Cm-BBX24, a zinc finger transcription factor gene, causes the changes of flowering time under non-floral-inductive long-day conditions in short-day-requiring chrysanthemums, and the regulation involves FT, which integrates photoperiod and GA biosynthesis pathways. Cm-BBX24 also regulates abiotic stress tolerance through influencing bioactive GA biosynthesis.
Abstract
Flowering time and an ability to tolerate abiotic stresses are important for plant growth and development. We characterized BBX24, a zinc finger transcription factor gene, from Chrysanthemum morifolium and found it to be associated with both flowering time and stress tolerance. Transgenic lines with suppressed expression of Cm-BBX24 (Cm-BBX24-RNAi) flowered earlier than wild-type plants and showed decreased tolerance to freezing and drought stresses. Global expression analysis revealed that genes associated with both photoperiod and gibberellin (GA) biosynthesis pathways were upregulated in Cm-BBX24-RNAi lines, relative to the wild type. By contrast, genes that were upregulated in overexpressing lines (Cm-BBX24-OX), but downregulated in Cm-BBX24-RNAi lines (both relative to the wild type), included genes related to compatible solutes and carbohydrate metabolism, both of which are associated with abiotic stress. Cm-BBX24 expression was also influenced by daylength and GA4/7 application. Under long days, changes in endogenous GA1, GA4, GA19, and GA20 levels occurred in young leaves of transgenic lines, relative to the wild type. Regulation of flowering involves the FLOWERING TIME gene, which integrates photoperiod and GA biosynthesis pathways. We postulate that Cm-BBX24 plays a dual role, modulating both flowering time and abiotic stress tolerance in chrysanthemum, at least in part by influencing GA biosynthesis.
INTRODUCTION
Flowering at the appropriate time of year is essential for successful reproduction and also has commercial significance for crops and ornamental plants. Flowering time is determined by external environmental cues and endogenous developmental signals (Lang 1965; Fornara et al., 2010). Plants have been shown to utilize various and often interconnecting flowering mechanisms, including photoperiod, vernalization, gibberellin (GA) biosynthesis, and aging pathways (Song et al., 2013). Outputs from these pathways are integrated by a set of common downstream flowering-time integrators, such as FLOWERING LOCUS T (FT) and SUPPRESSOR OF OVEREXPRESSION OF CO1 (SOC1), whose expression leads to the induction of floral meristem identity genes, including LEAFY (LFY) and APETALA1 and, consequently, flowering (Song et al., 2013).
Plants can be classified as long-day (LD), short-day (SD), or day-neutral based on daylength requirements for flowering (Lang, 1965; Srikanth and Schmid, 2011). A core component of the photoperiod pathway is CONSTANS (CO), a gene that upregulates the immediate downstream genes FT and SOC1 (Samach et al., 2000). CO is classified as a B-box protein and was the first such protein identified from Arabidopsis thaliana (Putterill et al., 1995). The B-box is a zinc finger binding domain consisting of conserved Cys and His residues, and proteins with one or two B-boxes in the N-terminal region are all classed as transcription factors and termed BBX, according to the nomenclature proposed for Arabidopsis (Khanna et al., 2009). In Arabidopsis, the BBX family has 32 members and is divided into five structural groups, based on the number and sequence features of the B-box domain and whether the protein also contains a CCT domain (Khanna et al., 2009). Structure group I has six members (CO, also known as BBX1, and BBX2 to 6), each of which contains two B-box domains and a CCT domain. Structure group II has seven members (BBX7 to 13), which are structurally similar to group I but have differences in their second B-box domain. Structure group III has four members (BBX14 to 17) characterized by having only one B-box domain and a CCT domain. BBX2 to 17 are all CO-LIKE (COL) proteins. Structure group IV has eight members (BBX18 to 25), which contain two B-box domains but have no CCT domain. Lastly, structure group V has seven members (BBX26 to 32), each of which has just a single B-box domain (Khanna et al., 2009).
It has been demonstrated that some members of structure groups I and III act to regulate flowering time. Of these, CO (BBX1) is the best-characterized BBX member, acting as a transcription factor in the photoperiodic pathway’s regulation of flowering time (Putterill et al., 1995). Additionally, COL genes are known to be associated with the positive or negative regulation of flowering time. For example, COL5 (BBX6) can induce flowering in Arabidopsis (Hassidim et al., 2009), while COL3 (BBX4) expression delays flowering (Cheng and Wang, 2005). In rice (Oryza sativa), Heading date 1, the ortholog of Arabidopsis CO, shortens time to heading (Yano et al., 2000), while Os-CO3 delays flowering (Kim et al., 2008). Arabidopsis plants with a mutation in the eip6 (bbx32) gene, which encodes a member of structure group V, display earlier flowering and increased expression of flowering time and floral organ identity genes, while plants overexpressing EMF1-Interacting Protein 6 (BBX32) show late-flowering phenotypes (Park et al., 2011). However, less is known about BBXs from structure group IV, with regard to their ability to modulate other flowering-related pathways.
Induction of flowering and shortening of time to flowering is also promoted in many plant species by application of GAs, as was initially demonstrated by application of GA3 to Arabidopsis (Langridge, 1957), and it has subsequently been shown that changes in GA biosynthesis and signaling are involved in the regulation of flowering in Arabidopsis and Lolium temulentum (Srikanth and Schmid, 2011). Gibberellin A12, an early GA in the biosynthesis pathway, is converted to bioactive GA4 via GA15, GA24, and GA9 (in the early nonhydroxylation pathway) or to bioactive GA1 via GA53, GA44, GA19, and GA20 (in the early C-13-hydroxylation pathway; Yamaguchi, 2008). Two 2-oxoglutarate–dependent dioxygenases GA 20-oxidase (GA20ox) and GA 3-oxidase (GA3ox) catalyze the formation of these bioactive GAs (Olszewski et al., 2002).
A mechanistic basis for the interaction between the photoperiodic and GA biosynthesis pathways is hinted at by the convergence of both pathways on the promotion of FT and SOC1 transcription in the meristem of Arabidopsis (Moon et al., 2003; Porri et al., 2012). In the LD plant Arabidopsis, mutations that inhibit the GA biosynthesis pathway or increase the degradation of GAs can delay flowering in SD (Fornara et al. 2010). Conversely, application of florigenic GA structures or overexpression of GA biosynthesis pathway genes leads to early flowering phenotypes in Arabidopsis by activating SOC1 and its target gene LFY (Blázquez et al., 1998; Moon et al., 2003). However, to date, little is known about the possible regulation of flowering time by members of the BBX gene family and specifically in association with the GA biosynthesis pathway, especially for SD-requiring plant species.
In addition to flowering time, another crucial factor that can limit plant growth and productivity are environmental stresses, such as those induced by low temperature, drought, and salinity. These abiotic stresses can induce physiological and biochemical changes in plants, changes that are apparent as a broad range of adaptive responses (Hirayama and Shinozaki, 2010). Many of these responses have been associated with transcriptional regulation, and it appears that various transcription factors play a causal role in the tolerance of plants to abiotic stresses. Well-characterized examples include members of the Dehydration-Responsive Element Binding (DREB) protein, NAM ATAF CUC2, MYB, MYC, WRKY, and zinc finger protein transcription factor classes. These transcription factors can variously function as transcriptional activators or repressors and govern downstream gene expression either cooperatively or independently in stress signaling (Hirayama and Shinozaki, 2010).
Several reports have also described the involvements of BBX proteins in abiotic stress tolerance. In Arabidopsis, the transcripts of BBX1-3, BBX6-9, BBX11, BBX13-14, BBX16, BBX29, BBX31, and structure group IV members BBX18-19, BBX22, BBX24 (STO), and BBX25 (STH1/STH) are all induced by low temperature (Winter et al., 2007). Similarly, in banana (Musa nana), the abundance of Ma-COL1 transcripts, from structure group I, increases in response to chilling (J. Chen et al., 2012), as does grapevine (Vitis vinifera) ZFPL, which is an ortholog of Arabidopsis BBX32 from structure group V, whose overexpression can enhance cold tolerance (Takuhara et al., 2011). Another BBX protein associated with abiotic stress is BBX24, which was initially described as a protein complementing the salt-sensitive phenotype of yeast calcineurin null mutants cna1, cna2, and cnb1, and a yeast calcium channel-deficient mutant, cch1 (Lippuner et al., 1996). In Arabidopsis seedlings, the overexpression of BBX24 (STO) enhances root growth on a high salt medium (Nagaoka and Takano, 2003). However, to date, other than these few examples, there is limited information concerning possible regulatory mechanisms of abiotic stress tolerance being associated with BBX family members.
Chrysanthemum (Chrysanthemum morifolium), a commercially important ornamental plant worldwide, is a plant in which SD hastens time to flowering. The annual commercial production of flowering chrysanthemum plants depends mainly on artificial regulation of daylength (Cockshull, 1985). Abiotic stresses, such as low temperature and drought, can be major impediments to the growth, development, and ornamental traits of chrysanthemum and can thus limit productivity. Elucidation of the molecular mechanisms of time to flowering and abiotic stress tolerance is therefore of considerable importance. In this study, we report that Cm-BBX24, a zinc finger transcription factor gene from chrysanthemum, plays dual roles in the regulation of flowering time and abiotic stress tolerance: the former by influencing the expression of genes related to the photoperiod pathway and the biosynthesis of endogenous GAs and the latter (abiotic stress tolerance) by being associated with a range of stress response mechanisms.
RESULTS
Isolation of Chrysanthemum BBX24
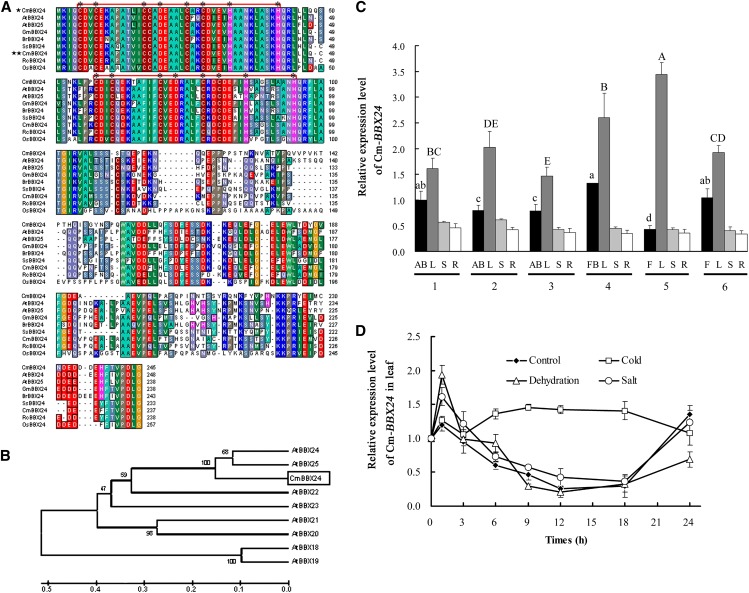
To investigate the function of BBX genes in chrysanthemum, we identified a BBX homolog with a predicted open reading frame (ORF) of 738 bp that encodes a deduced protein of 245 amino acids. A BLASTP search showed that the deduced polypeptide has similarity to BBX proteins from a range of plant species and contains all of the characteristics of structure group IV members of the BBX family: a highly conserved double B-box domain in the N terminus but no CCT domain in the C terminus (Figure 1A). Phylogenetic analysis of BBX proteins from a range of plant species revealed the predicted protein to cluster with structure group IV BBX homologs from Arabidopsis and that this gene is a homolog of At-BBX24 and At-BBX25 (Figure 1B), with the greatest similarity to At-BBX24. Accordingly, we designated the gene as Cm-BBX24, following the nomenclature system suggested by Khanna et al. (2009).
Figure 1.
Comparison of Deduced Amino Acid Sequences of Cm-BBX24 with BBXs from Other Plant Species and Expression Patterns of Cm-BBX24 in Chrysanthemum.
(A) Alignment of deduced amino acid sequences of Cm-BBX24 with other plant BBXs, with conserved amino acids shaded in different colors. The stars before Cm-BBX24 indicate the deduced protein sequence from chrysanthemum and melon (Cucumis melo), respectively. Red lines indicate two conserved B-box domains (B1 and B2), and the asterisks indicate the Zn2+-ligating conserved Cys, His, and Asp residues present in the B-box.
(B) Phylogenetic analysis of Cm-BBX24 and eight other structure group IV BBXs from Arabidopsis. Bootstrap values indicate the divergence of each branch and the scale indicates branch length.
