Magnesium (Mg2+) is an essential mineral for plant growth and development. This study identifies a transporter responsible for absorbing magnesium ion from the soil into the roots. This transporter is expressed in the roots only under low-Mg2+ conditions and thus represents a mechanism for low-Mg2+ sensing in plants.
Abstract
Although magnesium (Mg2+) is the most abundant divalent cation in plant cells, little is known about the mechanism of Mg2+ uptake by plant roots. Here, we report a key function of Magnesium Transport6 (MGT6)/Mitochondrial RNA Splicing2-4 in Mg2+ uptake and low-Mg2+ tolerance in Arabidopsis thaliana. MGT6 is expressed mainly in plant aerial tissues when Mg2+ levels are high in the soil or growth medium. Its expression is highly induced in the roots during Mg2+ deficiency, suggesting a role for MGT6 in response to the low-Mg2+ status in roots. Silencing of MGT6 in transgenic plants by RNA interference (RNAi) resulted in growth retardation under the low-Mg2+ condition, and the phenotype was restored to normal growth after RNAi plants were transferred to Mg2+-sufficient medium. RNAi plants contained lower levels of Mg2+ compared with wild-type plants under low Mg2+ but not under Mg2+-sufficient conditions. Further analysis indicated that MGT6 was localized in the plasma membrane and played a key role in Mg2+ uptake by roots under Mg2+ limitation. We conclude that MGT6 mediates Mg2+ uptake in roots and is required for plant adaptation to a low-Mg2+ environment.
INTRODUCTION
Magnesium (Mg2+) is an essential macronutrient and the most abundant free divalent cation that is required for important functions related to chlorophyll synthesis, enzyme activation, and membrane stability in plants (Knoop et al., 2005). Although the importance of Mg2+ in plant growth and development has been well recognized, the molecular mechanisms of Mg2+ uptake, translocation, and homeostasis remain poorly understood.
The CorA family of Mg2+ transporters, originally discovered in bacteria, is the most extensively studied Mg2+ transport system, which exists widely in bacteria, fungi, animals, and plants, mediating both the influx and efflux of Mg2+ (Lusk and Kennedy, 1969; Snavely et al., 1989a, 1989b; Schock et al., 2000; Li et al., 2001). CorA proteins are characterized by the GMN motif (for three conserved amino acid residues) that is essential for Mg2+ transport (Szegedy and Maguire, 1999). A crystal structure of the CorA Mg2+ transporter from Thermotoga maritima has been solved, identifying CorA as a pentameric cone-shaped channel (Eshaghi et al., 2006; Lunin et al., 2006; Maguire, 2006; Payandeh and Pai, 2006).
A CorA-like gene family in Arabidopsis thaliana, namely, MGT/MRS2, encodes ∼10 putative Mg2+ transport proteins (Schock et al., 2000; Li et al., 2001). Two independent groups simultaneously identified this gene family in Arabidopsis (Schock et al., 2000; Li et al., 2001). Schock et al. (2000) showed that one of the family members complemented the yeast mrs2 mutant and named the gene family MRS2 (Mitochondrial RNA Splicing2). Li et al. (2001) found that two members complemented bacterial and yeast mutants defective in Mg2+ transport and named the gene family MGT (Magnesium Transport). We refer to this family as MGT for simplicity unless studies using different terms are discussed. The Mg2+ transport activities of several MGTs have been characterized in heterologous systems, including bacteria or yeast mutants lacking Mg2+ transport capacity (Schock et al., 2000; Li et al., 2001, 2008; Mao et al., 2008; Chen et al., 2009; Gebert et al., 2009). In these systems, members of the MGT family vary in their affinities for Mg2+ transport, including high-affinity (MGT1 and MGT10), low-affinity (MGT7 and MGT9), and dual-affinity (MGT5) members (Schock et al., 2000; Li et al., 2001, 2008; Drummond et al., 2006; Mao et al., 2008; Chen et al., 2009; Gebert et al., 2009).
Members of the MGT family are expressed in a broad range of plant tissues (Li et al., 2001, 2008; Mao et al., 2008; Chen et al., 2009; Gebert et al., 2009), and the proteins are located to the plasma membrane or in intracellular organelles, including mitochondria, plastid, and tonoplast (Li et al., 2001, 2008; Drummond et al., 2006; Gebert et al., 2009; Conn et al., 2011), indicating their roles in Mg2+ transport in and out of the cell and in exchange among the compartments of the cell.
MGT1 is localized in the plasma membrane (Li et al., 2001) and is expressed in the root hair and elongation zones, in the vascular tissues, and in trichomes of adult leaves (Gebert et al., 2009), suggesting a role in Mg2+ uptake in the root or translocation in the above tissues. MGT10 is localized in the chloroplast envelope membrane system and is expressed in the rosette leaves and vascular tissues of cauline leaves (Gebert et al., 2009), indicating a possible function in Mg2+ translocation into chloroplasts (Drummond et al., 2006). MGT2 and MGT3 are tonoplast localized and possibly involved in Mg2+ homeostasis in mesophyll cells (Conn et al., 2011), consistent with their expression in both rosette and cauline leaves (Gebert et al., 2009). MGT5 is localized in the mitochondria and plays an essential role in pollen development (Li et al., 2008). The loss-of-function mutant mgt7/mrs2-7 displayed a growth retardation phenotype under low Mg2+ (Gebert et al., 2009). MGT7 is localized in the endomembrane system and may be involved in the regulation of Mg2+ compartmentalization in the cell and thereby the regulation of Mg2+ homeostasis (Gebert et al., 2009). MGT9 is essential for male gametophyte development and male fertility (Chen et al., 2009), in line with its predominant expression in anther (Chen et al., 2009; Gebert et al., 2009). Despite functional links of MGT members to various processes, none of them has been shown to play a role in a specific Mg2+ transport process in plants. In particular, MGT members responsible for Mg2+ uptake from soil have not been identified, leaving the mechanism of Mg2+ acquisition by plant roots unknown. Here, we demonstrate that MGT6 functions as a plasma membrane Mg2+ transporter, mediating Mg2+ uptake in the roots especially when plants encounter a low-Mg2+ environment.
RESULTS
MGT6 Displays Low-Affinity Mg2+ Transport in a Bacterial Model
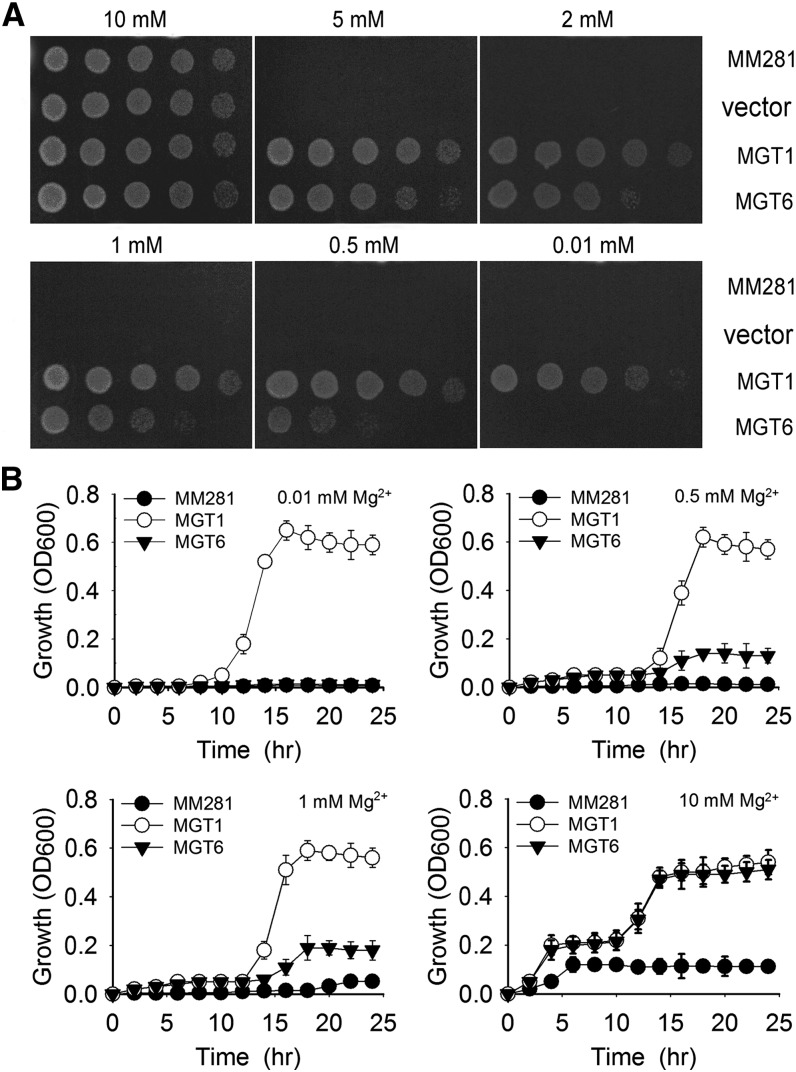
To determine whether MGT6 encodes a Mg2+ transporter, we expressed it in the Salmonella typhimurium mutant strain MM281, which lacks the Mg2+ transport systems CorA, MgtA, and MgtB (Maguire, 1992) and requires 10 mM Mg2+ in the medium for its normal growth. Our experiments showed that the negative controls (MM281 and MM281 transformed with empty pTrc99A vector) did not grow on the medium containing less than 10 mM Mg2+. As a positive control, MM281 transformed with MGT1 grew well on medium containing 0.01 mM Mg2+. MM281 transformed with MGT6 grew on the medium containing 0.5 mM Mg2+, suggesting that MGT6 is capable of mediating Mg2+ uptake, but with lower affinity than MGT1 (Figure 1A). The growth curve analysis in liquid medium was also consistent with the complementation results observed on agar plates (Figure 1B).
Figure 1.
Complementation of the MM281 Mutant by MGT6.
(A) Growth of bacterial strains on N-minimal medium containing 0.01, 0.05, 1, 2, 5, or 10 mM MgSO4. The strains are MM281, MM281 cells transformed with the pTrc99A vector only (vector), MGT1 cDNA in the pTrc99A vector (MGT1), and MGT6 cDNA in pTrc99A vector (MGT6). From left to right is a 10-fold dilution series of bacterial cultures.
(B) Bacterial growth curves plotted from OD600 values in liquid cultures. Bacterial cells from the strains described in (A) were grown in N-minimal liquid medium containing increasing concentrations of MgCl2 from 0.01 to 10 mM. Growth of the cultures was monitored every 2 h by OD600 readings for 24 h. Data are average values of three independent experiments and are presented as means ± se.
