Abstract
Context:
Genetic and environmental factors play an essential role in the pathogenesis of Graves' Disease (GD). Children with GD have less exposure time to environmental factors and therefore are believed to harbor stronger genetic susceptibility than adults.
Objective:
The aim of the study was to identify susceptibility loci that predispose to GD in patients with young-age-of-onset (YAO) GD.
Setting and Design:
One hundred six patients with YAO GD (onset <30 y) and 855 healthy subjects were studied. Cases and controls were genotyped using the Illumina Infinium Immunochip, designed to genotype 196,524 polymorphisms. Case control association analyses were performed using the PLINK computer package. Ingenuity Pathway Analysis program (QIAGEN) was used to carry out pathway analyses.
Results:
Immunochip genetic association analysis identified 30 single-nucleotide polymorphisms in several genes that were significantly associated with YAO GD, including major histocompatibility complex class I and class II genes, BTNL2, NOTCH4, TNFAIP3, and CXCR4. Candidate gene analysis revealed that most of the genes previously shown to be associated with adult-onset GD were also associated with YAO GD. Pathway analysis demonstrated that antigen presentation, T-helper cell differentiation, and B cell development were the major pathways contributing to the pathogenesis of YAO GD.
Conclusions:
Genetic analysis identified novel susceptibility loci in YAO GD adding a new dimension to the understanding of GD etiology.
Graves' disease (GD) is one of the most common autoimmune endocrine disorders with a prevalence in the United States of approximately 0.5–1% (1). GD is a classic antibody-mediated autoimmune disease, but it is unique in that the pathogenic antibodies stimulate the TSH receptor (2). Clinically, GD is characterized by hyperthyroidism, diffuse goiter, and in some patients is associated with complications including ophthalmopathy and dermopathy (2). Biochemically, the hallmark of GD is the production of TSH receptor (TSHR) –stimulating antibodies (2). The pathogenesis of GD involves breakdown of central and peripheral tolerance, infiltration of the thyroid with thyroid-directed T cells that escaped tolerance, and activation of B cells to secrete TSHR stimulating antibodies (2).
What triggers this cascade of events is still unknown, but accumulating data point to an interaction between susceptibility genes (1) and environmental triggers such as viral infection, diet, and iodine (3). During the past 2 decades, several GD-susceptibility genes have been identified and confirmed through linkage and association studies including HLA-DR, CTLA-4, PTPN22, CD40, CD25, thyroglobulin (Tg), and the TSHR gene (reviewed in Ref. 1). Moreover, in a subset analysis, we have shown a unique genetic susceptibility in young-age-of-onset (YAO) GD (age of onset [AO] < 30 y) (4). The goal of the present study was to dissect this unique genetic susceptibility found in YAO GD. We used the Immunochip (Illumina Infinium) approach, analyzing approximately 9000 genes in a cohort of YAO GD patients and healthy controls.
Materials and Methods
Study subjects
The project was approved by the Icahn School of Medicine and the Boston's Children's Hospital Institutional Review Boards. One hundred six Caucasian patients with GD (AO < 30 y) and 855 healthy Caucasian patients were studied. Detailed characteristics of the subjects enrolled onto our study are included as Supplemental Materials and Methods.
Genotyping and Principal Component Analysis
Genotyping and principal component analysis (PCA) of the cases and controls were performed as described previously (5). Briefly, DNA samples were genotyped using the Immunochip, designed to genotype 196,524 polymorphisms. Quality-control testing was performed to ensure high call rate for the variants on the Immunochip. More information on the genotyping and the PCA analysis are available as Supplemental Material.
Association analysis
Association analyses were performed using the program PLINK, which utilizes the Cochran-Armitage trend test (http://pngu.mgh.harvard.edu/purcell/plink/) (6). P < 1×10−5 was considered significant. Detailed information on the association analysis is available as Supplemental Materials and Methods.
