Abstract
Transposable elements (TEs) and satellite DNAs (satDNAs) are abundant components of most eukaryotic genomes studied so far and their impact on evolution has been the focus of several studies. A number of studies linked TEs with satDNAs, but the nature of their evolutionary relationships remains unclear. During in silico analyses of the Drosophila virilis assembled genome, we found a novel DNA transposon we named Tetris based on its modular structure and diversity of rearranged forms. We aimed to characterize Tetris and investigate its role in generating satDNAs. Data mining and sequence analysis showed that Tetris is apparently nonautonomous, with a structure similar to foldback elements, and present in D. virilis and D. americana. Herein, we show that Tetris shares the final portions of its terminal inverted repeats (TIRs) with DAIBAM, a previously described miniature inverted transposable element implicated in the generation of chromosome inversions. Both elements are likely to be mobilized by the same autonomous TE. Tetris TIRs contain approximately 220-bp internal tandem repeats that we have named TIR-220. We also found TIR-220 repeats making up longer (kb-size) satDNA-like arrays. Using bioinformatic, phylogenetic and cytogenomic tools, we demonstrated that Tetris has contributed to shaping the genomes of D. virilis and D. americana, providing internal tandem repeats that served as building blocks for the amplification of satDNA arrays. The β-heterochromatic genomic environment seemed to have favored such amplification. Our results imply for the first time a role for foldback elements in generating satDNAs.
Keywords: repetitive DNA, transposable elements, satellite DNA, fiber-FISH, β-heterochromatin, polytene chromosomes
Introduction
Repetitive DNAs are major components of most eukaryotic genomes studied so far, comprising more than 50% of the total DNA in many organisms, such as humans, maize, and the red flour beetle (Wang et al. 2008; Schnable et al. 2009; de Koning et al. 2011). They include many types of elements that can be classified based on their genomic organization as tandem or interspersed repeats.
Satellite DNAs (satDNAs) are the most abundant class of tandem repeats. They consist of repeat units typically organized as large arrays (up to several megabases), oriented in a head-to-tail fashion, and located in heterochromatic regions of chromosomes near centromeres and telomeres (Charlesworth et al. 1994; Plohl et al. 2008). Interspersed repeats are mainly represented by transposable elements (TEs), which consist of repeats that are able to proliferate and move throughout a genome (Kidwell 2002). TEs are classified in a hierarchical system of many levels. The first level is that of class and divides TEs into two groups based on the existence of an RNA intermediate step in transposition. Lower levels of classification include subclass, order, and superfamily (Wicker et al. 2007).
Several studies show that TEs have a more dispersed chromosome distribution compared with satDNAs, but both tend to accumulate preferentially in heterochromatic regions (Pimpinelli et al. 1995; Heslop-Harrison and Schwarzacher 2011). TEs and satDNAs are among the most rapidly evolving genomic elements and account for most of the variation seen in genome size and architecture among eukaryotes (Devos et al. 2002; Bosco et al. 2007).
The finding that some satDNAs share sequence similarity to TEs suggests an evolutionary link between these two types of repetitive DNAs. However, this issue was investigated only in a few studies. For example, data reviewed by Wong and Choo (2004) suggest that centromeric satDNAs may originate from whole or internal parts of TEs, probably by unequal crossing over (UCO). Most of such cases described so far involve retroelements from the gypsy superfamily in plants (Cheng and Murata 2003; Macas et al. 2009; Sharma et al. 2013). Studies of this phenomenon in animal species have evidenced no generalities. Examples include the SGM DNA transposons in Drosophila guanche (Miller et al. 2000), a Tc1-like element in the frog Rana esculenta (Pontecorvo et al. 2000), and the L1 retrotransposon in cetaceans (Kapitonov et al. 1998). Gaffney et al. (2003) also described a miniature inverted transposable element (MITE)-like interspersed repeat in Crassostrea virginica showing similarity to satDNA from several mollusk species. The identification of additional cases, specially using data from sequenced animal genomes, may contribute for a better understanding of the origin of satDNAs from TEs, the factors and mechanisms related to such phenomenon and its importance for genome evolution.
Drosophila virilis (virilis group; Drosophila subgenus) is one of the Drosophila species whose sequenced genome is publicly available (Drosophila 12 Genomes Consortium 2007). This species represents a particularly interesting model for the study of repetitive DNAs because it has one of the largest genomes (∼400 Mb) and one of the highest heterochromatic contents (∼50%) within the Drosophila genus (Mahan and Beck 1986; Bosco et al. 2007). As for most shotgun-sequenced eukaryote genomes, the D. virilis assembled genome mainly covers the euchromatic fraction. This hinders the characterization of repetitive DNA and requires the simultaneous employment of other methodologies.
More than 800 TE families have been identified in the sequenced genome of D. virilis, most of them long terminal repeat (LTR) or non-LTR retrotransposons (Feschotte et al. 2009). Together, they represent about 14% of the D. virilis genome (Drosophila 12 Genomes Consortium 2007). In spite of the large number of identified TEs, very little is known about their characteristics, such as detailed structure, copy number, genomic distribution, and association with other genetic elements. The recent availability of the sequence data for the genome of D. americana (Fonseca et al. 2013), another member of the virilis species subgroup (Morales-Hojas et al. 2011), further helps to study the repetitive DNA fraction in D. virilis, because it offers a phylogenetically close genome for comparisons.
In this study, we discovered a novel DNA transposon in D. virilis that we named Tetris. The name is a reference to the classic game TETRIS and alludes to the characteristics of the transposon, for example, its modular structure and diversity of rearranged forms. We used a combination of bioinformatic and experimental approaches in order to characterize Tetris and to discuss its role in generating a new satDNA in D. virilis. Our results imply for the first time a role for foldback DNA transposons in the generation of satDNAs.
Materials and Methods
Identification of Tetris in D. virilis and Sequence Analysis
We serendipitously found Tetris in the assembled genome of D. virilis while looking for tandemly repetitive DNA sequences using the Tandem Repeats Finder software (Benson 1999). The BLAST tool was used for searching sequences similar to Tetris in the 21 sequenced genomes of Drosophila available in FlyBase (http://flybase.org, last accessed May 31, 2014) and in the D. americana genome (available at http://cracs.fc.up.pt/∼nf/dame/, last accessed May 31, 2014). Hits with e values smaller than 10−5 and covering at least 60% of the query sequence were selected for further analysis. BLAST searches for annotated sequences were performed against the nucleotide collection (nr/nt) database in GenBank.
Sequence alignments were performed with Muscle (Edgar 2004) implemented in MEGA5 (Tamura et al. 2011) with default setup. MEGA5 was also used for the estimation of genetic distances (p distance), AT content, and repeat length analysis. The evolutionary relationships among sequences were inferred by neighbor-joining (NJ) trees using the implemented option in MEGA5 and the proportion of nucleotide differences (p distance).
