Summary
Previously, extracellular vesicle production in Gram-positive bacteria was dismissed due to the absence of an outer membrane, where Gram-negative vesicles originate, and the difficulty in envisioning how such a process could occur through the cell wall. However, recent work has shown that Gram-positive bacteria produce extracellular vesicles and that the vesicles are biologically active. In this study, we show that Bacillus subtilis produces extracellular vesicles similar in size and morphology to other bacteria, characterized vesicles using a variety of techniques, provide evidence that these vesicles are actively produced by cells, show differences in vesicle production between strains, and identified a mechanism for such differences based on vesicle disruption. We found that in wild strains of B. subtilis, surfactin disrupted vesicles while in laboratory strains harboring a mutation in the gene sfp, vesicles accumulated in the culture supernatant. Surfactin not only lysed B. subtilis vesicles, but also vesicles from Bacillus anthracis, indicating a mechanism that crossed species boundaries. To our knowledge, this is the first time a gene and a mechanism has been identified in the active disruption of extracellular vesicles and subsequent release of vesicular cargo in Gram-positive bacteria. We also identify a new mechanism of action for surfactin.
Introduction
In recent years, several laboratories have reported that bacteria, fungi, and parasites produced extracellular vesicles, which in some instances contain proteins associated with virulence. The most extensive studies of extracellular vesicle production in bacteria have focused on the outer membrane vesicles (OMV) produced by Gram-negative bacteria (Kuehn; Kesty, 2005; Ellis; Kuehn, 2010; Kulp; Kuehn, 2010). OMVs can release materials in a concentrated and protected manner, and therefore are potent virulence factors of pathogenic Gram-negative bacteria. These vesicles contain toxins, DNA, immunomodulatory compounds, communication factors, and adhesins and have been associated with cytotoxicity, bacterial attachment, intercellular DNA transfer, and invasion (Dorward et al., 1989; Kuehn; Kesty, 2005; Ellis; Kuehn, 2010; Maldonado et al., 2011). Until recently, vesicle production by Gram-positive bacteria was overlooked due to the inference that differences in the cell wall structure of Gram-negative and Gram-positive bacteria precluded their release. Unlike Gram-negative bacteria, the cell wall of Gram-positive bacteria consists of primarily peptidoglycan and does not have an outer membrane. However, the discovery of extracellular vesicles in fungi, which also have large cell walls, catalyzed the search of vesicular structures in Gram-positive bacteria (Rodrigues et al., 2007).
The first evidence for vesicles in Gram-positive bacterial cells was provided by Dorward and Garon but they were unable to establish function (Dorward; Garon, 1990). More recently, vesicle production was described in Mycobacterium ulcerans, Staphylococcus aureus, Mycobacterium tuberculosis, and Bacillus anthracis (Marsollier et al., 2007; Lee et al., 2009; Rivera et al., 2010; Gurung et al., 2011; Hong et al., 2011; Prados-Rosales et al., 2011; Thay et al., 2013). M. tuberculosis vesicles contain compounds that modulate TLR2 responses while those of B. anthracis contain anthrax toxin components, indicating that Gram-positive membrane vesicles are potentially involved in pathogenesis (Rivera et al., 2010; Prados-Rosales et al., 2011). This observation suggested that Gram-positive cells, like Gram-negative bacteria, utilized vesicle secretion to deliver concentrated amounts of toxins and other compounds into the extracellular space (Marsollier et al., 2007; Lee et al., 2009; Rivera et al., 2010; Prados-Rosales et al., 2011; Hong et al., 2011; Gurung et al., 2011; Thay et al., 2013). The disruption of B. anthracis vesicles by serum was identified as a potential mechanism for vesicle cargo release, as it is important to understand what happens to vesicles and their contents after release from the cell (Wolf et al., 2012). Recently, vesicle production has also been described for Streptococcus pneumonia and pneumococcal vesicles are used to release pneumolysin (Olaya-Abril et al., 2014).
In this work, we explored vesicle production in the Gram-positive, endospore-forming bacterium, Bacillus subtilis. B. subtilis is considered the model system for Gram-positive bacteria and is therefore an excellent organism to study the genetic and cellular machinery involved in Gram-positive vesiculogenesis. Two common strains of B. subtilis studied were the 3610 environmental strain, which forms robust biofilms and the laboratory strain 168, which has attenuated biofilm production. Both strains produced varying amounts of recoverable vesicles under different conditions. Environmental strains of B. subtilis are often not easily genetically manipulated; therefore domesticated lab strain 168 was created to be easily transformable and used for genetic studies in Gram-positive bacteria (Earl et al., 2007). It is hypothesized that both strains 168 and 3610 are descendants of B. subtilis Marburg strain and vary from each other by only 22 SNPs (Zeigler et al.,2008). These limited genetic differences and ability to isolate differing amounts of vesicles make these two strains ideal to study the mechanism of vesicle production in B. subtilis. McLoon et al identified five of these genetic differences between strains 168 and 3610 that are involved in biofilm formation (mutations in 168 in the genes sfp, epsC, swrA, PdegQ, and lack of the plasmid-borne gene rapP) (McLoon et al., 2011). Further study of these two strains has led us to identify the gene, sfp, which affects the amount of vesicles recovered in B. subtilis. Lab strains, including strain 168, harbor a mutation in sfp resulting in a truncated, nonfunctional protein, whereas environmental strains, like strain 3610, harbor a wild type copy of this gene (Nakano et al., 1988).
Sfp is a 4′-phosphopantetheinyl transferase which transfers a phosphopantetheine group from coenzyme A to a peptidyl carrier protein during biosynthesis of the lipopeptide, surfactin, and the siderophore, bacillibactin, in B. subtilis (Nakano et al., 1988; Quadri et al., 1998; May et al., 2001). We found that strains harboring a functional sfp gene produced very few recoverable vesicles whereas strains with nonfunctional copies of sfp, produced a massive amount of recoverable vesicles. To determine the mechanism behind this difference in vesicle quantity, and how sfp was affecting it, we explored the effect of surfactin on vesicles. We found that vesicles from B. subtilis and B. anthracis were indeed disrupted by the presence of surfactin. And this was not surprising considering surfactin can perturb lipid membranes (Bernheimer; Avigad, 1970; Heerklotz; Seelig, 2001; Carrillo et al., 2003). This data suggests that the production of surfactin serves as a mechanism for vesicular cargo release in B. subtilis, as well as the ability to disrupt vesicles from B. anthracis, indicating a mechanism that reaches across species barriers. This work sets the stage for dissecting the function of vesiculogenesis in a genetically tractable Gram-positive bacterium.
Results
Bacillus subtilis Produces Extracellular Vesicles
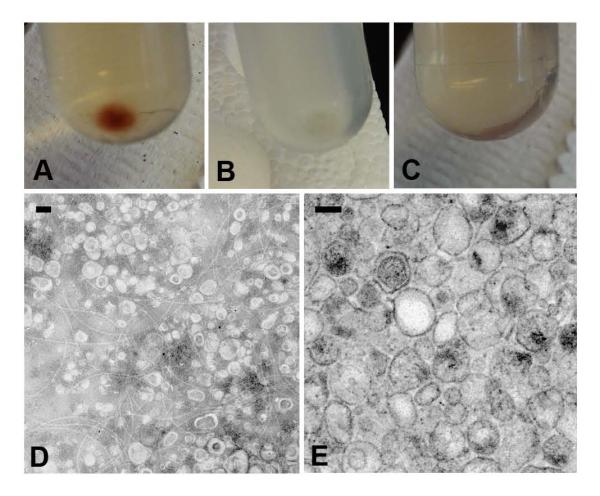
We recovered vesicles from the culture supernatants of Bacillus subtilis lab strain 168, wild type strain 3610, AM373, RL3090, AD3610, SL3610, and RL2663 (Table 1). The morphology and color of the vesicle pellet from each strain was the first indication that there were differences in vesicle preparations among strains. Centrifugation of strain 168 culture supernatants produced a large, reddish-brown vesicle pellet whereas centrifugation of strain 3610 culture supernatant and 168 heat-killed preparations produced only small, clear pellets (Fig. 1A-C). A massive amount of vesicles from strain 168 were visualized by both negative and positive staining transmission electron microscopy (TEM) (Figure 1D-E). In contrast, very few vesicles were identified in electron micrographs of identical preparations of vesicles from strain 3610. Due to the abundance of vesicles in the supernatant of strain 168, most of the subsequent characterization was performed on vesicles isolated from this strain.
Table 1. Strains used in this study.
| Strain | Relevant Characteristics | Vesicle Production | Source or Reference |
|---|---|---|---|
| 168 | B. subtilis lab strain sfp− | High | ATCC #23857 |
| PY79 | 168 prototrophic derivative | N/A | Losick Lab |
| 3610 | B. subtilis environmental strain sfp+ | Low | Losick Lab |
| AM373 | 168 sfp+ | Low | (McLoon et al., 2011) |
| RL3090 | 168 sfp+ (mls linked to sfp) | Low | (McLoon et al., 2011) |
| AD3610 | 3610 sfp::kan | High | AD, Losick Lab |
| SL3610 | AD3610 sfp::kan amyE::sfp-spec | Low | L.B., S.L., Losick Lab |
| RL2663 | 3610 srfAA::spec | High | (Kearns; Losick, 2003) |
| Sterne 34F2 | B. anthracis Sterne pXO1+ pXO2− | Medium | Alex Hoffmaster, CDC |
Figure 1. Bacillus subtilis produces extracellular vesicles.
Vesicles were isolated and pelleted by ultracentrifugation from strains (A) 168, (B) 3610, and (C) heat-killed 168. Vesicles from strain 168 were visualized by (D) negative staining TEM and (E) embedded TEM. Scale bars 100 nm.
