Abstract
There has been a surge of diagnosis of autism spectrum disorders (ASD) over the past decade. While large, high powered genome screening studies of children with ASD have identified numerous genetic risk factors, research efforts to understanding how each of these risk factors contributes to the development autism has met with limited success. Revealing the mechanisms by which these genetic risk factors affect brain development and predispose a child to autism requires mechanistic understanding of the neurobiological changes underlying this devastating group of developmental disorders at multifaceted molecular, cellular and system levels. It has been increasingly clear that the normal trajectory of neurodevelopment is compromised in autism, in multiple domains as much as aberrant neuronal production, growth, functional maturation, patterned connectivity, and balanced excitation and inhibition of brain networks. Many autism risk factors identified in humans have been now reconstituted in experimental mouse models to allow mechanistic interrogation of the biological role of the risk gene. Studies utilizing these mouse models have revealed that underlying the enormous heterogeneity of perturbed cellular events, mechanisms directing synaptic and circuit assembly may provide a unifying explanation for the pathophysiological changes and behavioral endophenotypes seen in autism, although synaptic perturbations are far from being the only alterations relevant for ASD. In this review, we discuss synaptic and circuit abnormalities obtained from several prevalent mouse models, particularly those reflecting syndromic forms of ASD that are caused by single gene perturbations. These compiled results reveal that ASD risk genes contribute to proper signaling of the developing gene networks that maintain synaptic and circuit homeostasis, which is fundamental to normal brain development.
Keywords: autism spectrum disorders, development, risk genes, synapse, circuits, behavior, neurodevelopmental disorders
Introduction
In his 1943 milestone paper “Autistic Disturbances of Affective Contact” (Kanner, 1943), Leo Kanner described 11 children affected by a psychiatric condition that “differs so markedly and uniquely from anything reported so far.” In one case, he observed the boy patient “was happiest when left alone, almost never cried to go with his mother, did not seem to notice his father’s homecomings, and was indifferent to visiting relatives,” and, referring to this patient, “He seems to be self-satisfied. He has no apparent affection when petted. He does not observe the fact that anyone comes or goes, and never seems glad to see father or mother or any playmate. He seems almost to draw into his shell and live within himself.” Later, in his second year, the patient “developed a mania for spinning blocks and pans and other round objects.” These 11 cases all feature what Kanner later believed what all these patients had in common: the “autistic aloneness” and the “obsessive insistence on the preservation of sameness.”
Over the next seven decades, the definition of autism has evolved to incorporate a clinically heterogeneous group of disorders, collectively termed ‘autism spectrum disorders (ASD)’. ASD shares the common feature of marked impairment in social interactions, verbal and non-verbal communications, and repetitive behavior and restricted interests. Historically, DSM-IV-TR (published in 2000) defined ASD as a heterogeneous group of conditions that include autistic disorder, Rett syndrome, Asperger’s syndrome, and pervasive developmental disorder-not otherwise specified (PDD-NOS). However, radical revisions are made in the DSM-V (2013) (Baker, 2013). ASD is now defined in terms of two categories, “persistent impairment in reciprocal social communication and social interaction,” and “restricted, repetitive patterns of behavior,” with both conditions present from early childhood. Asperger's disorder, PDD-NOS, and other subcategories such as atypical autism are eliminated (although not without contentions) from ASD according to DSM-V. The new ASD diagnosis also requires specifying whether there is intellectual disability, language impairment or associated known medical or genetic conditions.
ASD is a collection of “pervasive developmental disorders.” Pervasive because it involves a broad range of developmental abnormalities with many brain regions believed to be affected. Clinical signs of ASD are frequently manifested before three years of age and recent studies indicate that abnormal social, communication and play behavior may present at as early as 14 months of age (Landa et al., 2007). The fact that ASD is a clinical syndromic diagnosis underscores the lack of a better alternative or unifying brain pathophysiology shared by this group of disorders. Adding to the phenotypic complexity is that ASD symptomatology is on a graded continuum, and is often co-morbid with other behavioral and neurological abnormalities, including varying degrees of intellectual disability, hyperactivity, epilepsy, abnormalities in sensory processing, gastrointestinal symptoms, sleep disturbances, obsessive-compulsive disorder, anxiety, and aggression. This phenotypic spectrum, although not necessary for ASD diagnosis, further complicates the study of its etiology.
Genetics plays an important role in ASD pathogenesis. Epidemiological twin studies suggest that a substantial proportion of the ASD risk is heritable, with monozygotic twins showing 60%–90% concordance in ASD diagnoses (Steffenburg et al., 1989; Bailey et al., 1995; Hallmayer et al., 2011). This level of heritability far exceeds the estimated heritability of other complex polygenic diseases, such as heart disease, cancer, or other psychiatric conditions such as schizophrenia and major depression. Therefore, defining the underlying genetic etiology of ASD has been the major focus of the field in the past a few decades. On the other hand, studies into the genetic basis for ASD have been hampered by inherent problems shared by many complex genetic disorders, such as multiple gene effects, gene-gene interactions, environmental factors, gene–environment interactions and variable penetrance for each individual gene. These studies revealed a remarkable heterogeneity of ASD genetic structure, amounting to hundreds of different genes that may contribute to this disorder (Banerjee-Basu and Packer, 2010; Miles, 2011). These genes are very heterogenous, involving de novo mutations, common and rare variants, chromosome abnormalities, copy number variations, as well as the monogenic, syndromic disorders that display autistic features (Abrahams and Geschwind, 2008; Walsh et al., 2008; Pinto et al., 2010). Despite this genetic heterogeneity, ASD patients with these distinct genetic etiologies share a common triad symptomatology, indicative that different genetic abnormalities may be converging on a discrete set of neurons or circuits in the brain that mediate these behavioral symptoms. Therefore, although the diverse genetic nature of ASD poses a daunting task for ascertaining the contributions of each individual risk genes, it also provides a unique opportunity to study the shared etiological mechanisms.
One of these shared mechanisms involves the normal function of the neuronal synapse, a sub-micron-scale structure that connects neurons into functional networks capable of computational outputs. Many well-established autism risk genes encode proteins that functionally converge at the excitatory and inhibitory synapses. These proteins participate in synaptic signaling networks that transforming patterned synaptic activity reflecting an individual’s experience into circuits of defined cell types and connectivity. These circuits and their constituent cell types may also be the key substrates onto which specific neurological symptoms can be mapped. Indeed, an intriguing and enduring question is which specific neuronal or circuitry type disruptions are sufficient to reproduce the specific symptoms of ASD. Understanding these affected brain circuits would help identify novel avenues for treatment and/or behavioral interventions for ASD. For instance, for genetic abnormalities that share a common synaptic or circuit level pathology, that circuit, rather than a particular risk gene, may be potentially targeted for more effective intervention.
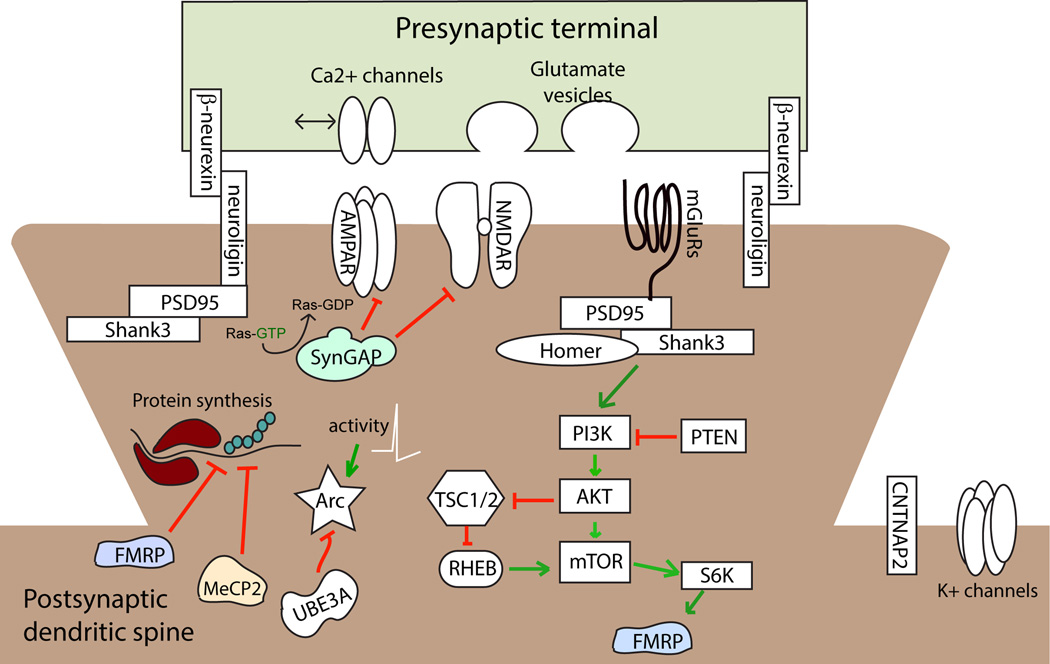
In this review, we first discuss some pathophysiological findings at the synaptic and neural circuit level in autism patients, and then provide mechanistic insights on the pathogenesis gained from mutant mice modeling various human genetic conditions. The notion that ASD represents both ‘synaptopathies’ and ‘circuilopathies’ (Zoghbi and Bear, 2012) is now supported by substantial experimental evidence. We discuss a few rare syndromic disorders that are associated with ASD. These disorders are characterized by highly penetrant gene mutations that disrupt synaptic and circuit function in mouse genetic models. These single gene mutation disorders associated with ASD include fragile X syndrome (FMR1 gene mutation), Rett syndrome (MECP2), Angelman syndrome (UBE3A), the Phelan-McDermid syndrome (SHANK3) as well as rare mutations of CNTNAP2, SYNGAP1, and the neuroligin and neurexin gene families. These gene encode proteins that either contribute to synapse formation and maintenance, or regulate transcription and translation of a myriad of other genes known to affect synapse structure and function. Although an exhaustive overview of a growing list of ASD genes is beyond the scope of this review, we highlight the synaptic and circuit abnormalities caused by these critical regulators of the synapse. The simplified molecular context in which these ASD risk gene products are functioning is sketched in Fig. 1. Literature discussed here supports the model that fundamental circuit abnormalities unify the etiology of ASD arising from diverse genetic factors. Moreover, despite the complexity of genes associated with ASD, shared deficits in synaptic function contribute to the pathogenesis of these disorders. Therefore, therapeutic interventions focused on correcting or compensating for these synaptic or circuit dysfunctions may restore neural systems to their normal operating potentials and alleviate neurologic symptoms of the disorder.
Figure 1.
Schematic depiction of several prevalent ASD risk genes associated with syndromic disorders, and their molecular interactions at the excitatory synapse.
It is also important to note that while we focus here on the role of genetic risk factors implicated in ASD on synapse and circuit dysfunction, accumulating evidence reveals that environmental factors, such as maternal immune activation, and metabolic conditions, also contribute to the biological basis of autism. It may well be the case that atypical gene-environment interactions lead to the pathophysiology in later emerging neural circuits responsible for communication and social competency. Readers are directed to recent studies and review articles for these additional topics on environmental effects (Parker-Athill and Tan, 2010; Hsiao, 2013; Hsiao et al., 2013; Pierre, 2013).
