Summary
The CtIP protein is known to function in 5′ strand resection during homologous recombination similar to the budding yeast Sae2 protein, although its role in this process is unclear. Here we characterize recombinant human CtIP and find that it exhibits 5′ flap endonuclease activity on branched DNA structures, independent of the MRN complex. Phosphorylation of CtIP at known ATM-dependent sites and other sites is essential for its catalytic activity, although the S327 and T847 phosphorylation sites are dispensable. A catalytic mutant of CtIP that is deficient in endonuclease activity exhibits wild-type levels of homologous recombination at restriction enzyme-generated breaks but is deficient in processing topoisomerase adducts and radiation-induced breaks in human cells, suggesting that the nuclease activity of CtIP is specifically required for the removal of DNA adducts at sites of DNA breaks.
Introduction
Double-strand breaks (DSBs) in chromosomal DNA can be caused by external agents or by internal sources of DNA damage such as reactive oxygen species or the process of replication. Eukaryotic cells respond very rapidly to DSBs with the initiation of both DNA repair as well as cell cycle checkpoint arrest (Ciccia and Elledge, 2010). The Mre11/Rad50/Nbs1(Xrs2) (MRN) complex plays a central role in coordinating these events, through activation of the ATM protein kinase at sites of DSBs and also in performing the initiating steps of homologous recombination (HR) (Stracker and Petrini, 2011). Recent studies in budding yeast indicate that MRX, together with the Sae2 endonuclease, carry out short-range processing of DSBs to resect ends and also help recruit the long-range endo- and exonucleases that perform long-range 5′ strand resection (Mimitou and Symington, 2009; Paull, 2010).
The Sae2 protein shows little evolutionary conservation in primary sequence but has functional orthologs in other species that also act in promoting 5′ strand resection (You and Bailis, 2010). The mammalian ortholog is CtIP, the CtBP (carboxy-terminal binding protein)-interacting protein, which binds to the Brca1 tumor suppressor and to the cell cycle regulator Rb (retinoblastoma protein). CtIP has been shown to promote DNA end resection in mammalian cells (Helmink et al., 2011; Huertas and Jackson, 2009; Sartori et al., 2007; You et al., 2009), in chicken DT40 cells (Nakamura et al., 2010; Yun and Hiom, 2009), and in nematodes and plants (Penkner et al., 2007; Uanschou et al., 2007).
The role of Sae2 in DSB repair in budding yeast was first recognized through its role in meiosis, where it is essential for the processing of covalent Spo11 intermediates (McKee and Kleckner, 1997; Prinz et al., 1997). This meiosis-specific function is also conserved in S. pombe and in higher organisms (Hartsuiker et al., 2009a; Penkner et al., 2007; Uanschou et al., 2007). Spo11 is a putative topoisomerase that forms intermediates with DNA through a covalent tyrosine linkage (Keeney et al., 1997). Topoisomerase I and II also form covalent intermediates, which are stabilized by drugs used for cancer therapy, including derivatives of camptothecin and etoposide. Eukaryotic cells deleted or depleted for Sae2/CtIP orthologs show a pronounced sensitivity to these chemotherapeutic agents (Hartsuiker et al., 2009b; Huertas and Jackson, 2009; Nakamura et al., 2010; Quennet et al., 2011; Sartori et al., 2007; Wang et al., 2013b), suggesting that the processing of covalent protein-DNA intermediates may be a conserved function for this enzyme.
HR in eukaryotic cells is regulated during the cell cycle to occur most efficiently during the S and G2 phases when sister chromatids are present. Sae2 and CtIP are among the primary targets of this regulation, which occurs through phosphorylation by cyclin-dependent kinases (CDKs) and by ATM and ATR (Fu et al., 2014; Li et al., 2000; Peterson et al., 2012; Wang et al., 2013a; You and Bailis, 2010).
CtIP appears to be essential in vertebrates, and even haploinsufficiency generates genomic instability and higher rates of tumorigenesis (Chen et al., 2005; Nakamura et al., 2010). Conversely, CtIP also contributes to translocations through its role in alternative end-joining pathways (Lee-Theilen et al., 2011; Zhang and Jasin, 2011), a role also conserved with Sae2 in S. cerevisiae (Lee and Lee, 2007). Recently, mutations in CtIP were also identified as the causative factors in the congenital microcephaly disorders Jawad and Seckel syndromes (Qvist et al., 2011).
Despite the large amount of information currently available about CtIP, it is unknown if the vertebrate protein acts as a nuclease in a manner similar to Sae2 and how the complex phosphorylation patterns affect CtIP function. To address these questions, we expressed and purified recombinant human CtIP from insect cells and evaluated its activities in vitro. We find that CtIP is a 5′ flap endonuclease that recognizes and cleaves branched DNA structures, and identify a CtIP mutant that is deficient in nuclease activity but proficient for DNA binding. CtIP-deficient cells expressing this mutant show defects in the survival of radiation-induced damage and topoisomerase poisons, but exhibit normal resection of unmodified DNA breaks. These findings establish an enzymatic role for CtIP and illuminate how this enzyme functions in DNA DSB repair.
Results
CtIP is a 5′ flap endonuclease
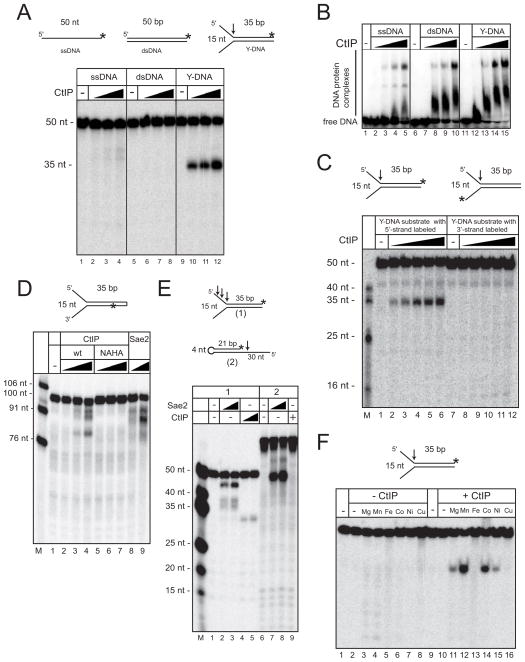
The functional similarity between CtIP in mammalian cells and Sae2 in yeast suggests that CtIP might also act as an endonuclease. To address this question, we purified human recombinant CtIP to homogeneity from insect cells (Fig. S1A) and tested its activity to cleave different deoxyoligonucleotide substrates. We found that CtIP cleaves a branched, Y-shaped DNA structure at the base of the 5′ flap, but has no activity on single-stranded (ss) or double-stranded (ds) DNA (Fig. 1A). Gel mobility shift experiments showed that the protein binds to all these structures, although CtIP exhibits a higher affinity for dsDNA and Y structures compared to ssDNA (Fig. 1B). The cleavage pattern suggests that CtIP recognizes ss/ds junctions in DNA, so structures containing 5′ overhangs and 3′ overhangs were tested (Fig. S1B). Neither of these was cleaved by CtIP, nor was the 3′ flap in the Y structure (Fig. 1C), thus the nuclease activity of human CtIP is specific for the 5′ strand in a branched, Y-shaped structure.
Figure 1. CtIP is a 5′ strand flap endonuclease.
(A) Structures of DNA substrates (top): ssDNA, and dsDNA, Y-structure with 15 nt branch. Each substrate contains the same top strand, labeled with [32P] at the 3′ end (asterisk). Nuclease assays were performed with human wt CtIP (50, 100, and 200 nM) in 5 mM MgCl2, and products were separated by denaturing polyacrylamide gel electrophoresis (PAGE). The arrow shows the position of CtIP-mediated cleavage. (B) Gel mobility shift assays were performed with DNA substrates as in (A) with human CtIP (25, 50, 100, or 200 nM), separated by native PAGE. (C) Nuclease assays as in (A) using Y structures labeled on the 3′ end of the top strand or the bottom strand as shown. (D) Nuclease assays with wt CtIP (50, 100, or 200 nM) or wt recombinant Sae2 (1.5 or 6 nM) as in (A) with 5 mM MgCl2 and a Y-structure DNA containing a hairpin and an internal [32P] label as indicated. (E) Nuclease assays with wt CtIP (100 or 200 nM) or wt recombinant Sae2 (3 or 6 nM) in 5 mM MgCl2 and a Y-structure DNA (1) or a hairpin with a 5′ overhang (2). (F) Nuclease assays with wt CtIP (200 nM) on Y-structure DNA as in (A) but with various metals (6 mM Mg2+ or 2 mM other metals, and 1 mM EDTA) as indicated.
Hairpin structures were also tested as substrates for CtIP, since we previously showed that recombinant Sae2 cleaves ssDNA adjacent to hairpins (Lengsfeld et al., 2007). On a hairpin with both 5′ and 3′ flaps, CtIP and Sae2 generated similar products, not cutting the hairpin itself but cleaving within the single-stranded DNA tails (Fig. 1D). However, on a Y structure, Sae2 generates a different cleavage pattern compared to CtIP, cutting at several positions within the 5′ flap as well as at the base (Fig. 1E). In addition, CtIP failed to cut a hairpin with an adjacent ssDNA overhang, whereas this is a preferred substrate for Sae2 (Fig. 1E) (Lengsfeld et al., 2007). Overall, CtIP and Sae2 share some substrate preferences and both cleave 5′ flaps in a branched structure, but CtIP does not exhibit obvious hairpin specificity.
We also investigated the metal requirements of recombinant CtIP and found that, although it is active in the presence of Mg2+, Ni2+, and Co2+ (all assays in Fig. 1 were performed in 5 mM MgCl2), it exhibits 4 to 8-fold higher activity in manganese compared to Mn2+ (Fig. 1F). In contrast, Sae2 does not exhibit a preference for Mn2+ (Fig. S1C).
Figure 4. CtIP nuclease activity is required for DNA end processing and damage survival.
