Abstract
A diverse family of protein 4.1R isoforms is encoded by a complex gene on human chromosome 1. Although the prototypical 80-kDa 4.1R in mature erythrocytes is a key component of the erythroid membrane skeleton that regulates erythrocyte morphology and mechanical stability, little is known about 4.1R function in nucleated cells. Using gene knockout technology, we have generated mice with complete deficiency of all 4.1R protein isoforms. These 4.1R-null mice were viable, with moderate hemolytic anemia but no gross abnormalities. Erythrocytes from these mice exhibited abnormal morphology, lowered membrane stability, and reduced expression of other skeletal proteins including spectrin and ankyrin, suggesting that loss of 4.1R compromises membrane skeleton assembly in erythroid progenitors. Platelet morphology and function were essentially normal, indicating that 4.1R deficiency may have less impact on other hematopoietic lineages. Nonerythroid 4.1R expression patterns, viewed using histochemical staining for lacZ reporter activity incorporated into the targeted gene, revealed focal expression in specific neurons in the brain and in select cells of other major organs, challenging the view that 4.1R expression is widespread among nonerythroid cells. The 4.1R knockout mice represent a valuable animal model for exploring 4.1R function in nonerythroid cells and for determining pathophysiological sequelae to 4.1R deficiency.
Introduction
Protein 4.1R is best known as an ∼80-kDa structural protein in red blood cells that assembles into the peripheral membrane skeleton and plays an important role in determining red cell mechanical properties in the circulation. By interacting with components of the spectrin-based membrane skeleton, as well as with integral proteins in the overlying lipid bilayer, 4.1R is critical to both the structural integrity of the skeleton and its attachment to the membrane. Deficiency of 4.1R results in red cells with morphological abnormalities and unstable membranes due to the assembly of a mechanically compromised membrane skeleton (1, 2). Such defects are manifested by the appearance of circulating cells with elliptical morphology and also by the presence of fragmented cells due to the decreased membrane mechanical stability. In severe cases, 4.1R deficiency leads to hemolytic anemia.
In contrast to the relatively simple pattern of 4.1R expression in red cells and the well-understood role of 4.1R in the red cell membrane skeleton, the structure and function of 4.1R in nonerythroid cells has not been investigated in detail. While the red cell membrane skeleton provides a valuable paradigm for understanding 4.1R function in nucleated cells, available data suggests that the activities of 4.1R (or closely related 4.1 homologues) in such cells may be more complex. Erythroid progenitor cells (3) and nonerythroid cells of various types express a wide array of 4.1-immunoreactive proteins that localize to diverse subcellular sites. For example, 4.1 polypeptides ranging from 30 to 210 kDa have been reported in various tissues of rats (4) and chickens (5, 6). Immunofluorescence studies have demonstrated that 4.1 localizes not only to peripheral membrane sites, but also to other intracellular compartments, including the nucleus and centrosomes (7–11). To date, specific functional studies of these 4.1 isoforms have been hampered by the lack of precise molecular characterization of their structural identity.
Molecular analyses have revealed that the cellular complement of 4.1 isoforms has a complex genetic origin, involving both multiple alternative splicing pathways and multiple genes. In the prototypical erythroid 4.1R gene, alternative splicing regulates expression of alternative translation initiation sites in exon 2 (AUG1) and exon 4 (AUG2), thus controlling expression of 135-kDa and 80-kDa isoforms of the protein (12, 13). Multiple internal coding exons can also be alternatively spliced (13–15), generating an array of products potentially displaying different protein interaction domains and/or different subcellular localization signals. Interestingly, several 4.1R pre-mRNA splicing events exhibit developmental switches in their expression patterns, implying that distinct 4.1R functions are critical at different stages of differentiation. Clear examples of 4.1R splicing switches have been documented in erythroid (3, 13) and mammary epithelial cells (16). Finally, superimposed on this array of 4.1R polypeptides is the recent finding that multiple, closely related 4.1R gene homologues are also expressed in nucleated cells (17–20). The relative contributions of 4.1R versus the novel 4.1 protein(s) to nonerythroid cell biology remain to be determined.
Until now, the phenotype of a true 4.1R-null mutation has not been possible to investigate directly. Human patients with erythroid 4.1 deficiency have isoform-specific mutations that block selectively the expression of 80-kDa isoforms of the protein initiated at AUG2 (12, 21, 22), while sparing expression of higher molecular weight isoforms originating at upstream AUG1. Because AUG1 is expressed in nonerythroid cells and in early erythroid progenitors, but switched off (by alternative splicing) during erythropoiesis, AUG2 mutations lead to essentially complete 4.1R deficiency in erythrocytes, but only partial deficiency in other cell types. As one approach to characterizing 4.1R functions in nonerythroid cells, we used gene-targeting techniques to create a strain of mice with complete 4.1R deficiency. This paper describes the phenotypic characterization of cells in the hematopoietic lineage of these 4.1R-null mice. Concomitant with the primary 4.1R deficiency in erythrocytes, reduced membrane assembly of other key skeletal proteins was observed, including glycophorin C and p55 (also reduced or absent in human 4.1R deficiency), as well as spectrin (not previously reported to be abnormal in human 4.1R-deficient erythrocytes). In addition, by incorporating a lacZ transcriptional reporter into the targeted 4.1R locus, we have investigated 4.1R promoter activity in nonerythroid cells. The surprisingly focal expression pattern observed by this approach challenges the prevailing view that 4.1R is universally expressed among mammalian tissues, and suggests that 4.1R knockout mice represent a potent animal model for exploring nonerythroid function.
Methods
Construction of the 4.1R targeting vector.
Genomic clones encompassing the 5′ end of the mouse 4.1R gene spanning exons 2–4 and substantial flanking sequences were isolated and characterized. Appropriate fragments of the gene were excised and linked to a β-geo selection/reporter cassette to construct a targeting vector. This vector contained a 10-kb Sse8387I–BamHI fragment ∼2 kb upstream of exon 2 as the long arm and a 2-kb KpnI–EcoRV fragment located ∼0.5 kb downstream of exon 4 as the short arm. Both arms were cloned into Bluescript SK+(II), separated by a synthetic polylinker sequence into which was inserted a 7-kb Sal fragment of pGT1.8Iresβgeo (a gift of W.C. Skarnes, University of Edinburgh, Edinburgh, United Kingdom) to generate the final 21.5-kb targeting vector, PFmβ-geor. The β-geo cassette (containing the lacZ/neo fusion gene) by itself lacks a transcriptional promoter, but the coding sequence in this fragment is preceded by a splice acceptor sequence and a ribosomal binding site. Functional expression of neomycin resistance is thus enhanced when integration into a transcribed gene downstream of an active promoter occurs, thus selecting against the majority of nonhomologous recombination events in the transfected cell population and enriching for homologous recombination into the 4.1R gene. The β-geo cassette also included an SV40 polyadenylation signal at the 3′ end. These features were designed to incorporate the following critical elements into the targeted 4.1R gene: (a) deletion of exons 2–4, containing the known translation initiation sites, to block 4.1R protein expression; (b) expression of the β-galactosidase activity to serve as a reporter for the upstream promoter(s) of the 4.1R gene; and (c) abrogation of expression of the downstream 4.1 exons.
Generation of mice lacking protein 4.1R.
