Abstract
Since thiols can undergo nitrosation and the cell membrane is rich in thiol-containing proteins, we considered the possibility that membrane surface thiols may regulate cellular entry of NO. Recently, protein disulfide isomerase (PDI), a protein that catalyzes thio–disulfide exchange reactions, has been found on the cell-surface membrane. We hypothesized that cell-surface PDI reacts with NO, catalyzes S-nitrosation reactions, and facilitates NO transfer from the extracellular to intracellular compartment. We observed that PDI catalyzes the S-nitrosothiol–dependent oxidation of the heme group of myoglobin (15-fold increase in the rate of oxidation compared with control), and that NO reduces the activity of PDI by 73.1 ± 21.8% (P < 0.005). To assess the role of PDI in the cellular action of NO, we inhibited human erythroleukemia (HEL) cell-surface PDI expression using an antisense phosphorothioate oligodeoxynucleotide directed against PDI mRNA. This oligodeoxynucleotide decreased cell-surface PDI content by 74.1 ± 9.3% and PDI folding activity by 46.6 ± 3.5% compared with untreated or “scrambled” phosphorothioate oligodeoxynucleotide–treated cells (P < 0.0001). This decrease in cell-surface PDI was associated with a significant decrease in cyclic guanosine monophosphate (cGMP) generation after S-nitrosothiol exposure (65.4 ± 26.7% reduction compared with control; P < 0.05), with no effect on cyclic adenosine monophosphate (cAMP) generation after prostaglandin E1 exposure. These data demonstrate that the cellular entry of NO involves a transnitrosation mechanism catalyzed by cell-surface PDI. These observations suggest a unique mechanism by which extracellular NO gains access to the intracellular environment.
Introduction
Protein disulfide isomerase (PDI) is a homodimeric enzyme that is primarily located within the endoplasmic reticulum (ER) (1, 2) where it assists in the folding of newly synthesized proteins (3). The ability of PDI to catalyze protein disulfide bond formation, reduction, or isomerization (4) is a function of the redox state of the protein substrate and of the thiol redox state of the local microenvironment. The catalytic site of PDI contains a CGHC sequence found in both subunits of the homodimer. Disulfide exchange essentially involves breakage and formation of protein disulfide bonds by thiol–disulfide interchange involving the two active site cysteines, leading to an accumulation of the set of disulfides consistent with the most stable conformational state for a polypeptide. The enzyme shows little specificity in its protein substrates and acts on a wide variety of proteins ranging from small, single-domain proteins to large oligomers with multiple domains. In addition to its isomerase activity, PDI also manifests chaperone-like activity (5) and is also an essential subunit of prolyl-4-hydroxylase (6, 7) and of the microsomal triglyceride transfer protein complex (8).
The location and role of PDI within the ER have been well established (5, 9). In addition, PDI has recently been found on the plasma membrane of several cell types (10, 11), including megakaryocytes and platelets (12, 13). In certain cases, surface-associated PDI is reported to be secreted from cells; e.g., rat exocrine pancreatic cells transport PDI to the plasma membrane and secrete it into the acinar lumen (10). Additionally, activated platelets have been reported to release PDI (12).
Presently, several functions have been attributed to cell-surface PDI. In human thyroid cells, surface PDI is responsible for the shedding of the ectodomain of thyrotropin (14). On the platelet surface, PDI has been shown to catalyze the isomerization of disulfides in surface-associated thrombospondin 1, subsequently altering its binding to neutrophil cathepsin (15). Surface PDI has also been shown to reduce disulfide bonds established between a ligand and a cell-surface receptor. Specifically, Ryser and colleagues (16) have shown that activation and translocation of receptor-bound diphtheria toxin, as well as entry of receptor-bound human immunodeficiency virus, require the presence of cell-surface PDI.
Nitric oxide (NO) is a heterodiatomic free radical with a wide range of physiological actions. Its expanding range of functions includes neurotransmission, regulation of platelet function, blood pressure control, and a role in the immune system's ability to kill tumor cells and intracellular parasites. More specifically, vascular NO induces vasorelaxation and inhibition of platelet function, in part via soluble guanylyl cyclase activation. NO is transported in the vasculature in the form of S-nitrosothiols involving either low–molecular weight thiols or serum albumin (17).
Unlike most small signaling molecules that depend upon interactions with specific receptors, transporters, or channels for their entry into cells, NO is currently thought to diffuse freely from its source of synthesis across effector cell membranes nonspecifically. Because of its relatively low reactivity compared with other biological free radicals, small size, and rapid diffusibility, it is presumed that the key to its regulation lies at the level of synthesis. Furthermore, in contrast to other biological free radicals such as superoxide anion, it has been suggested that the charge neutrality of NO may facilitate its diffusion across cell membranes (18).
However, because thiols can liberate NO from S-nitrosothiols, and under certain conditions can react with nitrosonium equivalents, it is possible that cellular entry of NO may be regulated by transnitrosation reactions with thiols at the level of the plasma membrane. Cell membranes contain a significant thiol pool (19–21), and surface PDI is one member of this pool. There are several reasons why we hypothesized that PDI is a likely candidate for specific reactivity toward NO on the cell surface. Firstly, the active site of PDI includes two cysteinyl residues (hence, two thiol groups), one of which has an anomalously low pK (22), rendering the resulting thiolate anion susceptible for reaction with nitrosonium ion or facilitating its nucleophilic attack of an –S-NO bond. Secondly, the reducing environment on the extracellular surface of cells, which is believed to be maintained by PDI, is crucial for the formation of an –S-NO bond. Thirdly, PDI is involved in thiol–disulfide exchange reactions, which are mechanistically akin to thiol–S-nitrosothiol exchange reactions. And finally, transfer of nitrosonium ions from one thiolate ion to another (transnitrosation) may be an important mechanism for the biological effects of S-nitrosothiols (RSNO). These reactions occur rapidly in vitro and in vivo under physiological conditions and are involved in the biological functions of NO in vivo (23).
