This work shows that plants derive the ring of ubiquinone from tyrosine and phenylalanine via two distinct pathways. A trans-cinnamate 4-hydroxylase, a peroxisomal p-coumarate-CoA ligase, and the peroxisomal transporter PXA1 were identified in the phenylalanine branch.
Abstract
It is not known how plants make the benzenoid ring of ubiquinone, a vital respiratory cofactor. Here, we demonstrate that Arabidopsis thaliana uses for that purpose two separate biosynthetic branches stemming from phenylalanine and tyrosine. Gene network modeling and characterization of T-DNA mutants indicated that acyl-activating enzyme encoded by At4g19010 contributes to the biosynthesis of ubiquinone specifically from phenylalanine. CoA ligase assays verified that At4g19010 prefers para-coumarate, ferulate, and caffeate as substrates. Feeding experiments demonstrated that the at4g19010 knockout cannot use para-coumarate for ubiquinone biosynthesis and that the supply of 4-hydroxybenzoate, the side-chain shortened version of para-coumarate, can bypass this blockage. Furthermore, a trans-cinnamate 4-hydroxylase mutant, which is impaired in the conversion of trans-cinnamate into para-coumarate, displayed similar defects in ubiquinone biosynthesis to that of the at4g19010 knockout. Green fluorescent protein fusion experiments demonstrated that At4g19010 occurs in peroxisomes, resulting in an elaborate biosynthetic architecture where phenylpropanoid intermediates have to be transported from the cytosol to peroxisomes and then to mitochondria where ubiquinone is assembled. Collectively, these results demonstrate that At4g19010 activates the propyl side chain of para-coumarate for its subsequent β-oxidative shortening. Evidence is shown that the peroxisomal ABCD transporter (PXA1) plays a critical role in this branch.
INTRODUCTION
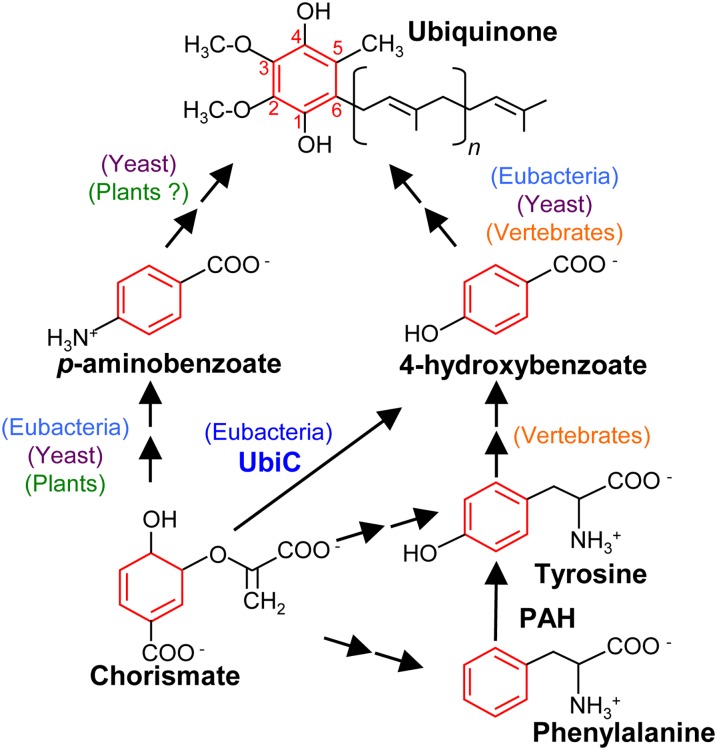
Ubiquinone (coenzyme Q) is a prenylated benzoquinone that serves as a vital electron and proton carrier in the respiratory chain of mitochondria and some bacteria (Nowicka and Kruk, 2010). It is made up of a redox active benzoquinone ring and a prenyl side chain (Figure 1). In plants as in fungi and vertebrates, it is now established that the synthesis of the prenyl moiety, its conjugation to the benzenoid ring, and the subsequent methylation of the latter on carbon 5 (Figure 1) all take place in mitochondria (Avelange-Macherel and Joyard, 1998; Okada et al., 2004; Ducluzeau et al., 2012).
Figure 1.
The Structure of Ubiquinone and the Metabolic Origin of Its Benzenoid Ring in E. coli, Yeast, and Vertebrates.
Note that UbiC (chorismate-pyruvate lyase) occurs exclusively in eubacteria and that PAH (phenylalanine-4-hydroxylase) appears to be lacking in angiosperms.
In contrast, the origin of the benzenoid backbone is not known (Kawamukai, 2009). Indeed, not only have the cognate enzymes yet to be identified, but what is known about the biosynthesis of the ring of ubiquinone in non-plant organisms delineates a puzzling evolutionary pattern. For instance, in Escherichia coli, the ring of ubiquinone is made from 4-hydroxybenzoate, which comes from the direct aromatization of chorismate catalyzed by chorismate pyruvate-lyase or UbiC (Figure 1; Nichols and Green, 1992). However, this enzyme appears to be restricted to the eubacterial lineage (Clarke, 2000; Kawamukai, 2009; Pfaff et al., 2014).
The yeast Saccharomyces cerevisiae is able to derive the ring of ubiquinone from the folate precursor para-aminobenzoate (pABA), which is prenylated and then deaminated (Figure 1; Pierrel et al., 2010). Yeast and other fungi can also make the ring of ubiquinone from 4-hydroxybenzoate, although none of the steps in this alternative pathway are known and the relative contribution of the pABA and 4-hydroxybenzoate branches is difficult to estimate (Law et al., 1971; Marbois et al., 2010). Vertebrates cannot synthesize pABA, but early radiotracer studies have shown that mammals incorporate the ring of phenylalanine and tyrosine into the benzenoid moiety of ubiquinone and that 4-hydroxybenzoate is a likely intermediate (Figure 1; Olson et al., 1963). Since vertebrates are able to convert phenylalanine into tyrosine, these two amino acids most likely belong to a single linear biosynthetic branch leading to 4-hydroxybenzoate (Figure 1; Olson et al., 1963; Clarke, 2000). As is the case for yeast, the vertebrate enzymes involved in the biosynthesis of 4-hydroxybenzoate have not been identified.
Flowering plants a priori do not convert phenylalanine into tyrosine, as they appear to lack phenylalanine-4-hydroxylase (Pribat et al., 2010). However, plants do synthesize pABA (Quinlivan et al., 2003), suggesting that a pABA route toward ubiquinone could operate in the green lineage as it does in fungi (Figure 1). There is also the possibility that plants have evolved the capacity to synthesize the ring of ubiquinone via an entirely different pathway from those of other eukaryotes.
In this study, we show that Arabidopsis thaliana derives the ring of ubiquinone from that of phenylalanine and tyrosine via two nonintersecting routes. We focused on the phenylalanine pathway, and combining gene network modeling, isotopic labeling, and reverse genetics identified two enzymes and one transporter involved in the formation of the benzenoid ring of ubiquinone from the metabolism of phenylpropanoids.
RESULTS
Arabidopsis Derives the Benzenoid Moiety of Ubiquinone from Phenylalanine and Tyrosine, Not from pABA
When axenic cultures of Arabidopsis were given identical doses of uniformly 13C-labeled phenylalanine (Phe-[13C9]) or tyrosine (Tyr-[13C9;15N]), ultraperformance liquid chromatography–electrospray ionization–tandem mass spectrometry analysis of the corresponding extracts readily detected the ion pairs characteristic of ubiquinone-[Ring-13C6] (Supplemental Figure 1). The isotopic enrichment of the benzoquinone ring was twice as high with Phe-[13C9] as with Tyr-[13C9;15N] in both shoots and roots (Table 1). Importantly, 13C isotopes of ubiquinone heavier than those labeled at the six ring positions were not detected, indicating that only the aromatic moiety of Phe-[13C9] and Tyr-[13C9;15N] had been incorporated into ubiquinone. Because angiosperms appear to be unable to convert phenylalanine into tyrosine (Pribat et al., 2010), we inferred that the incorporation of these two aromatic amino acids into ubiquinone might occur, unlike in vertebrates, via two independent pathways. In stark contrast with yeast, Arabidopsis did not incorporate pABA into ubiquinone (Table 1).
Table 1. Ubiquinone-[Ring13C6] Levels in Arabidopsis.
| Ubiquinone-[Ring13C6 ], pmole⋅g−1 FW |
|||
|---|---|---|---|
| Phe-[13C9] | Tyr-[13C9;15N] | pABA-[13C6] | |
| Shoots | 13 ± 2 | 6 ± 1 | n.d. |
| Roots | 120 ± 21 | 54 ± 13 | n.d. |
Data are means of three biological replicates ± se. n.d., not detected; FW, fresh weight.