DNA Extraction, PCR Amplification, Cloning, and Sequencing
Genomic DNA was extracted from a pool of individuals from the D. virilis strain 15010-1051.51 (Santiago, Chile). PCR amplification was carried out with two sets of primers: Primers terminal inverted repeat (TIR)-220-F (CACGATTTATCAATCATTTTGC) and TIR-220-R (CTCTATATGCACAACTACCGTGC) were designed to amplify the TIR-220 tandem repeats, whereas primers TIR-FD (flanking domain)-F (TCGAGCCCTTATTTCTTAGC) and TIR-FD-R (AACTGCGCTATTAAGTATGAC) were designed to amplify a part of the TIR-FD domain. The position of each primer is indicated in figure 1B. PCR products were purified and cloned using the pGEM-T-Easy cloning kit (Promega). Recombinant plasmids were sequenced on the ABI3130 platform (Life Technologies).
Fig. 1.—
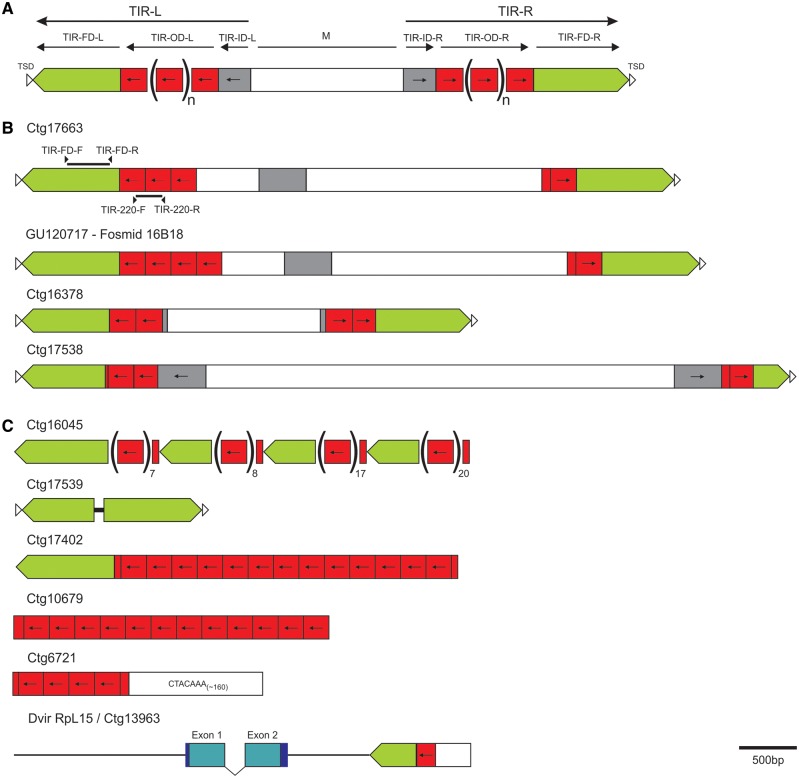
Schematic representation of the foldback structure and examples of the main types of variation found in the analyzed Tetris copies. (A) Schematic representation of foldback elements (adapted from Casals et al. 2005). M, middle domain. L and R refer to the left and right extremities of the transposon, respectively. (B) Schematic representation of the structurally complete copies of Tetris found in the Drosophila virilis assembled genome plus one hit from Fosmid 16B18 (accession: GU120717). Arrowheads in opposite orientation and solid black bars on top of the Ctg 17663 scheme represent the primers used to amplify the two fragments used as probes for the FISH experiments. (C) Main types of arrangements found between the Tetris and its TIR-220 repeats in the D. virilis genome, including one partial copy of Tetris next to the short heterochromatic gene RpL15. Green boxes represent Tetris’ TIR. Red boxes indicate the tandem repeats (TIR-220), and the grey boxes represent the Tetris’ TIR-ID.
Fluorescence In Situ Hybridization
Neuroblasts and salivary glands of third instar larvae from D. virilis (strain 15010-1051.51) and D. americana (strain W11) were used to obtain metaphase and polytene chromosome preparations, respectively (Baimai 1977; Ashburner 1989). Extended DNA fibers were obtained from adult flies and slides containing chromosomes or DNA fibers were prepared for fluorescence in situ hybridization (FISH) as described by Kuhn et al. (2008). The probes used for FISH were obtained from recombinant plasmids labeled with digoxigenin 11-dUTP or biotin 11-dUTP by nick translation with the DIG-nick and Biotin-nick translation mix (Roche Applied Science), respectively. Chromosomes were denatured by incubation in 0.07 M NaOH for 3 min and DNA fibers were denatured by incubation in 70% formamide/2× SSC at 80 °C for 3 min. The hybridization mix consisting of 100–200 ng of each probe in 50% formamide/2× SSC and water to a final volume of 40 µl per slide was denatured for 10 min at 80 °C and applied onto the slides. Hybridizations were performed in a moist chamber at 37 °C for 16–20 h. Posthybridization washes consisted of two baths in 2× SSC at 37 °C for 5 min. The probes were immunodetected with antidigoxigenin—FITC and neutravidin—Rhodamine (Roche Applied Science). Chromosomes and DNA fibers were counterstained with DAPI (4′,6-diamidino-2-phenylindole) in antifade reagent (SlowFade; Invitrogen) and analyzed under an Axio Imager A2 epifluorescence microscope equipped with the AxiocamMRm camera (Zeiss). Images were captured with Axiovision (Zeiss) and edited in Adobe Photoshop.
Dot-Blot of TIR-FD and TIR-220 in D. virilis and D. americana
In order to detect and verify the abundance of sequences similar to TIR-220 and TIR-FD, genomic DNA of D. virilis and D. americana was dot-blotted onto a positively charged nylon membrane (Roche) and hybridized with cloned TIR-220 and TIR-FD probes separately. Control DNA DIG-labeled (Roche) and the probe plasmids were also dot-blotted as positive controls. DNA from D. buzzatii was used as a negative control. Positive signals were visualized using chemoluminiscent CDP-Star (Roche) and images acquired using the ChemiDoc XRS system and Quantity ONE 4.7 software (Bio-Rad) at the Laboratory of Analysis and Photodocumentation of the Universitat Autònoma de Barcelona.
Results
Characterization of Tetris
After genome searches for repetitive elements in the D. virilis assembled genome, we serendipitously found a TE with a structure very similar to that of foldback elements (see fig. 1A). Foldbacks are a group of DNA transposons first described in D. melanogaster (Potter et al. 1980) and later found in several other organisms, including rye, Arabidopsis, the sea urchin and Chironomus thumi (Lieberman et al. 1983; Hankeln and Schmidt 1990; Windsor and Waddell 2000; Alves et al. 2005). We named this element Tetris. Sequence analysis of Tetris revealed long TIRs (up to 1,735 bp) made up of three possible domains: A most external flanking domain (FD) (TIR-FD) with up to 866 bp, an intermediate outer domain (TIR-OD) with a variable number of approximately 220-bp tandem repeats (named TIR-220), and an inner domain (TIR-ID). The TIRs are separated by a middle domain (M) with variable composition and size and no apparent protein-coding capacity. We found only three “structurally complete” copies of Tetris (i.e., copies with conserved structure and identifiable target site duplications) in the D. virilis assembled genome (fig. 1B; supplementary table S3, Supplementary Material online). These three copies lack conserved open reading frames (ORFs) and showed 8–9 bp target site duplications (TSDs). Additional analysis of four rearranged copies of Tetris revealed TSDs between 9 and 10 bp (supplementary table S1, Supplementary Material online).
