Abstract
Affinity maturation of antibodies requires a unique process of targeted mutation that allows changes to accumulate in the antibody genes while the rest of the genome is protected from off-target mutations that can be oncogenic. This targeting requires that the same deamination event be repaired either by a mutagenic or a high-fidelity pathway depending on the genomic location. We have previously shown that the BRCT domain of the DNA-damage sensor PARP-1 is required for mutagenic repair occurring in the context of IgH and IgL diversification in the chicken B cell line DT40. Here we show that immunoprecipitation of the BRCT domain of PARP-1 pulls down Ku70 and the DNA–PK complex although the BRCT domain of PARP-1 does not bind DNA, suggesting that this interaction is not DNA dependent. Through sequencing the IgL variable region in PARP-1−/− cells that also lack Ku70 or Lig4, we show that Ku70 or Lig4 deficiency restores GCV to PARP-1−/− cells and conclude that the mechanism by which PARP-1 is promoting mutagenic repair is by inhibiting high-fidelity repair which would otherwise be mediated by Ku70 and Lig4.
Keywords: PARP-1, DNA–PK, DNA repair, Gene conversion
1. Introduction
Antibody diversity is generated by a an ingenious program of V(D)J recombination to form a huge variety of B cell receptors, followed by somatic hypermutation to generate extremely high affinity antibodies. V(D)J recombination is mediated by the Rag proteins, which generate controlled breaks at specific RSS sequences and recombine the DNA ends to form a functional gene [1]. This process is dependent on DNA–PK and Lig4 among other proteins [2,3]. Mechanisms of targeting of mutations to Ig loci during somatic hypermutation and gene conversion are just starting to unfold. AID targets these genes for deamination by an unknown mechanism. The resulting lesion is either excised by UNG or recognized by MMR pathways that recruit a mutagenic polymerase Polη, rather than a high fidelity polymerase for repair. The abasic site generated by UNG can be filled in by Rev1 or bound by an AP lyase which generates a DNA backbone break. This break stimulates gene conversion (GCV) in organisms with templates for homologous repair, such as chicken.
Recently, we have shown that PARP-1 has a distinct role in promoting vertebrate somatic IgH and IgL gene diversification through GCV by inhibiting high-fidelity repair of AID-induced lesions thus directing repair through mutagenic pathways [4]. This role was found to be dependent on PARP-1's BRCT domain, which is not required for high-fidelity base excision repair [4]. Gene conversion is thought to be a process similar to homologous recombination repair and indeed, it is dependent on some of the same proteins (XRCC2/XRCC3). However, gene conversion at hypermutating Ig loci results in mutagenic repair rather than high fidelity repair, due to the presence of homologous upstream pseudogenes with ~90% identity to the variable region. Whether gene conversion at mutating Ig loci should be considered a unique form of mutagenic DNA repair or an unusual instance of high fidelity homologous recombination is an open question. It is notable that when XRCC2 or XRCC3 are knocked out of the DT40 system, the deamination which would have been repaired by gene conversion are diverted to other mutagenic repair pathways, not high fidelity repair, while PARP-1−/− results in high fidelity repair of the deamination. This suggests that PARP-1 is not mediating gene conversion so much as preventing high fidelity repair of the lesion, placing gene conversion in the realm of mutagenic repair options.
Since BRCT domains are generally phosphoprotein interacting domains, we hypothesized that the mechanism underlying PARP-1's role in promoting mutagenic repair involves BRCT domain-mediated interactions that impair high-fidelity repair, thus allowing alternative, mutagenic repair pathways to predominate at diversifying loci [4]. To test this hypothesis, we used the isolated PARP-1 BRCT domain as a functional and affinity probe of BRCT-mediated PARP-1 interactions occurring during IgL diversification in DT40 cells. We observe that overexpression of the isolated BRCT domain of PARP-1 inhibits gene conversion, and that immunoprecipitation using the isolated BRCT domain pulled down the entire DNA–PK complex, suggesting that PARP-1's role in gene conversion is due to modulation of DNA–PK complex function through BRCT domain-mediated interactions. Further supporting this model, we also observed that knocking out Ku70 or Lig4 rescues gene conversion in PARP-1−/− DT40s. Taken together, our data support a model in which PARP-1 and the DNA–PK complex act in competing repair pathways at diversifying Ig loci. Additionally, that a major role of PARP-1 in promoting gene conversion can be recapitulated by downregulation of Ku70/Lig4 activity, suggesting that PARP-1 may interact directly or indirectly with the Ku70/Lig4 complex in order to block its activity at the DNA lesion, thus allowing slower, mutagenic repair pathways such as gene conversion to predominate.
