This work shows that the cucumber anthracnose fungus Colletotrichum orbiculare accumulates its virulence effectors in the plant–pathogen interfacial region around the neck of primary biotrophic hyphae developed in host cells. It also describes that the pathogen secretes the effectors toward the interfacial region via SEC22-dependent traffic and SEC4-mediated exocytosis.
Abstract
The hemibiotrophic pathogen Colletotrichum orbiculare develops biotrophic hyphae inside cucumber (Cucumis sativus) cells via appressorial penetration; later, the pathogen switches to necrotrophy. C. orbiculare also expresses specific effectors at different stages. Here, we found that virulence-related effectors of C. orbiculare accumulate in a pathogen–host biotrophic interface. Fluorescence-tagged effectors accumulated in a ring-like region around the neck of the biotrophic primary hyphae. Fluorescence imaging of cellular components and transmission electron microscopy showed that the ring-like signals of the effectors localized at the pathogen–plant interface. Effector accumulation at the interface required induction of its expression during the early biotrophic phase, suggesting that transcriptional regulation may link to effector localization. We also investigated the route of effector secretion to the interface. An exocytosis-related component, the Rab GTPase SEC4, localized to the necks of biotrophic primary hyphae adjacent to the interface, thereby suggesting focal effector secretion. Disruption of SEC4 in C. orbiculare reduced virulence and impaired effector delivery to the ring signal interface. Disruption of the v-SNARE SEC22 also reduced effector delivery. These findings suggest that biotrophy-expressed effectors are secreted, via the endoplasmic reticulum-to-Golgi route and subsequent exocytosis, toward the interface generated between C. orbiculare and the host cell.
INTRODUCTION
Hemibiotrophic fungal pathogens initially grow inside living hosts by maintaining the viability of their host cells (biotrophy) and later kill the host cells to obtain nutrients (necrotrophy). The ascomycete genus Colletotrichum comprises over 600 species that infect a wide range of plant species, including important cultivars, and many members of Colletotrichum have a hemibiotrophic lifestyle (Perfect et al., 1999; O’Connell et al., 2012; Gan et al., 2013). Colletotrichum fungi develop specialized infection structures called appressoria, which are darkly pigmented with melanin (Kubo and Furusawa, 1991; Kubo and Takano, 2013).
The penetration peg emerging from melanized appressoria of Colletotrichum species develops bulbous biotrophic hyphae in living host cells before switching to a necrotrophic mode (Perfect et al., 1999). To establish biotrophy in host plants, pathogens need to secrete an arsenal of effector proteins that manipulate their host environment, particularly by suppressing plant immune responses, such as hypersensitive cell death (Bozkurt et al., 2012; Dou and Zhou, 2012; Rafiqi et al., 2012). Subsequently, pathogens kill the plant cells, probably by secreting toxins, and degrade host tissues by using lytic enzymes (Kleemann et al., 2012; O’Connell et al., 2012; Gan et al., 2013).
Recent genome analyses identified numerous effector candidate genes in the hemibiotrophic Colletotrichum fungi C. higginsianum (pathogen of Brassica spp), C. graminicola (pathogen of maize [Zea mays]), C. orbiculare (pathogen of cucumber [Cucumis sativus]), and C. gloeosporioides (pathogen of strawberry [Fragaria spp]) (O’Connell et al., 2012; Gan et al., 2013). Comparative analyses of the effector candidate genes suggested that each Colletotrichum species has a large set of unique effector genes. Thus, each set of unique effectors in the Colletotrichum species likely has crucial roles in the manipulation of each host plant’s condition. Importantly, comprehensive expression analyses of C. higginsianum and C. orbiculare also detected major changes in the expression of effector candidate genes during the biotrophic and necrotrophic stages, which suggested a correlation between sequential effector expression and stage transitions (Kleemann et al., 2012; O’Connell et al., 2012; Gan et al., 2013). C. higginsianum expresses a set of effectors, including cell death suppressors, during its biotrophic stages, but it expresses genes encoding toxin-like proteins during the switch from biotrophy to necrotrophy, indicating the importance of the coordinated expression of antagonistic effectors for hemibiotrophy (Kleemann et al., 2012).
Fluorescent tagging of several effectors detected a clear, punctate signal at the basal penetration pore of C. higginsianum appressoria infecting cells of Arabidopsis thaliana (Kleemann et al., 2012). This finding, together with transmission electron microscopy (TEM) immunogold labeling, suggests that the effectors are not secreted uniformly from the appressorium surface in C. higginsianum (Kleemann et al., 2012). Instead, they are secreted focally from appressorial penetration pores before the invasion of the host, suggesting that the penetration pore is an interface for effector delivery between C. higginsianum and Arabidopsis. After effectors are secreted, they may act outside the host cells (called apoplastic effectors) or be translocated into plant cells, where they have intracellular effects (called cytoplasmic effectors).
Furthermore, in the rice blast fungus Magnaporthe oryzae, effector proteins accumulate in an interfacial structure named the biotrophic interfacial complex (BIC), which can be visualized as a strong dot signal using fluorescently labeled effectors (Mosquera et al., 2009; Khang et al., 2010; Yi and Valent, 2013). BIC development is coupled to hyphal differentiation in the transition from filamentous to bulbous invasive hyphal growth. In addition, several BIC-localized effector proteins translocate into the rice (Oryza sativa) cytoplasm, which suggests that BIC may be the site for effector translocation during rice–M. oryzae interactions (Mosquera et al., 2009; Khang et al., 2010). Recently work also reported that M. oryzae has two distinct secretion systems of effectors, a conventional pathway for apoplastic effectors and a BIC-related secretion pathway for cytoplasmic effectors (Giraldo et al., 2013; Giraldo and Valent, 2013).
However, no BIC-like structures were detected during C. higginsianum–Arabidopsis interactions; instead, multiple punctate signals of the fluorescently labeled effectors, called interfacial bodies, were observed randomly on the biotrophic hyphal surface (Kleemann et al., 2012; Giraldo and Valent, 2013). Also, between the biotrophic powdery mildew fungus Golovinomyces orontii and its host Arabidopsis, the biogenesis of a specialized interface (called the haustorial complex), distinct from the BIC, was observed by TEM where the extrahaustorial matrix outside the haustorial wall contained putative exosome vesicles, which suggested the existence of a portal into host cells to allow effector translocation (Micali et al., 2011). These findings suggest the diversity of biotrophic interfaces generated between fungal pathogens and plants, probably resulting from specific coevolution in each set of pathogen and host.
Here, we report a biotrophic interface where effectors are preferentially delivered in C. orbiculare–host cucumber interactions. Recently, we identified three distinct effectors of C. orbiculare: NIS1, DN3, and MC69 (Saitoh et al., 2012; Yoshino et al., 2012). NIS1 is preferentially expressed during the biotrophic stage and is recognized by Nicotiana benthamiana. Recognition is followed by a plant cell death response, implying that NIS1 has potential avirulence effects. C. orbiculare can infect N. benthamiana in addition to cucumber; however, the deletion of NIS1 produced no apparent effect on C. orbiculare virulence on N. benthamiana, suggesting possible suppression of NIS1 recognition and/or the subsequent defense response (Yoshino et al., 2012). Consistent with this idea, we have identified the effector Co-DN3 of C. orbiculare (the ortholog of C. gloeosporioides Cg-DN3) as a factor that can suppress NIS1-induced cell death (Stephenson et al., 2000; Yoshino et al., 2012). MC69 is a small secreted protein essential for the pathogenicity of M. oryzae (Saitoh et al., 2012), and interestingly, we found that Co-MC69 (the C. orbiculare ortholog of M. oryzae MC69) is also required for the successful infection of C. orbiculare (Saitoh et al., 2012).
In this study, we found that during host infection, three C. orbiculare effectors, NIS1, DN3, and MC69, localize to a characteristic ring at the neck region of biotrophic primary hyphae in the interfacial region outside the fungal cell, representing the presence of an effector-associated biotrophic interface. Effector accumulation to the ring signal interface largely depends on induced expression of the effectors during the early biotrophic phase and requires SEC4 Rab GTPase-dependent exocytosis via the SEC22 v-SNARE–dependent endoplasmic reticulum (ER)-to-Golgi route.
RESULTS
C. orbiculare Effectors Tagged with Fluorescent Proteins Exhibit a Ring Signal in Host Infection Sites
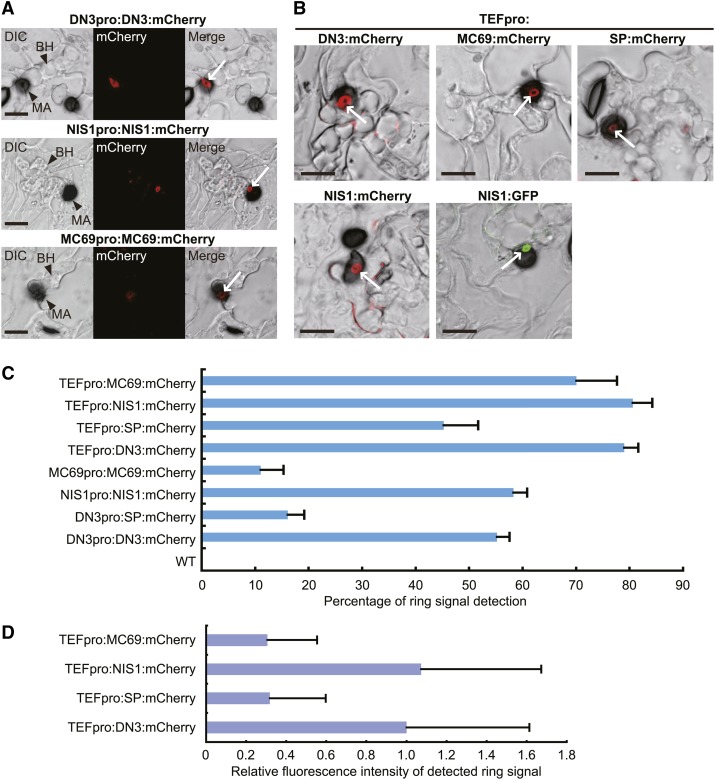
We have identified and characterized three virulence-related effector genes of C. orbiculare: DN3, MC69, and NIS1 (Saitoh et al., 2012; Yoshino et al., 2012). Here, we investigated the dynamics of these effectors during infection by C. orbiculare by tagging each effector with the fluorescent protein mCherry. Reporter strains of C. orbiculare were generated that expressed each effector tagged with mCherry under the control of the corresponding native promoter. Subsequently, the fungal strains generated were inoculated on host cucumber plants. Notably, fluorescent signals of the tagged effectors were detected as a unique ring-shaped signal around the appressorial invasion site at 4 d postinoculation (dpi), when the pathogen formed biotrophic invasive hyphae (Figure 1A), although there was a difference between MC69:mCherry and the other two tagged effectors. In detail, we detected strong ring signals in the reporter lines expressing DN3:mCherry or NIS1:mCherry and we detected weaker signal intensity from MC69:mCherry, all driven by respective native promoters. The frequencies of ring signal detection with DN3:mCherry and NIS1:mCherry (55 and 58%, respectively) were also significantly higher than that with MC69:mCherry (11%) (Figure 1C). This result suggested that the expression of MC69 was relatively lower during the early biotrophic phase. However, the green fluorescent protein (GFP)–based promoter assay of MC69 and DN3 suggested that MC69 was strongly expressed during early biotrophy, the same as DN3 (Supplemental Figure 1).
Figure 1.
Fluorescent Protein–Tagged Effectors Accumulated around the Penetration Site to Form a Ring Signal.
(A) Localization pattern of C. orbiculare effectors under the control of the corresponding native promoter. C. orbiculare strains carrying each effector gene tagged with mCherry were incubated on cucumber cotyledons for 4 d. Tagged DN3, NIS1, and MC69 accumulated around the appressorial penetration site to form a ring signal, although DN3 and NIS1 exhibit preferential accumulation compared with MC69. The ring signals are indicated by white arrows. BH, biotrophic hypha; DIC, differential interference contrast; MA, melanized appressorium. Bars = 10 μm.
(B) Localization pattern of C. orbiculare effectors under the control of the TEF promoter. C. orbiculare strains carrying each effector gene tagged with mCherry or GFP were incubated on cucumber for 4 d. The ring signals are indicated by white arrows. SP, signal peptide of DN3. Bars = 10 μm.
(C) The ring signal ratio of mCherry fluorescence for appressoria that formed biotrophic hyphae. Each reporter line was inoculated on cucumber cotyledons, and the inoculated plants were incubated for 4 d. At least 70 appressoria that formed biotrophic hyphae were investigated for each plant sample. The mean and sd were calculated from three independent plant samples. WT indicates the wild-type strain without mCherry.
(D) Quantification of the fluorescence intensity of the detected ring signal in each reporter line. Each reporter line was inoculated on cucumber cotyledons, and the inoculated plants were incubated for 4 d. At least 50 detected ring signals were investigated for each sample. The mean fluorescence intensity value of DN3:mCherry was normalized to 1.0.
In contrast with the detection of ring signal around the appressorial invasion sites, no ring signal was detected in the absence of biotrophic hyphae; instead, tiny dot signals of effectors tagged with mCherry were detected at the base of appressoria without invasion (Supplemental Figure 2), which was consistent with a previous report that effectors accumulated in the appressorial pore of C. higginsianum before invasion (Kleemann et al., 2012). Thus, the ring signal of effector:mCherry is associated with the formation of biotrophic hyphae.