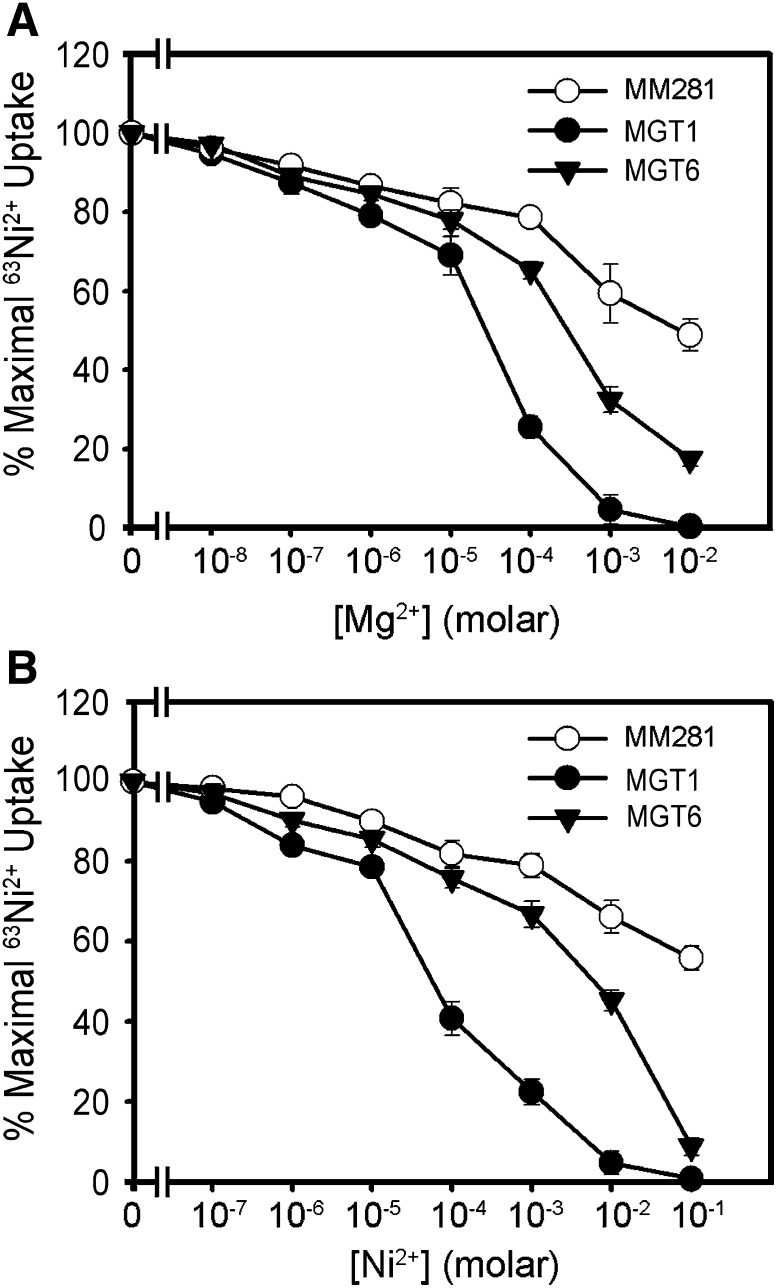
To examine the transport capacity of MGT6, we performed Mg2+ uptake experiments using 63Ni2+ as a tracer based on a previously described procedure (Snavely et al., 1989a, 1989b, 1991; Li et al., 2001). Studies have demonstrated that Mg2+ and Ni2+ can use the same transport system with similar kinetics in the cell (Smith et al., 1998; Szegedy and Maguire, 1999). Inhibition of 63Ni2+ tracer uptake by nonradioactive Mg2+ or other cations represents the uptake efficiency of these cations. We compared MM281 cells and MM281 cells harboring MGT1 or MGT6 cDNA in the tracer-uptake assays. In the MM281 cells expressing MGT1, inhibition of tracer uptake occurred in the presence of 0.001 to 0.01 mM Mg2+ or 0.01 to 0.1 mM Ni2+. Similar inhibition took place in concentrations of 0.1 to 1 mM Mg2+ or 1 to 10 mM Ni2+ in MGT6-expressing MM281 cells, suggesting that MGT6 functions as a low-affinity Mg2+ transporter in the Salmonella expression system (Figure 2).
Figure 2.
Ni2+ and Mg2+ Transport Measured by the Inhibition of 63Ni2+ Uptake in MM281.
(A) Mg2+ inhibition of 63Ni2+ uptake.
(B) Ni2+ inhibition of 63Ni2+uptake.
Transport assays were performed using MM281 cells or MM281 cells transformed with MGT1 or MGT6. Tracer uptake is presented as the percentage of the maximal uptake by each strain. Uptake was measured for 3 min with triplicates at each cation concentration. Data were collected from three independent experiments and are presented as means ± se.
Mg2+ Deficiency Alters the Expression Pattern of MGT6 in Arabidopsis Roots
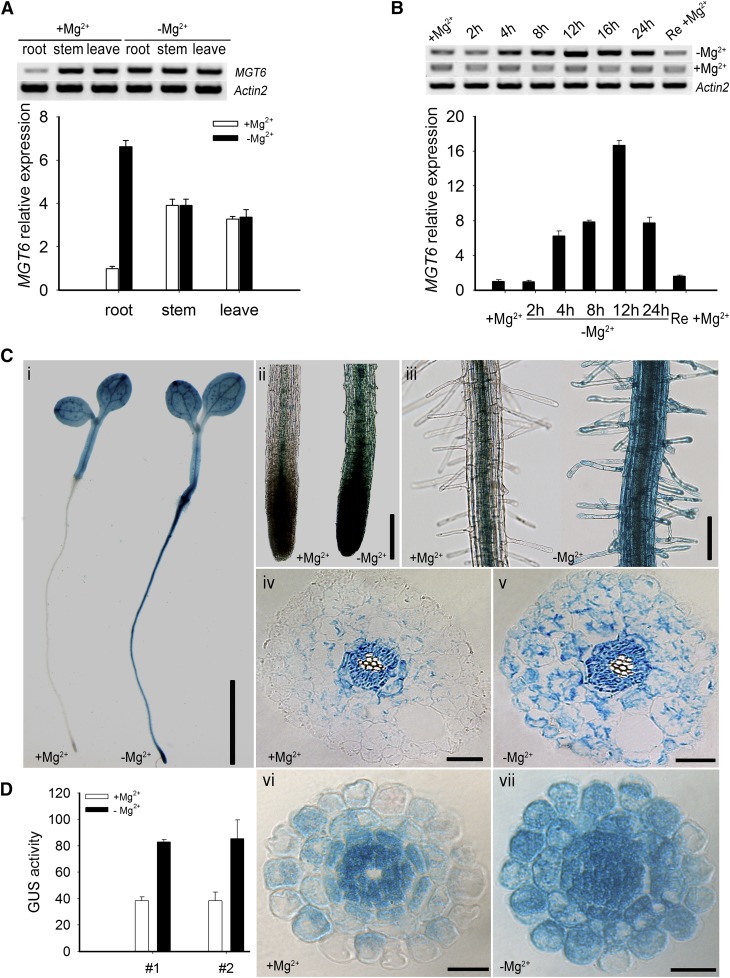
RT-PCR and quantitative RT-PCR (qRT-PCR) analysis showed that MGT6 transcripts were detected in roots, stems, and leaves (Figure 3A). Interestingly, the expression of MGT6 in the roots was significantly up-regulated when seedlings were transferred from the Mg2+-sufficient to the Mg2+-deficient condition (Figure 3A). Further qRT-PCR analysis showed that MGT6 transcript in the roots was induced to the peak level in 12 h and then decreased (Figure 3B). The expression of MGT6 in roots declined to the basal level within 12 h after the Mg2+-starved plants were transferred back to the Mg2+-sufficient medium (Figure 3B). The qRT-PCR data showed that MGT6 transcript levels in the roots under Mg2+-deficient conditions were 8-fold higher than in the control plants grown under Mg2+-sufficient conditions (Figure 3B). These results demonstrated Mg2+-dependent regulation of MGT6 mRNA accumulation in Arabidopsis roots.
Figure 3.
Expression of MGT6 Is Induced by Low-Mg2+ Conditions.
(A) RT-PCR products (top panel) and qRT-PCR analyses (bottom panel) showing MGT6 mRNA accumulation in roots, stems, and leaves. Wild-type plants (Columbia-0) were grown on MS solid medium containing 3 mM Mg2+ for 1 week and transferred to MS solid medium containing either 0 mM Mg2+ (−Mg2+) or 3 mM Mg2+ (+Mg2+) for 12 h. Significant differences between 0 and 3 mM samples are indicated in root tissue (P < 0.005 by Student’s t test). No significant difference was found in stem and leaf tissues (P > 0.05 by Student’s t test). Data are means of three replicates of one experiment. The experiment was repeated three times with similar results. Error bars represent sd.
(B) Time course of MGT6 mRNA accumulation in roots as shown by gel analysis of PCR products (top panel) and real-time PCR analyses (bottom panel). RNA samples were isolated from roots of plants grown in MS medium for 1 week and transferred to 0 mM Mg2+ (−Mg2+) or 3 mM Mg2+ (+Mg2+) for the indicated times. The right column represents the transcription level of the 12-h Mg2+-deprived plants resupplied with 3 mM Mg2+ (Re +Mg2+). Data are means of three replicates from one experiment. Similar results were obtained in three other experiments. Error bars represent sd.
(C) Histochemical analysis of MGT6 promoter–driven GUS reporter expression in transgenic Arabidopsis plants. The transgenic plants were grown as described in (A), and GUS activities (indicated in blue) were examined using histochemical staining. (i) GUS staining of 1-week-old plants (line 1) grown in minimal medium with 3 mM (+Mg2+) or 0 mM (−Mg2+) MgCl2. (ii) Enlarged images of the root tips in (i). (iii) Enlarged images of the root hair zones in (i). (iv) and (v) Cross section images of the elongation zone in (i). (vi) and (vii) Cross section images of the elongation zone in GUS-stained 1-week-old plants (line 2). Bar in (i) = 4 mm, bar in (ii) = 10 mm, bar in (iii) = 5 mm, and bars in (iv) to (vii) = 50 μm.
(D) Quantification of GUS activity in the cortex region of transgenic Arabidopsis plants containing the MGT6 promoter-GUS construct. Image J software was used to quantify the GUS signals in the root cortex areas of two transgenic lines (lines 1 and 2) treated with Mg2+-sufficient or Mg2+-deficient conditions. Three plants from each line were used for the analysis in one experiment. The experiment was repeated three times with similar results. Error bars represent sd.
To analyze MGT6 gene expression in more detail, we constructed a reporter gene using the MGT6 promoter to drive the expression of the β‑glucuronidase (GUS) gene and produced transgenic plants containing this construct. We screened and identified 20 independent transgenic lines containing ProMGT6:GUS. GUS activity was detected in the cotyledon, leaves, stems, and roots (Figures 3Ci, 3Cii, and 3Ciii; Supplemental Figure 1). In addition, GUS signal was enhanced in roots when transferred to Mg2+-deficient conditions, consistent with the qRT-PCR data. We chose three independent transgenic lines (lines 1, 2, and 3) to compare the GUS signals in the roots of seedlings grown under Mg2+-deficient and Mg2+-sufficient conditions and found ∼6-fold higher GUS activity in the roots under the Mg2+-deficient condition, as determined by Image J software (Supplemental Figure 1). We analyzed two lines (lines 1 and 2) in more detail by examination of root cross sections. The results showed that MGT6-GUS was expressed mainly in vascular tissues of roots under normal conditions and was strongly induced by Mg2+ deficiency in cortex cells and epidermal cells, including root hairs (Figure 3C). We quantified the GUS signals in the root cortex by Image J software in three plants from each line treated under Mg2+-sufficient or Mg2+-deficient conditions. There was a 2-fold increase in GUS signal in the cortex under Mg2+-deficient conditions. This induction pattern suggests a potential role of MGT6 in Mg2+ uptake in response to the low-Mg2+ stress in the roots.
