Abstract
Peptide signals have emerged as an important class of regulators in cell-to-cell communication in plants. Several families of small, secreted proteins with a conserved C-terminal Pro-rich motif have been identified as functional peptide signals in Arabidopsis thaliana. These proteins are presumed to be trimmed proteolytically and undergo posttranslational modifications, such as hydroxylation of Pro residues and glycosylation, to form mature, bioactive signals. Identification and matching of such ligands with their respective receptors remains a major challenge since the genes encoding them often show redundancy and low expression restricted to a few cells or particular developmental stages. To overcome these difficulties, we propose the use of ectopic expression of receptor genes in suitable plant cells like Nicotiana benthamiana for testing ligand candidates in receptor output assays and in binding studies. As an example, we used the IDA peptide HAE/HSL2 receptor signaling system known to regulate floral organ abscission. We demonstrate that the oxidative burst response can be employed as readout for receptor activation by synthetic peptides and that a new, highly sensitive, nonradioactive labeling approach can be used to reveal a direct correlation between peptide activity and receptor affinity. We suggest that these approaches will be of broad value for the field of ligand-receptor studies in plants.
CHALLENGES IN MATCHING LIGAND-RECEPTOR PAIRS
Over the last decade, increasing numbers of secreted peptides have been shown to influence a variety of developmental processes in plants, including meristem size, root growth, stomatal differentiation, and organ abscission (Butenko and Aalen, 2012). The Arabidopsis thaliana genome encodes about a thousand small proteins that could function as peptide signals and more than 600 plasma membrane-bound receptor-type proteins that could act as receptors for peptide ligands (Shiu and Bleecker, 2001; Lease and Walker, 2006). So far, however, less than a dozen peptide ligand-receptor pairs have been identified. Identification has mainly been by genetic interaction studies, and direct physical interaction has been demonstrated experimentally for only a few of these pairs (Murphy et al., 2012). Thus, the singling out of matching pairs and confirmation of physical interaction is needed for the vast majority of these putative peptide ligands and their respective receptors. Identification of these regulatory pairs is crucial to advance our understanding of cell-to-cell communication during plant development.
Genetic evidence for ligand-receptor pairs usually arises from the observation that the encoding genes have corresponding expression patterns and that mutations in such genes are associated with similar or identical changes in phenotypes. Genetic interaction subsequently can be corroborated if a scorable overexpression phenotype of a peptide-encoding gene is lost in the receptor mutant background. However, these phenotypes must be interpreted with caution. Similar, but not identical, phenotypes are indicative for missing signaling components, as exemplified by studies of the CLAVATA3 (CLV3) ligand, belonging to the CLV3/ENDOSPERM SURROUNDING REGION (CLE) peptide family, and its signaling partner CLV1 (Müller et al., 2008; Kinoshita et al., 2010). Conversely, constitutive overexpression of signal precursors or application of large doses of a synthetic version of the peptide or a related family member can give phenotypes resulting from unspecific signaling through a non-native receptor, as observed when overexpressing INFLORESCENCE DEFCIENT IN ABSCISSION-LIKE5 (IDL5), a protein in the presumed INFLORESCENCE DEFCIENT IN ABSCISSION (IDA) peptide family. Constitutive overexpression of IDA results in precocious floral abscission, as does overexpression of IDL5, even if this gene is not expressed in floral abscission zones (AZs) (Stenvik et al., 2006, 2008). Similarly, exogenous application or overexpression of many different CLE peptides rescues the clv3 phenotype, indicating that receptors of CLV3 are able to recognize a certain degree of CLE sequence variation (Kinoshita et al., 2007; Lee and Torii, 2012).
Unfortunately, the majority of the Arabidopsis receptor-like kinases (RLKs) do not have a known function despite the systematic effort to identify aberrant phenotypes of single T-DNA insertions, and often higher order mutants of homologous genes are required to obtain informative phenotypes (Lehti-Shiu et al., 2009; Butenko and Aalen, 2012; Li and Tax, 2013). Furthermore, due to the small size of peptide ligands and their corresponding open reading frames, the publicly available T-DNA insertion collections are far from complete with respect to insertions in all of the putative peptide ligand encoding genes, and many peptide ligand genes may remain unannotated since they fall below the cutoff values for open reading frames in gene prediction programs (Lease and Walker, 2006). In addition, peptide encoding genes often belong to gene families comprising several members, and genetic redundancy might preclude approaches based on genetics alone (Amano et al., 2007). Due to these shortcomings, only a few peptide-receptor pairs have been identified with the aid of mutant studies to date (Murphy et al., 2012).
A more direct approach consists of straightforward purification and identification of peptide signals. This approach depends on biological activity present in plant extracts that can be monitored quantitatively with bioassays and has allowed the purification and identification of peptide ligands regulating developmental pathways, such as PHYTOSULFOKINE (PSK) and TRACHEARY ELEMENT DIFFERENTIATION INHIBITORY FACTOR (TDIF), as well as innate immune response, such as Pep1, systemin, and rapid alkalinization factor (RALF) (Pearce et al., 1991, 2001; Matsubayashi and Sakagami, 1996; Huffaker et al., 2006; Ito et al., 2006). In turn, identified peptide signals can then be used to screen for mutants showing insensitivity to the signal in order to identify the corresponding receptors. Examples for successful application of this approach include the TDIF receptor PHLOEM INTERCALATED WITH XYLEM/TRD (Hirakawa et al., 2008) and the RLK FERONIA that binds RALF (Haruta et al., 2014).
Unfortunately, many peptide signals may not show a scorable phenotype in tissue culture or cell suspensions due to natural low expression levels restricted to a few specific cells in the plant of both receptors and peptide ligands. Thus, there is a need for the establishment of novel assays suitable for detection of peptide signaling through RLKs. Such assays are additionally important since most plant peptide signals identified so far are derived from larger precursor proteins. For genes encoding such precursors identified genetically or in silico, a rapid cellular bioassay would be advantageous for delineation of the bioactive part of the protein and for testing the importance of peptide trimming and posttranslational modifications that may be predicted from comparison to known, structurally related signals (Amano et al., 2007; Ohyama et al., 2008).
While genetic approaches are important to identify candidates that may form ligand-receptor pairs, additional methods are required to uncover physical interaction of ligands with their receptors. Here, innovative methods developed for human cell cultures may prove useful to identify plant peptide-receptor pairs (Uebler and Dresselhaus, 2014). Elucidation of the structural requirements of peptides is important to establish labeled ligands as probes for direct physical interactions in biochemical binding assays. Classical methods of labeling ligands made use of radiolabeling with 125I, 35S, or 3H in order to obtain the high specific activity required to detect receptor molecules occurring in low abundance on the surface of cells. Apart from the inconvenience of working with radioactive material, the radiolabeling of peptides is expensive and probes can have a relatively short half-life.
