SUMMARY
Melanoma is an invasive malignancy with a high frequency of blood-borne metastases, but circulating tumor cells (CTCs) have not been readily isolated. We adapted microfluidic CTC capture to a tamoxifen-driven B-RAF/PTEN mouse melanoma model. CTCs were detected in all tumor-bearing mice, rapidly declining after B-RAF inhibitor treatment. CTCs were shed early from localized tumors and a short course of B-RAF inhibition following surgical resection was sufficient to dramatically suppress distant metastases. The large number of CTCs in melanoma-bearing mice enabled comparison of RNA sequencing profiles with the matched primary tumor. A mouse melanoma CTC-derived signature correlated with invasiveness and cellular motility in human melanoma. In patients with metastatic melanoma, CTCs were detected in smaller numbers in patients with metastatic melanoma and declined with successful B-RAF targeted therapy. Together, the capture of CTCs and their molecular characterization provide insight into the hematogenous spread of melanoma.
INTRODUCTION
Recent advances in the treatment of metastatic melanoma have altered the outlook in this previously refractory cancer. In patients with metastatic B-RAF mutant melanoma, dramatic albeit transient responses to B-RAF-MAPK blockade require timely monitoring of drug response so as to adjust treatment options (Flaherty et al., 2012). The identification of effective treatments for metastatic disease suggests that these drugs may be applied even more successfully in patients with earlier disease, such as Stage II and III, where microscopic distant metastases may be eradicated by targeted therapies (Balch et al., 2009). However, identifying high-risk patients who should receive such postoperative “adjuvant” therapy is challenging using the current clinical staging and would benefit from more reliable indicators of tumor invasiveness and recurrence risk. For patients with advanced melanoma as well as those with localized disease, the analysis of circulating tumor cells (CTCs) may therefore provide a novel biomarker to guide therapeutic decisions.
In addition to diagnostic applications, the detailed molecular characterization of melanoma cells circulating in the bloodstream may yield new insights into the process of melanoma metastasis (Liu et al., 2011; Ramskold et al., 2012). However, there are significant challenges: CTCs are rare even in patients with advanced cancer. Moreover, melanomas do not express the classical epithelial cell surface marker EpCAM, which has formed the basis for most CTC isolation strategies (Yu et al., 2011). Some melanoma-specific cell surface epitopes have been proposed for CTC enrichment (Khoja et al., 2013) and the large size of tumor cells within primary melanomas has led to the application of filtering strategies to isolate melanoma CTCs (De Giorgi et al., 2010), although recent studies have suggested that melanoma CTCs may span a wide range of cell size (Ozkumur et al., 2013). Given the difficulty in isolating whole melanoma CTCs, RT-PCR-based measurements of blood-derived melanoma-specific transcripts have also been employed. In patients with locally invasive tumors, positive PCR signals in such assays are associated with a poor prognosis and an increased risk of distant metastasis (Hoshimoto et al., 2012). Taken together, multiple approaches suggest the presence of melanoma cells in the bloodstream of patients with various stages of disease, but a robust cell capture platform is essential for their efficient detection and molecular characterization.
Here, we adapted a microfluidic platform, the HBCTC-Chip (Stott et al., 2010), to capture melanoma CTCs using panels of antibodies against melanoma-specific cell surface markers, followed by immunofluorescence (IF) staining for melanoma antigens and optimized on-chip imaging. We applied this HBCTC-Chip to a robust, inducible B-RAF-PTEN-driven mouse melanoma model (Dankort et al., 2009), in which we validated capture of bona fide melanoma CTCs, monitored their response to targeted therapy and the timing of CTC generation by localized lesions. The large number of CTCs generated in the mouse melanoma model, together with the consistent genetic background, made it possible to compare RNA sequencing profiles of matched primary tumors, metastatic lesions and CTCs, generating a melanoma CTC signature that identified high-risk subsets in human melanoma-derived specimens. Based on its validation in the mouse model, we applied the melanoma HBCTC-Chip to a pilot cohort of human samples.
Results
Microfluidic isolation of CTCs and their response to B-RAF targeted therapy in a mouse melanoma model
Given the considerable heterogeneity of human melanoma, we sought to adapt the HBCTC-Chip to capture cells from a genetically-engineered mouse model of melanoma (Dankort et al., 2009). In this model, subcutaneous injection of tamoxifen at the flank of the animal leads to activation of oncogenic B-RAFV600E, coincident with deletion of PTEN within melanocytes. Melanomas are formed at the injection site with 100% penetrance within 3 weeks, with distant cutaneous metastases appearing at 6-7 weeks (Figure 1A). Following testing B-RAF/PTEN-driven mouse tumors for expression of multiple lineage-specific markers, we selected antibodies against the cell surface epitopes CSPG4 and MCAM for CTC capture, and antibody against the melanoma marker S100 for staining and imaging of captured CTCs (Figure S1A-D). We collected and processed blood specimens from tumor-bearing mice (Yu et al., 2012), passing blood through anti-CSPG4 and anti-MCAM functionalized HBCTC-Chips.
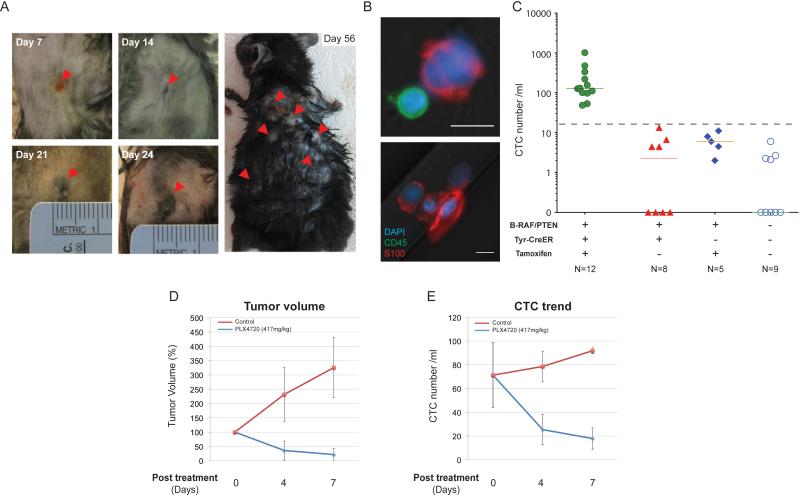
Figure 1. Identification of CTCs in the B-RAFCA/+/PTENflox/flox mouse melanoma model.