Thus, the specific objectives of the present study were to (a) test the ability of PDI to catalyze transnitrosation, (b) study the effect of NO on the thiol–disulfide exchange activity of PDI, and (c) examine the effect of inhibition of cell-surface PDI expression on the intracellular bioavailability of extracellular NO. To address these objectives, we used the human erythroleukemia (HEL) cell line and measured RNase folding activity and NO-dependent cyclic guanosine monophosphate (GMP) accumulation in cells in which PDI mRNA translation is suppressed.
Methods
Reagents and solutions.
RNase T1 was a generous gift of C.N. Pace (Texas A&M University Health Science Center, College Station, Texas, USA) and glutathionyl-RNase T1 (GS-RNase T1) was prepared as described previously (24). PDI was obtained from Pierce Chemical Co. (Rockford, Illinois, USA). Rabbit anti-PDI antibody was obtained from StressGen Biotechnologies Corp. (Victoria, British Columbia, Canada). Antisense (5′-GGCAGCGAGACTCCGAACACGGTA-3′) and “scrambled” (5′-GATGGCACAAGCCTCAGAGCGACGG-3′) phosphorothioate oligodeoxynucleotides, as well as oligofectin F (lipofectin F), were obtained from Sequitur Inc. (Natick, Massachusetts, USA). 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB), reduced and oxidized glutathione (GSH and GSSG, respectively), dithiothreitol (DTT), and lactate dehydrogenase assay kits were obtained from Sigma Chemical Co. (St. Louis, Missouri, USA). Cyclic GMP and cyclic AMP enzyme immunoassay kits were purchased from Cayman Chemical Co. (Ann Arbor, Michigan, USA). Mercurous chloride and sodium nitrite were purchased from Fisher Scientific Co. (Fair Lawn, New Jersey, USA) Sulfanilamide, ammonium sulfamate, and N-(1-naphthyl)ethylenediamine dihydrochloride were obtained from Aldrich Chemical Co. (Milwaukee, Wisconsin, USA). Activated thiol Sepharose 4B was obtained from Pharmacia Biotech Inc. (Piscataway, New Jersey, USA). PBS (10 mM NaH2PO4, 150 mM NaCl; pH 7.4) and Tris-buffered saline (TBS) (10 mM Trizma base, 150 mM NaCl; pH 8.0) were prepared as stock solutions. S-nitroso-BSA (SNO-serum albumin) was prepared as described previously (23).
Preparation of S-nitroso-glutathionyl–Sepharose 4B.
In a previous study, we described and characterized S-nitroso-glutathionyl–Sepharose 4B (SNO-4B) as a very stable and potent RSNO (25). SNO-4B was prepared as previously described (25) and used as a cell-impermeant RSNO. Briefly, freeze-dried, activated thiol Sepharose 4B (4B) was swollen and washed twice with PBS using a vacuum funnel. Free thiols on the Sepharose 4B were generated by suspending the 4B in TBS (1 g 4B per 5 ml TBS; pH 8.0) containing 1% DTT, 0.3 M sodium bicarbonate, and 1 mM EDTA for 40 min while continuously mixing at room temperature. The 4B suspension was then transferred to the vacuum funnel and washed with 0.1 M acetic acid solution containing 0.5 M NaCl and 1 mM EDTA, using a total volume of 400 ml of washing solution for each gram of 4B. The beads were subsequently washed and suspended in PBS. The free thiol level was determined using a modified DTNB assay (26). S-nitrosation of the free thiols on 4B was achieved with NaNO2 in the presence of 0.5 N HCl. The resulting SNO-4B was then washed thoroughly with PBS, and suspended in PBS at concentrations as desired for a given experiment.
Thiol content determination.
The sulfhydryl content of all of the modified forms of RNase T1, SNO-4B beads, and cell membranes was estimated by DTNB titration in 0.3 M Tris-HCl and 1 mM EDTA (pH 7.5) containing 6 M guanidinium chloride. The formation of the 2-nitro-5-thiobenzoate dianion was measured as an increase in absorbance at 412 nm (ελ=412 = 13,600 M–1 cm–1).
Vicinal thiol (defined in cellular and biochemical terms as cysteinyl side chains sufficiently contiguous to engage in redox reactions with each other) (27, 28) content of HEL cells was measured using phenylarsine oxide (PAO). Cells were collected, washed, and suspended in PBS (∼10,000 cells/μl), then treated with a range of concentrations of PAO (0, 10, 100, 200, and 500 μM) for 10 min. The cells were subsequently washed and resuspended in PBS for determination of the membrane vicinal and total thiol content using the DTNB assay as described above with reduced glutathione (GSH) as the standard (26). Total cell thiols were determined without PAO treatment.
Preparation of GS-RNase T1.
GS-RNase T1 was prepared as described by Ruoppolo and colleagues (24). Briefly, RNase T1 (10 mg/ml) in 0.1 M Tris HCl, 1 mM EDTA, and 6 M guanidinium chloride was reduced by incubation with reduced DTT (mol DTT/mol –S-S- = 50:1) for 2 h at 37°C under a nitrogen atmosphere at pH 8.5. After the addition of 0.2 vol of 1 M HCl, the protein was gel filtered on a Sephadex G-25 column, equilibrated, and eluted with 0.01 M HCl. The protein fraction was then recovered, tested for –SH content, and lyophilized.
The fully reduced protein (2 mg/ml) was treated with GSSG (neutralized with Trizma-base) in 0.5 M Tris-HCl and 1 mM EDTA (pH 8.5) containing 6 M guanidinium chloride at room temperature for 3–10 h under a nitrogen atmosphere (mol GSSG/mol protein = 1,000:1). The reaction mixture was acidified with 0.2 vol 1 M HCl, desalted on a Sephadex G-25 column, and eluted with 0.01 M HCl. The protein fraction was recovered, tested for SH content, and lyophilized. The samples were stored at –20°C before use.
RNase folding analysis.
