Abstract
Objective
To measure the performance characteristics of an immunochromatographic rapid antigen test for respiratory syncytial virus (RSV) and determine how its interpretation should be contextualized in patients presenting to the emergency department (ED) with bronchiolitis.
Design
Diagnostic accuracy study of a rapid RSV test.
Setting
County hospital emergency department.
Intervention
We took paired nasal samples from consecutively enrolled infants with bronchiolitis and tested them with a rapid immunochromatographic antigen test and reverse transcriptase polymerase chain reaction gold standard.
Outcome measures
Sensitivity, specificity, likelihood ratios, predictive values, evidence of spectrum bias and clinical characteristics of the patients. Using these we constructed a graphical contextual model to show how the results of RSV antigen tests from infants presenting within 24 hours should influence interpretation of subsequent antigen tests.
Results
We analyzed 607 patients. The sensitivity and specificity for immunochromatographic testing was 79.4% (95% CI 73.9%, 84.2%) and 67.1% (95% CI 61.9%, 72%) respectively. We found little evidence of spectrum bias. In our contextual model the best predictor of a positive RT-PCR test was a positive antigen test OR 5.47 (95%CI 3.65, 8.18) and the number of other infants having positive tests within 24 hours OR 1.48 (95%CI 1.26, 1.72) per infant. Increasing numbers presenting to the ED with bronchiolitis in a given day increases the probability of RSV infection.
Conclusion
The RSV antigen test we examined had modest performance characteristics. The results of the antigen test should be interpreted in the context of the results of previous tests.
Keywords: infant, respiratory syncytial virus, antigen testing, polymerase chain reaction, predictive value
Introduction
Respiratory syncytial virus (RSV) infection is ubiquitous. Most children have serological evidence of RSV infection by two years.1,2 RSV is the commonest cause of bronchiolitis,3,4 and one of the commonest causes of infant hospitalization.5,6
Diagnostic confirmation of respiratory syncytial virus (RSV) infection greatly decreases the risk of coexisting bacterial illness, particularly in the presence of bronchiolitis.7–10 Clinicians can often safely limit or omit testing and treating for bacterial infection if RSV is confirmed. Similarly although any infection can induce central apnea, RSV is the commonest cause.11 Consequently RSV testing is common in emergency departments (ED). As RSV-specific anti-viral agents become available emergency physicians will come under pressure to prescribe them, and therefore to test for RSV.
Clinical laboratories typically rely on easily-performed rapid immunochromatographic antigen (RA) tests to detect RSV. The results are available in approximately twenty minutes and are reported as positive, negative, or rarely equivocal. To ascertain if this apparent simplicity of interpretation is justified we asked three questions:
First, what is the independently-measured sensitivity and specificity of an RA RSV test using polymerase chain reaction (RT-PCR) as the reference standard? Independent assessment rather than relying on manufacturer supplied data is important as others have failed to reproduce manufacturer claimed performance in other tests.12–14
Second, how does this diagnostic performance affect a test result given local RSV prevalence? For clinicians the pertinent question is, if the patient tests positive for RSV what is the probability that this patient in fact has RSV? The probability that a positive test is truly positive is related to how prevalent RSV is in infants at the time of testing.
Third, should clinical context modify interpretation of rapid RSV test results? We defined clinical context as the physical examination findings and severity of illness in the patient being tested.
Overall context incorporates both the effects of local prevalence and clinical findings. In this study, overall context incorporates the results of antigen tests performed on other infants who presented with bronchiolitis 24 hours of the infant being tested, rather than over days or weeks. This approach accounts for both the recognized seasonality of RSV, and the practical problem of never being quite sure where one is in a given season until after the season has ended.
Methods
Study design
We conducted a prospective diagnostic accuracy study of a rapid immunochromatographic RSV antigen test. We defined RT-PCR as the gold standard. Our outcomes were sensitivity, specificity, the effect of prevalence, clinical findings, and overall context on predictive values for the rapid immunochromatographic antigen test.
Setting
Emergency Department in a county teaching hospital.
Participants
We included patients who had a clinical diagnosis of bronchiolitis, and in whom the clinician ordered a rapid RSV test. We defined bronchiolitis operationally as evidence (e.g. wheezing, chest wall retractions) of lower airway obstruction following a period of upper respiratory tract symptoms up to the eighteenth month of life. Research assistants identified potential subjects by real time triage note review and observation of waiting patients. Eligibility was determined by mid-level providers, faculty and selected resident physicians. We obtained written, informed consent from the parent/guardian.
Patients were excluded if the clinician did not intend to order a rapid RSV test, consent was refused, RSV testing was obtained prior to screening, or if an alternative transport or collection system to that specified was used.
Outcome measures
Our primary outcome measures were: RSV antigen test results, positive or negative (we repeated equivocal tests), and RT-PCR RSV results positive or negative. These were used to calculate antigen test characteristics. Secondary outcomes included the presence of an additional or alternative etiology by RT-PCR, specifically human metapneumovirus (HMV) or influenza (INF) A or B.
Sample collection
We collected samples for antigen testing using saline anterior nasal aspirates as described by Hall.15 These specimens were immediately sent to our clinical laboratory for testing. We obtained paired samples for reverse transcriptase polymerase chain reaction (RT-PCR) testing with nasal mid-turbinate nylon flocked swabs placed in universal transport medium-room temperature (UTM-RT), (Copan Diagnostics, Murrieta, CA). The UTM-RT-swab specimens were frozen to −20°C prior to being shipped on dry ice for batch testing. This collection and transport system has been described in detail elsewhere.16 The order of swab or wash was not specified. Each collection system was performed on opposite nostrils.
Specimen testing
We performed immunochromatographic testing using Directigen RSV, (Becton Dickinson & Company, Sparks, MD) in accordance with the manufacturer’s instructions in the hospital clinical laboratory by certified laboratory technicians.17 This test uses antibodies against the fusion (F) and nucleocapsid proteins. RT-PCR testing was performed in batches at a remote research laboratory. The laboratory performing the RT-PCR was blinded to the antigen test results. The RT-PCR assay is described in the Appendix.
