SUMMARY
FtsN is a bitopic membrane protein and the last essential component to localize to the Escherichia coli cell division machinery, or divisome. The periplasmic SPOR domain of FtsN was previously shown to localize to the divisome in a self-enhancing manner, relying on the essential activity of FtsN and the peptidoglycan synthesis and degradation activities of FtsI and amidases, respectively. Because FtsN has a known or purported role in stimulation of these activities, it follows that FtsN initially localizes to division sites in a SPOR-independent manner. Here, we show that the cytoplasmic and transmembrane domains of FtsN (FtsNCyto-TM) facilitated localization of FtsN independently of its SPOR domain but dependent on the early cell division protein FtsA. In addition, SPOR-independent localization preceded SPOR-dependent localization, providing a mechanism for the initial localization of FtsN. In support of the role of FtsNCyto-TM in FtsN function, a variant of FtsN lacking the cytoplasmic domain localized to the divisome but failed to complement an ftsN deletion unless it was overproduced. Simultaneous removal of both domains abolished localization and complementation. These data support a model in which FtsA-FtsN interaction recruits FtsN to the divisome, where it can then stimulate the peptidoglycan remodeling activities required for SPOR-dependent localization.
Keywords: cell division, cytokinesis, FtsA
INTRODUCTION
Division of a single cell into two equal daughter cells is a fundamental process shared among eukaryotic and prokaryotic organisms. Yet, despite the conservation of this developmental stage, we continue to lack a complete understanding of the mechanisms driving cytokinesis. To further characterize this process, the essential proteins and protein-protein interactions that participate in cell division must be further investigated. At present, nearly a dozen essential cell division proteins have been identified in the model bacterium Escherichia coli (Rico et al., 2013). These proteins comprise a division machine or “divisome” that assembles at midcell into a ring-shaped structure prior to cytokinesis.
The components of the E. coli divisome are incorporated in two overall stages, with some proteins localizing early to midcell and others localizing late (Aarsman et al., 2005). FtsZ, a homolog of eukaryotic tubulin, is the first essential cell division protein to localize at midcell (Erickson, 1997). FtsZ marks the division site by polymerizing into a ring structure (Z-ring) that serves as a scaffold for the remaining cell division proteins (Bi and Lutkenhaus, 1991). FtsA and ZipA are recruited to the division site soon after polymerization of FtsZ and tether the Z-ring to the inner membrane (Ma et al., 1996, Raychaudhuri, 1999, Pichoff and Lutkenhaus, 2002, Pichoff and Lutkenhaus, 2005). Once the early divisome proteins are assembled at midcell, the late cell division proteins are recruited at a defined later stage (Aarsman et al., 2005) in a linear order of dependency (FtsK → FtsQBL complex → FtsW → FtsI → FtsN). The order of essential cell division protein recruitment is based on several pioneering studies in which one or more divisome proteins were inactivated and the localization of downstream cell division proteins was monitored [reviewed in (Goehring and Beckwith, 2005) and (Vicente and Rico, 2006)].
Despite the established order of dependency, recent studies have suggested that recruitment of many cell division proteins is more complicated. FtsN, for example, fails to localize to division sites in the absence of FtsA even after all other essential cell division proteins, including FtsI, are artificially targeted to midcell (Goehring et al., 2005). FtsN is a transmembrane protein with a small cytoplasmic tail, a single transmembrane segment, and a relatively large periplasmic domain (Dai et al., 1993). Because FtsN is an essential cell division protein, FtsN depletion causes cell filamentation and eventual death (Dai et al., 1993). Although the essential function of FtsN is unknown, FtsN appears to have a role in divisome stabilization, as removal of FtsN leads to disassembly of divisome components (Rico et al., 2010). Domain swap experiments showed that the cytoplasmic and transmembrane domains of FtsN can be replaced by analogous domains from other proteins and still complement an ftsN null mutant (Dai et al., 1996, Addinall et al., 1997, Goehring et al., 2007, Gerding et al., 2009). The domain of FtsN essential for its function was pinpointed to a three-alpha helix segment in the membrane-proximal region of the periplasmic domain. In addition, the C-terminal region of FtsN harbors a SPOR domain involved in binding peptidoglycan (Ursinus et al., 2004), which is also present in several other E. coli proteins (Gerding et al., 2009, Arends et al., 2010).
FtsN localizes to the divisome most efficiently via its periplasmic SPOR domain. This localization requires three activities: the peptidoglycan synthesis activity of penicillin binding protein FtsI (PBP3); the essential activity of FtsN, as accomplished by FtsN itself or by its suppressor, FtsA(E124A); and the activity of at least one periplasmic murein amidase (Gerding et al., 2009). Although SPOR-mediated localization of FtsN is dependent upon prior activation of these proteins, FtsN has a demonstrated role in the recruitment of amidases and an implied role in FtsI stimulation, suggesting that its targeting to the divisome is self-reinforced. Specifically, the recruitment of amidases AmiB and AmiC to division sites is dependent on FtsN, as is the localization of NlpD, the cognate activator of AmiC (Peters et al., 2011). FtsN has also been shown to activate PBP1b and is suspected of activating FtsI (Muller et al., 2007). If FtsN is involved in the initial localization and stimulation of these proteins, how does FtsN get to the divisome in the first place?
Here, we provide evidence that the previously established interaction between FtsA and FtsN (Karimova et al., 2005, Corbin et al., 2004, Busiek et al., 2012) facilitates midcell localization of FtsN. We also show that this SPOR-independent means of FtsN localization precedes SPOR-dependent midcell recruitment. Together, these results provide a mechanism for the initial localization of FtsN that is needed for recruitment or stimulation of FtsI, FtsN, and amidase activities and subsequent SPOR-dependent localization of FtsN.
RESULTS
The cytoplasmic domain of FtsN contributes to its localization to the divisome independently of native FtsN
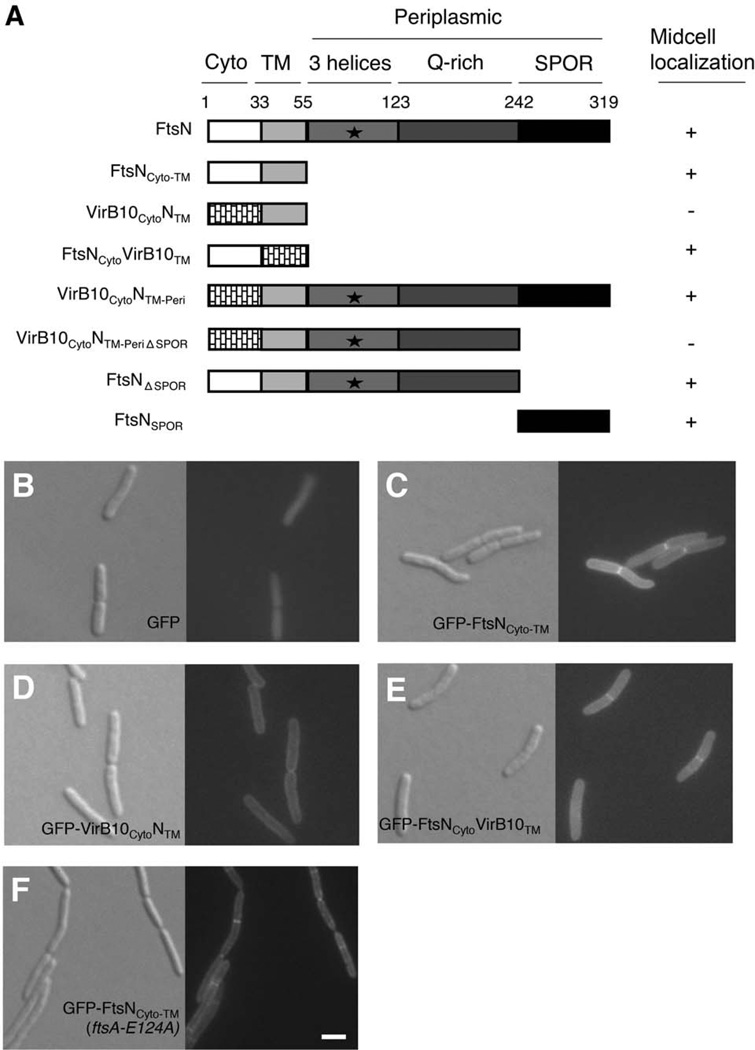
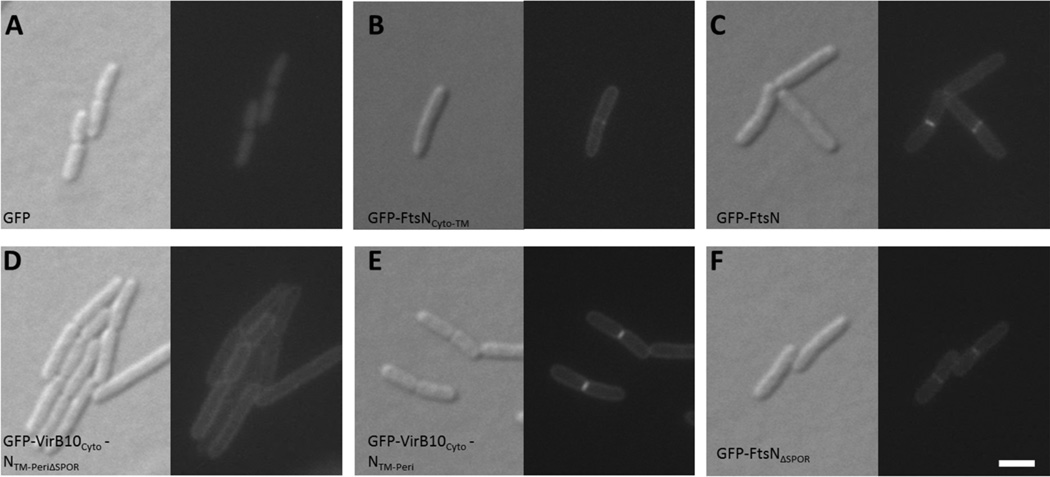
Previously, our laboratory provided in vitro evidence that proteins FtsA and FtsN interact, and that the first 55 residues of FtsN, including the short cytoplasmic tail and transmembrane domain (FtsNCyto-TM) are sufficient for this interaction (Busiek et al., 2012). We also found that strong overproduction of FtsNCyto-TM caused moderate filamentation of cells (data not shown), which prompted us to ask if the cytoplasmic and transmembrane domains of FtsN can localize to division sites without the aid of the known divisome targeting determinants in the periplasmic domain. To observe the localization of FtsNCyto-TM, we fused green fluorescent protein (GFP) to the amino terminus of FtsNCyto-TM (Fig. 1A) and expressed the fusion at uninduced levels from plasmid pDSW207, which has a weakened trc promoter with leaky expression. Although GFP itself localized diffusely throughout the cell (Fig. 1B), GFP-FtsNCyto-TM localized specifically to division sites and the membrane (Fig. 1C). The ability of FtsN to localize weakly in the absence of the SPOR domain was also noted when observing GFP fusions to FtsN1–243, FtsN1–105, and FtsN1–90 (Gerding et al., 2009).
