Abstract
Low-cost detection of pathogens and biomolecules at the point-of-care promises to revolutionize medicine through more individualized monitoring and increased accessibility to diagnostics in remote and resource-limited areas. While many approaches to biosensing are still limited by expensive components or inadequate portability, we present here an ELISA-inspired lab-on-a-chip strategy for biological detection based on liposome tagging and ion-release impedance spectroscopy. Ion-encapsulating dipalmitoylphosphatidylcholine (DPPC) liposomes can be functionalized with antibodies and are stable in deionized water yet permeabilized for ion release upon heating, making them ideal reporters for electrical biosensing of surface-immobilized antigens. We demonstrate the quantification of these liposomes by real-time impedance measurements, as well as the qualitative detection of viruses as a proof-of-concept toward a portable platform for viral load determination which can be applied broadly to the detection of pathogens and other biomolecules.
Keywords: Impedance sensing, Point-of-care, Liposomes, Microfluidics
1 Introduction
The point-of-care detection of biological entities will play an essential role in the changing face of health care delivery by making the detection of disease and monitoring of treatment more ubiquitous in developed nations and more accessible in developing nations. Sensing platforms for biomolecular entities such as cardiac enzymes, cancer biomarkers, toxins, bacteria and viruses can dramatically improve clinical outcomes by enabling earlier diagnosis and personalized monitoring. In particular, populations residing in remote and resource-limited settings stand to benefit significantly from low-cost portable technologies capable of detecting the mediators of major communicable diseases.
As an example, 34 million people worldwide are living with human immunodeficiency virus (HIV), the virus that causes acquired immune deficiency syndrome (AIDS), and many face barriers to accessing diagnostic testing necessary for effective treatment. A critical need exists for a low-cost HIV detection assay for the point-of-care which could provide (i) a qualitative assessment of plasma viremia, especially during the early weeks of infection prior to seroconversion, and (ii) a quantitative assessment of plasma viral load as a tool for informing treatment strategies in individuals with established HIV-positive status, especially during initial viremia when antibodies are undetectable but plasma viral load is in the range of 106 copies/ml (Ho 1996).
Several micro- and nanotechnology solutions have been described in attempts toward point-of-care platforms for HIV/AIDS diagnostic applications (Damhorst et al. 2013). These approaches have often included assays for the p24 viral capsid protein (Kim et al. 2008; Lee et al. 2004; Parpia et al. 2010; Tang et al. 2007; Tang and Hewlett 2010) and viral enzyme reverse transcriptase activity (Ekstrand et al. 1996). Others have attempted isothermal amplification of viral nucleic acid on point-of-care platforms (Lee et al. 2010; Tang et al. 2010) or miniaturized polymerase chain reaction (PCR) (Lee et al. 2008; Shen et al. 2011; Tanriverdi et al. 2010). Few approaches have attempted detection of whole virus particles for a point-of-care measurement, although strategies for the separation of HIV from whole blood or plasma for diagnostic assays by microfluidic immunoaffinity chromatography (Kim et al. 2009; Wang et al. 2010, 2012) and superparamagnetic nanoparticles (Chen et al. 2010) have been reported and may lend themselves well to a whole particle detection approach. Many of the approaches reported to date, however, are not fully portable and automated, rely on expensive detection instrumentation, or do not demonstrate the sensitivity necessary to rival state-of-the-art nucleic acid amplification assays, thus a ubiquitous platform for point-of-care HIV viral load detection is yet to emerge.
The concept of lysate impedance spectroscopy was introduced by Cheng, et al. for the detection of human CD4+ T lymphocytes (Cheng et al. 2007). Cells were captured in a microfluidic channel and lysed by the addition of low-conductivity hypotonic solution and impedance changes detected by surface electrodes were showed to be indicative of cell number down to just 20 cells μl−1. Others have applied impedance spectroscopy more broadly for the detection of bacterial growth (Gómez et al. 2002; Gómez-sjöberg et al. 2005), binding of an analyte to a recognition site on electrodes (Lisdat and Schäfer 2008), and, recently, the qualitative detection of HIV from viral lysis at high concentrations (Shafiee et al. 2013).
The concept of releasing liposome contents as a biosensor component has been reported in the past; however, the materials encapsulated by the liposomes in these cases were either fluorescent molecules or redox couples (Connelly et al. 2012; Edwards et al. 2012; Kwakye et al. 2006; Liu and Boyd 2012; Nugen et al. 2009; Zaytseva et al. 2005a, b). Our ion-encapsulating liposomes represent a simpler approach which would better accommodate the requirements for portable sensing platforms.
Here we report an ELISA-inspired approach to biological detection consisting entirely of on-chip electrical impedance sensing and apply it to HIV detection. At the core of this detection strategy is a micron-sized, antibody-functionalized liposome particle encapsulating concentrated phosphate buffered saline (PBS). These liposome tags can be quantified by impedance spectroscopy upon ion release in low conductivity media and are also shown to specifically bind surface-immobilized virus particles captured in a microfluidic channel. Here we characterize dipalmitoylphosphatidylcholine (DPPC) liposome particles, show quantification by this electrical sensing method, and demonstrate an ELISA-like approach to virus capture, liposome tagging and sensing.
2 Modelling of impedance spectroscopy
2.1 Conductivity changes from liposome ion release
The change in impedance sensed by our system is the result of ion release into the surrounding media from the permeabilization of liposomes. The average diameter of liposomes used in this report (see Fig. 1) was determined to be 3.7 μm, which corresponds to an average volume of 26.5 fL per liposome. Each liposome encapsulated 10X PBS containing 1.37M NaCl and 0.027 M KCl. Each liposome therefore contributes approximately 3.6×10−14 moles of NaCl to the surrounding media upon complete release of its contents. Assuming that the beginning solution is true deionized water (conductivity of 0.055 MΩ−1 cm−1) (Omega Engineering 2013), 33,000 liposomes (~5,000 liposomes per μl) would produce a 0.18 mM change in NaCl in the 6.628 μl volume fluidic channel. Assuming the effects on conductivity of NaCl in 10X PBS dominates over the effects of KCl, this change in ion concentration can be calculated to produce a change per liposome per microliter of approximately 4.27 GΩ−1 cm−1 (CSM; Technical Manual 2010). It has been shown previously that change in capacitance from ion release into solution is several orders of magnitude lower than the change in bulk conductance, thus the effect on conductance dominates (Cheng et al. 2007).