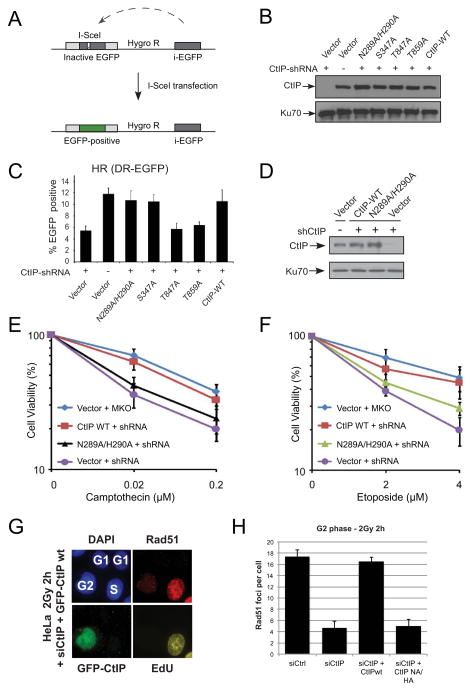
(A) Schematic representation of an eGFP-based restriction enzyme induced HR repair reporter assay in U2OS cells (Wang et al., 2012). Expression of I-SceI endonuclease induces a DSB; repair via HR results in expression of green fluorescent eGFP protein. (B) Expression of wt, NA/HA, S347A, T847A, or T859A proteins in U2OS cells with CtIP depleted was analyzed by western blot for CtIP protein, using Ku70 for normalization. (C) U2OS cells from (B) carrying the HR reporter and expressing HA-CtIP wt or indicated mutants, with CtIP depleted by shRNA, were induced with I-SceI and assayed for EGFP-positive events. Error bars indicate standard error from at least 3 independent experiments. (D) Expression of wt or NA/HA proteins in U2OS cells with CtIP depleted was analyzed by western blot for CtIP, using Ku70 for normalization. U2OS cells from (D) were analyzed for cell survival after Camptothecin (CPT) (E) or etoposide (F) mediated DNA damage in U2OS cells expressing HA-CtIP WT or indicated mutants, with CtIP shRNA knockdown or mock control (MKO). (G) Endogenous CtIP was depleted in HeLa cells by siRNA and cells were transfected with GFP-tagged wt or NA/HA CtIP plasmids 48 h before 2 Gy irradiation. Cells were incubated with EdU 30 min prior to IR and during the entire repair period. All EdU-positive S-phase cells were excluded from the analysis and only EdU-negative G2-irradiated cells were analyzed. (H) Rad51 foci were enumerated in GFP-positive G2 cells at 2 h after 2 Gy. Error bars indicate standard error from 3 independent experiments. Also see Fig. S3.
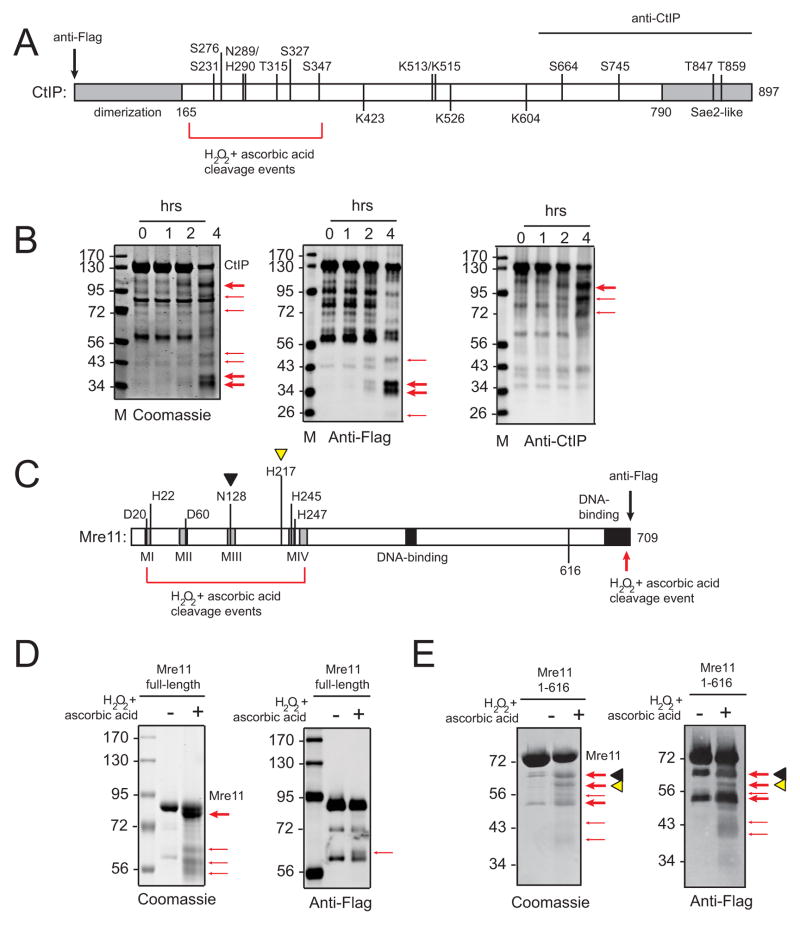
CtIP contains an N-terminal dimerization domain, a conserved C-terminus with limited similarity to Sae2 and Ctp1 orthologs, and several identified phosphorylation and acetylation sites (Fig. 2A). In order to identify amino acid residues responsible for CtIP nuclease activity, we performed a limited sequence similarity search based on the active site of Mre11, considering that this enzyme also shows 5′ strand endonucleolytic activity and also exhibits increased activity with Mn2+ (Hopkins and Paull, 2008; Paull and Gellert, 1998). This search revealed two conserved amino acid residues in CtIP, N289 and H290, which are conserved as an NH or DH sequence in all mammalian CtIP orthologs (Fig. S2A). Mutation of these residues resulted in a mutant (N289A/H290A, NA/HA) that showed an impaired nuclease activity but fully retained the ability to bind DNA, comparable to that of the wild-type (wt) protein (Fig. 2B, C), and the ability to bind to MRN (Fig. S2B). The H290N mutation, in theory equivalent to H129N in Mre11 (Moreau et al., 1999), was also nuclease-deficient. Quantification of wt and NA/HA CtIP activity in the presence of Mg2+ compared to Mn2+ is shown in Fig. 2D.
Figure 2. Regulation of CtIP nuclease activity by conserved amino acids and phosphorylation.
(A) Schematic diagram of the CtIP protein showing known features and phosphorylation sites relevant to this study: S231, S276, T315, and S347 (see Table S1). (B) Nuclease assays with Y-structure as in Fig. 1A, using wt, NA/HA, or H290N CtIP proteins at 25, 50, 100, or 200 nM. (C) Gel mobility shift assays with a 249 bp dsDNA substrate internally labeled with [32P], using wt or NA/HA CtIP at 6.25, 12.5, 25, 50, 100, or 200 nM. Reactions were separated by native PAGE. (D) Quantitation of CtIP endonuclease assays performed as in (B) with 200 nM wt or NA/HA proteins in 5 mM MgCl2 or 1 mM MnCl2 as indicated. The average of 3 experiments is shown with error bars indicating standard deviation. (E, F) Nuclease assays with a Y-structure as in Fig. 1A, using wt and mutant proteins at 100 or 200 nM. (G) Nuclease assays with a Y-structure as in Fig. 1A, using wt and mutant proteins at 50, 100 or 200 nM.
Effects of Post-translational Modifications on CtIP Activity
CtIP is known to be a target of CDK cell cycle-dependent phosphorylation as well as DNA damage-induced phosphorylation by the ATM and ATR kinases (Huertas and Jackson, 2009; Li et al., 2000; Peterson et al., 2012; Sartori et al., 2007; Wang et al., 2013b; Yu and Chen, 2004), although it is not clear if all the relevant sites have been identified. We analyzed human CtIP expressed in insect cells and human cells, both in the presence and absence of DNA damage, and identified 36 phosphorylation sites and two acetylation sites (Table S1), including novel and previously reported residues. Here we found that mutation of the known S327 and T847 CDK sites to alanine generates mutants that are still active in nuclease activity in vitro (Fig. 2E). One of the novel modifications identified by mass spectrometry was S347, a site that matches an SP/TP CDK target consensus. In contrast to the previously characterized CDK sites, we found that the S347A mutation blocks endonuclease activity, while a phosphomimic form S347D exhibits higher activity than the wt protein (Fig. 2E and S2C), suggesting that phosphorylation of this residue directly impacts CtIP activity.
A recent study identified two phosphorylation sites in CtIP that affect the binding of Pin1, a prolyl isomerase (Steger et al., 2013) and showed that mutation of T315 and S276 phosphorylation sites inhibited Pin1-dependent isomerization, ubiquitination, and subsequent degradation of CtIP. We also tested the T315A and S276A mutants of CtIP and found that both of the single mutants are inactive for nuclease activity (Fig. 2E and Fig. S2D). Whether this is due to loss of phosphorylation (both sites are phosphorylated in CtIP isolated from insect cells or human cells, Table S1) or to a loss of proline isomerization will require further investigation.
Many other phosphorylation sites were analyzed by mutation, but the only other residues that have an effect on CtIP endonuclease activity in vitro are ones which match an ATM/ATR SQ/TQ consensus. Of the 8 putative SQ/TQ phosphorylation sites in CtIP, we observed S745 and S231 phosphorylation repeatedly in damage-treated samples by mass spectrometry (Table S1). Mutation of S231 to alanine had a strong inhibitory effect on CtIP activity and, like the S347 site, a phosphomimic aspartate at this position increased CtIP activity over the wt level (Fig. 2F). The S664 and S745 mutations identified as ATM-dependent sites in CtIP (Li et al., 2000) in combination with S231 (S231A/S664A/S745A) strongly reduce the activity of the mutant protein to an undetectable level (Fig. 2F). None of the phosphorylation-based mutations altered the interactions between CtIP and DNA (Fig. S2E). Overall, we have found that CtIP is the target of many phosphorylation events but only a small number of these affect its endonuclease activity in vitro (see also Fig. S2F). These residues include both SP/TP and SQ/TQ phosphorylation sites in CtIP, and the strongest effects are seen with novel sites and sites known to be phosphorylated by ATM. The phosphorylation sites that appear to be the most important for CtIP nuclease activity are concentrated in an N-terminal domain, from a.a. 231 to 347.