FIG. 1. The cytoplasmic domain of FtsN contributes to midcell localization independently of native FtsN.
(A) Domain organization of FtsN and engineered constructs used in this study. Midcell localization of each GFP-tagged construct in a wild-type strain is indicated by a (+) symbol; diffuse or peripheral membrane localization is noted by a (−) symbol. The essential region of FtsN (aa71–105; Gerding et al. 2009) is represented by a black star. (B–F) DIC (left) and fluorescence (right) images of cells grown to low OD600 at 37°C in the absence of IPTG. Cells shown in panels B–E are wild-type WM1074 harboring plasmids pWM1088 (pDSW207-gfp) (B), pWM4528 (pDSW207-gfp-ftsNCyto-TM) (C), pWM4610 (pDSW207-gfp-virB10CytoNTM) (D), or pWM4611 (pDSW207-gfp-ftsNCytovirB10TM) (E). Cells shown in (F) lack the native ftsN allele but harbor a single chromosomal copy of ftsA-E124A (WM3303 background) as well as plasmid pWM4528 (pDSW207-gfp-ftsNCyto-TM). Scale bar = 4 µm.
To narrow down the segment of FtsNCyto-TM required for midcell localization, we replaced the cytoplasmic and transmembrane segments of FtsNCyto-TM with the corresponding domains of the unrelated Agrobacterium tumefaciens protein VirB10. VirB10 is a key component of the Type IV secretion system and has a bitopic membrane topology similar to FtsN (Garza and Christie, 2013). GFP-VirB10CytoNTM localized uniformly around the membrane but failed to localize to division sites (Fig. 1D), whereas GFP-FtsNCytoVirB10TM localized clearly to midcell (Fig. 1E). These results indicate that the FtsN cytoplasmic domain is sufficient to promote midcell localization of GFP-FtsNCyto-TM. However, GFP-FtsNCyto alone did not localize to division sites (data not shown), suggesting that the transmembrane domain of VirB10 facilitates midcell localization of GFP-FtsNCytoVirB10TM, possibly through the weak dimeric activity or membrane association of VirB10TM (Garza and Christie, 2013). Consistent with this idea, the cytoplasmic domain of FtsN alone fails to interact with FtsA unless it is fused to a dimerization motif, such as a leucine zipper (Busiek et al., 2012).
Because self-interaction of FtsN was previously reported (Karimova et al., 2005, Alexeeva et al., 2010), we wanted to rule out the possibility that GFP-FtsNCyto-TM localizes to division sites through its interaction with native FtsN. If midcell localization of GFP-FtsNCyto-TM is indeed dependent on interaction with previously localized FtsN, then loss of FtsN should result in delocalization of GFP-FtsNCyto-TM. Although FtsN is an essential cell division protein, cells can survive the loss of ftsN when a single amino acid mutation in FtsA (FtsA-E124A) is present (Bernard et al., 2007). Using an ftsN deletion strain carrying a chromosomal ftsA-E124A allele (WM3302), we observed localization of GFP-FtsNCyto-TM at division sites in 87% of cells, indicating that GFP-FtsNCyto-TM is efficiently recruited to the divisome independently of FtsN (Fig. 1F).
Localization of GFP-FtsNCyto-TM to midcell is dependent on FtsA
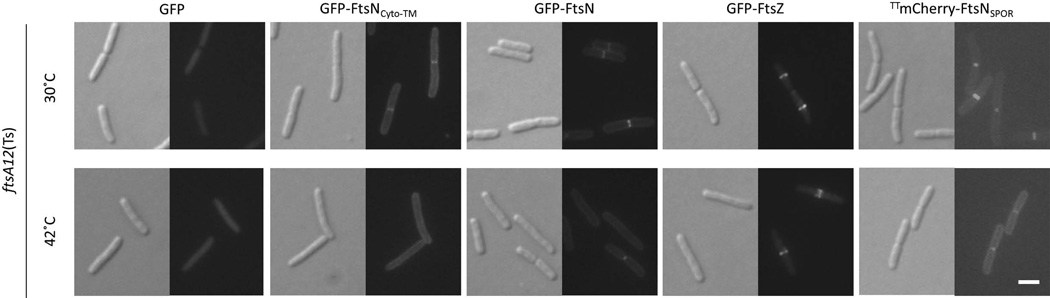
Because the amino terminus of FtsN interacts with cell division protein FtsA, we hypothesized that FtsA recruits GFP-FtsNCyto-TM directly to division sites. Using the ftsA12(Ts) strain WM1115, we compared the localization of GFP-FtsNCyto-TM at the permissive temperature of 30°C to the pattern of localization at the non-permissive temperature of 42°C. Because FtsA12 delocalizes from division sites within 5 minutes at 42°C (Fig. S1), we visualized cells 5 minutes after the temperature shift. GFP-FtsNCyto-TM formed midcell rings at the permissive temperature as expected, but delocalized within the 5 minute temperature shift (Fig. 2). Similar results were seen when we monitored the localization of GFP-tagged FtsNΔSPOR, a construct that retains the cytoplasmic and transmembrane domains of FtsN but lacks the SPOR domain (Fig. 1A and data not shown). It is unlikely that thermal instability or other non-specific factors caused GFP-FtsNCyto-TM to delocalize, as GFP-FtsNCyto-TM continued to form fluorescent rings at midcell after 5 or 30 minutes at 42°C in WM1074, the wild-type parent of WM1115 (Fig. S2).
FIG. 2. Localization of GFP-FtsNCyto-TM to midcell is dependent on FtsA.
DIC (left) and fluorescence (right) images of ftsA12(Ts) strains harboring plasmids pWM1088 (pDSW207-gfp), pWM4528 (pDSW207-gfp-ftsNCyto-TM), pWM1152 (pDSW207-gfp-ftsN), pWM3775 (pDSW207-gfp-ftsZ), or pWM4740 (pKG116-TTmCherry-ftsNSPOR). Cells were grown to low OD600 without IPTG (strains containing pDSW207 derivatives) or with 1 µM sodium salicylate (strain containing pKG116 derivative) at 30°C then shifted to 42°C for 5 minutes. Scale bar = 4 µm.
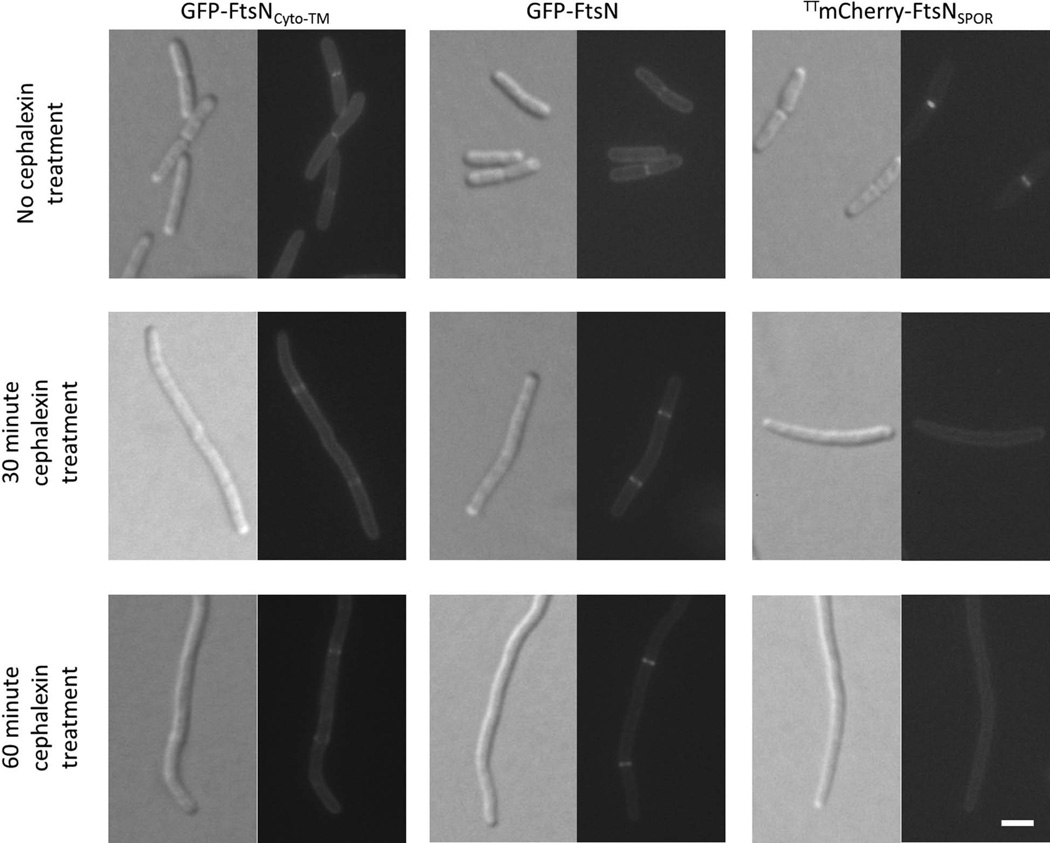
Localization of FtsN’s SPOR domain requires the peptidoglycan synthesis activity of FtsI (Gerding et al., 2009). To understand if localization of GFP-FtsNCyto-TM has a similar requirement, we treated cells with cephalexin, an inhibitor of FtsI activity, and visualized cells for altered localization. As a negative control, we observed the localization of the isolated SPOR domain (FtsNSPOR) after cephalexin treatment. Because the SPOR domain is periplasmic, we tagged FtsNSPOR with Tat-targeted mCherry (TTmCherry-FtsNSPOR). TTmCherry harbors the signal sequence of TorA, a Tat-targeted substrate that allows efficient export of FtsNSPOR to the periplasm (Thomas et al., 2001). As expected, TTmCherry-FtsNSPOR delocalized within 30 minutes of cephalexin treatment, confirming that SPOR-dependent localization of FtsN requires the peptidoglycan synthesis activity of FtsI. Unlike TTmCherry-FtsNSPOR, GFP-FtsN and GFP-FtsNCyto-TM continued to localize after cephalexin treatment, demonstrating that the cytoplasmic and transmembrane domains of FtsN have different requirements for localization than the SPOR domain (Fig. 3). Although rapid cell lysis has been described during cephalexin treatment of E. coli cells under specific conditions (Chung et al., 2009), we did not observe lysis using our standard growth conditions. Additionally, we tested whether FtsQ or FtsI are needed for localization of GFP-FtsNCyto-TM. Both FtsQ and FtsI interacted with FtsN in previously published in vivo studies, and could potentially recruit GFP-FtsNCyto-TM to midcell (Karimova et al., 2005). However, GFP-FtsNCyto-TM continued to localize at potential division sites after thermoinactivation of ftsQ(Ts) or ftsI(Ts) at 42°C (Fig. S3 and Table S2), suggesting that neither FtsQ nor FtsI promotes SPOR-independent localization of FtsN.