Purified GS-RNase T1 was used to evaluate the folding activity of free or cell-surface PDI (24, 29). The conformational change that occurs in RNase during folding was monitored spectrofluorometrically. GS-RNase T1 (6 μM) was incubated with purified PDI or HEL cell-surface PDI in the presence of 6 mM GSSG and 6 μM GSH in PBS for various times, after which intrinsic GS-RNase T1 fluorescence was monitored; in the case of cell-surface PDI experiments, cells were removed from the incubation mixture by centrifugation (8,000 g for 3 min) before recording the fluorescence spectra. The emission spectra were recorded in 1-cm2 cuvettes in a spectrofluorometer at 18°C (LS-5B; Perkin-Elmer Corp., Norwalk, Connecticut, USA). The intrinsic fluorescence emission spectra of GS-RNase T1 were recorded upon excitation at 278 nm (10-nm wavelength increments, 5-nm excitation and emission slit widths), and then the emission maxima and peak amplitude were determined.
NO-dependent oxymyoglobin oxidation assay.
The role of PDI in transnitrosation was assessed using the conversion of oxymyoglobin to metmyoglobin by NO. Myoglobin (Mb) was chosen rather than hemoglobin because it does not contain any free cysteinyl residues, which could potentially complicate the kinetics of NO transfer with the heme iron group in the reaction. NO release from the NO donor and the subsequent oxidative reaction (Mb-Fe2+-O2 + NO → Mb-Fe2+-O2NO → Mb-Fe3+ + NO3) with the heme moiety of myoglobin was monitored spectrophotometrically (30). Absorbance decreases at 542 nm were followed over time and are indicative of metmyoglobin formation. The reaction mixture contained 100 mM potassium phosphate (pH 7.4), 10 μM oxymyoglobin, 1 mM EDTA, and 30 μM or 3 mM GSNO with and without 0.3 μM or 3 μM PDI, respectively. All solutions were prepared with Chelex-treated water. The extinction coefficient of oxymyoglobin at pH 7.0 is 13.9 mM–1 cm–1 at 580 nm, and that of GSNO is 0.015 mM–1 cm–1 at 545 nm. All measurements were performed using a diode array spectrophotometer (8452A; Hewlett Packard Co., Palo Alto, California, USA).
Transfection of phosphorothioate oligodeoxynucleotides into HEL cells.
The RNA sequence for PDI was obtained from the National Center for Biotechnology Information protein database library for human PDI cDNA. The sequence of the β subunit of prolyl 4-hydroxylase (6), which is identical to PDI, was used to select the antisense phosphorothioate oligodeoxynucleotide sequence (F5). The control, scrambled sequence (CA) was composed of identical nucleotides as the antisense, except that the former are randomly arranged. HEL cells were washed once with Opti-MEM media (GIBCO BRL, Grand Island, New York, USA); then oligofectin F (16.5 μl) was added to 500 μl of media. Next, the F5 and CA were added to the Opti-MEM/oligofectin F lipid mixture to final concentrations of 200 nM, and the suspension was mixed for 10 min by repeatedly inverting the tube. The mixture was then placed in a petri dish containing 5 ml HEL cell suspension (10,000 cells/ml). The cells were incubated for 24 h at 37°C. After transfection, the cells were gently washed and resuspended in PBS until ready for use.
Immunofluorescent staining of HEL cel -surface PDI.
HEL cells were washed with PBS (pH 7.4) ultracentrifuged for 5 min at room temperature, and resuspended in PBS containing 4% paraformaldehyde. After a 1-h fixation period at 4°C, the cells were sedimented, resuspended in PBS, and then allowed to settle under unit gravity. The loosely sedimented cells were placed on poly-L-lysine–coated glass slides and allowed to dry at room temperature. The slides were then incubated for 2 h with a blocking buffer containing dehydrated milk, 5% Tween-20, and Fc antibody fragments (10 mg/ml). The slides were washed three times with PBS and then treated with rabbit anti-PDI IgG for 1 h. After the primary antibody incubation, the slides were again washed three times with PBS for 20 min per wash, incubated with the secondary, FITC-labeled goat anti–rabbit antibody for 1 h, and subsequently washed three times with PBS for 20 min per wash. The slides were allowed to dry at room temperature and sealed with SlowFade (Fisher Scientific Co., Fair Lawn, New Jersey, USA) under No. 1 coverslips (Fisher Scientific Co.). Once dried and properly sealed, the slides were examined by confocal microscopy. To ensure that the binding of the primary antibody was not a consequence of Fc receptor binding, we performed studies on HEL cells using Fc fragments alone. We observed no fluorescence when Fc fragments were incubated with the HEL cells in place of the primary antibody.
Confocal scanning microscopy.
The HEL cells were analyzed with a Leica upright confocal laser scanning microscope (Leica AG, Heerbrugg, Switzerland) equipped with an argon ion laser (output power of 250 mW) and two photomultiplier tubes. The apertures were set at the minimum size for optimal signal detection. The typical z-series of images are composed of sequential en face optical sections in the x–y plane, which lies at right angles to the vertical axis of the tissue. When analyzing the cells, confocal optical sections were scanned at 1.0-μm intervals in the 512- × 512-pixel format.
Protein concentration determination.
Protein concentrations were determined by the bicinchoninic acid assay (Pierce Chemical Co.) with BSA as the standard.
PDI determination by fluorescence immunoassay.
HEL cells were washed with PBS (pH 7.4) ultracentrifuged for 5 min at room temperature, and resuspended in PBS. The cells were then incubated for 1 h with a blocking buffer containing 1% filtered dehydrated milk and Fc antibody fragments (10 mg/ml). The cells were sedimented three times, resuspended in PBS, and then treated with rabbit anti-PDI IgG for 45 min. After the primary antibody incubation, the cells were again washed three times with PBS, incubated with the secondary, FITC-labeled goat anti–rabbit antibody for 45 min, and subsequently washed two times with PBS. The cells were counted using a Coulter counter (Coulter Immunology, Hialeah, Florida, USA). A final wash was then performed using PBS with 1% Triton X-100. The lysed, solubilized cell suspension was sedimented by ultracentrifugation, the supernatant removed, and the fluorescence intensity measured at an emission wavelength of 515 nm with an excitation wavelength of 494 nm.