Candidate gene analysis
Genes that have been previously reported to be associated with GD were analyzed separately, taking the nominal cutoff (P < .05) as significant. These candidate genes included TSHR, Tg, CD40, CTLA-4, IL23R, CD25, FOXP3, FCRL3, PTPN22, and PTPN2 (7–9). All candidate genes were genotyped using the Immunochip platform except for Tg and FOXP3 (See Supplemental Materials and Methods).
Pathway analysis
Pathway analysis was performed using the Ingenuity Pathway Analysis (IPA) system version 8.6 (http://www.ingenuity.com/). One thousand single nucleotide polymorphism (SNP) that reached the highest statistical significance were used in our analysis. The probability that a pathway was significantly enriched when compared with the genome database was computed using Fisher's exact test. P < .05 was considered significant. More information on the pathway analysis is included in Supplemental Materials and Methods.
Results
Immunochip analysis
Among the 106 Caucasian patients with GD analyzed, the average age at diagnosis was 18.9 ± 7.0 y (range, 2–30 y); there were 83 female and 23 male patients. Thirty six patients (34%) had Graves' ophthalmopathy (GO), 40 (38%) had no evidence of GO, and in 30 patients (28%), the GO status could not be confirmed. Of the 36 patients with GO 7 (19%) had mild GO and 29 (81%) had moderate-to-severe GO as defined by European Group on Graves' Orbitopathy. Of the 106 GD patients, one was removed for “missingness” (missing genotypes) and therefore 105 patients were used in the final analysis. The average age of healthy controls was 37 ± 12.6 y.
To avoid effects of population stratification we performed a PCA analysis on our cases and controls. Both patients and controls were North American Caucasians of European ancestry. PCA demonstrated that our Caucasian YAO GD patients and Caucasian controls were ethnically matched (Supplemental Figure 1) with an inflation factor (λ) of 1.0, demonstrating very close matching of our patients and controls.
The quantile-quantile plot, which illustrates results of the association analyses for all the tested SNPs, showed that multiple SNPs in several loci were significantly associated with YAO GD (Supplemental Figure 2). Association analysis results of the Immunochip SNPs in YAO GD compared with health controls are summarized in the Manhattan plot shown in Figure 1. The strongest association was seen at the major histocompatibility complex (MHC) (HLA) locus; however, other non-MHC loci also showed significant associations. To reduce the likelihood of false-positive results, we only considered genes that had at least two SNPs with a P value less than 1×10−5 as significant. We identified 30 SNPs within eight genes that met those criteria. These included MHC class I and class II genes, as well as four additional genes, BTNL2, NOTCH4, TNFAIP3, and CXCR4 (Table 1).
Figure 1.
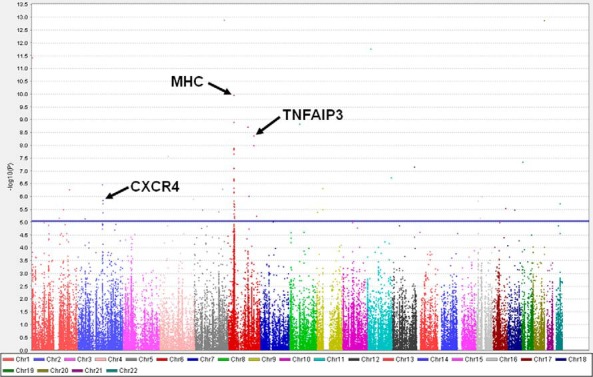
Manhattan plot of Immunochip association analysis data comparing patients with YAO GD (n = 105) to healthy controls (n = 855). Variants on the immunochip (154,847 including 6628 HLA variants) that passed the quality control testing were sorted by chromosomal location (x-axis) and were plotted against the −log10(P value) (y-axis), with the height of each point corresponding to the strength of association with disease. The blue line indicated a P value < 1×10−5. SNPs that showed association with YAO GD with P < 1×10−5 are above this line.
Table 1.