Gene Network Reconstruction Identifies a Functional Link between the Biosynthesis of Ubiquinone and a 4-Coumarate-CoA Ligase-Like in Arabidopsis
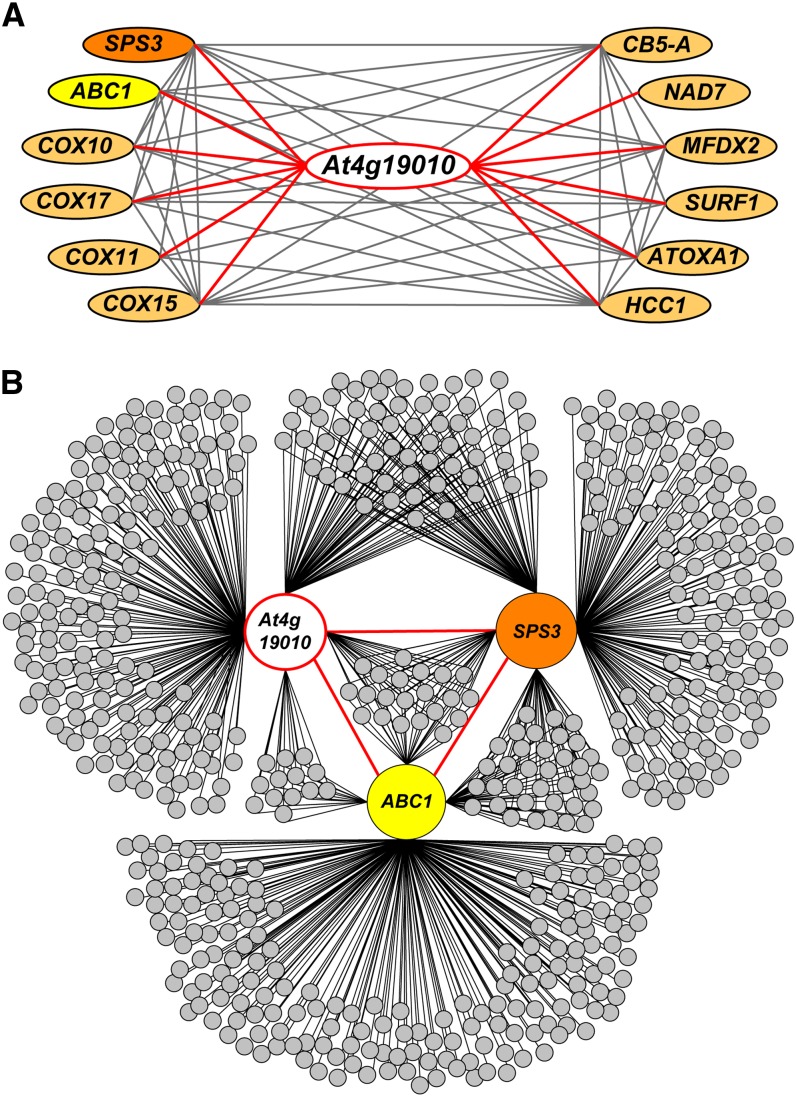
Mining the ATTED-II coexpression database (Obayashi et al., 2009) with genes involved in the assembly of the respiratory chain as queries detected gene At4g19010 as a remarkable node interactor (Figure 2A). The cognate 57-kD protein features a conserved domain (cd05904) found in 4-coumarate-CoA ligases, and previous studies confirmed that recombinant At4g19010 displays CoA ligase activity with some hydroxycinnamate derivatives as substrates (Costa et al., 2005; Kienow et al., 2008). Two of At4g19010’s strongest interactors, solanesyl-diphosphate synthase3 (SPS3; At2g34630), a COQ1/IspB ortholog, and ABC transporter1 (ABC1; At4g01660), a COQ8/UbiB ortholog (Figure 2A), encode for enzymes involved in the biosynthesis of ubiquinone (Cardazzo et al., 1998; Ducluzeau et al., 2012; Block et al., 2013). Expanding the network reconstruction to the top 250 genes that coexpress with At4g19010, SPS3, and ABC1 revealed that At4g19010 shares 38% of its interactors with SPS3 and/or ABC1 (Figure 2B). Of those, 65% are shared uniquely with SPS3, 13% uniquely with ABC1, and 22% with both SPS3 and ABC1 (Figure 2B). It thus appears that the function of At4g19010 is intimately connected to the mitochondrial electron and proton transport chain and more specifically to ubiquinone biosynthesis. It should also be noted that At4g19010 displays a C-terminal SerArgLeu tripeptide typifying a peroxisomal targeting signal-1; we will come back to this point later.
Figure 2.
Gene Functional Network of At4g19010.
(A) Interaction network reconstituted from the top 4.5% elements of the 22,263 expressed loci of the ATTED-II database that coexpress with the respiratory genes SPS3, ABC1, COX10, COX17, COX11, COX15, CB5-A, NAD7, MFDX2, SURF1, OXA1, and HCC1.
(B) Reconstruction of the top 250 coexpressors of At4g19010, SPS3, and ABC1. The annotated lists of these genes and the correlation ranks of the latter are given in Supplemental Data Set 1.
At4g19010 Contributes to the Formation of the Ring of Ubiquinone from Phenylalanine, but Not from Tyrosine
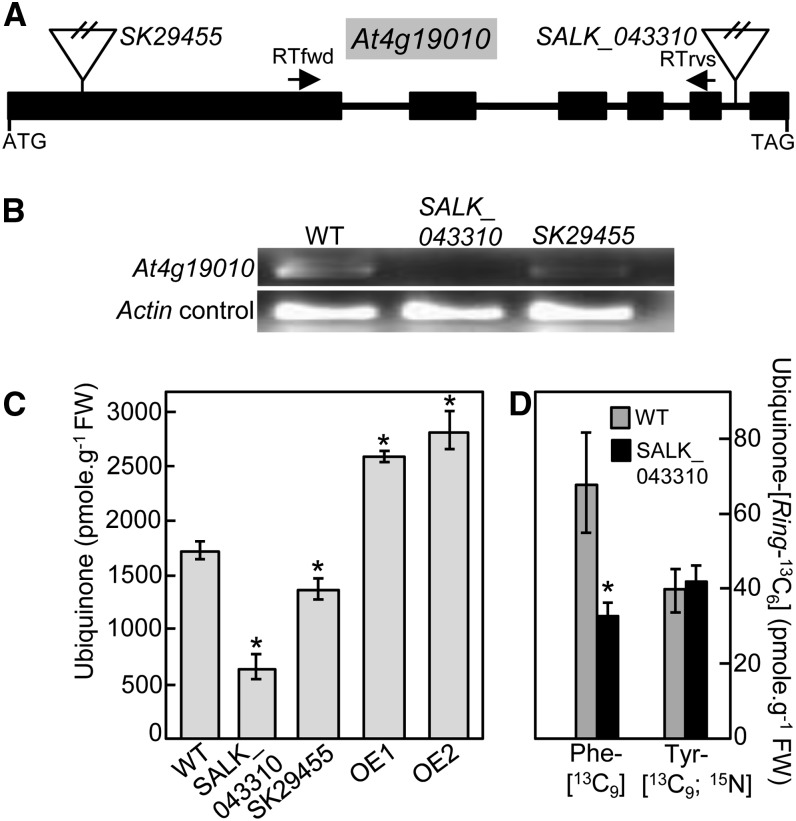
Two mutants corresponding to T-DNA insertions located in the first exon (SK_29455) and fifth intron (SALK_043310) of At4g19010, respectively, were identified using the T-DNA Express gene mapping tool of the SALK Institute (Figure 3A) and confirmed by DNA genotyping. RT-PCR analyses demonstrated that the insertion in line SK_29455 resulted in a knockdown mutation, while that of line SALK_043310 corresponded to a genuine knockout (Figure 3B). Both T-DNA lines were visually indistinguishable from wild-type plants. However, quantification of ubiquinone showed that its levels correlated with the expression of At4g19010; lines SK_29455 and SALK_043310 displaying 21 and 62% decrease in ubiquinone content compared with wild-type plants, respectively (Figure 3C). Similarly, overexpression of At4g19010 cDNA in two independent Arabidopsis transgenic lines resulted in the accumulation of ubiquinone to ∼155% of wild-type content (Figure 3C). Furthermore, when Phe-[13C9] was fed to the at4g19010 knockout, the isotopic enrichment of ubiquinone-[Ring-13C6] was half of that found in the wild-type control (Figure 3D). On the other hand, no statistically significant difference in 13C isotope incorporation was observed between knockout and wild-type plants when Tyr-[13C9;15N] was given to the plants (Figure 3D). All together these results demonstrate that At4g19010 is involved in the biosynthesis of the benzenoid moiety of ubiquinone via the phenylalanine route.
Figure 3.
At4g19010 Is Involved in the Biosynthesis of the Ring of Ubiquinone from Phenylalanine.
(A) Structure of the At4g19010 locus and location of the SALK_043310 and SK29455 insertions. Boxes and lines represent exons and introns, respectively. RTfwd and RTrvs indicate the location of the RT-PCR primers.
(B) RT-PCR analyses of RNA abundance in wild-type and at4g19010 plants showing that SALK_043310 is a knockout mutant and that SK29455 is a knockdown mutant.