We also performed RT-PCR testing for three other pathogens: HMV, INF A and INF B on all specimens. All RT-PCR tests had a minimum detection threshold of 100 genomic copies. We did this rather than attempting direct immunoflourescence (DFA) or viral culture as a “tiebreaker.” DFA is a highly microscopist dependent technique. The relative fragility of the RSV virus would result in a falsely low detection rate because viral culture was not immediately available.
Sample size calculation
Assuming half the subjects would truly have RSV, an RA test sensitivity of 90% +/− 5%, a power of 90% and a significance level of 5% we would have required 154 patients.
Anticipating the multivariate models contemplated by our study questions we followed Long’s guideline of at least 500 subjects and anticipated requiring three bronchiolitis seasons to achieve this.18
Calculation of diagnostic accuracy
We calculated sensitivity, specificity, predictive values and likelihood ratios using the diagt command in Stata.19 Spectrum bias occurs when supposedly stable test characteristics, such as sensitivity and specificity, actually differ between groups. For example; echocardiography may detect central pulmonary emboli but not smaller peripheral ones. We sought spectrum bias by repeating these calculations for the following subgroups: age < 2 months, increased work of breathing, hypoxia (SaO2<92%), and National Children’s Hospital severity of bronchiolitis (NCH-SoB) score. The NCH-SoB score is a validated ordinal regression model which classifies bronchiolitis as mild, moderate, or severe.20 These subgroups were chosen a priori for their clinical relevance.
Discordant Results
We addressed discordant RT-PCR and immunochromatographic RSV tests by comparing the prevalence of alternative etiologies. The alternative etiologies we tested for were human metapneumovirus, (HMV) and influenza (INF) A and B. We compared the prevalence of an alternative etiology when the immunochromatographic RSV test was positive but the RT-PCR negative, with the prevalence of an alternative etiology when the immunochromatographic RSV test was negative but the RT-PCR was positive using Fisher’s exact test. Since there are no conserved epitopes between these viruses more alternative diagnoses among infants who had negative RT-PCR tests in the face of a positive RSV antigen test would reassure us of our criterion reference’s performance.
Estimating the effects of prevalence on test performance
We used the sensitivity and specificity calculated on the whole sample to recalculate and graph the positive and negative predictive values for all possible prevalence. We used likelihood ratios to construct a Fagan nomogram.21 The Fagan nomogram allows users to calculate the post-test probability that an infant has RSV given baseline prevalence and the test result.
Estimating the effect of prior test results
In practice clinicians rarely know what local prevalence is, especially for RSV whose seasons vary by year and location. We used logistic regression to model the effect of the antigen test and prior antigen tests on RT-PCR proven RSV. The first two models only additional positive or negative prior tests to allow for a clear one-dimensional depiction of the process. The third model allowed combinations of positive and negative tests within 24 hours. We created a two panel color coded graph to allow visualization of these effects; one panel for the interpretation of prior results given a new positive antigen test and one panel for a new negative antigen test.
Estimating the effect of clinical characteristics on test performance
We performed univariate analysis on clinical characteristics that we thought may influence test performance. Those with a p value of <0.1 were eligible for inclusion in the model. Rather than relying on automated forward or backward stepwise regression procedures we manually selected plausible candidate variables and combination of variables. We incorporated these variables into the models described above and compared model fit and diagnostics. The institutional review board of Kern Medical Center approved this study.
Results
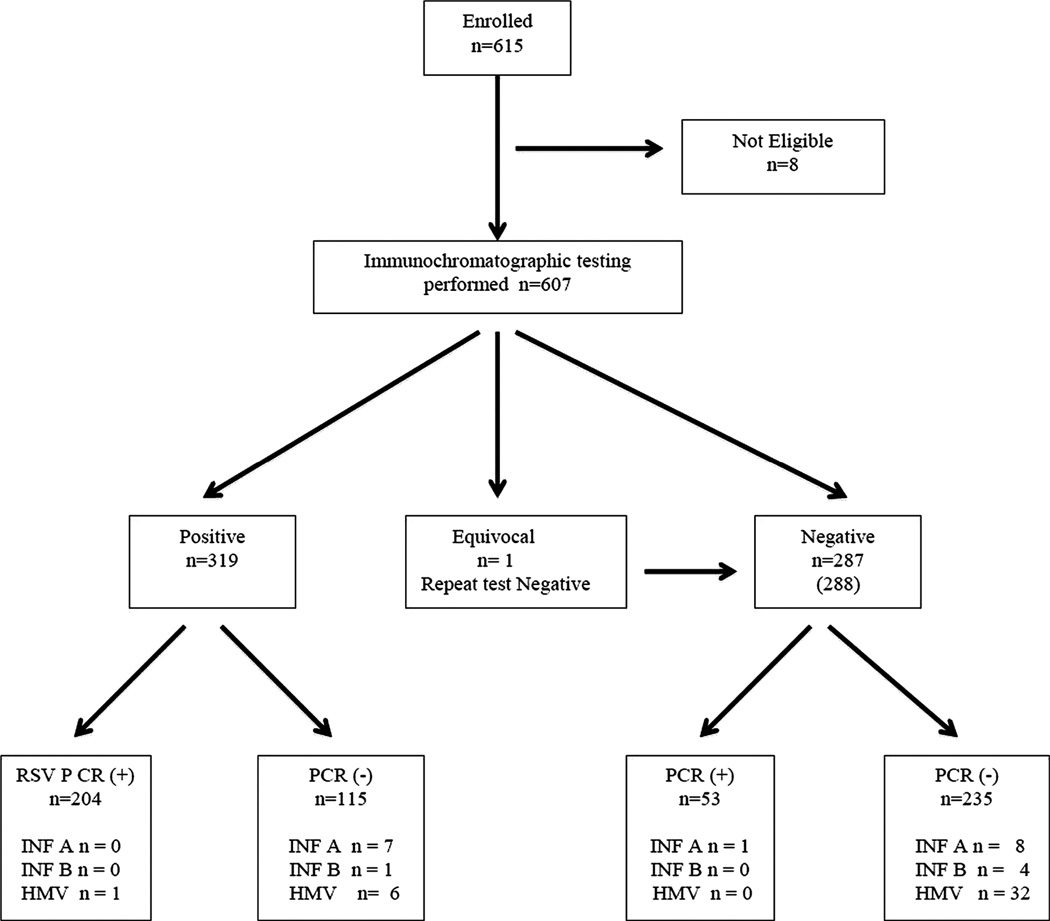
We enrolled 615 patients. Although it was theoretically possible to enroll patients in summer as a practical matter none were. Eight (1.3%) patients were excluded for lack of paired antigen/swab testing. Of the remaining 607 patients, 346 (57.0%) were male. Most had moderately severe bronchiolitis. The patients are described in Table 1. Figure 1 shows the antigen and RT-PCR results in standards for the reporting of diagnostic accuracy studies (STARD) format available at www.stard-statement.org.