When we constitutively and highly expressed each mCherry-fused effector under the control of the Aureobasidium pullulans TRANSLATION ELONGATION FACTOR (TEF) promoter (Vanden Wymelenberg et al., 1997), we detected increased ring signals with all of the tagged effectors (Figures 1B and 1C). However, the fluorescence intensity of the detected ring signals of MC69:mCherry was still lower than that of DN3:mCherry and NIS1:mCherry ring signals (Figures 1B and 1D). In the expression with the TEF promoter, we frequently found punctate effector:mCherry signals on biotrophic hyphae (Figure 1B). We also generated C. orbiculare reporter lines that expressed NIS1 fused to GFP under the control of the TEF promoter. As a result, we detected the ring signal of NIS1:GFP at 4 dpi, similar to the ring signal of NIS1:mCherry (Figure 1B), demonstrating that the ring signal was not specific to mCherry fusions and that it was likely an intrinsic phenomenon.
We further investigated ring signals in the expression of mCherry carrying the signal peptide of DN3, named SP:mCherry. When cucumber cotyledons were inoculated with C. orbiculare expressing SP:mCherry under the control of the TEF promoter, the frequency of the ring signal detection in infection sites was significantly reduced compared with the strain expressing DN3:mCherry with the TEF promoter (Figures 1B and 1C), although the mCherry signal in preincubated conidia of the SP:mCherry strain was similar to that in preincubated conidia of the DN3:mCherry strain (Supplemental Figure 3). The fluorescence intensity of the detected ring signal of SP:mCherry was also strongly reduced compared with that of DN3:mCherry, which was similar to the case of MC69:mCherry (Figures 1B and 1D). Consistent with these results, when SP:mCherry was expressed under the control of the DN3 promoter, the frequency of the ring signal detection was also severely reduced (less than 20%) compared with the expression of DN3:mCherry with the DN3 promoter (Figure 1C).
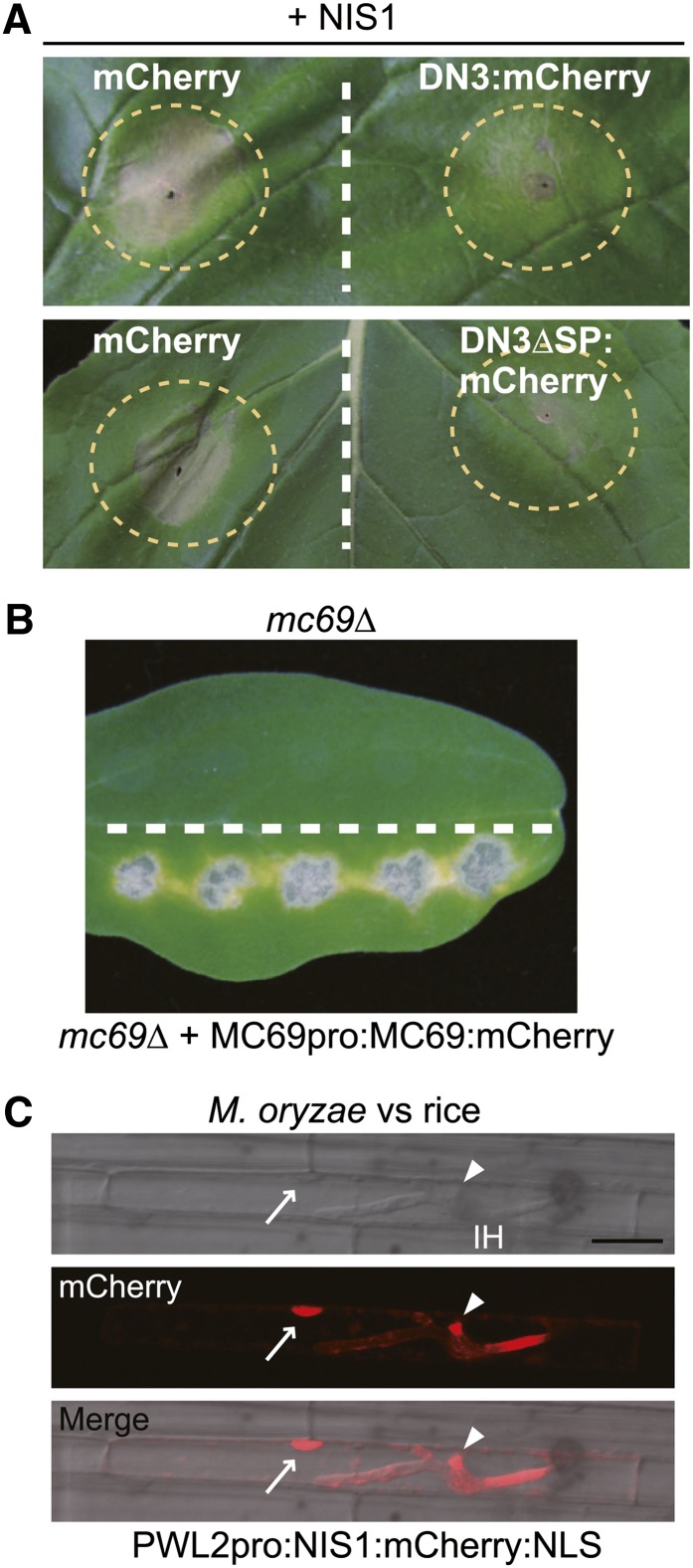
We also investigated the functionality of the effector genes fused to mCherry. As mentioned earlier, DN3 suppresses NIS1-induced cell death in N. benthamiana (Yoshino et al., 2012). We found that the expression of DN3:mCherry also suppressed NIS1-induced cell death, thereby indicating that the addition of mCherry to DN3 did not abolish its functionality (Figure 2A). DN3 lacking its signal peptide (DN3ΔSP) also suppresses NIS1-induced cell death (Yoshino et al., 2012), which strongly suggests that DN3 functions as a cytoplasmic effector. We also found that DN3ΔSP fused to mCherry suppressed NIS1-induced cell death (Figure 2A). In contrast with DN3, MC69 is regarded as a putative apoplastic effector because M. oryzae MC69 (Mo-MC69 shares 64% identity on the amino acid level with Co-MC69) fused to mCherry was localized to the BIC but was not translocated into host rice cells (Saitoh et al., 2012). We found that introduction of the Co-MC69:mCherry gene enabled the mc69Δ mutant to develop lesions on cucumber cotyledons (Figure 2B), thereby suggesting that the MC69:mCherry is functional.
Figure 2.
Functionality of the Effector:mCherry Fusion Proteins.
(A) Cell death suppression assay using DN3:mCherry and DN3ΔSP:mCherry. Agroinfiltration sites that expressed mCherry (control), DN3:mCherry, or DN3ΔSP:mCherry were challenged with Agrobacterium expressing NIS1 in N. benthamiana. The infiltration sites are represented by dashed circles. Strong cell death induced by NIS1 was observed in infiltration sites that expressed mCherry, whereas it was suppressed in the DN3:mCherry and DN3ΔSP:mCherry sites. The photograph was taken at 5 d after the infiltration challenge.
(B) Pathogenicity assay using the mc69Δ mutant that expressed Co-MC69:mCherry under the control of the native promoter. The mc69Δ strain was inoculated onto the upper half of the cucumber cotyledon as a control. The tested strain was inoculated onto the lower half. The inoculated cotyledons were incubated for 7 d.
(C) NIS1:mCherry:NLS was secreted by M. oryzae and translocated into rice cells. The M. oryzae strain that carried PWL2pro:NIS1:mCherry:NLS was inoculated onto rice leaf sheath, incubated for 32 h, and observed by confocal laser-scanning microscopy. The NIS1:mCherry:NLS signal localized to the BIC (arrowheads) and nucleus (arrows) of the invaded rice leaf sheath cell. IH, invasive hypha. Bar = 20 μm.
It was unclear whether NIS1 acts as an apoplastic or a cytoplasmic effector. To address this point, we generated an M. oryzae strain that expressed an NIS1:mCherry construct that carried a modified nuclear localization signal (NLS) of Simian virus 40, expressed the fusion under the control of the PWL2 promoter (Khang et al., 2010; Saitoh et al., 2012), and inoculated this strain onto rice leaf sheath samples. The NIS1:mCherry:NLS signal was found in both the BIC and the nuclei of invaded rice cells, thereby indicating that NIS1:mCherry:NLS secreted from the M. oryzae strain is translocated into rice cells (Figure 2C). This finding suggests that (1) NIS1 likely acts as a cytoplasmic effector, at least in rice cells, and (2) the addition of mCherry to NIS1 did not abolish its functionality with respect to host translocation. Collectively, these findings support the functionality of each effector tagged with mCherry, so we conclude that their ring-shaped signal is an intrinsic characteristic.
The Ring Signal of the Effectors Is Located in a Pathogen–Host Interface around the Biotrophic Hyphal Neck beneath Appressoria
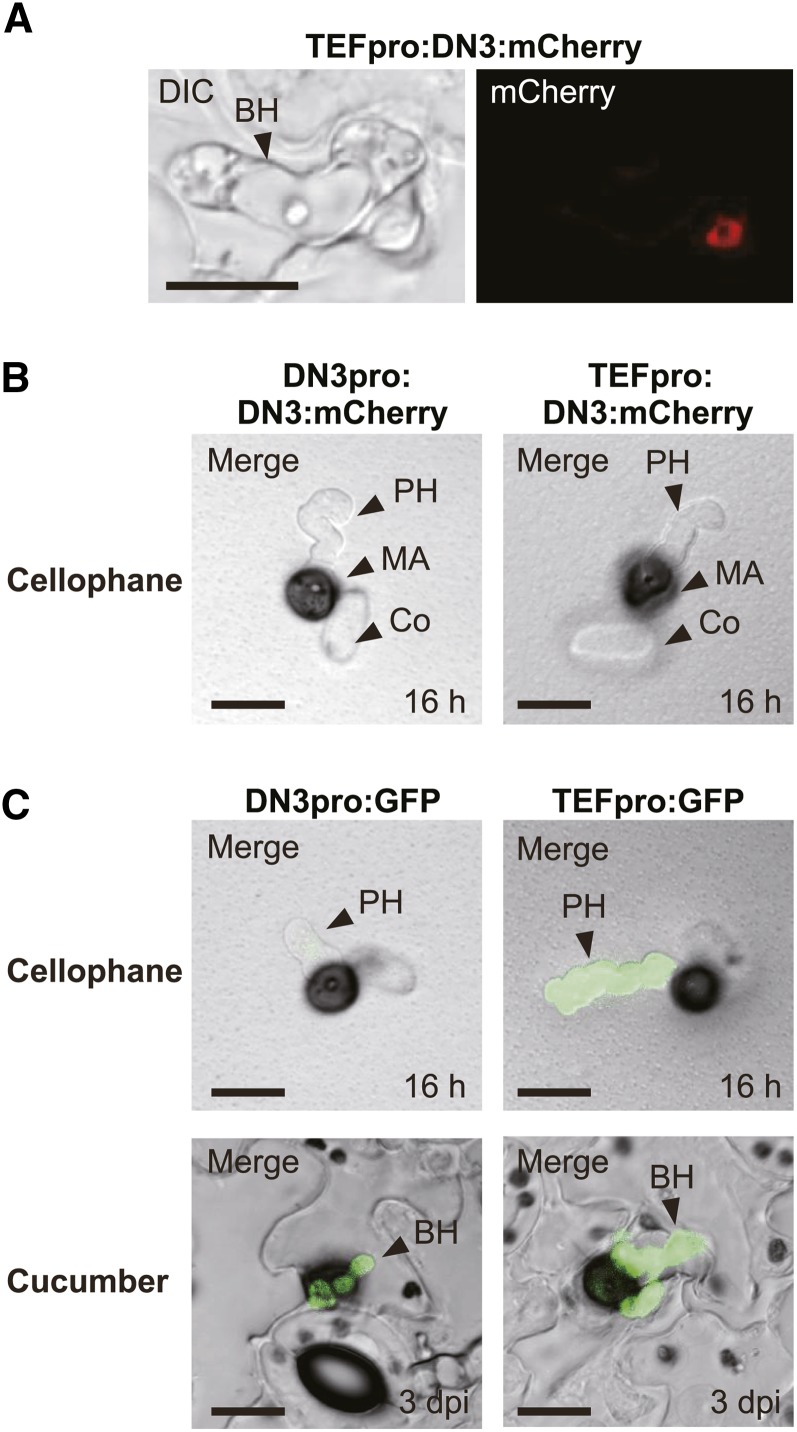
The effector ring signal was detected in appressorial invasion sites, but the precise location of this ring signal was not certain. The ring signal could have been located around the appressorial base or the biotrophic primary hyphal neck. Thus, we addressed this issue by inoculating cucumber cotyledons with the strains expressing DN3:mCherry and removed the appressoria after biotrophic colonization to determine whether the DN3:mCherry signal was present or absent after the removal of appressoria. We clearly observed the ring signal of DN3:mCherry after the removal, which suggested that the effector accumulated around the biotrophic hyphal neck but not the appressorial base (Figure 3A). When the strains with DN3pro:DN3:mCherry were incubated on cellophane membrane (an artificial, penetrable substratum) (Figure 3B), DN3:mCherry ring signals were not detected around the penetration sites. A GFP-based promoter assay indicated that the promoter activity of DN3 was much lower on cellophane than that on the host plant (Figure 3C), which suggested that the absence of the ring signal might be due to the lower expression level of the reporter gene. However, when DN3:mCherry was expressed under the control of the TEF promoter, we still did not find ring signals around appressorial penetration sites on cellophane (Figure 3B), although the TEF promoter exhibited its strong activity on cellophane (Figure 3C). This finding strongly suggested that the ring signal specifically developed around the neck of biotrophic primary hyphae.
Figure 3.
The Ring Signal of the Effectors Localized at the Biotrophic Hyphal Neck beneath the Appressorium.