(C) Total ubiquinone (reduced and oxidized) levels in the leaves of wild-type plants, SALK_043310 knockout and SK29455 knockdown T-DNA mutants, and At4g19010 overexpressing line OE1 and OE2. FW, fresh weight.
(D) Total ubiquinone-[Ring-13C6] levels in axenically grown wild-type and SALK_043310 Arabidopsis plants fed with 250 μM doses of Phe-[13C9] or Tyr-[13C9;15N] for 3 h.
Data in (C) and (D) are means of four biological replicates ± se. Asterisk annotations indicate significant difference from the wild type as determined by Fisher’s test (P < α = 0.05) from an analysis of variance.
In Vitro Activities of At4g19010
Previous investigations of the in vitro substrate preference of acyl activating enzymes reported conflicting data for At4g19010: one study mentioning marginal activity with caffeate and ferulate only (Costa et al., 2005), while a later study showed that the enzyme possessed conspicuous activity with t-cinnamate, p-coumarate, caffeate, and ferulate as well as with various aliphatic acids (Kienow et al., 2008). To clarify this point, an N-terminally 6xhis-tagged version of At4g19010 was codon-optimized for expression in E. coli, purified, and assayed using a standard spectrophotometric method with CoA, ATP, and t-cinnamate, p-coumarate, ferulate, caffeate, sinapate, 4-hydroxybenzoate, or acetate as substrates. CoA ligase activity was readily detected with t-cinnamate, p-coumarate, ferulate, and caffeate, allowing further investigation of At4g19010’s kinetic properties. Km, kcat, and kcat/Km values determined with p-coumarate, ferulate, and caffeate were similar (Table 2). The catalytic efficiency of the enzyme with t-cinnamate was only 20 to 35% of that measured with p-coumarate, ferulate, or caffeate, owing to a lower kcat value with this substrate (Table 2). Overall, the kinetic parameters of At4g19010 determined with t-cinnamate, p-coumarate, ferulate, and caffeate were similar to those previously reported for plant p-coumarate and t-cinnamate-CoA ligases (Ehlting et al., 1999; Hamberger and Hahlbrock, 2004; Klempien et al., 2012). Notably, as is usually observed for p-coumarate-CoA ligases (Ehlting et al., 1999; Hamberger and Hahlbrock, 2004), no activity was detected with sinapate, the ring of which is methoxylated on C5, while t-cinnamate, p-coumarate, ferulate, and caffeate are unsubstituted at this position (Table 2). Anecdotal catalytic efficiency was found with 4-hydroxybenzoate, while no activity could be detected with acetate (Table 2). These data thus verify that recombinant At4g19010 is predominantly active with t-cinnamate, p-coumarate, ferulate, and caffeate, although they say nothing about which of these substrates might actually play a role in ubiquinone biosynthesis in vivo. Assays with plant extracts did not show any statistically significant difference in CoA ligase activity between wild-type and at4g19010 knockout plants (Supplemental Table 1).
Table 2. Kinetic Properties of Recombinant At4g19010.
| Km (μM) | kcat (s−1) | kcat/Km (s−1⋅mM−1) | |
|---|---|---|---|
| t-Cinnamate | 112 ± 37 | 0.7 ± 0.2 | 6 ± 2 |
| p-Coumarate | 125 ± 25 | 2.1 ± 0.4 | 17 ± 3.4 |
| Ferulate | 113 ± 45 | 3.6 ± 1.4 | 32 ± 13 |
| Caffeate | 85 ± 12 | 2.4 ± 0.3 | 28 ± 4 |
| Sinapate | n.d. | n.d. | n.d. |
| 4-Hydroxybenzoate | 67 ± 30 | 0.01 ± 0.01 | 0.15 ± 0.07 |
| Acetate | n.d. | n.d. | n.d. |
Data are means of six experimental replicates from two enzyme purifications ± se. n.d., not determined, for no activity was detected at any of the substrate concentrations tested.
t-Cinnamate, p-Coumarate, and 4-Hydroxybenzoate Serve as Ubiquinone Precursors
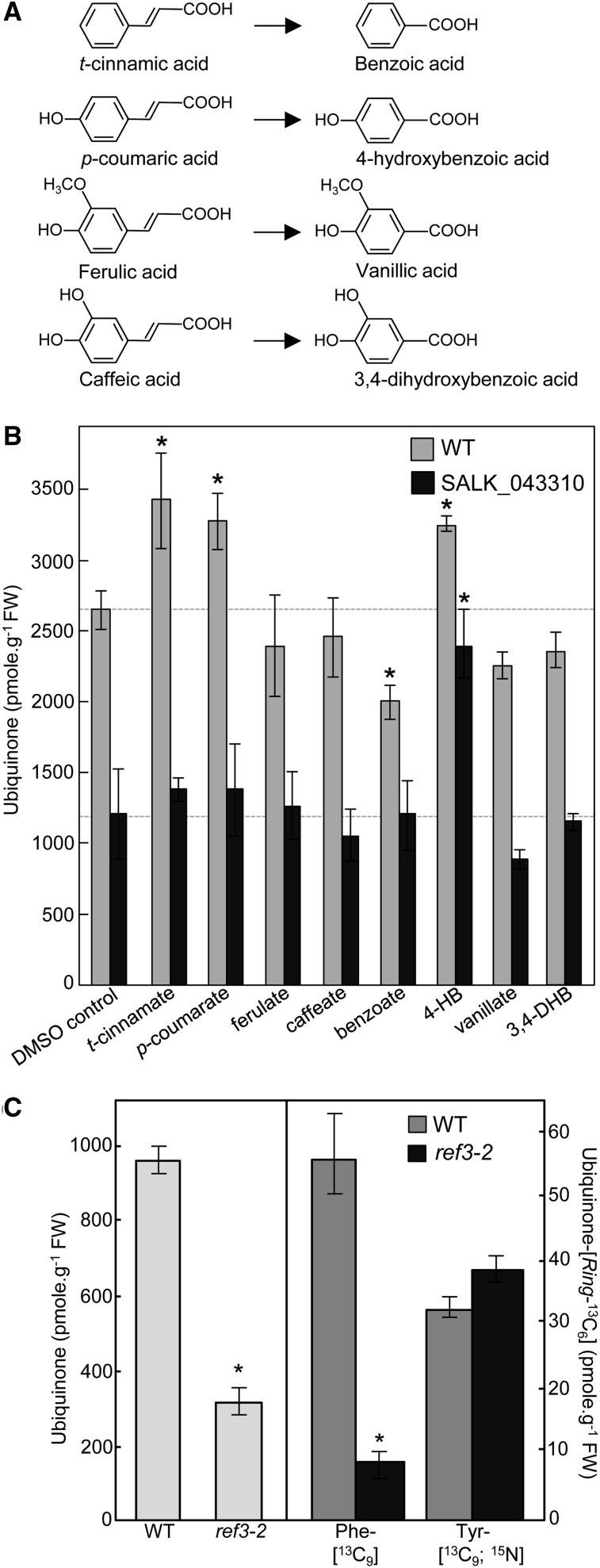
To gain some insight into the architecture of the metabolic branch of which At4g19010 belongs to, ubiquinone levels were determined in wild-type and at4g19010 knockout plants fed with t-cinnamate (the product of phenylalanine ammonia lyase), p-coumarate, ferulate, or caffeate, or with their corresponding shortened aliphatic side-chain versions, i.e., benzoate, 4-hydroxybenzoate, vanillate, or 3,4-dihydroxybenzoate, respectively (Figure 4A). Feeding wild-type plants with t-cinnamate, p-coumarate, and 4-hydroxybenzoate increased ubiquinone content by ∼30% compared with the DMSO control, whereas only 4-hydroxybenzoate restored ubiquinone biosynthesis in the at4g19010 knockout (Figure 4B). Because t-cinnamate is the immediate precursor of p-coumarate (Schilmiller et al., 2009) and plants are able to derive 4-hydroxybenzoate from p-coumarate (Löscher and Heide, 1994), our results strongly suggest that t-cinnamate, p-coumarate, and 4-hydroxybenzoate are used as precursors of ubiquinone via the same metabolic branch. That ubiquinone biosynthesis in the at4g19010 knockout is rescued exclusively by 4-hydroxybenzoate would therefore indicate that the loss of function of At4g19010 creates a metabolic blockage upstream of 4-hydroxybenzoate and downstream of t-cinnamate and p-coumarate. Agreeing with such a model, an Arabidopsis reduced epidermal fluorescence3-2 (ref3-2) leaky mutant that displays reduced cinnamate 4-hydroxylase activity and is thus impaired in its ability to convert t-cinnamate into p-coumarate (Schilmiller et al., 2009) contained only 40% of wild-type ubiquinone level (Figure 4C) and displayed a 75% decrease in isotopic enrichment of ubiquinone-[Ring-13C6] from Phe-[13C9] (Figure 4C). As observed for the at4g19010 knockout, the incorporation of Tyr-[13C9;15N] into ubiquinone in the ref3-2 mutant remained intact (Figure 4C). Taken together, these results are consistent with At4g19010 operating as a CoA ligase that activates the propyl side chain of p-coumarate for its subsequent β-oxidative shortening, leading to 4-hydroxybenzoate.