MGT6 Is Localized to the Plasma Membrane
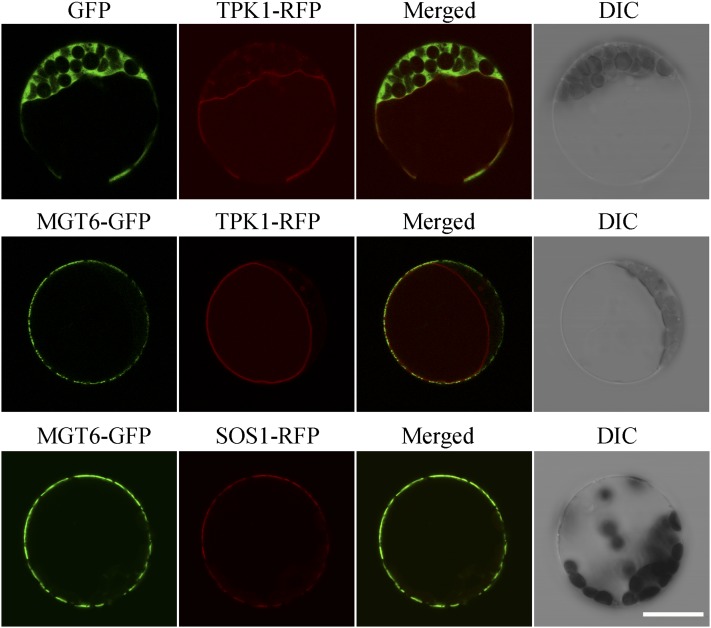
In addition to the expression pattern, the subcellular localization of a transporter provides important clues in understanding its in vivo function. To determine the subcellular location of MGT6, a green fluorescent protein (GFP) reporter construct was generated to express a fusion protein of GFP fused to the C terminus of MGT6. Expression was driven by the cauliflower mosaic virus 35S promoter. The same vector with GFP only was used as a control. The GFP-only control and the MGT6-GFP fusion construct were introduced into Arabidopsis mesophyll protoplasts separately, and GFP signals were examined using confocal microscopy. In all cases examined, GFP signals from the GFP control construct were detected in all cytoplasmic areas of the cell except for areas covered by chloroplasts or the central vacuole, whereas GFP signals from the MGT6-GFP construct were specifically associated with the cell surface area (Figure 4), showing that MGT6 is likely located to the plasma membrane. To confirm the plasma membrane localization of MGT6, we cotransformed the MGT6-GFP construct with a construct containing a known plasma membrane protein fused to red fluorescent protein (SOS1-RFP) (Shi et al., 2002) into the protoplasts from Arabidopsis mesophyll cells. We found that GFP signals produced by the MGT6-GFP fusion overlapped well with RFP signals generated by SOS1-RFP, indicating that MGT6 is localized to the plasma membrane. Because areas of plasma membrane often overlap with the tonoplast of the central vacuole, we decided to examine the localization further by introducing a construct containing a known tonoplast protein fused to RFP (TPK1-RFP) (Latz et al., 2007) as a tonoplast marker. When MGT6-GFP and TPK1-RFP were cotransformed into protoplasts from Arabidopsis mesophyll cells, we found that GFP signals produced by the MGT6-GFP fusion and RFP signals generated by the TPK1-RFP fusion were well separated, especially in areas where the plasma membrane and tonoplast were separated by chloroplasts, further confirming that MGT6 is localized to the plasma membrane (Figure 4). We also coexpressed the MGT6-GFP and SOS1-RFP or TPK1-RFP fusions in protoplasts isolated from Arabidopsis suspension culture cells that do not contain chloroplasts. The results also indicated that MGT6-GFP signals overlapped with SOS1-RFP signals but were separated from TPK1-RFP signals (Supplemental Figure 2). Thus, we concluded that MGT6 is localized to the plasma membrane in plant cells.
Figure 4.
Plasma Membrane Localization of MGT6.
The GFP coding sequence was fused to the C terminus of the MGT6 coding region in the pRTL2 vector and then transformed into Arabidopsis mesophyll protoplasts. GFP alone in the same vector was used as a control. Signals from GFP, TPK1-RFP, and SOS1-RFP were imaged using a Leica TCS-SL confocal laser scanning microscope (Leica Microsystems). Columns 1 to 4 show GFP signals, TPK1-RFP (or SOS1-RFP) signals, merged images of GFP and RFP signals, and bright-field differential interference contrast (DIC), respectively. Bar = 20 μm.
MGT6 Is Required for Growth under Low-Mg2+ Conditions
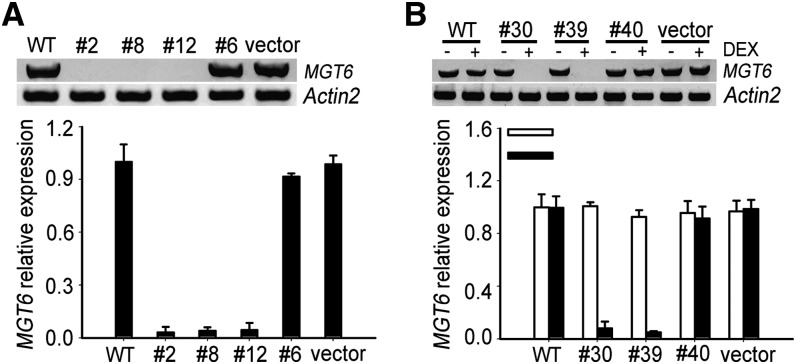
Because a T-DNA insertion mutant is not available for the MGT6 gene, we used an RNA interference (RNAi) strategy to silence the gene and address the function of MGT6 in planta. Two RNAi procedures were taken: one was constitutive RNAi driven by the 35S promoter, and the other was inducible RNAi using the dexamethasone (DEX)-inducible expression system. We screened 40 transgenic lines in each case, and the MGT6 transcript levels in the transgenic lines were determined by RT-PCR and qRT-PCR. In the constitutive RNAi group, we chose three severely suppressed lines (lines 2, 8, and 12) and one line (line 6) with a wild-type level of RNA as a control. In the DEX-inducible lines, we chose two highly suppressed lines (lines 30 and 39) and one control (line 40). We also included two empty vector–transformed lines and wild-type Columbia-0 plants as controls (Figures 5A and 5B). To exclude the possibility that the RNAi construct affected the expression of other closely related MGT genes, the expression of all other MGT genes was also analyzed (Supplemental Figure 3). Such analysis indicated that the RNAi constructs specifically reduced the level of MGT6 transcripts without affecting other MGT genes.
Figure 5.
MGT6 Expression Levels in MGT6 RNAi Plants.
(A) MGT6 transcript levels in constitutive RNAi plants.
(B) MGT6 transcript levels in inducible RNAi plants.
MGT6 transcript levels in MGT6 RNAi plants were determined by gel analysis of PCR products (top panels) and real-time RT-PCR (bottom panels). Lines 2, 8, 12, and 6 are constitutive RNAi plants, and lines 30, 39, and 40 are DEX-inducible RNAi plants. Significant differences were found between the RNAi plants and the controls (P < 0.005 by Student’s t test). Data are means of three replicates from one experiment. The experiment was repeated three times with similar results. Error bars represent sd.
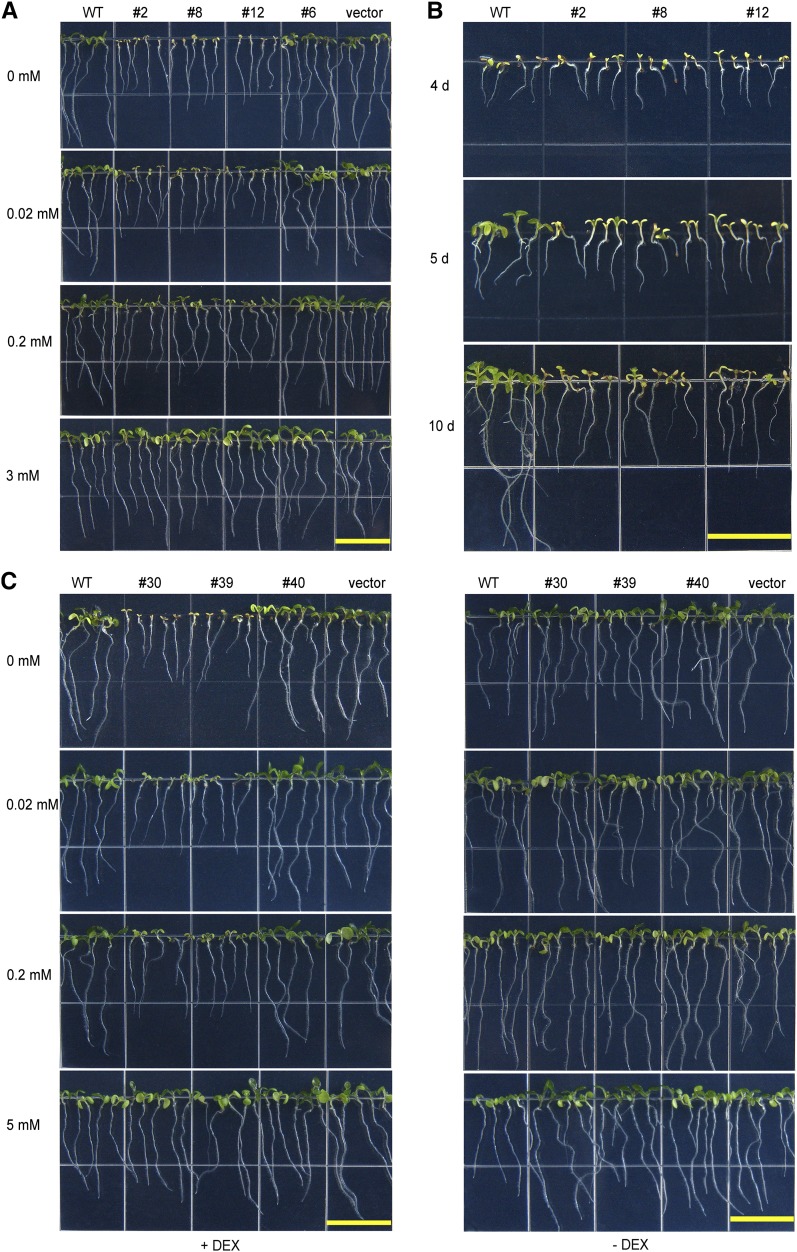
We examined the growth of the constitutive RNAi lines under Mg2+-sufficient and Mg2+-deficient conditions. When planted on plates containing Mg2+-sufficient medium (3 mM Mg2+), all transgenic plants and wild-type plants germinated and grew well without discernible differences (Figure 6A). When grown on medium with 0 mM Mg2+, however, the difference between the MGT6 RNAi plants and their controls was clearly visible within 5 d after germination. The RNAi plants showed stunted stature and pale cotyledons, and this phenotype became more evident as plants grew older (Figure 6B). The differences between the RNAi and control plants became smaller and gradually disappeared when sufficient levels of Mg2+ were added to the medium (Figure 6A). The inducible RNAi plants (lines 30 and 39), like the MGT6 constitutive RNAi plants (lines 2, 8, and 12), showed similar stunted growth when grown on medium containing low Mg2+ in the presence of DEX. Before RNAi was induced by DEX, however, the RNAi transgenic plants were indistinguishable from the control plants. All plants grew well on medium containing 5 mM Mg2+ (Figure 6C).