BLAST searches using the three complete sequences of Tetris as queries against the 21 Drosophila sequenced genomes available at FlyBase showed no significant hits, except for those observed in D. virilis in this study.
BLAST searches using the Tetris sequences against the nucleotide collection available in GenBank retrieved several hits from fosmids mapped on chromosome 6 (Müller element F, also known as the “dot” chromosome) of D. virilis (Leung et al. 2010). Most of these sequences consist of small fragments or rearranged copies of Tetris. One of them (from fosmid 16B18; accession: GU120717) showed an allelic copy of the Tetris element present in contig 17663 from the D. virilis assembled genome (fig. 1B). These two copies are nearly identical, except for the fact that the copy from the assembled genome has three TIR-220 repeats in the left TIR and the copy from the sequenced fosmid has four TIR-220 repeats. Sequence comparisons and phylogenetic analysis allowed us to infer that this array size variation was due to the duplication of the second TIR-220 repeat in the fosmid copy (supplementary figs. S1 and S2, Supplementary Material online).
Besides the fosmids from D. virilis, BLAST searches in GenBank with Tetris as query retrieved significant hits in D. americana, another member of the virilis group. These hits correspond to an element that was found in the breakpoints of three chromosomal inversions and that was tentatively classified as a MITE and named DAIBAM (Evans et al. 2007; Fonseca et al. 2012). The similarity between Tetris and DAIBAM is high at both TIRs (left and right) of Tetris (95% and 89% identities) and actually extends over most of DAIBAM sequence (95%) (fig. 2). When self-aligned, DAIBAM copies show only short TIRs (up to ∼240 bp) and no further identifiable features. The extent of similarity between DAIBAM and Tetris indicates that DAIBAM probably originated from an element similar to Tetris by deletion (fig. 2).
Fig. 2.—
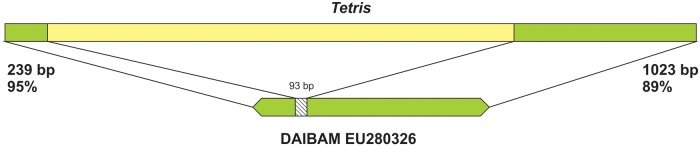
Schematic comparison between Tetris (Ctg 16378) and DAIBAM (EU280326). Regions with high sequence identity between the elements are shown in green. The DAIBAM region that could not be confidently aligned with Tetris is shown in the striped bar.
Characterization of TIR-220 Repeats
The TIRs from Tetris include an outer domain (OD) (TIR-OD) with a variable number of approximately 220-bp tandem repeats that we named TIR-220. BLAST searches using a TIR-220 consensus sequence against the D. virilis assembled genome retrieved 610 TIR-220 repeats distributed along 69 contigs. In 25 contigs, TIR-220 repeats are associated with adjacent TIR-FD segments of Tetris (n = 202), 39 contigs comprise exclusively TIR-220 repeats (n = 395), and five contigs encompass TIR-220 repeats neighboring other genomic elements (n = 13) but without the TIR-FD (supplementary table S2, Supplementary Material online).
A PCR experiment using primers with opposite orientations designed to amplify solely TIR-220 repeats resulted in a “ladder-like” pattern of amplicons representing multimers (up to seven) of approximately 220 bp, confirming the tandem organization of TIR-220 repeats seen in the assembled genome of D. virilis (supplementary fig. S3, Supplementary Material online).
Sequence analysis of 482 copies of TIR-220 repeats showed that they are AT-rich (∼70% on average) and highly homogeneous, displaying an overall similarity of approximately 94% (±0.4%) and an average repeat length of 218 bp. When analyzed separately, the TIR-220 repeats located inside structurally complete Tetris elements revealed a mean nucleotide similarity of 87% (±1.3%), whereas the rest of the TIR-220 repeats presented an average similarity of 94%. This result indicates that TIR-220 repeats are more homogeneous within long than within short arrays inside Tetris. Such pattern would be expected if a small number of copies within Tetris suffered expansion generating large TIR-220 arrays.
We found 27 complete out of 60 identified copies of TIR-220 in the D. americana assembled genome, but only as part of short arrays (up to three copies), typical for the TIRs of Tetris.
Phylogeny of TIR-220 Repeats
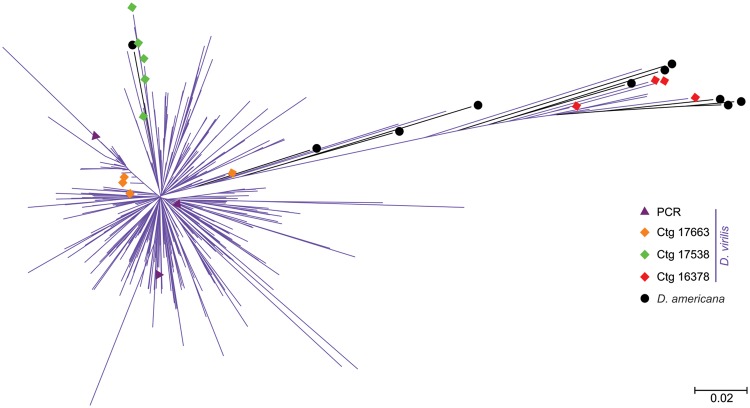
An NJ tree with all sampled TIR-220 repeats from D. virilis and D. americana revealed two main groups of sequences (fig. 3). The first one, on the left part of the tree, is mainly comprised short branches of TIR-220 repeats from D. virilis and a single TIR-220 copy from D. americana. The right side of the tree shows long branches leading to the repeats that are located inside one structurally complete copy of Tetris from D. virilis and to most repeats from D. americana.
Fig. 3.—
NJ tree containing TIR-220 repeats extracted from Drosophila virilis and D. americana assembled genomes. Purple branches with no symbols at the tips represent the bulk of D. virilis amplified copies. Repeats belonging to the three structurally complete copies of Tetris are shown in orange (Ctg 17663), red (Ctg 16378), and green (Ctg 17538) diamonds. Purple triangles represent the three D. virilis TIR-220 repeats obtained by PCR and sequenced in this work. Drosophila americana TIR-220 repeats are shown in black circles and branches. The tree was estimated using the NJ algorithm and the p-distance substitution method.
The NJ tree showed no consistent clustering of TIR-220 repeats from single contigs, indicating that adjacent TIR-220 repeats are as similar to each other as they are to distant repeats present in the same or different arrays. However, in two of the three complete Tetris elements of D. virilis (Ctg 17538 and 16378) the TIR-220 repeats were grouped together, indicating the possible action of homogenization mechanisms (e.g., UCO and gene conversion). The TIR-220 sequences obtained by PCR are dispersed throughout the left side of the tree, indicating that they belonged to the larger and more abundant TIR-220 arrays (fig. 3).
Chromosome Location of Tetris and TIR-220-bp Repeats
The karyotype of D. virilis consists of five pairs of large acrocentric chromosomes (Muller elements A–E) and a very small pair of microchromosomes (Muller element F; also known as dot chromosomes) and is thought to retain the ancestral condition inferred for the Drosophila genus (Clayton and Guest 1986). The karyotype of D. americana shows a centromeric fusion of chromosomes 2 and 3, and a polymorphic centromeric fusion of chromosomes X and 4 (Caletka and McAllister 2004).