TOOLS FOR STUDYING LIGAND-RECEPTOR INTERACTIONS
In this Perspective, we propose to overcome the various impediments in studying ligand-receptor interactions summarized above using the following strategies: (1) to increase the amount of receptor of interest with respect to the number of cells expressing the receptor and the amount of receptor molecules per cell by enhanced ectopic expression in suitable plant cells like those of Nicotiana benthamiana leaves; (2) to establish a cellular bioassay suitable for quantitative assessment of the receptor output to test and optimize natural and synthetic ligand candidates predicted from in silico analysis; (3) to use optimized ligand peptide to produce a labeled probe for interaction studies with receptor proteins. In particular, as an alternative to radiolabeling, we propose the use of acridinium-esters as luminogenic probes that allow detection down to the lower fmol and attmol ranges with standard luminometers.
We exemplify these strategies and approaches with the peptide ligand IDA that signals through the two leucine-rich repeat receptor-like kinases HAESA (HAE) and HAESA-LIKE2 (HSL2) to control floral organ abscission in Arabidopsis (Butenko et al., 2003; Cho et al., 2008; Stenvik et al., 2008). Mutations in either the gene encoding the peptide signal or both of the receptor genes lead to plants exhibiting defects in floral abscission (Cho et al., 2008; Stenvik et al., 2008). Constitutive overexpression of IDA results in detachment of organs not normally shed in Arabidopsis in addition to precocious floral abscission, a phenotype not observed in the hae hsl2 mutant background (Cho et al., 2008; Stenvik et al., 2008). All genetic evidence is consistent with IDA as the source of a signal that acts by stimulating the receptors HAE and HSL2. However, the structure of the active signal molecule and whether it interacts physically with HAE, HSL2, or both receptors is not known.
OXIDATIVE BURST, A BIOASSAY USED TO STUDY RECEPTORS IN PATHOGEN DEFENSE, CAN BE EMPLOYED TO MEASURE OUTPUT OF HSL2
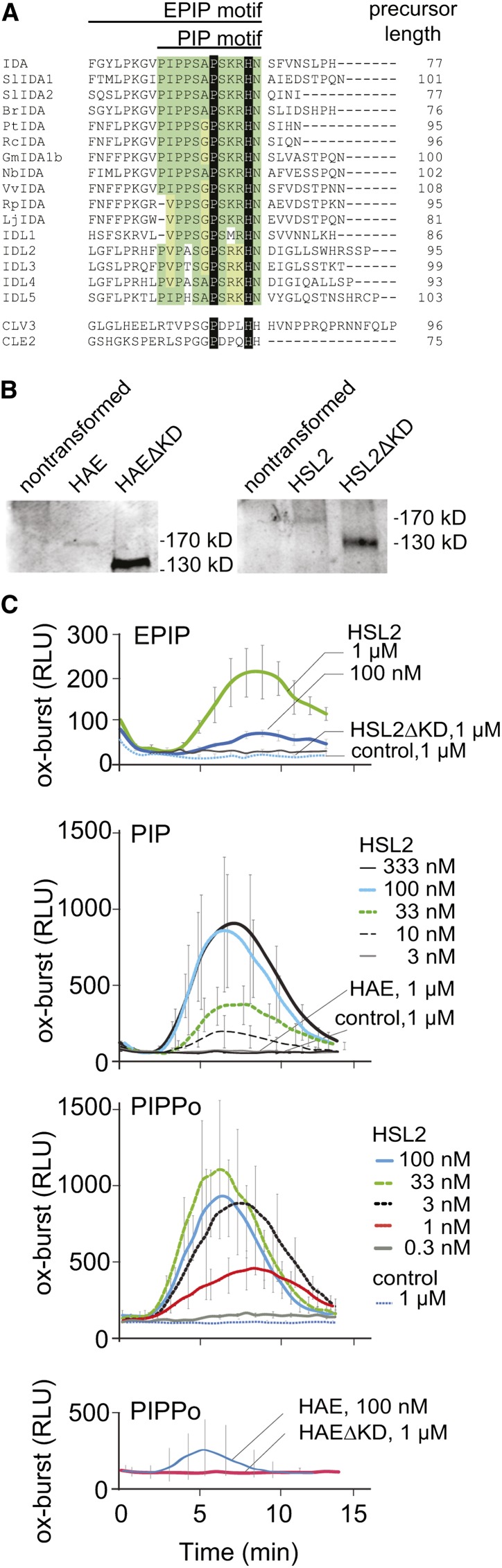
Release of reactive oxygen species (ROS) as an oxidative burst (ox-burst) was first documented as output of plant cells treated with elicitors or microbe-associated molecular patterns (Apostol et al., 1989) and was implicated in defense against pathogens and also in intercellular signaling (Lamb and Dixon, 1997). Earlier reports suggested ROS to be important regulators of plant development through their role in regulating cell expansion (Gapper and Dolan, 2006). Because cell elongation is an intrinsic step of floral organ abscission (Shi et al., 2011), we tested whether the HAE and HSL2 RLKs could trigger induction of ROS by expressing them in leaf tissue of N. benthamiana. The functional part of IDA resides within the C-terminal part of the protein and comprises a 20–amino acid Pro-rich motif, the extended PIP (EPIP) (Figure 1A), which is sufficient to rescue the abscission defect of the ida mutant in a HAE/HSL2-dependent manner (Stenvik et al., 2008). By employing a luminol-dependent assay, a significant ox-burst was detected after treatment of N. benthamiana leaf tissue expressing HSL2 with concentrations exceeding 100 nM EPIP (Figures 1B and 1C, Table 1). As expected, no ox-burst response was observed in N. benthamiana leaves expressing HSL2 without kinase domain (HSL2ΔKD) (Figures 1B and 1C). Interestingly, no ox-burst was seen for leaves expressing HAE, despite expression levels similar to HSL2 (Figure 1B). While the synthetic EPIP peptide reproducibly induced an ox-burst by signaling through the HSL2 receptor, it did so only at relatively high concentrations exceeding concentrations reported for bioactivity of other known peptide ligands like PSK, CLV3, and Pep1 (Matsubayashi et al., 1999; Huffaker et al., 2006; Ohyama et al., 2009). This, and the lack of ox-burst observed in HAE expressing leaves, suggested that the peptide in use was suboptimal.
Figure 1.
Ox-Burst by the IDA-HAE/HSL2 Signaling Module.