(C) Expression of Cm-BBX24 in different tissues at various developmental periods. 1, young plant; 2, mature plant before differentiation; 3, initial period of flower bud differentiation; 4, visible capitulum; 5, initial flowering; 6, flowering; AB, apical buds; L, leaves; S, stems; R, roots; FB, flower buds; F, flowers. Ubiquitin was used as the control. Error bars indicate sd (n = 3). Significant differences were analyzed by Duncan’s multiple range test (P < 0.05).
(D) Expression of Cm-BBX24 under cold, dehydration and salt stress conditions. Error bars indicate sd (n = 3).
[See online article for color version of this figure.]
To investigate the potential role of Cm-BBX24 in regulating time to flowering, we evaluated its expression in different organs, including apical buds, flower buds, leaves, stems, and roots at different vegetative and reproductive growth stages, by quantitative RT-PCR (qRT-PCR). Cm-BBX24 transcripts were detected in all organs and at all developmental stages tested, and expression in leaves and apical buds, including flower buds, showed significant changes during development, with relatively high transcript abundance in young plants, a decrease in mature plants prior to flower bud differentiation, and a further decrease during the initial period of flower bud differentiation, i.e., prior to the time when the capitulum first becomes visible (Figure 1C). These results suggest that during plant development a temporal decrease of Cm-BBX24 expression in leaves and apical buds might be associated with the beginning of flower bud differentiation.
To characterize the regulation of Cm-BBX24 expression, we isolated ∼2.7 kb of genomic DNA sequence upstream of the Cm-BBX24 coding sequence. Analysis of putative cis-regulatory elements using the PLACE database (Supplemental Figure 1A) revealed that the promoter contains predicted flowering-related cis-elements, such as CARG, and motifs that are associated with plant hormones (GARE/CARE, GAs; ABRE, abscisic acid), light (T-BOX, GT-1, circadian, and CCA1), and abiotic stress–related (DRE, MYC, and MYB) signaling. The presence of various potential cis-elements in the upstream region of Cm-BBX24 suggested that the gene is regulated by multiple external environmental and internal hormonal cues. In addition, β-glucuronidase activity of the Cm-BBX24 promoter in Arabidopsis was significantly enhanced by treatment with GA4/7, cold, and mannitol and was slightly induced by abscisic acid and NaCl treatments (Supplemental Figure 1B).
We then investigated whether the expression of the Cm-BBX24 in leaves is under the regulation of a circadian clock. The results showed that transcript levels of Cm-BBX24 oscillated, with a peak occurring at about ZT3 (Zeitgeber time 3 from light), followed by a second peak ∼24 h later (ZT27) and a third peak ∼48 h later (Supplemental Figure 2A). This indicates that Cm-BBX24 exhibits a robust free-running circadian rhythm.
The possible involvement of Cm-BBX24 in abiotic stress responses was assessed by exposing young plants to dehydration, low temperature (4°C), and salt conditions. Under dehydrating conditions, Cm-BBX24 transcripts showed a rapid and transient induction in leaves (Figure 1D), amounting to an ∼1.6-fold increase at the 1-h time point, relative to untreated control plants. Low-temperature treatment also induced expression after 3 h, with further increased expression maintained through 18 h. Salt stress conditions elicited a slight induction of expression in leaves at the 1-h time point. These results indicated that gene transcription was inducible by both low temperature and dehydration.
We further assessed the expression of Cm-BBX24 in chrysanthemum wild-type plants under different daylengths for 30 d. Under LD, expression of Cm-BBX24 remained unchanged and at a relatively high level. By contrast, under SD, where time to flowering is substantially reduced, expression substantially decreased from 3 d, and remained at a relatively low level (Supplemental Figure 2B). Thus, expression of Cm-BBX24 is responsive to daylength.
Structural Features of Cm-BBX24
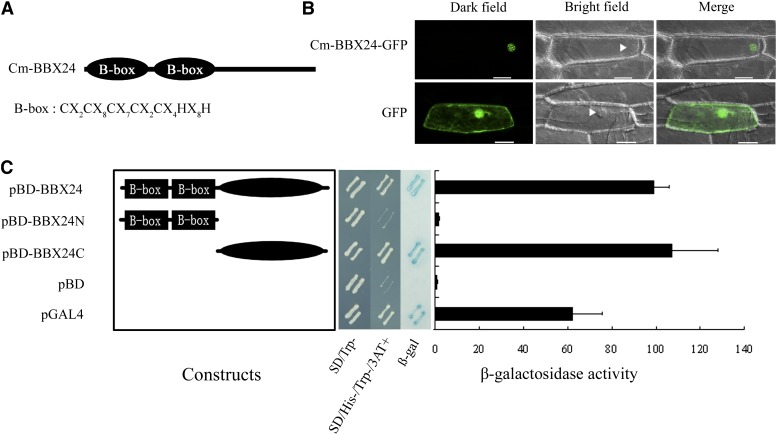
Cm-BBX24 contains two B-box domains in the N terminus and no CCT domain (Figure 2A). The B-box is a zinc binding domain, consisting of conserved Cys and His residues. To obtain supporting evidence that Cm-BBX24 functions as a transcription factor, we determined the subcellular localization of a fusion protein comprising Cm-BBX24 fused to green fluorescent protein (Cm-BBX24-GFP) following transient expression in onion epidermal cells. The Cm-BBX24-GFP fusion protein localized predominantly in the nucleus, while the GFP protein alone was detected in both cytoplasm and nucleus, as expected (Figure 2B).
Figure 2.
Protein Structure, Subcellular Localization, and Transactivation of Cm-BBX24.
(A) Schematic diagram of the protein structures.
(B) Subcellular localization in onion epidermal cells. Arrowheads indicate nuclei. Images are shown as dark field (left), bright field (middle), and merged (right). Bars = 50 μm.
(C) Transactivation activity analysis in yeast cells. Left panel: Diagram of the three different constructs of Cm-BBX24. The pBD and pGAL4 plasmids were used as the negative and positive controls, respectively. Middle panel: transactivation activity. SD-Trp, SD medium without tryptophan; SD/His-/Trp-/3AT+ medium, SD medium without histidine and tryptophan, but with 10 mM 3-AT; β-Gal, β-galactosidase activity. Right panel: Quantitative assay of β-galactosidase activity. Error bars indicate sd (n = 3).
We then performed a transactivation assay in yeast. The sequences encoding the ORF, N-terminal, and C-terminal regions of Cm-BBX24 were individually inserted into the expression vector pBD-GAL4, and each of the different constructs was transformed into the yeast strain YRG-2 containing the His3 and LacZ reporter genes. The pGAL4 transcription factor and pBD vector were used as positive and negative controls, respectively. Yeast growth on the His-deficient medium, LacZ staining, and the relative quantitative assay of β-galactosidase activity (Figure 2C) indicated that Cm-BBX24 plays a role as a transcriptional activator and that the activation domain is present in the C-terminal region.
Growth and Development of Cm-BBX24–Overexpressing or RNA Interference–Suppressed Chrysanthemum Lines, and Cm-BBX24 Overexpression in Arabidopsis
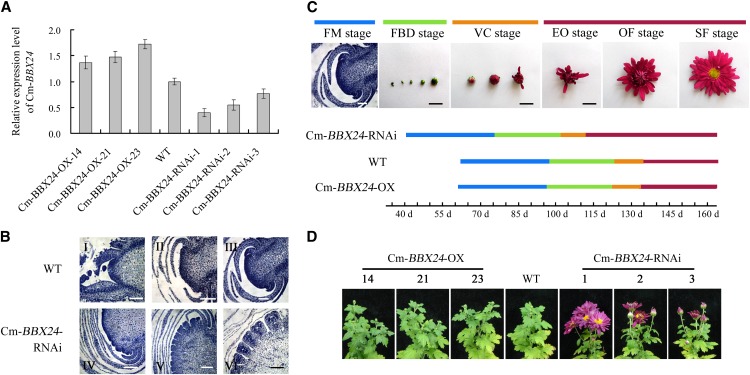
To establish whether Cm-BBX24 plays a role in regulating flowering time, we overexpressed (OX) and suppressed (RNA interference [RNAi]) Cm-BBX24 in chrysanthemum, obtaining 15 T0 clones of Cm-BBX24-OX and 11 T0 clones of Cm-BBX24-RNAi. We also evaluated the expression of all five members of chrysanthemum BBX structure group IV and confirmed that only Cm-BBX24 was silenced in Cm-BBX24-RNAi plants (Supplemental Figure 3). We selected three representative transformants each for the Cm-BBX24-OX and Cm-BBX24-RNAi lines to assess their growth and development (Figure 3A). No obvious differences were observed in plant height, crown diameter, leaf numbers, flower diameter, and number of flowers in any of the transgenic lines, compared with the wild type. However, the time to flowering was significantly different in the Cm-BBX24-RNAi plants, relative to the wild-type or Cm-BBX24-OX plants (Figures 3B to 3D). At 55 d after transplanting, the Cm-BBX24-RNAi plants held under LD were already exhibiting differentiation of involucral primordia, while wild-type plants showed no such differentiation. Moreover, at 65 d after transplanting, the Cm-BBX24-RNAi plants showed differentiation of floret primordia, while wild-type plants had just initiated involucral primordium differentiation, and at 75 d after planting, the Cm-BBX24-RNAi plants had entered stage of petal primordium differentiation, while the wild-type plants were at the stage of floret primordium and floret preprimordium formation (Figure 3B). Subsequently, flower buds emerged and bloomed in the Cm-BBX24-RNAi plants in advance of the wild-type plants after ∼20 d. No significant difference of flowering time was observed between Cm-BBX24-OX and wild-type plants (Figures 3C and 3D). All of the above observations were made on plants growing under LD.
Figure 3.
Phenotypic Characterization of the Cm-BBX24–Overexpressing (Cm-BBX24-OX) or RNAi-Suppressed (Cm-BBX24-RNAi) Chrysanthemum Plants.
(A) Expression of Cm-BBX24 in the wild-type and transgenic plants at ZT3, determined by qRT-PCR. Cm-BBX24-OX-14, -21, and -23 correspond to three independent Cm-BBX24-OX lines, and Cm-BBX24-RNAi-1, -2, and -3 correspond to three independent Cm-BBX24 RNAi lines.
(B) Inflorescence differentiation in Cm-BBX24-RNAi and the wild type at various stages. I, before flower bud differentiation; II, involucral primordium differentiation; III, before floret primordium differentiation; IV, involucral primordium differentiation; V, floret primordium formation; VI, petal primordium differentiation. Bars = 100 μm.
(C) Flower bud differentiation in the transgenic lines and the wild type. FM stage, floral meristem stage (bars = 100 µm); FBD stage, flower bud development stage; VC stage, visible color stage; EO stage, earlier opening stage; OF stage, opened flower stage; SF stage, senescing flower stage. Bars = 1 cm.
(D) Generative phenotype of wild-type, Cm-BBX24-OX, and Cm-BBX24-RNAi chrysanthemum lines at the reproductive stage. Plants were photographed 117 d after transplanting.
Chrysanthemum is classed as a SD-requiring plant and, as such, there might be insufficient potential to delay flowering time in Cm-BBX24-OX plants. To test this hypothesis, we overexpressed Cm-BBX24 in Arabidopsis, which is a LD-requiring plant. Compared with Arabidopsis wild-type plants, Cm-BBX24-OX lines grown under LD showed no obvious differences in phenotypes, such as size and color rosette leaves and plant height, but there was a significant delay in flowering time, and at the bolting stage the transgenic Arabidopsis plants had many more rosette leaves than the control plants (Supplemental Figure 4). These results indicate that Cm-BBX24 has the capacity to regulate flowering time in both chrysanthemum and Arabidopsis.
Abiotic Stress Tolerance in Cm-BBX24-OX and Cm-BBX24-RNAi Chrysanthemum
To assess the influence of Cm-BBX24 expression on abiotic stress tolerance in chrysanthemum, we defined specific phenotypes according to the severity of the stress-induced injury. When the injury is relatively mild, the apical shoots can recover growth quickly, when the injury is moderate the apical shoots cannot recover growth, but the lateral shoots grow out instead. When the injury is severe the aerial parts die, but the basal shoots can still grow out. However, when the injury is very severe, plants cannot recover and die.