Fig. 1.
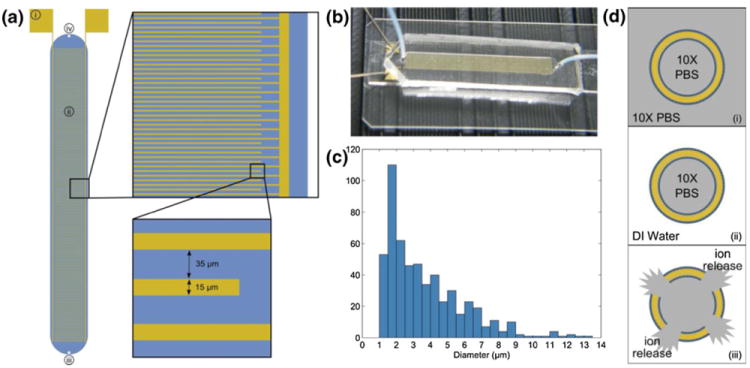
(a) Schematic of interdigitated electrode impedance sensing device. Each electrode finger is 15 μm wide with a 35 μm gap between electrodes. Labeled components are (i) gold contact pads where micromanipulator probes contact the device, (ii) sensing region/fluidic chamber, (iii) fluidic inlet port, (iv) fluidic outlet port. The blue region represents the fluidic chamber defined by the PDMS cover. (b) Photograph of device with PDMS cover mounted on a microscope slide. Micromanipulator probes can be seen in contact with the device. (c) Histogram of liposome diameters as determined by optical microscopy (mean diameter=3.682 μm, standard deviation=2.262 μm) and analysis with ImageJ software. (d) Illustration of liposome sensing concept: (i) liposomes are hydrated in 10X PBS, producing ion-encapsulating particles, (ii) the external media is replaced with low-conductivity DI water, (iii) ion release is triggered and impedance change is measured in order to quantify liposomes present in the sensing device
2.2 Modelling impedance spectra
Electrodes in solution can be modelled by the equivalent circuit in Fig. 4d. In this simplified model, Cdi is the dielectric capacitance resulting from all materials surrounding the electrodes, Rsol is the solution resistance, Rser is the series resistance of the electrodes and wires, and Zpara is the parasitic impedance, which is measured from the open circuit (Cheng et al. 2007; Gómez et al. 2002; Gómez-sjöberg et al. 2005). Zdl, the interfacial or Warburg resistance, is described by
| (1) |
Fig. 4.
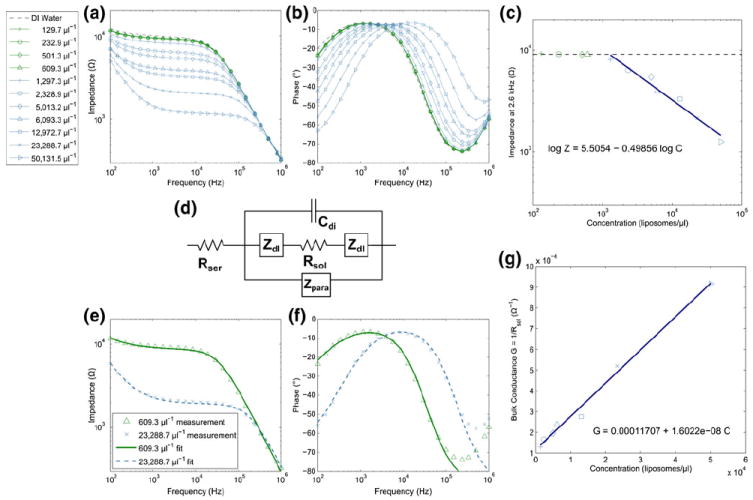
(a–b) Ion-release impedance spectroscopy measurements from several heat-permeabilized liposome samples at concentrations ranging from 129.7 μl−1 to 50,131.5 μl−1 and (c) impedance measured at 2.6 kHz versus liposome concentration with fits to measurements above the limit of detection, approximately 1,000 μl−1. (d) Simplified circuit model of electrodes in solution which is fitted to experimental data, accounting for the dielectric capacitance Cdi, solution resistance Rsol, series resistance Rser, parasitic impedance Zpara, and interfacial impedance Zdl. (e–f) Sample fits of the simplified circuit model to measured data for high and low concentrations. (g) Bulk conductance calculated from values for Rsol extracted from fitting the experimental data are plotted versus liposome concentration and fitted with a straight line. The slope of this line represents the sensitivity of the device
Where j = √(−1), and the parameters n and B depend on the properties of the electrolytes and electrodes (Cheng et al. 2007; Gómez et al. 2002; Gómez-sjöberg et al. 2005). This model will be used to fit the impedance spectra of liposome ion release in the interdigitated electrode device described here.
3 Materials and methods
3.1 Reagents
1,2-dipalmitoyl-sn-glycero-3-phosphocholine (DPPC) and 1-palmitoyl-2-{12-[(7-nitro-2-1,3-benzoxadiazol-4-yl)amino]dodecanoyl}-sn-glycero-3-phosphocholine (NBD PC) were purchased from Avanti Polar Lipids (Alabaster, AL, USA). Protein A was purchased from ProSpec (East Brunswick, NJ, USA). Phosphate buffered saline (10X solution) was purchased from Fisher Scientific (Hampton, NH, USA) and Lonza (Basel, Switzerland). Sucrose, dextrose and Triton X-100, deoxycholic acid, sodium azide palmitic acid N-hydroxysuccinimide (pal-NHS), and rhodamine B isothiocyanate were purchased from Sigma-Aldrich (St. Louis, MO, USA). Calcein was purchased from Fisher Scientific (Hampton, NH, USA). Anit-gp120 goat IgG was purchased from Abcam (Cambridge, MA, USA). Virus buffer consisted of Tris, NaCl, and EDTA purchased from Sigma-Aldrich (St. Louis, MO, USA). HIV-1 strain IIIB was purchased from Advanced Biotechnologies Inc. (Columbia, MD, USA).
3.2 Preparation of liposomes
DPPC was dissolved in chloroform and dried in a round bottom flask by rotary evaporation. The lipid film was hydrated with 10X PBS at 50 °C to create liposomes at a lipid concentration of 1 mg/ml. This mixture was then sonicated for 15 min in an ice bath.