Given the propensity of lipids to organize into vesicle-like structures, considerable effort was devoted to establishing that recovered vesicles were not artifacts of broken cells or a result of self-assembly by extracellular lipids. Consistent with a requirement for cell viability in vesicle production, we observed very few vesicles by negative staining TEM when heat-killed 168 cells were added to bacterial media, allowed to incubate at 37° C for 18 h in a shaker, and then processed for vesicle preparation under the same protocol (data not shown).
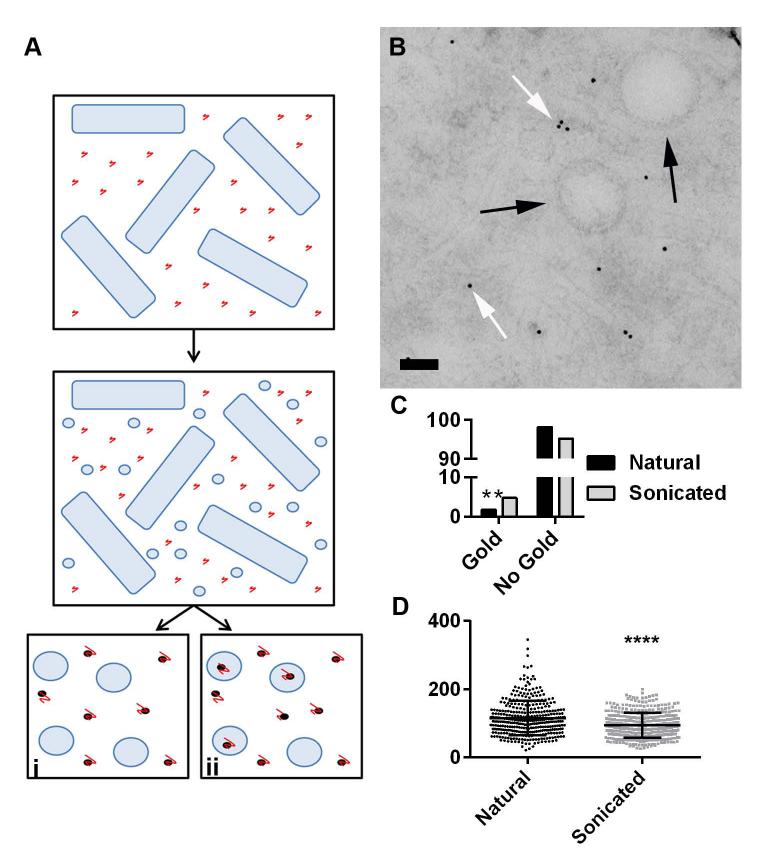
To exclude the possibility that the recovered vesicles were the result of spontaneous extracellular assembly of secreted lipids we performed an experiment where the fungal polysaccharide glucuronoxylomannan (GXM), of Cryptococcus neoformans, was added to the bacterial culture and then its presence and absence inside vesicles was measured by immunogold. To ensure that the GXM was small enough in size to become encapsulated inside vesicles it was fractionated to less than 10 kDa in size by ultrafiltration through selective molecular mass membranes. This size retains reactivity with specific monoclonal antibodies (mAbs). We reasoned that if vesicles were forming spontaneously from lipids, they would contain far more GXM than vesicles assembled by the bacterial cell (for a scheme of the experimental design see Figure 2A), and that was precisely what was measured. As a control, we sonicated vesicles isolated from cultures incubated with GXM, expecting that these would contain more GXM, since subsequent self-assembly in the presence of GXM would result in trapping of the polysaccharide in newly formed vesicles. TEM images of the natural vesicle preparations showed the presence of GXM in the sample (Figure 2B). In accordance to predictions, significantly more GXM was inside vesicles that reformed after sonication compared to natural vesicles produced from live bacterial cultures (6.9% and 2.9%, respectively) (Figure 2C). These results provided strong support for the notion that vesicles are produced by the cell and are not the result of spontaneous organization of lipids in the media. Sonicated vesicle diameter was also significantly smaller and varied less than that of natural vesicles, indicating another difference between B. subtilis-produced vesicles and those that reformed from the aggregation of lipids (94.54 ± 1.869 and 115.4 ± 2.69) (Figure 2D). We conclude that B. subtilis, like other Gram-positive bacteria, actively produce extracellular vesicles (Marsollier et al., 2007; Lee et al., 2009; Rivera et al., 2010; Prados-Rosales et al., 2011).
Figure 2. Vesicles are produced directly by the cell not by aggregation of lipids.
Strain 168 cells were grown in the presence of GXM and vesicles were isolated and left natural or sonicated. (A) Schema of experimental design. Vesicles were predicted to either (i) be gold-free if they were formed by cellular processes at the cell membrane or (ii) encapsulate gold particles if they formed in the presence of GXM (e.g. spontaneous formation in supernatant containing GXM). Immunogold was used to delineate the presence of GXM in individual vesicles at ultrastructural scales. In the schema, black dots represent gold particles and red lines, GXM. (B) Representative micrograph showing immunogold staining of GXM in a vesicle preparation. Note that all gold particles occur outside of vesicles. (C) Percentage of natural and sonication-derived vesicles in the presence of GXM that contained gold particles. Sonicated vesicles contained significantly more gold than natural vesicles (4.8 and 1.8%, n=564 and n=712, p<0.01 Chi2) and demonstrated a significantly smaller (D) diameter and standard deviation (94.54 ± 1.869, n=392 and 115.4 ± 2.69, n=355, p<0.0001 unpaired t-test, n=3) than natural vesicles. Black arrows indicate vesicles and white arrow indicates GXM. Scale bars 100 nm.
Physical Characterization of B. subtilis Vesicles
Vesicle preparations from planktonic B. subtilis cultures were studied using dynamic light scattering (DLS) to determine the approximate size of the vesicles. DLS revealed two populations of approximately 50 and 150-250 nm in preparations from strain 168 (Figure 3B). This size distribution is similar to that reported for vesicles produced by other bacteria (Kuehn; Kesty, 2005). DLS analysis of vesicle preparations from B. subtilis strain 3610 under the same conditions revealed less distinct distributions and the presence of larger structures, which may be flagella given their abundance in TEM micrographs.
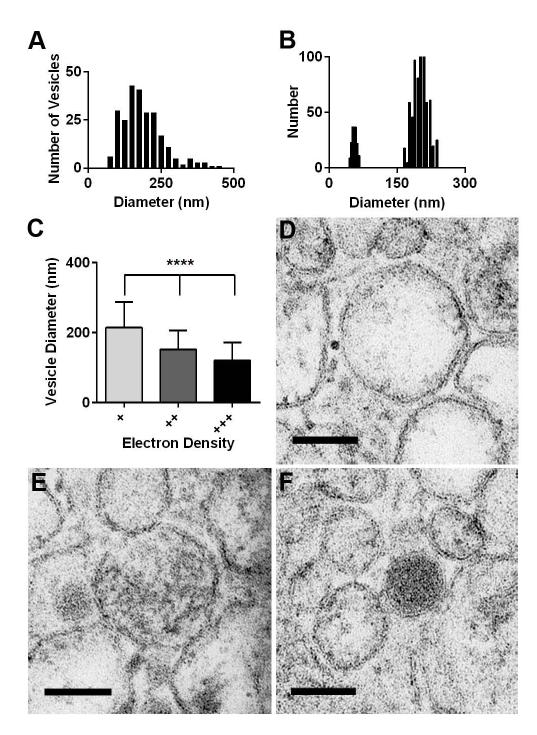
Figure 3. B. subtilis vesicles are heterogeneous in diameter and density.
(A) Vesicle diameters measured from TEM images had a mean of 174.4 nm after applying 1.27 correction factor. (B) Dynamic light scattering indicated vesicle diameters having two peaks approximately 50 and 150-200 nm. (C) Vesicle diameters and standard deviations were measured and mean densities scored of (D) least electron density (+) (214.5 ± 73.28 nm, n=85), (E) medium electron density (++) (152.2 nm ± 53.95, n=137), and (F) most electron dense (+++) (120.4 ± 51.14 nm, n=105) from strain 168 (p<0.0001, one-way ANOVA). Scale bars 100 nm.
Analysis of strain 168 supernatant pellets by TEM with negative staining revealed large numbers of vesicles (Figure 1D) whereas very few vesicles were recovered from strain 3610 when isolated from the same culture conditions. Images of strain 168 vesicles obtained by embedded TEM were measured using ImageJ software, and the result was a mean diameter of 137.7 nm (Figure 1E). However, because vesicles are spherical, and TEM represents transverse sections, a 1.27 correction factor for these 2D measurements was utilized as suggested by Kong et al to estimate the actual equatorial dimensions (Kong et al., 2005). With the 1.27 correction factor, the mean diameter of vesicles was 174.4 nm (Figure 3A). It is noteworthy that when DLS and TEM measurements are compared, TEM tends to underestimate size while DLS overestimates the size of samples (Egelhaaf et al., 1996; Hong et al., 2011). TEM images of strain 168 vesicles indicated that some vesicles were more electron dense than others. Smaller vesicles were significantly more electron dense than larger vesicles (Figure 3C). Vesicle diameters were categorized visually in three groups based on electron density with mean diameters of 214.5 nm for least dense (+), 152.2 nm for medium dense (++), and 120.4 nm for most dense (+++) (Fig. 3D-F).