(A) The effector ring signal was localized at the biotrophic invasive hyphal neck. Cucumber cotyledons were inoculated with a C. orbiculare strain that carried TEFpro:DN3:mCherry and incubated for 4 d. Then, appressoria were removed by brushing the surface of cucumber cotyledons infected with the pathogen. BH, biotrophic hypha; DIC, differential interference contrast. Bar = 10 μm.
(B) The signal of DN3:mCherry expressed under the control of the DN3 or TEF promoter was undetectable in C. orbiculare cells during penetration of the cellophane membrane. C. orbiculare strains that carried DN3pro:DN3:mCherry or TEFpro:DN3:mCherry were incubated for 16 h. Co, conidium; MA, melanized appressorium; PH, pseudobiotrophic hypha. Bars = 10 μm.
(C) GFP-based promoter assays of the DN3 promoter and the TEF promoter in C. orbiculare, which were incubated on cellophane and cucumber. C. orbiculare strains that carried the GFP gene with the DN3 or TEF promoter were incubated on cellophane for 16 h or on cucumber for 3 d. Bars = 10 μm.
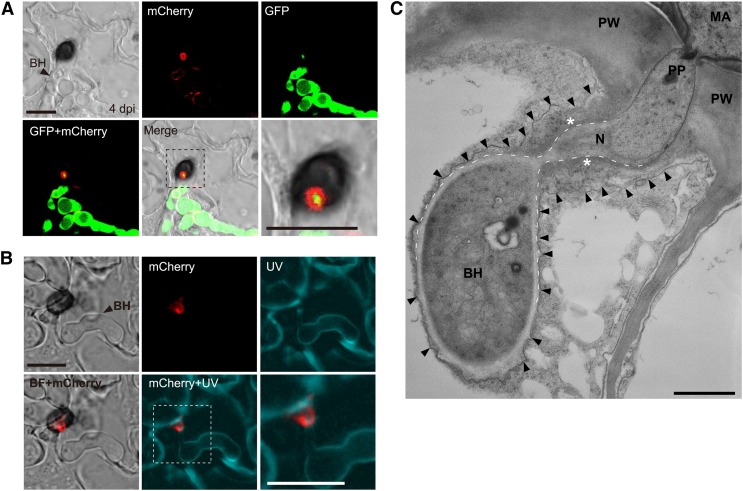
To investigate the location of the effector ring further, we visualized the fungal cytoplasm and the DN3 at the same time by expressing GFP and DN3:mCherry under the control of the TEF promoter (Figure 4A). A strong GFP signal was detected in the cavity of the DN3:mCherry fluorescent ring. However, the GFP signal does not overlap with the ring of DN3:mCherry, suggesting that the effector ring signal likely locates outside of the fungal cell. The fungal cell wall is composed primarily of polysaccharides, including chitin (Latgé, 2007). Thus, we also visualized the cell wall of C. orbiculare expressing DN3:mCherry using Calcofluor White M2R, which stains chitin. Calcofluor White M2R clearly visualized the cell walls of invasive fungal hyphae, but no chitin signal was detected outside the DN3:mCherry ring (Figure 4B). Furthermore, the TEM analysis showed that biotrophic primary hyphae were tightly surrounded by plant-derived membrane, whereas the neck regions of biotrophic hyphae were not (i.e., the TEM analysis detected a relatively wide space between the neck region of biotrophic hyphae and the plant-derived membrane, which is considered to include the interfacial region where the effector:mCherry accumulates as the ring signal) (Figure 4C). Also, the interfacial regions detected by TEM, which were close to the surface of the neck region, were regions of high electron density (Figure 4C). Overall, these results indicate that the effector ring forms outside the neck of biotrophic primary hyphae developed by appressoria and locates in a pathogen–plant interface. Consistent with this, the ring signal of DN3:mCherry was not detected around penetration hyphae on cellophane as shown in Figure 3B, which might be due to diffusion of the secreted DN3:mCherry from the appressorium site in the absence of the interfacial region and the plant-derived membrane.
Figure 4.
The Effectors Localized at the Pathogen–Host Interface around the Biotrophic Primary Hyphal Neck.
(A) Visualization of fungal cytoplasm by GFP. A C. orbiculare strain that carried both TEFpro:GFP and TEFpro:DN3:mCherry constructs was incubated for 4 d on cucumber. The bottom right panel shows a zoomed image of the dashed box. BH, biotrophic hypha. Bars = 10 μm.
(B) Visualization of the fungal cell wall using Calcofluor White M2R. A C. orbiculare strain with TEFpro:DN3:mCherry was incubated for 4 d on cucumber. The infected cucumber sample was treated with Calcofluor White M2R for 5 min before UV excitation. The bottom right panel shows a zoomed image of the dashed box. BF, bright field. Bars = 10 μm.
(C) TEM analysis of the biotrophic hypha that developed from the appressorium of C. orbiculare. The C. orbiculare wild-type strain was inoculated on cucumber cotyledons. The arrowheads represent the plant-derived membrane, and the dashed lines represent the surface of the biotrophic hypha around the neck. The asterisks indicate the putative interfacial region with high electron density, related to the effector ring signal, around the biotrophic hyphal neck. MA, melanized appressorium; N, neck of the biotrophic hypha; PP, penetration peg; PW, plant cell wall. Bar = 1 μm.
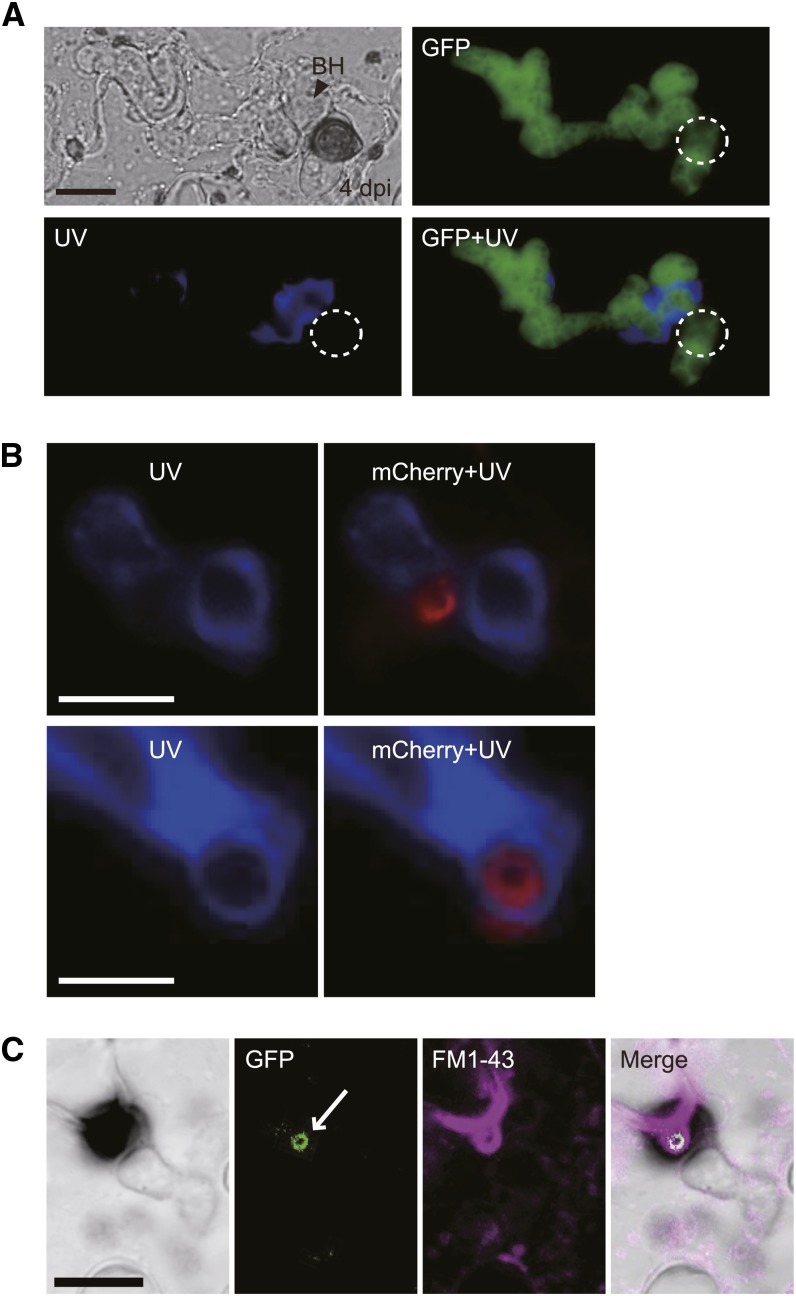
The plant cell wall apposition, called papilla, is known to be formed beneath appressorial penetration trial sites. Papilla is regarded as an inducible structural barrier in plant defense and contains callose as a major constituent that can be stained by aniline blue fluorochrome (Hückelhoven, 2007). Consistent with this, we generally detected the typical deposition of papillary callose beneath the appressorial penetration site at 1 dpi during C. orbiculare–cucumber interactions (Supplemental Figure 4). At this time, C. orbiculare appressoria did not develop biotrophic primary hyphae. At 4 dpi, however, the papillary callose signal was hardly detectable, whereas callose was deposited randomly around C. orbiculare biotrophic hyphae expressing cytoplasmic GFP (Figure 5A). The simultaneous observation of callose and effector ring also demonstrated that callose does not colocalize with the effector ring, whereas callose was often found around biotrophic hyphae (Figure 5B). These findings indicate that the effector ring develops de novo during the formation of biotrophic hyphae. Also, FM1-43 staining of cotyledons inoculated with C. orbiculare expressing NIS1:GFP suggested that the ring signal interface is associated with concentrated regions of FM1-43–stained materials, implying the presence of membranous components around the ring signal interface (Figure 5C).
Figure 5.
The Ring Signal Interface Did Not Include Papillary Callose and Was Associated with Membrane Components.
(A) Visualization of callose deposition during the biotrophic invasive stage. C. orbiculare with TEFpro:GFP was incubated for 4 d on cucumber. Staining was performed using aniline blue fluorochrome. The dashed circles represent the position of the appressorium. BH, biotrophic hypha. Bar = 10 μm.
(B) Simultaneous observation of the effector ring and callose deposition during the biotrophic invasive stage. C. orbiculare with TEFpro:DN3:mCherry was incubated for 4 d on cucumber. Callose staining was performed with aniline blue fluorochrome. Bars = 5 μm.
(C) Staining of the membrane components using FM1-43. C. orbiculare with TEFpro:NIS1:GFP was incubated for 4 d on cucumber. The infected cucumber samples were treated rapidly with FM1-43 before observation in distilled water. The white arrow indicates the GFP-labeled ring signal interface. Bar = 10 μm.
Recruitment of the Effector to the Ring Signal Interface Depends on Its Expression during Early Biotrophy
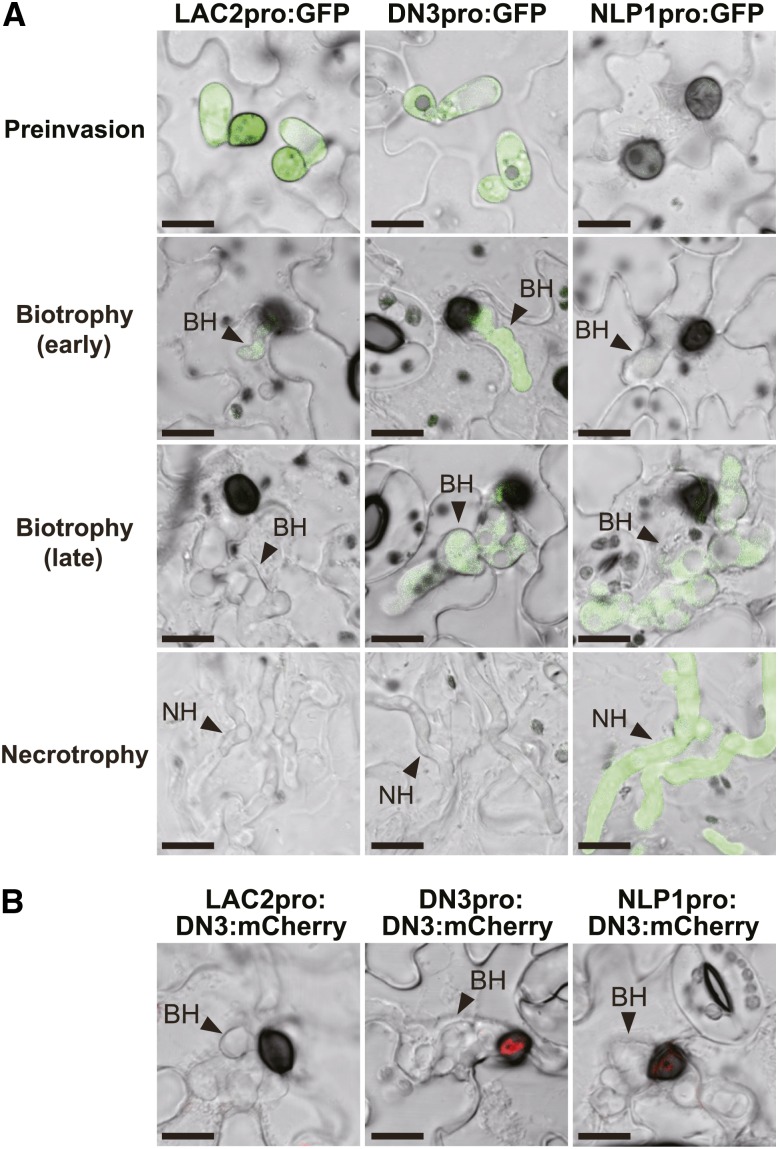
As described earlier, DN3:mCherry and NIS1:mCherry strongly localized to the ring signal interface when they were expressed under the control of their own promoters (Figures 1A and 1C). GFP-based promoter assays of DN3 and NIS1 indicated that both genes show preferential expression during the biotrophic phase (Figure 3C; Supplemental Figure 1) (Yoshino et al., 2012). These findings suggest that the preferential expression of the effector genes may be important for the localization of the effectors to the interface. To test this hypothesis, we expressed DN3:mCherry under the control of the promoters of two Colletotrichum secretion protein genes, Laccase2 (LAC2) (Lin et al., 2012) and C. orbiculare Necrosis- and ethylene-inducing peptide1-like protein1 (NLP1) (Gijzen and Nürnberger, 2006), which have distinct expression patterns compared with DN3. LAC2 encodes a secreted laccase, which is involved with appressorial melanization (Lin et al., 2012), and its expression is strongly induced during appressorium maturation, whereas it declines rapidly after appressorial invasion (Figure 6A; Lin et al., 2012). We found three NLP homologs in the C. orbiculare genome, Cob_11657, Cob_09815, and Cob_00931 (Gan et al., 2013), which we designated as NLP1, NLP2, and NLP3, respectively (Supplemental Figure 5). Co-NLP1 shares high sequence similarity with Ch-NLP1 from C. higginsianum (Kleemann et al., 2012) and is predicted to be expressed specifically during necrotrophy (Gan et al., 2013). Our GFP-based promoter assay confirmed that C. orbiculare NLP1 was preferentially expressed in necrotrophic invasive hyphae (Figure 6A).