In order to investigate the genomic location of both TIR-220 repeats within structurally complete copies of Tetris and expanded TIR-220 arrays, we performed double FISH experiments in metaphase and polytene chromosomes of D. virilis and metaphase chromosomes of D. americana. Probes for FISH were prepared from two regions of the Tetris element: One specific for the TIR-FD region and another specific for the TIR-220 repeats (fig. 1B).
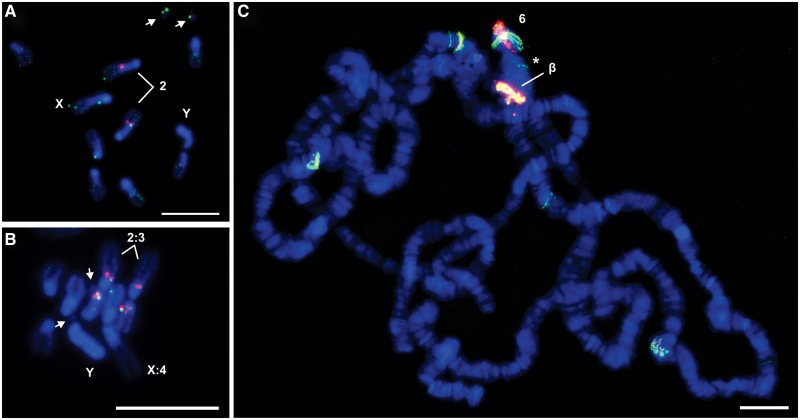
FISH in the mitotic chromosome spreads of D. virilis produced colocalizing hybridization signals of both the probes in the distal extremity of the pericentromeric heterochromatin of chromosome 2 (Müller element E), identified as the biggest acrocentric autosome (fig. 4A). In this specific region, the signals corresponding to TIR-220 repeats were seemingly brighter than the signals produced by TIR-FD, suggesting overabundance of TIR-220 repeats. The two probes also colocalized in the small dot chromosomes, showing similar signal intensities. Small hybridization signals with the TIR-FD probe were distributed throughout all chromosomes, including the Y. The absence of such ubiquitous distribution of TIR-220 signals may be explained by the presence of Tetris elements containing partial or no TIR-220 repeats and therefore difficult to detect (see examples in fig. 1).
Fig. 4.—
FISH of TIR-FD (green) and TIR-220 (red) probes hybridized on (A) Drosophila virilis metaphase, (B) D. americana metaphase, and (C) D. virilis polytene chromosomes. Arrows in (A) and (B) indicate the chromosome 6 (the dot chromosome). The chromocenter of the polytene chromosomes is indicated with an asterisk in (C). The “β” indicates the chromosome 2 β-heterochromatin and the number 6 is placed next to the polytene arm corresponding to the chromosome 6. Bars in (A) and (B) correspond to 10 µm, and in (C), to 5 µm.
FISH in D. americana mitotic chromosomes with the same two probes produced colocalizing hybridization signals in the distal extremity of the pericentromeric heterochromatin of chromosomes 2 and 3 (Müller elements E and D respectively; fig. 4B). As observed for D. virilis, signals corresponding to TIR-220 repeats were seemingly brighter than the signals produced by TIR-FD. No hybridization signals were detected in the dot chromosomes of D. americana.
FISH experiments on polytene chromosomes of D. virilis using the same probes produced strong colocalizing hybridization signals with similar intensities in the proximal region of chromosome 2. This region consists of β-heterochromatin, an intermediate zone located between the heterochromatin and the euchromatin that, unlike α-heterochromatin, is replicated during polytenization. In the dot chromosome, strong colocalizing hybridization signals were observed in a region that extends from the end of the chromocenter to the most distal regions of the chromosome arm. A few euchromatic signals of TIR-FD with or without TIR-220 were also observed (fig. 4C).
The analysis of the assembled genome of D. virilis showed that some Tetris elements are present in contigs assigned to chromosomes 2, 3, and X. Interestingly, in two contigs (one of them is shown in fig. 1C) we found TIR-220 repeats ending abruptly with the Satellite I sequences of D. virilis (5′-ACAAACT-3′) (supplementary table S2, Supplementary Material online), known to comprise 20% of the genome and located in the tightly packed α-heterochromatin from all chromosomes (Gall and Atherton 1974) (fig. 1C). Such configuration further indicates that Tetris elements with expanded arrays accumulated close to the α-heterochromatin of chromosome 2.
Long-Range Organization of Tetris and TIR-220 Repeats
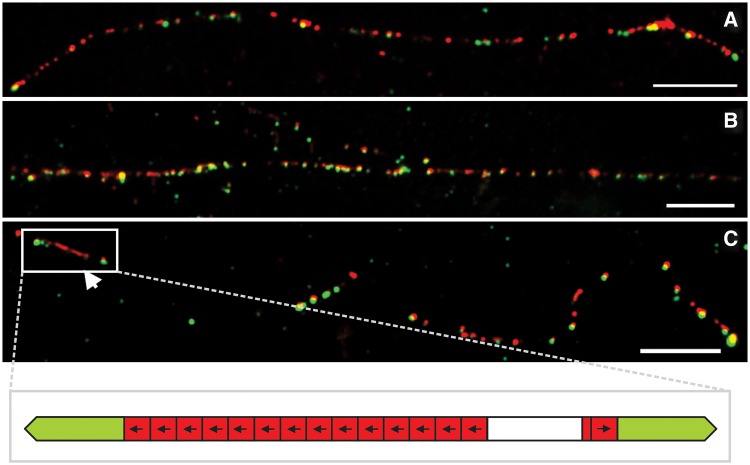
High-resolution FISH on DNA fibers (fiber-FISH) with the two probes described above showed several cases where multiple copies of Tetris are located in a genomic region corresponding to less than 100 kb (fig. 5A and B). Such regions may correspond to those characterized by the strong hybridization signals observed in the metaphase and polytene chromosomes. The fiber-FISH results confirm the variety of rearrangements between Tetris and TIR-220 that we found in our in silico analyses of the D. virilis assembled genome (fig. 1C) and indicate a similar scenario for D. americana. In some fibers, it was possible to visualize TIR-FD hybridization signals associated with a variable number of TIR-220 repeats, including the presence of long arrays of TIR-220 repeats flanked by two TIR-FD (fig. 5C). No array composed solely by TIR-220 was detected by fiber-FISH. The presence of isolated signals generated only by the TIR-FD probe indicates the presence of Tetris elements containing none or partial TIR-220 repeats.
Fig. 5.—
FISH of TIR-220 and TIR-FD probes onto extended DNA fibers of Drosophila virilis. Long DNA fibers with several Tetris hybridization signals including some expanded TIR-220 arrays are shown in (A) for D. americana and in (B) for D. virilis. Arrowhead in (C) indicates a long array of TIR-220 within Tetris from D. virilis. A representation of the hybridization signals highlighted in (C) is shown in the bottom row. The bars correspond to 10 kb (assuming 10 µm = 29 kb; Schwarzacher and Heslop-Harrison 2000).
