This work investigates the proteome changes during the onset of postharvest physiological deterioration (PPD) in cassava, an important root crop for food security in the tropics. The time-course proteomic study was instrumental in the identification of gene candidates to delay PPD in cassava roots.
Abstract
Cassava (Manihot esculenta) is the most important root crop in the tropics, but rapid postharvest physiological deterioration (PPD) of the root is a major constraint to commercial cassava production. We established a reliable method for image-based PPD symptom quantification and used label-free quantitative proteomics to generate an extensive cassava root and PPD proteome. Over 2600 unique proteins were identified in the cassava root, and nearly 300 proteins showed significant abundance regulation during PPD. We identified protein abundance modulation in pathways associated with oxidative stress, phenylpropanoid biosynthesis (including scopoletin), the glutathione cycle, fatty acid α-oxidation, folate transformation, and the sulfate reduction II pathway. Increasing protein abundances and enzymatic activities of glutathione-associated enzymes, including glutathione reductases, glutaredoxins, and glutathione S-transferases, indicated a key role for ascorbate/glutathione cycles. Based on combined proteomics data, enzymatic activities, and lipid peroxidation assays, we identified glutathione peroxidase as a candidate for reducing PPD. Transgenic cassava overexpressing a cytosolic glutathione peroxidase in storage roots showed delayed PPD and reduced lipid peroxidation as well as decreased H2O2 accumulation. Quantitative proteomics data from ethene and phenylpropanoid pathways indicate additional gene candidates to further delay PPD. Cassava root proteomics data are available at www.pep2pro.ethz.ch for easy access and comparison with other proteomics data.
INTRODUCTION
Cassava (Manihot esculenta) is the most important staple crop consumed by food-insecure populations in sub-Saharan Africa (Lobell et al., 2008). It is mostly produced for its starchy roots, but the leaves are also part of the diet in several African regions (Achidi et al., 2005). Efforts have been made to increase and secure sustainable cassava production through improving agronomic practices and reducing biotic and abiotic stresses (El-Sharkawy, 2006; Fermont et al., 2009). The potential of cassava as a food and industrial crop, however, is still limited because of rapid postharvest physiological deterioration (PPD) of the root (Wenham, 1995; Sayre et al., 2011). PPD is induced by mechanical damage during harvesting and handling operations (Booth, 1976; Rickard, 1985), and progression depends on cassava genotypes and storage conditions (Sanchez et al., 2006). The blue-black discoloration of the vascular parenchyma that develops during PPD and that is followed by a general discoloration of the storage parenchyma, as well as physiological and biochemical changes, ultimately render the roots unpalatable (Beeching et al., 1998).
Initial studies of PPD focused on gene expression changes (Han et al., 2001; Huang et al., 2001; Reilly et al., 2001, 2004, 2007). However, changes in protein accumulation, interactions, modifications, and activities during PPD development are equally important but currently not well understood. Earlier investigations revealed that PPD is associated with changes in the activities of enzymes such as catalase, superoxide dismutase, phenylalanine ammonia lyase (PAL), and peroxidase (Tanaka et al., 1983; Rickard, 1985; Reilly et al., 2001, 2004).
Mass spectrometry (MS)–based proteomics offers a new approach to discover proteins and pathways associated with crop physiological and stress responses (Vanderschuren et al., 2013). The identification of protein candidates generally requires quantitative data, because their precise modulation rather than presence/absence is involved in the regulation of physiological processes. Cassava proteomics studies have so far mostly used two-dimensional gel electrophoresis (Sheffield et al., 2006; Baba et al., 2008; Mitprasat et al., 2011). The number of proteins identified in those studies was limited by the method as well as the lack of a fully annotated cassava genome. To increase proteome coverage, we previously reported an iTRAQ-based matrix-assisted laser desorption/ionization time-of-flight approach using the Viridiplantae protein database to identify 1387 nonredundant protein groups (Owiti et al., 2011). With the recent release and annotation of the cassava genome (Prochnik et al., 2012), it is now possible to expand cassava proteome coverage and better characterize its modulation during PPD.
Here, we analyzed the proteome of cassava roots at harvest and during the onset of PPD. We identified several proteins in pathways that were regulated during the onset of PPD and extended the number of proteins that likely have a role during root deterioration. Protein identifiers and protein abundance ratios were integrated into the pep2pro database (Baerenfaller et al., 2011; Hirsch-Hoffmann et al., 2012) to enable a comprehensive cassava proteome data analysis. Based on proteomics data, enzymatic assays, and lipid peroxidation, we identified GLUTATHIONE PEROXIDASE (GPX) as a candidate gene for delayed PPD and demonstrated that transgenic overexpression of an Arabidopsis thaliana GPX in cassava storage root could delay PPD.
RESULTS
Properties of the Samples Used for Analysis
Cassava storage roots of the model cv 60444 that is routinely used for transformation (Bull et al., 2009) were harvested in the greenhouse and sliced. Root slices were collected for characterization of the root proteome at harvest (0 h). Additional root slices were incubated for 6, 12, and 24 h at 28°C in the dark and imaged prior to flash freezing and sample storage at −80°C. Image analysis of the root slices was used to establish a standardized quantitative measure of PPD progression in cassava roots and to ensure that the material used for analysis showed significant and reproducible differences during PPD progression. The image-based analysis we developed relies on the gray value distribution. Because PPD symptoms develop by continuous darkening of root tissues commencing from the epidermis, their identification by standard histogram thresholding (Raju and Neelima, 2012) is not appropriate as in the case of a bimodal histogram. We observed that the gray value histogram broadened with PPD symptoms spreading from the epidermis through the root tissue. Using the PPD imaging method, we confirmed that the root sample replicates collected for proteomics analysis showed homogenous and reproducible PPD progression at the selected time points (Supplemental Figure 1). The small variation between replicates demonstrates that our PPD assay is a reliable method to investigate PPD progression.
Proteomics Workflow for the Identification of Cassava Root Proteins
Previously, we used detergent to extend proteome analysis to the nonsoluble protein fraction of cassava roots (Owiti et al., 2011), which substantially increased proteome coverage. Here, we used SDS to extract total protein from cassava roots during PPD progression. Three independent biological replicates of each time point were analyzed, resulting in a total of 702,738 tandem mass spectra (MS/MS) (Figure 1).
Figure 1.
Experimental and Bioinformatics Workflow of the Proteome Analysis.
Total proteins were extracted from roots, separated by gel electrophoresis, digested with trypsin, and analyzed by liquid chromatography–MS/MS using an LTQ-Orbitrap device. The workflow references Supplemental Data Sets 1 to 5, corresponding to the analysis steps.
Selected spectra were searched against the cassava proteome database 4.1, which comprises 30,666 proteins from 34,151 transcripts (Prochnik et al., 2012). We identified 4121 proteins when peptides with conflicting assignments were taken into account, and identified proteins were not grouped (Supplemental Data Set 1). To reduce redundancy in protein identification, conflicting peptides were removed from the initial data set and the remaining identified proteins were compiled into protein groups, resulting in 2632 nonredundant proteins (Supplemental Data Set 2).
The identified proteins represent 8.6% of the protein-coding loci currently annotated in the cassava genome. In Arabidopsis, Baerenfaller and colleagues (2011) detected 41.6% of the genome-encoded proteins predicted in TAIR10 in root samples. The number of proteins expressed in cassava roots is not known; however, proteome coverage could be increased in the future by improving the cassava genome annotation, protein fractionation and enrichment strategies, and the sensitivity of MS/MS instruments. Low-abundance proteins as well as proteins whose expression is induced under specific conditions probably represent an important fraction of currently undetected proteins in cassava roots.
Identification of Abundant Proteins in Cassava Storage Roots
Cassava roots have a low protein content and do not express vegetative storage proteins that typically accumulate in vegetative tissues as a temporary reservoir of amino acids (Fujiwara et al., 2002; Stupak et al., 2006; Montagnac et al., 2009). As expected, our proteomics analysis did not reveal proteins related to known root and tuber storage proteins (Supplemental Data Sets 1 and 2) (Shewry, 2003). To generate a ranking of the proteins based on their abundance, we used the normalized volumes of the peptides calculated by the Progenesis software for the top-three protein quantification (T3PQ) method (Silva et al., 2006; Grossmann et al., 2010). Nearly half of the detected proteins could not be ranked because their identification and quantification were based on less than three peptides (Supplemental Data Set 3). The most abundant proteins were heat shock proteins, reactive oxygen species (ROS)–scavenging enzymes, as well as several starch-related enzymes, consistent with the physiological role of the cassava root. We also detected the most abundant proteins previously reported from a 2D gel analysis of tuberous cassava roots (Sheffield et al., 2006). They are also ranked among the most abundant proteins on our list (Supplemental Data Set 3).
