Abstract
Recently, microRNAs (miRs) have been implicated in bone formation and homeostasis. We previously reported that Dicer generated miRs have pivotal roles in differentiation and activity of osteoclasts. However, recent studies have demonstrated that Dicer is implicated in production of endogenous small interfering RNAs, non-canonical miRs, and other small RNAs in mammals. Hence, a challenging question is the extent to which expression of canonical miRs are obligatory for osteoclastic control of bone metabolism. DiGeorge syndrome critical region gene 8 (DGCR8) is exclusively related with expression of miRs by a canonical processing pathway together with the nuclear RNase III enzyme Drosha. Osteoclast-specific deletion of DGCR8 led to impaired osteoclastic development and bone resorption so that bone development was significantly retarded. In culture, the expression levels of osteoclastic phenotype-related genes and proteins were remarkably inhibited during osteoclastogenesis in DGCR8-deficiency. Thus, we have identified that DGCR8-dependent miRs are indispensable for osteoclastic control of bone metabolism.
Keywords: DGCR8, microRNA, osteoclast, Dicer
Introduction
MicroRNAs (miRs) are small, non-coding RNAs that posttranscriptionally control gene expression by inhibiting protein translation or inducing mRNA destabilization [Yang et al., 2011]. MiRs are characterized by specific maturation processes defined by canonical and non-canonical biogenesis pathways [Yang et al., 2011]. Canonical miR biogenesis requires the Microprocessor complex, which is comprised of Drosha and DiGeorge syndrome critical region gene 8 (DGCR8), to generate precursor-miRs, and Dicer, which is the cytosolic RNase III enzyme, to form mature miRs [Yang et al., 2011]. In contrast, non-canonical pathways are DGCR8 independent and Dicer dependent processes [Yang et al., 2011]. For instance, mirtrons, which are short hairpin introns, are spliced and debranched from mRNA transcripts directly and their maturation is produced by Dicer, escaping the Microprocessor complex [Berezikov et al., 2007]. In addition, Dicer is related to generation of another class of small RNA such as endogenous small interfering RNAs, small nucleolar RNAs, and transfer RNAs [Yang et al., 2011]. Hence, the conclusion of our previous study that miRs play critical roles in osteoclastic control of bone metabolism because myeloid-osteoclast line-age-specific Dicer deficient mice produced osteopetrosis caused by decreased osteoclastic development and bone resorption [Sugatani et al., 2009] is now questionable. To address this issue, we created osteoclast-specific DGCR8 deficient mice because DGCR8 is exclusively associated with the canonical miR pathway, except for miR-451 biogenesis, which is in a Dicer independent and DGCR8 dependent non-canonical pathway, to produce canonical miRs and does not generate other small RNAs [Yang et al., 2011]. Here, we demonstrate that the DGCR8 mutant mice have osteopetrosis caused by significantly decreased osteoclast development and bone resorption, showing that DGCR8-dependent miRs have critical roles in differentiation and activity of osteoclasts.
Materials & Methods
Generation of the osteoclast-specific DGCR8 deficient mice
DGCR8 floxed (DGCR8f/f) mice were obtained from Dr. Elaine Fuchs (Howard Hughes Medical Institute, Rockefeller University, New York, NY) [Yi et al., 2009]. DGCR8f/f mice were crossed with ctsk-Cre mice [Nakamura et al., 2007] to generate DGCR8wt/f with Cre mice. The wild-type mice (DGCR8f/f without Cre) and the mutant mice (DGCR8f/f with Cre) were obtained by crossing DGCR8wt/f with Cre male mice and DGCR8f/f without Cre female mice. All animals were housed under pathogen-free conditions according to the guidelines of the Division of Comparative Medicine, Washington University School of Medicine. The animal ethics committee approved all experiments.