Understanding ASD brain pathophysiology with mechanistic studies in animal models
Limited knowledge of the neural circuits subserving human cognitive, social, and communication competency, as well as the practical difficulties of studying the living human brain have posed major obstacles to our understanding on ASD pathophysiology. Although rapid progress has been made over the last decades in functional imaging technologies, our ability to interrogate human brain structure and function is still met with major limitations in spatial, temporal and functional resolution. It is important to note that much knowledge on autism etiology is obtained through translating clinically-defined autism genes into biological mechanisms. A better mechanistic understanding of these biological mechanisms, in turn, helps identify potential therapeutic targets and translate into successful novel treatment or interventions of this disorder. For autism, and many other genetic neurological disorders, the conventional path from gene discovery to novel treatment strategy often begins with molecular genetic studies in diagnosed patients to identify a genetic variation for the disorder. In the event that a single high penetrance gene causes diseases with autistic traits, such as fragile X syndrome and Rett syndrome, it is possible to create a mouse model with the same genetic disruption. This is based on the reasonable assumption that gene function and disruption in the fundamental neuronal functions are likely shared between the mouse and humans. In this context, mice models offer a unique advantage for mechanistic studies because ASD candidate genes identified through human genetic studies can be reconstituted in another organism with considerable physiologic and behavioral sophistications.
Ideally, ASD models should be based on a known genetic cause (construct validity), resemble key aspects of the human conditions (face validity), and responds to treatments that are effective in the modeled disease (predictive validity) (Moy and Nadler, 2008; Nestler and Hyman, 2010; Silverman et al., 2010). Various mouse models carrying genetic mutations for ASD risk genes have been developed, and most of these mice have established validities in modeling the single gene syndromic disorders associated with ASD traits. Additionally, robust phenotypes in mouse models hold great promise as translational tools for interrogating the pathophysiological mechanisms resulting from genetic mutations, and for identifying novel effective treatments. Because the diagnostic criteria for autism are behavioral, and single genes or proteins do not encode specific behaviors, but instead encode biological functions, these mice may serve as model animals for neurobiological investigations, not for the disease per se. It is important, therefore, to emphasize that these single gene mouse models are useful for understanding fundamental changes in neural structure-function relationships, yet these ‘ASD models’ do not manifest the entirety of phenotypes relevant to ASD. After all, to quote Norbert Wiener (Rosenblueth and Wiener, 1945), ‘The best material model of a cat is another, or preferably the same, cat.’
Impaired brain circuit connectivity as a pathological hallmark of autism
Recent genetic findings, coupled with anatomical and functional imaging studies, suggest a potential unifying model for autism brain pathology. It is now generally agreed upon that deficits in brain circuits and function underlie autism, and virtually all neurodevelopmental disorders. These deficits unfold as an individual develops early in life. The specificity of brain circuits that are affected and their differential ontogeny may determine the presentation of clinical phenotype and disease progression. This connectivity-based etiological hypothesis is supported by numerous mechanistic studies using mouse models (discussed below in this review) and many functional and structural imaging studies in autism patients, and may readily explain the complex disease phenotypes underlying the social and communication components of the diagnosis.
The signs and symptoms of ASD often appear before three years of life, during which social, emotional and cognitive skills are developing rapidly (Walsh et al., 2008). This period correlates with a phase of profound development of the brain architecture, including the generation of new neurons, dendritic growth, synaptogenesis, neural circuit formation and experience-dependent remodeling (Clowry et al., 2010). Studies on postmortem brain have indicated altered neuron production and migration, disrupted balance of excitatory and inhibitory synapses, and faulty assembly of microcircuits in multiple brain regions of ASD patients (Rubenstein and Merzenich, 2003; Geschwind and Levitt, 2007). These structural findings are consistent with an emerging hypothesis that neural pathophysiology of individuals with autism is characterized by hyperconnectivity in local circuits and hypoconnectivity between brain regions (Just et al., 2004; Courchesne and Pierce, 2005; Geschwind and Levitt, 2007). One current model proposes that ASD may reflect a ‘developmental brain disconnection’ syndrome (Geschwind and Levitt, 2007) arising from dysregulation of gene expression that results in the failure of different parts of brain to communicate and form connections appropriately in response to experiences and sensory input. This hypothesis is supported by the fact that many genes associated with ASD are known to play important roles in synapse formation and neuronal connectivity (Comery et al., 1997; Zoghbi, 2003; Clement et al., 2012). However, how altered synaptic connections resulting from defective genes contribute to the abnormalities in specific neural circuits, particularly those underlie the socio-cognitive impairments characteristic of these disorders, remains poorly understood.
Numerous functional and structural imaging studies in ASD patients are also consistent with this hypothesis (Just et al., 2004; Geschwind and Levitt, 2007; Kana et al., 2009; Sahyoun et al., 2010; Hong et al., 2011; Shukla et al., 2011b). These imaging studies indicate alterations in both local brain regions and long-range connectivity among different functional brain regions. For instance, communications between higher-order association areas of the brain involving the frontal lobe are partially impaired during development. ASD patients display impaired synchronous activity in language processing areas during a semantic comprehension task that involves the fronto-parietal areas (Just et al., 2004; Just et al., 2007). Similar compromised synchrony in ASD patients has been reported during social processing tasks (Kana et al., 2009). In addition, ASD patients exhibit reduced functional connectivity with frontal cortical regions while performing a face recognition task or visuospatial and linguistic reasoning (Sahyoun et al., 2010).
Anatomical evidence gained from studying ASD brain also support the hypo-functioning in long-range circuits. Reduced corpus callosum volume has been a variable anatomical phenotype among ASD patients, and the severity of the reduction of corpus callosum volume correlates with the degree of disrupted connectivity and behavioral phenotypes as well (Just et al., 2007; Hong et al., 2011; Shukla et al., 2011a; Shukla et al., 2011b; Thomas et al., 2011). Diffusion tensor imaging (DTI) of the ASD brain also reveals deficits in most major long-range fiber tracts, suggesting a potential global deficit in functional connectivity among different brain regions (Shukla et al., 2011b). In contrast to the hypo-connectivity of long range connections in ASD brain, local circuit connectivity may be normal or even hyper-functional in ASD (Casanova et al., 2002; Rubenstein and Merzenich, 2003; Courchesne and Pierce, 2005; Geschwind and Levitt, 2007; Rubenstein, 2010). A recent fMRI study demonstrated that increased local frontal lobe connectivity is associated with variation of an ASD risk allele (CNTNAP2 gene) that may predispose carriers to language endophenotypes (Scott-Van Zeeland et al., 2010). Other relevant findings have shown that approximately 20% of idiopathic autism patients show macrocephaly (Fombonne et al., 1999; Lainhart, 2003). The brain enlargement seen in ASD are postulated as resultant of mechanisms leading to excess synaptic connections, either due to an initial overproduction of synapses or a failure in experience-dependent synaptic pruning (Eigsti and Shapiro, 2003; Piggot et al., 2009). Other possibilities, such as excessive gliogenesis, neurogenesis, and decreased apoptosis are less frequently invoked, although they are biologically plausible.
The hypothesis of altered synaptic connectivity in ASD brain is further supported by the observations in post-mortem ASD brain tissue, which displayed atypical minicolumn structure in several regions of cortex (Casanova et al., 2002; Casanova et al., 2010). Dendritic spines, protrusions containing excitatory synapses, are altered in postmortem ASD human brain tissue. Spine density is increased on apical dendrites of pyramidal neurons from cortical layers in frontal, temporal and parietal cortices of ASD brain (Hutsler and Zhang, 2010). Similar differences in spine density have been found in individuals with diseases comorbid with ASD, such as in fragile X syndrome (FXS). Increased dendritic spine density in FXS brain has been found, with elongated and tortuous spine morphologies (Irwin et al., 2001; Pfeiffer and Huber, 2007), indicative of altered connectivity and function. Such findings are consistent with the postulated hyper-connectivity in local brain regions in ASD.
Synaptic and circuit dysfunction indicated in rare syndromic forms of ASD
Fragile X syndrome protein FMRP
The model that altered synaptic functions could underlie ASD pathophysiology was perhaps best supported by the phenotypic overlap between ASD, fragile X syndrome and Rett syndrome (RTT) (Zoghbi, 2003; Belmonte and Bourgeron, 2006). Fragile X syndrome (FXS), the most common form of inherited intellectual disability, accounts for about 3% of all cases of ASD. The human FMR1 gene encodes fragile X mental retardation protein (FMRP). FMRP localizes to the somatodendritic compartment and regulates protein synthesis by binding to mRNA to primarily function as a transcriptional repressor. FMRP regulates the expression of a variety of genes, many of which are activity-dependent. Fmr1 mutant mice exhibited increased rate of protein synthesis (Qin et al., 2005; Osterweil et al., 2010), which can be corrected through partial inhibition of the metabotropic glutamate receptor mGluR5 (Dölen et al., 2007; Osterweil et al., 2013). Fmr1 mutant mice also display enhanced mGluR5-dependent long-term potentiation (mGluR5-LTP), and a large spectrum of ASD related phenotypes that can be corrected by suppressing mGluR5 activity (Dölen et al., 2007).
Studies of post mortem brain tissue obtained from FXS patients revealed dendritic spine abnormalities (Irwin et al., 2000; Irwin et al., 2001). Increased dendritic protrusions on apical and basal dendrites in multiple cortical regions were observed, raising the possibility that loss of FMRP leads to stunted spine maturation or impaired elimination. However, several studies examining cortical dendritic spines in Fmr1 mutant mice and wild-type controls observed similar spine density, spine length, and spine head size, yet spines in Fmr1 mutant mice were less stable, consistent with a delay in the developmental downregulation of spine turnover and a persistent overproduction of transient spines (Cruz-Martín et al., 2010; Pan et al., 2010; Padmashri et al., 2013). Consistent with these anatomical findings, extensive studies have shown that Fmr1 mutant mice display altered synaptic plasticity at glutamatergic synapses. These deficits include exaggerated mGluR-dependent hippocampal LTP (Huber et al., 2002) and reduced cortical LTP and GluR1 expression (Li et al., 2002). Loss of FMRP leads to impairments in N-methyl-d-aspartate receptor (NMDAR)-dependent synaptic plasticity in the dentate gyrus (Bostrom et al., 2013), and a downregulation of tonic GABAA receptor currents in the subicular pyramidal cells (Curia et al., 2009). In basal lateral amygdala, the number of inhibitory synapses is reduced, as is the frequency and amplitude of inhibitory postsynaptic currents (Olmos-Serrano et al., 2010). This finding is especially intriguing as this region is involved in the modulation of anxiety state and social avoidance responses, both of which are disrupted in FXS and ASD. In addition, the principal neurons in Fmr1 knockout mice are hyperexcitable, and the homeostatic excitatory/inhibitory (E/I) balance was disrupted. Notably, loss of Fmr1 in mice leads to developmental deficits in the functional laminar cortical connectivity (Bureau et al., 2008), an imbalance of neocortical excitation and inhibition favoring increased excitability (Gibson et al., 2008), increased synchrony of neuronal network activity in somatosensory cortex (Gonçalves et al., 2013) and abnormal experience-dependent plasticity in visual cortex (Dölen et al., 2007). Thus, Fmr1 is capable of profoundly shaping cortical circuits, many associated with the etiology of ASD.