To create fluorescent NBD-liposomes, NBD-PC lipids were dissolved with DPPC at 1 mol%. Liposomes were then formed by the same hydration and sonication processes described above.
3.3 Devices
Planar interdigitated electrode chips were fabricated on glass with standard photolithography and gold lift-off technique. A polydimethylsiloxane (PDMS) cover 4 mm × 34 mm × 50 μm (6.628 μl total volume) was cured on a master mold of SU-8 photoresist patterned on a 4” silicon wafer using standard photolithography technique. Inlet and outlet ports were punched in each PDMS cover, which were then aligned with the electrodes and bonded after oxygen plasma activation in a barrel etcher. A schematic and photograph of the device are provided in Fig. 1.
3.4 Liposome stability characterization
NBD-liposomes were prepared in 10X PBS as described above. Initially, four samples were each added to deionized (DI) water, centrifuged at 3220 rcf for 10 min to pellet the liposomes, and the supernatant was aspirated. The pellet was then re-suspended in DI water and the process was repeated to achieve thorough washing. After the second aspiration of supernatant, each pellet was re-suspended in 10X PBS, 1X PBS, sugar solution, or DI water. Each sample was wellmixed by pipetting up and down to ensure even resuspension of the pellet and placed on a sample rotisserie to prevent settling. Liposome number was assessed regularly for each sample by a Guava EasyCyt Plus flow cytometer (EMD Millipore, Billerica, MA, USA) over the course of several hours.
To consider the effects of detergent on liposome stability, NBD-liposomes were washed as described above and resuspended in DI water or DI water containing 1 % v/v Triton X-100. Samples were placed on a sample rotisserie and analysed by flow cytometry over the course of several hours.
To consider the effects of heating on similar samples, NBD-liposomes were washed and re-suspended in DI water or 1 % v/v Triton X. Each sample was aliquoted into several vials which were independently heated to temperatures ranging from 30 °C to 99 °C. After heating and cooling to room temperature, liposomes were analysed in a flow cytometer to determine liposome number.
3.5 Liposome permeability characterization
Liposomes were prepared in 10X PBS containing 50 mM calcein and washed twice by adding an equal volume of DI water, centrifugation and aspiration of supernatant. After the second wash, liposomes were re-suspended in 10X PBS, 1X PBS, sugar solution, or DI water and added as eight replicates of each sample in 0.2 ml PCR reaction tubes. This reaction strip was placed in a Mastercycler ep realplex real time PCR system (Eppendorf, Hauppauge, NY, USA) for temperature analysis. The sample temperature was increased from 25 °C to 95 °C over 30 min and 520 nm emission was monitored throughout. Control samples containing 10X PBS without calcein were also analysed in the same assay.
3.6 Heat-induced liposome permeabilization for impedance spectroscopy
NBD-liposomes were washed and re-suspended in DI water in four different concentrations. Serial dilutions of each of these liposome suspensions were performed in DI water to achieve the desired range of liposome number. A dilution within the range of the flow cytometer was analysed to determine liposome concentration for each of the four sample sets. Samples were heated to 50 °C for 15 min and allowed to cool to room temperature. Each sample was then injected into the interdigitated electrode chip for impedance spectroscopy analysis.
3.7 Impedance spectroscopy measurements
Impedance magnitude and phase angle were determined using an Agilent 4284 LCR meter (Agilent Technologies Inc., Palo Alto, CA, USA). Micromanipulator probes were used to connect the microfabricated device to the LCR meter and the measurement process was automated by MATLAB (MathWorks, Natick, MA, USA). Impedance spectra were measured for frequencies between 100 Hz and 1 MHz with an increase factor of 1.5 and amplitude of 250 mV.
3.8 Real-time impedance monitoring of ion release from liposomes
NBD-liposomes were prepared and washed as described previously. However, to ensure complete elimination of ionic contamination from liposome samples, each wash was performed in a 50 ml centrifuge tube and repeated twice for a total of three washes. Liposomes were then suspended at various solutions and a portion of the sample was immediately analysed by flow cytometry to determine liposome number. Within 1 min of re-suspension in DI water, the liposome sample was injected into the interdigitated electrode device. The device was set on a heating stage and held at 23 °C for 30 s before being heated to 50 °C and held for severalminutes. Impedance was monitored throughout at 2 kHz and 10 kHz.
3.9 Preparation of antibody-functionalized liposomes
Protein A was hydrophobically modified with hexadecyl chains for anchoring on the liposome surface. Briefly, 1 mg of protein A was reacted with 0.1 mg of pal-NHS in a solution of 0.3 % deoxycholic acid and 0.1 % sodium azide in 1X PBS according to the procedure described by Kim and Peacock (Kim and Peacock 1993). After reacting for 24 h at room temperature, palmitated protein A was purified by centrifuge filter (MWCO 3 kDa, Millipore).
Protein A was adsorbed to liposomes by mixing 5 μg of palmitate-modified protein A for every 1 mg of lipid. The mixture was incubated for 12 h at room temperature, followed by addition of DI water to pellet the liposomes upon centrifugation for 10 min at 3220 rcf to remove protein A that was not adsorbed.
To verify adsorption of protein A, the protein was fluorescently labeled with the amine-reactive rhodamine B isothiocyanate, followed by purification by centrifugal filtration and dialysis to remove unreacted rhodamine. This fluorescent palmitated protein A was incubated with liposomes, which were then purified by centrifugation as described. Fluorescent intensity of the supernatant and re-suspended liposomes was recorded with a Tecan Infinite 200 PRO (Tecan AG, Switzerland), indicating that 40 % of the protein A initially added remained adsorbed to the liposomes.
Liposomes with protein Awere then mixed with 18 μg/ml anti-gp120 IgG for 4 h on a sample rotisserie.