Figure 6.
Growth Retardation of MGT6 RNAi Seedlings under Mg2+-Deficient Conditions.
(A) Growth retardation of the transgenic plants harboring constitutive MGT6 RNAi constructs under Mg2+-deficient conditions. Wild-type and MGT6 RNAi seeds were plated on medium containing 0, 0.02, 0.2, or 3 mM Mg2+ and photographed after 7 d. Lines 2, 8, and 12, but not line 6, displayed growth retardation under the Mg2+-deficient condition. This is consistent with the RNA levels in these lines (Figure 5A). “Vector” represents plants transformed with the empty vector pART27.
(B) Time course of growth retardation of the constitutive MGT6 RNAi lines under Mg2+-deficient conditions. Wild-type and RNAi (lines 2, 8, and 12) seeds were plated on Mg2+-free MS medium and photographed after growing for 4, 5, or 10 d.
(C) Growth retardation of transgenic plants containing the DEX-inducible MGT6 RNAi construct under Mg2+-deficient conditions. Seeds were plated on MS medium containing 0, 0.02, 0.2, or 5 mM Mg2+ in the presence (+) or absence (−) of 6 μM DEX. Plants were photographed after 7 d. Lines 30 and 39 showed defects consistent with the RNA levels of MGT6 in these plants (Figure 5B). “Vector” represents plants transformed with the empty vector pTA7002.
Bars = 1.5 cm.
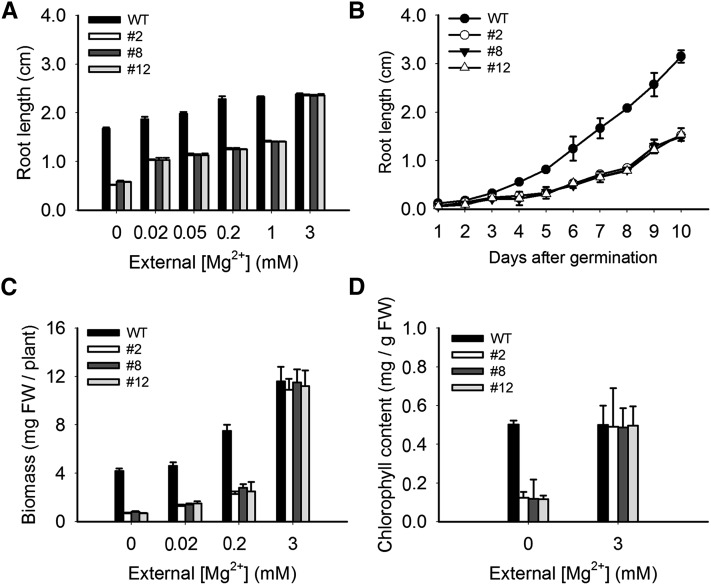
As a more quantitative measure, we assayed root growth. MGT6 RNAi plants were more severely inhibited compared with the controls when planted on medium containing less than 3 mM Mg2+ (Figures 6A, 6B, 7A, and 7B). Consistent with the observed size difference, the fresh weights of the RNAi-suppressed transgenic plants were significantly lower compared with the control (Figure 7C). In addition, we measured the chlorophyll content of 10-d-old seedlings grown under the 0 mM Mg2+ condition and found that the RNAi plants had significantly lower levels of chlorophyll than the control (Figure 7D). We also examined the plants under hydroponic growth conditions and found that RNAi plants (lines 2 and 8) showed low-Mg2+-related growth retardation (Supplemental Figure 4).
Figure 7.
Phenotypic Analyses of MGT6 RNAi Lines under Mg2+-Deficient Conditions.
(A) Root growth of wild-type and RNAi plants under various Mg2+ concentrations. Root lengths of 7-d-old seedlings were measured (n = 20 for each data point). Three independent experiments were performed, and values represent means ± se.
(B) Time course of root growth in wild-type and RNAi plants under the Mg2+-deficient condition. Root length of the seedlings was measured every day for 10 d after germination (n = 20 for each data point). Three independent experiments were performed, and values represent means ± se.
(C) Biomass of wild-type and RNAi plants. Growth conditions were as described in Figure 6A. Three independent experiments were performed, and values represent means ± se. Significant differences were found between the wild type and RNAi lines (lines 2, 8, and12) (P < 0.005 by Student’s t test). FW, fresh weight.
(D) Chlorophyll contents of wild-type and RNAi plants. The chlorophyll contents of the seedlings were measured after 10 d of growth as described in Figure 6A. Three independent experiments were performed, and values represent means ± se. Significant differences were found compared with the control.
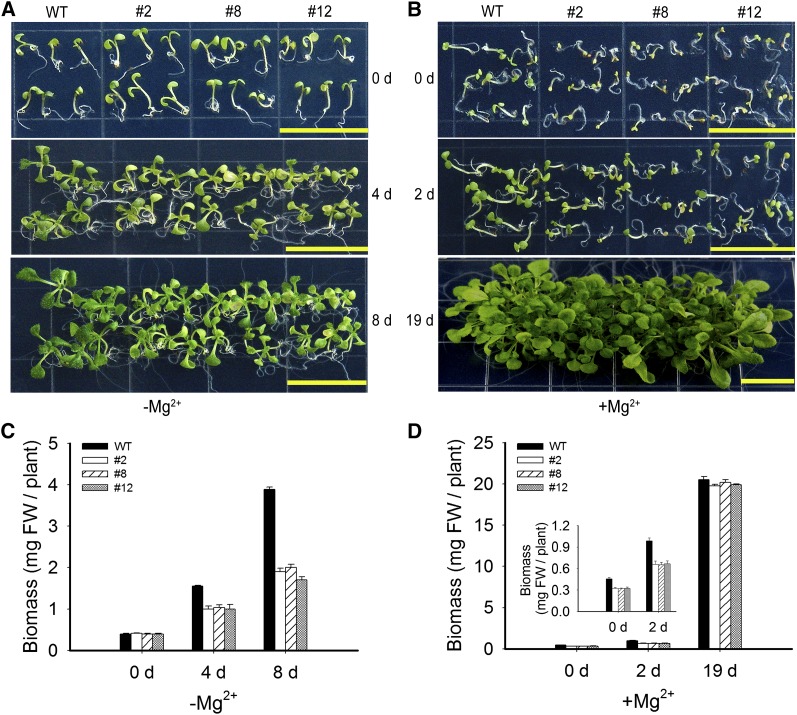
Our data above suggest that silencing MGT6 did not appear to affect seed germination or the initial phase of seedling establishment but rather affected further plant growth under the Mg2+-deficient condition. To examine this further, we grew wild-type and RNAi plants (lines 2, 8, and 12) on Mg2+-sufficient medium for 4 d and then transferred the seedlings to the Mg2+-deficient medium and monitored plant growth for a number of days after transfer. The RNAi plants began to turn yellow 4 d after transfer to the low-Mg2+ medium and became visibly smaller than the wild-type control 8 d after the transfer. The fresh weight of the RNAi plants was ∼25% less than that of the wild-type control 4 d after transfer and 50% less than the control 8 d after transfer (Figures 8A and 8C).
Figure 8.
Recovery of RNAi Plants after Mg2+ Replenishment.
(A) Postgermination Mg2+ deficiency assay. The wild-type and RNAi (lines 2, 8, and 12) plants were first grown in solid medium containing 3 mM Mg2+ for 4 d and then transferred to Mg2+-deficient (−Mg2+) MS plates. Plants were photographed immediately after transfer (0 d) and after growing for 4 and 8 d. Bars = 1.5 cm.
(B) Recovery of RNAi plants after Mg2+ replenishment. The wild-type and RNAi (lines 2, 8, and 12) plants were first grown in the Mg2+-free medium for 4 d, when the MGT6 RNAi plants displayed the growth retardation phenotype as shown in Figure 6. Then the plants were transferred to medium containing 3 mM Mg2+ (+Mg2+) and grown for a number of days. Photographs were taken just after transfer (0 d) and after growing for 2 and 19 d. Bars = 1.5 cm.
(C) Changes in the fresh weight (FW) of plants after transfer to Mg2+-deficient conditions. The control and RNAi plants (lines 2, 8, and12) were first grown in the solid medium containing 3 mM Mg2+ for 4 d and then transferred to Mg2+-deficient (−Mg2+) medium for the number of days indicated. Data are means of 10 replicates from one experiment. The experiment was repeated three times with similar results. Error bars represent sd. Significant differences were found between the wild type and the RNAi lines after 8 d of growth (P < 0.005 by Student’s t test).
(D) Changes in the fresh weight of plants after transfer to Mg2+ replenishment conditions. The wild-type and RNAi (lines 2, 8, and 12) plants were first grown in the Mg2+-free medium for 4 d and then transferred to medium containing 3 mM Mg2+ (+Mg2+) and grown for the number of days indicated. Data are means of 10 replicates from one experiment. The experiment was repeated three times with similar results. Error bars represent sd. The RNAi transgenic plants were indistinguishable from the control plants after 19 d of growth.
In a parallel experiment, we tested whether a sufficient Mg2+ supply would restore seedling growth in the RNAi plants. The RNAi transgenic plants (lines 2, 8, and 12) and wild-type plants were first germinated and grown on Mg2+-deficient medium for 4 d, which is when the growth retardation and pale cotyledon became evident with the RNAi plants. The plants were then transferred to the Mg2+-sufficient medium. Soon after transfer (2 d), the cotyledons and leaves of the RNAi plants became greener, although the fresh weight of the RNAi plants was 34% less than that of the wild-type control. The RNAi plants were essentially indistinguishable from the control plants 2 weeks later, and the fresh weights of the RNAi and control plants were also similar (Figures 8B and 8D).
The above results suggest that silencing the MGT6 gene affected plant growth under low-Mg2+ conditions, possibly due to changes in Mg2+ accumulation in the transgenic plants.
