Abstract
Promoter DNA methylation may reflect the interaction between genetic backgrounds and environmental factors in the development of metabolic disorders, including type 2 diabetes (T2D). Calcium/calmodulin-dependent protein kinase 1D (CAMK1D), cryptochrome 2 (CRY2) and calmodulin 2 (CALM2) genes have been identified to be associated with a risk of T2D. Therefore, the aim of the present study was to investigate the contribution of promoter DNA methylation of these genes to the risk of T2D. Using bisulfite pyrosequencing technology, the DNA methylation levels of the CpG dinucleotides within the CAMK1D, CRY2 and CALM2 gene promoters were measured in 48 patients with T2D and 48 age- and gender-matched healthy controls. The results demonstrated that the promoters of these three genes were hypomethylated in the peripheral blood of all the subjects, and DNA methylation of these three genes did not contribute to the risk of T2D.
Keywords: type 2 diabetes, calcium/calmodulin-dependent protein kinase 1D, cryptochrome 2, calmodulin 2, DNA methylation, promoter
Introduction
Type 2 diabetes (T2D) is a major public health problem in numerous countries (1,2). Genetic and environmental factors are hypothesized to contribute to the initiation and development of the disease (3). Genetic association studies based on genome-wide association and meta-analyses have revealed a number of susceptible genetic polymorphisms associated with the pathogenesis of T2D (2,4–7). However, a number of these polymorphisms are not functional variants and are most likely cotransmitted with the causative polymorphisms in large linkage disequilibrium blocks, and do not directly control the expression of T2D susceptibility genes (8). Evaluating the impact of these genetic-environmental interactions and predicting the personal onset risk of developing T2D remains a challenge.
Epigenetic regulation is sensitive to a number of environmental factors and may provide a bridge between the environment and genetic background (1). As a crucial mechanism in epigenetic regulation, DNA methylation regulates gene expression by transferring a methyl group to cytosine nucleotides via DNA methyltransferase (1). Unlike genetic polymorphisms, DNA methylation levels may alter during numerous processes, including development, tissue differentiation and aging (1). In a monozygotic twins study, the older twins showed more distinct changes according to DNA methylation profiles in the epigenome (9,10). Environmental factors, including nutrition and lifestyle, may influence DNA methylation in mammals. For example, the promoter methylation of certain genes was increased in human primary muscle cells due to the exposure of the free fatty acids palmitate (10). A series of genes in primary metabolic processes were reported to show a differential methylation status in skeletal muscle in patients with T2D compared with normal glucose-tolerant individuals (11). Furthermore, dysregulation of promoter methylation has been found in pancreatic islets from patients with T2D (1). Elucidating the association between promoter methylation and the pathogenesis of T2D is a promising field in the research of metabolic disorders (1,10).
The calcium/calmodulin-dependent protein kinase 1D (CAMK1D) gene encodes a member of the Ca2+/calmodulin-dependent protein kinase 1 subfamily of serine/threonine kinases that plays an important role in the regulation of granulocyte function via the chemokine signaling transduction pathway (12,13). Single nucleotide polymorphisms (SNPs) in CAMK1D have been found to be associated with the susceptibility of developing T2D in an east Asian population (14). However, no significant association between the CAMK1D SNP and T2D was found in a study based on European populations (15). Thus, the role of the CAMK1D gene in the pathophysiology of T2D remains unclear (13). The calmodulin 2 (CALM2) gene encodes a member of the human calmodulin family, and polymorphisms in this gene have been associated with dialysis survival in T2D-associated renal disease (16). The cryptochrome 2 (CRY2) gene is a circadian signal gene, whose genetic variation has been associated with T2D and metabolic characteristics (17,18). A recent study investigating monozygotic twins found that the methylation status of CRY2 in subcutaneous adipose tissue differed between the twin with T2D and the healthy twin (18). Further investigations into the association between promoter methylation of the CRY2 gene and T2D in the peripheral blood should be conducted.
