Nuclei change their shape and size during cell growth in Arabidopsis thaliana. This work identifies a component of the plant nuclear envelope, KAKU4, which affects nuclear shape and size, possibly by regulating nuclear envelope expansion.
Abstract
In animals, the nuclear lamina is a fibrillar meshwork on the inner surface of the nuclear envelope, composed of coiled-coil lamin proteins and lamin binding membrane proteins. Plants also have a meshwork on the inner surface of the nuclear envelope, but little is known about its composition other than the presence of members of the CROWDED NUCLEI (CRWN) protein family, possible plant lamin analogs. Here, we describe a candidate lamina component, based on two Arabidopsis thaliana mutants (kaku2 and kaku4) with aberrant nuclear morphology. The responsible gene in kaku2 encodes CRWN1, and the responsible gene in kaku4 encodes a plant-specific protein of unknown function (KAKU4) that physically interacts with CRWN1 and its homolog CRWN4. Immunogold labeling revealed that KAKU4 localizes at the inner nuclear membrane. KAKU4 deforms the nuclear envelope in a dose-dependent manner, in association with nuclear membrane invagination and stack formation. The KAKU4-dependent nuclear envelope deformation was enhanced by overaccumulation of CRWN1, although KAKU4 can deform the nuclear envelope even in the absence of CRWN1 and/or CRWN4. Together, these results suggest that plants have evolved a unique lamina-like structure to modulate nuclear shape and size.
INTRODUCTION
The cell nucleus is enclosed by the nuclear envelope (NE), which consists of the outer and inner membranes, nuclear pore complexes, and the nuclear lamina beneath the inner membrane (Stuurman et al., 1998). The nuclear lamina is a fibrillar meshwork that provides not only structural integrity to the NE but also anchoring sites for chromatin domains, signaling molecules, and transcription factors to support a variety of cellular functions (Burke and Stewart, 2013). Major components of the nuclear lamina are the coiled-coil type V intermediate filament proteins called lamins. Lamin genes are widely conserved in animals (Stuurman et al., 1998; Dittmer and Misteli, 2011). Apart from the lamins in the nuclear scaffold, the nuclear lamina of mammals also contains a number of lamin binding membrane proteins (Gruenbaum et al., 2005), which play various roles in the nucleus (Schirmer and Foisner, 2007; Wilson and Foisner, 2010). Defects in the nuclear lamina components cause tissue-specific diseases in association with morphological changes of nuclei (Gruenbaum et al., 2005; Dauer and Worman, 2009). The lamina components in mammals have attracted a great deal of attention over the years because of their linkage to severe genetic diseases.
Despite extensive studies of the nuclear lamina components in animals, plants have no intermediate filament proteins (Goldberg, 2013) and Arabidopsis thaliana has no lamin homologs (Dittmer and Misteli, 2011). In tobacco (Nicotiana tabacum) cultured cells, a meshwork structure similar to the fibrillar lamin meshwork of Xenopus laevis was found beneath the inner nuclear membrane (Fiserova et al., 2009). A nuclear matrix constituent protein (NMCP1) was found as a peripheral framework component of nuclei in carrot (Daucus carota) cells (Masuda et al., 1997) and was recently reported to be localized underneath the inner nuclear membranes in onion (Allium cepa) cells (Ciska et al., 2013). NMCP1 is a coiled-coil protein that belongs to the NMCP family, which occurs widely in land plants, including mosses (Ciska et al., 2013; Ciska and Moreno Diaz de la Espina, 2013).
Arabidopsis has four NMCP homologs called CROWDED NUCLEI (CRWN) (Wang et al., 2013), formerly named LITTLE NUCLEI (Dittmer et al., 2007; Ciska et al., 2013; Ciska and Moreno Diaz de la Espina, 2013; Sakamoto and Takagi, 2013). Arabidopsis mutants crwn1 and crwn4 displayed aberrantly small and spherical nuclei in mature epidermal cells (Dittmer et al., 2007; Sakamoto and Takagi, 2013). CRWN1 and CRWN4 might be required for nuclear growth during cell development because the nucleus changes from spherical to spindle-shaped during development of epidermal cells of Arabidopsis (Chytilova et al., 1999). Similar nuclear shape phenotypes have been found in Arabidopsis mutants with defects of NE proteins. These include nup136 lacking the nuclear pore complex component NUCLEOPORIN136 (Nup136) (Tamura et al., 2010), the double mutant sun1 sun2 defective in the nuclear membrane proteins SAD1/UNC-84 (SUN) DOMAIN PROTEINs (Oda and Fukuda, 2011; Zhou et al., 2012), and the triple mutant wip1 wip2 wip3 lacking the outer nuclear membrane proteins WPP-domain interacting proteins (WIPs) (Zhou et al., 2012). Deficiency of myosin XI-i, which links the outer nuclear membrane to the cytoskeletal actin filaments, causes small nuclei in mature epidermal cells of Arabidopsis tissues (Tamura et al., 2013). Together, these findings implied that NE components act as nuclear shape determinants and are important for nuclear growth.
In animals, nuclear shape is determined by several proteins, including the nuclear pore complex components Nup35 (also known as Nup53) (Hawryluk-Gara et al., 2005) and Nup153 (Bastos et al., 1996; Zhou and Panté, 2010), and the inner nuclear membrane proteins LBR (Ellenberg et al., 1997) and Kugelkern (Brandt et al., 2006; Polychronidou et al., 2010), as well as by the actin binding, NE-associated protein Nesprin-2 Giant (Lüke et al., 2008). Nuclear shapes in mammals are also controlled by the nuclear lamina components, including the lamin proteins (Sullivan et al., 1999; Goldman et al., 2004; Prüfert et al., 2004, 2005; Lammerding et al., 2006; Volkova et al., 2011) and the lamin binding protein Titin (Zastrow et al., 2006). Although many factors affect nuclear morphology (Walters et al., 2012), the nuclear lamina seems to be the major contributor to maintenance of the nuclear shape (Webster et al., 2009). Hence, focusing on nuclear shape would make it possible to identify lamina components.
Nuclei in differentiated epidermal cells of Arabidopsis have a characteristic spindle shape (Chytilova et al., 1999; Jovtchev et al., 2006), which makes it easy to find mutants that have a defective nuclear shape. In this study, we isolated two ethyl methanesulfonate mutant alleles with defects in nuclear shape and size and designated them as kaku4 and kaku2, after the Japanese word for nucleus. The responsible genes encode plant-specific proteins, KAKU4 with unknown function and CRWN1 with coiled-coil domains, which interact with each other at the NE. Our findings suggest that KAKU4 modulates nuclear morphology by regulating the surface area of the nucleus.
RESULTS
The Plant-Unique Protein KAKU4 Is Required to Maintain the Spindle Shape of Nuclei in Epidermal Cells
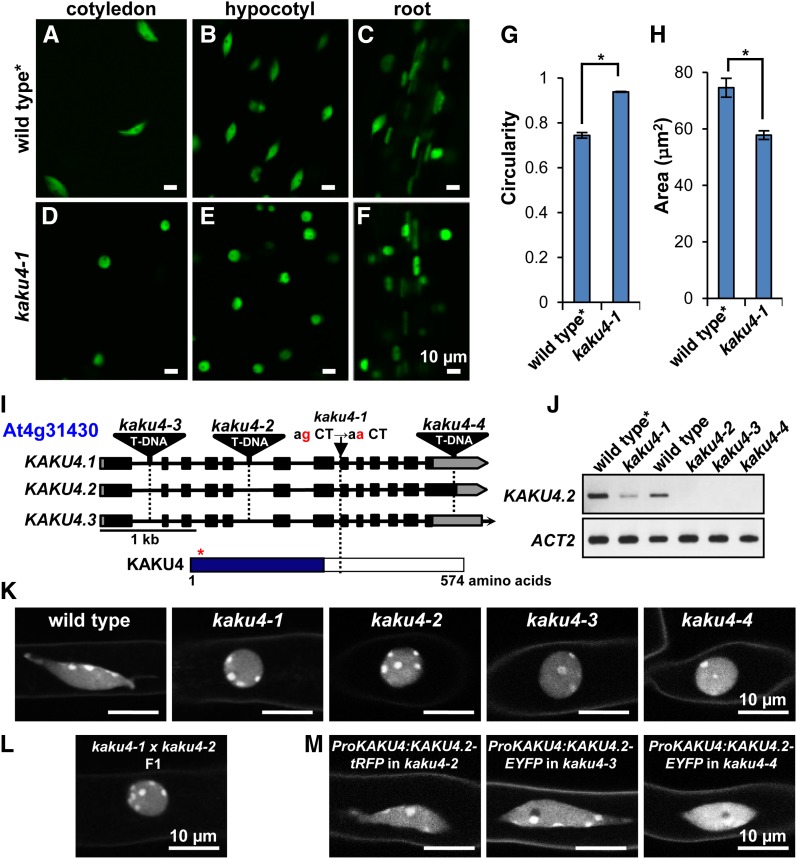
To identify genes that affect nuclear morphology, we conducted a mutant screen for mutants with altered morphology of nuclei. To mark the nuclei, we used a transgenic Arabidopsis line that expresses the nuclear marker Nup50a-GFP (green fluorescent protein) (Tamura et al., 2010). These plants show the wild-type morphology of spindle-shaped nuclei in the epidermal cells of cotyledons, hypocotyls, and roots of seedlings (Figures 1A to 1C). From an ethyl methanesulfonate–mutagenized pool of plants from this transgenic line (Tamura et al., 2013), we isolated two mutants with aberrant nuclear morphology: kaku4-1 and kaku2. The kaku4-1 nuclei were nearly spherical in various tissues (Figures 1D to 1F), with a significantly higher circularity index (defined as 4πA/P2, where A and P are the cross-sectional area and perimeter of the nucleus, respectively) (Figure 1G) and a significantly smaller area (Figure 1H) than the nuclei of the parental line, indicating that kaku4-1 nuclei are smaller and less elongated than nuclei of the parental line. kaku4 mutant plants exhibited no significant defect in their growth (Supplemental Figure 1).