MGT6 Functions in Mg2+ Accumulation in Plants
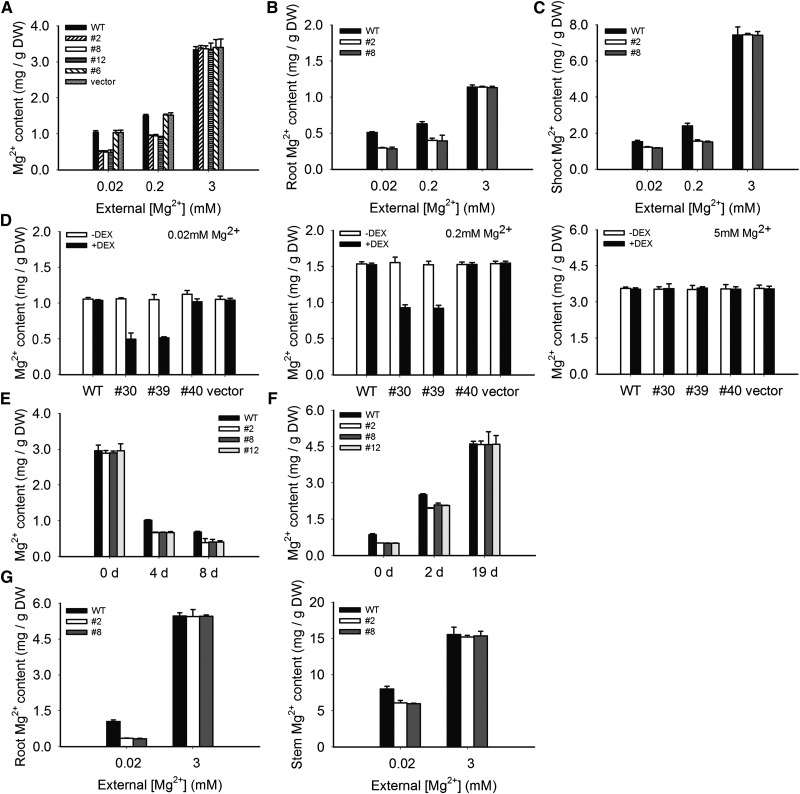
To test whether the growth retardation of MGT6 RNAi plants in low-Mg2+ medium results from a lower level of Mg2+ in plants, we measured Mg2+ content in the RNAi-suppressed lines and the control plants (including the wild type, the RNAi lines without suppression, and the empty vector–transformed plants).
We grew the constitutive RNAi lines, the inducible RNAi lines, and the control plants on medium containing 3, 0.2, or 0.02 mM Mg2+ for 10 d. The inducible RNAi plants were divided into two groups: one grown on medium with DEX and the other without. The seedlings were collected and the Mg2+ contents were determined by inductively coupled plasma–atomic emission spectrometry (ICP-AES). As shown in Figure 9A, Mg2+ content was similar among different plants grown in 3 mM Mg2+. As the Mg2+ level in the medium became lower, all plants accumulated less Mg2+, as expected. However, the RNAi plants were affected more severely. For example, under 0.2 mM Mg2+, the RNAi plants (constitutive line 2) contained ∼40% less Mg2+ compared with the control plants. Under 0.02 mM Mg2+, the RNAi plants accumulated 50% less Mg2+ than the controls.
Figure 9.
Mg2+ Contents in Wild-Type and RNAi Plants under Different External Mg2+ Levels.
(A) Mg2+ contents in whole seedlings of the wild type and constitutive RNAi lines 2, 8, and 12. Plants were grown on medium containing 0.02, 0.2, or 3 mM Mg2+ and subjected to Mg2+ measurements as described in Methods.
(B) Mg2+ contents in the roots.
(C) Mg2+ contents in the shoots.
(D) Mg2+ contents in whole seedlings of the wild type and inducible RNAi lines 30 and 39. Plants were grown on agar medium containing 0.02, 0.2, or 5 mM Mg2+.
(E) Changes in Mg2+ contents in Arabidopsis seedlings after transfer to Mg2+-deficient conditions. The wild-type and RNAi (lines 2, 8, and 12) plants were first grown in medium containing 3 mM Mg2+ for 4 d and then transferred to Mg2+-free medium for up to 8 d.
(F) Changes in Mg2+ contents in plants after transfer from Mg2+-deficient medium to Mg2+-rich medium. The wild-type and RNAi (lines 2, 8, and 12) plants were first grown in Mg2+-free solid medium for 4 d, when the MGT6 RNAi plants displayed the growth retardation phenotype. The plants were then transferred to medium containing 3 mM Mg2+ for up to 19 d.
(G) Mg2+ contents in plant tissues grown under hydroponic conditions. The wild-type and RNAi plants were first grown for 2 weeks on medium containing 0.02 and 3 mM Mg2+, respectively, and then transferred to a hydroponic solution with the equivalent Mg2+ concentrations for 2 weeks.
Mg2+ contents were determined by ICP-AES. The experiment was repeated three times with similar results. Error bars represent se. Significant differences were found between the wild type and the RNAi lines (P < 0.01 by Student’s t test). DW, dry weight.
The other two constitutive RNAi lines (lines 8 and 12) showed similar Mg2+ accumulation patterns to line 2 in that they accumulated, respectively, 36.6 and 36.5% less Mg2+ compared with the controls under 0.2 mM Mg2+, and they showed 53.0 and 53.5% reductions under 0.02 mM Mg2+ (Figure 9A).
Using the same approach, we also measured the Mg2+ content in the inducible RNAi line (line 30) in comparison with the control plants (wild-type plants, line 40, and vector-transformed plants). In the presence of various Mg2+ levels (5, 0.2, or 0.02 mM) in the medium without DEX induction, there was no difference in Mg2+ content among these plants (Figure 9D). When DEX was applied, the Mg2+ content in the inducible RNAi line (line 30) and the control plants (wild-type plants, line 40, and vector-transformed plants) did not differ in the presence of 5 mM Mg2+ but differed significantly under 0.2 or 0.02 mM Mg2+. Compared with the wild-type plants, the Mg2+ content in an inducible RNAi plant (line 30) was reduced by 38.8 and 51.9%, respectively, under 0.2 and 0.02 mM Mg2+ (P < 0.01). Analysis of another inducible RNAi line (line 39) showed similar results (Figure 9D).
When plants were grown on the Mg2+-sufficient medium for 4 d and then transferred to the Mg2+-deficient medium, the Mg2+ content decreased in both RNAi and control plants during the time course on low-Mg2+ medium (Figure 9E). However, the RNAi plants (lines 2, 8, and 12) displayed a more drastic decrease in their Mg2+ content compared with the controls. Four days after the transfer, plants from RNAi line 2 appeared pale green, and the Mg2+ content in these plants dropped from 2.89 mg/g dry weight (before transfer) to 0.87 mg/g dry weight, which represented an additional 34% decrease compared with the wild-type plants (P < 0.01) (Figure 9E). Eight days after the transfer, the Mg2+ content dropped to only 0.39 mg/g dry weight, a 42% greater decrease than in the wild-type plants (P < 0.01) (Figure 9F). The analyses of the other two RNAi lines (lines 8 and 12) also showed similar results to line 2 (Figure 9E).
In the Mg2+-replenishing experiment, the RNAi transgenic plants (lines 2, 8, and 12) and the wild type were first germinated and grown on Mg2+-deficient medium (0 mM) for 4 d and then transferred to Mg2+-sufficient medium (3 mM Mg2+). On day 2 after transfer, the Mg2+ content of line 2 increased from 0.52 to 1.96 mg/g dry weight, and in the wild-type plants, it increased from 0.86 to 2.51 mg/g dry weight (Figure 9F), along with the visual changes in the color of cotyledons. On day 19 after transfer, Mg2+ content in line 2 and wild-type plants became similar (4.59 mg/g dry weight for RNAi and 4.61 mg/g dry weight for the wild type) (Figure 9F), consistent with an indistinguishable phenotype.
We conclude from the above data that silencing of MGT6 impaired Mg2+ accumulation in the plants under the low-Mg2+ condition. As the wild-type plants were similar to the empty vector–transformed plants in phenotype and Mg2+ accumulation, we used only the wild-type plants as a control in the later experiments.
MGT6 Is a Key Transporter for Mg2+ Uptake in Plants and Acts Specifically under Low Mg2+
Reduced Mg2+ levels in the RNAi plants, together with the fact that MGT6 is a Mg2+ transporter, suggest defects in Mg2+ acquisition by the RNAi plants. We further determined if either Mg2+ uptake by roots or Mg2+ translocation from roots to shoots was impaired in the RNAi plants. First, we measured the Mg2+ contents in roots and shoots separately to examine the distribution of Mg2+ in the plants. As shown in Figures 9B and 9C, when grown on low-Mg2+ medium, the RNAi plants (lines 2 and 8) contained less Mg2+ in both roots and shoots compared with the wild type. To examine the transport process in adult plants, we grew the plants in the hydroponic system. The wild type and RNAi lines (lines 2 and 8) were grown on Mg2+-deficient plates for 2 weeks and then transferred to Mg2+-deficient hydroponic solution for another 2 weeks before roots and stems were collected separately for the determination of Mg2+ contents. Figure 9G showed, again, that Mg2+ contents in both roots and stems were lower in the RNAi plants than in the wild type.
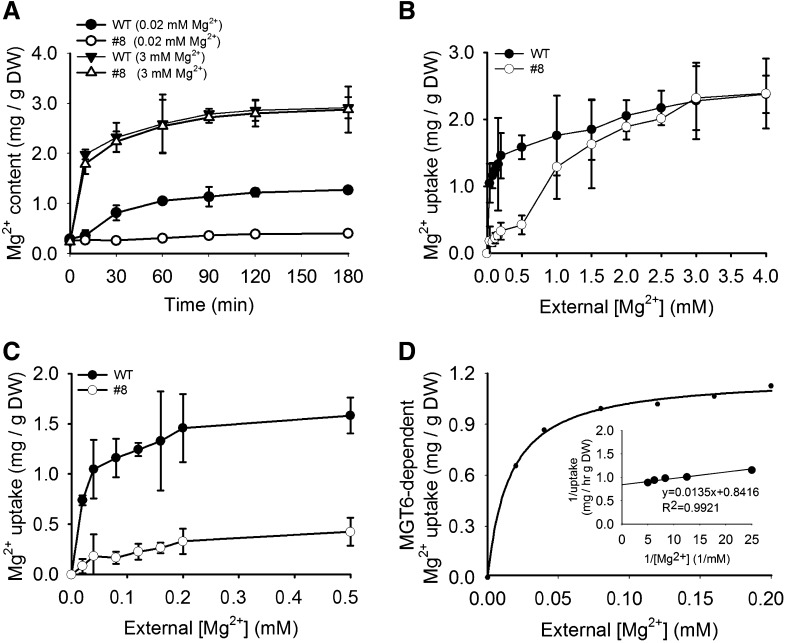
The above results suggest that MGT6 may function in Mg2+ uptake from the external environment into the roots when external Mg2+ level is low. To test this hypothesis, we performed Mg2+ uptake assays using wild-type and MGT6 RNAi plants. The kinetics of Mg2+ uptake in roots was measured by ICP-AES and 63Ni2+ tracer uptake analyses. For the Mg2+ uptake assay, two MGT6 RNAi lines (lines 8 and 2) and wild-type plants were first grown on soil for 2 weeks, then transferred to hydroponic medium with 3 mM Mg2+ for 1 week, and subsequently transferred to hydroponic medium containing 0 mM Mg2+ for another 1 week to induce the Mg2+ acquisition system. For the time-dependent Mg2+ uptake assay, Mg2+-starved plants were incubated in the medium containing 0.02 or 3 mM Mg2+ for 10, 30, 60, 90, 120, or 180 min (Figure 10A), and the roots were collected for Mg2+ content measurements using inductively coupled plasma analysis. For the concentration-dependent Mg2+ uptake assay, Mg2+-starved plants were transferred to medium containing various Mg2+ concentrations (from 0 to 4.0 mM) for 60 min, and roots were collected for Mg2+ content measurements (Figure 10B).