Aberrant gene methylation has been identified not only in diseased tissues, but also in human peripheral tissue, including blood lymph cells (8). Detecting the promoter methylation levels of T2D-associated genes in the peripheral blood may be beneficial for the identification of novel diabetic biomarkers with preventative and/or diagnostic values. In the present study, the methylation levels of three candidate genes (CAMK1D, CRY2 and CALM2) in patients with T2D and non-diabetic individuals were investigated to determine the value of this epigenetic marker, with the aim of providing further understanding into the disease etiology of T2D.
Materials and methods
Subjects
A total of 48 patients with T2D and 48 age- and gender-matched healthy controls were recruited from the Affiliated Hospital of Ningbo University (Ningbo, China). The characteristics of the individuals are shown in Table I. All the individuals were of Han Chinese origin and had lived in the Ningbo area for at least three generations. Patients with T2D were recruited if their plasma glucose levels were >7.0 mmol/l at fasting or >11.1 mmol/l at 2 h following glucose load (World Health Organization) (19). Healthy individuals were recruited according to the standard of fasting blood glucose of <6.1 mmol/l. None of the controls had a family history of T2D in first-degree relatives or had received any medication. Subjects were excluded from the study if they had hypertension, coronary heart disease, renal inadequacy, drug abuse or any other serious diseases. The study was approved by the Ethics Committee of Ningbo University and written informed consent was obtained from all the subjects. Blood samples were collected in 3.2% citrate sodium-treated tubes and stored at −80°C for DNA extraction.
Table I.
Characteristics of the subjects (n=96).
| Characteristics | Value | Range | T2D | Controls | P-value |
|---|---|---|---|---|---|
| Age (years) | 59.2±7.5 | 35–69 | 59.2±7.5 | 59.2±7.5 | 1.000 |
| Gender (M/F) | 48/48 | - | - | - | - |
| BMI (kg/m2) | 23.71±3.28 | 17.15–42.96 | 24.17±4.18 | 23.18±1.64 | 0.146 |
| Total cholesterol (mmol/l) | 5.19±0.96 | 2.95–7.90 | 5.34±0.83 | 5.05±1.06 | 0.140 |
| Total triglycerides (mmol/l) | 1.60±1.36 | 0.40–9.92 | 1.90±1.69 | 1.31±0.82 | 0.034 |
| Glucose (mmol/l) | 6.76±2.65 | 4.38–22.84 | 8.31±2.91 | 5.22±0.92 | 0.000 |
| ALT (IU/l) | 21.5±15.9 | 5.0–99.0 | 25.1±18.5 | 18.0±12.1 | 0.028 |
| Uric acid (μmol/l) | 294.9±81.1 | 132.0–531.0 | 289.3±70.5 | 300.6±90.9 | 0.499 |
| CALM2 methylation (%) | |||||
| CpG1 | 0.96±0.38 | 0–2 | 1.02±0.40 | 0.88±0.33 | 0.099 |
| CpG2 | 2.01±0.50 | 0–3 | 2.18±0.39 | 1.79±0.55 | 0.001 |
| CpG3 | 0.92±0.48 | 0–2 | 1.00±0.53 | 0.82±0.39 | 0.087 |
| CpG4 | 1.40±0.86 | 0–5 | 1.11±0.58 | 1.79±1.02 | 0.001 |
| CRY2 methylation (%) | |||||
| CpG1 | 1.25±0.53 | 0–2 | 1.13±0.41 | 1.44±0.65 | 0.022 |
| CpG2 | 0.80±0.41 | 0–1 | 0.79±0.41 | 0.80±0.41 | 0.961 |
| CpG3 | 1.61±0.63 | 0–3 | 1.64±0.63 | 1.56±0.65 | 0.621 |
| CpG4 | 0.98±0.42 | 0–3 | 1.05±0.39 | 0.88±0.44 | 0.110 |
| CpG5 | 1.14±3.32 | 0–27 | 0.64±0.54 | 1.92±5.24 | 0.134 |
| CAMK1D methylation (%) | |||||
| CpG1 | 1.04±0.29 | 0–3 | 1.04±0.20 | 1.04±0.36 | 1.000 |
| CpG2 | 0.75±0.48 | 0–2 | 0.90±0.31 | 0.60±0.57 | 0.003 |
| CpG3 | 1.53±0.54 | 1–3 | 1.75±0.48 | 1.31±0.51 | 0.000 |
| CpG4 | 1.04±0.29 | 0–2 | 1.10±0.31 | 0.98±0.25 | 0.032 |
| CpG5 | 1.02±0.20 | 0–2 | 1.04±0.20 | 1.00±0.21 | 0.320 |
| CpG6 | 0.96±0.32 | 0–2 | 1.02±0.14 | 0.90±0.43 | 0.057 |
| CpG7 | 1.04±0.29 | 0–2 | 1.06±0.38 | 1.02±0.14 | 0.480 |
| CpG8 | 0.92±0.45 | 0–3 | 1.02±0.25 | 0.81±0.57 | 0.023 |
| CpG9 | 0.82±0.54 | 0–3 | 1.00±0.41 | 0.65±0.60 | 0.001 |
Results are expressed as the mean ± standard error.