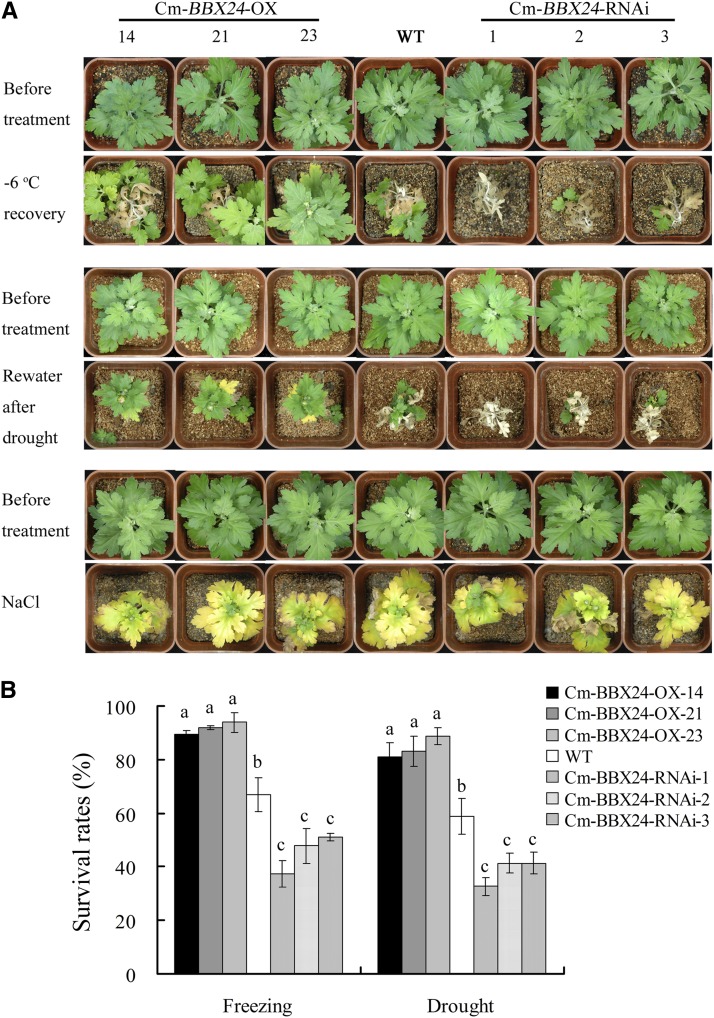
To evaluate freezing tolerance, plants were treated at −6°C for 8 h. After a subsequent 30-d recovery under normal growing conditions, wild-type plants displayed moderate damage, while almost all the Cm-BBX24-OX plants showed only mild damage and maintained a vigorous growth. By contrast, the Cm-BBX24-RNAi plants suffered serious damage (Figure 4A). In one experiment, 71% of the wild-type plants survived, with 67% depending on the outgrowth of lateral or basal shoots, i.e., there was no recovery of apical shoots. Less than 52% of the Cm-BBX24-RNAi plants survived, and they had weak growth, with all surviving plants depending on the outgrowth of lateral or basal shoots. However, more than 88% of the Cm-BBX24-OX plants survived and exhibited strong growth, and plants from Cm-BBX24-OX line 23 recovered with continued apical shoot formation (Figures 4A and 4B).
Figure 4.
Abiotic Stress Tolerance of Cm-BBX24-OX and Cm-BBX24-RNAi Chrysanthemum Lines.
(A) Abiotic stress tolerance of Cm-BBX24-OX and Cm-BBX24-RNAi lines. Cm-BBX24-OX-14, -21, and -23 are three independent T0 lines, as are Cm-BBX24-RNAi-1, -2, and -3.
(B) The survival rates of Cm-BBX24-OX and Cm-BBX24-RNAi lines grown under abiotic stress conditions. Three independent experiments were performed and error bars indicate sd. Significant differences were determined by Duncan’s multiple range test (P < 0.05).
In the drought stress tolerance test, involving 23 d without watering followed by a 30-d recovery period, almost all the Cm-BBX24-OX plants showed mild damage with subsequent vigorous growth, while the Cm-BBX24-RNAi plants were seriously damaged. In one experiment, 54% of wild-type plants survived, but for the Cm-BBX24-RNAi plants, survival rate was only 39%, and almost all of the surviving plants showed only weak outgrowth of the lateral or basal shoots. However, more than 77% of the Cm-BBX24-OX plants survived, and half of those plants recovered quickly with continuous growth of their apical shoots (Figures 4A and 4B).
For the salt stress tolerance study, plants were treated with NaCl for 28 d, after which time all the leaves displayed symptoms of damage, with no obvious differences being observed between the wild-type, Cm-BBX24-RNAi, and Cm-BBX24-OX plants (Figure 4A).
Collectively, these results provide evidence that Cm-BBX24 overexpression can enhance tolerance by chrysanthemum of both low temperatures and drought. By contrast, there was no evidence of improved tolerance to salt stress.
Global Expression Analysis of Cm-BBX24 Downstream-Associated Transcripts Related to Flowering Time and Abiotic Stress Tolerance
To better understand the regulatory mechanisms of Cm-BBX24–mediated regulation of flowering time and abiotic stress tolerance, we performed a large-scale screen of genes that are differentially expressed between wild-type and Cm-BBX24-OX or RNAi chrysanthemum plants using an RNA sequencing (RNA-seq) approach. The raw sequence reads were deposited into the National Center for Biotechnology Information SRA database under the accession numbers SRP038981 and SRA091277. The assembled transcripts that were differentially expressed in Cm-BBX24 transgenic chrysanthemum plants are listed in Supplemental Data Set 1.
In the context of the regulation of flowering time by Cm-BBX24, we focused on the transcripts that were differentially expressed between Cm-BBX24-RNAi and wild-type plants. Based on their annotation, we selected nine transcripts that were upregulated 2-fold and seven that were upregulated 1.5-fold in Cm-BBX24-RNAi plants compared with wild-type plants (Supplemental Figure 5). The corresponding genes are predicted to be components of the photoperiod pathway, GA biosynthesis genes, GA signaling–related genes, MADS box transcription factors, etc. (Table 1). Of these genes, PRR5, GI, and CO (COL) encode proteins related to the photoperiod pathway, while the genes associated with the GA pathway are predicted to play a role in biosynthesis or signal transduction, such as those encoding GA3-oxidase, GA20-oxidase, gibberellin-regulated protein (GRP), GRAS, and flowering promoting factor-like 1 (FPF1). We also observed that FT and SOC1, which are known flowering integrators, were differentially expressed between Cm-BBX24-RNAi and wild-type chrysanthemum lines (Table 1). Collectively, the expression data suggest that Cm-BBX24 effects on flowering time may occur by influencing the regulation of expression for genes that are related to both the photoperiod and GA pathways.
Table 1. Upregulated Transcripts Related to Flowering Time in Cm-BBX24-RNAi Plants.
| Gene | Annotation | Fold Change |
|
|---|---|---|---|
| OX/WT | RNAi/WT | ||
| Photoperiod pathway | |||
| UN052290 | GIGANTEA | 1.14 | 1.62 |
| UN055998 | GIGANTEA | 0.95 | 1.53 |
| UN005597 | PRR5 | 0.65 | 1.88 |
| UN052854 | CONSTANS | 1.02 | 2.11 |
| UN064661 | CONSTANS-like | 0.73 | 1.70 |
| GA biosynthesis and signaling | |||
| UN031599 | Gibberellin 20-oxidase | 0.19 | 3.38 |
| UN049353 | Gibberellin 3-oxidase2 | 0.62 | 2.71 |
| UN035573 | GRP | 1.05 | 2.01 |
| UN030255 | GAI, RGA, and SCARECROW (GRAS) | 1.37 | 2.99 |
| UN052024 | FPF1 | 1.68 | 3.56 |
| Flowering integrators | |||
| UN060780 | SOC1 | 1.07 | 1.61 |
| UN052931 | FT | 1.14 | 1.51 |
| MADS box genes | |||
| UN014190 | MADS box transcription factor CDM111 | 1.04 | 13.00 |
| UN001497 | MIKC MADS box transcription factor | 1.75 | 7.60 |
| UN059046 | MADS box transcription factor CDM41 | 0.68 | 1.83 |
| AP2-EREBP genes | |||
| UN033682 | Transcription factor AP2-EREBP | 0.33 | 11.64 |
Significant differences were determined with P < 0.05 and expression ratio ≥ 1.5. WT, the wild type.
In terms of abiotic stress tolerance, we focused on transcripts that were upregulated in Cm-BBX24-OX plants but downregulated in Cm-BBX24-RNAi plants, both relative to wild-type chrysanthemum. Of the 43 assembled transcripts in this category, 36 have functional annotations based on sequence similarity, while seven have no assigned function (Supplemental Figure 5). The putative proteins corresponding to the assembled transcripts were further classified into two groups: regulatory proteins and functional proteins (Supplemental Table 1). The former category includes five predicted transcription factors (e.g., WRKY and MYB), two signaling proteins (e.g., calmodulin), and four protein kinases (e.g., a receptor kinase). We note that most of these classes of proteins have previously been reported as being stress responsive (Seki et al., 2002). The category of functional proteins includes LEA and dehydration response proteins, compatible solute-related proteins (e.g., lectins), carbohydrate metabolism–related proteins (e.g., UDP-glycosyltransferase), and oxidation-reduction process-related proteins (e.g., cytochrome oxidase subunit). These proteins have also previously been reported to act either directly or indirectly in abiotic stress responses (Seki et al., 2002).
To validate the data from the RNA-seq digital expression analysis, we performed qRT-PCR assays of seven abiotic response- and flowering time–related genes. The results showed that although there were some anomalous quantitative differences, the trends of gene expression changes detected by the two different approaches were generally consistent (Supplemental Table 2), thereby confirming the validity of the RNA-seq data.
Involvement of GAs in Cm-BBX24–Influenced Flowering Time and Abiotic Stress Tolerance
As described above, some of the transcripts related to GA biosynthesis genes and GA signaling genes were downregulated in the Cm-BBX24-OX lines and upregulated in Cm-BBX24-RNAi lines (Table 1). The qRT-PCR analyses confirmed that expression of GA20ox and GA3ox was increased to 3.34- and 2.92-fold in Cm-BBX24 transgenic chrysanthemum plants relative to expression levels seen for wild-type plants (Supplemental Table 2). We also determined expression of genes related to GA biosynthesis, including GA20ox (1 to 5), GA3ox (1 and 2), and GA2ox (GA 2-oxidases) (1 to 4, 6, and 7) in Cm-BBX24-OX Arabidopsis plants. Relative to wild-type plants, expression of GA20ox2 and GA3ox1 was clearly downregulated in Cm-BBX24-OX Arabidopsis, whereas expression of all the GA2ox genes tested showed no differences (Supplemental Figures 6A and 6B).
We then quantified the levels of endogenous GAs in young leaves of the transgenic chrysanthemum lines and compared these GA levels to those found for wild-type plants grown under different daylengths. As a general tendency, bioactive GA1 was much more abundant than GA4 in both transgenic and wild-type plants under either LD or SD (Table 2). Under LD, the Cm-BBX24-RNAi plants had concentrations of GA1 and GA4 that were 2.3 and 1.8 times greater, respectively, than levels found for wild-type plant leaves. For GA19 and GA20, which are precursors of GA1, their levels in young leaves were 2.1 and 1.5 times greater than found for the wild type (Table 2). Other GAs quantified by the stable isotope dilution method either showed no changes or were not detectable. Under SD, GA1 and GA4 increased slightly, relative to wild-type young leaves, but no obvious changes in GA19 and GA20 levels were apparent in Cm-BBX24-RNAi plants compared with wild-type plants (Table 2). For Cm-BBX24-OX plants compared with the wild type, of the GAs detected, GA1 and GA4 contents showed a substantial decrease under LD, though these two bioactive GAs remained relatively high under SD (Table 2). These results suggest that Cm-BBX24 expression results in changes in the steady state levels of GA4 and GA1 concentrations in the young leaves, and notably the two precursors of GA1, GA19, and GA20 also showed similar increases. It should be noted, though, that these changes in GA concentration in the young leaves occurred only under LD, where flowering is appreciably delayed.