2. Materials and methods
2.1. PCR and sequencing of IgL and IgH
The variable regions of IgL and IgH were amplified with Accuprime Pfx and blunt end Topo cloned. Single colonies were picked and sequenced using the M13 reverse primer. The primers used for PCR were IgL-F: CAGGAGCTCGCGGGGCCGTCACTGATTGCCG, IgL-R: GCGCAAGCTTCCCCAGCCTGCCGCCAAGTCCAAG.
2.2. Categorization of mutations
Sequences were aligned using Phred and Phrap and viewed in Consed. High quality base discrepancies were noted and subjected to further analysis. As the total mutation rate was much lower than 1 mutation/read, tracks of multiple mutations in a read were scored as gene conversion events. Single mutations for which no donor template could be identified (using blastn against NCBI gi 118098819) were scored as point mutations. To avoid difficulties in interpretation due to the presence of ambiguous mutations, which match the pseudogene templates but occur in isolation, results are reported when these mutations were either counted as GCV events or as point mutations.
2.3. Tissue culture
DT40 cells were cultured in RPMI with 10% FBS, 1% CS, Pen/Strep, and b-ME at 41°. PARP-1−/− cells and the WT parent cell line were a generous gift from S. Takeda. Cell lines reconstituted with human PARP-1 or the BRCT domain were generated by electroporation of PARP-1 cDNA in the 5/TO vector (Invitrogen) at 550 V, 25 μF in 4 mm cuvettes using the GenePulser from Bio-Rad. For experiments involving treatment with PARP inhibitors, 25 μM AZD2281 was included in the media. Cells were allowed to accumulate mutations for a period of three to four weeks. There were no notable differences in generation time and all cells were split 1:16 every other day, except for Lig4-deficient cells which grew more slowly and were split 1:10 every other day.
2.4. MMS survival assays
Cells were exposed to MMS at the indicated concentrations for 1 h at 37°. They were then washed 2× in fresh media and resuspended in 3 mL media. 450 μL of 3% agar was added and 1 mL was plated in triplicate. Plates were grown for 3–4 days at 41° before colonies were counted.
2.5. Coimmunoprecipitation
Ramos cells were grown to a density of 5 × 105 cells/mL in 50 mL media. Cells were pelleted, washed, and incubated with anti-FLAG (M2) sepharose beads (Sigma) overnight at 4°. The pellets were washed 3× with IP buffer and 3× with TBS then digested for analysis by mass spectrometry.
2.6. Sample digestion
Alkylation, reduction, and tryptic digestion was performed by adding 100 μL of 100 mM ammonium bicarbonate/0.1% PPS/5 mM dithiothreitol (Protein Discovery Labs, Sigma) directly to the affinity resin, incubating for 1 h at 70 °C, followed by reduction with 15 mM iodoacetamide. Alkylated samples were then digested using (1:50 w/w) of modified porcine trypsin (V5111, Promega). The digestion proceeded at 37 °C overnight at which point digestion was quenched with HCL (200 mM), incubated at 37 °C for 45 min, and centrifuged for 10 min to remove PPS prior to LCMS application.
2.7. Online LCMS analysis
LCMS analysis was performed on an LTQ-Velos (Thermo Scientific) inline with an Eksigent 1D plus nano-LC (Eksigent). 20 μL of each digest was loaded onto a reverse phase trap (5 μm, 200 Å, Magic; Michrom Bioresources) with 100% buffer A and washed for 15 min prior to separation on a microcapillary column. The microcapillary column was constructed by slurry packing 10 cm of C18 material (2.7 μm, 90 Å, HALO, Michrom Bioresources) into a 75 μm ID Picofrit (New Objective). Buffer solutions were made from LC–MS grade water, acetonitrile, and formic acid (Fisher) and were comprised of 5% acetonitrile/0.1% acetic acid (Buffer A) and 100% acetonitrile/0.1% acetic acid (Buffer B). A 30 min gradient from 0% to 5% B over 1 min and 5% to 35% buffer B over 29 min was used at a flow rate of 0.3 μL/min. Full MS/MS spectra were acquired in profile mode, with a mass range of 400–2000 with the following dynamic exclusion: Repeat count = 2, Repeat duration = 60 s, and Exclusion list size = 500. Spectra were collected using normalized collision energy of 30% and an isolation window of 3 Da.
