SUMMARY
Recurrent mutations in histone modifying enzymes imply key roles in tumorigenesis yet their functional relevance is largely unknown. Here we show that JARID1B, encoding a histone H3 lysine 4 (H3K4) demethylase, is frequently amplified and overexpressed in luminal breast tumors and a somatic mutation in a basal-like breast cancer results in the gain of unique chromatin binding and luminal expression and splicing patterns. Downregulation of JARID1B in luminal cells induces basal genes expression and growth arrest, which is rescued by TGFβ pathway inhibitors. Integrated JARID1B chromatin binding, H3K4 methylation, and expression profiles suggest a key function for JARID1B in luminal cell-specific expression programs. High luminal JARID1B activity is associated with poor outcome in patients with hormone receptor positive breast tumors.
INTRODUCTION
Histone lysine methylation is important for chromatin organization and the regulation of gene expression (Dawson and Kouzarides, 2012). Systemic sequencing of human cancer genomes identified numerous alterations in genes encoding histone modifying enzymes including somatic mutations of KDM6A, KDM5C, and KDM5B (also called JARID1B, PLU-1), in multiple cancer types (You and Jones, 2012). However, the functional and clinical relevance of these mutations are still poorly understood.
In breast cancer, genes encoding transcriptional regulators (e.g., FOXA1, GATA3) are among the most frequently mutated genes, what suggests a key role for perturbed epigenetic programs in tumorigenesis (Polyak and Metzger Filho, 2012). Several of these genes are mutated in a breast tumor subtype-specific manner (TCGA, 2012). Breast tumors are classified based on the expression of estrogen receptor (ER), progesterone receptor (PR), and HER2 into ER+, HER2+, and ER−PR−HER2− (triple negative breast cancer, TNBC) disease. Based on gene expression profiles, breast tumors are characterized as luminal or basal-like (Perou et al., 2000; Sørlie et al., 2001). HER2+ and ER+ tumors typically have luminal features whereas TNBCs show significant but not complete overlap with the basal-like subtype. In general many more recurrent mutations are present in luminal than in basal-like breast tumors (Polyak and Metzger Filho, 2012). The high frequency of mutations in GATA3 and FOXA1 transcription factors with essential roles in luminal epithelial differentiation (Bernardo et al., 2010; Kouros-Mehr et al., 2006) in luminal breast tumors suggests that perturbed differentiation programs are important for tumor development.
Increasing evidence indicates that histone methyltransferases (HMTs) and demethylases (HDMs) play key roles in regulating cellular differentiation states. KDM5B, which encodes the histone H3 lysine4 (H3K4) demethylase JARID1B (Christensen et al., 2007; Iwase et al., 2007; Yamane et al., 2007), was first identified as a gene whose expression was reversibly regulated by HER2 signaling in breast cancer cells (Lu et al., 1999). Overexpression of JARID1B in HeLa cells results in loss of tri-, di-, and mono-methyl H3K4, but does not affect H3K9, H3K27 or H3K36 methylation.
JARID1B is overexpressed in prostate carcinomas where it associates with the androgen receptor and regulates its transcriptional activity (Xiang et al., 2007). Overexpression and tumor growth-promoting effects of JARID1B have also been described in bladder and lung cancers (Hayami et al., 2010). In contrast, the expression of JARID1B is downregulated in melanoma, where it has been implicated as a tumor suppressor inhibiting cell proliferation in an RB-dependent manner (Roesch et al., 2006; Roesch et al., 2008). However, JARID1B also plays a role in melanoma tumor maintenance and in metastatic progression based on xenotransplant assays in immunodeficient mice (Roesch et al., 2010).
JARID1B regulates the proliferation and differentiation of embryonic stem cells (ESCs) (Dey et al., 2008; Frankenberg et al., 2007; Kidder et al., 2013; Schmitz et al., 2011; Xie et al., 2011; Yamane et al., 2007; You and Jones, 2012) and it is involved in murine mammary gland development (Catchpole et al., 2011). JARID1B overexpression in ESCs decreases the expression of differentiated cell-specific genes and increases proliferation (Dey et al., 2008). The targeting of key transcriptional regulators of progenitor cell function may also play a role in JARID1B’s tumor-promoting effects, a hypothesis that has not been tested experimentally. Despite the importance of JARID1B in normal development and in tumorigenesis, its mechanism of action is still poorly defined.
RESULTS
JARID1B is amplified and overexpressed in breast tumors
We identified JARID1B as a gene with common copy number gain based on our SNP (Single Nucleotide Polymorphism) array analysis of breast tumors and cancer cell lines (Nikolsky et al., 2008), which we also confirmed by qPCR and FISH (fluorescent in situ hybridization) (Figure 1A and S1A and data not shown). Analysis of the METABRIC dataset (Curtis et al., 2012) showed that JARID1B copy number gain is associated with increased transcript levels (Figure 1B), especially in luminal subtypes (compare Figure 1C and S1B). JARID1B mRNA levels are also the highest in luminal A and HER2+ tumors both when using the PAM50 (Parker et al., 2009) and the IC10 (Curtis et al., 2012) classification (Figure 1D–E) and somewhat higher in ER+ than in ER− cases (Figure S1C). JARID1B protein levels displayed a similar trend in breast cancer cell lines (Figure S1D).
Figure 1. JARID1B is a luminal lineage-specific oncogene in breast cancer.
(A) A representative image of metaphase FISH analysis of MCF7 luminal ER+ breast cancer cells using JARID1B BAC (green) and chromosome 1 centromeric (red) probes. (B, C) Correlation between JARID1B gene expression (mRNA) and copy number in all tumors (n=1,944) (B) and luminal A (n=711) subset (C) based on the analysis of breast tumors in the METABRIC dataset. r indicates linear correlation coefficient. (D, E) Associations between JARID1B expression and PAM50 (n=1,944) (D) and 10 different breast tumor (integrative clusters, IC10) (n=1,980) (E) subtypes in the METABRIC dataset. Small colored rectangles in panel E indicate the composition of IC10 clusters according to PAM50 subtype. The differences in JARID1B mRNA levels among breast tumor subtypes are statistically significant (see Supplemental Experimental Procedures for details). (F) shRNA clones identified as hits in the cellular viability screen in the indicated cell lines. Colors indicate luminal (blue), basal-like (red), and HER2+ (pink) breast cell lines, respectively. Numbers indicate fraction (%) of viable cells compared to control. Red shading indicates growth inhibition above cut off (75%). See also Figure S1.
Loss of JARID1B inhibits breast cancer cell growth
To investigate the functional relevance of JARID1B overexpression in breast cancer, first we performed a lentiviral shRNA screen for cellular viability in a panel of luminal and basal-like breast cancer cell lines. Both in the primary and secondary screens shRNAs against JARID1B had the most pronounced growth inhibitory effect in ER+ luminal breast cancer cells (MCF7 and T-47D) based on the significance of decrease in viable cells (i.e., z-score in the primary and % decrease of viability in the secondary screen) and the number of independent shRNA clones that were hits in these assays (Figure 1F and S1E–F). Similar results were obtained using JARID1B-targeting siRNAs (Figure S1G–I) and the decrease in cell viability was rescued by the exogenous expression of siRNA-resistant JARID1B cDNAs (Figure S1J–K). We did not detect significant apoptosis in any of the cell lines after siJARID1B transfection, consistent with prior reports (Yamane et al., 2007). The downregulation of JARID1B did not affect the protein levels of the closely related JARID1A, and it only slightly increased global histone H3K4me3 levels in the MCF7 and T-47D luminal breast cancer cell lines with no obvious change in H3K4me1/2 or H3K27me3 (Figure S1L–M). These results support the hypothesis that JARID1B is a target of the 1q32 amplicon and suggested that JARID1B function might particularly be important in ER+ luminal breast cancer cells.