Figure 6.
Accumulation of the Effectors at the Ring Signal Interface Was Dependent on Preferential Gene Expression during Early Biotrophy.
(A) GFP-based promoter assay using the LAC2, DN3, and NLP1 promoters in the C. orbiculare infection process. C. orbiculare strains that carried the GFP gene with each promoter were incubated on cucumber for 6 h (preinvasion), 3 d (early biotrophy), 4 d (late biotrophy), and 5 d (necrotrophy). BH, biotrophic hypha; NH, necrotrophic hypha. Bars = 10 μm.
(B) Promoter-exchange experiment with DN3:mCherry. Each C. orbiculare strain expressed DN3:mCherry under the control of the LAC2, DN3, or NLP1 promoter. Bars = 10 μm.
We generated C. orbiculare strains that expressed DN3:mCherry under the control of LAC2 and NLP1 promoters, which we designated as the LAC2pro:DN3:mCherry strain and the NLP1pro:DN3:mCherry strain, respectively (Figure 6B). The ring-shaped mCherry signal was undetectable around the biotrophic hyphal neck of the LAC2pro:DN3:mCherry strain (Figure 6B), excluding the possibility that DN3:mCherry produced in appressorial cells before invasion was later secreted toward the interface around the hyphal neck. The NLP1pro:DN3:mCherry strain also failed to exhibit the strong ring signal of DN3:mCherry, although DN3:mCherry accumulated weakly at the interface (Figure 6B). These results indicate that the accumulation of the effector in the ring signal interface depends on the preferential expression of the effector gene during the early biotrophic stage, thereby highlighting the coupling of the transcriptional regulation of the effector with the mechanism of delivery to the ring signal interface.
Preferential Localization of SEC4 and Actin Cytoskeleton to the Biotrophic Hyphal Neck
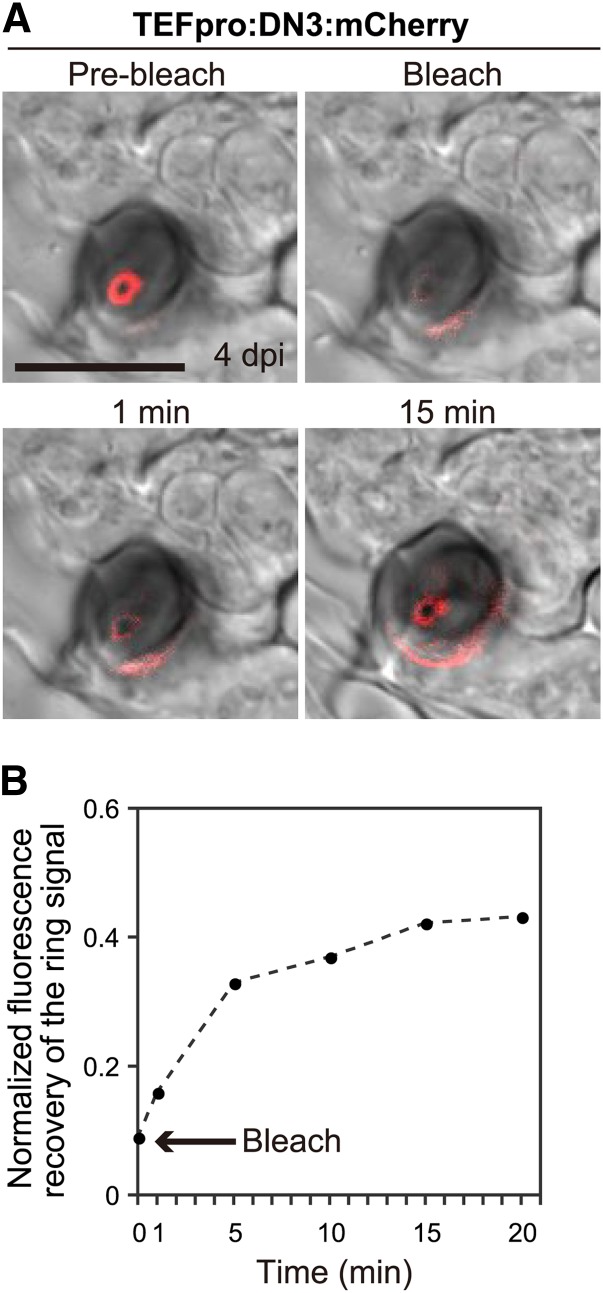
To test whether the preferential accumulation of the effector:mCherry constructs in the ring signal interface was attributable to their continuous secretion to the interface, we performed a fluorescence recovery after photobleaching (FRAP) analysis, which was directed at the ring signal interface. We selectively photobleached the ring signal, which was labeled with DN3:mCherry expressed under the control of the TEF promoter, and monitored the fluorescence recovery over time. After almost total elimination of the signal, we found that the fluorescence recovered after 5 min and this recovery was reproducible, although the fluorescence did not recover fully after further incubation (Figure 7). The samples were prepared by peeling away the surface layers from inoculated cotyledons, so we assumed that further incubation exacerbated the damage caused by peeling. This analysis suggests that the effector proteins are secreted continuously from the pathogen to the ring signal interface.
Figure 7.
Effectors Are Secreted to the Ring Signal Interface Continuously.
(A) FRAP analysis indicated the continuous secretion of DN3:mCherry to the ring signal interface. The fluorescence at the interface at 4 dpi (Pre-bleach) was photobleached (Bleach) and allowed to recover over time. Bar = 10 μm.
(B) Plot showing the normalized fluorescence intensity of the effector ring. The arrow indicates the time point when bleaching occurred. The fluorescence intensity of the prebleached effector ring was normalized to 1.0.
We next investigated whether the effectors are secreted toward the ring signal interface from the biotrophic hyphal neck beneath the appressoria. We identified the C. orbiculare orthologs of two exocytosis-related components (named SEC4 and EXO70) (Supplemental Figure 6) and fluorescently labeled these components in C. orbiculare by expressing the corresponding GFP fusions. EXO70 is a core component of the exocyst complex, and SEC4 is a Rab GTPase required for exocytosis (Guo et al., 1999; Munson and Novick, 2006). As a result, we found that GFP:SEC4 and EXO70:GFP were frequently localized at the tip of vegetative mycelia, although GFP:SEC4 signal was stronger than that of EXO70:GFP and also exhibited subapical localization (Supplemental Figure 7A). This was consistent with previous studies of these exocytosis-related components in other filamentous fungi (Jones and Sudbery, 2010; Giraldo et al., 2013), supporting the roles of SEC4 and EXO70 for exocytosis in C. orbiculare.
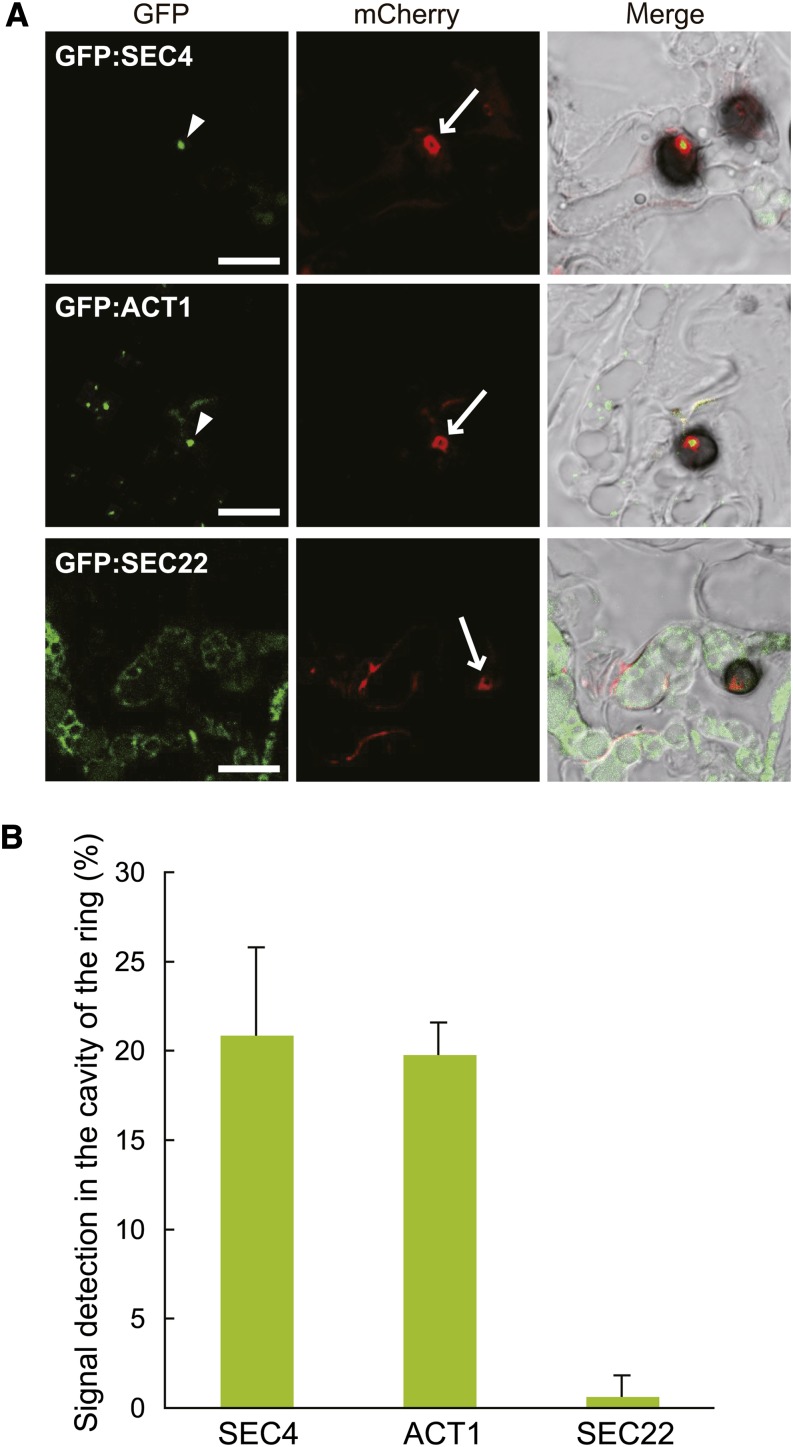
We visualized the ring signal interface and each exocytosis-related component simultaneously by generating C. orbiculare strains expressing NIS1:mCherry and each component tagged with GFP. Notably, we found that SEC4 frequently localized to the neck of the biotrophic hyphae under appressoria (corresponding to the cavity region of ring-shaped signal) (Figure 8A). The signal of EXO70:GFP, however, was hardly observed in biotrophic hyphae including the neck region (Supplemental Figure 7B), presumably due to the lower expression level of the fusion protein for in planta detection. Also, we investigated the localization of the C. orbiculare ortholog of the v-SNARE protein SEC22, which is involved with transport between the ER and the Golgi apparatus in other organisms (Jahn and Scheller, 2006) (Supplemental Figure 6). The expression of GFP:SEC22 and DN3:mCherry indicated that GFP:SEC22 did not localize to the neck region of biotrophic hyphae (Figure 8A), whereas GFP:SEC22 exhibited the canonical fluorescence pattern of the perinuclear ER network in other regions of biotrophic hyphae, which was consistent with a report from another fungus (Kuratsu et al., 2007). In the quantification analysis, the frequency of SEC4 localization to the neck of the biotrophic hyphae was 20.9% ± 4.9% whereas that of SEC22 was 2.0% ± 0.7% (Figure 8B), suggesting preferential localization of SEC4 to the biotrophic hyphal neck.
Figure 8.
SEC4 and Actin Localized to the Cavity Region of the Effector Ring.
(A) Subcellular localization of GFP:SEC4, GFP:ACT1, and GFP:SEC22 in C. orbiculare cells during the biotrophic invasive stage. C. orbiculare strains that carried combinations of the effector:mCherry gene and each GFP-tagged test gene (TEFpro:NIS1:mCherry × SCD1pro:GFP:SEC4, TEFpro:NIS1:mCherry × SCD1pro:GFP:ACT1, and TEFpro:DN3:mCherry × SCD1pro:GFP:SEC22, respectively) were inoculated onto cucumber cotyledons, and the inoculated plants were analyzed at 4 dpi. SEC4 and actin (arrowheads) preferentially localized to the cavity region of the effector ring signal (which is equivalent to the neck region of a biotrophic primary hypha). The arrows indicate the effector ring signal. Bars = 10 μm.