Genes/Loci That Were Significantly Associated with YAO GD, Containing at Least Two SNPs with P < 1×10−5
| Gene | Chromosome | Locus | Most Strongly Associated SNP | P Value | Odds Ratio | No. SNPs (P < 1×10−5) | Gene Function |
|---|---|---|---|---|---|---|---|
| HLA-C | 6 | MHCa | rs3132486 | 2.3 × 10− 7 | 3 | 2 | Antigen presentation |
| HLA-DQA1 | 6 | MHC | rs9273012 | 1.1 × 10−10 | 3.1 | 8 | Antigen presentation |
| HLA-DQB2 | 6 | MHC | rs6457680 | 3.4 × 10−6 | 2.1 | 5 | Antigen presentation |
| HCG22 | 6 | MHC | rs1265061 | 1.3 × 10−8 | 2.6 | 3 | HLA complex group 22 |
| BTNL2 | 6 | MHC | rs17577980 | 6.9 × 10−7 | 2.8 | 2 | Negative regulator of T cell proliferation |
| NOTCH4 | 6 | MHC | rs424232 | 1.3 × 10−8 | 2.6 | 5 | Regulator of cell-fate determination |
| TNFAIP3 | 6 | 6q23 | rs78242606 | 4.2 × 10−9 | 12 | 2 | Involved in cytokine mediated immune and inflammatory responses |
| CXCR4 | 2 | 2q22 | rs4954573 | 1.4 × 10−6 | 2.1 | 3 | Chemokine receptor |
The MHC locus is on chromosome 6p21.
Candidate gene analysis
Several genes have been previously reported to be associated with GD regardless of AO, and these have been tested separately for association with YAO GD. With the exception of FOXP3 and PTPN22, all known GD susceptibility genes showed association with YAO GD. These included TSHR (P = 1.9×10−3), Tg (P = 8.5×10−3), CD40 (P = 1×10−2), CTLA-4 (P = 9×10−4), IL23R (P = 1×10−5), CD25 (P = 6.4×10−4), FCRL3 (P = 1×10−2), and PTPN2 (P = 2.2×10−4) (Supplemental Table 1).
Pathway analysis
We did a pathway analysis to provide insight into mechanisms underlying the etiology of YAO GD. Several pathways were found to be significantly associated with YAO GD (Supplemental Table 2). The most strongly associated pathways were antigen presentation and T-helper cell differentiation, consistent with accumulating evidence that the immunological synapse harbors most of the susceptibility genes for GD. Of interest, CD28 and CTLA-4 signaling pathways, also involved in antigen presentation to T helper cells, showed significant associations. Similarly, B-cell development and IL-4 signaling pathways showed significant associations with YAO GD, consistent with the fact that GD is an antibody-mediated autoimmune disease.
Discussion
A strong hereditary component in the pathogenesis of GD has long been recognized (1). The inheritance of GD is complex, involving multiple genes with variable penetrances; environmental and epigenetic factors also play a critical role in the etiology of GD (1). One approach to identifying key genes that predispose to disease is to study children and patients with YAO of disease. The main advantage of studying patients with YAO of disease is that they have had less exposure time to environmental triggers and therefore the genetic contribution to their disease is expected to be stronger. Indeed, our previous studies suggested unique genetic susceptibility to YAO GD (4). Moreover, studies by Segni et al (10) have shown distinctive genetic susceptibility to juvenile autoimmune thyroid diseases. In a follow-up study by Segni and Badenhoop (11) on 91 juvenile Hashimoto's thyroiditis patients and 12 juvenile GD patients, they found an increased transmission of the DQ2 allele by fathers, but not by mothers of both juvenile GD and Hashimoto's thyroiditis patients, suggesting paternal imprinting of the risk alleles.
In several autoimmune disorders, unique genes predisposing to YAO disease have been identified. A study of type 1 diabetes patients reported that polymorphisms in the IL-2 and RNLS genes were associated with a younger age at diagnosis (12). The Arg620Trp variant in the PTPN22 gene was also found to be associated with YAO type 1 diabetes (13). Other groups reported associations between PDCD1 polymorphisms and childhood-onset systemic lupus erythematosus (14).