FIG. 3. Localization of GFP-tagged FtsNCyto-TM and FtsN is not dependent on FtsI activity.
DIC (left) and fluorescence (right) images of wild-type (WM1074) strains harboring plasmids pWM4528 (pDSW207-gfp-ftsNCyto-TM), pWM1152 (pDSW207-gfp-ftsN), or pWM4740 (pKG116-TTmCherry-ftsNSPOR). Cells were grown to low OD600 without IPTG (strains containing pDSW207 derivatives) or with 1 µM sodium salicylate (strain containing pKG116 derivative) at 37°C. Cultures were back-diluted 1:1 and imaged at 30 minutes and 60 minutes after addition of cephalexin to inhibit FtsI activity. Cultures were also back-diluted after the 30-minute time point to maintain a low OD600. Scale bar = 4 µm.
We then asked whether full length FtsN depended on FtsA for its localization. Like GFP-FtsNCyto-TM, GFP-FtsN localized at the permissive temperature in the ftsA12(Ts) strain (Fig. 2). Localization of GFP-FtsN at midcell decreased in frequency after thermoinactivation at 42°C (Table 2), but weak fluorescent rings could still be observed at division sites at 42°C, perhaps via SPOR-dependent localization. To test this hypothesis, we observed the localization of TTmCherry-FtsNSPOR in the ftsA12(Ts) strain at the non-permissive temperature. TTmCherry-FtsNSPOR remained at midcell after thermoinactivation of ftsA12(Ts) (Fig. 2), supporting the idea that the SPOR domain of FtsN localizes to the divisome independently of FtsA. Not surprisingly, GFP-tagged FtsZ, which does not depend solely on FtsA for localization (Pichoff and Lutkenhaus, 2002), remained largely unaffected by the temperature shift, forming fluorescent rings at 42°C. Overall, the failure of GFP-FtsNCyto-TM to form fluorescent rings in the ftsA12(Ts) strain at the non-permissive temperature suggests that localization of GFP-FtsNCyto-TM to division sites is dependent on proper localization of FtsA.
TABLE 2. Frequency of division site localization of GFP-tagged and TTmCherry-tagged division proteins in the ftsA12(Ts) strain.
Cells were grown as described in Figure 2 and imaged live.
| Plasmid | Fusion | Total # cells |
% Cells with rings at 30°C |
% Cells with rings after 5 minutes at 42°C |
|---|---|---|---|---|
| pWM1088 | GFP | 233 | 0 | 0 |
| pWM4528 | GFP-FtsNCyto-TM | 211 | 63 | 0 |
| pWM1152 | GFP-FtsN | 595 | 67 | 15 |
| pWM3775 | GFP-FtsZ | 241 | 94 | 97 |
| pWM4740 | TTmCherry-FtsNSPOR | 408 | 88 | 39 |
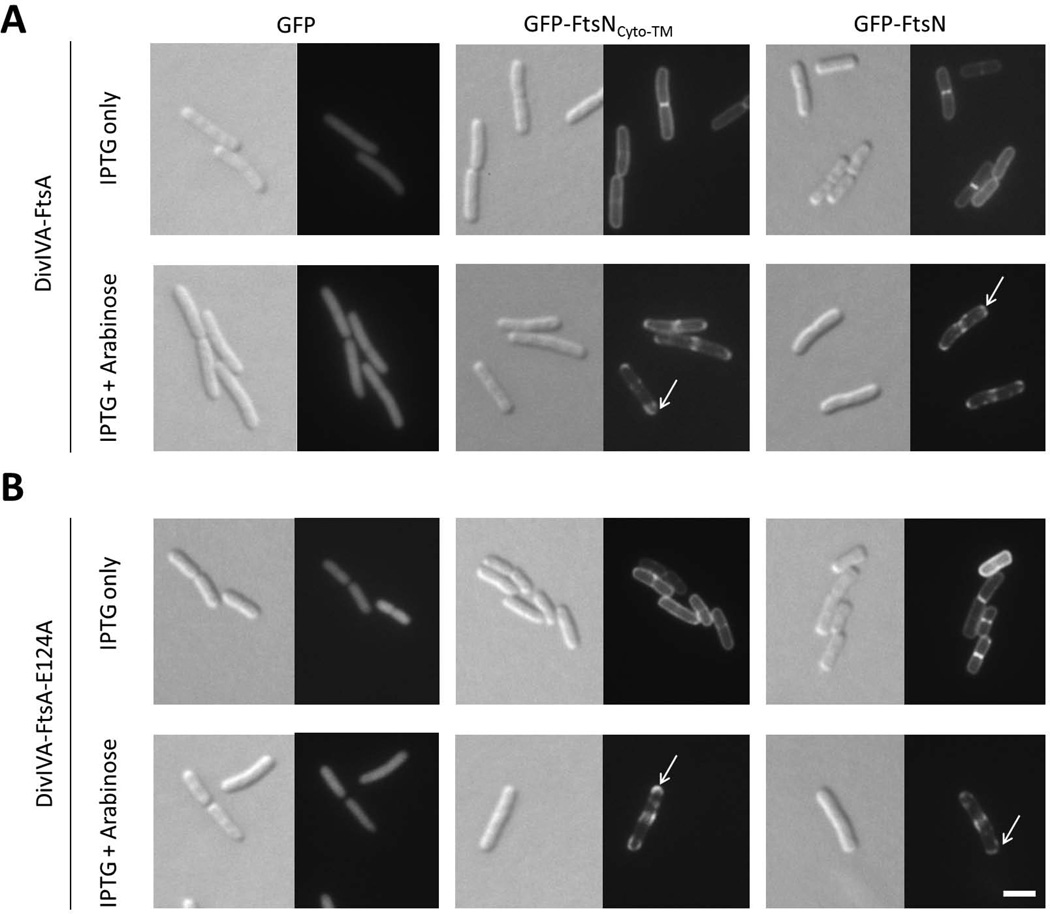
To test whether FtsA is sufficient to recruit FtsNCyto-TM, we turned to our polar recruitment assay, which previously showed for the first time that FtsA and full-length FtsN could interact (Corbin et al., 2004). Briefly, a bait protein is fused to the Bacillus subtilis protein DivIVA that preferentially localizes to areas of curvature within any cell, which in E. coli are the cell poles and division septa (Edwards et al., 2000). If the co-produced GFP-tagged prey protein is a divisome protein and interacts with the bait protein, the prey will relocalize from solely the septum to the cell poles as well as the septum. This change in fluorescence localization is an easy visual readout (Ding et al., 2002). For the current experiment, bait protein FtsA was fused to DivIVA (pBAD33-divIVA-ftsA) and prey protein FtsNCyto-TM was fused to GFP (pDSW207-gfp-ftsNCyto-TM). Upon induction of gfp-ftsNCyto-TM expression only, GFP-FtsNCyto-TM localized to cell division sites but did not accumulate at poles (Fig. 4A, top middle panel). When expression of both gfp-ftsNCyto-TM and divIVA-ftsA was induced, however, GFP-FtsNCyto-TM localized not only at division sites, but also the cell poles, indicating that DivIVA-FtsA can efficiently recruit GFP-FtsNCyto-TM to cell poles (Fig. 4A, bottom middle panel, arrow). Additionally, we confirmed that DivIVA-FtsA does not affect the diffuse localization of unfused GFP but does recruit GFP-FtsN to cell poles as reported in Corbin et al. (2004) (Fig. 4A, left and right panels, arrow).
FIG. 4. DivIVA-FtsA and DivIVA-FtsA-E124A recruit GFP-FtsNCyto-TM to cell poles.
Images are of wild-type cells containing pWM1088 (pDSW207-gfp), pWM4528 (pDSW207-gfp-ftsNCyto-TM), or pWM1152 (pDSW207-gfp-ftsN) plus either pWM1806 (pBAD33-divIVA-ftsA) (A) or pWM4637 (pBAD33-divIVA-ftsA-E124A) (B). Cells were grown to low OD600 at 30°C and then induced for two hours with either 40 µM IPTG only (to induce expression from pDSW207 plasmids) or 40 µM IPTG plus 0.2% arabinose (to induce expression from both plasmids). White arrows indicate examples of polar localization of GFP-tagged constructs. Scale bar = 4 µm.
Because GFP-FtsNCyto-TM localized to midcell in a ΔftsN ftsA-E124A strain that lacks the wild-type ftsA allele (Fig. 1F), we asked whether the FtsA with the E124A lesion can interact with FtsN by investigating whether DivIVA-FtsA-E124A could recruit GFP-FtsNCyto-TM to cell poles. Like DivIVA-FtsA, DivIVA-FtsA-E124A recruited both GFP-FtsNCyto-TM and GFP-FtsN to sites of cell curvature (Fig. 4B, arrows). Based on the delocalization of GFP-FtsNCyto-TM upon thermoinactivation of FtsA12(Ts) (Fig. 2) and the polar recruitment of GFP-FtsNCyto-TM by DivIVA-FtsA and DivIVA-FtsA-E124A (Fig. 4), we conclude that FtsA is likely necessary and sufficient for recruitment of GFP-FtsNCyto-TM to division sites.