The pep2pro Database as a Repository of Quantitative Cassava Proteomics Data
The protein identifications of this study were loaded into the pep2pro database (Baerenfaller et al., 2011; Hirsch-Hoffmann et al., 2012) and are available at www.pep2pro.ethz.ch. We provide information on all proteins and their identified peptides as well as visualization of the proteogenomic mapping of the peptides onto the annotated cassava genome sequence. We also provide the scores for the peptide spectrum assignment and display the spectra in a Spectrum Viewer. For visualizing quantitative information, the normalized abundance of each protein across the different time points is shown together with the P value and the fold change of the quantitative analysis.
Regulated Proteins and Pathways during PPD Progression and Target Gene Identification
MS peak intensity was used to identify 293 proteins regulated during PPD progression (Supplemental Data Set 4). This list of proteins was subsequently used for the further characterization and interpretation of pathways activated during PPD progression.
Protein Categories Overrepresentation
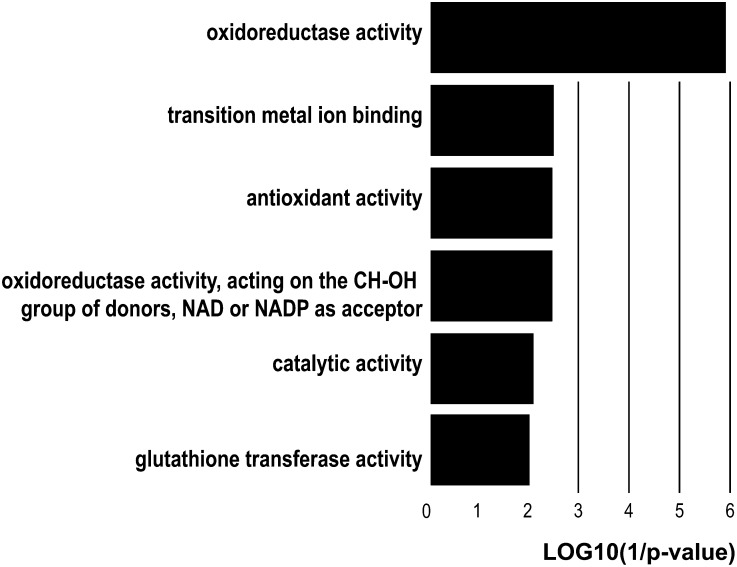
Overrepresentation of protein categories from biological process and molecular function was investigated during PPD progression using the list of regulated proteins (Supplemental Data Set 4). Proteins involved in phenylpropanoid biosynthetic and metabolic processes were enriched in the fraction of modulated proteins (Supplemental Figure 2). Other biological processes overrepresented during PPD progression include lipid and sulfur metabolic processes, response to biotic stresses, as well as toxin catabolic process. Analysis of overrepresented molecular function categories revealed that the majority accounted for antioxidant and redox activities (i.e., oxidoreductase activity, antioxidant activity, and glutathione transferase activity) (Figure 2), suggesting that oxidative stress is a key process that induces PPD and facilitates PPD progression (Reilly et al., 2004, 2007). We used the AraCyc database (Mueller et al., 2003) and the list of modulated cassava proteins to identify and analyze pathways with at least two enzymes that were significantly regulated during the onset and progression of PPD.
Figure 2.
Molecular Function Categories Overrepresented in the Proteins Regulated during PPD.
Category overrepresentation was performed with Arabidopsis identifiers and determined relative to the background set of all identified proteins for which an Arabidopsis identifier was available to take into account the bias introduced by the extraction and detection methods.
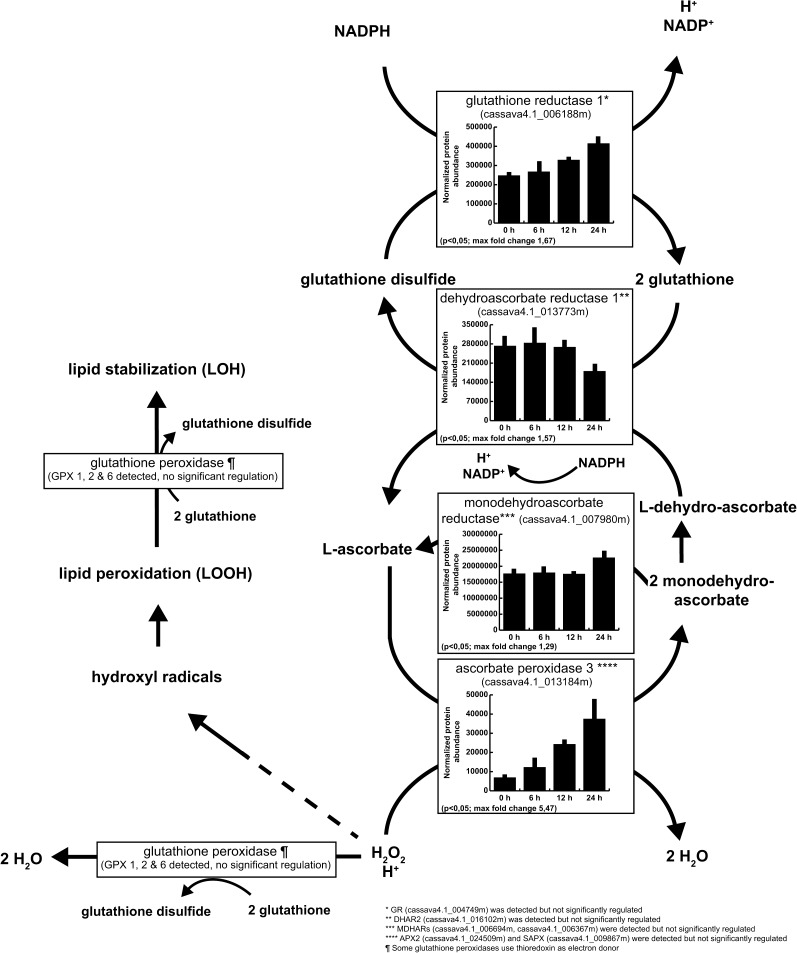
Ascorbate/Glutathione Cycles Are Pivotal to PPD
Both superoxide and H2O2 accumulate in cassava roots during PPD (Buschmann et al., 2000a; Reilly et al., 2004). Enzymes in the ascorbate/glutathione cycle, which scavenges H2O2, were modulated during PPD (Figure 3). Notably, the ASCORBATE PEROXIDASE3 (APX3) protein level was strongly upregulated 6 h after harvest. We also detected APX2, which was reported to be upregulated at the mRNA level (Reilly et al., 2007), but its levels did not change significantly during PPD progression. Other peroxidases that were regulated at the transcript level during PPD (i.e., PER12 and PX3) (Reilly et al., 2001, 2007) had corresponding protein level changes (i.e., cassava4.1_010796m and cassava4.1_011662m; Supplemental Data Set 4). Glutathione dehydrogenase ascorbate reductase (DHAR) and monodehydroascorbate reductase (MDHAR) levels were not significantly changed or even slightly downregulated at 6 and 12 h after harvest. Similarly, the level of glutathione dehydrogenase was significantly reduced 24 h after harvest. While the increase of APX abundance does not correlate with increased APX activities during early PPD progression (Owiti et al., 2011) (Supplemental Figure 3), our study also revealed that levels of enzymes involved in the reduction of ascorbate and dehydroascorbate are not significantly upregulated.
Figure 3.
Detection and Regulation of Proteins Involved in the Ascorbate/Glutathione Cycle.
Regulated proteins are selected based on the Progenesis ANOVA analysis (P < 0.05), and regulation is represented graphically using normalized protein abundance and sd (n = 3).