Figure 4.
t-Cinnamate, p-Coumarate, and 4-Hydroxybenzoate Serve as Precursors of Ubiquinone in Arabidopsis.
(A) Structures of t-cinnamic, p-coumaric, ferulic and caffeic acids, and their corresponding shortened aliphatic side-chain versions.
(B) Total ubiquinone (reduced and oxidized) levels in axenically grown wild-type and SALK_043310 Arabidopsis plants fed for 24 h with 200 μM t-cinnamate, p-coumarate, ferulate, caffeate, benzoate, 4-hydroxybenzoate (4-HB), vanillate, or 3,4-dihydroxybenzoate (3,4-DHB). Data are means of four biological replicates ± se. Asterisk annotations indicate significant difference from the corresponding DMSO control as determined by Fisher’s test (P < α = 0.05) from an analysis of variance. FW, fresh weight.
(C) Left panel, total ubiquinone levels in the leaves of the wild type and cinnamate-4 hydroxylase mutant (ref3-2) Arabidopsis plants. Right panel, total ubiquinone-[Ring-13C6] levels in axenically grown wild-type and ref3-2 Arabidopsis plants fed with 250 μM doses of Phe-[13C9] or Tyr-[13C9;15N] for 3 h. Data are means of four biological replicates ± se. Asterisk annotations indicate significant difference from the wild type as determined by Fisher’s test (P < α = 0.05) from an analysis of variance.
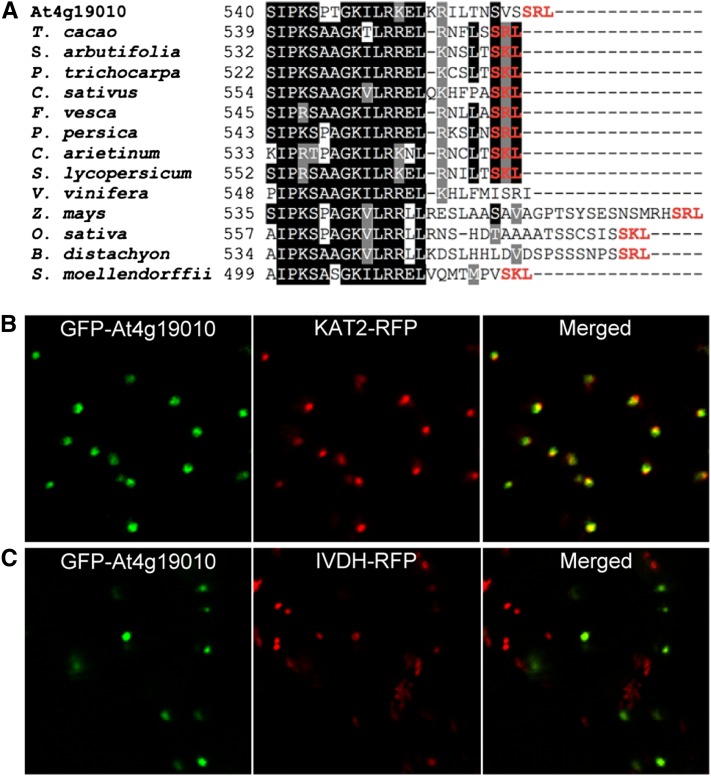
At4g19010 Is Localized in Peroxisomes
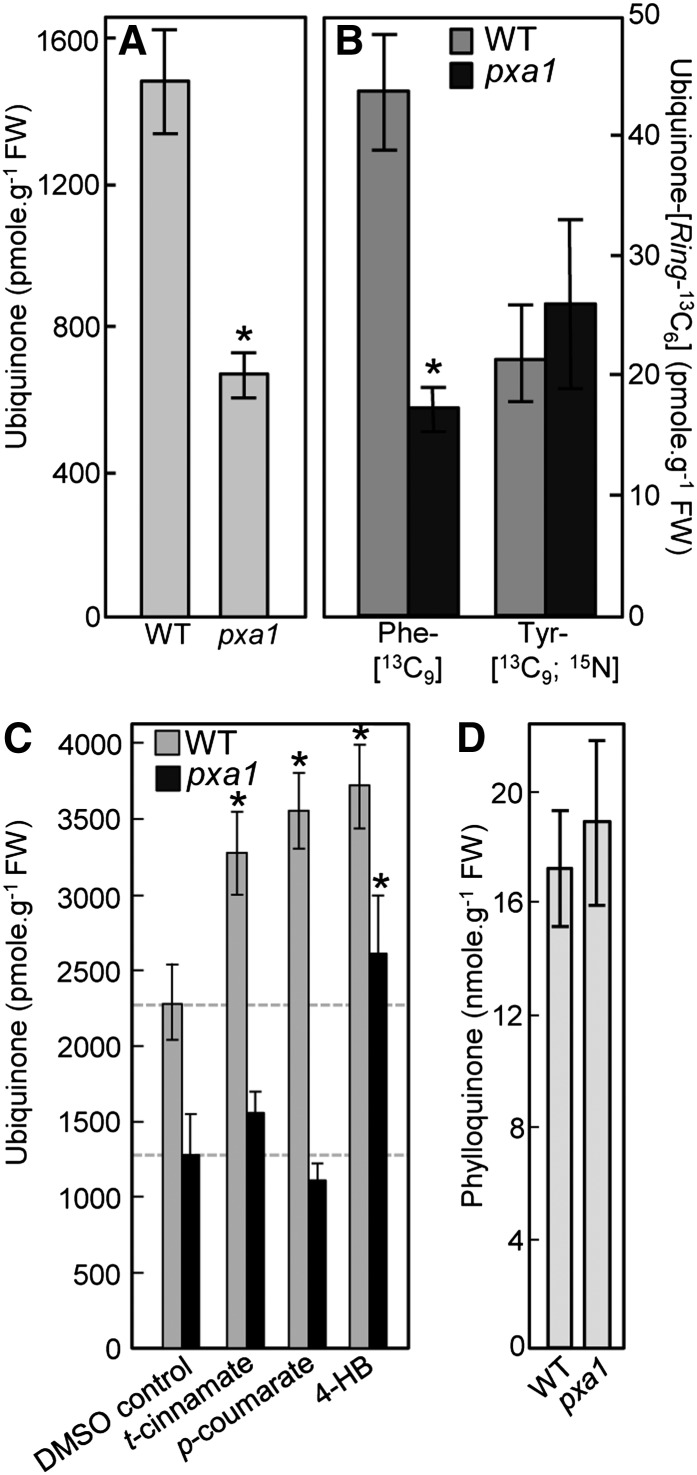
Homology searches across terrestrial plant lineages showed that the putative orthologs of At4g19010 display the hallmark stringent conservation of an authentic peroxisomal targeting sequence-1 (Figure 5A). Coexpression experiments in Nicotiana benthamiana mesophyll cells confirmed that the green pseudocolor of the green fluorescent protein (GFP) fused to the N terminus of At4g19010 strictly colocalized with the red pseudocolor of a red fluorescent protein (RFP)–tagged fragment of 3-keto-acyl-CoA thiolase 2 in peroxisomes (Figure 5B). Control coexpression of the GFP-At4g19010 fusion protein with RFP-tagged isovaleryl-CoA dehydrogenase verified that the fluorescence of GFP was not associated with mitochondria (Figure 5C). That At4g19010 is localized in peroxisomes, while cinnamate-4-hydroxylase is bound to the endomembrane system in the cytosol (Achnine et al., 2004), implies that p-coumarate might be transported into peroxisomes. Reexamination of the At419010/SPS3/ABC1 functional network indentified the peroxisome ABC subfamily D transporter 1 or PXA1 (Zolman et al., 2001) as one of the interacting genes shared between At4g19010 and SPS3 (rank 214 and 291, respectively; Supplemental Data Set 1). Quinone analysis showed that the pxa1 knockout contained 55% less ubiquinone than the wild-type control and that as observed for the at4g19010 and ref3-2 mutants, such a loss was exclusively attributable to a decrease in biosynthetic flux from phenylalanine (Figures 6A and 6B). Feeding experiments showed that of t-cinnamate, p-coumarate, and 4-hydroxybenzoate, only the latter rescued ubiquinone biosynthesis in the pxa1 knockout, demonstrating that here again the metabolic blockage occurred between p-coumarate and 4-hydroxybenzoate (Figure 6C). Further verifying that the impact of the pxa1 mutation on quinone metabolism is specific to ubiquinone biosynthesis, no statistically significant difference in the levels of phylloquinone, the naphthoquinone ring of which is made in part in peroxisomes (Reumann, 2013), was observed between the pxa1 mutant and the wild-type control (Figure 6D).
Figure 5.
Subcellular Localization of At4g19010.