Relative Abundances of TIR-FD and TIR-220 in D. virilis and D. americana
To gain further insight on Tetris abundance in D. virilis and D. americana genomes, we performed dot-blot hybridizations using the same TIR-FD and TIR-220 probes. For both species, the results indicated a marked overabundance of the TIR-220 sequences when compared with the TIR-FD probe, thus supporting the amplification of the former within Tetris. The abundance of TIR-220 in D. americana as evidenced by the dot-blot seems in contrast with the low copy number found in the assembled genome.
Discussion
Tetris Is a Foldback TE
In this work, we identified a TE in the assembled genome of D. virilis with structural similarities to foldback elements, which has been named Tetris. However, we found only three “structurally complete” copies of Tetris, that is, copies with conserved structure and identifiable target site duplications (fig. 1B), all of them lacking conserved ORFs and therefore defective or nonautonomous. Despite our efforts, autonomous copies encoding the transposase that mobilizes Tetris have not been found, perhaps because there are no complete copies in the sequenced D. virilis genome.
It has been suggested that the foldback structure may evolve independently from different lineages of TEs by the loss of the transposase gene and the elongation of TIRs (Marzo et al. 2008). However, most foldback elements seem to belong to the Mutator superfamily (Feschotte and Pritham 2007; Marquez and Pritham 2010; Gross and Williamson 2011), although there are exceptions (Marzo et al. 2008; Badal et al. 2013). The lack of a complete copy encoding the transposase prevents a definite classification of Tetris within the order of TIR transposons (Wicker et al. 2007). Nevertheless, the size of TSDs is related to the transposase that catalyzes the cut-and-paste reaction of TIR transposons and is an important clue that differentiates many of the TE superfamilies (Feschotte and Pritham 2007; Yuan and Wessler 2011). Based on the verified Tetris TSDs (8–10 bp) and from previous suggestions that foldback elements belong to the Mutator superfamily, we suggest that Tetris belongs to the Mutator superfamily.
DAIBAM Is Closely Related to Tetris: Implications for Chromosome Inversions
It has been shown that Drosophila chromosome inversions may be generated by ectopic recombination between copies of the same transposon (Evans et al. 2007; Delprat et al. 2009; Fonseca et al. 2012; Rius et al. 2013). In D. buzzatii, for instance, three polymorphic inversions, 2j, 2q7 and 2z3, were generated by the transposon Galileo that belongs to the P superfamily (Delprat et al. 2009). Likewise, BuT5, a MITE associated with the P element, generated two inversions fixed in the repleta group, 2s and 2x3 (Rius et al. 2013). Finally, a MITE named DAIBAM was involved in the origin of three chromosome inversions present in the D. virilis phylad, Xa fixed in D. virilis, and 4a and 5a polymorphic in D. americana (Evans et al. 2007; Fonseca et al. 2012). Herein, we show that the previously studied copies of DAIBAM are related to Tetris and present a high similarity at both ends of the element. Similarity between terminal sequences of a transposon and a MITE has been previously found and seemingly indicates that the autonomous element is responsible for the MITE origin and amplification because these sequences are known to be important for transposition (Rius et al. 2013). However, the available information on DAIBAM (Fonseca et al. 2012) is very limited and a more thorough characterization of its copies in the D. americana genome would be desirable. In addition, the complete element encoding the transposase that mobilizes Tetris and DAIBAM was not found. The characterization of such canonical copies would be also important for a definite classification of these elements.
Origin of TIR-220 Repeats and First Stages of Copy Number Variation
The TIR from Tetris is composed of a most external FD (TIR-FD), an intermediate outer domain with a variable number of tandem repeats (TIR-220), and an ID (TIR-ID). A careful comparison of the TIR-FD and TIR-220 domains revealed that they share a stretch of 37 bp with high similarity (∼80%) (fig. 6). This result suggests that the TIR-220 repeats may have originated as a duplication of a part of the original TIR-FD sequence. The foldback-like transposon Galileo of D. buzzatti also possesses internal tandem repeats within the TIRs that contain additional transposase binding sites (Marzo et al. 2013). These secondary binding sites may facilitate recognition by the THAP (Thanatos-associated protein) domain of the Galileo transposase. Increasing the number of tandem repeats containing the THAP recognition sequence may also lead to an increased transposition efficiency (Potter 1982; Cheng et al. 2000; Marzo et al. 2013). We suggest a similar scenario for the evolution of TIR-220 repeats from D. virilis. Initial stages of formation and expansion of the TIR-220 repeats may have increased the transposition efficiency of Tetris through the advantages explained above. However, larger arrays of TIR-220 may have disrupted the transposition of the Tetris element, leaving the door opened for the formation of satDNA arrays. Further studies will be needed to evaluate these suggestions.
Fig. 6.—
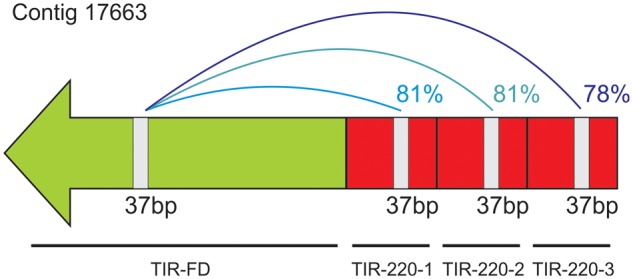
Sequence identity between a 37-bp stretch shared by the TIR-FD and TIR-220 domains of Tetris. The nucleotide identity values are given on the right side of the connection lines.
UCO has long been considered as an important mechanism leading to the expansion or contraction of tandemly repeated DNAs, such as satDNAs (Smith 1976). In addition, rolling-circle replication of extrachromosomal repeats followed by reintegration in the genome may be another powerful mechanism for array expansion (Cohen and Segal 2009). Although there are several examples in the literature showing the participation of UCO in array size variation (Martin et al. 2003; Alkan et al. 2004), the participation of rolling-circle replication remains hypothetical. In this study, we found two allelic copies of Tetris that are nearly identical, except for the fact that one allele has three TIR-220 repeats and the other has four (fig. 1B; supplementary figs. S2 and S3, Supplementary Material online). We showed, by sequence comparisons and phylogenetic analyses, that this variation resulted from the duplication of the second TIR-220 copy present in one of the alleles. This duplication could be the outcome of an event of UCO between TIR-220 repeats. Nevertheless, other mechanisms may have also participated in the expansion of TIR-220 repeats inside Tetris.
Large TIR-220 satDNA Arrays Originated Inside Tetris
During the analysis of the D. virilis assembled genome, we also found TIR-220 repeats as part of long and homogeneous satDNA arrays made of up to 66 copies (see supplementary table S2, Supplementary Material online). By a combination of sequence analysis and FISH on chromosomes and DNA fibers, we showed that TIR-220 repeats have undergone tandem amplification within Tetris. Such amplification may have resulted in the disruption of Tetris thus facilitating the expansion of large satDNA arrays.
Previous studies showed that TEs may provide different substrates for satDNA emergence. A few studies suggested that the first steps of satDNA formation could involve UCO promoted by ectopic recombination between TEs (Wong and Choo 2004). Consequently, some satDNAs share high degrees of sequence similarity to parts of the original TEs. Examples showing this phenomenon include the centromeric retroelements of maize (Sharma et al. 2013), the L1 retroelement of cetaceans (Kapitonov et al. 1998), the Ty3/gypsy-like retroelement from pea (Macas et al. 2009), and the SGM transposon in the D. obscura species group (Miller et al. 2000).