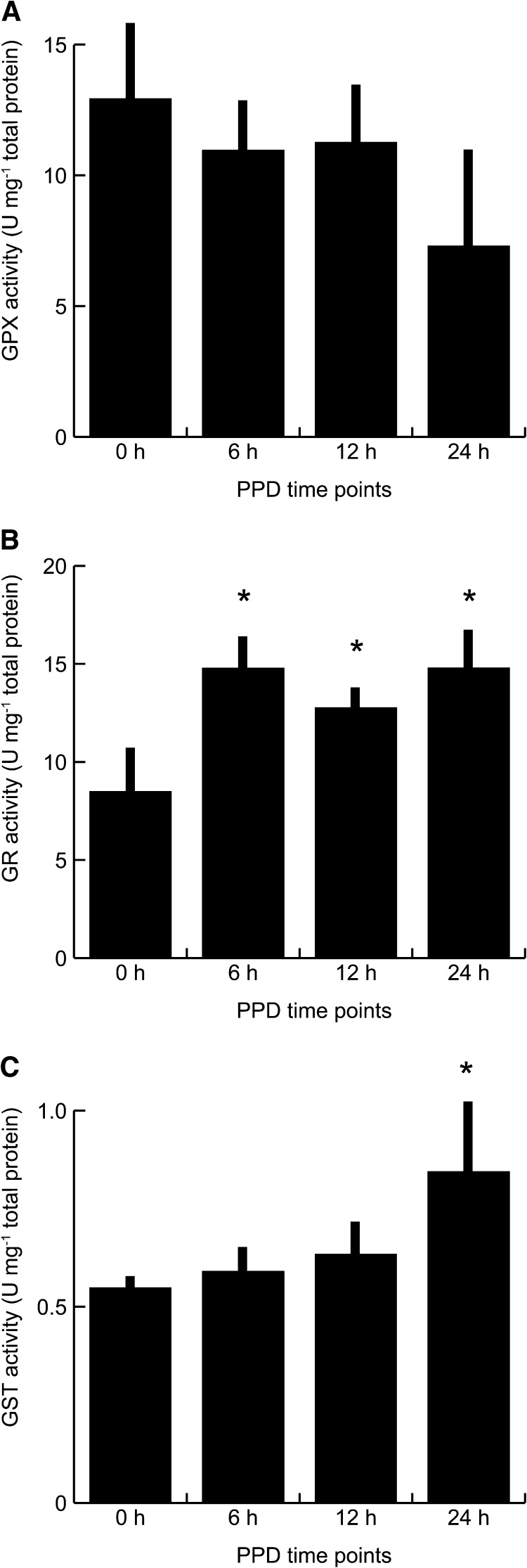
Enzymes that use glutathione to detoxify H2O2 (i.e., GPX1, GPX2, and GPX6) were detected, but their abundances and total GPX activity were not significantly changed during PPD progression (Figure 4A). However, glutathione disulfide reductase (GR) accumulated during PPD (Supplemental Data Set 4) and had increased activities at 6, 12, and 24 h after harvest (Figure 4B). GR converts GSSG into GSH to maintain GSH/GSSG homeostasis. Because GPX activity is stable (glutathione redox reactions I) and DHAR abundance is decreased during PPD, other ROS-scavenging pathways using GSH might also modulate the GSH/GSSG ratio and therefore require increased GR activities.
Figure 4.
Enzymatic Activities in Protein Fractions from Collected Time Points.
Protein fractions were GPX (A), GR (B), and GST (C). Means ± sd of three biological replicates are shown (Student’s t test, *P < 0.05).
In addition to DHAR, we detected four glutathione transferases belonging to the two large plant-specific ϕ and τ classes that were among the proteins that showed the strongest upregulation during PPD. The difference of glutathione S-transferase (GST) abundances correlated with a significant change in GST activity only 24 h after harvest (Figure 4C). The lack of significant regulation at 6 and 12 h might be due to stable and high abundances of other GSTs contributing to the overall GST activity measured in our assays.
Glutathione can also be conjugated spontaneously to formaldehyde or by GSTs to methylglyoxal in the formaldehyde oxidation II and methylglyoxal degradation I pathways. The two enzymes from the glutathione-dependent formaldehyde oxidation II pathway, which detoxifies formaldehyde, were detected, but only S-formylglutathione hydrolase (cassava4.1_009729m) was found to be downregulated at 24 h after harvest. The hydroxyacylglutathione hydrolase (cassava4.1_011683m), which catalyzes the hydrolysis of S-lactoylglutathione, displayed a similar abundance pattern (Supplemental Data Set 4).
Increased GPX Activity in Transgenic Cassava Root Significantly Reduces PPD Symptoms
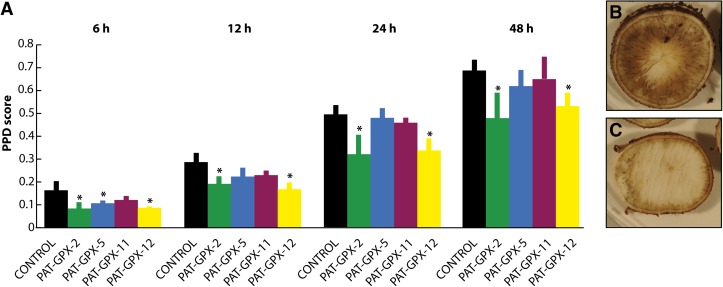
Based on our observation that lipid peroxidation occurs during PPD (Supplemental Figure 4) and both GPX protein level and activity (Figures 3 and 4A) are not altered during PPD, we hypothesized that increasing GPX activity in cassava roots could delay PPD onset. Transgenic cassava plants overexpressing an Arabidopsis cytosolic GPX in storage roots were generated (Supplemental Figure 5), and four transgenic lines were assessed for PPD onset using the PPD scoring method. We found that the appearance of PPD symptoms in the inner section of cassava storage roots was significantly delayed in the transgenic lines at 6 h after harvest (Figure 5). The reduced PPD symptom score was maintained until 48 h after harvest in the PAT-GPX2 and PAT-GPX12 lines. Enhanced GPX activity in transgenic cassava appeared to limit lipid peroxidation (Supplemental Figure 6A) and lower H2O2 accumulation during PPD onset (Supplemental Figure 6B).
Figure 5.
PPD Score Analysis of Wild-Type and Transgenic Cassava Storage Roots.
(A) Comparison of the PPD scores of the control line and transgenic PAT-GPX lines at 6, 12, 24, and 48 h after harvest (Student’s t test, *P < 0.05).
(B) Root slice of the control line at 24 h after harvest.
(C) Root slice of the PAT-GPX2 line at 24 h after harvest.
[See online article for color version of this figure.]
Other Enzymatic Pathways Regulated during PPD
Ethene Biosynthesis
Early studies of PPD identified the emission of ethene in cassava roots that peaked 18 h after harvest (Hirose et al., 1984), which correlates with the upregulation of 1-aminocyclopropane-1-carboxylate (ACC) oxidase mRNA 12 h after harvest (Reilly et al., 2007). ACC oxidase catalyzes the final step of ethene synthesis (Wang et al., 2002). Ethene biosynthesis using l-Met as substrate requires two additional enzymes, S-adenosyl-l-methionine (SAM) synthetase and ACC synthase. Both SAM synthetase and ACC oxidase showed significant and coordinated upregulation consistent with increased ethene synthesis during PPD progression (Supplemental Figure 7).
Phenylpropanoid Biosynthesis
Phenylpropanoids are produced in plants during biotic and abiotic stress (reviewed in Vogt, 2010). The coumarin scopoletin accumulates to high levels during PPD progression (Tanaka et al., 1983; Wheatley and Schwabe, 1985; Buschmann et al., 2000b; Bayoumi et al., 2010). Enzymes involved in the final steps of scopoletin biosynthesis were significantly upregulated 6 h after harvest (Supplemental Figure 8), including 4-coumarate-CoA ligase and caffeoyl-CoA O-methyltransferase enzymes, which convert 4-coumarate into coumaroyl-CoA and caffeoyl-CoA into feruoyl-CoA, respectively. 2-Oxoglutarate–dependent dioxygenase, which was recently identified as a pivotal enzyme converting feruoyl-CoA into 6′-hydroxyferuoyl-CoA during scopoletin synthesis (Kai et al., 2008), was upregulated during PPD progression only 12 h after harvest. PAL, which initiates phenylpropanoid synthesis, also had increased activity during PPD (Tanaka et al., 1983). We found that both PAL1 and PAL2 abundances were regulated, with up to an 8-fold increase at 24 h after harvest (Supplemental Figure 9). Analysis of PAL activity in the cassava root extracts used for the proteomics study revealed a significant increase in PAL activity at 12 and 24 h after harvest (Supplemental Figure 10), but PAL activity and protein levels did not strictly correlate.