Loss of JARID1B-induced gene expression changes
To further explore JARID1B function, we performed RNA-seq of luminal and basal-like breast cancer cell lines transfected with control or JARID1B-targeting siRNAs and identified genes with significant expression changes. Basal-like and luminal breast cancer cell lines clustered together based on RNA-seq profiles with the exception of the JARID1B-mutant HCC2157 basal-like line that clustered with luminal lines (Figure 2A). The cell lines showed variable number of genes with significantly altered expression after siJARID1B transfection (Figure S2A–B), thus, we selected the top 200 differentially expressed genes in each cell line for comparison (Table S1). Clustering of the samples based on the expression of all significantly [≥1.5 robust fold-change estimate (Feng et al., 2012)] differentially expressed transcripts (Figure S2A) or using the union of top 200 differentially expressed genes (Figure 2B) showed some luminal and basal differences. In luminal cells more genes were up than downregulated following siJARID1B transfection whereas the opposite was observed in basal-like cells (Figure S2B). The differences in expression levels were also confirmed by quantitative RT-PCR for selected genes in MCF7 cells (Figure 2C).
Figure 2. The effect of JARID1B on gene expression patterns.
(A, B) Hierarchical clustering of breast cancer cell lines by log2(RPKM) based on similarities in the expression pattern of all transcripts (A) and by GFold value of the top 200 genes differentially expressed after siJARID1B (B). Gene expression index is measured by the logarithm of gene’s RPKM value. The genes chosen for clustering in panel (B) are the union of top 200 differentially expressed genes after siJARID1B transfection in each cell line. (C) Measurement of expression changes for selected genes by quantitative RT-PCR. (D) Changes in the expression of basal/stem cell-related genes in siJARID1B-transfected MCF7 cells. (E) Clustering of breast cancer gene expression data from multiple different cohorts based on the top 200 differentially expressed genes with JARID1B promoter binding in the MCF7 cell line following transfection of siJARID1B. Change of expression level in MCF7 after siJARID1B transfection is indicated to the left (Cell-line expr), followed by the difference between luminal A and basal-like samples from all cohorts (Basal-lumA). (F, G) Rescue from JARID1B siRNA-induced growth inhibition by treatment with LY2109761. (F) or by downregulation of SMAD4 or TGFBR2 by siRNAs (G). In all panels, *, p<0.05; **, p<0.001; error bars mark STDEV. See also Figure S2 and Table S1.
TGFβ signaling in JARID1B-loss-mediated growth arrest
To explore the functional relevance of gene expression changes following downregulation of JARID1B, we performed network and pathway analyses of the top 200 differentially expressed genes using the Metacore suit (Bessarabova et al., 2011). This approach identified a prominent upregulation of basal/stem cell genes in siJARID1B-transfected luminal cells (Figures 2D–E and S2C) and significant enrichment for the TGFβ signaling pathway (Figure S2D). Correlating with this, the top 200 differentially expressed genes in siJARID1B transfected MCF7 cells showed distinct patterns of expression in luminal and basal-like primary breast tumors (Figures 2E and S2C), with the up genes in general having higher expression in basal-like tumors, demonstrating the physiologic relevance of these gene signatures in patient samples. Interestingly, in basal-like cells enrichment for the TGFβ signaling pathway was observed for the genes downregulated after siJARID1B transfection (Figure S2D) again suggesting luminal-basal differences in JARID1B’s function.
To further investigate the role of TGFβ signaling in JARID1B loss-mediated growth arrest, we first analyzed phosphoSMAD2, a key downstream target of TGFβ receptor kinases, and found increased levels in siJARID1B-transfected MCF7 cells (Figure S2E). To determine whether the activation of TGFβ signaling is required for JARID1B loss-mediated growth arrest, we downregulated key components of the TGFβ pathway (i.e., SMAD4 and TGFBR2) by siRNAs (Figure S2F) or inhibited TGFβ receptor kinase activity using a small molecule inhibitor (SMI). Treatment of MCF7 and T-47D luminal breast cancer cells with a TGFBR kinase inhibitor (LY2109761) rescued the siJARID1B-induced growth arrest in a dose-dependent manner whereas no effect was seen in SUM159PT basal-like cells (Figure 2F and data not shown). Downregulation of SMAD4 or TGFBR2 by siRNAs had similar effects (Figure 2G and data not shown). These results suggest that JARID1B expression in luminal breast cancer cells is required for the repression of basal cell-specific genes, including those important for TGFβ signaling, and the loss of this repression following decreased JARID1B leads to growth arrest. In contrast, in basal-like breast cancer cells JARID1B loss leads to decreased TGFβ pathway activity, but this may not play a role in JARID1B loss-mediated growth arrest in this cell type.
Genomic targets of JARID1B
To identify the genomic targets of JARID1B we performed ChIP-seq in luminal and basal-like breast cancer cells. ChIP was optimized and the specificity of the JARID1B antibody was validated by ChIP-qPCR that showed significant decrease in signal at known JARID1B-binding regions in siJARID1B-expressing MCF7 cells (Figure S3A–C). The number of JARID1B peaks varied among cell lines; thus, we used the top 10,000 peaks in each cell line for comparisons among samples. Clustering of the samples based on JARID1B peaks showed luminal and basal patterns (Figure 3A). Interestingly, the JARID1B-mutant HCC2157 basal-like cell line showed higher relatedness to luminal cells and had numerous unique peaks. The number and distribution of JARID1B peaks relative to transcription start site (TSS) varied among cell lines with luminal cells in general having more promoter and basal-like cells more non-promoter peaks (Figure 3B), and intriguingly >90% of the ~9,891 HCC2157 unique peaks were located in non-promoter regions (Table S2). Clustering of the samples using promoter and non-promoter peaks demonstrated that luminal-basal differences in JARID1B binding are more significant at non-promoter regions and again highlighted the uniqueness of HCC2157 cells (Figure 3C).
Figure 3. JARID1B binding patterns in breast tumor subtypes.
(A) Heatmap and hierarchical clustering of JARID1B genomic binding peaks (±3kb around peak summit) in luminal (blue) and basal-like (red) breast cancer cell lines. The JARID1B-mutant HCC2157 cell line is marked with an asterisk and the green rectangle highlights unique peaks. (B) Pie chart of top 10k JARID1B binding peaks location in relation to its distance from TSS. (C) Hierarchical clustering of breast cancer cell lines considering JARID1B binding signals only in non-promoter (left panel) and only in promoter (right panel) regions. Both clusters are significant in terms of p value (using pvclus). (D) Ratio of H3K4me3/H3K4me2 signal at JARID1B peak regions in MCF7 cells transfected with siControl and siJARID1B. P-value for Pearson correlation coefficient of siJARID1B and siControl H3K4me3 signals in JARID1B region (correlation value=0.98999, n=91,992, p value=0). (E) Union of JARID1B, H3K4me3, H3K4me2, and H3K4me1 binding peaks in MCF7 and SUM159PT cell lines. (F) Examples of peak maps and ChIP-qPCR for selected genes. Black bar and a-f indicates primers used for qPCR in the right panel. (G) Examples of H3K4me3 ChIP-qPCR for selected genes in control and siJARID1B transfected cells untreated or treated with LY2109761. (H) Heatmap of JARID1B signal around TSS of all RefSeq genes in breast cancer cell lines. Each row is a gene promoter region centered around TSS. K-mean clustering (k=2) is applied to all genes promoters. Histone marks and gene expression columns are plotted according to the order of genes from the clustering. Gene expression value is the logarithm of RPKM for each gene. In all panels, *, p<0.05; **, p<0.001; error bars mark STDEV. See also Figures S3 and Table S2.