n=86 (46 patients with T2D vs. 40 control patients).
T2D, type 2 diabetes; BMI, body mass index; ALT, alanine aminotransferase; CALM2, calmodulin 2; CRY2, cryptochrome 2; CAMK1D, calcium/calmodulin-dependent protein kinase 1D; M, male; F, female.
Phenotype collection
Blood samples were collected from the antecubital vein into vacutainer tubes containing ethylene diamine tetraacetic acid following fasting for 12 h overnight. Plasma levels of cholesterol, triglyceride, alanine aminotransferase (ALT), uric acid and glucose concentrations were enzymatically measured using the CX7 Analyzer (Beckman Coulter, Inc., Fullerton, CA, USA).
DNA methylation assay
Human genomic DNA was isolated from peripheral blood samples using a nucleic acid extraction automatic analyzer (Lab-Aid 820; Xiamen, Fujian, China). DNA was quantified using the PicoGreen double strand DNA Quantification kit (Molecular Probes, Inc., Eugene, OR, USA). Bisulfite pyrosequencing technology was used to determine the CpG dinucleotide methylation levels of fragments within the promoters of the CAMK1D, CRY2 and CALM2 genes (Fig. 1). The pyrosequencing assays involved sodium bisulfite DNA conversion chemistry, polymerase chain reaction (PCR) amplification and sequencing by synthesis assay of the target sequence. Sodium bisulfite preferentially deaminates unmethylated cytosine residues to thymines (following PCR amplification), while methyl-cytosines remain unmodified. PCR primers were selected using PyroMark Assay Design software v2.0.1.15 and the amplification primers for the CAMK1D, CRY2 and CALM2 gene promoters are shown in Table II.
Figure 1.
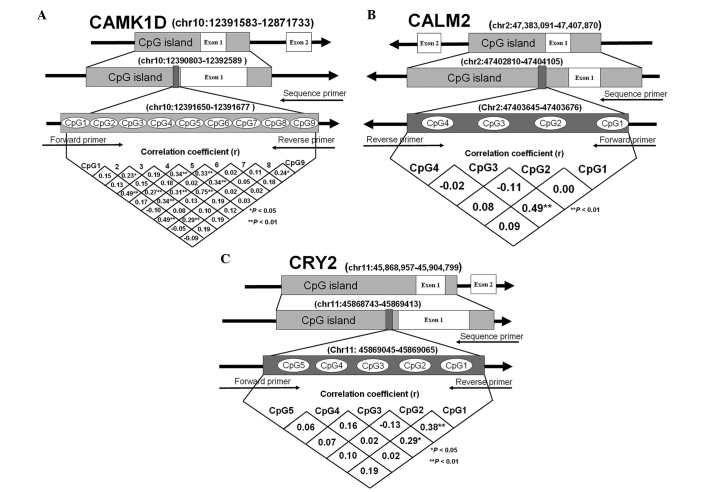
CpG islands within the CAMK1D, CRY2 and CALM2 promoters. The diagrams indicate the correlation coefficients, and (A) nine CpG dinucleotides within CAMK1D, (B) four CpG dinucleotides within CALM2 and (C) five CpG dinucleotides within CRY2. *P<0.05, **P<0.01. P-values were calculated by Pearson test which analyzed the correlation of the methylation levels among the tested CpGs. CALM2, calmodulin 2; CRY2, cryptochrome 2; CAMK1D, calcium/calmodulin-dependent protein kinase 1D.