(A) Representative images of melanoma induced by focal tamoxifen injection. Arrowheads show tumor progression at the injection site from day 7 to day 24 (left) and cutaneous metastasis at day 56 after tumor induction (right). (B) Upper panel, representative image of a mouse melanoma CTC adjacent to a leukocyte. Lower panel, a cluster of four CTCs. DAPI, blue; CD45, green; S100, red; scale bars, 10μm. (C) Quantification of CTCs from a cohort of tumor-bearing (green, N=12) and control mice (red, genotype-matched tumor-free mice, N=8; blue, Tyr-CreER- mice received tamoxifen injection, N=5; open circles, syngeneic C57BL/6 mice, N=9). Solid lines, median CTC counts. Dashed line, threshold of ≥14 S100+ cells/ ml. Tam, tamoxifen. Concordant change in (D) tumor volume and (E) CTCs in tumor-bearing mice fed with control chow or PLX4720-containing chow are shown (PLX4720, blue; control, red) (Mean value, error bars represent standard deviation). (See also Figure S1)
Captured cells were stained for S100 and the common leukocyte marker CD45 and CTCs were defined as S100+/CD45− (Figure 1B). As for all fluorescence-based CTC imaging, we set a baseline for positive scoring using tumor-free controls (Yu et al., 2012): under the established imaging conditions, a median of two S100+/CD45− cells/ml of blood (mean 3; range 0-13 cells/ml) was observed in non-tumor bearing mice (Tyr-CreER+ mice without tamoxifen treatment, N=8; Tyr-CreER- mice with tamoxifen injection, N=5; syngeneic C57BL/6 mice, N=9). All of 12 melanoma-bearing mice (100%) scored positive for CTCs using a threshold of ≥ 14 S100+/CD45− cells/ml, with a median 114 cells/ml (mean 232; range 48-1015 cells/ml) (Figure 1C). The CTCs were significantly larger than leukocytes (median size 14 um, range 13-18um), and CTC clusters of 2-4 cells were also recovered (Figure 1B) as reported in other cancer types (Stott et al., 2010).
The CTC count rose progressively with increasing tumor burden following tumor induction (Figure S1E). Oral administration of the selective B-RAF inhibitor PLX4720 resulted in a concordant reduction in both the size of the primary tumor and the number of CTCs. A >50% drop in CTC numbers was evident as early as 4 days after initiation of PLX4720 therapy and these continued to decline progressively over the 14 day treatment period (Figure 1D and E, Figure S1F-H).
Prevention of metastases from early melanomas by post-resection administration of B-RAF inhibitor
The efficient generation and capture of CTCs in a mouse melanoma model, coupled with our ability to monitor the earliest stages tumor formation in the skin, made it possible to test the timeline of CTC generation and seek histopathological correlates within the primary tumor. For these experiments we chose to apply minute amounts of the active tamoxifen metabolite, 4-hydroxytamoxifen (4-HT), topically to the outer ear, thus avoiding skin trauma associated with subcutaneous injection and generating a small initial lesion, amenable to detailed histopathological analysis. A darkening of the outer ear, consistent with melanocyte proliferation, was grossly evident 17-21 days after topical application of 4-HT (Figure 2A), with rising CTC numbers detectable in the circulation as early as 7 days (Figure 2B). Histopathological features analyzed in the primary melanoma lesions included tumor thickness, tumor ulceration, tumor cell mitoses and fat invasion (Table S1). Consistent with well-established predictors of clinical outcome (Carrillo et al., 2002), the thickness of the primary lesion was most highly correlated with increasing generation of CTCs (Figure 2C).
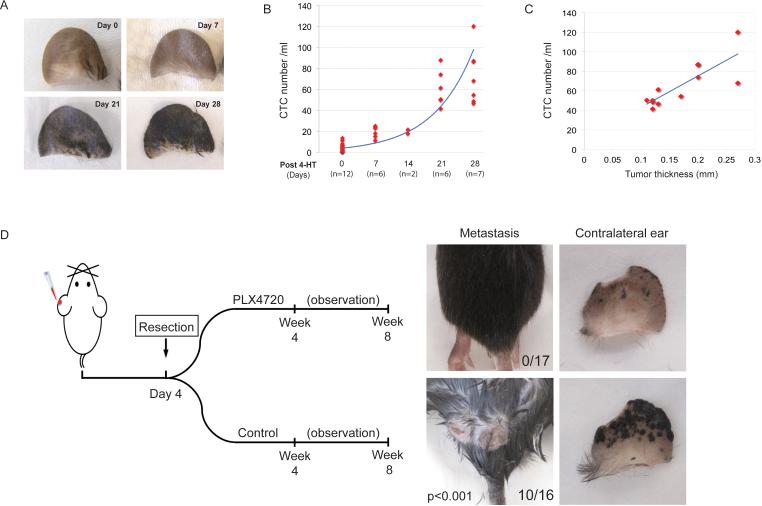
Figure 2. Distant metastasis is associated with the presence of CTCs and is prevented by adjuvant B-RAF inhibition.