Fatty Acid α-Oxidation
α-DOX1 (cassava4.1_003891m), a fatty acid dioxygenase catalyzing the primary oxygenation of fatty acids (De León et al., 2002), was more than 15-fold upregulated 24 h after harvest. This was paralleled by the upregulation of an aldehyde dehydrogenase (cassava4.1_005092m) (Supplemental Figure 11) during PPD progression. Upregulation of both enzymes at 24 h might be indicative of fatty acid degradation through α-oxidation.
Folate Transformation
Several proteins involved in one-carbon metabolism accumulated during PPD progression (Supplemental Figure 12). The upregulation of Gly hydroxymethyltransferase (cassava4.1_006924m) and 5,10-methylenetetrahydrofolate reductase (cassava4.1_003870m), combined with the downregulation of methylenetetrahydrofolate dehydrogenase (cassava4.1_012783m), indicate that the folate transformation pathway is directed toward the production of 5′-methyltetrahydrofolate, which is the most reduced form of tetrahydrofolate. The use of 5′-methyltetrahydrofolate to convert homocysteine to Met represents the largest anabolic flux of one-carbon units (Hanson and Roje, 2001).
The upregulation of two Met synthases (cassava4.1_002156m and cassava4.1_006508) (Supplemental Figure 13) supports the hypothesis that Met biosynthesis is altered during PPD.
Sulfate Reduction II Pathway (Assimilatory)
A sulfate adenylyltransferase (APS2; cassava4.1_006467m), which converts sulfate into adenosine 5′-phosphosulfate, increased more than 20-fold 6 h after harvest and further increased to over 60-fold 12 and 24 h after harvest (Supplemental Figure 14). Sulfate adenylyltransferase catalyzes the first reaction of the sulfate reduction II pathway (Saito, 2004). The adenylyl-sulfate reductase detected in our study did not show significant abundance regulation, whereas sulfite reductase, which converts sulfite into hydrogen sulfide, was also upregulated during PPD progression. Modulation of the sulfate assimilation proteins suggests an increase of amino acids (i.e., Cys and Met) and glutathione biosynthesis during PPD.
DISCUSSION
We have significantly expanded the cassava root and PPD proteome to more than 2600 nonredundant proteins using a combination of high-accuracy label-free MS/MS and the cassava genome database. This data set provides a useful basis for further characterizing the cassava root proteome in different conditions relevant to cassava root physiology and trait improvement. The cassava root proteomics data that are available on the pep2pro web interface will allow comparison with other proteomics data to uncover common responses in model and crop plants.
An analysis of overrepresented biological process and molecular function categories shows that the phenylpropanoid pathway and proteins with antioxidant and redox activities are pivotal to PPD. The quantitative proteomics data indicate that quantities of DHAR and MDHAR enzymes are not altered at 6 and 12 h after harvest. DHAR is significantly downregulated at 24 h after harvest; therefore, DHAR activity might be limiting for maintenance of the ascorbate pool. This is supported by a previous observation that ascorbate supplementation can significantly delay PPD onset in cassava (Reilly et al., 2001). Altering DHAR expression levels could be instrumental to maintaining an ascorbate pool, as suggested by previous studies in transgenic tobacco (Nicotiana tabacum) (Kwon et al., 2003; Yin et al., 2010). Our data reveal that GR accumulates during PPD and correlate with increased GR activity, suggesting that the ascorbate/glutathione pathway might be more constrained by the availability of ascorbate than glutathione. The abundance of GluCys ligase (cassava4.1_005513m) and glutathione synthetase (cassava4.1_004677m), which catalyze the formation of glutathione from l-Glu and l-Cys, respectively, were not changed.
Our study also confirms that APX activity is not increased up to 24 h after harvest. The limited ascorbate availability and the nonactivation of peroxidases could explain the accumulation of H2O2 observed in cassava storage roots undergoing PPD (Reilly et al., 2004; Xu et al., 2013). We consistently observed an initial increase of H2O2 followed by a reduction at 48 h after harvest (Supplemental Figure 6B), which could indicate a limited capacity of the ascorbate/glutathione pathways to scavenge ROS. Recent work using transgenic cassava storage roots overexpressing catalase and superoxide dismutase demonstrates that limiting the accumulation of ROS independently from the ascorbate pool can effectively reduce PPD onset (Xu et al., 2013). Our measurement of detected GPX by quantitative proteomics reveals an absence of regulation during PPD (Figure 3), which is consistent with an overall decreasing activity of GSH-dependent GPX activity (Figure 4). Previous work in Arabidopsis and cassava suggests that GPX genes are regulated by stresses, including PPD, at the transcriptional level (Rodriguez Milla et al., 2003; Reilly et al., 2007). Our work shows that the transcriptional regulation does not lead to increased abundances of GPX protein during PPD. Transgenic cassava expressing At-GPX2 under the control of the patatin promoter shows an increased GSH-dependent GPX activity in storage roots, demonstrating that At-GPX2 can have GSH-dependent activity in heterologous systems in addition to its thioredoxin-dependent activity (Iqbal et al., 2006). The reduction of malondialdehyde (MDA) accumulation in transgenic PAT-GPX cassava storage roots during PPD (Supplemental Figure 6A) correlates with a significant decrease in PPD symptoms (Figure 5). Previous studies using transgenic tobacco overexpressing GPX enzymes already provided evidence that resistance to abiotic stresses correlates with decreased MDA accumulation (Roxas et al., 2000; Yoshimura et al., 2004).
Together, our proteomics data establish that the levels of enzymes in the phenylpropanoid pathway involved in feruloyl-CoA production are generally increased. However, further biochemical analysis will be necessary to determine if this overall modulation of the phenylpropanoid pathway is directed primarily toward an increased production of scopoletin and its glucosylated derivative scopolin that accumulate during PPD (Wheatley and Schwabe, 1985; Buschmann et al., 2000b). Previous studies demonstrated the antioxidant activity of free scopoletin as well as the phenylpropanoid caffeoyl quinate (chlorogenic acid), which is produced by p-coumaroyl shikimate/quinate 3′-hydroxylase (Chong et al., 1999; Niggeweg et al., 2004; Rommens et al., 2008). Thus, phenylpropanoid pathway modulation might also reflect a response to ROS production during PPD (Reilly et al., 2004). Enzymes of the phenylpropanoid pathway and the related suberin pathway up to feruloyl-CoA were generally upregulated (Supplemental Figures 9 and 15). However, the levels of cinnamoyl-CoA reductase, which converts feruloyl-CoA into coniferyl aldehyde, were not significantly changed during PPD progression (Supplemental Figure 15). Coniferyl aldehyde is a precursor of monolignols that are the building blocks of lignin (Vanholme et al., 2010). Enzymes leading to the formation of sinapyl alcohol, a precursor of primary monolignols (Dixon et al., 2001), were either not detected (i.e., coniferyl aldehyde 5-hydroxylase and caffeate O-methyltransferase) or downregulated (cinnamyl alcohol dehydrogenase). Similarly, tyramine N-feruloyltransferase, which converts feruloyl-CoA into N-feruloyltyramine, the major monomer in suberized potato (Solanum tuberosum; King and Calhoun, 2005), was detected, but its abundance was not changed during PPD progression. Our data indicate that the synthesis of lignin and suberin might be limited in cassava storage roots after harvest.
Our quantitative proteomics data also suggest an activation of the ethene pathway during PPD. It remains unclear if the rate-limiting enzyme ACC synthase (Wang et al., 2002) is also upregulated during PPD progression to convert SAM into ACC, because we did not detect ACC synthase in our proteomics study. Previous proteomics studies also failed to detect or quantify ACC synthase in Arabidopsis (Baerenfaller et al., 2011), suggesting that it is a low-abundance enzyme in plants (Kende, 1993; Wang et al., 2002).
In plants, Met is the only known precursor for ethene biosynthesis. Interestingly, proteins involved in the SAM cycle appeared to be positively regulated during PPD (Supplemental Figure 13). Activation of the Met pathway is consistent with the increased biosynthesis of ethene (Supplemental Figure 7) but could also contribute to protein synthesis during PPD (Beeching et al., 1998). The significant increase of three Met adenosyltransferases (Supplemental Figure 13) might reflect the conversion of l-Met to SAM, a donor for methylation reactions.
