About 60% of the nucleotide diversity was lost during domestication of the common bean. The whole pattern of gene expression has also been affected, with changes in the patterns of coexpression among genes and 18% reduction in the overall diversity of gene expression. About 9% of the genes were selected during domestication, which is associated with further reduced diversity of expression.
Abstract
Using RNA sequencing technology and de novo transcriptome assembly, we compared representative sets of wild and domesticated accessions of common bean (Phaseolus vulgaris) from Mesoamerica. RNA was extracted at the first true-leaf stage, and de novo assembly was used to develop a reference transcriptome; the final data set consists of ∼190,000 single nucleotide polymorphisms from 27,243 contigs in expressed genomic regions. A drastic reduction in nucleotide diversity (∼60%) is evident for the domesticated form, compared with the wild form, and almost 50% of the contigs that are polymorphic were brought to fixation by domestication. In parallel, the effects of domestication decreased the diversity of gene expression (18%). While the coexpression networks for the wild and domesticated accessions demonstrate similar seminal network properties, they show distinct community structures that are enriched for different molecular functions. After simulating the demographic dynamics during domestication, we found that 9% of the genes were actively selected during domestication. We also show that selection induced a further reduction in the diversity of gene expression (26%) and was associated with 5-fold enrichment of differentially expressed genes. While there is substantial evidence of positive selection associated with domestication, in a few cases, this selection has increased the nucleotide diversity in the domesticated pool at target loci associated with abiotic stress responses, flowering time, and morphology.
INTRODUCTION
Plant domestication has long stimulated scientific interest. As stated by Charles Darwin, domestication can be considered a giant evolutionary experiment (Darwin, 1875), while from a plant-breeding perspective, understanding domestication is key to the development of breeding strategies and the identification of useful genetic variants.
Selection related to domestication has modified a number of traits that now distinguish the modern crops from their wild forms. These common features of many crop species contribute collectively to the “domestication syndrome” (Gepts and Papa, 2002), and they include the size, shape, and color of the plant organs used by humans (e.g., of seeds, fruit, and leaves) and seed dispersal (e.g., shattering, dormancy). Indeed, while increased seed and fruit size has been the most impressive change from the wild to the domesticated forms, the loss of seed dispersal mechanisms represents a major factor that has reduced the fitness of domesticated plants in the wild environment and has thus prevented these plants from reproducing outside the agro-ecosystem.
The population genetics model of domestication predicts a reduction in diversity and increased divergence between wild and domesticated populations due to demographic factors affecting the whole genome and selection at target loci (Glémin and Bataillon, 2009). Allogamous species, such as maize (Zea mays), are generally characterized by a lower genetic bottleneck effect compared with autogamous species like the common bean (Phaseolus vulgaris) (Bitocchi et al., 2013). In particular, resequencing data have confirmed that in autogamous species, such as soybean (Glycine max) and rice (Oryza sativa ssp japonica) (Lam et al., 2010; Xu et al., 2012), a reduction in diversity has arisen as an effect of domestication, as also reported for the silkworm (Bombyx) and for mammalian species (Xia et al., 2009; Vonholdt et al., 2010; Lippold et al., 2011).
Signatures of selection during domestication have been reported for 2 to 4% of genes expressed in maize (Wright et al., 2005) and for 7.6% of the maize genome (Hufford et al., 2012). This finding suggests a prominent role for the combined effects of selection, drift, and reduction of effective recombination at loci linked to the selection targets. A strong hitchhiking effect (Smith and Haigh, 1974) has also been suggested for rice (Lu et al., 2006) and common bean (Papa et al., 2007), which supports the concept that domestication has had larger effects compared with those that can be explained solely by effects of selection.
Techniques such as next-generation sequencing offer a unique opportunity to scan the genome not only to obtain genotypic information, but also to analyze the molecular phenotype of the whole genome through the analysis of the transcriptome, the metabolome, and the proteome. Recent studies have reported major changes in the maize transcriptome expression, but without any reduction in the expression diversity of genes (Hufford et al., 2012; Swanson-Wagner et al., 2012). There is a need to extend these studies to other crop species to better establish the genome-wide consequences of domestication.
Here, we focused on the domestication process of the common bean in Mesoamerica, with the main aims of (1) describing the genome-wide molecular changes due to domestication using RNA sequencing (RNA-seq) technology and (2) identifying the molecular variants that are responsible for the phenotypic variations that constitute the basis of the domestication process within the common bean genome.
For P. vulgaris (2n=2x=22), at least two domestication events have occurred, in Mesoamerica and in the Andes (reviewed in Bitocchi et al., 2013). The two parallel domestications and the domestication of an additional four closely related Phaseolus species render the common bean a unique system in which to study domestication and crop evolution.
RESULTS
Transcriptome Sequencing and Assembly
To capture most of the allelic diversity observed for molecular markers in a cohort made up of 10 Mesoamerican wild (MW) genotypes and eight Mesoamerican domesticated (MD) genotypes, with one wild and two domesticated Andean genotypes as controls, we choose to use an approach based on de novo assembly of transcriptome from RNA-seq data. To maximize the information content provided by the data set, a reference transcriptome was built from a hypercore collection of the four most divergent wild genotypes in our cohort (three from Mesoamerica and one from the Andes). This approach was preferred over a reference-based/hybrid method as, given the well-established genetic divergence of the Andean and Mesoamerican gene pool, the use of the P. vulgaris reference genome derived from an Andean genotype (G19833) might lead to a loss of informative markers.
To minimize expression differences due to sampling errors, RNAs were extracted from the first trifoliate leaf, fully expanded and at stationary phase. On average, 38 × 106 paired-end reads (100 bp ×2) per sample were generated (Supplemental Table 1).
The transcriptome of each of the four members of the hypercore collection was assembled de novo using Trinity (Grabherr et al., 2011). Overall, each sample yielded from 55,069 to 70,826 clusters of contigs, as defined by the Chrysalis module of Trinity, with each cluster ideally representing a single gene. The longest contig out of each cluster was chosen as representative sequence, and redundancies among the four genotypes were collapsed with CD-HIT-EST (Li and Godzik, 2006; Supplemental Table 1). The set of resulting 124,166 sequences, comprising genes shared across all four members of the hypercore collection and genotype-specific genes, was thus used as the reference transcriptome for all subsequent analyses.
Comparing the sequences of each genotype with the reference transcriptome, we identified 284,812 high-quality homozygous single nucleotide polymorphisms (SNPs) on 43,789 contigs (see Methods). Contigs with only heterozygous SNPs or indels were not further considered. Excluding positions missing in more than three Mesoamerican genotypes (see Methods) and filtering for homozygous biallelic SNPs only, the final data set was further reduced to 188,107 SNPs on 27,243 contigs. When considering only the Mesoamerican accessions, the polymorphic contigs decreased to 26,141. Twenty-five of these contigs were fixed for alternative allelic states in the MW and MD populations (Table 1).