In this work, we show a different way by which TEs may provide templates for satDNA formation. In the case of the Tetris element of D. virilis, tandem repeat amplification involved repeats already present as part of the TE structure. It remains to be investigated if the peculiar repetitive structure of foldback-like elements makes them particularly prone to participate in the emergence of new satDNAs.
Tetris Is Present in the virilis Subgroup
We found Tetris in two Drosophila species from the virilis group, that is, D. virilis and D. americana, suggesting that it was already present at least in the genome of the last common ancestor of the virilis subgroup. In the sequenced genome of D. americana, we found TIR-220 repeats organized only in short arrays made up of three repeats at most. Although this result suggested that no expansion of TIR-220 repeats took place in the D. americana genome, fiber-FISH and dot-blot analysis indicated otherwise.
Fiber-FISH results for D. americana were much alike those obtained for D. virilis, showing clusters of Tetris insertions with expanded internal TIR-220 arrays. Moreover, the dot-blot analysis revealed a similar abundance of TIR-220 in D. americana and in D. virilis (supplementary fig. S4, Supplementary Material online), suggesting that amplification of TIR-220 repeats also occurred in the first species. The much lower TIR-220 copy number in the D. americana assembled genome is probably a consequence of the shorter read lengths produced by next generation sequencers, what poses even bigger challenges to the assemblage of repetitive regions.
Based on our data, TIR-FD and TIR-220 repeats account for 0.08% of the assembled genome of D. virilis, but only half of its estimated genome size has been assembled so far. Additionally, repetitive DNAs represent a challenge for the assembly of genomes sequenced by the whole-genome shotgun methodology, such as that of D. virilis (Drosophila 12 Genomes Consortium 2007). In fact, most of the heterochromatic sequences from the Drosophila sequenced genomes remain unassembled and therefore the actual contribution of Tetris for the genome size of D. virilis may be higher.
The Amplification of TIR-220 Occurred in the β-Heterochromatin
Multiple copies of Tetris were found scattered along all chromosomes of D. virilis and D. americana, but with a much higher density in some sites such as in the dot chromosome and β-heterochromatin of chromosome 2 of D. virilis and in the β-heterochromatin chromosomes 2 and 3 of D. americana. Such a distribution may indicate that part of the euchromatin of the dot chromosome shares common sequences with the β-heterochromatin of chromosome 2. This situation is similar to that found on D. melanogaster, in which the β-heterochromatin of the X chromosome and part of the euchromatin of the dot chromosome are composed of a shared mosaic of middle repetitive elements (Miklos et al. 1988). Interestingly, Slawson et al. (2006) also found an enrichment of repetitive elements in the dot chromosomes of D. virilis compared with the euchromatic parts of the remaining chromosomes. The heterochromatic accumulation of TEs is probably a result of the lower levels of recombination in this region. Once inserted in the heterochromatin, TEs are less prone to generate unviable rearrangements through ectopic recombination, what ultimately reduces the effectiveness of purifying selection upon these sequences (Petrov et al. 2011). Nevertheless, the mainly euchromatic D. virilis dot chromosome was shown to recombine at meiosis, although at a lower level. Some authors suggested that there might be an adaptive advantage in maintaining a high density of repetitive elements in the dot chromosome (Slawson et al. 2006). The dot chromosomes of D. americana seem to be devoid of Tetris.
The β-heterochromatin of chromosome 2 of D. virilis and chromosomes 2 and 3 of D. americana were the sites where major events of amplification of TIR-220 repeats took place. A more detailed study of the β-heterochromatin of D. melanogaster chromosomes revealed several interesting features including a high density of TEs and a high proportion of rearranged and mostly defective TEs (Vaury et al. 1989). In particular, sources of TE rearrangements in the β-heterochromatin include tandem amplifications and nested insertions (Vaury et al. 1989; Kuhn and Heslop-Harrison 2011). Such properties of β-heterochromatin add valuable and testable information related to the potential origin of satDNAs in this region.
Conclusions
In this study, we showed by a combination of bioinformatic and experimental data that a foldback element, named here as Tetris, has contributed to shaping the genomes of D. virilis and D. americana by providing internal tandem repeats that acted as “seeds” for the amplification of satDNA arrays. The β-heterochromatin genomic environment associated with the high density of Tetris copies seemed to have favored such amplification. There is increasing evidence for the participation of different families of TEs in the origin of other types of repetitive DNAs, such as microsatellites (e.g., Wilder and Hollocher 2001; Smýkal et al. 2009), minisatellites (e.g., Inukai 2004), and satDNAs (e.g., Macas et al. 2009). Our results imply for the first time a role for foldback elements in the generation of satDNAs.
In Drosophila, transposons have been shown to generate chromosome inversions by ectopic recombination between copies of the same family, as is the case of the transposon Galileo in D. buzzatti (Delprat et al. 2009). In D. americana, a TE classified as MITE and named DAIBAM was found to be implicated in the generation of three chromosome inversions of the virilis phylad (Fonseca et al. 2012). Interestingly, we showed in this work that DAIBAM is related to Tetris by the sequence similarity present between their ends, a region known to be important for transposition. DAIBAM and Tetris are likely to be mobilized by elements of the same family and DAIBAM could have been generated from an element similar to Tetris by deletion of internal sequences.
Supplementary Material
Supplementary tables S1–S3 and figures S1–S4 are available at Genome Biology and Evolution online (http://www.gbe.oxfordjournals.org/).
Acknowledgments
The authors would like to thank Claudia Carareto (Universidade Estadual Júlio Mesquita Filho, Brazil) and Jorge Vieira (Instituto de Biologia Molecular e Celular, Portugal) for providing the Drosophila virilis and D. americana stocks, respectively. They are also grateful for the valuable comments of Laila Alves Nahum (FIOCRUZ - Centro de Pesquisas René Rachou, Brazil) on an early draft of the paper. Finally, they thank the three anonymous reviewers for their valuable comments and suggestions. This work was supported by “Conselho Nacional de Desenvolvimento Científico e Tecnológico” (grant number 471921/2011-4) and Pró-Reitoria de Pesquisa da Universidade Federal de Minas Gerais to G.C.S.K., Fundação de Amparo à Pesquisa de Minas Gerais, “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior,” and grant BFU2011-30476 from Ministerio de Ciencia e Innovación (Spain) to A.R.