Figure 1.
KAKU4 Is Required to Maintain Nuclear Shape in Arabidopsis.
(A) to (F) Nuclei in cotyledon epidermal cells, hypocotyl epidermal cells, and root cells from 1-week-old seedlings were visualized in the wild type and kaku4-1 by expressing Nup50a-GFP. Wild type*, a transgenic plant expressing Nup50a-GFP.
(G) and (H) Nuclear circularity indices (G) and nuclear areas (H) of hypocotyl epidermal cells in 1-week-old seedlings. Thirty independent plants were examined. The circularity indices and nuclear areas of three nuclei in each plant were quantified. Means ± standard errors for n = 30. Asterisks indicate a significant difference from wild type (Student’s t test, *P < 0.05).
(I) Schematic representation of the KAKU4 gene (At4g31430) and the predicted protein KAKU4. The positions of the kaku4-1 mutation and of each T-DNA insertion in three kaku4 mutant alleles are shown. This gene has three splicing variants, KAKU4.1, KAKU4.2, and KAKU4.3. Closed boxes, exons; solid lines, introns; gray boxes, putative untranslated regions; red asterisk, the position of putative nuclear localization signal; blue box, the region conserved among seed-bearing plants. See Supplemental Figures 2 and 3 for details.
(J) RT-PCR of KAKU4 transcript in kaku4 mutant alleles. Actin2 (ACT2) was used as a loading control.
(K) to (M) Nuclei stained with Hoechst 33342 in root hair cells for kaku4 mutant alleles (K), for an allelism test between kaku4-1 and kaku4-2 (L), and for complementation tests of the kaku4 phenotype by expressing KAKU4-tRFP or KAKU4-EYFP under the control of the endogenous promoter in three kaku4 mutant alleles (M). See Supplemental Figure 4 for wide fields of view.
The kaku4-1 mutant has a point mutation at the splicing donor site in the seventh intron of the At4g31430 gene (Figure 1I), causing 22 bp in the exon to be spliced out, thereby generating a premature stop codon (Supplemental Figures 2A to 2C). All three T-DNA tagged alleles (kaku4-2, kaku4-3, and kaku4-4) (Figure 1I), which are null mutants (Figure 1J), had spherical nuclei like those of kaku4-1 (Figure 1K). The first filial (F1) progeny from crossing kaku4-1 with kaku4-2 also had spherical nuclei (Figure 1L). These data demonstrate that the aberrant nuclear morphology of kaku4 mutant plants is caused by mutations in the KAKU4 gene (At4g31430).
The TAIR database (http://www.Arabidopsis.org/) has three splicing variants (At4g31430.1, At4g31430.2, and At4g31430.3), which we designated as KAKU4.1, KAKU4.2, and KAKU4.3, respectively (Figure 1I; Supplemental Figure 2A). The predicted KAKU4.2 product differs slightly from the product encoded by KAKU4.1 and KAKU4.3. Only KAKU4.2 cDNA was detected in seedlings (Supplemental Figures 2D and 2E). Thus, we focused on KAKU4.2, which encodes the unknown protein KAKU4 (Figure 1I). KAKU4 has one predicted nuclear localization signal (Figure 1I; Supplemental Figure 3) but no typical transmembrane domains. Additionally, KAKU4 has an Arg- and Gly-rich region in the C-terminal tail (Supplemental Figure 3, blue line), which is similar to sequences that bind to tudor domains for RNA metabolism (Selenko et al., 2001). The N-terminal half of KAKU4 occurs widely in seed-bearing plants (Supplemental Figure 3) but not in the fern Selaginella moellendorffi or the moss Physcomitrella patens. Thus, KAKU4 is unique to seed-bearing plants.
KAKU4 Localizes at the Inner Nuclear Membrane
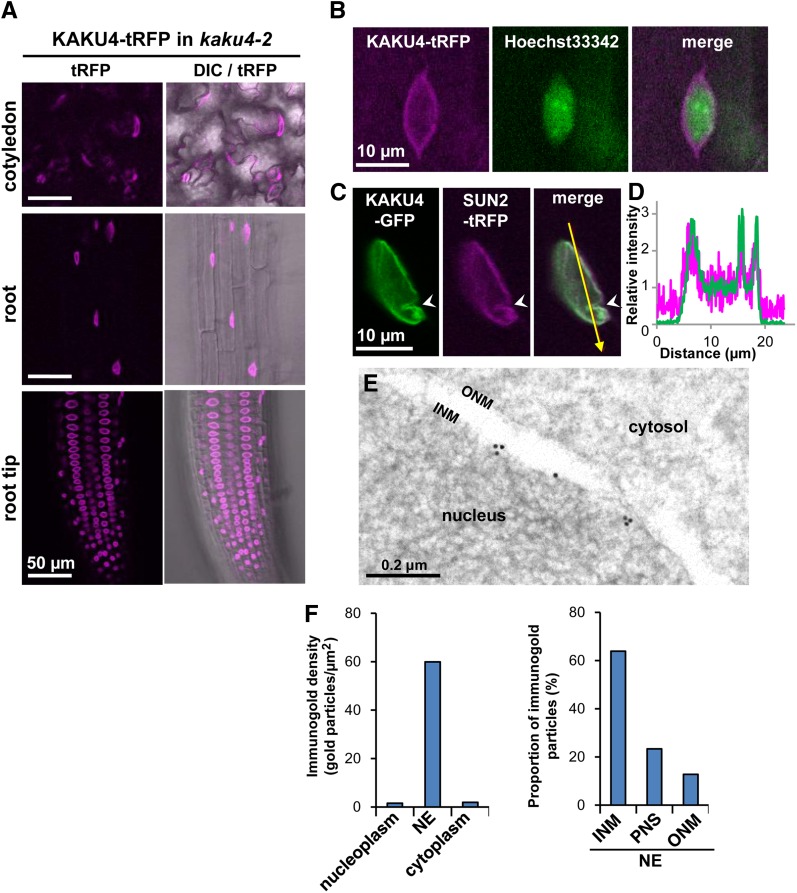
We examined the subcellular localization of KAKU4 by expressing fluorescent protein fusions with KAKU4. The aberrantly spherical nuclear morphology of three kaku4 mutant alleles was rescued by expressing either KAKU4-tRFP (tag red fluorescent protein) or KAKU4-EYFP (enhanced yellow fluorescent protein) under the control of the endogenous promoter (Figure 1M; Supplemental Figure 4), suggesting that the KAKU4 fusion proteins are functional and behave similarly to KAKU4. In these complemented plants, fluorescent signals were detected on the NE, regardless of the nuclear shape, in every tissue examined (Figure 2A). Consistent with this localization, both KAKU4-tRFP (Figure 2B) and KAKU4-EYFP (Supplemental Figures 5A to 5C and Supplemental Movie 1) were predominantly detected at the peripheral parts of nuclei stained with Hoechst in the root hair cells. The transgenic plants that expressed KAKU4-GFP together with the inner nuclear membrane marker SUN2-tRFP clearly showed the colocalization of these proteins (Figures 2C and 2D). A GFP fusion with the variant of KAKU4 that lacks the C-terminal Arg- and Gly-rich region of KAKU4, as encoded by the splicing variant KAKU4.1, also localized at the peripheral parts of nuclei of the root tip cells (Supplemental Figure 5D). Immunoelectron microscopy was done with root tip cells of the complemented plant (KAKU4-EYFP in kaku4-3) (Figure 2E; Supplemental Figure 5E). Quantitative analysis shows that gold particles were predominantly detected around the inner nuclear membrane (Figure 2F). These results suggest that KAKU4 localizes at the inner nuclear membrane.
Figure 2.
KAKU4 Localizes on the Nucleoplasmic Side of the Inner Nuclear Membrane.
(A) Fluorescence images showing subcellular localization of KAKU4 in cotyledon epidermal cells, root cells, and root tip cells from kaku4-2 seedlings stably expressing KAKU4-tRFP under the control of the endogenous promoter. DIC, differential interference contrast. See Supplemental Movie 1 for a Z-stack image and Supplemental Figure 5B.
(B) Fluorescence images of the root hair cell of a kaku4-2 seedling stably expressing KAKU4-tRFP under the control of the endogenous promoter. The nucleus was counterstained with Hoechst 33342. See Supplemental Figure 5A for the localization of KAKU4-EYFP.
(C) Fluorescence images of the root cell of a seedling that stably expressed both KAKU4-GFP and SUN2-tRFP, each under the control of the 35S promoter. The arrowheads indicate an invagination of the NE.