The Rett syndrome protein MECP2
Rett syndrome (RTT) is another well-studied monogenic neurodevelopmental disorder with autistic features and deficits in synapse function. Mutations in the X-linked human MECP2 gene, which encodes the methylcytosine binding protein MeCP2, cause RTT in the majority of the cases (Amir et al., 1999; Chahrour and Zoghbi, 2007; Neul et al., 2008). MeCP2 acts as a transcriptional repressor of thousands of genes, and is required for developmental as well as adult neuronal functions (Guy et al., 2001; McGraw et al., 2011). Mutations in MECP2 gene cause a broad spectrum of neuropsychiatric conditions associated with distinct brain pathological changes. RTT patients exhibit a decrease in the size of cortical minicolumns (Casanova et al., 2003), and cortical pyramidal neurons show reduced dendritic branching patterns and a smaller soma. In addition, dendritic spines in frontal cortex neurons are shorter and more sparse than normal (Belichenko et al., 1994).
Partially overlapping behavioral phenotypes are observed in mice either lacking Mecp2 (loss of function mutation), or overexpressing MECP2 (transgene overexpression to model the human MECP2 gene duplication). These findings support the model that MeCP2 normally functions within a narrow dynamic range. Loss of MeCP2 appears to influence local functional cortical circuits by altering the E/I balance. MeCP2 is regulated by neuronal activity through calcium/calmodulin-dependent protein kinase II (CaMKII) phosphorylation (Zhou et al., 2006). MeCP2 mutant mice have reduced LTP and impaired synaptic plasticity (Asaka et al., 2006; Moretti et al., 2006). In cultured hippocampal neurons that form autapses, lack of MeCP2 leads to reduced spontaneous excitatory postsynaptic currents (EPSCs), most likely reflecting decreased synapse numbers in the mutant neurons through cell autonomous mechanisms (Chao et al., 2010). Mecp2 mutant mice also show a decrease in the AKT/mTOR signaling prior to symptom onset (Ricciardi et al., 2011). It is noteworthy that genetic and pharmacological rescue of MeCP2 function in adult mutants reverses the RTT-like features in mice (Guy et al., 2001; Giacometti et al., 2007). These findings provide hope that RTT symptoms can be potentially reversed in patients as well by re-introducing functional MeCP2 in the developed mature brain.
The Angelman syndrome protein UBE3A
The human UBE3A gene encodes the E3 ubiquitin ligase E6-AP. Due to genetic imprinting of the paternal UBE3A gene allele, mutation of the maternal UBE3A allele alone leads to complete loss-of-function of UBE3A protein in neurons. This mutation results in Angelman syndrome (AS), a debilitating neurodevelopmental disorder characterized by motor dysfunction, severe mental retardation, speech impairment, seizures, and a high prevalence of autism (Kishino et al., 1997; Matsuura et al., 1997). AS is typically (in ~75% of cases) caused by the chromosomal deletions in the 15q11-q13 region. In a small number (~15%) of cases, the maternal UBE3A allele alone is mutated (Jiang et al., 1998a).
Studies of mice harboring a targeted deletion of the Ube3a gene support the role of this gene in AS and autism. Ube3a mutant mice inheriting the mutations through the maternal germ line display salient features of AS (Jiang et al., 1998b). However, how Ube3a loss of function leads to AS and autism brain pathophysiology is poorly understood. In particular, what proteins are the critical targets of UBE3A, which neural circuits are affected by Ube3a deletion, and how these relate to the behavioral endophenotypes in Ube3a mutant mice are unclear at present.
Specific synaptic and neuronal deficits have been described in the Ube3a mutant mice. Ube3a maternal deficiency mice display impaired LTP in the hippocampal CA1 region (Weeber et al., 2003). This deficit can be rescued by elevating CaMKII kinase activity by mutating a regulatory phosphorylation site (Thr305) (van Woerden et al., 2007). The density dendritic spines and strength of excitatory synapses are reduced in the visual cortex of Ube3a maternal deficient mice (Wallace et al., 2012). However, these mice also exhibit a more pronounced decrease in inhibitory neurotransmission, yielding a disrupted E/I balance favoring excitation. This aberrant E/I balance may account for the observed deficit in experience-dependent visual cortical plasticity (Yashiro et al., 2009).
UBE3A protein localizes both to the neuronal soma and synaptic sites. Levels of the protein appear to be regulated by neuronal activity as well (Greer et al., 2010). Among many potential Ube3a downstream targets, Arc is one of the best characterized. Ube3a maternal deficient mice have increased expression of Arc that correlates with greater internalization of the AMPA receptor submits and reduced excitatory synaptic transmission (Greer et al., 2010). In addition to deficits in cortical plasticity, Ube3a maternal deficient mice also exhibit behavioral deficits that are correlated with abnormal dopamine signaling (Riday et al., 2012). These mutant mice exhibited increased dopamine release in the mesolimbic pathway and decreased dopamine release at nigrostriatal pathway, revealing that Ube3a may have specific and different roles that are cell-type specific within subcortical circuits.
Interestingly, maternal duplication of 15q11-q13 is also observed in ~1–3% of nonsyndromic cases of ASD (Cook et al., 1998; Abrahams and Geschwind, 2008). A mouse mutant has been generated that models this chromosomal duplication of the human 15q11–13 region (Nakatani et al., 2009). Mice with a paternal duplication of this syntenic region exhibit significant behavioral deficits including abnormal social interactions, inflexible behavior and increased anxiety. Synaptic abnormalities include altered levels of MBII52, a post-transcriptional regulator of the G-protein-coupled serotonin 2c receptor (5-HT2cR). Disrupted post-transcriptional editing of 5-HT2cR pre-mRNA is known to lead to amino acid substitutions that affect synaptic receptor physiology and serotonin-guided behavior. This is consistent with the observation that primary neurons isolated from 15q11–13 paternal duplication mice display a significantly higher 5-HT2cR-mediated intracellular calcium increase compared to wild-type neurons (Nakatani et al., 2009). Therefore, synaptic deficits observed in genetic abnormalities involving the 15q11–13 region may arise from either increased or decreased gene dosage.
Phelan-McDermid syndrome protein SHANK3
The SH3 and ankyrin-domain-containing protein (Shank) family of proteins, comprising Shank1, 2, and 3, are core components of the postsynaptic density (PSD), and interact with the PSD 95 binding protein SAPAP and Homer (Sheng and Kim, 2000). Shank proteins contain several protein–protein interaction domains. Mutations of Shank3 cause multiple neurological phenotypes, including the Phelan-McDermid syndrome (PMS) (Bonaglia et al., 2006). Shank proteins bind to Homer as part of a macromolecular postsynaptic complex (PSD-95/SAPAP/Shank/Homer) (Hayashi et al., 2009). Shank3 may also be a cytoplasmic binding partner for the neuroligins (Meyer et al., 2004).
Shank3 point mutations, truncations, and disruption by balanced chromosome translocation all has been reported in ASD patients. SHANK3 deletions have been reported in patients with autism, intellectual disability and schizophrenia (Bonaglia et al., 2006; Durand et al., 2007; Gauthier et al., 2009), while SHANK3 duplication has been found in Asperger’s patients and patients with intellectual disability (Durand et al., 2007; Okamoto et al., 2007).
Shank3 promotes synaptogenesis by recruiting essential glutamatergic components to the synapse (Roussignol et al., 2005), resulting in a greater number of functional synapses and increased excitatory signaling. Knockdown of Shank3 in hippocampal neurons results in reduced number of dendritic spines. In developing neurons, Shank3 localizes to the tip of actin filaments and participates in growth cone motility by enhancing actin polymerization (Durand et al., 2011). Mutation that truncate Shank3 disrupt dendritic spine morphology and growth cone motility, whereas inherited mutations (R12C and R300C) results in impaired spine induction and maturation. Mice mutant for Shank3B display multiple neurological deficits including compulsive and repetitive behaviors (excessive grooming), avoidance of social interactions, poor perception of social novelty, increased anxiety and self-injurious behavior, that are associated with deficits in corticostriatal circuitry (Peça et al., 2011). These mice also have altered levels of several components of the PSD, including Homer, PSD93, SAPAP3, and multiple glutamate receptor subunits: GluR2, NR2A, and NR2B. A number of additional Shank3 mutant mouse strains have been generated to represent distinct rare mutantions linked to ASD. One such mutant disrupts the major isoforms of the Shank3 gene by deleting the ankryn-repeat encoding exons 4–9. These mutant mice also display deficits in synaptic plasticity, molecular composition of the post-synaptic density, and abnormal social behavior (Wang et al., 2011).
Neurexin and neuroligin genes
Neurexins and neuroligins are trans-synaptic cell adhesion molecules that physically organize the adjoining presynaptic and postsynaptic specializations (Südhof, 2008). There are three human neurexin genes (NRXN1, NRXN2, NRXN3), each generates transcripts under the control of two separate promoters for two isforms of neurexins, the longer form α-neurexins and shorter β-neurexins. In comparison, neuroligins are encoded by four human genes NLGN1, 2, 3 and 4, and also undergo alternative splicing.
Binding between neurexins and neuroligins recruits Shank3 to the postsynaptic site and stabilizes the synaptic contact, thus facilitating excitatory synapse formation and synaptic transmission. Mutations of both NRXN1 and the X-linked NLGN3 and NLGN4 genes have been reported in patients with ASD (Jamain et al., 2003; Kim et al., 2008). For example, a large deletion of NRXN1 promoter and exon 1–5 has been reported in a patient showing dysmorphic features, cognitive impairment and autistic features (Zahir et al., 2008). Mutant mice lacking any one of the Nrxn genes fail to thrive after birth and die at different times postnatally. Electrophysiologic recordings from acute brain slices of these Nrxn mutant mice reveal impaired synaptic function including decreased spontaneous and evoked neurotransmitter release. Deleting all three isoforms of the α-neurexins (Nrxn 1, 2 and 3) results in embryonic lethality and impaired NMDA receptor function (Kattenstroth et al., 2004). Notably, triple knockout of Ngln 1, 2 and 3 also die perinatally from respiratory failure, which may be due to defective glutamatergic and GABAergic/glycinergic transmission in brain stem respiratory centers (Varoqueaux et al., 2006). Thus, neurexins are critical for the proper synaptic circuit formation and function.
Missense and nonsense mutations in both neuroligin 3 and neuroligin 4 have been identified in a subset of human patients with ASD (Jamain et al., 2003; Laumonnier et al., 2004). Mutant mice containing the point mutant R452C exhibit enhanced inhibitory synaptic transmission and impaired social interactions but no apparent defect in excitatory synaptic transmission (Tabuchi et al., 2007). Interestingly, these R452C mutant mice also display enhanced spatial learning, indicating that the R451C mutation may represent a gain-of-function mutation.
The tuberous sclerosis complex gene TSC1 and TSC2
Tuberous sclerosis complex (TSC) is a rare multi-system genetic disorder that is inherited in an autosomal dominant pattern for a review, see (Leung and Robson, 2007). TSC causes benign tumors, termed harmatomas, in the brain and other organs including skin, eye, heart and kidney. A constellation of neurological phenotypes are present but quite variable and may include intellectual disability, seizures, developmental delays, lung and kidney disease, skin abnormalities, and behavioral problems. TSC is caused by a mutation of either human TSC1 or TSC2 genes (Crino et al., 2006; Curatolo et al., 2008). The TSC1 gene encodes the protein hamartin and TSC2 gene encodes protein Tuberin. Both proteins negatively regulate the mammalian target of rapamycin (mTOR) pathway by forming a heterodimeric complex with mTOR and thereby inhibit numerous intracellular signals that promote cell growth and proliferation. TSC patients typically have heterozygote germline mutations in TSC1/2. Seizures are common neurological symptoms in TSC patients while ASD and intellectual disability are seen in ~50% cases (Baker et al., 1998).