(A) Alignment of the C-terminal regions of At4g19010 and its orthologs in cacao tree (Theobroma cacao), willow (Salix arbutifolia), poplar (Populus trichocarpa), cucumber (Cucumis sativus), strawberry (Fragaria vesca), peach (Prunus persica), chickpea (Cicer arietinum), tomato (Solanum lycopersicum), grape vine (Vitis vinifera), maize (Zea mays), rice (Oryza sativa), Brachypodium distachyon, and Selaginella moellendorffii. The tripeptides corresponding to consensus peroxisomal targeting sequence-1 are highlighted in red. Identical residues are shaded in black and similar ones in gray. Dashes symbolize gaps introduced to maximize alignment.
(B) Left panel, green pseudocolor of GFP fused to the N terminus of At4g19010; center panel, red pseudocolor of peroxisomal marker RFP-tagged 3-keto-acyl-CoA thiolase 2 (KAT2); right panel, overlay.
(C) Left panel, green pseudocolor of GFP fused to the N terminus of At4g19010; center panel, red pseudocolor of mitochondrial marker RFP-tagged isovaeryl-CoA dehydrogenase (IVDH); right panel, overlay.
Figure 6.
The Peroxisomal Transporter PXA1 Is Involved in Ubiquinone Biosynthesis from Phenylalanine.
(A) Total ubiquinone (reduced and oxidized) levels in the leaves of wild-type and pxa1 knockout Arabidopsis plants. Data are means of four biological replicates ± se. Asterisk annotations indicate significant difference from the wild type as determined by Fisher’s test (P < α = 0.05) from ANOVA. FW, fresh weight.
(B) Total ubiquinone-[Ring-13C6] levels in axenically grown wild-type and pxa1 knockout Arabidopsis plants fed with 100 μM doses of Phe-[13C9] or Tyr-[13C9;15N] for 3 h. Data are means of four biological replicates ± se. Asterisk annotations indicate significant difference from the wild type as determined by Fisher’s test (P < α = 0.05) from ANOVA.
(C) Total ubiquinone levels in axenically grown wild-type and pxa1 knockout Arabidopsis plants fed for 24 h with 200 μM t-cinnamate, p-coumarate, or 4-hydroxybenzoate (4-HB). Data are means of three to five biological replicates ± se. Asterisk annotations indicate significant difference from the corresponding DMSO control as determined by Fisher’s test (P < α = 0.05) from ANOVA.
(D) Phylloquinone levels in the leaves of wild-type pxa1 Arabidopsis plants. Data are means of three biological replicates ± se. Wild-type and pxa1 values are not significantly different as determined by Fisher's test from ANOVA (P > 0.4).
DISCUSSION
Plant Peroxisomes Contribute to the Biosynthesis of Ubiquinone
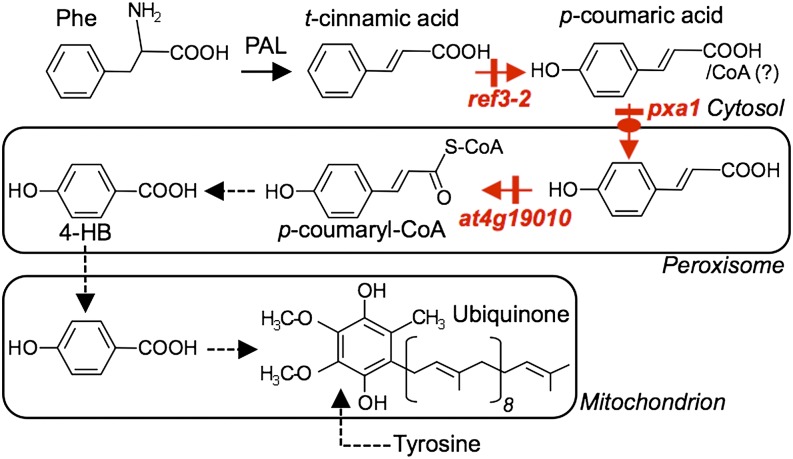
Despite the role of ubiquinone as a crucial cofactor for respiration, the metabolic origin of its benzenoid ring has long remained enigmatic in most eukaryotes (Clarke, 2000; Kawamukai, 2009). One might assume that since plants synthesize pABA de novo (Quinlivan et al., 2003), they could use this metabolite for the biosynthesis of ubiquinone as yeast does (Marbois et al., 2010; Pierrel et al., 2010). Our data show that this is not so, most probably because, unlike yeast, plant cells sequester pABA as a glucose ester conjugate and thus have a minute pool of the free acid form (Quinlivan et al., 2003). Collectively, our data demonstrate that plants have evolved instead the capacity to derive the benzenoid moiety of ubiquinone from the β-oxidative metabolism of phenylpropanoids, adding de facto respiration to the core cellular processes that depend upon plant peroxisomes. Such an arrangement results in an elaborate biosynthetic architecture, in which intermediates are trafficked between the cytosol, where p-coumarate is made (Achnine et al., 2004), peroxisomes, where this study shows that p-coumarate is activated by ligation with CoA for its subsequent shortening into 4-hydroxybenzoate, and mitochondria, where the latter is prenylated and modified (Avelange-Macherel and Joyard, 1998; Okada et al., 2004; Ducluzeau et al., 2012). A proposed scheme of the cognate metabolic branches and their respective compartmentation is shown in Figure 7. A compelling parallel can actually be drawn between our finding that the peroxisome transporter PXA1 participates in this trafficking and the earlier report that PXA1 is required for the β-oxidative activation of the protoherbicide 2,4-dichlorophenoxybutyric acid into 2,4-D (Footitt et al., 2002). Indeed, while wild-type Arabidopsis is sensitive to both 2,4-dichlorophenoxybutyric acid and 2,4-D, the pxa1 knockout is unable to β-oxidize 2,4-dichlorophenoxybutyric acid and is sensitive only to 2,4-D (Footitt et al., 2002). Similarly, our data show that ubiquinone biosynthesis in the pxa1 knockout is restored by 4-hydroxybenzoate, but not by its parent compound p-coumarate. Furthermore, the recent demonstration that PXA1 displays acyl-CoA thioesterase activity (De Marcos Lousa et al., 2013), raises the question of whether the cytosolic pool of p-coumaryl-CoA contributes to ubiquinone biosynthesis.
Figure 7.
Architecture of 4-Hydroxybenzoic Acid Biosynthesis from Phenylalanine in Relation to Ubiquinone Biosynthesis.
The Arabidopsis mutants investigated in this study are indicated in red. Dashed arrows denote multiple steps. 4-HB, 4-hydroxybenzoic acid; PAL, phenylalanine ammonia lyase.
The Function of At4g19010 in Ubiquinone Biosynthesis Is That of a p-Coumarate-CoA Ligase
At4g19010 belongs to clade V of the plant superfamily of acyl-activating enzymes, the respective cellular functions of which have been so far particularly difficult to attribute due to their general lack of substrate specificity in vitro (Kienow et al., 2008; Shockey and Browse, 2011). In fact, besides At4g19010, only one out of eight clade V Arabidopsis members, At1g20510, which encodes for 3′-oxo-2-(2’-[Z]-pentenyl) cyclopentane-1-octanoic acid CoA ligase 1 (OPCL1) of the jasmonate biosynthetic pathway, has a known physiological role (Koo et al., 2006). In vitro, OPCL1 is as active with C8-C18 aliphatic acids as it is with 3′-oxo-2-(2’-[Z]-pentenyl) cyclopentane-1-octanoic acid, its authentic in vivo substrate (Kienow et al., 2008). Similarly, At1g20500, another class V member, displays as much activity with sinapate as it does with the structurally unrelated jasmonate precursor 3′-oxo-2-(2’-[Z]-pentenyl) cyclopentane-1-hexanoic acid (Kienow et al., 2008). Therefore, it is clear that the in vivo function of class V acyl activating enzymes cannot be inferred from their substrate preference alone. In this study, we present three lines of evidence indicating that the role of At4g19010 in ubiquinone biosynthesis is that of a p-coumarate-CoA ligase. First, the usage of p-coumarate as a precursor for ubiquinone biosynthesis is blocked in the at4g19010 knockout. Second, this blockage can be bypassed exclusively by the exogenous supply of 4-hydroxybenzoate, the β-oxidized product of p-coumarate. Last, recombinant At4g19010 catalyzes the conversion of p-coumarate into p-coumaroyl-CoA with similar catalytic efficiency to that of previously characterized p-coumarate-CoA ligases, while as is generally observed for this class of enzymes, At4g19010 is not active with sinapate (Ehlting et al., 1999; Hamberger and Hahlbrock, 2004).
β-Oxidation of Phenylpropanoids Is a Common Mechanism in the Biosynthetic Pathways of Benzenoid-Derived Compounds
The β-oxidative shortening of p-coumarate is reminiscent of that of t-cinnamate, which undergoes a similar route in the biosynthetic pathways of benzoyloxyglucosinolates, xanthone phytoalexin, and benzenoid floral volatiles (Kliebenstein et al., 2007; Colquhoun et al., 2012; Gaid et al., 2012; Klempien et al., 2012; Lee et al., 2012; Bussell et al., 2014). The conversion of p-coumaryl-CoA into 4-hydroxybenzoate requires four yet to be identified steps, i.e., those corresponding to the hydration, dehydrogenation, and thiolation of the β-oxidative cycle followed by the final hydrolysis of the 4-hydroxybenzoyl-CoA thioester. A prime Arabidopsis candidate for the hydration and dehydrogenation steps is the bifunctional peroxisomal enzyme AIM1 (Richmond and Bleecker, 1999), the petunia (Petunia hybrida) ortholog of which was recently shown to participate in the β-oxidative shortening of cinnamoyl-CoA into benzoic acid and to be also active with p-coumaroyl-CoA (Qualley et al., 2012). Similarly, peroxisomal thiolases and CoA thioesterases that act on benzenoid substrates have recently been identified in plants (Van Moerkercke et al., 2009; Widhalm et al., 2012). Plant peroxisomes thus appear to contain the necessary core enzymatic machinery to complete the β-oxidation of p-coumaroyl-CoA to 4-hydroxybenzoate.