JARID1B binding pattern overlaps with H3K4me3 and active genes
JARID1B is a histone H3K4 demethylase and based on in vitro assays it can demethylate all three H3K4 methylated forms (i.e., mono, di, and trimethyl) albeit with varying efficiency and with the highest affinity for H3K4me3 (Christensen et al., 2007; Iwase et al., 2007; Kristensen et al., 2012; Yamane et al., 2007). Thus, we investigated whether JARID1B chromatin binding and changes in JARID1B levels influence H3K4 methylation patterns in breast cancer cells. Correlating with its HDM function, we detected increased H3K4me3/me2 signal ratios following downregulation of JARID1B in MCF7 cells (Figure 3D). However, despite its H3K4me3/2 HDM activity, we found that JARID1B binding overlapped with high H3K4me3 and H3K4me2 signal in both luminal and basal-like breast cancer cells (Figures 3E and S3D), potentially due to the high affinity of its PHD domains to H3K4me3 as reported for JARID1A (Wang et al., 2009). We validated increased H3K4me3/me2 levels after siJARID1B expression for several genes with clearly overlapping JARID1B and H3K4me3/me2 peaks in their promoter region and increased expression in siJARID1B-transfected MCF7 cells (Figures 3F–G and S3B–C). Interestingly, the increase in H3K4me3 signal in siJARID1B-transfected cells was abrogated by treatment with a TGFBR kinase inhibitor (Figure 3G).
H3K4me3 and H3K4me2 mark transcriptionally active genes (Ram et al., 2011; Zhang and Pugh, 2011). In line with this, genes with JARID1B promoter binding were highly expressed in all cell lines analyzed (Figure 3H). These data suggest that JARID1B chromatin binding by itself may not be sufficient for HDM function or that JARID1B is fine-tuning the levels of genes already destined to be expressed in the cells. To further explore these associations, we analyzed JARID1B binding and H3K4me3/me2 enrichment patterns of genes up or down-regulated in siJARID1B-transfected MCF7 cells. We found that the promoter region of the differentially expressed genes was significantly enriched for JARID1B binding and also for H3K4me3 and H3K4me2 (Figure 4A) indicating a clear and strong association between JARID1B promoter binding and downstream gene expression changes in siJARID1B-transfected cells.
Figure 4. Integrated view of JARID1B activity.
(A) Integrated view of siJARID1B differential genes in MCF7 cells and its association with JARID1B, H3K4me2 and H3K4me3 promoter binding signal. Each column is a gene, ranking by the differential gene GFold value comparing siJARID1B with siControl. (B) Two of the highest scoring consensus sequence motifs in top 5,000 JARID1B peaks in MCF7 cells. z-score −87.5043 (top) and −87.3197 (bottom), p value=1e-30. (C) Box plots depict average beta values of DNA methylation for all genes, and genes up or down regulated after expression of siJARID1B. *** marks p<0.001, detailed statistical analysis is included in Supplementary Experimental Procedures. (D) Integrated view of differential gene expression (G-Fold>1, 947 genes), JARID1B, FOXA1, GATA3, ER, and TFAP2C binding at promoters and enhancers in luminal (MCF7 and T-47D combined) and basal (SUM159PT and MDA-MB-231 combined) breast cancer cell lines. Wilcoxon test for basal and luminal JARID1B signal in promoter (W=430557937, p value<2.2e-16) and in enhancer (W=511285604, p value<2.2e-16). See also Figure S4.
To investigate the sequence-specificity of JARID1B binding, we performed motif analysis of JARID1B binding sites and identified highly CG-rich sequences (Figures 4B and S4A), confirming the results of in vitro binding assays (Scibetta et al., 2007). The presence of CpG nucleotides in these consensus motifs raised the possibility that JARID1B binding might be influenced by DNA methylation. To explore this, we analyzed potential associations between DNA methylation and JARID1B targets (see Supplemental Experimental Procedures for details). Baseline DNA methylation was higher in genes that were upregulated in siJARID-transfected luminal MCF7 and T-47D cells, whereas the opposite was observed in some basal-like cells (Figure 4C). However, these differences might simply reflect the association of JARID1B with H3K4m3/me2-enriched regions as unmethylated H3K4 has been linked to de novo DNA methylation (Ooi et al., 2007). Correlating with this, JARID1B-binding sites had lower level of DNA methylation than other regions analyzed and this was also associated with differences in H3K4me3 enrichment (Figure S4B–C). Thus, DNA methylation might influence JARID1B binding and its effect on gene expression, but delineating these relationships will require further studies.
JARID1B binding is enriched in luminal-high genes
To further investigate JARID1B chromatin binding patterns, we analyzed associations between JARID1B binding and genes differentially expressed between luminal and basal-like breast cancer cells. Despite its overall binding to actively transcribed genes in all cell types, JARID1B showed significant enrichment in the promoter and enhancer regions of luminal-high genes in luminal cells with a much less pronounced pattern observed for basal-high genes in basal cells (Figure 4D). These results imply potential cooperation between JARID1B and luminal lineage-specific transcription factors (TFs). We investigated this by integrating ChIP-seq data for such TFs in breast cancer cells with our results. We found that luminal-high genes in luminal cells were enriched for genomic targets of ER, GATA3, FOXA1, and TFAP2C transcription factors with known roles in luminal differentiation (Kouros-Mehr et al., 2008; Siegel and Muller, 2010) (Figure 4D). We also found a significant overlap between JARID1B promoter binding genes and genes differentially expressed three hours after estradiol treatment in MCF7 cells (Figure S4D). However, since JARID1B peaks are enriched in highly expressed genes in luminal cells and ER is a key transcriptional activator in MCF7 cells, it is not unexpected to see a significant overlap. Thus, this observation by itself does not prove a functionally relevant link between JARID1B and ER targets.
In contrast, promoters and enhancers of basal-high genes with JARID1B binding did not show enrichment for any TFs with available ChIP-seq data (data not shown). Thus, the combined activation of luminal-specific and repression of basal-specific genes might be required for the establishment of luminal cell-specific gene expression programs and associated phenotypes, whereas basal-specific patterns might represent default states or regulated by not-yet characterized TFs.
Clinical relevance of JARID1B activity in breast cancer
To investigate the clinical relevance of JARID1B copy number gain and overexpression in breast cancer, we analyzed molecular data from 1,980 breast carcinomas (the METABRIC cohort) (Curtis et al., 2012). JARID1B expression level varied with respect to PAM50 subtype and the 10 Integrative clusters (Figure 1D–E). To explore the activity of JARID1B in a more comprehensive manner, we used PARADIGM (Vaske et al., 2010) to create signatures mirroring JARID1B activity in MCF7 and SUM159PT cell lines based on gene expression, copy-number and JARID1B ChIP-seq data (MCF7 and SUM159PT-index). The two JARID1B activity indexes were calculated for each METABRIC sample by extracting gene expression and copy number features included in the signature. For each index, samples were split into three groups; negative, zero or positive JARID1B activity, with distinct clinico-pathological features (Table S3). The three JARID1B MCF7-index groups showed different outcomes where patients with ER+ tumors and a positive JARID1B activity score had a shorter disease-specific survival than patients with ER+ tumors and negative JARID1B activity (Figure 5A). Positive JARID1B activity had a hazard ratio of 1.9 (95%CI 1.1–3.3, p=0.029) in ER+ tumors compared to negative activity after correcting for known prognostic covariates in a Cox regression model (Table S4). The differences in survival depending on JARID1B activity remained significant in ER+ tumors treated with endocrine therapy alone (Figure 5A). The JARID1B SUM159PT-index also showed a significant difference in survival for patients with ER+ tumors, but here zero activity indicated worse prognosis (Figure 5B). These data again highlight that JARID1B activity is distinct between luminal and basal breast cancer cells, and that positive luminal JARID1B activity is associated with worse outcome in luminal ER+ tumors implying oncogenic function.
Figure 5. Clinical relevance of JARID1B activity in breast tumors.
Associations between MCF7 (A) and SUM159PT (B) PARADIGM JARID1B activity indices and disease-specific survival in breast cancer patients with ER+ and ER− tumors. Red, green, and blue colors indicate positive, negative, and zero JARID1B activity. “end treat” indicates endocrine treatment only. See also Tables S3-S4.