The importance of TSC1/2 in brain development is illustrated by the effects of mutating these genes in mice. Homozygous silencing mutations of either Tsc1 or Tsc2 results in embryonic lethality (Onda et al., 1999; Ma et al., 2014). Tsc1 null mice display liver and neural tube defects, while Tsc2 null mice have defects in the formation of the heart, neural tube and motor systems. Homozygote mutants for either gene die by embryonic day 12. In heterozygote Tsc1+/− mice, no spontaneous seizures or cerebral lesions are present, and dendritic spine number and branching appear normal. However, Tsc1+/− mice have impaired hippocampus-dependent learning tasks and social and cognitive deficits. Thus, haploinsufficiency for the Tsc1 genes leads to aberrations in neuronal function and results in impaired learning and social behavior (Goorden et al., 2007). Signaling by both Tsc1 and Tsc2 is also required for the expression of specific forms of hippocampal synaptic plasticity as well as the maintenance of normal excitatory synaptic strength. Single neuron deletion of a conditional Tsc1 allele in mouse CA1 neurons with Cre recombinase does not significantly affect spine density, morphology, or presynaptic release probability, but AMPA and NMDA receptor-mediated EPSCs and miniature spontaneous EPSC frequency are increased (Bateup et al., 2011). In addition, the protein synthesis-independent form of NMDA receptor-mediated LTD is normal, whereas the protein synthesis-dependent hippocampal mGluR-LTD is abolished by loss of Tsc1.
The Tsc1-mTOR pathway has been shown to promote network activity by repressing inhibitory synapses formation on excitatory neurons (Bateup et al., 2013). Deregulated mTOR activity causes weakened inhibition onto Tsc1 knockout neurons, alters E/I balance, results in hippocampal hyper-excitability. Mice with conditional knockout of Tsc1 in the hippocampus (CaMKIIα-cre, Tsc1fx/fx) display increased seizure severity that leads to premature death. Interestingly, Tsc1 expression is also required in astrocytes, but how Tsc1 operates in these glial cells to maintain normal brain function is not yet clear (Uhlmann et al., 2002).
In comparison, Tsc2+/− mice also have no overt neuropathology or seizures, but also display deficits in learning and memory (Ehninger et al., 2008). Increased hippocampal mTOR signaling in these mice correlates an enhancement of the late phase LTP in the CA1 region of the hippocampus, and deficits in hippocampal-dependent learning. Interesting, treatment of adult mice with the mTOR inhibitor rapamycin rescues not only the abnormal synaptic plasticity, but also the behavioral deficits in the Tsc2+/− mice (Ehninger et al., 2008).
It is noteworthy that some ASD gene may indirectly contribute to the impaired excitatory and inhibitory synaptic transmission and altered network activity through alterations of protein synthesis at the synapse. These genes include, but not limited to TSC1/2, FMR1, and UBE3A. Autism relevant pathogenesis induced by these genes may be a result of loss of normal constraints on activity-induced protein synthesis. In the case of FXS, this may be due to reduced inhibition of FMRP-regulated mRNA levels; in TSC, this might be due to de-repression of mTOR protein kinase acticity, which stimulates mRNA translation and leads to growth-related protein synthesis; in AS, there may be increased synaptic protein content due to reduced UBE3A-mediated protein ubiquitination and degradation. It has been proposed that childhood synaptic disorders, such as ASD, intellectual disability and seizures may be due to impaired regulation of protein abundance at certain neuronal compartments, and aberrant synaptic protein synthesis may represent one possible pathway leading to autistic phenotypes (Kelleher and Bear, 2008).
The human CNTNAP2 gene
The contactin-associated protein-like 2 protein (CNTNAP2) is encoded by the human CNTNAP2 gene. Genetic association and linkage studies all support the role of both common and rare variants of CNTNAP2 in ASD (Alarcón et al., 2008; Arking et al., 2008; Vernes et al., 2008). For example, a rare recessive mutation in CNTNAP2 results in a syndromic form of ASD termed cortical dysplasia-focal epilepsy syndrome (Strauss et al., 2006). CNTNAP2 is a member of the neurexin superfamily and is involved in neuron-glia interactions and clustering of potassium channels in axons in the vertebrate nervous system (Poliak et al., 2003).
CNTNAP2 protein plays additional important roles in brain development and neural circuit formation, including cell migration, axon guidance, and synaptic transmission velocity (related to the potassium channel function). These are critical events that when disturbed could lead to dysfunction of synapses and circuits that are involved in ASD (Poliak et al., 1999; Poliak et al., 2003; Peñagarikano et al., 2011). In the developing human brain, CNTNAP2 is expressed in structures known for the established roles in ASD pathophysiology, including frontal lobe, striatum, and thalamus, (Alarcón et al., 2008). Common genetic variants in CNTNAP2 are correlated with deficits of frontal lobe connectivity (Scott-Van Zeeland et al., 2010).
Cntnap2 mutant mice exhibit epilepsy, neuronal migration defects and some core autism-related behavior deficits (Peñagarikano et al., 2011). The laminar position of some cortical projection neurons is aberrant in Cntnap2 knockout mice. The number of cortical GABAergic interneurons is also reduced. These mutant mice also have less synchronous electrical activity of cortical networks. Critically, Cntnap2 mutant mice exhibit abnormal ultrasonic communications, as well as hyperactivity, seizures and repetitive behaviors. Cntnap2 knockout mice also demonstrated a predictive validity for modeling autism, such that the repetitive and restrictive behaviors can be partially corrected by risperidone, an FDA approved drug to treat ASD-related symptoms.
SYNGAP1
The human SYNGAP1 gene encodes a synaptic GTPase-activating protein (SynGAP) for Ras-like GTPases. SynGAP is selectively expressed in the brain and is highly enriched at excitatory synapses and dendritic spines (Chen et al., 1998; Kim et al., 1998). SYNGAP1 is a complex gene with multiple isoforms generated by various transcriptional start sites and alternative splicing of the C terminus (Chen et al., 1998). The SynGAP protein is a component of the PSD macromolecular complex and interacts with the PDZ domains of PSD-95 and SAP102. SynGAP can either stimulate or inhibit excitatory synapse function in an isoform-dependent manner (Rumbaugh et al., 2006; McMahon et al., 2012). SynGAP negatively regulates Ras activity at excitatory synapses by stimulating the GTPase activity of Ras, which may be involved in the modulation of excitatory synaptic transmission by neurotrophins and NMDA receptors (Kim et al., 1998; Komiyama et al., 2002; Krapivinsky et al., 2004).
De novo autosomal dominant truncating mutations in the SYNGAP1 gene have been reported to occur in ~3% of nonsyndromic intellectual disability cases (Hamdan et al., 2009; Hamdan et al., 2011; Writzl and Knegt, 2013). In two studies, patients with identified SYNGAP1 haploinsufficiency all show varying degree of nonsyndromic form of intellectual disability, and some of these patients were on the ASD spectrum (Pinto et al., 2010; Hamdan et al., 2011). Therefore, SYNGAP1 de novo mutations are highly penetrant and disruptive to normal brain function. Adult mice heterozygous for a null mutation of SynGAP1 exhibit only modest impairment in synaptic plasticity but some cognitive impairment (Komiyama et al., 2002). However, a recent study examining the developmental trajectory of SynGAP1 mutant mice identified that heterozygous SynGAP mutant mice develop excitatory synapses earlier than controls and this precocious maturation increases excitability in the developing hippocampus during a critical period when behavioral abnormalities are emerging. In contrast, SYNGAP1 mutations at adult stage had minimal impact on synapse function. Thus, the timeline of synapse maturation in early life and associated excitatory-inhibitory balance with neural circuitry are critical determinants of normal cognition and behavior.
Concluding remarks
To gain a better understanding of ASD brain pathophysiology and devise interventions and treatments for ASD requires translation of the identified genetic risk factors into biological mechanisms, particularly those mechanisms that are shared by multiple genetic risk factors. The discovery of genetic causes in several syndromic form ASD discussed here have improved our understanding of the molecular pathways operating at the synapse, and how disruption of these genes impairs proper synapse development and function. Knowledge in these areas can be largely attributed to the fact that single gene disorders can be readily modeled in mice, thus enabling biological mechanistic studies.
The evidence discussed here support the model that the wide phenotypic spectrum of ASD may derive from altered synaptic connection and/or impaired balance of excitatory/inhibitory local networks (‘synaptolopathies’ Zoghbi and Bear, 2012). Although currently more than 250 ASD risk factors have been discovered (Miles, 2011), and no doubt more awaits discovery, these disparate genetic risks may result in the disruption of a defined number of key cell types or circuits. Therefore, while both syndromic and idiopathic ASD may disrupt any one (or more) of many components in a number of signaling pathways that regulate neuronal cell specification, migration, gene transcriptions, synaptic protein synthesis, activity-dependent plasticity, and maintaining the delicate balance of local network excitatory/inhibitory network, by understanding how to modulate these synapses, cell types, or circuits, we may be able to compensate for preceding disruptions to neural circuitry independent of fully understanding the original insult (Bourgeron, 2009). Thus, ascertaining the synaptic and circuit substrates shared by ASD risk genes can be leveraged for the design of effective therapeutic interventions, a concept that will no doubt be a perennial theme in the field of autism research for the forthcoming decades.
Footnotes
Compliance with ethics guidelines
Aaron McGee, Guohui Li, Zhongming Lu and Shenfeng Qiu declare that they have no conflict of interest.
The manuscript is a review article and does not involve a research protocol requiring approval by the relevant institutional review board or ethics committee.