The cytoplasmic and SPOR domains of FtsN share overlapping roles in localization of FtsN
Although the FtsA-FtsNCyto-TM interaction is capable of recruiting FtsNCyto-TM to specific cellular locations, the periplasmic SPOR domain is thought to be the major localization determinant of FtsN (Gerding et al., 2009). To elucidate the relative contributions of the cytoplasmic and SPOR domains to midcell localization of FtsN, we visualized cells producing GFP fusions to FtsN variants that lacked the native cytoplasmic domain (VirB10CytoNTM-Peri), the SPOR domain (FtsNΔSPOR), or both domains (VirB10CytoNTM-PeriΔSPOR) and quantified the occurrence of midcell localization at non-constricting and constricting sites. We had also attempted to truncate the first 20 residues of the cytoplasmic domain of FtsN (FtsNΔ1–20), but the construct was unstable (data not shown). Additionally, we mutated residues 16–19 that are involved in FtsA-FtsN interaction in vitro (Busiek et al., 2012) but, like FtsNΔ1–20, this protein was also unstable (data not shown). Although GFP-VirB10CytoNTM-Peri and GFP-FtsNΔSPOR localized to potential division sites, the fusion lacking both domains (GFP-VirB10CytoNTM-PeriΔSPOR) failed to localize (Fig. 5).
FIG. 5. Removal of both cytoplasmic and SPOR domains of FtsN abolishes midcell localization.
DIC (left) and fluorescence (right) images of wild-type strains containing plasmid pWM1088 (pDSW207-gfp) (A), pWM4528 (pDSW207-gfp-ftsNCyto-TM) (B), pWM1152 (pDSW207-gfp-ftsN) (C), pWM4694 (pDSW207-gfp-virB10CytoNTM-PeriΔSPOR) (D), pWM4696 (pDSW207-gfp-virB10CytoNTM-Peri) (E), or pWM4693 (pDSW207-gfp-ftsNΔSPOR) (F). Cultures were grown to low OD600 at 30°C without inducer before visualization. Scale bar = 4 µm.
These results indicate that localization of FtsN requires either the cytoplasmic or SPOR domain for localization to potential division sites and that loss of both domains prevents recruitment of FtsN. Furthermore, the majority of fluorescent GFP-VirB10CytoNTM-Peri rings were located at division sites that were visibly constricting (64%), whereas the majority of fluorescent GFP-FtsNΔSPOR rings were located at non-constricting sites (62%; Table 3). These results imply that on average, GFP-VirB10CytoNTM-Peri localizes to midcell during a later stage of cytokinesis than GFP-FtsNΔSPOR.
TABLE 3. Frequency of division site localization of GFP-tagged FtsN variants.
Cells were grown as described in Figure 4 and imaged live. Visible constriction of cells was determined using DIC images and FM4-64 staining of cell membrane.
| Plasmid | Fusion | Total # cells |
% Cells with rings |
% Cells without rings |
% Rings at no constrictions |
% Rings at constrictions |
|---|---|---|---|---|---|---|
| pWM1152 | GFP-FtsN | 130 | 72 | 28 | 57 | 43 |
| pWM4528 | GFP-FtsNCyto-TM | 165 | 56 | 44 | 58 | 42 |
| pWM4693 | GFP-FtsNΔSPOR | 139 | 60 | 40 | 62 | 38 |
| pWM4696 | GFP-VirB10CytoNTM-Peri | 130 | 64 | 36 | 36 | 64 |
| pWM4694 | GFP-VirB10CytoNTM-PeriΔSPOR | 142 | 0 | 100 | NA | NA |
GFP-FtsNCyto-TM localizes to midcell prior to TTmCherry-FtsNSPOR
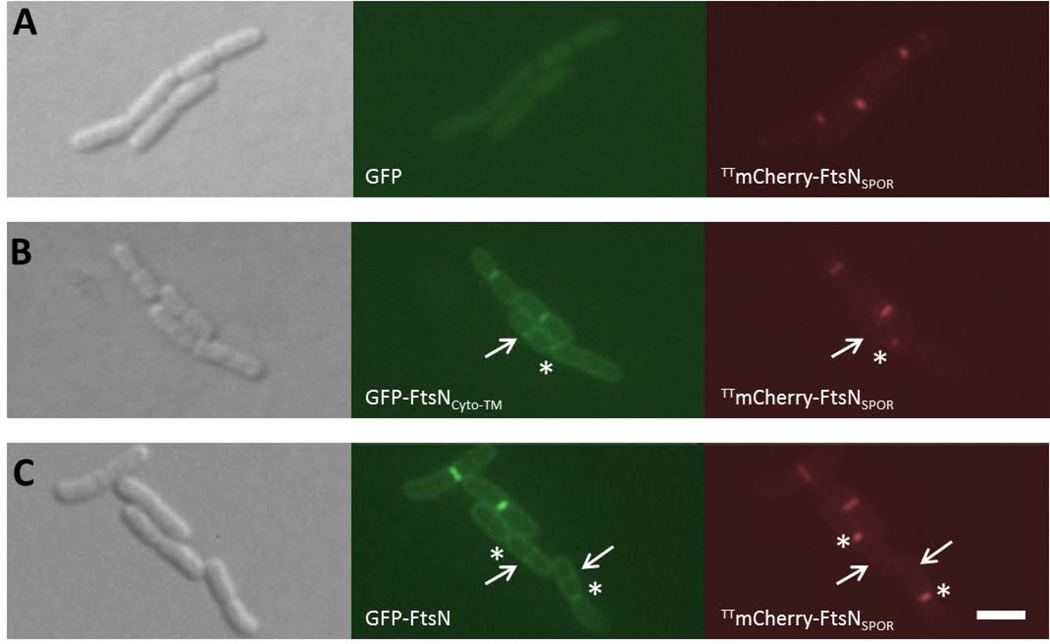
To test directly the hypothesis that SPOR-independent localization of FtsN precedes SPOR-dependent localization, we separately co-produced GFP-FtsNCyto-TM and Tat-targeted mCherry (TTmCherry) fused to FtsNSPOR in the same cell and compared their localization. The Tat-targeting sequence was fused to the mCherry-FtsNSPOR chimera to facilitate export of the construct to the periplasm (Thomas et al., 2001). Although we observed frequent co-localization of GFP-FtsNCyto-TM and TTmCherry-FtsNSPOR as expected, GFP-FtsNCyto-TM and TTmCherry-FtsNSPOR also localized to division sites independent of one another. Specifically, rings of GFP-FtsNCyto-TM were present at quarter sites of cells in the late stages of cytokinesis (denoted by arrows in Fig. 6), consistent with GFP-FtsNCyto-TM localizing to nascent division sites. Conversely, TTmCherry-FtsNSPOR lingered at deep constrictions (denoted by asterisks in Fig. 6) and was often not detectable at the nascent division sites that contained GFP-FtsNCyto-TM (arrows).
Fig. 6. GFP-FtsNCyto-TM localizes to division sites prior to TTmCherry-FtsNSPOR.
DIC (left), GFP (middle), and mCherry (right) filter sets were used to capture images of cells grown to low OD600 at 30°C in the absence of inducers. All strains were wild-type background and contained plasmid pWM4740 (pKG116-TTmCherry-ftsNSPOR) plus either plasmid pWM1088 (pDSW207-gfp) (A), pWM4528 (pDSW207-gfp-ftsNCyto-TM) (B), or pWM1152 (pDSW207-gfp-ftsN) (C). Arrows denote division site localization of GFP-tagged proteins only and asterisks mark localization of TTmCherry-FtsNSPOR only. Scale bar = 4 µm.
We quantified the difference in localization between the constructs by measuring the frequency of division site localization at constricting and non-constricting sites. In cells co-producing both fusions, the vast majority of GFP-FtsNCyto-TM rings (83%) were located at non-constricting sites, whereas the majority of TTmCherry-FtsNSPOR rings were located at constrictions (62%; Table 4). We observed similar localization patterns when GFP-FtsN was co-produced with TTmCherry-FtsNSPOR, although a larger proportion of GFP-FtsN rings were located at constricting sites. The higher percentage of GFP-FtsN rings at constricting sites compared to GFP-FtsNCyto-TM is expected, as GFP-FtsN also contains the SPOR domain that, according to the above data, localizes preferentially to constricting sites. Together, these data support the hypothesis that SPOR-independent localization of FtsN precedes SPOR-dependent localization.
TABLE 4. Distinct localization biases of two FtsN domains.