Table II.
Primers for the promoter CpG methylation analysis.
| Gene | Sequence |
|---|---|
| CALM2 | |
| Forward primer | 5′-AGGAGGAGTTGTTGGAGAATATGA-3′ |
| Reverse primer | 5′-biotin- ACTACCCCCCTAACCCCTCT-3′ |
| Sequencing primer | 5′-GTTTTGAGTGTTTAGGTAAGG-3′ |
| CRY2 | |
| Forward primer | 5′-GGGGTGGTTGGAGTAGTTTGG-3′ |
| Reverse primer | 5′-biotin-AATCCCCTCACCTCCATC-3′ |
| Sequencing primer | 5′-GGAGTAGTTTGGATAGTTA-3′ |
| CAMK1D | |
| Forward primer | 5′-GGAGGTAAGAAAGTAGTAGAAAGTGA-3′ |
| Reverse primer | 5′-biotin-CCTCCTCTACAATTTCCTCTT-3′ |
| Sequencing primer | 5′-GAGTTAGGGAGGGAT-3′ |
CALM2, calmodulin 2; CRY2, cryptochrome 2; CAMK1D, calcium/calmodulin-dependent protein kinase 1D.
Statistical analysis
Pearson’s χ2 test was used to compare the categorical variables, while mean group differences for continuous variables were compared using the Student’s t-test. Pearson’s correlation analysis was applied to determine the associations between the methylation status and metabolic features of the subjects. P<0.05 was considered to indicate a statistically significant difference. All statistical analyses were performed using PASW Statistics 18.0 software (SPSS, Inc., Chicago, IL, USA).
Results
Low methylation levels within the CAMK1D, CRY2 and CALM2 promoters
A total of 48 patients with T2D and 48 age- and gender-matched controls were recruited for the association study, and the methylation levels of three genes (CAMK1D, CRY2 and CALM2) were investigated. In total, four CpG dinucleotides within the CALM2 gene promoter, five CpG dinucleotides within the CRY2 gene promoter and nine CpG dinucleotides within the CAMK1D gene promoter were identified. The correlations between the DNA methylation levels among all the CpGs are shown in Fig. 1 and the characteristics of the subjects are shown in Table I. As shown in Table I, the promoters of the three genes in the peripheral blood exhibited low methylation levels for all the subjects. The DNA methylation information of all the CpGs within the CAMK1D, CRY2 and CALM2 promoters is shown in Fig. 2.
Figure 2.
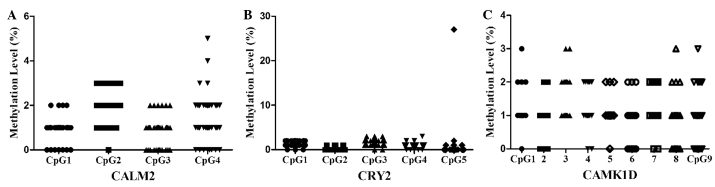
DNA methylation of all the CpGs within the CAMK1D, CRY2 and CALM2 promoters. Methylation levels of each dinucleotide of the (A) four CpGs within CALM2, (B) five CpGs within CRY2 and (C) nine CpGs within CAMK1D. CALM2, calmodulin 2; CRY2, Cryptochrome 2; CAMK1D, calcium/calmodulin-dependent protein kinase 1D.
Statistical analysis of clinical phenotypes
No statistically significant differences in clinical phenotypes, including age, body mass index, total cholesterol and uric acid, were observed between the T2D and controls patients (Table I; P>0.05). However, the glucose level in patients with T2D was significantly higher compared with the control patients (Table I; P<0.001), and a similar result was observed in the breakdown analysis by gender. The levels of ALT and total triglycerides were higher in patients with T2D (Table I; P=0.028 and P=0.034, respectively). However, in the breakdown analysis by gender, no differential level of total triglycerides was observed. Notably, higher levels of ALT in female patients with T2D were observed (Table III; P=0.019).