3.10 Virus detection by heat-induced liposome permeabilization
Interdigitated electrode devices were infused with 25 μl of 0.3 mg/ml anti-gp120 IgG in PBS and incubated at 4 °C overnight. Excess antibody was rinsed from the fluidic chamber with 100 μl PBS flowing at 10 μl min−1. Following this rinse, 50 μl buffer containing 6.7e11 virus particles per μl or buffer without viruses was infused into each device at 5 μl min−1 and incubated for approximately 4 h. Following incubation, each device was rinsed with 100 μl PBS at 10-μl min−1 to remove the buffer and unbound viruses. Subsequently, 100 μl of 6,537±70 μl−1 liposomes functionalized with anti-gp120 IgG were injected at a rate of 10 μl min−1 followed immediately by rinsing with 200 μl DI water at 20 μl min−1 to rinse away unbound liposomes and lower the conductivity of the bulk solution. Each device was then placed on a heating stage and real-time impedance was measured during heating as described above.
4 Results
4.1 Liposome stability characterization
To determine the stability of liposomes in various media, flow cytometry measurements were recorded over several hours with NBD-liposomes. Three replicates each of liposomes suspended in isotonic (10X PBS) and hypotonic (1X PBS, sugar solution, DI water) media were measured to assess the effects of various osmotic forces. Sugar solution was chosen because it has been used in the past as an approach to maintaining lymphocyte stability when a low-conductivity medium is desired for impedance measurements of cell lysate (Cheng et al. 2007). The 8.5 % sucrose/0.3 % dextrose solution used in these measurements would have approximately the same osmolarity as cytosol or 1X PBS. A sugar solution which matches the osmolarity of 10X PBS is not practical (too viscous) and therefore was not investigated.
Flow cytometry analysis of liposomes in the four media considered showed little to no decrease in both PBS samples and some depletion in both sugar and DI water over approximately 14 h. To analyse the counting data for each sample, each replicate was fitted separately and the three fits were averaged. This averaged fit is presented in Fig. 2a. The slope of the fits showed depletion rates of approximately 0.0905 % min−1, 0.0569 % min−1, 0.0089 % min−1, and 0.0125 % min−1 for DI water, sugar solution, 1X PBS and 10X PBS respectively (Fig. 2a, inset).
Fig. 2.
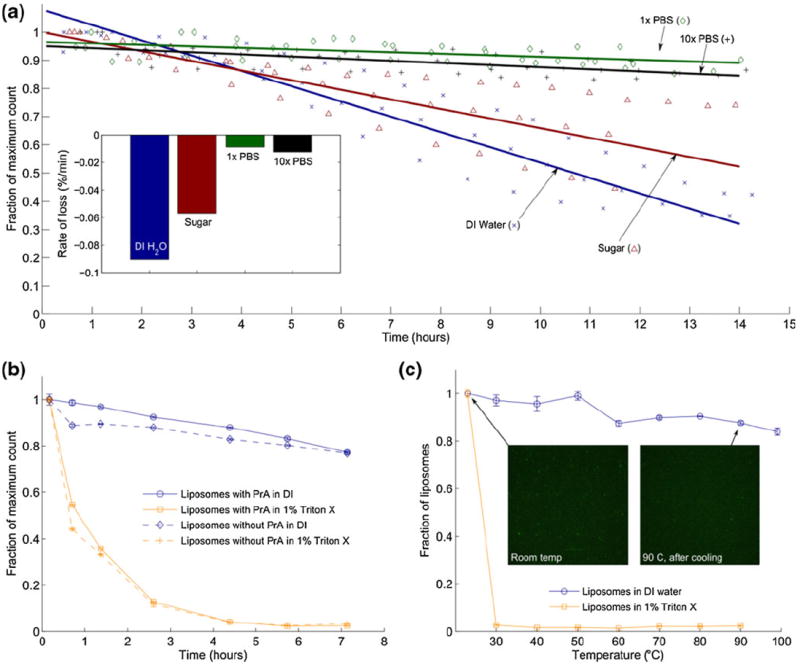
Liposomes in various conditions were counted by flow cytometry to characterize stability. (a) Liposome count was measured over several hours in DI water, sugar solution, 1X PBS, and 10X PBS. The measurement was repeated for each condition for a total of three samples. Each repeat was fitted with a linear best fit. The average of the three fits is shown. (inset) Bar chart of slopes determined from averaged fits of three repeats of each condition. (b) Liposomes with and without protein A were suspended in DI water and 1 % Triton X. Counting over time shows that detergent accelerates liposome depletion. (c) Similarly, liposomes were suspended in DI water and 1 % Triton X, heated to various temperatures and counted with a flow cytometer after cooling to room temperature. This data suggests that heating accelerates the effects of detergent but does not compromise the overall structure of liposomes in DI, although additional data presented in this paper indicates that heating above ~41 °C promotes the release of liposome contents. Fluorescence microscopy images of samples at room temperature and after cooling from 90° confirm flow cytometry measurements by demonstrating that liposome particles are still present after extreme heating
In order to compare the stability of liposomes in DI water with a common liposome permeabilization/lysis reagent, Triton X-100, a separate flow-cytometry measurement was performed with liposomes in DI water and 1 % Triton X. In this case, liposomes with and without membrane-integrated protein A were also compared to determine if liposome functionalization would compromise stability. The results, presented in Fig. 2b, show that liposomes are compromised in the presence of detergent, showing rapid depletion in the first hour and nearly 100 % depletion of liposomes after 5 h. The presence of adsorbed protein A showed little effect.
Upon initial consideration of heating as a chemical-free permeabilization technique for liposome impedance sensing, liposomes in DI water and 1 % Triton X were also analysed by flow cytometry after heating to various temperatures. Counting data in Fig. 2c shows that the destabilizing effects on the liposomes of detergent were accelerated by the addition of heat to the solution. A proposed mechanism explaining the behaviour of liposomes upon heating and in the presence of detergent is provided in the section 5 of this paper.
4.2 Effects of heat on liposome stability
A DNA melting curve assay proved to be the ideal measurement for characterizing the effects of heating on liposome stability and to determine the critical temperature, which we define as the temperature at which the lipid bilayer is partially compromised and encapsulated contents are released into the surrounding media. An ideal reporter molecule for this measurement was calcein, a fluorescent dye which selfquenches at millimolar concentrations (Chen et al. 2004; Hwang et al. 2001).
Liposomes were prepared in concentrated calcein and removed from free calcein in surrounding media by centrifugation and re-suspension of the liposome pellet. Calcein encapsulated within liposomes exhibited very little emission at 520 nm. Upon heating in the thermocycler, however, a sharp increase in fluorescence is observed in the range of 40–43 °C, indicating the release of liposome contents as calcein dissociates and self-quenching subsides. A plot of raw fluorescence is presented in Fig. 3a, with the first derivative with respect to temperature, dI/dT, presented in Fig. 3b as reported by the instrument’s data analysis software.