2.8. Data analysis for peptide and protein identification
Peptide and protein identification was conducted using the SPIRE analysis pipeline to search MS/MS spectra (http://proteinspire.com). Searches were conducted against the Human IPI FASTA using the X!Tandem search engine [5] with a 2.5 Da mass error, a variable modification for methionine oxidation (16@M) and a fixed modification for iodoacetamide (57.02@C) along with the default search parameters. A randomly reshuffled version of the database was appended for error estimation.
The search results were processed with the LIPS (logistic identification of peptide sequences) model [6] to generate peptide spectra scores. Peptide identification probabilities and FDRs were calculated based on the reshuffled matches using an isotonic regression model [7,8]. A 90% certainty was used as the basis for spectra identifications. A recently introduced approach was used to estimate the protein identification FDR from individual peptide identification probabilities [8,9].
2.9. Western blot
Coimmunoprecipitation was performed as described above, but rather than submit the final sample to mass spectroscopic analysis, western blot was performed using the antibody sc-5309 (Ku70).
3. Results
3.1. Parameters of PARP-1 function in GCV: PARP-1 catalytic activity is not required, BRCT domain can bind its target independent of full length PARP-1 protein
We wished to clarify the parameters of the role of PARP-1 in GCV. We previously established that DNA binding of PARP-1 is required to mediate GCV, and the automodification domain is not. Presumably, the DNA binding domain is required to target PARP-1 to the site of DNA repair, but the signal generated by self-ADP ribosylation is not required. However, the requirement for the BRCT domain does not present a simple explanation, as BRCT domains are generally phosphoprotein interaction domains without intrinsic activity. Rather, this suggests a few possible mechanisms by which PARP-1 could mediate GCV. PARP-1 could be binding/recruiting another protein via its BRCT domain, binding could induce a conformational change in the target protein, or binding could simply result in steric hinderance which impedes function. To address these questions, we asked whether the BRCT domain could interact with its partner in isolation, or if it required the rest of the PARP-1 protein, including the DNA binding domain. Additionally, we wanted to investigate whether PARP-1 catalytic activity was required to mediate this effect.
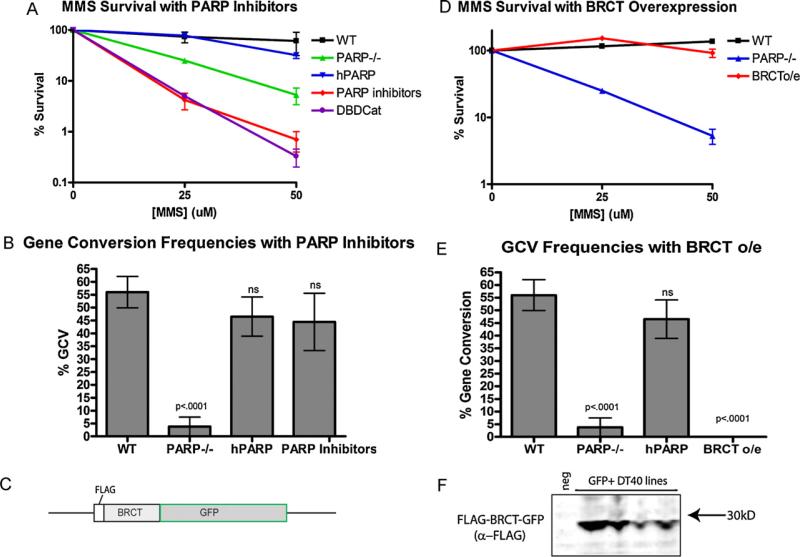
To assess the role of PARP-1 catalytic activity, we cultured DT40 cells in the PARP inhibitor AZD2281, which inhibits catalytic activity without impairing the zinc finger or BRCT domains. We then PCR amplified, cloned, and sequenced the IgL variable region to assess the accumulation of mutations (complete sequences are available in supplemental Figure S1). Surprisingly, we found that culturing DT40 cells in sufficient PARP inhibitor to render a phenotype identical to PARP-1 knockouts in an MMS survival assay of base excision repair (BER) (Fig. 1a), had no detrimental effect on gene conversion (Fig. 1b).
Fig. 1.
PARP inhibitors impair survival in response to challenge with MMS, but do not impair gene conversion, while overexpression of the BRCT domain of PARP-1 has the opposite effect. (A) Survival of indicated cell lines in response to MMS induced damage. Error bars indicate the SEM of three experiments. (B) Frequency of gene conversion events in respective cell lines. Error bars indicate the SEM of 2–4 cell lines cultured independently. (C) Schematic of BRCT overexpression construct. (D) Survival of indicated cell lines in response to MMS induced damage. Error bars indicate the SEM of three experiments. (E) Frequency of gene conversion events in respective cell lines. Error bars indicate the SEM of 2–4 cell lines cultured independently. (F) Western blot showing expression of the BRCT overexpression construct in multiple independent DT40 cell lines which fluoresced green, stained with anti-FLAG (Sigma).
