Abstract
The carcinoembryonic antigen (CEA) gene family belongs to the immunoglobulin (Ig) superfamily and codes for a vast number of glycoproteins that differ greatly both in amino acid composition and function. The CEA family is divided into two groups, the carcinoembryonic antigen-related cell adhesion molecules (CEACAMs) and the pregnancy-specific glycoproteins. The CEA family members are implicated in pleiotropic (patho)physiological functions including cell–cell adhesion, pregnancy, immunity, neovascularization, regulation of insulin homeostasis, and carcinogenesis. In general, the CEA-encoded proteins are composed of an extracellular region with Ig variable and constant-like domains and a cytoplasmic region containing signaling motifs. Of particular interest, the well-studied human and mouse CEA genes are arranged in clusters in a single chromosome. Taking into account this characteristic, we made an effort to reconstruct the evolutionary history of the CEA gene family. Toward this end, the publicly available genomes were searched extensively for CEA homologs. The domain organization of the retrieved protein sequences was analyzed, and, subsequently, comprehensive phylogenetic analyses of the entire length CEA homologous proteins were performed. A series of evolutionarily conserved amino acid residues, functionally important, were identified. The relative positioning of these residues on the modeled tertiary structure of novel CEA protein domains revealed that they are, also, spatially conserved. Furthermore, the chromosomal arrangement of CEA genes was examined, and it was found that the CEA genes are preserved in terms of position, transcriptional orientation, and number in all species under investigation.
Keywords: CEA, CEACAM, PSG, immunoglobulins, phylogeny, gene duplication
Introduction
The carcinoembryonic antigen (CEA) gene family, which belongs to the immunoglobulin (Ig) gene superfamily, comprised an exceptionally diverse array of highly glycosylated glycoproteins (Paxton et al. 1987; Zhou et al. 2001). The CEA family is broadly divided into two groups, the CEA-related cell adhesion molecules (CEACAMs) and the pregnancy-specific glycoproteins (PSGs) (Hammarstrom 1999). In humans, based on our current phylogenetic analysis, the CEA family consists of 35 genes, 21 of which are protein coding, arranged in contiguous clusters in chromosome 19 in the region 19q13.2-19q13.4 (Hammarstrom 1999).
The CEA-encoded proteins have varying length and domain organization, which probably reflects their functional divergence. All currently reported CEA-encoded proteins consist of at least one Ig variable (IgV)-like domain, followed by a varying number of Ig constant (IgC)-like domains (Brummendorf and Rathjen 1995). The core structure of these domains, the Ig-like fold, is characterized by two β-sheets (faces) that cross over each other. The IgV-like domain, approximately 110 amino acids long, contains a conserved basic (arginine) and an acidic (aspartate) amino acid, which are proposed to stabilize the Ig-like fold via an intradomain salt bridge. The CFG-face of the IgV-like domain (named after the C-F-G strands it is composed of) mediates homotypic and heterotypic cell–cell adhesion (Taheri et al. 2000). The IgC-like domain, contains two conserved cysteine residues, that occupy the corresponding positions of arginine and aspartate, stabilize the Ig-like conformation by forming a disulphide bridge (Williams and Barclay 1988; Bork et al. 1994).
CEACAM genes are expressed in a wide variety of cell types including epithelial, endothelial, and immune cells such as leukocytes and dendritic cells, whereas PSGs are expressed exclusively in the placental trophoblasts (Hammarstrom 1999). CEACAMs are either inserted into the cell membrane via a transmembrane (TM) domain or they are linked to the membrane via semipenetrating glycosylphosphatidylinositol (GPI) anchorage (Naghibalhossaini et al. 2007). The latter type of membrane anchorage has been detected only in primates thus far. The membrane-bound CEACAMs possess a C-terminal cytoplasmic domain, which may contain motifs associated with signal transduction (Hammarstrom 1999).
Members of the CEA family are implicated in diverse physiological and pathological functions (Obrink 1997; Kuespert et al. 2006). For instance, CEACAMs play a vital role during embryonic development where cell–cell adhesion is necessary to integrate the cells into functional organs (Kuespert et al. 2006). Members of the CEACAM group also serve as receptors of several bacterial and viral pathogens, such as the murine hepatitis virus, Haemophilus influenza, Neisseria meningitides, and N. gonorrhea, which bind CEACAM proteins via their N-terminal IgV-like domain (Bos et al. 1999; Virji et al. 1999, 2000; Villullas et al. 2007). PSGs are secreted proteins from fetal trophoblasts, which are proposed to regulate the maternal–fetal interactions during pregnancy (Hau et al. 1985; Ha et al. 2010). Of particular note, CEA play an important role in carcinogenesis (Scorilas et al. 2003; Michaelidou et al. 2013). The prototypic member of this family, human CEA (henceforth referred to as CEACAM5), was discovered by Gold and Freedman (1965) in the mid-1960s in the blood of patients with colon cancer. CEACAM5 is consistently overexpressed in various malignancies frequently associated with poor patients’ clinical outcome and reduced overall survival (Chevinsky 1991). These properties have made CEACAM5 a prominent clinical cancer biomarker, widely used in early diagnosis, effective prognosis, and monitoring of colon cancer, as well as other types of cancers (Gaglia et al. 1988; Ballesta et al. 1995).
In this study, we made an effort to reconstruct the evolutionary history of CEA gene family and identify conserved amino acids that may play important role in the overall structure and function of CEA proteins. To this direction, the fully sequenced and the nearly complete sequenced genomes were searched for CEA homologs. Members of the CEA family were identified in diverse taxa covering an evolutionary range from cartilaginous fishes to human. Subsequently, comprehensive phylogenetic analyses were performed employing the maximum likelihood (ML) and the neighbor-net methods. The genomic arrangement of the identified CEA-related genes was also analyzed, and it was shown that in different species, these genes are arranged in contiguous clusters with conserved position and orientation of transcription. On the basis of both the phylogenetic and syntenic analyses, we identified eight conserved gene clusters. Furthermore, the protein domain organization of the CEA homologs was examined and amino acid conservation patterns were identified. The three-dimensional (3D) structure of domains from species of the basal taxonomy was predicted with homology modeling, and the evolutionarily conserved amino acids were mapped onto these structures.
Materials and Methods
Sequence Database Searching
The names or accession numbers of the characterized CEA reported in literature were used initially to retrieve their corresponding sequences from the publicly available nonredundant sequence databases ENSEMBL (Flicek et al. 2013), National Center for Biotechnology Information (NCBI)’s RefSeq (Pruitt et al. 2012), and UniProtKB (Magrane and UniProt Consortium 2011). To obtain more putative CEA homologs, these sequences were used subsequently as probes to perform extensive reciprocal BLASTp and tBLASTn (Altschul et al. 1990) searches of genomes with high coverage (>6×) and low coverage (2×). This process was reiterated until convergence, that is, no novel putative CEA sequences could be detected. The longer known transcript was selected. The partial or ambiguous sequences were not included in the subsequent steps of the study. The Translate program (http://web.expasy.org/translate/, last accessed May 27, 2014) was used to translate nucleotide sequences.
Motifs Construction
Representative CEA peptide sequences were aligned with MAFFT v.7 (Katoh and Standley 2013, 2014) and edited with Utopia suite’s CINEMA alignment editor (Pettifer et al. 2009). Sequence motifs were excised from the multiple sequence alignments, manually edited for insertions or gaps. They were submitted to WebLogo3 (Crooks et al. 2004) with default options, to generate consensus sequences.