Literature Cited
- Alkan C, Eichler EE, Bailey JA, Sahinalp SC, Tüzün E. The role of unequal crossover in alpha-satellite DNA evolution: a computational analysis. J Comput Biol. 2004;11(5):933–944. doi: 10.1089/cmb.2004.11.933. [DOI] [PubMed] [Google Scholar]
- Alves E, Ballesteros I, Linacero R, Vázquez AM. RYS1, a foldback transposon, is activated by tissue culture and shows preferential insertion points into the rye genome. Theor Appl Genet. 2005;111:431–436. doi: 10.1007/s00122-005-2013-9. [DOI] [PubMed] [Google Scholar]
- Ashburner M. Drosophila: a laboratory handbook. Cold Spring Harbor (NY): Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Badal M, Xamena N, Cabré O. FB-NOF is a non-autonomous transposable element, expressed in Drosophila melanogaster and present only in the melanogaster group. Gene. 2013;526(2):459–463. doi: 10.1016/j.gene.2013.04.082. [DOI] [PubMed] [Google Scholar]
- Baimai V. Chromosomal polymorphisms of constitutive heterochromatin and inversions in Drosophila. Genetics. 1977;85(1):85–93. doi: 10.1093/genetics/85.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. Tandem Repeats Finder: a program to analyze DNA sequences. Nucleic Acids Res. 1999;27(2):573–580. doi: 10.1093/nar/27.2.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosco G, Campbell P, Leiva-Neto J, Markow T. Analysis of Drosophila species genome size and satellite DNA content reveals significant differences among strains as well as between species. Genetics. 2007;177:1277–1290. doi: 10.1534/genetics.107.075069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caletka BC, McAllister BF. A genealogical view of chromosomal evolution and species delimitation in the Drosophila virilis species subgroup. Mol Phylogenet Evol. 2004;33:664–670. doi: 10.1016/j.ympev.2004.08.007. [DOI] [PubMed] [Google Scholar]
- Casals F, Cáceres M, Manfrin MH, González J, Ruiz A. Molecular characterization and chromosomal distribution of Galileo, Kepler and Newton, three foldback transposable elements of the Drosophila buzzatti species complex. Genetics. 2005;169:2047–2059. doi: 10.1534/genetics.104.035048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charlesworth B, Sniegowski P, Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. doi: 10.1038/371215a0. [DOI] [PubMed] [Google Scholar]
- Cheng C, Tsuchimoto S, Ohtsubo H, Ohtsubo E. Tnr8, a foldback transposable element from rice. Genes Genet Syst. 2000;75:327–333. doi: 10.1266/ggs.75.327. [DOI] [PubMed] [Google Scholar]
- Cheng Z, Murata M. A centromeric tandem repeat family originating from a part of Ty3/gypsy-retroelement in wheat and its relatives. Genetics. 2003;164:665–672. doi: 10.1093/genetics/164.2.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton FE, Guest WC. Overview of chromosomal evolution in the family Drosophilidae. In: Ashburner M, editor. The genetics and biology of Drosophila. Waltham (MA): Academic Press; 1986. pp. 1–38. [Google Scholar]
- Cohen S, Segal D. Extrachromosomal circular DNA in eukaryotes: possible involvement in the plasticity of tandem repeats. Cytogenet Genome Res. 2009;124:327–338. doi: 10.1159/000218136. [DOI] [PubMed] [Google Scholar]
- de Koning APJ, Gu W, Castoe TA, Batzer MA, Pollock DD. Repetitive elements may comprise over two-thirds of the human genome. PLoS Genet. 2011;7(12):e1002384. doi: 10.1371/journal.pgen.1002384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delprat A, Negre B, Puig M, Ruiz A. The transposon Galileo generates natural chromosomal inversions in Drosophila by ectopic recombination. PLoS One. 2009;4(11):e7883. doi: 10.1371/journal.pone.0007883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devos KM, Brown JKM, Bennetzen JL. Genome size reduction through illegitimate recombination counteracts genome expansion in Arabidopsis. Genome Res. 2002;12:1075–1079. doi: 10.1101/gr.132102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drosophila 12 Genomes Consortium. Evolution of genes and genomes on the Drosophila phylogeny. Nature. 2007;450:203–218. doi: 10.1038/nature06341. [DOI] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AL, Mena PA, McAllister BF. Positive selection near an inversion breakpoint on the neo-X chromosome of Drosophila americana. Genetics. 2007;177:1303–1319. doi: 10.1534/genetics.107.073932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Keswani U, Ranganathan N, Guibotsy ML, Levine D. Exploring repetitive DNA landscapes using REPCLASS, a tool that automates the classification of transposable elements in eukaryotic genomes. Genome Biol Evol. 2009;1:205–220. doi: 10.1093/gbe/evp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca NA, et al. Drosophila americana as a model species for comparative studies on the molecular basis of phenotypic variation. Genome Biol Evol. 2013;5(4):661–679. doi: 10.1093/gbe/evt037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonseca NA, Vieira CP, Schlötterer C, Vieira J. The DAIBAM MITE element is involved in the origin of one fixed and two polymorphic Drosophila virilis phylad inversions. Fly. 2012;6(2):1–4. doi: 10.4161/fly.19423. [DOI] [PubMed] [Google Scholar]
- Gaffney PM, Pierce JC, Mackinley AG, Titchen DA, Glenn WK. Pearl, a novel family of putative transposable elements in bivalve mollusks. J Mol Evol. 2003;56:308–316. doi: 10.1007/s00239-002-2402-5. [DOI] [PubMed] [Google Scholar]
- Gall JG, Atherton DD. Satellite DNA sequences in Drosophila virilis. J Mol Biol. 1974;85:633–664. doi: 10.1016/0022-2836(74)90321-0. [DOI] [PubMed] [Google Scholar]
- Gross SM, Williamson VM. Tm1: a Mutator/Foldback transposable element family in root-knot nematodes. PLoS One. 2011;6(9):e24534. doi: 10.1371/journal.pone.0024534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankeln T, Schmidt ER. New foldback transposable element TFB1 found in histone genes of the midge Chironomus thummi. J Mol Biol. 1990;215:477–482. doi: 10.1016/S0022-2836(05)80159-7. [DOI] [PubMed] [Google Scholar]
- Heslop-Harrison JS, Schwarzacher T. Organization of the plant genome in chromosomes. Plant J. 2011;66:18–33. doi: 10.1111/j.1365-313X.2011.04544.x. [DOI] [PubMed] [Google Scholar]
- Inukai T. Role of transposable elements in the propagation of minisatellites in the rice genome. Mol Genet Genomics. 2004;271:220–227. doi: 10.1007/s00438-003-0973-5. [DOI] [PubMed] [Google Scholar]
- Kapitonov VV, Holquist GH, Jurka J. L1 repeat is a basic unit of heterochromatin satellites in cetaceans. Mol Biol Evol. 1998;15:611–612. doi: 10.1093/oxfordjournals.molbev.a025963. [DOI] [PubMed] [Google Scholar]
- Kidwell MG. Transposable elements and the evolution of genome size in eukaryotes. Genetica. 2002;115:49–63. doi: 10.1023/a:1016072014259. [DOI] [PubMed] [Google Scholar]
- Kuhn GCS, Franco FF, Manfrin MH, Moreira-Filho O, Sene FM. Low rates of homogenization of the DBC-150 satellite DNA family restricted to a single pair of microchromosomes in species from the Drosophila buzzatii cluster. Chromosome Res. 2008;16:307–324. doi: 10.1007/s10577-007-1138-x. [DOI] [PubMed] [Google Scholar]