(A) Tumor progression on 4-HT treated ear. (B) CTCs can be detected as early as day 7 after tumor induction and correspond to tumor progression. (C) CTC counts correlate with tumor thickness when thickness is greater than 0.1mm (N=12, R2=0.66). (D) Left panel, schematic diagram of the experimental procedures. Right panels, post-surgical administration of B-RAF inhibitor delays or prevents distant metastasis. After the affected ears were removed, the mice were fed with PLX4720-containing chow (N=17) or control chow (N=16) for 4 weeks. Subcutaneous metastases were observed in the control group (10 out of 16) but not in the PLX4720-fed group (0 out of 17). Tumors on the contralateral ear were also largely suppressed by PLX4720. (See also Figure S2)
Initiation of melanoma in the mouse ear was followed by its appearance in the contralateral ear at 4 weeks and disseminated cutaneous spread (to the torso) at 7-8 weeks. We could not determine whether the early contralateral ear involvement represented true blood-borne metastasis with a predilection for the ear, or more likely the effect of mouse grooming, with spread of 4-HT by contact to the contralateral ear. Consistent with the rapid generation of CTCs in the mouse ear model, resection of the primary tumor as early as four days following 4-HT application did not prevent the subsequent development of distant metastases (Figure 2D, S2). As a potential model for the adjuvant treatment of early stage melanoma, we tested whether resection of the primary tumor, followed by a short course of B-RAF inhibition, could delay or prevent the development of metastases. Of 16 mice treated with 4-HT followed by resection of the treated ear at day 4, 10 (63%) developed distant cutaneous metastases at two months. Remarkably, none of 17 mice (0%) that received four weeks of post-surgical B-RAF inhibitor developed cutaneous metastases at two months (P<0.001; Figure 2D). The appearance of tumors in the contralateral ear was significantly reduced but not completely abrogated by PLX4720 treatment. When compared to the time limited efficacy of B-RAF inhibition in the setting of metastatic melanoma, these observations suggest that a brief treatment with B-RAF inhibitor in the post-surgical “adjuvant” setting, when tumor burden is low, may have a dramatic impact on the risk of tumor recurrence.
RNA sequencing profile of mouse melanoma CTCs
The availability of matched primary melanoma, CTCs and metastatic deposits in a genetically defined mouse model made it possible to compare expression profiles for these sequential stages of cancer dissemination. As previously described (Yu et al., 2012), we applied single molecule next-generation RNA-Sequencing (HeliscopeTM Single Molecule Sequencer), so as to avoid the loss of signal associated with PCR-amplification of low quantity, heterogeneous templates. The median purity of CTCs enriched by HBCTC-Chip from tumor-bearing mice was 0.3%(range 0.073% to 3.85%, N=12) (Figure 1C), with higher purity achieved from mice with higher tumor burdens. We therefore selected another 5 mice with multiple metastatic lesions, obtained CTC-enriched populations, and subjected these to microfluidic in-line extraction of nucleic acids, followed by RNA-Sequencing. To allow digital subtraction of contaminating leukocyte transcripts from the CTC-enriched reads, each mouse-derived blood specimen was processed through paired HBCTC-Chips, one functionalized with antibodies to CSPG4 and MCAM (Capture-HBCTC-Chip), and the other mock-functionalized with matching IgG (Control-IgG-Chip), thereby achieving a more specific digital expression pattern for melanoma CTCs. Primary and metastatic melanoma deposits were also processed using the same single molecule sequencing platform (Figure 3A). While expression differences between matched primary and metastatic tumors were evident, none of these was shared across the four pairs that were successfully sequenced (Figure S3). We therefore did not identify consistent metastasis-specific transcripts in this mouse model.
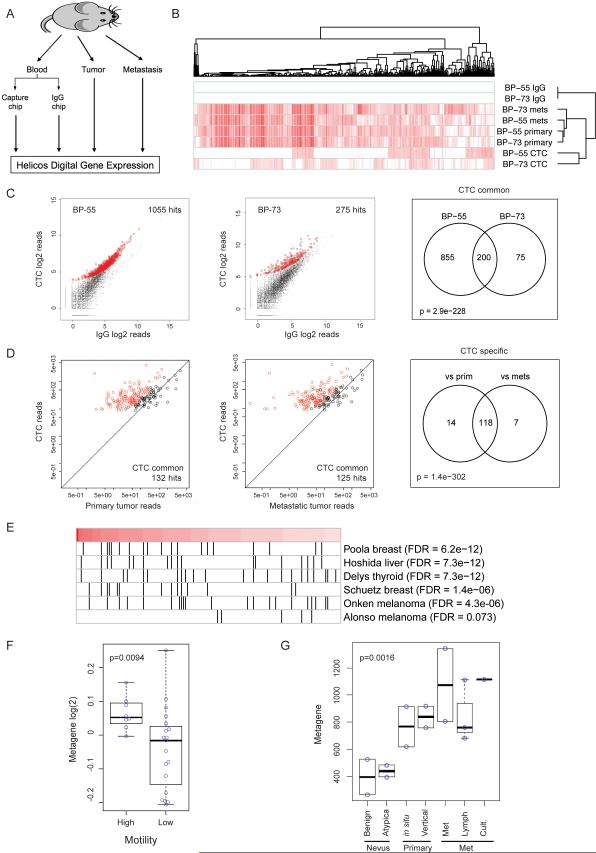
Figure 3. Digital Gene Expression (DGE) analysis of mouse CTCs.