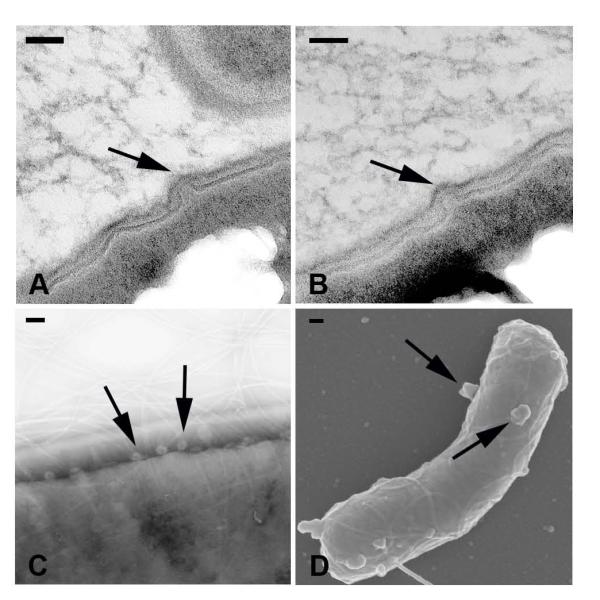
Spherical protrusions from bacterial cells with dimensions similar to vesicles were observed in electron micrographs of B. subtilis cells. TEM images of strain 168 cells showed rounded structures strongly suggestive for putative vesiculation in the cell membrane (Figure 4A-B). Structures similar to vesicles were also identified on strain 168 cells by negative staining TEM in various stages of release (Figure 4C). Finally, SEM images of strain 168 cells also showed multiple protrusions on cells consistent with the emergence of vesicles (Figure 4D). Although we recovered various quantities of vesicles from strains 168 and 3610, it is notable that the number of putative vesicle protrusions on cells was not significantly different (data not shown).
Figure 4. Structures suggestive of extracellular vesicles forming on B. subtilis cells.
Strain 168 cells were visualized by (A-B) embedded TEM, (C) negative staining EM, and (D) SEM. Arrows indicate protrusions suggestive of emerging vesicles; scale bars 100 nm.
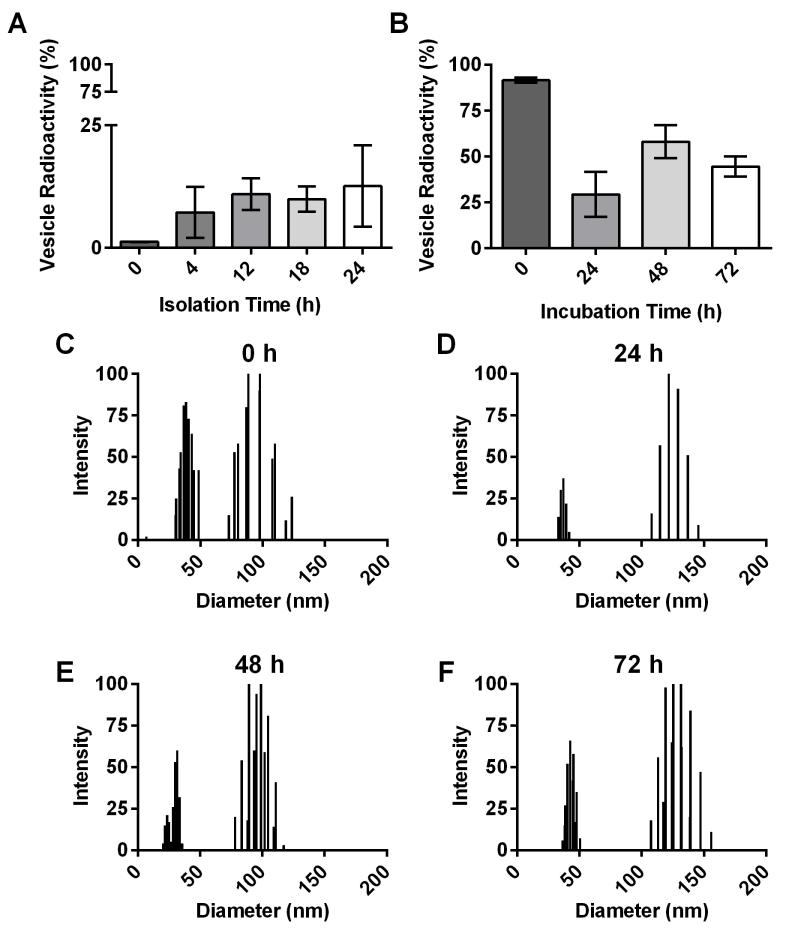
Bacterial cells were labeled with 14C palmitic acid (14C PA), which is incorporated into vesicles, to study stability, production over time, and to compare recoverable vesicle quantity among strains (Rivera et al., 2010; Prados-Rosales et al., 2011; Wolf et al., 2012). B. subtilis strain 168 vesicles were isolated after 0, 4, 12, 18, and 24 h of growth and radioactivity of the vesicle pellets were analyzed as fractions of the total culture. Strain 168 manifested increased vesicle production over time, which leveled off after 12 h of growth (Figure 5A). Stability of strain 168 vesicles in PBS was also measured by 14C PA labeling. Vesicle preparations were incubated for 0, 24, 48 and 72 h before being spun down, counted and the ratio of vesicle versus supernatant radioactivity was analyzed. Vesicle pellets contained high levels of radioactivity for 24 h but subsequently decreased, indicating that vesicles were stable for approximately 24 h (Figure 5B). DLS measurements of vesicle stability preparations from each time point showed consistent peaks of approximately 50 and 100-150 nm in diameter (Figure 5C-F).
Figure 5. Vesicle production increases and vesicle stability decreases over time.
Strain 168 cells were labeled with 14C PA and vesicles were isolated. (A) Vesicle pellet radioactivity as fraction of total sample isolated at 0, 4, 12, 18, and 24 h and (B) vesicle pellet stability in PBS compared to supernatant recovered at 0, 24, 48, and 72 h (n=4). DLS of vesicles incubated in PBS at (C-F) 0, 24, 48, and 72 h, respectively.
Chemical Composition of Vesicles
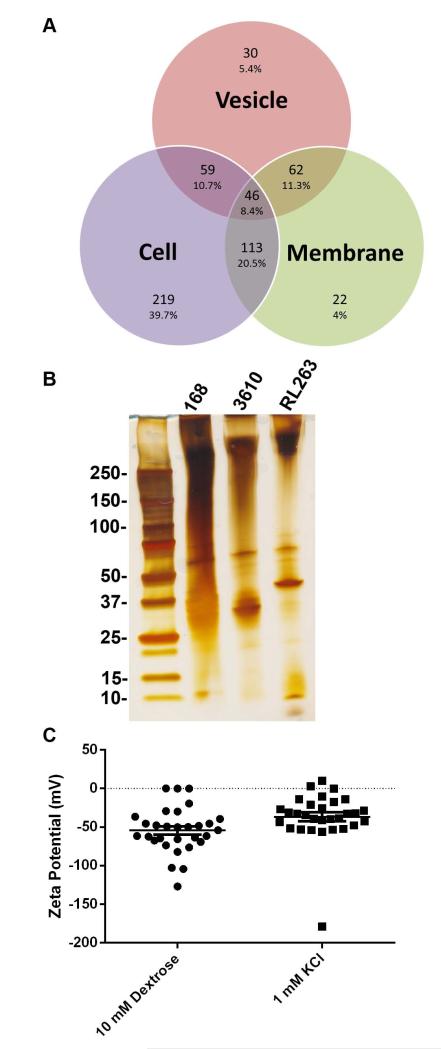
Proteomic analysis was performed on strain 168 vesicles, cell membrane fractions, and cells. B. subtilis 168 vesicles were enriched in 30 proteins and shared 46 with both the cell and cell membrane fractions (Figure 6A). Vesicles were enriched in lipoproteins and contained many siderophore-binding proteins (Table S2). SunI, the sublancin immunity protein, was found associated with the vesicles (Dubois et al., 2009). Most of the proteins in the vesicle preparation were membrane proteins, suggesting that the vesicles originated from the membrane of the bacterial cell (Table S2). SDS-PAGE gel of vesicle preparations from strains 168, 3610, and RL2663 exhibited similar profiles with only a few differing bands (Figure 6B).
Figure 6. Vesicles are enriched in specific proteins and have a negative zeta-potential.
Proteomic analysis of strain 168 (A) vesicles, cell membrane, and whole cells. (B) Vesicle preparations from strains 168, 3610, and RL2663 exhibit various banding patterns on silver stained SDS-PAGE gel. (C) The zeta-potential and standard deviation of vesicles in 10 mM dextrose and 1 mM KCl is −54.2917 +/− 9.24 and −36.724 +/− 10.2, respectively.
Vesicle Membrane Potential
The ζ potential of strain 168 vesicle preparations were determined to be −54.2917 +/− 9.24 and −36.724 +/− 10.2 in 10 mM Dextrose and 1 mM KCl, respectively (Figure 6C).