Chromosomal Localization
The chromosomal localization of the CEA genes was determined using the ENSEMBL GeneView (Flicek et al. 2013) and the NCBI MapViewer (Wolfsberg 2011).
Alignments and Phylogenetic Analyses
The full-length CEA amino acid sequences were aligned with MAFFT v.7. The resulting multiple sequence alignments were edited using CINEMA alignment editor (Pettifer et al. 2009). The trimmed alignments were then used to reconstruct phylogenetic trees by employing two separate methods. To obtain ML-based trees, the method implemented in the software package MEGA, version 5.2 (Tamura et al. 2011) was used. In this study, a distance-based tree (BIONJ) (Gascuel 1997) was used as seed, as well as the nearest-neighbor-interchange heuristic with five discrete gamma categories of evolutionary rates. The number of amino acid substitutions per position was estimated with the JTT model (Jones et al. 1992). Trees were also reconstructed employing the neighbor-net method (Bryant and Moulton 2004) implemented in SplitsTree v.4 (Huson 1998; Kloepper and Huson 2008), a distance-based method able to detect conflict between phylogenetic signals in the form of networks; the Ucorrected P model of substitution was used. For both methods, bootstrap analyses (200 pseudoreplicates) were conducted to evaluate the statistical significance of the reconstructed trees. The trees generated with the ML method were illustrated with Dendroscope v.3 (Huson and Scornavacca 2012).
Protein Domain Organization
The consensus boundaries of the individual protein domains in CEA proteins were determined from the full-length CEA amino acid sequences combining the outputs of the search engines available in SMART v.7 (Letunic et al. 2012), PFAM v.27 (Punta et al. 2012) and CDD v.3 (Derbyshire et al. 2012) and InterPro v.42 (Hunter et al. 2012) protein signature databases. The TM regions were predicted with the programs MINNOU (Cao et al. 2006) and PRED-TMR2 (Pasquier et al. 1999).
Homology Modeling
The 3D structures of the IgV-like domain of MedakaCea, FrogCea7, and LizardCeacam19, and the 3D structure of the IgC-like domain of the FrogCea2 and PlatypusCea1 (target proteins) were predicted by homology modeling. The X-ray crystal structures of the murine Ceacam1a (PDB ID: 1L6Z) (Tan et al. 2002) and the human CEACAM1 (PDB: 2GK2) (Fedarovich et al. 2006) were used as templates to model the IgC-like and IgV-like domain, respectively, with the modeling package Modeller (Sali et al. 1995). To remove any local constraints, the generated protein models were subjected to energy minimization using the Charmm27 forcefield, implemented in Gromacs v.4.5.5 (Hess et al. 2008). The quality of the final modeled protein structures was evaluated using Procheck (Laskowski et al. 1996) and ANOLEA (Melo et al. 1997). The protein models were illustrated with PyMol (DeLano 2002). Furthermore, the secondary structure of the TM domain of the sequences ZebrafishCea1, FrogCea7, LizardCeacam19, and HumanCEACAM1 was predicted using the bioinformatics tools described in Pavlopoulou and Michalopoulos (2011). The predicted TM helices were modeled, template free, as described above.
Results
Identification of CEA Homologs
In this study, we performed comprehensive and updated phylogenetic analyses of the CEA homologs in the available genomes of 33 species: Anolis carolinensis (lizard), Bos taurus (cow), Branchiostoma lancelatum (amphioxus), Callithrix jacchus (marmoset), Canis familiaris (dog), Ciona intestinalis (ascidia), Danio rerio (zebrafish), Dasypus novemcinctus (armadillo), Drosophila melanogaster (fruit fly), Equus caballus (horse), Gallus gallus (chicken), Homo sapiens (human), Latimeria chalumnae (coelacanth), Lepisosteus oculatus (spotted gar), Leucoraja erinacea (little skate), Loxodonta africana (elephant), Macaca mulatta (macaque), Microcebus murinus (mouse lemur), Monodelphis domestica (opossum), Mus musculus (mouse), Myotis lucifugus (microbat), Ornithorhynchus anatinus (platypus), Oryzias latipes (medaka), Otolemur garnettii (bushbaby), Pan troglodytes (chimpanzee), Pelodiscus sinensis (Chinese softshell turtle), Petromyzon marinus (lamprey), Pongo abelii (orangutan), Rattus norvegicus (rat); Taeniopygia guttata (zebra finch); Takifugu rubripes (pufferfish), Tupaia belangeri (tree shrew), and Xenopus tropicalis (frog). The genomes with high coverage were selected to avoid underestimation of the number of CEA genes, like in the case of low coverage genomes. Collectively, 207 CEA protein-encoding genes, 13 pseudogenes, and 1 expressed sequence tag (EST) sequence were identified in the genomes of 20 species representing diverse eukaryotic taxonomic divisions (according to the NCBI taxonomy database [Federhen 2012]) (supplementary table S1, Supplementary Material online) rimates (87), rodentia (47), perissodactyla (9), cetartiodactyla (6), carnivore (10), afrotheria (5), xenarthra (7), metatheria (15), proteotheria (5), sauria (2), amphibia (13) and teleosts (14), and chondrichthyes (1). Despite extensive database searches, CEA homologs were not detected in the complete and well-annotated genomes of aves, insects, and in lower vertebrates such as craniata, cephalochordata, and ascidia.
To prevent confusion, we used the revised nomenclature by Beauchemin et al. (1999), for human and rodent CEA sequences; for the rest, we used the names provided in the original references and the sequence databases. Regarding the newly identified sequences (e.g., primate CEA), they were named by virtue of homology to their closest related well-annotated human and mouse CEA genes. The distant homologs (e.g., frog and fish CEA), with no significant sequence similarity to the known CEA, were commenced by CEA followed by an ascending number depending on their order in the chromosome.
Conserved Structural Features of CEA Proteins
The CEA homologous proteins were found to differ greatly in their length and domain organization. On the basis of the combined output of the signature databases and the multiple alignment of CEA protein sequences, we determined the organization of the three major protein domains in the extracellular region of the CEA proteins, namely IgV-like, IgC-like, and TM, the immunoreceptor tyrosine-based activation motif (ITAM) and immunoreceptor tyrosine-based inhibition motif (ITIM) in the cytoplasmic region and the GPI anchors. Furthermore, consensus protein motifs were derived from the multiple alignment of sequences that correspond to the three extracellular domains and the cytoplasmic domain and a number of conserved amino acid residues were identified (figs. 1–4).
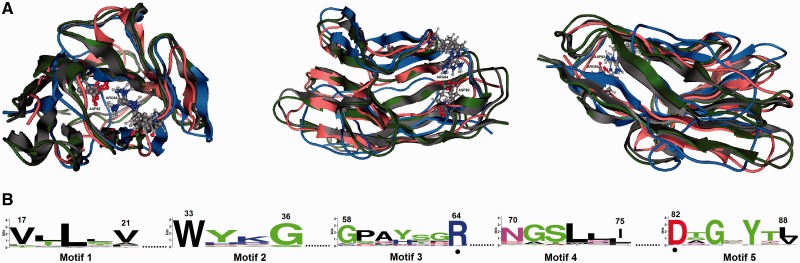
Fig. 1.—
IgV-like domain. (A) Homology models of the IgV-like domain of MedakaCea (pink), FrogCea7 (green), and LizardCeacam19 (blue) in cartoon representation superposed on the human CEACAM1 (PDB ID: 2GK2) (gray). The residues arginine and aspartate that form a salt bridge are shown as a ball-and-stick representation. (B) The conserved proteins motifs derived from the IgV-like domain. The amino acid residue numbers (according to human CEACAM1) are indicated. The invariant residues arginine and aspartate are indicated by dots. The letters, representing amino acid residues of the motif sequences, are piled one on top of another at every position in the sequences. The height of each letter is proportional to the frequency of the corresponding amino acid at that position; the letters are ordered, so the most frequent one is on the top. The height of the whole pile is normalized, so that it indicates the information content (measured in bits) in each position.