Table 2. Concentration of GAs in the Leaves of Cm-BBX24-OX/RNAi and Wild-Type Chrysanthemum Lines under SD and LD.
| Daylength | Lines |
GA Content (ng⋅g−1
FW) |
||||||
|---|---|---|---|---|---|---|---|---|
| GA1 | GA4 | GA19 | GA24 | GA20 | GA3 | GA8 | ||
| SD | Cm-BBX24-RNAi | 1.73 ± 0.17 a | 0.33 ± 0.04 a | 1.10 ± 0.16 a | 0.31 ± 0.07 a | 0.35 ± 0.03 a | 0.34 ± 0.05 a | 0.80 ± 0.23 a |
| Wild type | 1.64 ± 0.19 a | 0.22 ± 0.05 b | 0.94 ± 0.17 a | 0.29 ± 0.05 a | 0.36 ± 0.09 a | 0.38 ± 0.09 a | 0.60 ± 0.06 a | |
| Cm-BBX24-OX | 1.46 ± 0.13 a | 0.23 ± 0.04 b | 1.02 ± 0.14 a | 0.21 ± 0.06 a | 0.37 ± 0.06 a | 0.42 ± 0.09 a | 0.56 ± 0.23 a | |
| LD | Cm-BBX24-RNAi | 1.45 ± 0.26 a | 0.34 ± 0.03 a | 1.40 ± 0.04 a | nd | 0.43 ± 0.04 a | 0.21 ± 0.03 a | 1.04 ± 0.34 a |
| Wild type | 0.62 ± 0.11 b | 0.18 ± 0.01 b | 0.67 ± 0.07 b | nd | 0.28 ± 0.07 b | 0.19 ± 0.04 a | 0.69 ± 0.18 a | |
| Cm-BBX24-OX | 0.37 ± 0.08 c | 0.12 ± 0.01 c | 0.58 ± 0.05 b | nd | 0.32 ± 0.02 b | 0.20 ± 0.02 a | 0.94 ± 0.07 a | |
Cm-BBX24-OX, the wild type, Cm-BBX24-RNAi, and three different chrysanthemum lines. SD, 8-h SD; LD, 16-h LD. Significant differences were determined by Duncan’s multiple range test (P < 0.05, n = 3), and lowercase letters, a to c, indicate significant differences between wild-type and transgenic plants under SD or LD condition. nd, endogenous GA not detected, deuterated GA internal standard present. GA5, GA9, GA29, GA34, and GA51 were not detected.
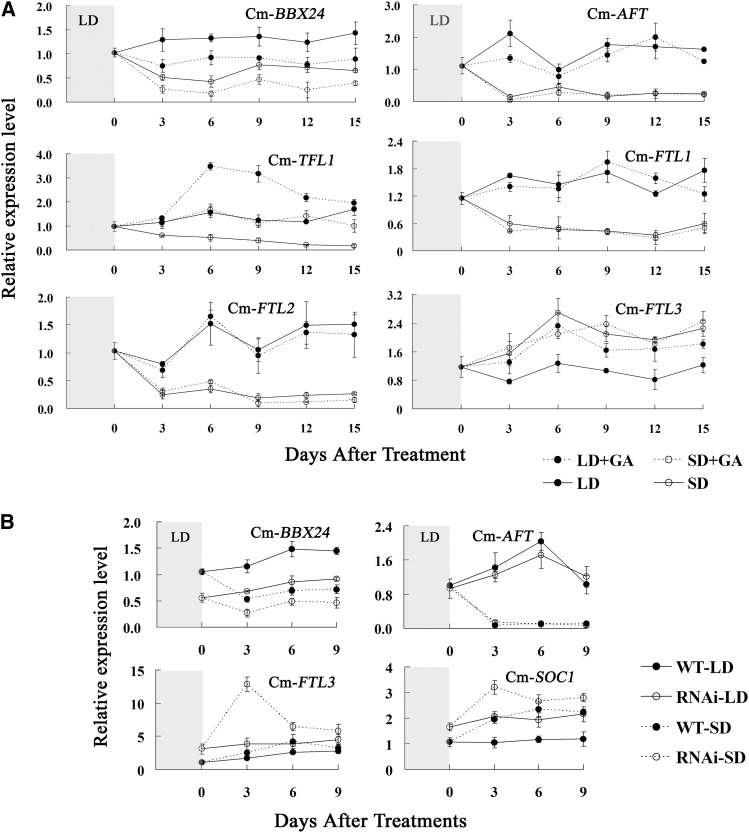
To establish whether Cm-BBX24 influences the floral integrator through the GA pathway, we determined expression patterns of Cm-BBX24 and five chrysanthemum FT genes with GA4/7 applications under the two different daylengths. The five FT genes were identified by querying our chrysanthemum RNA-seq database (Table 1), as well as data presented in a recent publication (Higuchi et al., 2013). Cm-BBX24 expression changed in response to daylength, and the expression patterns were consistent with that of the antiflorigen (e.g., Cm-AFT), and opposite from those of florigen (e.g. Cm-FTL3) under both SD and LD (Figure 5A). Exogenously applied GA4/7 suppressed the expression of Cm-BBX24 under either LD or SD, whereas the expression of the five Cm-FTs showed varying patterns in response to the GA4/7 treatment. We note that clear suppression of Cm-BBX24 expression by the GA treatment is consistent with that of antiflorigen (e.g., Cm-AFT) but is opposite to that of florigen (e.g., Cm-FTL3 and Cm-TFL1) under LD (Figure 5A). It should also be noted that applied bioactive GAs act as potent florigens in chrysanthemum, both in this study (Figure 5A) as well as in a previous study (Pharis, 1972).
Figure 5.
Expression Profiles of Cm-BBX24 and Flowering-Time Integrators under Different Daylengths or following GA4/7 Applications for Chrysanthemum Plants.
(A) Expression profiles of Cm-BBX24 and Cm-FTs under different daylengths or following GA4/7 treatments. The gray area indicates LD, and 0 indicates the time at which the plants were transferred to SD. The data were normalized with Ubiquitin expression. Error bars indicate sd (n = 3).
(B) Expression profiles of Cm-BBX24 and flowering-time integrators under different daylengths in Cm-BBX24-RNAi plants. RNAi, Cm-BBX24-RNAi.
We then analyzed the consequences of Cm-BBX24 suppression on the expression patterns of flowering-time integrators in chrysanthemum. Under either LD or SD, expression of Cm-FTL3 and Cm-SOC1 was much higher in the Cm-BBX24-RNAi lines than in wild-type plants, whereas expression of Cm-AFT showed essentially no difference (Figure 5B). We also evaluated the expression of potential target genes of Cm-BBX24, including GI, CO, FT, and SOC1 in the Arabidopsis Cm-BBX24 transgenic plants. Relative to wild-type plants, expression of all the tested genes related to the photoperiod pathway was clearly downregulated in Cm-BBX24-OX Arabidopsis (Supplemental Figure 6A).
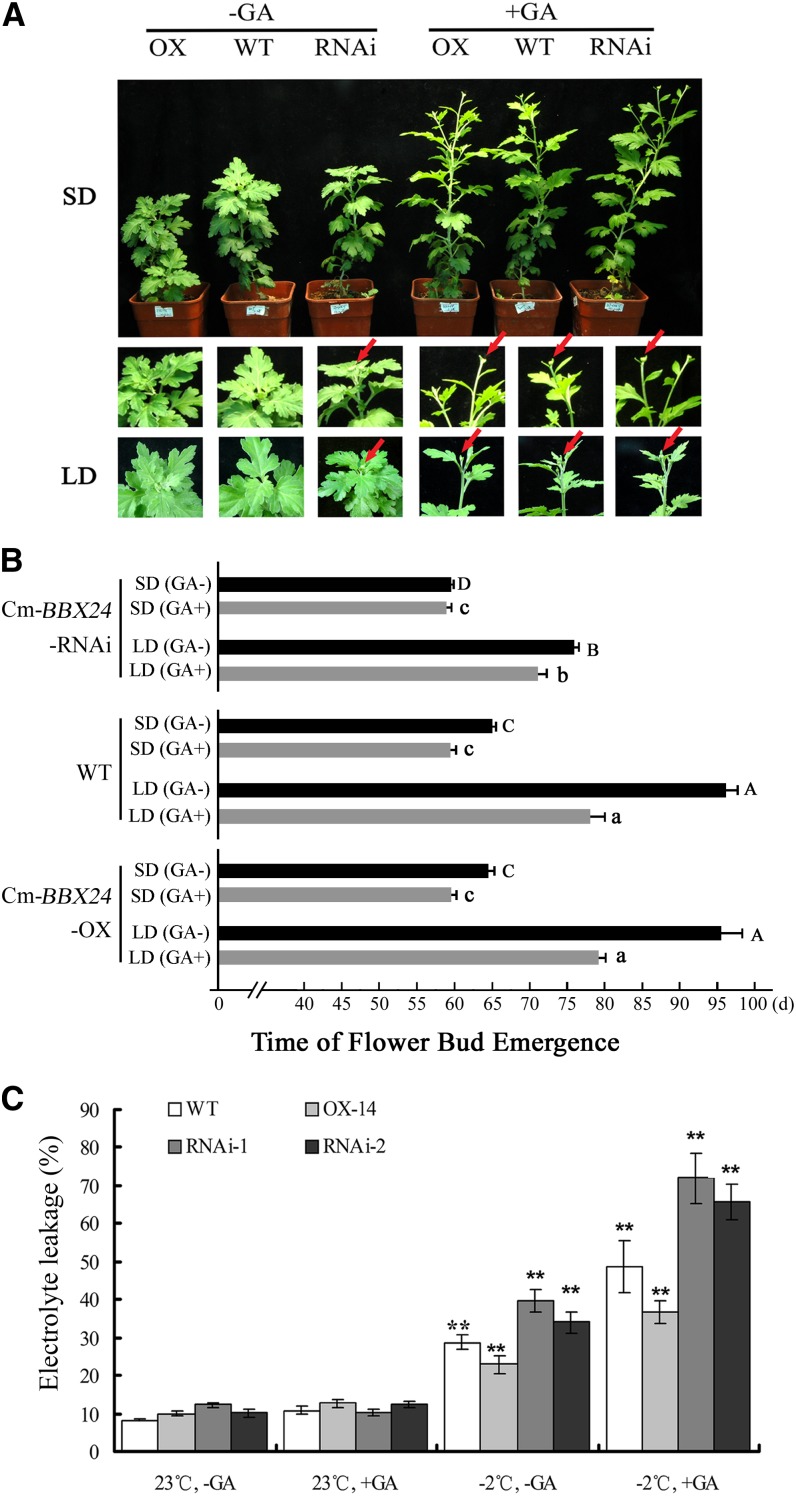
We further tested the effect of exogenous GA4/7 on flowering time of chrysanthemum. In one experiment under LD, without GA treatment, the blossom time was 76 and 96 d in Cm-BBX24-RNAi and wild-type plants, respectively, a 20-d advance in flowering time. With GA4/7 treatment, the flowering time was 72 and 78 d in Cm-BBX24-RNAi and wild-type plants, respectively, an advance of only 6 d by the transgenic line under LD, though an 18-d advance was seen for the wild type. Under SD, the GA4/7 applications yielded a flowering time for all three genotypes that was approximately the same (Figures 6A and 6B). GA4/7 application to the Cm-BBX24-OX Arabidopsis plants under LD rescued the late-flowering phenotype of the transgenic line, but there was no significant difference in leaf numbers between the transgenic and wild-type plants (Supplemental Figures 6D and 6E). These results indicate that the GA4/7 treatment countered the suppressing effect of Cm-BBX24 overexpression.
Figure 6.
Effect of GA4/7 Treatment on Flowering Time and Electrolyte Leakage in Transgenic Cm-BBX24 Chrysanthemum Plants.
(A) Flower bud emergence of wild-type and Cm-BBX24-OX/RNAi chrysanthemum plants treated with GA4/7 under LD or SD. The plants were photographed 65 and 85 d after transplanting under SD and LD, respectively. Red arrows indicate the emergence of visible flower buds.
(B) Effect of GA4/7 application on flower bud emergence time in wild-type and Cm-BBX24-OX/RNAi transgenic chrysanthemum lines. The columns indicate the duration of the vegetative growth period of different plants with or without GA4/7 treatments under LD or SD. 0 d represents the transplanting time, and GA treatment was performed at 30 d. Three independent experiments were performed, and error bars indicate sd (n = 3). Significant differences were determined by Duncan’s multiple range test (P < 0.05), and capital letters (A to D) and small letters (a to c) indicate significant differences in comparisons among control plants (no GA4/7 treatment) and plant given the GA4/7 spray treatments, respectively.
(C) Effect of GA4/7 treatment on electrolyte leakage of chrysanthemum. OX-14, a line of Cm-BBX24-OX; RNAi-1 and RNAi-2, two independent lines of Cm-BBX24-RNAi. 23°C, -GA (normal growth conditions); 23°C, +GA (normal growth conditions with an exogenous GA4/7); -2°C -GA (freezing treatment); -2°C +GA (freezing with an exogenous GA4/7). Three independent experiments were performed, and error bars indicate sd (n = 3). Significant differences were determined by Duncan’s multiple range test (P < 0.05), and asterisks indicate significant differences in comparisons among control plants (no GA4/7 treatment) or plants given the GA4/7 spray treatments.