Fig. 3.
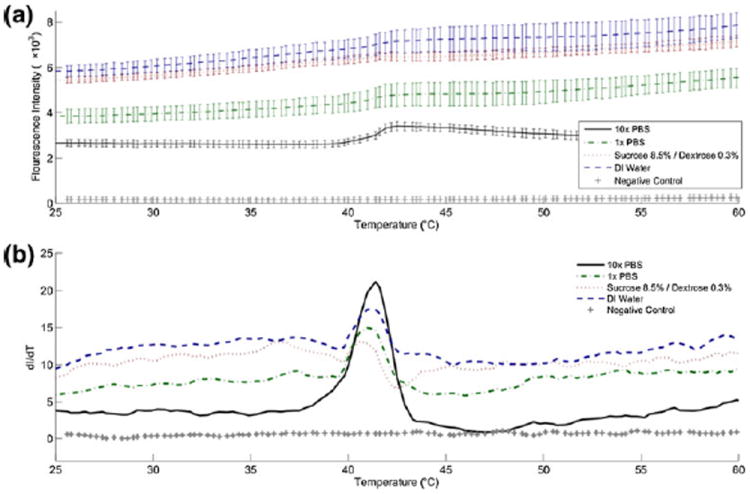
Liposomes encapsulating calcein were prepared, re-suspended in various media and analyzed with a melting curve program in a realtime PCR instrument. The sample temperature was increased from 25 °C to 95 °C over 30 min and at 520 nm emission was monitored throughout. (a) Raw fluorescence intensity and (b) the rate of change dI/dT versus temperature show that a dramatic increase in fluorescence intensity occurs at approximately 41°, indicating the release of calcein from within liposomes. This agrees with the lipid transition temperature cited by the product literature
4.3 Liposome quantification by ion-release impedance spectroscopy
The quantification of liposomes by impedance spectroscopy was demonstrated by suspension of liposomes at various concentrations in DI water and off-chip heating to promote release of encapsulated ions. After cooling to room temperature, each sample was injected into the fluidic chamber and impedance spectra were measured. The measured impedance depends on the concentration of ions in the fluid. We assume the average liposome size is consistent between measurements and that all liposomes encapsulate 10X PBS, which allows for impedance to be directly related to liposome number. Figures 4a–b show the impedance magnitude and phase angle versus AC frequency for each sample. In Fig. 4c, the impedance recorded at 2.6 kHz is plotted versus liposome concentration as measured by a flow cytometer. A lower limit of detection was determined to be at a liposome concentration of approximately 103 μl−1 and impedance values measured at concentrations above this were fitted with a straight line on log-log axes.
4.4 Impedance modelling
The simplified circuit model in Fig. 4d was used to fit measurements displayed in Figs. 4a–b. The parasitic impedance Zpara was determined by fitting the impedance data measured with the device prior to adding any fluid and was then held constant for the other fits. Examples of fits with the measured data are provided in Figs. 4e–f. The bulk conductance Gsol = 1/Rsol can be determined from the parameters extracted for each measurement and are plotted in Fig. 4g versus liposome concentration (Cheng et al. 2007). The slope of the fit to conductivity data gives the sensitivity of the interdigitated electrode chip, determined in this case to be 16.022 GΩ−1 (liposomes per μl)−1 (Cheng et al. 2007).
4.5 Real-time impedance monitoring and quantification
Ion release from liposomes was monitored in real time and the resulting impedance trace could be used to determine liposome concentration as described above. 10X PBS-containing liposomes in DI water were injected into the interdigitated electrode device which was placed on a small heating stage. The temperature was held constant near room temperature for 30 s and quickly increased to 50 °C where it was held constant until the measurement completed. Because conductivity increases with temperature, the impedance trace shows an initial steady decrease as a result of heating of the device; however, a second, sharper decrease is observed as the device reaches critical temperature and the encapsulated ions are released. Sample traces of this behavior are presented as the normalized impedance over time in Fig. 5a and the time derivative of normalized impedance in Fig. 5c. Figures 5b and d show two methods for quantifying liposomes from this data: by fitting the normalized impedance after 200 s (Fig. 5b) and by determining the maximum rate of change (Fig. 5d). Once again, the signals produced by samples below concentrations of 103 ml−1 appear commensurate with measurements of DI water and are not included in fitting of the data.
Fig. 5.
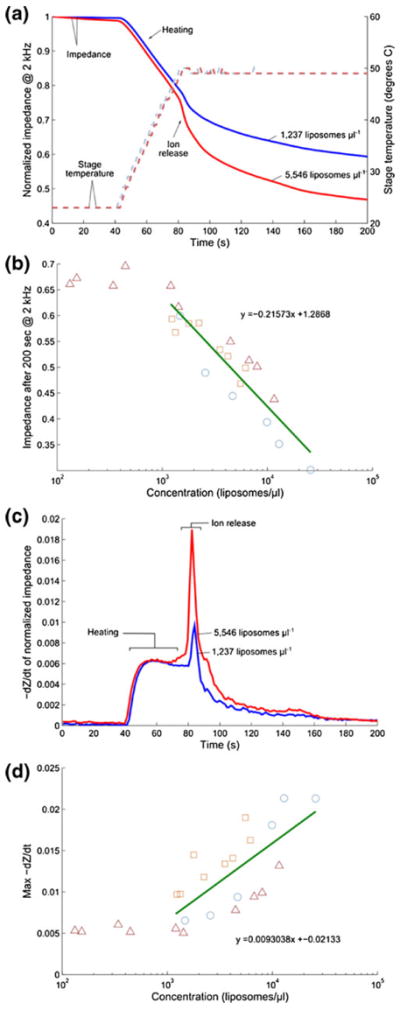
Real-time impedance detection of ion release from liposomes by heating of interdigitated electrode device. (a) Sample data for liposomes at two different concentrations is shown, including the normalized impedance and the stage temperature for 1,237 μl−1 and 5,546 μl−1 samples. The actual device temperature lags behind the stage temperature by a few seconds because it is insulated by a glass microscope slide. (b) Shows the normalized impedance value at 200 s for all samples versus liposome concentration with linear best fit (on a semilog axis). Measurements indicated by each unique symbol/color combination were recorded from the same batch of liposomes. Three different batches of liposomes were prepared and measured in total. (c) The rate of normalized impedance decrease, −dZ/dt, is plotted for the same two representative samples. (d) Summarizes all measurements with the same symbol/color scheme and linear best fit (on a semilog axis). Only liposome concentrations above the lower limit of detection (103 μl−1) are fitted
4.6 Virus detection
As a proof-of-concept toward liposome ion-release impedance spectroscopy for virus detection, several devices were functionalized with anti-gp120 antibody and exposed to 6.7e11 μl−1 HIV to be compared with controls. After prolonged exposure to the virus, the devices were rinsed and 100 μl of IgG-functionalized liposomes (6,537±70 μl−1) in PBS were injected through the device, followed immediately by rinsing with 200 μl DI water. A schematic of this ELISA-like assay is depicted in Fig. 6a. The same heating and real-time impedance monitoring protocols as described above were followed for analysis of these devices, and the normalized impedance after 200 s for devices prepared with immobilized viruses is compared with a control treated with virus-free buffer. Figure 6b shows the normalized impedance data and Fig. 6c summarizes data from four separate devices for each case, showing a significantly larger change in impedance after 200 s for virus-containing devices, representing specific capture of liposomes on immobilized HIV.