In addition, our quantitative proteomics data reveal significant regulation of other pathways. For example, the abundances of several enzymes involved in geranylgeranyl diphosphate biosynthesis II are upregulated during PPD. Our quantitative proteomics analysis also reveals the upregulation of the two key enzymes involved in the ent-kaurene biosynthesis pathway, ent-copalyl diphosphate synthase (cassava4.1_001987m) and ent-kaurene synthase (cassava4.1_001987m), which converts geranylgeranyl diphosphate into ent-kaurene (Sun and Kamiya, 1997). A combination of the phytohormones jasmonic acid and ethene was previously shown to have a synergistic role in positively regulating kauralexin accumulation, which is preceded by an increase of the ent-copalyl diphosphate synthase An2 transcript in maize (Zea mays; Schmelz et al., 2011). It remains to be elucidated whether the upregulation of ent-kaurene biosynthesis proteins during PPD has a biological role or is a consequence of ethene accumulation.
Our work demonstrates that combining proteomics analysis with functional validation of protein candidates facilitates the improvement of orphan crops such as cassava. Based on our quantitative proteomics data, we suggest that reducing ethene biosynthesis and increasing enzymes involved in suberization and lignification could further delay PPD onset in cassava roots.
METHODS
Plant Material
Stem-propagated 10-month-old plants of cassava (Manihot esculenta cv 60444) were grown in a greenhouse (16 h of light, 60% humidity, day/night temperatures of 26/17°C), harvested at 10 am, and roots were sliced into 5-mm-thick slices. The root slices were placed on Petri dishes containing filter papers presoaked with 2 mL of distilled water to prevent drying. The sliced roots were then randomized and stored at a constant temperature of 28°C in the dark. For each time point, three replicates consisting of two root slices each were sampled. Slices were taken and frozen in liquid nitrogen after incubation for 0, 6, 12, and 24 h.
Plant Transformation
The transformation vector was designed using pCAMBIA 1301 as a backbone vector. The cauliflower mosaic virus 35S promoter was substituted with a gene fragment containing a 1.0-kb class I patatin promoter from potato (Solanum tuberosum) followed by a 500-bp Arabidopsis thaliana cytosolic GPX. The cDNA encoding cytosolic At-GPX2 (AT2G31570.1) was amplified by RT-PCR from Arabidopsis total RNA using primers 5′-ATGGCGGATGAATCTCCAAAGTC-3′ and 5′-TTAAGAAGAGGCCTGTCCCAAC-3′ and cloned downstream of the patatin promoter to generate the PAT-GPX transformation vector. Transgenic cassava plants were generated according to an optimized transformation procedure (Bull et al., 2009). Transgenic plant analysis and characterization were performed according to procedures described previously (Vanderschuren et al., 2009).
Image Analysis
Root slices were photographed under standard light conditions with a Nikon D700 camera. Image analysis was performed using the PPD Symptom Score Software written in MatLab (The Mathworks) (Supplemental File 1). Photographs of root probes were first interactively marked with a polygon in the image. The software then converted the color image into a gray value image and determined its histogram. To obtain a robust PPD score that is insensitive to changing illumination conditions, and outliers, we used the width of a binary histogram (Kunttu et al., 2003) to calculate the number of occurring gray values. The range of the gray value interval, covering 95% of the occurring gray values, was normalized with the 97.5% quantile to generate a PPD score, which ranges in theory from 0 to 1. Because the root slice method triggers rapid and homogenous PPD symptoms, we observed saturation of the PPD symptom score between 48 and 72 h after harvest. The inner 50% PPD value was automatically calculated based on the gray values occurring in the inner 50% surface area of the initially selected root area.
Protein Extraction
Root slices were ground in liquid nitrogen with a mortar and pestle. Total proteins were extracted from root powder with an SDS extraction buffer (4% [w/v] SDS, 40 mM Tris base, and 2× EDTA-free protease inhibitor [Roche] in a 1:2 ratio [w/v]). Protein extracts were ultracentrifuged at 100,000g for 45 min to remove contaminants, and proteins were quantitated with the BCA Protein Assay Kit (Pierce).
For enzymatic assays, ground tissue powder was mixed in a 3:4 ratio (v/v) with an extraction buffer containing 20 mM HEPES buffer, pH 8.0, 1% polyvinylpyrrolidone, and 1× Complete EDTA-free protease inhibitor (Roche). In the case of the APX assay, 10 mM ascorbic acid was included in the extraction buffer (Amako et al., 1994). For the GPX assay, fresh protein samples were extracted from an independent PPD assay experiment.
For MS, equal amounts of proteins (180 µg) per sample were subjected to 1D SDS-PAGE. After electrophoresis, the gel was stained with Coomassie Brilliant Blue R 250. Each lane, corresponding to one replicate, was cut into five equal gel slices longitudinally, and each gel slice was diced into small pieces. The gel pieces were destained completely and washed prior to an in-gel tryptic digestion according to Shevchenko et al. (1996) with an overnight incubation at 30°C. Peptides were eluted and purified using Sep-Pak reverse-phase cartridges (Waters).
Mass Spectrometry
Samples were analyzed on an LTQ-Orbitrap mass spectrometer (Thermo Fischer Scientific) coupled to an Eksigent-Nano-HPLC system (Eksigent Technologies). Solvent composition at the two channels was 0.2% formic acid and 1% acetonitrile for channel A and 0.2% formic acid and 80% acetonitrile for channel B. Peptides were loaded on a self-made tip column (75 μm × 80 mm) packed with reverse-phase C18 material (AQ, 3 μm, 200 Å; Bischoff) and eluted with a flow rate of 200 nL/min by a gradient from 3 to 15% B in 5 min, 40% B in 50 min, and 97% B in 56 min.
Full-scan MS spectra (300 to 2000 m/z) were acquired with a resolution of 60,000 at 400 m/z after accumulation to a target value of 500,000. Collision-induced dissociation MS/MS spectra were recorded in a data-dependent manner in the ion trap from the five most intense signals above a threshold of 500, using a normalized collision energy of 28% and an activation time of 30 ms. Charge state screening was enabled, and singly charged states were rejected. Precursor masses already selected for MS/MS were excluded for further selection for 60 s, and the exclusion window was set to 20 ppm. The size of the exclusion list was set to a maximum of 500 entries.
Label-Free Proteomics Analysis
Raw data of gel slices with equal molecular weight were loaded together into the commercial software package Progenesis LCMS version 4.0 (Nonlinear Dynamics), a software tool developed for label-free quantification of liquid chromatography–MS data. For data loading, the option High Mass Accuracy Instrument was selected. Liquid chromatography–MS data were normalized and aligned according to the manufacturer’s specifications. In the aligning step, three to five vectors along the retention time gradient were manually seeded to give the automatic alignment a good starting point.
For the annotation of the master map, Mascot generic files (.mgf file format) generated with Progenesis LCMS (using up to five tandem mass spectra for each sequenced feature with the top 200 fragment ion peaks and the deisotoping and charge deconvolution option from Progenesis LCMS) were searched against the cassava 4.1 protein database with known mass spectrometry contaminants attached using the Mascot 2.3 search engine. Parameters for precursor tolerance and fragment ion tolerance were set to ±10 ppm and ±0.6 D, respectively. Carbamidomethylation of Cys was set as fixed modification, and oxidation of Met was set as variable. The Mascot results were exported and filtered. Only rank 1 assignments and assignments matching the term bold red (significant and assigned to the best protein match) were kept. The Mascot ion score cutoff was set at 25, which resulted in a peptide false discovery rate < 1%. For proteins identified with single peptide spectrum match, the annotated spectra are provided in Supplemental Data Set 5. The three biological replicates were grouped for quantification analysis. The Progenesis experiments were first performed individually for each molecular weight fraction and subsequently combined within the software using the combine-fractions approach. Peptide features confidently assigned to proteins are stacked for quantification. The statistical analysis was then performed for each protein on the stacks of normalized feature volumes. The ANOVA statistical test was applied to the selected PPD time points using data from the three biological replicates, and proteins with significant regulation were identified (P < 0.05). We also applied a minimum 1.5-fold change cutoff to the list of regulated proteins to identify proteins with differential regulation. All regulated proteins were identified with at least two peptide spectrum matches.
Cassava proteins were ranked with the T3PQ method, which is based on the linearity between the average of the three most intense MS signals of different tryptic peptides of a given protein and its abundance (Silva et al., 2006; Grossmann et al., 2010).
pep2pro Database
Data import into pep2pro was based on the peptide and protein output files generated with Progenesis (Nonlinear Dynamics). The protein output file contained the quantitative abundance data for each protein, which are graphically represented on the pep2pro website. For each protein, the peptide sequences and corresponding ion scores were retrieved from the peptide output file, and the sample spectra shown on the pep2pro website were read out from the Mascot output file. The display of the cassava proteome data on the pep2pro website relied on the peptide-protein assignment from the Progenesis output, and the peptide and ambiguity filters, which are the basis of the uploading procedure for other search algorithms, were not applied.