Vesicles are Constituents of Biofilms
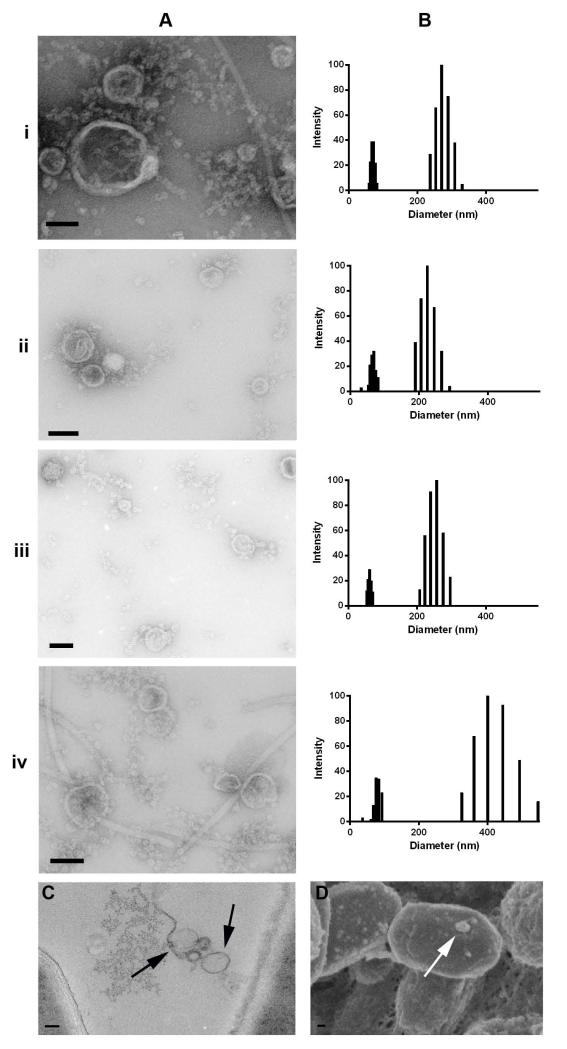
Negative staining TEM revealed the presence of vesicle-like structures in B. subtilis biofilms. After 4 and 8 d of growth, vesicles were isolated from disrupted B. subtilis strains 168 (i-ii) and 3610 (iii-iv) biofilm pellicles. Despite the previously mentioned differences in both recovered vesicle quantity and biofilm growth between the two strains, similar amounts of vesicles were isolated from all cultures and visualized using electron microscopy with negative staining (Figure 7A). The size distribution of the vesicles isolated from strains 168 and 3610 biofilms was determined by DLS, which revealed the presence of two distinct size populations across all biofilm vesicle samples with measurements comparable to those described for supernatant vesicles from planktonic cultures (Figure 7B).
Figure 7. Vesicles are found in B. subtilis biofilms.
Vesicles isolated from disrupted strains 168 (i-ii) and 3610 (iii-iv) biofilms at 4 and 8 days, respectively. (A) Negatively stained electron micrographs and (B) DLS size distribution of biofilm vesicles. (C) Embedded TEM and (D) SEM micrographs of strain 3610 whole biofilms show the presence of vesicles. Black arrow indicates vesicles, white arrow indicates vesicle protrusions. Scale bars 70 nm.
Samples of intact B. subtilis strains 168 and 3610 biofilm were further visualized using TEM and SEM. Embedded biofilm TEM images of strain 3610 biofilm showed vesicles as part of the biofilm matrix (Figure 7C). For SEM, biofilm samples were prepared with ferrocyanide in a process that ultimately peels back the outer layer of the biofilm to allow for viewing of the internal biofilm constituents. SEM images generated from the ferrocyanide-treated samples indicated the presence of putative vesicular events on cells within the B. subtilis strain 3610 biofilm (Figure 7D).
Vesicle Quantification
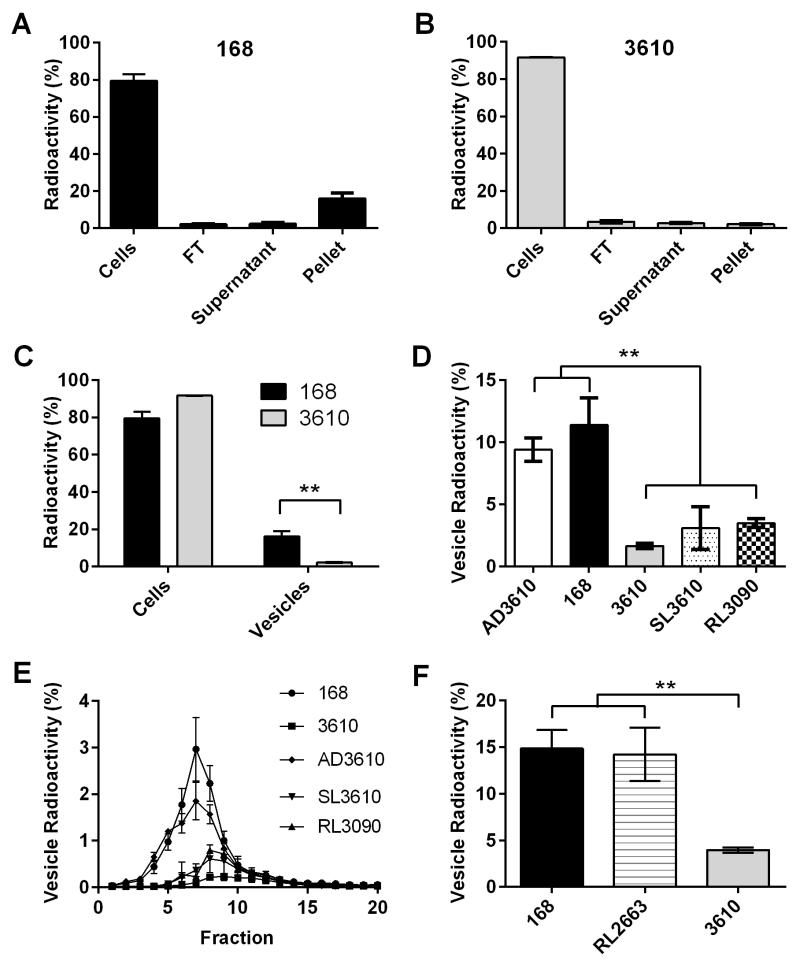
A dramatic difference in vesicle quantity between strains 168 and 3610 was apparent in TEM micrographs. To further explore this difference, we utilized metabolic labeling to gain more quantifiable information about vesicles from various strains. Cells from strains 168 and 3610 were labeled with 14C PA and then we measured radioactive counts in cells, flow through (FT), supernatant, and vesicle pellets, with each measurement determined relative to the total sample (Figure 8A-B). Strain 168 had significantly more radioactivity incorporated into the vesicle pellet than strain 3610, even though both strains took up a similar amount of radioactivity in the initial labeling step (Figure 8C). This data, along with the DLS and electron micrographs, provides further support for the observation that the laboratory strain 168 produces more recoverable vesicles than environmental strain 3610.
Figure 8. Nonfunctional sfp gene results in large quantities of recovered vesicles.
Cells from strains (A) 168 and (B) 3610 were labeled with 14C PA and the radioactivity of each fraction measured (cells, FT, supernatant, and vesicle pellet). (C) Strain 168 vesicle pellet contained significantly more radioactivity than that of strain 3610 (p<0.01 t-test, n=3). Vesicle pellets of B. subtilis strains 168 and AD3610 harboring (D) nonfunctional sfp, contain significantly higher radioactivity in the vesicle pellet than that of functional sfp strains 3610, RL3090, and SL3610 (p<0.01 one-way ANOVA, n=3). (E) Density gradient of 14C PA labeled vesicles indicating the location and density of vesicles from each strain. (F) Vesicle pellets from strains 168 and RL2663 contain significantly more radioactivity than that of the surfactin-producing strain 3610 (p<0.01 one-way ANOVA, n=3).
Metabolic labeling and density gradient centrifugation was utilized to compare vesicle quantity and density among various B. subtilis strains (Table 1). Cultures of each strain were pulsed with 14C PA and vesicles were isolated. Cells, FT, and supernatant fractions of the labeled samples were also recovered and counted; all fraction radioactivities are represented as a percentage of the whole sample.
Strains 168 and AD3610 showed a significantly higher percentage of radioactivity levels in the vesicle pellet than pellets of strains 3610, SL3610, and RL3090 (Figure 8D). This indicates that supernatants from strains 168 and AD3610 contain a larger quantity of recoverable vesicles than 3610, SL3610, and RL3090. Strains 168 and AD3610 have a nonfunctional sfp gene, whereas strains 3610, SL3610, and RL3090 harbor fully functional copies of the gene.
The vesicle preparations were subjected to an Optiprep density gradient and subsequently 0.5 ml fractions were recovered (Figure 8E). Fractions containing the majority of radioactivity were 6, 7, and 8, and the presence of vesicles in these fractions was confirmed by TEM. Outer membrane vesicles from Pseudomonas aeruginosa were also found in similar fractions (Tashiro et al., 2010).
Disruption of Vesicles by Surfactin
We found that we were able to recover large amounts of vesicles from strains that were unable to produce surfactin due to a mutation in sfp and very few vesicles from strains with a functional sfp gene. This association suggested to us the possibility that vesicle quantities and surfactin expression were linked. The gene, sfp, is involved in the biosynthesis of the lipopeptide antibiotic, surfactin, in B. subtilis. Because surfactin is known to disrupt membranes, we sought to determine if the presence of surfactin in these sfp+ strains resulted in lower amounts of recoverable vesicles. Strain RL2663 (3610 background) has a functional sfp gene but a mutation in srfAA, an important module of surfactin biosynthesis, rendering the strain unable to produce surfactin (Kearns; Losick, 2003). We were able to recover similar amounts of vesicle pellet radioactivity from strain RL2663 as strain 168 (Figure 8F), showing the same phenotype for two mutations in the surfactin pathway. We also confirmed the large amount of vesicles by TEM (data not shown). This result suggests that the presence of surfactin disrupts vesicles, leading to less recoverable reactivity in the vesicle pellet fraction.