The targeting construct was introduced into R1 embryonic stem (ES) cells (23) by electroporation using 25 μg of Sse8387I-linearized PFmβ-geor. Transfected cells were cultured for 2 days and then subjected to selection using G418 (160 μg/ml) for 8–9 days. A total of 200 neomycin-resistant colonies were screened by Southern blot analysis with a 0.4-kb probe derived from just upstream of exon 5; this probe hybridizes to a 16-kb BamHI fragment in normal mouse genomic DNA and a 10-kb BamHI fragment in the targeted variant (Fig. 1). Three positive clones identified by this approach were further confirmed by nested PCR using primers specific for the β-geo cassette and exon 5 of the 4.1R gene. The first set of primers consisted of neoprobe-f4 (5′-TCTGGATTCATCGACTGTGG-3′) and neoprobe-r4 (5′-AATGTTAAGTAAAATCCCACCTCC-3′); the second primer set consisted of neoprobe-f5 (5′-GCTATCAGGACATAGCGTTGG-3′) and neoprobe-r5 (5′-CCCTTCTTTCTCCATCTCTATCC-3′).
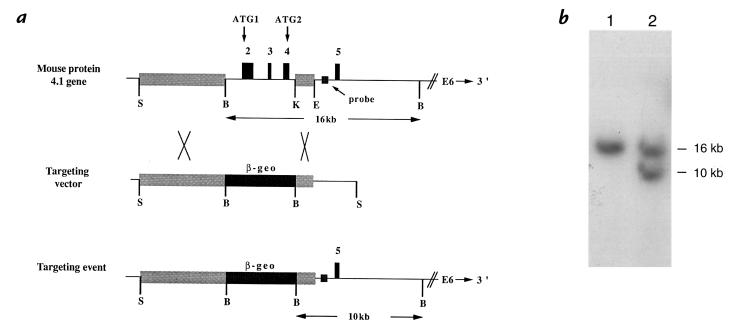
Figure 1.
Targeted disruption of the 4.1R gene. (a) Targeting vector. Top panel depicts a portion of the normal mouse 4.1R gene. Shown are the exon 2–4 region targeted for deletion and the flanking regions upstream (10-kb Sse837I–BamHI fragment) and downstream (2-kb KpnI–EcoRV fragment) that constitute the long arm and short arm of the targeting vector (middle panel). The lower panel represents the structure of the correctly targeted 4.1R gene in which the β-geo cassette has replaced exons 2–4. The targeted 4.1R gene retains the putative upstream promoter and all sequences downstream of exon 5. The intron probe adjacent to exon 5 hybridized to a 16-kb BamHI band in the normal gene and a 10-kb band in the targeted gene. (b) Southern blot showing a successful targeting event in ES cells. In lane 1, control ES cell DNA shows the normal 16-kb BamHI band detected with the intron 4 probe; in lane 2, targeted cell DNA shows the normal 16-kb band and the targeted 10-kb band. ES, embryonic stem.
Targeted ES cells were injected into C57Bl/6J blastocysts to generate chimeric mice.
Chimeric males having agouti coat color were bred with C57Bl/6J × DBA F1 hybrids, and germline transmission in agouti pups was confirmed by Southern blot analysis. Genotyping was performed by multiplex PCR to identify mice inheriting the targeted 4.1R gene and to distinguish heterozygous mice from homozygous targeted animals. Subsequently, chimeric males that exhibited germline transmission were bred with 129/SvEv (Taconic Farms, Germantown, New York, USA) females to maintain the targeted 4.1R allele in a genetically inbred background. Primers specific for neo confirmed the presence or absence of the β-geo cassette (neo-f: 5′-GATGGATTGCACGCAGGT-3′; neo-r 5′ GGCAGGAGCAAGGTGAGA-3′); primers specific for exon 4 (PFm4-f: 5′-GCTCAGGAAGAACACAGAGAGG-3′; PFm4-r: 5′-CATTCGTAGACCGTGTCATCC-3′) or exon 2 (lib-f: 5′-AAGAAACTCAGCTGGAGGAGG-3′; lib-r: 5′-TCCAAGTTCTATCTCCTTCTGACC-3′) tested for the presence or absence of the deleted exon 2–4 region of the 4.1 gene.
Gene expression analysis of protein 4.1R-deficient mice.
Expression of 4.1R mRNA was assayed by standard Northern blot analysis. RNA was isolated from mouse tissues by a phenol/guanidinium thiocyanate technique using the RNA STAT-60 kit according to manufacturer's instructions (Tel-Test Inc., Friendswood, Texas, USA). Then 20 μg of total RNA was electrophoresed in a denaturing (formamide) 1.2% agarose gel, transferred to a nylon membrane (Hybond; Amersham Life Sciences Inc., Arlington Heights, Illinois, USA), and hybridized to either a COOH-terminal probe (representing exons 18–21 of the 4.1R gene) or a full-length probe (spanning exons 2–21). These probes should hybridize to a 6-kb mRNA in mice containing an intact 4.1R gene, but should not hybridize to RNA from a homozygous 4.1R knockout mouse. The control β-actin probe (CLONTECH Laboratories Inc., Palo Alto, California, USA) hybridizes to an ∼2-kb band in most tissues and a slightly smaller band in heart and skeletal muscle.
Expression of 4.1R protein was assayed by Western blot analysis.
Red blood cell ghost proteins were resolved in SDS-polyacrylamide gels (24, 25), electrotransferred to nitrocellulose, and labeled with anti–human 4.1R antibody 24-3 (9), anti-p55 (gift of Athar Chishti, St. Elizabeth's Medical Center, Boston, Massachusetts, USA), or antibody to the cytoplasmic domain of glycophorin C (gift of Marion Reid, New York Blood Center, New York, New York, USA). The complex was detected by enhanced chemiluminescence using anti–rabbit antibody conjugated to horseradish peroxidase.
Histochemical detection of lacZ expression in tissues.
Various tissues were removed from 4.1R targeted (–/–) and wild-type (+/+) mice, embedded in OCT (Miles Inc., Elkart, Indiana, USA) and frozen in isopentane cooled at –70°C. Tissue sections (5–10 μm) were cut on a cryostat; then slides were air dried and immediately frozen until ready for fixation and staining. Fixation was carried out for 5 min in a solution of 0.25% glutaraldehyde in PBS. After rinsing thoroughly, the sections were incubated overnight at 37°C in 1 mg/ml solution of 5-bromo-4-chloro-3-indolyl-β-galactosidase (X-gal) in 2 mM MgCl2, 0.1 M PO4 buffer, 0.01% (wt/vol) sodium deoxycholate, 0.02% (wt/vol) Nonidet P-40, 5 mM K3Fe(CN)6, and 5 mM K4Fe(CN)6. The sections were then rinsed four times in PBS and counterstained with hematoxylin and eosin. After dehydrating the sections in 70%, 96%, and 100% ethanol, they were passed through xylene and mounted with Permount. Photographs were taken on a Nikon photomicroscope using Kodak-64 tungsten slide film.
Histology
Tissues (spleen, liver, kidney, heart, brain, intestine, stomach, colon, and testis) were fixed in Bouin's solution for 24 h, processed, and placed in paraffin. Sections (5 μM) were stained with hematoxylin and eosin for routine pathological examination or with Prussian blue for the detection of iron (26).
Hematologic analysis
Red cell analysis.
Red cell indices and reticulocyte counts were determined using the automated hematology analyzer H*3 (Bayer Diagnostics, Tarrytown, New York, USA). In this system, volume and hemoglobin concentration of individual red cells and reticulocytes are measured by laser light scattering (27). From the measured values of volume and hemoglobin concentration of each cell, histograms of volume, hemoglobin content, and hemoglobin concentration distribution of reticulocytes and mature red cells are constructed (28).
Osmotic gradient ektacytometry.