Plants Possess a Second Route to Synthesize the Ring of Ubiquinone
It is clear from our data that phenylalanine supplies approximately half of ubiquinone biosynthetic flux and that plants can also use tyrosine to make the ring of ubiquinone. That the at4g19010, ref3-2, and pxa1 mutants have fully retained the capacity to incorporate the benzenoid skeleton of tyrosine into ubiquinone indicates that this alternate route does not intersect with that originating from phenylalanine. Interestingly, the biosynthesis of terpenoid benzoquinones in plastids provides an evolutionary instance of tyrosine serving as a ring precursor via a non-β-oxidative route (DellaPenna and Pogson, 2006). In this pathway, tyrosine is first deaminated into 4-hydroxyphenylpyruvate, which is then decarboxylated and hydroxylated with concomitant migration of the side chain, yielding homogentisate (Supplemental Figure 2). The loss of a second carbonyl occurs simultaneously with the prenylation of homogentisate (Supplemental Figure 2). Future investigations will examine the possible occurrence of metabolic branch-points from 4-hydroxyphenylpyruvate and homogentisate toward ubiquinone.
METHODS
Bioinformatics
Protein and gene sequences were retrieved from the National Center for Biotechnology Information database (http://www.ncbi.nlm.nih.gov). Protein sequences were aligned using MultAlin (http://multalin.toulouse.inra.fr/multalin/). Orthology was determined using phylogeny.fr (http://www.phylogeny.fr). Gene networks were reconstructed by mining coexpressors from the ATTED-II database (http://atted.jp) using Arabidopsis thaliana respiratory genes as queries. Coexpressing genes were then aggregated using GeneVenn (http://genevenn.sourceforge.net).
Chemicals and Reagents
p-Aminobenzoic acid-[13C6] was from IsoSciences, l-phenylalanine-[13C9], and l-tyrosine-[13C9;15N] were from Cambridge Isotope Laboratories. t-Cinnamic and benzoic acids and their hydroxyl/methoxy derivatives were from Sigma-Aldrich. Unless mentioned otherwise, other reagents were from Fisher Scientific.
Plant Material and Growth Conditions
Arabidopsis SALK_043310 (Alonso et al., 2003) and SK_29455 (Robinson et al., 2009) insertion lines, as well as the pxa1 ethyl methanesulfonate mutant (Zolman et al., 2001; stock number CS3950), were obtained from the ABRC. The ref3-2 mutant was generated and identified in the laboratory of one of the coauthors (C.C.) as previously described (Schilmiller et al., 2009). Seeds were germinated on Murashige and Skoog solid medium, transferred to soil, and then grown at 22°C in 16-h days (110 μE m−2 s−1) for 4 weeks. For feeding experiments, plants were germinated and grown in Murashige and Skoog liquid medium containing sucrose (10 g/liter) on an orbital shaker (60 rpm) in 10-h days (200 μE m−2 s−1) at 22°C.
RT-PCR Analyses
Total leaf RNA was extracted using the RNeasy plant mini kit (Qiagen). cDNAs were then prepared from total RNA (500 ng) using the ImProm-II reverse transcription system (Promega). PCR amplification was performed on a 3Prime thermal cycler (Techne) using the following gene-specific primer pairs: At4g19010, 5′-GGAGTTTGTGGTGAAGTGTTT-3′ and 5′-CAAGGGATGTGAAACAAGAAC-3′; actin control, 5′-CTAAGCTCTCAAGATCAAAGGC-3′ and 5′-TTAACATTGCAAAGAGTTTCAAGG-3′.
Preparation of Recombinant At4g19010 and Plant Extracts
Full-length At4g19010 cDNA (ABRC clone G60335) was codon optimized (GenScript) and subcloned using Gateway technology into expression vector pDEST17 (Invitrogen) for N-terminal fusion with a 6xHis tag. This construct was introduced in Escherichia coli BL21 Star (DE3) pLysS (Invitrogen). Protein expression was induced in Luria-Bertani medium with 0.5 mM isopropyl β-d-1-thiogalactopyranoside for 16 h at 20°C. Cells were harvested by centrifugation and disrupted in 50 mM NaH2PO4 (pH 8.0), 300 mM NaCl, 10 mM imidazole, and 0.1% (v/v) Triton X-100 using 0.1-mm Zirconia/silica beads in a MiniBeadbeater (BioSpec Products). The extract was cleared by centrifugation, and the recombinant protein was purified under native conditions with Ni-NTA His-Bind resin (Novagen) following the manufacturer's recommendations. The isolated enzyme was desalted on a PD-10 column equilibrated in 100 mM KH2PO4 (pH 8.0), 10% (v/v) glycerol, and 5 mM MgCl2. The desalted enzyme was frozen in liquid N2 and stored at −80°C. Negative controls consisting of untransformed BL21 Star (DE3) pLysS cells were processed in parallel. For the preparation of plant extracts, Arabidopsis leaves (1 g) were ground into a fine powder using a mortar and pestle under liquid N2. The powder was then thawed in 5 mL of 100 mM KH2PO4 (pH 8.0), 10% (v/v) glycerol, 5 mM MgCl2, 1% (v/v) of β-mercaptoethanol, and 5% (w/v) of cross-linked poly vinylpyrrolidone. Extracts were centrifuged (10,000g for 10 min at 4°C) to pellet the cell debris and the supernatants immediately desalted on a PD-10 column equilibrated in 100 mM KH2PO4 (pH 8.0), 10% (v/v) glycerol, and 5 mM MgCl2. Desalted fractions were dispensed into 500 μL aliquots, flash frozen in liquid N2, and stored at −80°C. Protein concentration was quantified by the method of Bradford (Bradford, 1976) using BSA as a standard.
Enzyme Assays
Assays (1 mL) contained 100 mM KH2PO4 (pH 8.0), 10% (v/v) glycerol, 5 mM MgCl2, 2.5 mM ATP, 0.5 mM CoA, 0 to 300 μM of organic acid, and 0 to 25 μg of protein. Reactions were performed for 30 to 60 min at 30°C. The increase in absorbance due to the formation of the CoA thioesters was monitored on an Agilent 8453 diode array spectrophotometer using the following wavelengths and molar extinction coefficients: 311 nm and 22,000 M−1 cm−1 for t-cinnamoyl-CoA, 333 nm and 21,000 M−1 cm−1 for p-coumaroyl-CoA, 345 nm and 19,000 M−1 cm−1 for feruloyl-CoA, 346 nm and 18,000 M−1 cm−1 for caffeoyl-CoA, 352 nm and 20,000 M−1 cm−1 for sinapoyl-CoA, 330 nm and 24,000 M−1 cm−1 for 4-hydroxybenzoyl-CoA, and 232 nm and 4500 M−1 cm−1 for acetyl-CoA (Dawson et al., 1986; Lee et al., 1997; Egland and Harwood, 2000).
Terpenoid Quinones Analyses
For ultraperformance liquid chromatography–electrospray ionization–tandem mass spectrometry analysis, ubiquinone was extracted as previously described (Ducluzeau et al., 2012), except that the reduction step with NaBH4 was omitted. Reduced and oxidized ubiquinones were separated on an Eclipse Plus C18 column (Agilent Technologies) held at 40°C at a flow rate of 0.2 mL min−1 with a binary gradient system consisting of solvent A, methanol:isopropanol (80:20) + 10 mM ammonium acetate, and solvent B, isopropanol:hexane (60:40) + 10 mM ammonium acetate. An 8-min gradient was generated starting with 30% solvent B for 3 min and increased to 60% solvent B over 2 min and held at 60% for 1 min before returning to 30% B at 6.6 min. The eluate was directed to a TurboV ion source of the 4000 QTRAP triple quadrupole mass spectrometer operated in positive mode at +5000 V. The temperature was held at 200°C, nebulizing gas (GS1) was set at 16 p.s.i., focusing gas (GS2) at 20 p.s.i., curtain gas at 20 p.s.i., and the interface heater was engaged. Declustering potential (40 V), collision energy (34 V), entrance potential (10 V), and collision exit potential (11 V) were optimized using a standard of ubiquinone-10. Reduced and oxidized ubiquinones were measured by Multiple Reaction Monitoring using dwell times of 50 ms and the following ion pairs: ubiquinone-9 (812.8/197.1), ubiquinol-9 (814.8/197.1), ubiquinone-10 (880.8/197.1), ubiquinol-10 (882.8/197.1), ubiquinone-9-[13C6] (818.8/203.1), ubiquinol-9-[13C6] (820.8/203.1), ubiquinone-10-[13C6] (886.8/203.1), and ubiquinol-10-[13C6] (888.8/203.1). Retention times were 3.1 min for ubiquinol-9, 3.6 min for ubiquinol-10, 4.1 min for ubiquinone-9, and 5 min for ubiquinone-10. HPLC-diode array spectrophotometry analysis of total ubiquinones was performed as previously described (Ducluzeau et al., 2012). Phylloquinone levels were determined on the leaves of Arabidopsis plants grown on Murashige and Skoog medium containing sucrose using the HPLC-fluorescence method of Widhalm et al. (2012).