While sequencing and comparison with reference sequence make it clear which mutagenic pathway (point mutation or gene conversion) resulted in most of the mutations analyzed, a small proportion of mutations generated will match the pseudogene sequences, but occur in isolation, thus we cannot know for certain if those mutations arose as a gene conversion event that changed only one nucleotide or a point mutation which by coincidence matched the pseudogenes. As there is some evidence to suggest that many of these ambiguous mutations are gene conversion events [10], our analyses above are based on this assumption. However, to confirm the robustness of the data in view of these ambiguities, we also analyzed the data when all ambiguous mutations are scored as point mutations instead of gene conversion events and found that the alternative analysis does not alter the conclusions (Supplemental Figure S2). From this evidence, we conclude that the role of PARP-1 in GCV is not dependent on the catalytic activity of PARP-1, which is a major distinction between the role of PARP-1 in GCV and the previously known roles of PARP-1 in high fidelity BER.
That the role of PARP-1 in preventing high fidelity repair and promoting mutagenic repair at the site of AID-induced lesions is dependent on its BRCT domain but not catalytic activity led us to hypothesize that an isolated PARP-1 BRCT domain may be able to bind its target and mediate an effect, even in the absence of the rest of the PARP-1 protein. To test this hypothesis, we created a construct for the overexpression of the BRCT domain of PARP-1 in WT DT40 cells in which we fused the FLAG tag, the BRCT domain of PARP-1 (aa 384–479) and GFP (Fig. 1c). We expressed this construct in WT DT40 cells (Fig. 1f) and evaluated whether overexpression of the isolated BRCT domain, with no DNA binding domain or catalytic activity could influence gene conversion. Remarkably, BRCT domain overexpression nearly completely abolished gene conversion at the IgL locus, while not having any effect on BER (Fig. 1d and e). These data suggest that the isolated BRCT domain is able to bind its target, but without the rest of the protein, has the opposite effect, either by binding without corresponding function, analogous to the effect of a blocking antibody, or sequestration of a required component away from the site of action. To differentiate between these hypotheses, we sought to identify the proteins interacting with the BRCT domain of PARP-1.
3.2. PARP-1 competes with the DNA–PK complex in the same pathway
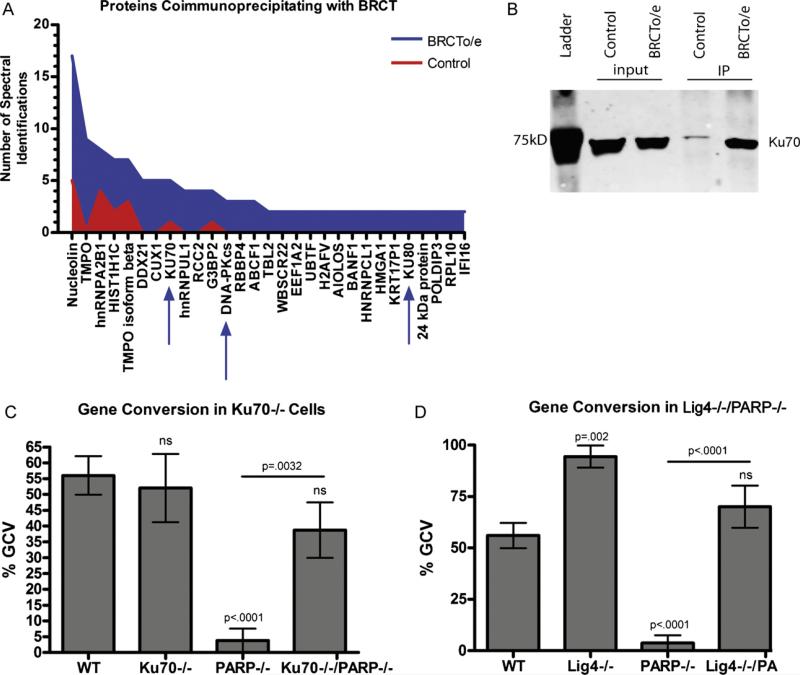
Since human PARP-1 complements chicken PARP function in the DT40 cells, and because human proteins are far better annotated than chicken, we used the human PARP-1 BRCT domain as bait for interacting proteins in human RAMOS B-cells, and analyzed the bound proteins by mass spectrometry. Of the 363 proteins identified, only 30 (including the tagged BRCT domain of PARP-1) are upregulated ≥2-fold in the positive over the negative sample, and of these, 15 are known to be involved with DNA regulation or repair (Fig. 2a). The most notable aspect of the mass spectrometry profile of bound proteins is that the entire DNA–PK complex was pulled down, along with hnRNPB1, which has been shown to bind and inhibit DNA–PK activity [11], and Aiolos, a B cell transcription factor, although without an established role in DNA repair (Supplemental Table S1). To confirm the interaction with the DNA–PK complex, we repeated the immunoprecipitation and confirmed the interaction with Ku70 by immunoblot (Fig. 2b).