Table 1. Demographic and clinical characteristics of the patients studied.
NCH-SOB, National Children’s Hospital severity of bronchiolitis scale. SD standard deviation, IQR interquartile range. All vital signs refer to the first set obtained at triage.
| Patient characteristics |
RSV n =257 |
Non RSV n=350 |
Total n=607 |
|---|---|---|---|
| Age, median (IQR) | 3.7 (5.4) | 4.3 (7.0) | 4.1 ( 6.3) |
| Age <2 month | 82 (32%) | 84 (24%) | 166(27%) |
| HR Mean (SD) | 154 (23) | 156 (23) | 155 (23) |
| Tachycardic | 55(21%) | 81 (23%) | 136 (22%) |
| Temp | 99.9 (2.0) | 99.9 (1.9) | 99.9 (1.6) |
| Febrile | 86(33%) | 115 (33%) | 201 (33%) |
| Respiratory Rate , mean (SD) | 46 (13) | 44 (13) | 45(13) |
| Rate >60 | 35 (14%) | 36 (10%) | 71 (12%) |
| Work of Breathing* | |||
| Normal /Mild | 117 (46%) | 124 (36%) | 241(40%) |
| Moderate/severe | 137 (54%) | 222 (64%) | 359(60%) |
| SaO2,mean (SD) | 97% (4) | 98% (3) | 98(3) |
| Hypoxic | 18 (7%) | 15 (4%) | 33(5%) |
| NCH-SoB(19) | |||
| Mild | 36 (16%) | 52 (17%) | 88 (16%) |
| Moderate | 171 (75%) | 231 (74%) | 402 (74%) |
| Severe | 21 (9%) | 29 (9%) | 50 (9%) |
total less than 607 because of incomplete data sheets. Total percentage may not 100 because of rounding.
Figure 1.
Patient flow and PCR and antigen results
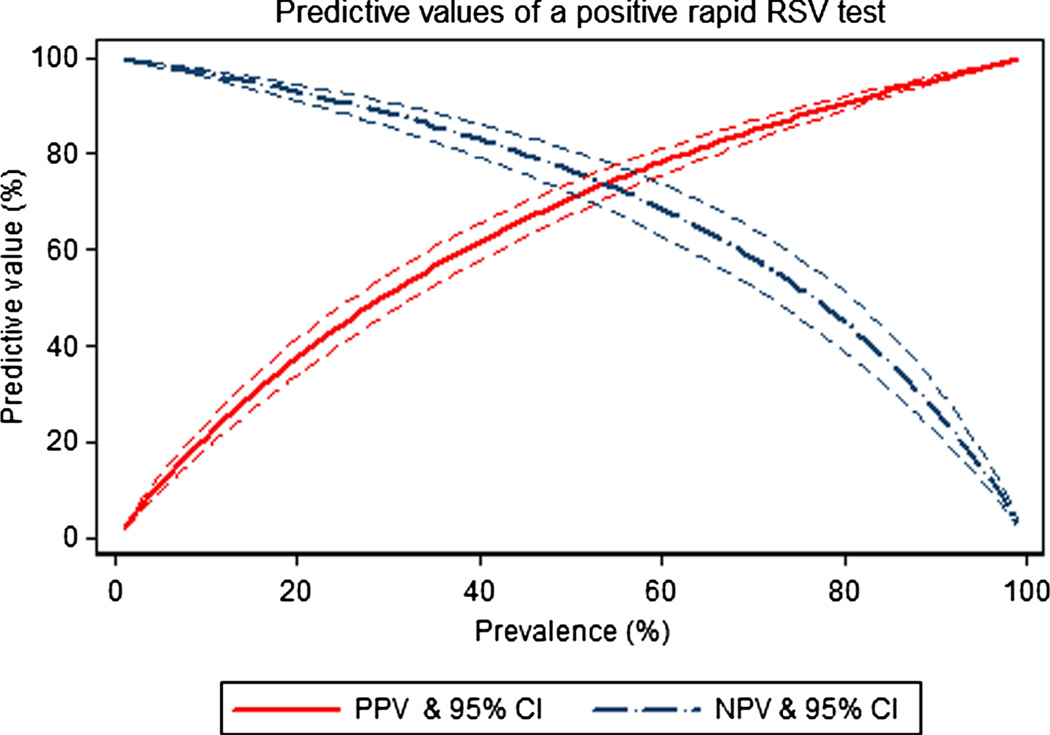
The sensitivity and specificity for antigen testing were 79.4% (95% CI 73.9%, 84.2%) and 67.1% (95% CI 61.9%, 72%) respectively. The area under the receiver operating curve (ROC) was 0.73. Predictive values and their confidence intervals for all possible prevalence are shown in Figure 2. This should not be confused with the more familiar ROC. Whereas the ROC allows the user to determine optimum sensitivity and specificity cut points for a numeric test Figure 2 shows what the predictive value is for positive and negative tests for all possible prevalence. Figure 2 also shows that the discriminating ability of the antigen test peaks when the prevalence of RSV is 53.5%.
Figure 2.