Subcellular Localization and Overexpression of At4g19010 in Arabidopsis
Full-length At4g19010 cDNA was subcloned into pK7WGF2 (Karimi et al., 2002) using Gateway technology, resulting in an in-frame fusion of GFP to the N terminus of At4g19010. The KAT2-eqFP611 peroxisomal marker cassette, encoding for a fragment (residues 1 to 99) of Arabidopsis 3-keto-acyl-CoA thiolase 2 fused to the N-terminal end of RFP under the control of the 35S promoter (Forner and Binder, 2007), was subcloned into the EcoRI and PstI sites of binary vector PZP212 (Hajdukiewicz et al., 1994). Vector pLN3639 allowed expression of the IVDH mitochondrial marker consisting of the N-terminal region of isovaleryl-CoA dehydrogenase fused to the N-terminal end of RFP under the control of the 35S promoter (Block et al., 2010). The 35S:GFP-At4g19010 construct was electroporated into Agrobacterium tumefaciens C58C1 for subsequent coinfiltration into the leaves of Nicotiana benthamiana with a second strain harboring either PZP212-35S:KAT2-eqFP611 or pLN3639. Epidermal cells were imaged by confocal microscopy 48 h later using a Nikon A1 laser scanning microscope. GFP, RFP, and chlorophyll were excited and collected sequentially. Excitation wavelengths were 488 nm for GFP, 561 nm for RFP, and 641 nm for chlorophyll. Emissions were collected from 500 to 550 nm for GFP, 570 to 620 nm for RFP, and 663 to 738 nm for chlorophyll.
Stable Overexpression of At4g19010 in Arabidopsis
Full-length At4g19010 cDNA was subcloned under the control of the 35S promoter in pB2GW7 (Karimi et al., 2002) using Gateway technology. This construct was introduced into Arabidopsis plants ecotype Columbia-0 using the floral dip method (Clough and Bent, 1998), and the resulting transgenics were selected on soil by spraying 100 mg L−1 of BASTA.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL libraries under the following accession numbers: At2g34630 (SPS3), At4g01660 (ABC1), At2g44520 (COX10), At3g15352 (COX17), At1g02410 (COX11), At5g56090 (COX15), At1g26340 (CB5-A), AtMg00510 (NAD7), At4g21090 (MFDX2), At3g17910 (SURF1), At5g62050 (OXA1), and At3g08950 (HCC1). At4g19010 (AAO64847.1) and its orthologs EOY25276.1 (T. cacao), AGO89326.1 (S. arbutifolia), XP_002304364.1 (P. trichocarpa), XP_004144410.1 (C. sativus), XP_004301643.1 (F. vesca), EMJ05131.1 (P. persica), XP_004512448.1 (C. arietinum), XP_004245937.1 (S. lycopersicum), XP_002270360.1 (V. vinifera), DAA46521.1 (Z. mays), EAZ05563.1 (O. sativa), EAZ05563.1 (B. distachyon), and XP_002985111.1 (S. moellendorffii).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Extracted Counts of Ion Pairs for Multiple Reaction Monitorings of Ubiquinone Isotopes.
Supplemental Figure 2. Formation of the Ring of Plastid Prenyl Benzoquinones from Tyrosine.
Supplemental Table 1. CoA Ligase Activity in Arabidopsis Extracts.
Supplemental Data Set 1. Correlation Ranks and Functional Annotations of the Top 500 Coexpressors of At4g19010, SPS3, and ABC1.
Supplementary Material
Acknowledgments
This work was made possible in part by National Science Foundation Grants MCB-0918258, MCB-1148968 (to G.J.B.), and MCB-0919987 (to N.D.) and by the Gordon and Betty Moore Foundation through Grant GBMF2550.02 to the Life Sciences Research Foundation (to J.R.W.). We thank Doriane LeBourse for assistance with T-DNA mutant screening and subcloning of At4g19010 cDNA.
AUTHOR CONTRIBUTIONS
A.B., J.R.W., R.E.C., and G.J.B. designed and performed the research, analyzed the data, and wrote the article. A.F., Y.W., and C.E. performed the research and analyzed the data. S.A.M., E.B.C., C.C., and N.D. analyzed the data.
Glossary
- pABA
para-aminobenzoate
Footnotes
Online version contains Web-only data.
References
- Achnine L., Blancaflor E.B., Rasmussen S., Dixon R.A. (2004). Colocalization of L-phenylalanine ammonia-lyase and cinnamate 4-hydroxylase for metabolic channeling in phenylpropanoid biosynthesis. Plant Cell 16: 3098–3109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso J.M., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301: 653–657 [DOI] [PubMed] [Google Scholar]
- Avelange-Macherel M.-H., Joyard J. (1998). Cloning and functional expression of AtCOQ3, the Arabidopsis homologue of the yeast COQ3 gene, encoding a methyltransferase from plant mitochondria involved in ubiquinone biosynthesis. Plant J. 14: 203–213 [DOI] [PubMed] [Google Scholar]
- Block A., Fristedt R., Rogers S., Kumar J., Barnes B., Barnes J., Elowsky C.G., Wamboldt Y., Mackenzie S.A., Redding K., Merchant S.S., Basset G.J. (2013). Functional modeling identifies paralogous solanesyl-diphosphate synthases that assemble the side chain of plastoquinone-9 in plastids. J. Biol. Chem. 288: 27594–27606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Block A., Guo M., Li G., Elowsky C., Clemente T.E., Alfano J.R. (2010). The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell. Microbiol. 12: 318–330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M.M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Bussell J.D., Reichelt M., Wiszniewski A.A., Gershenzon J., Smith S.M. (2014). Peroxisomal ATP-binding cassette transporter COMATOSE and the multifunctional protein abnormal INFLORESCENCE MERISTEM are required for the production of benzoylated metabolites in Arabidopsis seeds. Plant Physiol. 164: 48–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardazzo B., Hamel P., Sakamoto W., Wintz H., Dujardin G. (1998). Isolation of an Arabidopsis thaliana cDNA by complementation of a yeast abc1 deletion mutant deficient in complex III respiratory activity. Gene 221: 117–125 [DOI] [PubMed] [Google Scholar]
- Clarke C.F. (2000). New advances in coenzyme Q biosynthesis. Protoplasma 213: 134–147 [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Colquhoun T.A., Marciniak D.M., Wedde A.E., Kim J.Y., Schwieterman M.L., Levin L.A., Van Moerkercke A., Schuurink R.C., Clark D.G. (2012). A peroxisomally localized acyl-activating enzyme is required for volatile benzenoid formation in a Petunia×hybrida cv. ‘Mitchell Diploid’ flower. J. Exp. Bot. 63: 4821–4833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa M.A., et al. (2005). Characterization in vitro and in vivo of the putative multigene 4-coumarate:CoA ligase network in Arabidopsis: syringyl lignin and sinapate/sinapyl alcohol derivative formation. Phytochemistry 66: 2072–2091 [DOI] [PubMed] [Google Scholar]
- Dawson, R.M.C., Elliot, D.C., Elliot, W.H., and Jones, K.M. (1986). Data for Biochemical Research. (New York: Oxford University Press). [Google Scholar]
- DellaPenna D., Pogson B.J. (2006). Vitamin synthesis in plants: tocopherols and carotenoids. Annu. Rev. Plant Biol. 57: 711–738 [DOI] [PubMed] [Google Scholar]
- De Marcos Lousa C., van Roermund C.W., Postis V.L., Dietrich D., Kerr I.D., Wanders R.J., Baldwin S.A., Baker A., Theodoulou F.L. (2013). Intrinsic acyl-CoA thioesterase activity of a peroxisomal ATP binding cassette transporter is required for transport and metabolism of fatty acids. Proc. Natl. Acad. Sci. USA 110: 1279–1284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ducluzeau A.-L., Wamboldt Y., Elowsky C.G., Mackenzie S.A., Schuurink R.C., Basset G.J.C. (2012). Gene network reconstruction identifies the authentic trans-prenyl diphosphate synthase that makes the solanesyl moiety of ubiquinone-9 in Arabidopsis. Plant J. 69: 366–375 [DOI] [PubMed] [Google Scholar]
- Egland P.G., Harwood C.S. (2000). HbaR, a 4-hydroxybenzoate sensor and FNR-CRP superfamily member, regulates anaerobic 4-hydroxybenzoate degradation by Rhodopseudomonas palustris. J. Bacteriol. 182: 100–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehlting J., Büttner D., Wang Q., Douglas C.J., Somssich I.E., Kombrink E. (1999). Three 4-coumarate:coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J. 19: 9–20 [DOI] [PubMed] [Google Scholar]