Data analysis.
The data are expressed as the mean ± SD for individual experiments. One- and two-way ANOVA was performed followed by multiple comparison analysis as appropriate. The results of any analysis were considered to be significant when P ≤ 0.05.
Results
Membrane thiol content.
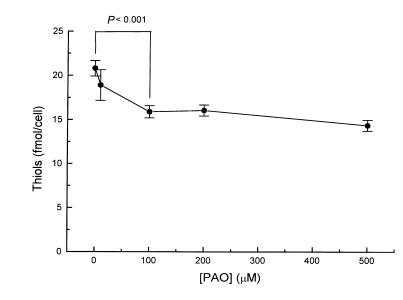
The total free thiol content on the HEL cell surface, as detected with DTNB (which does not enter cells under these conditions as determined by absorbance measurements of cytosolic extracts), was 21.0 ± 0.9 fmol/cell (n = 3). To quantify the fraction of the total cell-surface thiol pool that is vicinal dithiol in conformation, we exposed cells to PAO before the DTNB reaction. Pretreatment of cells with increasing concentrations of PAO, a membrane vicinal thiol blocker, led to a dose-dependent decrease in total membrane thiol content. As shown in Fig. 1, PAO maximally decreased the thiol level by 23.6 ± 3.8% at 100 μM PAO (n = 3). Higher concentrations did not produce any greater decrease in measurable thiols.
Figure 1.
Quantification of cell-surface thiols. HEL cells were incubated for 10 min with increasing concentrations of PAO, and the remaining free surface thiols were measured by the DTNB assay. We observed that each cell contains 20 ± 0.9 fmol of thiols in which 23.6 ± 3.8% are vicinal dithiols. DTNB, 5,5′-dithiobis(2-nitrobenzoic acid); HEL, human erythroleukemia; PAO, phenylarsine oxide.
Cell-surface PDI pool.
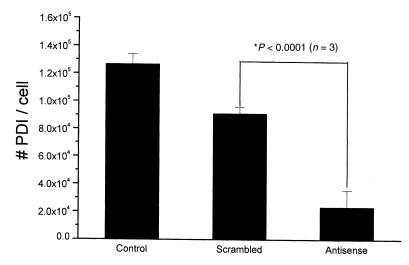
We next performed immunolocalization studies to determine whether or not PDI is present on the surface of HEL cells. Figure 2 shows the results of immunofluorescent staining of HEL cells using confocal microscopy. Control cells, cells treated with oligofectin F, and cells treated with scrambled antisense phosphorothioates against PDI mRNA all showed fluorescence staining localized to the periphery of the plasmalemmal rim of the cells (Fig. 2a). Transfection of cells with an antisense phosphorothioate against PDI mRNA, however, led to a marked reduction in peripheral immunofluorescence (Fig. 2b). This treatment did not appreciably affect cell viability (>95% trypan blue exclusion in both antisense and scrambled phosphorothioate–treated cells and no significant LDH release). In addition, Western blot analysis showed a significant reduction in total cellular PDI content (Fig. 3). Cell-surface PDI content was also quantified by fluorescence immunoassay and found to be reduced by 74.1 ± 9.2% with antisense compared with scrambled phosphorothioate treatment (P < 0.001) (Fig. 4).
Figure 2.
Suppression of cell-surface PDI expression. HEL cells were treated with an IgG polyclonal antibody against PDI and a secondary FITC-labeled anti–rabbit antibody. We observed under confocal microscopy a significant decrease in cell-surface PDI expression when HEL cells were preincubated with antisense phosphorothioate against PDI mRNA for 24 h (a) compared with the scrambled control (b). PDI, protein disulfide isomerase.
Figure 3.
Effects of antisense phosphorothioate oligodeoxynucleotides on PDI content. The total PDI content of HEL cells was assessed by immunoblotting analysis. We observe a significant decrease in total PDI (lowest band, ∼57 kDa) in the HEL cells preincubated with antisense phosphorothioates against PDI mRNA compared with controls (scrambled phosphorothioates).
Figure 4.
Suppression of cell-surface PDI expression. Antisense phosphorothioate blockade of PDI translation suppressed the expression of cell-surface PDI by 74.1 ± 9.2% compared with scrambled phosphorothioates, as detected by fluorescent-labeled anti-PDI antibody.
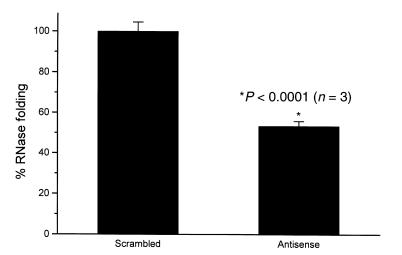
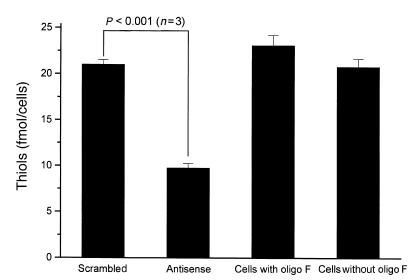
The functional consequences of this decrease in cell-surface PDI were next determined. There was a 46.6 ± 3.5% reduction in cell-surface PDI folding activity using RNase T1 as a substrate (P < 0.001) (Fig. 5). In addition, there was a 53.6 ± 2.3% reduction in total free thiols on the cell surface (21 ± 0.5 fmol/cell vs. 9.8 ± 0.5 fmol/cell), which supports the view that cell-surface PDI maintains the thiol redox state of the surface thiol pool (Fig. 6).
Figure 5.