Fig. 2.
Proteins which bind to the BRCT domain of PARP-1 were identified by mass spectrometry, confirmed by immunoblot, and validated by KO cell lines. (A) Graphical representation of the number of spectral identifications of proteins which were enriched in the BRCT overexpressing (BRCT o/e) cell line and the parent cell lines as a control. Proteins were coimmunoprecipitated with the BRCT construct by anti-FLAG antibodies bound to sepharose beads. (B) Western blot to verify enrichment of proteins identified by mass spectrometry. The BRCT domain was immunoprecipitated with anti-FLAG antibody and proteins which coimmunoprecipitated were probed with antibody to Ku70. (C) Frequency of gene conversion events in Ku70−/− cell lines. Error bars indicate the SEM of 2–4 cell lines cultured independently. (D) Frequency of gene conversion events in Lig4−/− cell lines. Error bars indicate the SEM of 2–4 cell lines cultured independently.
The pulldown of the complete DNA–PK complex with the BRCT domain of PARP-1 combined with existing evidence that PARP-1 can bind DNA–PK [12,13] led us to investigate whether PARP-1 function during mutagenic repair might be mediated through inhibition of DNA–PK complex activity, the major high fidelity repair pathway in humans. To test this, we looked at PARP−/− cells that were also deficient in Ku70, the DNA-binding subunit of DNA–PK. If the requisite function of PARP-1 in GCV were primarily inhibition of DNA–PK activity at the DNA lesion, then the knockout of Ku70 should rescue GCV in PARP-1−/− cells, whereas if PARP-1 is actively promoting GCV, the double knockout cells would still be GCV-deficient. Sequencing of IgL in Ku−/−/PARP-1−/− DT40s revealed that gene conversion was fully rescued (Fig. 2c), which suggests that the function of the BRCT domain of PARP-1 is likely the inhibition of Ku70 function. To further test this hypothesis, we also examined the phenotype of Lig4−/−/PARP-1−/− cells and found that Lig4−/− similarly rescues the PARP−/− phenotype (Fig. 2d). Additionally, the Lig4−/− cells that were PARP-1-sufficient had even higher levels of GCV than WT, suggesting that Lig4 plays a role in the repair of AID-induced lesions during SHM/GCV as well. Taken together, our data suggest that the mechanism by which PARP-1 is promoting mutagenic repair at mutating IgL loci is inhibition of the high-fidelity repair pathway mediated by Ku70 binding, and recruitment of Lig4.
4. Discussion and conclusions
Hochegger et al. suggested that PARP-1 and Ku70 compete for repair of damaged DNA [14]. They used a model in which DNA double strand breaks were either generated genome-wide through irradiation or at a site targeted by the homing endonuclease ISceI. They proposed that PARP-1 is competing with Ku70 to favor high fidelity repair by homologous recombination rather than NHEJ, which may be mutagenic. However, subsequent studies have suggested that while PARP-1 is probably competing with Ku for repair of breaks, PARP-1 is actually promoting alternative NHEJ [15,16], which is the more mutagenic pathway of NHEJ, whereas classic NHEJ is more likely to repair a clean break without introducing mutations [17–19]. Here we looked at an unusual instance of DNA damage, generated by targeted deamination resulting in a single-strand break and repaired in a manner that promotes the introduction of mutations and again find evidence that PARP-1 is competing with Ku for repair of the lesion, and in this case, PARP-1 promotes mutagenic repair mediated by entirely different mechanisms of SHM and GCV. An consistent interpretation across all these studies is that PARP-1 inhibits the high fidelity repair mediated by Ku, allowing alternate, slower repair pathways to predominate. As PARP-1 does not have intrinsic DNA repair capabilities, it follows that the resulting repair pathway is dependent on context and the local environment.
