This work compares the genomes of four Brassicaceae species to examine the patterns of gene gains and losses following whole-genome duplication, finding that retained genes showed substantial divergence in sequence, expression, function, and network connectivity. This information was used to establish a statistical learning model for predicting whether a duplicate would be retained postpolyploidization.
Abstract
Polyploidization events are frequent among flowering plants, and the duplicate genes produced via such events contribute significantly to plant evolution. We sequenced the genome of wild radish (Raphanus raphanistrum), a Brassicaceae species that experienced a whole-genome triplication event prior to diverging from Brassica rapa. Despite substantial gene gains in these two species compared with Arabidopsis thaliana and Arabidopsis lyrata, ∼70% of the orthologous groups experienced gene losses in R. raphanistrum and B. rapa, with most of the losses occurring prior to their divergence. The retained duplicates show substantial divergence in sequence and expression. Based on comparison of A. thaliana and R. raphanistrum ortholog floral expression levels, retained radish duplicates diverged primarily via maintenance of ancestral expression level in one copy and reduction of expression level in others. In addition, retained duplicates differed significantly from genes that reverted to singleton state in function, sequence composition, expression patterns, network connectivity, and rates of evolution. Using these properties, we established a statistical learning model for predicting whether a duplicate would be retained postpolyploidization. Overall, our study provides new insights into the processes of plant duplicate loss, retention, and functional divergence and highlights the need for further understanding factors controlling duplicate gene fate.
INTRODUCTION
All angiosperms are polyploids or have experienced a polyploidization event in their recent evolutionary history (Ramsey and Schemske, 1998; Jiao et al., 2011), resulting in multiplication of their gene content. These duplicated genes may remain functionally redundant briefly but will eventually be retained due to new function gains (Ohno, 1970) or due to partitioning of ancestral functions (Force et al., 1999) or will be lost through deletion or other processes leading to pseudogenization (Li et al., 1981). Aside from these mechanisms, the retention of duplicate genes may also be attributed to the selection for balanced gene drive/gene balance (Freeling and Thomas, 2006; Birchler and Veitia, 2007), functional buffering (Chapman et al., 2006), dosage selection (Conant and Wolfe, 2008), and/or escape from adaptive conflict (Des Marais and Rausher, 2008; reviewed in Edger and Pires, 2009; Innan and Kondrashov, 2010). The retention of duplicates, especially those derived from polyploidization, is correlated with certain gene functions (Blanc and Wolfe, 2004; Hanada et al., 2008), gene complexity (Chapman et al., 2006; Jiang et al., 2013), levels of gene expression (Pál et al., 2001), parental genome dominance (Chang et al., 2010; Schnable et al., 2011), and network connectivity (Thomas et al., 2006). Despite correlations of these features with duplicate retention, it remains unclear to what extent these features may explain duplicate retention. This issue can be addressed in greater detail in Brassicaceae due to the close evolutionary relationship between species in the Brassiceae tribe, including wild radish (Raphanus raphanistrum) and Brassica rapa, and those in the Arabidopsis genus (diverged 43 million years ago [mya]; Beilstein et al., 2010). Also, a broad range of molecular data in Arabidopsis thaliana can be used to infer the potential roles of Brassiceae duplicates. In addition, there is a recent hexaploidization event in the Brassiceae lineage (Lagercrantz and Lydiate, 1996), allowing a closer look at the patterns of duplicate loss and retention.
In Brassicaceae, studies of duplicate genes in A. thaliana suggest three rounds of whole-genome duplication (WGD) occurred after its lineage diverged from the monocot lineage. The most recent WGD event (α) occurred 50 to 65 mya (Bowers et al., 2003; Beilstein et al., 2010), prior to the divergence of species in the Brassicaceae family. Notably, a further hexaploidization event (hereafter referred to as the α’ whole-genome triplication [WGT] event) occurred recently in the common ancestor of Brassica and Raphanus (Lagercrantz and Lydiate, 1996; Lysak et al., 2005; Yang et al., 2006; Town et al., 2006; Wang et al., 2011). Among Brassiceae species, much of the knowledge about the evolution of α’ duplicates is derived from species in the Brassica genus (Wang et al., 2011). Since the α’ WGT, >50% of the Brassica duplicates may have been lost via deletion and pseudogenization, some of which has occurred in a biased fashion (Wang et al., 2011; Tang et al., 2012). These findings provide a baseline understanding of duplicate evolution post WGT and raise additional questions regarding rate of pseudogenization of duplicate genes and patterns of expression divergence.
R. raphanistrum is native to the Mediterranean region and is a close relative of the cultivated radish (Raphanus sativus). The wild radish has evolved a weedy form that is a global agricultural pest (Warwick and Francis, 2005), and it is also a model for the study of ecology and evolution (Sahli et al., 2008; Conner et al., 2009). Thus, availability of genomic and transcriptomic resources for Raphanus will contribute to a better understanding of the molecular basis and evolutionary characteristics of weediness and aid in improvement of cultivated radish. In addition, these resources enable comparative genomic and transcriptomic analyses between Raphanus, Brassica, and Arabidopsis species to understand evolution of duplicate genes post α’ WGT. In this study, we report the draft assembly and annotation of the Raphanus genome and ask four major questions. First, what are the patterns of gene loss and retention post α’ WGT in Brassica and Raphanus? Second, how may pseudogenes in Brassica and Raphanus genomes provide information on gene death post triplication? Third, what is the extent of duplicate gene expression divergence? Finally, can we computationally predict which genes would be retained or lost after duplication? Our results suggest that the patterns of evolution of α’ duplicates are similar in Brassica and Raphanus and that the retention process possesses biases that can be exploited for computationally predicting whether a duplicate would be lost or retained post polyploidization.
RESULTS AND DISCUSSION
Sequencing and Assembly of the Wild Radish Genome
As the first step in generating a draft assembly for the R. raphanistrum (referred to as Raphanus) genome, we estimated the genome size of Raphanus using flow cytometry. The estimated size of 515 Mb is comparable to genome size estimates of related species, including Brassica (529 Mb), Brassica oleraceae (696 Mb), and R. sativus (573 Mb) (Johnston et al., 2005). We sequenced the genome of a 5th generation inbred plant, and the reads were assembled with a hybrid approach (Supplemental Figure 1). The final assembly size was 254 Mb, representing 49.3% of the estimated genome size, with an N50 of 10.1 kb (Table 1). This is comparable to the draft B. rapa (referred to as Brassica) genome where the assembly is 283.8 Mb, or 53.7% of the estimated genome size, despite its significantly better sequencing coverage of 72× (Wang et al., 2011). The size of the euchromatic space in Brassica is estimated to be ∼220 Mb (Mun et al., 2009). In addition, ∼30% of all Brassica chromosomes are composed of centromeric repeats that occupy ∼50% of all heterochromatic domains (Lim et al., 2007). Assuming that most of this heterochromatin consists of repetitive, nongenic regions and Raphanus is similar to Brassica in its heterochromatin content, it is likely that we captured most of the nonrepetitive genome space in our Raphanus assembly.
Table 1. Comparison between Raphanus and Brassica Assemblies.
| R. raphanistrum Contigs | B. rapa Contigs | B. rapa Scaffolds | |
|---|---|---|---|
| Assembly size | 254.6 Mb | 264.1 Mb | 283.8 Mb |
| Number of contigs (>100 bp) | 68,331 | 60,521 | 40,549 |
| N50 | 10.1 kb | 27.3 kb | 1.9 Mb |
| Coverage of core eukaryotic genesa | 97.2% | NA | 100.0% |
| % Unique transcripts mapping to assemblyb | 94.2% | NA | 96.9% |
NA, not available.
Conservation of 248 core eukaryotic Genes was determined using the core eukaryotic gene mapping approach program (Parra et al., 2007).
163,862 Raphanus and 213,105 Brassica ESTs from NCBI dbEST were assembled into 106,052 and 85,508 unique transcripts (see Methods).
Using the MAKER annotation pipeline (Cantarel et al., 2008), we predicted 38,174 protein coding genes in the Raphanus assembly (Supplemental Figures 2A and 2B). The coverage of the gene space in our Raphanus and the published Brassica assembly was further assessed using ESTs and using the core eukaryotic gene mapping approach (Parra et al., 2007). We found that 96.9 and 94.2% of the Brassica and Raphanus unique ESTs could be mapped to their respective assemblies (Table 1). In addition, the Brassica and Raphanus assemblies contained complete matches for 248 (100%) and 241 (97.2%) core eukaryotic genes, respectively (Table 1). These observations suggest that the Raphanus assembly is less complete and more fragmented than Brassica. However, a significant proportion of the gene space in Raphanus is covered in the draft assembly. For further comparisons of the gene space across species, we employed a combination of similarity and synteny-based approaches to define orthologous groups (OGs) between A. thaliana, Arabidopsis lyrata, Brassica, and Raphanus protein-coding genes (Supplemental Figure 3).