Together, these results suggest that Cm-BBX24 influences flowering, at least in part, via the GA pathway under LD, i.e., under a daylength where flowering is appreciably delayed.
Changes in GA biosynthesis are known to be associated with abiotic stress responses in some plant species (Achard et al., 2008). Here, Cm-BBX24 transgenic chrysanthemum lines, relative to wild-type plants, showed clear changes in expression of several GA biosynthesis genes (GA20ox and GA3ox) and in young leaf GA concentration (Tables 1 and 2). However, the expression levels of GA2ox and concentration of GA8, the product of GA2ox activity, showed no apparent difference (Supplemental Figure 6C; Table 2). Therefore, we speculate that changed abiotic stress tolerance in Cm-BBX24 transgenic plants may be related to the influence of increased levels of bioactive GAs, rather than by not inactivation via GA2ox action.
To further investigate the speculation, we tested the influence on cold tolerance of GA4/7 applied to young chrysanthemum leaves. Under normal temperatures, GA4/7 treatment had no effect on electrolyte leakage, but following a −2°C low-temperature treatment, the electrolyte leakage of all plants increased significantly, and the GA4/7 treatment exacerbated the low temperature-induced damage: The GA4/7 treatment increased leakage by 70% in wild-type plants, but only 59% in Cm-BBX24-OX plants. By contrast, electrolyte leakage was 82 and 93% in the two Cm-BBX24-RNAi lines. Thus, exogenously applied GA4/7 reduced the tolerance to low temperatures and did so particularly for the Cm-BBX24-RNAi transgenic plants (Figure 6C).
DISCUSSION
Involvement of Cm-BBX24 in the Regulation of Flowering Time
It has been established that CO encodes a putative zinc finger transcription factor and that its temporal and spatial regulation is key to the daylength-dependent induction or promotion of flowering (Putterill et al., 1995). In terms of the relationship between the structures and functions of BBX proteins, a previous analysis of seven Arabidopsis co mutant alleles demonstrated that mutations within either the B-box region or the CCT domain influence flowering time, underlining the importance of these two conserved regions in the regulation of flowering time by CO (Robson et al., 2001). However, it is worth noting that BBX32, a member of structure group V, and some COLs (COL6-8, 16/BBX14-17), belonging to structure group III, can also influence flowering time, even though they encode proteins with just a single B-box domain. In this study we demonstrate that chrysanthemum BBX24, a member of structure group IV, influences flowering time, even though it does not contain a CCT domain. Collectively, our data suggest that the first B-box domain in the BBX proteins plays a causal role in influencing flowering time.
As components of the photoperiod pathway, GI, PRR5, CO, FT, and SOC1 are thought to be important in the control of flowering time. For example, GI is an essential component of the daylength control of flowering, and in Arabidopsis, gi mutants show a late-flowering phenotype and reduced accumulation of CO transcripts (Fowler et al., 1999). In this study, we used RNA-seq-based transcript profiling to show that the expression levels of a range of genes with homology to known components of flowering regulatory pathways, including GI, PRR5, CO, FT, and SOC1, were upregulated in Cm-BBX24-RNAi transgenic chrysanthemum, relative to wild-type plants (Table 1). These findings suggest that Cm-BBX24 influences flowering time in part by modulating the photoperiod-induced pathway at the transcriptional level.
GA biosynthesis genes and GA downstream-signaling responses are involved in the regulation of flowering time (Mouradov et al., 2002). The GA3ox protein catalyzes the conversion of precursor 3-deoxy GAs to their bioactive forms and therefore plays a direct roles in determining the levels of bioactive GAs in plants (Pimenta Lange and Lange, 2006). However, the bioactive GA1 and GA4 can be deactivated through oxidation by GA2ox to the inactive GA34 and GA8, respectively, and GA2ox may also play a regulatory role in determining bioactive GA levels (Hedden and Phillips, 2000; Eriksson et al., 2006). In addition, GRP, an important component of the GA signaling pathway, has been reported to participate in regulating flowering time in Arabidopsis (Zhang et al., 2009). GRAS genes also function in GA signal transduction and thus can play a role in regulation of flowering (Dill and Sun, 2001). Additionally, the FPF1 functions in the GA-dependent signaling pathway in Arabidopsis (Kania et al., 1997).
Chrysanthemum, a SD-requiring plant, has long been used as a model in classical physiological experiments to study the effect of daylength and low-temperature vernalization on flowering. In its natural setting, chrysanthemum flower bud initiation occurs eventually in all cultivars, even when grown in LD, but this may take many months in unvernalized shoots of cold-requiring cultivars (Cockshull, 1985). Flowering time of chrysanthemum is also influenced by several plant hormones (Cockshull, 1985), and among them GAs have been documented as promoting chrysanthemum flowering under LD (Pharis, 1972; Sumitomo et al., 2009). In this study, the Cm-BBX24-RNAi chrysanthemum clearly exhibited early flowering in LD, and RNA-seq analyses revealed that some key GA biosynthesis genes, such as GA20ox and GA3ox, as well as those genes associated with downstream GA signaling, such as a GRP, GRAS, and FPF1, were upregulated in Cm-BBX24-RNAi plants, relative to wild-type plants (Table 1).
The influence of GAs on flowering time depends on daylength. For LD-requiring plants, such as the ‘Ceres’ cultivar of the grass L. temulentum, GAs are the only plant hormones known to substitute for the single LD required for flowering (Evans and King, 1985). In L. temulentum, GA5 is the primary LD-induced flowering stimulus with GA1 and GA4 being secondary, late-acting LD stimuli for inflorescence differentiation. Gibberellin A6 is also a LD-induced florigenic GA (King et al., 2001, 2003; reviewed in King, 2012). In the LD-requiring Arabidopsis, GA4 is the key bioactive GA in both growth and (likely) the regulation of flowering (Eriksson et al., 2006). Applied GAs have a long history of being able to replace the LD requirement and can also substitute for low temperatures in some cold-requiring plants (Pharis and King, 1985). Additionally, overexpression of GA20ox or GA3 application can promote early flowering in Arabidopsis (Coles et al., 1999). Studies with spinach (Spinacia oleracea) further revealed that GA53, a precursor of GA19, is converted only to GA19 in noninductive SD, but is rapidly converted to GA20 when plants are transferred from SD to LD (reviewed in Pharis and King, 1985). It seems likely that LD-induced changes in GA20ox expression are involved in these conversion processes (Olszewski et al., 2002).
In chrysanthemum, inflorescence development (flowering) can eventually occur under noninductive LD, and exogenous application of GA3, GA4/7, or a combination of GA5 with the cytokinin benzyladenine can appreciably shorten the time to flowering, with GA concentration being a limiting factor for inflorescence development and the GA5 plus benzyladenine treatment having the strongest effect (Pharis, 1972). We also observed that suppression of Cm-BBX24 significantly increased the levels of both bioactive GA1 and GA4, as well as the concentration of GA19 and GA20 (Table 2), and that these changes in endogenous GA concentration were more apparent under LD than SD. Additionally, GA4/7 application rescued the early flowering phenotype of chrysanthemum that was created by a reduction in Cm-BBX24 expression (Figure 6). We therefore hypothesize that under LD, the observed earlier flowering in Cm-BBX24-RNAi chrysanthemum lines is partially due to an increase in steady state levels of bioactive GA1 and GA4, the former involving the GA19→ GA20→ GA1 pathway.
We noted that compared with wild-type plants, BBX24-OX chrysanthemum lines did not exhibit delayed flowering, whereas a delay was seen when Cm-BBX24 was overexpressed in Arabidopsis (Supplemental Figure 4). Nevertheless, possible target genes of Cm-BBX24 related to GA biosynthesis pathways were clearly downregulated in Cm-BBX24-OX transgenic Arabidopsis, relative to the wild-type controls (Supplemental Figure 6B). Thus, we postulate that Cm-BBX24 acts, under LD, by regulating GA biosynthesis and/or signaling pathways.
FT and SOC1 are known as integrators of photoperiodic and GA pathways (Moon et al., 2003; Song et al., 2013). For the LD plant L. temulentum, the regulation of floral apex induction appears to be mediated by LD-induced changes in endogenous GA5 (and possible also GA6) in the apex, as well as by FT (King et al., 2006). In Arabidopsis, flowering under noninductive SD also appears to be GA dependent, as treatment with exogenous GAs activates the transcription of SOC1 and LFY (Blázquez et al., 1998; Moon et al., 2003).
For chrysanthemum, a SD plant, responses of different FT genes to daylength differed. FTL3 is a key regulator of the photoperiodic flowering pathway in chrysanthemum (Oda et al., 2012) and Cs-FTL3 signaling from the leaves to the shoot tip has been reported to be affected by environmental factors, including high temperature (Nakano et al., 2013). In addition, a recent analysis revealed that the gated induction system of a systemic floral inhibitor, Cs-ATF1, an antiflorigen, determines obligate SD flowering in chrysanthemum (Higuchi et al., 2013).
In this study, we characterized the expression patterns of Cm-BBX24 and five FT genes and performed a correlation analysis with daylength and GA4/7 treatment of chrysanthemum. Our results indicated that Cm-BBX24 is expressed at high levels under LD and was suppressed by GA4/7 application. Furthermore, the expression patterns of Cm-BBX24 are consistent with that of the antiflorigen (Cm-AFT). In addition, silencing of Cm-BBX24 in chrysanthemum clearly induced expression levels of Cm-FTL3 and Cm-SOC1 (Figure 5B). We therefore propose that Cm-BBX24 suppresses flowering by influencing both the photoperiod and GA pathways and that under LD it is Cm-BBX24 that modulates flowering, mainly through effects on the GA pathway.
Involvement of Cm-BBX24 in the Regulation of Abiotic Stress Tolerance
Since we determined that Cm-BBX24 expression can confer abiotic stress tolerance, especially to drought and freezing, we focused on the possible downstream genes that are upregulated in Cm-BBX24-OX lines, but downregulated in Cm-BBX24-RNAi lines. Among the 36 assembled annotated transcripts that fell into this category, seven encode regulatory proteins, including members of the WRKY, MYB, and zinc finger families, which are known to function as part of a large regulatory network that senses and responds to different environmental stimuli (Hirayama and Shinozaki, 2010). In addition, two transcripts encode proteins that are related to signaling mediated by calcium ions, which are regarded as important secondary messengers in eliciting responses to abiotic stresses (Kader and Lindberg, 2010). In addition, among the five genes encoding protein kinases, a receptor-like protein kinase was identified, a homolog of which was previously reported to be stress inducible (Hong et al., 1997).
The group of differentially expressed genes also includes those annotated as encoding functional proteins. One subset comprises proteins associated with compatible solutes, such as lectins and a homolog of tuber agglutinin. Lectins are carbohydrate binding proteins that specifically recognize diverse sugar structures and can participate in abiotic stress regulation (Jiang et al., 2010). A second group comprises genes encoding carbohydrate metabolism-related proteins. Of these, trehalose-6-phosphate synthase, UDP-glucose glucosyltransferase, and β-amylases have been reported to be regulated by drought, cold, or high-salinity stress (Seki et al., 2002), and drought stress can increase β-amylase activity (Todaka et al., 2000). Overexpression of the Arabidopsis TREHALOSE-6-PHOSPHATE SYNTHASE (TPS) gene conferred dehydration tolerance and delayed flower development in transgenic plants (Avonce et al., 2004), and overexpression of the yeast TPS gene improved tolerance to abiotic stresses in tomato (Solanum lycopersicum) (Cortina and Culianez-Macia, 2005), suggesting that these genes play similar roles in chrysanthemum.
In terms of correlations between GA biosynthesis and abiotic stress tolerance, it has been reported that overexpression of an endogenous GA20ox in citrus plants resulted in increased of GA content and thus decreased in abiotic stress tolerance (Huerta et al., 2008). This was accompanied by global upregulation of genes involved in photosynthetic and carbon utilization, downregulation of genes associated with protein biosynthesis and ribosome biogenesis, and the differential expression of numerous genes related to abiotic stress responses (Huerta et al., 2008). In Arabidopsis, endogenous GA4 content of the shoot is negatively correlated with expression of CBF1/DREB1b, and elevated CBF expression reduces the accumulation of bioactive GAs through expression levels of GA2ox in cold-induced plants (Achard et al., 2008). CBFs are considered to function as integrators of phytochrome and GA signaling during cold acclimation, and such integration can lead to the activation of the CBF regulon and subsequent upregulation of COR gene and GA2ox expression, the latter resulting in a dwarf phenotype, coupled with increased freezing tolerance and enhanced photosynthetic performance (Kurepin et al., 2013).