A plot of positive and negative predictive value with CIs for the immunochromatographic RSV test related to prevalence. The red continuous curve shows how the positive predictive value increases with prevalence. The blue dot dashed line shows how the negative predictive value decreases with prevalence. The dashed lines represent the 95% CIs. The CIs for test performance are expected to approach zero as the true prevalence approaches zero or 100%. The two lines intersect at the prevalence where the discriminatory ability of the test is maximal.
Table 2 shows the performance of the antigen test in various subgroups. The antigen test may perform slightly better with moderately severe bronchiolitis and increased work of breathing, but overall we found little evidence of spectrum bias.
Table 2.
Effect of patient characteristics on observed sensitivity and specificity of the immunochromatographic RSV test. NCH-SOB, National Children’s Hospital severity of bronchiolitis scale. All vital signs refer to the first set obtained at triage.
| Sensitivity (95% CI) | Specificity (95% CI) | |
|---|---|---|
| Age | ||
| < 2 months | 78.3 (71.4, 84.2) | 69.5 (63.6, 75.0) |
| ≥ 2 months | 81.7 (71.6, 89.4) | 59.5 (48.3, 70.1) |
| Respiratory rate | ||
| > 60 | 79.7 (73.8, 84.8) | 69.7 (64.3, 74.8) |
| ≤ 60 | 77.1 (59.9, 89.6) | 44.4 (27.9, 61.9) |
| Pulse oximetry | ||
| Sa O2 >92% | 79.5 (73.8, 84.4) | 67.7 (62.5, 72.7) |
| Sa O2≤92% | 77.8 (52.4, 93.6) | 53.3 (26.6, 78.7) |
| Work of breathing | ||
| Normal/Mild | 76.9 (68.2, 84.2) | 62.9 (53.8, 71.4) |
| Moderate/severe | 81.8 (74.3, 87.8) | 69.8 (63.3, 75.8) |
| Heart Rate | ||
| <98th centile | 79.7 (73.5, 85.0) | 66.9 (60.9, 72.5) |
| ≥98th centile | 78.2 (65.0, 88.2) | 67.9 (56.6, 77.8) |
| Rectal temperature | ||
| <38°C | 77.8 (62.9, 88.8) | 67.9 (56.8, 77.6) |
| ≥38°C | 79.7 (73.7, 84.9) | 66.9 (60.9, 72.5) |
| NCH-SoB | ||
| Mild | 69.4 (51.9, 83.7) | 55.8 (41.3, 69.5) |
| Moderate | 81.8 (75.3, 87.3) | 70.1 (63.8, 76.0) |
| Severe | 71.4 (47.8, 88.7) | 62.1 (42.3, 79.3) |
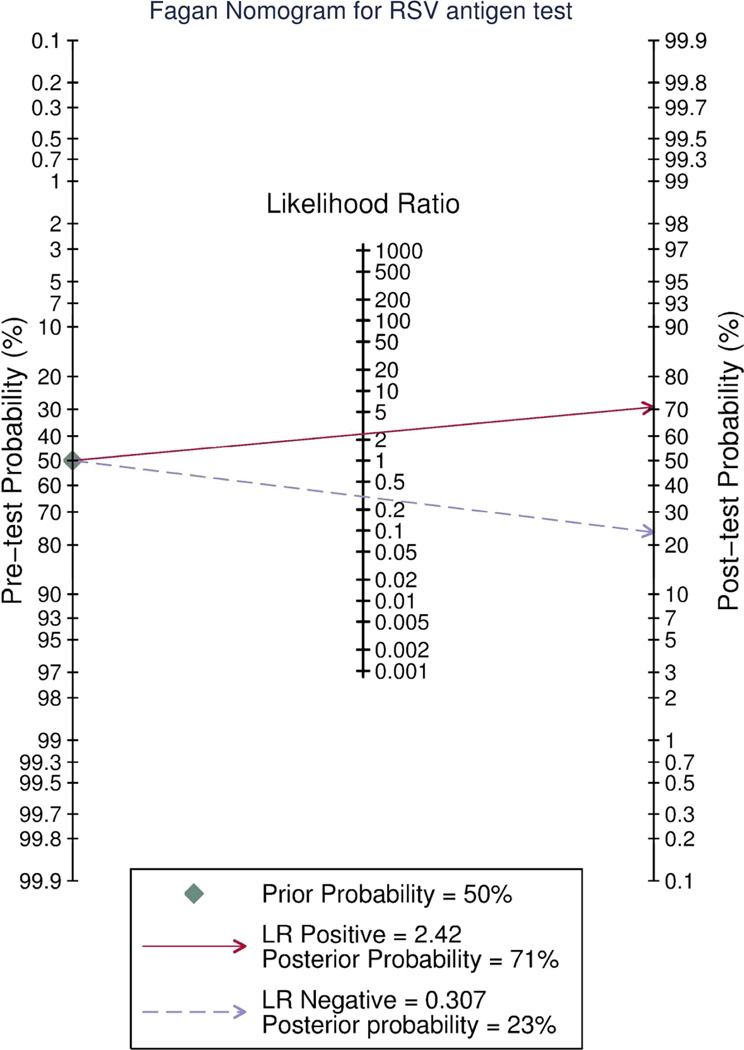
The Fagan nomogram (Figure 3) shows how the antigen test result changes the post-test probability of RSV being present. Drawing a line from the pretest probability through the positive (for a positive test) or negative (for a negative test) likelihood ratios on the central axis will show the post-test probability of RT-PCR proven RSV. When pretest probability is uncertain local prevalence is often substituted. For illustrative purposes we have drawn the lines where the pretest probability is 50%.
Figure 3.
The Fagan nomogram allows the user obtain a Bayesian estimate of the probability of a positive or negative immunochromatographic RSV test being truly positive or negative. To use this graph the reader must first estimate the pretest probability that the patient truly hasRSV. If the antigen test is positive the user draws a straight line from the pre-test probability line through the likelihood ratio (LR) positive and continues the line through the post-test probability line to read off the post test probability that the patient has RSV. If the antigen test is negative the reader draws the line from the pre-test probability through the LR negative through the post test probability line to read off the post-test probability that the patient has RSV. EDs that treat very few infants even during bronchiolitis season may rely on this rather than the more complex model in figure 5.