Most of the variants exhibiting association with YAO GD were located within the MHC locus (Table 1), demonstrating that this locus confers much of the genetic susceptibility to YAO GD. One of the MHC-locus genes we found to be associated with YAO GD was the BTNL2 gene, which was also reported to influence the AO of vitiligo (15). The BTNL2 gene is a ligand expressed on antigen-presenting cells that suppresses T-cell proliferation. Recent studies suggested that BTNL2 expression and binding to the T-cell receptor play a major role in the differentiation of T cells into Treg cells (16). Taken together, these data suggest that variants in BTNL2 may impair Treg function, thereby predisposing individuals to autoimmunity at a young age. Indeed, the BTNL2 gene was reported to be associated with several other autoimmune diseases in addition to vitiligo (15), including ulcerative colitis (17) and rheumatoid arthritis (18), albeit an association with AO was reported only for vitiligo. Because the BTNL2 gene is located within the MHC class II region, we cannot rule out that the association of BTNL2 with YAO GD is due to linkage disequilibrium with DR3 (19).
In addition to screening for new genes, we analyzed known GD susceptibility genes for association with YAO GD. Analysis of known GD genes demonstrated that most of them were also associated with YAO GD, including TSHR, Tg, CD40, CTLA-4, IL-23R, and CD25 (Supplemental Table 1). Our IPA pathway analysis showed that pathways associated with antigen presentation to T-helper cells and with B-cell development are the major pathways contributing to the pathogenesis of YAO GD, in keeping with reports that antigen presentation pathway genes are associated with GD regardless of AO (1). Therefore, the genetic susceptibility to YAO GD may be explained by two possible mechanisms: 1) all genes predisposing to YAO GD also predispose to GD in general, but their effect is more pronounced in patients that develop the disease at a young age; 2) some genes predispose to GD regardless of AO whereas other genes predispose uniquely to YAO GD (eg, BTNL2).
Although the subset of YAO GD patients was homogeneous, the main limitation of our study was the small size of our cohort. Therefore, our findings must be replicated in a larger group of patients. Furthermore, because we performed a pathway analysis selecting only the top-scoring 1000 SNPs, we cannot exclude that if we expanded the subset of SNPs used in the IPA, additional pathways would be identified as contributing to YAO GD.
In summary, analysis of YAO GD demonstrated that in addition to the known GD-susceptibility genes, unique genes contribute to YAO GD. The mechanisms by which these genes contribute to the AO of GD are not known but perturbation of the immunological synapse seems to be a key mechanism.
Acknowledgments
This work was supported in part by Grants DK061659, DK067555, and DK073681 from NIDDK. In addition, this material is based upon work supported in part by the Department of Veterans Affairs.
Disclosure Summary: The authors have nothing to disclose.
Footnotes
- AO
- age of onset
- GD
- Graves' Disease
- GO
- Graves' ophthalmopathy
- IPA
- Ingenuity Pathway Analysis
- MHC
- major histocompatibility complex
- PCA
- Principal Component Analysis
- SNP
- single nucleotide polymorphism
- TSHR
- TSH receptor
- YAO
- young age of onset.