References
- Abrahams BS, Geschwind DH. Advances in autism genetics: on the threshold of a new neurobiology. Nat Rev Genet. 2008;9(5):341–355. doi: 10.1038/nrg2346. PMID:18414403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alarcón M, Abrahams BS, Stone JL, Duvall JA, Perederiy JV, Bomar JM, Sebat J, Wigler M, Martin CL, Ledbetter DH, Nelson SF, Cantor RM, Geschwind DH. Linkage, association, and gene-expression analyses identify CNTNAP2 as an autism-susceptibility gene. Am J Hum Genet. 2008;82(1):150–159. doi: 10.1016/j.ajhg.2007.09.005. PMID:18179893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amir RE, Van den Veyver IB, Wan M, Tran CQ, Francke U, Zoghbi HY. Rett syndrome is caused by mutations in X-linked MECP2, encoding methyl-CpG-binding protein 2. Nat Genet. 1999;23(2):185–188. doi: 10.1038/13810. PMID:10508514. [DOI] [PubMed] [Google Scholar]
- Arking DE, Cutler DJ, Brune CW, Teslovich TM, West K, Ikeda M, Rea A, Guy M, Lin S, Cook EH, Jr, Chakravarti A. A common genetic variant in the neurexin superfamily member CNTNAP2 increases familial risk of autism. Am J Hum Genet. 2008;82(1):160–164. doi: 10.1016/j.ajhg.2007.09.015. PMID:18179894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asaka Y, Jugloff DG, Zhang L, Eubanks JH, Fitzsimonds RM. Hippocampal synaptic plasticity is impaired in the Mecp2-null mouse model of Rett syndrome. Neurobiol Dis. 2006;21(1):217–227. doi: 10.1016/j.nbd.2005.07.005. PMID:16087343. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, Bolton P, Simonoff E, Yuzda E, Rutter M. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25(1):63–77. doi: 10.1017/s0033291700028099. PMID:7792363. [DOI] [PubMed] [Google Scholar]
- Baker JP. Autism at 70—redrawing the boundaries. N Engl J Med. 2013;369(12):1089–1091. doi: 10.1056/NEJMp1306380. PMID:24047057. [DOI] [PubMed] [Google Scholar]
- Baker P, Piven J, Sato Y. Autism and tuberous sclerosis complex: prevalence and clinical features. J Autism Dev Disord. 1998;28(4):279–285. doi: 10.1023/a:1026004501631. PMID:9711484. [DOI] [PubMed] [Google Scholar]
- Banerjee-Basu S, Packer A. SFARI gene: an evolving database for the autism research community. Dis Model Mech. 2010;3(3–4):133–135. doi: 10.1242/dmm.005439. PMID:20212079. [DOI] [PubMed] [Google Scholar]
- Bateup HS, Johnson CA, Denefrio CL, Saulnier JL, Kornacker K, Sabatini BL. Excitatory/inhibitory synaptic imbalance leads to hippocampal hyperexcitability in mouse models of tuberous sclerosis. Neuron. 2013;78(3):510–522. doi: 10.1016/j.neuron.2013.03.017. PMID:23664616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateup HS, Takasaki KT, Saulnier JL, Denefrio CL, Sabatini BL. Loss of Tsc1in vivo impairs hippocampal mGluR-LTD and increases excitatory synaptic function. J Neurosci. 2011;31(24):8862–8869. doi: 10.1523/JNEUROSCI.1617-11.2011. PMID:21677170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belichenko PV, Oldfors A, Hagberg B, Dahlström A. Rett syndrome: 3-D confocal microscopy of cortical pyramidal dendrites and afferents. Neuroreport. 1994;5(12):1509–1513. PMID:7948850. [PubMed] [Google Scholar]
- Belmonte MK, Bourgeron T. Fragile X syndrome and autism at the intersection of genetic and neural networks. Nat Neurosci. 2006;9(10):1221–1225. doi: 10.1038/nn1765. PMID:17001341. [DOI] [PubMed] [Google Scholar]
- Bonaglia MC, Giorda R, Mani E, Aceti G, Anderlid BM, Baroncini A, Pramparo T, Zuffardi O. Identification of a recurrent breakpoint within the SHANK3 gene in the 22q13.3 deletion syndrome. J Med Genet. 2006;43(10):822–828. doi: 10.1136/jmg.2005.038604. PMID:16284256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bostrom CA, Majaess NM, Morch K, White E, Eadie BD, Christie BR. Rescue of NMDAR-dependent synaptic plasticity in fmr1 knock-out mice. Cereb Cortex. 2013 doi: 10.1093/cercor/bht237. [DOI] [PubMed] [Google Scholar]
- Bourgeron T. A synaptic trek to autism. Curr Opin Neurobiol. 2009;19(2):231–234. doi: 10.1016/j.conb.2009.06.003. PMID:19545994. [DOI] [PubMed] [Google Scholar]
- Bureau I, Shepherd GM, Svoboda K. Circuit and plasticity defects in the developing somatosensory cortex of FMR1 knock-out mice. J Neurosci. 2008;28(20):5178–5188. doi: 10.1523/JNEUROSCI.1076-08.2008. PMID:18480274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden D, Switala A, Roy E. Rett syndrome as a minicolumnopathy. Clin Neuropathol. 2003;22(4):163–168. PMID:12908751. [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden DP, Switala AE, Roy E. Minicolumnar pathology in autism. Neurology. 2002;58(3):428–432. doi: 10.1212/wnl.58.3.428. PMID:11839843. [DOI] [PubMed] [Google Scholar]
- Casanova MF, El-Baz A, Vanbogaert E, Narahari P, Switala A. A topographic study of minicolumnar core width by lamina comparison between autistic subjects and controls: possible minicolumnar disruption due to an anatomical element in-common to multiple laminae. Brain Pathol. 2010;20(2):451–458. doi: 10.1111/j.1750-3639.2009.00319.x. PMID:19725830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chahrour M, Zoghbi HY. The story of Rett syndrome: from clinic to neurobiology. Neuron. 2007;56(3):422–437. doi: 10.1016/j.neuron.2007.10.001. PMID:17988628. [DOI] [PubMed] [Google Scholar]
- Chao HT, Chen H, Samaco RC, Xue M, Chahrour M, Yoo J, Neul JL, Gong S, Lu HC, Heintz N, Ekker M, Rubenstein JL, Noebels JL, Rosenmund C, Zoghbi HY. Dysfunction in GABA signalling mediates autism-like stereotypies and Rett syndrome phenotypes. Nature. 2010;468(7321):263–269. doi: 10.1038/nature09582. PMID:21068835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen HJ, Rojas-Soto M, Oguni A, Kennedy MB. A synaptic Ras-GTPase activating protein (p135 SynGAP) inhibited by CaM kinase II. Neuron. 1998;20(5):895–904. doi: 10.1016/s0896-6273(00)80471-7. PMID:9620694. [DOI] [PubMed] [Google Scholar]
- Clement JP, Aceti M, Creson TK, Ozkan ED, Shi Y, Reish NJ, Almonte AG, Miller BH, Wiltgen BJ, Miller CA, Xu X, Rumbaugh G. Pathogenic SYNGAP1 mutations impair cognitive development by disrupting maturation of dendritic spine synapses. Cell. 2012;151(4):709–723. doi: 10.1016/j.cell.2012.08.045. PMID:23141534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clowry G, Molnár Z, Rakic P. Renewed focus on the developing human neocortex. J Anat. 2010;217(4):276–288. doi: 10.1111/j.1469-7580.2010.01281.x. PMID:20979582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comery TA, Harris JB, Willems PJ, Oostra BA, Irwin SA, Weiler IJ, Greenough WT. Abnormal dendritic spines in fragile X knockout mice: maturation and pruning deficits. Proc Natl Acad Sci USA. 1997;94(10):5401–5404. doi: 10.1073/pnas.94.10.5401. PMID:9144249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cook EH, Jr, Courchesne RY, Cox NJ, Lord C, Gonen D, Guter SJ, Lincoln A, Nix K, Haas R, Leventhal BL, Courchesne E. Linkage-disequilibrium mapping of autistic disorder, with 15q11-13 markers. Am J Hum Genet. 1998;62(5):1077–1083. doi: 10.1086/301832. PMID:9545402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courchesne E, Pierce K. Why the frontal cortex in autism might be talking only to itself: local over-connectivity but long-distance disconnection. Curr Opin Neurobiol. 2005;15(2):225–230. doi: 10.1016/j.conb.2005.03.001. PMID:15831407. [DOI] [PubMed] [Google Scholar]
- Crino PB, Nathanson KL, Henske EP. The tuberous sclerosis complex. N Engl J Med. 2006;355(13):1345–1356. doi: 10.1056/NEJMra055323. PMID:17005952. [DOI] [PubMed] [Google Scholar]
- Cruz-Martín A, Crespo M, Portera-Cailliau C. Delayed stabilization of dendritic spines in fragile X mice. J Neurosci. 2010;30(23):7793–7803. doi: 10.1523/JNEUROSCI.0577-10.2010. PMID:20534828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Curatolo P, Bombardieri R, Jozwiak S. Tuberous sclerosis. Lancet. 2008;372(9639):657–668. doi: 10.1016/S0140-6736(08)61279-9. PMID:18722871. [DOI] [PubMed] [Google Scholar]
- Curia G, Papouin T, Séguéla P, Avoli M. Downregulation of tonic GABAergic inhibition in a mouse model of fragile X syndrome. Cereb Cortex. 2009;19(7):1515–1520. doi: 10.1093/cercor/bhn159. PMID:18787232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dölen G, Osterweil E, Rao BS, Smith GB, Auerbach BD, Chattarji S, Bear MF. Correction of fragile X syndrome in mice. Neuron. 2007;56(6):955–962. doi: 10.1016/j.neuron.2007.12.001. PMID:18093519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Betancur C, Boeckers TM, Bockmann J, Chaste P, Fauchereau F, Nygren G, Rastam M, Gillberg IC, Anckarsäter H, Sponheim E, Goubran-Botros H, Delorme R, Chabane N, Mouren-Simeoni MC, de Mas P, Bieth E, Rogé B, Héron D, Burglen L, Gillberg C, Leboyer M, Bourgeron T. Mutations in the gene encoding the synaptic scaffolding protein SHANK3 are associated with autism spectrum disorders. Nat Genet. 2007;39(1):25–27. doi: 10.1038/ng1933. PMID:17173049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand CM, Perroy J, Loll F, Perrais D, Fagni L, Bourgeron T, Montcouquiol M, Sans N. SHANK3 mutations identified in autism lead to modification of dendritic spine morphology via an actin-dependent mechanism. Mol Psychiatry. 2011;17(1):71–84. doi: 10.1038/mp.2011.57. PMID:21606927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehninger D, Han S, Shilyansky C, Zhou Y, Li W, Kwiatkowski DJ, Ramesh V, Silva AJ. Reversal of learning deficits in a Tsc2+/− mouse model of tuberous sclerosis. Nat Med. 2008;14(8):843–848. doi: 10.1038/nm1788. PMID:18568033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eigsti IM, Shapiro T. A systems neuroscience approach to autism: biological, cognitive, and clinical perspectives. Ment Retard Dev Disabil Res Rev. 2003;9(3):205–215. doi: 10.1002/mrdd.10081. PMID:12953300. [DOI] [PubMed] [Google Scholar]
- Fombonne E, Rogé B, Claverie J, Courty S, Frémolle J. Microcephaly and macrocephaly in autism. J Autism Dev Disord. 1999;29(2):113–119. doi: 10.1023/a:1023036509476. PMID:10382131. [DOI] [PubMed] [Google Scholar]
- Gauthier J, Spiegelman D, Piton A, Lafrenière RG, Laurent S, St-Onge J, Lapointe L, Hamdan FF, Cossette P, Mottron L, Fombonne E, Joober R, Marineau C, Drapeau P, Rouleau GA. Novel de novo SHANK3 mutation in autistic patients. Am J Med Genet B Neuropsychiatr Genet. 2009;150B(3):421–424. doi: 10.1002/ajmg.b.30822. PMID:18615476. [DOI] [PubMed] [Google Scholar]