Microcomputer tomography (μCT) and bone histomorphometry analysis
Micro-CT analysis of femurs from wild-type (wt) and mutant mice (12-week-old) was performed at the Ohio State University at Department of Veterinary Clinical Sciences using a high-resolution μCT system (Siemens). Images were reconstructed with IRW (Siemens) and analyzed with bone morphometric software (Ratoc System Engineering). In the trabecular bone analysis, volume/total volume (BV/TV), trabecular thickness (Tb.Th), trabecular number (Tb.N), and trabecular spacing (Tb.Sp) were examined in the secondary trabecular bone area of distal femurs. In cortical bone analysis, cortical wall thickness (C.Th) was analyzed by axial images at femur mid shafts. Bone histomorphometric analysis of femurs from wt and mutant mice (12-week-old) was conducted at the University of Kentucky at Bone Diagnostic and Research Laboratory. All bone samples were dehydrated, and embedded in methylmethacrylate. Serial sections of 4- and 7-μm thicknesses were cut with a microm microtome and the sections were stained with the modified Masson-Goldner trichrome stain. Histomorphometric parameters of bone were evaluated at standardized sites in cancellous bone using the semiautomatic method (Osteoplan II, Kontron, Germany) [Malluche et al., 1982].
Cell culture and infection
Macrophage colony-stimulating factor (M-CSF)-dependent DGCR8f/f bone marrow macrophages (BMMs) were prepared from the femurs and tibias of 4-to-6-week-old DGCR8f/f mice and Cre-containing retroviruses were infected into the cells as described previously [Sugatani et al., 2011]. After two days blasticidin selection (2 μg/mL) (Invitrogen), the infected cells were expanded and harvested for following assays.
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was performed with the imprint ChIP assay kit (Sigma) according to the manufacturer’s suggestions using antibodies against c-Fos (Santa Cruz), nuclear factor of activated T-cells, cytoplasmic 1 (NFATc1) (Abcam), and normal IgG (Sigma). The purified DNA was analyzed by PCR using primers that detect sequences containing the mouse NFATc1 promoter [Sugatani et al., 2011] and the mouse cathepsin K promoter [Miyauchi et al., 2010].
RNA isolation, reverse transcription and real-time PCR (qRT-PCR) analysis
M-CSF-dependent BMMs were prepared from the femurs and tibias of 4-to-6 week old wt and mutant male mice and cultured with 50 ng/ml M-CSF (Peprotech) and 100 ng/ml RANKL (Peprotech) for three days. Total RNA with small RNA (< 200 nucleotides) was isolated using the mirVana™ miRNA isolation kit (Life technologies) according to the manufacturer’s instructions. For qRT-PCR, cDNA was generated using the 1st strand cDNA synthesis system kit (OriGene) following the manufacturer’s suggestion. QRT-PCR was performed using the OriGene qSTAR SYBR Green Kit on an Mx4000 multiplex quantitative PCR System (Stratagene). The relative expression for c-Fos, NFATc1, and cathepsin K mRNA were estimated from triplicate qRT-PCR reactions following normalization to the β2 microglobulin. All primers were purchased from the OriGene.
Immunoblotting
To visualize NFATc1 (Thermo Scientific), cathepsin K (Santa Cruz), and α-tubulin (Santa Cruz), whole-cell lysates were prepared by the RIPA buffer (Thermo Scientific). Proteins were resolved by SDS-PAGE, electroblotted to polyvinylidene difluoride membrane (Millipore), blocked in 1 X TBST (Cell Signaling), and probed with primary antibodies (1:1000). Following incubation with anti-mouse IgG HRP-linked antibody (1:2000) (Cell Signaling) or anti-rabbit IgG HRP-linked antibody (1:2000) (Cell Signaling) was detected using enhanced chemiluminescence (Thermo Scientific).
Osteoclastic bone-resorbing activity assay
Quantification of bone resorption by osteoclasts in vitro was measured by the OsteoLyse assay kit (Lonza) according to the manufacturer’s instructions.