Figure 10.
Mg2+ Uptake in Wild-Type and RNAi Plants.
(A) Time course of Mg2+ uptake in the roots of wild-type and RNAi (line 8) plants. Plants were first grown on soil for 2 weeks and then transferred to hydroponic medium with 3 mM Mg2+ for 1 week, followed by growth in 0 mM Mg2+ hydroponic medium for another 1 week. The Mg2+-starved plants were then transferred to hydroponic medium containing 0.02 or 3 mM Mg2+ and incubated for the indicated times. Roots were harvested subsequently, and Mg2+ contents in the roots were determined by ICP-AES.
(B) and (C) Concentration-dependent Mg2+ uptake rates in roots of wild-type and RNAi (line 8) plants. Plants were cultured as described in (A). The Mg2+-starved plants were transferred to a hydroponic medium containing various Mg2+ concentrations for 1 h before roots were harvested, and Mg2+ contents were determined by ICP-AES.
(D) MGT6-dependent Mg2+ uptake. The value for MGT6-dependent Mg2+ uptake was calculated by subtracting Mg2+ uptake of RNAi plant roots from that of the wild-type roots. The data were fitted to a Michaelis-Menten function by nonlinear least-squares analysis.
Three independent experiments were performed, and values represent means ± se. DW, dry weight.
The time course of Mg2+ uptake showed that MGT6 RNAi line 8 and the wild-type plants displayed similar Mg2+ accumulation patterns under 3 mM Mg2+ (Figure 10A). Under Mg2+-deficient conditions (0.02 mM Mg2+), the Mg2+ content in the MGT6 RNAi plants (line 8) did not change significantly (from 0.26 to 0.31 mg/g dry weight) after 60 min of uptake, whereas the wild-type plants showed a strong increase (from 0.29 to 1.25 mg/g dry weight) (Figure 10A). The uptake represents a net increase in Mg2+ content. For example, the uptake of wild-type plants under 0.02 mM Mg2+ was 1.25 to 0.29, or 0.96 mg·g−1 dry weight·h−1. These results suggest that under a low-Mg2+ environment, the majority of Mg2+ uptake in the wild-type plants is mediated by MGT6. Disruption of MGT6 function by RNAi essentially eliminated this uptake activity.
The concentration-dependent Mg2+ uptake assays indicated that the Mg2+ uptake rate in RNAi line 8 was much lower than that in the wild-type plants when external Mg2+ concentrations were in the micromolar range (Figures 10B and 10C). However, under 3 mM Mg2+, the RNAi and wild-type plants showed similar rates of Mg2+ uptake (Figure 10B). We then calculated the rate of MGT6-dependent uptake in the micromolar range of external Mg2+ by subtracting the Mg2+ uptake (mg/g dry weight) of At MGT6 RNAi roots from that of wild-type roots. Using the Michaelis-Menten function, we calculated apparent Km and Vmax values of ∼16.04 μM and 1.19 mg·g−1 dry weight·h−1, respectively (Figure 10D).
Analysis of another independent RNAi line (line 2) showed results similar to those of RNAi line 8 in Mg2+ uptake kinetics, and the apparent Km and Vmax values of MGT6-dependent Mg2+ uptake were 19.54 μM and 1.23 mg·g−1 dry weight·h−1, respectively (Supplemental Figure 5).
To assess Mg2+ uptake in MGT6 RNAi plants using another procedure, we conducted 63Ni2+ isotope tracer uptake analysis, as Mg2+ and Ni2+ can use the same transport system with similar kinetics in the cell (Li et al., 2001). The Mg2+-starved seedlings were transferred into 20 mL of hydroponic medium supplemented with the indicated levels of MgSO4, 100 μM NiCl2, and 10 μCi 63Ni2+. The time courses of root Mg2+ uptake under 3 and 0.02 mM Mg2+ were determined by measuring the level of 63Ni2+ radioactivity in the plants using a liquid scintillation counter. Under 3 mM Mg2+, the root Mg2+ uptake in RNAi line 8 is comparable to that in the wild-type control at the time points of 30, 60, 90, and 120 min (Supplemental Figure 6B). However, at 0.02 mM Mg2+, the root Mg2+ uptake in RNAi line 8 is significantly lower than that in the wild-type control (Supplemental Figure 6A). These data further support that the MGT6 RNAi plants are defective in Mg2+ uptake under the low-Mg2+ condition.
From the data above, we conclude that MGT6-mediated Mg2+ uptake accounts for the majority of Mg2+ acquisition in roots under submillimolar Mg2+ and is required for plant adaptation to the low-Mg2+ environment.
DISCUSSION
Mg2+ is an essential cation that, among other functions in plants, is a core component of chlorophyll. Like other mineral nutrients, Mg2+ enters plants through root epidermal cells (in particular, root hairs) that possess specific transporters. Whereas the MGT/MRS2-type transporters are generally considered as the major family of proteins that mediate Mg2+ transport in plants, none of them has been shown to mediate Mg2+ uptake from the external environment. Our study here provides direct evidence that MGT6 plays such a role, particularly when the external Mg2+ level is in the submillimolar range, which is the prevailing condition in the soil. Therefore, we expect that MGT6 plays a key role under low-Mg2+ conditions and that other Mg2+ transporters, including other MGT members and nonspecific cation channels (Demidchik et al., 2002; Guo et al., 2010), may function in Mg2+ uptake in Mg2+-rich soils (where [Mg2+] is in the millimolar range).
After Mg2+ enters plant roots, a portion of the Mg2+ will be distributed into various cell types in the roots through both apoplast and symplast transport. Within each cell type, Mg2+ is required in a number of compartments, including essentially all organelles. After satisfying the needs of the roots, the rest of the Mg2+ will move up to shoots through xylem mass flow. The transport routes will include Mg2+ movement from epidermal cells to the cortex and endodermal cells, to the stele, and export of Mg2+ from xylem parenchyma cells into the xylem (Shaul, 2002). Based on the current understanding of the MGT-type transporters, we hypothesize the following MGT-mediated Mg2+ transport processes.
For Mg2+ uptake from the soil, several MGT members may be relevant; these include MGT6, MGT1, MGT7, MGT9, MGT2, and MGT3, which are expressed in Arabidopsis roots (Chen et al., 2009; Gebert et al., 2009). Two of them (MGT1 and MGT6) are located in the plasma membrane and expressed in the epidermal cells and, therefore, can potentially function in Mg2+ uptake from the soil. Although MGT1 has not been shown to play a role in this process, MGT6 does function in this first step of Mg2+ acquisition. MGT6 is expressed in the epidermal cells only when external Mg2+ is in the submillimolar range, and it is believed that Mg2+ concentrations in soils vary from 125 μM to 8.5 mM (Conn et al., 2011). This will make the MGT6-mediated transport a routine mechanism for Mg2+ uptake when plants encounter submillimolar Mg2+ in the soils.
As an MGT1 loss-of-function mutant did not display a growth phenotype under a broad range of Mg2+ levels, we speculate that MGT1 may function redundantly with other Mg2+ transporters such as MGT2 and MGT3, as the atmgt1/2/3 triple mutant and the atmgt1/2 double mutant showed growth retardation phenotypes under Mg2+ limitation (Lenz et al., 2013). However, it is not known where and how these transporters may work within plants. It is also shown that MGT members, like the bacterial CorA protein (Maguire, 2006), can physically interact with each other, suggesting the possibility that MGT members form a heteropentamer as a functional Mg2+ transporter (Schmitz et al., 2013). The fact that most of the MGT members can function alone in the bacterial MM281 system indicates that they may function as homooligomers as well. After Mg2+ gets into root cells, some will be transported into organelles such as the endoplasmic reticulum by MGT7 or the vacuole by MGT2, as it is also expressed in roots and in leaves (Gebert et al., 2009). For Mg2+ translocation toward the xylem, vascular tissue–expressed MGT members, including MGT3, MGT4, MGT6, and MGT9 (Gebert et al., 2009), may function in Mg2+ loading from xylem parenchyma cells to the xylem and unloading from xylem to tissues in shoots. When Mg2+ is transported up into leaf tissues through the xylem sap, four MGT members may participate in the regulation of Mg2+ homeostasis. The chloroplast-localized MGT10 may be involved in Mg2+ loading into the photosynthetic organelle (Drummond et al., 2006). The tonoplast-located MGT2 and MGT3 may play a role in Mg2+ equilibrium between the cytosol and the vacuole (Conn et al., 2011). Again, MGT7 may distribute Mg2+ into the endoplasmic reticulum (Mao et al., 2008; Gebert et al., 2009). There are three anther-expressed MGT members, MGT5, MGT9, and MGT6, which could be responsible for the Mg2+ uptake and homeostasis in the male gametophyte, whereas two ovule-expressed MGT members, MGT4 and MGT9, may be responsible for the Mg2+ uptake and homeostasis during ovule development or seed formation. Among all these processes, perhaps the best characterized is the MGT6-mediated Mg2+ uptake reported here, whereas in most other processes, further work is needed to examine the detailed mechanism of the transport.
METHODS
Plant Materials and Growth Conditions
Arabidopsis thaliana (ecotype Columbia-0) plants were used in this study unless otherwise indicated. Plant growth conditions and Agrobacterium tumefaciens–mediated transformation were performed as described previously (Clough and Bent, 1998). The transgenic lines were selected on 0.5× MS medium (Sigma-Aldrich) containing 50 μg/mL kanamycin (for pBI101- and pART27-based vectors) or 30 μg/mL hygromycin B (for pCAMBIA 1300– and pTA7002-based vectors). Hoagland hydroponic cultivation of Arabidopsis plants was performed according to an earlier procedure (Tocquin et al., 2003) with modification in the concentrations of Mg2+ as specified in the figure legends. Nutrient solutions were replaced once per week or as needed by experimental treatments.
RT-PCR and qRT-PCR
Total RNA was extracted using the TRIzol reagent (Invitrogen) following the manufacturer’s instructions. Samples were treated with DNase (Promega) to eliminate genomic DNA contamination. Two micrograms of DNA-free RNA was used for reverse transcription by Moloney murine leukemia virus reverse transcriptase (Promega) with anchored oligo(dT18). qRT-PCR was performed on an Bio-Rad C1000 Thermal Cycler using Power SYBR Green (Qiagen; CFX96 system). All primer pairs for expression assays or subcloning are listed in Supplemental Table 1. For qRT-PCR, the reaction solution included 1 μL of cDNA template (every cDNA template was diluted 20 times), 1 μL of reverse primer, 1 μL of forward primer, 12.5 μL of 2× SYBR Green Master Mix, and 9.5 μL of RNA-free water (total of 25 μL). qRT-PCR was performed as follows: 94°C for 4 min followed by 40 cycles of 94°C for 15 s, 60°C for 30 s, and 72°C for 30 s. Primer specificity was confirmed by analysis of the melting curves. The 2−ΔΔCT method (Livak and Schmittgen, 2001) was used to quantify the value of every sample using Actin2 as an internal reference.
