Abstract
A small 12-kb haplotype upstream of the AKT1 gene has been found to be associated with insulin resistance phenotypes. We sought to define the functional consequences of the three component polymorphic loci (rs1130214, rs10141867, rs33925946) on AKT1 and the upstream ZBTB42 gene. 5′ RACE analysis of AKT1 transcripts in human skeletal muscle biopsies showed the predominant promoter to be 2.5 kb upstream of exon 2, and distinct from those promoters previously reported in rat. We then studied the effect of each of the three haplotype polymorphisms in transcriptional reporter assays in muscle, bone, and fat cell culture models, and found that each modulated enhancer and repressor activity are in a cell-specific and differentiation-specific manner. Our results in promoter assays are consistent with the human phenotype data; we found an anabolic effect on muscle and bone with increased mRNA expression of AKT1, and catabolic effect on fat with decreased expression. To test the hypothesis that rs10141867 affects transcription levels of the novel zinc finger protein ZBTB42 in vivo, we developed the allele-specific expression assay using Taqman technology to test for allelic differences within heterozygotes. The allele containing the derived polymorphism (haplotype H2) showed a 1.75-fold increase in expression in human skeletal muscle. Our data show a particularly complex effect of the component polymorphisms of a single haplotype on cells and tissues, suggesting that the coordination of different tissue-specific effects may have driven selection for the H2 haplotype. In light of the recent abundance of SNP association studies, our approach can serve as a method for exploring the biological function of polymorphisms that show significant genotype/phenotype associations.
Introduction
AKT1/PKBα is part of a family of protein kinases that includes three members (AKT1/PKBα, AKT2/PKBβ, and AKT3/PKBγ) believed to share some functional redundancy (Cheng et al. 1992; Jones et al. 1991a, b; Konishi et al. 1995; Peng et al. 2003; Vanhaesebroeck and Alessi 2000; Yang et al. 2005). AKT1 is the most extensively studied of this family. AKT1 is a key part of the IGF1/insulin-signaling pathway, is activated by both intracellular (altered receptor tyrosine kinase, Src and Ras) and extracellular (growth factors, cytokines, stress) stimuli, and regulates a variety of cellular processes, including cell growth, survival, differentiation, and metabolism (Alessi et al. 1996; Burgering and Coffer 1995; Cross et al. 1995; Liu et al. 1998; Shaw et al. 1998). Murine knockouts for the AKT1 gene show stunted growth, but otherwise appear normal (Chen et al. 2001).
As a result of its position in a key signaling pathway, AKT1 has been postulated to have two major roles: regulated induction of muscle cell hypertrophy and atrophy and as a key mediator in the insulin-signaling pathway. Animal experiments have provided evidence that AKT1 plays a significant role in muscle hypertrophy and atrophy (Bodine et al. 2001; Chen et al. 2001; Stitt et al. 2004). Specifically, AKT1 phosphorylation state dictates downstream induction of mTOR, leading to increased protein synthesis and promoting muscle hypertrophy (Brunn et al. 1997; Lin and Lawrence 1996). In addition, AKT1 phosphorylates (and thus inactivates) the FOXO family of transcription factors, which prevents the induction of the atrophy signals (Sandri et al. 2004; Stitt et al. 2004).
The location of AKT1 in the insulin-signaling pathway has led to examination of its potential role in glucose homeostasis and the development of insulin resistance. Ueki et al. (1998) showed that expression of a constitutively active form of AKT1 induced glucose uptake, glycogen synthesis, and protein synthesis in cultured L6 myotubes. Basal (i.e. without insulin action) glucose uptake and basal glycogen synthesis were reduced in cells in which AKT1 was knocked down with siRNA (Bouzakri et al. 2006). Krook et al. (1998) found evidence for a link between AKT1 and insulin resistance when they reported that AKT1 activity in response to maximal insulin was markedly reduced in skeletal muscle from subjects with T2D (66% of control levels, p < 0.01) when compared to that of control muscles. Similar results were found in adipocytes, with AKT1 activity in response to insulin impaired by ~50% in adipocytes from T2D subjects when compared to controls (Rondinone et al. 1999).