Reduction in cell-surface PDI folding activity with antisense treatment. Refolding of GS-RNase T1 was performed at 25°C in PBS (pH 7.4). RNase folding activity was measured as extent of folding at 45 min; 100% activity corresponds to the maximal folding by control HEL cells. GS-RNase T1 (6 μM final concentration) was added to incubation mixtures containing 10,000 cells/μl. In antisense-treated cells, folding activity decreased by 46.6 ± 3.5% compared with control. GS-RNase, glutathionyl-RNase T1.
Figure 6.
PDI maintains the redox state of cell-surface thiols. HEL cells were treated with an antisense phosphorothioate against PDI mRNA. We observed a 53.6 ± 2.3% decrease in surface thiols, as detected by the DTNB assay.
Interaction of PDI and NO.
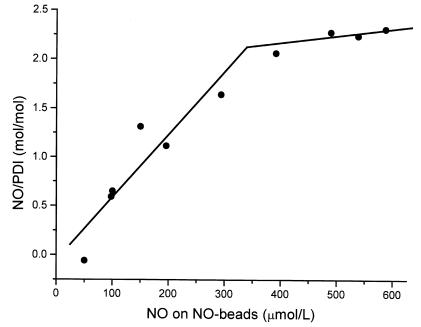
To determine the maximal stoichiometry between nitrosonium equivalents and PDI under steady-state conditions, we incubated PDI with increasing concentrations of SNO-4B beads, providing a 50–600-fold molar excess of NO equivalents over PDI, and determined the concentration of NO bound to PDI (30). PDI appeared saturated with NO at a ratio of two moles of NO per mole of PDI monomer (Fig. 7), and this reaction involved an –S-NO intermediate presumably at the active site thiol, as demonstrated by a commensurate loss of DTNB-detectable thiol on PDI (data not shown). We also examined the effect of S-nitrosation on the folding activity of cell-surface PDI to determine whether or not S-nitrosation modulated PDI function. After treatment of HEL cells with increasing concentrations of SNO-4B, PDI folding activity decreased by 17.2 ± 21.1% and 70.1 ± 21.8% (at 50 and 100 μM –S-NO, or 50 and 100 nmol NO equivalents/103 beads, respectively) (P < 0.005).
Figure 7.
PDI interacts with NO. PDI (1 μM) was incubated for 10 min with an increasing concentration of SNO-4B. Using the Saville assay, we found that NO binds to PDI in a dose-dependent manner with an NO/PDI ratio of 2:1 at saturation. SNO-4B, S-nitroso-glutathionyl–Sepharose 4B.
Additionally, because PDI's active site thiols may engage in oxidation reactions with RSNO, we reasoned that PDI may also catalyze transnitrosation reactions. To test this hypothesis, we used horse heart oxymyoglobin as a means to monitor the release of NO from GS-NO, following the rate of decrease in absorbance at 542 nm. PDI caused a 15-fold increase in the rate of oxidation of myoglobin by GS-NO compared with enzyme-free control (t1/2 = 1 min vs. t1/2 = 15 min; n = 3, P < 0.05); this effect was eliminated by heat denaturation of PDI under N2, which produced approximately 10% oxidation of protein thiols. Thus, PDI appears to catalyze transnitrosation reactions under physiologically relevant conditions.
Role of PDI in NO-induced cyclic GMP production in HEL cells.
As a measure of the potential physiological relevance of cell-surface PDI–dependent transnitrosation reactions, we investigated the role of cell-surface PDI on the NO-induced increase in intracellular cyclic GMP (Fig. 8). HEL cells that had been transfected with antisense or scrambled phosphorothioates were incubated with 50 μM SNO-4B for 5 minutes at 25°C, and cell cGMP content was measured by ELISA. The NO-induced cGMP increase was reduced by 65.4 ± 26.7% (2.41 ± 0.21 pmol/105 cells vs. 0.51 ± 0.41 pmol/105 cells; P < 0.05) when cell-surface PDI expression was inhibited with PDI antisense phosphorothioates compared with scrambled control. We performed a similar set of experiments (under identical conditions, except for the addition of 1 mM DTPA to the incubation solution) using 1 μM SNO-serum albumin (a physiologically relevant concentration) (23). We observed that the NO-induced cGMP increase was reduced under these conditions by 77.0 ± 14.8% (1.92 ± 0.11 pmol/105 cells vs. 0.44 ± 0.08 pmol/105 cells; P < 0.05) by inhibition of cell-surface PDI expression.
Figure 8.
PDI, NO, and soluble guanylyl cyclase activation. To determine the role of PDI in the transfer of NO into HEL cells, we measure intracellular cyclic GMP levels. We observed a 65.4 ± 26.7% decrease in cyclic GMP levels in antisense phosphorothioate–treated cells incubated with 10 μM SNO-4B for 10 min compared with scrambled phosphorothioate–treated controls.
To determine the specificity of the impairment of guanylyl cyclase activation, we performed an additional set of control experiments. We first examined the consequences of suppression of cell-surface PDI expression on prostaglandin E1–stimulated cyclic AMP formation in HEL cells and found no difference compared with control cells: 212 pmol cyclic AMP/105 cells for cells treated with PDI antisense phosphorothioates and 189 pmol cyclic AMP/105 cells for control cells, each treated with 10 μM prostaglandin E1 for 5 min. These results suggest that the effect of suppressing PDI expression on the cell surface is a consequence of alterations in the mechanism of access of NO to the cell cytosol and not a consequence of a generalized abnormality in protein folding in the endoplasmic reticulum that affects other related proteins or enzymes. These results are consistent with the uniform viability of cells treated with antisense phosphorothioates, and with the selective loss of PDI from the cell surface with adequate concentrations retained in the endoplasmic reticulum. The KDEL sequence in PDI, in fact, ensures preferential retention of PDI in the endoplasmic reticulum unless and until a critical threshold concentration is reached that facilitates translocation to the plasma membrane; under the conditions of antisense treatment used in these experiments, this threshold is not exceeded.