- Geschwind DH, Levitt P. Autism spectrum disorders: developmental disconnection syndromes. Curr Opin Neurobiol. 2007;17(1):103–111. doi: 10.1016/j.conb.2007.01.009. PMID:17275283. [DOI] [PubMed] [Google Scholar]
- Giacometti E, Luikenhuis S, Beard C, Jaenisch R. Partial rescue of MeCP2 deficiency by postnatal activation of MeCP2. Proc Natl Acad Sci USA. 2007;104(6):1931–1936. doi: 10.1073/pnas.0610593104. PMID:17267601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson JR, Bartley AF, Hays SA, Huber KM. Imbalance of neocortical excitation and inhibition and altered UP states reflect network hyperexcitability in the mouse model of fragile X syndrome. J Neurophysiol. 2008;100(5):2615–2626. doi: 10.1152/jn.90752.2008. PMID:18784272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonçalves JT, Anstey JE, Golshani P, Portera-Cailliau C. Circuit level defects in the developing neocortex of Fragile X mice. Nat Neurosci. 2013;16(7):903–909. doi: 10.1038/nn.3415. PMID:23727819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goorden SM, van Woerden GM, van der Weerd L, Cheadle JP, Elgersma Y. Cognitive deficits in Tsc1+/− mice in the absence of cerebral lesions and seizures. Ann Neurol. 2007;62(6):648–655. doi: 10.1002/ana.21317. PMID:18067135. [DOI] [PubMed] [Google Scholar]
- Greer PL, Hanayama R, Bloodgood BL, Mardinly AR, Lipton DM, Flavell SW, Kim TK, Griffith EC, Waldon Z, Maehr R, Ploegh HL, Chowdhury S, Worley PF, Steen J, Greenberg ME. The Angelman Syndrome protein Ube3A regulates synapse development by ubiquitinating arc. Cell. 2010;140(5):704–716. doi: 10.1016/j.cell.2010.01.026. PMID:20211139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy J, Hendrich B, Holmes M, Martin JE, Bird A. A mouse Mecp2-null mutation causes neurological symptoms that mimic Rett syndrome. Nat Genet. 2001;27(3):322–326. doi: 10.1038/85899. PMID:11242117. [DOI] [PubMed] [Google Scholar]
- Hallmayer J, Cleveland S, Torres A, Phillips J, Cohen B, Torigoe T, Miller J, Fedele A, Collins J, Smith K, Lotspeich L, Croen LA, Ozonoff S, Lajonchere C, Grether JK, Risch N. Genetic heritability and shared environmental factors among twin pairs with autism. Arch Gen Psychiatry. 2011;68(11):1095–1102. doi: 10.1001/archgenpsychiatry.2011.76. PMID:21727249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamdan FF, Daoud H, Piton A, Gauthier J, Dobrzeniecka S, Krebs MO, Joober R, Lacaille JC, Nadeau A, Milunsky JM, Wang Z, Carmant L, Mottron L, Beauchamp MH, Rouleau GA, Michaud JL. De novo SYNGAP1 mutations in nonsyndromic intellectual disability and autism. Biol Psychiatry. 2011;69(9):898–901. doi: 10.1016/j.biopsych.2010.11.015. PMID:21237447. [DOI] [PubMed] [Google Scholar]
- Hamdan FF, Gauthier J, Spiegelman D, Noreau A, Yang Y, Pellerin S, Dobrzeniecka S, Côté M, Perreau-Linck E, Carmant L, D’Anjou G, Fombonne E, Addington AM, Rapoport JL, Delisi LE, Krebs MO, Mouaffak F, Joober R, Mottron L, Drapeau P, Marineau C, Lafrenière RG, Lacaille JC, Rouleau GA, Michaud JL the Synapse to Disease Group. Mutations in SYNGAP1 in autosomal nonsyndromic mental retardation. N Engl J Med. 2009;360(6):599–605. doi: 10.1056/NEJMoa0805392. PMID:19196676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi MK, Tang C, Verpelli C, Narayanan R, Stearns MH, Xu RM, Li H, Sala C, Hayashi Y. The postsynaptic density proteins Homer and Shank form a polymeric network structure. Cell. 2009;137(1):159–171. doi: 10.1016/j.cell.2009.01.050. PMID:19345194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S, Ke X, Tang T, Hang Y, Chu K, Huang H, Ruan Z, Lu Z, Tao G, Liu Y. Detecting abnormalities of corpus callosum connectivity in autism using magnetic resonance imaging and diffusion tensor tractography. Psychiatry Res. 2011;194(3):333–339. doi: 10.1016/j.pscychresns.2011.03.009. PMID:22047729. [DOI] [PubMed] [Google Scholar]
- Hsiao EY. Immune dysregulation in autism spectrum disorder. Int Rev Neurobiol. 2013;113:269–302. doi: 10.1016/B978-0-12-418700-9.00009-5. PMID:24290389. [DOI] [PubMed] [Google Scholar]
- Hsiao EY, McBride SW, Hsien S, Sharon G, Hyde ER, McCue T, Codelli JA, Chow J, Reisman SE, Petrosino JF, Patterson PH, Mazmanian SK. Microbiota modulate behavioral and physiological abnormalities associated with neurodevelopmental disorders. Cell. 2013;155(7):1451–1463. doi: 10.1016/j.cell.2013.11.024. PMID:24315484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber KM, Gallagher SM, Warren ST, Bear MF. Altered synaptic plasticity in a mouse model of fragile X mental retardation. Proc Natl Acad Sci USA. 2002;99(11):7746–7750. doi: 10.1073/pnas.122205699. PMID:12032354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hutsler JJ, Zhang H. Increased dendritic spine densities on cortical projection neurons in autism spectrum disorders. Brain Res. 2010;1309:83–94. doi: 10.1016/j.brainres.2009.09.120. PMID:19896929. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Galvez R, Greenough WT. Dendritic spine structural anomalies in fragile-X mental retardation syndrome. Cereb Cortex. 2000;10(10):1038–1044. doi: 10.1093/cercor/10.10.1038. PMID:11007554. [DOI] [PubMed] [Google Scholar]
- Irwin SA, Patel B, Idupulapati M, Harris JB, Crisostomo RA, Larsen BP, Kooy F, Willems PJ, Cras P, Kozlowski PB, Swain RA, Weiler IJ, Greenough WT. Abnormal dendritic spine characteristics in the temporal and visual cortices of patients with fragile-X syndrome: a quantitative examination. Am J Med Genet. 2001;98(2):161–167. doi: 10.1002/1096-8628(20010115)98:2<161::aid-ajmg1025>3.0.co;2-b. PMID:11223852. [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Betancur C, Råstam M, Colineaux C, Gillberg IC, Soderstrom H, Giros B, Leboyer M, Gillberg C, Bourgeron T, Gillberg C, Råstam M, Gillberg C, Nydén A, Söderström H, Leboyer M, Betancur C, Philippe A, Giros B, Colineaux C, Cohen D, Chabane N, Mouren-Siméoni MC, Brice A, Sponheim E, Spurkland I, Skjeldal OH, Coleman M, Pearl PL, Cohen IL, Tsiouris J, Zappella M, Menchetti G, Pompella A, Aschauer H, Van Maldergem L the Paris Autism Research International Sibpair Study. Mutations of the X-linked genes encoding neuroligins NLGN3 and NLGN4 are associated with autism. Nat Genet. 2003;34(1):27–29. doi: 10.1038/ng1136. PMID:12669065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Y, Tsai TF, Bressler J, Beaudet AL. Imprinting in Angelman and Prader-Willi syndromes. Curr Opin Genet Dev. 1998a;8(3):334–342. doi: 10.1016/s0959-437x(98)80091-9. PMID:9691003. [DOI] [PubMed] [Google Scholar]
- Jiang YH, Armstrong D, Albrecht U, Atkins CM, Noebels JL, Eichele G, Sweatt JD, Beaudet AL. Mutation of the Angelman ubiquitin ligase in mice causes increased cytoplasmic p53 and deficits of contextual learning and long-term potentiation. Neuron. 1998b;21(4):799–811. doi: 10.1016/s0896-6273(00)80596-6. PMID:9808466. [DOI] [PubMed] [Google Scholar]
- Just MA, Cherkassky VL, Keller TA, Kana RK, Minshew NJ. Functional and anatomical cortical underconnectivity in autism: evidence from an FMRI study of an executive function task and corpus callosum morphometry. Cereb Cortex. 2007;17(4):951–961. doi: 10.1093/cercor/bhl006. PMID:16772313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Just MA, Cherkassky VL, Keller TA, Minshew NJ. Cortical activation and synchronization during sentence comprehension in high-functioning autism: evidence of underconnectivity. Brain. 2004;127(Pt 8):1811–1821. doi: 10.1093/brain/awh199. PMID:15215213. [DOI] [PubMed] [Google Scholar]
- Kana RK, Keller TA, Cherkassky VL, Minshew NJ, Just MA. Atypical frontal-posterior synchronization of Theory of Mind regions in autism during mental state attribution. Soc Neurosci. 2009;4(2):135–152. doi: 10.1080/17470910802198510. PMID:18633829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanner L. Autistic disturbances of affective contact. Nervous Child. 1943;2:217–250. [PubMed] [Google Scholar]
- Kattenstroth G, Tantalaki E, Südhof TC, Gottmann K, Missler M. Postsynaptic N-methyl-D-aspartate receptor function requires alpha-neurexins. Proc Natl Acad Sci USA. 2004;101(8):2607–2612. doi: 10.1073/pnas.0308626100. PMID:14983056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleher RJ, 3rd, Bear MF. The autistic neuron: troubled translation? Cell. 2008;135(3):401–406. doi: 10.1016/j.cell.2008.10.017. PMID:18984149. [DOI] [PubMed] [Google Scholar]
- Kim HG, Kishikawa S, Higgins AW, Seong IS, Donovan DJ, Shen Y, Lally E, Weiss LA, Najm J, Kutsche K, Descartes M, Holt L, Braddock S, Troxell R, Kaplan L, Volkmar F, Klin A, Tsatsanis K, Harris DJ, Noens I, Pauls DL, Daly MJ, MacDonald ME, Morton CC, Quade BJ, Gusella JF. Disruption of neurexin 1 associated with autism spectrum disorder. Am J Hum Genet. 2008;82(1):199–207. doi: 10.1016/j.ajhg.2007.09.011. PMID:18179900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JH, Liao D, Lau LF, Huganir RL. SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron. 1998;20(4):683–691. doi: 10.1016/s0896-6273(00)81008-9. PMID:9581761. [DOI] [PubMed] [Google Scholar]
- Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat Genet. 1997;15(1):70–73. doi: 10.1038/ng0197-70. PMID:8988171. [DOI] [PubMed] [Google Scholar]
- Komiyama NH, Watabe AM, Carlisle HJ, Porter K, Charlesworth P, Monti J, Strathdee DJ, O’Carroll CM, Martin SJ, Morris RG, O’Dell TJ, Grant SG. SynGAP regulates ERK/MAPK signaling, synaptic plasticity, and learning in the complex with postsynaptic density 95 and NMDA receptor. J Neurosci. 2002;22(22):9721–9732. doi: 10.1523/JNEUROSCI.22-22-09721.2002. PMID:12427827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapivinsky G, Medina I, Krapivinsky L, Gapon S, Clapham DE. SynGAP-MUPP1-CaMKII synaptic complexes regulate p38 MAP kinase activity and NMDA receptor-dependent synaptic AMPA receptor potentiation. Neuron. 2004;43(4):563–574. doi: 10.1016/j.neuron.2004.08.003. PMID:15312654. [DOI] [PubMed] [Google Scholar]
- Lainhart JE. Increased rate of head growth during infancy in autism. JAMA. 2003;290(3):393–394. doi: 10.1001/jama.290.3.393. PMID:12865381. [DOI] [PubMed] [Google Scholar]