Cells were grown as described in Figure 5 and imaged live. Visible constriction of cells was determined using DIC images of each cell.
| Plasmids | Fusion | Total # cells |
% GFP rings at no constrictions |
% GFP rings at constrictions |
% mCherry rings at no constrictions |
% mCherry rings at constrictions |
|---|---|---|---|---|---|---|
| pWM4528 + pWM4740 | GFP-FtsNCyto-TM + TTmCherry-FtsNSPOR | 67 | 83 | 16 | 38 | 62 |
| pWM1152 + pWM4740 | GFP-FtsN + TTmCherry-FtsNSPOR | 70 | 59 | 41 | 35 | 65 |
Inactivation of cytoplasmic or SPOR domains inhibits the ability of FtsN to function in cell division
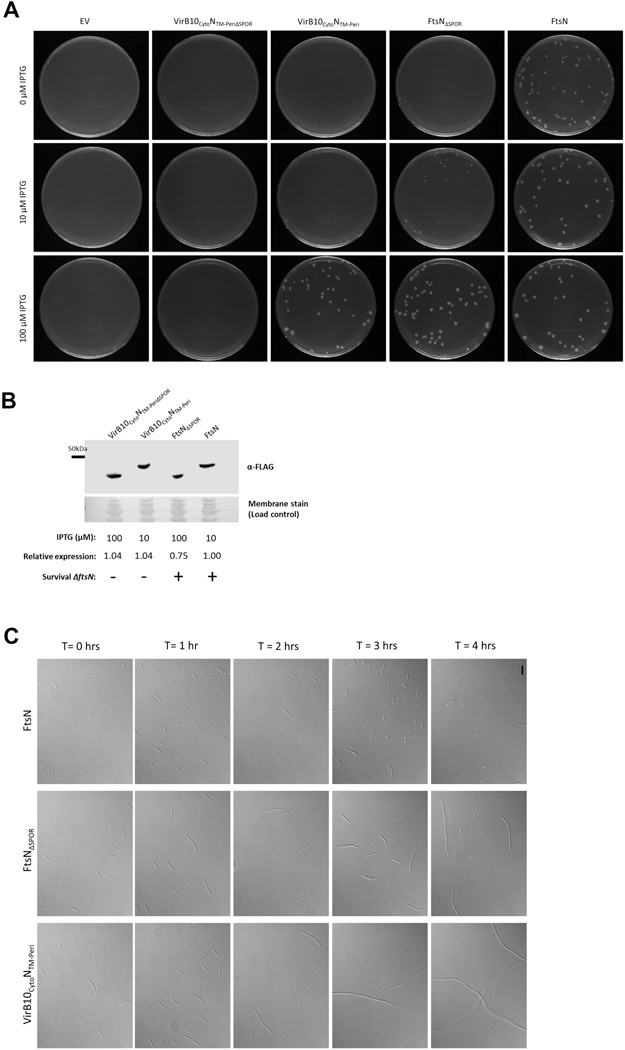
To explore the relative physiological importance of SPOR-independent and SPOR-dependent means of FtsN localization, we attempted to create strains carrying a single ftsN allele that lacked the cytoplasmic, SPOR, or both domains. To engineer these strains, we introduced the ΔftsN::kan allele by P1 phage transduction into wild-type cells harboring plasmids pDSW210-flag-virB10CytoNTM-Peri, -ftsNΔSPOR, or -virB10CytoNTM-PeriΔSPOR. Strains carrying pDSW210-flag or pDSW210-flag-ftsN were transduced as negative and positive controls, respectively. In the absence of inducer, leaky expression of ftsN from the weakened trc promoter of pDSW210 complemented the loss of the chromosomal ftsN allele (Fig. 7A, right column), whereas no transductants were found with empty pDSW210 vector at any IPTG concentration (Fig. 7A, left column). Induced expression of flag-ftsN also conferred survival in the absence of chromosomal ftsN at 10 µM and 100 µM IPTG, suggesting that higher levels of ftsN expression did not inhibit growth significantly as has been shown previously (Aarsman et al., 2005).
FIG. 7. The cytoplasmic and SPOR domains are both needed for full FtsN function in cell division.
(A) ΔftsN::kan P1 transduction of wild-type strains containing pWM2784 (pDSW210-flag), pWM4582 (pDSW210-flag-virB10CytoNTM-Peri), or pWM3157 (pDSW210-flag-ftsN). Transductants were plated onto LB medium containing 0, 10, or 100 µM IPTG. (B) α-FLAG Western blot analysis of FLAG-VirB10CytoNTM-PeriΔSPOR, FLAG-VirB10CytoNTM-Peri, FLAG-FtsNΔSPOR, and FLAG-FtsN produced from ftsN(Ts) strain WM4028. Samples were collected after strains were grown without IPTG to low OD600 at 30°C and subsequently shifted to 42°C at various levels of IPTG induction for 4 hours. The ability of the FtsN constructs to survive P1 transduction at the various levels of IPTG induction are represented by (+) and (−) symbols. (C) DIC images of transductants isolated in (A). Cultures were grown in the presence of 100 µM IPTG to low-mid OD600 at 37°C and visualized (T = 0 hours). Cells were then washed and grown in the absence of IPTG for four hours (T = 1, 2, 3, and 4 hours) and back-diluted as needed to prevent entry into stationary phase. Scale bar = 12 µm.
The various ftsN constructs were similarly tested for growth in the absence of chromosomal ftsN. None of the constructs could complement the loss of the native ftsN allele in the absence of inducer. However, in the presence of 10 µM IPTG, FLAG-FtsNΔSPOR partially complemented the loss of the native allele as evidenced by the small colonies formed on this plate. FLAG-FtsNΔSPOR completely complemented upon full induction of the plasmid (100 µM IPTG). Although FLAG-VirB10CytoNTM-Peri did not complement at 0 µM or 10 µM IPTG, full induction of flag-virB10CytoNTM-Peri conferred survival. FLAG-VirB10CytoNTM-PeriΔSPOR was unable to complement the loss of ftsN at any level of induction, consistent with the fluorescence microscopy results that showed no midcell localization of GFP-VirB10CytoNTM-PeriΔSPOR(Fig. 5). The lack of complementation by FLAG-VirB10CytoNTM-PeriΔSPOR was not due to instability of the protein, because it was present at equal or greater levels than FLAG-FtsNΔSPOR or FLAG-FtsN, both of which were capable of complementing the loss of native FtsN (Fig. 7B). Importantly, anti-FtsN Western blot analysis showed that FLAG-FtsN was produced at a level equivalent to endogenous FtsN when induced at 10 µM IPTG (data not shown), indicating that each construct shown in Fig. 7B was produced at a level similar to native FtsN. Notably, a strain synthesizing FLAG-VirB10CytoNTM-Peri failed to survive the deletion of ftsN at levels of protein equivalent to or higher than FLAG-FtsNΔSPOR (Fig. 7A, 10 µM IPTG; compare protein levels at 10 and 100 µM IPTG in Fig. 7B), supporting the idea that SPOR-independent localization of FtsN plays an important role in proper localization of FtsN.
To confirm the transduction data, we also tested the effects of depleting FtsN in a strain carrying the same plasmids used in the transduction experiments. Consistent with our transduction data, FLAG-VirB10CytoNTM-PeriΔSPOR failed to complement the loss of ftsN at any level of induction and FLAG-FtsNΔSPOR began complementing at a lower level of induction than FLAG-VirB10CytoNTM-Peri (Fig. S4). Also consistent with the transduction data, FLAG-FtsN complemented both in the absence and presence of inducer. Although substitution of the cytoplasmic and transmembrane regions or deletion of the SPOR domain did not result in cell death in previous publications (Dai et al., 1996, Addinall et al., 1997, Goehring et al., 2007, Gerding et al., 2009), all of these studies relied on overproduction of the mutated constructs. Our results are consistent with these previous results in that FLAG-VirB10CytoNTM-Peri and FLAG-FtsNΔSPOR could complement the loss of ftsN, but only when their expression was induced.
To ensure that the cell viability defects of the FtsN constructs were caused by inhibition of cell division and not a general problem with growth, we observed the morphology of the transduction survivors upon full IPTG induction of the plasmids and for several generations after depletion of inducer. As expected, ΔftsN::kan cells producing FLAG-FtsN did not form filaments prior to or after depletion of IPTG (Fig. 7C, right panels). Transductants lacking chromosomal ftsN but producing FLAG-FtsNΔSPOR, on the other hand, formed moderate filaments upon depletion of the inducer (Fig. 7C, middle panels), consistent with the results in Fig. 7A–B and Fig. S4. Also consistent with the transduction and spot dilution data, ΔftsN::kan cells producing FLAG-VirB10CytoNTM-Peri were more affected by the lack of IPTG, forming long filaments, inferring that cell division was severely compromised. The impacts on cell division among the mutant FtsN strains at low levels of induction are compatible with the idea that these proteins localize poorly. These results suggest that although the cytoplasmic and SPOR domains have overlapping roles in localization, both domains are needed for efficient localization of FtsN to the divisome.
DISCUSSION
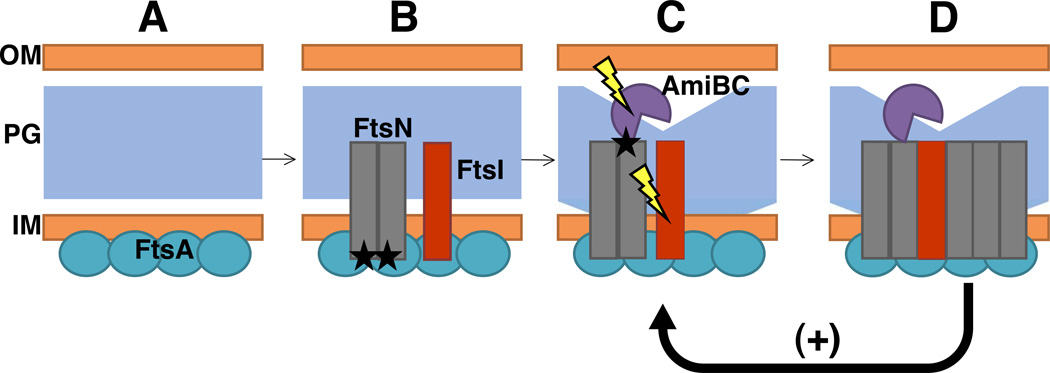
The data presented here support a model in which interaction between FtsA and FtsN weakly recruits FtsN to midcell by its amino-terminus following incorporation of FtsA into the divisome (Fig. 8). After its recruitment to the divisome, FtsN is poised to recruit amidases and stimulate the activity of FtsI. The activity of FtsN, amidases, and FtsI attracts additional FtsN molecules via the SPOR domain and reinforces a positive feedback loop. A similar model of self-enhanced recruitment of FtsN was first proposed by Gerding et al. (2009), but the mechanism of initial FtsN localization was unknown at the time.
Fig. 8. Proposed role of FtsA-FtsN interactions in cell division.
Following midcell localization of FtsA (A), interaction between FtsA and the amino-terminus of FtsN weakly recruits FtsN to midcell in a SPOR-independent manner (B; black asterisks represent recruitment). The small fraction of FtsN localized at midcell recruits amidases AmiB and AmiC and stimulates the peptidoglycan synthesis activity of FtsI. Activities of amidases AmiB and AmiC are stimulated by their cognate activators including FtsN-dependent NlpD (not shown) to degrade peptidoglycan (C; lightning bolt graphic represents stimulation of activity). Peptidoglycan remodeling at midcell exposes denuded glycan strands which, in turn, promote SPOR-dependent localization of FtsN (D). Additional FtsN molecules at the division site stimulate further peptidoglycan remodeling in a positive feedback loop, represented by a “+” symbol. OM=outer membrane, PG=peptidoglycan, IM=inner membrane.