Statistical evaluations
All the numeral data in the results were presented as mean values ± standard deviations (SD). Statistical analysis was performed based on Student’s t-test or Welch’s t-test according to the results of F-test. Difference was judged to be statistically significant when p values were < 0.05.
Results and Discussion
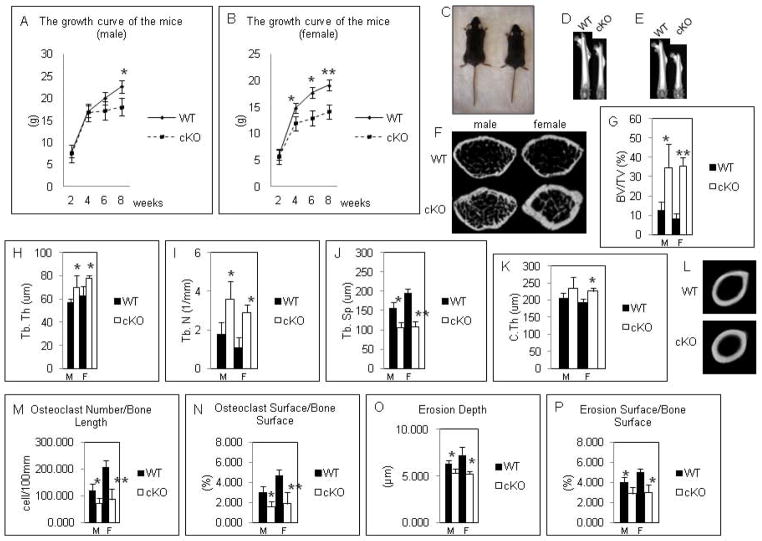
We created osteoclast-specific DGCR8 deficient mice using DGCR8 floxed mice with ctsk-Cre knock-in mice. The mutant mice were born normally without exhibiting any significant abnormalities in skeletal patterning (data not shown). In male mice, the growth of the mutant mice was significantly retarded compared to wt mice by 8 weeks after birth (Fig. 1A). In contrast, in female mutant mice, it was markedly impaired compared to wt mice after 4 weeks after birth (Fig. 1B and C). 3D uCT analysis showed that the anatomical length of femurs of mutant mice were decidedly shorter than wt of that in male (Fig. 1D) and female (Fig. 1E). In addition, the cross-section of distal metaphysis of femurs by uCT analysis revealed increased the trabecular and cortical bone mass in mutant mice (Fig. 1F). In particular, it was strikingly salient in female mutant mice (Fig. 1F). Consistent with these results, uCT analysis showed significantly increased the trabecular bone mass, trabecular thickness, and trabecular numbers and significantly decreased trabecular space in mutant mice compared to wt mice (Fig. 1G–J). On the other hand, the cortical thickness of mid shaft of femurs was significantly increased in female mutant mice, but not in male mutant mice (Fig. 1K and L). Bone morphometric analysis showed significantly impaired osteoclastogenesis and osteoclastic bone resorption activity in mutant mice (Fig. 1M–P). In contrast, osteoblastogenesis and its function had no difference between wt and mutant mice (data not shown). These data indicate that DGCR8 mutant mice produced osteopetrotic phenotypes caused by impaired osteoclastic development and bone resorption activity, suggesting that DGCR8-dependent canonical miRs may play critical roles in bone homeostasis implicated in osteoclast function. In addition, some data gave us the gender differences; however, we were not able to find the causes in this study.
Figure 1. Skeletal phenotypes of DGCR8f/f;ctsk-Cre mice.