Table III.
Characteristics of the subjects by gender.
| Characteristics | T2D | Controls | P-value |
|---|---|---|---|
| Males (n=48) | |||
| Age (years) | 59.1±8.7 | 59.1±8.7 | |
| BMI (kg/m2)a | 24.92±5.17 | 23.10±1.21 | 0.124 |
| Total cholesterol (mmol/l) | 5.06±0.74 | 4.86±1.11 | 0.464 |
| Total triglycerides (mmol/l) | 1.81±1.56 | 1.38±0.90 | 0.254 |
| Glucose (mmol/l) | 8.59±3.49 | 4.94±0.34 | 0.000 |
| ALT (IU/l) | 30.4±23.8 | 21.1±15.9 | 0.119 |
| Uric acid (μmol/l) | 304.7±70.6 | 346.5±82.7 | 0.066 |
| Females (n=48) | |||
| Age (years) | 59.4±6.4 | 59.4±6.4 | |
| BMI (kg/m2)b | 23.49±2.97 | 23.25±1.93 | 0.747 |
| Total cholesterol (mmol/l) | 5.62±0.83 | 5.24±1.00 | 0.161 |
| Total triglycerides (mmol/l) | 1.98±1.85 | 1.23±0.75 | 0.071 |
| Glucose (mmol/l) | 8.04±2.22 | 5.49±1.20 | 0.000 |
| ALT (IU/l) | 19.8±8.6 | 14.8±4.8 | 0.019 |
| Uric acid (μmol/l) | 273.9±68.3 | 254.6±75.0 | 0.357 |
n=39 (22 patients with T2D vs. 17 control patients);
n=47 (24 patients with T2D vs. 23 control patients).
T2D, type 2 diabetes; BMI, body mass index; ALT, alanine aminotransferase; SE, standard error.
Discussion
Epigenetic regulation is involved in numerous types of human diseases, and epigenetic biomarkers have been reported to be associated with pathological processes, of which a number have potential value as laboratory diagnostic tools (20–23). DNA methylation, as one of the most studied epigenetic regulatory mechanisms, is essential in the interaction between genetic and environmental factors (10,20). DNA methylation usually occurs in the CpG islands of functional gene promoter regions (20), and this type of epigenetic modification was originally considered to be stable in vitro and function as a tissue-specific feature in disease (10,24,25). A number of disease-associated DNA methylation variations have been identified in a diverse range of tissues, including the peripheral blood (8,10,20,26).
T2D is a major metabolic disease, which physiological and pathological states may be regulated by epigenetic modifications that control gene expression (10). Hundreds of differential DNA methylation CpGs of genes promoters have been identified in genomic DNA isolated from pancreatic islets using Infinium methylation assay technology (1). These disease-specific methylation signatures also exist in human accessible peripheral tissues (8). A prospective study found that significant hypomethylation of young individuals was associated with a higher risk of developing T2D in later life (8). In the present study, the DNA promoter methylation levels of three T2D candidate genes (CALM2, CRY2 and CAMK1D) in the peripheral blood were investigated, providing further information regarding DNA promoter methylation regulation in T2D.
Low levels of DNA promoter methylation were identified in the three genes in the healthy subjects and patients with T2D (Table I). The majority of the methylation correlation coefficients of the CpGs within these genes were also not significant (Fig. 1). Two reasons may explain these observations. In epigenetics, not all candidate genes are directly regulated by DNA methylation, with other epigenetic factors, including non-coding RNA and histone variants, also involved in the regulation of gene expression (20,23). Since T2D is a polygenic disease, the low DNA promoter methylation levels of CALM2, CRY2 and CAMK1D genes may indicate that DNA promoter methylation has no direct effect on gene function. In addition, the methylation status was only investigated in the peripheral blood, and DNA promoter methylation modification has been demonstrated to exhibit a tissue-specific methylation pattern (25). The insulin-2 gene, an additional candidate gene for diabetes, was found to be unmethylated in β cells, but methylated in other tissues (25). The identification of DNA methylation profiling in pancreatic islets revealed that differences in the methylation status of certain candidate genes between T2D and non-diabetic patients were not present in blood cells (1). The two aforementioned reasons may explain the low DNA methylation levels of CALM2, CRY2 and CAMK1D observed in the present study.