CtIP is acetylated in human cells (Kaidi et al., 2010), but mutation of the known acetylated lysine residues to arginine (K432R/K526R/K604R) did not change the activity of CtIP in vitro (Fig. 2G). In contrast, lysine residues in a previously identified DNA-binding domain (K513 and K515) (You et al., 2009) are important for nuclease activity (Fig. 2G). Paradoxically, this mutant appears to bind DNA normally (Fig. S2E) but it could be that contributions from other domains in the full-length protein mask the DNA binding deficiency.
The C-terminus of CtIP is truncated in two rare genetic disorders in humans, Seckel and Jawad syndromes, where patient cells exhibit defects in ATR-mediated signaling of replication damage (Qvist et al., 2011). Truncation of CtIP at a.a. 790 to mimic the human truncation (L790X), which also removes the domain implicated in MRN binding (Sartori et al., 2007; Yuan and Chen, 2009), reduces the activity of the protein to levels similar to the NA/HA mutant (Fig. 2G). Thus it is likely that this domain has multiple roles, one of which is to facilitate nuclease activity of CtIP. It is important to note that the CtIP endonuclease activity demonstrated here occurs in the absence of MRN, although it could be that MRN facilitates the functions of CtIP in a cellular context.
Oxidative cleavage identifies the active site domain in CtIP
To determine if the N289/H290 residues define part of the active site of CtIP, we tested Fe2+/EDTA-dependent hydroxyl radical cleavage of the protein backbone; however, this did not yield any protein cleavage products. Nevertheless, incubation of CtIP with ascorbic acid and H2O2 alone, without Fe2+/EDTA, generated distinct protein fragments (Fig. 3). Using antibodies specific for either the N-terminus (anti-Flag) or the C-terminus (anti-CtIP, epitope from a.a. 620–897), we determined from the size of the fragments (approximately 26, 34, 36, and 43 kD) that the major cleavage sites are between a.a. 180 and 350. Efficient cleavage of polypeptide chains by Cu2+ or by Co2+ bound to a protein in the presence of ascorbic acid and H2O2 has been described previously (Andberg et al., 2007; Sereikaite et al., 2006). The cleavage of CtIP by H2O2 and ascorbate is blocked by EDTA (data not shown), so it is likely that a transition metal pre-existing in the active site is reduced by ascorbic acid and generates hydroxyl radicals at the site of the bound metal which cut the protein backbone. The estimated region of cleavage includes N289/H290 and the N181/R185/E267/E268 residues identified by Wu and colleagues (Wang et al., 2014), consistent with these residues composing part of an active site, although structural data will be necessary to conclusively demonstrate this.
Figure 3. CtIP and Mre11 have N-terminal oxidation-sensitive metal binding sites.
(A) Schematic diagram of CtIP showing known features as in Fig. 2 but also indicating regions of recognition by Flag and CtIP antibodies and domain where oxidation takes place (see main text). (B) Human CtIP was incubated with ascorbic acid and H2O2; reactions were stopped at the indicated time points, separated by SDS-PAGE, and either stained with coomassie blue or transferred to a membrane and blotted with anti-Flag or anti-CtIP antibodies as indicated. Red arrows indicate predominant cleavage products. (C) Schematic diagram of Mre11 showing known features and indicating the amino acids that directly contact metal ions in the crystal structure of human Mre11 (Park et al., 2011). (D) Full-length human Mre11 was incubated with ascorbic acid and H2O2; reactions were separated by SDS-PAGE and stained with coomassie blue or probed for the C-terminal Flag epitope as indicated. Red arrows indicate cleavage products with the bold arrow indicating the predominant product. (E) Truncated Mre11 (a.a. 1 to 616) was treated as in (D). Bands marked with black or yellow triangles were identified by N-terminal sequencing as C-terminal fragments starting with a.a. 132 and a.a. 218, respectively (see Table S2).
Considering the proposed similarity between CtIP and Mre11 in transition metal utilization, we also incubated recombinant human Mre11 with H2O2 and ascorbic acid. Inclusion of Fe2+/EDTA also failed to generate cleavage products for Mre11 (data not shown); however, H2O2 and ascorbic acid treatment alone produced several protein fragments. The most prominent is very close to the full-length size, and arises from a cleavage event close to the C-terminus, based on the observation that the C-terminal Flag epitope is lost. The cleavage event is very likely within a stretch of 5 consecutive aspartate residues at a.a. 691–695, based on the size of the fragment and the fact that poly-aspartate is well-known to chelate metal (Stair and Holcombe, 2007). There are also cleavage events in the N-terminus of Mre11, one of which is consistent with a cut in nuclease motif III (see Flag-tagged band), consistent with a transition metal bound to this motif.
To clarify the locations of the cleavage sites, we performed the H2O2/ascorbic acid reaction with a truncation mutant of Mre11 lacking residues 617 to 709 but still containing a Flag epitope at the C-terminus (Fig. 3E). This truncation did not yield the C-terminal cleavage product but did show several oxidation sites in the N-terminus. N-terminal sequence was obtained by Edman sequencing for two of these cleavage products, showing that the most N-terminal cut site is at a.a.131 and the second is at a.a. 217, both coincident with the metal ion in the position of Mn2+ II in the human Mre11 structure (Park et al., 2011) (N128-D131 and H217)(Park et al., 2011). This site was also identified as the high-affinity metal-binding site in the structurally related lambda phosphatase protein (White et al., 2001). By comparison, the location of the cleavage sites in CtIP is consistent with the presence of one or more transition metal ions contacting 4 sites within the N-terminal a.a. 180–350 nuclease domain. Both the CtIP NA/HA mutant and an Mre11 catalytic mutant were tested for oxidative cleavage and the patterns appeared similar to the wt proteins (data not shown); however, previous studies with Mre11 and lambda phosphatase have also shown that single catalytic mutations in these proteins do not block metal binding (Arthur et al., 2004; White et al., 2001).
CtIP but not its nuclease activity is required for the resection of restriction enzyme induced double-strand breaks
To test whether CtIP is required for the repair of DSBs in cells, we utilized a reporter assay in human U2OS cells in which induced expression of I-SceI generates a DSB that is subsequently repaired by gene conversion with a homologous sequence on the same chromosome (Fig. 4A) (Wang et al., 2012). In this system, successful HR repair results in expression of full-length eGFP protein. CtIP levels were depleted in U2OS cells using siRNA, which then were transfected either an empty vector or with constructs expressing wt or mutant alleles (Fig. 4B). Measurement of eGFP levels after I-SceI induction showed that wt, NA/HA, and S347A alleles supported HR and DNA repair comparably in comparison to the control transfection, whereas the previously characterized T847A and T859A mutants were similar to the uncomplemented cells (Fig. 4C).
We also examined the resection of DSBs by utilizing 293T cells that conditionally translocate the AsiSI restriction enzyme into the nucleus (Iacovoni et al., 2010). Endogenous CtIP was depleted in these cells, followed by expression of either wt or NA/HA CtIP. ssDNA at two of the AsiSI sites was measured by quantitative PCR (Zhou et al., 2014) which showed that CtIP is required for the production of ssDNA at these DSB sites, but that both the wt and NA/HA mutant complement this deficiency (Fig. S3A). These results, together with the in vitro analysis of the recombinant protein, suggest that CtIP is required for the resection of breaks induced by restriction enzyme cleavage in human cells but its nuclease activity is dispensable for this process.
To determine if CtIP acts directly to promote DSB resection, we used a reconstituted system with Dna2, BLM helicase, RPA, and the Ku70/80. MRN has previously been shown to stimulate Dna2/BLM activity in vitro by promoting the initiation of resection in the presence of RPA, which limits Dna2 resection to the 5′ strand (Cejka et al., 2010; Nimonkar et al., 2011; Niu et al., 2010). We have also shown that recombinant yeast Sae2 can promote the activity of Exo1 both in a MRX-dependent and independent manner (Nicolette et al., 2010). In both this system and the reaction with yeast proteins we have found that MRN(X) has a unique ability to overcome the inhibitory effects of the Ku70/80 on Exo1 and Dna2 activity in vitro (Nicolette et al., 2010; Yang et al., 2013). Here we find that CtIP increases close-range resection by Dna2 and BLM ~3-fold in a manner that is strongly dependent on MRN (Fig. S3C). There was no significant difference between the activities of the wt and mutant CtIP proteins, consistent with the ability of this mutant to promote resection of clean DSBs in cells.
CtIP nuclease activity is required for cell survival of topoisomerase and ionizing radiation-induced DNA damage
Vertebrate CtIP, fission yeast Ctp1, and budding yeast Sae2 have been implicated in resection of topoisomerase induced DSBs (Deng et al., 2005; Hartsuiker et al., 2009a; Hartsuiker et al., 2009b; Nakamura et al., 2010; Neale et al., 2005; Rothenberg et al., 2009; Sartori et al., 2007; Wang et al., 2013a). We reasoned that CtIP nuclease activity could be critical for resection in situations where a protein adduct such as a topoisomerase must be removed to facilitate repair. To test this we depleted CtIP in U2OS cells using siRNA and transfected with either an empty vector or with the vectors expressing wt or NA/HA mutant CtIP alleles (Fig. 4D). Cells were treated with either camptothecin (CPT) or etoposide and their survival was assessed. We found that the absence of CtIP significantly sensitizes cells to DSBs generated by either topoisomerase I (CPT) or topoisomerase II (etoposide) (Fig. 4E, F). Expression of mutant NA/HA CtIP did not completely suppress the sensitivity to the drug, while expression of wt protein significantly improved cell survival, close to the level of cells without CtIP depletion. These results suggest that both CtIP and its nuclease activity are important for the resolution of topoisomerase-induced DSBs.