(D) Fluorescence intensity profile of the area indicated by the arrow in (C). The x axis is the distance from the starting point of the arrow.
(E) Immunogold analysis of the root tip of a kaku4-3 seedling expressing KAKU4-EYFP (see Figure 1M) with an anti-GFP antibody. INM, inner nuclear membrane; ONM, outer nuclear membrane. Also see Supplemental Figure 4D.
(F) Immunogold densities for KAKU4-EYFP in nucleoplasm, NE, and cytoplasm were calculated with total 397 gold particles of 19 electron micrographs (left). Proportion of the gold particles on inner nuclear membrane (INM), perinuclear space (PNS), and outer nuclear membrane (ONM) was calculated with total 501 gold particles of 47 electron micrographs (right).
KAKU4 and CRWN1 Interact with Each Other and Localize Independently at the NE
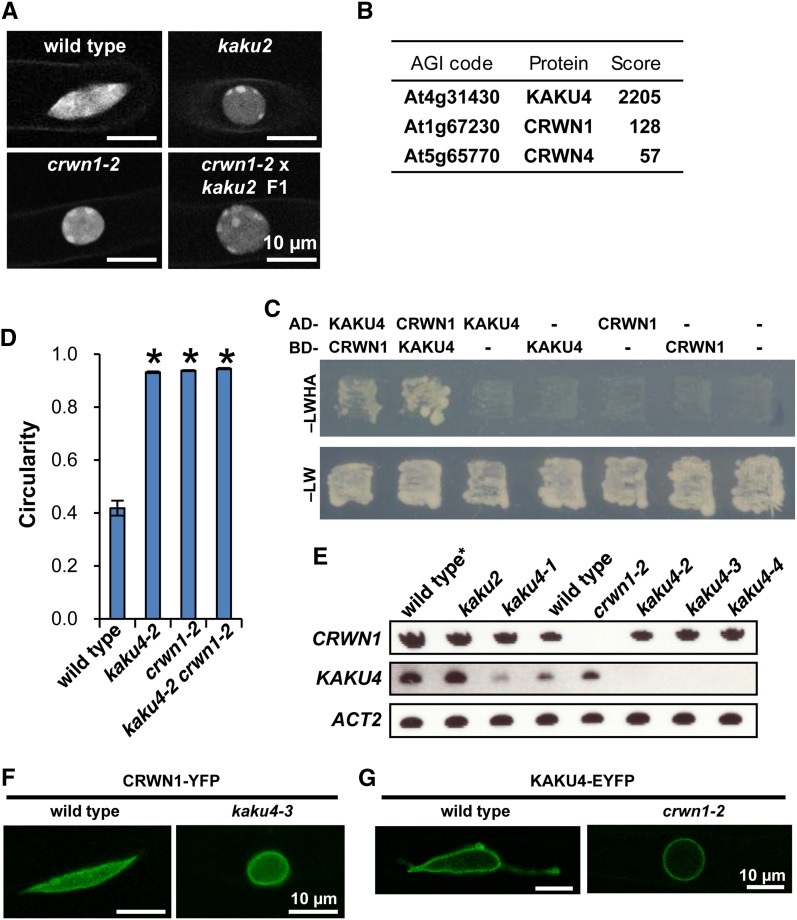
A second kaku mutant, kaku2, which has aberrant nuclear morphology, has a point mutation in the sixth exon of CRWN1 (Supplemental Figures 6A to 6J). F1 progeny obtained by crossing kaku2 with crwn1-2 (a null mutant) had spherical nuclei like those of the parental lines (Figure 3A), indicating that the responsible gene for the kaku2 phenotype is CRWN1.
Figure 3.
KAKU4 Interacts with CRWN1.
(A) Allelism test between kaku2 and crwn1-2. Fluorescence images of nuclei stained with Hoechst 33342 in root hair cells. See Supplemental Figure 6 for details of kaku2 mutant.
(B) LTQ-Orbitrap mass spectrometry of the anti-GFP antibody pull-down fraction of plants expressing KAKU4-GFP–identified CRWN1 and CRWN4. Arabidopsis Genome Initiative (AGI) codes and annotations are from the TAIR database (http://www.Arabidopsis.org). Scores were calculated using the Mascot program (Matrix Science). See Supplemental Figure 7 for raw data.
(C) Yeast two-hybrid analysis of the strain expressing a fusion protein containing the GAL4 DNA binding domain (BD) and a fusion protein containing the GAL4 activation domain (AD). Transformants were incubated on SD/–Leu/–Trp/–His/–Ade medium supplemented with X-α-Gal and Aureobasidin A (–LWHA) or SD/–Leu/–Trp medium (–LW). Negative controls with empty vector (-) were also included.
(D) Nuclear circularity indices of nuclei stained with Hoechst 33342 in root hair cells of seedlings. Fifteen independent plants were examined. The circularity indices of three nuclei in each plant were quantified. Means ± standard errors for n = 15. Asterisks indicate a significant difference from the wild type (Student’s t test, P < 0.05). Also see Supplemental Figure 6K.
(E) RT-PCR of CRWN1, KAKU4, and ACT2 transcripts in crwn1 (kaku2 and crwn1-2) and kaku4 mutants. Wild type*, a transgenic plant expressing Nup50a-GFP.
(F) Fluorescence images of root cells of wild-type and kaku4-3 seedlings that stably expressed CRWN1-YFP under the control of the 35S promoter.
(G) Fluorescence images of root hair cells of wild-type and crwn1-2 seedlings that stably expressed KAKU4-EYFP under the control of the endogenous promoter.
To examine the interactions of KAKU4, we conducted pull-down assays and examined its interaction partners. KAKU4-GFP expressed in whole seedlings was pulled down with anti-GFP antibody, and the pull-down product was analyzed by mass spectrometry. Intriguingly, two CRWN proteins (CRWN1 and CRWN4) were pulled down together with KAKU4 (Figure 3B; Supplemental Figure 7). The interaction between KAKU4 and CRWN1 was supported by yeast two-hybrid assay (Figure 3C). Among the four CRWN genes in the Arabidopsis genome, the other two CRWN gene products (CRWN2 and CRWN3) were not detected in mass spectrometry. The absence of CRWN2 and CRWN3 in the pull-down fraction, together with the fact that neither crwn2 nor crwn3 mutants exhibited drastic abnormalities in nuclear shape (Sakamoto and Takagi, 2013), suggest that CRWN1 and CRWN4 are the predominant factors maintaining nuclear morphology.
We next examined whether KAKU4 and CRWN1 act independently or together. The nuclei of root hairs in a kaku4-2 crwn1-2 double mutant were just as spherical as those in the parental lines kaku4-2 and crwn1-2 (Figure 3D; Supplemental Figure 6K), indicating that combining the kaku4-2 and crwn1-2 mutations had neither synergistic nor additive effects on nuclear shape. Additionally, transcription of CRWN1 was not affected by mutation of KAKU4, and vice versa (Figure 3E). Next, we examined the effect of deficiency of KAKU4 on the localization of CRWN1 and the effect of deficiency of CRWN1 on the localization of KAKU4 at the NE. Stably expressed CRWN1-YFP under the 35S promoter localized at the NE in kaku4-3 as it did in the wild type (Figure 3F) and, in the reverse case, KAKU4-EYFP localized at the NE in crwn1-2 (Figure 3G). Notably, expressing CRWN1 did not rescue the spherical nuclear phenotype of kaku4-3 and expressing KAKU4 did not rescue the spherical nuclear phenotype of crwn1-2 (Figures 3F and 3G). These results suggest that KAKU4 and CRWN1 localize at the NE independently of each other.