There is also evidence that AKT1 regulates heart size. Several studies have shown that overexpression of an activated form of AKT1 induced cardiac hypertrophy via an increase in cardiomyocyte size (Condorelli et al. 2002; Matsui et al. 2002; Shiojima et al. 2005). In addition, mouse AKT1 knockout models failed to show the normal cardiac hypertrophy in response to exercise training, revealing further evidence for the role of AKT1 in cardiac growth and size (DeBosch et al. 2006).
It is clear that AKT1 plays a role in a variety of conditions; therefore, understanding polymorphic variation in this gene may offer insight into disease states as well as normal human phenotypic variation. Although several studies have shown associations between AKT1 polymorphisms and various phenotypes (Bajestan et al. 2006; Emamian et al. 2004; Ikeda et al. 2004; Ludlow et al.2007; Riska et al. 2007; Schwab et al. 2005), the literature examining the functional role of AKT1 polymorphisms in vivo is very scarce. We have recently conducted a SNP discovery program on the AKT1 gene, and found genotype/phenotype associations with fasting glucose and insulin levels as well as predisposition for the development of metabolic syndrome in human populations (J. Devaney et al., submitted). We determined that there are no common polymorphisms (minor allele frequency greater than 10%) within the protein-coding sequence of the AKT1 gene; however, there were a series of polymorphisms in introns, and upstream of the gene. Particularly important was a 12-kb region immediately upstream of the AKT1 gene that showed only two common haplotypes, defined by three core polymorphisms in tight linkage disequilibrium (rs1130214, rs10141867, rs33925946). To define the functional significance of these polymorphisms, we sought to determine their placement relative to the transcriptional start site and promoter of the AKT1 gene in human skeletal muscle. We also sought to determine the functional significance of the H2 haplotype polymorphisms, which have shown associations with human metabolic phenotypes.
We characterized the novel protein ZBTB42, which contains SNP rs10141867, one of the SNPs of haplotype H2, in its coding sequence (S. Devaney and E. Hoffman, submitted). We showed that this protein is highly expressed in human skeletal muscle where it localizes to the nuclei. To define the functional significance of SNP rs10141867 on transcriptional activity of ZBTB42, we designed an assay to test the allele-specific expression of this coding sequence SNP.
Patients, materials, and methods
Patient materials
A human muscle biopsy used for mRNA studies (5′ RACE) was from previously existing pathological specimens from a molecular diagnostics program, where the sample was deidentified and included a consent form for use of the specimen for research purposes approved by the CNMC IRB (#2405). The muscle showed no evidence of degeneration or regeneration.
5′ RACE
5′ Rapid Amplification of cDNA Ends (RACE) was performed on total RNA isolated from a human muscle biopsy. The RACE technique was performed using the SMART RACE cDNA Amplification Kit (Clontech) according to manufacturer’s recommendations, using an AKT1-specific anti-sense primer (5′-TGTACTCCCCTCG TTTGTGCAGCCAACC-3′). Following RACE, the resultant cDNA was PCR-amplified (Clontech Advantage GC 2 PCR Kit) using a forward primer specific to the Clontech 5′ RACE adaptor sequence and a nested reverse primer specific for AKT1 (5′-CTTCACAATAGCCACGTCGC-3′). The PCR products were gel-purified using Qiagen’s QIAquick Gel Extraction Kit and cloned into a TOPO TA cloning plasmid (Invitrogen). The plasmids were then grown up in One Shot Chemically Competent E. coli (Invitrogen) and plated on LB plates containing 50 μg/ml ampicillin. Plasmids were isolated from ampicillin-resistant colonies using Qiagen’s Plasmid Mini Kit, and inserts sequenced via automated sequencing (Applied Biosystems 3100 Genetic Analyzer). Sequence data were aligned and analyzed using Sequencher software, version 4.1.4 (Gene Codes Corporation).