Ionizing radiation (IR) also can generate protein-DNA adducts as well as “dirty” ends on broken DNA which show a requirement for CtIP-dependent repair (Huertas and Jackson, 2009). To investigate if the nuclease activity of CtIP is required for its resection function following IR exposure, we measured Rad51 foci formation as described previously (Beucher et al., 2009). We transfected HeLa cells with siRNA-resistant wt or mutant GFP-CtIP constructs following depletion of endogenous CtIP by siRNA (Fig. 4G and Fig. S3D). CtIP depletion strongly diminished the level of Rad51 foci measured in G2-phase cells at 2 h post 2 Gy (Fig. 4H), a time when Rad51 foci formation is maximal in wt cells (Beucher et al., 2009). Quantitation of Rad51 foci in transfected G2 cells (GFP+) showed that wt CtIP restored the level of Rad51 foci formation to that of control cells whereas transfection with the CtIP NA/HA mutant did not. Examination of RPA foci in these cells also showed a striking deficiency with the NA/HA mutant, confirming a direct effect on DSB resection after IR treatment (Fig. S3E). The NA/HA still is competent for DSB localization after IR damage, visualizing foci in transfected cells (Fig. S3F). These data suggest that the nuclease activity of CtIP is required for efficient DNA end-resection of breaks caused by ionizing radiation in G2 phase.
CtIP is required to open hairpin-sealed coding ends
CtIP has been reported to resect DNA coding ends generated by RAG recombinase during V(D)J recombination. (Helmink et al., 2011) CtIP-mediated resection of coding ends was observed in the absence of histone H2AX, which protects coding ends from nucleases other than the Artemis enzyme that normally processes these intermediates. To test whether CtIP nuclease activity is important in this context, CtIP levels were reduced by shRNA expression in mouse Artemis−/−/H2AX−/− DELCJ Abl Pre-B cells, then either the wt or NA/HA mutant version was expressed. G1 cell cycle phase arrest and RAG cleavage was induced by adding STI 571 v-abl inhibitor. Analysis of the coding ends revealed that wt CtIP was able to resect hairpin-sealed ends but the NA/HA nuclease-deficient mutant was deficient in this activity (Fig. S3G).
Discussion
DNA end processing is required to generate a 3′ strand for single-strand annealing and HR. The extent of resection depends on the type of DNA damage, the cell cycle phase, and the availability of a homologous donor sequence (Symington and Gautier, 2011). Efficient resection is known to require CtIP, a binding partner of MRN and BRCA1 and a distant ortholog of the Sae2 endonuclease in budding yeast. Here we show that recombinant human CtIP exhibits endonuclease activity and is specific for the 5′ strand of a Y-shaped DNA substrate, similar to Sae2 (Lengsfeld et al., 2007). This activity is required for the survival of camptothecin-induced topoisomerase damage, ionizing radiation damage, and for the processing of hairpin-capped intermediates in G1 phase cells, establishing the importance of CtIP catalytic activity in DNA repair.
Recombinant human CtIP exhibits 5′ flap endonuclease activity in vitro, like Sae2 from S. cerevisiae (Lengsfeld et al., 2007). This specificity is consistent with a role in 5′ end processing, although we find that this nuclease activity is not required for resection of restriction enzyme-generated DSB ends in vitro or in human cells. Since the NA/HA catalytic mutant does exhibit defects in survival of more complex forms of DNA damage, we propose that the nuclease activity is only essential when adducts are present on the DNA.
CtIP catalytic activity is clearly specific for 5′ strands at a branched DNA structure, which suggests a mechanism for the removal of 5′ adducts such as those generated by topoisomerase II and Spo11, but it is difficult to imagine how it would remove a 3′ adduct created by topoisomerase I. Sae2 and CtIP have been shown to be required for cell survival after treatment with topoisomerase I poisons such as CPT (Deng et al., 2005; Nakamura et al., 2010; Sartori et al., 2007). One possibility is that a subset of topoisomerase I adducts may form at sites of secondary structure in DNA, for instance cruciforms or hairpins in ssDNA intermediates, and that cleavage of these branched structures could effectively remove the secondary structure as well as the protein adduct (Fig. S3H). There is already evidence for Sae2-dependent removal of Spo11 and topoisomerase II with an attached oligonucleotide in both budding and fission yeasts (Hartsuiker et al., 2009a; Hartsuiker et al., 2009b; Milman et al., 2009; Neale et al., 2005), and the work by Wu et al also establishes a role for the catalytic activity of CtIP in processing secondary structures formed at inverted repeat sequences (Wang et al., 2014). How exactly the Mre11 and Sae2 nuclease activities are coordinated at sites of protein-DNA adducts and the nature of the lesion recognized in vivo are important questions for further study.
While Sae2 and CtIP are clearly related in function and both stimulate resection and act as 5′ flap endonucleases in vitro, there are some obvious differences between these proteins. Sae2 exhibits specificity for hairpins, cleaving ssDNA in overhangs adjacent to these structures, while CtIP does not appear to have this property unless the hairpin is adjacent to a branched DNA structure. The two proteins are very different in size and are not obviously related in primary sequence, apart from the short identity in the C-terminus of both proteins that is not part of the nuclease domain of CtIP. Lastly, CtIP nuclease activity is highest in manganese, and to a lesser extent cobalt, while Sae2 does not exhibit this preference. Based on these differences, we are considering two possibilities: 1) Sae2 and CtIP may not derive from a common ancestor but have evolved to promote similar functions in DNA repair, or 2) Sae2 and CtIP are related but have diverged so significantly that their similarities are nearly unidentifiable in the absence of structural analysis. In support of the second hypothesis, both proteins show extremely high rates of positive selection, particularly in the N-termini, which is also observed in other proteins that affect NHEJ and may reflect ongoing antagonistic relationships with viruses and other mobile elements (Demogines et al.; Sawyer and Malik, 2006).
CtIP does not share significant homology with Mre11; however, we have uncovered here a similarity between the enzymes in their preference for manganese, a nuclease motif that in Mre11 is part of a network of histidine and aspartate residues in the active site that coordinate metal ions, and their ability to be oxidatively cleaved in the presence of ascorbate and peroxide. The use of the oxidative cleavage method to identify known metal-binding sites in Mre11 validates this procedure as a powerful method to locate potential active sites in other proteins. While it is not obvious that CtIP contains the other nuclease motifs that are conserved in Mre11, it could be that there is a similarity in transition metal coordination that could extend to the structural organization of the active site.
Regulation of CtIP activity by phosphorylation
Here we identified 36 phosphorylation sites in CtIP that include previously known and novel sites. Our results with recombinant protein in vitro do not support a direct dependence of nuclease activity on the phosphorylation sites that have previously been implicated in CtIP activity (S327, T847, and T859), suggesting that these modifications either promote CtIP function in an indirect way or alter its activities at break sites in a manner that is not directly related to its nuclease activity.
In contrast, we found that the putative CDK-targeted residues S276, T315, and S347 are very important for CtIP nuclease activity, as are the ATM-targeted residues S231, S664 and S745. The importance of the ATM phosphorylation events for CtIP may explain a recent report showing that ATM activity is essential for repair involving topoisomerase adducts in human cells (Alagoz et al., 2013).
In summary, we have demonstrated that human CtIP is a 5′ flap endonuclease and that this activity is required in some contexts for the efficient function of CtIP. These findings suggest that its role in DNA repair occurs through the catalytic activity of CtIP on DNA adducts, as well as a non-catalytic mechanism by which CtIP promotes the activity of other nucleases, as we have previously shown for Sae2 (Nicolette et al., 2010). CtIP is the target of a large number of post-translational modifications, a small subset of which directly regulate its nuclease activity whereas others affect interactions between CtIP, MRN, and Brca1. We do not yet understand the function of some of these modifications, but it is clear that CtIP receives input from both the DNA damage responses kinases (ATM and ATR) in the regulation of DNA end resection that is used to coordinate DNA repair with cell cycle progression.
Experimental Procedures
Cloning and Protein expression
CtIP oligonucleotide cleavage assay
Nuclease assays were performed with the internally labeled Y-hairpin or with [32P-cordycepin]-labeled oligonucleotides (sequences in Supplementary Experimental Procedures). DNA substrates (0.125 nM) were incubated with CtIP in nuclease buffer (25 mM MOPS pH 7.0, 65 mM NaCl, 1 mM DTT, 5 mM MgCl2, 0.1 mg/mL BSA) at 30°C (or at 37°C, where indicated) for 2 hours. Reactions were stopped by adding 2 μL of stop solution (0.5% SDS, 20 mM EDTA pH 8.0, 5 μM TP2622 oligonucleotide), lyophilized, resuspended in formamide loading buffer, resolved on a 20% acrylamide/urea gel at constant wattage (40 W) for 2.5 hours, and analyzed by phosphorimager (GE). For CtIP oligonucleotide cleavage assays in the presence of different divalent metals the method was modified by substituting TCEP for DTT and adding 1 mM EDTA along with 6 mM Mg2+, or 2 mM Mn2+, Fe2+, Co2+, Ni2+, Cu2+. For the experiments with hairpin DNA substrates, the denaturing gels contained 10% polyacrylamide as well as 20% formamide.
DNA binding assay
Gel mobility shift assays in Fig. 1B were performed with azido-modified [32P-cordycepin]-labeled oligonucleotides TP771, by itself (ssDNA) or annealed to TP828 (dsDNA) or to P3795 (Y-DNA). DNA substrates (0.125 nM) were incubated with CtIP in DNA binding buffer (25 mM MOPS pH 7.0, 1 mM DTT, 10 mM EDTA pH 8.0, 0.1 mg/mL BSA, 65 mM NaCl) on ice for 20 minutes, UV crosslinked, and were resolved on acrylamide/agarose composite gels (0.5% acrylamide, 0.5% agarose)(Suh et al., 2005) using 0.5X TBE as a running buffer. Gels were dried and analyzed by phosphorimager (GE). Gel mobility shift assays in Fig. 2C and in Fig. S2E were performed with a 249 bp dsDNA substrate, internally labeled with [32P]. DNA substrates (0.2 nM) were incubated with CtIP in binding buffer (25 mM MOPS pH 7.0, 1 mM DTT, 10 mM EDTA pH 8.0, 0.1 mg/mL BSA, 30 mM NaCl) on ice for 20 minutes, and were separated in 1% agarose gels using 0.5X TBE as a running buffer and analyzed as described above.