Timing the Speciation and Polyploidization Events in Brassicaceae
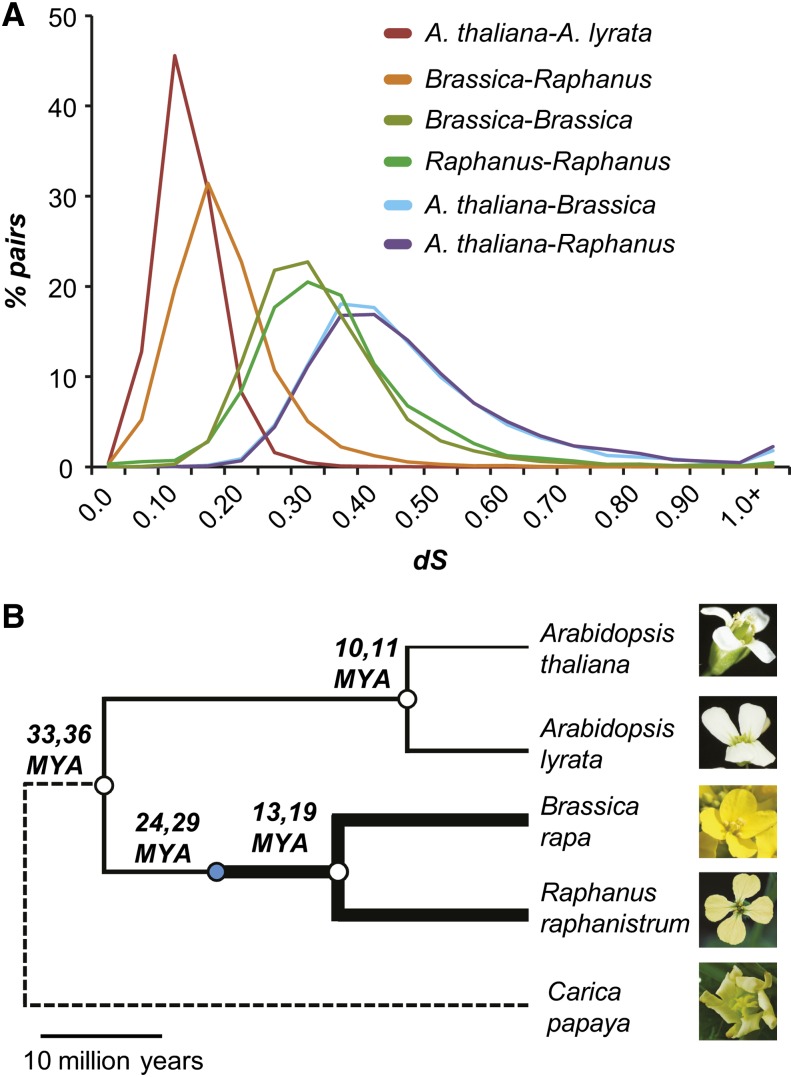
To understand the patterns of duplicate gene evolution before and after the genome triplication event, it is important to know the Brassica-Raphanus speciation time in relation to the timing of the α’ WGT and A. thaliana–Brassica speciation events. Previous studies have suggested a broad range of timings for speciation and the WGT events in the Brassicaceae family (Supplemental Figure 4A), and some of these estimates have been revised based on availability of new data (Beilstein et al., 2010). Due to inconsistent times and methodological differences between these studies, we used the most recent published data to reestimate the timing of the α’ WGT event and the timing of the Brassica-Raphanus speciation event. Using two dating methods, the first based on synonymous substitution rate (dS) and the second based on Bayesian approximation, the median divergence times between Brassica and Raphanus were estimated to be 13 and 19 mya, respectively (Figure 1A). These estimates are older than the predicted divergence of A. thaliana and A. lyrata (10 and 11 mya) and more recent than the divergence time between the A. thaliana–Brassica lineages (32 and 36 mya) (Figure 1B; Supplemental Table 1).
Figure 1.
Synonymous Substitution Rate (dS) and Relationships between Brassicaceae Species.
(A) dS between ortholog pairs and between paralogs derived from α’ WGT among Brassicaceae species.
(B) Timing of polyploidization (blue circle) and speciation (open circles) events. The first and second numbers corresponding to each event are estimated based on the dS and Bayesian dating approaches, respectively. Thickness of the solid lines corresponds to the genome size. (The image for A. lyrata is used with permission, ©Ya-Long Guo, Max Planck Institute for Developmental Biology.)
These estimates are significantly older than some of the previous estimates (13 to 24 mya for A. thaliana–Brassica split) for two reasons. First, the Bayesian prior and the lower limit for the divergence time between A. thaliana and Brassica/Raphanus lineages we used is based on the most recent fossil data (Beilstein et al., 2010). The second reason is that we used a lower but more recent neutral substitution rate estimate (Ossowski et al., 2010) than some of the earlier studies (Supplemental Figure 4B). Using α’ WGT-derived Brassica and Raphanus duplicates, we estimated that the WGT event took place 24 and 29 mya (Figure 1B). Our estimates are in agreement with some previous studies, which estimated the A. thaliana–Brassica split to have occurred 33 to 43 mya and the WGT event 22 to 29 mya (Town et al., 2006; Beilstein et al., 2010; Couvreur et al., 2010). Taken together, our results suggest that the polyploidization event likely occurred 3 to 12 million years after the separation of the A. thaliana–Brassica lineages and that the Raphanus genus may have been diverging from Brassica for a longer time than previously estimated (Yang et al., 2002; Lysak et al., 2005). In addition, the α’ duplicates may have 5 to 16 mya of shared ancestry, followed by 13 to 19 mya of independent evolution in Brassica and Raphanus.
Patterns of Loss and Retention of Duplicate Genes Post α’ WGT
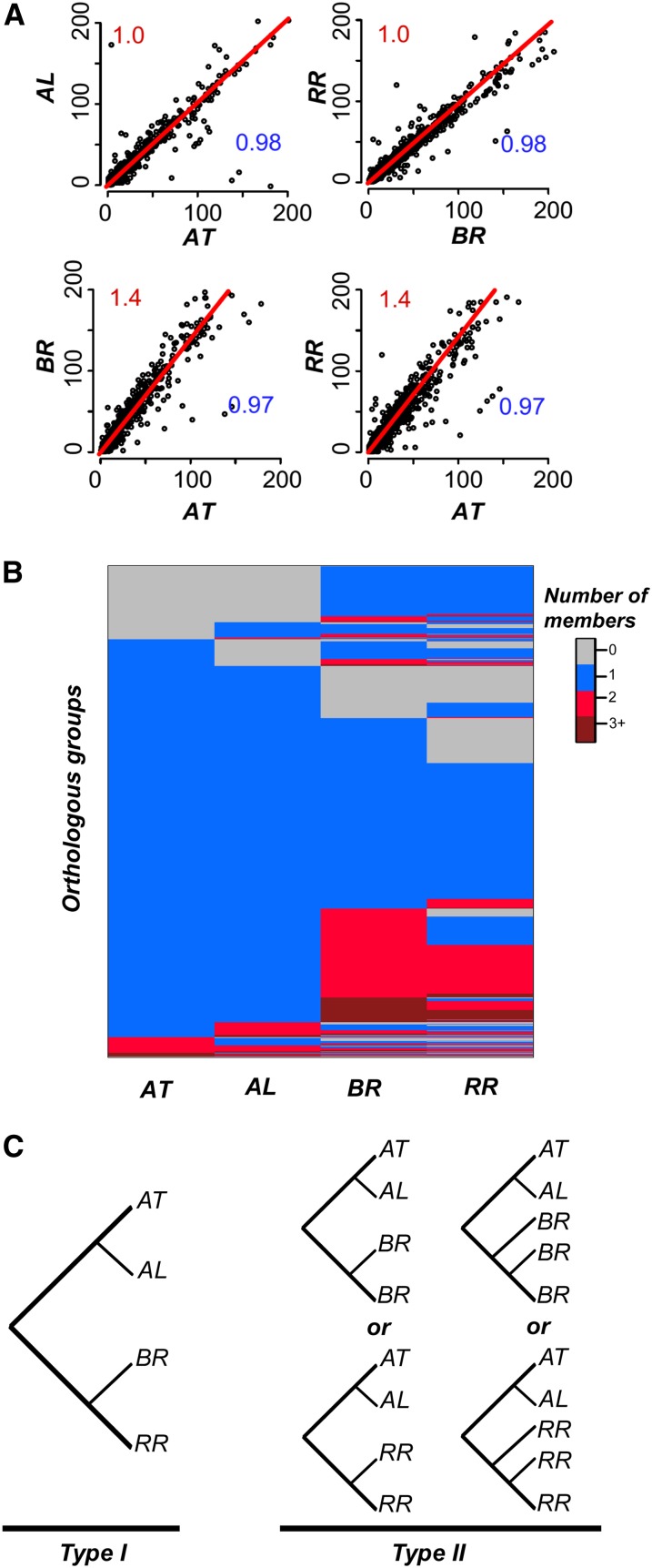
A. thaliana and A. lyrata have 27,416 and 32,670 annotated protein-coding genes, respectively. Assuming that the common ancestor of A. thaliana/A. lyrata/Brassica/Raphanus had ∼30,000 genes, the α’ event should have created ∼90,000 genes. Considering that there are 41,174 Brassica and 38,174 Raphanus annotated genes, only ∼42 to 45% genes from the ancestral hexaploid have been retained. The extent of gene loss is evident at the protein domain level because there are, on average, 1.4 times more domain family members in both Brassica and Raphanus versus Arabidopsis species instead of the expected three times more (Figure 2A). Next, we examined the patterns of duplicate gene retention at the level of OGs, where each OG specifies one ancestral gene that existed prior to the divergence of the Brassiceae species examined. We identified 16,567 OGs containing high-confidence Brassica and Raphanus genes derived from the α’ WGT event (Supplemental Figure 3). Based on these OG definitions, both the Brassica and Raphanus lineages have experienced gene losses in ∼70% of the OGs, returning them to a singleton or complete gene loss state (Figure 2B). Among 10,521 and 8871 OGs returned to a singleton state in Brassica and Raphanus, respectively, 6235 (70.3%) OGs overlap, which is significantly higher than random expectation (Fisher’s exact test P < 1e-16).