Plasmid Constructs
The MGT6 cDNA was amplified by RT-PCR using the primer pair MGT6-F and MGT6-R and ligated to the pMD18-T vector. For the constructs used in functional complementation and tracer uptake assays in MM281 cells, the MGT6 cDNA fragment was amplified using the primers MGT6-FC and MGT6-RC and ligated to the pTrc99A vector. For subcellular localization analysis, the MGT6 cDNA was amplified without its stop codon using the primer pair MGT6-G1F and MGT6-G1R, subsequently digested with EcoRI and NcoI, respectively, and then inserted into the multicloning sites in the plant binary vector pRTL2 (Carrington et al., 1990) to make the 35S:MGT6-GFP fusion construct. To make the ProMGT6:GUS construct for histochemical analysis, a 1.5-kb fragment upstream of the ATG starting codon of MGT6 was PCR amplified using the primers MGT6-P1F and MGT6-P1R and cloned into the pMD18-T vector, then subcloned into the pBI101.2 vector using the primers MGT6-P2F and MGT6-P2R (Datla et al., 1992).
For RNAi constructs, a 500-bp fragment in the coding region of MGT6 was amplified and subcloned into the pKANNIBAL (CSIRO) vector as described previously (Wesley et al., 2001). This fragment was chosen because it exhibited low nucleotide sequence identity (<40%) to the corresponding regions in other MGT genes. The RNAi cassette assembled in pKANNIBAL was transferred into the binary vector pART27 (CSIRO) as a NotI fragment, yielding the final RNAi construct pART27-MGT6 to produce the transgenic plants used in this study. The primers used were as follows: MGT6sense-F and MGT6sense-R, introducing a XhoI site and a EcoRI site, and MGT6anti-F and MGT6anti-R, introducing a XbaI site and a HindIII site. For the MGT6-inducible RNAi construct, the small fragment from pKANNIBAL-MGT6 digested with XhoI and SpeI was subcloned into pTA7002 (Aoyama and Chua, 1997), giving rise to pTA7002-MGT6. All primer pairs for expression assays or subcloning are listed in Supplemental Table 1.
Salmonella typhimurium Mutant Complementation and Tracer Uptake Assays
For the complementation experiment, MGT6 cDNA was inserted into pTrc99A (Li et al., 2001) plasmid vector, and pTrc99A-MGT6 was transformed into MM281 cells by electroporation. Cells were plated onto Luria-Bertani (LB) medium supplemented with 10 mM Mg2+ and appropriate antibiotics (50 μg/mL ampicillin and 34 μg/mL chloramphenicol) and incubated at 37°C overnight. The transformants were confirmed by PCR amplification of both the vector and MGT6 coding sequence. Individual transformants were grown in LB liquid medium containing 10 mM Mg2+, the same antibiotics, and 0.05 mM isopropyl β-d-1-thiogalactopyranoside for the induction of MGT6 expression. MM281 and MM281 transformed with empty pTrc99A vector were used as negative controls, and MM281-MGT1 was used as a positive control. The cultures were adjusted to 1.0 OD600, diluted in a 10-fold series, and spotted (2 μL) onto N-minimal medium [5 mM KC1, 7.5 mM (NH4)2SO4, 0.5 mM K2SO4, 1 mM KH2PO4, 0.1 mM Tris-HCl, pH 7.4, and 0.2% Glc with required amino acids at 0.10 g/L] supplemented with different concentrations of MgSO4 (0.01, 0.5, 1, 2, 5, and 10 mM) and the antibiotics. The growth of different strains was scored after incubation at 37°C for 48 h. The growth rate of the four strains in liquid medium was also monitored. After growing in LB liquid medium to log phase (OD600 = 0.6 to 0.8), the cells were harvested by centrifugation (5000g, 10 min), washed twice with distilled water to remove traces of Mg2+, and resuspended in distilled water. N-minimal liquid medium was prepared with increasing concentrations of MgSO4 (0.01, 0.5, 1, and 10 mM). Cells were then added to a final OD600 of 0.001 to 0.002. The growth of the cultures was monitored over 26 h, and the OD600 was plotted as a function of growth time.
Uptake of 63Ni2+ was performed as described previously (Snavely et al., 1989b; Smith et al., 1998; Li et al., 2001). 63Ni2+ uptake was assayed using MM281-MGT6 for MGT6, mutant strain MM281 as a negative control, and MM281-MGT1 for MGT1 as a positive control. Briefly, bacterial cultures of different strains were grown overnight in LB broth containing 10 mM Mg2+ and the appropriate antibiotics. Cells were washed with N-minimal medium without added Mg2+ and then diluted 1:5 in N-minimal medium containing 1 mM Mg2+ and appropriate antibiotics. After a 3-h subculture, cells were collected by centrifugation at 1000g for 15 min and washed twice with ice-cold Mg2+-free N-minimal medium. The washed cells were resuspended in the same medium and adjusted to 1.0 OD600. For a standard assay, uptake was initiated by adding 0.2 mL of cells to 0.8 mL of N-minimal medium containing 100 μM NiCl2 and 0.5 μCi of 63Ni2+. Typically, uptake was stopped at 5 min by adding 1 mL of ice-cold washing buffer. Cells were washed four times with 1.5 mL of ice-cold washing buffer, and 63Ni2+ activity associated with cells was determined by a scintillation counter (model A210001; Packard Instruments). The relative tracer uptake was standardized against the maximal value and presented as a percentage of maximal uptake.
Histochemical GUS Analysis
Transformed Arabidopsis lines carrying the promoter-GUS fusion were selected based on kanamycin resistance. The promoter activity was reflected by GUS activity visualized in the plant tissues using 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid as a substrate according to published protocols (Jefferson et al., 1987). Briefly, 1-week-old seedlings were incubated in GUS staining solution [10 mg/mL 5-bromo-4-chloro-3-indolyl-β-d-glucuronic acid, 0.1 M K3(Fe(CN)6), 0.1 M K4Fe(CN)6·3H2O, 1 M Na2EDTA, and 0.2 M Na3PO4, pH 7.0] at 37°C for 12 h. Then the plant tissues were decolorized for 30 min in each of the gradient ethanol solutions (30, 40, 50, 60, and 70%) and incubated in 75% ethanol at 37°C overnight. The samples were examined and photographed with an Olympus SZX12 microscope equipped with a camera. Quantification of GUS activity was performed using the Image J software provided by the microscope manufacturer.
Subcellular Localization of MGT6
Arabidopsis mesophyll protoplasts were prepared from 4-week-old rosette leaf tissues by digesting leaf slices with Cellulase R-10 and Macerozyme R-10 (Yakult Pharmaceutical) as described previously (Sheen, 2001). The protoplasts were resuspended in W5 medium (154 mM NaCl, 125 mM CaCl2, 5 mM KCl, and 2 mM MES, pH 5.7) and transfected separately or in combination with 20 μg of recombinant plasmid DNA representing each of the constructs (GFP-pRTL2, MGT6-GFP-pRTL2, TPK1-RFP-pA7, and SOS1-RFP-pA7) using a polyethylene glycol–mediated transformation protocol (Sheen, 2001). The transformed protoplasts were incubated in the dark at 23°C for 16 h before confocal imaging analysis.
Arabidopsis suspension culture cells were subcultured in Murashige and Skoog (MS) medium once per week by transferring 5 mL of suspension cells into 45 mL of fresh MS medium at 27°C in the dark condition on a rotating platform at 130 rpm. After a 3-d subculture, cells were collected by centrifugation at 100g for 2 min at room temperature and digested with Cellulase R-10 and Macerozyme R-10 until 95% of the cells became protoplasts. After centrifugation at room temperature for 15 min at 100g using a swinging bucket rotor, protoplasts floating to the top of the tubes were collected (Miao and Jiang, 2007) and transformed by the same methods described above. GFP signals in protoplasts were imaged using a confocal microscope (Leica TCS-SL) with excitation at 488 nm, and the emission signal was collected between 498 and 539 nm. The fluorescence signals of TPK1-RFP and SOS1-RFP were observed with excitation at 543 nm, and the emission signal was collected between 590 and 651 nm.
Measurement of the Mg2+ Content in Arabidopsis
Wild-type and MGT6 RNAi plants were grown on half-strength MS agar medium on plates or Hoagland solution for hydroponic culture containing various Mg2+ concentrations for different times. The seedlings were then collected, rinsed with distilled water, dried at 70°C for 48 h, and weighed. The samples were digested with 1 N HNO3 and then boiled for 30 min. Mg2+ contents were measured by ICP-AES (Varian 715-ES) at the 285.2-nm wavelength.
Mg2+ Uptake Assay
Wild-type and RNAi plants were grown in the soil for 2 weeks and then transferred to the hydroponic culture with 3 mM Mg2+ for 1 week. The plants were further transferred to hydroponic medium containing 0 mM Mg2+ for another 1 week and then were incubated in hydroponic medium containing different Mg2+ concentrations as indicated in Figure 10. To plot the time course of Mg2+ uptake, Mg2+-starved plants were transferred to medium containing 0.02 or 3 mM Mg2+, and roots were collected separately at different times (0, 10, 30, 60, 90, 120, and 180 min). For concentration-dependent Mg2+ accumulation assays, Mg2+-starved plants were incubated for 60 min in medium containing various Mg2+ concentrations (0.02, 0.04, 0.08, 0.12, 0.16, 0.2, 0.5, 1.0, 1.5, 2.0, 2.5, 3.0, and 4.0 mM), and roots were collected. Tissue samples were rinsed three times in 50 mL of ice-cold Mg2+-free medium and then dried at 70°C for 48 h and weighed. The samples were ground and dissolved with 1 N HNO3 and then boiled for 30 min. Measurement of Mg2+ contents was performed by ICP-AES (Varian 715-ES). The Mg2+ uptake rate (mg∙g−1 dry weight∙h−1) of the plants for each incubation was calculated by subtracting the Mg2+ content of the Mg2+-starved plants from that at the Mg2+ concentration incubated.
To measure Mg2+ uptake using another procedure, we performed 63Ni2+ tracer uptake assays. The plant materials and treatment procedures were similar to those for the Mg2+ uptake assay, except that 100 μM NiCl2 and 10 μCi of 63Ni2+ were added to a 20-mL uptake solution containing Mg2+-free medium supplemented with the indicated levels of MgSO4. The time course of Mg2+ uptake at 20 μM or 3 mM Mg2+ was performed for 0, 30, 60, 90, and 120 min. The radioactivity was measured as described for the Salmonella mutant MM281 tracer uptake assays.