In this study, we also noted that under LD, GA20ox and GA3ox were significantly upregulated in Cm-BBX24-RNAi transgenic lines and significantly downregulated in Cm-BBX24-OX lines, both relative to wild-type chrysanthemum (Supplemental Table 2). By contrast, GA2ox showed no difference in expression in our transgenic lines (Supplemental Figure 6C). These results suggest that Cm-BBX24 regulates bioactive GA levels mainly through influencing GA biosynthesis rather than inactivation. Therefore, we propose that Cm-BBX24 acts independently of CBF1/DREB1b.
It has been reported that some flowering-associated genes function to regulate flowering time and are also involved in the abiotic stress response. GI, a circadian clock gene, is induced by cold stress and appears to be involved in mediating the cold stress response, possibly by positively regulating freezing tolerance via a CBF-independent pathway (Cao et al., 2005). Plants with loss-of-function mutations in GI and CO exhibited abnormal drought-escape phenotypes. The peak mRNA levels of GI and Flavin binding Kelch domain F box protein 1 and the mRNA levels of CO and FT were shown to change under drought stress (Han et al., 2013). Pseudo-RRs (PRRs), clock-associated genes that regulate flowering time positively, are also implicated in abiotic stress responses, where they function as negative regulators (Nakamichi et al., 2009). In Arabidopsis, a decreased level of SOC1 results in derepression of cold-inducible genes, which appears to enable plants to respond more effectively to low temperature stress (Seo et al., 2009). In Pharbitis nil, the expression of FT2, one of two homologs of the floral pathway integrator gene, FT, was induced by low temperature (Yamada and Takeno, 2014).
Our results imply that Cm-BBX24 may be an important regulator of the crosstalk between flowering time and abiotic stress responses regulation in chrysanthemum. If so, the ability to influence flowering time and abiotic stress responses via the same set of genes may provide an evolutionary advantage.
In summary, Cm-BBX24 encodes a zinc finger transcription factor that may play a pivotal role in flowering time regulation and abiotic stress responses. Cm-BBX24 possibly acts as a repressor of flowering by negatively regulating the expression of the photoperiod flowering pathway genes GI, PRR5, CO, FT, and SOC1, as well as the GA biosynthesis genes GA20ox and GA3ox. It also appears to be involved in regulating the response of plants to two abiotic stresses by influencing a subset of genes that mediate stress responses and GA biosynthesis (Supplemental Figure 7). Our results suggest a conserved mechanism for regulating flowering time and abiotic stress responses in plants, and we propose that the regulation of Cm-BBX24 may be a common feature in these two distinct responses. Future investigation to determine a more precise mechanism by which Cm-BBX24 acts in the two intersecting pathways will provide a better understanding of the modulation of reproductive development in plants under abiotic stress conditions.
METHODS
Plant Materials and Treatments
A popular ground-cover chrysanthemum cultivar (Chrysanthemum morifolium cv Fall Color) with pink flowers was used as the wild-type control in this study. Chrysanthemum plants were propagated by in vitro culture for 50 d, then transplanted into 9-cm-diameter pots containing a mixture of 1:1 (v/v) of peat and vermiculite and transferred to a controlled temperature culture room with normal conditions (23 ± 1°C, 40% relative humidity, and 100 μmol⋅m−2⋅s−1 illumination by fluorescent lamps) under a LD cycle (16 h light/8 h dark). It should be noted that the fluorescent lamps produced a far-red light irradiance of only ∼3% of the total irradiance. When the plants had seven to nine fully expanded leaves they were root-washed and placed in distilled water for 24 h for subsequent experiments.
Cm-BBX24 Gene Isolation
Plants were transferred to a 4°C chamber for 6 h. Total RNA samples were extracted from whole plants using the Trizol reagent (Invitrogen) according to the manufacturer’s instructions. Using the DREB1A regulon cDNA library obtained by suppression subtractive hybridization (Ma et al., 2010), 5′- and 3′-rapid amplification of cDNA ends was performed to isolate the 5′ and the 3′ cDNA ends of Cm-BBX24 using the SMARTer RACE cDNA amplification kit (Clontech) according to the manufacturer’s instructions. The full length of Cm-BBX24 was obtained using gene-specific primers (Supplemental Table 3). The resulting PCR product was purified and cloned into the pGEM-T Easy Vector (Promega) for sequencing. Multiple sequence alignments were constructed using the ClustalW (http://www.ch.embnet.org/software/ClustalW.html) programs and BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html).
Phylogenetic Analysis
The protein sequences of Cm-BBX24 and 8 structure group IV BBXs from Arabidopsis thaliana were multiply aligned using MUSCLE algorithm (gap open, 22.9; gap extend, 0; hydrophobicity multiplier, 1.5; clustering method, upgmb). The alignment used for the analysis is available as Supplemental Data Set 2. A phylogenetic analysis was conducted by MEGA version 4 (Tamura et al., 2007) using the neighbor-joining method with 1000 bootstrap replications.
Abiotic Stress Treatment of Chrysanthemum
For the freezing treatments, plants were transferred to a −6°C chamber for 8 h and then returned to 23°C for 30 d. For drought treatments, the plants were given adequate water and then water was withheld for 23 d and rewatered regularly and allowed to recover for 30 d. For the salt treatment, plants were transferred to a container filled with 400 mM NaCl solution for 4 weeks. The initial height of the solution was noted and maintained each day. Survival rates were recorded after the treatments finished and the plants were photographed to record their phenotypes.
Chrysanthemum Transformation
To construct the overexpression vector, the ORF sequence of Cm-BBX24 was amplified using a pair of primers with XbaI and SmaI sites (Supplemental Table 3), and the resulting PCR product was digested with XbaI and SmaI and then cloned into the pBIG vector (Becker, 1990). To construct the RNAi vector, 361-bp sense and antisense fragments of Cm-BBX24 containing XhoI/EcoRI and XbaI/HindIII sites were amplified and the two PCR products were digested with the corresponding restriction enzymes, as above, and directionally inserted into the two sides of the Pdk Intron in the pHANNIBAL vector step-by-step to form intron-containing hairpin RNA constructs. The intron-containing hairpin RNA construct with promoter and terminator was then subcloned into the binary vector pART27 (Wesley et al., 2001). The overexpression and RNAi plasmids were separately introduced into Agrobacterium tumefaciens strain LBA4404 and transformed into chrysanthemum by Agrobacterium-mediated transformation (Hong et al., 2006).
Phenotypic Characterization of Transgenic Chrysanthemum
The plants were considered to be entering the flowering stage when at least 50% of the ray flowers on at least one inflorescence were reflexed (Blanchard and Runkle, 2009). The times of first visible flower buds and first flower opening were recorded. For observation of flower bud differentiation, stem apices of Cm-BBX24-OX, wild-type, and Cm-BBX24-RNAi plants were isolated, paraffin embedded, and sectioned.
RNA-seq Analysis
Total RNA samples were extracted from the aerial parts of 4-week-old Cm-BBX24–overexpressing plants (line Cm-BBX24-OX-23), Cm-BBX24 RNAi plants (line Cm-BBX24-RNAi-1), and wild-type plants, grown under normal conditions, using the RNeasy plant mini kit (Qiagen). Triplicate samples, comprising three independent plants of each line, were taken at ZT3. The RNA sequencing library was prepared as described by Zhong et al. (2011), and sequencing was performed on an Illumina HiSequation 2000 in the Genomics Facility of the Institute of Biotechnology, Cornell University (http://www.biotech.cornell.edu/).
RNA-seq Data Processing, de Novo Assembly, and Annotation
RNA-seq reads were first processed with a custom R script based on the ShortRead package to trim low quality (Q value < 20) nucleotides on both ends and to clip the adapter and barcode sequences from the 3′ end. The resulting reads with lengths <40 bp or containing more than two ambiguous (‘‘N’’) nucleotides were discarded. The RNA-seq reads were then aligned to GenBank virus (version 186) and the rRNA sequence databases using BWA and default parameters. Reads mapped to these two databases were discarded. The resulting high-quality cleaned reads were assembled de novo into contigs using Trinity with strand specific option “–SS_lib_type” set to “F” and “min_kmer_cov” set to 2 (Grabherr et al., 2011). To remove the redundancy of Trinity-generated contigs, they were further assembled de novo using iAssembler with minimum percent identify (-p) set to 99 (Zheng et al., 2011).
The resulting unique transcripts were screened by BLAST against the GenBank nonredundant (nr), UniProt (Swiss-Prot and TrEMBL), and Arabidopsis protein databases with a cutoff E-value of 1e-5. Gene Ontology terms were assigned to the chrysanthemum assembled transcripts based on the Gene Ontology terms annotated to their corresponding homologs in the UniProt database.
qRT-PCR Analysis
For the spatiotemporal expression analysis of Cm-BBX24, leaves, stems, roots, shoot apices at the vegetative growth stage and flower bud differentiation stage, flower buds, and flowers were taken from wild-type chrysanthemum grown under LD (16 h light/8 h dark). Three replicate samples were taken.
For the abiotic stress induction analysis of Cm-BBX24, the root-washed chrysanthemum plants were exposed for fixed times to the following conditions: control (distilled water treatment under LD and normal growth conditions); cold (chilling at 4°C in a growth chamber); dehydration (air drying on filter paper at room temperature); and salt (125 mM NaCl treatment under normal growth conditions). Shoots were then harvested and stored at −80°C prior to RNA extraction. The treatments were started at ZT3, and three replicate samples were taken.
To analyze the effect of daylength and GA4/7 application on the expression patterns of Cm-BBX24 and Cm-FT, we transferred wild-type chrysanthemum plants grown under LD for 2 weeks, to SD (8 h light/16 h dark) or kept under LD for 15 d, with or without a 100 μM GA4+7 treatment once every 3 d. Samples were collected at ZT3 with 3-d intervals and three replicate samples were taken. To detect correlations in the expression of Cm-BBX24 and floral integrators under different daylengths, Cm-BBX24-RNAi and wild-type chrysanthemum plants grown under LD for 2 weeks were transferred to SD or kept under LD for 9 d. Samples were collected at ZT3 with 3-d intervals, and three replicate samples were taken.
Total RNA samples were extracted from the above sample stock using the Trizol agent, as above, and cDNAs were synthesized from 1 μg total RNA using M-MLV reverse transcriptase (Promega). qRT-PCR reactions (20 μL volume containing 2 μL cDNA as the template) were performed using the StepOne real-time PCR system (Applied Biosystems) in standard mode with the KAPA SYBR FAST Universal qRT-PCR kit (Kapa Biosystems). All reactions were performed in triplicate, and the chrysanthemum Ubiquitin gene was used as the internal control. PCR primers are listed in Supplemental Table 3.
Subcellular Localization Analysis of Cm-BBX24
A cDNA fragment containing the ORF of Cm-BBX24 (stop codon removed) was amplified using a pair of primers containing EcoRI or SalI sites (Supplemental Table 3). The PCR products were digested with EcoRI and SalI and then cloned into the pEZS-NL vector (D. Ehrhardt, http://deepgreen.stanford.edu), which resulted in the Cm-BBX24 coding sequence fused in frame with the 5′ terminus of a sequence encoding GFP, driven by the cauliflower mosaic virus 35S promoter. The vector pEZS-NL deleted Ala (10) linker containing 35S:GFP was used as a control. Plasmids were individually transformed into onion epidermal cells using a particle bombardment method. Transformed onion epidermal cells were cultured on Murashige and Skoog media in dark conditions for 24 to 30 h at 22°C and were observed using a confocal laser scanning microscope (Nikon C1 Plus).