Figure 2.
Patterns of α' Duplicate Evolution.
(A) Comparison of PFAM domain family sizes between species pairs. Each dot corresponds to the number of genes possessing a particular PFAM domain. The numbers in red and blue indicate the slope of the best fit line (red line) and the R2 value, respectively.
(B) Comparison of OG sizes between the four species. Each row indicates the number of genes from each of the four species (column) in an OG.
(C) Schematic representations of Type I and Type II OGs. AT, Arabidopsis thaliana; AL, Arabidopsis lyrata; BR, Brassica rapa; RR, Raphanus raphanistrum.
The presence of such common singletons may be due to common gene losses in the ancestral branch leading up to the Brassica-Raphanus last common ancestor or independent, parallel losses in these two lineages post speciation. To distinguish between these two possibilities, information from the Brassica homoeologous blocks (Wang et al., 2011) was jointly analyzed with Brassica-Raphanus ortholog assignments. In the previous study, 27,774 Brassica genes were assigned to 22,546 orthologous and homoeologous relationships (Wang et al., 2011), of which 21,170 (76.6%) genes in 19,036 relationships are common to our stringently defined set of homologs. Each homoeologous relationship represents one ancestral gene prior to genome triplication; thus, at least 35,938 (19,036 times 3 minus 21,170) and up to 49,000 (30,000 times 3 minus 41,000) genes were lost from the Brassica genome since WGT. Of the 21,170 retained Brassica genes, 2912 (13.7%) genes do not have Raphanus orthologs. The likely explanation is that independent losses of these Raphanus orthologs took place post Brassica-Raphanus speciation. Based on this relaxed definition of independent loss, we estimate that as many as 86% of the losses may have occurred in the shared lineage of both species. Because the criterion for calling independent losses is relaxed, this estimate of losses in the shared lineage may be considered an upper limit.
To address the question of whether OGs with retained duplicates have distinct properties from those with singleton genes, we classified the OGs into three types (Figure 2C). The type I OGs (2534) contain only one member from A. thaliana and A. lyrata and one member from Brassica and Raphanus, excluding OGs containing tandem or segmental duplicates. Type II OGs contain one member each from A. thaliana and A. lyrata and two or three members from Brassica or Raphanus. Type III consists of the remaining OGs (9331). Brassica and Raphanus genes in type I and type II OGs are referred to as singletons and retained duplicates, respectively. We found that more of the retained duplicates tend be involved in biotic and abiotic stress response, hormonal signaling, development, as well as regulation of transcription, compared with singletons (Supplemental Figure 5A). In contrast, singletons were enriched in processes such as DNA repair, cell division, metabolic processes, as well as RNA modification and processing (Supplemental Figure 5B). These results are consistent with previous findings in Brassica and other flowering plants (Wang et al., 2011; De Smet et al., 2013).
Our findings indicate that a large percentage of OGs (∼70%) experienced losses in Brassica and Raphanus, returning them to a singleton state with significant functional bias. Such a behavior may be expected as the polyploid became diploidized over the past 27 million years. To better understand the process of gene loss, we identified and analyzed pseudogenes in the Brassica and Raphanus genomes.
Pseudogenization of Duplicate Genes
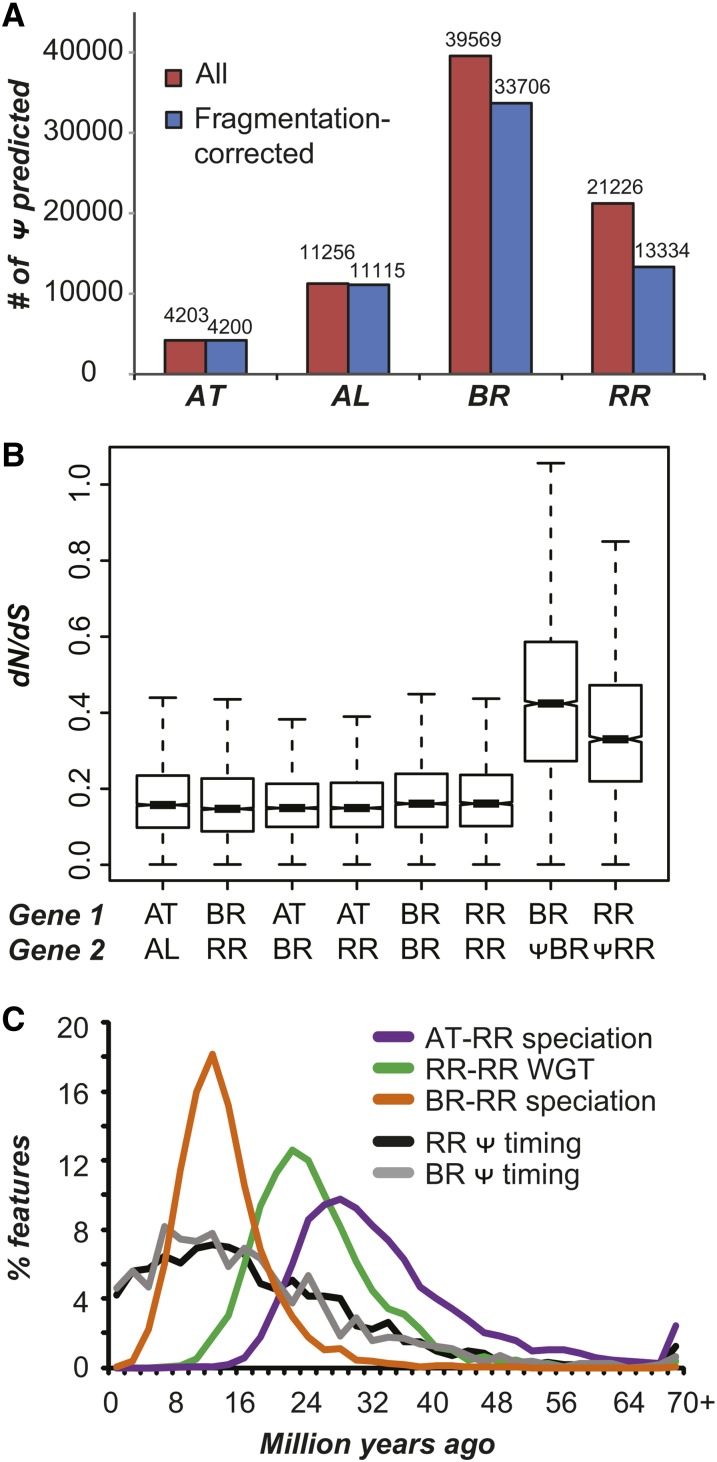
Studies on synthetic and naturally occurring recent polyploids suggest that newly formed polyploids may undergo rapid genomic arrangements and chromosomal losses in the first few generations (Shaked et al., 2001; Tian et al., 2010; Matsushita et al., 2012; Chester et al., 2012), which can result in instantaneous loss of several thousand genes from the genome via deletion. Another mode of gene loss is through accumulation of substitutions and/or small indels in the gene body leading to pseudogenization. We identified 39,659 Brassica and 21,226 Raphanus pseudogenes that are fragments of their paralogs (>80% covered only <50% of the paralog length) and/or contain premature stops/frameshifts (Figure 3A; Supplemental Figures 6 and 7A). To assess the error rate in misclassifying a gene as a pseudogene, four analyses were conducted (Supplemental Figure 6). The predicted pseudogenes have significantly higher dN/dS values compared with functional ortholog and paralog pairs (Kolmogorov–Smirnov test P < 1e-15; Figure 3B). Although some pseudogenes had dN/dS values comparable to functional duplicate genes, these pseudogenes contain in-frame stops and/or frameshifts or are short fragments (Supplemental Figure 7A), suggesting that they are not simply false positives but may have been created recently.
Figure 3.
Patterns of Pseudogenization in Brassicaceae Species.
(A) Number of pseudogenes (Ψ) predicted in each species, before (red) and after (blue) correcting for the fragmented nature of the genomic assemblies.
(B) Evolutionary rates (dN/dS) of orthologs between A. thaliana (AT), A. lyrata (AL), Brassica (BR), and Raphanus (RR) and between paralogs in BR and in RR. The paralog rates were calculated between pairs of annotated, presumably functional paralogs and between functional gene-pseudogene pairs.
(C) Timing of pseudogenization (black and gray lines) compared with the timing of other events.