(A) The growth curve of the male wt and mutant mice. cKO: conditional knockout. (B) The growth curve of the female wt and mutant mice. cKO: conditional knockout. (C) The picture of 12-week-old female wt (left) and mutant mice (right). (D) 3D-uCT images of the anatomical length of femurs of male wt and mutant mice (8 weeks). (E) 3D-uCT images of the anatomical length of femurs of female wt and mutant mice (12 weeks). (F) The cross-section of distal metaphysis (12 weeks). cKO: conditional knockout. (G)–(K) uCT analysis. The parameters are measured in the distal femurs of 12-week-old wt and mutant mice. The data are expressed as the means ± S.D. of five or six mice of each genotype. BV/TV, bone volume; Tb.Th, trabecular thickness; Tb.N, trabecular number; Tb.Sp, trabecular separation; C.Th, cortical bone thickness. M: male, F: female, cKO: conditional knockout. (L) uCT images of femoral mid shaft. cKO: conditional knockout. (M)–(P) Bone histomorphometric analysis. The parameters were measured in the distal femurs of 12-week-old wt and mutant mice. The data are expressed as the means ± S.D. of five or six mice of each genotype. cKO: conditional knockout. *p < 0.05, **p < 0.01. The data are expressed as the means ± five or six mice of each genotype.
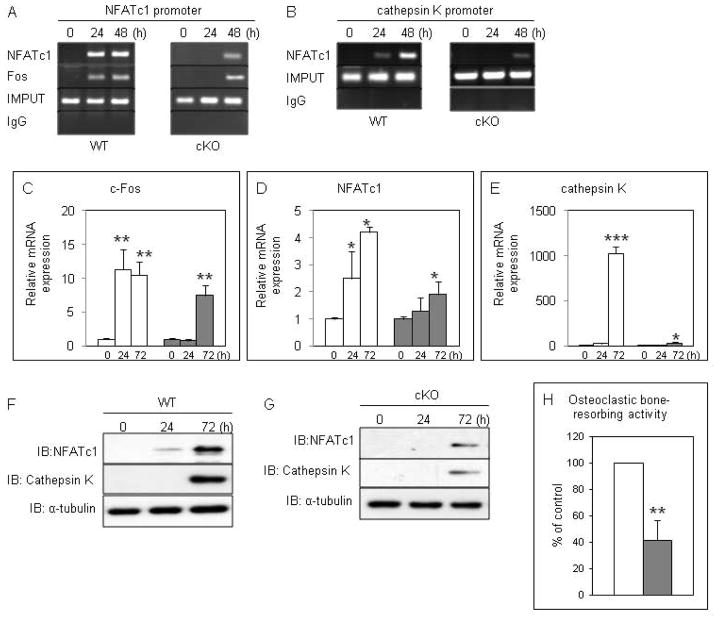
We have demonstrated that RANKL-induced osteoclastogenesis is significantly impaired in DGCR8-deficient BMMs, even though RANKL induces TRAP-positive osteoclasts from DGCR8f/f BMMs [Sugatani et al., 2011]. Thereby, we next asked whether the transcription factors required in osteoclastogenesis, such as c-Fos and NFATc1, are recruited to the NFATc1 or cathepsin K promoter in DGCR8-null cells using ChiP analysis. ChIP analysis showed that NFATc1 and c-Fos were recruited to the NFATc1 promoter by 24 and 48 h after RANKL stimulation in ex vivo DGCR8f/f BMMs (wt cells) (Fig. 2A). However, the binding of these factors to the NFATc1 promoter was observed by 48 h after RANKL treatment by a loss of DGCR8 protein (Fig. 2A). Moreover, NFATc1 was slightly recruited to the cathepsin K promoter by 24 and was strikingly bound to the promoter by 48 h after RANKL stimulation in wt cells (Fig. 2B). In contrast, the binding of NFATc1 to the cathepsin K promoter was drastically attenuated by a loss of DGCR8 protein (Fig. 2B). Consistent with the results of the ChIP analysis, the mRNA expression levels of the osteoclastic phenotype-related genes were markedly inhibited during RANKL-induced DGCR8-deficient osteoclastogenesis (Fig. 2C–E). QRT-PCR analysis showed that c-Fos and NFATc1 expression were markedly increased by 24 h and 72 h and cathepsin K expression was drastically elevated by 72 h after RANKL stimulation in wt cells (Fig. 2C–E). In contrast, those mRNA expression levels were significantly up-regulated by 72 h after RANKL stimulation in DGCR8-deficient osteoclastogenesis (Fig. 2C–E). In particular, cathepsin K mRNA expression levels were dramatically suppressed by 72 h after RANKL stimulation in DGCR8-null cells compared with wt cells (Fig. 2E). These data of Chip assay and qRT-PCR analysis can be attributed to inhibition of expression of the transcription factor proteins important for osteoclastogenesis by loss of DGCR8 during osteoclastic development as shown by Fig. 2F and G. Immunoblotting analysis showed RANKL strongly enhanced the protein expression levels of NFATc1 and cathepsin K by 72 h in wt cells (Fig. 2F). However, the effect was obviously attenuated by a loss of DGCR8 protein (Fig. 2G). Consistent with these findings, osteoclastic bone-resorbing activity was significantly decreased in DGCR8-deficient osteoclasts (Fig. 2H), suggesting that miRs generated by DGCR8 play critical roles for osteoclastic development and activity.