Transactivation Analysis of Cm-BBX24 in Yeast Cells
The ORF (1 to 245 amino acids), B-box domain at the N terminus (1 to 95 amino acids), and the C terminus (96 to 245 amino acids) of the Cm-BBX24 gene were amplified using three pairs of primers with EcoRI and SalI sites. The primers are listed in Supplemental Table 3. The PCR products were digested with EcoRI and SalI and were then cloned into the DNA binding domain vector pBD-GAL4 Cam (Clontech) to yield the expression vectors pBD-Cm-BBX24, pBD-Cm-BBX24N, and pBD-Cm-BBX24C. pBD-GAL4 and pGAL4 were used as negative and positive controls, respectively. The constructs were transformed into yeast (Saccharomyces cerevisiae) strain YRG-2 containing the reporter genes LacZ and HIS3, according to the method described in the Yeast Protocols Handbook PT3024-1 (Clontech). The yeast transformants were incubated at 30°C for 3 d on SD deleting tryptophan medium with or without histidine. The transformed yeast cells were then transferred to filter paper and incubated at 30°C for 3 to 8 h in the presence of X-gal to assess the presence of β-galactosidase by observing blue color formation. A quantitative β-galactosidase assay was also performed also using o-nitrophenyl β-d-galactopyranoside, according to the method described in the Yeast Protocols Handbook PT3024-1. Three transformants were selected and subjected to the β-galactosidase assay.
The Determination of GA Content in Transgenic Chrysanthemum Leaves
Cm-BBX24-OX/RNAi and wild-type plants were grown under LD for 40 d and then were transferred to SD or kept in LD for 3 d. Three replicate leaf samples were taken from each line, and each leaf sample was from top three fully opened leaves, and harvest time was 3 h after the lights were turned on. Leaf tissue GA concentrations (expressed as ng/g fresh weight [FW] of leaf tissue extracted) were analyzed by the stable isotope dilution method as described by M.L. Chen et al. (2012).
Each sample (100 mg FW) was extracted with 1.0 mL 80% methanol (v/v) at 4°C for 12 h. As internal standards, [2H2] GA1 (1.0 ng/g), [2H2] GA3 (1.0 ng/g), [2H2] GA4 (1.0 ng/g), [2H2] GA5 (1.0 ng/g), [2H2] GA8 (1.0 ng/g), [2H2] GA19 (1.0 ng/g), [2H2] GA20 (1.0 ng/g), [2H2] GA24 (1.0 ng/g), [2H2] GA34 (1.0 ng/g), [2H2] GA51 (1.0 ng/g), and [2H2] GA53 (1.0 ng/g) were added to the samples before grinding. After centrifugation (10,000g, 4°C, 20 min), the supernatants were collected and then passed through the tandem SPE cartridges containing C18 sorbent (50 mg) and SAX sorbent (200 mg). After sample loading, the C18 cartridge was removed and the SAX cartridge was rinsed with 2 mL methanol/water (20/80, v/v). Three milliliters of acetonitrile (ACN) with 1% formic acid (v/v) was applied to dilute the targeted acidic phytohormones, and the diluents were evaporated and then redissolved in 100 μL water. The resulting solution was then acidified with 10 μL formic acid and extracted with ether (2 × 1 mL). The ether phase was combined, dried under nitrogen gas, and reconstituted in 100 μL ACN. Subsequently, 10 μL triethylamine (20 μmol/mL) and 10 μL 3-bromoactonyltrimethylammonium bromide (20 μmol/mL) were added. The reaction solution was vortexed for 30 min at 35°C and evaporated under nitrogen gas and then redissolved in 200 μL water/ACN (90/10, v/v) for further analysis (M.L. Chen et al., 2012).
GA Treatment of Chrysanthemum
For flower bud observation, wild-type and Cm-BBX24-OX/RNAi plants were grown under LD for 30 d and then transferred to SD or kept under LD before spraying with 100 μM GA4/7. The GA4/7 was dissolved in absolute ethanol, and a 1000-fold dilution was used for the spray application experiment. The same concentration of ethanol solution was used as a mock control. The GA4/7 treatment was performed twice per week until the flower buds emerged.
Electrolyte Leakage
Chrysanthemum plants were grown at 23°C under LD and then sprayed three times with 100 μM GA4+7, once every 3 d. Plants with/without GA treatment were then transferred to −2°C chamber for 48 h. Leaves were harvested from the middle of the stem, and electrolyte leakage was determined according to the method of Leopold et al. (1981).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL databases under the following accession numbers: Cm-BBX24, C. morifolium, KF385866; the raw sequence reads of RNA-seq, C. morifolium, SRP038981 and SRA091277; At-BBX24, Arabidopsis, NP172094, At1g06040; At-BBX25, Arabidopsis, NP565722, At2g31380; Gm-BBX24, Glycine max, ABB29467; Br-BBX24, Brassica rapa, ABV89657; Ss-BBX24, Solanum sogarandinum, ABC25454; Cm-BBX24, Cucumis melo, ADN34088; Rc-BBX24, Ricinus communis, XP002534139; Os-BBX24, Oryza sativa, CAH67738; At-BBX18, Arabidopsis, NP565507, At2g21320; At-BBX19, Arabidopsis, NP195607, At4g38960; At-BBX20, Arabidopsis, NP195618, At4g39070; At-BBX21, Arabidopsis, NP177686, At1g75540; AtBBX22, Arabidopsis, NP565183, At1g78600; AtBBX23, Arabidopsis, NP192762, At4g10240; Cm-Ubiquitin, C. morifolium, EU862325; Cs-AFT, C. seticuspe, AB839766; Cs-TFL1, C. seticuspe, AB839767; Cs-FTL1, C. seticuspe, AB545936; Cs-FTL2, C. seticuspe, AB679271; Cs-FTL3, C. seticuspe, AB679272; GI, Arabidopsis, NM102124, At1g22770; CO, Arabidopsis, NM001036810, At5g15840; FT, Arabidopsis, NM1052, At1G65480; SOC1, Arabidopsis, NM130128, At2g45660; GA20ox1, Arabidopsis, NM118674, At4g25420; GA20ox2, Arabidopsis, NM124560, At5g51810; GA20ox3, Arabidopsis, NM120802, At5g07200; GA20ox4, Arabidopsis, NM104778, At1g60980; GA20ox5, Arabidopsis, NM103535, At1g44090; GA3ox1, Arabidopsis, NM101424, At1g15550; GA3ox2, Arabidopsis, NM106683, At1g80340; GA2ox1, Arabidopsis, NM106491, At1g78440; GA2ox2, Arabidopsis, NM001036035, At1g30040; GA2ox3, Arabidopsis, NM129007, At2g34555; GA2ox4, Arabidopsis, NM103695, At1g47990; GA2ox6, Arabidopsis, NM100121, AT1g02400; GA2ox7, Arabidopsis, NM103976, At1g50960; GA2ox8, Arabidopsis, NM101973, At1g21200.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Activities of the Cm-BBX24 Promoter in Response to Hormone Treatments and Abiotic Stresses in Transgenic Arabidopsis Plants.
Supplemental Figure 2. Relative Expression of Cm-BBX24 Transcripts under the Control of the Circadian Clock and Different Daylengths.
Supplemental Figure 3. Comparison of the Expression of Cm-BBX24 and Other Members of BBX Structure Group IV in Cm-BBX24-RNAi and Wild-Type Chrysanthemum Plants.
Supplemental Figure 4. Flowering Time of Cm-BBX24–Overexpressing Arabidopsis Plants.
Supplemental Figure 5. The Number of Assembled Transcripts Differentially Expressed between Cm-BBX24-OX, Cm-BBX24-RNAi, and Wild-Type Chrysanthemum Plants.
Supplemental Figure 6. Expression of the Photoperiod Pathway and GA Biosynthesis Pathway-Relevant Genes and Effect of GA4/7 Treatment on Flowering Time in Transgenic Arabidopsis and Chrysanthemum Plants.
Supplemental Figure 7. Schematic Model Describing the Involvement of Cm-BBX24 in Flowering and Abiotic Stress Tolerance in Chrysanthemum.
Supplemental Table 1. Annotation of Genes Upregulated in Cm-BBX24-OX Plants but Downregulated in Cm-BBX24-RNAi Plants.
Supplemental Table 2. Differentially Expressed Genes in Cm-BBX24-OX/RNAi and Wild-Type Chrysanthemum Plants by qRT-PCR Analysis.
Supplemental Table 3. Primers Used for Gene Isolation, qRT-PCR Analysis, and Vector Construction.
Supplemental Data Set 1. List of Assembled Transcripts Upregulated or Downregulated (>2.0-Fold, Adjusted P < 0.05) in Cm-BBX24 Transgenic Chrysanthemum Plants, Relative to Wild-Type Plants.
Supplemental Data Set 2. Text File of the Alignment Used for the Phylogenetic Analysis of Group IV BBXs Shown in Figure 1B.
Supplementary Material
Acknowledgments
We acknowledge PlantScribe (www.plantscribe.com) for careful editing of this article. This work was supported by the National Natural Science Foundation of China (Grants 31372094, 31171990, and 91217309).
AUTHOR CONTRIBUTIONS
Y.Y. performed the experiments and wrote the article. C.M., Y.X., Q.W., M.I., H.L., L.C., and M.W. helped with experiments. S.G. and Z.F. analyzed the data. B.H. and J.G. conceived and designed the experiments and wrote the article. All authors read and approved the final article.