Table 1. Number of SNPs and Contigs Identified in This Study.
| Total number of biallelic SNPs | 188,107 |
| Total number of contigs | 27,243 |
| Total number of monomorphic contigs in Mesoamerican sample | 1,102 |
| Total number of polymorphic contigs in Mesoamerican sample | 26,141 |
| Number of contigs monomorphic in both MW and MD populations, except for alternative alleles | 25 |
| Shared polymorphic contigs between MW and MD | 13,411 |
| Number of contigs monomorphic in MD, but polymorphic in MW | 12,014 |
| Number of contigs monomorphic in MW, but polymorphic in MD | 691 |
Population Structure, and Diversity and Expression Analysis
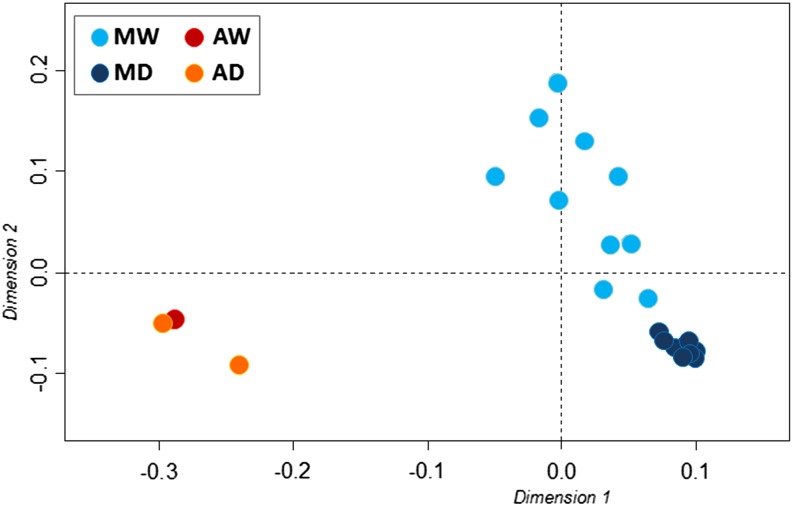
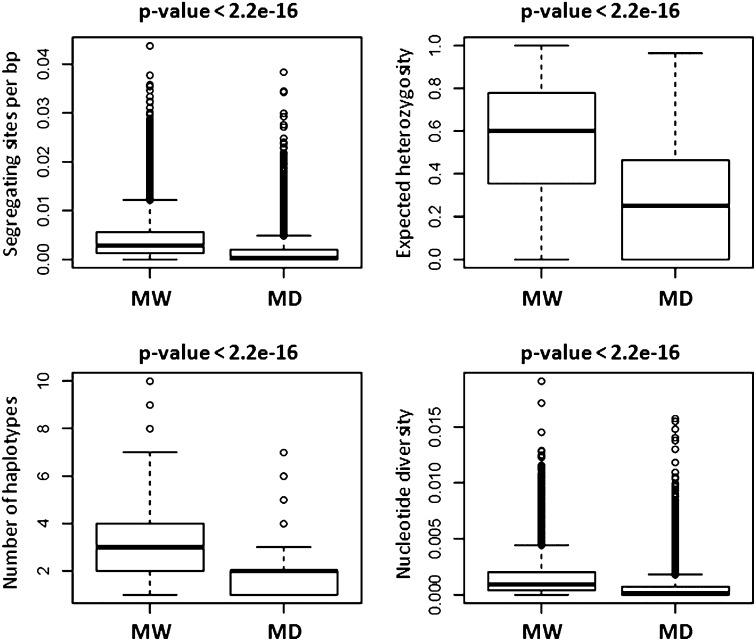
Multidimensional scaling analysis (Figure 1) reproduced the known genetic structure of the common bean populations: The Mesoamerican and Andean pools were separated, as were the MW and MD forms. The analysis also revealed that compared with MD, MW is characterized by a higher diversity. This agrees with all of the estimated statistics (e.g., S, nH, π, θ, and He) (Table 2; Supplemental Figure 1), with ∼60% loss of diversity in the MD population (Table 2). Moreover, almost half of the contigs that were polymorphic in Mesoamerica (46%) were monomorphic in MD (Table 1; Supplemental Figure 1). The difference between the genetic variation within MW and MD was highly significant for all of the indices (Wilcoxon test, P ≤ 2.2 ×10−16; Figure 2).
Figure 1.
Multidimensional Scaling Analysis Representing the Genetic Relationships among the 21 Common Bean Genotypes.
Table 2. Diversity Estimates in the MW and MD Accessions Computed Considering the Polymorphic Contigs in the Entire Mesoamerican Sample (26,141 Contigs) and Loss of Nucleotide Diversity Estimates.
| Diversity Estimate | MW | MD | |
|---|---|---|---|
| N | 10 | 8 | |
| S | 153,971 | 56,053 | |
| S1 | 5.9 | 2.1 | |
| He | 0.57 | 0.25 | |
| nH | 3.5 | 1.8 | |
| π | 2.11 | 0.85 | |
| θ | 2.08 | 0.83 | |
| ΔH | 0.56 | ||
| ΔH1 | 0.56 | ||
| Lπ | 0.60 | ||
| Lπ1 | 0.58 | ||
| Lθ | 0.60 | ||
| Lθ1 | 0.58 | ||
| ϕST | 0.15 |
N, sample size; S, total number of segregating sites; S1, mean number of segregating sites per contig; He, average expected heterozygosity (Nei, 1978); nH, mean number of haplotypes per contig; π, θ, averaged estimates of nucleotide diversity (Tajima, 1983; Watterson, 1975; respectively); ΔH, Lπ, and Lθ, loss of nucleotide diversity, from averaged He, π, and θ, for (MW – MD) comparison; ΔH1, Lπ1, and Lθ1, loss of nucleotide diversity, from averaged ΔH, Lπ, and Lθ of each contig, for (MW – MD) comparison; averaged ϕST.
Figure 2.
Within-Population Genetic Diversity Comparison between MW and MD Populations.
Box plots of the number of segregating sites (per base pair), the expected heterozygosity, the number of haplotypes, and the nucleotide diversity in the MW versus the MD population, evaluated over all of the contigs. The statistical significance was computed with the Wilcoxon signed rank test for paired data (P value: above each box plot).
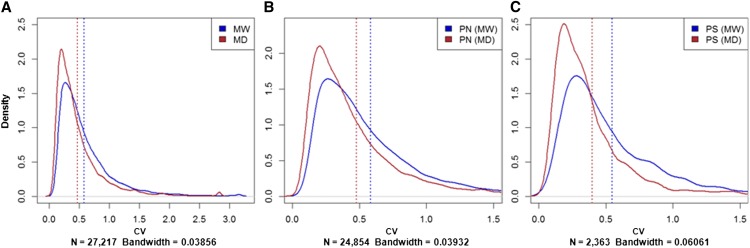
We tested the differential expression of MW versus MD considering the different individual genotypes within groups as replicates. Out of 27,243 contigs, 198 (0.7%) were differentially expressed when comparing MW and MD (Supplemental Data Set 1A), and 146 of them (74%) were downregulated in MD. Moreover, the log2 fold change in the level of transcription shifted significantly toward negative values (mean, −0.09; median, −0.02; skewness of distribution, −3.49), which indicates an abundance of downregulation in MD, with the mean log2 fold change significantly smaller than 0 (Wilcoxon two-sided test: P ≤ 2e−16). The coefficient of variation (CV) of gene expression was higher in MW (0.57) than in MD (0.47), with an 18% loss of expression diversity (Figure 3A, Table 3).
Figure 3.
Estimated Density Functions for the Coefficients of Variation in MD and MW as a Reference.
Comparison of the density functions of the CVs considering: all contigs (n = 27,217 CVs of finite value in MW; 27,114 CVs of finite value in MD) (A), subdivision in PN contigs (n = 24,854 CVs of finite value in MW; 24,759 CVs of finite value in MD) (B), and PS contigs (n = 2363 CVs of finite value in MW; 2355 CVs of finite value in MD) (C), where N denotes the number of contigs with CVs of finite values in MW as a reference.
Table 3. Coefficients of Variation of P. vulgaris Expression for the MW and MD Forms.
LCV, loss of expression diversity, calculated as LCV = 1 − (CVMD/CVMW); L’CV, loss of expression diversity, calculated as the mean of the single contig LCV.
Gene Coexpression Networks
A total of 10,616 contigs were selected for the network-based analysis (Supplemental Figure 2 and Supplemental Methods 1). The selection avoided potentially noisy or invariant gene expression profiles, which would lead to the inclusion of spurious edges in the extracted networks. The introduction of systematic bias due to this selection strategy was examined, and no bias associated with the contig variations in gene expression was found (Supplemental Figure 3).