- Kuhn GCS, Heslop-Harrison JS. Characterization and genomic organization of PERI, a repetitive DNA in the Drosophila buzzatii cluster related to DINE-1 transposable elements and highly abundant in the sex chromosomes. Cytogenet Genome Res. 2011;132:79–88. doi: 10.1159/000320921. [DOI] [PubMed] [Google Scholar]
- Leung W, et al. Evolution of a distinct genomic domain in Drosophila: comparative analysis of the dot chromosome in Drosophila melanogaster and Drosophila virilis. Genetics. 2010;185:1519–1534. doi: 10.1534/genetics.110.116129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman D, et al. An unusual transposon with long terminal inverted repeats in the sea urchin Strongylocentrotus purpuratus. Nature. 1983;306:342–347. doi: 10.1038/306342a0. [DOI] [PubMed] [Google Scholar]
- Macas J, Koblížková A, Navrátilová A, Neumann P. Hypervariable 3′ UTR region of plant LTR-retrotransposons as a source of novel satellite repeats. Gene. 2009;448:198–206. doi: 10.1016/j.gene.2009.06.014. [DOI] [PubMed] [Google Scholar]
- Mahan JT, Beck ML. Heterochromatin in mitotic chromosomes of the virilis species group of Drosophila. Genetica. 1986;68:113–118. [Google Scholar]
- Marquez CP, Pritham EJ. Phantom, a new subclass of Mutator DNA transposons found in insect viruses and widely distributed in animals. Genetics. 2010;185:1507–1517. doi: 10.1534/genetics.110.116673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin MP, Bashirova A, Traherne J, Trowsdale J, Carrington M. Cutting edge: expansion of the KIR locus by unequal crossing over. J Immunol. 2003;171:2192–2195. doi: 10.4049/jimmunol.171.5.2192. [DOI] [PubMed] [Google Scholar]
- Marzo M, Liu D, Ruiz A, Chalmers R. Identification of multiple binding sites for the THAP domain of the Galileo transposase in the long terminal inverted-repeats. Gene. 2013;525:84–91. doi: 10.1016/j.gene.2013.04.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzo M, Puig M, Ruiz A. The foldback-like element Galileo belongs to the P superfamily of DNA transposons and is widespread within the Drosophila genus. Proc Natl Acad Sci U S A. 2008;115(8):2957–2962. doi: 10.1073/pnas.0712110105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miklos GLG, Yamamoto M, Davies J, Pirrota V. Microcloning reveals a high frequency of repetitive sequences characteristic of chromosome 4 and the β-heterochromatin of Drosophila melanogaster. Proc Natl Acad Sci U S A. 1988;88:2051–2055. doi: 10.1073/pnas.85.7.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller WJ, Nagel A, Bachmann J, Bachmann L. Evolutionary dynamics of the SGM transposon family in the Drosophila obscura species group. Mol Biol Evol. 2000;17(11):1597–1609. doi: 10.1093/oxfordjournals.molbev.a026259. [DOI] [PubMed] [Google Scholar]
- Morales-Hojas R, Reis M, Vieira CP, Vieira J. Resolving the phylogenetic relationships and evolutionary history of the Drosophila virilis group using multilocus data. Mol Phylogenet Evol. 2011;60:249–258. doi: 10.1016/j.ympev.2011.04.022. [DOI] [PubMed] [Google Scholar]
- Petrov DA, Fiston-Lavier A, Lipatov M, Lenkov K, González J. Population genomics of transposable elements in Drosophila melanogaster. Mol Biol Evol. 2011;28(5):1633–1644. doi: 10.1093/molbev/msq337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pimpinelli S, et al. Transposable elements are stable structural components of Drosophila melanogaster heterochromatin. Proc Natl Acad Sci U S A. 1995;92:3804–3808. doi: 10.1073/pnas.92.9.3804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plohl M, Luchetti A, Meštrović N, Mantovani B. Satellite DNAs between selfishness and functionality: structure, genomics and evolution of tandem repeats in centromeric (hetero)chromatin. Gene. 2008;409:72–82. doi: 10.1016/j.gene.2007.11.013. [DOI] [PubMed] [Google Scholar]
- Pontecorvo G, De Felice B, Carfagna M. A novel repeated sequence DNA originated from a Tc1-like transposon in water green frog Rana esculenta. Gene. 2000;261:205–210. doi: 10.1016/s0378-1119(00)00539-4. [DOI] [PubMed] [Google Scholar]
- Potter S, Truett M, Philips M, Maher A. Eukaryotic transposable genetic elements with inverted terminal repeats. Cell. 1980;20:639–647. doi: 10.1016/0092-8674(80)90310-4. [DOI] [PubMed] [Google Scholar]
- Potter SS. DNA sequence of a foldback transposable element in Drosophila. Nature. 1982;297:201–204. doi: 10.1038/297201a0. [DOI] [PubMed] [Google Scholar]
- Rius N, Delprat A, Ruiz A. A divergent P element and its associated MITE, BuT5, generate chromosomal inversions and are widespread within the Drosophila repleta species group. Genome Biol Evol. 2013;5(6):1127–1141. doi: 10.1093/gbe/evt076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable PS, et al. The B37 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1115. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- Schwarzacher T, Heslop-Harrison JS. Practical In Situ Hybridization. Oxford: BIOS Scientific Publishers Limited; 2000. [Google Scholar]
- Sharma A, Wolfgruber TK, Presting GG. Tandem repeats derived from centromeric retrotransposons. BMC Genomics. 2013;14:142. doi: 10.1186/1471-2164-14-142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slawson EE, et al. Comparison of dot chromosome sequences from D. melanogaster and D. virilis reveals an enrichment of DNA transposon sequences in heterochromatic domains. Genome Biol. 2006;7(2):R15. doi: 10.1186/gb-2006-7-2-r15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith GP. Evolution of repeated DNA sequences by unequal crossover. Science. 1976;191:528–535. doi: 10.1126/science.1251186. [DOI] [PubMed] [Google Scholar]
- Smýkal P, Kalendar R, Ford R, Macas J, Griga M. Evolutionary conserved lineage of Angela-family retrotransposons as a genome-wide microsatellite repeat dispersal agent. Heredity. 2009;103:157–167. doi: 10.1038/hdy.2009.45. [DOI] [PubMed] [Google Scholar]
- Tamura K, et al. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaury C, Bucheton A, Pelisson A. The β heterochromatic sequences flanking the I elements are themselves defective transposable elements. Chromosoma. 1989;98:215–224. doi: 10.1007/BF00329686. [DOI] [PubMed] [Google Scholar]
- Wang S, Lorenzen MD, Beeman RW, Brown SJ. Analysis of repetitive DNA distribution patterns in the Tribolium castaneum genome. Genome Biol. 2008;9:R61. doi: 10.1186/gb-2008-9-3-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker T, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8(12):973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- Wilder J, Hollocher H. Mobile elements and the genesis of microsatellites in Dipterans. Mol Biol Evol. 2001;18(3):384–392. doi: 10.1093/oxfordjournals.molbev.a003814. [DOI] [PubMed] [Google Scholar]
- Windsor AJ, Waddell CS. FARE, a new family of foldback transposons in Arabidopsis. Genetics. 2000;156(4):1983–1995. doi: 10.1093/genetics/156.4.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong LH, Choo KHA. Evolutionary dynamics of transposable elements at the centromere. Trends Genet. 2004;20(12):611–616. doi: 10.1016/j.tig.2004.09.011. [DOI] [PubMed] [Google Scholar]
- Yuan Y, Wessler SR. The catalytic domain of all eukaryotic cut-and-paste transposase superfamilies. Proc Natl Acad Sci U S A. 2011;108(19):7884–7889. doi: 10.1073/pnas.1104208108. [DOI] [PMC free article] [PubMed] [Google Scholar]