Glossary
- GA
gibberellin
- LD
long-day
- SD
short-day
- ORF
open reading frame
- qRT-PCR
quantitative RT-PCR
- RNA-seq
RNA sequencing
- FW
fresh weight
- ACN
acetonitrile
References
- Achard P., Gong F., Cheminant S., Alioua M., Hedden P., Genschik P. (2008). The cold-inducible CBF1 factor-dependent signaling pathway modulates the accumulation of the growth-repressing DELLA proteins via its effect on gibberellin metabolism. Plant Cell 20: 2117–2129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avonce N., Leyman B., Mascorro-Gallardo J.O., Van Dijck P., Thevelein J.M., Iturriaga G. (2004). The Arabidopsis trehalose-6-P synthase AtTPS1 gene is a regulator of glucose, abscisic acid, and stress signaling. Plant Physiol. 136: 3649–3659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker D. (1990). Binary vectors which allow the exchange of plant selectable markers and reporter genes. Nucleic Acids Res. 18: 203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanchard M.G., Runkle E.S. (2009). Use of a cyclic high-pressure sodium lamp to inhibit flowering of chrysanthemum and velvet sage. Sci. Hortic. (Amsterdam) 122: 448–454 [Google Scholar]
- Blázquez M.A., Green R., Nilsson O., Sussman M.R., Weigel D. (1998). Gibberellins promote flowering of Arabidopsis by activating the LEAFY promoter. Plant Cell 10: 791–800 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao S., Ye M., Jiang S. (2005). Involvement of GIGANTEA gene in the regulation of the cold stress response in Arabidopsis. Plant Cell Rep. 24: 683–690 [DOI] [PubMed] [Google Scholar]
- Chen J., Chen J.Y., Wang J.N., Kuang J.F., Shan W., Lu W.J. (2012). Molecular characterization and expression profiles of MaCOL1, a CONSTANS-like gene in banana fruit. Gene 496: 110–117 [DOI] [PubMed] [Google Scholar]
- Chen M.L., Fu X.M., Liu J.Q., Ye T.T., Hou S.Y., Huang Y.Q., Yuan B.F., Wu Y., Feng Y.Q. (2012). Highly sensitive and quantitative profiling of acidic phytohormones using derivatization approach coupled with nano-LC-ESI-Q-TOF-MS analysis. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 905: 67–74 [DOI] [PubMed] [Google Scholar]
- Cheng X.F., Wang Z.Y. (2005). Overexpression of COL9, a CONSTANS-LIKE gene, delays flowering by reducing expression of CO and FT in Arabidopsis thaliana. Plant J. 43: 758–768 [DOI] [PubMed] [Google Scholar]
- Cockshull, K.E. (1985). Chrysanthemum morifolium In CRC Handbook of Flowering, Vol. II, A.H. Halevy, ed (Boca Raton, FL: CRC), pp. 238–257. [Google Scholar]
- Coles J.P., Phillips A.L., Croker S.J., García-Lepe R., Lewis M.J., Hedden P. (1999). Modification of gibberellin production and plant development in Arabidopsis by sense and antisense expression of gibberellin 20-oxidase genes. Plant J. 17: 547–556 [DOI] [PubMed] [Google Scholar]
- Cortina C., Culianez-Macia F.A. (2005). Tomato abiotic stress enhanced tolerance by trehalose biosynthesis. Plant Sci. 169: 75–82 [Google Scholar]
- Dill A., Sun T.P. (2001). Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159: 777–785 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eriksson S., Böhlenius H., Moritz T., Nilsson O. (2006). GA4 is the active gibberellin in the regulation of LEAFY transcription and Arabidopsis floral initiation. Plant Cell 18: 2172–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans, L.T., and King, R.W. (1985). Lolium temulentum In CRC Handbook of Flowering, A.H. Halevy, ed (Boca Raton, FL: CRC Press), pp. 306–323. [Google Scholar]
- Fornara F., de Montaigu A., Coupland G. (2010). SnapShot: Control of flowering in Arabidopsis. Cell 141: 550–, e1–e2. [DOI] [PubMed] [Google Scholar]
- Fowler S., Lee K., Onouchi H., Samach A., Richardson K., Morris B., Coupland G., Putterill J. (1999). GIGANTEA: a circadian clock-controlled gene that regulates photoperiodic flowering in Arabidopsis and encodes a protein with several possible membrane-spanning domains. EMBO J. 18: 4679–4688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grabherr M.G., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y., Zhang X., Wang Y., Ming F. (2013). The suppression of WRKY44 by GIGANTEA-miR172 pathway is involved in drought response of Arabidopsis thaliana. PLoS ONE 8: e73541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassidim M., Harir Y., Yakir E., Kron I., Green R.M. (2009). Over-expression of CONSTANS-LIKE 5 can induce flowering in short-day grown Arabidopsis. Planta 230: 481–491 [DOI] [PubMed] [Google Scholar]
- Hedden P., Phillips A.L. (2000). Gibberellin metabolism: new insights revealed by the genes. Trends Plant Sci. 5: 523–530 [DOI] [PubMed] [Google Scholar]
- Higuchi Y., Narumi T., Oda A., Nakano Y., Sumitomo K., Fukai S., Hisamatsu T. (2013). The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proc. Natl. Acad. Sci. USA 110: 17137–17142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Shinozaki K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61: 1041–1052 [DOI] [PubMed] [Google Scholar]
- Hong B., Tong Z., Ma N., Kasuga M., Yamaguchi-Shinozaki K., Gao J.P. (2006). Expression of Arabidopsis DREB1A gene in transgenic chrysanthemum enhances tolerance to low temperature. J. Hortic. Sci. Biotechnol. 81: 1002–1008 [Google Scholar]
- Hong S.W., Jon J.H., Kwak J.M., Nam H.G. (1997). Identification of a receptor-like protein kinase gene rapidly induced by abscisic acid, dehydration, high salt, and cold treatments in Arabidopsis thaliana. Plant Physiol. 113: 1203–1212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerta L., Forment J., Gadea J., Fagoaga C., Peña L., Pérez-Amador M.A., García-Martínez J.L. (2008). Gene expression analysis in citrus reveals the role of gibberellins on photosynthesis and stress. Plant Cell Environ. 31: 1620–1633 [DOI] [PubMed] [Google Scholar]
- Jiang S.Y., Ma Z., Ramachandran S. (2010). Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol. Biol. 10: 79–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader M.A., Lindberg S. (2010). Cytosolic calcium and pH signaling in plants under salinity stress. Plant Signal. Behav. 5: 233–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kania T., Russenberger D., Peng S., Apel K., Melzer S. (1997). FPF1 promotes flowering in Arabidopsis. Plant Cell 9: 1327–1338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna R., Kronmiller B., Maszle D.R., Coupland G., Holm M., Mizuno T., Wu S.H. (2009). The Arabidopsis B-box zinc finger family. Plant Cell 21: 3416–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S.K., Yun C.H., Lee J.H., Jang Y.H., Park H.Y., Kim J.K. (2008). OsCO3, a CONSTANS-LIKE gene, controls flowering by negatively regulating the expression of FT-like genes under SD conditions in rice. Planta 228: 355–365 [DOI] [PubMed] [Google Scholar]
- King R.W. (2012). Mobile signals in day length_regulated flowering: gibberellins, flowering Locus T, and sucrose. Russ. J. Plant Physiol. 59: 479–490 [Google Scholar]
- King R.W., Evans L.T., Mander L.N., Moritz T., Pharis R.P., Twitchin B. (2003). Synthesis of gibberellin GA6 and its role in flowering of Lolium temulentum. Phytochemistry 62: 77–82 [DOI] [PubMed] [Google Scholar]
- King R.W., Moritz T., Evans L.T., Junttila O., Herlt A.J. (2001). Long-day induction of flowering in Lolium temulentum involves sequential increases in specific gibberellins at the shoot apex. Plant Physiol. 127: 624–632 [PMC free article] [PubMed] [Google Scholar]
- King R.W., Moritz T., Evans L.T., Martin J., Andersen C.H., Blundell C., Kardailsky I., Chandler P.M. (2006). Regulation of flowering in the long-day grass Lolium temulentum by gibberellins and the FLOWERING LOCUS T gene. Plant Physiol. 141: 498–507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurepin L.V., Dahal K.P., Savitch L.V., Singh J., Bode R., Ivanov A.G., Hurry V., Hüner N.P. (2013). Role of CBFs as integrators of chloroplast redox, phytochrome and plant hormone signaling during cold acclimation. Int. J. Mol. Sci. 14: 12729–12763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang, A. (1965). Physiology of flower initation. In Encyclopedia of Plant Physiology, W. Ruhland, ed (Berlin, Heidelberg, New York: Springer-Verlag), pp. 1380–1536. [Google Scholar]
- Langridge J. (1957). Effect of day-length and gibberellic acid on the flowering of Arabidopsis. Nature 180: 36–37 [Google Scholar]
- Leopold A.C., Musgrave M.E., Williams K.M. (1981). Solute leakage resulting from leaf desiccation. Plant Physiol. 68: 1222–1225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lippuner V., Cyert M.S., Gasser C.S. (1996). Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J. Biol. Chem. 271: 12859–12866 [DOI] [PubMed] [Google Scholar]
- Ma C., Hong B., Wang T., Yang Y.J., Tong Z., Zuo Z.R., Yamaguchi-Shinozaki K., Gao J.P. (2010). DREB1A regulon expression in rd29A:DREB1A transgenic chrysanthemum under low temperature or dehydration stress. J. Hortic. Sci. Biotechnol. 85: 503–510 [Google Scholar]
- Moon J., Suh S.S., Lee H., Choi K.R., Hong C.B., Paek N.C., Kim S.G., Lee I. (2003). The SOC1 MADS-box gene integrates vernalization and gibberellin signals for flowering in Arabidopsis. Plant J. 35: 613–623 [DOI] [PubMed] [Google Scholar]
- Mouradov A., Cremer F., Coupland G. (2002). Control of flowering time: interacting pathways as a basis for diversity. Plant Cell 14 (suppl.): 111–130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaoka S., Takano T. (2003). Salt tolerance-related protein STO binds to a Myb transcription factor homologue and confers salt tolerance in Arabidopsis. J. Exp. Bot. 54: 2231–2237 [DOI] [PubMed] [Google Scholar]
- Nakamichi N., Kusano M., Fukushima A., Kita M., Ito S., Yamashino T., Saito K., Sakakibara H., Mizuno T. (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50: 447–462 [DOI] [PubMed] [Google Scholar]
- Nakano Y., Higuchi Y., Sumitomo K., Hisamatsu T. (2013). Flowering retardation by high temperature in chrysanthemums: involvement of FLOWERING LOCUS T-like 3 gene repression. J. Exp. Bot. 64: 909–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olszewski N., Sun T.P., Gubler F. (2002). Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14 (suppl.): S61–S80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda A., Narumi T., Li T., Kando T., Higuchi Y., Sumitomo K., Fukai S., Hisamatsu T. (2012). CsFTL3, a chrysanthemum FLOWERING LOCUS T-like gene, is a key regulator of photoperiodic flowering in chrysanthemums. J. Exp. Bot. 63: 1461–1477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park H.Y., Lee S.Y., Seok H.Y., Kim S.H., Sung Z.R., Moon Y.H. (2011). EMF1 interacts with EIP1, EIP6 or EIP9 involved in the regulation of flowering time in Arabidopsis. Plant Cell Physiol. 52: 1376–1388 [DOI] [PubMed] [Google Scholar]
- Pharis R.P. (1972). Flowering of Chrysanthemum under non-inductive long days by gibberellins and N(6)-benzyladenine. Planta 105: 205–212 [DOI] [PubMed] [Google Scholar]
- Pharis R.P., King R.W. (1985). Gibberellins and reproductive deve in seed plants. Annu. Rev. Plant Physiol. 36: 517–568 [Google Scholar]
- Pimenta Lange M.J., Lange T. (2006). Gibberellin biosynthesis and the regulation of plant development. Plant Biol (Stuttg) 8: 281–290 [DOI] [PubMed] [Google Scholar]
- Porri A., Torti S., Romera-Branchat M., Coupland G. (2012). Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 139: 2198–2209 [DOI] [PubMed] [Google Scholar]
- Putterill J., Robson F., Lee K., Simon R., Coupland G. (1995). The CONSTANS gene of Arabidopsis promotes flowering and encodes a protein showing similarities to zinc finger transcription factors. Cell 80: 847–857 [DOI] [PubMed] [Google Scholar]
- Robson F., Costa M.M., Hepworth S.R., Vizir I., Piñeiro M., Reeves P.H., Putterill J., Coupland G. (2001). Functional importance of conserved domains in the flowering-time gene CONSTANS demonstrated by analysis of mutant alleles and transgenic plants. Plant J. 28: 619–631 [DOI] [PubMed] [Google Scholar]
- Samach A., Onouchi H., Gold S.E., Ditta G.S., Schwarz-Sommer Z., Yanofsky M.F., Coupland G. (2000). Distinct roles of CONSTANS target genes in reproductive development of Arabidopsis. Science 288: 1613–1616 [DOI] [PubMed] [Google Scholar]
- Seki M., et al. (2002). Monitoring the expression profiles of 7000 Arabidopsis genes under drought, cold and high-salinity stresses using a full-length cDNA microarray. Plant J. 31: 279–292 [DOI] [PubMed] [Google Scholar]
- Seo E., Lee H., Jeon J., Park H., Kim J., Noh Y.S., Lee I. (2009). Crosstalk between cold response and flowering in Arabidopsis is mediated through the flowering-time gene SOC1 and its upstream negative regulator FLC. Plant Cell 21: 3185–3197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y.H., Ito S., Imaizumi T. (2013). Flowering time regulation: photoperiod- and temperature-sensing in leaves. Trends Plant Sci. 18: 575–583 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikanth A., Schmid M. (2011). Regulation of flowering time: all roads lead to Rome. Cell. Mol. Life Sci. 68: 2013–2037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sumitomo K., Li, T., and Hisamatsu, T. (2009). Gibberellin promotes flowering of chrysanthemum by upregulating CmFL, a chrysanthemum FLORICAULA/LEAFY homologous gene. Plant Sci. 176: 643–649 [Google Scholar]
- Takuhara Y., Kobayashi M., Suzuki S. (2011). Low-temperature-induced transcription factors in grapevine enhance cold tolerance in transgenic Arabidopsis plants. J. Plant Physiol. 168: 967–975 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. (2007). MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24: 1596–1599 [DOI] [PubMed] [Google Scholar]
- Todaka D., Matsushima H., Morohashi Y. (2000). Water stress enhances β-amylase activity in cucumber cotyledons. J. Exp. Bot. 51: 739–745 [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
- Winter D., Vinegar B., Nahal H., Ammar R., Wilson G.V., Provart N.J. (2007). An “Electronic Fluorescent Pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2: e718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada M., Takeno K. (2014). Stress and salicylic acid induce the expression of PnFT2 in the regulation of the stress-induced flowering of Pharbitis nil. J. Plant Physiol. 171: 205–212 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59: 225–251 [DOI] [PubMed] [Google Scholar]
- Yano M., Katayose Y., Ashikari M., Yamanouchi U., Monna L., Fuse T., Baba T., Yamamoto K., Umehara Y., Nagamura Y., Sasaki T. (2000). Hd1, a major photoperiod sensitivity quantitative trait locus in rice, is closely related to the Arabidopsis flowering time gene CONSTANS. Plant Cell 12: 2473–2484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S., Yang C., Peng J., Sun S., Wang X. (2009). GASA5, a regulator of flowering time and stem growth in Arabidopsis thaliana. Plant Mol. Biol. 69: 745–759 [DOI] [PubMed] [Google Scholar]
- Zheng Y., Zhao L.J., Gao J.P., Fei Z.J. (2011). iAssembler: a package for de novo assembly of Roche-454/Sanger transcriptome sequences. BMC Bioinformatics 12: 453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong S.L., Joung J.G., Zheng Y., Chen Y.R., Liu B., Shao Y., Xiang J.Z., Fei Z.J., Giovannoni J.J. (2011). High-throughput illumina strand-specific RNA sequencing library preparation. Cold Spring Harb Protoc 2011: 940–949 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.