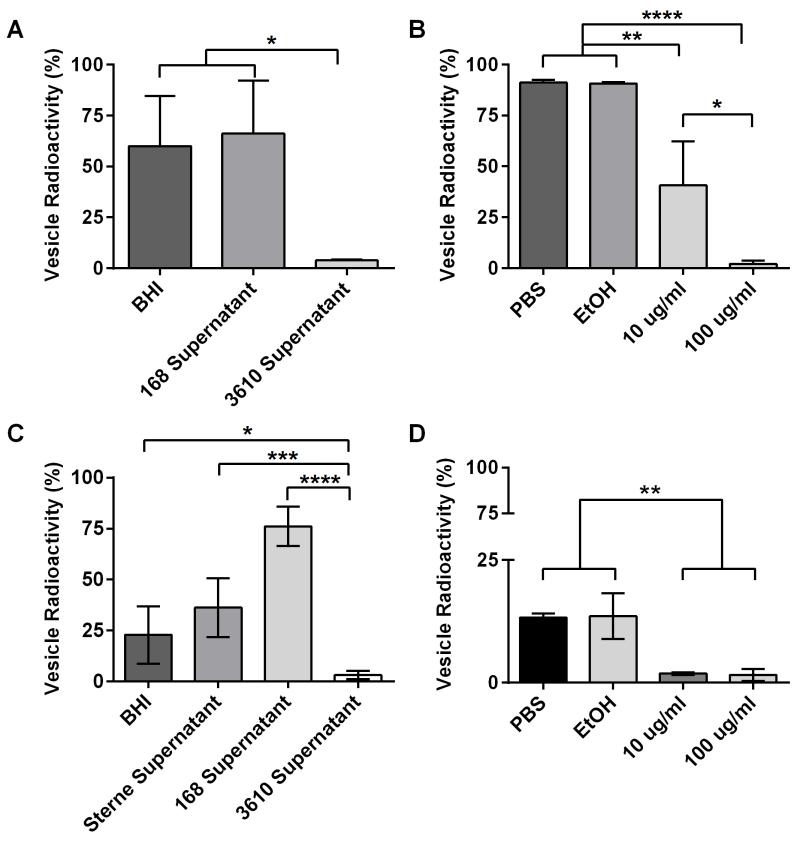
To ascertain whether surfactin action was indeed the mechanism behind the differences in recoverable vesicles among strains, we incubated 14C PA-labeled vesicles isolated from strain 168 in BHI media, strain 168 cell-free supernatant, strain 3610 cell-free supernatant, PBS, 10% EtOH, 10 μg ml-1 surfactin, and 100 μg ml-1 surfactin overnight at 37° C on a nutator and then recovered the vesicle pellet and spent supernatants. We found that recoverable vesicle radioactivity was significantly reduced when incubated in supernatant from strain 3610 (surfactin competent) but stable in strain 168 supernatant (surfactin deficient) (Figure 9A). We also recovered less radioactivity in the pellets of vesicles incubated with purified surfactin and the data showed a dose response (Figure 9B). These results indicate that the presence of surfactin de-stabilizes vesicles and is responsible for the inability to recover large amounts of vesicles from surfactin-producing strains of B. subtilis.
Figure 9. Surfactin production disrupts extracellular vesicles.
Cells from strain 168 and Sterne 34F2 were labeled with 14C PA and the radioactivity of vesicle pellets compared to radioactivity of the supernatant fractions was measured after 24 h incubation to determine disruption. (A) B. subtilis vesicle pellets incubated in media or strain 168 cell-free supernatant were stable and radioactivity remained high compared to radioactivity in the supernatant, whereas the radioactivity of vesicle pellets incubated in strain 3610 cell-free supernatant was almost completely lost (p<0.05 one-way ANOVA, n=3). (B) B. subtilis vesicle pellets incubated in PBS and 10% EtOH remained stable, whereas the radioactivity of vesicle pellets compared to supernatants diminished in 10 μg ml-1 and 100 μg ml-1 of surfactin, indicating vesicle disruption (p<0.05, p<0.01, p<0.0001 one-way ANOVA, n=3). (C) B. anthracis vesicle pellets from B. anthracis were more stable in media, Sterne 34F2 strain supernatant, and strain 168 supernatant compared to pellet radioactivity when incubated in strain 3610 supernatant (p<0.05, p<0.001, p<0.0001 one-way ANOVA, n=3). B. anthracis vesicles are incredibly stable in strain 168 cell-free supernatant. (D) B. anthracis vesicle pellet radioactivity was greatly reduced when incubated in pure surfactin and was more stable in PBS and 10% EtOH (p<0.01 one-way ANOVA, n=3).
To explore whether the mechanism of vesicle disruption by surfactin extended across bacterial species we studied the effect of surfactin on vesicles isolated from B. anthracis. We found that pellet radioactivity of 14C PA-labeled B. anthracis vesicles significantly decreased after incubation with strain 3610 supernatant and purified surfactin (Figure 9C-D). It is notable that vesicles from B. anthracis are much less stable under these conditions when compared to B. subtilis. B. anthracis vesicle instability was also reported previously (Wolf et al., 2012). We also performed the reverse experiments, incubating strain 168 in week-old B. anthracis Sterne, Pasteur, and DeltaT cell-free supernatants and did not see a significant change in pellet radioactivity compared to BHI and strain 168 supernatant controls (data not shown). This indicates B. anthracis does not actively produce a vesicle destabilizing molecule. This result extends the role of surfactin to disruption of vesicles from other bacterial species.
Discussion
The production of extracellular vesicles appears to be a common phenomenon in both Gram-positive and Gram-negative bacteria. Here, we established that B. subtilis produces vesicles, the quantity of vesicles varies with the strain, that the gene, sfp, influences vesicle recovery through the effect of its product surfactin, and surfactin disrupts vesicles produced by different species, suggesting a role for unloading vesicles. These results establish B. subtilis as a system for studying the physiology of Gram-positive vesicular production.
Any new description of vesiculogenesis by microorganisms must rule out the possibility that such vesicles are artifacts of lipid self-assembly and/or damaged membranes resealing into spherical structures. Consequently, we devoted considerable effort to establishing that natural vesicles were indeed produced from living cells. We conclude that B. subtilis produces membrane derived vesicles from several lines of evidence: 1) vesicles were recovered from live and not dead cells; 2) metabolic labeling with 14C PA resulted in the incorporation of radioactivity into vesicle preparations; 3) the vesicles described for B. subtilis had similar dimensions to those produced by other microorganisms; 4) vesicle preparations manifested similarities and differences in protein content to membranes consistent with both a common origin and a separate synthetic pathway; 5) electron microscopy revealed protrusions from cells with dimensions comparable to those of extracellular vesicles suggesting that both are the same; 6) differences in vesicle recovery among B. subtilis strains implied strain-related and -regulated differences in vesicle fate; 7) significantly reduced incorporation of GXM in naturally produced vesicles (Rodrigues et al.,2007; Marsollier et al.,2007; Lee et al.,2009; Rivera et al.,2010; Prados-Rosales et al.,2011; Maldonado et al.,2011). The latter experiment was designed to show that these vesicles were not forming spontaneously in media, which was assessed by taking advantage of a fungal polysaccharide for which mAbs suitable for immunogold labeling of vesicle preparations are available. We predicted that if vesicles were forming spontaneously by the self-assembly of free lipids that we would find a similarly large amount of vesicles containing gold particles in both natural and sonicated vesicles. This, however, was not the case, as sonicated vesicles contained significantly more GXM encapsulated within the vesicles. Additionally, these structures were smaller and less variable in size than natural vesicles recovered from bacterial supernatants. Furthermore, we noted diameter differences between sonicated vesicles that presumably had the opportunity to reassemble and naturally recovered vesicles, providing further evidence against our vesicle preparations being artifacts of the propensity of lipids for self-assembly in solution.
Vesicles were isolated from both planktonic cultures and bacterial biofilms. Biofilm is considered the primary state in which bacteria exist in the environment therefore it is perhaps not surprising that we identified extracellular vesicles within the matrix of B. subtilis biofilm. Vesicles isolated from all biofilm preparations were similar in size to planktonic vesicles, implying natural vesicle production by B. subtilis despite different growth conditions. Notably, vesicles isolated from strain 168 biofilm were free floating and non-adherent to other material whereas strain 3610 biofilm vesicles were often associated to flagella and other debris. This may reflect reduced production of exopolysaccharide by strain 168 due to a mutation in the epsC gene (McLoon et al., 2011).
Previously, Tashiro et al showed that OMVs with stronger ζ potentials were more likely to associate with cells in P. aeruginosa (Tashiro et al., 2010). B. subtilis cells have a strongly negative ζ potential with a magnitude similar to the vesicles recovered from the supernatant. This finding is consistent with notion that B. subtilis extracellular vesicles originate from the cell membrane and have the potential to interact with bacterial cells (Ahimou et al., 2001).
When isolating vesicles from the B. subtilis 168 lab and 3610 environmental strains, we noted that vesicles isolated from each strain under the same culture conditions resulted in different DLS profiles. Strain 168 produced two populations similar in size to vesicles from other organisms whereas strain 3610 showed signals not typically associated with vesicles and more consistent with the presence of contaminating structures. The apparent paucity of vesicle signatures in the DLS profile of vesicle preparation from strain 3610 most likely reflects flagella contamination, which could skew the DLS data. For strain 168, vesicles were so abundant relative to any contaminating flagella that they exhibited a profile like those reported for other vesicle preparations (Rivera et al., 2010). DLS results of sfp+ strains, SL3610 and RL3090, also demonstrated similar contaminant distributions to strain 3610 whereas strain AD3610 (sfp−) displayed two clean peaks like strain 168. The dimensions obtained from DLS data were consistent with measurements from TEM images, which also revealed that more vesicles could be recovered from strain 168 compared to strain 3610. Strain 168 vesicle preparations contained so many vesicles that they were identified in the entire EM field of view whereas in strain 3610 vesicle preparations, only an occasional vesicle was present in microscopic fields. It is notable that cells from all strains exhibited putative vesicular blebs on the cell membrane in SEM images even though we recovered various amounts of vesicles from the strains (data not shown). Although such blebs cannot be conclusively identified as vesicles based on sharing similar morphology and dimensions, their presence in the surface of wild type strains, and their relatively scarcity in the culture supernatants of those strains is consistent with, and supportive of the notion that they are made by wild type strains but disrupted by surfactin after release from cells.