References
- 1. Huber A, Menconi F, Corathers S, Jacobson EM, Tomer Y. Joint genetic susceptibility to type 1 diabetes and autoimmune thyroiditis: from epidemiology to mechanisms. Endocr Rev. 2008;29:697–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Davies TF. 2000 Graves' diseases: pathogenesis. In: Braverman LE, Utiger RD, eds. Werner and Ingbar's the thyroid: a fundamental and clinical text. 8th ed Philadelphia: Lippincott Williams & Wilkins; 518–530 [Google Scholar]
- 3. Tomer Y, Huber A. The etiology of autoimmune thyroid disease: a story of genes and environment. J Autoimmun. 2009;32:231–239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tomer Y, Menconi F, Davies TF, et al. Dissecting genetic heterogeneity in autoimmune thyroid diseases by subset analysis. J Autoimmun. 2007;29:69–77 [DOI] [PubMed] [Google Scholar]
- 5. Hasham A, Zhang W, Lotay V, et al. Genetic analysis of interferon induced thyroiditis (IIT): evidence for a key role for MHC and apoptosis related genes and pathways. J Autoimmun. 2013;44:61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Inoue N, Watanabe M, Yamada H, et al. 2012 associations between autoimmune thyroid disease prognosis and functional polymorphisms of susceptibility genes, CTLA4, PTPN22, CD40, FCRL3, and ZFAT, previously revealed in genome-wide association studies. J Clin Immunol. 2012;32:1243–1252 [DOI] [PubMed] [Google Scholar]
- 8. Ban Y, Tozaki T, Tobe T, et al. The regulatory T cell gene FOXP3 and genetic susceptibility to thyroid autoimmunity: An association analysis in Caucasian and Japanese cohorts. J Autoimmun. 2007;28:201–207 [DOI] [PubMed] [Google Scholar]
- 9. Tomer Y, Hasham A, Davies TF, et al. Fine mapping of loci linked to autoimmune thyroid disease identifies novel susceptibility genes. J Clin Endocrinol Metab. 2013;98:E144–E152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Segni M, Wood J, Pucarelli I, et al. Clustering of autoimmune thyroid diseases in children and adolescents: a study of 66 families. J Pediatr Endocrinol Metab. 2001;14 Suppl 5:1271–1275; discussion 1297–1298 [PubMed] [Google Scholar]
- 11. Segni M, Pani MA, Pasquino AM, Badenhoop K. Familial clustering of juvenile thyroid autoimmunity: higher risk is conferred by human leukocyte antigen DR3-DQ2 and thyroid peroxidase antibody status in fathers. J Clin Endocrinol Metab. 2002;87:3779–3782 [DOI] [PubMed] [Google Scholar]
- 12. Howson JM, Cooper JD, Smyth DJ, et al. Evidence of gene-gene interaction and age-at-diagnosis effects in type 1 diabetes. Diabetes. 2012;61:3012–3017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kordonouri O, Hartmann R, Badenhoop K, Kahles H, Ilonen J. PTPN22 1858T allele is associated with younger age at onset of type 1 diabetes and is not related to subsequent thyroid autoimmunity. Hum Immunol. 2010;71:731–732 [DOI] [PubMed] [Google Scholar]
- 14. Velázquez-Cruz R, Orozco L, Espinosa-Rosales F, et al. Association of PDCD1 polymorphisms with childhood-onset systemic lupus erythematosus. Eur J Hum Genet. 2007;15:336–341 [DOI] [PubMed] [Google Scholar]
- 15. Jin Y, Birlea SA, Fain PR, et al. Genome-wide analysis identifies a quantitative trait locus in the MHC class II region associated with generalized vitiligo age of onset. J Invest Dermatol. 2011;131:1308–1312 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Swanson RM, Gavin MA, Escobar SS, et al. Butyrophilin-like 2 modulates B7 costimulation to induce Foxp3 expression and regulatory T cell development in mature T cells. J Immunol. 2013;190:2027–2035 [DOI] [PubMed] [Google Scholar]
- 17. Pathan S, Gowdy RE, Cooney R, et al. Confirmation of the novel association at the BTNL2 locus with ulcerative colitis. Tissue Antigens. 2009;74:322–329 [DOI] [PubMed] [Google Scholar]
- 18. Mitsunaga S, Hosomichi K, Okudaira Y, Nakaoka, et al. Exome sequencing identifies novel rheumatoid arthritis-susceptible variants in the BTNL2. J Hum Genet. 2013;58:210–215 [DOI] [PubMed] [Google Scholar]
- 19. Simmonds MJ, Heward JM, Barrett JC, Franklyn JA, Gough SC. Association of the BTNL2 rs2076530 single nucleotide polymorphism with Graves' disease appears to be secondary to DRB1 exon 2 position beta74. Clin Endocrinol (Oxf). 2006;65:429–432 [DOI] [PubMed] [Google Scholar]