- Landa RJ, Holman KC, Garrett-Mayer E. Social and communication development in toddlers with early and later diagnosis of autism spectrum disorders. Arch Gen Psychiatry. 2007;64(7):853–864. doi: 10.1001/archpsyc.64.7.853. PMID:17606819. [DOI] [PubMed] [Google Scholar]
- Laumonnier F, Bonnet-Brilhault F, Gomot M, Blanc R, David A, Moizard MP, Raynaud M, Ronce N, Lemonnier E, Calvas P, Laudier B, Chelly J, Fryns JP, Ropers HH, Hamel BC, Andres C, Barthélémy C, Moraine C, Briault S. X-linked mental retardation and autism are associated with a mutation in the NLGN4 gene, a member of the neuroligin family. Am J Hum Genet. 2004;74(3):552–557. doi: 10.1086/382137. PMID:14963808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung AK, Robson WL. Tuberous sclerosis complex: a review. J Pediatr Health Care. 2007;21(2):108–114. doi: 10.1016/j.pedhc.2006.05.004. PMID:17321910. [DOI] [PubMed] [Google Scholar]
- Li J, Pelletier MR, Perez Velazquez JL, Carlen PL. Reduced cortical synaptic plasticity and GluR1 expression associated with fragile X mental retardation protein deficiency. Mol Cell Neurosci. 2002;19(2):138–151. doi: 10.1006/mcne.2001.1085. PMID:11860268. [DOI] [PubMed] [Google Scholar]
- Ma A, Wang L, Gao Y, Chang Z, Peng H, Zeng N, Gui YS, Tian X, Li X, Cai B, Zhang H, Xu KF. Tsc1 deficiency-mediated mTOR hyperactivation in vascular endothelial cells causes angiogenesis defects and embryonic lethality. Hum Mol Genet. 2014;23(3):693–705. doi: 10.1093/hmg/ddt456. PMID:24129405. [DOI] [PubMed] [Google Scholar]
- Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat Genet. 1997;15(1):74–77. doi: 10.1038/ng0197-74. PMID:8988172. [DOI] [PubMed] [Google Scholar]
- McGraw CM, Samaco RC, Zoghbi HY. Adult neural function requires MeCP2. Science. 2011;333(6039):186. doi: 10.1126/science.1206593. PMID:21636743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon AC, Barnett MW, O’Leary TS, Stoney PN, Collins MO, Papadia S, Choudhary JS, Komiyama NH, Grant SG, Hardingham GE, Wyllie DJ, Kind PC. SynGAP isoforms exert opposing effects on synaptic strength. Nat Commun. 2012;3:900. doi: 10.1038/ncomms1900. PMID:22692543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer G, Varoqueaux F, Neeb A, Oschlies M, Brose N. The complexity of PDZ domain-mediated interactions at glutamatergic synapses: a case study on neuroligin. Neuropharmacology. 2004;47(5):724–733. doi: 10.1016/j.neuropharm.2004.06.023. PMID:15458844. [DOI] [PubMed] [Google Scholar]
- Miles JH. Autism spectrum disorders—a genetics review. Genet Med. 2011;13(4):278–294. doi: 10.1097/GIM.0b013e3181ff67ba. PMID:21358411. [DOI] [PubMed] [Google Scholar]
- Moretti P, Levenson JM, Battaglia F, Atkinson R, Teague R, Antalffy B, Armstrong D, Arancio O, Sweatt JD, Zoghbi HY. Learning and memory and synaptic plasticity are impaired in a mouse model of Rett syndrome. J Neurosci. 2006;26(1):319–327. doi: 10.1523/JNEUROSCI.2623-05.2006. PMID:16399702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moy SS, Nadler JJ. Advances in behavioral genetics: mouse models of autism. Mol Psychiatry. 2008;13(1):4–26. doi: 10.1038/sj.mp.4002082. PMID:17848915. [DOI] [PubMed] [Google Scholar]
- Nakatani J, Tamada K, Hatanaka F, Ise S, Ohta H, Inoue K, Tomonaga S, Watanabe Y, Chung YJ, Banerjee R, Iwamoto K, Kato T, Okazawa M, Yamauchi K, Tanda K, Takao K, Miyakawa T, Bradley A, Takumi T. Abnormal behavior in a chromosome-engineered mouse model for human 15q11-13 duplication seen in autism. Cell. 2009;137(7):1235–1246. doi: 10.1016/j.cell.2009.04.024. PMID:19563756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nestler EJ, Hyman SE. Animal models of neuropsychiatric disorders. Nat Neurosci. 2010;13(10):1161–1169. doi: 10.1038/nn.2647. PMID:20877280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neul JL, Fang P, Barrish J, Lane J, Caeg EB, Smith EO, Zoghbi H, Percy A, Glaze DG. Specific mutations in methyl-CpG-binding protein 2 confer different severity in Rett syndrome. Neurology. 2008;70(16):1313–1321. doi: 10.1212/01.wnl.0000291011.54508.aa. PMID:18337588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamoto N, Kubota T, Nakamura Y, Murakami R, Nishikubo T, Tanaka I, Takahashi Y, Hayashi S, Imoto I, Inazawa J, Hosokai N, Kohsaka S, Uchino S. 22q13 Microduplication in two patients with common clinical manifestations: a recognizable syndrome? Am J Med Genet A. 2007;143A(23):2804–2809. doi: 10.1002/ajmg.a.31771. PMID:17975801. [DOI] [PubMed] [Google Scholar]
- Olmos-Serrano JL, Paluszkiewicz SM, Martin BS, Kaufmann WE, Corbin JG, Huntsman MM. Defective GABAergic neurotransmission and pharmacological rescue of neuronal hyperexcitability in the amygdala in a mouse model of fragile X syndrome. J Neurosci. 2010;30(29):9929–9938. doi: 10.1523/JNEUROSCI.1714-10.2010. PMID:20660275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onda H, Lueck A, Marks PW, Warren HB, Kwiatkowski DJ. Tsc2(+/−) mice develop tumors in multiple sites that express gelsolin and are influenced by genetic background. J Clin Invest. 1999;104(6):687–695. doi: 10.1172/JCI7319. PMID:10491404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterweil EK, Chuang SC, Chubykin AA, Sidorov M, Bianchi R, Wong RK, Bear MF. Lovastatin corrects excess protein synthesis and prevents epileptogenesis in a mouse model of fragile X syndrome. Neuron. 2013;77(2):243–250. doi: 10.1016/j.neuron.2012.01.034. PMID:23352161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterweil EK, Krueger DD, Reinhold K, Bear MF. Hypersensitivity to mGluR5 and ERK1/2 leads to excessive protein synthesis in the hippocampus of a mouse model of fragile X syndrome. J Neurosci. 2010;30(46):15616–15627. doi: 10.1523/JNEUROSCI.3888-10.2010. PMID:21084617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Padmashri R, Reiner BC, Suresh A, Spartz E, Dunaevsky A. Altered structural and functional synaptic plasticity with motor skill learning in a mouse model of fragile X syndrome. J Neurosci. 2013;33(50):19715–19723. doi: 10.1523/JNEUROSCI.2514-13.2013. PMID:24336735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan F, Aldridge GM, Greenough WT, Gan WB. Dendritic spine instability and insensitivity to modulation by sensory experience in a mouse model of fragile X syndrome. Proc Natl Acad Sci USA. 2010;107(41):17768–17773. doi: 10.1073/pnas.1012496107. PMID:20861447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker-Athill EC, Tan J. Maternal immune activation and autism spectrum disorder: interleukin-6 signaling as a key mechanistic pathway. Neurosignals. 2010;18(2):113–128. doi: 10.1159/000319828. PMID:20924155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peça J, Feliciano C, Ting JT, Wang W, Wells MF, Venkatraman TN, Lascola CD, Fu Z, Feng G. Shank3 mutant mice display autistic-like behaviours and striatal dysfunction. Nature. 2011;472(7344):437–442. doi: 10.1038/nature09965. PMID:21423165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peñagarikano O, Abrahams BS, Herman EI, Winden KD, Gdalyahu A, Dong H, Sonnenblick LI, Gruver R, Almajano J, Bragin A, Golshani P, Trachtenberg JT, Peles E, Geschwind DH. Absence of CNTNAP2 leads to epilepsy, neuronal migration abnormalities, and core autism-related deficits. Cell. 2011;147(1):235–246. doi: 10.1016/j.cell.2011.08.040. PMID:21962519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeiffer BE, Huber KM. Fragile X mental retardation protein induces synapse loss through acute postsynaptic translational regulation. J Neurosci. 2007;27(12):3120–3130. doi: 10.1523/JNEUROSCI.0054-07.2007. PMID:17376973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre G. Neurodegenerative disorders and metabolic disease. Arch Dis Child. 2013;98(8):618–624. doi: 10.1136/archdischild-2012-302840. PMID:23698595. [DOI] [PubMed] [Google Scholar]
- Piggot J, Shirinyan D, Shemmassian S, Vazirian S, Alarcón M. Neural systems approaches to the neurogenetics of autism spectrum disorders. Neuroscience. 2009;164(1):247–256. doi: 10.1016/j.neuroscience.2009.05.054. PMID:19482063. [DOI] [PubMed] [Google Scholar]
- Pinto D, Pagnamenta AT, Klei L, Anney R, Merico D, Regan R, Conroy J, Magalhaes TR, Correia C, Abrahams BS, Almeida J, Bacchelli E, Bader GD, Bailey AJ, Baird G, Battaglia A, Berney T, Bolshakova N, Bölte S, Bolton PF, Bourgeron T, Brennan S, Brian J, Bryson SE, Carson AR, Casallo G, Casey J, Chung BH, Cochrane L, Corsello C, Crawford EL, Crossett A, Cytrynbaum C, Dawson G, de Jonge M, Delorme R, Drmic I, Duketis E, Duque F, Estes A, Farrar P, Fernandez BA, Folstein SE, Fombonne E, Freitag CM, Gilbert J, Gillberg C, Glessner JT, Goldberg J, Green A, Green J, Guter SJ, Hakonarson H, Heron EA, Hill M, Holt R, Howe JL, Hughes G, Hus V, Igliozzi R, Kim C, Klauck SM, Kolevzon A, Korvatska O, Kustanovich V, Lajonchere CM, Lamb JA, Laskawiec M, Leboyer M, Le Couteur A, Leventhal BL, Lionel AC, Liu XQ, Lord C, Lotspeich L, Lund SC, Maestrini E, Mahoney W, Mantoulan C, Marshall CR, McConachie H, McDougle CJ, McGrath J, McMahon WM, Merikangas A, Migita O, Minshew NJ, Mirza GK, Munson J, Nelson SF, Noakes C, Noor A, Nygren G, Oliveira G, Papanikolaou K, Parr JR, Parrini B, Paton T, Pickles A, Pilorge M, Piven J, Ponting CP, Posey DJ, Poustka A, Poustka F, Prasad A, Ragoussis J, Renshaw K, Rickaby J, Roberts W, Roeder K, Roge B, Rutter ML, Bierut LJ, Rice JP, Salt J, Sansom K, Sato D, Segurado R, Sequeira AF, Senman L, Shah N, Sheffield VC, Soorya L, Sousa I, Stein O, Sykes N, Stoppioni V, Strawbridge C, Tancredi R, Tansey K, Thiruvahindrapduram B, Thompson AP, Thomson S, Tryfon A, Tsiantis J, Van Engeland H, Vincent JB, Volkmar F, Wallace S, Wang K, Wang Z, Wassink TH, Webber C, Weksberg R, Wing K, Wittemeyer K, Wood S, Wu J, Yaspan BL, Zurawiecki D, Zwaigenbaum L, Buxbaum JD, Cantor RM, Cook EH, Coon H, Cuccaro ML, Devlin B, Ennis S, Gallagher L, Geschwind DH, Gill M, Haines JL, Hallmayer J, Miller J, Monaco AP, Nurnberger JI, Jr, Paterson AD, Pericak-Vance MA, Schellenberg GD, Szatmari P, Vicente AM, Vieland VJ, Wijsman EM, Scherer SW, Sutcliffe JS, Betancur C. Functional impact of global rare copy number variation in autism spectrum disorders. Nature. 2010;466(7304):368–372. doi: 10.1038/nature09146. PMID:20531469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poliak S, Gollan L, Martinez R, Custer A, Einheber S, Salzer JL, Trimmer JS, Shrager P, Peles E. Caspr2, a new member of the neurexin superfamily, is localized at the juxtaparanodes of myelinated axons and associates with K+ channels. Neuron. 1999;24(4):1037–1047. doi: 10.1016/s0896-6273(00)81049-1. PMID:10624965. [DOI] [PubMed] [Google Scholar]