The ability of strain 168 to be easily genetically manipulated as well as to produce large quantities of vesicles makes it an ideal model strain to study the genetics and mechanisms of vesicle production in Gram-positive bacteria. Further, mutations in sfp, epsC, swr,and PdegQ, as well as the lack of the plasmid-borne gene rapP in strain 168 were identified as genetic differences between strain 168 and strain 3610 (McLoon et al., 2011). We further investigated the effect of the sfp gene on vesiculogenesis. Strain AM373 (168 harboring wild type copies of aforementioned genes) showed decreased amounts of recoverable vesicles compared to the background 168 strain by TEM and metabolic labeling (data not shown). This was evidence that the lack of a functional copy of one or more of these genes could be responsible for high vesicle recovery in the 168 strain. We investigated the candidate gene, sfp, further for its effects on vesicle production in B. subtilis. When a functional copy of sfp was restored into strain 168, the quantity of recoverable vesicles was greatly reduced (RL3090). Furthermore, when sfp was disrupted in strain 3610 (AD3610), vesicle recovery levels increased similar to strain 168. Strain SL3610, a sfp complement strain of AD3610, again showed abrogated vesicle recovery. These results suggest that sfp is an important factor in the amount of vesicles recovered between wild strains and lab strains of B. subtilis and further reinforces the authenticity of vesiculogenesis by the Gram-positive bacterium. We note that genetic regulation of vesicular production has recently been reported in mycobacteria (Rath et al., 2013).
Sfp protein is required for surfactin biosynthesis; therefore strains lacking sfp cannot produce surfactin. Surfactin has been shown to disrupt lipid membranes and so we investigated surfactin as the mechanism behind recovering variable amounts of vesicles between wild and domestic strains of B. subtilis. We were able to recover similar amounts of vesicles from strain RL2663 as from strain 168. RL2663 has a functional sfp gene but a mutation in srfAA, which is, also required for surfactin synthesis. This data led us to believe that surfactin production affected the amount of recoverable vesicles. We confirmed that vesicle incubation in pure surfactin and cell-free supernatant from surfactin-producing strain 3610 leads to less recoverable radioactivity in the vesicle pellet compared to incubation in surfactin-nonproducing strain 168 supernatant, PBS, BHI broth, and 10% EtOH indicative of vesicle disruption. We also determined that strain 3610 supernatant and pure surfactin disrupted vesicles from B. anthracis, suggesting that wild B. subtilis can utilize surfactin to disrupt extracellular vesicles produced by other species. The high stability of B. anthracis vesicles in strain 168 supernatant, which corresponds to depleted BHI media, may be due to the cells utilizing one or more destabilizing molecules in the media but this difference was not pursued further. Since vesicles have been implicated in such critical functions as iron transport, it is conceivable that surfactin-mediated disruption of vesicles from other species provides a competitive advantage to B. subtilis (Rath et al., 2013). In summary, our data implies that environmental strains produce vesicles that are disrupted by surfactin and provides a new function for this lipopeptide in promoting release of vesicular cargo into the extracellular space.
In conclusion, we have characterized extracellular vesicles produced by Bacillus subtilis. These vesicles are not only similar in size and morphology to other bacterial vesicles but we were also able to describe vesiculogenesis in various strains. In addition, the ability to recover a large amount of vesicles from strain 168 makes it an efficient model for the study of vesiculogenesis in Gram-positive bacteria. We have identified a gene, sfp, which is nonfunctional in most laboratory strains, that indirectly affects the quantity of vesicles recoverable from cell cultures. The lipopeptide surfactin, which requires functional sfp, disrupts vesicles in the extracellular milieu to release vesicle cargo. To our knowledge, this is the first time a mechanism for self-and foreign-vesicle cargo release has been reported in Gram-positive bacteria. The occurrence of vesicular production in B. subtilis combined with the ability to induce release of vesicular content across species boundaries provides a potentially outstanding platform for the dissection of this enigmatic process in a model organism.
Experimental Procedures
Bacterial Strains
Bacillus subtilis 168 strain was obtained from ATCC #23857, and B. subtilis strains NCIB# 3610, AMA373, RL3090, RL2663, and AD3610 were gifts from Richard Losick (Boston, MA) (Table 1). Strain AD3610 was constructed using techniques previously described (Yasbin; Young, 1974; Gryczan et al., 1978; Wach, 1996). Briefly, PCR was utilized to disrupt the sfp gene with a Kan cassette (Table S1). The construct was transformed into strain PY79 and moved into strain 3610 by phage transduction. Strain SL3610 was constructed by ectopic integration of sfp-spec into the AmyE locus of AD3610 by using pDG1730 and In-Fusion HD Cloning Plus kit (Clontech) (Table S1). Transformation was performed as stated above. The lab strain of B. anthracis Sterne 34F2 (pXO1+, pXO2−) used in stability assays was obtained from Alex Hoffmaster at the Center for Disease Control (Atlanta, GA). All strains were grown and maintained from 25% glycerol −80°C frozen stock on brain heart infusion (Difco) agar or broth at 37°C. Antibiotics used include 0.5 μg ml-1 erythromycin and 2.5 μg ml-1 lincomycin (mls), 100 μg ml-1 spectinomycin, and 10 μg ml-1 kanamycin.
Vesicle Isolation
Vesicles were obtained with minor modifications of the method used previously for the isolation of B. anthracis vesicles (Rivera et al., 2010). Briefly, bacterial cultures were spun for 20 min at 4°C at 15,000 × g to remove cells. The resulting supernatant was then filtered through a 0.22 um Milipore Express Plus Membrane and the filtrate was concentrated using Amicon ultrafiltration system (Millipore) with 100 kDa filter. The concentrate was then further filtered through a 0.8 um syringe filter (Corning) to remove larger cellular debris or aggregated material. The filtered supernatant was then centrifuged at 195,000 × g for 1 h at 4°C and washed with phosphate buffered saline (PBS). The vesicle pellet was resuspended in PBS or water. Heat-killed culture vesicles were prepared by autoclaving an overnight culture. The cells were then spun for 20 min at 4°C at 15,000 × g and washed with PBS. The dead cells were then suspended in fresh media and incubated overnight with shaking. Vesicles were then isolated in parallel with live cultures.
Vesicle Size by Dynamic Light Scattering (DLS)
The size distribution and diameter of vesicles resuspended in PBS or water was measured using 90Plus/BI-MAS Multi Angle Particle Sizing analyzer (Brookhaven Instruments Corp.), as previously described (Eisenman et al., 2009).
Transmission Electron Microscopy (TEM)
Vesicles were visualized by TEM using negative and positive staining techniques. For negative staining, vesicles were adhered to 300-mesh carbon and Formvar coated Copper grids for 2 min, stained with 1% phosphotungstic acid and viewed on a JEOL 100CXII or 1200EX at 80kV. For embedded TEM, vesicles were fixed with 2.5% glutaraldehyde and 2% paraformaldehyde in 0.1 M sodium cacodylate buffer. Samples were then postfixed with 1% osmium tetroxide followed by 2% uranyl acetate en-bloc staining. The samples were dehydrated through a graded series of ethanol and embedded in LX112 resin (LADD Research Industries, Burlington VT). Pellicle biofilms were fixed for 24 h with 2.5% gluteraldehyde, 0.2 M sodium cacodylate, 0.4 M sucrose, and 10mM MgCl2 (pH 7.4) in a 1:1 ratio with the MSgg culture media. Pieces of each fixed biofilm were processed in suspension on a rotator. The biofilm samples were postfixed with 1% osmium tetroxide, 0.7% potassium ferrocyanide for 1 h followed by 1% uranyl acetate. The biofilm samples were dehydrated through a graded series of ethanol and then embedded in LX112 resin (LADD Research Industries, Burlington VT). Ultrathin (80 nm) sections of all samples were cut on a Reichert Ultracut UCT, stained with uranyl acetate followed by lead citrate.
Scanning Electron Microscopy (SEM)
Cells were fixed in 5% glutaraldehyde, 0.2 M sodium Cacodylate, 0.4 M Sucrose, 10mM MgCl2 pH 7.4 and mixed 1:1 with suspension. The biofilm samples were postfixed for 30 min with 1% osmium tetroxide, 0.7% potassium ferrocyanide, 0.1 M sodium cacodylate, 0.2 M sucrose, and 5 mM MgCl2 (pH 7.4). Samples were dehydrated through a graded series of ethanol and critical point dried using liquid carbon dioxide in a Tousimis Samdri 795 Critical Point Drier (Rockville, MD). The samples were sputter coated with gold-palladium in a Denton Vacuum Desk-2 Sputter Coater (Cherry Hill, NJ). Prepared cells were then examined in a Zeiss Supra Field Emission Scanning Electron Microscope (Carl Zeiss Microscopy, LLC North America), using an accelerating voltage of 5 KV.
Glucuronoxylomannan Immunogold TEM
The polysaccharide, glucuronoxylomannan (GXM), was isolated from Cryptococcus neoformans and fractionated to less than 10 kDa in size by filtration through Amicon Ultra Centrifugal Filters (Millipore). GXM was added to a culture of strain 168 at inoculation and incubated at 37°C for 18 h. Vesicles were then isolated and either sonicated (20 sec sonication and 30 sec incubation on ice, repeated 4 times) or not (Sonic Dismembrator Model 100; Fisher Scientific). Vesicles were fixed with 2% paraformaldehyde, 0.1 % glutaraldehyde in 0.1 M Phosphate buffer, followed by dehydration with a graded series of ethanol, and a progressive lowering of the temperature to −20° in a RMC FS 7500. Samples were then embedded in Lowicryl HM-20 monostep resin (Electron Microscopy Sciences) and polymerized using UV light. Ultrathin sections were cut on a Reichert Ultracut UCT.