Discussion
In these studies, we have shown that the cell-surface thiol content of HEL cells is ∼21 fmol/cell, of which vicinal thiols comprise 23.6%. PDI contributes to this cell-surface vicinal thiol population but only accounts for ∼0.025% of the total pool. Using PDI antisense phosphorothioates, we reduced cell-surface PDI content significantly without altering cell viability. Antisense-treated cells also exhibited a marked reduction in surface thiol content compared with control cells, thereby demonstrating that PDI plays a significant role in maintaining the redox state of cell-surface thiols. We further demonstrated that PDI effectively undergoes S-nitrosation and also catalyzes transnitrosation. Inhibition of cell-surface PDI expression by antisense phosphorothioates directed against PDI mRNA significantly decreases in a selective manner the RSNO-induced cyclic GMP increase in HEL cells. These studies demonstrate a novel mechanism by which NO entry into cells is regulated, and consequently suggest another mechanism by which the bioactivity of NO may be modulated.
In this study, we successfully used antisense technology to suppress the expression of cell-surface PDI. In the past, many inhibitors of PDI have been used, but most lack either specificity or efficacy in inhibiting PDI folding activity. For example, bacitracin has been used as a putative selective PDI inhibitor to demonstrate the presence of PDI on the plasma membrane of Chinese hamster ovary cells. However, the high concentrations (–3 mM) required for its inhibitory action call into question its specificity as an inhibitor. Several PDI antibodies have been generated, and although they were specific in binding to PDI, few have demonstrated significant inhibitory activity toward the enzyme. Because PDI is >90% homologous to the T3 receptor (32, 33), it has been hypothesized that T3 might be used as a specific PDI inhibitor. At equilibrium, T3 binds to PDI at two binding sites, a high-affinity site (Kd = 21 nM) and a very low-affinity site (Kd = 100 μM). Despite the high-affinity site, however, T3 is not an effective inhibitor of PDI activity. Recent molecular modeling studies by us using the coordinates of the PDI active site amino acid residues derived from nuclear magnetic resonance (NMR) studies of the PDI active site failed to demonstrate an energetically favorable association of T3 with the active site (data not shown).
To decrease the presence of cell-surface PDI selectively, we therefore constructed three antisense phosphorothioates, with their accompanying scrambled sequences as controls, against the mRNA structure of human PDI. Only one sequence (5′-GGCAGCGAGACTCCGAACACGGTA-3′) exhibited marked inhibition of PDI expression; the other two did not (5′-CTCTGCCGTCAGCTCCTCCGATTCG-3′ and 5′-CGTGCGTTCCCCGTTGTAATCAATG-3′). With the active sequence, we were able to suppress cell-surface PDI expression significantly. Because PDI is important for protein folding, we questioned whether this suppression might cause the cells to die. Cell viability assays demonstrated that the cells remained viable under our transfection conditions, arguing that only a fraction of total PDI is required for cell survival. It has been shown that in yeast, PDI is a necessary protein for cell viability (34, 35); a PDI-null strain could not be rescued by overexpression of thioredoxin. However, when a mutant of thioredoxin in which the active site sequence (CGPC) was mutated to the PDI sequence (CGHC), the yeast were able to survive (36) even though the mutant thioredoxin had only 10% of the folding activity of PDI in vitro (37). These studies confirm our findings that minimal levels of PDI are sufficient for cell survival.
The importance of cell-surface thiols in many cellular functions has been amply demonstrated in several cell types. Cell-surface thiols are critical determinants of lymphocyte activation and proliferation (38). The cell-surface thiol redox state is also an important regulator of the ability of natural killer lymphocytes to bind to and lyse target tumor cells (39). The thiol redox state of platelet-surface glycoproteins is also an essential determinant of activation and aggregation responses (40). Owing to the role of PDI in maintaining the thiol redox state of cell-surface proteins, the cell surface–bound pool of this enzyme is essential for the surface-dependent functions of many cell types. Data presented here support this view in that suppressing cell-surface expression of PDI leads both to a significant reduction in the reduced thiol pool on the surface of HEL cells and to changes in the access of extracellular NO to the intracellular environment where it activates guanylyl cyclase.
Most investigators have assumed that NO traverses cell membranes freely, because of its relative lipophilicity and modest reactivity (compared with other biologically relevant free radicals, such as superoxide anion and hydroxyl radical). Yet, the natural occurrence of RSNOs and the importance of transnitrosation reactions in the transfer, bioactivity, and metabolism of nitric oxide (23) suggest that cell-surface thiols may engage in reactions that regulate the transfer of NO from the extracellular to intracellular environments. The data presented here support this view and suggest that cell-surface PDI catalyzes this transmembrane transfer of nitric oxide, facilitating its entry into cells from an RSNO source and, thereby, promoting its intracellular bioactivity (activation of guanylyl cyclase).
The precise chemical mechanism by which thiols liberate NO from S-nitrosothiols under physiological conditions remains controversial. The homolytic cleavage of the –S-N bond is unlikely even if a disulfide bond were to result from the reaction between two thiyl radicals. Superoxide anion can release NO from S-nitrosothiols; however, it is doubtful that this reaction mechanism is biologically significant. The most likely mechanism involves the reductive release of NO with the consequent formation of the disulfide (41): 2RSNO + 2R′SH → 2NO· + 2RSH + R′SSR′.
Another possible mechanism proceeds via the formation of a disulfide radical anion, which is a very strong reducing agent (42): RSNO + RSH → RS·–SR + NO· + H+.
The mechanism(s) by which PDI catalyzes transnitrosation is as yet unknown. Possibilities include direct transnitrosation (i.e., transfer from an RSNO donor to a membrane or transmembrane R′SH acceptor, RSNO + R′SH ⇔ RSH + R′SNO); transnitros(yl)ation (i.e., transfer from an RSNO to non-heme iron); or maintenance of the specific cell surface–reduced thiol pool(s) to which direct (i.e., non-catalyzed) transnitrosation occurs from the extracellular RSNO donor. In addition, whether or not cell-surface PDI catalyzes the transfer of NO from an intracellular RSNO pool to the extracellular surface or to extracellular R′SH acceptor(s) is as yet unknown. These possible mechanistic issues are the subject of ongoing investigation.