(A) Alignment of the C-terminal region of IDA, IDL1-5 from Arabidopsis, and IDA orthologs from other species, CLV3 and CLE2. Sequence similarities within the PIP domain of the IDA peptides are highlighted in green. The invariant proline, hydroxyprolinated in CLV3, CLE2, and presumably also IDA, and the His conserved in all these signal peptides are indicated in black.
(B) Protein gel blot of N. benthamiana leaves expressing Halo-tagged HAE and HSL2 as full-length constructs or as kinase truncated versions (ΔKD), respectively. Blots developed with α-Halo antibody stain single bands that migrate with molecular weights that are higher than those anticipated by the polypeptide chains alone. This is likely due to glycosylation of the receptors, as previously reported for HAE (Jinn et al., 2000).
(C) IDA triggers the production of ROS through HSL2 in N. benthamiana. Oxidative burst was measured in leaf pieces of untransformed N. benthamiana (control) or leaves transiently expressing the HSL2 receptor or HSL2ΔKD, HAE, or HAEΔKD as indicated. Leaf pieces were exposed to various concentration of the EPIP peptide of IDA (top panel), the PIP peptide (second panel from top), or the peptide PIPPo, PIP with hydroxylation of the conserved Pro at position 7 (bottom two panels). Ox-burst by the luminol-based assay was monitored over time as relative light units (RLU). Error bars indicate sd of n = 3 replicates.
Table 1. Peptide Sequences, Activity, and Affinity to the HSL2 Receptor.
| Peptide | Amino Acid Sequence | EC50 (nM) | IC50 (nM) |
|---|---|---|---|
| EPIP | FGYLPKGVPIPPSAPSKRHN | 333 | >10,000 |
| PIP | PIPPSAPSKRHN | 33 | >10,000 |
| PIP-o4 | PIPoSAPSKRHN | 33 | ND |
| PIPPo | PIPPSAoSKRHN | 1 | 100 |
| PIP-o1;7 | oIPPSAoSKRHN | 1 | ND |
| PIP-o3;7 | PIoPSAoSKRHN | 1 | 33 |
| PIPPo-∆N | PIPPSAoSKRH | >10,000 | >10,000 |
| VPIPPo | VPIPPSAoSKRHN | 1 | 33 |
| acri-PIPPo | acr-VPIPPSAoSKRHN | 100 | ND |
| CLV3* | RTVoSGoNPLHHH | >10,000 | ND |
| [Ara3]CLV3* | [Ara3]-RTVoSGoNPLHHH | >10,000 | ND |
Amino acid sequences of peptides used in this study are indicated in single-letter amino acid abbreviations with “o” for hydroxyproline. EC50, concentrations required to stimulate half-maximal oxidative burst in N. benthamiana leaves expressing HSL2; IC50, concentration required for 50% reduction of acri-PIPPo binding to HSL2; >10,000, no half-maximal effect on ox-burst or inhibition of binding even at 10 µM of peptides applied; ND, not determined. Asterisks indicate that the same results were obtained for HAE-expressing tissue.
The lack of ox-burst in untransformed N. benthamiana upon treatment with IDA peptides (Figures 1B and 1C) indicates that the predicted endogenous orthologs of HAE and HSL2 are not sufficiently expressed in leaf tissue to respond to the IDA peptides (http://solgenomics.net/organism/Nicotiana_benthamiana/genome). The EPIP domain is well conserved between the IDL proteins of Arabidopsis. Except for residues 2 and 3, the EPIP domains of putative IDA orthologs in N. benthamiana are identical to IDA EPIP of Arabidopsis (Figure 1A). Considering IDA peptides of dicot plant species in general, the PIP motif, comprising the 12 C-terminal amino acids of the EPIP domain, is close to 100% conserved in IDA homologs (Figure 1A). Exposure of the dodecapeptide PIP to HSL2 expressed in N. benthamiana leaves was significantly more effective than EPIP and elicited an ox-burst with a half maximal effective concentration (EC50) of 33 nM for PIP compared with 333 nM for EPIP (Figure 1C, Table 1), indicating that proper trimming is one of the modifications required for a fully active ligand of HSL2. Nevertheless, despite the increase in activity of the PIP peptide compared with EPIP, no response to the PIP peptide was observed in HAE-expressing leaf tissue or in untransformed control leaves (Figure 1C).
HYDROXYLATION OF PROLINES AFFECTS SIGNAL ACTIVITY
C-terminal motifs of proteins encompassing peptide signals, including IDA/IDL, CLV3/CLE, GOLVEN/ROOT GROWTH FACTOR/CLE-LIKE, and C-TERMINALLY ENCODED PEPTIDE, are Pro rich (Butenko and Aalen, 2012), and conserved Pro of mature, active CLV3/CLE peptides have been shown to be hydroxylated (Kondo et al., 2006; Ohyama et al., 2008; Hirakawa et al., 2010). We therefore tested the effect of substituting hydroxyprolines for Pro at different positions of the PIP peptide (Table 1). Hydroxylation of Pro at position 4 (PIP-o4) had no effect on peptide activity (Table 1), while a peptide named PIPPo with modification of the invariant Pro at position 7 (Figure 1A) increased the activity 30-fold over that observed for PIP (Figure 1C, Table 1). Peptides with additional hydroxylations on either the Pro at position 1 or 3 (PIP-o1;7 or PIP-o3;7) had the same activity as PIPPo and induced an ox-burst with an EC50 of 1 nM (Table 1).
Interestingly, PIP and CLE peptides have a Pro in corresponding positions (Figure 1A). Two of the Pro of CLV3 and CLE2 become hydroxylated during export of the precursor peptides to the apoplastic space (Kondo et al., 2006; Ohyama et al., 2009), and for these peptides, the activity and receptor affinity is increased when the hydroxyprolines are further glycosylated (Ohyama et al., 2009; Shinohara and Matsubayashi, 2013). The hydroxyproline in PIPPo corresponds to this relevant hydroxyproline in CLV3/CLE2. Despite the similarities between the CLE domain of CLV3 and PIP (Stenvik et al., 2008), neither the glycosylated version nor the hydroxyprolinated version of the mature CLV3 peptide (Shinohara and Matsubayashi, 2013) gave an ox-burst response in HSL2- or HAE-expressing tissue (Table 1).