Fig. 6.
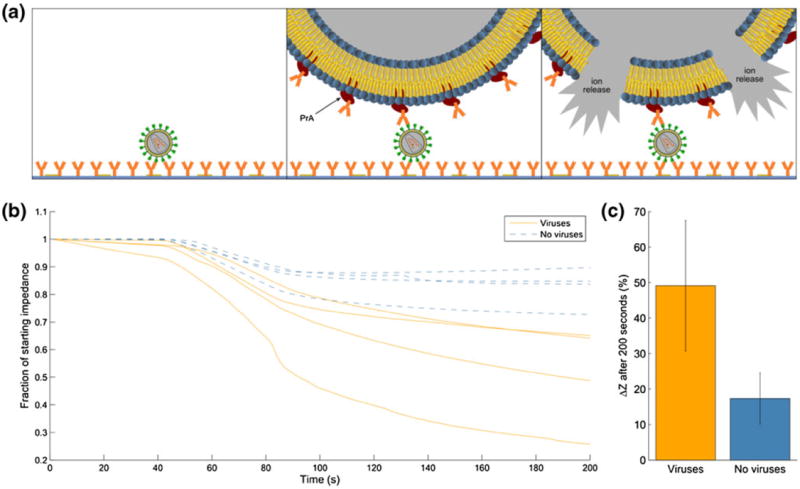
Virus sensing by liposome tagging and ion-release impedance spectroscopy. (a) Concept of detection: (1) Virus is immobilized by antibody binding on the surface of the microfluidic channel. (2) Liposomes, functionalized with protein A (PrA) which binds the Fc region of IgG, bind specifically to immobilized viruses and background media is replaced by DI water. (3) Device is heated to trigger the release of encapsulated ions. (b) Normalized impedance data measured at 2 kHz for four repeats of virus-containing devices and controls. (c) Summary of average impedance change after 200 s for virus-containing devices (mean=49.11 %, standard deviation=18.37 %, n=4) versus control (mean=17.34 %, standard deviation=7.18 %, n=4). The averages are confirmed by two-tailed t-test to be significantly different with 95 % confidence (t=3.22)
5 Discussion
This paper describes progress toward an ELISA-like liposome-based impedance sensing device for biological sensing at the point-of-care. We begin with the characterization of liposome particles containing 10X PBS which are the basis for electrical sensing, providing a simple and low-cost solution to detection of submicron particles. Liposome stability over time is compared in four different media, demonstrating that although liposome depletion rate is greatest in DI water as the result of osmotic forces, the loss of particles is still minimal over a short time span. In our assay, the exposure of liposomes to pure DI water is anticipated to be 10 min or less, thus this minimal depletion rate is inconsequential. This is a marked difference from the effects of DI water on human T lymphocytes where it was used by Cheng, et al. to promote rapid lysis of human cells (Cheng et al. 2007).
Liposome stability in DI water was also compared to stability in the presence of the detergent Triton X, a reagent which is used ubiquitously to restructure lipid membranes. We believe that the detergent disrupts the liposomes by inserting itself into the lipid bilayers, thus increasing permeability. At a constant temperature, rupture of the entire liposome population appears to occur over several hours and appears to be accelerated by heating the sample.
While heating characterization described in Fig. 3 demonstrates that liposomes release encapsulated materials above a temperature of approximately 41°, consistent with the lipid phase transition temperature of DPPC, the counting of liposomes inDIwater as shown in Fig. 2c indicates that even upon heating the sample near boiling point (99 °C), the liposome population is not significantly decreased upon cooling and counting in the flow cytometer. This phenomenon can likely be explained by the self-repair properties of the lipids. Above the transition temperature, the kinetic energy of these molecules is enough to overcome the hydrophobic interactions which stabilize the bilayer structure, thus explaining the release of encapsulated ions. Flow cytometry analysis, however, is performed after the sample has been cooled again. The data in Fig. 2c is explained by the presence or absence of detergent – without Triton X, DPPC lipids are free to re-stabilize their ordered bilayer structures, while reformation of liposomes is inhibited by Triton X, which favors the formation of micelles and may sequester or destabilize DPPC lipids.
In the quantification of human T lymphocytes by lysate impedance spectroscopy (Cheng et al. 2007), the introduction of DI water is used to trigger lysis of cells and release of contents for interrogation. In this measurement, liposomes are more stable in DI water, likely as a result of the absence of membrane proteins, particularly aquaporins, which would allow for the diffusion of water molecules into the cell driven by the osmostic gradient.
DI water is therefore not a suitable technique for triggering liposome permeabilization. Furthermore, chemical-based permeabilization is not preferred for this platform because the introduction of a reagent by flow necessarily results in the displacement of an equal volume of fluid from the microfluidic chamber and thus may result in the loss of lipids or ions and complication of the impedance measurement. Heating is therefore an ideal technique for stimulating the release of encapsulated ions from liposomes in the fluidic chamber as it allows for interrogation of the chamber without displacing its contents.