Osmotic gradient ektacytometry was used to quantitate cellular deformability, which is regulated by surface area, surface-area-to-volume ratio, membrane deformability, and state of hydration of red cells (29). Osmotic gradient ektacytometry was performed on fresh blood samples. Briefly, red cells were continuously mixed with a 4% polyvinylpyrrolidone solution of gradually increasing osmolality (from 60 to 450 mOsmol), and the deformability index is recorded as a function of osmolality at a constant applied shear stress of 170 dynes/cm2 using an ektacytometer (Bayer Diagnostics). The osmolality at which the deformability index reaches a minimum in the hypotonic region of the gradient (Omin) provides a measure of the average surface-area-to-volume ratio of the cells being studied. The maximum value of the deformability index attained at physiologically relevant osmolality (DImax) is determined by the mean surface area of red cells.
Blood chemistry
Serum blood chemistry analysis was performed according to standard procedures by a commercial laboratory (Consolidated Veterinary Diagnostics Inc., West Sacramento, California, USA).
Platelet analysis
Platelet counts and mean platelet volume.
Platelet counts and mean platelet volume (MPV) were determined by analysis of diluted citrated whole blood in a Serono Diagnostics System 9018cp analyzer (Serono Diagnostics, Allentown, Pennsylvania, USA) set for analyzing mouse cells.
Morphological and functional analysis.
Platelet aggregation studies were performed as described previously (30). In brief, ∼0.5 ml blood was collected in acid-citrate dextrose or sodium citrate from the retrobulbar venous plexus of mice anesthetized with methoxyflurane. Platelet-rich plasma (PRP) was collected following low-speed centrifugation, and the remaining buffy coat (200 μl) was mixed with an equal volume of modified Tyrode's buffer (HBMT) and centrifuged to obtain platelet-rich buffer (PRB). In one of three experiments, 200 μl of HBMT was also added to the erythrocyte pellet and centrifuged for PRB as above. The PRP and PRB samples were then combined, platelet counts adjusted with HBMT, and aggregation conducted in an aggregometer (Chrono-Log Corp., Havertown, Pennsylvania, USA) using human thrombin (1 U/ml; Sigma Chemical Co., St. Louis, Missouri, USA), ADP (20 μM; Chrono-Log Corp.), and collagen (10 μg/ml) as agonists. For analysis of clot retraction, PRP/PRB mixtures were adjusted to platelet counts of 263,000–288,000/μl, and 200-μl aliquots were added to siliconized glass aggregometer cuvettes. To initiate coagulation and activate platelets, 80 μl of 50 mM CaCl2 was added, and after 1 h at 37°C the clots were observed for retraction onto wood sticks. To assay for platelet microparticles, PRP/PRB (5 μl) in HBMT (45 μl) was reacted with FITC-labeled hamster anti–mouse platelet GPIIIa (CD41; PharMingen, San Diego, California, USA) (30 μl) and analyzed by flow cytometry (FACScan; Becton Dickinson Immunocytometry Systems, San Jose, California, USA) as described previously (31). Samples were analyzed before and after treatment with ionophore A23817 (25 μM).
Platelet adhesion to collagen and fibrinogen was studied as described previously (32). Briefly, polystyrene “popout” microtiter wells (Costar 2580; Corning-Costar Corp., Cambridge, Massachusetts, USA) were coated with 100 μl of 0.5% BSA in 0.15 M NaCl, 0.01M Tris/HCl (pH 7.4), lathyritic rat skin collagen (33 μg/ml), or purified human fibrinogen (33 μg/ml), for 1 h at 37°C. Wells were blocked with 0.5% BSA, washed in HBMT, and 50 μl of citrated PRP/PRB in HBMT (washed once in HBMT/0.1 vol acid-citrate dextrose; platelet count 78,000–82,000/μl) added to each well for a 90-min incubation at room temperature. After washing, 0.2 ml of 1.5% glutaraldehyde/2% paraformaldehyde in 0.1 M cacodylate was added, and samples were prepared for scanning electron microscopy by critical point drying and spatter coating with gold palladium.
Platelets were analyzed by transmission electron microscopy. In brief, citrated whole blood was centrifuged at 700 g for 20 min at room temperature, the plasma removed, and 3% glutaraldehyde in 0.1 M cacodylate was dripped down the side and incubated for 2 h at room temperature. The buffy coat was removed and cut into 1–2-mm cubes that were further fixed with 3% glutaraldehyde in 0.1 M cacodylate overnight at 4°C. After washing in cacodylate and embedding in Epon, sections (60 nm) were stained with uranyl acetate and lead citrate and viewed with a JEOL JEM 1000× microscope (JEOL USA Inc., Cranford, New Jersey, USA).
Results
Targeted disruption of the protein 4.1R gene.
The mouse protein 4.1R gene consists of at least 25 exons (33) extending over 200–300 kb. Expression of this large gene is characterized by complex alternative RNA splicing pathways that lead to tissue-specific synthesis of various 4.1R protein isoforms. A complete knockout of 4.1R transcription was not feasible due to insufficient information concerning the precise localization of 4.1R transcriptional promoter(s). Instead, a knockout strategy was designed to eliminate expression of all known 4.1R isoforms by interfering with translation initiation. Two alternative translation initiation sites are known to be encoded in exons 2 (high-molecular-weight 4.1 isoforms) and 4 (80-kDa isoforms), respectively. Consequently, targeted disruption of the 4.1R gene was accomplished by deletion of exons 2–4.
The targeting vector used in these studies is shown in Fig. 1. Exons 2–4 of the protein 4.1R gene were replaced with a β-geo cassette encoding neomycin resistance and β-galactosidase activity. The disrupted 4.1R gene was thus expected to use its normal upstream promoter element(s) to drive expression of a truncated transcript containing the normal 5′ end of 4.1R mRNA fused to the β-geo cassette; expression of downstream 4.1R exons should be abrogated by translation and transcription terminators incorporated into the β-geo cassette. This strategy had the added advantage that the β-galactosidase moiety could be used as a transcriptional reporter in recipient mice bearing the targeting construct.
Using the targeting vector, homologous recombination in ES cells was used to disrupt one allele of the 4.1R gene. Southern blot analysis showed that the parental ES cells exhibited only the expected 16-kb BamHI fragment when hybridized with an intron 4 probe (Fig. 1b, lane 1), whereas the targeted ES cells were heterozygous for the original 16-kb and the targeted 10-kb alleles (Fig. 1b, lane 2). Additional control experiments employing PCR reactions extending from the β-geo cassette into the surrounding sequence of the 4.1R gene verified the structure of the targeted gene (data not shown). These ES cells were used subsequently for blastocyst injections to generate a strain of mice exhibiting germline transmission of the targeted 4.1R allele.
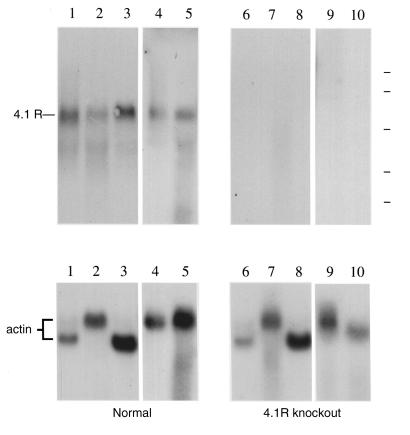
Breeding experiments revealed that 4.1R–/– mice were fertile and viable. To confirm that 4.1R expression was indeed knocked out in such animals, Northern blot analysis of RNA was performed. Using a short 4.1R hybridization probe derived from COOH-terminal coding sequences, we detected an ∼6-kb band in RNA from normal mouse tissues (Fig. 2, upper left), but not in RNA from comparable tissues of the knockout (Fig. 2, upper right). Similar results were obtained with a long 2-kb probe spanning coding exons 2–21, ruling out the possibility that cryptic promoters or splice patterns might yield partial 4.1R expression (data not shown). In contrast, hybridization of the same filters with a positive control actin probe yielded signals of equivalent intensity in both normal and 4.1R knockout mice (Fig. 2, lower). These results confirmed that the expression of downstream portions of the gene was reduced to below detectable levels. Disruption of the 4.1R gene thus produced a 4.1R-null phenotype.