- Poliak S, Salomon D, Elhanany H, Sabanay H, Kiernan B, Pevny L, Stewart CL, Xu X, Chiu SY, Shrager P, Furley AJ, Peles E. Juxtaparanodal clustering of Shaker-like K+ channels in myelinated axons depends on Caspr2 and TAG-1. J Cell Biol. 2003;162(6):1149–1160. doi: 10.1083/jcb.200305018. PMID:12963709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin M, Kang J, Burlin TV, Jiang C, Smith CB. Postadolescent changes in regional cerebral protein synthesis: an in vivo study in the FMR1 null mouse. J Neurosci. 2005;25(20):5087–5095. doi: 10.1523/JNEUROSCI.0093-05.2005. PMID:15901791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricciardi S, Boggio EM, Grosso S, Lonetti G, Forlani G, Stefanelli G, Calcagno E, Morello N, Landsberger N, Biffo S, Pizzorusso T, Giustetto M, Broccoli V. Reduced AKT/mTOR signaling and protein synthesis dysregulation in a Rett syndrome animal model. Hum Mol Genet. 2011;20(6):1182–1196. doi: 10.1093/hmg/ddq563. PMID:21212100. [DOI] [PubMed] [Google Scholar]
- Riday TT, Dankoski EC, Krouse MC, Fish EW, Walsh PL, Han JE, Hodge CW, Wightman RM, Philpot BD, Malanga CJ. Pathway-specific dopaminergic deficits in a mouse model of Angelman syndrome. J Clin Invest. 2012;122(12):4544–4554. doi: 10.1172/JCI61888. PMID:23143301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenblueth AWN, Wiener N. The role of models in science. Philos Sci. 1945;12(4):316–321. [Google Scholar]
- Roussignol G, Ango F, Romorini S, Tu JC, Sala C, Worley PF, Bockaert J, Fagni L. Shank expression is sufficient to induce functional dendritic spine synapses in aspiny neurons. J Neurosci. 2005;25(14):3560–3570. doi: 10.1523/JNEUROSCI.4354-04.2005. PMID:15814786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubenstein JL. Three hypotheses for developmental defects that may underlie some forms of autism spectrum disorder. Curr Opin Neurol. 2010;23(2):118–123. doi: 10.1097/WCO.0b013e328336eb13. PMID:20087182. [DOI] [PubMed] [Google Scholar]
- Rubenstein JL, Merzenich MM. Model of autism: increased ratio of excitation/inhibition in key neural systems. Genes Brain Behav. 2003;2(5):255–267. doi: 10.1034/j.1601-183x.2003.00037.x. PMID:14606691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rumbaugh G, Adams JP, Kim JH, Huganir RL. SynGAP regulates synaptic strength and mitogen-activated protein kinases in cultured neurons. Proc Natl Acad Sci USA. 2006;103(12):4344–4351. doi: 10.1073/pnas.0600084103. PMID:16537406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahyoun CP, Belliveau JW, Soulières I, Schwartz S, Mody M. Neuroimaging of the functional and structural networks underlying visuospatial vs. linguistic reasoning in high-functioning autism. Neuropsychologia. 2010;48(1):86–95. doi: 10.1016/j.neuropsychologia.2009.08.013. PMID:19698726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott-Van Zeeland AA, Abrahams BS, Alvarez-Retuerto AI, Sonnenblick LI, Rudie JD, Ghahremani D, Mumford JA, Poldrack RA, Dapretto M, Geschwind DH, Bookheimer SY. Altered functional connectivity in frontal lobe circuits is associated with variation in the autism risk gene CNTNAP2. Sci Transl Med. 2010;2(56):56ra80. doi: 10.1126/scitranslmed.3001344. PMID:21048216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheng M, Kim E. The Shank family of scaffold proteins. J Cell Sci. 2000;113(Pt 11):1851–1856. doi: 10.1242/jcs.113.11.1851. PMID:10806096. [DOI] [PubMed] [Google Scholar]
- Shukla DK, Keehn B, Müller RA. Tract-specific analyses of diffusion tensor imaging show widespread white matter compromise in autism spectrum disorder. J Child Psychol Psychiatry. 2011a;52(3):286–295. doi: 10.1111/j.1469-7610.2010.02342.x. PMID:21073464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shukla DK, Keehn B, Smylie DM, Müller RA. Microstructural abnormalities of short-distance white matter tracts in autism spectrum disorder. Neuropsychologia. 2011b;49(5):1378–1382. doi: 10.1016/j.neuropsychologia.2011.02.022. PMID:21333661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman JL, Yang M, Lord C, Crawley JN. Behavioural phenotyping assays for mouse models of autism. Nat Rev Neurosci. 2010;11(7):490–502. doi: 10.1038/nrn2851. PMID:20559336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steffenburg S, Gillberg C, Hellgren L, Andersson L, Gillberg IC, Jakobsson G, Bohman M. A twin study of autism in Denmark, Finland, Iceland, Norway and Sweden. J Child Psychol Psychiatry. 1989;30(3):405–416. doi: 10.1111/j.1469-7610.1989.tb00254.x. PMID:2745591. [DOI] [PubMed] [Google Scholar]
- Strauss KA, Puffenberger EG, Huentelman MJ, Gottlieb S, Dobrin SE, Parod JM, Stephan DA, Morton DH. Recessive symptomatic focal epilepsy and mutant contactin-associated protein-like 2. N Engl J Med. 2006;354(13):1370–1377. doi: 10.1056/NEJMoa052773. PMID:16571880. [DOI] [PubMed] [Google Scholar]
- Südhof TC. Neuroligins and neurexins link synaptic function to cognitive disease. Nature. 2008;455(7215):903–911. doi: 10.1038/nature07456. PMID:18923512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabuchi K, Blundell J, Etherton MR, Hammer RE, Liu X, Powell CM, Südhof TC. A neuroligin-3 mutation implicated in autism increases inhibitory synaptic transmission in mice. Science. 2007;318(5847):71–76. doi: 10.1126/science.1146221. PMID:17823315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas C, Humphreys K, Jung KJ, Minshew N, Behrmann M. The anatomy of the callosal and visual-association pathways in high-functioning autism: a DTI tractography study. Cortex. 2011;47(7):863–873. doi: 10.1016/j.cortex.2010.07.006. PMID:20832784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uhlmann EJ, Wong M, Baldwin RL, Bajenaru ML, Onda H, Kwiatkowski DJ, Yamada K, Gutmann DH. Astrocyte-specific TSC1 conditional knockout mice exhibit abnormal neuronal organization and seizures. Ann Neurol. 2002;52(3):285–296. doi: 10.1002/ana.10283. PMID:12205640. [DOI] [PubMed] [Google Scholar]
- van Woerden GM, Harris KD, Hojjati MR, Gustin RM, Qiu S, de Avila Freire R, Jiang YH, Elgersma Y, Weeber EJ. Rescue of neurological deficits in a mouse model for Angelman syndrome by reduction of αCaMKII inhibitory phosphorylation. Nat Neurosci. 2007;10(3):280–282. doi: 10.1038/nn1845. PMID:17259980. [DOI] [PubMed] [Google Scholar]
- Varoqueaux F, Aramuni G, Rawson RL, Mohrmann R, Missler M, Gottmann K, Zhang W, Südhof TC, Brose N. Neuroligins determine synapse maturation and function. Neuron. 2006;51(6):741–754. doi: 10.1016/j.neuron.2006.09.003. PMID:16982420. [DOI] [PubMed] [Google Scholar]
- Vernes SC, Newbury DF, Abrahams BS, Winchester L, Nicod J, Groszer M, Alarcón M, Oliver PL, Davies KE, Geschwind DH, Monaco AP, Fisher SE. A functional genetic link between distinct developmental language disorders. N Engl J Med. 2008;359(22):2337–2345. doi: 10.1056/NEJMoa0802828. PMID:18987363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallace ML, Burette AC, Weinberg RJ, Philpot BD. Maternal loss of Ube3a produces an excitatory/inhibitory imbalance through neuron type-specific synaptic defects. Neuron. 2012;74(5):793–800. doi: 10.1016/j.neuron.2012.03.036. PMID:22681684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walsh CA, Morrow EM, Rubenstein JL. Autism and brain development. Cell. 2008;135(3):396–400. doi: 10.1016/j.cell.2008.10.015. PMID:18984148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, McCoy PA, Rodriguiz RM, Pan Y, Je HS, Roberts AC, Kim CJ, Berrios J, Colvin JS, Bousquet-Moore D, Lorenzo I, Wu G, Weinberg RJ, Ehlers MD, Philpot BD, Beaudet AL, Wetsel WC, Jiang YH. Synaptic dysfunction and abnormal behaviors in mice lacking major isoforms of Shank3. Hum Mol Genet. 2011;20(15):3093–3108. doi: 10.1093/hmg/ddr212. PMID:21558424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeber EJ, Jiang YH, Elgersma Y, Varga AW, Carrasquillo Y, Brown SE, Christian JM, Mirnikjoo B, Silva A, Beaudet AL, Sweatt JD. Derangements of hippocampal calcium/calmodulin-dependent protein kinase II in a mouse model for Angelman mental retardation syndrome. J Neurosci. 2003;23(7):2634–2644. doi: 10.1523/JNEUROSCI.23-07-02634.2003. PMID:12684449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Writzl K, Knegt AC. 6p21.3 microdeletion involving the SYNGAP1 gene in a patient with intellectual disability, seizures, and severe speech impairment. Am J Med Genet A. 2013;161A(7):1682–1685. doi: 10.1002/ajmg.a.35930. PMID:23687080. [DOI] [PubMed] [Google Scholar]
- Yashiro K, Riday TT, Condon KH, Roberts AC, Bernardo DR, Prakash R, Weinberg RJ, Ehlers MD, Philpot BD. Ube3a is required for experience-dependent maturation of the neocortex. Nat Neurosci. 2009;12(6):777–783. doi: 10.1038/nn.2327. PMID:19430469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahir FR, Baross A, Delaney AD, Eydoux P, Fernandes ND, Pugh T, Marra MA, Friedman JM. A patient with vertebral, cognitive and behavioural abnormalities and a de novo deletion of NRXN1α. J Med Genet. 2008;45(4):239–243. doi: 10.1136/jmg.2007.054437. PMID:18057082. [DOI] [PubMed] [Google Scholar]
- Zhou Z, Hong EJ, Cohen S, Zhao WN, Ho HY, Schmidt L, Chen WG, Lin Y, Savner E, Griffith EC, Hu L, Steen JA, Weitz CJ, Greenberg ME. Brain-specific phosphorylation of MeCP2 regulates activity-dependent Bdnf transcription, dendritic growth, and spine maturation. Neuron. 2006;52(2):255–269. doi: 10.1016/j.neuron.2006.09.037. PMID:17046689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY. Postnatal neurodevelopmental disorders: meeting at the synapse? Science. 2003;302(5646):826–830. doi: 10.1126/science.1089071. PMID:14593168. [DOI] [PubMed] [Google Scholar]
- Zoghbi HY, Bear MF. Synaptic dysfunction in neurodevelopmental disorders associated with autism and intellectual disabilities. Cold Spring Harb Perspect Biol. 2012;4(3):4. doi: 10.1101/cshperspect.a009886. PMID:22258914. [DOI] [PMC free article] [PubMed] [Google Scholar]