Gene Ontology Categories and Pathway Representations
The assignment of protein and transcript functional categories was based on the TAIR Gene Ontology categories from aspect biological process (ATH_GO_GOSLIM_20110301.txt), excluding annotations inferred from electronic annotation (Gene Ontology evidence code IEA). The assignment was performed using the elim algorithm from the topGO package (Alexa et al., 2006).
Quantitative proteomics data for the four PPD time points were submitted to the Plant Metabolic Network (Mueller et al., 2003) to visualize pathways modulated during PPD. Proteins with the highest peptide coverage were selected for graphical representation when multiple homologs appeared regulated.
Assays
PAL Activity
PAL activity was assayed in a PAL assay buffer (0.1 M Tris-HCl buffer, pH 8.9, and 15 mM l-Phe). The reaction was followed at 290 nm for 1 h at 30°C, with readings at 5-min intervals, and the molar extinction coefficient of t-cinnamic acid (i.e., 1.0 × 104 M−1 cm−1) was used for calculation (Rubery and Fosket, 1969). One unit of PAL activity was defined as the amount of enzyme needed to produce 1 nmol of t-cinnamic acid per min.
APX Activity
APX activity was assayed in an APX assay buffer (50 mM potassium phosphate buffer, pH 7.0, 0.5 mM ascorbic acid, 0.2 mM H2O2, and 0.1 mM EDTA). The reaction was followed at 290 nm for 3 min at 25°C, with readings at 30-s intervals, and the molar extinction coefficient of ascorbic acid (2.8 mM−1 cm−1) was used for calculation (Amako et al., 1994). One unit of APX activity was defined as the amount of enzyme oxidizing 1 μmol of ascorbic acid per min at 25°C.
GR Activity
GR activity was assayed in a GR reaction buffer (25 mM sodium phosphate buffer, pH 7.8, 5 mM GSSG, and 1.2 mM NADPH). The reaction was followed at 340 nm for 5 min at 25°C, with readings at 30-s intervals, and the molar extinction coefficient of NADPH (6.22 mM−1 cm−1) was used for calculation (Foyer and Halliwell, 1976). One unit of GR activity was defined as the amount of enzyme oxidizing 1 nmol of NADPH per min at 25°C.
GPX Activity
GPX activity was assayed in a GPX assay buffer (50 mM potassium phosphate buffer, pH 7.0, 1 mM GSH, 1 mM EDTA, 1 mM NaN3, 0.2 mM NADPH, and 0.35 units/mL GR [Sigma-Aldrich]). Samples were preincubated at 30°C to deplete cellular GSSG, and the reaction was initiated by adding 0.25 mM H2O2 using a molar extinction coefficient (6.22 mM−1 cm−1) for calculation. One unit of GPX activity was defined as the amount of enzyme oxidizing 1 nmol of NADPH per min.
GST Activity
GST activity was assayed in a GST assay buffer (0.1 M potassium phosphate buffer, pH 6.5, 10 mM GSH, and 10 mM 1-chloro-2,4-dinitrobenzene [CDNB]). The reaction was followed at 340 nm for 10 min at 25°C, with readings at 30-s intervals, and a molar extinction coefficient of CDNB (9.6 mM−1 cm−1) was used for calculation (Loscalzo and Freedman, 1986). One unit of GST activity was defined as the amount of enzyme needed to conjugate 1 nmol of CDNB per min.
Determination of H2O2 Content and Lipid Peroxidation
H2O2 content was measured spectrophotometrically with potassium iodide according to standard methods established for plant tissues (Velikova et al., 2000). Lipid peroxidation was estimated by the level of MDA production using the thiobarbituric acid method according to previously described protocols (Hodges et al., 1999; Chen and Gallie, 2006).
Accession Numbers
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the PRIDE partner repository (Vizcaíno et al., 2013) with the data set identifier PXD000587.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Symptom Scores of Root Samples Used in the Proteomics Study.
Supplemental Figure 2. Biological Process Categories Overrepresentation in the Proteins Regulated during Postharvest Physiological Deterioration.
Supplemental Figure 3. Ascorbate Peroxidase Enzymatic Activities in Protein Fractions from Collected Time Points.
Supplemental Figure 4. MDA Content of Cassava Root Slices from Collected Time Points.
Supplemental Figure 5. Characterization of Transgenic PAT-GPX Cassava Lines.
Supplemental Figure 6. Characterization of Control and Transgenic Cassava Storage Roots during PPD.
Supplemental Figure 7. Detection and Regulation of Proteins Involved in Ethene Biosynthesis.
Supplemental Figure 8. Detection and Regulation of Proteins Involved in Scopoletin Biosynthesis.
Supplemental Figure 9. Detection and Regulation of Proteins Involved in Phenylpropanoid Biosynthesis.
Supplemental Figure 10. Enzymatic Activities of PAL in Protein Fractions from Collected Time Points.
Supplemental Figure 11. Detection and Regulation of Proteins Involved in the Fatty Acid α-Oxidation Pathway.
Supplemental Figure 12. Detection and Regulation of Proteins Involved in Folate Transformation II.
Supplemental Figure 13. Detection and Regulation of Proteins Involved in SAM Cycle II.
Supplemental Figure 14. Detection and Regulation of Proteins Involved in Sulfate Reduction II.
Supplemental Figure 15. Detection and Regulation of Proteins Involved in Suberin Biosynthesis.
The following materials have been deposited in the DRYAD repository under accession number http://dx.doi.org/10.5061/dryad.6f48r.
Supplemental Data Set 1. Proteins Identified in Cassava Samples from Four PPD Time Points.
Supplemental Data Set 2. Proteins Identified with No Conflicting Peptides in Cassava Samples from Four PPD Time Points.
Supplemental Data Set 3. T3PQ-Based Protein Abundances of Identified Proteins in Cassava Roots at 0 h.
Supplemental Data Set 4. Label-Free MS1-Based Quantitative Data and Statistical Analysis of Cassava Proteins Detected in Cassava Samples from Four PPD Time Points.
Supplemental Data Set 5. List of Proteins Identified with Single Peptide Spectrum Match and Their Corresponding Spectra.
Supplemental File 1. PPD Symptom Score Software.
Supplementary Material
Acknowledgments
We thank Judith Owiti for initiating the research project on PPD and Irene Zurkirchen (Eidgenössisch Technische Hochschule Zurich) for special care of the cassava plants. We thank the Institute of Complex Systems Biomechanik, Forschungszentrum Jülich, for providing initial access to and training on the MatLab package to N.K. This work was supported by ETH Zurich and by the Bill & Melinda Gates Foundation (BioCassava Plus Program Phase 1 trainee fellowship to E.N.).
AUTHOR CONTRIBUTIONS
H.V., E.N., and J.S.P. designed research. H.V., E.N., J.S.P., and P.N. performed research. H.V., E.N., J.S.P., K.B., and J.G. analyzed the data. N.K. and M.H.-H. contributed new computational tools. H.V. wrote the article. W.G. edited the article.
Glossary
- PPD
postharvest physiological deterioration
- MS/MS
tandem mass spectrometry
- ROS
reactive oxygen species
- MS
mass spectrometry
- ACC
1-aminocyclopropane-1-carboxylate
- SAM
S-adenosyl-l-methionine
- CDNB
1-chloro-2,4-dinitrobenzene
- MDA
malondialdehyde
- T3PQ
top-three protein quantification
Footnotes
Some figures in this article are displayed in color online but in black and white in the print edition.