(B) Quantification of the GFP signal ratio in the cavity region of the effector ring. At least 50 effector ring cavities were investigated in each plant sample. The mean and sd were calculated from three independent plant samples.
The actin cytoskeleton has a crucial role for vesicle transport in fungal exocytosis (Steinberg, 2007; Berepiki et al., 2011); thus, we also investigated the intracellular localization of the actin cytoskeleton in C. orbiculare. The expression of GFP-fused ACT1, encoding an actin, demonstrated the apical localization of GFP:ACT1 in the C. orbiculare vegetative mycelia, and cytochalasin E impaired its localization (Supplemental Figures 6 and 7C) (Taheri-Talesh et al., 2008). We also detected a strong signal of GFP:ACT1 at the base of the mature appressorium before penetration (Supplemental Figure 7D), consistent with previous reports (Srinivasan et al., 1996; Dagdas et al., 2012), showing that GFP:ACT1 correctly visualized the actin cytoskeleton of C. orbiculare. We detected the preferential localization of GFP:ACT1 in the cavity of the effector ring, although GFP:ACT1 also produced a punctate pattern around the edge of the biotrophic invasive hyphae (Figure 8). These findings support the idea that the neck region of primary biotrophic hyphae, corresponding to the cavity of the ring signal, has a role as an effector secretion point focused on the interface.
Delivery of the Effectors to the Ring Signal Interface Depends on SEC4-Mediated Exocytosis
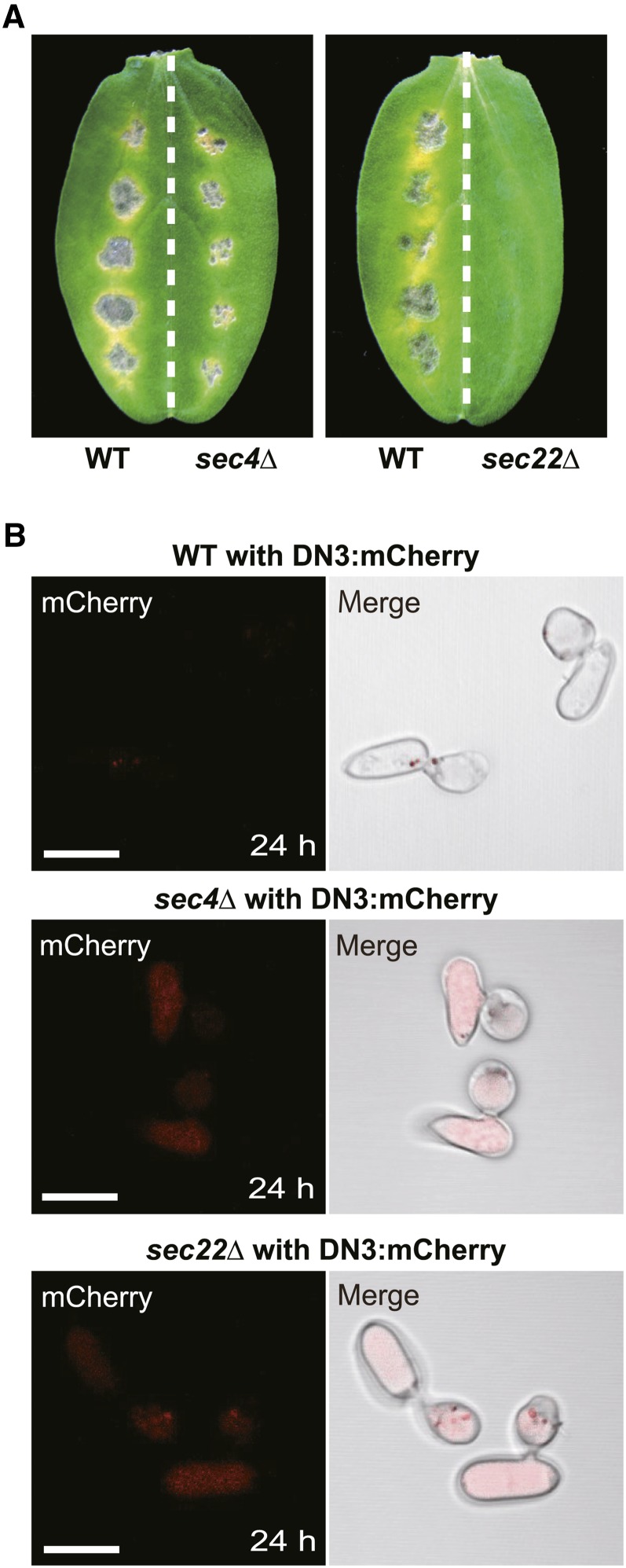
To assess the roles of Co-SEC4 during effector secretion to the interface, we investigated effector secretion in the absence of SEC4 via targeted gene disruption in C. orbiculare. We successfully isolated the sec4Δ strains of C. orbiculare, which exhibited growth retardation and reduced conidiation (Supplemental Figures 8 and 9). The sec4Δ mutants exhibited severe reduction in virulence, although the mutants developed small lesions on cucumber cotyledons (Figure 9A). However, the conidia of the sec4Δ mutants germinated normally and developed melanized appressoria. The melanized appressoria of the sec4Δ mutants penetrated cellophane (Supplemental Figure 8). These results suggest that the sec4Δ mutants retained the basic capacity for appressorial penetration but had a specific defect in appressorium-mediated host invasion.
Figure 9.
Effects of SEC4 and SEC22 Disruptions on Pathogenicity and Effector Secretion.
(A) Pathogenicity assays of the sec4 and sec22 null mutants (sec4Δ and sec22Δ) on host cucumber. The wild-type strain was inoculated onto the left halves of the cucumber cotyledons as a positive control. The sec4Δ and sec22Δ strains were inoculated onto the right halves. The inoculated plants were incubated for 7 d.
(B) Effector localization patterns of the wild-type, sec4Δ, and sec22Δ strains with TEFpro:DN3:mCherry on a glass surface. Each strain was incubated in the presence of the melanin biosynthesis inhibitor carpropamid on glass for 24 h. Bars = 10 μm.
To investigate the status of effector secretion in the absence of SEC4, we generated a sec4Δ strain that expressed DN3:mCherry under the control of the TEF promoter. After incubation for 24 h on a glass surface, the DN3:mCherry signal was barely observed in the appressoria of the wild-type reporter strains (Figure 9B). By contrast, a strong mCherry signal was detected inside the conidia and appressoria of sec4Δ background strains (Figure 9B). These findings indicate that the deletion of SEC4 resulted in the intracellular accumulation of DN3:mCherry, which suggests that SEC4-mediated exocytosis is involved with secretion of the expressed DN3:mCherry during the prepenetration stages in C. orbiculare.
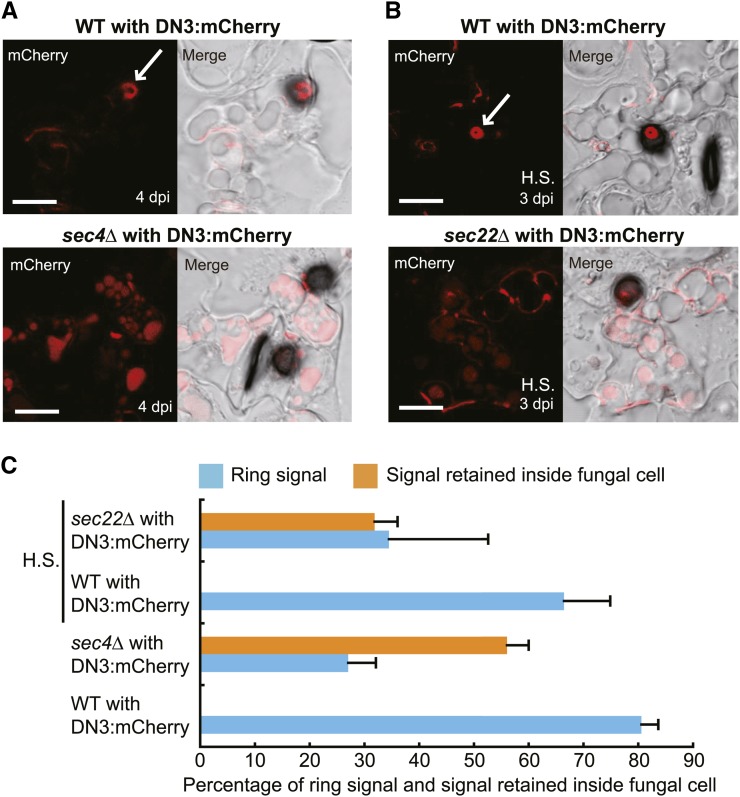
Importantly, we also found that the sec4Δ mutant had a defect in the secretion of effector to the ring signal interface. The sec4Δ reporter strain formed fewer invasive hyphae on cucumber cotyledons (22.1% ± 3.0%) compared with the wild-type reporter strain (56.8% ± 4.7%) at 4 dpi. We evaluated the presence or absence of the DN3:mCherry ring signal around the biotrophic hyphal neck and found that the sec4Δ mutant had a reduced capacity to deliver the effector to the ring signal interface (27.0% ± 5.1%) compared with the parental wild-type strains (80.6% ± 3.0%) (Figures 10A and 10C). Furthermore, the effector fluorescence of DN3:mCherry was retained inside the biotrophic hyphae of the sec4Δ strains as multiple small vesicles and vacuole-like puncta, which were not observed in the wild-type background (Figures 10A and 10C). In addition to the preferential localization of SEC4 to the biotrophic hyphal neck beneath appressoria, these findings further suggest that DN3:mCherry is secreted to the ring signal interface from the biotrophic hyphal neck via SEC4-mediated exocytosis.
Figure 10.
The sec4Δ and sec22Δ Mutants of C. orbiculare Reduced Effector Delivery to the Ring Signal Interface.
(A) Effector localization pattern during the biotrophic stage for wild-type and sec4Δ strains with TEFpro:DN3:mCherry. Each strain was incubated on cucumber for 4 d. The white arrow indicates the mCherry-labeled ring signal interface. Bars = 10 μm.
(B) Effector localization pattern during the biotrophic stage for wild-type and sec22Δ strains with TEFpro:DN3:mCherry. The cucumber cotyledons were pretreated by heat shock (H.S.) to enable the host invasion of the sec22Δ mutant. Each strain was incubated on cucumber for 3 d. Bars = 10 μm.
(C) Quantitative analysis of the effects of SEC4 and SEC22 disruptions on effector delivery to the ring signal interface. Each genotype with TEFpro:DN3:mCherry was incubated on cucumber for 4 d (no heat shock) or 3 d (heat shock). At least 74 biotrophic hyphae were investigated to determine the presence ratio of the effector ring signal. At least 50 mature biotrophic hyphae were investigated for each plant sample to determine the ratio of effector retention inside biotrophic hyphae. The mean and sd were calculated from three independent plant samples.
SEC22-Mediated Traffic Is Involved in Effector Delivery toward the Interface
We further investigated the intracellular traffic route of the effectors for delivery to the interface. SEC22 is a v-SNARE protein, which is known to be important for transport between the ER and the Golgi apparatus (Jahn and Scheller, 2006). Consistent with this, we detected the signal of GFP:SEC22 in the putative perinuclear ER of C. orbiculare cells (Figure 8A) (Kuratsu et al., 2007). To investigate the roles of ER-to-Golgi traffic for the effector delivery, we assessed the effector delivery in the absence of SEC22. The sec22Δ strains, generated by targeted gene disruption, are nonpathogenic on cucumber (Figure 9A; Supplemental Figure 8). However, the sec22Δ strains developed melanized appressoria, and appressoria of sec22Δ strains penetrated cellophane normally, which is similar to the phenotype of the sec4Δ strains (Supplemental Figure 8). Furthermore, when the sec22Δ strain expressing DN3:mCherry was incubated on glass, mCherry signals were detected inside the conidia and appressoria, consistent with the phenotype of the sec4Δ strain (Figure 9B). The effector:mCherry signal was also retained inside the penetration hyphae (pseudobiotrophic hyphae) of the mutant on cellophane (Supplemental Figure 10).
In contrast with the sec4 strains, which retain the ability to produce lesions on cucumber to some degree, we found that the sec22Δ mutants were completely defective in lesion formation (Figure 9A). The plant immunity of cucumber against C. orbiculare could be partially compromised by heat-shock pretreatment (Takano et al., 2006; Tanaka et al., 2007), although its molecular background has not been fully elucidated. When the sec22Δ mutant was inoculated on cucumber cotyledons with the heat-shock pretreatment, we found that sec22Δ mutants developed smaller lesions (Supplemental Figure 8). Thus, we inoculated the sec22Δ strain carrying TEFpro:DN3:mCherry on heat-treated cucumber cotyledons and found that appressoria of the sec22Δ strains developed biotrophic hyphae, although the formation ratio was still quite low (4.0% ± 1.3% of appressoria) at 3 dpi compared with the wild type (60.4% ± 9.3% of appressoria) in the same inoculation condition. We evaluated the Co-DN3:mCherry signal at the neck of the detected biotrophic hyphae of the sec22Δ mutant and found that the effector delivery to the ring signal interface in the mutant was lower (34.5% ± 18.1%) compared with that of the wild type (66.5% ± 8.4%) (Figures 10B and 10C). Furthermore, effector fluorescence was retained inside the biotrophic hyphae of the sec22Δ background strain, which was similar to the sec4Δ background strain (Figures 10B and 10C). These results suggest that SEC22-mediated intracellular traffic between the ER and the Golgi is involved in the effector secretion toward the ring signal interface.