While PARP-1 and DNA–PK have previously been implicated in other steps of the antibody diversification process, V(D)J recombination and CSR, PARP-1 has only recently been shown to also play a role in somatic hypermutation and gene conversion. That this role is rescued by concurrent knockout of Ku70 or Lig4, suggests that DNA–PK and PARP-1 are in antagonistic roles in this process. We previously showed that PARP-1 is critical to promote mutagenic repair [4]. Here we provide evidence that PARP-1 and Ku70 are competing for repair of the AID-induced lesions generated during immunoglobulin gene conversion. If PARP-1 is present to bind the break, it inhibits high fidelity repair in a fashion that is dependent on its BRCT domain. If Ku70 binds in the absence of PARP-1, it promotes high-fidelity repair in a Lig4-dependent manner, which results in a GCV-deficient phenotype. There is already evidence for a role for DNA–PK in inhibiting DT40 GCV [20,21]. In these papers, the authors conclude that since DNA–PK is responding to the lesion, that GCV is mediated through a DSB intermediate. However, there is also evidence that GCV does not require a DSB intermediate [22], and Nakahara et al. provide a convincing mechanism by which the MRN complex may stimulate GCV from a SSB [23]. The explanation for this discrepancy may lie in recent findings that Ku70 and Ku80 have activity at lesions other than DSBs. Ku70/Ku80 have been shown to bind abasic sites [24], and possess AP lyase activity [25], which may provide an explanation for how these components of the DNA–PK complex could be involved in SHM/GCV in the absence of a DSB. Alternatively, it is also possible that a DSB is generated as an intermediate step of repair, but if the end of the lesion is protected from exonuclease activity or other means of recognition as a DSB (perhaps by PARP-1 itself) if would be difficult to establish experimentally, while still allowing repair by a mechanism that involves DNA–PK.
Two different models could explain the capacity of the isolated PARP-1 BRCT domain to prevent high-fidelity repair mediated by Ku70 and Lig4 at AID-mediated DNA lesions. The observation that overexpression of the BRCT domain is able to suppress gene conversion in the context of endogenous full length PARP-1 is most simply explained by the isolated PARP-1 BRCT domain binding to Ku, thus preventing full length PARP-1 from binding and inhibiting Ku function. A second possibility is that the PARP-1 BRCT domain normally recruits a 3rd protein that acts to inhibit Ku70/Lig4 function. In this model, the isolated PARP-1 BRCT domain, which does not contain a DNA-binding domain, would sequester this 3rd protein away from the break, resulting in uninhibited, and thus high frequency, high fidelity repair. While the inhibition in either case would be independent of PARP-1 catalytic activity, it could be dependent on a conformational change which requires full-length PARP-1 or DNA-binding, or simple steric hinderance.
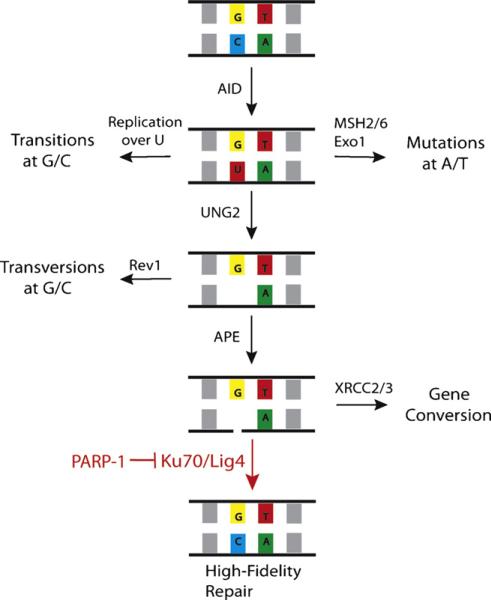
Taken together, our evidence suggests that SHM/GCV is an ordered, sequential process. Once a backbone break is generated by an AP lyase, GCV and high fidelity repair (HFR) are in competition for the repair of the lesion, with PARP-1 being critical for inhibiting HFR and allowing mutagenic repair [4]. Here we provide a mechanism by which PARP-1 is inhibiting HFR by showing that in the absence of Ku70 or Lig4, mutagenic repair predominates even in the absence of PARP-1, suggesting that the role of PARP-1 is to inhibit Ku70 function and the recruitment of Lig4 (Fig. 3). In systems with more active MMR pathways, such as mice and humans, the uracil is recognized before UNG excision. Since PARP-1 functions in the UNG-dependent repair pathways, in the absence of PARP-1, there would not be a defect in mutations arising at A/T. In the same vein, even in the absence of PARP-1, the abasic site can be filled in by trans-lesion polymerases before APE1 nicks the backbone. Since many of the deaminations may be repaired before the SSB stage in these systems, even if some of the deaminations which would have resulted in a G/C mutations undergo high fidelity repair in the absence of PARP-1, this would be a small portion of the total mutations and therefore harder to detect than in the DT40 system where most of the lesions are repaired by GCV after generation of a SSB.