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Achidi A.U., Ajayi O.A., Bokanga M., Maziya-Dixon B. (2005). The use of cassava leaves as food in Africa. Ecol. Food Nutr. 44: 423–435 [Google Scholar]
- Alexa A., Rahnenführer J., Lengauer T. (2006). Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics 22: 1600–1607 [DOI] [PubMed] [Google Scholar]
- Amako K., Chen G.X., Asada K. (1994). Separate assays specific for ascorbate peroxidase and guaiacol peroxidase and for the chloroplastic and cytosolic isozymes of ascorbate peroxidase in plants. Plant Cell Physiol. 35: 497–504 [Google Scholar]
- Baba A.I., Nogueira F.C.S., Pinheiro C.B., Brasil J.N., Jereissati E.S., Juca T.L., Soares A.A., Santos M.F., Domont G.B., Campos F.A.P. (2008). Proteome analysis of secondary somatic embryogenesis in cassava (Manihot esculenta). Plant Sci. 175: 717–723 [Google Scholar]
- Baerenfaller K., Hirsch-Hoffmann M., Svozil J., Hull R., Russenberger D., Bischof S., Lu Q., Gruissem W., Baginsky S. (2011). pep2pro: A new tool for comprehensive proteome data analysis to reveal information about organ-specific proteomes in Arabidopsis thaliana. Integr. Biol. (Camb.) 3: 225–237 [DOI] [PubMed] [Google Scholar]
- Bayoumi S.A., Rowan M.G., Beeching J.R., Blagbrough I.S. (2010). Constituents and secondary metabolite natural products in fresh and deteriorated cassava roots. Phytochemistry 71: 598–604 [DOI] [PubMed] [Google Scholar]
- Beeching J.R., Han Y.H., Gomez-Vasquez R., Day R.C., Cooper R.M. (1998). Wound and defense responses in cassava as related to post-harvest physiological deterioration. Recent Adv. Phytochem. 32: 231–248 [Google Scholar]
- Booth R.H. (1976). Storage of fresh cassava (Manihot esculenta). 1. Post-harvest deterioration and its control. Exp. Agric. 12: 103 [Google Scholar]
- Bull S.E., Owiti J.A., Niklaus M., Beeching J.R., Gruissem W., Vanderschuren H. (2009). Agrobacterium-mediated transformation of friable embryogenic calli and regeneration of transgenic cassava. Nat. Protoc. 4: 1845–1854 [DOI] [PubMed] [Google Scholar]
- Buschmann H., Reilly K., Rodriguez M.X., Tohme J., Beeching J.R. (2000a). Hydrogen peroxide and flavan-3-ols in storage roots of cassava (Manihot esculenta Crantz) during postharvest deterioration. J. Agric. Food Chem. 48: 5522–5529 [DOI] [PubMed] [Google Scholar]
- Buschmann H., Rodriguez M.X., Tohme J., Beeching J.R. (2000b). Accumulation of hydroxycoumarins during post-harvest deterioration of tuberous roots of cassava (Manihot esculenta Crantz). Ann. Bot. (Lond.) 86: 1153–1160 [Google Scholar]
- Chen Z., Gallie D.R. (2006). Dehydroascorbate reductase affects leaf growth, development, and function. Plant Physiol. 142: 775–787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong J., Baltz R., Fritig B., Saindrenan P. (1999). An early salicylic acid-, pathogen- and elicitor-inducible tobacco glucosyltransferase: Role in compartmentalization of phenolics and H2O2 metabolism. FEBS Lett. 458: 204–208 [DOI] [PubMed] [Google Scholar]
- De León I.P., Sanz A., Hamberg M., Castresana C. (2002). Involvement of the Arabidopsis alpha-DOX1 fatty acid dioxygenase in protection against oxidative stress and cell death. Plant J. 29: 61–62 [DOI] [PubMed] [Google Scholar]
- Dixon R.A., Chen F., Guo D., Parvathi K. (2001). The biosynthesis of monolignols: A “metabolic grid”, or independent pathways to guaiacyl and syringyl units? Phytochemistry 57: 1069–1084 [DOI] [PubMed] [Google Scholar]
- El-Sharkawy M.A. (2006). International research on cassava photosynthesis, productivity, eco-physiology, and responses to environmental stresses in the tropics. Photosynthetica 44: 481–512 [Google Scholar]
- Fermont A.M., van Asten P.J.A., Tittonell P., van Wijk M.T., Giller K.E. (2009). Closing the cassava yield gap: An analysis from smallholder farms in East Africa. Field Crops Res. 112: 24–36 [Google Scholar]
- Foyer C.H., Halliwell B. (1976). The presence of glutathione and glutathione reductase in chloroplasts: A proposed role in ascorbic acid metabolism. Planta 133: 21–25 [DOI] [PubMed] [Google Scholar]
- Fujiwara T., Nambara E., Yamagishi K., Goto D.B., Naito S. (2002). Storage proteins. The Arabidopsis Book 1: e0020, /10.1199/tab.0020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossmann J., Roschitzki B., Panse C., Fortes C., Barkow-Oesterreicher S., Rutishauser D., Schlapbach R. (2010). Implementation and evaluation of relative and absolute quantification in shotgun proteomics with label-free methods. J. Proteomics 73: 1740–1746 [DOI] [PubMed] [Google Scholar]
- Han Y.H., Gomez-Vasquez R., Reilly K., Li H.Y., Tohme J., Cooper R.M., Beeching J.R. (2001). Hydroxyproline-rich glycoproteins expressed during stress responses in cassava. Euphytica 120: 59–70 [Google Scholar]
- Hanson A.D., Roje S. (2001). One-carbon metabolism in higher plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52: 119–137 [DOI] [PubMed] [Google Scholar]
- Hirose S., Data E.S., Tanaka Y., Uritani I. (1984). Physiological deterioration and ethylene production in cassava roots after harvest, in relation with pruning treatment. Jpn. J. Crop Sci. 53: 282–289 [Google Scholar]
- Hirsch-Hoffmann M., Gruissem W., Baerenfaller K. (2012). pep2pro: The high-throughput proteomics data processing, analysis, and visualization tool. Front. Plant Sci. 3: 123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodges D.M., DeLong J.M., Forney C.F., Prange R.K. (1999). Improving the thiobarbituric acid-reactive-substances assay for estimating lipid peroxidation in plant tissues containing anthocyanin and other interfering compounds. Planta 207: 604–611 [DOI] [PubMed] [Google Scholar]
- Huang J., Bachem C., Jacobsen E., Visser R.G.F. (2001). Molecular analysis of differentially expressed genes during postharvest deterioration in cassava (Manihot esculenta Crantz) tuberous roots. Euphytica 120: 85–93 [Google Scholar]
- Iqbal A., Yabuta Y., Takeda T., Nakano Y., Shigeoka S. (2006). Hydroperoxide reduction by thioredoxin-specific glutathione peroxidase isoenzymes of Arabidopsis thaliana. FEBS J. 273: 5589–5597 [DOI] [PubMed] [Google Scholar]
- Kai K., Mizutani M., Kawamura N., Yamamoto R., Tamai M., Yamaguchi H., Sakata K., Shimizu B. (2008). Scopoletin is biosynthesized via ortho-hydroxylation of feruloyl CoA by a 2-oxoglutarate-dependent dioxygenase in Arabidopsis thaliana. Plant J. 55: 989–999 [DOI] [PubMed] [Google Scholar]
- Kende H. (1993). Ethylene biosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 44: 283–307 [Google Scholar]
- King R.R., Calhoun L.A. (2005). Characterization of cross-linked hydroxycinnamic acid amides isolated from potato common scab lesions. Phytochemistry 66: 2468–2473 [DOI] [PubMed] [Google Scholar]
- Kunttu, L., Lepisto, L., Rauhamaa, J., and Visa, A. (2003). Binary histogram in image classification for retrieval purposes. In Journal of WSCG, No. 1, WSCG'2003, February 3-7, 2003 (Plzen, Czech Republic: Copyright UNION Agency, Science Press), pp. 269–273. [Google Scholar]
- Kwon S.Y., Choi S.M., Ahn Y.O., Lee H.S., Lee H.B., Park Y.M., Kwak S.S. (2003). Enhanced stress-tolerance of transgenic tobacco plants expressing a human dehydroascorbate reductase gene. J. Plant Physiol. 160: 347–353 [DOI] [PubMed] [Google Scholar]
- Lobell D.B., Burke M.B., Tebaldi C., Mastrandrea M.D., Falcon W.P., Naylor R.L. (2008). Prioritizing climate change adaptation needs for food security in 2030. Science 319: 607–610 [DOI] [PubMed] [Google Scholar]
- Loscalzo J., Freedman J. (1986). Purification and characterization of human platelet glutathione-S-transferase. Blood 67: 1595–1599 [PubMed] [Google Scholar]
- Mitprasat M., Roytrakul S., Jiemsup S., Boonseng O., Yokthongwattana K. (2011). Leaf proteomic analysis in cassava (Manihot esculenta, Crantz) during plant development, from planting of stem cutting to storage root formation. Planta 233: 1209–1221 [DOI] [PubMed] [Google Scholar]