SPOR-dependent localization of cell division proteins appears to be a common mechanism for division site recruitment: the SPOR domain of E. coli cell division proteins FtsN, DamX, DedD, and RlpA directs all four of these proteins to midcell (Gerding et al., 2009, Arends et al., 2010). Although SPOR-dependent localization of FtsN is supported by our data, we also highlight the need for the amino-terminus of FtsN in its recruitment to midcell. The requirement for both cytoplasmic and periplasmic domains for midcell localization has already been shown for the essential E. coli cell division protein FtsQ. Similar to FtsN, the cytoplasmic domain of FtsQ was originally described as a dispensable portion of the protein (Goehring et al., 2007). However, as with the experiments performed with FtsN proteins that lacked the cytoplasmic domain, these studies were performed at induced levels of the protein (Guzman et al., 1997). It was later shown that the cytoplasmic domain of FtsQ is not dispensable and, in addition to its periplasmic localization determinant, it has a role in midcell localization of the protein (Goehring et al., 2007). The cytoplasmic and periplasmic domains are not the only domains of the bitopic division proteins that contribute to localization. The localization of FtsI, for example, requires its transmembrane region (Wissel et al., 2005). Although the transmembrane domain of FtsN is not directly involved in SPOR-independent localization (Fig. 1), our results suggest that putative dimerization of this domain (Busiek et al., 2012) is important for its interaction with FtsA and, hence, FtsA-dependent recruitment of FtsN.
The immediate delocalization of GFP-FtsNCyto-TM from division sites upon shifting the ftsA12(Ts) strain to the non-permissive temperature (Fig. 2) strongly suggests that SPOR-independent localization of GFP-FtsNCyto-TM is mediated by its interaction with FtsA. This conclusion is further supported by the efficient recruitment of GFP-FtsNCyto-TM to poles by DivIVA-FtsA and DivIVA-FtsA-E124A (Fig. 4) as well as previously published evidence that FtsA and FtsN interact directly (Busiek et al., 2012). FtsA-dependent localization of FtsN may also explain why FtsN fails to localize to midcell in the absence of FtsA, despite the recruitment of all other essential cell division proteins by a ZapA-FtsQ fusion (Goehring et al., 2005).
The involvement of FtsA in the localization of FtsN also provides a potential explanation for the ability of FtsN to suppress the ftsA12(Ts) strain at 37°C (but not at 42°C) when present in a multicopy pBR322 derivative plasmid (Dai et al., 1993). Overproduction of FtsN from the plasmid might partially overcome the need for SPOR-independent localization of FtsN, similar to how overproduction of VirB10CytoNTM-Peri is capable of complementing the lack of wild-type FtsN (Fig. 7A, 7C, S4). Overproduction of FtsN or VirB10CytoNTM-Peri enriches these proteins in the membrane and likely increases the probability that these proteins will interact with other divisome components. Although we believe that multicopy expression of ftsN may bypass the need for FtsA-mediated localization, we do not yet understand how the cytoplasmic and transmembrane domains of FtsN can partially suppress the loss of ftsK or ftsQ (Geissler and Margolin, 2005, Goehring et al., 2007).
The disparate enrichment of GFP-FtsNΔSPOR at non-constricting sites and GFP-VirB10CytoNTM-Peri at constricting sites initially suggested to us that SPOR-independent localization of FtsN precedes SPOR-dependent localization of FtsN (Table 3). We confirmed this timing by observing cells that co-produced GFP-tagged FtsNCyto-TM and TTmCherry-tagged FtsNSPOR (Fig. 6). Intriguingly, while TTmCherry-FtsNSPOR seemed to persist at deep constrictions, GFP-FtsNCyto-TM appeared to abandon these sites for nascent division sites (as shown by the loss of GFP-FtsNCyto-TM fluorescence at deep constrictions in Fig. 6). This observation suggests that whereas the amino-terminus of FtsN facilitates initial localization of FtsN, this interaction may weaken despite the continuous presence of FtsA at the septum until later stages of cytokinesis. This may be because at later stages, FtsNCyto-TM now faces increasing competition with other proteins for its binding site within subdomain 1c of FtsA, including potentially another molecule of FtsA (Pichoff et al., 2012). Another possibility is that FtsA itself begins to leave the divisome prior to septum completion, although a very recent study suggests that FtsA persists until septum closure (Soderstrom et al.). In any case, in this model, the main responsibility of maintaining FtsN at the growing septum until completion would then fall to the SPOR domain. The anchoring of FtsN to old division sites by the SPOR domain would prevent premature abandonment by FtsN as FtsA redeploys to newly assembled Z-rings. This built-in regulation of FtsN localization is likely an important feature of FtsN, as substitution or deletion of either localization determinant reduced cell division and viability (Fig. 7a, 7c, and S4).
While two of our previous publications have explored if and how FtsA and FtsN interact (Corbin et al., 2004, Busiek et al., 2012), this study addresses the more interesting question of why these early and late proteins contact one another. At least one physiological role of FtsA-FtsN interaction is efficient recruitment of FtsN to the divisome, although additional functions of this interaction may also be uncovered. One interesting possibility is that once recruited, FtsN may transduce a signal to FtsA that triggers initiation of Z-ring constriction, but further studies will be needed to address this prospect (Lutkenhaus, 2009, Busiek et al., 2012, Corbin et al., 2004). We also speculate that the role of other early-late cell division protein interactions (Corbin et al., 2004, Karimova et al., 2005) may be to facilitate more robust recruitment of these proteins to the divisome and/or to fine-tune the timing of their localization. This study provides the first evidence for a physiological role of FtsA-FtsN interaction under physiological conditions.
EXPERIMENTAL PROCEDURES
Strains, growth media, and microscopy
Escherichia coli strains and plasmids used in this study are listed in Table 1. All cultures were grown in Luria-Bertani (LB) medium at 37°C, unless otherwise indicated. LB medium was supplemented with ampicillin (Fisher Scientific; 50 µg ml−1), kanamycin (Sigma-Aldrich; 50 µg ml−1), chloramphenicol (Acros Organics; 10 µg ml−1), and glucose (Sigma-Aldrich; 1%) as needed. Gene expression from the trc promoter in pDSW207 and pDSW210 vectors was induced with isopropyl-ß-D-galactopyranoside (IPTG) at a final concentration of 10–100 µM. The araBAD promoter in pBAD33 vectors was induced at a final concentration of 0.2% l-arabinose and the nahR promoter in pKG116 vectors was induced with 1.0 µM sodium salicylate. Wild type strain WM1074 (TX3772, a lac- derivative of MG1655) was used for expression from all plasmids except where noted. To inhibit FtsI activity, cultures were treated with cephalexin (Sigma-Aldrich) at a final concentration of 200 µg mL−1 for 30 or 60 minutes. The ftsN depletion strain WM4028 was created by transducing strain WM1074 carrying pWM2964 (a temperature sensitive depletion plasmid encoding ftsN) with P1 phage carrying ΔftsN::kan, selecting for KanR colonies, and screening for colonies that could grow at 30°C but not at 42°C. Similarly, strain WM3302 was constructed by transducing strain WM2935 (W3110 Tn10 ftsA-E124A) with ΔftsN::kan P1 lysate and selecting for KanR survivors.
TABLE 1. Strains and plasmids used in this study.
| Strain or plasmid | Genotype or description | Source or reference |
|---|---|---|
| E. coli strains | ||
| WM1074 | MG1655 lacU169 (TX3772) | Laboratory collection |
| WM1115 | TX3772 leu::Tn10 ftsA12(Ts) | Geissler et al. (2003) |
| WM2935 | W3110 (WT strain) leu::Tn10 ftsA-E124A | Shiomi and Margolin (2008) |
| WM3302 | WM2935 ΔftsN::kan | This study |
| WM4028 | WM1074 ΔftsN::kan pWM2964 | This study |
| Plasmids | ||
| Vectors | ||
| pBAD33 | Derivative of pDHB60 with araBAD promoter (CmR) | Guzman et al. (1995) |
| pDSW210 | Ptrc-gfp pBR322 derivative with weak promoter (AmpR) | Weiss et al. (1999) |
| pDSW207 | Ptrc-gfp pBR322 derivative with strong promoter (AmpR) | Weiss et al. (1999) |
| pKG116 | pACYC184 derivative containing the nahG promoter (CmR) | J.S. Parkinson |
| divIVA fusions | ||
| pWM1806 | divIVA-ftsA in pBAD33 | Corbin et al. (2004) |
| pWM4637 | divIVA-ftsA-E124A in pBAD33 | This study |
| flag fusions | ||
| pWM2784 | flag epitope inserted between EcoRI and SacI sites of pDSW210 | Shiomi and Margolin (2007) |
| pWM3157 | flag-ftsN in pDSW210 | This study |
| pWM4582 | flag-virB10Cyto(aa2-32)-ftsNTM-Peri(aa29-319) in pDSW210 | This study |
| pWM4612 | flag-virB10Cyto(aa2-32)-ftsNTM-PeriΔSPOR(aa29-242) in pDSW210 | This study |
| pWM4613 | flag-ftsNΔSPOR(aa2-242) in pDSW210 | This study |
| gfp fusions | ||
| pWM1088 | gfp in pDSW207 | Weiss et al. (1999) |
| pWM1152 | gfp-ftsN in pDSW207 | Corbin et al. (2004) |
| pWM3775 | gfp-ftsZ in pDSW207 | This study |
| pWM4528 | gfp-ftsNCyto-TM(aa2-55) in pDSW207 | This study |
| pWM4610 | gfp-virB10Cyto(aa2-32)-ftsNTM(aa29-55) in pDSW207 | This study |
| pWM4611 | gfp-ftsNCyto(aa2-33)-virB10TM(aa33-50) in pDSW207 | This study |
| pWM4693 | gfp-ftsNΔSPOR(aa2-242) in pDSW207 | This study |
| pWM4694 | gfp-virB10Cyto(aa2-32)-ftsNTM-PeriΔSPOR(aa29-242) in pDSW207 | This study |
| pWM4696 | gfp-virB10Cyto(aa2-32)-ftsNTM-Peri(aa29-319) in pDSW207 | This study |
| Other plasmids | ||
| pWM4740 | TT(torA signal sequence)mCherry-ftsNSPOR(aa241-319) in pKG116 | This study |
| pWM2964 | pKD123 (Ts ftsN plasmid) | Dai et al. (1993) |
Strains were observed using an Olympus BX60 microscope and UPlanSApo 100X oil immersion objective. Differential interference contrast (DIC) and fluorescence images were captured on a Hamamatsu Digital Camera (Model C8484) using HCImageLive (Hamamatsu) software. Membranes were stained as needed to assist visualization by incubating 1 mL cultures with 0.5 µg mL−1 FM4–64 stain for 15 minutes at room temperature.