Acknowledgments
The authors wish to thank Stephanie Tribuna for expert technical assistance, and J. Kemmink (European Molecular Biology Laboratory, Heidelberg, Germany) for kindly providing the PDI active site. This work was supported, in part, by National Institutes of Health grants HL53919, HL48743, and HL53993; by a Merit Review Award from the US Veterans Administration; and by a grant from NitroMed Inc. (Bedford, Massachusetts, USA).
References
- 1.Lambert N, Freedman R. The latency of rat liver microsomal protein disulfide-isomerase. Biochem J. 1985;228:635–645. doi: 10.1042/bj2280635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Koch G. Reticuloplasmins: a novel group of proteins in the endoplasmic reticulum. J Cell Sci. 1985;87:491–492. doi: 10.1242/jcs.87.4.491. [DOI] [PubMed] [Google Scholar]
- 3.Creighton TE, Hillson D, Freedman R. Catalysis by protein-disulphide isomerase of the unfolding and refolding of proteins with disulphide bonds. J Mol Biol. 1980;142:43–62. doi: 10.1016/0022-2836(80)90205-3. [DOI] [PubMed] [Google Scholar]
- 4.Gilbert H. Molecular and cellular aspects of thiol-disulfide exchange. Adv Enzymol Relat Areas Mol Biol. 1990;63:69–172. doi: 10.1002/9780470123096.ch2. [DOI] [PubMed] [Google Scholar]
- 5.Gilbert H. Protein disulfide isomerase and assisted protein folding. J Biol Chem. 1997;272:29399–29402. doi: 10.1074/jbc.272.47.29399. [DOI] [PubMed] [Google Scholar]
- 6.Pihlajaniemi T, et al. Molecular cloning of the beta-subunit of human prolyl 4-hydroxylase. This subunit and protein disulphide isomerase are products of the same gene. EMBO J. 1987;6:643–649. doi: 10.1002/j.1460-2075.1987.tb04803.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koivu J, et al. A single polypeptide acts both as the beta subunit of prolyl 4-hydroxylase and as a protein disulfide-isomerase. J Biol Chem. 1987;262:6447–6449. [PubMed] [Google Scholar]
- 8.Wetterau J, Combs K, Spinner S, Joiner B. Protein disulfide isomerase is a component of the microsomal triglyceride transfer protein complex. J Biol Chem. 1990;265:9801–9807. [PubMed] [Google Scholar]
- 9.Freedman R, Hirst T, Tuite M. Protein disulfide isomerase: building bridges in protein folding. Trends Biochem Sci. 1994;19:331–336. doi: 10.1016/0968-0004(94)90072-8. [DOI] [PubMed] [Google Scholar]
- 10.Akagi S, et al. Localization of protein disulfide isomerase on plasma membranes of rat exocrine pancreatic cells. J Histochem Cytochem. 1988;36:1069–1074. doi: 10.1177/36.8.3292644. [DOI] [PubMed] [Google Scholar]
- 11.Kroning H, Kahne T, Ittenson A, Franke A, Ansorge S. Thiol-proteindisulfide-oxidoreductase (proteindisulfide isomerase): a new plasma membrane constituent of mature human B lymphocytes. Scand J Immunol. 1994;39:346–350. doi: 10.1111/j.1365-3083.1994.tb03384.x. [DOI] [PubMed] [Google Scholar]
- 12.Chen K, Lin Y, Detwiler T. Protein disulfide isomerase activity is released by activated platelets. Blood. 1992;79: 2226–2228. [PubMed] [Google Scholar]
- 13.Essex D, Chen K, Swiatkowska M. Localization of protein disulfide isomerase to the external surface of the platelet plasma membrane. Blood. 1995;86:2168–2173. [PubMed] [Google Scholar]
- 14.Couet J, et al. Cell surface protein disulfide-isomerase is involved in the shedding of human thyrotropin receptor ectodomain. Biochemistry. 1996;35:14800–14805. doi: 10.1021/bi961359w. [DOI] [PubMed] [Google Scholar]
- 15.Hotchkiss K, Chesterman C, Hogg P. Catalysis of disulfide isomerization in thrombospondin 1 by protein disulfide isomerase. Biochemistry. 1996;35:9761–9767. doi: 10.1021/bi9603938. [DOI] [PubMed] [Google Scholar]
- 16.Ryser HJ, Levy EM, Mandel R, DiSciullo GJ. Inhibition of human immunodeficiency virus infection by agents that interfere with thiol-disulfide interchange upon virus-receptor interaction. Proc Natl Acad Sci USA. 1994;91:4559–4563. doi: 10.1073/pnas.91.10.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Stamler J, Singel D, Loscalzo J. Biochemistry of nitric oxide and its redox-activated forms. Science. 1992;258:1898–1902. doi: 10.1126/science.1281928. [DOI] [PubMed] [Google Scholar]
- 18.Goretski J, Hollocher T. Trapping of nitric oxide produced during denitrification by extracellular hemoglobin. J Biol Chem. 1988;263:2316–2323. [PubMed] [Google Scholar]
- 19.Ammon H, Hagele R, Youssif N, Eujen R, El-Amri N. A possible role of intracellular and membrane thiols of rat pancreatic islets in calcium uptake and insulin release. Endocrinology. 1983;112:720–726. doi: 10.1210/endo-112-2-720. [DOI] [PubMed] [Google Scholar]
- 20.Rank B, Hebbel R, Carlsson J. Oxidation of membrane thiols in sickle erythrocytes. Prog Clin Biol Res. 1984;165:473–477. [PubMed] [Google Scholar]
- 21.Redelman D, Hudig D. Cell surface thiols, methylation, and complement-like components are involved in the early events of CML whereas proteases participate in the later, Ca++-dependent events. Adv Exp Med Biol. 1985;184:527–534. doi: 10.1007/978-1-4684-8326-0_34. [DOI] [PubMed] [Google Scholar]