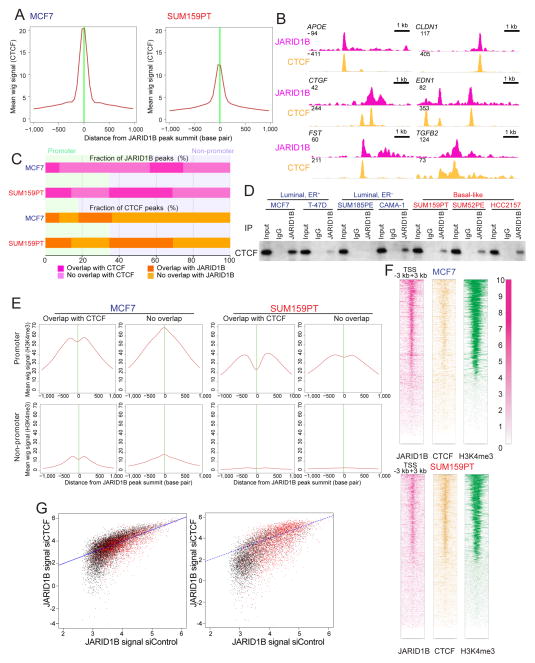
JARID1B and CTCF interaction
To identify potential JARID1B interacting proteins that may modulate its function in breast cancer cells, we conducted a DNA binding site motif analysis on JARID1B ChIP-seq peaks and identified CTCF as the top scoring factor (z-score −81.1077, −10*log(pval): 690.7755279) both in luminal and basal breast cancer cells (Table S5). We verified overlap with CTCF by ChIP-seq in both cell lines (Figure 6A–B) using ENCODE data for MCF7 (Lee et al., 2012) and our data in SUM159PT cells. Overall a relatively higher fraction of JARID1B overlaps with CTCF in SUM159PT compared to MCF7 cells both in promoter and non-promoter regions (Figure 6C). However, in MCF7 cells the ratio of overlapping sites was much smaller in promoter compared to non-promoter regions, whereas no such difference was detected in SUM159PT cells. Because the binding of CTCF is inhibited by DNA methylation (Shukla et al., 2011), we analyzed the level of DNA methylation of CTCF binding sites in relation to transcription start site (TSS) and JARID1B binding. CTCF binding sites had low level of DNA methylation, which was even lower at CTCF-JARID1B overlapping sites (Figure S5A). These differences were significant both in MCF7 and SUM159 cells but were more pronounced at non-promoters in MCF7 cells.
Figure 6. JARID1B-CTCF interaction.
(A) CTCF enrichment around JARID1B peak summits in both MCF7 and SUM159PT cells. (B) Examples of JARID1B-CTCF overlap in MCF7 cells based on visualization of ChIP-seq data for the indicated genes. (C) Bar graphs depicting JARID1B and CTCF overlap at promoter and non-promoter sites in MCF7 and SUM159PT cell lines. The differences between JARID1B-only and JARID1B-CTCF overlapping sites in non-promoter and promoter regions are statistically significant both in MCF7 and SUM159PT cells (p<2.2e-16, Fisher exact test). (D) Immunoblots showing the immunoprecipitation of JARID1B and CTCF in breast cancer cell lines. (E) H3K4me3 signal profile around JARID1B peak summit in MCF7 and SUM159PT cell lines. (F) JARID1B, CTCF, and H3K4me3 signal around all promoters in MCF7 and SUM159PT cell lines. Gene promoters are ranked by the average signal of H3K4me3. The JARID1B and CTCF signal are significantly different between MCF7 and SUM159PT cells (p value<2.2e-16, Wilcoxon test). (G) Changes in JARID1B chromatin binding in promoter peaks (left panel) and non-promoter peaks (right panel) following transfection of siCTCF in MCF7 cells. Red dots mark JARID1B-CTCF overlapping sites. See also Figure S5 and Table S5.
To determine if the overlap of JARID1B and CTCF binding sites may indicate that the two proteins are present in the same complex, we performed JARID1B immunoprecipitation followed by CTCF immunoblot in a panel of breast cancer cell lines. We detected variable levels of CTCF protein in JARID1B immunoprecipitates in all cell lines tested except in SUM185PE, suggesting that the two proteins may interact in most breast cancer cells (Figure 6D).
To explore potential associations between JARID1B-CTCF and H3K4 methylation status, we analyzed H3K4me3 signal in relation to JARID1B-CTCF overlap. The level H3K4me3 signal at sites of JARID1B-CTCF overlap was significantly lower in MCF7 luminal cells (Figure S5B) and this was most pronounced in promoters (Figure 6E). In line with this, we detected significantly (p value < 2.2e-16, Wilcoxon test) stronger JARID1B and weaker CTCF signal at promoters in MCF7 luminal cells compared to the SUM159PT basal-like cells (Figure 6F). The non-overlapping sites had higher H3K4me3 signal in MCF7 cells whereas in SUM159PT cells even these sites showed somewhat lower values implying that the JARID1B-CTCF interaction may have a more pronounced effect in luminal compared to basal breast cancer cells. These observations are intriguing in light of recurrent CTCF mutations in ER+ breast cancer (Ellis et al., 2012; 2012) implying subtype-specific role in tumorigenesis.
CTCF is known to have many different biological functions and can have opposing effects on gene expression depending on the location of its binding site relative to TSS (Filippova, 2008; Fiorentino and Giordano, 2012). Thus, to further explore the potential relevance of CTCF-JARID1B interaction in luminal breast cancer cells, we analyzed JARID1B chromatin binding and gene expression patterns in MCF7 cells following transfection with siCTCF. JARID1B peak signal significantly decreased in siCTCF-transfected cells (Figure 6G) and this decrease was more significant (p value < 2.2e-16 Wilcox test) in the JARID1B-CTCF overlapping versus non-overlapping sites in the promoter regions whereas the opposite was observed in non-promoters (Figure S5C). Coincidentally with a decrease in CTCF and JARID1B promoter binding, we observed an increase in global H3K4me3 levels in MCF7 cells (Figure S5D). In contrast, in SUM185PT cells that were negative for JARID1B-CTCF complexes by immunoprecipitation, H3K4me3 decreased after siCTCF (Figure S5D) and this was accompanied by increased JARID1B chromatin binding both in promoter and non-promoter areas (Figure S5E). These results imply that CTCF may modulate JARID1B’s chromatin binding function and HDM activity. However, many of these associations might be due to global chromatin changes induced by decreased CTCF levels and may not reflect the functional relevance of CTCF-JARID1B interaction; dissecting the underlying mechanisms requires further studies.
JARID1B mutant with luminal gain-of-function
Lastly, we investigated the potential functional relevance of the K1435R JARID1B heterozygous missense mutation in the HCC2157 basal-like breast cancer cell line (Figure 7A). Gene expression and JARID1B ChIP-seq data revealed patterns unique to HCC2157 cells that made it more similar to luminal cells (Figures 2A and 3A) although this cell line is ER−PR−HER2− and classified as a basal-like breast cancer. Indeed, many of the genes associated with HCC2157-unique JARID1B peaks showed high expression in luminal breast tumors and cell lines (Figure 7B–C). The SPDEF transcription factor with known luminal functions was among the HCC2157-unique JARID1B peaks and both SPEDF and its targets showed elevated levels in HCC2157 cells (Table S2). These data imply a gain of JARID1B binding to luminal-specific genes leading to their increased expression in this basal-like breast cancer cell line potentially due to the JARID1B mutation. To test this experimentally, we exogenously expressed Myc epitope-tagged wild type (WT) and mutant JARID1B in SUM159PT basal breast cancer cells and assessed JARID1B binding and gene expression patterns. We found that the K1435R mutant displayed greater enrichment for HCC2157-unique JARID1B peaks (Figure 7D) and this was accompanied with modest differences in the expression of some genes nearest to these peaks (Figure S6A). The relatively modest effects on gene expression are not surprising since the exogenous transfection and overexpression of WT and mutant JARID1B in SUM159PT basal cells may not fully reproduce the physiologic differences in their function; protein levels might be higher than that of endogenous and some transcription factors required for the expression of luminal genes may be lacking in SUM159PT cells. Thus, precise understanding of mutant JARID1B function in HCC2157 cells would require the analysis of cells following the selective deletion of mutant or wild type JARID1B alleles.