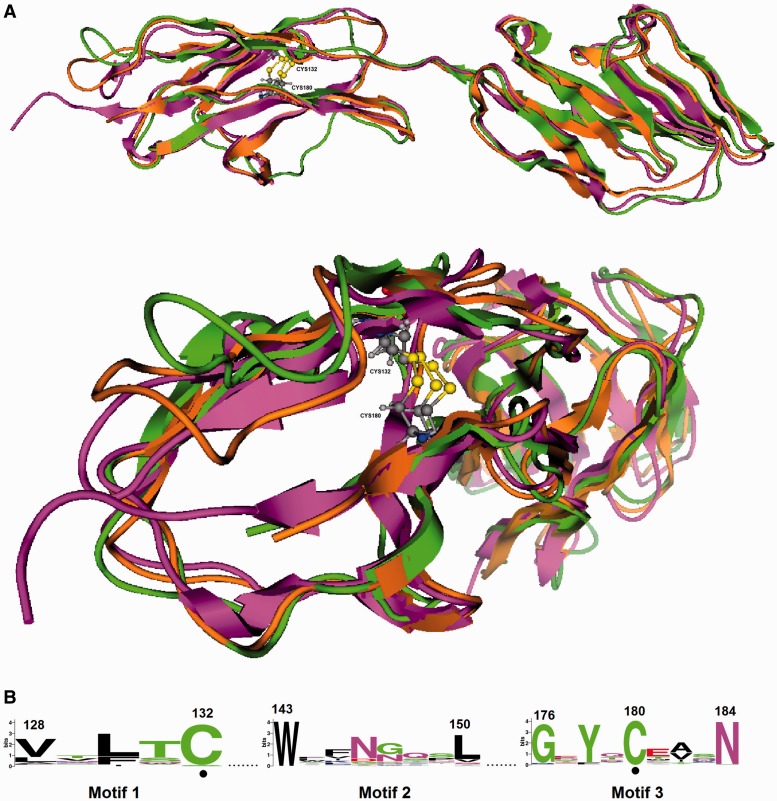
Fig. 2.—
IgC-like domain. (A) Modeled protein structures of the IgC-like domain of FrogCea2 (green) and PlatypusCea1 (brown) superimposed onto the murine Ceacam1a (PDB ID: 1L6Z) (purple). The cysteines involved in the formation of the disulfide bridge are shown as a ball-and-stick representation, and the disulfide bridges are indicated by yellow lines. (B) The conserved proteins motifs are detected in the IgC-like domain, numbered according to murine Ceacam1a. The invariant cysteine residues are denoted by dots.
Fig. 3.—
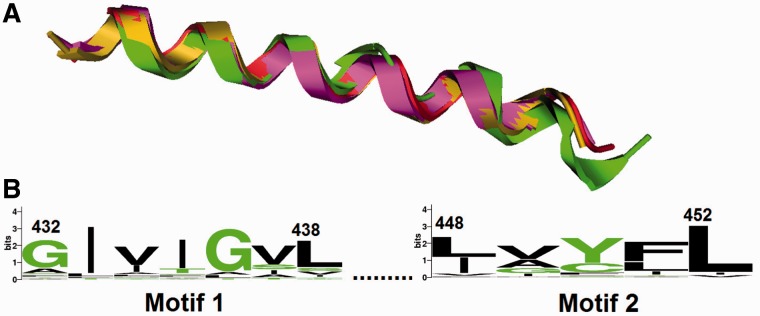
TM domain. (A) Modeled TM helices of HumanCEACAM1 (purple), LizardCeacam19 (gold), FrogCea7 (green), and ZebrafishCea1 (red). (B) The conserved TM protein; the amino acid numbering is based on HumanCEACAM1.
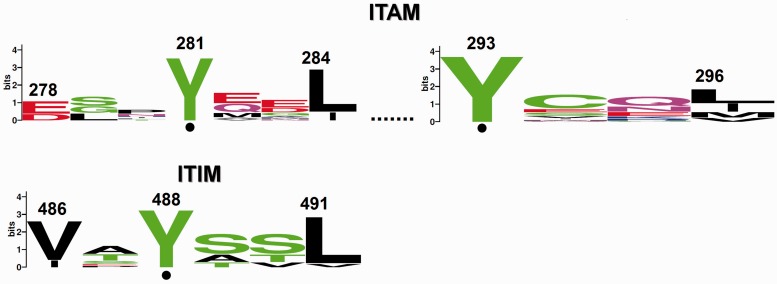
Fig. 4.—
Cytoplasmic domain. ITAM and ITIM. The invariant tyrosine residues are indicated by dots, numbered according to HumanCEACAM19 and MouseCeacam1, respectively.
Given that the 3D structure of a protein is more conserved than its corresponding amino acid sequence, an effort was made to map the position of these residues to the tertiary structure of representative CEA domains. Toward this end, the 3D structure of the IgV-like (fig. 1) and IgC-like (fig. 2) domains from putative, evolutionarily diverse CEA proteins were predicted with homology modeling using the resolved crystal structures of the IgV-like domain of the human CEACAM1 (PDB ID: 2GK2) and the murine Ceacam1a (PDB ID: 1L6Z) as templates, respectively.
As shown in the superimposed structures of the MedakaCea, FrogCea7, and LizardCeacam19 IgV-like domains (fig. 1), the major secondary structures are conserved. The residues arginine (R64) and aspartate (D82) that form a salt bridge are also conserved in the modeled protein structures. Also, the amino acid asparagine (N70), suggested to be involved in glycosylation, was found to be highly conserved.
As shown in figure 2, the modeled 3D structures of the IgC-like domain of FrogCea2 and PlatypusCea1 superimposed onto the N-terminus of the murine Ceacam1a exhibit notable similarity in their secondary structure elements. The two invariant cysteine residues, which are involved in the formation of the disulfide bridge, were found to be spatially conserved in the IgC-like domain of the CEA homologs (fig. 2).
The TM domains of the homologous CEA proteins were predicted to adopt an α-helical conformation (fig. 3). Two prime signature motifs were also identified in the TM domain.
In the cytoplasmic region, consensus ITAM and ITIM were identified, where the tyrosine residue is invariant (fig. 4). Phosphorylation of ITAM/ITIM initiates of terminates, respectively, signal transduction pathways implicated in cellular proliferation (Beauchemin et al. 1997) or regulation of immune response.