DISCUSSION
The expression of effector genes changes greatly during the transition between biotrophy and necrotrophy in C. higginsianum and C. orbiculare (Kleemann et al., 2012; O’Connell et al., 2012; Gan et al., 2013), suggesting that pathogens need to secrete their effectors at the correct times to ensure successful host infection. Interestingly, there are several examples of pathogens that have developed unique interfaces where secreted effectors are localized preferentially. This suggests that pathogens need to control the secretion of effectors at the spatial level as well as at the temporal level.
In this study, we discovered that the ring-shaped signal of the effector:mCherry is located around the neck of biotrophic primary hyphae of C. orbiculare when the pathogen infects its host, cucumber. Our multifaceted cytological studies by fluorescence microscopy, including FRAP, revealed the presence of a unique biotrophic interface actively accumulating effectors that develops during C. orbiculare–host cucumber interactions. Consistent with this, TEM analysis also showed the presence of an interfacial space between the biotrophic hyphal neck and plant-derived membrane, whereas the other surface region of the biotrophic hypha was tightly surrounded by a plant-derived membrane regarded as the extrainvasive hyphal membrane (Yi and Valent, 2013). The interfacial space around the biotrophic hyphal neck possesses high-electron-density components. Taking these results together, we conclude that C. orbiculare develops a unique pathogen–plant interface that locates around the neck region of biotrophic primary hyphae beneath appressoria in host cucumber infection. Our findings further support the diversity of host–pathogen biotrophic interfaces, which was probably established via specific coevolution between pathogen and host. FM1-43 staining assays suggested that the ring signal interface is likely surrounded by concentrated regions of membrane components, probably derived from cucumber (Figure 5C). Recent work showed that the BIC of M. oryzae associates with concentrated regions of the plant plasma membrane and ER, based on analyses of transgenic rice plants that expressed a fluorescently labeled plasma membrane marker (Lti6B-GFP) or an ER marker (HDEL-GFP) (Giraldo et al., 2013).
A key question is the role of the ring signal interface during host plant infection by C. orbiculare. Because the interface develops de novo during the formation of biotrophic hyphae, it might have a role in biotrophy, such as the maintenance of the biotrophic phase until the switch to necrotrophy. In the promoter-exchange experiments with DN3 using the LAC2 and NLP1 promoters, we found that the preferential expression of DN3 during the early biotrophic phase was critical for the accumulation of DN3:mCherry in the ring signal interface. Thus, the biotrophy-expressed effectors are programmed for effective delivery to the interface, which further supports an association between the ring signal interface and biotrophy.
It has been proposed that BIC of M. oryzae provides an entrance for cytoplasmic effectors into host rice cells, based on effector translocation experiments using effector:mCherry:NLS (Khang et al., 2010). However, one M. oryzae effector, AVR1-CO39, was translocated into rice cells independently of the interfacial structure (Ribot et al., 2013). The analysis of DN3 lacking its signal peptide suggested that DN3 acts as a cytoplasmic effector (Yoshino et al., 2012) (Figure 2A). The analysis using M. oryzae transgenic lines expressing NIS1:mCherry:NLS showed that NIS1 can be delivered to the host by M. oryzae (Figure 2C). This may suggest that NIS1 acts as a cytoplasmic effector in C. orbiculare, although we do not know if similar delivery mechanisms operate in these pathogens. Also, NIS1 lacking its signal peptide (NIS1∆SP) failed to induce cell death in N. benthamiana, which might be related to the reduced accumulation of NIS1∆SP in comparison with intact NIS1 in N. benthamiana (Yoshino et al., 2012). Together with the finding that NIS1:mCherry and DN3:mCherry accumulated in the ring signal interface when expressed under the control of their own promoters, we consider that NIS1 and Co-DN3 are programmed to localize in the ring signal interface, possibly for their efficient translocation into host plant cells. However, we preliminarily found that DN3:mCherry:NLS and NIS1:mCherry:NLS expressed by C. orbiculare exhibited the ring-shaped signal around the hyphal neck, but they did not accumulate in nuclei of epidermal cells of cucumber cotyledon. We speculate that the intensity of the mCherry signal in nuclei of cucumber cells might have been below the detection limit, because their translocation efficiency to epidermal cells in cucumber cotyledon might have been lower than that to rice leaf sheath cells.
It has been reported that the apoplastic effectors BAS4, BAS113, and SLP1 of M. oryzae accumulated inside the extrainvasive hyphal matrix surrounding invasive hyphae, whereas cytoplasmic effectors such as PWL2 and BAS1 of M. oryzae highly accumulated in BIC (Mosquera et al., 2009; Khang et al., 2010; Giraldo et al., 2013). By contrast, the effector MC69 of M. oryzae (Mo-MC69) fused to mCherry:NLS preferentially accumulates in BIC, although the signal of Mo-MC69:mCherry:NLS in BIC is relatively lower than that of the tested cytoplasmic effectors (Saitoh et al., 2012). However, Mo-MC69:mCherry:NLS was not translocated to rice leaf sheath cells, whereas the tested cytoplasmic effectors fused to mCherry:NLS were translocated there (Saitoh et al., 2012). Thus, we consider that Mo-MC69 is not a cytoplasmic effector, although its detailed localization and function have yet to be elucidated.
Interestingly, we also found that the ring signal ratio and the signal intensity of Co-MC69:mCherry were significantly lower than those of DN3 and NIS1 when they were expressed under the control of their own promoters (Figure 1). The results of the promoter assay of C. orbiculare MC69 suggested that MC69 was expressed during early biotrophy in the same way as DN3 (Supplemental Figure 1). Thus, we consider that the timing of gene expression is critical for effector recruitment to the interface around the biotrophic hyphal neck, but putative cytoplasmic effectors DN3 and NIS1 might possess structural features that facilitate their preferential accumulation in the ring signal interface compared with Co-MC69, which is consistent with the previous finding of Mo-MC69 accumulation in the BIC (Saitoh et al., 2012). Supporting this idea, we also found that mCherry having the signal peptide of DN3 (SP:mCherry) did not form ring signal efficiently, in contrast with DN3:mCherry (Figures 1B to 1D). Therefore, we consider that DN3:mCherry and NIS1:mCherry preferentially accumulate in the ring signal interface around the neck of biotrophic primary hyphae, which is probably due to their efficient targeting to the ring signal interface. Alternatively, their stability might be specifically enhanced inside the interface, although the signal intensity of NIS1:mCherry and DN3:mCherry inside fungal cells is similar to that of SP:mCherry and MC69:mCherry (Supplemental Figure 3).
We detected preferential localization of SEC4 to the fungal cellular region in the cavity of the ring signal interface (corresponding to the biotrophic hyphal neck), which suggested SEC4-mediated exocytosis in this cellular region. Consistent with this idea, it was previously shown that Colletotrichum lindemuthianum SEC4, named CLPT1 (which has 100% amino acid sequence identity with C. orbiculare SEC4), restored the defect in the yeast sec4 mutant, and expression of the dominant negative form of CLPT1 blocks vesicle transport in C. lindemuthianum by preventing the interaction of the wild-type CLPT1 with a transport component required for targeting and fusion of the vesicles with the plasma membrane (Siriputthaiwan et al., 2005), thereby strongly suggesting a function of CLPT1 in exocytosis. We revealed that the sec4∆ mutants exhibited reduced effector delivery into the interface, which was accompanied by the accumulation of the effector:mCherry fluorescence inside biotrophic hyphae of the mutant. These findings suggest the SEC4-mediated exocytosis of effectors in the biotrophic hyphal neck to the interface (Figure 10). The finding that the sec4Δ strains reduced pathogenicity also suggests the importance of the SEC4-mediated exocytosis of effectors in host infection of C. orbiculare (Figure 9A). However, when the labeled effectors were overexpressed by the TEF promoter (Figure 1B), we frequently detected the effector signals around other regions of biotrophic hyphae, possibly equivalent to interfacial bodies reported in C. higginsianum (Kleemann et al., 2012), implying effector secretion from the corresponding regions of biotrophic hyphae in addition to the neck region. We also found the tiny dot signal of the effector:mCherry fluorescence at the base of C. orbiculare appressoria that did not develop biotrophic primary hyphae (Supplemental Figure 2). This finding is consistent with the previous report that effectors are focally secreted from appressorial penetration pores of C. higginsianum before host invasion (Kleemann et al., 2012). These results suggest that C. orbiculare and C. higginsianum, in part, share spatial regulatory machineries of effector proteins. However, there was no description of the ring signal of the effector:mCherry fusions in the previous studies on the infection sites of C. higginsianum on Arabidopsis (Kleemann et al., 2012); furthermore, it seems that there is no interfacial wide space at the primary hyphal neck in a reported TEM image of C. higginsianum (Kleemann et al., 2012). Therefore, we consider that the development of a biotrophic interface at the primary hyphal neck is unique to C. orbiculare and is not the case for C. higginsianum.
In this study, we also performed a functional analysis of SEC22 in C. orbiculare. It has been reported that the SEC22 ortholog of M. oryzae can restore defects in the yeast sec22 mutant (Song et al., 2010). Because Co-SEC22 exhibits striking sequence similarity to Mo-SEC22 (80% identity), it is plausible that M. oryzae SEC22 and C. orbiculare SEC22 are functional orthologs of the yeast SEC22 involved in transport between the ER and the Golgi apparatus (Jahn and Scheller, 2006). The sec22Δ mutants exhibited reductions in effector secretion toward the ring signal interface, indicating the importance of SEC22-meidated intracellular traffic for effector secretion to the interface. Yeast sec22 mutants have multiple small vesicles and accumulated ER structures, indicating the impaired fusion of ER-derived transport vesicles with the Golgi apparatus (Kaiser and Schekman, 1990). Similarly, we also found that the sec22Δ mutants accumulated multiple vesicle-like structures and vacuole-like structures labeled with DN3:mCherry inside biotrophic hyphae. Furthermore, we found that the signal of GFP:SEC22 was localized in the putative perinuclear ER of C. orbiculare cells. Collectively, these data suggest that a set of effectors including DN3 is secreted toward the interface via SEC22-dependent ER-to-Golgi traffic inside C. orbiculare cells.
The sec22Δ mutants failed to develop any lesions on cucumber cotyledons, whereas the sec4Δ mutants retained the ability to develop lesions, although lesion development was clearly reduced. In Saccharomyces cerevisiae, SEC22 functions in both anterograde and retrograde traffic between the ER and the Golgi apparatus (Kaiser and Schekman, 1990; Spang and Schekman, 1998; Liu and Barlowe, 2002; Jahn and Scheller, 2006). We thus consider that some additive defects of the sec22Δ mutants, probably not directly linked to protein secretion, may explain why the sec22 mutant exhibited a more severe phenotype than the sec4 mutant during fungal pathogenesis. It is also notable that the sec22Δ mutants did not completely lose the ability to secrete the effectors to the interface. We consider that the deletion of SEC22 does not knock out ER-to-Golgi trafficking completely in C. orbiculare, because the complete block of ER-to-Golgi trafficking is assumed to result in lethality (Liu and Barlowe, 2002). In yeast, a sec22 mutant survives because Ykt6p, which is a related R-SNARE protein that operates in later stages of the secretory pathway, is upregulated and functionally substitutes for Sec22p, whereas the sec22 ykt6 double mutant is lethal (Liu and Barlowe, 2002). Thus, we assume that other factors (possibly the ortholog of YKT6) may function during ER-to-Golgi trafficking, including effector secretion, in the absence of Co-SEC22 in C. orbiculare, although we cannot completely exclude the possibility of an unconventional Golgi-independent secretory pathway for effectors.
The conventional ER-to-Golgi secretion pathway is involved in the secretion of a set of apoplastic effectors (BAS4, BAS113, and SLP1) in M. oryzae, whereas the pathway is unlikely to be essential for the secretion of BIC-associated cytoplasmic effectors, according to assays conducted using brefeldin A (Giraldo et al., 2013). The cytoplasmic effectors, but not BAS4, require the exocyst components EXO70 and SEC5 for their efficient secretion (Giraldo et al., 2013). By contrast, SEC22 is required for the secretion of the putative cytoplasmic effector DN3 to the ring signal interface, which implies the involvement of the conventional ER-to-Golgi secretion pathway in the secretion of a set of cytoplasmic effectors to the interface that develops between C. orbiculare and cucumber. The BIC forms at the tip of filamentous primary hyphae, whereas the ring signal interface investigated in this study develops at the neck of biotrophic bulbous hyphae beneath appressoria. Thus, although the ring signal interface is considered to be functionally related to the biotrophic phase in a similar way to the Magnaporthe BIC, the features of the ring signal interface, including the effector secretion route to the interface, seem to be quite different from those of the BIC. It is necessary to study (1) the dynamics of more effectors and (2) the roles of other traffic components for further understanding of the effector delivery of C. orbiculare to the interface in host infection.
It will be interesting to investigate whether similar interfaces are developed by other fungal pathogens, especially a group of Colletotrichum species that are closely related to C. orbiculare (e.g., Colletotrichum trifolii and C. lindemuthianum). This would provide insights into evolutionary aspects of the interface establishment. We assume that effector-related biotrophic interfaces will be discovered in many plant–fungal pathogen interactions in the future, which might further support the diversity of interfaces and contribute to a deeper understanding of the nature of fungal pathogens, especially their adaptations to host plants.