To determine whether pseudogenization is still ongoing among α’ duplicates, we estimated the timing of pseudogenization for the pseudogenes derived from the α’ WGT event. First, we stringently defined 2268 Brassica and 1261 Raphanus pseudogenes as derived from α’ WGT because they are located in homoeologous regions with their annotated, presumably functional paralogs. Given that we see ∼40,000 genes in each genome, ∼50,000 genes may have been lost from the neopolyploid ancestor, which may have had ∼90,000 genes (assuming the common ancestors of the four Brassiceae species had 30,000 genes). Thus, our prediction of 1000 to 2000 α’ WGT derived pseudogenes is an underestimate. To estimate timing of pseudogenization, a method was used assuming that the two duplicate genes experience the same degree of selective constraint before pseudogenization, and the pseudogenized copy evolves neutrally (see Methods; Supplemental Figure 7B) (Chou et al., 2002). The number of pseudogenized duplicates is higher after α’ WGT, but we do not see a sharp increase in pseudogenization immediately after the WGT event. Instead, we find a gradual pattern of pseudogenization wherein some pseudogenes were formed very recently (Figure 3C). The choice of criteria for defining α’ derived pseudogenes did not significantly affect this pattern (Supplemental Figures 7D to 7G). These results suggest that pseudogenization is ongoing even 27 million years after WGT.
Our results also suggest that there was no peak of pseudogenization soon after α’ WGT. However, this analysis has two caveats. First, because we can detect only ∼2000 α’ WGT pseudogenes, we may have missed older pseudogenes that have degraded beyond recognition. Second, related to the first issue, our analysis is limited to gene loss via pseudogenization and that gene loss via whole-gene deletion may have a different profile, contributing differently to overall gene loss compared with pseudogenization. Hence, the relative rates of loss via deletion versus pseudogenization need to be further studied.
Sequence Divergence of Duplicate Genes Post α’ WGT
Although a large proportion of the triplicated gene content has been lost, ∼15% of the duplicates are still retained. Over the past 27 million years, these retained duplicates may have subfunctionalized or neofunctionalized via sequence or expression divergence. To detect sequence level divergence, we performed a relative rates test on the protein sequences using an amino acid substitution model and found that 13.1 and 18.7% of the Brassica and Raphanus gene pairs, respectively, experienced asymmetric evolution (Supplemental Figure 8A). The asymmetrically evolving duplicate gene pairs were also found to have a 1.5× higher dN/dS ratio than duplicate pairs evolving at a uniform rate (Kolmogorov–Smirnov test P < 1e-15) (Supplemental Figure 8B), which is similar to observations in yeast (Fares et al., 2006), cotton (Gossypium hirsutum), and A. thaliana (Cronn et al., 1999; Blanc and Wolfe, 2004).
These results suggest that functional divergence in almost a fifth of the duplicate gene pairs may have occurred via asymmetric sequence divergence. It is likely that this is an underestimate because the power to detect asymmetry is reduced for older duplicates, shorter sequences, and asymmetry with small effect sizes (Seoighe and Scheffler, 2005). Also, instances of asymmetry on both branches at different times, which effectively negate each other, or phenomena such as gene conversion, will be hidden from our analysis. Nevertheless, our results suggest a substantial degree of sequence divergence that may significantly impact gene function. It has been suggested that there is a burst of protein sequence evolution immediately after a WGD event, and the genes that evolve fast initially tend to experience a more relaxed selective regime than the slow evolving ones, for a long time after the WGD event (Scannell and Wolfe, 2008). Such an accelerated rate of protein evolution, which leads to a rapid accumulation of independent mutations in the duplicate gene copies may set the stage for asymmetric sequence evolution of duplicates over time.
Expression Divergence of Duplicate Genes Post α’ WGT
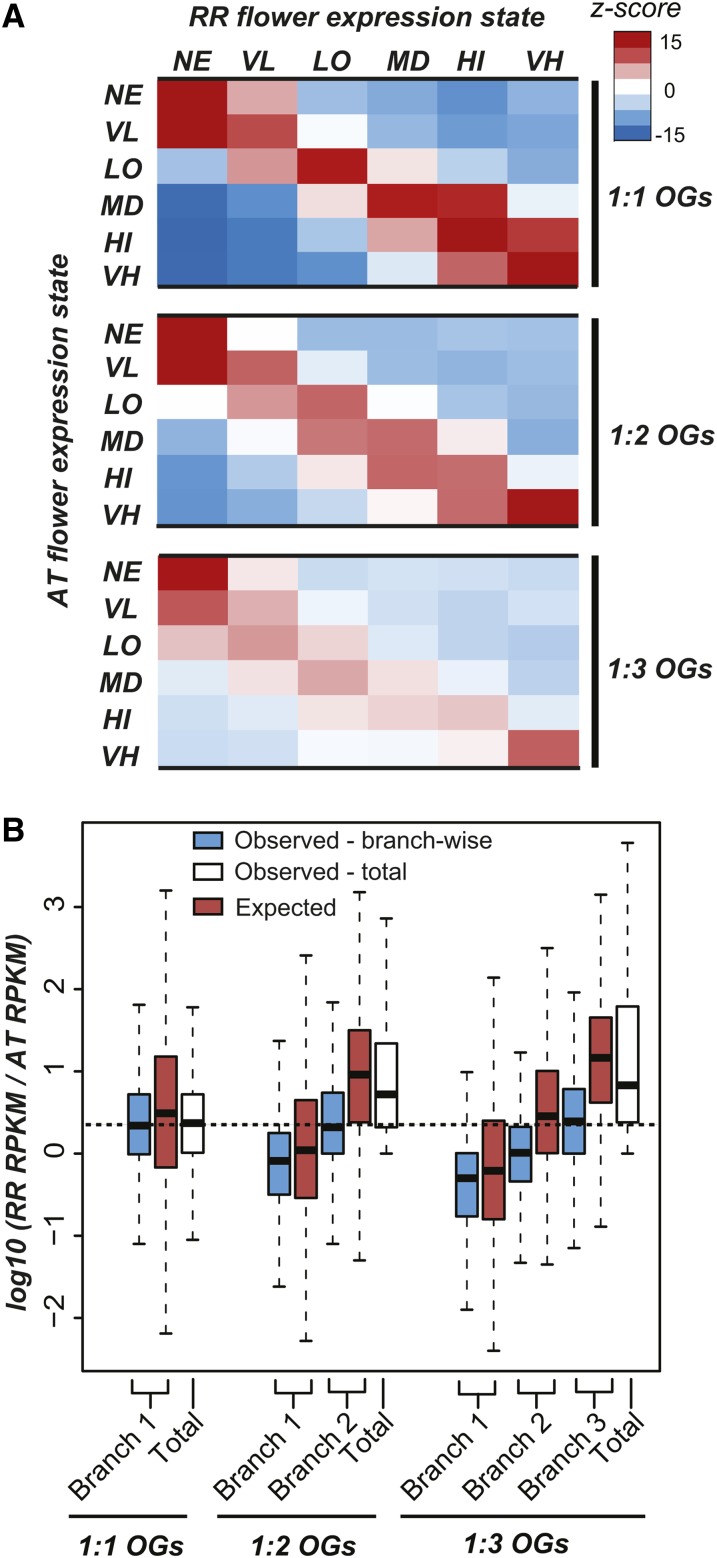
Duplicate genes may diverge not only in sequence but also expression (Conant and Wolfe, 2008). To understand the extent of expression divergence in α’ duplicates, we used gene expression data from A. thaliana flowers and Raphanus floral buds and asked (1) if Raphanus genes show signatures of expression level divergence when compared with their A. thaliana orthologs and (2) whether the expression patterns are different between 1:1, 1:2, and 1:3 A. thaliana:Raphanus OGs. Based on the expression distribution of A. thaliana and Raphanus genes, we partitioned their expression levels into five states, very low (0 to 20%), low (20 to 40%), medium (40 to 60%), high (60 to 80%), and very high (80 to 100%), as well as a sixth “not expressed” state, and examined transitions between states for pairwise A. thaliana:Raphanus comparisons. Defining A. thaliana and Raphanus orthologs with the same expression state as conserved, 36.6% Raphanus genes in 1:1 OGs have a conserved expression state, which is significantly higher than randomly expected (Figure 4A; z-scores range from 10 to 35 among blocks along the diagonal). On the other hand, the degree of expression state conservation drops substantially among Raphanus retained duplicates in 1:2 and 1:3 OGs compared with 1:1 OGs. Assuming that the A. thaliana ortholog expression level represents the ancestral expression level, there are apparently significantly more transitions from a higher expression state to a lower one among Raphanus retained duplicates in 1:2 and 1:3 OGs (Figure 4A).
Figure 4.
Expression Divergence of α' Duplicates.
(A) Z-scores of % overlaps between A. thaliana (AT) and Raphanus (RR) expression states compared with fitted distributions of % randomly expected overlaps (10,000 trials). NE, not expressed; VL, very low; LO, low; MD, medium; HI, high; VH, very high. Red, overrepresentation; blue, underrepresentation.
(B) Observed and expected distributions of reads per kilobase of transcript per million mapped reads (RPKM) ratios between RR and AT orthologs in the three OG types. The horizontal dotted line indicates the baseline according to the observed ratio in the 1:1 OG type. The branchwise observed values (blue) were calculated first by sorting orthologs in an OG based on their expression levels. Orthologs with lower expression levels also have smaller branch number designations. The expected values (red) were obtained by randomly shuffling the association between AT and RR orthologs for each OG type. The observed totals over all branches (white) were calculated using the sum of the RR ortholog RPKM values in an OG.