Liposomes were initially prepared containing 1X PBS, however, 10X PBS was later employed as a means of amplifying the signal produced by liposome permeabilization and is used in all the data presented here. The lower limit of detection determined by the data in Fig. 4 shows the same signal as the deionized water we obtained for our experiments. It is evident that the DI water that was available to us did contain some small amount of ionic contamination and we anticipate that a higher sensitivity would be possible if DI water containing fewer ions were available. We believe that this was the case for the similar measurements performed with human T lymphocytes in (Cheng et al. 2007) where a lower limit of detection is demonstrated.
We have demonstrated that the sensing device used in these measurements can be modelled with a simple circuit consisting of a capacitor, Cdi, in parallel with parasitic impedance Zpara and the series combination of solution resistance Rsol and interfacial impedance Zdl. From this model, the value Rsol is used to determine the bulk solution conductance, Gsol, which is related to the solution conductivity σ by the equation:
| (2) |
Where A is the solution cross-sectional areas between electrodes, L is the spacing between electrodes, and m is the number of electrodes (Cheng et al. 2007). For our device, the value of mA/L is 133.7 cm and this value can be used to determine a measured conductivity change of 0.12 GΩ−1-cm−1 (liposomes per μl)−1. This differs by one order of magnitude from our predicted conductivity change of 4.27-GΩ−1 cm−1 (liposomes per μl)−1, but can be attributed to the behaviour of ions in bulk as described by (Cheng et al. 2007).
During real-time monitoring of liposome permeabilization, two methods for quantification are demonstrated (Fig. 5): normalized impedance after 200 s and max −dZ/dt. It is still to be determined which method is a more consistent and sensitive method for detection. An improved technique which minimizes the fluctuation of device temperature during heating and heats the device more slowly may result in a lower baseline in the time derivative and thus allow for a lower limit of detection. This technique is to be optimized in future studies.
Additionally, the liposomes described in this report exhibited significant variation in diameter. We anticipate that the decrease in impedance magnitude and −dZ/dt would correlate better if liposome size was uniform. Several microfluidic techniques which produce highly uniform liposome particles have been described (deMello and Van Swaay 2012; Teh et al. 2011). These formation methods can be pursued in future work for a more accurate liposome-based measurement.
The accuracy and sensitivity of this device may also be improved by employing larger liposomes. We found that the lower limit of detection was approximately 1,000 liposomes/μl in our current measurements (average liposome diameter = 3.7 μm). Because the impedance change detected depends on the total volume of 10X PBS released from all liposomes on the device, larger liposomes would enable the sensing of fewer total particles. Furthermore, because the volume of a sphere scales with the radius cubed, a liposome with twice the radius of those used in our measurements would have eight times the volume. In other words, only one eighth of the number of liposomes per microliter would be needed to produce the same impedance change and fewer liposomes per microliter could be detected. As an example, a 10 μm diameter liposome has 19.7 times the volume of a 3.7 μm diameter liposome. We would expect, therefore, to be able to detect approximately 50 liposomes/μl if 10 μm liposomes were used.
Finally, the qualitative sensing of viruses based on impedance change that we demonstrated is only a proof-ofconcept. The time of incubation with virus sample was prolonged in our current study to ensure ample time for virus immobilization. We believe, however, that efficient and rapid capture of viruses can be achieved with optimized device geometry, antibody immobilization methods, and improved protocols for virus capture. We are now pursuing the quantitative detection of viruses after high-efficiency immunocapture from whole blood or plasma from HIV-positive individuals.
6 Conclusions
In conclusion, we have developed an electrical sensing technique for the detection of biological entities after tagging with ion-encapsulating liposome particles and ion-release impedance spectroscopy measurements. Our sensing approach eliminates the need for bulky and expensive optical equipment and, because liposome permeabilization occurs rapidly above the critical temperature and only small volumes of reagents are required, is well-suited to become a point-of-care diagnostic tool. Additionally, because the strategy for specific recognition is based exclusively on antigen-antibody interaction, the platform can be easily adapted for the detection of a variety of pathogens, proteins and other biomolecules from biological fluids.
Acknowledgments
We would like to thank Mehmet Toner at Massachusetts General Hospital, William Rodriguez and Marta Fernandez Suarez at Daktari Diagnostics, Inc., and Joshua Wood and Brian Dorvel at the University of Illinois for helpful discussions. Partial support was provided by the Illinois Distinguished Fellowship (to GLD) at the University of Illinois atUrbana-Champaign. This work was also supported by theNational Institutes of Health (1R01 HL109192 to H.J.K. and Chemistry-Biology Interface Training Program 5T32GM070421 to C.E.S.). The devices were fabricated and tested at the Micro and Nanotechnology Laboratory at the University of Illinois at Urbana-Champaign (www.mntl.illinois.edu).
Contributor Information
Gregory L. Damhorst, Micro and Nanotechnology Laboratory, University of Illinois at Urbana-Champaign, 208 N Wright St, Urbana, IL 61801, USA Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, USA.
Cartney E. Smith, Department of Chemical and Biomolecular Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, USA
Eric M. Salm, Micro and Nanotechnology Laboratory, University of Illinois at Urbana-Champaign, 208 N Wright St, Urbana, IL 61801, USA Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, USA.
Magdalena M. Sobieraj, Micro and Nanotechnology Laboratory, University of Illinois at Urbana-Champaign, 208 N Wright St, Urbana, IL 61801, USA
Hengkan Ni, Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, USA.
Hyunjoon Kong, Department of Chemical and Biomolecular Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, USA.
Rashid Bashir, Email: rbashir@illinois.edu, Micro and Nanotechnology Laboratory, University of Illinois at Urbana-Champaign, 208 N Wright St, Urbana, IL 61801, USA; Department of Bioengineering, University of Illinois at Urbana-Champaign, Urbana, IL, USA; Department of Electrical and Computer Engineering, University of Illinois at Urbana-Champaign, Urbana, IL, USA.