In conclusion, low methylation levels of CALM2, CRY2 and CAMK1D were observed in the peripheral blood of the healthy controls and T2D patients. These observations indicate that DNA methylation may not be the primary regulatory mechanism of CALM2, CRY2 and CAMK1D in T2D, and these methylation loci may not be regarded as biomarkers for T2D. The present study provides further information with regard to the epigenetic mechanisms and may provide reference value in genetic-based pharmacological development for future T2D treatments.
Acknowledgements
The study was supported by grants from the National Natural Science Foundation of China (nos. 31100919 and 81371469), the Natural Science Foundation of Zhejiang Province (no. LR13H020003) and the K.C. Wong Magna Fund in Ningbo University, Ningbo Social Development Research Projects (no. 2012C50032).
Abbreviations
- T2D
type 2 diabetes
- CAMK1D
calcium/calmodulin-dependent protein kinase 1D
- CRY2
cryptochrome 2
- CALM2
calmodulin 2
- CI
confidence interval
- ALT
alanine aminotransferase
- SNP
single nucleotide polymorphism
- PCR
polymerase chain reaction
References
- 1.Volkmar M, Dedeurwaerder S, Cunha DA, et al. DNA methylation profiling identifies epigenetic dysregulation in pancreatic islets from type 2 diabetic patients. EMBO J. 2012;31:1405–1426. doi: 10.1038/emboj.2011.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Omori S, Tanaka Y, Takahashi A, et al. Association of CDKAL1, IGF2BP2, CDKN2A/B, HHEX, SLC30A8, and KCNJ11 with susceptibility to type 2 diabetes in a Japanese population. Diabetes. 2008;57:791–795. doi: 10.2337/db07-0979. [DOI] [PubMed] [Google Scholar]
- 3.Gu T, Horová E, Möllsten A, et al. IGF2BP2 and IGF2 genetic effects in diabetes and diabetic nephropathy. J Diabetes Complications. 2012;26:393–398. doi: 10.1016/j.jdiacomp.2012.05.012. [DOI] [PubMed] [Google Scholar]
- 4.Kwak SH, Kim SH, Cho YM, et al. A genome-wide association study of gestational diabetes mellitus in Korean women. Diabetes. 2012;61:531–541. doi: 10.2337/db11-1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Grarup N, Rose CS, Andersson EA, et al. Studies of association of variants near the HHEX, CDKN2A/B, and IGF2BP2 genes with type 2 diabetes and impaired insulin release in 10,705 Danish subjects: validation and extension of genome-wide association studies. Diabetes. 2007;56:3105–3111. doi: 10.2337/db07-0856. [DOI] [PubMed] [Google Scholar]
- 6.Jia H, Yu L, Jiang Z, Ji Q. Association between IGF2BP2 rs4402960 polymorphism and risk of type 2 diabetes mellitus: a meta-analysis. Arch Med Res. 2011;42:361–367. doi: 10.1016/j.arcmed.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 7.Wu J, Wu J, Zhou Y, et al. Quantitative assessment of the variation in IGF2BP2 gene and type 2 diabetes risk. Acta Diabetol. 2012;49(Suppl 1):S87–S97. doi: 10.1007/s00592-011-0336-3. [DOI] [PubMed] [Google Scholar]
- 8.Toperoff G, Aran D, Kark JD, et al. Genome-wide survey reveals predisposing diabetes type 2-related DNA methylation variations in human peripheral blood. Hum Mol Genet. 2012;21:371–383. doi: 10.1093/hmg/ddr472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fraga MF, Ballestar E, Paz MF, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc Natl Acad Sci USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Barres R, Zierath JR. DNA methylation in metabolic disorders. Am J Clin Nutr. 2011;93:897S–900S. doi: 10.3945/ajcn.110.001933. [DOI] [PubMed] [Google Scholar]