- 22.Kortemme TN, Darby NJ, Creighton TE. Electrostatic interactions in the active site of the N-terminal thioredoxin-like domain of the protein disulfide isomerase. Biochemistry. 1996;35:14503–14511. doi: 10.1021/bi9617724. [DOI] [PubMed] [Google Scholar]
- 23.Scharfstein J, et al. In vivo transfer of NO between a plasma protein-bound reservoir and low molecular weight thiols. J Clin Invest. 1994;94:1432–1439. doi: 10.1172/JCI117480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ruoppolo M, Freedman R. Protein-S-S-glutathione mixed disulfides as models of unfolded proteins. Biochemistry. 1994;33:7654–7662. doi: 10.1021/bi00190a020. [DOI] [PubMed] [Google Scholar]
- 25.Liu Z, Rudd MA, Freedman JE, Loscalzo J. S-transnitrosation reactions are involved in the metabolic fate and biological actions of nitric oxide. J Pharm Exp Ther. 1998;284:526–534. [PubMed] [Google Scholar]
- 26.Albano E, Poli G, Chiarpotti E, Biasi F, Dianzani M. Paracetamol-stimulated lipid peroxidation in isolated rat and mouse hepatocytes. Chem Biol Interact. 1983;47:249–263. doi: 10.1016/0009-2797(83)90161-8. [DOI] [PubMed] [Google Scholar]
- 27.Frost SC, Lane MD. Evidence for the involvement of vicinal sulfhydryl groups in insulin-activated hexose transport by 3T3-L1 adipocytes. J Biol Chem. 1985;260:2646–2652. [PubMed] [Google Scholar]
- 28.Gitler C, Mogyoros M, Kalef E. Labeling of protein vicinal dithiols: role of protein-S2 to protein-(SH)2 conversion in metabolic regulation and oxidative stress. Methods Enzymol. 1994;233:403–415. doi: 10.1016/s0076-6879(94)33047-6. [DOI] [PubMed] [Google Scholar]
- 29.Ruoppolo M, Freedman R. Refolding by disulfide isomerization: the mixed disulfide between ribonuclease T1 and glutathione as a model refolding substrate. Biochemistry. 1995;34:9380–9388. doi: 10.1021/bi00029a014. [DOI] [PubMed] [Google Scholar]
- 30.Doyle MP, Hoekstra JW. Oxidation of nitrogen oxides by bound dioxygen in hemoproteins. J Inorg Chem. 1981;14:351–358. doi: 10.1016/s0162-0134(00)80291-3. [DOI] [PubMed] [Google Scholar]
- 31.Saville B. A scheme for the colorimetric determination of microgram amounts of thiols. Analyst. 1958;83:670–672. [Google Scholar]
- 32.Yamauchi K, et al. Sequence of membrane-associated thyroide hormone binding protein from bovine liver: its identity with protein disulfide isomerase. Biochem Biophys Res Commun. 1987;146:1485–1492. doi: 10.1016/0006-291x(87)90817-5. [DOI] [PubMed] [Google Scholar]
- 33.Horuichi R, et al. Purification and characterization of 55-kDa protein with 3,5,3′-triiodo-l-thyronine-binding activity and protein disulfide-isomerase activity from beef liver membrane. Eur J Biochem. 1989;183:529–538. doi: 10.1111/j.1432-1033.1989.tb21081.x. [DOI] [PubMed] [Google Scholar]
- 34.Farquhar R, et al. Protein disulfide isomerase is essential for viability in Saccharomyces cerevisiae. Gene. 1991;108:81–89. doi: 10.1016/0378-1119(91)90490-3. [DOI] [PubMed] [Google Scholar]
- 35.LaMantia M, Lennarz W. The essential function of yeast protein disulfide isomerase does not reside in its isomerase activity. Cell. 1993;74:899–908. doi: 10.1016/0092-8674(93)90469-7. [DOI] [PubMed] [Google Scholar]
- 36.Chivers P, Laboissiere M, Raines R. The CXXC motif: imperatives for the formation of native disulfide bonds in the cell. EMBO J. 1996;15:2659–2667. [PMC free article] [PubMed] [Google Scholar]
- 37.Lundstrom J, Krause G, Holmgren A. A Pro to His mutation in active site of thioredoxin increases its disulfide-isomerase activity 10-fold. New refolding systems for reduced or randomly oxidized ribonuclease. J Biol Chem. 1992;167:9047–9052. [PubMed] [Google Scholar]
- 38.Lawrence DA, Song R, Weber P. Surface thiols of human lymphocytes and their changes after in vitro and in vivo activation. J Leukoc Biol. 1996;60:611–618. doi: 10.1002/jlb.60.5.611. [DOI] [PubMed] [Google Scholar]
- 39.Ristow S, Starkey J, Stanford D, Davis W, Brooks C. Cell surface thiols, but not intracellular glutathione, are essential for cytolysis by a cloned murine natural killer cell line. Immunol Invest. 1985;14:401–414. doi: 10.3109/08820138509047608. [DOI] [PubMed] [Google Scholar]
- 40.Pasche B, Ouimet H, Francis S, Loscalzo J. Structural changes in platelet glycoprotein IIb/IIIa by plasmin: determinants and functional consequences. Blood. 1994;83:404–414. [PubMed] [Google Scholar]
- 41.Crow JP, Beckman JS. Reactions between nitric oxide, superoxide, and peroxynitrite: footprints of peroxynitrite in vivo. Adv Pharmacol. 1995;34:17–43. doi: 10.1016/s1054-3589(08)61079-0. [DOI] [PubMed] [Google Scholar]
- 42.Koppenol WH. The centennial of the Fenton reaction. Free Radic Biol Med. 1993;15:645–651. doi: 10.1016/0891-5849(93)90168-t. [DOI] [PubMed] [Google Scholar]