(A) Schematic diagram of the DGE experiment. RNA was extracted from Chip-enriched cells, matched primary and metastatic tumors and subjected to Helicos DGE analysis. (B) Unsupervised clustering of transcripts enriched in CTCs, primary and metastatic tumors from the two mice (BP-55 and BP-73) demonstrating distinct expression profiles of CTCs from those of the primary tumor and metastasis. (C) Genes upregulated in CTC-enriched population compared with mock IgG-enriched cells. There were 1055 (left) and 275 (middle) upregulated genes in samples BP-55 and BP-73, respectively, with an overlap of 200 genes (right, “CTC-common”). Of these 200 genes, 132 were specifically overexpressed in CTCs compared with the primary tumor (D, left) and 125 were overexpressed in CTCs compared with the metastasis (D, middle), with 118 genes found to be in common (D, right). (E) Gene Set Enrichment Analysis (GSEA) on the 132 CTC-specific genes identified a number of gene sets related to cancer. Red bar, the 132 genes ranked by expression level from high (left, dark red) to low (right, light red); black vertical lines, overlap of the 132 genes with known gene sets. Poola breast, Poola invasive breast cancer genes (Poola et al., 2005); Hoshida liver, Hoshida liver carcinoma genes (Hoshida et al., 2009); Delys thyroid, Delys papillary thyroid cancer genes (Delys et al., 2007); Schuetz breast, Schuetz ductal invasive breast cancer genes (Schuetz et al., 2006); Onken melanoma, Onken aggressive uveal melanoma genes (Onken et al., 2006); Alonso melanoma, Alonso metastatic melanoma genes (Alonso et al., 2007). (F) The 132 CTC-specific gene signature is associated with increased motility in melanoma cell lines (Jeffs et al., 2009). (G) The signature also showed a progressive increase from benign or atypical nevus to primary and metastatic lesions. Benign, benign nevus; Atypica, atypical nevus; in situ, melanoma in situ; Vertical, melanoma vertical growth phase; Met, metastatic melanoma; Lymph, melanoma lymph node metastasis; Cult., metastatic melanoma short-term culture (Smith et al., 2005). (See also Figure S3)
Of five mice analyzed, two (BP-55 and BP-73) had CTC-derived RNA of sufficient quality to generate data for detailed digital gene expression profiling. Unsupervised clustering of the 1000 genes with the highest standard deviation suggested that the CTCs express a set of transcripts that are under-represented in both the primary and metastatic tumors (Figure 3B). Mice BP-55 and BP-73 respectively had 1,055 and 275 transcripts that were overexpressed (FDR<1e-05 and >2-fold) in CTC-enriched cells, compared with mock-enriched controls, of which 200 transcripts were shared between the two specimens (“CTC-common”) (Figure 3C). Gene Ontology analysis applied to the 200 genes identified melanosome-associated genes (Table S2) reflecting CTC's melanocytic origin. Of these 200 “CTC-common” transcripts, 132 were specifically overexpressed in CTCs compared to the primary tumor reads (“CTC-specific”) and 125 were overexpressed compared with metastatic tumor reads, with 118 of these transcripts in common (Figure 3D).
Gene Set Enrichment Analysis (GSEA) applied to the 132 “CTC-specific” genes upregulated in CTCs versus primary tumor identified a signature previously implicated in the non-canonical WNT, TGF-β and BMP signaling (Hoshida et al., 2009) (Figure 3E, Figure S3C-F), as well as other gene sets implicated in tumor invasiveness in both melanomas and breast cancers (Onken et al., 2006; Poola et al., 2005; Schuetz et al., 2006). Also identified were gene sets driven by hyperactivation of the B-RAFV600E-MAPK pathway in papillary thyroid cancer (Delys et al., 2007) and Epithelial-to-Mesenchymal Transition, EMT (Alonso et al., 2007) (Figure 3E).
We applied the mouse melanoma CTC-derived 132 metagene signature to expression datasets of human melanoma specimens: the signature readily identified melanoma cell lines that were ranked as having high cell motility score (Jeffs et al., 2009) (Figure 3F). It also showed progressively increased expression in melanocytic lesions, ranging from benign or atypical nevi to primary and metastatic melanomas (Smith et al., 2005) (Figure 3G). Thus, a set of transcripts upregulated during CTC generation in a mouse melanoma model may represent pathways that denote invasive properties in human melanoma.
Microfluidic isolation of CTCs from patients with metastatic melanoma
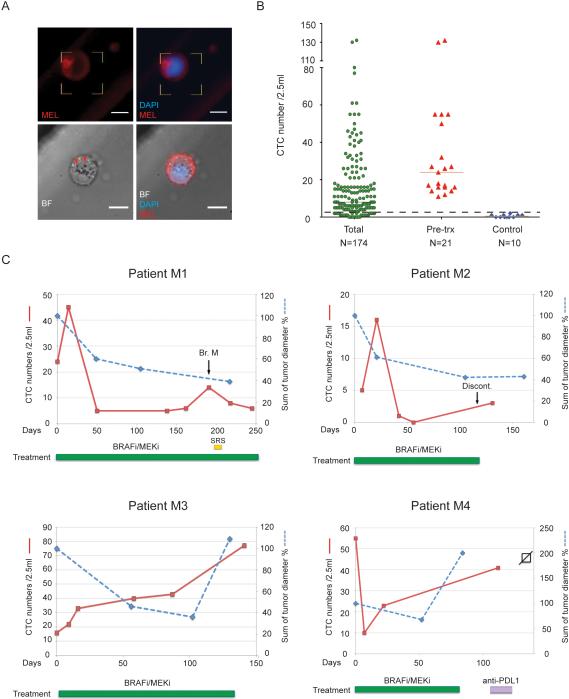
The successful isolation of mouse melanoma CTCs raised the possibility of capturing such cells from patient-derived blood specimens. Due to the highly heterogeneous expression of lineage markers in human melanomas (Herlyn and Koprowski, 1988), we screened 11 human melanoma cell lines for expression of 42 melanoma antigens (Figure S4). A cocktail of 12 antibodies against melanoma cell surface epitopes identifying all 11 melanoma cell lines but reacting minimally to normal blood cells, was selected, demonstrating capture of >90% melanoma cells (Figure S4). A second cocktail of 4 distinct antibodies against melanoma epitopes were chosen to stain captured cells (“melanoma stain”; Figure 4A, S4, Table S4). As for mouse CTCs, an imaging threshold was set using blood specimens from healthy donors: a median of one “melanoma stain” +/CD45− cell/2.5 ml blood was observed in control specimens (range 0-2 cells/2.5ml; N=10), leading us to define ≥3 cells/2.5ml as a positive CTC score. CTCs above this threshold were observed in 138 of 174 samples (79%) from 41 patients at various stages of treatment (median 8 cells/2.5ml; range 0-132). Significantly higher numbers of CTCs (P<0.0001) were identified from pre-treatment samples (median 24 CTCs/2.5ml; range 11 to 132; N=21) (Figure 4B). Captured melanoma CTCs exhibited a wide range in cell size (11-19 um diameter) and CTC clusters (≥2 cells) were identified 3 out of 174 samples (Figure S5F). While the relatively low purity of human CTCs captured using this assay (median 0.07%, range 0-0.77%) precluded the molecular analysis performed in the mouse model, it allowed for monitoring of patients undergoing therapeutic interventions. In four patients with metastatic BRAFV600E melanoma receiving targeted anti-B-RAF therapies, CTC numbers were generally correlated with clinical evaluation of disease status, including two patients experiencing relatively durable responses (M1 and M2) and two patients who had only transient benefit (M3 and M4) (Figure 4C).