Posttranslational modifications known from well-studied mature signal peptides such as CLV3 can provide a lead for predicting important modifications on related signal peptides. For peptides with a Pro-rich motif, hydroxylation of the conserved, centrally placed Pro can be proposed to occur and to have a positive effect on signal activity. Comparing CLV3 and IDA indicates that the pattern of pro-peptide processing also seems to be conserved. Similar to removal of the C-terminal residue of the mature CLV3 (mCLV3) (Kondo et al., 2006), activity of IDA was abolished by removal of the C-terminal Asn residue in PIPPo, as observed for peptide PIPPo-ΔΝ (Table 1). In summary, using an in silico approach to identify highly conserved amino acid residues to known peptides may provide a good starting point for determination of the length and residue composition of novel bioactive peptides.
DIFFERENT HSL RECEPTORS HAVE DIFFERENT AFFINITIES OR SPECIFICITIES FOR IDA-DERIVED SIGNALING MOLECULES
Interestingly, the highly active PIPPo peptide was also capable of inducing an ox-burst in N. benthamiana leaves transiently expressing HAE, albeit only when applied at a 100-fold higher peptide concentration than that required for HSL2 (Figure 1C). Thus, while hydroxylated PIPPo peptide efficiently activates HSL2, further modifications may be needed for efficient activation of HAE or HAE may require an additional coreceptor. So far, the structure of the naturally occurring IDA peptide has not been identified. However, hydroxylated Pro of CLV3, CLE2, and PSY1 have been found to be glycosylated in planta (Amano et al., 2007; Ohyama et al., 2009), making it likely that the mature IDA peptide is glycosylated. Hydroxylation of the central Pro of CLV3 did not change peptide activity (Kondo et al., 2006), but the structural impact on peptide conformation of attaching a triarabinoside branch to this hydroxyproline increased activity and receptor affinity (Ohyama et al., 2009; Shinohara and Matsubayashi, 2013). Similarly, full biological activity of the IDA peptide might depend on further modification of hydroxyprolines by glycosylation. The affinity of HAE for IDA ligands might depend more strongly on these presumptive modifications than that of HSL2.
PEPTIDE LIGANDS LABELED WITH CHEMILUMINESCENT ACRIDINIUM ARE SENSITIVE PROBES SUITABLE FOR LIGAND-RECEPTOR INTERACTION STUDIES
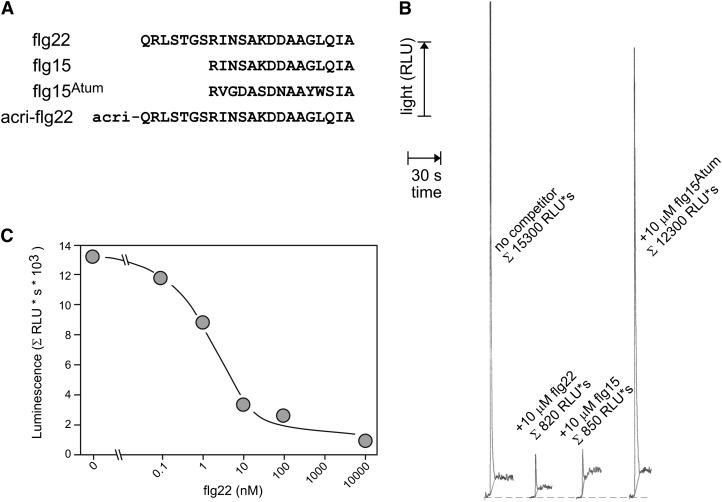
Individual membrane receptor proteins are usually of very low abundance and make up only a limited fraction of membrane proteins or total cellular proteins. Ligand binding studies with such receptors require methods that can detect minute amounts of ligands bound specifically to the binding sites. We investigated whether chemiluminescent labels like acridinium-esters could serve as a nonradioactive and cost-effective alternative to radiolabeling of ligands. Acridinium-esters rapidly and stoichiometrically decompose in the presence of H2O2 under emission of light that can be used to quantify this label down to attomolar amounts in a luminometer (Joss and Towbin, 1994). To establish the usefulness of acridinium-esters, we first tested it for the well-established interaction of flg22 with FLS2 on tomato (Solanum lycopersicum) cells (Meindl et al., 2000; Chinchilla et al., 2006). Indeed, as easily detectable in a luminometer, a significant amount of acri-flg22 (Figure 2A) remained bound to the cells after extensive washing (Figure 2B). This binding was specific and could be competed by adding an excess of biologically active ligands like flg22 and flg15 but not by the inactive analog flg15Atum to the binding assays (Figure 2).
Figure 2.
Acridinium Conjugate of flg22 as a Sensitive Probe to Detect Specific Binding Sites on Tomato Cells.
(A) Peptides used in the experiment. Acri-flg22 is flg22 with an acridinium-ester conjugated to its N terminus, and flg22 and flg15 are potent agonistic ligands for the FLS2 receptor, while flg15Atum has no affinity for this receptor in tomato cells (Meindl. et al., 2000).
(B) Binding of Acri-flg22 to intact tomato cells. Aliquots of cells were treated for 5 min with 30 pM Acri-flg22 and a high molar excess of unlabeled flg22, flg15, or flg15Atum as indicated. After extensive washing, acridinium bound to cells was measured by tracing light emission (relative light units [RLU]) induced by applying 20 mM H2O2 in 0.1 M NaOH.
(C) Binding competition by different concentration of unlabeled flg22. Cells were treated as in (B) and acri-flg22 bound to cells was quantified by integrating the flash of light emitted after application of 20 mM H2O2 in 0.1 M NaOH.
To test whether chemiluminescent labeling would be of help in studying the interaction of IDA-derived peptides with HSL2 and HAE, we synthesized a PIPPo peptide conjugated to acridinium. Attempts to conjugate the acridinium ester to the N terminus of PIPPo failed, but labeling was achieved after N-terminal prolongation of the peptide with a Val residue (VPIPPo) (Table 1). While the prolongation by Val did not change the specific activity of the peptide (Table 1), the final product, termed acri-PIPPo, was somewhat less active than unlabeled VPIPPo and PIPPo, inducing an ox-burst with an EC50 of 100 nM (Table 1).