Figure 2.
Analysis of 4.1R mRNA expression by Northern blot analysis. RNA isolated from tissues of a normal control mouse (left) and a homozygous 4.1 knockout mouse (right). Upper panel shows hybridization to a 4.1R cDNA probe representing sequences in exons 18–21, while lower panel shows hybridization to a control actin cDNA probe. Essentially identical results were obtained using a probe that spans exons 2–21. Source of RNA: lanes 1 and 6, heart; lanes 2 and 7, lung; lanes 3 and 8, skeletal muscle; lanes 4 and 9, brain; lanes 5 and 10, spleen. Position of RNA size standards of 9.4, 7.5, 4.4, 2.4, and 1.35 kb is indicated at the top right by the dashed lines.
Erythroid phenotype of 4.1R knockout mice.
In humans, red cell deficiency of 80-kDa 4.1R leads to secondary deficiency of the 4.1 interacting proteins glycophorin C and p55 (34) and to production of morphologically and mechanically abnormal red cells. The nature of mutation in these human patients is such that erythroid progenitors are known to, or are expected to, retain expression of 135-kDa isoforms of 4.1R and these may fulfill a critical role in early erythroid development (3, 12, 22). In contrast, erythropoiesis in knockout mice must proceed in the complete absence of 4.1R and might therefore lead to more severe phenotypic consequences.
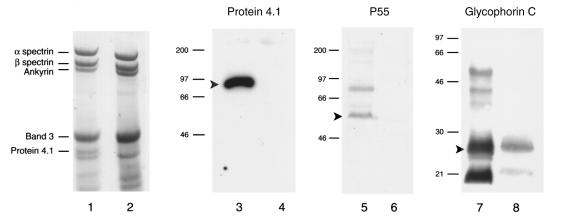
As a first step to characterize the phenotype of 4.1R knockout red cells, red cell membrane–associated proteins were examined by polyacrylamide gel electrophoresis. As shown in Fig. 3, Coomassie blue staining of red cell proteins revealed a protein band of 80 kDa migrating at the expected position of 4.1R in normal mouse red cells (lane 1), but not in red cells from the 4.1R knockout mouse (lane 2). Immunoblot analysis with anti-4.1R antibody confirmed the presence of this 80-kDa 4.1R band in normal (lane 3), but not knockout, red cells (lane 4). Further analysis of the Coomassie blue–stained gel by densitometry showed that the amounts of α- and β-spectrin were reduced by ∼30% in knockout mice compared with normal controls. These results, although unexpected because spectrin deficiency has not been reported previously in human red cells with 4.1 deficiency, were consistently observed in multiple experiments from several mice.
Figure 3.
Distribution of HETE compounds in plaques retrieved from asymptomatic (18 stable plaques) and symptomatic (12 unstable plaques) patients. Each HETE, with concentration expressed on a per-mole-of-lipid-phosphorus basis, taken separately was significantly more abundant in unstable, compared with stable, plaques (P < 0.01).
The abundance of other red cell proteins was next examined by immunoblot analysis using specific antibodies. Interestingly, the 4.1R deficiency was accompanied by apparently complete loss of p55 (lanes 5 and 6), as well as a dramatic reduction in the level of glycophorin C (to ∼16% of normal) (lanes 7 and 8). The coordinate loss or reduction in the levels of p55 and glycophorin C has also been observed in red cells from human patients with 4.1R deficiency (34).
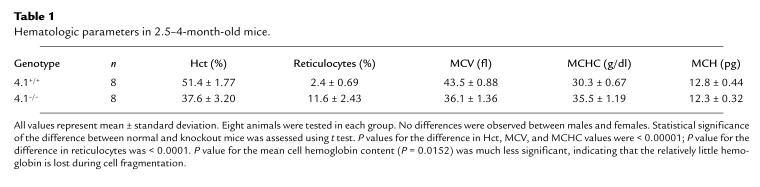
Hematological analysis of 4.1R-deficient mice revealed several significant abnormalities in red cell parameters. In contrast to the uniform biconcave disk morphology of normal mouse red cells (Fig. 4a, left), 4.1-deficient mouse red cells exhibited morphologic heterogeneity with numerous fragmented cells (Fig. 4a, right). Interestingly, while human 4.1R-deficient red cells also tend to fragment, they exhibit an elliptical morphology in contrast to the more spherocytic appearance of mouse 4.1R-deficient red cells (which have the additional spectrin deficiency not seen in the human red cells). Red cell parameters in knockout animals were also examined using an automated hematology analyzer. In particular, knockout mice had moderate hemolytic anemia with decreased hematocrits (37% vs. 51% in normal mice), coupled with increased reticulocyte counts (11% vs. 2.5% in normal mice). 4.1R-deficient erythrocytes also exhibited decreased mean cell volume and increased mean cell hemoglobin concentration, consistent with cell fragmentation due to a mechanically compromised membrane skeleton (Table 1). Finally, although early erythropoiesis was not examined directly, the observed increase in reticulocyte levels represents an appropriate response to the hemolytic anemia, suggesting that there is no gross dysfunction in the erythropoietic response in these animals even in the complete absence of 4.1R expression. The spleens of knockout mice were enlarged eight- to ninefold, consistent with increased erythropoietic activity. Regarding the phenotype of heterozygous (+/–) mice, hematological parameters such as mean cell volume and mean cell hemoglobin concentration were essentially normal.
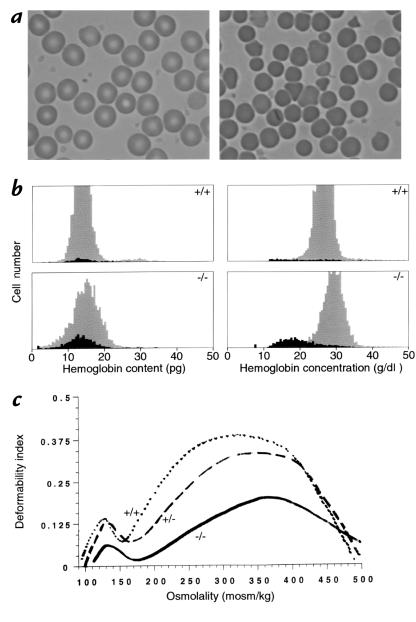
Figure 4.
Erythroid parameters. (a) Red cell morphology in blood from normal (left) and knockout (right) mice. Normal cells at left show the typical biconcave disk morphology, while homozygous 4.1-deficient red cells at right exhibit a more heterogeneous morphology marked by the presence of numerous cell fragments. Red cells from heterozygous mice were morphologically normal. (b) Red cell indices. Cell hemoglobin content and cell hemoglobin concentration histograms of whole blood from normal (upper panels) and knockout (lower panels) mice. The dark areas of the histogram represent reticulocytes, and the gray areas represent mature red cells. Significant numbers of fragmented red cells with decreased hemoglobin content (< 10 pg) can be seen in knockout mice. Cell dehydration is also a feature of knockout mice red cells, as indicated by the shift in hemoglobin concentration from reticulocytes (black area in lower right panel) to mature red cells (gray area in lower right panel). (c). Osmotic gradient deformability profiles of red cells from normal (+/+), heterozygous (+/–), and homozygous (–/–) 4.1R-deficient mice. The maximum value of the deformability index, a direct measure of red cell surface area, is reduced in (+/–) red cells and more dramatically reduced in (–/–) red cells.