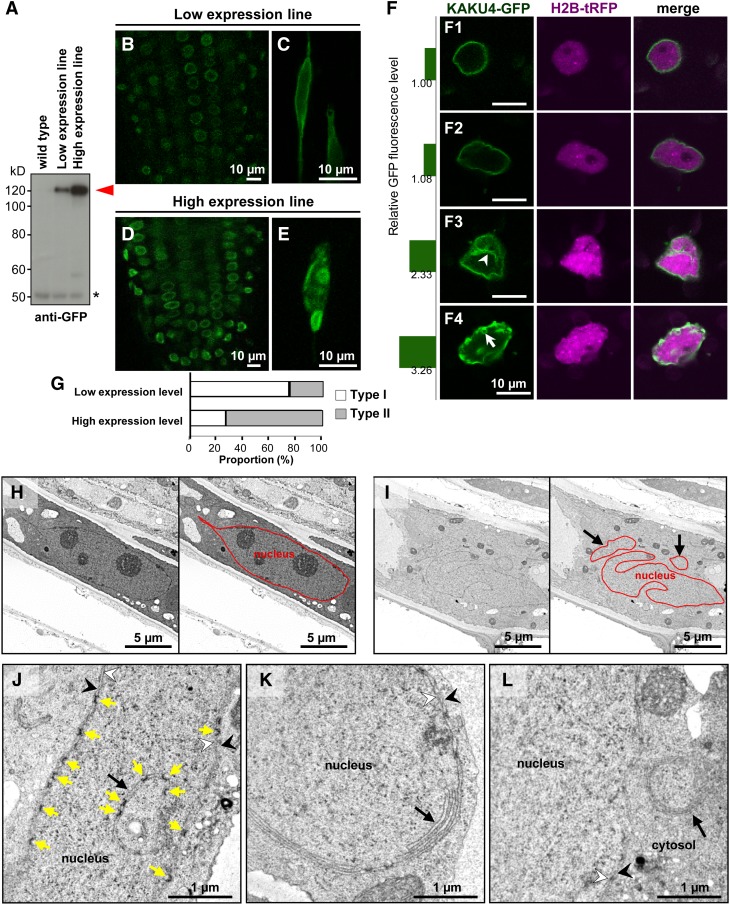
High Levels of KAKU4 Induce Growth and Deformation of Nuclear Membranes
To gain insight into the function of KAKU4, we examined the effect of KAKU4 expression levels on NE architecture. We isolated two stable transformant lines of Arabidopsis with different expression levels of KAKU4-GFP (Figure 4A). In both of these lines, KAKU4-GFP localized at the NE (Supplemental Figures 8A and 8B). In contrast to the normal structure of the GFP-labeled NE in the low-expression line (Figures 4B and 4C), in the high-expression line, the NE was deformed in all plant tissues examined except the meristematic tissues (Figures 4D and 4E). The deformations included ring-like structures, indentations, bleb-like structures, and nonuniform KAKU4-GFP distribution (Supplemental Figures 8C to 8I). The possibility that overexpressing the GFP moiety causes NE deformation can be eliminated because overexpressing FLAG-tagged KAKU4 also induced NE deformation, and overexpressing the GFP fusion with the inner nuclear membrane protein SAD1/UNC-84 DOMAIN PROTEIN2 (SUN2) did not (Supplemental Figure 8J). These data suggest that high levels of KAKU4 induce deformation of the NE.
Figure 4.
KAKU4-GFP Induces Nuclear Membrane Growth.
(A) Immunoblots with anti-GFP antibody of seedlings from wild-type plants and two transgenic lines expressing KAKU4-GFP at different levels under the control of the 35S promoter. Red arrowhead, full-length KAKU4-GFP; asterisk, a nonspecific signal.
(B) to (E) Fluorescence images of the root tips ([B] and [D]) and root cells ([C] and [E]) of seedlings from two transgenic KAKU4-GFP lines in (A). Also see Supplemental Figures 8A to 8I.
(F) Fluorescence images of N. tabacum cells transiently expressing KAKU4-GFP and HISTONE2B-tRFP (H2B-tRFP), each under the control of the 35S promoter. The numbers on the left side of the panels are relative fluorescence intensity. Arrowhead, invagination of the NE; arrow, a ring-like structure. Also see Supplemental Figure 8K and Supplemental Movie 2.
(G) According to the degree of deformation of the NE, the nuclei were classified into two types: Type I nuclei have normal NE and Type II nuclei have deformed NE. Proportions of Type I and Type II nuclei for the cells with high expression levels of KAKU4-GFP and for the cells with low expression levels of KAKU4-GFP were calculated. Expression level of KAKU4-GFP was determined by GFP fluorescence intensity (n = 328 nuclei).
(H) to (L) Electron micrographs of nuclei in root cells of seedlings of a wild-type plant (H) and a transgenic line overexpressing KAKU4-GFP ([I] to [L]). NEs in (H) and (I) are traced by red lines in the right panels. The black arrows indicate irregular patterns of the NE including regions enclosed by NE in (I), irregular invagination in (J), stacked nuclear membranes in (K), and an extranuclear membrane whorl in the cytoplasm near the nucleus in (L). The yellow arrows in (J) indicate nuclear pores. White arrowhead, inner nuclear membrane; black arrowhead, outer nuclear membrane.
The relationship between the KAKU4 levels and degree of deformation of the NE was examined by transiently expressing KAKU4-GFP in the N. tabacum leaves, which have predominantly spherical and oval nuclei. The nuclei with the low KAKU4-GFP levels were uniformly labeled with fluorescence at their periphery (Figure 4F, 1 and 2), while the nuclei with 2- to 3-fold higher levels of KAKU4-GFP were irregularly deformed in association with the invaginated NE structures (Figure 4F, 3) and ring-like NE structures (Figure 4F, 4). According to the degree of deformation of the NE, we classified the nuclei having normal NE into Type I and the nuclei having deformed NE into Type II. The proportion of Type II nuclei was noticeably higher in cells with high expression levels of KAKU4-GFP than in the cells with low expression levels (Figure 4G). Similar NE deformations were induced by overexpressing KAKU4-GFP in leaves of another Nicotiana species, N. benthamiana (Supplemental Figures 8K and 8L and Supplemental Movie 2). Hence, the deformation degree of the NE is positively correlated with the KAKU4 expression levels.
Ultrastructure analysis clearly showed that the NE of the high expression line of KAKU4-GFP was irregularly invaginated in root cells (Figures 4I), in contrast to the relatively smooth NE in the wild type (Figure 4H). Notably, the invaginated NE contained nuclear pores (Figure 4J). Stacked nuclear membranes without nuclear pores (Figure 4K) and extranuclear membrane whorls (Figure 4L) were also found in the high expression line. Taken together, these findings suggest that high amounts of KAKU4 induce growth of the nuclear membrane, which results in the formation of extensive nuclear membrane invaginations and stacks.
Enhancement of NE Deformation by KAKU4 and CRWN1
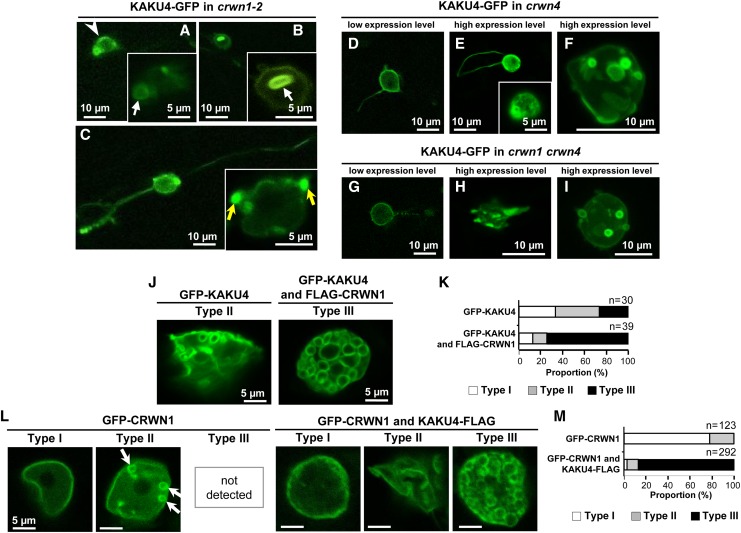
To determine the effects of a deficiency of CRWN1 on the KAKU4-mediated deformation of the NE, KAKU4-GFP was stably expressed under the 35S promoter in crwn1-2 mutants. In root cells, KAKU4-GFP often localized unevenly at the NE (Figure 5A, arrowhead). The nuclei frequently had ring-like and bleb-like structures and were typically spherical with long tails labeled by KAKU4-GFP (Figures 5A to 5C). On the whole, the NE deformation patterns in crwn1-2 were similar to those in the presence of CRWN1, suggesting that KAKU4 has an ability to deform the NE in the absence of CRWN1. Next, we examined the effects of a deficiency of CRWN4, a CRWN1 homolog, which was identified as a candidate for a KAKU4-interacting protein (Figure 3B). Similar expression-dependent deformation patterns of the NE were caused by transiently expressing KAKU4-GFP in the crwn4 mutant cells (Figures 5D to 5F) and the crwn1 crwn4 double mutant cells (Figures 5G to 5I). All of the single mutants (crwn1 and crwn4) and the double mutant crwn1 crwn4 have small spherical nuclei. Taken together, these results suggest that KAKU4 can deform the NE even in the absence of CRWN1 and/or CRWN4.
Figure 5.
Overexpression of KAKU4 and CRWN1 Induces NE Deformations
(A) to (C) Fluorescence images of root cells of crwn1-2 seedlings that stably expressed KAKU4-GFP under the control of the 35S promoter. Arrowhead, uneven fluorescence distribution; white arrows, ring-like structures; yellow arrows, bleb-like structures.
(D) to (F) Fluorescence images of cotyledon epidermal cells of crwn4 seedlings that transiently expressed KAKU4-GFP under the control of the 35S promoter. A Z-stack confocal image is reconstituted with 12 series of optical 1.0-μm sections (F).
(G) to (I) Fluorescence images of cotyledon epidermal cells of crwn1 crwn4 seedlings that transiently expressed KAKU4-GFP under the control of the 35S promoter. A Z-stack confocal image is reconstituted with 19 series of optical 1.0-μm sections (I).
(J) Fluorescence images of epidermal leaf cells of N. tabacum that transiently expressed GFP-KAKU4 without or with FLAG-CRWN1, each under the control of the 35S promoter. The nuclei are separated into three types: Type I, nuclei with normal NE; Type II, nuclei with moderately deformed NE; Type III, nuclei with severely deformed NE resulting in numerous ring structures.
(K) Proportions of Type I, Type II, and Type III nuclei in N. tabacum expressing GFP-KAKU4 without or with FLAG-CRWN1.