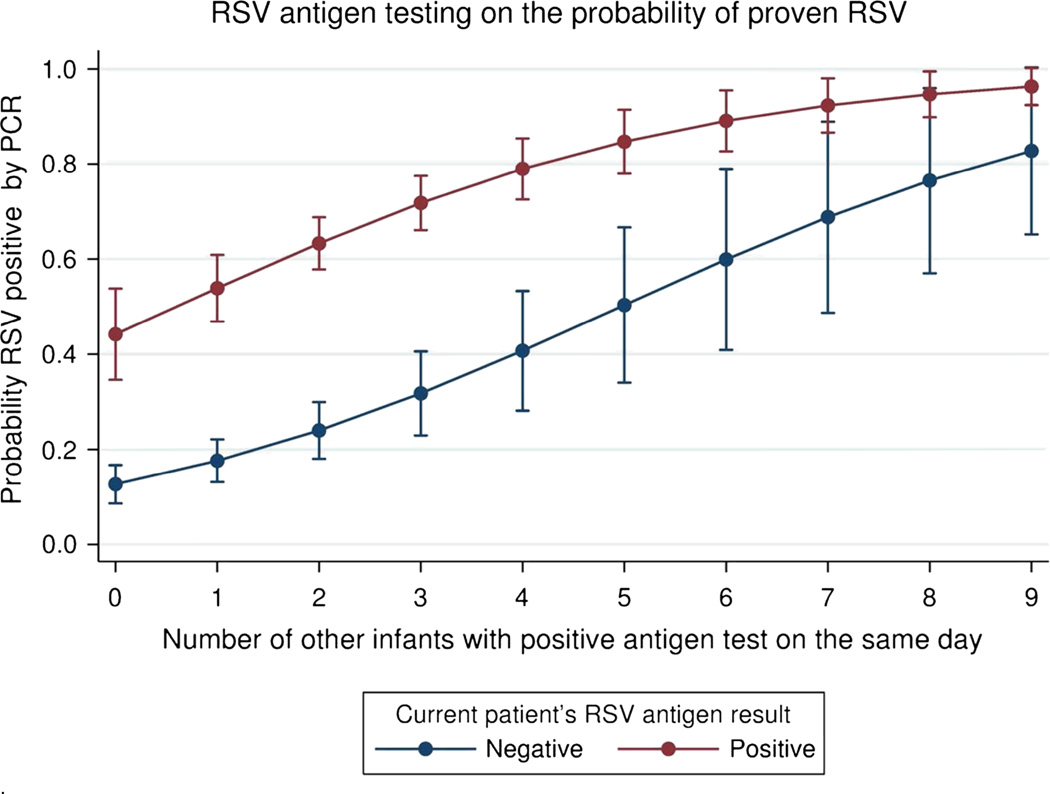
Clinical factors added little to our contextual multivariate models. Figure 4 is a graphical representation of the most parsimonious model that included the infants own RSV antigen test result and a count of the number of other infants who had positive immunochromatographic tests that day. This model showed that the best predictor of a positive RT-PCR test was a positive antigen test OR 5.47 (95%CI 3.65, 8.18) and the number of other infants having positive tests within one day OR 1.48 (95%CI 1.26, 1.72).
Figure 4.
This graph shows the marginal effect on probability of PCR proven RSV based both on the infants own antigen test result and the number of other children who tested positive within 24 hours of the index case. This graph demonstrates the concept that as cases of bronchiolitis increase does the probability that RSV is the aetiology. This model does not take into account the effect of prior negative test results. For clinical application figure 5 should be used.
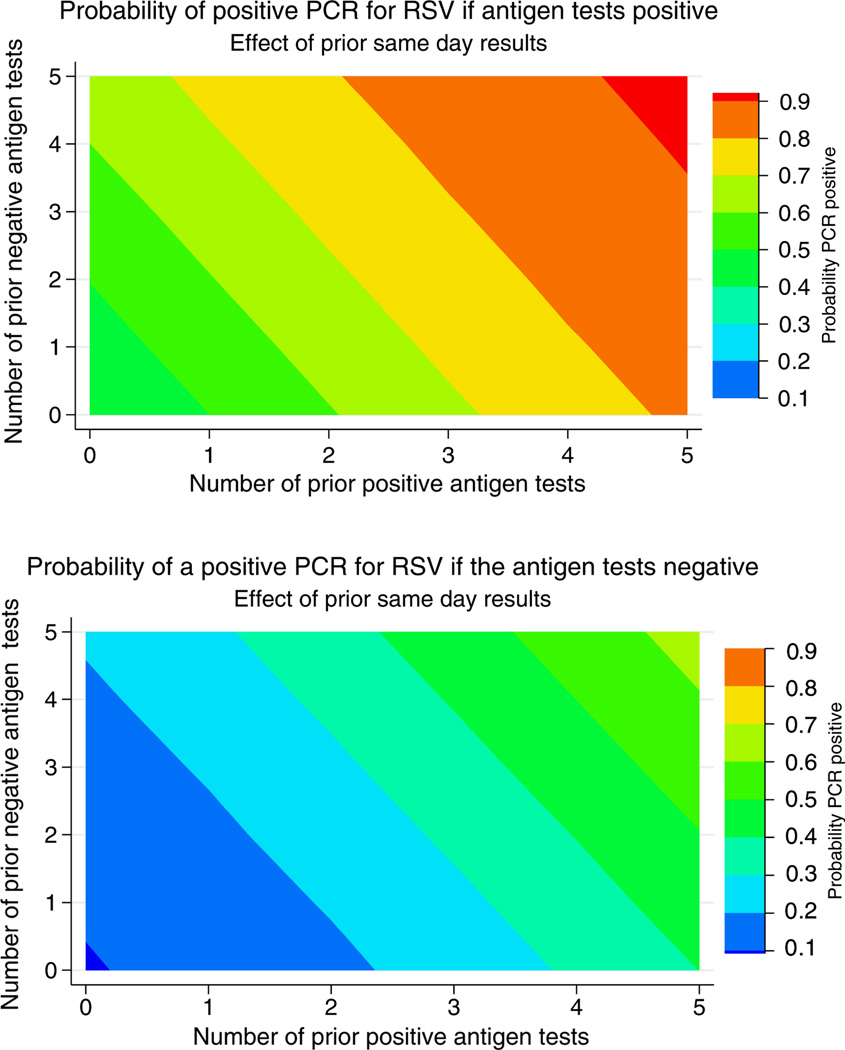
Figure 5 takes this further showing the marginal probabilities of a positive RSV RT-PCR test given a current positive (upper pane) or negative (lower pane) antigen test and the number of previous positive and negative antigen tests within 24 hours. The user picks the pane depending on whether the current test is positive or negative and then draws lines from the number of prior negative tests (the y-axis) and from the number of prior positives (x-axis). The color legend shows the probability of RT-PCR proven RSV. This also shows that as large numbers of children arrive at the ED with bronchiolitis, the probability that RSV will be the cause rises even if many of the earlier children had negative antigen tests.