Table 1.
Hematologic parameters in 2.5–4-month-old mice.
To obtain a more quantitative assessment of red cell fragmentation, the variation in hemoglobin content of individual red cells from normal and 4.1 knockout mice was examined. Normal red cells exhibited a fairly narrow range of hemoglobin content (Fig. 4b, top left), while homozygous knockout red cells had a broader distribution of hemoglobin content that included cells with lower than normal hemoglobin content (Fig. 4b, bottom left). This fraction represented the fragmented red cells in the circulation of knockout mice. Measurements of hemoglobin concentration of individual cells revealed that in 4.1 deficiency, the hemoglobin concentration in mature red cells was markedly increased compared with that of reticulocytes, implying that cell dehydration occurred in red cells as a result of fragmentation during their life span in the circulation (Fig. 4b, bottom right).
Red cells were also examined by ektacytometry to assess their deformability characteristics. Osmotic deformability profiles of red cells from wild-type, heterozygous, and homozygous 4.1R-deficient mice are shown in Fig. 4c. The maximum value of the deformability index (DImax), a direct measure of red cell membrane surface, was slightly reduced for red cells from heterozygous mice and markedly reduced for red cells from homozygous mice. The marked reduction in DImax seen for red cells from homozygous mice attests to the significant reduction in membrane surface area of these cells as a consequence of membrane fragmentation in vivo. The hypotonic osmolality at which deformability index reached a minimum (Omin) was elevated for red cells from both heterozygous and homozygous mice, implying that the increased osmotic fragility of these red cells is a result of membrane surface area loss.
Platelet phenotype of 4.1R knockout mice.
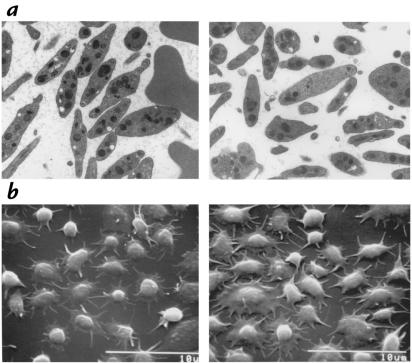
4.1R deficiency in platelets of the knockout mouse was confirmed by immunoblot experiments, which revealed a weak expression of an ∼75-kDa immunoreactive band in normal platelets (35, 36), but not knockout platelets (results not shown). The same antibody yielded strong staining of ∼80-kDa protein 4.1 in erythrocytes, consistent with a much lower expression in platelets than in erythrocytes. To determine whether 4.1R deficiency affected other hematopoietic cells, we analyzed platelet morphology and function. No abnormalities were observed in the number of peripheral blood platelets in 4.1R knockout mice, but the mean platelet volumes were slightly elevated (6.4, 5.9, and 5.7 fl vs. 5.2, 5.3, and 4.6 fl in wild-type mice). Platelet morphology, examined either in Wright-stained blood smears (data not shown) or in transmission electron micrographs of buffy coat fractions (Fig. 5a), also appeared normal. A number of parameters of platelet function were also tested in the 4.1R-deficient animals. Clot retraction and platelet adherence to collagen and fibrinogen appeared normal by phase-contrast microscopy. 4.1R-deficient mice had normal numbers of platelet microparticles and were able to make normal numbers of additional microparticles in response to stimulation with ionophore A23187. Likewise, scanning electron micrographs of platelets adhered to collagen or fibrinogen were similar for normal or 4.1R-deficient mice (Fig. 5b). Platelet aggregation in response to thrombin (1 U/ml) was tested on three separate occasions; the initial slopes of aggregation were the same for wild-type and 4.1-deficient mouse platelets on all three occasions, but in two of three experiments, the aggregation in the 4.1-deficient mice was reversible. Similarly, in one of the two experiments using adenosine diphosphate (ADP) the 4.1-deficient mouse platelets demonstrated reversible aggregation, even though the initial slope of aggregation was similar to that of wild-type. Finally, in response to collagen, the 4.1R-deficient mouse platelets underwent aggregation, but the lag phase was increased and the slope was slightly decreased. These data raise the possibility that 4.1R may play a role in irreversible platelet aggregation, which is plausible because platelet 4.1 has been demonstrated to associate with the platelet cytoskeleton after thrombin stimulation (36); it is equally likely, however, that ADP released from 4.1R-deficient erythrocytes during platelet preparation results in desensitization to ADP, with the resultant changes in aggregation response. From these data we conclude that, in contrast to the markedly abnormal erythrocytes in 4.1 knockout mice, platelet morphology and function appear essentially normal.
Figure 5.
Platelet parameters. (a) Transmission electron micrographs of buffy coat fractions of normal (left) and 4.1R-deficient (right) blood. (b) Scanning electron micrographs of platelets adhered to fibrinogen.
Histological findings of 4.1R–/– mice.
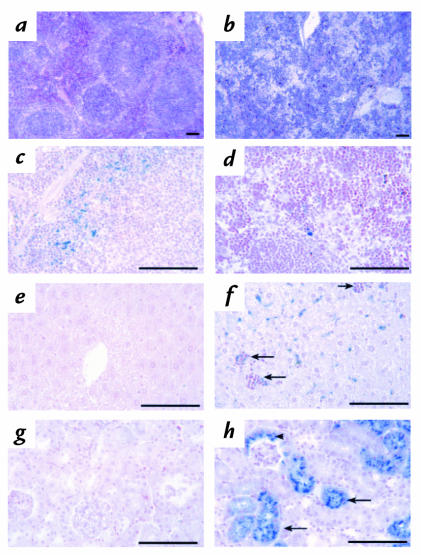
A routine microscopic examination of the major organs of 4.1R–/– mice was performed to determine whether 4.1R deficiency might produce any nonhematologic phenotype. Light microscopic examination of spleen, liver, and kidney sections revealed evidence of accelerated erythropoiesis and increased red cell destruction in 4.1R–/– mice. Such results are typical of other moderately to severely anemic mutants and are therefore not necessarily indicative of primary defects in these organs. As shown in Fig. 6, a and b, splenic sections stained with hematoxylin and eosin revealed red pulp areas that were dramatically expanded and filled with erythroid precursor cells, effacing much of the normal nodular architecture of the spleen. This finding is consistent with the splenomegaly noted above. Prussian blue staining revealed that iron deposition was reduced in 4.1R–/– spleen compared with normal spleen, presumably reflecting increased use by the rapidly proliferating erythroid precursors (Fig. 6, c and d). Increased erythropoiesis was also apparent in 4.1R–/– liver where hematopoietic foci were frequent (Fig. 6, e and f). Evidence of increased red cell destruction was seen by increased deposition of iron in 4.1R–/– liver (Fig. 6, e and f). Marked iron deposition was detected in 4.1R–/– kidney proximal convoluted tubules, indicating that extensive intravascular hemolysis occurred in the absence of protein 4.1R (Fig. 6, g and h). All other organs examined appeared grossly normal at the light microscopic level in 4.1R–/– mice.
Figure 6.