Immunogold labeling was performed by fixing the samples in 4% paraformaldehyde (Electron Microscopy Sciences), 0.1% glutaraldehyde (Polysciences), and buffered in 0.1 M sodium cacodylate (pH 7.2). Samples were incubated with the primary monoclonal antibody (mAb) 18B7 against GXM diluted in incubation buffer overnight at 4°C. The sample was washed with BSA-C (Electron Microscopy Sciences) and incubated with 10 nm immunogold-conjugated secondary antibody (donkey anti-mouse) (Electron Microscopy Sciences) for 2 h at room temperature. Samples were washed again and postfixed with 2% gluteraldehyde/PBS. Finally, samples were stained with 4% uranyl acetate and imaged using JEOL 100CXII transmission electron microscope at 80kv. Controls included preparations with no primary antibody as well as preparations with an irrelevant IgG antibody.
Metabolic labeling with C-14 Palmitic Acid
B. subtilis and B. anthracis cells were labeled with radioactivity as previously described for C. neoformans with minor modifications (Wolf et al., 2012). Briefly, an overnight culture of bacteria was diluted to a suspension with an optical density at 600 nm (OD600) of ~0.1. 1-2 μCi of 14C PA was added to aliquots of 15-30 ml. The samples were incubated at 37°C until OD600 doubled to ~0.2 and the cells were then pelleted, washed, and resuspended in fresh BHI. Aliquots of 1 ml were added to 14 ml BHI in PETG flasks (Thermo Scientific) and grown overnight at 37°C. Overnight cultures were normalized by OD600 and vesicles were harvested by centrifuging cells for 20 min at 15,000 × g at 4 °C and filtering the supernatant with a 0.8 μm syringe filter (Corning) and the resultant supernatant was concentrated with Amicon Ultra Centrifugal Filters 100 kDa (Millipore). The concentrate was spun at 195,000 × g for 1 h at 4 °C, and washed with PBS. The vesicle pellet was then resuspended in PBS for dynamic light scattering and scintillation counting (CPM). Controls included B. subtilis incubated without 14C PA, cells incubated with 150 mM NaN3, and cultures without B. subtilis cells. Vesicle production over time was studied by counting radioactivity of vesicles isolated at 0, 4, 12, and 18 h. Samples measured include cell pellets, supernatant, concentrate FT, concentrate supernatant, and vesicle pellet and each fraction represented as a ratio of the total sample. Scintillation counting was done in a Packard Bioscience Tricarb A2900 liquid scintillation counter. The position of vesicle fractions was determined by resuspending labeled vesicles in 30% Optiprep Density Gradient Medium (Sigma) and layering 2 ml of 25, 20, 15, and 10% Optiprep above the vesicle sample. The gradient was spun for 16 h at 140,000 × g at 4°C. 0.5 ml fractions were taken from the top of the gradient and counted.
Vesicle Stability
Vesicle stability over time, incubated in PBS, was measured by incubating 14C PA-labeled vesicle pellets at room temperature for 0, 24, 48 and 72 h in PBS followed by ultracentrifugation and counting of supernatants and pellets. Vesicle stability was measured in 10 μg ml-1 and 100 μg ml-1 surfactin (resuspended in EtOH at 10 mg ml-1) (Sigma), BHI media, PBS, 10% EtOH, and cell-free supernatants from B. subtilis strains 168 and 3610 week old cultures. B. anthracis stability was measured similarly to B. subtilis but with the added condition of cell-free supernatant from B. anthracis strain Sterne 34F2. Vesicle stability was measured by incubating 14C PA-labeled vesicle pellets resuspended in the various conditions at 37 °C on a nutator and after 24 h, samples were ultracentrifuged and radioactivity of supernatants and pellets were counted.
ζ Potential Measurements
The ζ potential of vesicles in water with 10mM dextrose and 1mM KCl was calculated using a ζ potential analyzer (ZetaPlus; Brookhaven Instruments Corp.).
Protein Characterization
Vesicles were isolated and purified by density gradient centrifugation using OptiPrep and vesicle containing fractions were dialyzed overnight against PBS at 4°C with one buffer change. Cells were grown for 18 h at 37°, spun down and washed with PBS. Cell membrane fractions were isolated as previously described (Petrackova et al., 2010). All samples were then lyophilized and analyzed.
Proteins were precipitated from vesicle suspensions, membrane fractions, and bacterial lysates by adding six volumes of ice-cold acetone. After incubation overnight, the acetone was removed and the pellets dissolved in 25 mM ammonium bicarbonate. Proteins were reduced with 10 mM DTT at room temperature for 1 h and the cysteine residues were subsequently alkylated with 10 mM iodoacetamide at room temperature for 1h in the dark. Protein digestion was carried out with trypsin (Promega, 1:20, w/w) at 37°C overnight. The peptide mixtures from in-solution tryptic digestions were analyzed using nanoflow LC-MS/MS. The peptides were loaded onto a 0.3 × 5 mm C18 precolumn (New Objective) and then eluted with a linear gradient of 2-90% acetonitrile in 0.1% aqueous solution of formic acid. The gradient was delivered over 120 min by a nanoLC-1Dplus (Eksigent) at a flow-rate of 200 nl/min, through a 75-μm × 15 cm fused silica capillary C18 HPLC PepMap column (LC Packings) to a stainless steel nano-bore emitter (Proxeon). The peptides were scanned and fragmented with an LTQ XL linear ion trap mass spectrometer (ThermoFinnigan) operated in data-dependent and MS/MS switching mode using the three most intense precursors detected in a survey scan from 400 to 1600 amu (three μscans). Normalized collision energy was set to 35%, and dynamic exclusion was applied during 3 min periods to avoid repetitive fragmentation of ions. Generated .raw files were converted to .dta files for Mascot database. A database containing the NCBI B. subtilis (4176 sequences, as of January 20, 2012) was searched using Mascot Software (version 2.3, Matrix Science) for protein identification. Search criteria included trypsin specificity with one missed cleavage allowed, and methionine oxidation as variable modification. A minimum precursor and fragment-ion mass accuracy of 1.2 and 0.3 Da, respectively, and a requirement of at least one bold red peptide (i.e. highest scoring peptide matching to protein with highest total score). Cut-off values for Mascot scores of peptides and proteins were set to 45 (p < 0.05) and 51 (p < 0.01), respectively, for considering them as being accurate identifications. Proteins identified with only one peptide were inspected manually. The false positive rate (1,23%) was calculated searching the same spectra against the NCBI B. subtilis decoy database, using the version updated January 20, 2012. A combined list of proteins identified in all replicates was condensed in order to remove redundant IDs such as orthologous sequences, redundant database entries, and indistinguishable isoforms based on observed peptide coverage.
Vesicle preparation protein content from strains 168, 3610, and RL2663 was normalized with Pierce BCA Protein Assay Kit (Thermo Scientific) and 7.5 ug of protein was run on 4-20% Mini-PROTEAN® TGX Stain-Free Gel (Bio-Rad) and silver stained using Pierce Silver Stain Kit (Thermo Scientific).
Biofilm Assay and Vesicle Isolation
Biofilms were grown as described in McLoon et al. with modifications (McLoon et al., 2011). Bacteria were grown from frozen stock on BHI agar overnight at 37°C. A single colony was then inoculated into BHI broth for 4 h at 37°C with shaking. The culture was inoculated (1:1000) into MSgg media (100 mM MOPS, pH 7, 5 mM potassium phosphate, pH 7, 700 μM CaCl2, 2 mM MgCl2, 50 μM FeCl3, 50 μM MnCl2, 1 μM ZnCl2, 2 μM thiamine, 50 μg ml-1 tryptophan, 50 μg ml-1 phenylalanine, and 50 μg ml-1 threonine, 0.5% glycerol and 0.5% glutamate). Biofilms were grown at 30°C for either 4 or 8 d without shaking. After the desired time, the biofilm culture was disrupted and centrifuged at 15,000 × g for 15 min to remove cellular debris. The resulting supernatant was filtered through a 0.22 um Milipore Express Plus Membrane and then concentrated using Amicon Ultra Centrifugal Filters 100 kDa (Millipore). The resultant concentrate was centrifuged at 195,000 × g at 4°C and washed with PBS. The vesicle pellet was resuspended in PBS or water. Whole biofilm pellicles were submitted to TEM and SEM preparations.
Statistics
Statistics were performed in Prism (GraphPad Software). Error bars indicate mean and standard deviation. Comparisons between ratio of natural and sonicated vesicles containing gold particles was done using Chi2 test. The diameters of natural and sonicated vesicles were compared using an unpaired t-test. One-way ANOVA was performed comparing vesicle density and diameter. Vesicle pellet radioactivity between strains 168 and 3610 was analyzed with an unpaired t-test. A one-way ANOVA was utilized when comparing vesicle pellet radioactivity among strains. One-way ANOVA was also used to compare stability of pellet radioactivity after incubation in various conditions. Significance was indicated as follows: p<0.05 *, p<0.01**, p<0.001*** p<0.0001****. Error bars represent triplicates and each graph is representative of one experiment done in duplicate or triplicate.
Supplementary Material
Acknowledgements
We are grateful to Dr. Richard Losick, Sara Leiman, and Aaron DeLoughery for various B. subtilis strains and assistance in the construction of SL3610. We thank Geoffrey Perumal, Benjamin Clark, and Juan Jimenez for assistance with the electron microscopy. The data in this paper were from a thesis that was submitted by LB in partial fulfillment of the requirements for the degree of doctor of philosophy in the Sue Golding Graduate Division of Medical Science, Albert Einstein College of Medicine, Yeshiva University, Bronx, NY.
Financial Disclosure
AC was supported by NIH grants HL059842, AI033774, AI033142, AI052733 and Center for AIDS Research at Albert Einstein College of Medicine. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Author Contributions
The author(s) have made the following declarations about their contributions: Conceived and designed the experiments: LB AC JLLG. Performed the experiments: LB AK PCS. Analyzed the data: LB AK PCS JLLG. Contributed reagents/materials/analysis tools: LB AK PCS JLLG AC. Wrote the paper: LB AK AC.