Figure 5.
The upper pane shows theprobability of a positive RSV PCR test in an infant with bronchiolitis if the RSV antigen test is positiveconditioned on the results of prior RSV antigen tests performed on other infants with bronchiolitis within 24 hours of the case being tested. The lowerpanel shows this if the antigen test is negative. To use this graph: (1) Pick the top panel if the RSV antigen test is positive or the lower panel if the antigen test is negative. (2) Draw a horizontal line from the y-axis at the number of prior negative antigen tests and a vertical line from the x-axis at the number of prior positive antigen tests. The colour where these lines intersect gives the probability that the patient’s PCR test would be positive.
Discussion
RSV antigen testing is used for diagnostic certainty. Such certainty is sometimes useful and can eliminate the need for more invasive tests. However, we found that antigen test interpretation is not as simple as positive or negative. The antigen test had substantially lower sensitivity and specificity than that reported by the manufacturer. We also found that RA test interpretation is best contextualised, that is interpreted in light of the presentation of other infants to the ED within 24 h. This approach is more useful to practicing clinicians than expecting them to know their community’s prevalence of RSV.
The number of positive antigen tests that were negative on RT-PCR testing was striking. The antigen test kits were from several batches and were correctly performed by certified technicians. The RT-PCR detection threshold was 100 copies and carried out in a commercial laboratory. All the RT-PCR samples passed internal quality controls. Consequently, although disappointing, the results are likely correct. The resultant sensitivity and specificity results contrast with those obtained by some who have found better performances for antigen tests.15 Conversely, others have found even poorer performance for RA RSV testing than we did.25
Two processes may explain the differences between studies such as ours and those showing better test performance. First, in the early stages of test development, it is reasonable to evaluate test performance in groups of patients at very high and very low risk for the target condition (eg, a pregnancy test might be evaluated in men and women on a maternity ward. Any positives in men or negatives in the maternity ward patients allow the test to be discarded without additional expenditure of resources). Although a useful first step, this initial testing is insufficient. A diagnostic test must be able to detect the target condition where both the group with and without the condition are in other respects similar. Bronchiolitis is such a condition. The antigen test seemed calibrated to have maximal discriminatory ability when the RSV positive bronchiolitis rate approximated 50%. A second cause for these differences may be subtle differences in study design and blinding. For example, one protocol for a rapid streptococcal test required that tonsillar exudates first be swabbed with the antigen swab being investigated and then for the confirmatory swab for culture. Other differences include choices of gold standard; this is particularly important if culture is relied upon for fragile viruses or microscopist dependent techniques are used. Finally, regulatory agencies may dictate the gold standard used. This can invite designs that lead to better results than clinically oriented investigators using RT-PCR subsequently find. Although our results contrast with those of some, our results for the antigen test performance are similar to those obtained by other independent investigators of similar tests.13,14,26,27
Our graphical presentation of contextual diagnostic test modelling is novel and facilitates rapid interpretation. Consider the following clinical vignette.
Clinicians A and B are in different EDs. Both are evaluating a febrile 3-month old infant with bronchiolitis. Both are considering an intervention where RSV status matters, say a new RSV drug or a urinalysis. Clinician A has seen two other patients whose RSV antigen tests were negative and one whose test was positive. The current patient is RSV positive. Referring to figure 5, this patient’s probability of actually having RSV is 60–70%. Clinician B has seen five patients whose RSV antigen tests were negative and two in whom it was positive. The current patient’s RSV antigen test is negative. Referring to figure 5, this patient’s probability of actually having RSV is 40–50%. Clinician A’s management may now hinge on the relative costs of the intervention. Clinician B is little better off than if he had flipped a coin.
This also suggests a role for hospital laboratories; rather than reporting RSV antigen tests as positive or negative, the results of tests from the previous 24 h could be leveraged to provide a probability estimate of what the RT-PCR result would be.
Contextualised test interpretation by incorporating results from previous patients has been shown to improve the performance of diagnostic algorithms for meningitis and pertussis.28,29 Generally, these have been presented as modifications to pre-existing rules for the end user. For example, in the case of meningitis, a previously described rule was modified to include a variable ‘Were there four or more cases of viral meningitis in the preceding 10 days?’ This requires the treating doctor to know how many cases of viral meningitis were diagnosed in his department in the preceding 10 days.28 Our graphical presentation allows the clinician estimate the risk based on the data to hand and is a logical progression from prior contextual constructs.