(L) Fluorescence images of epidermal leaf cells of N. tabacum that transiently expressed GFP-CRWN1 without or with KAKU4-FLAG, each under the control of the 35S promoter. Arrows indicate small ring structures.
(M) Proportions of Type I, Type II, and Type III nuclei in N. tabacum expressing GFP-CRWN1 without or with KAKU4-FLAG.
Co-overexpression of FLAG-CRWN1 with GFP-KAKU4 has a more severe impact on NE shape in N. tabacum leaves (Figure 5J). The nuclei were filled with numerous NE ring structures (Figure 5J). In this study, we separated the nuclei into three types. Type I nuclei had normal NE, Type II nuclei had moderately deformed NE, and Type III nuclei had severely deformed NE and were filled with numerous ring structures (Figure 5L). The proportion of the Type III nuclei was noticeably higher in the cells overexpressing both GFP-KAKU4 and FLAG-CRWN1 than in the cells overexpressing GFP-KAKU4 alone (Figure 5K). On the other hand, overexpressing GFP-CRWN1 alone caused the Type II nuclei to have punctate structures and ring structures (Figure 5L), but not the Type III nuclei. By contrast, co-overexpression of KAKU4-FLAG with GFP-CRWN1 frequently generated Type III nuclei filled with ring structures (Figure 5L) and made ∼90% of nuclei into Type III (Figure 5M). These results clearly demonstrate that high levels of CRWN1 dramatically enhance NE deformation in the presence of high levels of KAKU4. Taken together, these results suggest that KAKU4 and CRWN1 have an ability to deform the NE independently of each other and that the total amounts of KAKU4 and CRWN1 determine the degree of NE deformation.
DISCUSSION
KAKU4 Is a Putative Component of the Lamina-Like Structure in Plants
Here, we provide evidence that KAKU4 localizes at the inner nuclear membrane in plant tissues and interacts with CRWN1, which is a putative lamin analog, although it has little sequence similarity with lamins (Meier and Brkljacic, 2010; Evans et al., 2011). These results suggest that KAKU4 acts as a component of the nuclear lamina-like structure in plants. KAKU4 is a protein specific to seed-bearing plants, suggesting that these plants might have evolved a unique lamina-like structure to modulate nuclear shape and size.
Animals have intermediate filament protein lamins, but plants and single-celled eukaryotes have no lamin proteins (Meier and Brkljacic, 2010; Dittmer and Misteli, 2011). In single-celled eukaryotes, coiled-coil proteins that are functionally homologous to lamins have been reported as lamina components. These include NE81 in Dictyostelium (Krüger et al., 2012) and NUP-1 in Trypanosoma (DuBois et al., 2012), neither of which resemble lamins at the amino acid sequence level. These results, together with our findings, indicate that eukaryotic organisms have highly diverse lamina components. The diversity of lamina components might be contrasted with the high similarities of other NE proteins, such as the nuclear pore complex (Tamura and Hara-Nishimura, 2013) and nuclear membrane protein SUN (Graumann and Evans, 2010; Starr and Fridolfsson, 2010), between plants, animals and yeast.
Despite the high diversity among different organisms, lamina proteins regulate a variety of fundamental nuclear processes in these organisms, including DNA replication, transcription, and chromatin organization. Thus, deficiency of a lamina component can cause genetic diseases and inhibit growth (Dauer and Worman, 2009). Wang et al. (2013) reported their inability to isolate a null mutant lacking all four CRWN genes (Wang et al., 2013). By contrast, KAKU4 is not essential, which suggests that plants might have acquired nonessential components of the lamina-like structure.
Possible Interaction of the Lamina Complex KAKU4-CRWN1 with Other NE Components to Maintain Nuclear Morphology
The Arabidopsis mutant lacking plant-specific myosin XI-i has a similar aberrant nuclear shape to kaku4 and crwn1 mutants (Tamura et al., 2013). Myosin XI-i functions in nuclear movement along actin filaments by interacting with the nucleoplasm through the SUN-WIP-WIT bridge, which links the inner and outer nuclear membranes (Tamura et al., 2013). Accordingly, the double mutant sun1 sun2 (Oda and Fukuda, 2011; Zhou et al., 2012), the double mutant wit1 wit2 (Tamura et al., 2013), and the triple mutant wip1 wip2 wip3 (Zhou et al., 2012) also have the same phenotype as kaku4 and crwn1. Both Arabidopsis SUN1 and SUN2 are homologous to animal SUN-domain proteins (Graumann et al., 2010), some of which interact with lamin proteins (Starr and Fridolfsson, 2010). This raises the possibility that KAKU4 and CRWN1 interact with the SUN-WIP-WIT bridge. Additionally, deficiency of the nuclear pore protein Nup136 causes a similar aberrant nuclear shape as kaku4 and crwn1 mutations (Tamura et al., 2010); Nup136 is a functional homolog of animal Nup153 that physically associates with lamin (Smythe et al., 2000). KAKU4 and CRWN1 might maintain nuclear morphology by interacting with the nucleocytoplasmic linker and Nup136 to mechanically support the NE.
How Does KAKU4 Modulate Nuclear Morphology?
Both KAKU4 and CRWN1 might control nuclear growth during cell differentiation by regulating nuclear shape and size. A deficiency of KAKU4 impairs expansion of the NE and thereby inhibits growth of the nucleus. By contrast, when excess amounts of KAKU4 are present, KAKU4 was unevenly concentrated on certain parts of the NE. In these regions, the nuclear membrane might elongate to form invaginations and protrusions of the NE, resulting in stable ring-like and tail structures. Overexpression of other proteins can also cause NE deformations in other eukaryotic cells. These include Saccharomyces cerevisiae Esc1 (Hattier et al., 2007), Drosophila melanogaster Kugelkern (Brandt et al., 2006; Polychronidou et al., 2010), human lamins (Volkova et al., 2011), human LBR (Ellenberg et al., 1997), X. laevis LBR (Ma et al., 2007), and mammalian Nopp140 (Isaac et al., 1998, 2001; Kittur et al., 2007). A common feature of these proteins is that they localize on the nucleoplasmic side of the inner membrane, suggesting that putative lamina components have an immense influence on the shape of the NE. These data from animal and yeast cells support the idea that KAKU4 can affect the shape of the NE by acting as a lamina-like structure in plants.
METHODS
Plant Materials
Arabidopsis thaliana (Columbia-0) was used as the wild type. T-DNA insertion mutants (SALK_041774 [crwn1-2], SALK_076754 [kaku4-2], SALK_010298 [kaku4-3], and SAIL_711_E09 [kaku4-4]) were obtained from the ABRC at Ohio State University. The seeds of crwn4 (SALK_079288) and crwn1 crwn4 (crwn1, SALK_016800; crwn4, SALK_079288) were kindly provided by Yuki Sakamoto and Shingo Takagi (Osaka University). The seeds of transgenic plants expressing CRWN1-YFP (Dittmer et al., 2007) were kindly provided by Eric J. Richards (Cornell University). A kaku4-3 line expressing CRWN1-YFP was produced by crossing the CRWN1-YFP line with kaku4-3. A crwn1-2 line expressing KAKU4-EYFP and a crwn1-2 line expressing KAKU4-GFP were also produced by crossing. A transgenic Arabidopsis line expressing Nup50a-GFP was mutagenized with ethyl methanesulfonate to obtain a mutant pool (Tamura et al., 2010). Two mutants (kaku2 and kaku4-1) were isolated from the pool. Arabidopsis seeds were germinated and grown at 22°C under continuous light (35 µmol m−2 s−1). Tobacco seeds (Nicotiana tabacum cv Samsun NN and Nicotiana benthamiana) (Hatsugai et al., 2004) were sown in culture soil (Sumirin) and grown at 22°C under long-day conditions (16 h light/8 h dark).
Isolation of kaku2 and kaku4-1 Mutants and Map-Based Cloning of the KAKU2 and KAKU4 Genes
The kaku2 and kaku4-1 mutants were isolated as described previously (Tamura et al., 2013). Each of the kaku2 and kaku4-1 mutants was backcrossed with a wild-type plant at least once. Map-based cloning was performed as described previously (Tamura et al. 2005).
Confocal Laser Scanning Microscopy
Fluorescence images, with the exception of those presented in Supplemental Figure 4, were obtained using a confocal laser scanning microscope (LSM510 META or LSM780; Carl Zeiss). The 405-nm line of a blue diode laser, the 488-nm line of a 40-mW Ar/Kr laser, and the 544-nm line of a 1-mW He/Ne laser were used to excite Hoechst 33342, GFP/YFP, and RFP, respectively. Images were acquired with a ×63 1.2–numerical aperture (NA) water immersion objective (C-Apochromat, 441777-9970-000; Carl Zeiss), a ×40 0.95-NA dry objective (Plan-Apochromat, 440654-9902-000; Carl Zeiss), or a ×20 0.80-NA dry objective (Plan-Apochromat, 440640-9903-000; Carl Zeiss). Data were exported as TIFF files and processed using Adobe Photoshop Elements 9.0 (Adobe Systems) or ImageJ 1.45s (National Institutes of Health).
Fluorescence Microscopy
Fluorescence images in Supplemental Figure 4 were obtained using an Axioskop2 plus microscope (Carl Zeiss) and a high-sensitivity cooled CCD color camera VB-7010 (Keyence).