Our findings indicate that Raphanus genes in 1:2 and 1:3 OGs have experienced higher degrees of expression level divergence since the WGT event, while those in 1:1 OGs tend to have conserved expression levels in flowers. In addition, most instances of expression divergence between Raphanus and A. thaliana genes in 1:2 and 1:3 OGs are in the form of state transitions to lower expression levels in one or more of the Raphanus branches (Figure 4A). Assuming expression level increases and reductions are equally likely among Raphanus paralogs in 1:2 and 1:3 OGs (red box plots, Figure 4B), significantly more cases of expression level reduction are observed than randomly expected (Kolmogorov–Smirnov test P < 1e-15 in all comparisons) (blue box plots, Figure 4B). More importantly, despite expression divergence, the Raphanus copies with the highest expression levels within OGs (branch 2 in 1:2 and branch 3 in 1:3 OGs) appear to maintain the ancestral expression level. This inference is made because the ratio between the highest expressing Raphanus duplicate to its A. thaliana ortholog in both 1:2 and 1:3 OGs (blue boxes, Figure 4B) is similar to the median ratio in 1:1 OGs (horizontal dotted line, Figure 4B). Thus, after genome triplication, one duplicate likely maintains the ancestral level of expression while the other retained copies have reduced expression in a particular tissue. Such expression differentiation may be reflective of functional differentiation occurring in the retained duplicates through sub- or neofunctionalization.
We also found that the sum of expression levels among the retained Raphanus duplicates in 1:2 and 1:3 OGs was higher than the expression level of their A. thaliana orthologs (unfilled box plots, Figure 4B). Assuming the expression of the A. thaliana ortholog is similar to the ancestral level, these results suggest that the total expression level of all duplicates in an OG may not be subjected to strong selection to match the expression level in the ancestral gene. This finding, however, does not rule out the possibility that dosage balance is important because the balance may occur at posttranscriptional and posttranslational levels. For example, retained duplicates may possess different efficacies of performing the same function (Nowak et al., 1997), and dosage balance can be established at the level of protein activity. We also note that our results are obtained from analyzing only floral tissues in two species from two different studies and that the assumed preduplication ancestral expression level may not be the same as the expression level of the A. thaliana ortholog. Although comparing between 1:1, 1:2, and 1:3 orthologs may reduce the influence of cross-species/cross-study biases, a more stringent definition of ancestral expression state based on data for all four species under the same conditions in multiple developmentally similar tissues will provide a more complete picture of duplicate expression evolution.
Informative Features Correlated with α and α’ Duplicate Retention
Our results so far indicate that duplicates in ∼15% of the OGs may have been retained post α’ WGT. Such retained duplicates may exhibit functional (Supplemental Figures 5A and 5B) or other biases (Pál et al., 2001; Chapman et al., 2006; Schnable et al., 2011). One unanswered question is whether some of these features are better predictors of duplicate retention than others. To address these questions, we first examined five types of gene features, including GO-Slim classification, sequence-related features, expression-related features, network-related features, and conservation-related features (see Methods; Supplemental Table 2). For each feature, we asked if the feature values of retained duplicates were significantly different from those of singletons. In addition, we compared the properties of α’ retained duplicates and singletons against those derived from the α WGD event (Bowers et al., 2003). Because the general trends in Brassica and Raphanus are essentially the same, in all subsequent discussions we discuss the joint results of both species.
With some exceptions, most features are consistently over- or underrepresented among retained duplicates between the α’ WGT and α WGD events (Figure 5A). For example, among biological functions, retained duplicates are most strongly enriched in GO-Slim categories related to transcriptional regulation, stress response, signal transduction, and transport for both polyploidization events (Figure 5A). Compared with singleton genes, duplicates retained after the α’ WGT event tend to have larger gene sizes (P < 1e-9), higher GC3 content (P < 1e-21), higher expression levels (P < 1e-25), and broader expression profiles (P < 1e-3). They also tend to be responsive to biotic and abiotic stresses (P < 1e-7 and P < 1e-4, respectively) and have greater network connectivity (P < 1e-21). In addition, compared with singletons, retained duplicates tend to have homologs in a higher number of land plant genomes (P < 1e-45) and lower dN/dS values compared with their A. thaliana orthologs (P < 1e-24). The observation that genes with greater network connectivity and with signal transduction and regulatory functions are retained may indicate a tendency to maintain dosage balance among certain genes, as per the predictions of the gene balance hypothesis (Freeling and Thomas, 2006). Biased retention of genes possessing these properties may lead to conservation and subsequent functional divergence of duplicated gene modules through time, in turn resulting in increased morphological and physiological diversity in polyploid lineages (Freeling and Thomas, 2006).
Figure 5.
Comparison of Features between Retained Duplicates and Singletons.
(A) Features with overrepresented (red) or underrepresented (blue) numbers of retained duplicates according to multiple testing corrected Fisher’s exact test P values (Q-values). The value distributions of some features were divided into four quartiles (shades of gray). Names of certain GO-Slim categories marked with an asterisk have been abbreviated as noted in Supplemental Methods.
(B) The AUC-ROC (Receiver Operating Characteristic) for the α WGD (blue) and α' WGT (red) duplicate retention prediction models using all features in (A).
(C) Comparison of the SVM weight of the α WGD and the α' WGT models. Informative features (|weight| > 0.05) in a consistent direction between the α and the α' models are colored blue while those in opposite direction are colored red. Numbers correspond to feature IDs noted in (A).
Predicting Duplicate Gene Retention
The enrichment analyses indicate that some features are significantly different between retained duplicates and singletons, many of which are consistent between α and α’ events. Two questions remain. The first is regarding the relative importance of these features in differentiating retained duplicates from singletons. Second, our results are consistent with a recent study investigating gene retention across multiple plant WGD events (Jiang et al., 2013), raising the question whether a predictive, unifying model for the process of gene retention can be generated computationally by combining multiple gene properties. To address these questions, we considered all features (Supplemental Table 2) and generated predictive models for the α WGD and the α’ WGT events using a machine learning algorithm, Support Vector Machine (SVM; see Methods). The model performance was evaluated using Area Under Curve (AUC) where a perfect model will have an AUC of 1 and a random model will have an AUC of 0.5.
For the model predicting α’ duplicate retention using all features (the full model), the average AUC is 0.73, which is significantly better than the model constructed with randomized data (average AUC = 0.51; Figure 5B) or using single sets of features (the individual models, average AUC = 0.56; Supplemental Figure 9B). The results are similar for α duplicates, although, compared with random guesses, the performance in classifying α duplicates (average AUC = 0.75) is slightly better than predicting α’ duplicates (Supplemental Figure 9A), likely because GO-Slim, expression features, and network features for the Brassica and Raphanus genes were inferred from their A. thaliana orthologs. We also found that excluding one feature set at a time from the full model (“leave-one-out” models) did not significantly affect the model performance (average AUC = 0.72; Supplemental Figure 9B). Thus, combining multiple features into a single model allows for a better classification of retained duplicates from singletons than models based on random guesses or single features. Next, we asked whether models generated based on training data of the α’ event can be used to predict retention of α duplicates and vice versa. The model trained on the α’ data set generated an average AUC of 0.61 when used to classify α duplicates, while the model trained on the α data set generated an average AUC of 0.67 for α’ duplicates. While both AUCs are better than the individual models and random guesses, the performance of these models is significantly worse than the models trained and used to predict retained duplicates from the same event, suggesting the presence of unique properties of retained duplicates associated with each WGD event. This is consistent with the results of enrichment tests, which showed variable degrees of over- and underrepresentation for different feature types (Figure 5A). In addition, some features have positive SVM weights (associated with better prediction of duplicates) for the α’ duplicates but negative weights (associated with better predictions of singletons) for the α duplicates (Figure 5C), indicating divergent properties between α and α’ events.
Overall, these observations suggest that a three-feature set, including sequence-related features, Gene Ontology, and conservation-related features, allows us to generate a reasonable model for predicting gene retention. We find that models constructed using additional features do not perform better (average AUC α = 0.75, compared with average AUC α = 0.73 in models using the three-feature set). Thus, additional features, such as interaction partners and expression profile, depending on the WGD event under study, may or may not lead to further improvement. In addition, there is an issue of overfitting as the number of parameters in the model increases. Because the three-feature sets can be readily obtained in sequenced species (perhaps with the exception of Gene Ontology categories that are inferred mostly based on conservation), the machine learning approach can be broadly applied to model gene retention across various polyploidization events in a quantitative manner. More importantly, the model performance provides a measure of our current state of understanding regarding factors affecting duplicate retention. Based on our findings, there are additional factors beyond the three-feature set that might contribute to predicting duplicate gene retention, but these factors have yet to be modeled and/or discovered.