Syntenic Mapping of CEA Homologous Genes
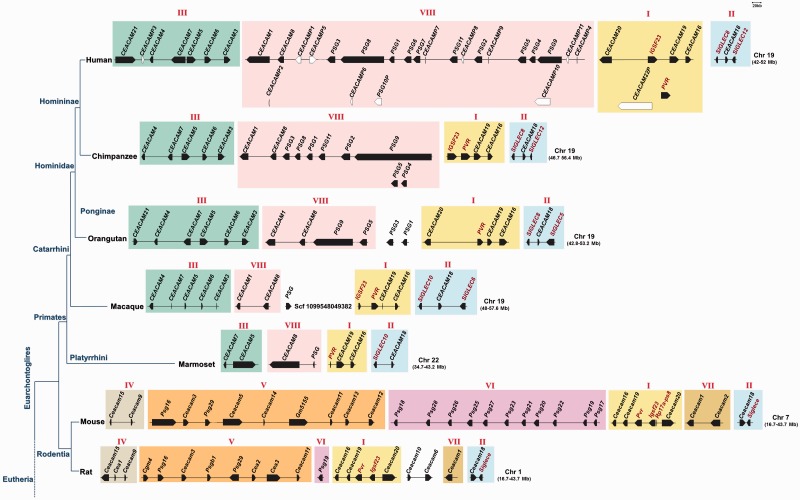
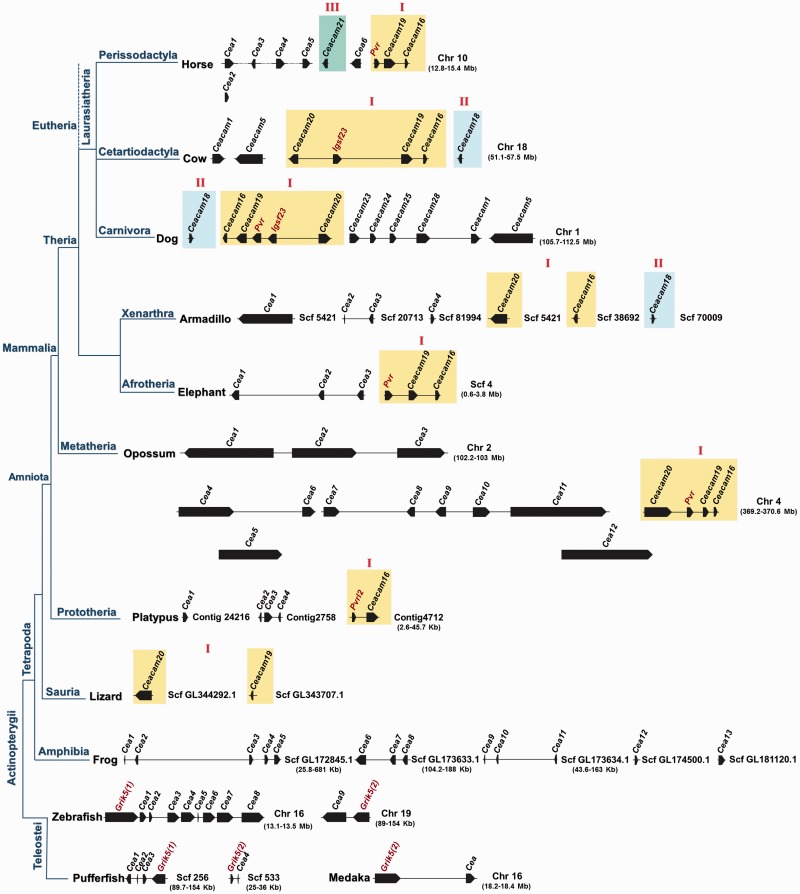
The chromosomal arrangement of the CEA homologous genes found in the genomes of all species under study was investigated. As shown in figure 5, the homologous CEA genes are arranged in clusters, with conserved sequential order, transcriptional orientation, number, and flanking genes, in all species under investigation—at least in the high-coverage genomes. We identified eight (I–VIII) conserved gene clusters in our study, which are indicated by roman numerals and numbers according to the order of their appearance in the evolutionary timetable (fig. 5). The “ancestral” Cluster I appeared first in the common ancestor of extant amniotes and contains the genes CEACAM20, CEACAM19, and CEACAM16. Cluster II was emerged in the common ancestor of extant eutherians for the first time and contains a single gene, CEACAM18. Cluster III appeared in the common ancestor of euarchontoglires and laurasiatheria for the first time (CEACAM21). Subsequent duplications of CEACAM21 have apparently given rise to CEACAM3–7 in the primate lineage. Clusters IV–VII are restricted to the glire lineage and more specifically to rodents. Cluster VIII is primate specific because it was detected only in primates. Of particular note, the CEA homologs of the New World monkeys are localized on chromosome 19. Because of incomplete genomic studies, the CEA genes of several organisms were detected in chromosomal fragments. Therefore, in this study, a CEA member was considered to be absent both if the gene was not detected and the CEA genes that flank it in the prototypic human and mouse sequential order are detected in the same chromosome, scaffold, or contig (fig. 5).
Fig. 5.—
Schematic depiction of the chromosomal arrangement of CEA genes. The orientation of transcription and approximate position and size of CEA genes are indicated. The genomic boundaries of each chromosome/scaffold/contig are shown in parentheses. The CEA protein encoding genes are shown as filled arrowheads, and the CEA pseudogenes are indicated by open arrowheads. The non-CEA genes flanking the CEA genes are shown in dark red. The CEA gene clusters are indicated by roman numerals and different coloration. An NCBI-derived cladogram illustrating the evolutionary relationships of the taxa under study is shown on the left. Chr: chromosome; Scf: scaffold.
Phylogenetic Analyses
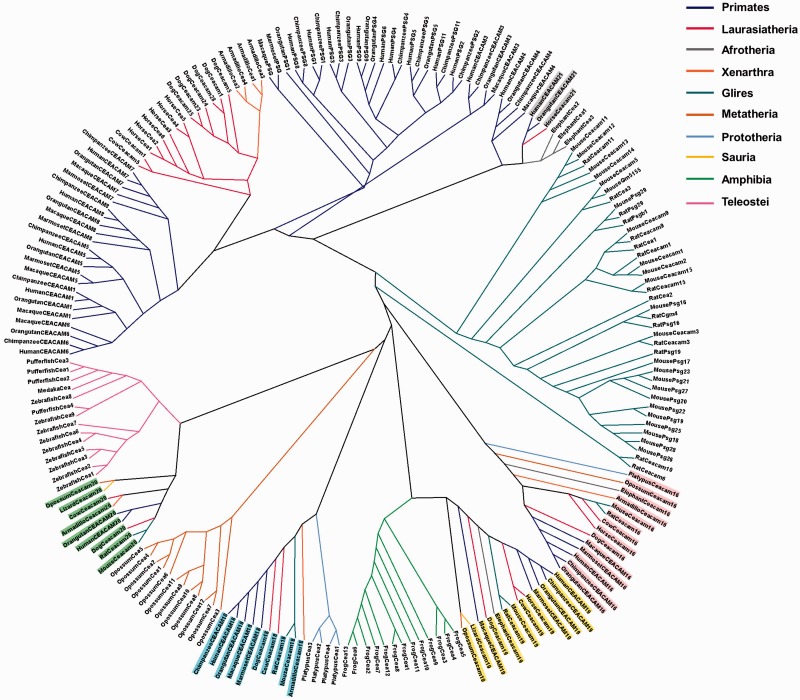
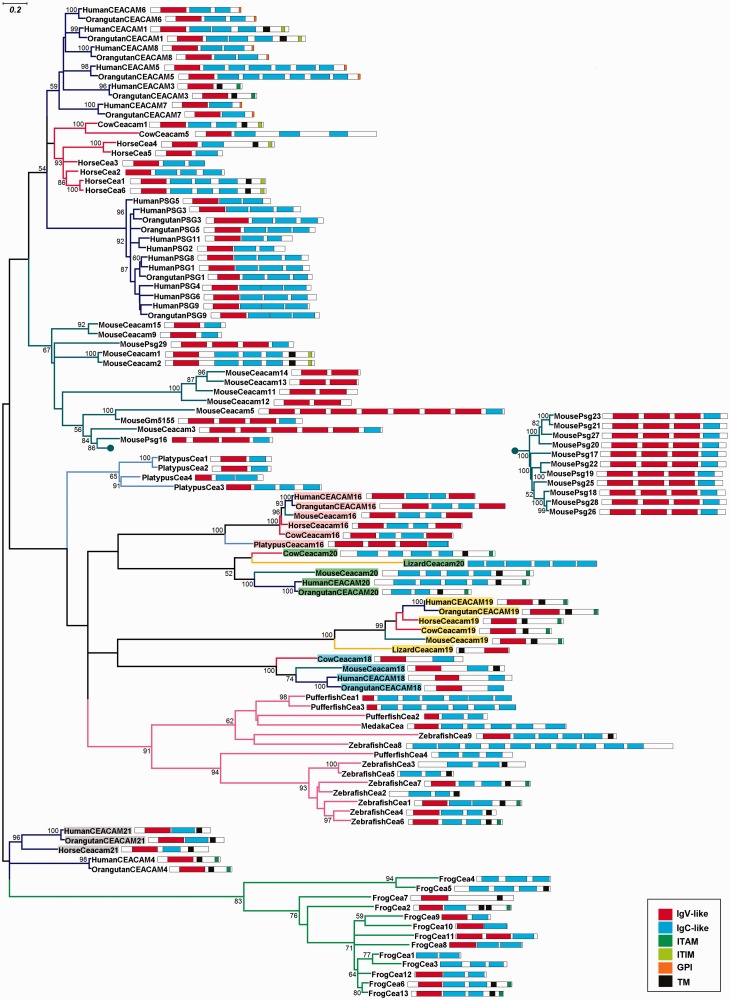
To investigate the evolutionary relationships among CEA, comprehensive phylogenetic analyses based on the entire length protein sequences of all species under study were conducted. Two different methods for phylogenetic reconstruction, ML and neighbor-net, were employed to resolve better the evolutionary relationships. The trees generated with both methods are congruent as their overall topology is similar (fig. 6 and supplementary fig. S1, Supplementary Material online). Representative CEA sequences of selected species with complete or almost complete genomes were selected for more accurate phylogenetic analysis, using both tree construction methods (fig. 7 and supplementary fig. S2, Supplementary Material online). The low support values (below 50) in some nodes suggest alternative branching patterns.
Fig. 6.—
ML radial cladogram of CEA proteins. The sequences are represented by the species name and the CEA protein name. The branches are colored according to the eukaryotic taxa. CEACAM16, CEACAM18, CEACAM19, CEACAM20, and CEACAM21are highlighted by different shading.
Fig. 7.—
ML phylogram of representative CEA proteins. For clarity, the mouse Psg clade (Cluster VI) is condensed and shown separately. Bootstrap values greater than 50% are indicated at the nodes. The branch lengths depict evolutionary distance. The domain organization of CEA proteins is presented on the right of the corresponding sequences. The domain legends are shown in the figure inset. The scale bar at the upper left indicates the length of amino acid substitutions per position. The conventions are the same as in figure 6.