- 11.Barrès R, Osler ME, Yan J, et al. Non-CpG methylation of the PGC-1alpha promoter through DNMT3B controls mitochondrial density. Cell Metab. 2009;10:189–198. doi: 10.1016/j.cmet.2009.07.011. [DOI] [PubMed] [Google Scholar]
- 12.Verploegen S, Ulfman L, van Deutekom HW, et al. Characterization of the role of CaMKI-like kinase (CKLiK) in human granulocyte function. Blood. 2005;106:1076–1083. doi: 10.1182/blood-2004-09-3755. [DOI] [PubMed] [Google Scholar]
- 13.Shu XO, Long J, Cai Q, et al. Identification of new genetic risk variants for type 2 diabetes. PLoS Genet. 2010;6:e1001127. doi: 10.1371/journal.pgen.1001127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Imamura M, Iwata M, Maegawa H, et al. Genetic variants at CDC123/CAMK1D and SPRY2 are associated with susceptibility to type 2 diabetes in the Japanese population. Diabetologia. 2011;54:3071–3077. doi: 10.1007/s00125-011-2293-3. [DOI] [PubMed] [Google Scholar]
- 15.Schleinitz D, Tönjes A, Böttcher Y, et al. Lack of significant effects of the type 2 diabetes susceptibility loci JAZF1, CDC123/CAMK1D, NOTCH2, ADAMTS9, THADA, and TSPAN8/LGR5 on diabetes and quantitative metabolic traits. Horm Metab Res. 2010;42:14–22. doi: 10.1055/s-0029-1233480. [DOI] [PubMed] [Google Scholar]
- 16.Murea M, Lu L, Ma L, et al. Genome-wide association scan for survival on dialysis in African-Americans with type 2 diabetes. Am J Nephrol. 2011;33:502–509. doi: 10.1159/000327985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–116. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ribel-Madsen R, Fraga MF, Jacobsen S, et al. Genome-wide analysis of DNA methylation differences in muscle and fat from monozygotic twins discordant for type 2 diabetes. PLoS One. 2012;7:e51302. doi: 10.1371/journal.pone.0051302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alberti KG, Zimmet PZ. Definition, diagnosis and classification of diabetes mellitus and its complications. Part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet Med. 1998;15:539–553. doi: 10.1002/(SICI)1096-9136(199807)15:7<539::AID-DIA668>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 20.García-Giménez JL, Sanchis-Gomar F, Lippi G, et al. Epigenetic biomarkers: A new perspective in laboratory diagnostics. Clin Chim Acta. 2012;413:1576–1582. doi: 10.1016/j.cca.2012.05.021. [DOI] [PubMed] [Google Scholar]
- 21.Zampetaki A, Kiechl S, Drozdov I, et al. Plasma microRNA profiling reveals loss of endothelial miR-126 and other microRNAs in type 2 diabetes. Circ Res. 2010;107:810–817. doi: 10.1161/CIRCRESAHA.110.226357. [DOI] [PubMed] [Google Scholar]
- 22.Zhao C, Dong J, Jiang T, et al. Early second-trimester serum miRNA profiling predicts gestational diabetes mellitus. PLoS One. 2011;6:e23925. doi: 10.1371/journal.pone.0023925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dehwah MA, Xu A, Huang Q. MicroRNAs and type 2 diabetes/obesity. J Genet Genomics. 2012;39:11–18. doi: 10.1016/j.jgg.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 24.Poy MN, Eliasson L, Krutzfeldt J, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 25.Husseiny MI, Kuroda A, Kaye AN, Nair I, Kandeel F, Ferreri K. Development of a quantitative methylation-specific polymerase chain reaction method for monitoring beta cell death in type 1 diabetes. PLoS One. 2012;7:e47942. doi: 10.1371/journal.pone.0047942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kong L, Zhu J, Han W, et al. Significance of serum microRNAs in pre-diabetes and newly diagnosed type 2 diabetes: a clinical study. Acta Diabetol. 2011;48:61–69. doi: 10.1007/s00592-010-0226-0. [DOI] [PubMed] [Google Scholar]