In vitro resection assays
Resection reactions were performed with 0.135 nM DNA linearized plasmid DNA (pNO1; ~ 4.5 kb) and Dna2, BLM helicase, MRN, Ku70/80, and RPA as described previously (Yang et al., 2013) but with CtIP wt and mutant proteins added as indicated. Reactions contained 25 mM MOPS pH 7.0, 1 mM DTT, 5 mM MgCl2, 1 mM ATP, and 30 mM NaCl. The reactions were incubated at 37 °C for 60 min. and stopped with 0.1% SDS and 10 mM EDTA. 33% of the reaction was reserved for qPCR analysis while the remainder was separated on a native agarose gel. The gel was stained with SYBR green (Invitrogen) and imaged using a Typhoon imager (GE), and then transferred to a nylon membrane with non-denaturing transfer. After UV crosslinking of the DNA to the membrane, it was probed with an RNA probe specific for the 3′ strand of a 1 kb region at one end of the linearized DNA, as described previously (Hopkins and Paull, 2008). The level of ssDNA produced during the reaction was also quantified by real-time PCR as described previously (Nicolette et al., 2010).
Site-specific proteolytic cleavage in ascorbate and peroxide
CtIP (1.5 μM) was incubated with 250 mM NaCl, 50 mM MOPS pH 7, 5% glycerol, 100 mM sodium ascorbate, and 10 mM H2O2 in a 20 μL reaction at 4 °C for 1, 2, or 4 hrs. Reactions were stopped by adding 5 μL of 5X SDS loading buffer supplemented with 50 mM EDTA. Samples were kept on ice until the time-course was completed, boiled for 15 minutes, and resolved on a 4–12% gradient polyacrylamide gel. The gel was either stained with Coomassie blue or transferred to a PVDF membrane and probed for the N-terminal Flag-tag (M2 anti-Flag antibody, Sigma) or the C-terminus of CtIP (#61141, Active Motif) as described in Figs. 3A and 3B.
Mre11 (5 μM) was incubated in 80 mM NaCl, 25 mM MOPS pH 7, 20 mM Tris pH 8, 8% glycerol, 5 mM H2O2, and 5 mM sodium ascorbate, at room temperature for 10 min. The oxidative cleavage was stopped by addition of SDS-PAGE loading buffer containing beta-mercaptoethanol. The cleavage products were separated by 10% SDS-PAGE and stained with Coomassie blue or transferred to a PVDF membrane and probed for the C-terminal Flag tag (M2 anti-Flag antibody, Sigma).
Mass Spectrometry
Cell Culture, Plasmid Construction, and Antibodies Used
Homologous recombination (HR) assay
The EGFP-HR DSB repair substrate and I-SceI-induced HR assay were described previously (Wang et al., 2012). U2OS cells carrying EGFP-HR were stably expressed with HA-CtIP-WT or HA-CtIP- NA/HA mutant, and silenced for endogenous CtIP by retroviral infection. Cells were transfected with I-SceI-IRES-dsRedNLS and after 72 h, collected for fluorescence-activated cell sorting (FACS) analysis of EGFP-positive events, using a BD Accuri C6 flow cytometer (Becton-Dickinson). The percentage of EGFP-positive cells is shown, with error bars representing standard deviation of three independent experiments.
Clonogenic Survival Assay
U2OS cells stably expressing HA-CtIP-WT, HA-CtIP- NA/HA or vector, with endogenous CtIP silenced, were treated with indicated amounts of camptothecin or etoposide for 1h, and cultured in complete media for 10–14 days as described (Sartori et al., 2007). Colonies were stained with a solution containing 0.5% crystal violet and 20% ethanol then counted, with the percentage of cell viability shown, error bars show standard deviation from three independent experiments.
Ionizing radiation (IR) assay
siRNA transfection of HeLa cells was carried out using HiPerFect Transfection Reagent (Qiagen) following the manufacturer’s instructions. 50 nM CtIP siRNA (Qiagen) was used: AAG CUA AAA CAG GAA CGA AUC. 24 h after incubation with CtIP siRNA, HeLa cells were transfected with MATra-A (IBA Bio TAGnology) following the manufacturer’s protocol to transfect various GFP-tagged siRNA-resistant CtIP plasmids. 48 h later, cells were irradiated with 2 Gy, fixed and stained for Rad51 and GFP. Immunofluorescence: Cells were grown on glass coverslips. EdU (10 μM) was added 0.5 h prior to IR to exclude S-phase cells from the analysis. Cells were fixed and stained as described (Quennet et al., 2011) and additionally stained with Click-it®EdU (Life technologies). Antibodies were rabbit-α-Rad51 at 1:15,000 (Abcam) and mouse-α-GFP at 1:200 (Roche). Cells were examined with a Zeiss microscope and Metafer software (Metasystems) to identify G1-, S- and G2-phase cells according to their DAPI and EdU signals (Lobrich et al., 2010). Control experiments confirmed that all cells detected with G2 DNA content are positive for the G2 marker CENP-F and all cells with G1 DNA content are negative. Foci enumeration in GFP-positive G2-phase cells was performed in a blinded manner.
Supplementary Material
Figure S1 (related to Figure 1).
(A) Recombinant CtIP proteins used in this study. Human CtIP was expressed in insect cells and purified as described in the Materials and Methods. Shown here are the wild-type and mutant proteins separated by SDS-PAGE and stained with coomassie blue. * C-terminal truncated CtIP.
(B) Nuclease assays as in Fig. 1A using 5′-overhang and 3′ overhang substrates and 50, 100, or 200 nM CtIP as shown.
(C) Nuclease assays as in Fig. 1D using a Y-structure with an internal [32P] label as shown but in the presence of 1 mM MnCl2. In comparison to Fig. 1D, CtIP activity is strongly stimulated by MnCl2 but Sae2 activity is not. See also Fig. 2D.
Figure S2 (related to Figure 2).
(A) Alignment of the CtIP N289/N290 region with nuclease motif III of Mre11.
(B) Recombinant hCtIP (75 nM wt or N289A/H290A as indicated) were incubated with recombinant wild-type hMRN (160 nM) in binding buffer (50 mM NaCL, 25 mM Tris pH 8, 10% glycerol, and 0.4% CHAPS) in a final volume of 200 μl. CtIP was immunoprecipitated from the reaction with anti-Flag antibody resin (hMRN in this case does not have a Flag tag), washed with washing buffer (100 mM NaCl, 25 mM Tris pH 8, 10% glycerol, and 0.1% CHAPS), separated on a 6% SDS polyacrylamide gel and probed for Flag-CtIP (top panel), Rad50 (middle panel), and Nbs1 (bottom panel) individually. hMRN binds to the anti-Flag antibody resin in a CtIP-dependent manner, indicating an interaction with recombinant wild-type and mutant hCtIP.
(C) Quantitation of CtIP endonuclease assays performed as in Fig. 1A with 200 nM wild-type, N289A/H290A, or S347D proteins as indicated. The average of 3 experiments is shown with error bars indicating standard deviation.
(D) The T315A and S276A mutants of CtIP are catalytically inactive. Nuclease assays as in Fig. 1A with wild-type, N289A/H290A (NAHA), T315A, or S276A proteins (50, 100, or 200 nM) using a Y-structure with a 3′ [32P] label as shown, in the presence of 1 mM MgCl2.
(E) The CtIP mutants S347D, S347A, T315A, S276A, N289A/H290A, K513A/K515A, and S231D proteins bind DNA similarly to wild-type CtIP. Wild-type and mutant proteins were incubated with 249 bp DNA internally labeled with [32P] and complexes were separated by native PAGE as in Fig. 2C. Concentrations of CtIP proteins were 12.5, 25, 50, 100, and 200 nm (first panel) and 6.25, 12.5, 25, 50, 100, and 200 nM (other panels).
(F) List of CtIP mutants exhibiting wild-type levels of activity
Figure S3 (related to Figure 4).
(A) Wild-type and N289A/H290A CtIP promote DNA end resection at AsiSI-induced breaks. Endogenous CtIP in ER-AsiSI-293T cells was depleted by shRNA treatment (KD) and transfected with vector (v), shRNA-resistant wild-type (wtCtIP) or N289A/H290A CtIP (CtIP(NA/HA)) plasmids 24 h before treatment with 600 nM 4-OHT, which induces nuclear translocation of the AsiSI enzyme (Iacovoni et al., 2010). After 4 hrs, cells were harvested, genomic DNA was prepared, and either digested with restriction enzymes to distinguish between single-stranded and double-stranded DNA, or mock-digested as described (Zhou and Paull, 2013). Quantitation of single-stranded DNA intermediates generated by resection at two selected AsiSI-induced DSBs (“DSB1” and “DSB2”) was performed by real-time PCR. The percentage of ssDNA at various distances from the two DSBs (335 nt, 1618 nt and 3500 nt from DSB1; 364 nt, 1754 nt and 3564 nt from DSB2) was measured. Background was examined by measuring %ssDNA at a site where there is no nearby AsiSI-induced break (“No DSB”). The average of two experiments is shown, with the qPCR performed in triplicate and the error bars showing standard deviation.
(B) Western blot of CtIP with actin as a loading control.
(C) Resection reactions with linearized plasmid DNA were performed with Dna2 (16 nM), BLM (10 nM), MRN (10 nM), Ku70/80 (100 nM), RPA (80 nM), and CtIP wild-type or nuclease-deficient (N289A/H290A) proteins (50 nM) as indicated. Reactions were separated in a 1% non-denaturing agarose gel and further analyzed by SYBR Green staining (top panel) for total DNA, by non-denaturing Southern hybridization (middle panel) using an RNA probe complementary to the 3′ end of DNA adjacent to one of the break sites, and by quantitative PCR (qPCR) using two sets of primers to measure ssDNA levels at sites located 29 nt or 1025 nt from the DNA end as described previously (Nicolette et al., 2010).
(D) Representative images of GFP-CtIP and Rad51 foci in HeLa cells after 2 Gy irradiation. Endogenous CtIP in HeLa cells was depleted by siRNA treatment and transfected with GFP-tagged wild-type or N289A/H290A (NA/HA) CtIP plasmids 48 h before irradiation. Cells were incubated with EdU 30 min prior to IR exposure (2Gy) and during the entire repair period. GFP-CtIP foci in G2 cells were measured 4 h after irradiation.