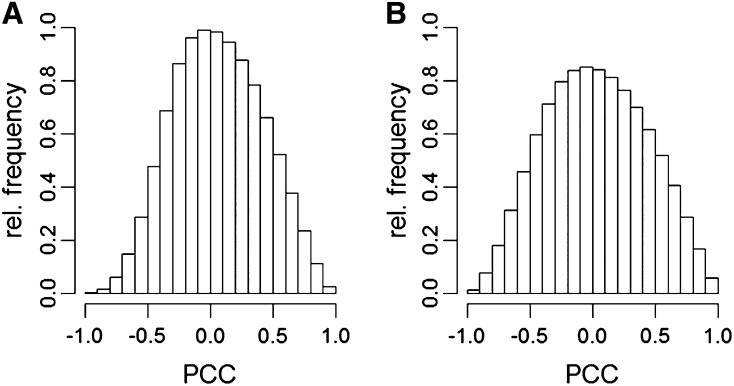
The correlations among gene expression profiles were based on the Pearson correlation coefficients (PCCs), which were similar for MW and MD, with both resembling normal distributions. In MD, the distribution was wider than in MW, with variance of 0.16 and 0.12, respectively (one-sided F-test, P < 2.2e−16) (Figure 4). This implied that there was a higher number of stronger correlations in MD than in MW.
Figure 4.
Distributions of the Pearson Correlation Coefficients Obtained from the Expression Profiles in the MW and MD Populations.
Distributions of the MW (A) and MD (B) populations resemble normal distributions, with the PCCs in MD showing greater values than those in MW.
Coexpression of the 10,616 contigs in MW and MD was also considered using network analysis: extraction of proximity networks and generation of relevance networks. These two networks gave very similar results (Supplemental Methods 1); here, we describe in detail only those from the proximity networks.
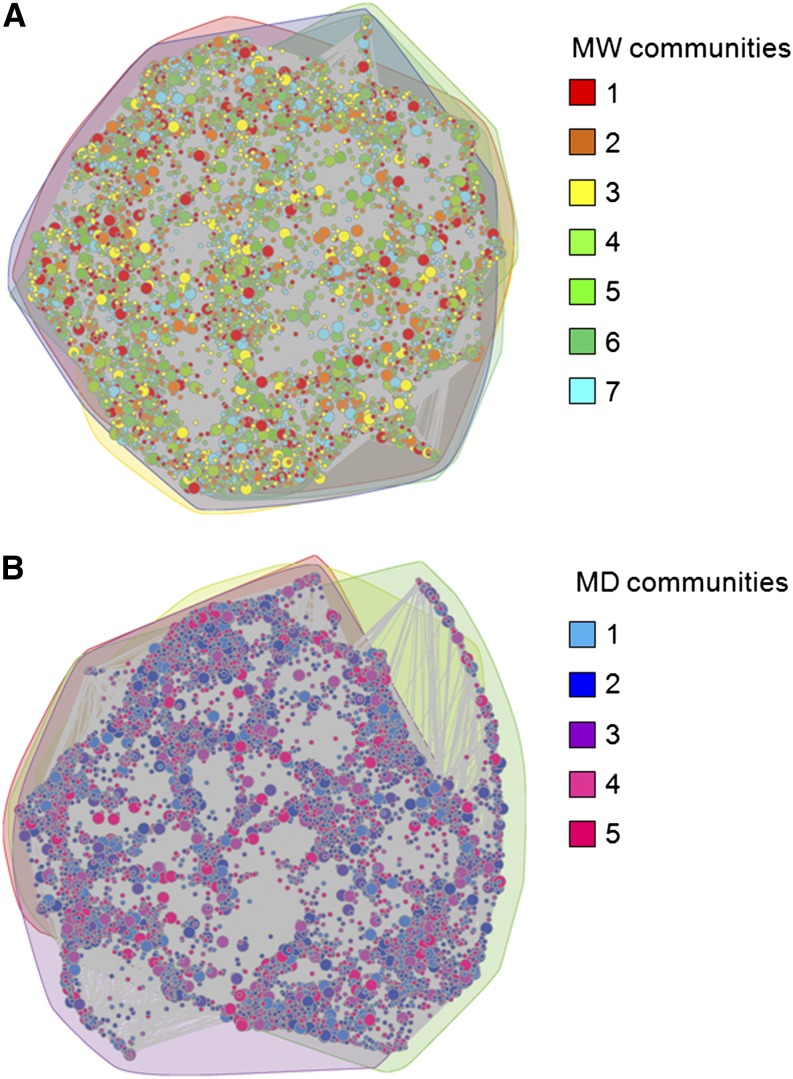
The seminal properties (Newman, 2003, 2012) of the proximity networks appeared similar in MW and MD. Indeed, only slight differences were observed for the density (0.0012 and 0.0013) and transitivity (0.12 and 0.14) of the MW and MD networks (respectively). The MW network contained seven communities that correspond to groups of genes with mutually correlated expression (Figure 5A), while the MD network contains five communities (Figure 5B). Comparing the MW and MD networks using an adjusted Rand index, it appeared that although they had relatively similar properties, their community structures were divergent. This was supported by the consideration of the Jaccard similarity coefficient (Supplemental Table 2).
Figure 5.
Proximity Networks of MW and MD and Their Community Structure.
Nodes are color-coded according to their participation in one of the seven and five communities in the MW (A) and MD (B) proximity networks, respectively, containing a single connected component. Nodes of bigger size correspond to contigs under selective pressure.
The enrichment gene function analysis (α = 0.01) indicated that eight of the 12 communities in the MW and MD networks were enriched for at least one gene function, with a mean of four and a maximum of seven gene functions over the communities (Supplemental Table 3 and Supplemental Data Sets 1B and 1C). Aside from having pronounced structural differences, these communities were also modular structures of largely different functions. For example, while there were communities in the MW and MD networks that were enriched for RNA regulation of transcription, the first and second MD communities were enriched in specific transcription factor families that are involved in floral development (e.g., MADS box), abiotic stress responses (e.g., b-ZIP), and several biological functions related to domestication (Supplemental Methods 2).
We next determined the intersection network, with the expectation that the two networks would share only a few edges (otherwise, their community structures would have shown greater similarities); indeed, these edges were incident on only 857 nodes.
Selection
A total of 2364 contigs (9% of those polymorphic) were identified as putatively under selection (PS) during domestication. This was revealed by simulation of the evolutionary dynamics of Mesoamerican and Andean wild beans with the assumption of absence of selection during domestication, considering the demographic details available from previous studies (Mamidi et al., 2011, 2013). These simulations reconstructed the distribution across the genome of summary statistics that describe processes of differential selection. The comparison between these distributions with those of real contigs (controlling for false positives) identified those contigs that were most likely affected by selection during domestication (directly, or due to hitchhiking). Most of the PS contigs (82%) were fixed in MD and polymorphic in MW, with 14.2% showing shared polymorphism between MD and MW. A small fraction (2.8%) was fixed in MW and polymorphic in MD. Finally, ∼1% was fixed both in MD and MW for alternative allelic states.
Contigs differentially expressed in MW compared with MD were highly enriched (about 5-fold) in PS compared with the putatively neutral (PN) contigs (2.75% versus 0.53%; Table 4), suggesting that selection was active for the expression pattern at already the first true leaf stage. In parallel, the loss of expression diversity due to domestication appeared significantly higher (P < 0.0001; χ2 test) for PS (26%; Table 3, Figure 3C) than PN (17%; Table 3, Figure 3B); this effect may be the outcome of direct selection or hitchhiking in regulatory regions (within or outside the exome).
Table 4. Enrichment of Differentially Expressed Contigs in the PS Contig Group.
The gene set enrichment analysis (GSEA) is presented in Supplemental Data Set 1D. Single enriched MapMan bins are not common to PS and PN sets, which further indicates that the two sets have the tendency to participate in different metabolic pathways or have different functions. Briefly, when ordered according to 5% statistical significance of the GSEA, the genes that have been annotated in Arabidopsis thaliana as involved in regulation of RNA transcription, synthesis of ribosomal proteins, RNA processing and regulation, and DNA repair are overrepresented in the PS contigs. By contrast, PN had a lower number of significantly overrepresented MapMan bins, which encoded proteins involved in, among other things, cofactor and vitamin metabolism and nucleotide metabolism.