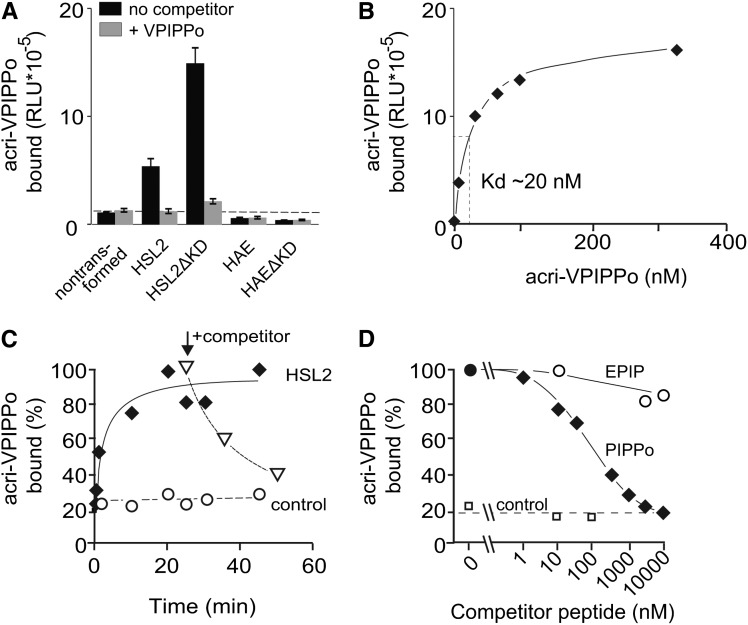
Acri-PIPPo was used to detect specific binding sites in the insoluble fractions of crude leaf preparations of N. benthamiana. Preparations from leaves expressing either HSL2 or HSL2ΔKD were found to bind significant amounts of acri-PIPPo that was clearly competed by unlabeled VPIPPo (Figure 3A).
Figure 3.
Binding of the IDA-Derived Peptide Acri-PIPPo to HSL2.
(A) Modified IDA peptides bind directly to the ectodomain of HSL2. Leaf material from N. benthamiana expressing HSL2, HAE, or the truncated version HSL2ΔKD or HAEΔKD lacking the kinase domain was incubated with 10 nM acri-PIPPo peptide in the presence or absence of 10 μM unlabeled VPIPPo as a competitor. Controls show binding to leaf material from nontransformed N. benthamiana. Bars and error bars show averages and standard deviations of n = 3 replicates. RLU, relative light units.
(B) Saturation of acri-PIPPo binding to the HSL2 receptor. Specific binding of acridinium-tagged VPIPPo was determined by subtracting binding observed with untransformed leaf material at the different concentrations from the values measured for the HSL2 expressing leaf material.
(C) Binding kinetics and reversibility of binding to the HSL2 receptor. Plant material from HSL2 expressing or nonexpressing (control) leaf samples were incubated with 10 nM acri-PIPPo. Reversibility of binding was tested by adding 10 μM of VPIPPo (indicated with an arrow) at 25 min. Binding is expressed as percentage of maximal binding in material expressing HSL2.
(D) HSL2-Halo expressing leaf material incubated with acri-PIPPo and increasing concentrations of EPIP and PIPPo as competitors. Leaf tissue not expressing the receptor was incubated with the acri-PIPPo peptide and different concentrations of PIPPo as a control to determine background signal. The signal detected from samples without competitor was defined as 100% binding.
HAEΔKD and HSL2ΔKD accumulated to higher levels than the full-length forms HAE and HSL2 in N. benthamiana leaves (Figure 1B). Correlating with the amount of ectodomain present, leaf tissues expressing HSL2ΔKD showed higher specific binding of acri-PIPPo than tissue expressing full-length HSL2 (Figure 3A). Binding assays with HSL2ΔKD demonstrated saturable binding with a dissociation constant (Kd) of ∼20 nM for acri-PIPPo (Figure 3B). Binding kinetics showed rapid association that could be reversed by the addition of unlabeled VPIPPo, implying reversibility of ligand binding (Figure 3C). Control samples with nontransformed leaves retained little of the acridinium-labeled peptide. Preparations from leaves expressing HAE or HAEΔKD only showed marginal binding, and this binding appeared nonspecific since it was not altered by the addition of an excess of unlabeled VPIPPo to the assays (Figure 3A), indicating that the very low ox-burst output in the presence of IDA peptides was due to low binding affinity of the HAE receptor.
The specificity of the acri-PIPPo binding to HSL2 was tested in competition experiments with acri-PIPPo and different concentrations of the unlabeled peptides EPIP, PIP, PIPPo, PIP-o3;7, PIPPo-ΔN, and VPIPPo, and half maximal inhibitory concentration (IC50) values were determined (Figure 3D, Table 1). In good correlation with their relative activity as inducers of ROS in N. benthamiana leaves, PIPPo and PIP-o3;7 efficiently competed binding with IC50 in the same range with values of 30 to 100 nM, while EPIP and PIP had IC50 values >10,000 nM. The IC50 value of 33 nM for VPIPPo confirmed that the addition of a Val to the PIPPo peptide did not interfere with its binding capacity toward HSL2. PIPPo-ΔΝ did not significantly inhibit binding of acri-PIPPo even at concentrations of 10,000 nΜ, indicating the Asn at the C terminus of the dodecapeptide PIPPo is essential for both binding and activation of HSL2.
It was shown previously that enzymatic activity in cauliflower (Brassica oleracea) protein extracts can process the IDA precursor protein, but an exact processing site was not determined (Stenvik et al., 2008). It remains to be tested whether trimming after His-Asn, marking the C-terminal end of the conserved part in all members of the IDL family, is occurring in vivo and is relevant for the biological activity of the ligand peptide.
PEPTIDE SIGNALS IN DEVELOPMENT AND DEFENSE: VARIATIONS OVER A THEME
Here, we investigated the signaling potential of IDA by its genetically identified receptor partners HAE and HSL2 through transient expression of the receptors in (heterologous) plant cells of N. benthamiana. We show that a peptide signal regulating a developmental process can trigger ox-burst, a characteristic response previously associated with innate immunity. During floral organ abscission, specialized AZ cells undergo cell wall remodeling prior to the shedding of the organ (Bleecker and Patterson, 1997), and an intrinsic step during this process is the expansion of the AZ cells that takes place prior to organ separation (Shi et al., 2011). ROS release is known to play a role during leaf expansion (Rodríguez et al., 2002), and the spatial regulation of ROS production is an important factor in controlling plant form. Furthermore, RALF, a peptide known to affect levels of ROS (Bedinger et al., 2010), binds FERONIA and mediates the inhibition of proton transport and cell expansion (Haruta et al., 2014), showing that a growth mediated signaling pathway may also intersect with the innate immune response (Kessler et al., 2010). FERONIA is a member of the CrRLK1L family of receptors of which several members have been implicated in regulating cell elongation, and it is therefore possible that other CrRLK1L members could be regulated by RALF-like peptides to modulate cell wall properties through intracellular Ca2+ increase and ROS production (Wolf and Höfte, 2014). It is possible that the ox-burst observed by HSL2 signaling could have a function in AZ cell expansion. Also, recent transcriptome analysis of AZs from wild-type and hae hsl2 mutants indicate that the IDA-HAE/HSL2 signaling module triggers cell wall degrading and cell wall remodeling (CWR) genes that are needed for the abscission process, but also genes commonly related to defense against bacteria and fungi (Niederhuth et al., 2013). Finally, the ox-burst observed could also be triggering the activation of pathogen defense genes because shedding exposes a fresh cell surface that could be highly susceptible to pathogen infection.