Hoechst 33342 Staining
Nuclei of 7-d-old seedlings were stained with a solution containing 1 μg/mL Hoechst 33342, 3.7% (w/v) paraformaldehyde, 10% (v/v) DMSO, 3% (v/v) Nonidet P-40, 50 mM PIPES-KOH (pH 7.0), 1 mM MgSO4, and 5 mM EGTA. The staining time (10 s to 10 min) depended on the tissue examined.
Quantitative Determination of Nuclear Shape and Size
Fluorescence images of nuclei labeled with Nup50a-GFP in hypocotyls of 10 7-d-old seedlings were obtained using a confocal laser scanning microscope. The images were exported as TIFF files and processed using the Analyze Particles function of ImageJ. The circularity index and area were calculated. We selected three measurement values in descending order per plant and regarded the mean of the three measurements as the value from a plant. Then, the means and standard errors of values from the 10 plants were calculated and a two-tailed homoscedastic Student’s t test was performed using Microsoft Excel. The circularity index was calculated using the equation 4πA/P2 (where A = area of nucleus and P = perimeter of nucleus) and indicates how closely each nucleus corresponds to a spherical shape (a perfect sphere has a circularity index of 1). Any deviation from a circular shape (e.g., elongated, lobulated, or spindle shaped) causes the index to decrease.
Fluorescence images of nuclei stained with Hoechst 33342 from three 7-d-old seedlings were obtained with a confocal laser scanning microscope, with three root hairs imaged per plant. Image processing and calculations were performed as described above. The mean of the three values from each plant was calculated. Then, the mean values and standard errors of the three plants were calculated.
Bioinformatics of the Arabidopsis Proteins
Information about the predicted gene structures and sequences (exons, introns, and untranslated regions) were obtained from TAIR (http://www.Arabidopsis.org). The domains of proteins were predicted using the normal mode of the SMART (Simple Modular Architecture Research Tool) program (http://smart.embl-heidelberg.de/).
RT-PCR Analysis
Total RNA was isolated from 6- to 7-d-old seedlings using the RNeasy plant mini kit (Qiagen). Total RNA was treated with DNaseI (Invitrogen) and subjected to first-strand cDNA synthesis using Ready-To-Go RT-PCR beads (GE Healthcare Bio-Science) and an oligo(dT)12-18 primer (Invitrogen). The primers used are shown in Supplemental Table 1.
Plasmid Construction
To construct ProKAKU4:KAKU4-EYFP/tRFP, a genomic fragment encompassing the 2-kb sequence upstream of the coding sequence (supposedly the endogenous promoter of KAKU4) and the entire coding sequence of KAKU4 (KAKU4.2) except for a stop codon was amplified by PCR and cloned into pENTR1A (Invitrogen) and then fused upstream of EYFP or TagRFP in a plant transformation vector (pGWB540 or pGWB559). To construct Pro35S:GFP- KAKU4 and Pro35S:KAKU4-GFP/FLAG, a genomic fragment containing the entire coding sequence of KAKU4 (KAKU4.2) except for a stop codon was cloned into pENTR1A (Invitrogen) and then fused downstream/upstream of sGFP or FLAG tag and downstream of the constitutive cauliflower mosaic virus promoter 35S in a plant transformation vector (pGWB406, pGWB405, or pGWB411). To construct Pro35S:KAKU4.1-GFP, a genomic fragment containing the entire coding sequence of KAKU4.1 except for a stop codon was cloned into pENTR1A (Invitrogen) and then fused upstream of sGFP tag and downstream of the promoter 35S in a plant transformation vector (pGWB405). To construct Pro35S:GFP/FLAG-CRWN1, a genomic fragment containing the entire coding sequence of CRWN1 was amplified, cloned into pENTR1A (Invitrogen), and then fused downstream of sGFP or FLAG tag in a plant transformation vector (pGWB406 or pGWB412). The construction of Pro35S:AtSUN2-GFP used in this study was described previously (Tamura et al., 2013). For the yeast two-hybrid assay, cDNA fragments of CRWN1 or KAKU4 were cloned into pENTR1A or pENTR/D-TOPO, respectively, and then fused into two vectors (pDEST-GBKT7 and pDEST-GADT7; Rossignol et al., 2007). To construct Pro35S:H2B-tRFP, the cDNA was cloned into the entry clone previously (Tamura et al., 2010) and the amplified region was fused upstream of TagRFP in a plant transformation vector (pGWB460). The primers used are shown in Supplemental Table 1.
Stable and Transient Expression of Fusion Proteins in Arabidopsis
For stable expression, Arabidopsis wild-type plants or kaku4 mutants were transformed by the floral dip method (Clough and Bent, 1998) with Agrobacterium tumefaciens (GV3101). For transient expression, each construct was introduced into wild-type, kaku4-2, crwn1-2, crwn4, or crwn1 crwn4 lines via particle bombardment. Seven-day-old seedlings were bombarded with 1-µm gold particles coated with two plasmids that carried KAKU4-GFP and mRFP-PTS1 (Tamura et al., 2005) using a Helios gene gun system (Bio-Rad Laboratories).
Transient Expression of Fusion Proteins in Tobacco
Constructs were introduced by agroinfiltration of true leaves (Sparkes et al., 2006). Leaves were infiltrated with an Agrobacterium suspension (optical density of 0.1). Two days later, the leaves were observed using a confocal laser scanning microscope.
Transmission Electron Microscopy and Immunogold Labeling
For transmission electron microscopy, the root tips of 7-d-old wild-type seedlings and those overexpressing KAKU4-GFP were used. The samples were fixed with 0.05 M cacodylate buffer (pH 7.4) containing 2% paraformaldehyde and 2% glutaraldehyde at 4°C overnight. Then, the samples were fixed with 0.05 M cacodylate buffer containing 0.5% tannic acid at 4°C for 2 h, rinsed four times with 0.05 M cacodylate buffer for 30 min each, and then postfixed with 0.05 M cacodylate buffer containing 2% osmium tetroxide at 4°C for 4 h. The samples were dehydrated through a graded ethanol series (50, 70, 90, and 100%) as follows: 50 and 70% for 30 min each at 4°C, 90% for 30 min at room temperature, and three changes of 100% for 24 h each at room temperature. The samples were infiltrated with propylene oxide (PO) twice for 30 min each time and incubated in a 70:30 mixture of PO and resin (Quetol-651; Nisshin EM) for 1 h. Then, the cap of the tube was opened and PO was evaporated overnight. The samples were transferred to fresh 100% resin and polymerized at 60°C for 48 h. The blocks were cut into 70-nm-thick sections using a diamond knife and an ultramicrotome (Ultracut UCT; Leica). The sections were placed on copper grids, stained with 2% uranyl acetate at room temperature for 15 min, rinsed with distilled water, and stained with lead stain solution (Sigma-Aldrich) at room temperature for 3 min. The grids were observed using a transmission electron microscope (JEM-1200 EX; JEOL) at an acceleration voltage of 80 kV. Digital images (2048 × 2048 pixels) were acquired using a CCD camera (VELETA; Olympus Soft Imaging Solutions).
For immunogold labeling and transmission electron microscopy, the root tips of 7-d-old kaku4-3 seedlings stably expressing KAKU4-EYFP were used. Samples sandwiched between copper disks were frozen in liquid propane at −175°C and then freeze substituted with acetone containing 1% tannic acid and 3% distilled water at −80°C for 48 h. The samples were kept at −20°C for 3 h and then at 4°C for 1 h. The samples were dehydrated in anhydrous ethanol three times at 4°C for 1 h each time. The samples were infiltrated with a 50:50 mixture of ethanol and resin (LR white; London Resin) at 4°C for 3 h. Then, the samples were incubated in 100% LR White three times at 4°C for 1 h each time, transferred to fresh 100% resin, and polymerized at 50°C overnight. The blocks were cut into 80-nm-thick sections using a diamond knife and an ultramicrotome (Ultracut UCT), and placed onto nickel grids. The grids were then incubated with the primary antibody (anti-GFP rabbit polyclonal antibody) diluted in PBS containing 1% BSA for 90 min at room temperature, rinsed with PBS containing 1% BSA three times for 1 min each time, and then placed onto drops of the secondary antibody conjugated to gold particles (15 nm) (Anti-Rabbit IgG; British BioCell International) for 1 h at room temperature. The grids were rinsed with PBS, placed in 0.1 M phosphate buffer containing 2% glutaraldehyde, dried, stained with 2% uranyl acetate for 15 min, rinsed with distilled water, and then incubated with lead stain solution (Sigma-Aldrich) at room temperature for 3 min. The grids were observed using a transmission electron microscope (JEM-1200 EX; JEOL) at an acceleration voltage of 80 kV. Digital images (2048 × 2048 pixels) were acquired with a CCD camera (VELETA; Olympus Soft Imaging Solutions).
SDS-PAGE and Immunoblot Analysis
Protein extracts from seedlings were subjected to SDS-PAGE, followed by immunoblot analysis. Immunoreactive signals were detected using the ECL detection system (GE Healthcare) and an anti-GFP antibody (JL-8; Clontech) (1:2500).