We observed that there was no shift of the CV toward higher/lower values for the PS contigs retained and those not included in the network analysis (Supplemental Figure 4). With respect to the position in the proximity networks, the PS contigs were underrepresented in the intersection network. We next tested if there was difference in the average centrality measures of PS and PN contigs, assessing their global position in the networks. We did not identify any statistically significant differences with respect to the average centrality measures of PS and PN contigs in both the MW and MD networks; however, a small, but statistically significant, difference was seen for the closeness centrality (Supplemental Table 4). At a local level, i.e., by focusing only on the immediate coexpressing partners of the PS contigs, we also found that the PS contigs showed significant, although small, assortativity in the MD network, which was not the case in the MW network (Supplemental Table 5). Qualitatively similar findings were obtained when the selection index was used instead of the partition of the contigs into the PS and PN classes. These data indicate that while the global position of the PS contigs in the coexpression network on average did not show differences with respect to the PN contigs, there were small local changes of coexpression patterns, as quantified by the assortativity, that might lead to tighter coexpression of the PS contigs in MD compared with MW. Associations between PS contigs and features of the expression network, like centrality indices and assortativity, were largely not significant. This might be due to our experimental system that is based on a specific developmental stage of the plant, which supports the view that only a fraction of the genes under selection had phenotypic effects associated to a differential fitness at this stage. Moreover, PS genes might be indirectly affected by selection due to hitchhiking. In addition, if domestication is considered a multitrait selection process, we have no reason to assume specific and common roles for all of the selected genes in the determination of the structure of the expression network.
Furthermore, function information allowed the investigation of whether a subset of contigs identified as PS is associated to the domestication process in other species. We focused on the 380 contigs with the highest selection index, including also the 23 PS contigs with an alternative allelic state between MW and MD as well as the 67 PS contigs monomorphic in MW and polymorphic in MD. Functional evaluation of these PS contigs is comprehensively discussed in Supplemental Methods 2.
The analysis revealed that several PS contigs are homologous to genes implicated in the process of domestication in other species or have functions associated with domestication, like light responses, signaling, plant development, and biotic and abiotic stress. For instance, the annotated PS contigs that showed greater genetic diversity in MW compared with MD included a sequence homolog to GIGANTEA (GI), which has a pivotal role in the photoperiodic response, as it regulates flowering in a circadian clock–controlled manner. In Arabidopsis, under long days, GI acts earlier in the pathway than CONSTANS (CO) and FLOWERING TIME (FT), by increasing the CO and FT mRNA abundance. CO and FT were targets of selection during domestication of crops such as rice (Takahashi and Shimamoto, 2011; Wu et al., 2013) and sunflower (Helianthus annuus; Blackman et al., 2011). In pea (Pisum sativum), Hecht et al. (2007) identified LATE BLOOMER1 (LATE1) as the pea ortholog of Arabidopsis GI and showed that LATE1 is necessary for the promotion of flowering, the production of a mobile flowering stimulus, and the induction of an FT homolog under long-day conditions. Another interesting example among the PS contigs with two alternative allelic states is the homolog of YABBY5 (YAB5), a transcription factor that is implicated in the regulation of seed shattering in cereal species, including sorghum (Sorghum bicolor), rice, and maize (Lin et al., 2012). A YAB-like transcription factor (FASCIATED) has also been shown to influence carpel number during flower and/or fruit development in tomato (Solanum lycopersicum; Cong et al., 2008).
Among the 67 PS contigs that show an increase in variability in MD is a homolog of K+ uptake transporter6 (KUP6). Osakabe et al. (2013) demonstrated that the KUP potassium transporter family has important roles in water stress responses and growth; moreover, KUP-type K+ transporters are induced by various stresses that have an osmotic component, and they specifically inhibit cell expansion, while enhancing drought tolerance.
DISCUSSION
This report describes the profound effect that domestication has imposed on the genome variation and gene expression patterns of common bean. About one out of 10 contigs was likely to have been affected by selection during domestication: Directional selection was the rule, but diversifying selection was also probably active, with contigs of the domesticated gene pool frequently having different levels of expression and different patterns of coexpression compared with the wild relatives. The practical implication for future crop improvement is that lot of variation at DNA sequences and regulatory regions is still available in the wild bean for crop breeding, but that to fully exploit the diversity of wild germplasm a substantial effort is needed to understand the complex relationship between the genotypic and phenotypic diversity in plant populations.
As highlighted in this study, in common bean, expressed genomic regions lost half of the wild bean nucleotide diversity during domestication in Mesoamerica. Compared with common bean, in maize, there was a smaller reduction in diversity at the nucleotide level (Hufford et al., 2012), which suggests that there was a smaller effect of domestication on the maize genetic diversity. The different mating systems between these two crops might help to explain these results. In autogamous species like common bean, self-fertilization is expected to reduce the effective population size, which will enhance the effects of genetic drift and increase the extent of linkage disequilibrium, leading to large genomic windows affected by genetic sweep (Glémin and Bataillon, 2009; Bitocchi et al., 2013) as also confirmed by the resequencing results in the autogamous soybean and rice (O. sativa, variety japonica) (Lam et al., 2010; Xu et al., 2012).
Our study also demonstrated that there was a drastic change because of domestication in the pattern and structure of gene expression over the entire set of genes. This was also found in maize, albeit with reduced intensity (Swanson-Wagner et al., 2012). Moreover, in common bean, we found that the reduction in sequence diversity also affects DNA regions implicated in the regulation of transcription, where ∼20% reduction in gene expression levels has been associated with domestication. In other words, here, we demonstrate that the loss of genetic variation has direct genome-wide phenotypic consequences on transcriptome diversity. These findings differ from the case of maize and its wild progenitor teosinte, where no reduction in the variation of gene expression was observed (Swanson-Wagner et al., 2012). It is particularly relevant that such different expression levels and patterns are observed at a developmental stage that is considered relatively important for domestication, even if the presence of larger leaves and seedlings is a hallmark trait of domestication (Gepts, 2002).
The occurrence in domesticated bean of mostly downregulated transcripts among those differentially expressed (74%) points to loss-of-function mutations, which are relatively frequent compared with gain-of-function changes, as a largely available source of variation that supports selection during rapid environmental changes (Olson, 1999). Such was the case of the transition from the wild to cultivated agro-ecosystems. In support of this, as first noted by Darwin (1859), in domesticated plants, the domestication traits have a recessive genetic nature (Lester, 1989). Moreover, a lower genome-wide gene expression level was found for domesticated compared with wild transcripts as if slightly deleterious mutations due to hitchhiking (mostly loss-of-function or with reduced expression) have been accumulated in the domesticated pool. This can be considered as the “cost of domestication.” The accumulation of loss-of-function (or reduced expression) mutations might also have been due to reduced effective recombination, which would have increased the frequency of deleterious mutations in the domesticated pool, with a negative influence on fitness, as suggested in rice (Lu et al., 2006).
About 10% of the contigs were affected by selection during domestication or were physically linked to the selected genes. This supports again the view that domestication had a relevant influence on the common bean genome. In the allogamous species maize, ∼2 to 4% of genes and ∼7.6% of the whole genome (Wright et al., 2005; Hufford et al., 2012, respectively) were detected as affected by selection during domestication. Similarly, in sunflower, which is also predominantly allogamous, ∼7.3% of genes show signatures of selection due to domestication (Chapman et al., 2008). These differences may be determined by a more relevant role of genetic hitchhiking in producing the observed results in P. vulgaris due to its autogamous mating system.
Most of the contigs affected by selection during domestication show reduced diversity in MD compared with MW, as would be expected following positive selection due to domestication. However, in a few cases, the opposite was observed: For instance, for 2.8% of the PS contigs, there was no diversity in MW, while there was diversity in MD. This can be taken as being due to diversifying selection in MD, with domestication increasing the level of functional diversity. The functional analysis of the drought-related KUP6 gene shows that it is significantly overexpressed in MD compared with MW, as if domestication has also increased the functional diversity of selected genes and not just increased the nucleotide diversity. Our data therefore indicate that in parallel with an overall reduction in diversity, domestication increased the functional diversity at target loci. This can be imputed to novel mutations (or those that exist at low frequencies) that were selected because of the crop expansion into new environments with unexpected biotic and abiotic stress or because of selection for traits that improved the use of the plant organs by humans (de Alencar Figueiredo et al., 2008). As such, the data contribute to resolving the Darwin paradox (Darwin, 1878; Glémin and Bataillon, 2009): Domestication is associated with an increased phenotypic diversity at target traits and a reduction of nucleotide variation.