(E) Wild-type CtIP but not N289A/H290A promotes resection of DSBs after radiation-induced DNA damage. Endogenous CtIP in HeLa cells was depleted by siRNA treatment and transfected with GFP-tagged wild-type or N289A/H290A CtIP plasmids 48 h before irradiation. pRPA foci were enumerated in GFP-positive G2 cells at 2 h after 2 Gy. Error bars indicate standard error from 4 independent experiments.
(F) Wild-type and N289A/H290A CtIP form foci after DNA damage. Endogenous CtIP in HeLa cells was depleted by siRNA treatment and transfected with GFP-tagged wild-type or N289A/H290A (NA/HA) CtIP plasmids 48 h before irradiation. Cells were incubated with EdU 30 min prior to IR exposure (2Gy) and during the entire repair period. GFP-CtIP foci in G2 cells were measured 4 h after irradiation. HeLa cells were pre-extracted 5 min with PBS/0.5% Triton, fixed 20 min in PBS/2.5% formaldehyde, permeabilised 20 min in PBS/1% FCS/0.5% Triton and blocked 20 min in PBS/1% FCS/5% BSA. Between the different steps the cells were washed with PBS/1%FCS. Antibodies used were mouse-α-GFP at 1:200 (Roche) and rabbit-α-γH2AX at 1:2,000 (Abcam).
(G) Left panel: CtIP was depleted by siRNA and either wild type or nuclease mutant CtIP was re-expressed in mouse Artemis−/−/H2AX−/− DELCJ Abl Pre-B Cells. G1 cell cycle phase arrest and Rag cleavage was induced by adding STI 571 v-abl inhibitor. Right panel: CtIP levels in cells transfected with the shRNA control (shNT), or shRNA directed against CtIP (shCtIP), with shRNA-resistant CtIP alleles expressing vector only (empty), wild-type CtIP (WT), or the N289A/H290A mutant as indicated, with GAPDH as a loading control.
(H) Schematic models of Topoisomerase I conjugates at a replication fork. Topoisomerase I forms 3′ conjugates with DNA, which can generate double-strand breaks during replication. In theory these could be on either the leading or lagging strand templates, but a previous study of the polarity of conjugates in human cells only found evidence for topo I cleavage complexes on the leading strand template (Strumberg et al., 2000), as shown in (1). In theory there could also be conjugates present on both strands (2), or the conjugate may form at the site of secondary structure in the template DNA (3), which has been observed in human cells at replication origins (Abdurashidova et al., 2007) and in some cases have been shown to form unusual secondary structures (Kusic et al., 2005). Note that there are a number of configurations of the adduct which give rise to double-strand break ends lacking a protein conjugate. Red arrows in (3) indicate potential sites of 5′ flap cleavage by CtIP that would contribute to the removal of the adduct. There may also be other events that promote DSB formation, including transcription and single-strand breaks near the Top1 cleavage site (Pommier et al., 2003).
Table S1 (separate Excel file; related to Figure 2).
List of all post-translational modifications identified in human CtIP expressed in human cells, with or without DNA damage (camptothecin treatment), or in Sf21 insect cells. For each modification the peptide mass and scores from Scaffold and Scaffold PTM analysis are shown.
Table S2 (related to Figure 3).
Mre11 C-terminal fragments generated by oxidative cleavage of the a.a. 1 to 616 truncated form of Mre11 were identified by N-terminal sequencing. The sequence identified is shown with the first residue of the fragment shown in parentheses, as well as the estimated molecular mass (including the C-terminal linker and Flag epitope). Mre11 runs on SDS-PAGE gels slightly above its predicted molecular weight. * P132 was not uniquely identified but is inferred to be the first residue since the subsequent TGADA sequence was uniquely identified. The estimated cleavage site is based on known residues that contact metal ions in human Mre11 (Park et al., 2011), which in the case of these products were located just N-terminal of the sequenced fragments.
Highlights.
human CtIP exhibits 5′ flap endonuclease activity
catalytic activity is dependent on phosphorylation
resection of “clean” breaks does not require catalytic activity
resection of ends with adducts requires catalytic activity
Acknowledgments
We thank members of the Paull laboratory for helpful comments and Wen Hwa Lee and Richard Baer for reagents. Studies in the Paull laboratory were supported by NIH CA094008; studies in the Wu laboratory were supported by NIH CA102361 and CA140972; studies in the Sleckman laboratory were supported by NIH AI074953.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Alagoz M, Chiang SC, Sharma A, El-Khamisy SF. ATM deficiency results in accumulation of DNA-topoisomerase I covalent intermediates in neural cells. PloS one. 2013;8:e58239. doi: 10.1371/journal.pone.0058239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andberg M, Jantti J, Heilimo S, Pihkala P, Paananen A, Koskinen AM, Soderlund H, Linder MB. Cleavage of recombinant proteins at poly-His sequences by Co(II) and Cu(II) Protein science: a publication of the Protein Society. 2007;16:1751–1761. doi: 10.1110/ps.072846407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur LM, Gustausson K, Hopfner KP, Carson CT, Stracker TH, Karcher A, Felton D, Weitzman MD, Tainer J, Carney JP. Structural and functional analysis of Mre11–3. Nucleic acids research. 2004;32:1886–1893. doi: 10.1093/nar/gkh343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beucher A, Birraux J, Tchouandong L, Barton O, Shibata A, Conrad S, Goodarzi AA, Krempler A, Jeggo PA, Lobrich M. ATM and Artemis promote homologous recombination of radiation-induced DNA double-strand breaks in G2. EMBO J. 2009;28:3413–3427. doi: 10.1038/emboj.2009.276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cejka P, Cannavo E, Polaczek P, Masuda-Sasa T, Pokharel S, Campbell JL, Kowalczykowski SC. DNA end resection by Dna2-Sgs1-RPA and its stimulation by Top3-Rmi1 and Mre11-Rad50-Xrs2. Nature. 2010;467:112–116. doi: 10.1038/nature09355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PL, Liu F, Cai S, Lin X, Li A, Chen Y, Gu B, Lee EY, Lee WH. Inactivation of CtIP leads to early embryonic lethality mediated by G1 restraint and to tumorigenesis by haploid insufficiency. Mol Cell Biol. 2005;25:3535–3542. doi: 10.1128/MCB.25.9.3535-3542.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciccia A, Elledge SJ. The DNA damage response: making it safe to play with knives. Mol Cell. 2010;40:179–204. doi: 10.1016/j.molcel.2010.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demogines A, East AM, Lee JH, Grossman SR, Sabeti PC, Paull TT, Sawyer SL. Ancient and recent adaptive evolution of primate non-homologous end joining genes. PLoS genetics. 6:e1001169. doi: 10.1371/journal.pgen.1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng C, Brown JA, You D, Brown JM. Multiple endonucleases function to repair covalent topoisomerase I complexes in Saccharomyces cerevisiae. Genetics. 2005;170:591–600. doi: 10.1534/genetics.104.028795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Q, Chow J, Bernstein KA, Makharashvili N, Arora S, Lee CF, Person MD, Rothstein R, Paull TT. Phosphorylation-regulated transitions in an oligomeric state control the activity of the sae2 DNA repair enzyme. Mol Cell Biol. 2014;34:778–793. doi: 10.1128/MCB.00963-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker E, Mizuno K, Molnar M, Kohli J, Ohta K, Carr AM. Ctp1CtIP and Rad32Mre11 nuclease activity are required for Rec12Spo11 removal, but Rec12Spo11 removal is dispensable for other MRN-dependent meiotic functions. Mol Cell Biol. 2009a;29:1671–1681. doi: 10.1128/MCB.01182-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartsuiker E, Neale MJ, Carr AM. Distinct requirements for the Rad32(Mre11) nuclease and Ctp1(CtIP) in the removal of covalently bound topoisomerase I and II from DNA. Mol Cell. 2009b;33:117–123. doi: 10.1016/j.molcel.2008.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmink BA, Tubbs AT, Dorsett Y, Bednarski JJ, Walker LM, Feng Z, Sharma GG, McKinnon PJ, Zhang J, Bassing CH, et al. H2AX prevents CtIP-mediated DNA end resection and aberrant repair in G1-phase lymphocytes. Nature. 2011;469:245–249. doi: 10.1038/nature09585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins B, Paull TT. The P. furiosus Mre11/Rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell. 2008;135:250–260. doi: 10.1016/j.cell.2008.09.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huertas P, Jackson SP. Human CtIP mediates cell cycle control of DNA end resection and double strand break repair. J Biol Chem. 2009;284:9558–9565. doi: 10.1074/jbc.M808906200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacovoni JS, Caron P, Lassadi I, Nicolas E, Massip L, Trouche D, Legube G. High-resolution profiling of gammaH2AX around DNA double strand breaks in the mammalian genome. EMBO J. 2010;29:1446–1457. doi: 10.1038/emboj.2010.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaidi A, Weinert BT, Choudhary C, Jackson SP. Human SIRT6 promotes DNA end resection through CtIP deacetylation. Science. 2010;329:1348–1353. doi: 10.1126/science.1192049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- Lee K, Lee SE. Saccharomyces cerevisiae Sae2- and Tel1-Dependent Single Strand DNA Formation at DNA Break Promotes Microhomology-Mediated End Joining. Genetics. 2007 doi: 10.1534/genetics.107.076539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee-Theilen M, Matthews AJ, Kelly D, Zheng S, Chaudhuri J. CtIP promotes microhomology-mediated alternative end joining during class-switch recombination. Nat Struct Mol Biol. 2011;18:75–79. doi: 10.1038/nsmb.1942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lengsfeld BM, Rattray AJ, Bhaskara V, Ghirlando R, Paull TT. Sae2 Is an Endonuclease that Processes Hairpin DNA Cooperatively with the Mre11/Rad50/Xrs2 Complex. Mol Cell. 2007;28:638–651. doi: 10.1016/j.molcel.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Ting NS, Zheng L, Chen PL, Ziv Y, Shiloh Y, Lee EY, Lee WH. Functional link of BRCA1 and ataxia telangiectasia gene product in DNA damage response. Nature. 2000;406:210–215. doi: 10.1038/35018134. [DOI] [PubMed] [Google Scholar]
- Lobrich M, Shibata A, Beucher A, Fisher A, Ensminger M, Goodarzi AA, Barton O, Jeggo PA. gammaH2AX foci analysis for monitoring DNA double-strand break repair: strengths, limitations and optimization. Cell cycle. 2010;9:662–669. doi: 10.4161/cc.9.4.10764. [DOI] [PubMed] [Google Scholar]