Figure 7. JARID1B gain of function mutation in HCC2157 cells.
(A) Schematic structure of JARID1B gene with exons, functional protein domains, and the location of somatic mutations identified in various human cancer types. (B) Heatmap of clustering of samples based on the expression of genes associated with HCC2157-unique peaks. (C) Heatmap showing the differential expression of a subset of genes (top 500 genes with higher expression in HCC2157 cells compared to other basal lines) associated with HCC2157-unique JARID1B peaks. “Cell line expr” indicates transcript levels in HCC2157 cells, Basal-lumA denotes difference in expression level between basal and luminal tumors, where blue indicates higher expression in the latter group. Green rectangle highlights luminal genes highly expressed in HCC2157 cells. (D) ChIP-qPCR analysis of enrichment for HCC2157-unique JARID1B peaks by Myc-tagged wild type and mutant (K1435R) JARID1B transfected into SUM159PT cells. * and ** asterisks indicate p<0.05 and p<0.001, respectively, whereas error bars mark STDEV. Inset shows immunoblot analysis of exogenously expressed Myc-tagged wild type and K1435R mutant JARID1B in SUM159PT cells. (E) Degree of enrichment of CTCF binding sites in JARID1B all peaks (left panel) and unique peaks (right panels) in the HCC2157 cell line. (F) Immunoblots showing the immunoprecipitation of CTCF in wild type or K1435R JARID1B mutant cells. (G) Patterns of H3K4me3 and H3K4me2 signal profile around common and HCC2157-unique JARID1B peak summit. See also Figure S6.
Intriguingly, ESRP1, encoding a key regulator of luminal splicing patterns (Warzecha and Carstens, 2012), was one of the genes more significantly enriched for mutant than wild type JARID1B and its expression increased the most significantly after mutant JARID1B transfection in SUM159PT cells (Figure S6A). ESRP1 is also one of the most significantly differentially expressed genes between luminal and basal breast cancer cells and the differences in its endogenous levels between HCC2157 and other basal-like cell lines were the largest among all genes analyzed (Figure S6A). These observations led us to explore the splicing patterns of genes with known luminal-basal cell-specific variants such as ENAH and CD44 (Shapiro et al., 2011) in our RNA-seq data. We found that the HCC2157 basal cell line expressed the luminal splice variants of all of these genes (Figure S6B) potentially due to high ESRP1 levels induced by mutant JARID1B. Correlating with this, transfection of mutant JARID1B in SUM159PT cells increased the expression of luminal splice variants for several genes (Figure S6C) further supporting the functional relevance of mutant JARID1B in HCC2157 cells.
Interestingly, HCC2157-unique JARID1B peaks did not overlap with CTCF in contrast to all JARID1B peaks detected in these cells (Figure 7E) implying the lack of co-occurrence of the two factors at the unique sites. We experimentally tested this hypothesis by performing immunoprecipitation for Myc-tagged wild type or mutant JARID1B followed by CTCF western. Lower levels of CTCF were detected in mutant JARID1B immunoprecipitates potentially implying that mutant JARID1B and CTCF may not be found in the same protein complexes (Figure 7F). These results further support the functional relevance of CTCF-JARID1B interaction and imply that somatic mutations in JARID1B (or potentially also in CTCF) may alter the composition of these complexes leading to altered JARID1B function. To explore this, we investigated H3K4me2 and H3K4me3 signal in HCC2157 cells at common and HCC2157-unique JARID1B peaks in promoter and non-promoter areas assuming that all or most of unique binding is due to the mutant protein. Unique JARID1B promoter peaks had much lower H3K4me2 and H3K4me3 signal compared to the common ones whereas non-promoters had about the same low signal for both unique and common sites (Figure 7G). Although there could be many potential explanations for these differences, these data suggest that mutant JARID1B-containing complexes may have increased HDM activity.
DISCUSSION
Perturbations of cell type-specific transcriptional programs play key roles in tumorigenesis, in particular, in breast cancer a significant fraction of recurrently mutated genes encode for transcription factors or epigenetic regulators (Polyak and Metzger Filho, 2012). Although therapeutic targeting of aberrant transcriptional programs has been challenging, the identification of BET bromodomain inhibitors as regulators of the MYC oncogene (Delmore et al., 2011; Filippakopoulos et al., 2010) have re-ignited interest in this area. Developing specific inhibitors of chromatin regulators is one of the most active areas of current drug discovery efforts. However, the validation and application of these inhibitors require the precise understanding of the biochemical and tumorigenic function of these epigenetic regulators. Here we describe a role of JARID1B in breast cancer and reveal that its chromatin binding and HDM activity might be modulated by associated proteins such as CTCF. Thus, its potential therapeutic targeting might require a more sophisticated approach than simple inhibition of enzymatic activity.
Our finding of a nearly complete overlap between JARID1B binding and H3K4me3 mark seems paradoxical as JARID1B is a H3K4me3 demethylase. However, similar observations have been made in two unrelated cell types, human leukemia (Ram et al., 2011) and mouse ESCs (Schmitz et al., 2011), thus, it appears to be a general phenomenon and it may reflect enzyme-substrate co-localization as reported for JARID1A (Lopez-Bigas et al., 2008). Correlating with the preferential binding of JARID1B to promoters enriched for H3K4me3 and active genes, downregulation of JARID1B had only modest effects on global H3K4me3 and transcript levels, especially in luminal breast cancer cells. Thus, it appears that JARID1B is not a strong transcriptional repressor, but rather it fine tunes cell type-specific histone H3K4 methylation and transcript levels.
Despite the association of JARID1B binding with actively transcribed genes both in luminal and basal-like breast cancer cells, the promoters and enhancers of luminal-high genes were significantly enriched for JARID1B binding, whereas no such enrichment is seen for basal-high genes. Thus, despite the fact that JARID1B binding is associated with active genes, there is still a significant luminal-basal difference. These results are in line with recent observations for other HDMs like KDM4B/C in ESCs (Das et al., 2013) and also the preferential effects of BET bromodomain inhibitors on BRD4 binding at “super-enhancers” and associated genes (Whyte et al., 2013). Thus, it appears that many chromatin regulators may have a relatively broad, low level binding at many sites implying non-specific action, but a subset of sites with the highest binding signal confer specificity.
We also found a colocalization between JARID1B and CTCF that appears to be specific for breast cancer cells (or epithelial cells) as it was not observed in hematopoietic and ES cells. Based on our data CTCF may modulate JARID1B binding and HDM activity, but neither of these appears to have a pronounced effect on gene transcription due to the complex regulation of Pol II at multiple different levels. Since CTCF with promoter binding pauses Pol II (Paredes et al., 2012; Shukla et al., 2011), JARID1B might be recruited to promoters to reduce H3K4me3 level to counteract Pol II accumulation (Li et al., 2007). Knockdown of CTCF, although releases Pol II pausing, dissociates JARID1B from the chromatin as well, creating muted expression changes.
The consensus JARID1B binding motif is CG-rich implying potential modulation by DNA methylation. Indeed, we found that JARID1B-bound promoters were less methylated and promoter regions of up and down-regulated genes after siJARID1B transfection displayed significant differences in DNA methylation levels. CTCF binding is also inhibited by DNA methylation (Shukla et al., 2011) and JARID1B-CTCF overlapping sites were even less methylated than those with CTCF-only binding. Based on our observations that CTCF may modulate JARID1B’s DNA binding and HDM activity, it is intriguing to speculate that the combinatorial effects of JARID1B-CTCF and regulators of DNA and H3K27 methylation work together to establish luminal epithelial differentiation programs. Correlating with this, CTCF and KDM6A mutations are commonly found in luminal breast cancer cells (Ciriello et al., 2013; Ellis et al., 2012).