References
- Ahimou F, Paquot M, Jacques P, Thonart P, Rouxhet PG. Influence of electrical properties on the evaluation of the surface hydrophobicity of bacillus subtilis. J Microbiol Methods. 2001;45:119–126. doi: 10.1016/s0167-7012(01)00240-8. [DOI] [PubMed] [Google Scholar]
- Bernheimer AW, Avigad LS. Nature and properties of a cytolytic agent produced by bacillus subtilis. J Gen Microbiol. 1970;61:361–369. doi: 10.1099/00221287-61-3-361. [DOI] [PubMed] [Google Scholar]
- Carrillo C, Teruel JA, Aranda FJ, Ortiz A. Molecular mechanism of membrane permeabilization by the peptide antibiotic surfactin. Biochim Biophys Acta. 2003;1611:91–97. doi: 10.1016/s0005-2736(03)00029-4. [DOI] [PubMed] [Google Scholar]
- Dorward DW, Garon CF, Judd RC. Export and intercellular transfer of DNA via membrane blebs of neisseria gonorrhoeae. Journal of Bacteriology. 1989;171:2499–2505. doi: 10.1128/jb.171.5.2499-2505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorward DW, Garon CF. DNA is packaged within membrane-derived vesicles of gram-negative but not gram-positive bacteria. Applied and Environmental Microbiology. 1990;56:1960–1962. doi: 10.1128/aem.56.6.1960-1962.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubois JY, Kouwen TR, Schurich AK, Reis CR, Ensing HT, Trip EN, et al. Immunity to the bacteriocin sublancin 168 is determined by the SunI (YolF) protein of bacillus subtilis. Antimicrob Agents Chemother. 2009;53:651–661. doi: 10.1128/AAC.01189-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl AM, Losick R, Kolter R. Bacillus subtilis genome diversity. J Bacteriol. 2007;189:1163–1170. doi: 10.1128/JB.01343-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egelhaaf S?, Wehrli E, Müller M, Adrian M, Schurtenberger P. Determination of the size distribution of lecithin liposomes: A comparative study using freeze fracture, cryoelectron microscopy and dynamic light scattering. J Microsc. 1996;184:214–228. [Google Scholar]
- Eisenman HC, Frases S, Nicola AM, Rodrigues ML, Casadevall A. Vesicle-associated melanization in cryptococcus neoformans. Microbiology. 2009;155:3860–3867. doi: 10.1099/mic.0.032854-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis TN, Kuehn MJ. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol Mol Biol Rev. 2010;74:81–94. doi: 10.1128/MMBR.00031-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gryczan TJ, Contente S, Dubnau D. Characterization of staphylococcus aureus plasmids introduced by transformation into bacillus subtilis. J Bacteriol. 1978;134:318–329. doi: 10.1128/jb.134.1.318-329.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gurung M, Moon DC, Choi CW, Lee JH, Bae YC, Kim J, et al. Staphylococcus aureus produces membrane-derived vesicles that induce host cell death. PLoS One. 2011;6:e27958. doi: 10.1371/journal.pone.0027958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heerklotz H, Seelig J. Detergent-like action of the antibiotic peptide surfactin on lipid membranes. Biophys J. 2001;81:1547–1554. doi: 10.1016/S0006-3495(01)75808-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S-, Kim M-, Lee E-, Kim JH, Kim Y-, Jeon SG, et al. Extracellular vesicles derived from staphylococcus aureus induce atopic dermatitis-like skin inflammation. Allergy. 2011;66:351–359. doi: 10.1111/j.1398-9995.2010.02483.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kearns DB, Losick R. Swarming motility in undomesticated bacillus subtilis. Mol Microbiol. 2003;49:581–590. doi: 10.1046/j.1365-2958.2003.03584.x. [DOI] [PubMed] [Google Scholar]
- Kong M, Bhattacharya RN, James C, Basu A. A statistical approach to estimate the 3D size distribution of spheres from 2D size distributions. Bulletin of the Geological Society of America. 2005;117:244–249. [Google Scholar]
- Kuehn MJ, Kesty NC. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 2005;19:2645–2655. doi: 10.1101/gad.1299905. [DOI] [PubMed] [Google Scholar]
- Kulp A, Kuehn MJ. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu Rev Microbiol. 2010;64:163–184. doi: 10.1146/annurev.micro.091208.073413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E, Choi D, Kim D, Kim J, Park JO, Kim S, et al. Gram-positive bacteria produce membrane vesicles: Proteomics-based characterization of staphylococcus aureus-derived membrane vesicles. Proteomics. 2009;9:5425–5436. doi: 10.1002/pmic.200900338. [DOI] [PubMed] [Google Scholar]
- Maldonado R, Wei R, Kachlany SC, Kazi M, Balashova NV. Cytotoxic effects of kingella kingae outer membrane vesicles on human cells. Microb Pathog. 2011;51:22–30. doi: 10.1016/j.micpath.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsollier L, Brodin P, Jackson M, Kordulakova J, Tafelmeyer P, Carbonnelle E, et al. Impact of mycobacterium ulcerans biofilm on transmissibility to ecological niches and buruli ulcer pathogenesis. PLoS Pathog. 2007;3:e62. doi: 10.1371/journal.ppat.0030062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JJ, Wendrich TM, Marahiel MA. The dhb operon of bacillus subtilisEncodes the biosynthetic template for the catecholic siderophore 2,3-dihydroxybenzoate-glycine-threonine trimeric ester bacillibactin. Journal of Biological Chemistry. 2001;276:7209–7217. doi: 10.1074/jbc.M009140200. [DOI] [PubMed] [Google Scholar]
- McLoon AL, Guttenplan SB, Kearns DB, Kolter R, Losick R. Tracing the domestication of a biofilm-forming bacterium. J Bacteriol. 2011;193:2027–2034. doi: 10.1128/JB.01542-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano MM, Marahiel MA, Zuber P. Identification of a genetic locus required for biosynthesis of the lipopeptide antibiotic surfactin in bacillus subtilis. J Bacteriol. 1988;170:5662–5668. doi: 10.1128/jb.170.12.5662-5668.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olaya-Abril A, Prados-Rosales R, McConnell MJ, Martin-Pena R, Gonzalez-Reyes JA, Jimenez-Munguia I, et al. Characterization of protective extracellular membrane-derived vesicles produced by streptococcus pneumoniae. J Proteomics. 2014 doi: 10.1016/j.jprot.2014.04.023. [DOI] [PubMed] [Google Scholar]
- Petrackova D, Semberova L, Halada P, Svoboda P, Svobodova J. Stress proteins in the cytoplasmic membrane fraction of bacillus subtilis. Folia Microbiol (Praha) 2010;55:427–434. doi: 10.1007/s12223-010-0072-z. [DOI] [PubMed] [Google Scholar]
- Prados-Rosales R, Baena A, Martinez LR, Luque-Garcia J, Kalscheuer R, Veeraraghavan U, et al. Mycobacteria release active membrane vesicles that modulate immune responses in a TLR2-dependent manner in mice. J Clin Invest. 2011;121:1471–1483. doi: 10.1172/JCI44261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quadri LE, Weinreb PH, Lei M, Nakano MM, Zuber P, Walsh CT. Characterization of sfp, a bacillus subtilis phosphopantetheinyl transferase for peptidyl carrier protein domains in peptide synthetases. Biochemistry. 1998:1585–95. doi: 10.1021/bi9719861. [DOI] [PubMed] [Google Scholar]
- Rath P, Huang C, Wang T, Wang T, Li H, Prados-Rosales R, et al. Genetic regulation of vesiculogenesis and immunomodulation in mycobacterium tuberculosis. Proc Natl Acad Sci U S A. 2013;110:E4790–7. doi: 10.1073/pnas.1320118110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera J, Cordero RJB, Nakouzi AS, Frases S, Nicola A, Casadevall A. Bacillus anthracis produces membrane-derived vesicles containing biologically active toxins. Proceedings of the National Academy of Sciences. 2010;107:19002–19007. doi: 10.1073/pnas.1008843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues ML, Nimrichter L, Oliveira DL, Frases S, Miranda K, Zaragoza O, et al. Vesicular polysaccharide export in cryptococcus neoformans is a eukaryotic solution to the problem of fungal trans-cell wall transport. Eukaryot Cell. 2007;6:48–59. doi: 10.1128/EC.00318-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tashiro Y, Ichikawa S, Shimizu M, Toyofuku M, Takaya N, Nakajima-Kambe T, et al. Variation of physiochemical properties and cell association activity of membrane vesicles with growth phase in pseudomonas aeruginosa. Appl Environ Microbiol. 2010;76:3732–3739. doi: 10.1128/AEM.02794-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thay B, Wai SN, Oscarsson J. Staphylococcus aureus alpha-toxin-dependent induction of host cell death by membrane-derived vesicles. PLoS One. 2013;8:e54661. doi: 10.1371/journal.pone.0054661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wach A. PCR-synthesis of marker cassettes with long flanking homology regions for gene disruptions in S. cerevisiae. Yeast. 1996;12:259–265. doi: 10.1002/(SICI)1097-0061(19960315)12:3%3C259::AID-YEA901%3E3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- Wolf JM, Rivera J, Casadevall A. Serum albumin disrupts cryptococcus neoformans and bacillus anthracis extracellular vesicles. Cell Microbiol. 2012;14:762–773. doi: 10.1111/j.1462-5822.2012.01757.x. [DOI] [PubMed] [Google Scholar]
- Yasbin RE, Young FE. Transduction in bacillus subtilis by bacteriophage SPP1. J Virol. 1974;14:1343–1348. doi: 10.1128/jvi.14.6.1343-1348.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeigler DR, Pragai Z, Rodriguez S, Chevreux B, Muffler A, Albert T, et al. The origins of 168, W23, and other bacillus subtilis legacy strains. J Bacteriol. 2008;190:6983–6995. doi: 10.1128/JB.00722-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.