Figure 4. Detection and characterization of circulating tumor cells in melanoma patients.
(A) Representative images of CTCs from patients with metastatic melanoma. CTCs are defined as being “melanoma stain (MEL)”+ and CD45−. Upper panels, IF image of a MEL+CD45− melanoma CTC. Lower panels, bright-field (BF) image and merged image IF and BF of a CTC reveals intracellular “granules” marked with arrowheads. MEL, CSPG4/MCAM/TYRP-1/αSMA antibody cocktail (upper panels) or CSPG4 (lower panels). DAPI, blue; MEL, red; scale bar, 10μm. (B) CTC counts of 174 samples from metastatic patients, 21 pre-treatment samples and 11 healthy donors. Median MEL(C/M/T/S)+/CD45− cell count is 8 cells/2.5ml for all patient samples, 24 cells/2.5ml for pre-treatment samples and 1 cells/2.5ml for controls. Solid lines, median CTC counts. Dashed line, threshold of ≥ 3 cells/2.5ml. (C) CTCs display dynamic changes in patients and correlate with therapy responses. Radiographic tumor measurement, blue curve; CTC counts, red curve. Br. M, brain metastasis; SRS, Stereotactic Radiosurgery; Discont., treatment discontinued; B-RAFi/MEKi, dabrafenib/trametinib (Patient M1, M3 and M4) or LGX818/MEK163 (Patient M2). (See also Figure S4)
Discussion
We have demonstrated the successful adaptation of a microfluidic device for melanoma CTC enrichment and applied it to a mouse model that recapitulates the biology of B-RAF-driven melanoma. The characterization of melanoma CTCs isolated in this model provides an initial insight into their invasive properties and sets the stage for potential future clinical applications. The mouse B-RAF/PTEN-driven melanoma model demonstrates a rise in CTC numbers as the primary tumor enlarges and metastases appear. Similarly, a brisk decline in CTCs is evident concomitant with tumor shrinkage in response to targeted therapy. Thus, in this genetically homogeneous model, CTC numbers are well correlated with tumor burden. In different human cancers, which exhibit considerable genetic heterogeneity, CTC numbers are notoriously variable across different individuals and they are poorly correlated with radiographic measurements of tumor burden (reviewed in Yu et al., 2011). However, longitudinal follow-up of individual patients does show a correlation with tumor response or progression. Such a trend is also evident in the melanoma patients studied here, and it suggests that once the detection platforms for human melanoma CTCs are optimized, monitoring the number of these cells during therapy may help guide treatment for metastatic cancer.
While most applications of CTC technologies have focused on monitoring advanced cancers, the highly invasive nature of cancer cells in this melanoma model made it possible to study their generation in the setting of early, localized disease. Indeed, in the mouse ear melanoma initiation model, we find that CTCs are shed very early into the circulation, at a time when the primary lesion is defined by an increase in pigmentation, preceding the appearance of a tumor. Resection of the ear at this early stage does not prevent the development of distant cutaneous metastases, confirming the rapid blood-borne dissemination of tumor cells. Remarkably, a relatively brief course of B-RAF inhibitor therapy is sufficient to suppress the development of distant cutaneous metastases. Conceptually, these experiments lend support to the use of anti-B-RAF therapies in the postoperative adjuvant setting, where the relatively low tumor burden may allow the drug to kill all residual disease and prevent recurrence at distant sites. Two ongoing clinical trials (NCT01682083, NCT01667419) are currently testing this hypothesis. However, identifying the subset of patients with stage II and III disease who are at high risk of tumor recurrence following resection of the primary lesion remains a major challenge, with histological hallmarks, such as tumor thickness, ulceration and mitotic rate, providing only incomplete patient stratification. Such patient stratification is essential, given BRAF inhibitor-related toxicity (Su et al., 2012). Optimization of human melanoma CTC capture platforms may allow detection of rare cells in patients with early disease, potentially providing a predictive measure of recurrence risk. In this context, it is of interest that the ability of primary melanoma explants to generate CTCs following implantation into immunodeficient mice appears to be predictive of their recurrence risk (Quintana et al., 2012).