References
- Chen T, McIntosh D, He Y, Kim J, Tirrell DA, Scherrer P, Fenske DB, Sandhu AP, Cullis PR. Mol Membr Biol. 2004;21:385. doi: 10.1080/09687860400010516. [DOI] [PubMed] [Google Scholar]
- Chen G, Alberts CJ, Rodriguez W, Toner M. Anal Chem. 2010;82:723. doi: 10.1021/ac9024522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng X, Liu Y, Irimia D, Demirci U, Yang L, Zamir L, Rodríguez WR, Toner M, Bashir R. Lab Chip. 2007;7:746. doi: 10.1039/b705082h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly JT, Kondapalli S, Skoupi M, Parker JSL, Kirby BJ, Baeumner AJ. Anal Bioanal Chem. 2012;402:315. doi: 10.1007/s00216-011-5381-9. [DOI] [PubMed] [Google Scholar]
- Damhorst GL, Watkins NN, Bashir R. IEEE Trans Bio-med Eng. 2013;60:715. doi: 10.1109/TBME.2013.2244894. [DOI] [PubMed] [Google Scholar]
- deMello A, van Swaay D. Lab Chip. 2012;13:752. doi: 10.1039/c2lc41121k. [DOI] [PubMed] [Google Scholar]
- Edwards KA, Bolduc OR, Baeumner AJ. Curr Opin Chem Biol. 2012;16:1. doi: 10.1016/j.cbpa.2012.05.182. [DOI] [PubMed] [Google Scholar]
- Ekstrand DH, Awad RJ, Källander CF, Gronowitz JS. Biotechnol Appl Biochem. 1996;23(Pt 2):95. [PubMed] [Google Scholar]
- Gómez R, Bashir R, Bhunia A. Sens Actuators B: Chem. 2002;86:198. [Google Scholar]
- Gómez-sjöberg R, Morisette DT, Bashir R, Member S. J Microelectromech Syst. 2005;14:829. [Google Scholar]
- Ho DD. Science. 1996;272:1124. doi: 10.1126/science.272.5265.1124. [DOI] [PubMed] [Google Scholar]
- Hwang M, Prud’homme R, Kohn J, Thomas J. Langmuir. 2001;17 [Google Scholar]
- Kim SA, Peacock JS. J Immunol Methods. 1993;158:57. doi: 10.1016/0022-1759(93)90258-9. [DOI] [PubMed] [Google Scholar]
- Kim E, Stanton J, Korber B. Nanomedicine. 2008;3 doi: 10.2217/17435889.3.3.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim Y-G, Moon S, Kuritzkes DR, Demirci U. Biosens Bioelectron. 2009;25:253. doi: 10.1016/j.bios.2009.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwakye S, Goral VN, Baeumner AJ. Biosens Bioelectron. 2006;21:2217. doi: 10.1016/j.bios.2005.11.017. [DOI] [PubMed] [Google Scholar]
- Lee K-B, Kim E-Y, Mirkin CA, Wolinsky SM. Nano Lett. 2004;4:1869. [Google Scholar]
- Lee SH, Kim S-W, Kang JY, Ahn CH. Lab Chip. 2008;8:2121. doi: 10.1039/b811131f. [DOI] [PubMed] [Google Scholar]
- Lee HH, Dineva MA, Chua YL, Ritchie AV, Ushiro-Lumb I, Wisniewski CA. J Infect Dis. 2010;201(Suppl):S65. doi: 10.1086/650385. [DOI] [PubMed] [Google Scholar]
- Lisdat F, Schäfer D. Anal Bioanal Chem. 2008;391:1555. doi: 10.1007/s00216-008-1970-7. [DOI] [PubMed] [Google Scholar]
- Liu Q, Boyd B. Analyst. 2013;138:391. doi: 10.1039/c2an36140j. [DOI] [PubMed] [Google Scholar]
- Nugen SR, Asiello PJ, Connelly JT, Baeumner AJ. Biosens Bioelectron. 2009;24:2428. doi: 10.1016/j.bios.2008.12.025. [DOI] [PubMed] [Google Scholar]
- Omega Engineering. Vol. 11. Omega Engineering, Inc.; 2013. [Google Scholar]
- Parpia ZA, Elghanian R, Nabatiyan A, Hardie DR, Kelso DM. JAIDS J Acquir Immune Defic Syndr. 2010;55:413. doi: 10.1097/QAI.0b013e3181f1afbc. [DOI] [PubMed] [Google Scholar]
- Shafiee H, Jahangir M, Inci F, Wang S, Willenbrecht RBM, Giguel FF, Tsibris AMN, Kuritzkes DR, Demirci U. Small. 2013;1 doi: 10.1002/smll.201202195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen F, Sun B, Kreutz JE, Davydova EK, Du W, Reddy PL, Joseph LJ, Ismagilov RF. J Am Chem Soc. 2011;133:17705. doi: 10.1021/ja2060116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Hewlett I. J Infect Dis. 2010;201(Suppl):S59. doi: 10.1086/650386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S, Zhao J, Storhoff J. JAIDS J Acquir Immune Defic Syndr. 2007;46:231. doi: 10.1097/QAI.0b013e31814a554b. [DOI] [PubMed] [Google Scholar]
- Tang W, Chow WHA, Li Y, Kong H, Tang Y-W, Lemieux B. J Infect Dis. 2010;201(Suppl):S46. doi: 10.1086/650388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanriverdi S, Chen L, Chen S. J Infect Dis. 2010;201(Suppl):S52. doi: 10.1086/650387. [DOI] [PubMed] [Google Scholar]
- Teh S-Y, Khnouf R, Fan H, Lee AP. Biomicrofluidics. 2011;5:44113. doi: 10.1063/1.3665221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Ip A, Xu F, Giguel FF, Moon S, Akay A, Kuritzkes DR, Demirci U. sensors, and command, control, communications, and intelligence (C3I) technologies for homeland security and homeland defense IX. 2010;7666:76661H. doi: 10.1117/12.853132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Esfahani M, Gurkan UA, Inci F, Kuritzkes DR, Demirci U. Lab Chip. 2012;12:1508. doi: 10.1039/c2lc20706k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woongjin Chemical Co Ltd. CSM Technical Manual. Seoul: Republic of Korea; 2010. p. 137. [Google Scholar]
- Zaytseva NV, Goral VN, Montagna RA, Baeumner AJ. Lab Chip. 2005a;5:805. doi: 10.1039/b503856a. [DOI] [PubMed] [Google Scholar]
- Zaytseva NV, Montagna RA, Baeumner AJ. Anal Chem. 2005b;77:7520. doi: 10.1021/ac0509206. [DOI] [PubMed] [Google Scholar]