Figure 2. DGCR8 deficiency impacts on RANKL-induced osteoclastogenesis.
(A) and (B) Shown are ChIP assays to study the association of NFATc1 and c-Fos with NFATc1 or cathepsin K promoters during osteoclastogenesis with RANKL (100 ng/ml) stimulation for indicated times. cKO: conditional knockout. (C)–(E) QRT-PCR analysis. Ex vivo DGCR8f/f and DGCR8-null BMMs were treated with RANKL (100 ng/ml) for 3 days. Total RNA was extracted at the indicated time points, and expression of c-Fos (C), NFATc1 (D) and cathepsin K (E) mRNA levels were measured by qRT-PCR analysis. The PCR products were normalized to β2M for each reaction. The data represent the means ± S.D. of three experiments in triplicate. wt cells: open bar, DGCR8-null cells: closed bar. *p < 0.05, **p < 0.01, ***p < 0.001. (F) and (G) Ex vivo DGCR8f/f (F) and DGCR8-null BMMs (G) were treated with RANKL (100 ng/ml) for 3 days, and whole-cell lysates were analyzed by immunoblotting with antibodies against NFATc1 and cathepsin K. α-tubulin is loading control. IB: immunoblotting. cKO: conditional knockout. (H) Osteoclasts bone-resorbing activity was measured using each cell culture supernatant by the OsteoLyse™ assay kit. The data represent the means ± S.D. of three experiments in duplicate. wt cells: open bar, DGCR8-null cells: closed bar. **p < 0.01.
We have demonstrated that miR-223 and miR-21, which are DGCR8-dependent canonical miRs, contribute to osteoclastogenesis [Sugatani et al., 2009; Sugatani et al., 2011; Lian et al., 2012]. The mechanism for miR-223 stimulating osteoclastogenesis is that PU.1 stimulates the transcription of primary-miR-223, which in turns mature miR-223 suppresses nuclear factor IA (NFIA) levels posttranscriptionally so that expression of M-CSF receptor is increased, which is critical for osteoclast differentiation and function [Sugatani et al., 2009; Lian et al., 2012]. A second molecular event for regulation of osteoclastogenesis is a positive feedback loop of c-Fos/miR-21/programmed cell death 4 (PDCD4) [Sugatani et al., 2011; Lian et al., 2012]. Since PDCD4 is a negative regulator for osteoclastogenesis [Sugatani et al., 2011; Lian et al., 2012], down-regulation of PDCD4 by miR-21, which is triggered by c-Fos, is indispensable for osteoclastic development [Sugatani et al., 2011; Lian et al., 2012]. Taken together, expression of miRs generated by DGCR8-dependent canonical pathway is critical for down-regulation of the suppressors of osteoclastogenesis, NFIA and PDCD4. Given that Dicer- and DGCR8-null mice both have an osteopetrotic phenotypes caused by impaired osteoclastic development and bone resorption activity [Sugatani et al., 2009; Mizoguchi et al., 2010], expression and function of canonical and non-canonical miRs including other small RNAs may have roles in bone remodeling and homeostasis. Aberrant expression and function of miRs and other small RNAs generated by impaired canonical and/or non-canonical pathways may contribute to bone diseases, such as osteoporosis.