The gene expression patterns correlated with melanoma aggressiveness have not been well defined, possibly reflecting the considerable heterogeneity in this disease (Schramm et al., 2012). In contrast, in the syngeneic mouse model, we were able to identify a small set of markers that were consistently upregulated in CTCs, compared with either primary or metastatic tumor. Despite the initiating B-RAF mutation triggering the melanoma model, CTCs demonstrated a further upregulation of B-RAF-MAPK activity (Figure 3E, “Delys Thyroid”). Consistent with a B-RAF-MAPK induced melanoma “dedifferentiation” phenotype (Frederick et al., 2013), expression of many melanocytic differentiation genes, including MITF, GP100 and Melan-A, were considerably lower in CTCs than in primary tumors (Figure S3G, X.L. and D.H., unpublished data). Even the “melanosome” genes expressed by CTCs were mainly genes whose products involve the maintenance of melanosome structures, e.g. ANXA2 and LAMP1, rather than the classic melanocytic differentiation genes (Table S2). The existence of melanoma stem cells remains controversial (Quintana et al., 2008), and we did not find any candidate melanoma stem cell-specific genes to be enriched in CTCs, including ABCB5 (Schatton et al., 2008), CD34 and CD271 (Boiko et al., 2010; Held et al., 2010). Interestingly, we did note significantly increased expression levels of two master transcription factors involving in stem cell reprogramming, Klf4 and Myc, in the melanoma CTC populations (Table S3).
Canonical Wnt signaling has been shown to contribute to metastatic dissemination in the B-RAF/PTEN mouse model (Damsky et al., 2011), but the most striking component of the CTC signature identified here was a similarity to a liver carcinoma gene set linked to non-canonical Wnt, TGF-β and BMP signaling pathways (Figure 3E, “Hoshida liver”, Figure S3C-F) (Hoshida et al., 2009). That signature also emerged in CTCs derived from a mouse model of pancreatic cancer, where it was correlated with anchorage independent survival signals (Yu et al., 2012). This raises the intriguing possibility that a set of conserved CTC-specific gene expression patterns may exist across different cancer types, linked to signaling pathways that regulate cell survival by normally adherent cells that travel in the bloodstream. When analyzed all together, the mouse melanoma CTC signature identified here shows a correlation with cellular invasiveness in human melanoma. If confirmed in larger clinical trials, such a CTC-derived expression signature may identify an aggressive subset of melanoma with a high risk of blood-borne metastatic spread and help guide the rational treatment of early disease.
While the HBCTC-Chip has proved effective in capturing melanoma CTCs in the BRAF/PTEN model, robust platforms to study human melanoma CTCs have proved illusive possibly due to heterogeneity and phenotypic plasticity in human melanomas (Hoek et al., 2008). Compared with the mouse, we found that a large number of antibodies were required to reliably capture human melanoma CTCs, resulting in complex chip functionalization protocols and elevated nonspecific binding by contaminating leukocytes. As such, detailed molecular analysis of human melanoma CTCs will require single cell isolation protocols, as well as non-tumor epitope driven CTC isolation platforms (Ozkumur et al., 2013) (see supplementary discussion). Nonetheless, the human melanoma-specific HBCTC-Chip designed here allowed enumeration of CTCs from patients with metastatic disease, with a dynamic range that enabled longitudinal monitoring of treatment response and disease progression. Indeed, within the parameters of periodic sampling that was not timed to specific treatment evaluations or therapeutic interventions, we observed a general concordance between CTC numbers and clinical status in patients undergoing B-RAF targeted therapies for metastatic melanoma. An unexpected finding in both human and mouse melanoma CTCs was the presence of CTC-clusters (Figure 1B and Figure S4F). Such groupings of CTCs in the circulation have been observed in patients with various epithelial cancers (Cho et al., 2012; Stott et al., 2010), and are likely a result of tumor fragments breaking off into the circulation. However, transformed from primary melanocytes that are thought to be single cells, melanoma cells do not form strong intercellular adhesions even although it has been reported that artificially generated melanoma microemboli did display increased tumorigenic potential than single melanoma cells (Fidler, 1973). Therefore, the origin and functional significance of melanoma CTC-clusters and their contribution to human melanoma metastasis remains to be defined.
In summary, the analysis of melanoma CTCs in a well defined and homogeneous mouse model, together with emerging data on human melanoma CTCs, are likely to provide important insights into the biological drivers of blood-borne invasion in this disease. The detection of CTCs in early invasive disease, as well as in established metastatic cancer, may help guide the application of increasingly powerful therapeutic regimens.
EXPERIMENTAL PROCEDURES
Detailed procedures and reagent information are in the Supplemental Experimental Procedures.
Supplementary Material
HIGHLIGHTS.