The frequent copy number gain and our ChIP-seq and functional data imply that JARID1B activity may particularly be important in luminal breast tumors that in general have better outcome than basal-like breast cancer. However, our data also shows that high luminal JARID1B activity in ER+ tumors may be associated with endocrine resistance. Furthermore, the expression of a luminal-gain-of-function JARID1B mutant (e.g., K1435R JARID1B in HCC2157 cells) aberrantly activating some luminal genes in basal breast tumors may result in less favorable clinical behavior. Correlating with this, gain of luminal features such as the increased expression of CD24 is commonly observed in treatment-resistant distant metastatic lesions (Shipitsin et al., 2007) and in chemotherapy-resistant TNBCs (Almendro et al., 2014). The JARID1B mutant in HCC2157 cells displayed gain of binding to genomic loci not detected in other cells expressing the WT protein. Mutant JARID1B also appears to be associated with different proteins at these unique sites and these complexes may have increased HDM activity compared to those with WT JARID1B. Thus, the mutation may perturb both chromatin binding and enzymatic activity. Somatic point mutations in JARID1B have not been commonly observed in breast and other tumor types. However, thus far all large-scale sequencing studies have focused on primary treatment-naïve tumors, which may not reveal mutations associated with metastatic progression and therapy resistance.
Together these data imply that JARID1B functions as an oncoprotein in breast cancer by driving a luminal transcriptional program. There can be many reasons why cancer cells with luminal expression patterns may have growth advantage. One possibility is that such cells may be more resistant to therapeutic interventions, as has been described in melanoma (Roesch et al., 2010; Roesch et al., 2013). Our data demonstrating enrichment for cancer cells with luminal features in neoadjuvant chemotherapy-resistant breast tumors supports this hypothesis (Almendro et al., 2014).
In summary, here we describe the detailed molecular characterization of the JARID1B H3K4 histone demethylase in breast cancer. Our data support a key role for JARID1B in luminal cell-specific gene expression programs and demonstrate that JARID1B is a luminal lineage-driving oncogene in breast cancer. Furthermore, as ER+ breast tumors with high JARID1B activity are associated with worse clinical outcome and resistance to endocrine therapy, therapeutic targeting of JARID1B may be a potential treatment strategy for this disease.
EXPERIMENTAL PROCEDURES
Detailed procedures are described in Supplemental Experimental Procedures.
Breast cancer cell lines
Breast cancer cell lines were obtained from ATCC or generously provided by Steve Ethier (SUM cell lines, University of Michigan) and cultured following the provider’s recommendations.
siRNA transfection, RNA isolation and qRT–PCR
siRNAs against JARID1B, CTCF, SMAD4, and TGFBR2 were purchased from Dharmacon in the form of ONTARGET-plus SMARTpool reagents (L-009899, L-020165, L-003902, and L-003930, respectively). Individual siRNAs against JARID1B (LQ-009899) were also purchased and used for the rescue experiments. Transfections of siRNA (15 nM) were performed using DharmaFECT transfection reagents (Dharmacon) according to the manufacturer’s protocol. Five days after transfection, total RNA was isolated from cells using RNeasy Mini Kit (Qiagen) following the manufacturer’s protocol. First-strand cDNA synthesis was carried out using random hexamer oligos and SuperScript II (Life Technologies). Then cDNA was analyzed by quantitative PCR (qPCR) using Power SYBR PCR Master Mix and 7500 Real Time PCR Systems (Applied Biosystems). Primer sequences are available upon request.
TGFBR (Transforming Growth Factor-Beta Receptor) inhibitor treatment
Cells were seeded in a 96-well plate, and siControl or siJARID1B were transfected. On the next day, TGFBR inhibitor (LY2109761) dissolved in DMSO was added into medium at 0, 1, 5, or 10 μM final concentration. DMSO concentration in culture medium is adjusted to 0.5% final concentration. Two days later, the culture medium was changed to fresh medium containing the inhibitor and 2 days later cell viability was assessed using CellTiter-Glo (Promega).
Immunoblotting and co-immunoprecipitation (co-IP)
Cell lysates were prepared 5 days after siRNA transfection. The proteins resolved in SDS – polyacrylamide gels (4–12%) were transferred electrophoretically for 2 hr at 4 °C to polyvinylidene difluoride membranes by using a Tris-glycine buffer system. The membranes were blocked with 5% milk powder in 0.1% Tween20 in PBS (PBS-T) for 1 hr at least, and then the antibodies were added with 2.5% milk in PBS-T. The antibodies used for immunoblotting were anti-Jarid1B (HPA027179) from Sigma, anti-Jarid1A (ab78322), anti-H3K4me3 (ab8580), anti-H3K4me1 (ab8895), anti-Histone H3 (ab1791) from Abcam, anti-H3K4me2 (07-030) from Millipore, and anti-CTCF (BD612148) from BD Biosciences. For co-IP, nuclear extract of cell-lines were prepared as for the ChIP experiments, sheared by passing through syringe needle, diluted, and treated with DNase I. The samples were incubated at 4°C overnight with the JARID1B antibody, and then precipitated with Dynabeads Protein G for 2 hr. Beads were washed with buffer containing 150 mM NaCl and 0.5% NP-40 three times, and then resuspended in SDS – polyacrylamide gel loading buffer.
Myc-JARID1B overexpression
N-terminal Myc-His-tagged human JARID1B (wild-type and K1435R) expression constructs in pcDNA3.1 were kindly provided by Dr. Yi Zhang (Harvard Medical School). 15 μg of JARID1B expression plasmids were transfected into cells cultured in a 10 cm dish by Lipofectamine LTX (Life Technologies). For immunoprecipitation, cells were harvested and lysed 4 days after transfection. Then exogenous JARID1B was probed with 2.5 μg of anti-Myc antibody (9E10, Millipore). For gene expression analysis, total RNA was prepared 4 days after transfection, and evaluated by qRT-PCR. For rescue experiments, 25 ng of JARID1B expression plasmids and 20 nM siRNA were co-transfected in 96-well plate by jetPRIME transfection reagent (Polyplus). Cell viability was examined five days after transfection.
Supplementary Material
HIGHLIGHTS.
Somatic genetic alterations of JARID1B in breast cancer
Importance of JARID1B for luminal gene expression programs
Missense mutation in JARID1B leads to gain of luminal genes expression
High luminal JARID1B activity is associated with poor outcome in ER+ tumors
SIGNIFICANCE.
Transcriptional regulators are recurrently mutated in breast cancer highlighting the importance of perturbed epigenetic programs in tumorigenesis. Here we show that JARID1B is commonly mutated in breast tumors and it modulates luminal and basal cell-specific expression programs. A heterozygous JARID1B mutation in basal-like cells leads to gain of binding to luminal genes. JARID1B binding overlaps with CTCF and the two proteins form a potentially functionally relevant complex as H3K4me3 signal is lower at JARID1B-CTCF-overlapping compared to non-overlapping sites. Our data suggest that JARID1B is a luminal lineage-driving oncogene that may represent a target in luminal breast tumors. However, the complexity of JARID1B’s functions suggests that its therapeutic targeting may require a more intricate approach than inhibition of enzymatic activity.
Acknowledgments
We thank members of our laboratories for their critical reading of this manuscript and useful discussions. We thank Dr. Jonathan Yingling (Eli Lilly) for providing the LY2109761 TGBR kinase inhibitor, Dr. P. Senthupathy (University of North Carolina) for providing the list of paused genes in MCF7 cells, and Chenfei Wang for his help with GEO submission. This research was supported by the National Cancer Institute CA080111 (K.P.) and GM099409 (X.S.L.), the Norwegian Cancer Society (H.G.R.), Radiumhospitalets Foundation (H.G.R.), Novartis Oncology (K.P.), and the Susan G. Komen Foundation (R.M. and K.P.).