Summary of Findings, Implications, and Unanswered Questions
In this study, we sequenced the genome of R. raphanistrum, a wild relative of the cultivated crops R. sativus and Brassica. This genome sequence, together with other sequenced Brassicaceae species (Arabidopsis Genome Initiative, 2000; Dassanayake et al., 2011; Hu et al., 2011; Wang et al., 2011; Cheng et al., 2013; Haudry et al., 2013; Slotte et al., 2013), makes Brassicaceae a highly desirable plant family for comparative genomic analyses. The 254-Mb assembly of Raphanus encompasses ∼49% of the estimated genome size, has an N50 of 10.1 kb, and includes a majority (38,174) of the genes in the Raphanus genome. We found that ∼60% of the genes in the neopolyploid ancestor of Brassica and Raphanus were lost since the WGT event; however, several thousand genes are still retained within the Brassica and Raphanus genomes and may contribute to evolutionary novelty. For example, a recent study showed that circadian rhythm regulated genes are preferentially retained in Brassica (Lou et al., 2012), suggesting the possibility of phenological changes in post α’ WGT species. In our study, retained duplicates were found to possess functions related to transcriptional regulation, stress regulation, and development. Retention of duplicates may lead to the immediate evolution of novel functions that can be adaptive and allow conquest of new ecological niches. Alternatively, the retention of these genes can be due to subfunctionalization (Force et al., 1999) and dosage balance (Birchler and Veitia, 2007), which may not involve the evolution of new functions in the short term but may pave a path toward eventual neofunctionalization (He and Zhang, 2005).
What are the properties of retained duplicates? Over the past decade, several studies have taken advantage of the increased availability of genome sequence data and comparative genomic tools to analyze the evolution of WGD-derived duplicate genes in multiple species, assessing features important for the loss and retention (Blanc and Wolfe, 2004; Schnable et al., 2011; Jiang et al., 2013). In this study, we confirmed the features assessed as important in earlier studies and determined their relative importance in distinguishing duplicates from singletons using machine learning. Our framework identifies features consistently correlated with gene loss/retention across the α and α’ duplicates. We found that although existing knowledge is useful for building predictive models of duplicate retention, the model performance is far from perfect, suggesting additional features are important for explaining the gene retention process. Examples of missing features may include subgenome bias (Schnable et al., 2011), importance of dosage balance (Freeling and Thomas, 2006; Birchler and Veitia, 2007), or simply random loss. In addition, a recent study suggests that retained duplicates from one WGD event have only a 50% chance of being retained after a subsequent WGD event (Schnable et al., 2012). Modeling using such additional features may help provide a more complete picture of gene retention and loss post WGD.
One counterintuitive finding in our analyses is that retained α and α’ duplicates tend to have lower evolutionary rates compared with singletons. This phenomenon has been noted before in plants (Chapman et al., 2006; Sémon and Wolfe, 2007; Jiang et al., 2013). One explanation is that retained duplicates may provide a buffering effect against perturbation of essential functions under certain circumstances (Nowak et al., 1997; Chapman et al., 2006), and selection for such buffering may constrain the rate of duplicate evolution (see Supplemental Methods for discussion). Another possibility is that, if duplicate genes were retained due to selection for maintaining proper dosage in macromolecular complexes, accumulation of nonsynonymous substitutions in any of the components in the complex may disturb the established stoichiometry, a phenomenon that might be selected against (Freeling and Thomas, 2006). Considering that retained duplicates tend to have higher network connectivity, broader and higher expression, and certain biological functions, these properties may lead to higher retention probability due to a need to maintain dosage among network, coexpression, and functional modules, respectively.
Our results also suggest a complex pattern of expression evolution between retained duplicates in Raphanus, where one of the triplicates tends to have a similar expression state as its A. thaliana ortholog, while other copies have reduced expression level. We found that the sums of Raphanus duplicate or triplicate expression levels are in general higher than their A. thaliana orthologs. This suggests that, at least at the transcriptional level, a “dosage imbalance” can persist for more than 20 million years after polyploidization. However, our study involves the transcriptome from only one organ, and expression divergence among retained duplicates needs to be investigated in more detail using transcriptomic data from more tissues/conditions. In recent years, genomic and transcriptomic data from multiple plant species, many of which have undergone recent or ancient polyploidization events, have been made available. Comparative analyses of pseudogenes and duplicate genes derived via WGD events and their expression patterns in these species will provide a comprehensive picture of the loss/retention/divergence process in plants.
METHODS
Estimating Genome Size
The procedure used to analyze nuclear DNA content in plant cells was modified from a previously published study (Arumuganathan and Earle, 1991). For flow cytometry, 50 mg fresh leaf tissue was sliced into 0.25- to 1-mm segments in a solution containing 10 mM MgSO4.7H2O, 50 mM KCl, 5 mM HEPES, pH 8.0, 3 mM DTT, 0.1 mg/mL propidium iodide, 1.5 mg/mL DNase free RNase (Roche), and 0.25% Triton X-100. The suspended nuclei were filtered and incubated at 37°C for 30 min. The sample nuclei was spiked with standard nuclei and analyzed with a FACScalibur flow cytometer (Becton-Dickinson). We used multiple DNA standards including Chicken Red blood cells (2.5 pg/2C), Glycine max (2.45 pg/2C), Oryza sativa cv Nipponbare (0.96 pg/2C), and Arabidopsis thaliana (0.36 pg/2C). For each sample, the propidium iodide fluorescence area signals (FL2-A) from 1000 nuclei were collected. The mean position of the G0/G1 (Nuclei) peak and the internal standard were determined by CellQuest (Becton-Dickinson).
Genome Sequencing, Assembly, and Annotation
Raphanus is an obligate outcrosser. To reduce the amount of heterozygosity in the genome, R. raphanistrum subspecies raphanistrum (weedy) from the Binghampton population in New York was inbred for five generations and sequenced using Illumina Genome Analyzer II. Sequence reads were preprocessed and assembled with a combination of ABySS 1.2.5 (Simpson et al., 2009), Newbler 2.5.3 (Margulies et al., 2005), the Celera Assembler 6.1 (Miller et al., 2008), and Minimus2 from AMOS 3.1.0 package (Sommer et al., 2007) (Supplemental Figure 1). The MAKER 2.10 pipeline (Cantarel et al., 2008) was used to annotate the Raphanus assembly as detailed in Supplemental Figure 2A. Functional annotations of gene models were obtained using BLAST2GO (Conesa et al., 2005). The assembled genome and annotations are available at http://radish.plantbiology.msu.edu.
EST Sequencing and Assembly
ESTs were sequenced from three R. sativus cultivars (convars sativus, caudatus, oleifera) and four R. raphanistrum populations (subspecies raphanistrum NY weedy, raphanistrum Central Spain, maritimus Coastal Spain, and landra France populations). Total RNA from whole seedlings of R. raphanistrum and R. sativus, buds, and anthers was pooled together. Double-strand cDNA was synthesized from pooled RNA using SMART technology (Clontech). The prepared cDNA was normalized by cDNA denaturation/reassociation, treatment by duplex-specific nuclease, and amplification of the normalized fraction by PCR. The normalized cDNA was then digested with SfiI, fractioned, directionally ligated into pDNR-LIB (Clontech), and electroporated into GC10-competent cells (Gene Choice). Sequences were generated from the 5′ and 3′ ends of clones. A total of 185.4 Mb and 310,844 ESTs were generated and deposited in National Center for Biotechnology Information (NCBI) dbEST.
For assembly, 163,862 Raphanus and 213,105 Brassica EST sequences were downloaded from NCBI dbEST and were assembled into 106,152 and 85,508 unique transcripts, respectively, using a modified version of the PlantGDB pipeline (http://www.plantgdb.org/prj/ESTCluster/PUT_procedure.php). Specifically, the ESTs were processed using a combination of Vmatch 2.1.7, TrimEST (EMBOSS package 6.4.0) and RepeatMasker 3.3.0 (coverage = 225 and divergence = 30). Unique transcripts ≤100 bp were removed and were mapped to their respective genomes using GMAP v2011-09-14 at 30% coverage and 90% identity thresholds. Among overlapping matches, only the longest ones were used for further analyses.
Orthology Inference
We determined orthologous groups between the four Brassicaceae species using a combination of two approaches: similarity based and synteny based. In the similarity-based approach, an all-against-all BLAST (Altschul et al., 1997) search was performed between protein sequences from eight species, and similar genes were assigned orthologous groups using multiple alignment followed by phylogenetic reconstruction. In the synteny-based approach, syntenic groups were first determined between the four species, and orthologous relationships were then defined among the syntenic groups using a phylogenetic approach (Supplemental Figure 3).
Pseudogene Identification
A modified version of a previously defined pseudogene pipeline (Zou et al., 2009) was used to predict pseudogenes in genomes of all four species under study (Supplemental Figure 6). The procedure first involves using protein sequences to search the genome. The matches are regarded as “pseudo-exons,” concatenated together and classified as pseudogenes. Using four different approaches, we confirmed that a significant majority of our pseudogene predictions were not false positives, i.e., real genes misclassified as pseudogenes. To account for false positive predictions as a result of the fragmentary nature of the Raphanus and Brassica genomes, we also eliminated pseudogene predictions lying close to contig ends.