The CEACAM16, CEACAM18, CEACAM19, and CEACAM20 homologs form their own distinct clades with representatives from almost every taxonomic division (figs. 6 and 7, supplementary figs. S1 and S2, Supplementary Material online). CEACAM19 and CEACAM20 appear to be the primordial genes of the CEA family (fig. 5) because they were found in the common ancestor of amniotes. However, neither CEACAM19 nor CEACAM20 orthologs were identified in prototheria (platypus) (fig. 5); this is probably due to incomplete genomic studies. CEACAM16 was detected for first time in prototheria, whereas CEACAM18 appeared later in evolution in the common ancestor of extant eutherians. CEACAM21 orthologs also forms a coherent clade (figs. 6 and 7, supplementary figs. S1 and S2, Supplementary Material online). Interestingly, CEACAM21, which was detected first in the order of perissodactyla and then again in the superfamily of apes (fig. 5), is restricted to three species, namely human, orangutan, and horse. This finding triggers the speculation that either a CEACAM21 gene may have existed in other species, which was deleted during the course of evolution, or CEACAM21 evolved independently within these three species.
CEACAM1, 3–8 were found only in primates (fig. 5). In particular, CEACAM5, CEACAM7, and CEACAM8 appeared in the common ancestor of New World monkeys (marmoset) for first time whereas CEACAM1, CEACAM3, CEACAM4, and CEACAM6 arose later in Old World monkeys (macaque) (fig. 5). CEACAM1 and CEACAM3–8 appear to form separate monophyletic branches (albeit moderately supported) (figs. 6 and 7, supplementary figs. S1 and S2, Supplementary Material online), leading to the suggestion that primate-specific CEACAM1, 3–8 gene duplications must have taken place. The domain organization of CEACAM1, 3–8 is also preserved among species (fig. 7). The GPI anchor was detected only in primates CEACAM5–8 corroborating, in this way, previous reports (Naghibalhossaini and Stanners 2004).
Based on the phylogenies (figs. 6 and 7, supplementary figs. S1 and S2, Supplementary Material online), the PSG genes of primates and the corresponding Psg genes of rodents form two different monophyletic branches, leading to the suggestion that PSG and Psg genes have likely expanded after the divergence of primates and rodents. Given that the PSG protein sequences of apes cluster with the corresponding PSGs of the fellow apes and not with the PSGs of their own species (e.g., human, chimpanzee, and orangutan PSG3) (figs. 6 and 7, supplementary figs. S1 and S2, Supplementary Material online) along with the observation that both their length and domain organization are different (fig. 7), we speculate that the PSGs of apes were derived from duplication events that have presumably preceded the speciation of apes. On the other hand, the PSGs of the New World monkeys form a subclade within the PSG clade (fig. 6 and supplementary fig. S1, Supplementary Material online), suggesting that they have rather evolved independently of those in apes.
As opposed to primate PSGs, a series of species-specific gene duplications must have occurred in rodents yielding 11 Psg paralogs (Cluster VI) (fig. 5) in mouse, which share significant sequence similarity (figs. 6 and 7 and supplementary figs. S1 and S2, Supplementary Material online). Regarding the domain organization, mouse Psg proteins harbor three IgV-like domains, whereas primate PSGs possess only one (fig. 7). In the rodent-specific Cluster VII (fig. 5), Ceacam1 and Ceacam2 are likely the products of a tandem gene duplication subsequent to the mouse–rat divergence because mouse Ceacam1 and Ceacam2 cluster together with high confidence (figs. 6 and 7, supplementary figs. S1 and S2, Supplementary Material online). However, this is not the case in the genes located in Clusters IV and V (fig. 5), which appear to have expanded prior to the rodent speciation (figs. 6 and 7, supplementary figs. S1 and S2, Supplementary Material online).
The Cea homologs that were identified in zebrafish and pufferfish are located in two different chromosomal fragments flanked by the Grik5 co-orthologs, Grik5(1) and Grik5(2) (fig. 5). Given that a whole-genome duplication occurred in teleost fishes subsequent to their divergence from nonteleost ray-finned fishes, approximately 320–400 Ma (Hoegg et al. 2004; Jaillon et al. 2004; Meyer and Van de Peer 2005; Kasahara et al. 2007), it would be reasonable to suggest that the zebrafish and pufferfish Cea are probably the products of this teleost-specific duplication.