Our study had several limitations. We did not use a third method to resolve discrepant results. Consequently, it is possible that, when discordant, RA tests were correct, and the RT-PCR results incorrect. However, the finding of significantly more alternative aetiologies when the RA test was positive but the RT-PCR negative for RSV than the converse supports our choice of gold standard. We did not test for other viruses, such as rhinoviruses or adenoviruses. This was based partly on resources and partly because rhinovirus in particular is prevalent regardless of clinical presentation. We did not test for bacterial infections. Our rate of RSV was lower than others; this likely reflects both our methodology which required diagnosis without knowledge of the antigen result and our relatively high prevalence of HMV. The sample collection methods for RA testing and RT-PCR were different, but reflect the preferred methods for each. Our results should not be extrapolated to other tests, collection and transport systems, the elderly or manifestations of RSV other than bronchiolitis. We confined our study to bronchiolitis to limit the breadth of patients our contextual model would have to account for; a broader model would have been a much more ambitious undertaking. Finally, we have not addressed the value of diagnostic certainty itself. This is a different and thorny topic. We have established only that diagnostic certainty cannot be established with a widely used antigen test, but that contextualised interpretation improves its performance.
Conclusion
The RA RSV antigen test we examined has modest performance characteristics and is best interpreted in the context of previous patients’ results. Increasing numbers of infants presenting to the ED with bronchiolitis in a given day increases the probability of RSV being present even if initial RA RSV tests are negative.
Acknowledgments
The authors acknowledge the input of the following: Nathan Kuppermann MD (1) Nicholas Kenyon MD (1) Nicholas Walker BS (4) Valerie Aguilar BS (4) and the Emergency medicine research assistants program at Kern Medical Center. PW had full access to all of the data in the study and takes responsibility for the integrity of the data and the accuracy of the data analysis.
Funding
This work was supported by Award Number 5K12HL108964-02 from the National Heart, Lung, and Blood Institute at the National Institutes for Health and by The Pediatric Emergency Medicine Research Foundation, Long Beach, CA. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Heart, Lung, and Blood Institute or the National Institutes of Health or The Pediatric Emergency Medicine Research Foundation.
Abbreviations
- RSV
respiratory syncytial virus
- INF
influenza
- HMV
human metapneumovirus
- RA
rapid immunochromatographic antigen
- RT-PCR
Polymerase chain reaction
- ED
Emergency department
- ROC
receiver operating curve
- OR
odds ratio
Appendix
The research laboratory is part of the research and development division of a commercial RT-PCR provider.
Viral RNA was extracted with the QIAamp Viral RNA purification kit (QIAgen, Valencia, CA). The extracted RNA was tested by reverse transcriptase real time RT-PCR for RSV A and RSV B. Reverse transcriptase by real-time RT-PCR was performed for HMV, Influenza A and Influenza B. All assays were optimized for specific genes and validated for sensitivity, specificity, interference, accuracy and precision. Sensitivity was determined by testing serial dilutions of RNA transcriptional run-off from each pathogen assay.
We cross-referenced the primers and probes in all assays by BLAST analysis against all DNA sequences deposited in the Entrez nr database. Analytical specificity was determined by testing for amplification in the presence of DNA and RNA extracted from 52 known bacterial, fungal, and viral pathogens purchased from ATCC. We tested for potential interference by adding 400ng of RNA which we had extracted from a patient sample that had tested negative for each pathogen in question.
We individually established stability tests for RSV A, RSV B, HMV, INF A and INF B. Each stability test was completed by spiking un-extracted pathogen purchased from ATCC into a previously tested negative sample. As there is no repository stock available for HMV, an artificial control was utilized. An oligonucleotide strand based upon Genbank accessioned sequences was synthesized by Integrated DNA Technologies, Inc (IDT) corresponding to the target sequence of the assay. This synthetic control was used to establish the fidelity of the designed assay for this sequence in the presence of other pathogenic species that are inherent to the sampling site. Stability testing was evaluated from Day 0 through Day 5 with storage at room temperature and 4°C. Each sample was extracted for RNA using the QIAamp viral RNA purification kit and then stored at -20°C until the final extraction time point. After the last time point was extracted, all samples were tested in triplicate for each assay.
Reproducibility of each assay was determined by testing a panel of nine positive samples of varying concentrations, nine negative samples, three positive controls and a negative template control using the appropriate reverse transcriptase RT-PCR method. The assays were tested by five different technicians on five different days and inter-operator precision was evaluated. The RT-PCR products from the first precision set of samples from each assay were saved and submitted for sequencing on the Genetic Analyzer 3130 (ABI) for accuracy validation. After the last time point was extracted, all samples were tested in triplicate for each assay.
Footnotes
COI: Neither the clinical laboratory nor the research laboratory teams was allowed a role in data analysis. Both laboratory teams were kept blinded to the other team's results. The MDL provides PCR testing on a commercial basis. Copan Innovation (Murrietta, CA) provided swabs and media. Neither company was allowed a role in study design, execution or analysis.