Footnotes
ACCESSION NUMBERS
All raw genomic data was deposited to GEO under accession number GSE46073. The accession number for the SNP array data is GSE19399.
Supplemental Information includes Supplemental Experimental Procedures, six figures, and five tables.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Almendro V, Cheng YK, Randles A, Itzkovitz S, Marusyk A, Ametller E, Gonzalez-Farre X, Munoz M, Russnes HG, Helland A, et al. Inference of Tumor Evolution during Chemotherapy by Computational Modeling and In Situ Analysis of Genetic and Phenotypic Cellular Diversity. Cell reports. 2014 doi: 10.1016/j.celrep.2013.12.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernardo GM, Lozada KL, Miedler JD, Harburg G, Hewitt SC, Mosley JD, Godwin AK, Korach KS, Visvader JE, Kaestner KH, et al. FOXA1 is an essential determinant of ERalpha expression and mammary ductal morphogenesis. Development. 2010;137:2045–2054. doi: 10.1242/dev.043299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bessarabova M, Pustovalova O, Shi W, Serebriyskaya T, Ishkin A, Polyak K, Velculescu VE, Nikolskaya T, Nikolsky Y. Functional synergies yet distinct modulators affected by genetic alterations in common human cancers. Cancer Res. 2011;71:3471–3481. doi: 10.1158/0008-5472.CAN-10-3038. [DOI] [PubMed] [Google Scholar]
- Catchpole S, Spencer-Dene B, Hall D, Santangelo S, Rosewell I, Guenatri M, Beatson R, Scibetta AG, Burchell JM, Taylor-Papadimitriou J. PLU-1/JARID1B/KDM5B is required for embryonic survival and contributes to cell proliferation in the mammary gland and in ER+ breast cancer cells. Int J Oncol. 2011;38:1267–1277. doi: 10.3892/ijo.2011.956. [DOI] [PubMed] [Google Scholar]
- Christensen J, Agger K, Cloos PA, Pasini D, Rose S, Sennels L, Rappsilber J, Hansen KH, Salcini AE, Helin K. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Ciriello G, Miller ML, Aksoy BA, Senbabaoglu Y, Schultz N, Sander C. Emerging landscape of oncogenic signatures across human cancers. Nat Genet. 2013;45:1127–1133. doi: 10.1038/ng.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, et al. The genomic and transcriptomic architecture of 2000 breast tumours reveals novel subgroups. Nature. 2012 doi: 10.1038/nature10983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das PP, Shao Z, Beyaz S, Apostolou E, Pinello L, Angeles AD, O’Brien K, Atsma JM, Fujiwara Y, Nguyen M, et al. Distinct and Combinatorial Functions of Jmjd2b/Kdm4b and Jmjd2c/Kdm4c in Mouse Embryonic Stem Cell Identity. Mol Cell. 2013 doi: 10.1016/j.molcel.2013.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150:12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- Delmore JE, Issa GC, Lemieux ME, Rahl PB, Shi J, Jacobs HM, Kastritis E, Gilpatrick T, Paranal RM, Qi J, et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell. 2011;146:904–917. doi: 10.1016/j.cell.2011.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dey BK, Stalker L, Schnerch A, Bhatia M, Taylor-Papidimitriou J, Wynder C. The histone demethylase KDM5b/JARID1b plays a role in cell fate decisions by blocking terminal differentiation. Mol Cell Biol. 2008;28:5312–5327. doi: 10.1128/MCB.00128-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis MJ, Ding L, Shen D, Luo J, Suman VJ, Wallis JW, Van Tine BA, Hoog J, Goiffon RJ, Goldstein TC, et al. Whole-genome analysis informs breast cancer response to aromatase inhibition. Nature. 2012;486:353–360. doi: 10.1038/nature11143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Meyer CA, Wang Q, Liu JS, Shirley Liu X, Zhang Y. GFOLD: a generalized fold change for ranking differentially expressed genes from RNA-seq data. Bioinformatics. 2012;28:2782–2788. doi: 10.1093/bioinformatics/bts515. [DOI] [PubMed] [Google Scholar]
- Filippakopoulos P, Qi J, Picaud S, Shen Y, Smith WB, Fedorov O, Morse EM, Keates T, Hickman TT, Felletar I, et al. Selective inhibition of BET bromodomains. Nature. 2010;468:1067–1073. doi: 10.1038/nature09504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filippova GN. Genetics and epigenetics of the multifunctional protein CTCF. Current topics in developmental biology. 2008;80:337–360. doi: 10.1016/S0070-2153(07)80009-3. [DOI] [PubMed] [Google Scholar]
- Fiorentino FP, Giordano A. The tumor suppressor role of CTCF. J Cell Physiol. 2012;227:479–492. doi: 10.1002/jcp.22780. [DOI] [PubMed] [Google Scholar]
- Frankenberg S, Smith L, Greenfield A, Zernicka-Goetz M. Novel gene expression patterns along the proximo-distal axis of the mouse embryo before gastrulation. BMC Dev Biol. 2007;7:8. doi: 10.1186/1471-213X-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayami S, Yoshimatsu M, Veerakumarasivam A, Unoki M, Iwai Y, Tsunoda T, Field HI, Kelly JD, Neal DE, Yamaue H, et al. Overexpression of the JmjC histone demethylase KDM5B in human carcinogenesis: involvement in the proliferation of cancer cells through the E2F/RB pathway. Molecular cancer. 2010;9:59. doi: 10.1186/1476-4598-9-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwase S, Lan F, Bayliss P, de la Torre-Ubieta L, Huarte M, Qi HH, Whetstine JR, Bonni A, Roberts TM, Shi Y. The X-linked mental retardation gene SMCX/JARID1C defines a family of histone H3 lysine 4 demethylases. Cell. 2007;128:1077–1088. doi: 10.1016/j.cell.2007.02.017. [DOI] [PubMed] [Google Scholar]
- Kidder BL, Hu G, Yu ZX, Liu C, Zhao K. Extended self-renewal and accelerated reprogramming in the absence of Kdm5b. Mol Cell Biol. 2013;33:4793–4810. doi: 10.1128/MCB.00692-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H, Kim JW, Bechis SK, Werb Z. GATA-3 and the regulation of the mammary luminal cell fate. Curr Opin Cell Biol. 2008;20:164–170. doi: 10.1016/j.ceb.2008.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouros-Mehr H, Slorach EM, Sternlicht MD, Werb Z. GATA-3 maintains the differentiation of the luminal cell fate in the mammary gland. Cell. 2006;127:1041–1055. doi: 10.1016/j.cell.2006.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kristensen LH, Nielsen AL, Helgstrand C, Lees M, Cloos P, Kastrup JS, Helin K, Olsen L, Gajhede M. Studies of H3K4me3 demethylation by KDM5B/Jarid1B/PLU1 reveals strong substrate recognition in vitro and identifies 2,4-pyridine-dicarboxylic acid as an in vitro and in cell inhibitor. FEBS J. 2012;279:1905–1914. doi: 10.1111/j.1742-4658.2012.08567.x. [DOI] [PubMed] [Google Scholar]
- Lee BK, Bhinge AA, Battenhouse A, McDaniell RM, Liu Z, Song L, Ni Y, Birney E, Lieb JD, Furey TS, et al. Cell-type specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res. 2012;22:9–24. doi: 10.1101/gr.127597.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- Lopez-Bigas N, Kisiel TA, Dewaal DC, Holmes KB, Volkert TL, Gupta S, Love J, Murray HL, Young RA, Benevolenskaya EV. Genome-wide analysis of the H3K4 histone demethylase RBP2 reveals a transcriptional program controlling differentiation. Mol Cell. 2008;31:520–530. doi: 10.1016/j.molcel.2008.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu PJ, Sundquist K, Baeckstrom D, Poulsom R, Hanby A, Meier-Ewert S, Jones T, Mitchell M, Pitha-Rowe P, Freemont P, Taylor-Papadimitriou J. A novel gene (PLU-1) containing highly conserved putative DNA/chromatin binding motifs is specifically up-regulated in breast cancer. J Biol Chem. 1999;274:15633–15645. doi: 10.1074/jbc.274.22.15633. [DOI] [PubMed] [Google Scholar]