According to the phylogenetic trees (figs. 6 and 7, supplementary figs. S1 and S2, Supplementary Material online), the teleost Cea protein sequences cluster in a well-supported monophyletic clade (with a bootstrap value of 91) (fig. 7). Therefore, the Cea genes detected in the contemporary teleost genomes must have been derived from a series of lineage-specific duplications, as in the case of amphibian, prototherian, metatherian, and specific therian CEA-related genes. This hypothesis is also supported by the relatively large evolutionary distances and the diverse domain organization of the proteins encoded by the above genes (fig. 7).
Discussion
Several experimental studies have focused on the expansion of CEA in specific species or taxa (Zhou et al. 2001; McLellan et al. 2005; Zebhauser et al. 2005; Weichselbaumer et al. 2011). In a more recent experimental effort, several CEA-related genes were also detected in vertebrates (Chang et al. 2013). The availability of a growing number of sequenced genomes enabled us to perform, for the first time, comprehensive phylogenetic and structural analyses of CEA. In this study, CEA members were identified in organisms from different taxonomic divisions, ranging from cartilaginous fishes to humans. An EST sequence was detected in little skate (L. erinacea), which was found to be a CEA homolog based on BLAST searches. This allowed us to trace the evolutionary origin of CEA approximately 450–420 Ma when chondrichthyes emerged (Venkatesh et al. 2014). A large number of CEA members were detected in teleosts, frog, platypus, opossum, elephant, armadillo, dog, and horse without any homologs from other species, suggesting lineage/species-specific gene amplification. PSG homologs were detected exclusively in the superorder of euarchontoglires (primates and rodents), which have hemochorial placentae (Carter and Enders 2004) and not in other mammalian orders with different type of placentation such as epitheliochorial or endotheliochorial (Zeiler et al. 2007). On the basis of this finding, we could suggest that PSGs have expanded after the radiation of euarchontoglires to perform functions related to the hemochorial mode of placentation.
Subsequently, phylogenetic reconstructions were performed with the entire length of the CEA-encoded proteins to include all the available evolutionary information that is present in the amino acid sequences. In this way, a series of sequentially and spatially conserved amino acids were also identified in the IgV- and IgC-like domains. The conservation of these residues across the diverse CEA family members suggests the importance of these residues in the overall structure and function of CEA.
In this study, the chromosomal arrangement of the CEA homologs in all species under investigation was examined. A prominent feature of the CEA gene family is that it consists of clusters of genes with conserved order and orientation, mapped to a single chromosome, in all species. On the basis of both syntenic and phylogenetic analyses, we identified a total of eight (I–VIII) conserved gene clusters, the first one appearing for first time in the common ancestor of amniotes. Moreover, the flanking non-CEA genes such as Grik5, PVR, SIGLECs, and IGSF23 are also conserved with the same order and position in all organisms under study. Given that shared synteny is likely associated with function (Wang et al. 2008), we suggest that these genes may have evolved along with CEA to complement CEA’s function.
The CEA gene family represents a notable example of gene duplication, a process suggested to be essential for the development of novel genes (Demuth et al. 2006). The extensive presence of duplicated genes such as kallikreins (Pavlopoulou et al. 2010), bitter taste receptor (T2R) genes, mammalian lysozyme gene family (Dong et al. 2009), genes encoding for keratin associated proteins (KRTAPs) (Wu et al. 2008), and the oxidative phosphorylation (OXPHOS) gene families (De Grassi et al. 2008), all of which are implicated in important physiological processes, points out the importance of this process. We assume that successive rounds of gene duplications, followed by deletions, inversions, translocations, and divergence have likely given rise to the CEA genes found in the contemporary genomes.
Of particular note, both ITAM and ITIM were detected together in human, orangutan, mouse, horse, and cow CEACAM proteins. This observation leads to the suggestion that evolutionary pressure could have applied to ITAM and ITIM, motifs exerting opposing signaling effects (activating vs. inhibitory), to coevolve (Kammerer and Zimmermann 2010). In particular, recognition of bacterial pathogens by CEACAM3 results to phosporylation of its ITAM by protein kinases of the Src family; in turn, a signal transduction pathway is initiated that leads to bacterial engulfment and killing (Hauck et al. 1998; McCaw et al. 2003). On the other hand, it was shown that the presence of the ITIM of CEACAM1 was essential to suppress adaptive immune response upon bacterial infection of the genus Neisseria (Boulton and Gray-Owen 2002). Moreover, CEACAM1, as opposed to CEACAM3, acts as a tumor suppressor, shown to inhibit the growth of prostate, colon, and breast tumors (Estrera et al. 2001; Volpert et al. 2002; Sappino et al. 2012). The ITIM could presumably account for CEACAM1’s tumor suppressive properties. In this study, an ITAM was also detected in the cytoplasmic tail of CEACAM19, which is overexpressed in several types of cancer (Scorilas et al. 2003; Michaelidou et al. 2013). The oncogenic potential of CEACAM19 may, at least partially, depend on the presence of the ITAM. Further experimental studies could probably verify the signaling regulatory role of ITAM/ITIM in various cellular activities.
We expect that the findings of our study could lay the foundation for the design of experimental studies directed toward the elucidation of the biochemical function of the putative CEA and CEA-encoding proteins, taking into consideration the identified protein patterns. The conserved amino acids, also, detected in the protein sequences could represent potential drug targets and should be considered in light of their exploitation in the design of therapeutic agents in anticancer research.
Supplementary Material
Supplementary table S1 and figures S1 and S2 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
This work was supported by the Commission of the European Community through the INsPiRE project (EU-FP7-REGPOT-2011-1, Proposal no: 284460).