Acknowledgments
We thank Dr. Elaine Fuchs (Howard Hughes Medical Institute, Rockefeller University, New York, NY) for the gift of DGCR8 flox mice. This work was supported by the National Institutes of Health (grant DK070790; K.A.H.).
Footnotes
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Contribution: T.S. designed and performed research, analyzed data, and contributed the primary draft of the manuscript; B.E.H and R.E.T. performed uCT analysis; H.H.M. conducted bone histomorphometric analysis; and K.A.H. revised and produced the final manuscript.
References
- Berezikov E, Chung WJ, Willis J, Cuppen E, Lai EC. Mammalian mirtron genes. Mol Cell. 2007;28:328–36. doi: 10.1016/j.molcel.2007.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian JB, Stein GS, van Wijnen AJ, Stein JL, Hassan MQ, Gaur T, Zhang Y. MicroRNA control of bone formation and homeostasis. Nat Rev Endocrinol. 2012;8:212–27. doi: 10.1038/nrendo.2011.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malluche HH, Sherman D, Meyer W, Massry SG. A new semiautomatic method for quantitative static and dynamic bone histology. Calcif Tissue Int. 1982;34:439–448. doi: 10.1007/BF02411282. [DOI] [PubMed] [Google Scholar]
- Miyauchi Y, Ninomiya K, Miyamoto H, Sakamoto A, Iwasaki R, Hoshi H, Miyamoto K, Hao W, Yoshida S, Morioka H, Chiba K, Kato S, Tokuhisa T, Saitou M, Toyama Y, Suda T, Miyamoto T. The Blimp1-Bcl6 axis is critical to regulate osteoclast differentiation and bone homeostasis. J Exp Med. 2010;207:751–62. doi: 10.1084/jem.20091957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizoguchi F, Izu Y, Hayata T, Hemmi H, Nakashima K, Nakamura T, Kato S, Miyasaka N, Ezura Y, Noda M. Osteoclast-specific Dicer gene deficiency suppresses osteoclastic bone resorption. J Cell Biochem. 2010;109:866–75. doi: 10.1002/jcb.22228. [DOI] [PubMed] [Google Scholar]
- Nakamura T, Imai Y, Matsumoto T, Sato S, Takeuchi K, Igarashi K, Harada Y, Azuma Y, Krust A, Yamamoto Y, Nishina H, Takeda S, Takayanagi H, Metzger D, Kanno J, Takaoka K, Martin TJ, Chambon P, Kato S. Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell. 2007;130:811–23. doi: 10.1016/j.cell.2007.07.025. [DOI] [PubMed] [Google Scholar]
- Sugatani T, Hruska KA. Impaired micro-RNA pathways diminish osteoclast differentiation and function. J Biol Chem. 2009;284:4667–78. doi: 10.1074/jbc.M805777200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O’Carroll D, Stoffel M, Tuschl T, Fuchs E. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci U S A. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugatani T, Vacher J, Hruska KA. A microRNA expression signature of osteoclastogenesis. Blood. 2011;117:3648–57. doi: 10.1182/blood-2010-10-311415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang JS, Lai EC. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43:892–903. doi: 10.1016/j.molcel.2011.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi R, Pasolli HA, Landthaler M, Hafner M, Ojo T, Sheridan R, Sander C, O’Carroll D, Stoffel M, Tuschl T, Fuchs E. DGCR8-dependent microRNA biogenesis is essential for skin development. Proc Natl Acad Sci U S A. 2009;106:498–502. doi: 10.1073/pnas.0810766105. [DOI] [PMC free article] [PubMed] [Google Scholar]