- Nikolsky Y, Sviridov E, Yao J, Dosymbekov D, Ustyansky V, Kaznacheev V, Dezso Z, Mulvey L, Macconaill LE, Winckler W, et al. Genome-wide functional synergy between amplified and mutated genes in human breast cancer. Cancer Res. 2008;68:9532–9540. doi: 10.1158/0008-5472.CAN-08-3082. [DOI] [PubMed] [Google Scholar]
- Ooi SK, Qiu C, Bernstein E, Li K, Jia D, Yang Z, Erdjument-Bromage H, Tempst P, Lin SP, Allis CD, et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature. 2007;448:714–717. doi: 10.1038/nature05987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paredes SH, Melgar MF, Sethupathy P. Promoter proximal CTCF binding is associated with an increase in the transcriptional pausing index. Bioinformatics. 2012 doi: 10.1093/bioinformatics/bts596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker JS, Mullins M, Cheang MCU, Leung S, Voduc D, Vickery T, Davies S, Fauron C, He X, Hu Z. Supervised risk predictor of breast cancer based on intrinsic subtypes. Journal of Clinical Oncology. 2009;27:1160. doi: 10.1200/JCO.2008.18.1370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–752. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- Polyak K, Metzger Filho O. SnapShot: breast cancer. Cancer Cell. 2012;22:562–562. e561. doi: 10.1016/j.ccr.2012.06.021. [DOI] [PubMed] [Google Scholar]
- Ram O, Goren A, Amit I, Shoresh N, Yosef N, Ernst J, Kellis M, Gymrek M, Issner R, Coyne M, et al. Combinatorial patterning of chromatin regulators uncovered by genome-wide location analysis in human cells. Cell. 2011;147:1628–1639. doi: 10.1016/j.cell.2011.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Becker B, Schneider-Brachert W, Hagen I, Landthaler M, Vogt T. Re-expression of the retinoblastoma-binding protein 2-homolog 1 reveals tumor-suppressive functions in highly metastatic melanoma cells. J Invest Dermatol. 2006;126:1850–1859. doi: 10.1038/sj.jid.5700324. [DOI] [PubMed] [Google Scholar]
- Roesch A, Fukunaga-Kalabis M, Schmidt EC, Zabierowski SE, Brafford PA, Vultur A, Basu D, Gimotty P, Vogt T, Herlyn M. A temporarily distinct subpopulation of slow-cycling melanoma cells is required for continuous tumor growth. Cell. 2010;141:583–594. doi: 10.1016/j.cell.2010.04.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roesch A, Mueller AM, Stempfl T, Moehle C, Landthaler M, Vogt T. RBP2-H1/JARID1B is a transcriptional regulator with a tumor suppressive potential in melanoma cells. Int J Cancer. 2008;122:1047–1057. doi: 10.1002/ijc.23211. [DOI] [PubMed] [Google Scholar]
- Roesch A, Vultur A, Bogeski I, Wang H, Zimmermann KM, Speicher D, Korbel C, Laschke MW, Gimotty PA, Philipp SE, et al. Overcoming intrinsic multidrug resistance in melanoma by blocking the mitochondrial respiratory chain of slow-cycling JARID1B(high) cells. Cancer Cell. 2013;23:811–825. doi: 10.1016/j.ccr.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz SU, Albert M, Malatesta M, Morey L, Johansen JV, Bak M, Tommerup N, Abarrategui I, Helin K. Jarid1b targets genes regulating development and is involved in neural differentiation. Embo J. 2011;30:4586–4600. doi: 10.1038/emboj.2011.383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scibetta AG, Santangelo S, Coleman J, Hall D, Chaplin T, Copier J, Catchpole S, Burchell J, Taylor-Papadimitriou J. Functional analysis of the transcription repressor PLU-1/JARID1B. Mol Cell Biol. 2007;27:7220–7235. doi: 10.1128/MCB.00274-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shapiro IM, Cheng AW, Flytzanis NC, Balsamo M, Condeelis JS, Oktay MH, Burge CB, Gertler FB. An EMT-driven alternative splicing program occurs in human breast cancer and modulates cellular phenotype. PLoS genetics. 2011;7:e1002218. doi: 10.1371/journal.pgen.1002218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shipitsin M, Campbell LL, Argani P, Weremowicz S, Bloushtain-Qimron N, Yao J, Nikolskaya T, Serebryiskaya T, Beroukhim R, Hu M, et al. Molecular definition of breast tumor heterogeneity. Cancer Cell. 2007;11:259–273. doi: 10.1016/j.ccr.2007.01.013. [DOI] [PubMed] [Google Scholar]
- Shukla S, Kavak E, Gregory M, Imashimizu M, Shutinoski B, Kashlev M, Oberdoerffer P, Sandberg R, Oberdoerffer S. CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature. 2011;479:74–79. doi: 10.1038/nature10442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siegel PM, Muller WJ. Transcription factor regulatory networks in mammary epithelial development and tumorigenesis. Oncogene. 2010;29:2753–2759. doi: 10.1038/onc.2010.43. [DOI] [PubMed] [Google Scholar]
- Sørlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70. doi: 10.1038/nature11412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D, Stuart JM. Inference of patient-specific pathway activities from multi-dimensional cancer genomics data using PARADIGM. Bioinformatics. 2010;26:i237–245. doi: 10.1093/bioinformatics/btq182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GG, Song J, Wang Z, Dormann HL, Casadio F, Li H, Luo JL, Patel DJ, Allis CD. Haematopoietic malignancies caused by dysregulation of a chromatin-binding PHD finger. Nature. 2009;459:847–851. doi: 10.1038/nature08036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warzecha CC, Carstens RP. Complex changes in alternative pre-mRNA splicing play a central role in the epithelial-to-mesenchymal transition (EMT) Semin Cancer Biol. 2012;22:417–427. doi: 10.1016/j.semcancer.2012.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte WA, Orlando DA, Hnisz D, Abraham BJ, Lin CY, Kagey MH, Rahl PB, Lee TI, Young RA. Master transcription factors and mediator establish super-enhancers at key cell identity genes. Cell. 2013;153:307–319. doi: 10.1016/j.cell.2013.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang Y, Zhu Z, Han G, Ye X, Xu B, Peng Z, Ma Y, Yu Y, Lin H, Chen AP, Chen CD. JARID1B is a histone H3 lysine 4 demethylase up-regulated in prostate cancer. Proc Natl Acad Sci U S A. 2007;104:19226–19231. doi: 10.1073/pnas.0700735104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie L, Pelz C, Wang W, Bashar A, Varlamova O, Shadle S, Impey S. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. Embo J. 2011;30:1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamane K, Tateishi K, Klose RJ, Fang J, Fabrizio LA, Erdjument-Bromage H, Taylor-Papadimitriou J, Tempst P, Zhang Y. PLU-1 is an H3K4 demethylase involved in transcriptional repression and breast cancer cell proliferation. Mol Cell. 2007;25:801–812. doi: 10.1016/j.molcel.2007.03.001. [DOI] [PubMed] [Google Scholar]
- You JS, Jones PA. Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell. 2012;22:9–20. doi: 10.1016/j.ccr.2012.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Pugh BF. High-resolution genome-wide mapping of the primary structure of chromatin. Cell. 2011;144:175–186. doi: 10.1016/j.cell.2011.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.