- Montagnac J.A., Davis C.R., Tanumihardjo S.A. (2009). Nutritional value of cassava for use as a staple food and recent advances for improvement. Comprehensive Reviews in Food Science and Food Safety 8: 181–194 [DOI] [PubMed] [Google Scholar]
- Mueller L.A., Zhang P., Rhee S.Y. (2003). AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 132: 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niggeweg R., Michael A.J., Martin C. (2004). Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 22: 746–754 [DOI] [PubMed] [Google Scholar]
- Owiti J., Grossmann J., Gehrig P., Dessimoz C., Laloi C., Hansen M.B., Gruissem W., Vanderschuren H. (2011). iTRAQ-based analysis of changes in the cassava root proteome reveals pathways associated with post-harvest physiological deterioration. Plant J. 67: 145–156 [DOI] [PubMed] [Google Scholar]
- Prochnik S., Marri P.R., Desany B., Rabinowicz P.D., Kodira C., Mohiuddin M., Rodriguez F., Fauquet C., Tohme J., Harkins T., Rokhsar D.S., Rounsley S. (2012). The cassava genome: Current progress, future directions. Trop. Plant Biol. 5: 88–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raju P.D.R., Neelima G. (2012). Image segmentation by using histogram tresholding. International Journal of Computer Science Engineering and Technology 2: 776–779 [Google Scholar]
- Reilly K., Bernal D., Cortés D.F., Gómez-Vásquez R., Tohme J., Beeching J.R. (2007). Towards identifying the full set of genes expressed during cassava post-harvest physiological deterioration. Plant Mol. Biol. 64: 187–203 [DOI] [PubMed] [Google Scholar]
- Reilly K., Gómez-Vásquez R., Buschmann H., Tohme J., Beeching J.R. (2004). Oxidative stress responses during cassava post-harvest physiological deterioration. Plant Mol. Biol. 56: 625–641 [DOI] [PubMed] [Google Scholar]
- Reilly K., Han Y., Tohme J., Beeching J.R. (2001). Isolation and characterisation of a cassava catalase expressed during post-harvest physiological deterioration. Biochim. Biophys. Acta 1518: 317–323 [DOI] [PubMed] [Google Scholar]
- Rickard J.E. (1985). Physiological deterioration of cassava roots. J. Sci. Food Agric. 36: 167–176 [Google Scholar]
- Rodriguez Milla M.A., Maurer A., Rodriguez Huete A., Gustafson J.P. (2003). Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 36: 602–615 [DOI] [PubMed] [Google Scholar]
- Rommens C.M., Richael C.M., Yan H., Navarre D.A., Ye J., Krucker M., Swords K. (2008). Engineered native pathways for high kaempferol and caffeoylquinate production in potato. Plant Biotechnol. J. 6: 870–886 [DOI] [PubMed] [Google Scholar]
- Roxas V.P., Lodhi S.A., Garrett D.K., Mahan J.R., Allen R.D. (2000). Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase. Plant Cell Physiol. 41: 1229–1234 [DOI] [PubMed] [Google Scholar]
- Rubery P.H., Fosket D.E. (1969). Changes in phenylalanine ammonia-lyase activity during xylem differentiation in Coleus and soybean. Planta 87: 54–62 [DOI] [PubMed] [Google Scholar]
- Saito K. (2004). Sulfur assimilatory metabolism: The long and smelling road. Plant Physiol. 136: 2443–2450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez T., Chavez A.L., Ceballos H., Rodriguez-Amaya D., Nestel P., Ishitani M. (2006). Reduction or delay of post-harvest physiological deterioration in cassava roots with higher carotenoid content. J. Sci. Food Agric. 86: 634–639 [Google Scholar]
- Sayre R., et al. (2011). The BioCassava Plus Program: Biofortification of cassava for sub-Saharan Africa. Annu. Rev. Plant Biol. 62: 251–272 [DOI] [PubMed] [Google Scholar]
- Schmelz E.A., Kaplan F., Huffaker A., Dafoe N.J., Vaughan M.M., Ni X., Rocca J.R., Alborn H.T., Teal P.E. (2011). Identity, regulation, and activity of inducible diterpenoid phytoalexins in maize. Proc. Natl. Acad. Sci. USA 108: 5455–5460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield J., Taylor N., Fauquet C., Chen S. (2006). The cassava (Manihot esculenta Crantz) root proteome: Protein identification and differential expression. Proteomics 6: 1588–1598 [DOI] [PubMed] [Google Scholar]
- Shevchenko A., Wilm M., Vorm O., Mann M. (1996). Mass spectrometric sequencing of proteins silver-stained polyacrylamide gels. Anal. Chem. 68: 850–858 [DOI] [PubMed] [Google Scholar]
- Shewry P.R. (2003). Tuber storage proteins. Ann. Bot. (Lond.) 91: 755–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva J.C., Gorenstein M.V., Li G.Z., Vissers J.P., Geromanos S.J. (2006). Absolute quantification of proteins by LCMSE: A virtue of parallel MS acquisition. Mol. Cell. Proteomics 5: 144–156 [DOI] [PubMed] [Google Scholar]
- Stupak M., Vanderschuren H., Gruissem W., Zhang P. (2006). Biotechnological approaches to cassava protein improvement. Trends Food Sci. Technol. 17: 634–641 [Google Scholar]
- Sun T.P., Kamiya Y. (1997). Regulation and cellular localization of ent-kaurene synthesis. Physiol. Plant. 101: 701–708 [Google Scholar]
- Tanaka Y., Data E.S., Hirose S., Taniguchi T., Uritani I. (1983). Biochemical changes in secondary metabolites in wounded and deteriorated cassava roots. Agric. Biol. Chem. 47: 693–700 [Google Scholar]
- Vanderschuren H., Alder A., Zhang P., Gruissem W. (2009). Dose-dependent RNAi-mediated geminivirus resistance in the tropical root crop cassava. Plant Mol. Biol. 70: 265–272 [DOI] [PubMed] [Google Scholar]
- Vanderschuren H., Lentz E., Zainuddin I., Gruissem W. (2013). Proteomics of model and crop plant species: Status, current limitations and strategic advances for crop improvement. J. Proteomics 93: 5–19 [DOI] [PubMed] [Google Scholar]
- Vanholme R., Demedts B., Morreel K., Ralph J., Boerjan W. (2010). Lignin biosynthesis and structure. Plant Physiol. 153: 895–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velikova V., Yordanov I., Edreva A. (2000). Oxidative stress and some antioxidant systems in acid rain-treated bean plants: Protective role of exogenous polyamines. Plant Sci. 151: 59–66 [Google Scholar]
- Vizcaíno J.A., et al. (2013). The PRoteomics IDEntifications (PRIDE) database and associated tools: Status in 2013. Nucleic Acids Res. 41: D1063–D1069 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogt T. (2010). Phenylpropanoid biosynthesis. Mol. Plant 3: 2–20 [DOI] [PubMed] [Google Scholar]
- Wang K.L., Li H., Ecker J.R. (2002). Ethylene biosynthesis and signaling networks. Plant Cell 14 (suppl.): S131–S151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenham, J.E. (1995). Postharvest Deterioration of Cassava: A Biotechnological Perspective. FAO Plant Production and Protection Paper 130. (Rome: FAO). [Google Scholar]
- Wheatley C.C., Schwabe W.W. (1985). Scopoletin involvement in post-harvest physiological deterioration of cassava root (Manihot esculenta Crantz). J. Exp. Bot. 36: 783–791 [Google Scholar]
- Xu J., Duan X., Yang J., Beeching J.R., Zhang P. (2013). Enhanced reactive oxygen species scavenging by overproduction of superoxide dismutase and catalase delays postharvest physiological deterioration of cassava storage roots. Plant Physiol. 161: 1517–1528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Wang S., Eltayeb A.E., Uddin M.I., Yamamoto Y., Tsuji W., Takeuchi Y., Tanaka K. (2010). Overexpression of dehydroascorbate reductase, but not monodehydroascorbate reductase, confers tolerance to aluminum stress in transgenic tobacco. Planta 231: 609–621 [DOI] [PubMed] [Google Scholar]
- Yoshimura K., Miyao K., Gaber A., Takeda T., Kanaboshi H., Miyasaka H., Shigeoka S. (2004). Enhancement of stress tolerance in transgenic tobacco plants overexpressing Chlamydomonas glutathione peroxidase in chloroplasts or cytosol. Plant J. 37: 21–33 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.