Pathological changes in 4.1R–/– tissues. Hematoxylin- and eosin-stained sections of (a) normal and (b) 4.1R–/– spleen sections. Note the effacement of the splenic nodules by the dramatic expansion of the red pulp areas in 4.1R–/– spleen. Prussian blue–stained (c) normal and (d) 4.1R–/– spleen. Iron deposition is decreased in 4.1R–/– spleen red pulp areas due to increased use by rapidly proliferating erythroid precursors. Prussian blue–stained (e) normal and (f) 4.1R–/– liver. Hematopoietic foci (arrows) are frequent and liver iron deposition increased in 4.1R–/– mice, reflecting increased destruction of 4.1-null red cells. Note that no iron deposition is seen in areas of proliferating erythroid precursors, i.e., liver hematopoietic foci (arrows). Prussian blue stains of (g) normal and (h) 4.1R–/– kidney. Note the extensive deposition of iron in 4.1R–/– proximal tubules (arrows) and Bowman's capsule (arrowhead), indicating severe intravascular hemolysis of 4.1R-null red cells. Bar, 100 μM.
Blood chemistry of 4.1R–/– mice.
A number of blood chemistry parameters were examined to gain possible insights into other tissue abnormalities in the 4.1R–/– mouse. Although most values were within normal limits, the levels of two enzymes were significantly elevated in the knockout animals. Creatine phosphokinase (CPK) was consistently increased about twofold in the blood of 4.1R–/– mice, regardless of age or sex of the animals. Likewise, serum glutamic-oxaloacetic transaminase (SGOT) was similarly elevated two- to threefold above normal levels. Both of these enzyme abnormalities could be consistent with a skeletal or cardiac muscle dysfunction, although no obvious muscle defects were observed histologically.
Activity of the 4.1R promoter in nonerythroid cells.
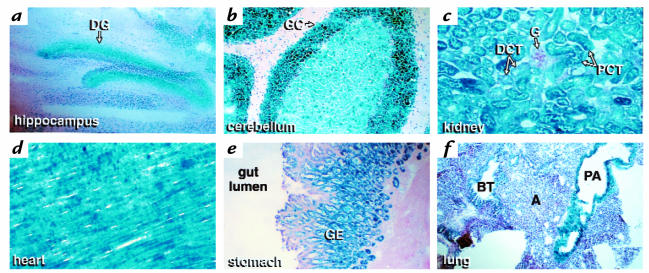
Previous studies using sensitive reverse transcriptase (RT)-PCR techniques have shown that 4.1R transcripts are expressed in a wide variety of nonerythroid tissues. However, such assays cannot resolve precisely which cell types within the tissue are actually expressing the 4.1R gene products. Because the targeted 4.1R gene contained a β-gal reporter inserted downstream of the gene's transcriptional promoter(s), we were able to obtain a high-resolution map of 4.1R promoter activity in various nonerythroid cells (Fig. 7). Very focal expression was observed in discrete cell populations of selective nonerythroid tissues throughout the body. For example, sagittal sections of the brain revealed robust expression in the granule cell layer (GC) of the cerebellum and the dentate gyrus (DG) of the hippocampus. In the kidney, β-galactosidase activity predominated in the proximal convoluted tubules (PCT), with negligible levels in the distal convoluted tubules (DCT) and no detectable activity in the glomeruli (G). Cardiac muscle exhibited uniform, high-level expression. In the stomach, expression was selectively localized to proliferating mucous cells in the gastric pits, parietal cells, and chief cells of the gastric epithelium (GE); β-galactosidase activity was negligible in differentiated mucous neck and mucous surface cells, and absent in the submucosal and muscular layers. In the lung, staining was confined to the pulmonary artery (PA) wall; the degree of expression corresponds to the caliber of the vessel wall, with smaller arterial branches and arterioles showing only a thin rim of β-galactosidase staining (data not shown). In contrast, negligible staining of the bronchial tree (BT) and alveoli (A) was observed. Identical patterns of expression were observed in tissues from 4.1R heterozygous mice, although the intensity of staining was decreased (data not shown).
Figure 7.
Expression of β-galactosidase activity encoded within the β-geo cassette of the targeted 4.1R gene. Frozen sections of the knockout mouse were fixed and stained with the chromogenic substrate 5-bromo-4-chloro-3-indolyl-β-galactosidase, as described in Methods. Expression was observed in the following tissues: (a) hippocampus (b) cerebellum (c) kidney (d) heart (e) stomach, and (f) lung. A, alveoli; BT, bronchiolar tree; DCT, distal convoluted tubule; DG, dentate gyrus; G, glomerulus; GC, granule cells of the cerebellum; GE, gastric epithelium; PA, pulmonary artery; PCT, proximal convoluted tubules.
Discussion
Using homologous recombination in mouse ES cells to create a specific targeting event in the protein 4.1R gene, we have created the first genetic model for complete 4.1R deficiency. The 4.1R-null mouse differs from previously described cases of 4.1R deficiency in humans, where the isoform-specific deficiency of 80-kDa 4.1R allows for continued expression of 135-kDa isoforms in erythroid progenitors and in nonerythroid cells. Indeed, this difference in availability of 4.1R in erythroid progenitors may influence the assembly of the membrane skeleton, because mouse 4.1R-deficient erythrocytes exhibited secondary deficiencies of spectrin and ankyrin that have not been observed in the human 4.1R-deficient individuals. The 4.1R–/– animal model thus provides a novel opportunity to explore the phenotypic consequences of complete 4.1R deficiency to the assembly of a mechanically stable erythrocyte membrane skeleton in erythroid progenitors.
The erythroid phenotype of 4.1R–/– mice was characterized by analysis of circulating red cells. 4.1R–/– red cells lacked detectable 4.1R protein expression and exhibited abnormal morphology and decreased deformability compared with control red cells with normal 4.1 content. Interestingly, in contrast to the elliptocytic phenotype of red cells from human patients with 4.1R deficiency, for unknown reasons (perhaps related to the partial spectrin deficiency), 4.1R-deficient mouse red cells had a more spherocytic appearance. Despite this morphological difference, both human and mouse red cells with 4.1R deficiency exhibit mechanically compromised erythrocyte membranes that fragment in the circulation, leading to decreased membrane surface area. Careful analysis of mature red cells from the 4.1R knockout mice showed, in addition, that they had elevated hemoglobin concentration, suggesting that cellular dehydration occurs in response to the loss of surface area. This observation is consistent with the hypothesis that red cells respond to loss of surface area by decreasing cell volume, in an attempt to maintain a requisite degree of deformability (37). Recent studies have shown that cell dehydration can also occur in human hereditary elliptocytosis (38).
To compensate for the moderate anemia arising from red cell destruction, 4.1R–/– animals had enlarged spleens and significantly increased reticulocyte counts. In general, the phenotype of knockout mouse erythrocytes, despite the complete deficiency of all 4.1R isoforms throughout erythroid differentiation, is quite similar to that observed in human patients with hereditary elliptocytosis due to selective 80-kDa 4.1R deficiency. No evidence of a more severe dysfunction of erythropoiesis was evident in these studies. In addition, few or no abnormalities in platelet morphology or function were detected, suggesting that 4.1R may not be an essential protein in platelets.