Our work presents relevant implications for the development of prebreeding strategies. Similarly to other studies, our findings support the need for wild germplasm for further crop improvements and calls for careful conservation of the wild populations. However, we also showed that the effect of domestication is pervasive throughout the genome in terms of expression patterns and diversity, probably because of the combination of linkage and pleiotropy. However, complex interactions within and among genes and their expression levels played an important role during the domestication of this species, suggesting that further genetic amelioration strongly requires new tools for genomics, molecular phenotyping, and phenomics. Moreover, our results suggest that the diversity in the domesticated pool (e.g., traditional landraces) that was originated by the fixation of useful mutations after domestication needs increased consideration as source of novel genetic variation for crop improvement.
METHODS
Sampling
On the basis of the molecular characterization of a wide and representative collection of Phaseolus vulgaris genotypes (Rossi et al., 2009; Nanni et al., 2011; Bitocchi et al., 2012, 2013; Desiderio et al., 2013) and with a focus on the Mesoamerican gene pool, 21 inbred genotypes (two cycles of single seed descent) were selected as the core collection to maximize the genetic diversity. The core included 10 MW genotypes, eight MD genotypes, and as controls, two domesticated and one wild Andean genotypes. With the aim also being to capture most of the allelic diversity observed for molecular markers, a further hypercore collection of four wild genotypes was built (three from Mesoamerica and one from the Andes). A complete list of the accessions used is reported in Supplemental Table 6.
The 21 individual genotypes were grown under greenhouse-controlled conditions (relative humidity, ∼70%; average night/day temperature, 25°C). To minimize expression differences that might be attributed to developmental disparity between individuals, the fully expanded first trifoliate leaf at stationary phase was collected and frozen for all genotypes.
RNA Extraction
Frozen plant tissues were ground in liquid nitrogen, and 100 mg ground tissue was used for RNA isolation using Spectrum Total RNA kits (Sigma-Aldrich). The RNA was then treated with RNase-Free DNase using the On-Column DNase I Digestion Set (Sigma-Aldrich). Qualitative and quantitative control was performed with a Nanodrop 2000 spectrophotometer (Thermo Scientific) and an RNA 7500 series II chip bioanalyzer (Agilent). Only RNA samples with an RNA integrity number >8.0 were used.
Library Preparation and Sequencing
For each of the 21 RNA samples, 3 µg was used for the construction of a nondirectional Illumina RNA-seq library, using TruSeq RNA sample preparation kits, v2 (Illumina), following the manufacturer’s instructions. Libraries were quantified using quantitative PCR, and quality control was performed with the DNA 1000 series II chip bioanalyzer (Agilent).
RNA-seq was performed with an Illumina HiSequation 1000 Sequencer using TruSeq SBS v3-HS kits (200 cycles) and TruSeq PE Cluster v3-cBot-HS kits (Illumina) generating 100-bp paired-end reads.
De Novo Transcriptome Assembly
Reads obtained from the sequencing of the four hypercore collection genotypes (Supplemental Table 1) were assembled de novo to obtain a common reference transcriptome. Each sample was assembled separately using Trinity version R2011-11-2 (Grabherr et al., 2011) using default parameters. To minimize the redundancy due to different transcript isoforms belonging to the same gene, a custom script was used to retain only the longest contig out of each trinity cluster as a representative of the cluster. The filtered contigs from the four assemblies were pooled together and redundancy among data sets was removed using CD-HIT-EST (Li and Godzik, 2006), with a 90% threshold on the contig identity. The contigs were compared with the sequences in the TAIR 10 protein database of Arabidopsis thaliana using BLASTX (Altschul et al., 1997), with an E-value <10E−2.
Variant Identification
RNA-seq reads of each of the 21 genotypes were mapped on the reference transcriptome using BWA version 0.6.2-r126 (Li and Durbin, 2009) using default parameters and with a minimum mapping quality threshold of q = 30 to minimize false variant calls due to misalignments or reads mapping to multiple positions in the transcriptome. The variants in the transcriptome sequence were identified using Samtools 0.1.18 (Li et al., 2009) and Varscan v2.2.8 (Koboldt et al., 2012), with a maximum P value of 0.01 and a minimum read depth of three reads in order not to penalize transcripts that are present in low abundance in the samples. Only positions of the transcriptome covered by at least three reads in all the 18 Mesoamerican genotypes analyzed were considered for variant calling. For all of the positions for which a homozygous SNP (percentage of reads supporting the alternate allele ≥75%) was called in at least one sample, we analyzed the samples with no SNP call in the same position using the following criteria: (1) if read depth ≥3 and percentage of reads supporting the reference base >75%, the reference base was called; (2) if read depth ≥3 and percentage of reads supporting an alternate allele already called in other samples (P value <0.01) for that position >75%, the alternate base was called; (3) if read depth ≥3 and percentage of reads supporting an alternate allele already called in the other samples (P value < 0.01) was between 25 and 75%, a heterozygous call was recorded. The positions for which a genotype was not detected in at least 15 of the Mesoamerican genotypes were removed, allowing a maximum of three missing data for each called SNP. Only biallelic homozygous SNPs were retained for the analyses.
RNA-seq Expression Analysis
Gene expression levels were based on TopHat2 (Kim et al., 2013) and HTSeq (Anders, 2010), with the default parameters. Differentially expressed contigs between the wild and domesticated genotypes (|logFC| >1; FDR <5%) were identified using DESeq version 1.6.1 (Anders and Huber, 2010). The experimental design contrasted two groups of accessions, as MW versus MD, using individual genotypes as replicates. The CV was calculated as the ratio between the sd and the mean number of fragments mapping on each contig, for each genotype, in both the MW and MD populations.
Population Genetics Analysis
Exploratory analysis of the genetic relationships among individuals was based on a metric multidimensional scaling, using the cmdscale command in the R statistical environment (http://www.r-project.org/; R Development Core Team, 2013). Genetic distances were computed as one minus the average fraction of nonshared alleles.
The following diversity statistics were computed using Arlequin 3.5 (Excoffier and Lischer, 2010): S, total and mean number of segregating sites; expected heterozygosity (He; Nei, 1978); nH, number of haplotypes; and π and ⊝, two measures of nucleotide diversity from Tajima (1983) and Watterson (1975), respectively, computed on SNPs within each contig. The divergence between the MW and MD forms was measured using ΦST (Excoffier et al., 1992).
To assess the reduction of diversity in MD versus MW, we used the statistical loss of diversity, as proposed by Vigouroux et al. (2002), and computed as [1 − (xMD/xMW)], where xMD and xMW are the diversities in the MD and MW populations, respectively, measured using three different statistics: He, π, and ⊝; the loss of diversity parameter ranges from zero to one, whereby zero indicates no loss of diversity and one indicates a total loss of diversity. The differences between the distributions of each genetic diversity statistic (S, He, nH, π, and ⊝) in MW and MD were statistically evaluated by the Wilcoxon signed ranked test for paired data.
Identification of Contigs Putatively under Selection
PS contigs in MD compared with MW were identified by computing two selection indices and testing their significance with a simulation approach. The selection indices were based on two between-groups and one within-groups genetic variation statistics, and their neutral distribution (assuming no selection) was based on coalescent simulations calibrated with previous demographic inferences on P. vulgaris divergence and domestication. Missing data were statistically imputed in the real data set before comparison to the simulated null distributions. All of these steps are individually described below.
Missing Data Imputation
A small fraction of the 188,107 SNPs had missing data. In particular, 2.73% of the total number of nucleotides was missing in the data set, with similar fractions in the different groups. We performed missing data imputation using the clustering algorithm implemented in fastPhase1.4 (Scheet and Stephens, 2006). This method does not require pedigree information and takes the population information into account. Individuals were assigned to three groups, according to their sampling origin: MW, MD, and Andean (due to the low number of Andean genotypes, all three of the individuals belonging to this area were grouped). Haplotype reconstruction was switched off, and the missing genotypes were imputed independently for each contig, setting the number of the cluster from one to 15. The complete set of fastPhase parameters was -KL1 -KU15 -Ki2 -H-4 -n -B -u.