Fig. 3.
A model for the roles of PARP-1, Ku70, and Lig4 in somatic hypermutation and gene conversion. Ku70 and Lig4 mediate high fidelity repair unless inhibited by PARP-1, which perpetuates the lesion and shunts to a mutagenic repair pathway.
The lessons we learn from studying DNA repair at hypermutating loci have important implications in understanding DNA repair more generally, as they illustrate how the same repair pathways which allow high-fidelity genome-wide repair may be co-opted to produce mutations depending on the particular circumstances and local environment of the break. Understanding how the decision between high-fidelity and mutagenic repair pathways is made may help us to better understand how to exploit DNA repair pathways to enhance chemotherapeutic responses of malignancies, and lead to approaches to controlling the decision between mutagenic and high-fidelity repair for genome engineering purposes.
Supplementary Material
Acknowledgements
We would like to thank Dr. Noritaki Adachi for use of the Lig4−/− and Lig4/PARP−/− DT40 cells.
Source of funding: Cancer Research Institute Predoctoral Emphasis Pathway in Tumor Immunology and NIH RL1CA133832 of the Northwest Genome Engineering Consortium.
Footnotes
Conflict of interest
None.
Appendix A. Supplementary data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.dnarep.2010.12.005.
References
- 1.Fugmann SD, Lee AI, Shockett PE, Villey IJ, Schatz DG. The RAG proteins and V(D)J recombination: complexes, ends, and transposition. Annu. Rev. Immunol. 2000;18:495–527. doi: 10.1146/annurev.immunol.18.1.495. [DOI] [PubMed] [Google Scholar]
- 2.Taccioli GE, Rathbun G, Oltz E, Stamato T, Jeggo PA, Alt FW. Impairment of V(D)J recombination in double-strand break repair mutants. Science. 1993;260:207–210. doi: 10.1126/science.8469973. [DOI] [PubMed] [Google Scholar]
- 3.Grawunder U, Zimmer D, Fugmann S, Schwarz K, Lieber MR. DNA ligase IV is essential for V(D)J recombination and DNA double-strand break repair in human precursor lymphocytes. Mol. Cell. 1998;2:477–484. doi: 10.1016/s1097-2765(00)80147-1. [DOI] [PubMed] [Google Scholar]
- 4.Paddock MN, Buelow BD, Takeda S, Scharenberg AM. The BRCT domain of PARP-1 is required for immunoglobulin gene conversion. PLoS Biol. 2010;8(7):e1000428. doi: 10.1371/journal.pbio.1000428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Craig R, Beavis RC. TANDEM: matching proteins with tandem mass spectra. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 6.Higdon R, Kolker N, Picone A, van Belle G, Kolker E. LIP index for peptide classification using MS/MS and SEQUEST search via logistic regression. OMICS. 2004;8:357–369. doi: 10.1089/omi.2004.8.357. [DOI] [PubMed] [Google Scholar]
- 7.Higdon R, Hogan JM, Kolker N, van Belle G, Kolker E. Experiment-specific estimation of peptide identification probabilities using a randomized database. OMICS. 2007;11:351–365. doi: 10.1089/omi.2007.0040. [DOI] [PubMed] [Google Scholar]
- 8.Hather G, Higdon R, Bauman A, von Haller PD, Kolker E. Estimating false discovery rates for peptide and protein identification using randomized databases. Proteomics. 2010;10(12):2369–2376. doi: 10.1002/pmic.200900619. [DOI] [PubMed] [Google Scholar]
- 9.Higdon R, Kolker E. A predictive model for identifying proteins by a single peptide match. Bioinformatics. 2007;23:277–280. doi: 10.1093/bioinformatics/btl595. [DOI] [PubMed] [Google Scholar]
- 10.Saberi A, Nakahara M, Sale JE, Kikuchi K, Arakawa H, Buerstedde JM, Yamamoto K, Takeda S, Sonoda E. The 9-1-1 DNA clamp is required for immunoglobulin gene conversion. Mol. Cell. Biol. 2008;28:6113–6122. doi: 10.1128/MCB.00156-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwanaga K, Sueoka N, Sato A, Hayashi S, Sueoka E. Heterogeneous nuclear ribonucleoprotein B1 protein impairs DNA repair mediated through the inhibition of DNA-dependent protein kinase activity. Biochem. Biophys. Res. Commun. 2005;333:888–895. doi: 10.1016/j.bbrc.2005.05.180. [DOI] [PubMed] [Google Scholar]