Red cells from the 4.1–/– mouse will be useful for biophysical assays probing the mechanical properties of the membrane skeleton. A number of previous studies have used rare human 4.1R-deficient red cells to explore the contribution of 4.1R to membrane stability and to investigate its role in linking various integral proteins to the skeletal network. In addition, functional differences among alternative isoforms of 4.1R have also been addressed in reconstitution experiments assaying the ability of recombinant 4.1R proteins to restore membrane stability to 4.1R-deficient red cell ghosts (2, 39–41). Indeed, we have previously demonstrated dramatic differences in the ability of different spliced forms of 4.1R to mechanically stabilize 4.1R-deficient membranes, leading to the conclusion that the peptide encoded by alternative exon 16 is critical for normal membrane stability (39, 41, 42). The ready availability of 4.1R-deficient red cells from 4.1–/– mice should facilitate further biophysical probing of the mechanical properties of red cell membranes.
A second important feature of the 4.1R gene-targeting event was the insertion of a lacZ transcriptional reporter into the modified 4.1R locus, which facilitated characterization of 4.1R mRNA expression throughout the body via a simple histochemical staining procedure. In nonerythroid cells, the observed focal expression of 4.1R provided a fundamentally new perception of 4.1R expression patterns. Previous assays had suggested that 4.1R RNA and protein, although abundant in erythroid cells, were also expressed almost universally among nonerythroid tissues (3–6, 14, 15, 43, 44). However, these results were derived in many cases from experiments with crude tissue extracts (where individual cell type differences might be missed) and/or with immunological probes of uncertain specificity (because antibodies raised against 4.1R from red cells may also cross-react with the recently reported nonerythroid 4.1 homologues). Both of these problems are largely overcome with the histochemical approach employed in this study, which offers both high specificity and high resolution. The histochemical results yielded intriguing patterns of expression: 4.1R in brain was detected in granular cells of the cerebellum and in the dentate gyrus within the hippocampus, but the vast majority of neuronal cells appeared negative by this assay. In other tissues, 4.1R promoter activity was prominent in cardiac muscle, in proximal convoluted tubules of the kidney, in pulmonary arteries (but not pulmonary veins) within the lung, and in select cells of the gastric epithelium. Importantly, the fidelity of this reporter system as an indicator of 4.1R promoter activity is supported by the identification of identical patterns of 4.1R gene expression in the brain using in situ hybridization with 4.1R-specific antisense RNA probes (45). The physiological relevance of the neuronal localizations is supported further by the preliminary observation that 4.1R-null mice exhibit neurobehavioral deficits that correlate with cerebellar and dentate functions, including coordination/balance and spatial learning, respectively (45). These data suggest that the 4.1R–/– mice should provide a unique opportunity to explore the phenotypic consequences of total 4.1R deficiency, not only during erythroid differentiation, but also with regard to the cell biology of selected nonerythroid cells.
In summary, the protein 4.1R gene is a complex genetic locus that encodes a large number of structurally diverse 4.1R protein isoforms via tissue-specific regulation of alternative pre-mRNA splicing. In addition to the well-documented role of 4.1R in the erythroid membrane skeleton and the recent evidence that 4.1R is required for normal cerebellar and dentate gyrus function, a number of other potential roles for 4.1-immunoreactive proteins have been proposed. Particularly intriguing are the observations that 4.1R may be involved in nuclear (7–9, 11) or centrosomal (10) function, although it is not yet clear how to reconcile these observations with the finding that 4.1R–/– mice have apparently normal viability and fertility. It is possible that one or more of the recently described 4.1 nonerythroid homologues (17–19) might compensate for the loss of 4.1R or that a 4.1R-deficient phenotype might be manifest only under specific conditions. Regardless, we expect that the 4.1R–/– mice will provide a useful model in which to explore the intricacies of 4.1R function encoded in the complex 4.1R locus.
Acknowledgments
The authors thank Lesley Scudder, Ronald E. Gordin, and Norman Katz of the Core Electron Microscopy Facility, as well as Heikki Vaananen of the Cell Imaging Laboratory, all at the Mount Sinai School of Medicine, and Phil Cooper of Lawrence Berkeley National Laboratory for his expert, professional handing of the mice used in this study. This work was supported by grants DK32094 (to N. Mohandas) and HL 55321 (to L. Peters) from the National Institutes of Health, and by the March of Dimes (to L. Peters).
References
- 1.Tchernia G, Mohandas N, Shohet SB. Deficiency of skeletal membrane protein 4.1 in homozygous hereditary elliptocytosis. J Clin Invest. 1981;68:454–460. doi: 10.1172/JCI110275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Takakuwa Y, Tchernia G, Rossi M, Benabadji M, Mohandas N. Restoration of normal membrane stability to unstable protein 4.1-deficient membranes by incorporation of purified protein 4.1. J Clin Invest. 1986;78:80–85. doi: 10.1172/JCI112577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chasis JA, et al. Differentiation-associated switches in protein 4.1 expression. Synthesis of multiple structural isoforms during normal human erythropoiesis. J Clin Invest. 1993;91:329–338. doi: 10.1172/JCI116189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Anderson RA, Correas I, Mazzucco C, Castle JD, Marchesi VT. Tissue-specific analogues of erythrocyte protein 4.1 retain functional domains. J Cell Biochem. 1988;37:269–284. doi: 10.1002/jcb.240370303. [DOI] [PubMed] [Google Scholar]
- 5.Granger BL, Lazarides E. Membrane skeletal protein 4.1 of avian erythrocytes is composed of multiple variants that exhibit tissue-specific expression. Cell. 1984;37:595–607. doi: 10.1016/0092-8674(84)90390-8. [DOI] [PubMed] [Google Scholar]
- 6.Granger BL, Lazarides E. Appearance of new variants of membrane skeletal protein 4.1 during terminal differentiation of avian erythroid and lenticular cells. Nature. 1985;313:238–241. doi: 10.1038/313238a0. [DOI] [PubMed] [Google Scholar]
- 7.Correas I. Characterization of isoforms of protein 4.1 present in the nucleus. Biochem J. 1991;279:581–585. doi: 10.1042/bj2790581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Carcer G, Lallena MJ, Correas I. Protein 4.1 is a component of the nuclear matrix of mammalian cells. Biochem J. 1995;312:871–877. doi: 10.1042/bj3120871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krauss SW, et al. Structural protein 4.1 in the nucleus of human cells: dynamic rearrangements during cell division. J Cell Biol. 1997;137:275–289. doi: 10.1083/jcb.137.2.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krauss SW, et al. Structural protein 4.1 is located in mammalian centrosomes. Proc Natl Acad Sci USA. 1997;94:7297–7302. doi: 10.1073/pnas.94.14.7297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mattagajasingh SN, Huang SC, Benz EJ., Jr Direct evidence for a nuclear localization and function of protein 4.1 in the nucleus: in vivo association with mitotic apparatus proteins. Blood. 1996;88(Suppl.):276a.. (Abstr.) [Google Scholar]