Coalescent Simulations of Domestication Models
Coalescent simulations were used to generate neutral distributions of summary statistics, assuming three likely domestication scenarios reconstructed in previous studies (Supplemental Figure 5). In particular, our models were based on the population histories and demographic parameters estimated by Mamidi et al. (2011, 2013).
In all of the models, the Mesoamerican and Andean gene pools originated from an ancestral population, and domestication then occurred independently in each group. Only in more recent times did the domesticated groups expand exponentially, and some hybridization between domesticated and wild forms took place. Models B1 and B2 included a bottleneck event in the Andean or in both groups, respectively.
For each parameter, we defined prior distributions (Supplemental Table 7) based on previous estimates of their uncertainty (Mamidi et al., 2011, 2013). For each model, 100,000 simulations were performed by sampling random parameter combinations from these distributions using the ABCsampler program from the ABCtoolbox package (Wegmann et al., 2010). In each simulation, we generated a sample for each population that was equal to the observed one (10 haploid individuals for MW, eight for MD, two for AD, and one for AW). The lengths of the DNA sequences were extracted from a distribution calibrated for the distribution of contig lengths in the real samples.
Selection Index
For each locus, we initially calculated three statistics that were likely to be affected by differential selection in MD compared with MW. First, the population differentiation statistic ΦST (Excoffier et al., 1992) between MW and MD was calculated, following the classical view that loci differently affected by natural selection in different populations can be detected as outliers in population comparisons (Lewontin and Krakauer, 1973). Second, we considered the locus-specific branch-length statistic (Shriver et al., 2004). This is based on the genetic distance between two populations, but it also includes a third reference group (Andean, in our case) to identify which of the two populations experienced the positive selective pressure.
Finally, we computed a third statistic as the ratio of (\SMW-SMD\)/(SMW+SMD), where SMW is the number of segregating sites in MW, and SMD is the number of segregating sites in MD. This statistic measures the absolute value of the difference between the genetic variation in the MW and MD forms, as standardized by their sum; it is intended to capture the relative change in genetic diversity due to selection (Pritchard et al., 2010). All of these statistics tend to increase with increasing evidence of selection.
ΦST and the number of segregating sites were computed using the command-line version of Arlequin 3.5 (Excoffier and Lischer, 2010). The statistics were normalized using the neutral distributions obtained by simulation.
The three statistics above were combined into two different indices. The first index was built as the sum of all of the above statistics, and it was computed for 26,116 contigs. Due to the different alleles fixed in the two groups, in 1127 contigs, the statistics based on the segregating sites were undetermined. For these contigs, we created a second index that was obtained by summing up only the standardized ΦST and the locus-specific branch length. The same procedure was followed in the simulated data to generate the distribution of both of these indices assuming that only the demographic processes, and not the selection processes, shaped the pattern of the genetic variation. P values for each contig were then computed as the fraction of the simulated indices larger that the real value, and corrected to account for false positives, following the approach of Benjamini and Hochberg (1995), implemented in the p.adjust function available in the R statistical environment. This approach was repeated for each of the three simulated models, and we obtained three different lists of corrected p values. We then identified positively selected contigs when the false discovery rate was <5%, in each of the three models.
Gene Annotation
A BLASTX (Altschul et al., 1997) analysis against protein databases of Arabidopsis TAIR 10 was performed for all of the transcripts. Moreover, for functional characterization of PS contigs or PN contigs, a MapMan GSEA was conducted. See Supplemental Methods 1 for further details.
Network Analysis of Gene Coexpression
We performed a network-based comparative analysis of the RNA-seq data in the MW and MD populations. The analysis was based on expression level estimates for the 27,243 contigs. First, the contigs were selected for subsequent network analyses to avoid inclusion of potentially noisy or invariant gene expression profiles. Altogether, we did not consider 581 contigs that showed zero expression levels in at least nine genotypes. For each of the remaining 26,662 contigs, two statistical tests were performed: the differences in the means and in the variance of expression levels between the MW and MD populations, based on ANOVA and on the F-test, respectively (Ho et al., 2008). A very loose level of significance (α = 0.1) was considered for both of these tests. These allowed the selection of a subset of contigs to be used for the network analysis. The Wilcoxon rank sum test with continuity correction (Bauer, 1972) was applied to the CVs computed for MW and MD for each chosen contig. To this end, we tested whether the strategy applied for contig selection introduced any systematic bias with respect to favoring contigs that vary strongly in both MW and MD populations, in comparison to the entire set of contigs we considered. The possibility of introducing a shift in CVs toward higher/lower values for the PS contigs retained and those excluded from subsequent analysis was also tested.
Correlations of gene expression profiles were estimated using PCCs. The MW and MD expression profiles for the selected contigs were subjected to network-based analysis following two procedures: (1) extraction of proximity networks and (2) generation of relevance networks; both of these were based on PCCs (Klie et al., 2012; Kleessen et al., 2013). For details of these procedures, see Supplemental Methods 1. In contrast to relevance networks, the extraction of the proximity network took into consideration the observation that genes are often activated as modules of a program to fulfill a particular function (Quackenbush, 2003).
For the proximity networks, the following properties (reviewed in Newman, 2003, 2012) were analyzed: number of edges (proportional to the density), degree distribution (a degree of a node is the number of nodes with which it shares edges; hence, the degree distribution is given for the probability distribution of degrees in the network), distribution of connected components (defined as maximal subnetworks in which any two nodes are connected by a path), radius (the smallest of all of the node eccentricities), transitivity, and community structure (where a network community is a set of nodes that have more edges between each other than with the rest of the network and can be regarded as modules with particular functions). The adjusted R and index (Hubert and Arabie, 1985) and Jaccard index (Jaccard, 1912) were used for comparisons of the community structures (partitions of nodes) of the MW and MD networks.
To determine whether the network communities have a biological signal, the enriched gene functions for each of the determined communities were investigated (significance level, α = 0.01). MapMan ontology and a test based on the hypergeometric function (Rivals et al., 2007) were used. Potential enrichment for PS contigs of each of the network communities identified was also investigated.
The intersection network composed of the edges shared between the MW and MD proximity networks was determined. Moreover, the potential overrepresentation or underrepresentation of PS contigs in the intersection network was investigated.
The position of PS contigs in the extracted proximity networks was determined by considering two attributes of the contigs: (1) as under selective pressure (both for binary attribution PS/PN, and for selection index); and (2) CV of their expression profiles. To test whether similar contigs tend to be neighbors with respect to these two attributes, we used the concept of weighted assortativity, which is equivalent to PCCs between the attributes of nodes and the attributes of all of their immediate neighbors (Newman, 2003). A value of 1 denotes a high degree of assortativity (i.e., similar is a neighbor of similar), 0 denotes no assortativity (i.e., complete dispersion), and −1 denotes dissortativity (i.e., similar is a neighbor of dissimilar).
We investigated whether the PS contigs tend to be more central than the rest of the contigs. To quantify the centrality of position in the network, we used several measures of node centrality in a network (Newman, 2003), including betweenness, closeness, degree, eigenvalue, page rank, eccentricity, Burt's constraint, and transitivity. We also used the latent centrality, which was obtained via principal component analysis and integrated the information from the eight centrality measures used. The correlation between the selection index and the node centrality measures was also investigated through PCC.
Finally, to examine the functional characterization of the PS contigs, GSEA (Rivals et al., 2007) based on MapMan ontology was conducted.
Accession Numbers
All data are available in the Sequence Reads Archive under accession number SRP028116. The Transcriptome Shotgun Assembly project has been deposited at DDBJ/EMBL/GenBank under accession number GAMK00000000. The version described in this article is the first version, GAMK01000000.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Diversity Estimates in MW and MD Populations.
Supplemental Figure 2. Network-Based Analysis: Results of the Contig Selection Strategy.