Genetic data show that IDA cannot signal in the absence of both HAE and HSL2 but that IDA can trigger abscission and promote lateral root emergence with either one or both of the receptors present (Stenvik et al., 2008; Kumpf et al., 2013). The functional equivalence of the two receptors in vivo seems at odds with their strong difference in affinity for the synthetic PIPPo observed in this work. However, a high local concentration of the IDA peptide in vivo might be sufficient to signal also through HAE alone. Furthermore, the lower affinity of HAE might be compensated by higher concentrations of HAE over HSL2 receptors in the cells of the responding tissues. Also, in analogy to CLV3 and CLE2, it is conceivable that the mature IDA peptide is posttranslationally glycosylated on a hydroxyproline and that this modification is more decisive for recognition by HAE than by HSL2. Experiments with chemically synthesized arabinosylated peptides such as described for CLV3 (Shinohara and Matsubayashi, 2013) or mature signal molecules isolated from plants overexpressing the IDA precursor will be required to test these possibilities.
Plant receptor kinases ectopically expressed in (heterologous) plant cells can be expected to be processed and localized correctly, unlike the situation in vitro. There is also a good chance that adaptors and coreceptors required for activation of downstream signaling are present in heterologous cells and allow a functional signal output upon stimulation with appropriate ligands. There are indications that signaling in defense and development have common features and, for instance, involve the same mitogen-activated protein kinase signaling components (Jonak et al., 2002; Betsuyaku et al., 2011). Hence, it is conceivable that elements of the stress response pathway, best studied after stimulation of pattern recognition receptors triggered by microbe-associated molecular patterns, can be co-opted by other signaling pathways. Kinases, including RLKs, can be subdivided into RD and non-RD kinases depending on the conservation of the amino acid residue preceding the core catalytic Asp residue in the kinase domain (Johnson et al., 1996; Nolen et al., 2004). Most of the RLKs functioning in innate immunity belong to the non-RD class (Dardick and Ronald, 2006). However, both HSL2 and HAE belong to the RD kinases, indicating that a broader range of RLKs might be able to activate ox-burst as an output to control developmental processes. We therefore anticipate that additional RD kinase plant receptors can activate ox-burst as a cellular output, as illustrated in a recent report on ROS production mediated by the receptor kinase FERONIA during rupture of the pollen tube in the fertilization process (Duan et al., 2014). A convenient and sensitive output assay such as the ox-burst assay is valuable to delineate the structural requirement for biologically active ligands. We suggest starting with a bioinformatic analysis for an initial delineation of peptide candidates based on the shortest stretch of conserved residues in a given peptide family, and, if comprising Pro residues, testing of these residues in their hydroxylated form.
Plant receptors are membrane proteins of low abundance in planta, with low expression restricted to a few cells and particular developmental stages. Transient overexpression in leaves of N. benthamiana can thus be used to accumulate plant receptor kinases or their extracellular binding domains in considerably higher amounts suitable for studying receptor ligand interaction. Together with methods for labeling of ligand candidates with tags that can be sensitively monitored, this should prove useful for the characterization of many other ligand-receptor pairs involved in regulation of important developmental and physiological processes in plants. Overall, ligand design in different peptide families seems to represent variation over a common theme. Hence, by recognizing nature’s reuse of the same modules and principles, we can hopefully move forward faster in our quest for understanding cell-to-cell communication in plants.
METHODS
Peptides and Conjugation with Acridinium
Peptides were synthesized using solid-phase technology with Fmoc-protected amino acids. Acridinium esters were conjugated to the N-terminal amino groups of the peptides on resin by coupling with N-hydroxysuccinimide activated acridinium esters (Cayman) before deprotection and purification of the peptides.
Transient Protein Expression in N. benthamiana Leaves
For transient expression, Agrobacterium tumefaciens carrying plasmids C58pGV2260 encoding HAE-Halo, HAE∆KD-Halo, HSL2-Halo, or HSL2∆KD-Halo under the 35S cauliflower mosaic virus promoter were infiltrated into Nicotiana benthamiana leaves according to Mueller et al. (2012). The Halo fusion constructs were made by amplifying the full-length and kinase-deleted coding region of HAE and HSL2 into the Halo-Tag vector (Promega). Accumulation of receptor constructs was detected by protein gel blot analysis using α-Halo antibody (Promega).
Oxidative Burst Measurements
Ox-burst experiments were performed as described (Felix et al., 1999; Albert et al., 2010) using a luminol-dependent assay (Keppler et al., 1989). Leaf pieces of N. benthamiana, cut 24 h after infiltration with the Agrobacteria and kept overnight floating on water, were placed in 96-well plates containing 100 μL water, 10 μg/mL horseradish peroxidase (Applichem), and 20 µM luminol. Peptides at given concentrations were added to the wells, and the emission of light was measured in an LB 960 microplate luminometer (Centro, Berthold Technologies) over a period of 15 to 20 min.
Receptor-Ligand Binding Assay
Frozen leaf tissue from N. benthamiana, 24 h after infiltration with Agrobacteria carrying constructs encoding HAE-Halo, HAE-∆KD-Halo, HSL2-Halo, or HSL2-∆KD-Halo, were ground to a powder and washed in binding buffer (25 mM MES, pH 6.0, 150 mM NaCl, 1 mM DTT, 3 µL/mL Proteinase Inhibitor mix [Sigma-Aldrich]) and collected by centrifugation (13,000g, 1 min, 4°C). The resulting pellet (P1) containing insoluble cell debris and membranes was resuspended in binding buffer (500 mg P1/mL). For standard assays, aliquots of 80 μL of this suspension was supplied with 10 nM of acri-PiPPo and different concentrations of competitor peptides as indicated. After incubation for 20 min at 4°C on a shaker ligand not bound to the tissue, aliquot was removed by washing pellets three times by quickly resuspending the pellets in 1 mL of binding buffer before immediate centrifugation (13,000g, 1 min, 4°C). Washed pellet was supplied with 50 μL 5 mM citric acid, and acridinium was measured by integrating light emission of the sample for 10 s in a single tube luminometer (FB12; Berthold) after injecting 100 μL of a solution with 20 mM H2O2 in 100 mM NaOH.
AUTHOR CONTRIBUTIONS
M.W., M.A., R.B.A., G.F., and M.A.B. designed the research. M.W., M.A., A.J., G.F., and M.A.B. performed the research. M.W., M.A., R.B.A., G.F., and M.A.B. analyzed the data. H.K. contributed reagents/materials/analysis tools. G.F. and M.A.B. wrote the article.