- 12.Chasis JA, et al. Differential use of protein 4.1 translation initiation sites during erythropoiesis: implications for a mutation-induced stage-specific deficiency of protein 4.1 during erythroid development. Blood. 1996;87:5324–5331. [PubMed] [Google Scholar]
- 13.Baklouti S, et al. Asynchronous regulation of splicing events within protein 4.1 pre-mRNA during erythroid differentiation. Blood. 1996;87:3934–3941. [PubMed] [Google Scholar]
- 14.Tang TK, Leto T, Marchesi VT, Benz EJ. Heterogeneity of mRNA and protein products arising from the protein 4.1 gene in erythroid and nonerythroid tissues. J Cell Biol. 1990;110:617–624. doi: 10.1083/jcb.110.3.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Conboy JG, Chan J, Chasis JA, Kan YW, Mohandas N. Tissue- and development-specific alternative RNA splicing regulates expression of multiple isoforms of erythroid membrane protein 4.1. J Biol Chem. 1991;266:8273–8280. [PubMed] [Google Scholar]
- 16.Schischmanoff PO, et al. Cell-shape dependent regulation of protein 4.1 alternative splicing in mammary epithelial cells. J Biol Chem. 1997;272:10254–10259. doi: 10.1074/jbc.272.15.10254. [DOI] [PubMed] [Google Scholar]
- 17.Parra M, et al. Cloning and characterization of 4.1-G, a new member of the skeletal protein 4.1 gene family. Genomics. 1998;49:298–306. doi: 10.1006/geno.1998.5265. [DOI] [PubMed] [Google Scholar]
- 18.Walensky LD, et al. Immunophilin FKBP13 interacts with a novel erythrocyte membrane cytoskeletal protein 4.1. J Cell Biol. 1998;273:143–153. doi: 10.1083/jcb.141.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Walensky LD, Blackshaw SS, Conboy JG, Mohandas N, Snyder SH. Molecular cloning of a novel neuron-specific homologue of the erythrocyte membrane skeletal protein 4.1. Soc Neurosci Abstr. 1997;23:1674. (Abstr.) [Google Scholar]
- 20.Parra M, et al. A family of distinct protein 4.1 genes is expressed in human tissues. Mol Biol Cell. 1997;8(Suppl.):177a.. (Abstr.) [Google Scholar]
- 21.Conboy JG, et al. An isoform-specific mutation in the protein 4.1 gene results in hereditary elliptocytosis and complete deficiency of protein 4.1 in erythrocytes but not in nonerythroid cells. J Clin Invest. 1993;91:77–82. doi: 10.1172/JCI116203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dalla Venezia N, et al. Homozygous 4.1(–) hereditary elliptocytosis associated with a point mutation in the downstream initiation codon of protein 4.1 gene. J Clin Invest. 1992;90:1713–1717. doi: 10.1172/JCI116044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc Natl Acad Sci USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 25.Fairbanks G, Steck TL, Wallach DF. Electrophoretic analysis of the major polypeptides of the human erythrocyte membrane. Biochemistry. 1971;10:2606–2617. doi: 10.1021/bi00789a030. [DOI] [PubMed] [Google Scholar]
- 26.Humason, G.L. 1979. Animal tissue techniques. W.H. Freeman Co. San Francisco, CA. 661 pp.
- 27.Mohandas N, et al. Accurate and independent measurement of volume and hemoglobin concentration of individual red cells by laser light scattering. Blood. 1986;68:506–513. [PubMed] [Google Scholar]
- 28.Brugnara C, et al. Effects of subcutaneous recombinant human erythropoietin in normal subjects: development of decreased reticulocyte hemoglobin content and iron-deficient erythropoiesis. J Lab Clin Med. 1994;123:660–667. [PubMed] [Google Scholar]
- 29.Clark MR, Mohandas N, Shohet SB. Osmotic gradient ektacytometry: comprehensive characterization of red cell volume and surface maintenance. Blood. 1983;61:899–910. [PubMed] [Google Scholar]
- 30.Coller BS, Peerschke EI, Scudder LE, Sullivan CA. A murine monoclonal antibody that completely blocks the binding of fibrinogen to platelets produces a thrombasthenic-like state in normal platelets and binds to glycoproteins IIb and/or IIa. J Clin Invest. 1983;72:325–338. doi: 10.1172/JCI110973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reverter JC, et al. Inhibition of platelet-mediated, tissue factor–induced thrombin generation by the mouse/human chimeric 7E3 antibody. Potential implications for the effect of c7E3 Fab treatment on acute thrombosis and “clinical restenosis.”. J Clin Invest. 1996;98:863–874. doi: 10.1172/JCI118859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Coller BS, Beer JH, Scudder LE, Steinberg MH. Collagen-platelet interactions: Evidence for a direct interaction of collagen with platelet GPIa/IIa and an indirect interaction with platelet GPIIb/IIa mediated by adhesive proteins. Blood. 1989;74:182–192. [PubMed] [Google Scholar]
- 33.Huang J-P, et al. Genomic structure of the locus encoding protein 4.1. J Biol Chem. 1993;268:3758–3766. [PubMed] [Google Scholar]
- 34.Alloisio N, et al. Evidence that red blood cell protein p55 may participate in the skeleton-membrane linkage that involves protein 4.1 and glycophorin C. Blood. 1993;82:1323–1327. [PubMed] [Google Scholar]
- 35.Cohen CM, Foley SF, Korsgren C. A protein immunologically related to erythrocyte band 4.1 is found on stress fibres of non-erythroid cells. Nature. 1982;299:648–650. doi: 10.1038/299648a0. [DOI] [PubMed] [Google Scholar]
- 36.Matsuoka Y, et al. Mediation of the physiological response of platelets by interactions of spectrin and protein 4.1 with the cytoskeleton. Biochem Biophys Res Commun. 1994;198:111–119. doi: 10.1006/bbrc.1994.1016. [DOI] [PubMed] [Google Scholar]
- 37.Waugh RE, Sarelius IH. Effects of lost surface area on red blood cells and red blood cell survival in mice. Am J Physiol. 1996;271:C1847–C1852. doi: 10.1152/ajpcell.1996.271.6.C1847. [DOI] [PubMed] [Google Scholar]
- 38.Silveira P, Cynober T, Dhermy D, Mohandas N, Tchernia G. Red blood cell abnormalities in hereditary elliptocytosis and their relevance to variable clinical expression. Am J Clin Pathol. 1997;108:391–399. doi: 10.1093/ajcp/108.4.391. [DOI] [PubMed] [Google Scholar]
- 39.Discher D, Parra M, Conboy JG, Mohandas N. Mechanochemistry of the alternatively spliced spectrin-actin binding domain in membrane skeletal protein 4.1. J Biol Chem. 1993;268:7186–7195. [PubMed] [Google Scholar]
- 40.Discher DE, et al. Mechanochemistry of protein 4.1's spectrin-actin binding domain: ternary complex associations, network integration, structural strengthening. J Cell Biol. 1995;130:897–907. doi: 10.1083/jcb.130.4.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schischmanoff PO, et al. Defining of the minimal domain of protein 4.1 involved in spectrin-actin binding. J Biol Chem. 1995;270:21243–21250. doi: 10.1074/jbc.270.36.21243. [DOI] [PubMed] [Google Scholar]
- 42.Horne WC, Huang SC, Becker PS, Tang TK, Benz EJ., Jr Tissue-specific alternative splicing of protein 4.1 inserts an exon necessary for formation of the ternary complex with erythrocyte spectrin and F-actin. Blood. 1993;82:2558–2563. [PubMed] [Google Scholar]
- 43.Conboy JG, Kan YW, Mohandas N. Molecular cloning of protein 4.1, a major structural element of the human erythrocyte membrane skeleton. Proc Natl Acad Sci USA. 1986;83:9512–9516. doi: 10.1073/pnas.83.24.9512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aster JC, Brinn K, Maisel H, Brewer GJ. Phosphorylation of the band 4.1-like proteins of the bovine lens. Biochem Biophys Res Commun. 1985;126:1166–1173. doi: 10.1016/0006-291x(85)90308-0. [DOI] [PubMed] [Google Scholar]
- 45.Walensky LD, et al. The discrete localization of erythrocyte protein 4.1 in mammalian brain correlates with coordination and learning deficits in knockout mice. Mol Biol Cell. 1997;8(Suppl.):275a.. (Abstr.) [Google Scholar]