Mass Spectrometry of the Pull-Down Product
Pull-down was performed using an anti-GFP antibody, transgenic seedlings expressing GFP (Mano et al., 1999) or KAKU4-GFP, and a µMACS Epitope Tag Protein Isolation Kit (Miltenyi Biotec). Whole seedlings of each transgenic Arabidopsis plant (∼0.5 g) were homogenized with liquid nitrogen on ice in 1.5 mL of lysis buffer (150 mM NaCl, 1% Triton X-100, and 50 mM Tris-HCl, pH 8.0). Homogenates were centrifuged at 20,400g for 10 min at 4°C to remove cellular debris. The supernatants (1 mL) were mixed with magnetic beads (50 µL) conjugated to an anti-GFP antibody (Miltenyi Biotec) and then incubated on ice for 30 min. The mixtures were applied to µ Columns (Miltenyi Biotec) in a magnetic field to capture the magnetic antigen-antibody complex. After extensive washing with wash buffers, immunoaffinity complexes were eluted with 50 μL of elution buffer (50 mM Tris-HCl, pH 6.8, 50 mM DTT, 1% SDS, 1 mM EDTA, 0.005% bromophenol blue, and 10% glycerol). Eluted proteins were resolved by SDS-PAGE and subjected to silver staining using the PlusOne Silver Staining Kit, Protein (GE Healthcare). The protein components of the immunoprecipitates were determined by mass spectrometry as described previously (Tamura et al., 2010).
Yeast Two-Hybrid Assay
Yeast two-hybrid assay was performed as described previously with minor modifications (Takagi et al., 2013). Saccharomyces cerevisiae Y2HGold strain was transformed using Frozen-EZ Yeast Transformation II (Zymo Research). Transformants were selected on SD/-Leu/-Trp plates, and the interactions were examined on SD/-Leu/-Trp/-His/-Ade plates supplemented with X-α-Gal (40 μg/mL) and Aureobasidin A (200 ng/mL).
Accession Numbers
Sequence data from this article can be found in the Arabidopsis Genome Initiative or GenBank/EMBL data libraries under the following accession numbers: ACT2 (At3g18780), CRWN1 (At1g67230), CRWN2 (At1g13220), CRWN3 (At1g68790), CRWN4 (At5g65770), H2B (At5g22880), KAKU4 (At4g31430), Myosin XI-i (At4g33200), Nup50a (At1g52380), Nup136 (At3g10650), SUN1 (At5g04990), SUN2 (At3g10730), WIP1 (At4g26455), WIP2 (At5g56210), WIP3 (At3g13360), WIT1 (At5g11390), and WIT2 (At1g68910).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. kaku4 Mutant Plants Grow Normally.
Supplemental Figure 2. KAKU4.2 mRNA Is Generated in Wild-type Plants, While Aberrantly Spliced KAKU4.2 mRNA Is Generated in the kaku4-1 Plant.
Supplemental Figure 3. Alignment of the Predicted Amino Acid Sequences of KAKU4 Homologs.
Supplemental Figure 4. Complementation of the Nuclear Shape Phenotypes of kaku4 Mutants by Stably Expressing Either KAKU4-tRFP or KAKU4-EYFP.
Supplemental Figure 5. KAKU4 Localizes at the NE.
Supplemental Figure 6. Nuclear Morphology of the kaku2 Mutant.
Supplemental Figure 7. Mass Spectrometry for KAKU4-Interacting Proteins.
Supplemental Figure 8. Overexpression of KAKU4 Induces Deformation of the NE.
Supplemental Table 1. DNA Sequences of the Primers Used in This Study.
Supplemental Movie 1. A Z-Stack Confocal Image of the Root Tip of a 1-Week-Old kaku4-3 Seedling Stably Expressing KAKU4-EYFP under the Control of the Endogenous KAKU4 Promoter.
Supplemental Movie 2. A Z-Stack Confocal Image Showing NE Growth and Ring-Like Structures.
Acknowledgments
We thank Tsuyoshi Nakagawa (Shimane University, Japan) for donating the Gateway vectors, Shoji Mano and Mikio Nishimura (National Institute for Basic Biology, Japan) for donating the Arabidopsis plants expressing GFP, Yuki Sakamoto and Shingo Takagi (Osaka University, Japan) for donating crwn4 and crwn1 crwn4, Eric J. Richards (Cornell University) for donating the Arabidopsis plants expressing CRWN1-YFP, and the ABRC for providing the T-DNA–tagged lines of Arabidopsis. This work was supported by a “Specially Promoted Research” Grant-in-Aid for Scientific Research to I.H-N. (22000014), by a Grant-in-Aid for Scientific Research to K.T. (26711017 and 25650096), by a Grant-in-Aid for JSPS Fellows to C.G. (25-1227) from the Japan Society for the Promotion of Science (JSPS), and in part by the Grant for Excellent Graduate Schools Program, MEXT, Japan.
AUTHOR CONTRIBUTIONS
C.G., K.T., T.S., and I.H-.N. designed the research. C.G. performed forward genetics, imaging analysis, biochemical analysis, and mass spectrometry. K.T. isolated kaku2 and kaku4 mutants. Y.F. performed mass spectrometry. C.G., K.T., and I.H-.N. wrote the article.
Glossary
- NE
nuclear envelope
- NA
numerical aperture
- PO
propylene oxide
Footnotes
Online version contains Web-only data.
References
- Bastos R., Lin A., Enarson M., Burke B. (1996). Targeting and function in mRNA export of nuclear pore complex protein Nup153. J. Cell Biol. 134: 1141–1156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandt A., et al. (2006). Developmental control of nuclear size and shape by Kugelkern and Kurzkern. Curr. Biol. 16: 543–552 [DOI] [PubMed] [Google Scholar]
- Burke B., Stewart C.L. (2013). The nuclear lamins: flexibility in function. Nat. Rev. Mol. Cell Biol. 14: 13–24 [DOI] [PubMed] [Google Scholar]
- Chytilova E., Macas J., Galbraith D.W. (1999). Green fluorescent protein targeted to the nucleus, a transgenic phenotype useful for studies in plant biology. Ann. Bot. (Lond.) 83: 645–654 [Google Scholar]
- Ciska M., Moreno Diaz de la Espina S. (2013). NMCP/LINC proteins: Putative lamin analogs in plants? Plant Signal. Behav. 8: e26669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciska M., Masuda K., Moreno Díaz de la Espina S. (2013). Lamin-like analogues in plants: the characterization of NMCP1 in Allium cepa. J. Exp. Bot. 64: 1553–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S.J., Bent A.F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Dauer W.T., Worman H.J. (2009). The nuclear envelope as a signaling node in development and disease. Dev. Cell 17: 626–638 [DOI] [PubMed] [Google Scholar]
- Dittmer T.A., Misteli T. (2011). The lamin protein family. Genome Biol. 12: 222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dittmer T.A., Stacey N.J., Sugimoto-Shirasu K., Richards E.J. (2007). LITTLE NUCLEI genes affecting nuclear morphology in Arabidopsis thaliana. Plant Cell 19: 2793–2803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DuBois K.N., et al. (2012). NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions. PLoS Biol. 10: e1001287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J., Siggia E.D., Moreira J.E., Smith C.L., Presley J.F., Worman H.J., Lippincott-Schwartz J. (1997). Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis. J. Cell Biol. 138: 1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans D.E., Shvedunova M., Graumann K. (2011). The nuclear envelope in the plant cell cycle: structure, function and regulation. Ann. Bot. (Lond.) 107: 1111–1118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiserova J., Kiseleva E., Goldberg M.W. (2009). Nuclear envelope and nuclear pore complex structure and organization in tobacco BY-2 cells. Plant J. 59: 243–255 [DOI] [PubMed] [Google Scholar]
- Goldberg, M.W. (2013). Nucleoskeleton in plants: The functional organization of filaments in the nucleus. In Annual Plant Reviews, Vol. 46: Plant Nuclear Structure, Genome Architecture and Gene Regulation, D.E. Evans, K. Graumann, and J.A. Bryant, eds (Oxford, UK: Wiley-Blackwell), pp. 93–122. [Google Scholar]
- Goldman R.D., Shumaker D.K., Erdos M.R., Eriksson M., Goldman A.E., Gordon L.B., Gruenbaum Y., Khuon S., Mendez M., Varga R., Collins F.S. (2004). Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 101: 8963–8968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K., Evans D.E. (2010). Plant SUN domain proteins: components of putative plant LINC complexes? Plant Signal. Behav. 5: 154–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graumann K., Runions J., Evans D.E. (2010). Characterization of SUN-domain proteins at the higher plant nuclear envelope. Plant J. 61: 134–144 [DOI] [PubMed] [Google Scholar]
- Gruenbaum Y., Margalit A., Goldman R.D., Shumaker D.K., Wilson K.L. (2005). The nuclear lamina comes of age. Nat. Rev. Mol. Cell Biol. 6: 21–31 [DOI] [PubMed] [Google Scholar]
- Hatsugai N., Kuroyanagi M., Yamada K., Meshi T., Tsuda S., Kondo M., Nishimura M., Hara-Nishimura I. (2004). A plant vacuolar protease, VPE, mediates virus-induced hypersensitive cell death. Science 305: 855–858 [DOI] [PubMed] [Google Scholar]