References
- 1.Harsten G, Prellner K, Löfgren B, Heldrup J, Kalm O, Kornfält R. Serum antibodies against respiratory tract viruses: A prospective three-year follow-up from birth. The Journal of Laryngology & Otology. 1989;103(10):904. doi: 10.1017/s0022215100110473. [DOI] [PubMed] [Google Scholar]
- 2.de Waal L, Koopman LP, van Benten IJ, et al. Moderate local and systemic respiratory syncytial virus-specific T-cell responses upon mild or subclinical RSV infection. Journal of medical virology. 2003;70(2):309–318. doi: 10.1002/jmv.10396. [DOI] [PubMed] [Google Scholar]
- 3.Mansbach JM, McAdam AJ, Clark S, et al. Prospective multicenter study of the viral etiology of bronchiolitis in the emergency department. Academic emergency medicine : official journal of the Society for Academic Emergency Medicine. 2008;15(2):111–118. doi: 10.1111/j.1553-2712.2007.00034.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Antunes H, Rodrigues H, Silva N, et al. Etiology of bronchiolitis in a hospitalized pediatric population: prospective multicenter study. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2010;48(2):134–136. doi: 10.1016/j.jcv.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Leader S, Kohlhase K. Respiratory syncytial virus-coded pediatric hospitalizations: 1997 to 1999. The Pediatric infectious disease journal. 2002;21(7) doi: 10.1097/00006454-200207000-00005. [DOI] [PubMed] [Google Scholar]
- 6.Stockman LJ, Curns AT, Anderson LJ, Fischer-Langley G. Respiratory Syncytial Virus-associated Hospitalizations Among Infants and Young Children in the United States: 1997–2006. The Pediatric infectious disease journal. 2012;31(1) doi: 10.1097/INF.0b013e31822e68e6. [DOI] [PubMed] [Google Scholar]
- 7.Hall CB, Powell KR, Schnabel KC, Gala CL, Pincus PH. Risk of secondary bacterial infection in infants hospitalized with respiratory syncytial viral infection. The Journal of pediatrics. 1988;113(2):266–271. [PubMed] [Google Scholar]
- 8.Purcell K, Fergie J. Concurrent serious bacterial infections in 912 infants and children hospitalized for treatment of respiratory syncytial virus lower respiratory tract infection. The Pediatric infectious disease journal. 2004;23(3):267–269. doi: 10.1097/01.inf.0000116759.21252.29. [DOI] [PubMed] [Google Scholar]
- 9.Levine DA, Platt SL, Dayan PS, et al. Risk of serious bacterial infection in young febrile infants with respiratory syncytial virus infections. Pediatrics. 2004;113(6):1728–1734. doi: 10.1542/peds.113.6.1728. [DOI] [PubMed] [Google Scholar]
- 10.Chee C, Walsh P, Kuan S, et al. Emergency Department Septic Screening in Respiratory Syncytial Virus (RSV) and Non-RSV Bronchiolitis. Western Journal of Emergency Medicine. 2010;11(1):60–67. [PMC free article] [PubMed] [Google Scholar]
- 11.Bruhn FW, Mokrohisky ST, McIntosh K. Apnea associated with respiratory syncytial virus infection in young infants. The Journal of pediatrics. 1977;90(3):382–386. doi: 10.1016/s0022-3476(77)80697-5. 3// [DOI] [PubMed] [Google Scholar]
- 12.Ginocchio CC, Zhang F, Manji R, et al. Evaluation of multiple test methods for the detection of the novel 2009 influenza A (H1N1) during the New York City outbreak. Journal of Clinical Virology. 2009;45(3):191–195. doi: 10.1016/j.jcv.2009.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Casiano-Colón AE, Hulbert BB, Mayer TK, Walsh EE, Falsey AR. Lack of sensitivity of rapid antigen tests for the diagnosis of respiratory syncytial virus infection in adults. Journal of Clinical Virology. 2003;28(2):169–174. doi: 10.1016/s1386-6532(03)00002-7. [DOI] [PubMed] [Google Scholar]
- 14.Halstead DC, Todd S, Fritch G. Evaluation of five methods for respiratory syncytial virus detection. Journal of clinical microbiology. 1990;28(5):1021–1025. doi: 10.1128/jcm.28.5.1021-1025.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hall CB, Douglas RG. Clinically Useful Method for the Isolation of Respiratory Syncytial Virus. Journal of Infectious Diseases. 1975;131(1):1–5. doi: 10.1093/infdis/131.1.1. [DOI] [PubMed] [Google Scholar]
- 16.Walsh P, Overmyer CL, Pham K, et al. Comparison of respiratory virus detection rates in infants and toddlers using flocked swabs, saline aspirates and saline aspirates mixed in UTM-RT. Journal of clinical microbiology. 2008 doi: 10.1128/JCM.00714-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Becton Dickinson. Co, Inventors. Package insert for Directigen RSV. 5,093,231; 5,135,8472004 US patent.
- 18.Long JS. Regression Models for Categorical and Limited Dependent Variables. Thousand Oaks, CA: Sage; 1997. [Google Scholar]
- 19.Seed P. DIAGT: Stata module to report summary statistics for diagnostic tests compared to true disease status. Boston College Department of Economics; 2001. [Google Scholar]
- 20.Walsh P, Rothenberg SJ, O'Doherty S, Hoey H, Healy R. A validated clinical model to predict the need for admission and length of stay in children with acute bronchiolitis. European Journal of Emergency Medicine. 2004;11(5):265. doi: 10.1097/00063110-200410000-00005. [DOI] [PubMed] [Google Scholar]
- 21.Fagan TJ. Nomogram for Bayes's Theorem. N Engl J Med. 1975;293(5):257–257. doi: 10.1056/NEJM197507312930513. [DOI] [PubMed] [Google Scholar]
- 22.Myers C, Wagner N, Kaiser L, Posfay-Barbe K, Gervaix A. Use of the Rapid Antigenic Test to Determine the Duration of Isolation in Infants Hospitalized for Respiratory Syncytial Virus Infections. Clinical Pediatrics. 2008 Jun 1;47(5):493–495. doi: 10.1177/0009922807310936. 2008. [DOI] [PubMed] [Google Scholar]
- 23.Rahman M, Vandermause MF, Kieke BA, Belongia EA. Performance of Binax NOW Flu A and B and direct fluorescent assay in comparison with a composite of viral culture or reverse transcription polymerase chain reaction for detection of influenza infection during the 2006 to 2007 season. Diagnostic microbiology and infectious disease. 2008;62(2):162–166. doi: 10.1016/j.diagmicrobio.2007.10.012. [DOI] [PubMed] [Google Scholar]
- 24.Hurt AC, Alexander R, Hibbert J, Deed N, Barr IG. Performance of six influenza rapid tests in detecting human influenza in clinical specimens. Journal of clinical virology : the official publication of the Pan American Society for Clinical Virology. 2007;39(2):132–135. doi: 10.1016/j.jcv.2007.03.002. [DOI] [PubMed] [Google Scholar]
- 25.Fine AM, Nigrovic LE, Reis BY, Cook EF, Mandl KD. Linking Surveillance to Action: Incorporation of Real-time Regional Data into a Medical Decision Rule. Journal of the American Medical Informatics Association. 2007;14(2):206–211. doi: 10.1197/jamia.M2253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fine AM, Reis BY, Nigrovic LE, et al. Use of population health data to refine diagnostic decision-making for pertussis. Journal of the American Medical Informatics Association. 2010;17(1):85–90. doi: 10.1197/jamia.M3061. [DOI] [PMC free article] [PubMed] [Google Scholar]