Plasmid constructions
Plasmids and primer sequences are listed in Table 1 and Table S1, respectively. Following the manufacturer’s guidelines, DNA was amplified using KAPA HiFi DNA polymerase from VWR (Radnor, PA) and primers from Sigma-Aldrich (St. Louis, MO) and Integrated DNA Technologies (IDT, Coralville, IA). Restriction and T4 DNA ligase enzymes were purchased from New England BioLabs, Inc. (NEB; Beverly, MA) and DNA cleanup kits were obtained from Promega (Madison, WI). Clones were sequenced by Genewiz (South Plainfield, NJ).
The polar recruitment assay vector pWM4637 (pBAD33-divIVA-ftsA-E124A) was constructed by amplifying the ftsA-E124A gene from template plasmid pWM2702 (pRR48-flag-ftsA-E124A) with primers 2051 and 2052. The PCR product and recipient vector pBAD33-divIVA were then digested with XbaI and PstI and ligated.
To create pWM4740 (pKG116-TTmCherry-ftsNSPOR), TTmCherry-ftsNSPOR was amplified from template pWM4682 (pDSW208-TTmCherry-ftsNSPOR) using primers 2063 and 2059. The insert was then cloned into vector pKG116 as an NsiI-XbaI fragment. To construct the original template plasmid pWM4682, the signal sequence of torA was amplified with primers 2056 and 2055 from pWM1487 (pBAD24-TTgfp) and the mCherry sequence was amplified with primers 2054 and 2057 from pWM4367 (pDSW208-ftsZ1–176-mCherry-ftsZ177–383). The torA signal sequence and mCherry were then fused using combinatorial PCR (primers 2056 and 2057) to create a TTmCherry fragment that was cloned into pDSW208 using EcoRI and BamHI restriction sites. The SPOR domain of ftsN was then amplified from template pWM1152 (pDSW207-ftsN) with primers 2058 and 2059 and cloned into pDSW208-TTmCherry as a BamHI-XbaI fragment.
All pWM2784 (pDSW210-flag) derivatives engineered for this study were cloned as XbaI-PstI fragments. To create plasmid pWM3157 (pDSW210-flag-ftsN), the ftsN insert was amplified with primers 1115 and 1116. Plasmid pWM4582 (pDSW210-flag-virB10CytoNTM-Peri) was created by amplifying the sequences encoding the cytoplasmic domain of virB10 with primers 1809 and 1804 and the transmembrane and periplasmic domains of ftsN with primers 1803 and 1116. The PCR products were then joined in a combinatorial PCR reaction with primers 1809 and 1116. To create plasmid pWM4612 (pDSW210-flag-virB10CytoNTM-PeriΔSPOR), the virB10CytoNTM-PeriΔSPOR sequence was amplified from template pWM4582 (pDSW210-flag-virB10CytoNTM-Peri) using forward primer 1809 and reverse primer 2049 to omit the SPOR sequence from the PCR product. The pDSW210-flag-ftsNΔSPOR plasmid pWM4613 was constructed by amplifying ftsNΔSPOR from template pWM3157 (pDSW210-flag-ftsN) using primers 1115 and 2049 to also omit the SPOR sequence.
Insertion of DNA sequences into the multiple cloning site of vector pDSW207 fuses GFP to the amino terminus of encoded proteins. The GFP-FtsZ fusion produced from pWM3775 (pDSW207-gfp-ftsZ) was created by subcloning ftsZ from pDSW209-gfp-ftsZ (pWM3439) into vector pDSW207 using SacI and HindIII restriction sites. All other pDSW207 plasmids were constructed using EcoRI and HindIII sites and all forward primers included a sequence to create an Asn-Asn-Asn linker between GFP and the protein of interest. pWM4528 (pDSW207-gfp-ftsNCyto-TM) was created by amplifying the cytoplasmic and transmembrane regions of ftsN from template pWM1152 (pDSW207-gfp-ftsN) with primers 112 and 2003. To construct pWM4610 (pDSW207-gfp-virB10CytoNTM), virB10CytoNTM was amplified from template pWM4582 (pDSW210-flag-virB10CytoNTM-Peri) with primers 2048 and 2003. pWM4611 (pDSW207-gfp-ftsNCytovirB10TM) was cloned by amplifying the cytoplasmic and transmembrane domains of pWM4432 (pKT25-flag-ftsNCytovirB10TMphoA) using primers 112 and 2047. For pWM4696 (pDSW207-gfp-virB10CytoNTM-Peri), the virB10CytoNTM-Peri sequence was amplified from pWM4582 (pDSW210-flag-virB10CytoNTM-Peri) using primers 2048 and 111. To create pDSW207-gfp-ftsNΔSPOR (pWM4693), ftsNΔSPOR was amplified from pWM4613 (pDSW210-flag-ftsN) using primers 112 and 2060. Finally, pWM4694 (pDSW207-gfp-virB10CytoNTM-PeriΔSPOR) was cloned by amplifying virB10CytoNTM-PeriΔSPOR from pWM4612 (210-flag-virB10CytoNTM-PeriΔSPOR) using primers 2048 and 2060.
Temperature-shift experiments with ftsA12(Ts)
Cultures of ftsA12(Ts) strain WM1115 expressing gfp (pWM1088), gfp-ftsNCyto-TM (pWM4528), gfp-ftsN (pWM1152), gfp-ftsZ (pWM3775), or TTmCherry-ftsNSPOR (pWM4740) were grown at 30°C to an optical density at 600nm (OD600) of 0.3–0.4 prior to temperature shift. Strains expressing gfp fusions from vector pDSW207 remained uninduced throughout the experiment, whereas the strain expressing TTmCherry-ftsNSPOR from pKG116 was grown in the presence of 1 µM sodium salicylate. Cultures were shifted to 42°C for 5 minutes. DIC and fluorescence images were obtained at each time point.
Polar recruitment assay
Cultures of strains carrying the pBAD33-divIVA-ftsA (pWM1806) or pBAD33-divIVA-ftsA-E124A (pWM4637) vector and pDSW207 plasmids expressing gfp (pWM1088), gfp-ftsNCyto-TM (pWM4528), or gfp-ftsN (pWM1152) were grown and induced as described previously (Corbin et al., 2004). Briefly, cultures were grown at 30°C to a low OD600 (~0.1) and induced for 1.5–2 hours with 40 µM IPTG only (inducing expression from pDSW207 plasmids) or 40 µM IPTG plus 0.2% arabinose (inducing expression from pDSW207 and pBAD33 plasmids).
ΔftsN::kan transductions and depletion experiments
To obtain transductants harboring the ΔftsN::kan allele, wild-type cells containing plasmid pWM2784 (pDSW210-flag), pWM4612 (pDSW210-flag-virB10CytoftsNTM-PeriΔSPOR), pWM4582 (pDSW210-flag-virB10CytoftsNTM-Peri), pWM4613 (pDSW210-flag-ftsNΔSPOR), or pWM3643 (pDSW210-flag-ftsN) were grown to low OD600 (~0.2) and transduced with a P1 phage lysate derived from ΔftsN::kan strain WM3303, which also carries the ftsA-E124A allele to permit survival in the absence of FtsN. Following a one hour outgrowth in media containing 10 mM sodium citrate, cells were plated onto media containing kanamycin and ampicillin to select for the plasmids and ΔftsN::kan, and with 0, 10, or 100 µM IPTG to induce expression from the pDSW210 vector. Survivors were picked from the plates containing 100 µM IPTG, grown at 37°C to an OD600 ≤0.5 in selective medium containing 100 µM IPTG, and visualized by DIC microscopy. Cells were then washed three times to remove IPTG and grown in the absence of inducer to decrease expression from the pDSW210 vector. Cultures were back-diluted as needed to maintain a low-mid OD600. Cells were visualized every hour following exposure to depletion conditions for a total of 4 hours.
To obtain samples for immunoblot analysis, plasmids pWM4612 (pDSW210-flag-virB10CytoftsNTM-PeriΔSPOR), pWM4582 (pDSW210-flag-virB10CytoftsNTM-Peri), pWM4613 (pDSW210-flag-ftsNΔSPOR), and pWM3643 (pDSW210-flag-ftsN) were transformed into ftsN depletion strain WM4028, grown at the permissive temperature of 30°C to low-mid OD600 in the absence of inducer, and shifted to 42°C in the presence of 10 or 100 µM IPTG for 4 hours. Cultures were continually back-diluted throughout the experiment to remain in logarithmic phase. Cultures were then normalized based on OD600 and boiled in sodium dodecyl sulfate (SDS) dye for 10 minutes prior to storage at −20°C.
Immunoblot analysis
Samples from the FtsN depletion strain were collected as described above and separated by SDS-PAGE, using equal volumes of normalized samples per lane. Using a wet apparatus, FLAG-tagged proteins were transferred onto a nitrocellulose membrane and stained with Swift membrane stain (G-Biosciences; St. Louis, MO) to show that similar levels of proteins in all lanes were loaded and transferred. The membrane was immunoblotted with mouse monoclonal anti-FLAG antibody (1:5,000) and goat anti-mouse secondary antibody conjugated to horseradish peroxidase (HRP; 1:10,000). HRP was detected using Western Lightning ECL Pro (PerkinElmer, Inc., Waltham, MA) and quantified using Image Studio Software (LI-COR; Lincoln, NE).
Supplementary Material
ACKNOWLEDGEMENTS
We thank past and present Margolin lab members for insightful comments and suggestions. We also thank Jennifer Herricks for sharing affinity purified α-FtsA antibody.