Supplemental Figure 3. Network-Based Analysis: Selection Strategy and Bias of the CVs.
Supplemental Figure 4. Network-Based Analysis: Selection Strategy and Bias of CVs in the PS Contigs.
Supplemental Figure 5. Demographic Models for the Mesoamerican and the Andean Populations Used in This Study.
Supplemental Table 1. Transcriptome Assembly Statistics for the Four P. vulgaris Reference Genotypes and for the Final Nonredundant Data Set.
Supplemental Table 2. Jaccard Similarity of the Community Structure in the MW and MD Proximity Networks.
Supplemental Table 3. Community Gene Function Enrichment: List of Selected Enriched Gene Functions for Each of the Communities Determined for the MW and MD Networks.
Supplemental Table 4. Difference in Mean Node Centralities of Contigs under Selective Pressure and the Rest of the Nodes in the MW and MD Networks.
Supplemental Table 5. Difference in Assortativity, Both Nominal and Based on the “Selection Index,” between the MW and MD Networks.
Supplemental Table 6. Accessions Used in This Study.
Supplemental Table 7. Demographic Parameters in the B0, B1, and B2 Models.
Supplemental Methods 1. Details on Network-Based Analysis and Gene-Set Enrichment Analysis Using MapMan.
Supplemental Methods 2. Details Regarding Gene Function Investigation.
Supplemental Data Set 1A. List of Contigs and Information about Results of Differential Expression and Selection Analyses.
Supplemental Data Set 1B. Community Gene Function Enrichment in MW: Full List of MapMan Bins Enriched in Each of the MW Communities at a Significance Level of α = 0.05.
Supplemental Data Set 1C. Community Gene Function Enrichment in MD: Full List of MapMan Bins Enriched in Each of the MD Communities at a Significance Level of α = 0.05.
Supplemental Data Set 1D. Gene function Enrichment of PS and PN Contigs: Overrepresentation of MapMan Bins in Both PS and PN Contigs by GSEA Analysis.
Supplementary Material
Acknowledgments
This work was supported by grants from the Italian Government (MIUR; Grant 20083PFSXA_001, PRIN Project 2008), the Università Politecnica delle Marche (2012–2013), and the Marche Region (Grant L.R.37/99 art. 2lett.I - PARDGR 247/10-DDPF98/CSI10). We thank L'Oréal Italia “Per le Donne e la Scienza” for fellowship support (to E. Bitocchi).
AUTHOR CONTRIBUTIONS
E. Bellucci, E. Bitocchi, L.N., and R.P. designed the project. E. Bellucci, E. Bitocchi, E. Biagetti, L.N., and R.P. managed the project. E. Bellucci, E. Bitocchi, L.N., and R.P. wrote the article. E. Bellucci, E. Bitocchi, A.F., A.B., E. Biagetti, S.K., D.R., M.R., G.A., E.A., S.A.J., L.N., A.R.F., Z.N., G.B., and R.P. contributed to the drafting and writing of the article. E. Bellucci and E. Biagetti performed greenhouse experiments and RNA extraction. A.F. and E. Biagetti developed library preparation and managed sequencing. A.F. and M.D. directed RNA-seq and bioinformatic analysis and performed differential expression analysis. A.F., L.V., and M.D. performed de novo transcriptome assembly. A.M. performed variant identification. A.F., E. Biagetti, A.M., and M.D. performed annotation. E. Bellucci, E. Bitocchi, A.B., E. Biagetti, A.P., G.B., and R.P. conducted genetic diversity analysis. A.B. and G.B. performed coalescence simulations. A.B., A.P., G.B., and R.P. conducted selection detection and population genetics analysis. S.K. and Z.N. conducted network-based analysis and GSEA. D.R., M.R., G.A. E.A., A.R.F., E. Bitocchi, and E. Biagetti performed gene function investigation. E. Bellucci, E. Bitocchi, D.R., G.B., and R.P. edited the article. All authors read and approved the article.
Glossary
- RNA-seq
RNA sequencing
- MW
Mesoamerican wild
- MD
Mesoamerican domesticated
- SNP
single nucleotide polymorphism
- CV
coefficient of variation
- PCC
Pearson correlation coefficient
- PS
putatively under selection
- PN
putatively neutral
- GSEA
gene set enrichment analysis
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders, S. (2010). HTSeq: analysing high-throughput sequencing data with Python. http://www-huber.embl.de/users/anders/HTSeq/doc/overview.html [DOI] [PMC free article] [PubMed]
- Anders S., Huber W. (2010). Differential expression analysis for sequence count data. Genome Biol. 11: R106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer D.F. (1972). Constructing confidence sets using rank statistics. J. Am. Stat. Assoc. 67: 687–690 [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B 57: 289–300 [Google Scholar]
- Bitocchi E., Nanni L., Bellucci E., Rossi M., Giardini A., Zeuli P.S., Logozzo G., Stougaard J., McClean P., Attene G., Papa R. (2012). Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proc. Natl. Acad. Sci. USA 109: E788–E796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitocchi E., et al. (2013). Molecular analysis of the parallel domestication of the common bean (Phaseolus vulgaris) in Mesoamerica and the Andes. New Phytol. 197: 300–313 [DOI] [PubMed] [Google Scholar]
- Blackman B.K., Rasmussen D.A., Strasburg J.L., Raduski A.R., Burke J.M., Knapp S.J., Michaels S.D., Rieseberg L.H. (2011). Contributions of flowering time genes to sunflower domestication and improvement. Genetics 187: 271–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman M.A., Pashley C.H., Wenzler J., Hvala J., Tang S., Knapp S.J., Burke J.M. (2008). A genomic scan for selection reveals candidates for genes involved in the evolution of cultivated sunflower (Helianthus annuus). Plant Cell 20: 2931–2945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cong B., Barrero L.S., Tanksley S.D. (2008). Regulatory change in YABBY-like transcription factor led to evolution of extreme fruit size during tomato domestication. Nat. Genet. 40: 800–804 [DOI] [PubMed] [Google Scholar]
- Darwin, C. (1859). On the Origins of Species by Means of Natural Selection. (London: John Murray). [Google Scholar]
- Darwin, C. (1875). The Variation of Animals and Plants under Domestication, 2nd ed. (London: John Murray). [Google Scholar]
- Darwin, C. (1878). The Effects of Cross and Self-Fertilization in the Vegetal Kingdom. (London: John Murray). [Google Scholar]
- de Alencar Figueiredo L.F., Calatayud C., Dupuits C., Billot C., Rami J.F., Brunel D., Perrier X., Courtois B., Deu M., Glaszmann J.C. (2008). Phylogeographic evidence of crop neodiversity in sorghum. Genetics 179: 997–1008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desiderio F., Bitocchi E., Bellucci E., Rau D., Rodriguez M., Attene G., Papa R., Nanni L. (2013). Chloroplast microsatellite diversity in Phaseolus vulgaris. Front. Plant Sci. 3: 312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L., Lischer H.E.L. (2010). Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 10: 564–567 [DOI] [PubMed] [Google Scholar]
- Excoffier L., Smouse P.E., Quattro J.M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131: 479–491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gepts P. (2002). A Comparison between crop domestication, classical plant breeding, and genetic engineering. Crop Sci. 42: 1780–1790 [Google Scholar]
- Gepts, P., and Papa, R. (2002). Evolution during domestication. In Encyclopedia of Life Sciences. (London: Macmillan Publishers, Nature Publishing Group), pp. 1–7. [Google Scholar]
- Glémin S., Bataillon T. (2009). A comparative view of the evolution of grasses under domestication. New Phytol. 183: 273–290 [DOI] [PubMed] [Google Scholar]
- Grabherr M.G., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hecht V., Knowles C.L., Vander Schoor J.K., Liew L.C., Jones S.E., Lambert M.J., Weller J.L. (2007). Pea LATE BLOOMER1 is a GIGANTEA ortholog with roles in photoperiodic flowering, deetiolation, and transcriptional regulation of circadian clock gene homologs. Plant Physiol. 144: 648–661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho J.W.K., Stefani M., dos Remedios C.G., Charleston M.A. (2008). Differential variability analysis of gene expression and its application to human diseases. Bioinformatics 24: i390–i398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubert L., Arabie P. (1985). Comparing partitions. J. Classif. 2: 193–218 [Google Scholar]