- Hattier T., Andrulis E.D., Tartakoff A.M. (2007). Immobility, inheritance and plasticity of shape of the yeast nucleus. BMC Cell Biol. 8: 47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawryluk-Gara L.A., Shibuya E.K., Wozniak R.W. (2005). Vertebrate Nup53 interacts with the nuclear lamina and is required for the assembly of a Nup93-containing complex. Mol. Biol. Cell 16: 2382–2394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaac C., Pollard J.W., Meier U.T. (2001). Intranuclear endoplasmic reticulum induced by Nopp140 mimics the nucleolar channel system of human endometrium. J. Cell Sci. 114: 4253–4264 [DOI] [PubMed] [Google Scholar]
- Isaac C., Yang Y.F., Meier U.T. (1998). Nopp140 functions as a molecular link between the nucleolus and the coiled bodies. J. Cell Biol. 142: 319–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovtchev G., Schubert V., Meister A., Barow M., Schubert I. (2006). Nuclear DNA content and nuclear and cell volume are positively correlated in angiosperms. Cytogenet. Genome Res. 114: 77–82 [DOI] [PubMed] [Google Scholar]
- Kittur N., Zapantis G., Aubuchon M., Santoro N., Bazett-Jones D.P., Meier U.T. (2007). The nucleolar channel system of human endometrium is related to endoplasmic reticulum and R-rings. Mol. Biol. Cell 18: 2296–2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krüger A., Batsios P., Baumann O., Luckert E., Schwarz H., Stick R., Meyer I., Gräf R. (2012). Characterization of NE81, the first lamin-like nucleoskeleton protein in a unicellular organism. Mol. Biol. Cell 23: 360–370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammerding J., Fong L.G., Ji J.Y., Reue K., Stewart C.L., Young S.G., Lee R.T. (2006). Lamins A and C but not lamin B1 regulate nuclear mechanics. J. Biol. Chem. 281: 25768–25780 [DOI] [PubMed] [Google Scholar]
- Lüke Y., et al. (2008). Nesprin-2 Giant (NUANCE) maintains nuclear envelope architecture and composition in skin. J. Cell Sci. 121: 1887–1898 [DOI] [PubMed] [Google Scholar]
- Ma Y., Cai S., Lv Q., Jiang Q., Zhang Q., Sodmergen, Zhai Z., Zhang C. (2007). Lamin B receptor plays a role in stimulating nuclear envelope production and targeting membrane vesicles to chromatin during nuclear envelope assembly through direct interaction with importin beta. J. Cell Sci. 120: 520–530 [DOI] [PubMed] [Google Scholar]
- Mano S., Hayashi M., Nishimura M. (1999). Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J. 17: 309–320 [DOI] [PubMed] [Google Scholar]
- Masuda K., Xu Z.J., Takahashi S., Ito A., Ono M., Nomura K., Inoue M. (1997). Peripheral framework of carrot cell nucleus contains a novel protein predicted to exhibit a long alpha-helical domain. Exp. Cell Res. 232: 173–181 [DOI] [PubMed] [Google Scholar]
- Meier I., Brkljacic J. (2010). The Arabidopsis nuclear pore and nuclear envelope. The Arabidopsis Book 8: e0139, /10.1199/tab.0139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda Y., Fukuda H. (2011). Dynamics of Arabidopsis SUN proteins during mitosis and their involvement in nuclear shaping. Plant J. 66: 629–641 [DOI] [PubMed] [Google Scholar]
- Polychronidou M., Hellwig A., Grosshans J. (2010). Farnesylated nuclear proteins Kugelkern and lamin Dm0 affect nuclear morphology by directly interacting with the nuclear membrane. Mol. Biol. Cell 21: 3409–3420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfert K., Vogel A., Krohne G. (2004). The lamin CxxM motif promotes nuclear membrane growth. J. Cell Sci. 117: 6105–6116 [DOI] [PubMed] [Google Scholar]
- Prüfert K., Alsheimer M., Benavente R., Krohne G. (2005). The myristoylation site of meiotic lamin C2 promotes local nuclear membrane growth and the formation of intranuclear membranes in somatic cultured cells. Eur. J. Cell Biol. 84: 637–646 [DOI] [PubMed] [Google Scholar]
- Rossignol P., Collier S., Bush M., Shaw P., Doonan J.H. (2007). Arabidopsis POT1A interacts with TERT-V(I8), an N-terminal splicing variant of telomerase. J. Cell Sci. 120: 3678–3687 [DOI] [PubMed] [Google Scholar]
- Sakamoto Y., Takagi S. (2013). LITTLE NUCLEI 1 and 4 regulate nuclear morphology in Arabidopsis thaliana. Plant Cell Physiol. 54: 622–633 [DOI] [PubMed] [Google Scholar]
- Schirmer E.C., Foisner R. (2007). Proteins that associate with lamins: many faces, many functions. Exp. Cell Res. 313: 2167–2179 [DOI] [PubMed] [Google Scholar]
- Selenko P., Sprangers R., Stier G., Bühler D., Fischer U., Sattler M. (2001). SMN tudor domain structure and its interaction with the Sm proteins. Nat. Struct. Biol. 8: 27–31 [DOI] [PubMed] [Google Scholar]
- Smythe C., Jenkins H.E., Hutchison C.J. (2000). Incorporation of the nuclear pore basket protein nup153 into nuclear pore structures is dependent upon lamina assembly: evidence from cell-free extracts of Xenopus eggs. EMBO J. 19: 3918–3931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sparkes I.A., Runions J., Kearns A., Hawes C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1: 2019–2025 [DOI] [PubMed] [Google Scholar]
- Starr D.A., Fridolfsson H.N. (2010). Interactions between nuclei and the cytoskeleton are mediated by SUN-KASH nuclear-envelope bridges. Annu. Rev. Cell Dev. Biol. 26: 421–444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuurman N., Heins S., Aebi U. (1998). Nuclear lamins: their structure, assembly, and interactions. J. Struct. Biol. 122: 42–66 [DOI] [PubMed] [Google Scholar]
- Sullivan T., Escalante-Alcalde D., Bhatt H., Anver M., Bhat N., Nagashima K., Stewart C.L., Burke B. (1999). Loss of A-type lamin expression compromises nuclear envelope integrity leading to muscular dystrophy. J. Cell Biol. 147: 913–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi J., Renna L., Takahashi H., Koumoto Y., Tamura K., Stefano G., Fukao Y., Kondo M., Nishimura M., Shimada T., Brandizzi F., Hara-Nishimura I. (2013). MAIGO5 functions in protein export from Golgi-associated endoplasmic reticulum exit sites in Arabidopsis. Plant Cell 25: 4658–4675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Hara-Nishimura I. (2013). The molecular architecture of the plant nuclear pore complex. J. Exp. Bot. 64: 823–832 [DOI] [PubMed] [Google Scholar]
- Tamura K., Fukao Y., Iwamoto M., Haraguchi T., Hara-Nishimura I. (2010). Identification and characterization of nuclear pore complex components in Arabidopsis thaliana. Plant Cell 22: 4084–4097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Iwabuchi K., Fukao Y., Kondo M., Okamoto K., Ueda H., Nishimura M., Hara-Nishimura I. (2013). Myosin XI-i links the nuclear membrane to the cytoskeleton to control nuclear movement and shape in Arabidopsis. Curr. Biol. 23: 1776–1781 [DOI] [PubMed] [Google Scholar]
- Tamura K., Shimada T., Kondo M., Nishimura M., Hara-Nishimura I. (2005). KATAMARI1/MURUS3 Is a novel golgi membrane protein that is required for endomembrane organization in Arabidopsis. Plant Cell 17: 1764–1776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volkova E.G., Kurchashova S.Y., Polyakov V.Y., Sheval E.V. (2011). Self-organization of cellular structures induced by the overexpression of nuclear envelope proteins: a correlative light and electron microscopy study. J. Electron Microsc. (Tokyo) 60: 57–71 [DOI] [PubMed] [Google Scholar]
- Walters A.D., Bommakanti A., Cohen-Fix O. (2012). Shaping the nucleus: factors and forces. J. Cell. Biochem. 113: 2813–2821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Dittmer T.A., Richards E.J. (2013). Arabidopsis CROWDED NUCLEI (CRWN) proteins are required for nuclear size control and heterochromatin organization. BMC Plant Biol. 13: 200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webster M., Witkin K.L., Cohen-Fix O. (2009). Sizing up the nucleus: nuclear shape, size and nuclear-envelope assembly. J. Cell Sci. 122: 1477–1486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson K.L., Foisner R. (2010). Lamin-binding proteins. Cold Spring Harb. Perspect. Biol. 2: a000554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zastrow M.S., Flaherty D.B., Benian G.M., Wilson K.L. (2006). Nuclear titin interacts with A- and B-type lamins in vitro and in vivo. J. Cell Sci. 119: 239–249 [DOI] [PubMed] [Google Scholar]
- Zhou L., Panté N. (2010). The nucleoporin Nup153 maintains nuclear envelope architecture and is required for cell migration in tumor cells. FEBS Lett. 584: 3013–3020 [DOI] [PubMed] [Google Scholar]
- Zhou X., Graumann K., Evans D.E., Meier I. (2012). Novel plant SUN-KASH bridges are involved in RanGAP anchoring and nuclear shape determination. J. Cell Biol. 196: 203–211 [DOI] [PMC free article] [PubMed] [Google Scholar]