Literature Cited
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Ballesta AM, Molina R, Filella X, Jo J, Gimenez N. Carcinoembryonic antigen in staging and follow-up of patients with solid tumors. Tumour Biol. 1995;16:32–41. doi: 10.1159/000217926. [DOI] [PubMed] [Google Scholar]
- Beauchemin N, et al. Association of biliary glycoprotein with protein tyrosine phosphatase SHP-1 in malignant colon epithelial cells. Oncogene. 1997;14:783–790. doi: 10.1038/sj.onc.1200888. [DOI] [PubMed] [Google Scholar]
- Beauchemin N, et al. Redefined nomenclature for members of the carcinoembryonic antigen family. Exp Cell Res. 1999;252:243–249. doi: 10.1006/excr.1999.4610. [DOI] [PubMed] [Google Scholar]
- Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- Bos MP, Hogan D, Belland RJ. Homologue scanning mutagenesis reveals CD66 receptor residues required for neisserial Opa protein binding. J Exp Med. 1999;190:331–340. doi: 10.1084/jem.190.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulton IC, Gray-Owen SD. Neisserial binding to CEACAM1 arrests the activation and proliferation of CD4+ T lymphocytes. Nat Immunol. 2002;3:229–236. doi: 10.1038/ni769. [DOI] [PubMed] [Google Scholar]
- Brummendorf T, Rathjen FG. Cell adhesion molecules 1: immunoglobulin superfamily. Protein Profile. 1995;2:963–1108. [PubMed] [Google Scholar]
- Bryant D, Moulton V. Neighbor-net: an agglomerative method for the construction of phylogenetic networks. Mol Biol Evol. 2004;21:255–265. doi: 10.1093/molbev/msh018. [DOI] [PubMed] [Google Scholar]
- Cao B, Porollo A, Adamczak R, Jarrell M, Meller J. Enhanced recognition of protein transmembrane domains with prediction-based structural profiles. Bioinformatics. 2006;22:303–309. doi: 10.1093/bioinformatics/bti784. [DOI] [PubMed] [Google Scholar]
- Carter AM, Enders AC. Comparative aspects of trophoblast development and placentation. Reprod Biol Endocrinol. 2004;2:46. doi: 10.1186/1477-7827-2-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CL, et al. Widespread divergence of the CEACAM/PSG genes in vertebrates and humans suggests sensitivity to selection. PLoS One. 2013;8:e61701. doi: 10.1371/journal.pone.0061701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chevinsky AH. CEA in tumors of other than colorectal origin. Semin Surg Oncol. 1991;7:162–166. doi: 10.1002/ssu.2980070309. [DOI] [PubMed] [Google Scholar]
- Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Grassi A, Lanave C, Saccone C. Genome duplication and gene-family evolution: the case of three OXPHOS gene families. Gene. 2008;421:1–6. doi: 10.1016/j.gene.2008.05.011. [DOI] [PubMed] [Google Scholar]
- DeLano WL. The PyMOL molecular graphics system. San Carlos (CA): DeLano Scientific; 2002. [Google Scholar]
- Demuth JP, De Bie T, Stajich JE, Cristianini N, Hahn MW. The evolution of mammalian gene families. PLoS One. 2006;1:e85. doi: 10.1371/journal.pone.0000085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derbyshire MK, Lanczycki CJ, Bryant SH, Marchler-Bauer A. Annotation of functional sites with the Conserved Domain Database. Database (Oxford) 2012;2012:bar058. doi: 10.1093/database/bar058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong D, Jones G, Zhang S. Dynamic evolution of bitter taste receptor genes in vertebrates. BMC Evol Biol. 2009;9:12. doi: 10.1186/1471-2148-9-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Estrera VT, Chen DT, Luo W, Hixson DC, Lin SH. Signal transduction by the CEACAM1 tumor suppressor. Phosphorylation of serine 503 is required for growth-inhibitory activity. J Biol Chem. 2001;276:15547–15553. doi: 10.1074/jbc.M008156200. [DOI] [PubMed] [Google Scholar]
- Fedarovich A, Tomberg J, Nicholas RA, Davies C. Structure of the N-terminal domain of human CEACAM1: binding target of the opacity proteins during invasion of Neisseria meningitidis and N. gonorrhoeae. Acta Crystallogr D Biol Crystallogr. 2006;62:971–979. doi: 10.1107/S0907444906020737. [DOI] [PubMed] [Google Scholar]
- Federhen S. The NCBI taxonomy database. Nucleic Acids Res. 2012;40:D136–D143. doi: 10.1093/nar/gkr1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flicek P, et al. Ensembl 2013. Nucleic Acids Res. 2013;41:D48–D55. doi: 10.1093/nar/gks1236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaglia P, et al. Prognostic value of CEA and ferritin assay in breast cancer: a multivariate analysis. Eur J Cancer Clin Oncol. 1988;24:1151–1155. doi: 10.1016/0277-5379(88)90121-6. [DOI] [PubMed] [Google Scholar]
- Gascuel O. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol. 1997;14:685–695. doi: 10.1093/oxfordjournals.molbev.a025808. [DOI] [PubMed] [Google Scholar]
- Gold P, Freedman SO. Specific carcinoembryonic antigens of the human digestive system. J Exp Med. 1965;122:467–481. doi: 10.1084/jem.122.3.467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ha CT, et al. Human pregnancy specific beta-1-glycoprotein 1 (PSG1) has a potential role in placental vascular morphogenesis. Biol Reprod. 2010;83:27–35. doi: 10.1095/biolreprod.109.082412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarstrom S. The carcinoembryonic antigen (CEA) family: structures, suggested functions and expression in normal and malignant tissues. Semin Cancer Biol. 1999;9:67–81. doi: 10.1006/scbi.1998.0119. [DOI] [PubMed] [Google Scholar]
- Hau J, Gidley-Baird AA, Westergaard JG, Teisner B. The effect on pregnancy of intrauterine administration of antibodies against two pregnancy-associated murine proteins: murine pregnancy-specific beta 1-glycoprotein and murine pregnancy-associated alpha 2-glycoprotein. Biomed Biochim Acta. 1985;44:1255–1259. [PubMed] [Google Scholar]
- Hauck CR, Meyer TF, Lang F, Gulbins E. CD66-mediated phagocytosis of Opa52 Neisseria gonorrhoeae requires a Src-like tyrosine kinase- and Rac1-dependent signalling pathway. EMBO J. 1998;17:443–454. doi: 10.1093/emboj/17.2.443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B, Kutzner C, van der Spoel D, Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J Chem Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol. 2004;59:190–203. doi: 10.1007/s00239-004-2613-z. [DOI] [PubMed] [Google Scholar]
- Hunter S, et al. InterPro in 2011: new developments in the family and domain prediction database. Nucleic Acids Res. 2012;40:D306–D312. doi: 10.1093/nar/gkr948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huson DH. SplitsTree: analyzing and visualizing evolutionary data. Bioinformatics. 1998;14:68–73. doi: 10.1093/bioinformatics/14.1.68. [DOI] [PubMed] [Google Scholar]
- Huson DH, Scornavacca C. Dendroscope 3: an interactive tool for rooted phylogenetic trees and networks. Syst Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- Jaillon O, et al. Genome duplication in the teleost fish Tetraodon nigroviridis reveals the early vertebrate proto-karyotype. Nature. 2004;431:946–957. doi: 10.1038/nature03025. [DOI] [PubMed] [Google Scholar]
- Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
- Kammerer R, Zimmermann W. Coevolution of activating and inhibitory receptors within mammalian carcinoembryonic antigen families. BMC Biol. 2010;8:12. doi: 10.1186/1741-7007-8-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kasahara M, et al. The medaka draft genome and insights into vertebrate genome evolution. Nature. 2007;447:714–719. doi: 10.1038/nature05846. [DOI] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Standley DM. MAFFT: iterative refinement and additional methods. Methods Mol Biol. 2014;1079:131–146. doi: 10.1007/978-1-62703-646-7_8. [DOI] [PubMed] [Google Scholar]
- Kloepper TH, Huson DH. Drawing explicit phylogenetic networks and their integration into SplitsTree. BMC Evol Biol. 2008;8:22. doi: 10.1186/1471-2148-8-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuespert K, Pils S, Hauck CR. CEACAMs: their role in physiology and pathophysiology. Curr Opin Cell Biol. 2006;18:565–571. doi: 10.1016/j.ceb.2006.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laskowski RA, Rullmannn JA, MacArthur MW, Kaptein R, Thornton JM. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR. 1996;8:477–486. doi: 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 2012;40:D302–D305. doi: 10.1093/nar/gkr931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magrane M, UniProt Consortium. 2011. UniProt Knowledgebase: a hub of integrated protein data. Database (Oxford) 2011:bar009. [DOI] [PMC free article] [PubMed]