This work was supported by NIGMS grant 61074.
REFERENCES
- Aarsman ME, Piette A, Fraipont C, Vinkenvleugel TM, Nguyen-Disteche M, den Blaauwen T. Maturation of the Escherichia coli divisome occurs in two steps. Mol Microbiol. 2005;55:1631–1645. doi: 10.1111/j.1365-2958.2005.04502.x. [DOI] [PubMed] [Google Scholar]
- Addinall SG, Cao C, Lutkenhaus J. FtsN, a late recruit to the septum in Escherichia coli. Mol Microbiol. 1997;25:303–309. doi: 10.1046/j.1365-2958.1997.4641833.x. [DOI] [PubMed] [Google Scholar]
- Alexeeva S, Gadella TW, Jr., Verheul J, Verhoeven GS, den Blaauwen T. Direct interactions of early and late assembling division proteins in Escherichia coli cells resolved by FRET. Mol Microbiol. 2010;77:384–398. doi: 10.1111/j.1365-2958.2010.07211.x. [DOI] [PubMed] [Google Scholar]
- Arends SJ, Williams K, Scott RJ, Rolong S, Popham DL, Weiss DS. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD, and RlpA. J Bacteriol. 2010;192:242–255. doi: 10.1128/JB.01244-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernard CS, Sadasivam M, Shiomi D, Margolin W. An altered FtsA can compensate for the loss of essential cell division protein FtsN in Escherichia coli. Mol Microbiol. 2007;64:1289–1305. doi: 10.1111/j.1365-2958.2007.05738.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E, Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991;354:161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Busiek KK, Eraso JM, Wang Y, Margolin W. The early divisome protein FtsA interacts directly through its 1c subdomain with the cytoplasmic domain of the late divisome protein FtsN. J Bacteriol. 2012;194:1989–2000. doi: 10.1128/JB.06683-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung HS, Yao Z, Goehring NW, Kishony R, Beckwith J, Kahne D. Rapid beta-lactam-induced lysis requires successful assembly of the cell division machinery. Proc Natl Acad Sci USA. 2009;106:21872–21877. doi: 10.1073/pnas.0911674106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corbin BD, Geissler B, Sadasivam M, Margolin W. Z-ring-independent interaction between a subdomain of FtsA and late septation proteins as revealed by a polar recruitment assay. J Bacteriol. 2004;186:7736–7744. doi: 10.1128/JB.186.22.7736-7744.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Xu Y, Lutkenhaus J. Cloning and characterization of ftsN an essential cell division gene in Escherichia coli isolated as a multicopy suppressor of ftsA12(Ts) J Bacteriol. 1993;175:3790–3797. doi: 10.1128/jb.175.12.3790-3797.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K, Xu Y, Lutkenhaus J. Topological characterization of the essential Escherichia coli cell division protein FtsN. J Bacteriol. 1996;178:1328–1334. doi: 10.1128/jb.178.5.1328-1334.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z, Zhao Z, Jakubowski SJ, Krishnamohan A, Margolin W, Christie PJ. A novel cytology-based, two-hybrid screen for bacteria applied to protein-protein interaction studies of a Type IV secretion system. J Bacteriol. 2002;184:5572–5582. doi: 10.1128/JB.184.20.5572-5582.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards DH, Thomaides HB, Errington J. Promiscuous targeting of Bacillus subtilis cell division protein DivIVA to division sites in Escherichia coli and fission yeast. EMBO J. 2000;19:2719–2727. doi: 10.1093/emboj/19.11.2719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erickson HP. FtsZ, a tubulin homologue in prokaryote division. Trends Cell Biol. 1997;7:362–367. doi: 10.1016/S0962-8924(97)01108-2. [DOI] [PubMed] [Google Scholar]
- Garza I, Christie PJ. A putative transmembrane leucine zipper of Agrobacterium VirB10 is essential for T-pilus biogenesis but not type IV secretion. J Bacteriol. 2013;195:3022–3034. doi: 10.1128/JB.00287-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Elraheb D, Margolin W. A gain of function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc Natl Acad Sci USA. 2003;100:4197–4202. doi: 10.1073/pnas.0635003100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissler B, Margolin W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol Microbiol. 2005;58:596–612. doi: 10.1111/j.1365-2958.2005.04858.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerding MA, Liu B, Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J Bacteriol. 2009;191:7383–7401. doi: 10.1128/JB.00811-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Beckwith J. Diverse paths to midcell: assembly of the bacterial cell division machinery. Curr Biol. 2005;15:514–526. doi: 10.1016/j.cub.2005.06.038. [DOI] [PubMed] [Google Scholar]
- Goehring NW, Gueiros-Filho F, Beckwith J. Premature targeting of a cell division protein to midcell allows dissection of divisome assembly in Escherichia coli. Genes Dev. 2005;19:127–137. doi: 10.1101/gad.1253805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goehring NW, Petrovska I, Boyd D, Beckwith J. Mutants, suppressors, and wrinkled colonies: Mutant alleles of the cell division gene ftsQ point to functional domains in FtsQ and a role for domain 1C of FtsA in divisome assembly. J Bacteriol. 2007;189:633–645. doi: 10.1128/JB.00991-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Belin D, Carson MJ, Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman LM, Weiss DS, Beckwith J. Domain-swapping analysis of FtsI, FtsL, and FtsQ, bitopic membrane proteins essential for cell division in Escherichia coli. J Bacteriol. 1997;179:5094–5103. doi: 10.1128/jb.179.16.5094-5103.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimova G, Dautin N, Ladant D. Interaction network among Escherichia coli membrane proteins involved in cell division as revealed by bacterial two-hybrid analysis. J Bacteriol. 2005;187:2233–2243. doi: 10.1128/JB.187.7.2233-2243.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lutkenhaus J. FtsN--trigger for septation. J Bacteriol. 2009;191:7381–7382. doi: 10.1128/JB.01100-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Ehrhardt DW, Margolin W. Colocalization of cell division proteins FtsZ and FtsA to cytoskeletal structures in living Escherichia coli cells by using green fluorescent protein. Proc Natl Acad Sci USA. 1996;93:12998–13003. doi: 10.1073/pnas.93.23.12998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller P, Ewers C, Bertsche U, Anstett M, Kallis T, Breukink E, Fraipont C, Terrak M, Nguyen-Disteche M, Vollmer W. The essential cell division protein FtsN interacts with the murein (peptidoglycan) synthase PBP1B in Escherichia coli. J Biol Chem. 2007;282:36394–36402. doi: 10.1074/jbc.M706390200. [DOI] [PubMed] [Google Scholar]
- Peters NT, Dinh T, Bernhardt TG. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J Bacteriol. 2011;193:4973–4983. doi: 10.1128/JB.00316-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 2002;21:685–693. doi: 10.1093/emboj/21.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichoff S, Lutkenhaus J. Tethering the Z ring to the membrane through a conserved membrane targeting sequence in FtsA. Mol Microbiol. 2005;55:1722–1734. doi: 10.1111/j.1365-2958.2005.04522.x. [DOI] [PubMed] [Google Scholar]
- Pichoff S, Shen B, Sullivan B, Lutkenhaus J. FtsA mutants impaired for self-interaction bypass ZipA suggesting a model in which FtsA's self-interaction competes with its ability to recruit downstream division proteins. Mol Microbiol. 2012;83:151–167. doi: 10.1111/j.1365-2958.2011.07923.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raychaudhuri D. ZipA is a MAP-Tau homolog and is essential for structural integrity of the cytokinetic FtsZ ring during bacterial cell division. EMBO J. 1999;18:2372–2383. doi: 10.1093/emboj/18.9.2372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rico AI, Garcia-Ovalle M, Palacios P, Casanova M, Vicente M. Role of Escherichia coli FtsN protein in the assembly and stability of the cell division ring. Mol Microbiol. 2010;76:760–771. doi: 10.1111/j.1365-2958.2010.07134.x. [DOI] [PubMed] [Google Scholar]
- Rico AI, Krupka M, Vicente M. In the beginning, Escherichia coli assembled the proto-ring: an initial phase of division. J Biol Chem. 2013;288:20830–20836. doi: 10.1074/jbc.R113.479519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Margolin W. Dimerization or oligomerization of the actin-like FtsA protein enhances the integrity of the cytokinetic Z ring. Mol Microbiol. 2007;66:1396–1415. doi: 10.1111/j.1365-2958.2007.05998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiomi D, Margolin W. Compensation for the loss of the conserved membrane targeting sequence of FtsA provides new insights into its function. Mol Microbiol. 2008;67:558–569. doi: 10.1111/j.1365-2958.2007.06085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Söderström B, Skoog K, Blom H, Weiss DS, von Heijne G, Daley DO. Disassembly of the divisome in Escherichia coli: Evidence that FtsZ dissociates before compartmentalization. Mol Microbiol. 2014;92:1–9. doi: 10.1111/mmi.12534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas JD, Daniel RA, Errington J, Robinson C. Export of active green fluorescent protein to the periplasm by the twin-arginine translocase (Tat) pathway in Escherichia coli. Mol Microbiol. 2001;39:47–53. doi: 10.1046/j.1365-2958.2001.02253.x. [DOI] [PubMed] [Google Scholar]
- Ursinus A, van den Ent F, Brechtel S, de Pedro M, Holtje JV, Löwe J, Vollmer W. Murein (peptidoglycan) binding property of the essential cell division protein FtsN from Escherichia coli. J Bacteriol. 2004;186:6728–6737. doi: 10.1128/JB.186.20.6728-6737.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vicente M, Rico AI. The order of the ring: assembly of Escherichia coli cell division components. Mol Microbiol. 2006;61:5–8. doi: 10.1111/j.1365-2958.2006.05233.x. [DOI] [PubMed] [Google Scholar]
- Weiss DS, Chen JC, Ghigo JM, Boyd D, Beckwith J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J Bacteriol. 1999;181:508–520. doi: 10.1128/jb.181.2.508-520.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wissel MC, Wendt JL, Mitchell CJ, Weiss DS. The transmembrane helix of the Escherichia coli division protein FtsI localizes to the septal ring. J Bacteriol. 2005;187:320–328. doi: 10.1128/JB.187.1.320-328.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.