Acknowledgments
We thank Ilonka Bock for technical assistance, Yoshikatsu Matsubayashi for providing chemically synthesized triarabinosylated CLV3 peptide (Shinohara and Matsubayashi, 2013), and Rüdiger Simon for constructive comments and suggestions to the article. This work was supported by Grants 13785/F20 (to M.A.B. and R.B.A.) and 348256/F20 (to M.W. and R.B.A.) from the Research Council of Norway; Grant 216856 (to M.A.B., M.A., M.W., G.F., and R.B.A.) from the Research Council of Norway and the Deutscher Akademischer Austausch Dienst; and Grants AL1426/1-1 (to M.A.) and FE 962/2-1 (to G.F. and A.J.) from the Deutsche Forschungsgemeinschaft.
Footnotes
Articles can be viewed online without a subscription.
References
- Albert M., Jehle A.K., Mueller K., Eisele C., Lipschis M., Felix G. (2010). Arabidopsis thaliana pattern recognition receptors for bacterial elongation factor Tu and flagellin can be combined to form functional chimeric receptors. J. Biol. Chem. 285: 19035–19042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano Y., Tsubouchi H., Shinohara H., Ogawa M., Matsubayashi Y. (2007). Tyrosine-sulfated glycopeptide involved in cellular proliferation and expansion in Arabidopsis. Proc. Natl. Acad. Sci. USA 104: 18333–18338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Apostol I., Heinstein P.F., Low P.S. (1989). Rapid stimulation of an oxidative burst during elicitation of cultured plant cells: role in defense and signal transduction. Plant Physiol. 90: 109–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bedinger P.A., Pearce G., Covey P.A. (2010). RALFs: peptide regulators of plant growth. Plant Signal. Behav. 5: 1342–1346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betsuyaku S., Takahashi F., Kinoshita A., Miwa H., Shinozaki K., Fukuda H., Sawa S. (2011). Mitogen-activated protein kinase regulated by the CLAVATA receptors contributes to shoot apical meristem homeostasis. Plant Cell Physiol. 52: 14–29 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleecker A.B., Patterson S.E. (1997). Last exit: senescence, abscission, and meristem arrest in Arabidopsis. Plant Cell 9: 1169–1179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butenko, M.A., and Aalen, R.B. (2012). Receptor ligands in development. In Receptor-Like Kinases in Plants: From Development to Defense, F. Tax and B. Kemmerling, eds (Berlin, Heidelberg: Springer-Verlag), pp. 195–226. [Google Scholar]
- Butenko M.A., Patterson S.E., Grini P.E., Stenvik G.-E., Amundsen S.S., Mandal A., Aalen R.B. (2003). Inflorescence deficient in abscission controls floral organ abscission in Arabidopsis and identifies a novel family of putative ligands in plants. Plant Cell 15: 2296–2307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chinchilla D., Bauer Z., Regenass M., Boller T., Felix G. (2006). The Arabidopsis receptor kinase FLS2 binds flg22 and determines the specificity of flagellin perception. Plant Cell 18: 465–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho S.K., Larue C.T., Chevalier D., Wang H., Jinn T.L., Zhang S., Walker J.C. (2008). Regulation of floral organ abscission in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 105: 15629–15634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardick C., Ronald P. (2006). Plant and animal pathogen recognition receptors signal through non-RD kinases. PLoS Pathog. 2: e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan Q., Kita D., Johnson E.A., Aggarwal M., Gates L., Wu H.M., Cheung A.Y. (2014). Reactive oxygen species mediate pollen tube rupture to release sperm for fertilization in Arabidopsis. Nat. Commun. 5: 3129. [DOI] [PubMed] [Google Scholar]
- Felix G., Duran J.D., Volko S., Boller T. (1999). Plants have a sensitive perception system for the most conserved domain of bacterial flagellin. Plant J. 18: 265–276 [DOI] [PubMed] [Google Scholar]
- Gapper C., Dolan L. (2006). Control of plant development by reactive oxygen species. Plant Physiol. 141: 341–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haruta M., Sabat G., Stecker K., Minkoff B.B., Sussman M.R. (2014). A peptide hormone and its receptor protein kinase regulate plant cell expansion. Science 343: 408–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Kondo Y., Fukuda H. (2010). TDIF peptide signaling regulates vascular stem cell proliferation via the WOX4 homeobox gene in Arabidopsis. Plant Cell 22: 2618–2629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirakawa Y., Shinohara H., Kondo Y., Inoue A., Nakanomyo I., Ogawa M., Sawa S., Ohashi-Ito K., Matsubayashi Y., Fukuda H. (2008). Non-cell-autonomous control of vascular stem cell fate by a CLE peptide/receptor system. Proc. Natl. Acad. Sci. USA 105: 15208–15213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffaker A., Pearce G., Ryan C.A. (2006). An endogenous peptide signal in Arabidopsis activates components of the innate immune response. Proc. Natl. Acad. Sci. USA 103: 10098–10103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito Y., Nakanomyo I., Motose H., Iwamoto K., Sawa S., Dohmae N., Fukuda H. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313: 842–845 [DOI] [PubMed] [Google Scholar]
- Jinn T.-L., Stone J.M., Walker J.C. (2000). HAESA, an Arabidopsis leucine-rich repeat receptor kinase, controls floral organ abscission. Genes Dev. 14: 108–117 [PMC free article] [PubMed] [Google Scholar]
- Johnson L.N., Noble M.E., Owen D.J. (1996). Active and inactive protein kinases: structural basis for regulation. Cell 85: 149–158 [DOI] [PubMed] [Google Scholar]
- Jonak C., Okrész L., Bögre L., Hirt H. (2002). Complexity, cross talk and integration of plant MAP kinase signalling. Curr. Opin. Plant Biol. 5: 415–424 [DOI] [PubMed] [Google Scholar]
- Joss U.R., Towbin H. (1994). Acridinium ester labelled cytokines: receptor binding studies with human interleukin-1 alpha, interleukin-1 beta and interferon-gamma. J. Biolumin. Chemilumin. 9: 21–28 [DOI] [PubMed] [Google Scholar]
- Keppler L.D., Baker C.J., Atkinson M.M. (1989). Active oxygen production during a bacteria-induced hypersensitive reaction in tobacco suspension cells. Phytopathology 79: 974–978 [Google Scholar]