Microfluidic isolation of circulating melanoma cells in a B-RAF driven mouse model
CTC numbers correlate with tumor response on B-RAF targeted therapies
CTCs are shed early and post resection B-RAF inhibition prevents metastasis in mouse
RNA-Sequencing of CTCs identifies a signature related to melanoma invasiveness
ACKNOWLEDGEMENT
The authors would like to thank C. Hart, A. McGovern and C. Koris and the Massachusetts General Hospital (MGH) clinical research coordinators for help with clinical studies, L. Libby for mouse studies, F. Ozsolak and P. Milos (Helicos) for RNA sequencing, J. Walsh, J. Lennerz and M. Ulman for microscopy expertise. This work was supported by grants from Stand Up To Cancer (D.A.H., M.T., S.M.), Howard Hughes Medical Institute (D.A.H.), Melanoma Research Foundation (X.L.), National Foundation for Cancer Research (D.A.H.), NIH CA129933 (D.A.H.) and NIBIB EB008047 (D.A.H, M.T.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Alonso SR, Tracey L, Ortiz P, Perez-Gomez B, Palacios J, Pollan M, Linares J, Serrano S, Saez-Castillo AI, Sanchez L, et al. A high-throughput study in melanoma identifies epithelial-mesenchymal transition as a major determinant of metastasis. Cancer Res. 2007;67:3450–3460. doi: 10.1158/0008-5472.CAN-06-3481. [DOI] [PubMed] [Google Scholar]
- Balch CM, Gershenwald JE, Soong SJ, Thompson JF, Atkins MB, Byrd DR, Buzaid AC, Cochran AJ, Coit DG, Ding S, et al. Final version of 2009 AJCC melanoma staging and classification. J Clin Oncol. 2009;27:6199–6206. doi: 10.1200/JCO.2009.23.4799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boiko AD, Razorenova OV, van de Rijn M, Swetter SM, Johnson DL, Ly DP, Butler PD, Yang GP, Joshua B, Kaplan MJ, et al. Human melanoma-initiating cells express neural crest nerve growth factor receptor CD271. Nature. 2010;466:133–137. doi: 10.1038/nature09161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrillo E, Prados J, Melguizo C, Marchal JA, Velez C, Serrano S, Boulaiz H, Merida JA, Aranega A. Reverse transcriptase-polymerase chain reaction detection of circulating tumor cells in patients with melanoma: correlation with clinical stage, tumor thickness and histological type. Pathol Int. 2002;52:294–299. doi: 10.1046/j.1440-1827.2002.01344.x. [DOI] [PubMed] [Google Scholar]
- Cho EH, Wendel M, Luttgen M, Yoshioka C, Marrinucci D, Lazar D, Schram E, Nieva J, Bazhenova L, Morgan A, et al. Characterization of circulating tumor cell aggregates identified in patients with epithelial tumors. Phys Biol. 2012;9:016001. doi: 10.1088/1478-3975/9/1/016001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Damsky WE, Curley DP, Santhanakrishnan M, Rosenbaum LE, Platt JT, Gould Rothberg BE, Taketo MM, Dankort D, Rimm DL, McMahon M, Bosenberg M. beta-catenin signaling controls metastasis in Braf-activated Pten-deficient melanomas. Cancer Cell. 2011;20:741–754. doi: 10.1016/j.ccr.2011.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dankort D, Curley DP, Cartlidge RA, Nelson B, Karnezis AN, Damsky WE, Jr., You MJ, DePinho RA, McMahon M, Bosenberg M. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet. 2009;41:544–552. doi: 10.1038/ng.356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Giorgi V, Pinzani P, Salvianti F, Panelos J, Paglierani M, Janowska A, Grazzini M, Wechsler J, Orlando C, Santucci M, et al. Application of a filtration- and isolation-by-size technique for the detection of circulating tumor cells in cutaneous melanoma. J Invest Dermatol. 2010;130:2440–2447. doi: 10.1038/jid.2010.141. [DOI] [PubMed] [Google Scholar]
- Delys L, Detours V, Franc B, Thomas G, Bogdanova T, Tronko M, Libert F, Dumont JE, Maenhaut C. Gene expression and the biological phenotype of papillary thyroid carcinomas. Oncogene. 2007;26:7894–7903. doi: 10.1038/sj.onc.1210588. [DOI] [PubMed] [Google Scholar]
- Fidler IJ. The relationship of embolic homogeneity, number, size and viability to the incidence of experimental metastasis. Eur J Cancer. 1973;9:223–227. doi: 10.1016/s0014-2964(73)80022-2. [DOI] [PubMed] [Google Scholar]
- Flaherty KT, Hodi FS, Fisher DE. From genes to drugs: targeted strategies for melanoma. Nat Rev Cancer. 2012;12:349–361. doi: 10.1038/nrc3218. [DOI] [PubMed] [Google Scholar]
- Frederick DT, Piris A, Cogdill AP, Cooper ZA, Lezcano C, Ferrone CR, Mitra D, Boni A, Newton LP, Liu C, et al. BRAF Inhibition Is Associated with Enhanced Melanoma Antigen Expression and a More Favorable Tumor Microenvironment in Patients with Metastatic Melanoma. Clin Cancer Res. 2013;19:1225–1231. doi: 10.1158/1078-0432.CCR-12-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held MA, Curley DP, Dankort D, McMahon M, Muthusamy V, Bosenberg MW. Characterization of melanoma cells capable of propagating tumors from a single cell. Cancer Res. 2010;70:388–397. doi: 10.1158/0008-5472.CAN-09-2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herlyn M, Koprowski H. Melanoma antigens: immunological and biological characterization and clinical significance. Annu Rev Immunol. 1988;6:283–308. doi: 10.1146/annurev.iy.06.040188.001435. [DOI] [PubMed] [Google Scholar]
- Hoek KS, Eichhoff OM, Schlegel NC, Dobbeling U, Kobert N, Schaerer L, Hemmi S, Dummer R. In vivo switching of human melanoma cells between proliferative and invasive states. Cancer Res. 2008;68:650–656. doi: 10.1158/0008-5472.CAN-07-2491. [DOI] [PubMed] [Google Scholar]