Accession Numbers
Sequence data from this article can be found in the GenBank/EMBL database and the Arabidopsis Genome Initiative database under the following accession numbers: MGT1, At1g80900, AC011713; MGT2, At1g16010, AC010924; MGT3, At3g03620, AC006836; MGT4, At3g19640, AP000417; MGT5, At4g28580, AL161573; MGT6, At3g58970, AL163527; MGT7, At5g09690, AL353994; MGT9, At5g64560, AB010076; and MGT10, At5g22830, AB005243.
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Quantitative Analysis of MGT6 Promoter Activity Represented by GUS Signal.
Supplemental Figure 2. Plasma Membrane Localization of MGT6-GFP.
Supplemental Figure 3. mRNA Levels of MGTs in MGT6 RNAi Plants.
Supplemental Figure 4. Phenotype of MGT6 RNAi Lines under the Mg2+-Deficient Hydroponic Condition.
Supplemental Figure 5. Mg2+ Uptake in Wild-Type and RNAi Plants.
Supplemental Figure 6. MGT6-Dependent Mg2+ Uptake in Roots Measured by 63Ni2+ Tracer Assay.
Supplemental Table 1. List of PCR Primers.
Supplementary Material
Acknowledgments
We thank Xinhuan Chen for assistance in the inductively coupled plasma assay and Langtao Xiao for assistance in the nickel isotope tracer uptake analysis. This work was supported by the National Science Foundation of China (Grants NSFC-31170229 and NSFC-31371244), the Hunan Natural Science Foundation (Grant 12JJ6021), a Key Project of the Hunan Education Commission (Grant 12A096), the Hunan Provincial Construct Program of the Key Discipline in Ecology, the Hunan Provincial Cooperative Innovation Center of Engineering, and New Products for Developmental Biology, the MOE 985 Project and IRT1020 of China, and the U.S. National Science Foundation (grant to S.L.).
AUTHOR CONTRIBUTIONS
D.M., D.L., and S.L. designed the experiments. D.M., J.C., L.T., Z.L., L.Y., R.T., J.L., and C.L. performed the experiments. D.M., Y.Y., J.S., D.L., L.C., and S.L. analyzed the data. D.M., D.L., and S.L. wrote the article.
Glossary
- qRT-PCR
quantitative RT-PCR
- RNAi
RNA interference
- DEX
dexamethosone
- ICP-AES
inductively coupled plasma–atomic emission spectrometry
- LB
Luria-Bertani
- MS
Murashige and Skoog
References
- Aoyama T., Chua N.H. (1997). A glucocorticoid-mediated transcriptional induction system in transgenic plants. Plant J. 11: 605–612 [DOI] [PubMed] [Google Scholar]
- Carrington J.C., Freed D.D., Oh C.S. (1990). Expression of potyviral polyproteins in transgenic plants reveals three proteolytic activities required for complete processing. EMBO J. 9: 1347–1353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Li L.G., Liu Z.H., Yuan Y.J., Guo L.L., Mao D.D., Tian L.F., Chen L.B., Luan S., Li D.P. (2009). Magnesium transporter AtMGT9 is essential for pollen development in Arabidopsis. Cell Res. 19: 887–898 [DOI] [PubMed] [Google Scholar]
- Clough S., Bent A. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Conn S.J., Conn V., Tyerman S.D., Kaiser B.N., Leigh R.A., Gilliham M. (2011). Magnesium transporters, MGT2/MRS2-1 and MGT3/MRS2-5, are important for magnesium partitioning within Arabidopsis thaliana mesophyll vacuoles. New Phytol. 190: 583–594 [DOI] [PubMed] [Google Scholar]
- Datla R.S., Hammerlindl J.K., Panchuk B., Pelcher L.E., Keller W. (1992). Modified binary plant transformation vectors with the wild-type gene encoding NPTII. Gene 122: 383–384 [DOI] [PubMed] [Google Scholar]
- Demidchik V., Davenport R.J., Tester M. (2002). Nonselective cation channels in plants. Annu. Rev. Plant Biol. 53: 67–107 [DOI] [PubMed] [Google Scholar]
- Drummond R.S.M., Tutone A., Li Y.C., Gardner R.C. (2006). A putative magnesium transporter AtMRS2-11 is localized to the plant chloroplast envelope membrane system. Plant Sci. 170: 78–89 [Google Scholar]
- Eshaghi S., Niegowski D., Kohl A., Martinez Molina D., Lesley S.A., Nordlund P. (2006). Crystal structure of a divalent metal ion transporter CorA at 2.9 angstrom resolution. Science 313: 354–357 [DOI] [PubMed] [Google Scholar]
- Gebert M., Meschenmoser K., Svidová S., Weghuber J., Schweyen R., Eifler K., Lenz H., Weyand K., Knoop V. (2009). A root-expressed magnesium transporter of the MRS2/MGT gene family in Arabidopsis thaliana allows for growth in low-Mg2+ environments. Plant Cell 21: 4018–4030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo K.M., Babourina O., Christopher D.A., Borsic T., Rengel Z. (2010). The cyclic nucleotide-gated channel AtCNGC10 transports Ca2+ and Mg2+ in Arabidopsis. Physiol. Plant. 139: 303–312 [DOI] [PubMed] [Google Scholar]
- Jefferson R.A., Kavanagh T.A., Bevan M.W. (1987). GUS fusions: β-Glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J. 6: 3901–3907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoop V., Groth-Malonek M., Gebert M., Eifler K., Weyand K. (2005). Transport of magnesium and other divalent cations: Evolution of the 2-TM-GxN proteins in the MIT superfamily. Mol. Genet. Genomics 274: 205–216 [DOI] [PubMed] [Google Scholar]
- Latz A., Becker D., Hekman M., Müller T., Beyhl D., Marten I., Eing C., Fischer A., Dunkel M., Bertl A., Rapp U.R., Hedrich R. (2007). TPK1, a Ca2+-regulated Arabidopsis vacuole two-pore K+ channel is activated by 14-3-3 proteins. Plant J. 52: 449–459 [DOI] [PubMed] [Google Scholar]
- Lenz H., Dombinov V., Dreistein J., Reinhard M.R., Gebert M., Knoop V. (2013). Magnesium deficiency phenotypes upon multiple knockout of Arabidopsis thaliana MRS2 clade B genes can be ameliorated by concomitantly reduced calcium supply. Plant Cell Physiol. 54: 1118–1131 [DOI] [PubMed] [Google Scholar]
- Li L., Tutone A.F., Drummond R.S., Gardner R.C., Luan S. (2001). A novel family of magnesium transport genes in Arabidopsis. Plant Cell 13: 2761–2775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L.G., Sokolov L.N., Yang Y.H., Li D.P., Ting J., Pandy G.K., Luan S. (2008). A mitochondrial magnesium transporter functions in Arabidopsis pollen development. Mol. Plant 1: 675–685 [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408 [DOI] [PubMed] [Google Scholar]
- Lunin V.V., Dobrovetsky E., Khutoreskaya G., Zhang R., Joachimiak A., Doyle D.A., Bochkarev A., Maguire M.E., Edwards A.M., Koth C.M. (2006). Crystal structure of the CorA Mg2+ transporter. Nature 440: 833–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusk J.E., Kennedy E.P. (1969). Magnesium transport in Escherichia coli. J. Biol. Chem. 244: 1653–1655 [PubMed] [Google Scholar]
- Maguire M.E. (1992). MgtA and MgtB: Prokaryotic P-type ATPases that mediate Mg2+ influx. J. Bioenerg. Biomembr. 24: 319–328 [DOI] [PubMed] [Google Scholar]
- Maguire M.E. (2006). The structure of CorA: A Mg2+-selective channel. Curr. Opin. Struct. Biol. 16: 432–438 [DOI] [PubMed] [Google Scholar]
- Mao D.D., Tian L.F., Li L.G., Chen J., Deng P.Y., Li D.P., Luan S. (2008). AtMGT7: An Arabidopsis gene encoding a low-affinity magnesium transporter. J. Integr. Plant Biol. 50: 1530–1538 [DOI] [PubMed] [Google Scholar]
- Miao Y.S., Jiang L.W. (2007). Transient expression of fluorescent fusion proteins in protoplasts of suspension cultured cells. Nat. Protoc. 2: 2348–2353 [DOI] [PubMed] [Google Scholar]
- Payandeh J., Pai E.F. (2006). A structural basis for Mg2+ homeostasis and the CorA translocation cycle. EMBO J. 25: 3762–3773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz J., Tierbach A., Lenz H., Meschenmoser K., Knoop V. (2013). Membrane protein interactions between different Arabidopsis thaliana MRS2-type magnesium transporters are highly permissive. Biochim. Biophys. Acta 1828: 2032–2040 [DOI] [PubMed] [Google Scholar]
- Schock I., Gregan J., Steinhauser S., Schweyen R., Brennicke A., Knoop V. (2000). A member of a novel Arabidopsis thaliana gene family of candidate Mg2+ ion transporters complements a yeast mitochondrial group II intron-splicing mutant. Plant J. 24: 489–501 [DOI] [PubMed] [Google Scholar]
- Shaul O. (2002). Magnesium transport and function in plants: The tip of the iceberg. Biometals 15: 309–323 [DOI] [PubMed] [Google Scholar]
- Sheen J. (2001). Signal transduction in maize and Arabidopsis mesophyll protoplasts. Plant Physiol. 127: 1466–1475 [PMC free article] [PubMed] [Google Scholar]
- Shi H.Z., Quintero F.J., Pardo J.M., Zhu J.K. (2002). The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell 14: 465–477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith R.L., Szegedy M.A., Kucharski L.M., Walker C., Wiet R.M., Redpath A., Kaczmarek M.T., Maguire M.E. (1998). The CorA Mg2+ transport protein of Salmonella typhimurium: Mutagenesis of conserved residues in the third membrane domain identifies a Mg2+ pore. J. Biol. Chem. 273: 28663–28669 [DOI] [PubMed] [Google Scholar]
- Snavely M.D., Florer J.B., Miller C.G., Maguire M.E. (1989a). Magnesium transport in Salmonella typhimurium: Expression of cloned genes for three distinct Mg2+ transport systems. J. Bacteriol. 171: 4752–4760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snavely M.D., Florer J.B., Miller C.G., Maguire M.E. (1989b). Magnesium transport in Salmonella typhimurium: 28Mg2+ transport by the CorA, MgtA, and MgtB systems. J. Bacteriol. 171: 4761–4766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snavely M.D., Gravina S.A., Cheung T.T., Miller C.G., Maguire M.E. (1991). Magnesium transport in Salmonella typhimurium: Regulation of mgtA and mgtB expression. J. Biol. Chem. 266: 824–829 [PubMed] [Google Scholar]
- Szegedy M.A., Maguire M.E. (1999). The CorA Mg2+ transport protein of Salmonella typhimurium: Mutagenesis of conserved residues in the second membrane domain. J. Biol. Chem. 274: 36973–36979 [DOI] [PubMed] [Google Scholar]
- Tocquin P., Corbesier L., Havelange A., Pieltain A., Kurtem E., Bernier G., Périlleux C. (2003). A novel high efficiency, low maintenance, hydroponic system for synchronous growth and flowering of Arabidopsis thaliana. BMC Plant Biol. 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wesley S.V., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27: 581–590 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.