- McKee AH, Kleckner N. A general method for identifying recessive diploid-specific mutations in Saccharomyces cerevisiae, its application to the isolation of mutants blocked at intermediate stages of meiotic prophase and characterization of a new gene SAE2. Genetics. 1997;146:797–816. doi: 10.1093/genetics/146.3.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milman N, Higuchi E, Smith GR. Meiotic DNA double-strand break repair requires two nucleases, MRN and Ctp1, to produce a single size class of Rec12 (Spo11)-oligonucleotide complexes. Mol Cell Biol. 2009;29:5998–6005. doi: 10.1128/MCB.01127-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimitou EP, Symington LS. DNA end resection: many nucleases make light work. DNA Repair. 2009;8:983–995. doi: 10.1016/j.dnarep.2009.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreau S, Ferguson JR, Symington LS. The nuclease activity of Mre11 is required for meiosis but not for mating type switching, end joining, or telomere maintenance. Mol Cell Biol. 1999;19:556–566. doi: 10.1128/mcb.19.1.556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kogame T, Oshiumi H, Shinohara A, Sumitomo Y, Agama K, Pommier Y, Tsutsui KM, Tsutsui K, Hartsuiker E, et al. Collaborative action of Brca1 and CtIP in elimination of covalent modifications from double-strand breaks to facilitate subsequent break repair. PLoS genetics. 2010;6:e1000828. doi: 10.1371/journal.pgen.1000828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neale MJ, Pan J, Keeney S. Endonucleolytic processing of covalent protein-linked DNA double-strand breaks. Nature. 2005;436:1053–1057. doi: 10.1038/nature03872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolette ML, Lee K, Guo Z, Rani M, Chow JM, Lee SE, Paull TT. Mre11-Rad50-Xrs2 and Sae2 promote 5′ strand resection of DNA double-strand breaks. Nat Struct Mol Biol. 2010;17:1478–1485. doi: 10.1038/nsmb.1957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nimonkar AV, Genschel J, Kinoshita E, Polaczek P, Campbell JL, Wyman C, Modrich P, Kowalczykowski SC. BLM-DNA2-RPA-MRN and EXO1-BLM-RPA-MRN constitute two DNA end resection machineries for human DNA break repair. Genes Dev. 2011;25:350–362. doi: 10.1101/gad.2003811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu H, Chung WH, Zhu Z, Kwon Y, Zhao W, Chi P, Prakash R, Seong C, Liu D, Lu L, et al. Mechanism of the ATP-dependent DNA end-resection machinery from Saccharomyces cerevisiae. Nature. 2010;467:108–111. doi: 10.1038/nature09318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park YB, Chae J, Kim YC, Cho Y. Crystal structure of human Mre11: understanding tumorigenic mutations. Structure. 2011;19:1591–1602. doi: 10.1016/j.str.2011.09.010. [DOI] [PubMed] [Google Scholar]
- Paull TT. Making the best of the loose ends: Mre11/Rad50 complexes and Sae2 promote DNA double-strand break resection. DNA Repair. 2010;9:1283–1291. doi: 10.1016/j.dnarep.2010.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paull TT, Gellert M. The 3′ to 5′ exonuclease activity of Mre 11 facilitates repair of DNA double-strand breaks. Mol Cell. 1998;1:969–979. doi: 10.1016/s1097-2765(00)80097-0. [DOI] [PubMed] [Google Scholar]
- Penkner A, Portik-Dobos Z, Tang L, Schnabel R, Novatchkova M, Jantsch V, Loidl J. A conserved function for a Caenorhabditis elegans Com1/Sae2/CtIP protein homolog in meiotic recombination. EMBO J. 2007;26:5071–5082. doi: 10.1038/sj.emboj.7601916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson SE, Li Y, Wu-Baer F, Chait BT, Baer R, Yan H, Gottesman ME, Gautier J. Activation of DSB Processing Requires Phosphorylation of CtIP by ATR. Mol Cell. 2012 doi: 10.1016/j.molcel.2012.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prinz S, Amon A, Klein F. Isolation of COM1, a new gene required to complete meiotic double-strand break-induced recombination in Saccharomyces cerevisiae. Genetics. 1997;146:781–795. doi: 10.1093/genetics/146.3.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quennet V, Beucher A, Barton O, Takeda S, Lobrich M. CtIP and MRN promote non-homologous end-joining of etoposide-induced DNA double-strand breaks in G1. Nucleic acids research. 2011;39:2144–2152. doi: 10.1093/nar/gkq1175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qvist P, Huertas P, Jimeno S, Nyegaard M, Hassan MJ, Jackson SP, Borglum AD. CtIP Mutations Cause Seckel and Jawad Syndromes. PLoS genetics. 2011;7:e1002310. doi: 10.1371/journal.pgen.1002310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenberg M, Kohli J, Ludin K. Ctp1 and the MRN-complex are required for endonucleolytic Rec12 removal with release of a single class of oligonucleotides in fission yeast. PLoS genetics. 2009;5:e1000722. doi: 10.1371/journal.pgen.1000722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sartori AA, Lukas C, Coates J, Mistrik M, Fu S, Bartek J, Baer R, Lukas J, Jackson SP. Human CtIP promotes DNA end resection. Nature. 2007;450:509–514. doi: 10.1038/nature06337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawyer SL, Malik HS. Positive selection of yeast nonhomologous end-joining genes and a retrotransposon conflict hypothesis. Proceedings of the National Academy of Sciences of the United States of America. 2006;103:17614–17619. doi: 10.1073/pnas.0605468103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sereikaite J, Jachno J, Santockyte R, Chmielevski P, Bumelis VA, Dienys G. Protein scission by metal ion-ascorbate system. The protein journal. 2006;25:369–378. doi: 10.1007/s10930-006-9014-7. [DOI] [PubMed] [Google Scholar]
- Stair JL, Holcombe JA. Metal binding characterization and conformational studies using Raman microscopy of resin-bound poly(aspartic acid) Analytical chemistry. 2007;79:1999–2006. doi: 10.1021/ac061602p. [DOI] [PubMed] [Google Scholar]
- Steger M, Murina O, Huhn D, Ferretti LP, Walser R, Hanggi K, Lafranchi L, Neugebauer C, Paliwal S, Janscak P, et al. Prolyl isomerase PIN1 regulates DNA double-strand break repair by counteracting DNA end resection. Mol Cell. 2013;50:333–343. doi: 10.1016/j.molcel.2013.03.023. [DOI] [PubMed] [Google Scholar]
- Stracker TH, Petrini JH. The MRE11 complex: starting from the ends. Nature reviews Molecular cell biology. 2011;12:90–103. doi: 10.1038/nrm3047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suh MH, Ye P, Datta AB, Zhang M, Fu J. An agarose-acrylamide composite native gel system suitable for separating ultra-large protein complexes. Analytical biochemistry. 2005;343:166–175. doi: 10.1016/j.ab.2005.05.016. [DOI] [PubMed] [Google Scholar]
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–271. doi: 10.1146/annurev-genet-110410-132435. [DOI] [PubMed] [Google Scholar]
- Uanschou C, Siwiec T, Pedrosa-Harand A, Kerzendorfer C, Sanchez-Moran E, Novatchkova M, Akimcheva S, Woglar A, Klein F, Schlogelhofer P. A novel plant gene essential for meiosis is related to the human CtIP and the yeast COM1/SAE2 gene. EMBO J. 2007;26:5061–5070. doi: 10.1038/sj.emboj.7601913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Li Y, Truong LN, Shi LZ, Hwang PY, He J, Do J, Cho MJ, Li H, Negrete A, et al. An end resection-independent CtIP endonuclease activity is required for maintaining genome stability at common fragile sites and inverted repeats. Molecular Cell. 2014 doi: 10.1016/j.molcel.2014.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shao Z, Shi LZ, Hwang PY, Truong LN, Berns MW, Chen DJ, Wu X. CtIP protein dimerization is critical for its recruitment to chromosomal DNA double-stranded breaks. J Biol Chem. 2012;287:21471–21480. doi: 10.1074/jbc.M112.355354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shi LZ, Wong CC, Han X, Hwang PY, Truong LN, Zhu Q, Shao Z, Chen DJ, Berns MW, et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS genetics. 2013a;9:e1003277. doi: 10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Shi LZ, Wong CCL, Han X, Hwang PY-H, Truong LN, Zhu Q, Shao Z, Chen DJ, Berns MW, et al. The interaction of CtIP and Nbs1 connects CDK and ATM to regulate HR-mediated double-strand break repair. PLoS genetics. 2013b doi: 10.1371/journal.pgen.1003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DJ, Reiter NJ, Sikkink RA, Yu L, Rusnak F. Identification of the high affinity Mn2+ binding site of bacteriophage lambda phosphoprotein phosphatase: effects of metal ligand mutations on electron paramagnetic resonance spectra and phosphatase activities. Biochemistry. 2001;40:8918–8929. doi: 10.1021/bi010637a. [DOI] [PubMed] [Google Scholar]
- Yang SH, Zhou R, Campbell J, Chen J, Ha T, Paull TT. The SOSS1 single-stranded DNA binding complex promotes DNA end resection in concert with Exo1. EMBO J. 2013;32:126–139. doi: 10.1038/emboj.2012.314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Bailis JM. DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints. Trends in cell biology. 2010;20:402–409. doi: 10.1016/j.tcb.2010.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- You Z, Shi LZ, Zhu Q, Wu P, Zhang YW, Basilio A, Tonnu N, Verma IM, Berns MW, Hunter T. CtIP links DNA double-strand break sensing to resection. Mol Cell. 2009;36:954–969. doi: 10.1016/j.molcel.2009.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu X, Chen J. DNA damage-induced cell cycle checkpoint control requires CtIP, a phosphorylation-dependent binding partner of BRCA1 C-terminal domains. Mol Cell Biol. 2004;24:9478–9486. doi: 10.1128/MCB.24.21.9478-9486.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Chen J. N terminus of CtIP is critical for homologous recombination-mediated double-strand break repair. J Biol Chem. 2009;284:31746–31752. doi: 10.1074/jbc.M109.023424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yun MH, Hiom K. CtIP-BRCA1 modulates the choice of DNA double-strand-break repair pathway throughout the cell cycle. Nature. 2009;459:460–463. doi: 10.1038/nature07955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Jasin M. An essential role for CtIP in chromosomal translocation formation through an alternative end-joining pathway. Nat Struct Mol Biol. 2011;18:80–84. doi: 10.1038/nsmb.1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Caron P, Legube G, Paull TT. Quantitation of DNA double-strand break resection intermediates in human cells. Nucleic acids research. 2014;42:e19. doi: 10.1093/nar/gkt1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 (related to Figure 1).