METHODS
Fungal Strains, Media, Transformation, and DNA Analysis
Colletotrichum orbiculare (syn. C. lagenarium) wild-type strain 104-T (MAFF240422) is stored at the Laboratory of Plant Pathology, Kyoto University. Cultures of all fungal isolates were maintained on 3.9% (w/v) PDA medium (BD Difco) at 24°C in the dark. Magnaporthe oryzae Sasa2, used as the wild-type strain, is stored at the Iwate Biotechnology Research Center. The transformation of C. orbiculare and M. oryzae was performed based on a previous report (Kimura et al., 2001). Hygromycin-resistant transformants (for C. orbiculare) or bialaphos-resistant transformants (for C. orbiculare and M. oryzae) were selected on plates with 100 μg/mL hygromycin B (Wako Pure Chemicals) or 250 μg/mL bialaphos (Wako Pure Chemicals). Restriction enzyme digestion, cloning, plasmid isolation, and gel electrophoresis were performed according to the manufacturers’ instructions and standard methods (Sambrook et al., 1989).
Plasmid Construction
All primers used in this study are listed in Supplemental Table 1. All plasmids carrying effector genes used for fluorescence observation were derived from pBAT (Kimura et al., 2001), a derivative of pCB1531 (Sweigard et al., 1997) that carries the bialaphos resistance (bar) and chloramphenicol acetyltransferase (cat) genes, which confer bialaphos and chloramphenicol resistance, respectively. Briefly, to generate effector:mCherry constructs under the control of the corresponding native promoters, NotI-EcoRI fragments of DN3pro:DN3:mCherry, NIS1pro:NIS1:mCherry, and MC69pro:MC69:mCherry were introduced into the NotI-EcoRI site of pBAT. The joint region between the effector genes and mCherry contained an XhoI site. For constitutive and strong expression of the effector:mCherry genes with the Aureobasidium pullulans TEF promoter (Vanden Wymelenberg et al., 1997), the XbaI-EcoRI fragments of DN3:mCherry, NIS1:mCherry, and MC69:mCherry were introduced into pBATTEFP (pBAT with the TEF promoter) (Asakura et al., 2009). The joint regions of mCherry with Co-DN3, NIS1, and Co-MC69 contained XhoI, BamHI, and XhoI sites, respectively. To generate the SP:mCherry construct with the TEF promoter, the XbaI-EcoRI fragment of mCherry fused with the signal peptide sequence of Co-DN3 was amplified and introduced into the corresponding site downstream of the TEF promoter in pBATTEFP. Then, the NotI-XbaI fragment of the TEF promoter was replaced with the fragment (2.4 kb) including the entire Co-DN3 promoter sequence, creating the expression construct of SP:mCherry under the control of the Co-DN3 native promoter. The TEFpro:NIS1:GFP (enhanced GFP) construct was created by introducing the XbaI-XbaI fragment of NIS1 into pBAFPGFPTEFP (Lin et al., 2012). To express a reporter GFP under the control of the Co-DN3 or Co-NLP1 promoter, NotI-XbaI fragments (2.4 and 2.0 kb, respectively) including the entire Co-DN3 and Co-NLP1 promoter sequences were introduced into the corresponding site in pBAFPGFP (Kojima et al., 2004). To express a reporter GFP under the control of the Co-MC69 promoter, the EcoRV-HindIII fragment of GFP was introduced into the corresponding site of pBAT, resulting in pBAT-eGFP. Then, the NotI-BamHI fragment (1.4 kb) of pBAT-CoMC69pro-mCherry (Saitoh et al., 2012) including the entire Co-MC69 promoter sequence was introduced into the corresponding site in pBAT-eGFP. To generate the CoDN3:mCherry construct under the control of the LAC2 promoter, the TEF promoter sequence (NotI-XbaI) in TEFpro:DN3:mCherry was replaced with the NotI-XbaI fragment of pBAFPGFPLAC2P (Lin et al., 2012) including the entire LAC2 promoter sequence. To generate the DN3:mCherry construct under the control of the Co-NLP1 promoter, the TEF promoter sequence (NotI-XbaI) in TEFpro:DN3:mCherry was replaced with the NotI-XbaI fragment (2.0 kb) including the entire Co-NLP1 promoter sequence. GFP-tagged exocytosis-related components and actin were expressed under the control of the constitutive scytalone dehydratase1 (SCD1) minimal promoter (Takano et al., 2001). All plasmids encoding the exocytosis-related components and actin that were used for fluorescence observation were derived from pCB16GFPSTSP (Yoshino et al., 2012) or pHYPT-eGFP, which are derivatives of pCB1636 (Sweigard et al., 1997) that carry the hygromycin phosphotransferase gene to confer hygromycin resistance. To generate pHYPT-eGFP, a NotI-BamHI fragment including the short promoter region of the melanin biosynthesis gene SCD1 (SCD1pro) of pBATP (Kimura et al., 2001) was introduced into the corresponding site in pCB16GFPSTSP, before the GFP gene was introduced into its EcoRV-HindIII site. The BamHI-PstI fragment of the Co-EXO70 gene was introduced into the corresponding site in pHYPT-eGFP to create the SCD1pro:EXO70:GFP construct. The BamHI-PstI, BamHI-BamHI, and BamHI-EcoRI fragments of Co-SEC4, Co-SEC22, and ACT1 were introduced into the corresponding site in pCB16GFPSTSP to generate the SCD1pro:GFP:SEC4, SCD1pro:GFP:SEC22, and SCD1pro:GFP:ACT1 constructs, respectively. To generate pBICP35-mCherry, the BamHI-EcoRI fragment of mCherry was introduced into the corresponding site in pBICP35 (Mori et al., 1991). Then, BamHI-BamHI fragments of Co-DN3 and Co-DN3∆SP were introduced into the corresponding site of the plasmid, resulting in pBICP35-DN3:mCherry and pBICP35-DN3∆SP:mCherry, respectively. To construct pCB-Ppwl2-NIS1-mCherry-NLS for the translocation assay of Co-NIS1 into rice (Oryza sativa) cells, a 0.5-kb NIS1 cDNA fragment was amplified from 35S-NIS1-HA (Yoshino et al., 2012) with the primers XCNU1 and BCNL1. The PCR product was digested with XbaI and BamHI and introduced into pCB-Ppwl2-mCherry-NLS (Saitoh et al., 2012), which resulted in pCB-Ppwl2-NIS1-mCherry-NLS.
Chemical Treatments
The chemical treatments used in this study were as follows. To test the effects of cytochalasin E (Sigma-Aldrich), cytochalasin E was added at a final concentration of 100 μg/mL. To stain the cell wall components or callose deposition, each sample was visualized using 0.1 mM Calcofluor White M2R solution (Cosmo Bio) or 0.01% aniline blue fluorochrome solution (Biosupplies) with 0.07 M K2HPO4 after incubation for 5 or 20 min, respectively. The observations of plasma membranes on pathogen infection sites of cucumber (Cucumis sativus) cotyledons were performed in distilled water immediately after a short treatment (<10 s) with 100 μM FM1-43 (Invitrogen).
Microscopy
For the appressorium formation and penetration assays, a conidial suspension (∼5 × 105 conidia/mL 0.1% yeast extract solution) was placed on a glass slide or cellophane membrane and incubated at 24°C. The 0.1% yeast extract solution was removed and replaced with water at 15 min after incubation. If necessary, the melanin biosynthesis inhibitor carpropamid (10 μg/mL) was added. To assess the fluorescent signals in the reporter strains of C. orbiculare during infection, reporter strain conidia were inoculated on the lower surface of cucumber cotyledons at 24°C. The epidermal layers of cucumber cotyledons were peeled away and mounted in water under cover slips. To observe the vegetative hyphae, selected mycelial blocks were mounted directly in water on slides. The detection of mCherry, FM1-43, and GFP fluorescence was achieved using an Olympus FluoView FV500 confocal microscope with a Nikon 60× PlanApo (1.4 numerical aperture) oil-immersion objective. The sets of dichroic mirror, beam splitter, and emission filter used were DM488/543/633, SDM630, and BA560-600 for mCherry and FM1-43 and DM488/543, SDM560, and BA505-525 for GFP. The observations using UV light were performed with an Olympus BX53 fluorescence microscope equipped with an Olympus DP72 camera using the imaging program Olympus cellSens. FRAP analysis was achieved using an Olympus FluoView FV1200 confocal microscope with a Nikon 60× PlanApo (1.35 numerical aperture) oil-immersion objective. Dichroic mirror DM405/473/559 and a variable barrier filter (600 to 640 nm) were used. mCherry fluorescence in rice leaf sheath inoculated with M. oryzae was observed using the Olympus Fluo View FV1000-D confocal laser-scanning microscope equipped with a multi argon laser, a HeNe G laser, and a 40× UPlanSApo (0.9 numerical aperture) objective lens. To quantify the fluorescence intensity of the detected ring signal, the images were analyzed by using the software ImageJ (rsb.info.nih.gov/ij/).
Pathogenicity Assays
In the pathogenicity assay of C. orbiculare on cucumber, 10-μL conidial suspensions (5 × 105 conidia/mL) were drop-inoculated onto detached cucumber cotyledons and incubated at 24°C. To heat shock the plants, the detached cucumber cotyledons were immersed in distilled water at 50°C for 50 s. The tested strains were then inoculated onto the heat-treated cotyledons and incubated at 24°C. A rice leaf sheath inoculation test of M. oryzae was performed according to the method described previously (Namai et al., 1996).
Gene Disruptions of SEC4 and SEC22 in C. orbiculare
The primers used for gene disruptions of SEC4 and SEC22 are listed in Supplemental Table 1. To generate the SEC4 gene disruption vector, the 2.0-kb upstream region and the 2.0-kb downstream region of the SEC4 gene were amplified from the C. orbiculare genome. The amplified products were digested with BamHI and EcoRI (for the upstream region) or with ApaI and KpnI (for the downstream region) and then inserted into the 5′ site (for the upstream region) or the 3′ site (for the downstream region) of the hygromycin-resistant gene HPH in pCB1636. To generate the SEC22 gene disruption vector, the 2.0-kb upstream region and the 2.0-kb downstream region of the SEC22 gene were amplified from the C. orbiculare genome. The amplified products were digested with NotI and EcoRI (for the upstream region) or with ApaI and KpnI (for the downstream region) and then inserted into the 5′ site (for the upstream region) and the 3′ site (for the downstream region) of HPH in pCB1636. To confirm SEC4 and SEC22 gene disruptions, we performed genomic PCR analysis using two primers containing the border sequences of the SEC4 and SEC22 genes.
Agrobacterium tumefaciens–Mediated Transient Expression in Nicotiana benthamiana
N. benthamiana plants (5 to 6 weeks old) were used for the agroinfection assays. Plants were grown in a controlled environment chamber at 25°C with 16 h of illumination per day. The constructs pBICP35-mCherry, pBICP35-CoDN3:mCherry, and pBICP35-CoDN3∆SP:mCherry were transformed into Agrobacterium strain GV3101 by electroporation. The construct pSfinx-NIS1 (Yoshino et al., 2012) was transformed into Agrobacterium strain GV3101 containing pSoup. The transformant cells were cultured and harvested by centrifugation and suspended in MMA induction buffer (1 L of MMA, 5 g of Murashige and Skoog salts, 1.95 g of MES, 20 g of Suc, and 200 μM acetosyringone, pH 5.6). At 1 d after infiltration of Agrobacterium carrying pBICP35-mCherry, pBICP35-CoDN3:mCherry, or pBICP35-CoDN3∆SP:mCherry, the infiltration sites were challenged with recombinant Agrobacterium carrying pSfinx-NIS1. All suspensions were incubated for 1 h prior to infiltration. NIS1-induced lesions were observed at 5 d after the infiltration.
TEM
To observe appressorial invasion sites between C. orbiculare and cucumber interactions by TEM, conidial suspensions (5 × 106 conidia/mL) were drop-inoculated onto detached cucumber cotyledons and incubated at 24°C. After 84 h of incubation, infected specimens were fixed with 2% glutaraldehyde in 0.05 mM potassium phosphate buffer, pH 7.0, for >2 h at 4°C and postfixed in 2% OsO4 in the same buffer for 2 h at 4°C. Afterward, these were dehydrated with a graded ethanol series and replaced in propyleneoxide. The samples were embedded in epoxy resin (Quetol 651; Nisshin EM). Ultrathin sections (80 to 100 nm) were stained with 2% uranyl acetate and lead stain solution (Sigma-Aldrich). TEM observation was performed using a JEM-1200EX device (JEOL) at 80 kV. Images were processed using Adobe Illustrator CS5 and Adobe Photoshop CS2.
Accession Numbers
Sequence data produced during this study can be found in the GenBank/EMBL data libraries under accession numbers AB778550 (Co-SEC4), AB778551 (Co-EXO70), AB778552 (Co-SEC22), and AB778553 (ACT1).
Supplemental Data
The following materials are available in the online version of this article.
Supplemental Figure 1. Gene Expression of DN3 and MC69 in Biotrophic Hyphae.
Supplemental Figure 2. The DN3:mCherry Signal Was Detected Frequently as a Dot Signal at the Bases of C. orbiculare Appressoria before Invasion.
Supplemental Figure 3. Cherry Fluorescence in Preincubated Conidia of the Wild Type and Each Reporter Strain.
Supplemental Figure 4. Typical Deposition of Papillary Callose beneath the Melanized Appressorium.
Supplemental Figure 5. Amino Acid Sequence Alignment of Three NLP Homologs of C. orbiculare and Ch-NLP1 of C. higginsianum.
Supplemental Figure 6. Amino Acid Sequence Alignments of SEC4, SEC22, EXO70, and Actin Orthologs of C. orbiculare with Corresponding Orthologs of Other Organisms.
Supplemental Figure 7. Localization of Exocytosis-Related Components and Actin in Vegetative Hyphae and Appressoria.
Supplemental Figure 8. Gene Disruptions of SEC4 and SEC22 in C. orbiculare.
Supplemental Figure 9. Vegetative Growth and Conidiation of sec4∆ and sec22∆ Mutants.