Timing of Speciation, Duplication, and Pseudogenization
Two approaches were used to determine the speciation and duplication time. First, using a lower limit of A. thaliana–Brassica divergence time of 30 mya (Beilstein et al., 2010) as well as a neutral substitution rate of 7*10−3 substitutions/site/million years (Ossowski et al., 2010), we performed Bayesian dating with a prior of 36 mya for the A. thaliana–Brassica divergence time (Town et al., 2006). In the second approach, we obtained divergence times based on dS and the neutral rate estimate indicated above (Supplemental Figures 4A and 4B). To estimate the timing of pseudogenization, we used a published approach (Supplemental Figure 7B) (Chou et al., 2002). All estimates ≤0 mya were discarded. To determine whether the timing was robust to the definition of α’ pseudogenes, we used four additional means of calling pseudogenes as α’ derived (Supplemental Figures 7D to 7G). We found no significant deviation from our proposed inferences.
RNA-seq Analyses
RNA-seq data from Arabidopsis flower (Jiao and Meyerowitz, 2010) was used. For Raphanus, 100 mg of buds of different sizes were used for RNA extraction using Qiagen RNEasy plant RNA mini kit and subsequent sequencing using Illumina Genome Analyzer using the standard library preparation protocol. The obtained 36-bp reads were quality filtered and mapped to the Raphanus genome as previously described using TopHat (Trapnell et al., 2012). Read counts per gene model were obtained using HT-Seq (http://www-huber.embl.de/users/anders/HTSeq/), and the reads per kilobase of transcript per million mapped reads value was obtained using custom scripts.
Classifying Retained Duplicates and Singletons with Machine Learning
We used an implementation of SVM (Joachims, 1999) to generate classifiers that allow distinguishing between retained duplicates and singletons. The feature sets used in this study are detailed in Supplemental Table 2. For expression-related features, we obtained data from previously published microarray (Kilian et al., 2007) and RNA-seq (Filichkin et al., 2010; Jiao and Meyerowitz, 2010; Moghe et al., 2013) expression data sets in A. thaliana. For network-related features, we analyzed data from AraNet, a probabilistic functional gene network (Lee et al., 2010). If the feature values could not be obtained from Brassica/Raphanus directly, they were inferred from the A. thaliana orthologs of the Brassica/Raphanus genes. See Supplemental Figure 9 and Supplemental Methods for more details.
Accession Numbers
Sequence data from this article can be found in the NCBI Sequence Read Archive under Bioproject PRJNA209513.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Sequencing and Assembly of the Raphanus Genome.
Supplemental Figure 2. Pipeline for Annotating the Raphanus Genome.
Supplemental Figure 3. Divergence Time Estimates.
Supplemental Figure 4. Pipeline for Defining Orthologous Groups between A. thaliana, A. lyrata, Brassica, and Raphanus.
Supplemental Figure 5. Gene Ontology Categories Enriched in Retained Duplicates and Singletons Derived from the α’ WGT Event.
Supplemental Figure 6. Pseudogene Annotation Pipeline.
Supplemental Figure 7. Patterns of Pseudogenization in Studied Species.
Supplemental Figure 8. Asymmetric Evolution of α’ Duplicates.
Supplemental Figure 9. Performance of Duplicate Retention Machine Learning Models.
Supplementary Material
Acknowledgments
We thank Cindy Mills and Melissa Lehti-Shiu for maintaining the radish plants, Melissa for reading the article, Barry Williams for discussion, Alicia Massa for helping isolate genomic DNA samples, Robin Buell for discussion on sequencing strategies, John Johnston for computational support, Guangxi Wu for assistance with orthology prediction, Alex Seddon for assistance with SVM analyses, and James Birchler and the reviewers for providing valuable comments allowing further improvement of the article. This work was partially supported by National Science Foundation Grants DBI-0638591 to J.K.C., C.D.T., S.-H.S., and Y.X., DEB-0919452 to J.K.C., I.D., and S.H.S., and IOS-1126998 to S.H.S. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the article. This is KBS contribution #1776.
AUTHOR CONTRIBUTIONS
G.D.M., S.H.S., I.D., C.D.T., and J.K.C. conceived and designed experiments. G.D.M., D.E.H., Y.X., H.T., and C.D.T. performed experiments. G.D.M. and S.-H.S. wrote the article.
Glossary
- mya
million years ago
- WGD
whole-genome duplication
- WGT
whole-genome triplication
- OG
orthologous group
- SVM
Support Vector Machine
- AUC
Area Under Curve
- NCBI
National Center for Biotechnology Information
Footnotes
Online version contains Web-only data.
Articles can be viewed online without a subscription.
References
- Altschul S.F., Madden T.L., Schäffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arabidopsis Genome Initiative (2000). Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Arumuganathan K., Earle E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9: 208–218 [Google Scholar]
- Beilstein M.A., Nagalingum N.S., Clements M.D., Manchester S.R., Mathews S. (2010). Dated molecular phylogenies indicate a Miocene origin for Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 107: 18724–18728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birchler J.A., Veitia R.A. (2007). The gene balance hypothesis: from classical genetics to modern genomics. Plant Cell 19: 395–402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanc G., Wolfe K.H. (2004). Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell 16: 1679–1691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowers J.E., Chapman B.A., Rong J., Paterson A.H. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422: 433–438 [DOI] [PubMed] [Google Scholar]
- Cantarel B.L., Korf I., Robb S.M.C., Parra G., Ross E., Moore B., Holt C., Sánchez Alvarado A., Yandell M. (2008). MAKER: an easy-to-use annotation pipeline designed for emerging model organism genomes. Genome Res. 18: 188–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang P.L., Dilkes B.P., McMahon M., Comai L., Nuzhdin S.V. (2010). Homoeolog-specific retention and use in allotetraploid Arabidopsis suecica depends on parent of origin and network partners. Genome Biol. 11: R125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman B.A., Bowers J.E., Feltus F.A., Paterson A.H. (2006). Buffering of crucial functions by paleologous duplicated genes may contribute cyclicality to angiosperm genome duplication. Proc. Natl. Acad. Sci. USA 103: 2730–2735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S., et al. (2013). The Tarenaya hassleriana genome provides insight into reproductive trait and genome evolution of crucifers. Plant Cell 25: 2813–2830 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chester M., Gallagher J.P., Symonds V.V., Cruz da Silva A.V., Mavrodiev E.V., Leitch A.R., Soltis P.S., Soltis D.E. (2012). Extensive chromosomal variation in a recently formed natural allopolyploid species, Tragopogon miscellus (Asteraceae). Proc. Natl. Acad. Sci. USA 109: 1176–1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou H.-H., Hayakawa T., Diaz S., Krings M., Indriati E., Leakey M., Paabo S., Satta Y., Takahata N., Varki A. (2002). Inactivation of CMP-N-acetylneuraminic acid hydroxylase occurred prior to brain expansion during human evolution. Proc. Natl. Acad. Sci. USA 99: 11736–11741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conant G.C., Wolfe K.H. (2008). Turning a hobby into a job: how duplicated genes find new functions. Nat. Rev. Genet. 9: 938–950 [DOI] [PubMed] [Google Scholar]
- Conesa A., Götz S., García-Gómez J.M., Terol J., Talón M., Robles M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21: 3674–3676 [DOI] [PubMed] [Google Scholar]
- Conner J.K., Sahli H.F., Karoly K. (2009). Tests of adaptation: functional studies of pollen removal and estimates of natural selection on anther position in wild radish. Ann. Bot. (Lond.) 103: 1547–1556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Couvreur T.L.P., Franzke A., Al-Shehbaz I.A., Bakker F.T., Koch M.A., Mummenhoff K. (2010). Molecular phylogenetics, temporal diversification, and principles of evolution in the mustard family (Brassicaceae). Mol. Biol. Evol. 27: 55–71 [DOI] [PubMed] [Google Scholar]
- Cronn R.C., Small R.L., Wendel J.F. (1999). Duplicated genes evolve independently after polyploid formation in cotton. Proc. Natl. Acad. Sci. USA 96: 14406–14411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassanayake M., Oh D.-H., Haas J.S., Hernandez A., Hong H., Ali S., Yun D.-J., Bressan R.A., Zhu J.-K., Bohnert H.J., Cheeseman J.M. (2011). The genome of the extremophile crucifer Thellungiella parvula. Nat. Genet. 43: 913–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Des Marais D.L., Rausher M.D. (2008). Escape from adaptive conflict after duplication in an anthocyanin pathway gene. Nature 454: 762–765 [DOI] [PubMed] [Google Scholar]
- De Smet R., Adams K.L., Vandepoele K., Van Montagu M.C.E., Maere S., Van de Peer Y. (2013). Convergent gene loss following gene and genome duplications creates single-copy families in flowering plants. Proc. Natl. Acad. Sci. USA 110: 2898–2903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edger P.P., Pires J.C. (2009). Gene and genome duplications: the impact of dosage-sensitivity on the fate of nuclear genes. Chromosome Res. 17: 699–717 [DOI] [PubMed] [Google Scholar]
- Fares M.A., Byrne K.P., Wolfe K.H. (2006). Rate asymmetry after genome duplication causes substantial long-branch attraction artifacts in the phylogeny of Saccharomyces species. Mol. Biol. Evol. 23: 245–253 [DOI] [PubMed] [Google Scholar]