- Footitt S., Slocombe S.P., Larner V., Kurup S., Wu Y., Larson T., Graham I., Baker A., Holdsworth M. (2002). Control of germination and lipid mobilization by COMATOSE, the Arabidopsis homologue of human ALDP. EMBO J. 21: 2912–2922 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner J., Binder S. (2007). The red fluorescent protein eqFP611: application in subcellular localization studies in higher plants. BMC Plant Biol. 7: 28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaid M.M., Sircar D., Müller A., Beuerle T., Liu B., Ernst L., Hänsch R., Beerhues L. (2012). Cinnamate:CoA ligase initiates the biosynthesis of a benzoate-derived xanthone phytoalexin in Hypericum calycinum cell cultures. Plant Physiol. 160: 1267–1280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajdukiewicz P., Svab Z., Maliga P. (1994). The small, versatile pPZP family of Agrobacterium binary vectors for plant transformation. Plant Mol. Biol. 25: 989–994 [DOI] [PubMed] [Google Scholar]
- Hamberger B., Hahlbrock K. (2004). The 4-coumarate:CoA ligase gene family in Arabidopsis thaliana comprises one rare, sinapate-activating and three commonly occurring isoenzymes. Proc. Natl. Acad. Sci. USA 101: 2209–2214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi M., Inzé D., Depicker A. (2002). GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 7: 193–195 [DOI] [PubMed] [Google Scholar]
- Kawamukai M. (2009). Biosynthesis and bioproduction of coenzyme Q10 by yeasts and other organisms. Biotechnol. Appl. Biochem. 53: 217–226 [DOI] [PubMed] [Google Scholar]
- Kienow L., Schneider K., Bartsch M., Stuible H.P., Weng H., Miersch O., Wasternack C., Kombrink E. (2008). Jasmonates meet fatty acids: functional analysis of a new acyl-coenzyme A synthetase family from Arabidopsis thaliana. J. Exp. Bot. 59: 403–419 [DOI] [PubMed] [Google Scholar]
- Klempien A., et al. (2012). Contribution of CoA ligases to benzenoid biosynthesis in petunia flowers. Plant Cell 24: 2015–2030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein D.J., D’Auria J.C., Behere A.S., Kim J.H., Gunderson K.L., Breen J.N., Lee G., Gershenzon J., Last R.L., Jander G. (2007). Characterization of seed-specific benzoyloxyglucosinolate mutations in Arabidopsis thaliana. Plant J. 51: 1062–1076 [DOI] [PubMed] [Google Scholar]
- Koo A.J.K., Chung H.S., Kobayashi Y., Howe G.A. (2006). Identification of a peroxisomal acyl-activating enzyme involved in the biosynthesis of jasmonic acid in Arabidopsis. J. Biol. Chem. 281: 33511–33520 [DOI] [PubMed] [Google Scholar]
- Law A.H., Threlfall D.R., Whistance G.R. (1971). Isoprenoid phenol and quinone precursors of ubiquinones and dihydroubiquinones [Ubiquinones (H2)] in fungi. Biochem. J. 123: 331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D., Meyer K., Chapple C., Douglas C.J. (1997). Antisense suppression of 4-coumarate:coenzyme A ligase activity in Arabidopsis leads to altered lignin subunit composition. Plant Cell 9: 1985–1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Kaminaga Y., Cooper B., Pichersky E., Dudareva N., Chapple C. (2012). Benzoylation and sinapoylation of glucosinolate R-groups in Arabidopsis. Plant J. 72: 411–422 [DOI] [PubMed] [Google Scholar]
- Löscher R., Heide L. (1994). Biosynthesis of p-hydroxybenzoate from p-coumarate and p-coumaroyl-CoA in cell-free extracts of Lithospermum erythrorhizon cell cultures. Plant Physiol. 106: 271–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marbois B., Xie L.X., Choi S., Hirano K., Hyman K., Clarke C.F. (2010). para-Aminobenzoic acid is a precursor in coenzyme Q6 biosynthesis in Saccharomyces cerevisiae. J. Biol. Chem. 285: 27827–27838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols B.P., Green J.M. (1992). Cloning and sequencing of Escherichia coli ubiC and purification of chorismate lyase. J. Bacteriol. 174: 5309–5316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowicka B., Kruk J. (2010). Occurrence, biosynthesis and function of isoprenoid quinones. Biochim. Biophys. Acta 1797: 1587–1605 [DOI] [PubMed] [Google Scholar]
- Obayashi T., Hayashi S., Saeki M., Ohta H., Kinoshita K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37: D987–D991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada K., Ohara K., Yazaki K., Nozaki K., Uchida N., Kawamukai M., Nojiri H., Yamane H. (2004). The AtPPT1 gene encoding 4-hydroxybenzoate polyprenyl diphosphate transferase in ubiquinone biosynthesis is required for embryo development in Arabidopsis thaliana. Plant Mol. Biol. 55: 567–577 [DOI] [PubMed] [Google Scholar]
- Olson R.E., Bentley R., Aiyar A.S., Dialameh G.H., Gold P.H., Ramsey V.G., Springer C.M. (1963). Benzoate derivatives as intermediates in the biosynthesis of coenzyme Q in the rat. J. Biol. Chem. 238: 3146–3148 [PubMed] [Google Scholar]
- Pierrel F., Hamelin O., Douki T., Kieffer-Jaquinod S., Mühlenhoff U., Ozeir M., Lill R., Fontecave M. (2010). Involvement of mitochondrial ferredoxin and para-aminobenzoic acid in yeast coenzyme Q biosynthesis. Chem. Biol. 17: 449–459 [DOI] [PubMed] [Google Scholar]
- Pribat A., et al. (2010). Nonflowering plants possess a unique folate-dependent phenylalanine hydroxylase that is localized in chloroplasts. Plant Cell 22: 3410–3422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qualley A.V., Widhalm J.R., Adebesin F., Kish C.M., Dudareva N. (2012). Completion of the core β-oxidative pathway of benzoic acid biosynthesis in plants. Proc. Natl. Acad. Sci. USA 109: 16383–16388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlivan E.P., Roje S., Basset G., Shachar-Hill Y., Gregory J.F., III, Hanson A.D. (2003). The folate precursor p-aminobenzoate is reversibly converted to its glucose ester in the plant cytosol. J. Biol. Chem. 278: 20731–20737 [DOI] [PubMed] [Google Scholar]
- Pfaff C., Glindemann N., Gruber J., Frentzen M., Sadre R. (2014). Chorismate pyruvate-lyase and 4-hydroxy-3-solanesylbenzoate decarboxylase are required for plastoquinone biosynthesis in the cyanobacterium Synechocystis sp. PCC6803. J. Biol. Chem. 289: 2675–2686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reumann S. (2013). Biosynthesis of vitamin k1 (phylloquinone) by plant peroxisomes and its integration into signaling molecule synthesis pathways. Subcell. Biochem. 69: 213–229 [DOI] [PubMed] [Google Scholar]
- Richmond T.A., Bleecker A.B. (1999). A defect in beta-oxidation causes abnormal inflorescence development in Arabidopsis. Plant Cell 11: 1911–1924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S.J., et al. (2009). An archived activation tagged population of Arabidopsis thaliana to facilitate forward genetics approaches. BMC Plant Biol. 9: 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilmiller A.L., Stout J., Weng J.K., Humphreys J., Ruegger M.O., Chapple C. (2009). Mutations in the cinnamate 4-hydroxylase gene impact metabolism, growth and development in Arabidopsis. Plant J. 60: 771–782 [DOI] [PubMed] [Google Scholar]
- Shockey J., Browse J. (2011). Genome-level and biochemical diversity of the acyl-activating enzyme superfamily in plants. Plant J. 66: 143–160 [DOI] [PubMed] [Google Scholar]
- Van Moerkercke A., Schauvinhold I., Pichersky E., Haring M.A., Schuurink R.C. (2009). A plant thiolase involved in benzoic acid biosynthesis and volatile benzenoid production. Plant J. 60: 292–302 [DOI] [PubMed] [Google Scholar]
- Widhalm J.R., Ducluzeau A.-L., Buller N.E., Elowsky C.G., Olsen L.J., Basset G.J.C. (2012). Phylloquinone (vitamin K(1)) biosynthesis in plants: two peroxisomal thioesterases of lactobacillales origin hydrolyze 1,4-dihydroxy-2-naphthoyl-coa. Plant J. 71: 205–215 [DOI] [PubMed] [Google Scholar]
- Zolman B.K., Silva I.D., Bartel B. (2001). The Arabidopsis pxa1 mutant is defective in an ATP-binding cassette transporter-like protein required for peroxisomal fatty acid β-oxidation. Plant Physiol. 127: 1266–1278 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.