Supplemental Figure 10. Retention of the Effector:mCherry Signal Inside Pseudobiotrophic Hyphae Developed in Cellophane.
Supplemental Table 1. Primers Used in This Study.
Supplementary Material
Acknowledgments
We thank John Andrews for kindly providing the plasmid pTEFEGFP and Shao Yu Lin, Eriko Oshiro, and Suthitar Singkaravanit for technical assistance. This work was supported by Grants-in-Aid for Scientific Research (Grants 23658040, 24248009, and 22780040) from the Ministry of Education, Culture, Sports, Science, and Technology of Japan; by the Programme for the Promotion of Basic and Applied Researches for Innovations in Bio-oriented Industries; and by the Science and Technology Research Promotion Program for Agriculture, Forestry, Fisheries, and Food Industry.
AUTHOR CONTRIBUTIONS
H.I. designed and performed the research, analyzed the data, and wrote the article. Y.T. designed the research, analyzed the data, and wrote the article. H.S. and R.T. designed and performed the research. H.M., K.A., A.H., and A.U. performed the research.
Glossary
- TEM
transmission electron microscopy
- BIC
biotrophic interfacial complex
- ER
endoplasmic reticulum
- dpi
days postinoculation
- FRAP
fluorescence recovery after photobleaching
Footnotes
Online version contains Web-only data.
References
- Asakura M., Ninomiya S., Sugimoto M., Oku M., Yamashita S., Okuno T., Sakai Y., Takano Y. (2009). Atg26-mediated pexophagy is required for host invasion by the plant pathogenic fungus Colletotrichum orbiculare. Plant Cell 21: 1291–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berepiki A., Lichius A., Read N.D. (2011). Actin organization and dynamics in filamentous fungi. Nat. Rev. Microbiol. 9: 876–887 [DOI] [PubMed] [Google Scholar]
- Bozkurt T.O., Schornack S., Banfield M.J., Kamoun S. (2012). Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15: 483–492 [DOI] [PubMed] [Google Scholar]
- Dagdas Y.F., Yoshino K., Dagdas G., Ryder L.S., Bielska E., Steinberg G., Talbot N.J. (2012). Septin-mediated plant cell invasion by the rice blast fungus, Magnaporthe oryzae. Science 336: 1590–1595 [DOI] [PubMed] [Google Scholar]
- Dou D., Zhou J.M. (2012). Phytopathogen effectors subverting host immunity: Different foes, similar battleground. Cell Host Microbe 12: 484–495 [DOI] [PubMed] [Google Scholar]
- Gan P., Ikeda K., Irieda H., Narusaka M., O’Connell R.J., Narusaka Y., Takano Y., Kubo Y., Shirasu K. (2013). Comparative genomic and transcriptomic analyses reveal the hemibiotrophic stage shift of Colletotrichum fungi. New Phytol. 197: 1236–1249 [DOI] [PubMed] [Google Scholar]
- Gijzen M., Nürnberger T. (2006). Nep1-like proteins from plant pathogens: Recruitment and diversification of the NPP1 domain across taxa. Phytochemistry 67: 1800–1807 [DOI] [PubMed] [Google Scholar]
- Giraldo M.C., Valent B. (2013). Filamentous plant pathogen effectors in action. Nat. Rev. Microbiol. 11: 800–814 [DOI] [PubMed] [Google Scholar]
- Giraldo M.C., Dagdas Y.F., Gupta Y.K., Mentlak T.A., Yi M., Martinez-Rocha A.L., Saitoh H., Terauchi R., Talbot N.J., Valent B. (2013). Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat. Commun. 4: 1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W., Roth D., Walch-Solimena C., Novick P. (1999). The exocyst is an effector for Sec4p, targeting secretory vesicles to sites of exocytosis. EMBO J. 18: 1071–1080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hückelhoven R. (2007). Cell wall-associated mechanisms of disease resistance and susceptibility. Annu. Rev. Phytopathol. 45: 101–127 [DOI] [PubMed] [Google Scholar]
- Jahn R., Scheller R.H. (2006). SNAREs—Engines for membrane fusion. Nat. Rev. Mol. Cell Biol. 7: 631–643 [DOI] [PubMed] [Google Scholar]
- Jones L.A., Sudbery P.E. (2010). Spitzenkorper, exocyst, and polarisome components in Candida albicans hyphae show different patterns of localization and have distinct dynamic properties. Eukaryot. Cell 9: 1455–1465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser C.A., Schekman R. (1990). Distinct sets of SEC genes govern transport vesicle formation and fusion early in the secretory pathway. Cell 61: 723–733 [DOI] [PubMed] [Google Scholar]
- Khang C.H., Berruyer R., Giraldo M.C., Kankanala P., Park S.-Y., Czymmek K., Kang S., Valent B. (2010). Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell 22: 1388–1403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura A., Takano Y., Furusawa I., Okuno T. (2001). Peroxisomal metabolic function is required for appressorium-mediated plant infection by Colletotrichum lagenarium. Plant Cell 13: 1945–1957 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleemann J., Rincon-Rivera L.J., Takahara H., Neumann U., Ver Loren van Themaat E., van der Does H.C., Hacquard S., Stüber K., Will I., Schmalenbach W., Schmelzer E., O’Connell R.J. (2012). Sequential delivery of host-induced virulence effectors by appressoria and intracellular hyphae of the phytopathogen Colletotrichum higginsianum. PLoS Pathog. 8: e1002643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima K., Takano Y., Yoshimi A., Tanaka C., Kikuchi T., Okuno T. (2004). Fungicide activity through activation of a fungal signalling pathway. Mol. Microbiol. 53: 1785–1796 [DOI] [PubMed] [Google Scholar]
- Kubo, Y., and Furusawa, I. (1991). Melanin biosynthesis: Prerequisite for successful invasion of the plant host by appressoria of Colletotrichum and Pyricularia In The Fungal Spore and Disease Initiation in Plants and Animals, G.T. Cole and H.C. Hoch, eds (New York: Plenum Publishing), pp. 205–217. [Google Scholar]
- Kubo Y., Takano Y. (2013). Dynamics of infection-related morphogenesis and pathogenesis in Colletotrichum orbiculare. J. Gen. Plant Pathol. 79: 233–242 [Google Scholar]
- Kuratsu M., Taura A., Shoji J.Y., Kikuchi S., Arioka M., Kitamoto K. (2007). Systematic analysis of SNARE localization in the filamentous fungus Aspergillus oryzae. Fungal Genet. Biol. 44: 1310–1323 [DOI] [PubMed] [Google Scholar]
- Latgé J.P. (2007). The cell wall: A carbohydrate armour for the fungal cell. Mol. Microbiol. 66: 279–290 [DOI] [PubMed] [Google Scholar]
- Lin S.Y., Okuda S., Ikeda K., Okuno T., Takano Y. (2012). LAC2 encoding a secreted laccase is involved in appressorial melanization and conidial pigmentation in Colletotrichum orbiculare. Mol. Plant Microbe Interact. 25: 1552–1561 [DOI] [PubMed] [Google Scholar]
- Liu Y., Barlowe C. (2002). Analysis of Sec22p in endoplasmic reticulum/Golgi transport reveals cellular redundancy in SNARE protein function. Mol. Biol. Cell 13: 3314–3324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Micali C.O., Neumann U., Grunewald D., Panstruga R., O’Connell R.J. (2011). Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell. Microbiol. 13: 210–226 [DOI] [PubMed] [Google Scholar]
- Mori M., Mise K., Kobayashi K., Okuno T., Furusawa I. (1991). Infectivity of plasmids containing brome mosaic virus cDNA linked to the cauliflower mosaic virus 35S RNA promoter. J. Gen. Virol. 72: 243–246 [DOI] [PubMed] [Google Scholar]
- Mosquera G., Giraldo M.C., Khang C.H., Coughlan S., Valent B. (2009). Interaction transcriptome analysis identifies Magnaporthe oryzae BAS1-4 as biotrophy-associated secreted proteins in rice blast disease. Plant Cell 21: 1273–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munson M., Novick P. (2006). The exocyst defrocked, a framework of rods revealed. Nat. Struct. Mol. Biol. 13: 577–581 [DOI] [PubMed] [Google Scholar]
- Namai T., Nukina M., Togashi J. (1996). Effects of two toxins and a derivative of one toxin produced by rice blast fungus on its infection to inner epidermal tissue of rice leaf sheath. Ann. Phytopathological Soc. Jpn. 62: 114–118 [Google Scholar]
- O’Connell R.J., et al. (2012). Lifestyle transitions in plant pathogenic Colletotrichum fungi deciphered by genome and transcriptome analyses. Nat. Genet. 44: 1060–1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect S.E., Hughes H.B., O’Connell R.J., Green J.R. (1999). Colletotrichum: A model genus for studies on pathology and fungal-plant interactions. Fungal Genet. Biol. 27: 186–198 [DOI] [PubMed] [Google Scholar]
- Rafiqi M., Ellis J.G., Ludowici V.A., Hardham A.R., Dodds P.N. (2012). Challenges and progress towards understanding the role of effectors in plant-fungal interactions. Curr. Opin. Plant Biol. 15: 477–482 [DOI] [PubMed] [Google Scholar]
- Ribot C., Césari S., Abidi I., Chalvon V., Bournaud C., Vallet J., Lebrun M.-H., Morel J.-B., Kroj T. (2013). The Magnaporthe oryzae effector AVR1-CO39 is translocated into rice cells independently of a fungal-derived machinery. Plant J. 74: 1–12 [DOI] [PubMed] [Google Scholar]
- Saitoh H., et al. (2012). Large-scale gene disruption in Magnaporthe oryzae identifies MC69, a secreted protein required for infection by monocot and dicot fungal pathogens. PLoS Pathog. 8: e1002711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook, J., Fritsch, E.F., and Maniatis, T. (1989). Molecular Cloning: A Laboratory Manual, 2nd ed. (Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press). [Google Scholar]
- Siriputthaiwan P., Jauneau A., Herbert C., Garcin D., Dumas B. (2005). Functional analysis of CLPT1, a Rab/GTPase required for protein secretion and pathogenesis in the plant fungal pathogen Colletotrichum lindemuthianum. J. Cell Sci. 118: 323–329 [DOI] [PubMed] [Google Scholar]
- Song W., Dou X., Qi Z., Wang Q., Zhang X., Zhang H., Guo M., Dong S., Zhang Z., Wang P., Zheng X. (2010). R-SNARE homolog MoSec22 is required for conidiogenesis, cell wall integrity, and pathogenesis of Magnaporthe oryzae. PLoS ONE 5: e13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spang A., Schekman R. (1998). Reconstitution of retrograde transport from the Golgi to the ER in vitro. J. Cell Biol. 143: 589–599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan S., Vargas M.M., Roberson R.W. (1996). Functional, organizational, and biochemical analysis of actin in hyphal tip cells of Allomyces macrogynus. Mycologia 88: 57–70 [Google Scholar]
- Steinberg G. (2007). Hyphal growth: A tale of motors, lipids, and the Spitzenkörper. Eukaryot. Cell 6: 351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stephenson S.A., Hatfield J., Rusu A.G., Maclean D.J., Manners J.M. (2000). CgDN3: An essential pathogenicity gene of Colletotrichum gloeosporioides necessary to avert a hypersensitive-like response in the host Stylosanthes guianensis. Mol. Plant Microbe Interact. 13: 929–941 [DOI] [PubMed] [Google Scholar]
- Sweigard J., Chumley F., Carroll A., Farrall L., Valent B. (1997). A series of vectors for fungal transformation. Fungal Genet. Newsl. 44: 52–55 [Google Scholar]
- Taheri-Talesh N., Horio T., Araujo-Bazán L., Dou X., Espeso E.A., Peñalva M.A., Osmani S.A., Oakley B.R. (2008). The tip growth apparatus of Aspergillus nidulans. Mol. Biol. Cell 19: 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takano Y., Oshiro E., Okuno T. (2001). Microtubule dynamics during infection-related morphogenesis of Colletotrichum lagenarium. Fungal Genet. Biol. 34: 107–121 [DOI] [PubMed] [Google Scholar]
- Takano Y., Takayanagi N., Hori H., Ikeuchi Y., Suzuki T., Kimura A., Okuno T. (2006). A gene involved in modifying transfer RNA is required for fungal pathogenicity and stress tolerance of Colletotrichum lagenarium. Mol. Microbiol. 60: 81–92 [DOI] [PubMed] [Google Scholar]
- Tanaka S., Yamada K., Yabumoto K., Fujii S., Huser A., Tsuji G., Koga H., Dohi K., Mori M., Shiraishi T., O’Connell R., Kubo Y. (2007). Saccharomyces cerevisiae SSD1 orthologues are essential for host infection by the ascomycete plant pathogens Colletotrichum lagenarium and Magnaporthe grisea. Mol. Microbiol. 64: 1332–1349 [DOI] [PubMed] [Google Scholar]
- Vanden Wymelenberg A.J., Cullen D., Spear R.N., Schoenike B., Andrews J.H. (1997). Expression of green fluorescent protein in Aureobasidium pullulans and quantification of the fungus on leaf surfaces. Biotechniques 23: 686–690 [DOI] [PubMed] [Google Scholar]
- Yi M., Valent B. (2013). Communication between filamentous pathogens and plants at the biotrophic interface. Annu. Rev. Phytopathol. 51: 587–611 [DOI] [PubMed] [Google Scholar]
- Yoshino K., Irieda H., Sugimoto F., Yoshioka H., Okuno T., Takano Y. (2012). Cell death of Nicotiana benthamiana is induced by secreted protein NIS1 of Colletotrichum orbiculare and is suppressed by a homologue of CgDN3. Mol. Plant Microbe Interact. 25: 625–636 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.