- 12.Galande S, Kohwi-Shigematsu T. Poly(ADP-ribose) polymerase and Ku autoantigen form a complex and synergistically bind to matrix attachment sequences. J. Biol. Chem. 1999;274:20521–20528. doi: 10.1074/jbc.274.29.20521. [DOI] [PubMed] [Google Scholar]
- 13.Ariumi Y, Masutani M, Copeland TD, Mimori T, Sugimura T, Shimotohno K, Ueda K, Hatanaka M, Noda M. Suppression of the poly(ADP-ribose) polymerase activity by DNA-dependent protein kinase in vitro. Oncogene. 1999;18:4616–4625. doi: 10.1038/sj.onc.1202823. [DOI] [PubMed] [Google Scholar]
- 14.Hochegger H, Dejsuphong D, Fukushima T, Morrison C, Sonoda E, Schreiber V, Zhao GY, Saberi A, Masutani M, Adachi N, Koyama H, de Murcia G, Takeda S. Parp-1 protects homologous recombination from interference by Ku and Ligase IV in vertebrate cells. EMBO J. 2006;25:1305–1314. doi: 10.1038/sj.emboj.7601015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Robert I, Dantzer F, Reina-San-Martin B. Parp1 facilitates alternative NHEJ, whereas Parp2 suppresses IgH/c-myc translocations during immunoglobulin class switch recombination. J. Exp. Med. 2009;206:1047–1056. doi: 10.1084/jem.20082468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang M, Wu W, Rosidi B, Zhang L, Wang H, Iliakis G. PARP-1, Ku compete for repair of DNA double strand breaks by distinct NHEJ pathways. Nucleic Acids Res. 2006;34:6170–6182. doi: 10.1093/nar/gkl840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guirouilh-Barbat J, Huck S, Bertrand P, Pirzio L, Desmaze C, Sabatier L, Lopez BS. Impact of the KU80 pathway on NHEJ-induced genome rearrangements in mammalian cells. Mol. Cell. 2004;14:611–623. doi: 10.1016/j.molcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- 18.Ferguson DO, Sekiguchi JM, Chang S, Frank KM, Gao Y, DePinho RA, Alt FW. The nonhomologous end-joining pathway of DNA repair is required for genomic stability and the suppression of translocations. Proc. Natl. Acad. Sci. U.S.A. 2000;97:6630–6633. doi: 10.1073/pnas.110152897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Difilippantonio MJ, Zhu J, Chen HT, Meffre E, Nussenzweig MC, Max EE, Ried T, Nussenzweig A. DNA repair protein Ku80 suppresses chromosomal aberrations and malignant transformation. Nature. 2000;404:510–514. doi: 10.1038/35006670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cook AJ, Raftery JM, Lau KK, Jessup A, Harris RS, Takeda S, Jolly CJ. DNA-dependent protein kinase inhibits AID-induced antibody gene conversion. PLoS Biol. 2007;5:e80. doi: 10.1371/journal.pbio.0050080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang ES, Martin A. NHEJ-deficient DT40 cells have increased levels of immunoglobulin gene conversion: evidence for a double strand break intermediate. Nucleic Acids Res. 2006;34:6345–6351. doi: 10.1093/nar/gkl830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ordinario EC, Yabuki M, Larson RP, Maizels N. Temporal regulation of Ig gene diversification revealed by single-cell imaging. J. Immunol. 2009;183:4545–4553. doi: 10.4049/jimmunol.0900673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakahara M, Sonoda E, Nojima K, Sale JE, Takenaka K, Kikuchi K, Taniguchi Y, Nakamura K, Sumitomo Y, Bree RT, Lowndes NF, Takeda S. Genetic evidence for single-strand lesions initiating Nbs1-dependent homologous recombination in diversification of Igv in chicken B lymphocytes. PLoS Genet. 2009;5:e1000356. doi: 10.1371/journal.pgen.1000356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ilina ES, Lavrik OI, Khodyreva SN. Ku antigen interacts with abasic sites. Biochim. Biophys. Acta. 2008;1784:1777–1785. doi: 10.1016/j.bbapap.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Roberts SA, Strande N, Burkhalter MD, Strom C, Havener JM, Hasty P, Ramsden DA. Ku is a 5′-dRP/AP lyase that excises nucleotide damage near broken ends. Nature. 2010;464:1214–1217. doi: 10.1038/nature08926. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.