- Kessler S.A., Shimosato-Asano H., Keinath N.F., Wuest S.E., Ingram G., Panstruga R., Grossniklaus U. (2010). Conserved molecular components for pollen tube reception and fungal invasion. Science 330: 968–971 [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Betsuyaku S., Osakabe Y., Mizuno S., Nagawa S., Stahl Y., Simon R., Yamaguchi-Shinozaki K., Fukuda H., Sawa S. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137: 3911–3920 [DOI] [PubMed] [Google Scholar]
- Kinoshita A., Nakamura Y., Sasaki E., Kyozuka J., Fukuda H., Sawa S. (2007). Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48: 1821–1825 [DOI] [PubMed] [Google Scholar]
- Kondo T., Sawa S., Kinoshita A., Mizuno S., Kakimoto T., Fukuda H., Sakagami Y. (2006). A plant peptide encoded by CLV3 identified by in situ MALDI-TOF MS analysis. Science 313: 845–848 [DOI] [PubMed] [Google Scholar]
- Kumpf R.P., Shi C.L., Larrieu A., Stø I.M., Butenko M.A., Péret B., Riiser E.S., Bennett M.J., Aalen R.B. (2013). Floral organ abscission peptide IDA and its HAE/HSL2 receptors control cell separation during lateral root emergence. Proc. Natl. Acad. Sci. USA 110: 5235–5240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C., Dixon R.A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48: 251–275 [DOI] [PubMed] [Google Scholar]
- Lease K.A., Walker J.C. (2006). The Arabidopsis unannotated secreted peptide database, a resource for plant peptidomics. Plant Physiol. 142: 831–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.S., Torii K.U. (2012). A tale of two systems: peptide ligand-receptor pairs in plant development. Cold Spring Harb. Symp. Quant. Biol. 77: 83–89 [DOI] [PubMed] [Google Scholar]
- Lehti-Shiu M.D., Zou C., Hanada K., Shiu S.-H. (2009). Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 150: 12–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Tax F.E. (2013). Receptor-like kinases: key regulators of plant development and defense. J. Integr. Plant Biol. 55: 1184–1187 [DOI] [PubMed] [Google Scholar]
- Matsubayashi Y., Sakagami Y. (1996). Phytosulfokine, sulfated peptides that induce the proliferation of single mesophyll cells of Asparagus officinalis L. Proc. Natl. Acad. Sci. USA 93: 7623–7627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsubayashi Y., Takagi L., Omura N., Morita A., Sakagami Y. (1999). The endogenous sulfated pentapeptide phytosulfokine-alpha stimulates tracheary element differentiation of isolated mesophyll cells of zinnia. Plant Physiol. 120: 1043–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meindl T., Boller T., Felix G. (2000). The bacterial elicitor flagellin activates its receptor in tomato cells according to the address-message concept. Plant Cell 12: 1783–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller K., Bittel P., Chinchilla D., Jehle A.K., Albert M., Boller T., Felix G. (2012). Chimeric FLS2 receptors reveal the basis for differential flagellin perception in Arabidopsis and tomato. Plant Cell 24: 2213–2224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller R., Bleckmann A., Simon R. (2008). The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy E., Smith S., De Smet I. (2012). Small signaling peptides in Arabidopsis development: how cells communicate over a short distance. Plant Cell 24: 3198–3217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niederhuth C.E., Patharkar O.R., Walker J.C. (2013). Transcriptional profiling of the Arabidopsis abscission mutant hae hsl2 by RNA-Seq. BMC Genomics 14: 37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nolen B., Taylor S., Ghosh G. (2004). Regulation of protein kinases; controlling activity through activation segment conformation. Mol. Cell 15: 661–675 [DOI] [PubMed] [Google Scholar]
- Ohyama K., Ogawa M., Matsubayashi Y. (2008). Identification of a biologically active, small, secreted peptide in Arabidopsis by in silico gene screening, followed by LC-MS-based structure analysis. Plant J. 55: 152–160 [DOI] [PubMed] [Google Scholar]
- Ohyama K., Shinohara H., Ogawa-Ohnishi M., Matsubayashi Y. (2009). A glycopeptide regulating stem cell fate in Arabidopsis thaliana. Nat. Chem. Biol. 5: 578–580 [DOI] [PubMed] [Google Scholar]
- Pearce G., Moura D.S., Stratmann J., Ryan C.A., Jr (2001). RALF, a 5-kDa ubiquitous polypeptide in plants, arrests root growth and development. Proc. Natl. Acad. Sci. USA 98: 12843–12847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pearce G., Strydom D., Johnson S., Ryan C.A. (1991). A polypeptide from tomato leaves induces wound-inducible proteinase inhibitor proteins. Science 253: 895–897 [DOI] [PubMed] [Google Scholar]
- Rodríguez A.A., Grunberg K.A., Taleisnik E.L. (2002). Reactive oxygen species in the elongation zone of maize leaves are necessary for leaf extension. Plant Physiol. 129: 1627–1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi C.-L., Stenvik G.-E., Vie A.K., Bones A.M., Pautot V., Proveniers M., Aalen R.B., Butenko M.A. (2011). Arabidopsis class I KNOTTED-like homeobox proteins act downstream in the IDA-HAE/HSL2 floral abscission signaling pathway. Plant Cell 23: 2553–2567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinohara H., Matsubayashi Y. (2013). Chemical synthesis of Arabidopsis CLV3 glycopeptide reveals the impact of hydroxyproline arabinosylation on peptide conformation and activity. Plant Cell Physiol. 54: 369–374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiu S.H., Bleecker A.B. (2001). Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc. Natl. Acad. Sci. USA 98: 10763–10768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik G.E., Butenko M.A., Urbanowicz B.R., Rose J.K., Aalen R.B. (2006). Overexpression of INFLORESCENCE DEFICIENT IN ABSCISSION activates cell separation in vestigial abscission zones in Arabidopsis. Plant Cell 18: 1467–1476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenvik G.E., Tandstad N.M., Guo Y., Shi C.L., Kristiansen W., Holmgren A., Clark S.E., Aalen R.B., Butenko M.A. (2008). The EPIP peptide of INFLORESCENCE DEFICIENT IN ABSCISSION is sufficient to induce abscission in arabidopsis through the receptor-like kinases HAESA and HAESA-LIKE2. Plant Cell 20: 1805–1817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uebler S., Dresselhaus T. (2014). Identifying plant cell-surface receptors: combining ‘classical’ techniques with novel methods. Biochem. Soc. Trans. 42: 395–400 [DOI] [PubMed] [Google Scholar]
- Wolf S., Höfte H. (2014). Growth control: A saga of cell walls, ROS, and peptide receptors. Plant Cell 26: 10.1105/tpc.114.125518. [DOI] [PMC free article] [PubMed] [Google Scholar]