- Filichkin S.A., Priest H.D., Givan S.A., Shen R., Bryant D.W., Fox S.E., Wong W.-K., Mockler T.C. (2010). Genome-wide mapping of alternative splicing in Arabidopsis thaliana. Genome Res. 20: 45–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Force A., Lynch M., Pickett F.B., Amores A., Yan Y.L., Postlethwait J. (1999). Preservation of duplicate genes by complementary, degenerative mutations. Genetics 151: 1531–1545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeling M., Thomas B.C. (2006). Gene-balanced duplications, like tetraploidy, provide predictable drive to increase morphological complexity. Genome Res. 16: 805–814 [DOI] [PubMed] [Google Scholar]
- Hanada K., Zou C., Lehti-Shiu M.D., Shinozaki K., Shiu S.-H. (2008). Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 148: 993–1003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haudry A., et al. (2013). An atlas of over 90,000 conserved noncoding sequences provides insight into crucifer regulatory regions. Nat. Genet. 45: 891–898 [DOI] [PubMed] [Google Scholar]
- He X., Zhang J. (2005). Rapid subfunctionalization accompanied by prolonged and substantial neofunctionalization in duplicate gene evolution. Genetics 169: 1157–1164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu T.T., et al. (2011). The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43: 476–481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Innan H., Kondrashov F. (2010). The evolution of gene duplications: classifying and distinguishing between models. Nat. Rev. Genet. 11: 97–108 [DOI] [PubMed] [Google Scholar]
- Jiang W.K., Liu Y.L., Xia E.H., Gao L.Z. (2013). Prevalent role of gene features in determining evolutionary fates of whole-genome duplication duplicated genes in flowering plants. Plant Physiol. 161: 1844–1861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., Meyerowitz E.M. (2010). Cell-type specific analysis of translating RNAs in developing flowers reveals new levels of control. Mol. Syst. Biol. 6: 419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y., et al. (2011). Ancestral polyploidy in seed plants and angiosperms. Nature 473: 97–100 [DOI] [PubMed] [Google Scholar]
- Joachims, T. (1999). Making Large-Scale Support Vector Machine Learning Practical. (Cambridge, MA: MIT Press). [Google Scholar]
- Johnston J.S., Pepper A.E., Hall A.E., Chen Z.J., Hodnett G., Drabek J., Lopez R., Price H.J. (2005). Evolution of genome size in Brassicaceae. Ann. Bot. (Lond.) 95: 229–235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilian J., Whitehead D., Horak J., Wanke D., Weinl S., Batistic O., D’Angelo C., Bornberg-Bauer E., Kudla J., Harter K. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50: 347–363 [DOI] [PubMed] [Google Scholar]
- Lagercrantz U., Lydiate D.J. (1996). Comparative genome mapping in Brassica. Genetics 144: 1903–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I., Ambaru B., Thakkar P., Marcotte E.M., Rhee S.Y. (2010). Rational association of genes with traits using a genome-scale gene network for Arabidopsis thaliana. Nat. Biotechnol. 28: 149–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W.H., Gojobori T., Nei M. (1981). Pseudogenes as a paradigm of neutral evolution. Nature 292: 237–239 [DOI] [PubMed] [Google Scholar]
- Lim K.-B., et al. (2007). Characterization of the centromere and peri-centromere retrotransposons in Brassica rapa and their distribution in related Brassica species. Plant J. 49: 173–183 [DOI] [PubMed] [Google Scholar]
- Lou P., Wu J., Cheng F., Cressman L.G., Wang X., McClung C.R. (2012). Preferential retention of circadian clock genes during diploidization following whole genome triplication in Brassica rapa. Plant Cell 24: 2415–2426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lysak M.A., Koch M.A., Pecinka A., Schubert I. (2005). Chromosome triplication found across the tribe Brassiceae. Genome Res. 15: 516–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margulies M., et al. (2005). Genome sequencing in microfabricated high-density picolitre reactors. Nature 437: 376–380 Erratum. Nature 441: 120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushita S.C., Tyagi A.P., Thornton G.M., Pires J.C., Madlung A. (2012). Allopolyploidization lays the foundation for evolution of distinct populations: evidence from analysis of synthetic Arabidopsis allohexaploids. Genetics 191: 535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.R., Delcher A.L., Koren S., Venter E., Walenz B.P., Brownley A., Johnson J., Li K., Mobarry C., Sutton G. (2008). Aggressive assembly of pyrosequencing reads with mates. Bioinformatics 24: 2818–2824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moghe G.D., Lehti-Shiu M.D., Seddon A.E., Yin S., Chen Y., Juntawong P., Brandizzi F., Bailey-Serres J., Shiu S.-H. (2013). Characteristics and significance of intergenic polyadenylated RNA transcription in Arabidopsis. Plant Physiol. 161: 210–224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mun J.-H., et al. (2009). Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 10: R111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak M.A., Boerlijst M.C., Cooke J., Smith J.M. (1997). Evolution of genetic redundancy. Nature 388: 167–171 [DOI] [PubMed] [Google Scholar]
- Ohno, S. (1970). Evolution by Gene Duplication. (New York: Springer-Verlag). [Google Scholar]
- Ossowski S., Schneeberger K., Lucas-Lledó J.I., Warthmann N., Clark R.M., Shaw R.G., Weigel D., Lynch M. (2010). The rate and molecular spectrum of spontaneous mutations in Arabidopsis thaliana. Science 327: 92–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pál C., Papp B., Hurst L.D. (2001). Highly expressed genes in yeast evolve slowly. Genetics 158: 927–931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra G., Bradnam K., Korf I. (2007). CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23: 1061–1067 [DOI] [PubMed] [Google Scholar]
- Ramsey J., Schemske D.W. (1998). Pathways, mechanisms, and rates of polyploid formation in flowering plants. Annu. Rev. Ecol. Syst. 29: 467–501 [Google Scholar]
- Sahli H.F., Conner J.K., Shaw F.H., Howe S., Lale A. (2008). Adaptive differentiation of quantitative traits in the globally distributed weed, wild radish (Raphanus raphanistrum). Genetics 180: 945–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scannell D.R., Wolfe K.H. (2008). A burst of protein sequence evolution and a prolonged period of asymmetric evolution follow gene duplication in yeast. Genome Res. 18: 137–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Springer N.M., Freeling M. (2011). Differentiation of the maize subgenomes by genome dominance and both ancient and ongoing gene loss. Proc. Natl. Acad. Sci. USA 108: 4069–4074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnable J.C., Wang X., Pires J.C., Freeling M. (2012). Escape from preferential retention following repeated whole genome duplications in plants. Front Plant Sci 3: 94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sémon M., Wolfe K.H. (2007). Consequences of genome duplication. Curr. Opin. Genet. Dev. 17: 505–512 [DOI] [PubMed] [Google Scholar]
- Seoighe, C., and Scheffler, K. (2005). Very low power to detect asymmetric divergence of duplicated genes. In Comparative Genomics, Lecture Notes in Computer Science, A. McLysaght and D.H. Huson, eds (Berlin, Heidelberg: Springer), pp. 142–152. [Google Scholar]
- Shaked H., Kashkush K., Ozkan H., Feldman M., Levy A.A. (2001). Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 13: 1749–1759 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J.M., Birol İ. (2009). ABySS: a parallel assembler for short read sequence data. Genome Res. 19: 1117–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slotte T., et al. (2013). The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45: 831–835 [DOI] [PubMed] [Google Scholar]
- Sommer D.D., Delcher A.L., Salzberg S.L., Pop M. (2007). Minimus: a fast, lightweight genome assembler. BMC Bioinformatics 8: 64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang H., Woodhouse M.R., Cheng F., Schnable J.C., Pedersen B.S., Conant G., Wang X., Freeling M., Pires J.C. (2012). Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics 190: 1563–1574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas B.C., Pedersen B., Freeling M. (2006). Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 16: 934–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian E., Jiang Y., Chen L., Zou J., Liu F., Meng J. (2010). Synthesis of a Brassica trigenomic allohexaploid (B. carinata × B. rapa) de novo and its stability in subsequent generations. Theor. Appl. Genet. 121: 1431–1440 [DOI] [PubMed] [Google Scholar]
- Town C.D., et al. (2006). Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy. Plant Cell 18: 1348–1359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Roberts A., Goff L., Pertea G., Kim D., Kelley D.R., Pimentel H., Salzberg S.L., Rinn J.L., Pachter L. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7: 562–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., et al. Brassica rapa Genome Sequencing Project Consortium (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43: 1035–1039 [DOI] [PubMed] [Google Scholar]
- Warwick S.I., Francis A. (2005). The biology of Canadian weeds. 132. Raphanus raphanistrum. L. Can. J. Plant Sci. 85: 709–733 [Google Scholar]
- Yang T.-J., et al. (2006). Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 18: 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Y.-W., Tai P.-Y., Chen Y., Li W.-H. (2002). A study of the phylogeny of Brassica rapa, B. nigra, Raphanus sativus, and their related genera using noncoding regions of chloroplast DNA. Mol. Phylogenet. Evol. 23: 268–275 [DOI] [PubMed] [Google Scholar]
- Zou C., Lehti-Shiu M.D., Thibaud-Nissen F., Prakash T., Buell C.R., Shiu S.-H. (2009). Evolutionary and expression signatures of pseudogenes in Arabidopsis and rice. Plant Physiol. 151: 3–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.