- McCaw SE, Schneider J, Liao EH, Zimmermann W, Gray-Owen SD. Immunoreceptor tyrosine-based activation motif phosphorylation during engulfment of Neisseria gonorrhoeae by the neutrophil-restricted CEACAM3 (CD66d) receptor. Mol Microbiol. 2003;49:623–637. doi: 10.1046/j.1365-2958.2003.03591.x. [DOI] [PubMed] [Google Scholar]
- McLellan AS, et al. Structure and evolution of the mouse pregnancy-specific glycoprotein (Psg) gene locus. BMC Genomics. 2005;6:4. doi: 10.1186/1471-2164-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melo F, Devos D, Depiereux E, Feytmans E. ANOLEA: a www server to assess protein structures. Proc Int Conf Intell Syst Mol Biol. 1997;5:187–190. [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD) Bioessays. 2005;27:937–945. doi: 10.1002/bies.20293. [DOI] [PubMed] [Google Scholar]
- Michaelidou K, Tzovaras A, Missitzis I, Ardavanis A, Scorilas A. The expression of the CEACAM19 gene, a novel member of the CEA family, is associated with breast cancer progression. Int J Oncol. 2013;42:1770–1777. doi: 10.3892/ijo.2013.1860. [DOI] [PubMed] [Google Scholar]
- Naghibalhossaini F, Stanners CP. Minimal mutations are required to effect a radical change in function in CEA family members of the Ig superfamily. J Cell Sci. 2004;117:761–769. doi: 10.1242/jcs.00903. [DOI] [PubMed] [Google Scholar]
- Naghibalhossaini F, Yoder AD, Tobi M, Stanners CP. Evolution of a tumorigenic property conferred by glycophosphatidyl-inositol membrane anchors of carcinoembryonic antigen gene family members during the primate radiation. Mol Biol Cell. 2007;18:1366–1374. doi: 10.1091/mbc.E06-10-0884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obrink B. CEA adhesion molecules: multifunctional proteins with signal-regulatory properties. Curr Opin Cell Biol. 1997;9:616–626. doi: 10.1016/S0955-0674(97)80114-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasquier C, Promponas VJ, Palaios GA, Hamodrakas JS, Hamodrakas SJ. A novel method for predicting transmembrane segments in proteins based on a statistical analysis of the SwissProt database: the PRED-TMR algorithm. Protein Eng. 1999;12:381–385. doi: 10.1093/protein/12.5.381. [DOI] [PubMed] [Google Scholar]
- Pavlopoulou A, Michalopoulos I. State-of-the-art bioinformatics protein structure prediction tools (Review) Int J Mol Med. 2011;28:295–310. doi: 10.3892/ijmm.2011.705. [DOI] [PubMed] [Google Scholar]
- Pavlopoulou A, Pampalakis G, Michalopoulos I, Sotiropoulou G. Evolutionary history of tissue kallikreins. PLoS One. 2010;5:e13781. doi: 10.1371/journal.pone.0013781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paxton RJ, Mooser G, Pande H, Lee TD, Shively JE. Sequence analysis of carcinoembryonic antigen: identification of glycosylation sites and homology with the immunoglobulin supergene family. Proc Natl Acad Sci U S A. 1987;84:920–924. doi: 10.1073/pnas.84.4.920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettifer S, et al. Visualising biological data: a semantic approach to tool and database integration. BMC Bioinformatics. 2009;10(Suppl 6):S19. doi: 10.1186/1471-2105-10-S6-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruitt KD, Tatusova T, Brown GR, Maglott DR. NCBI Reference Sequences (RefSeq): current status, new features and genome annotation policy. Nucleic Acids Res. 2012;40:D130–D135. doi: 10.1093/nar/gkr1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Punta M, et al. The Pfam protein families database. Nucleic Acids Res. 2012;40:D290–D301. doi: 10.1093/nar/gkr1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- Sappino AP, et al. The CEACAM1 tumor suppressor is an ATM and p53-regulated gene required for the induction of cellular senescence by DNA damage. Oncogenesis. 2012;1:e7. doi: 10.1038/oncsis.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorilas A, Chiang PM, Katsaros D, Yousef GM, Diamandis EP. Molecular characterization of a new gene, CEAL1, encoding for a carcinoembryonic antigen-like protein with a highly conserved domain of eukaryotic translation initiation factors. Gene. 2003;310:79–89. doi: 10.1016/s0378-1119(03)00499-2. [DOI] [PubMed] [Google Scholar]
- Taheri M, et al. Self recognition in the Ig superfamily. Identification of precise subdomains in carcinoembryonic antigen required for intercellular adhesion. J Biol Chem. 2000;275:26935–26943. doi: 10.1074/jbc.M909242199. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan K, et al. Crystal structure of murine sCEACAM1a[1,4]: a coronavirus receptor in the CEA family. EMBO J. 2002;21:2076–2086. doi: 10.1093/emboj/21.9.2076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, et al. Elephant shark genome provides unique insights into gnathostome evolution. Nature. 2014;505:174–179. doi: 10.1038/nature12826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villullas S, Hill DJ, Sessions RB, Rea J, Virji M. Mutational analysis of human CEACAM1: the potential of receptor polymorphism in increasing host susceptibility to bacterial infection. Cell Microbiol. 2007;9:329–346. doi: 10.1111/j.1462-5822.2006.00789.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virji M, et al. Critical determinants of host receptor targeting by Neisseria meningitidis and Neisseria gonorrhoeae: identification of Opa adhesiotopes on the N-domain of CD66 molecules. Mol Microbiol. 1999;34:538–551. doi: 10.1046/j.1365-2958.1999.01620.x. [DOI] [PubMed] [Google Scholar]
- Virji M, et al. Carcinoembryonic antigens are targeted by diverse strains of typable and non-typable Haemophilus influenzae. Mol Microbiol. 2000;36:784–795. doi: 10.1046/j.1365-2958.2000.01885.x. [DOI] [PubMed] [Google Scholar]
- Volpert O, et al. Inhibition of prostate tumor angiogenesis by the tumor suppressor CEACAM1. J Biol Chem. 2002;277:35696–35702. doi: 10.1074/jbc.M205319200. [DOI] [PubMed] [Google Scholar]
- Wang Y, et al. Sequencing and comparative analysis of a conserved syntenic segment in the Solanaceae. Genetics. 2008;180:391–408. doi: 10.1534/genetics.108.087981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichselbaumer M, et al. Phylogenetic discordance of human and canine carcinoembryonic antigen (CEA, CEACAM) families, but striking identity of the CEA receptors will impact comparative oncology studies. PLoS Curr. 2011;3:RRN1223. doi: 10.1371/currents.RRN1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams AF, Barclay AN. The immunoglobulin superfamily—domains for cell surface recognition. Annu Rev Immunol. 1988;6:381–405. doi: 10.1146/annurev.iy.06.040188.002121. [DOI] [PubMed] [Google Scholar]
- Wolfsberg TG. Using the NCBI Map Viewer to browse genomic sequence data. Curr Protoc Hum Genet. 2011 doi: 10.1002/0471142905.hg1805s69. Chapter 18:Unit18.5. [DOI] [PubMed] [Google Scholar]
- Wu DD, Irwin DM, Zhang YP. Molecular evolution of the keratin associated protein gene family in mammals, role in the evolution of mammalian hair. BMC Evol Biol. 2008;8:241. doi: 10.1186/1471-2148-8-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zebhauser R, et al. Identification of a novel group of evolutionarily conserved members within the rapidly diverging murine Cea family. Genomics. 2005;86:566–580. doi: 10.1016/j.ygeno.2005.07.008. [DOI] [PubMed] [Google Scholar]
- Zeiler M, Leiser R, Johnson GA, Tinneberg HR, Pfarrer C. Development of an in vitro model for bovine placentation: a comparison of the in vivo and in vitro expression of integrins and components of extracellular matrix in bovine placental cells. Cells Tissues Organs. 2007;186:229–242. doi: 10.1159/000107947. [DOI] [PubMed] [Google Scholar]
- Zhou GQ, Zhang Y, Hammarstrom S. The carcinoembryonic antigen (CEA) gene family in non-human primates. Gene. 2001;264:105–112. doi: 10.1016/s0378-1119(00)00595-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.