Plants don’t go into the light as a near-death experience—the plant’s complex response to light begins before the seedling even emerges. For example, in many plant species, red (R) light promotes germination and far-red (FR) light inhibits germination. Indeed, because chlorophyll absorbs R light, a high ratio of R:FR indicates direct sunlight and a low ratio indicates shade from other leaves; perception of this ratio conditions plant growth and development. Phytochromes play key roles in these (and other) plant responses to light, responses that involve signaling and substantial changes in the transcriptome (reviewed in Chen and Chory, 2011). After absorbing FR light, phytochrome A (phyA) moves to the nucleus, in association with its chaperone, FAR-RED ELONGATED HYPOCOTYL1 (FHY1), where it alters the transcriptome by regulating the stability of specific light-responsive transcription factors. In addition to indirectly altering the transcriptome, phyA and FHY1 also associate with the promoter of CHALCONE SYNTHASE (CHS), a key player in anthocyanin biosynthesis, to directly regulate its transcription.
To examine whether the direct association of phyA with target promoters functions as a general mechanism, Chen et al. (pages 1949–1966) use chromatin immunoprecipitation (ChIP) to identify phyA-associated genes and RNA-sequencing to identify phyA-regulated genes. phyA binds to DNA indirectly, by association with other DNA binding proteins; despite this, ChIP assays showed positive peaks in the CHS promoter, indicating that ChIP can identify phyA-associated genes. The ChIP and RNA-sequencing assays each identified about 3000 phyA-associated, or phyA-regulated genes, with 448 genes present in both sets (see figure). These putative direct targets of phyA include many genes that respond to other signals, including stress, defenses, and hormones, such as abscisic acid (ABA); the promoters of these genes also have an enrichment of cis-elements responsive to these signals. These direct targets are likely to be cotargeted by phyA in association with many known light- or hormone-responsive transcription factors, such as LONG HYPOCOTYL5 (HY5).
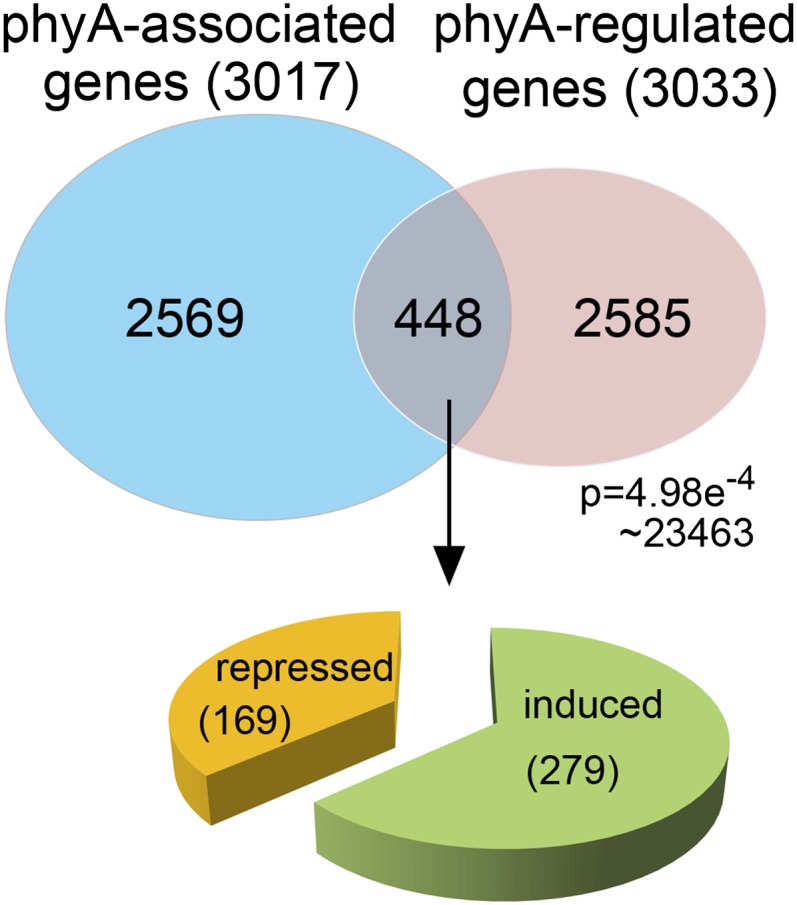
Identification of direct targets of phyA. The authors use ChIP to identify phyA-associated genes (blue) and RNA-sequencing to identify phyA-regulated genes (pink). The intersection of these two sets includes repressed and induced genes. (Reprinted from Chen et al. [2014], Figure 3.)
The authors next examined the ABA response as an example of how phyA affects other processes. Mutations that affected phyA signaling also were found to influence the inhibition of root elongation by ABA, indicating that, under FR light, phyA positively regulates ABA signaling. The authors also used ChIP and quantitative PCR to confirm the association of phyA with NAC019, which encodes an ABA-dependent transcription factor. Induction of NAC019 expression by FR light requires HY5, and direct binding assays showed that HY5 targets two G-box cis-elements in the NAC019 promoter. Luciferase reporter assays in Nicotiana benthamiana confirmed that HY5 activates NAC019, and coexpression of phyA and FHY1 increases NAC019 expression by increasing HY5 transcriptional activity. Moreover, in phyA, fhy1, and hy5 mutants, ABA cannot activate NAC019 expression.
Thus, phyA functions with HY5 to activate the NAC019 transcription factor and positively regulate the ABA response under FR light. This walk into the light response showed that plants may use phytochrome-mediated signaling to rapidly modulate multiple processes. However, how phyA affects these diverse processes and the significance of phyA association with so many genes that do not show phyA regulation remain unclear and intriguing subjects for future research.
References
- Chen F., Li B., Li G., Charron J.-B., Dai M., Shi X., Deng X.W. (2014). Arabidopsis phytochrome A directly targets numerous promoters for individualized modulation of genes in a wide range of pathways. Plant Cell 26: 1949–1966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M., Chory J. (2011). Phytochrome signaling mechanisms and the control of plant development. Trends Cell Biol. 21: 664–671 [DOI] [PMC free article] [PubMed] [Google Scholar]