- Hufford M.B., et al. (2012). Comparative population genomics of maize domestication and improvement. Nat. Genet. 44: 808–811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaccard P. (1912). The distribution of the flora in the alpine zone. New Phytol. 11: 37–50 [Google Scholar]
- Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleessen S., Klie S., Nikoloski Z. (2013). Data integration through proximity-based networks provides biological principles of organization across scales. Plant Cell 25: 1917–1927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klie S., Mutwil M., Persson S., Nikoloski Z. (2012). Inferring gene functions through dissection of relevance networks: interleaving the intra- and inter-species views. Mol. Biosyst. 8: 2233–2241 [DOI] [PubMed] [Google Scholar]
- Koboldt D.C., Zhang Q., Larson D.E., Shen D., McLellan M.D., Lin L., Miller C.A., Mardis E.R., Ding L., Wilson R.K. (2012). VarScan 2: somatic mutation and copy number alteration discovery in cancer by exome sequencing. Genome Res. 22: 568–576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam H.M., et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42: 1053–1059 [DOI] [PubMed] [Google Scholar]
- Lester R.N. (1989). Evolution under domestication involving disturbance of genic balance. Euphytica 44: 125–132 [Google Scholar]
- Lewontin R.C., Krakauer J. (1973). Distribution of gene frequency as a test of the theory of the selective neutrality of polymorphisms. Genetics 74: 175–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Durbin R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., 1000 Genome Project Data Processing Subgroup (2009). The sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25: 2078–2079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Godzik A. (2006). Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22: 1658–1659 [DOI] [PubMed] [Google Scholar]
- Lin Z., et al. (2012). Parallel domestication of the Shattering1 genes in cereals. Nat. Genet. 44: 720–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippold S., Knapp M., Kuznetsova T., Leonard J.A., Benecke N., Ludwig A., Rasmussen M., Cooper A., Weinstock J., Willerslev E., Shapiro B., Hofreiter M. (2011). Discovery of lost diversity of paternal horse lineages using ancient DNA. Nat. Commun. 2: 450. [DOI] [PubMed] [Google Scholar]
- Lu J., Tang T., Tang H., Huang J., Shi S., Wu C.I. (2006). The accumulation of deleterious mutations in rice genomes: a hypothesis on the cost of domestication. Trends Genet. 22: 126–131 [DOI] [PubMed] [Google Scholar]
- Mamidi S., Rossi M., Annam D., Moghaddam S., Lee R., Papa R., McClean P. (2011). Investigation of the domestication of common bean (Phaseolus vulgaris) using multilocus sequence data. Funct. Plant Biol. 38: 953–967 [DOI] [PubMed] [Google Scholar]
- Mamidi S., Rossi M., Moghaddam S.M., Annam D., Lee R., Papa R., McClean P.E. (2013). Demographic factors shaped diversity in the two gene pools of wild common bean Phaseolus vulgaris L. Heredity (Edinb.) 110: 267–276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanni L., Bitocchi E., Bellucci E., Rossi M., Rau D., Attene G., Gepts P., Papa R. (2011). Nucleotide diversity of a genomic sequence similar to SHATTERPROOF (PvSHP1) in domesticated and wild common bean (Phaseolus vulgaris L.). Theor. Appl. Genet. 123: 1341–1357 [DOI] [PubMed] [Google Scholar]
- Nei M. (1978). Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89: 583–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman M.E.J. (2003). The structure and function of complex networks. SIAM Rev. 45: 167–256 [Google Scholar]
- Newman M.E.J. (2012). Communities, modules and large-scale structure in networks. Nat. Phys. 8: 25–31 [Google Scholar]
- Olson M.V. (1999). When less is more: gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 64: 18–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osakabe Y., et al. (2013). Osmotic stress responses and plant growth controlled by potassium transporters in Arabidopsis. Plant Cell 25: 609–624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papa R., Bellucci E., Rossi M., Leonardi S., Rau D., Gepts P., Nanni L., Attene G. (2007). Tagging the signatures of domestication in common bean (Phaseolus vulgaris) by means of pooled DNA samples. Ann. Bot. (Lond.) 100: 1039–1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard J.K., Pickrell J.K., Coop G. (2010). The genetics of human adaptation: hard sweeps, soft sweeps, and polygenic adaptation. Curr. Biol. 20: R208–R215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quackenbush J. (2003). Genomics. Microarrays—guilt by association. Science 302: 240–241 [DOI] [PubMed] [Google Scholar]
- R Development Core Team (2013). R: A Language and Environment for Statistical Computing. (Vienna, Austria: R Foundation for Statistical Computing).
- Rivals I., Personnaz L., Taing L., Potier M.C. (2007). Enrichment or depletion of a GO category within a class of genes: which test? Bioinformatics 23: 401–407 [DOI] [PubMed] [Google Scholar]
- Rossi M., Bitocchi E., Bellucci E., Nanni L., Rau D., Attene G., Papa R. (2009). Linkage disequilibrium and population structure in wild and domesticated populations of Phaseolus vulgaris L. Evol. Appl. 2: 504–522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheet P., Stephens M. (2006). A fast and flexible statistical model for large-scale population genotype data: applications to inferring missing genotypes and haplotypic phase. Am. J. Hum. Genet. 78: 629–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shriver M.D., Kennedy G.C., Parra E.J., Lawson H.A., Sonpar V., Huang J., Akey J.M., Jones K.W. (2004). The genomic distribution of population substructure in four populations using 8,525 autosomal SNPs. Hum. Genomics 1: 274–286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J.M., Haigh J. (1974). The hitch-hiking effect of a favourable gene. Genet. Res. 23: 23–35 [PubMed] [Google Scholar]
- Swanson-Wagner R., Briskine R., Schaefer R., Hufford M.B., Ross-Ibarra J., Myers C.L., Tiffin P., Springer N.M. (2012). Reshaping of the maize transcriptome by domestication. Proc. Natl. Acad. Sci. USA 109: 11878–11883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tajima F. (1983). Evolutionary relationship of DNA sequences in finite populations. Genetics 105: 437–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi Y., Shimamoto K. (2011). Heading date 1 (Hd1), an ortholog of Arabidopsis CONSTANS, is a possible target of human selection during domestication to diversify flowering times of cultivated rice. Genes Genet. Syst. 86: 175–182 [DOI] [PubMed] [Google Scholar]
- Vigouroux Y., McMullen M., Hittinger C.T., Houchins K., Schulz L., Kresovich S., Matsuoka Y., Doebley J. (2002). Identifying genes of agronomic importance in maize by screening microsatellites for evidence of selection during domestication. Proc. Natl. Acad. Sci. USA 99: 9650–9655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonholdt B.M., et al. (2010). Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 464: 898–902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson G.A. (1975). On the number of segregating sites in genetical models without recombination. Theor. Popul. Biol. 7: 256–276 [DOI] [PubMed] [Google Scholar]
- Wegmann D., Leuenberger C., Neuenschwander S., Excoffier L. (2010). ABCtoolbox: a versatile toolkit for approximate Bayesian computations. BMC Bioinformatics 11: 116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S.I., Bi I.V., Schroeder S.G., Yamasaki M., Doebley J.F., McMullen M.D., Gaut B.S. (2005). The effects of artificial selection on the maize genome. Science 308: 1310–1314 [DOI] [PubMed] [Google Scholar]
- Wu W., et al. (2013). Association of functional nucleotide polymorphisms at DTH2 with the northward expansion of rice cultivation in Asia. Proc. Natl. Acad. Sci. USA 110: 2775–2780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia Q., et al. (2009). Complete resequencing of 40 genomes reveals domestication events and genes in silkworm (Bombyx). Science 326: 433–436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X., et al. (2012). Resequencing 50 accessions of cultivated and wild rice yields markers for identifying agronomically important genes. Nat. Biotechnol. 30: 105–111 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.