- Hoshida Y, Nijman SM, Kobayashi M, Chan JA, Brunet JP, Chiang DY, Villanueva A, Newell P, Ikeda K, Hashimoto M, et al. Integrative transcriptome analysis reveals common molecular subclasses of human hepatocellular carcinoma. Cancer Res. 2009;69:7385–7392. doi: 10.1158/0008-5472.CAN-09-1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshimoto S, Shingai T, Morton DL, Kuo C, Faries MB, Chong K, Elashoff D, Wang H-J, Elashoff RM, Hoon DSB. Association Between Circulating Tumor Cells and Prognosis in Patients With Stage III Melanoma With Sentinel Lymph Node Metastasis in a Phase III International Multicenter Trial. J Clin Oncol. 2012;30:3819–3826. doi: 10.1200/JCO.2011.40.0887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffs AR, Glover AC, Slobbe LJ, Wang L, He S, Hazlett JA, Awasthi A, Woolley AG, Marshall ES, Joseph WR, et al. A gene expression signature of invasive potential in metastatic melanoma cells. PLoS One. 2009;4:e8461. doi: 10.1371/journal.pone.0008461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoja L, Lorigan P, Zhou C, Lancashire M, Booth J, Cummings J, Califano R, Clack G, Hughes A, Dive C. Biomarker Utility of Circulating Tumor Cells in Metastatic Cutaneous Melanoma. J Invest Dermatol. 2013;133:1582–1590. doi: 10.1038/jid.2012.468. [DOI] [PubMed] [Google Scholar]
- Liu Z, Fusi A, Klopocki E, Schmittel A, Tinhofer I, Nonnenmacher A, Keilholz U. Negative enrichment by immunomagnetic nanobeads for unbiased characterization of circulating tumor cells from peripheral blood of cancer patients. J Transl Med. 2011;9:70. doi: 10.1186/1479-5876-9-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onken MD, Ehlers JP, Worley LA, Makita J, Yokota Y, Harbour JW. Functional Gene Expression Analysis Uncovers Phenotypic Switch in Aggressive Uveal Melanomas. Cancer Res. 2006;66:4602–4609. doi: 10.1158/0008-5472.CAN-05-4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozkumur E, Shah AM, Ciciliano JC, Emmink BL, Miyamoto DT, Brachtel E, Yu M, Chen PI, Morgan B, Trautwein J, et al. Inertial focusing for tumor antigen-dependent and -independent sorting of rare circulating tumor cells. Sci Transl Med. 2013;5:179, ra147. doi: 10.1126/scitranslmed.3005616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poola I, DeWitty RL, Marshalleck JJ, Bhatnagar R, Abraham J, Leffall LD. Identification of MMP-1 as a putative breast cancer predictive marker by global gene expression analysis. Nat Med. 2005;11:481–483. doi: 10.1038/nm1243. [DOI] [PubMed] [Google Scholar]
- Quintana E, Piskounova E, Shackleton M, Weinberg D, Eskiocak U, Fullen DR, Johnson TM, Morrison SJ. Human Melanoma Metastasis in NSG Mice Correlates with Clinical Outcome in Patients. Sci Transl Med. 2012;4:159ra149. doi: 10.1126/scitranslmed.3004599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quintana E, Shackleton M, Sabel MS, Fullen DR, Johnson TM, Morrison SJ. Efficient tumour formation by single human melanoma cells. Nature. 2008;456:593–598. doi: 10.1038/nature07567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Luo S, Wang YC, Li R, Deng Q, Faridani OR, Daniels GA, Khrebtukova I, Loring JF, Laurent LC, et al. Full-length mRNA-Seq from single-cell levels of RNA and individual circulating tumor cells. Nat Biotechnol. 2012;30:777–782. doi: 10.1038/nbt.2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schatton T, Murphy GF, Frank NY, Yamaura K, Waaga-Gasser AM, Gasser M, Zhan Q, Jordan S, Duncan LM, Weishaupt C, et al. Identification of cells initiating human melanomas. Nature. 2008;451:345–349. doi: 10.1038/nature06489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm SJ, Campain AE, Scolyer RA, Yang YH, Mann GJ. Review and cross-validation of gene expression signatures and melanoma prognosis. J Invest Dermatol. 2012;132:274–283. doi: 10.1038/jid.2011.305. [DOI] [PubMed] [Google Scholar]
- Schuetz CS, Bonin M, Clare SE, Nieselt K, Sotlar K, Walter M, Fehm T, Solomayer E, Riess O, Wallwiener D, et al. Progression-Specific Genes Identified by Expression Profiling of Matched Ductal Carcinomas In situ and Invasive Breast Tumors, Combining Laser Capture Microdissection and Oligonucleotide Microarray Analysis. Cancer Res. 2006;66:5278–5286. doi: 10.1158/0008-5472.CAN-05-4610. [DOI] [PubMed] [Google Scholar]
- Smith AP, Hoek K, Becker D. Whole-genome expression profiling of the melanoma progression pathway reveals marked molecular differences between nevi/melanoma in situ and advanced-stage melanomas. Cancer Biol Ther. 2005;4:1018–1029. doi: 10.4161/cbt.4.9.2165. [DOI] [PubMed] [Google Scholar]
- Stott SL, Hsu CH, Tsukrov DI, Yu M, Miyamoto DT, Waltman BA, Rothenberg SM, Shah AM, Smas ME, Korir GK, et al. Isolation of circulating tumor cells using a microvortex-generating herringbone-chip. Proc Natl Acad Sci U S A. 2010;107:18392–18397. doi: 10.1073/pnas.1012539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su F, Viros A, Milagre C, Trunzer K, Bollag G, Spleiss O, Reis-Filho JS, Kong X, Koya RC, Flaherty KT, et al. RAS mutations in cutaneous squamous-cell carcinomas in patients treated with BRAF inhibitors. N Engl J Med. 2012;366:207–215. doi: 10.1056/NEJMoa1105358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Stott S, Toner M, Maheswaran S, Haber DA. Circulating tumor cells: approaches to isolation and characterization. J Cell Biol. 2011;192:373–382. doi: 10.1083/jcb.201010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu M, Ting DT, Stott SL, Wittner BS, Ozsolak F, Paul S, Ciciliano JC, Smas ME, Winokur D, Gilman AJ, et al. RNA sequencing of pancreatic circulating tumour cells implicates WNT signalling in metastasis. Nature. 2012;487:510–513. doi: 10.1038/nature11217. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.