(A) Recombinant CtIP proteins used in this study. Human CtIP was expressed in insect cells and purified as described in the Materials and Methods. Shown here are the wild-type and mutant proteins separated by SDS-PAGE and stained with coomassie blue. * C-terminal truncated CtIP.
(B) Nuclease assays as in Fig. 1A using 5′-overhang and 3′ overhang substrates and 50, 100, or 200 nM CtIP as shown.
(C) Nuclease assays as in Fig. 1D using a Y-structure with an internal [32P] label as shown but in the presence of 1 mM MnCl2. In comparison to Fig. 1D, CtIP activity is strongly stimulated by MnCl2 but Sae2 activity is not. See also Fig. 2D.
Figure S2 (related to Figure 2).
(A) Alignment of the CtIP N289/N290 region with nuclease motif III of Mre11.
(B) Recombinant hCtIP (75 nM wt or N289A/H290A as indicated) were incubated with recombinant wild-type hMRN (160 nM) in binding buffer (50 mM NaCL, 25 mM Tris pH 8, 10% glycerol, and 0.4% CHAPS) in a final volume of 200 μl. CtIP was immunoprecipitated from the reaction with anti-Flag antibody resin (hMRN in this case does not have a Flag tag), washed with washing buffer (100 mM NaCl, 25 mM Tris pH 8, 10% glycerol, and 0.1% CHAPS), separated on a 6% SDS polyacrylamide gel and probed for Flag-CtIP (top panel), Rad50 (middle panel), and Nbs1 (bottom panel) individually. hMRN binds to the anti-Flag antibody resin in a CtIP-dependent manner, indicating an interaction with recombinant wild-type and mutant hCtIP.
(C) Quantitation of CtIP endonuclease assays performed as in Fig. 1A with 200 nM wild-type, N289A/H290A, or S347D proteins as indicated. The average of 3 experiments is shown with error bars indicating standard deviation.
(D) The T315A and S276A mutants of CtIP are catalytically inactive. Nuclease assays as in Fig. 1A with wild-type, N289A/H290A (NAHA), T315A, or S276A proteins (50, 100, or 200 nM) using a Y-structure with a 3′ [32P] label as shown, in the presence of 1 mM MgCl2.
(E) The CtIP mutants S347D, S347A, T315A, S276A, N289A/H290A, K513A/K515A, and S231D proteins bind DNA similarly to wild-type CtIP. Wild-type and mutant proteins were incubated with 249 bp DNA internally labeled with [32P] and complexes were separated by native PAGE as in Fig. 2C. Concentrations of CtIP proteins were 12.5, 25, 50, 100, and 200 nm (first panel) and 6.25, 12.5, 25, 50, 100, and 200 nM (other panels).
(F) List of CtIP mutants exhibiting wild-type levels of activity
Figure S3 (related to Figure 4).
(A) Wild-type and N289A/H290A CtIP promote DNA end resection at AsiSI-induced breaks. Endogenous CtIP in ER-AsiSI-293T cells was depleted by shRNA treatment (KD) and transfected with vector (v), shRNA-resistant wild-type (wtCtIP) or N289A/H290A CtIP (CtIP(NA/HA)) plasmids 24 h before treatment with 600 nM 4-OHT, which induces nuclear translocation of the AsiSI enzyme (Iacovoni et al., 2010). After 4 hrs, cells were harvested, genomic DNA was prepared, and either digested with restriction enzymes to distinguish between single-stranded and double-stranded DNA, or mock-digested as described (Zhou and Paull, 2013). Quantitation of single-stranded DNA intermediates generated by resection at two selected AsiSI-induced DSBs (“DSB1” and “DSB2”) was performed by real-time PCR. The percentage of ssDNA at various distances from the two DSBs (335 nt, 1618 nt and 3500 nt from DSB1; 364 nt, 1754 nt and 3564 nt from DSB2) was measured. Background was examined by measuring %ssDNA at a site where there is no nearby AsiSI-induced break (“No DSB”). The average of two experiments is shown, with the qPCR performed in triplicate and the error bars showing standard deviation.
(B) Western blot of CtIP with actin as a loading control.
(C) Resection reactions with linearized plasmid DNA were performed with Dna2 (16 nM), BLM (10 nM), MRN (10 nM), Ku70/80 (100 nM), RPA (80 nM), and CtIP wild-type or nuclease-deficient (N289A/H290A) proteins (50 nM) as indicated. Reactions were separated in a 1% non-denaturing agarose gel and further analyzed by SYBR Green staining (top panel) for total DNA, by non-denaturing Southern hybridization (middle panel) using an RNA probe complementary to the 3′ end of DNA adjacent to one of the break sites, and by quantitative PCR (qPCR) using two sets of primers to measure ssDNA levels at sites located 29 nt or 1025 nt from the DNA end as described previously (Nicolette et al., 2010).
(D) Representative images of GFP-CtIP and Rad51 foci in HeLa cells after 2 Gy irradiation. Endogenous CtIP in HeLa cells was depleted by siRNA treatment and transfected with GFP-tagged wild-type or N289A/H290A (NA/HA) CtIP plasmids 48 h before irradiation. Cells were incubated with EdU 30 min prior to IR exposure (2Gy) and during the entire repair period. GFP-CtIP foci in G2 cells were measured 4 h after irradiation.
(E) Wild-type CtIP but not N289A/H290A promotes resection of DSBs after radiation-induced DNA damage. Endogenous CtIP in HeLa cells was depleted by siRNA treatment and transfected with GFP-tagged wild-type or N289A/H290A CtIP plasmids 48 h before irradiation. pRPA foci were enumerated in GFP-positive G2 cells at 2 h after 2 Gy. Error bars indicate standard error from 4 independent experiments.
(F) Wild-type and N289A/H290A CtIP form foci after DNA damage. Endogenous CtIP in HeLa cells was depleted by siRNA treatment and transfected with GFP-tagged wild-type or N289A/H290A (NA/HA) CtIP plasmids 48 h before irradiation. Cells were incubated with EdU 30 min prior to IR exposure (2Gy) and during the entire repair period. GFP-CtIP foci in G2 cells were measured 4 h after irradiation. HeLa cells were pre-extracted 5 min with PBS/0.5% Triton, fixed 20 min in PBS/2.5% formaldehyde, permeabilised 20 min in PBS/1% FCS/0.5% Triton and blocked 20 min in PBS/1% FCS/5% BSA. Between the different steps the cells were washed with PBS/1%FCS. Antibodies used were mouse-α-GFP at 1:200 (Roche) and rabbit-α-γH2AX at 1:2,000 (Abcam).
(G) Left panel: CtIP was depleted by siRNA and either wild type or nuclease mutant CtIP was re-expressed in mouse Artemis−/−/H2AX−/− DELCJ Abl Pre-B Cells. G1 cell cycle phase arrest and Rag cleavage was induced by adding STI 571 v-abl inhibitor. Right panel: CtIP levels in cells transfected with the shRNA control (shNT), or shRNA directed against CtIP (shCtIP), with shRNA-resistant CtIP alleles expressing vector only (empty), wild-type CtIP (WT), or the N289A/H290A mutant as indicated, with GAPDH as a loading control.
(H) Schematic models of Topoisomerase I conjugates at a replication fork. Topoisomerase I forms 3′ conjugates with DNA, which can generate double-strand breaks during replication. In theory these could be on either the leading or lagging strand templates, but a previous study of the polarity of conjugates in human cells only found evidence for topo I cleavage complexes on the leading strand template (Strumberg et al., 2000), as shown in (1). In theory there could also be conjugates present on both strands (2), or the conjugate may form at the site of secondary structure in the template DNA (3), which has been observed in human cells at replication origins (Abdurashidova et al., 2007) and in some cases have been shown to form unusual secondary structures (Kusic et al., 2005). Note that there are a number of configurations of the adduct which give rise to double-strand break ends lacking a protein conjugate. Red arrows in (3) indicate potential sites of 5′ flap cleavage by CtIP that would contribute to the removal of the adduct. There may also be other events that promote DSB formation, including transcription and single-strand breaks near the Top1 cleavage site (Pommier et al., 2003).
Table S1 (separate Excel file; related to Figure 2).
List of all post-translational modifications identified in human CtIP expressed in human cells, with or without DNA damage (camptothecin treatment), or in Sf21 insect cells. For each modification the peptide mass and scores from Scaffold and Scaffold PTM analysis are shown.
Table S2 (related to Figure 3).
Mre11 C-terminal fragments generated by oxidative cleavage of the a.a. 1 to 616 truncated form of Mre11 were identified by N-terminal sequencing. The sequence identified is shown with the first residue of the fragment shown in parentheses, as well as the estimated molecular mass (including the C-terminal linker and Flag epitope). Mre11 runs on SDS-PAGE gels slightly above its predicted molecular weight. * P132 was not uniquely identified but is inferred to be the first residue since the subsequent TGADA sequence was uniquely identified. The estimated cleavage site is based on known residues that contact metal ions in human Mre11 (Park et al., 2011), which in the case of these products were located just N-terminal of the sequenced fragments.