Synthesis of complementary DNA from total RNA extracted from muscle biopsies of heterozygotes
Total RNA was extracted from human skeletal muscle biopsies. Complementary DNA was reverse transcribed from 1 lg of mRNA using a cDNA synthesis kit and oligo(dT) primers according to the manufacturer’s protocol (Invitrogen Corporation, Carlsbad, CA).
Promoter assays
Three distinct regions of the AKT1 gene were amplified from genomic DNA from an individual homozygous for H1 and an individual homozygous for H2 (see “Results”): region 1, +2,277 to +2,517 bp, from Genbank NM_001014431.1 containing +G2,347T (rs1130214); region 2, −6,163 to −5,956 bp, containing −C6,024T (rs10141867); region 3, −9,867 to −9,625 bp, containing −C9,756A (rs33925946). Each of the three regions was cloned into a pGL3-Promoter vector (Promega) to assay enhancer/repressor activity of the region. Three different murine cell lines were transfected, each in an undifferentiated (proliferating) state, and in a differentiated state: C2C12 myogenic cells (myoblasts, myotubes), 3T3-L1 adipocytes (undifferentiated, differentiated), MC3T3-E1 osteoblasts (undifferentiated, differentiated). C2C12 cells were cultured in growth medium (DMEM, 10% FBS, 1% Penicillin/Streptomycin) and transfected when they reached 70% confluence. Half the cells were harvested at 24 h post-transfection (myoblasts), while the other half were switched to a differentiation media (DMEM, 2% horse serum, 1× Insulin-Transferrin-Selenium-G supplement, 1% Penicillin/Streptomycin) at this time and then harvested 48 h post-induction of differentiation (myotubes). 3T3-L1 cells were cultured in growth medium (DMEM, 10% bovine calf serum, 1% Penicillin/Streptomycin) and transfected when they reached 70% confluence. Half the cells were harvested at 24 h post-transfection (undifferentiated), while the other half were allowed to grow for another 48 h before they were induced to differentiate (72 h post-transfection) by switching to a differentiation media (DMEM, 10% bovine calf serum, 0.5 mM isobutylmethylxanthine, 1 μg/ml insulin, 1 μM dexamethasone, 1% Penicillin/Streptomycin) and re-transfected at this time. The differentiated cells were then harvested 72 h post-induction of differentiation. MC3T3-E1 cells were cultured in growth medium (Alpha-MEM without ascorbic acid, 10% FBS, 1% Penicillin/Streptomycin) and transfected when they reached 70% confluence. Half the cells were harvested at 24 h post-transfection, while the other half were switched to a differentiation media (Alpha-MEM with ascorbic acid, 10% FBS, 10 mM beta-glycerol phosphate, 1% Penicillin/Streptomycin) at this time and then harvested 48 h post-induction of differentiation. Each undifferentiated and differentiated cell line was transfected at three separate times, with triplicate plates for each transfection and luciferase assay (total 9 values per construct and cell type and differentiation state). Three constructs for each region were transfected (vector alone, H1, H2). Thus, data from a total of 486 transfections were analyzed.
Transfection data were analyzed using repeated measures analysis of variance models with replicate and transfection effects (3 replicates for each of three independent transfection experiments; 9 transfections/construct total). All analyses except where noted were done using Stata Ver. 8.0 (College Station, TX).
ASE assay
The template (both cDNA and the corresponding genomic DNA) is mixed with 900 nM forward and reverse PCR primers, 200 nM fluorescent allele discrimination probes and TaqMan® Universal PCR Master Mix, No AmpErase® UNG [Applied Biosystems (ABI), Foster City, CA, USA] to a final volume of 25 μl. Samples are plated in triplicate. The amplification and probe release are done in the ABI 7900HT. Fluorescent readings are taken after each of the 44 amplification cycles.
Data analysis for ASE
Data analysis is done using Ct values, normalizing the cDNA to genomic DNA. The normalized ΔCt VIC is then set to 1, resulting in a fold-difference of Ct FAM in reference to VIC:
Fold-difference = 2(−ΔΔCt).
Results
Mapping AKT1 exon 1 in human skeletal muscle
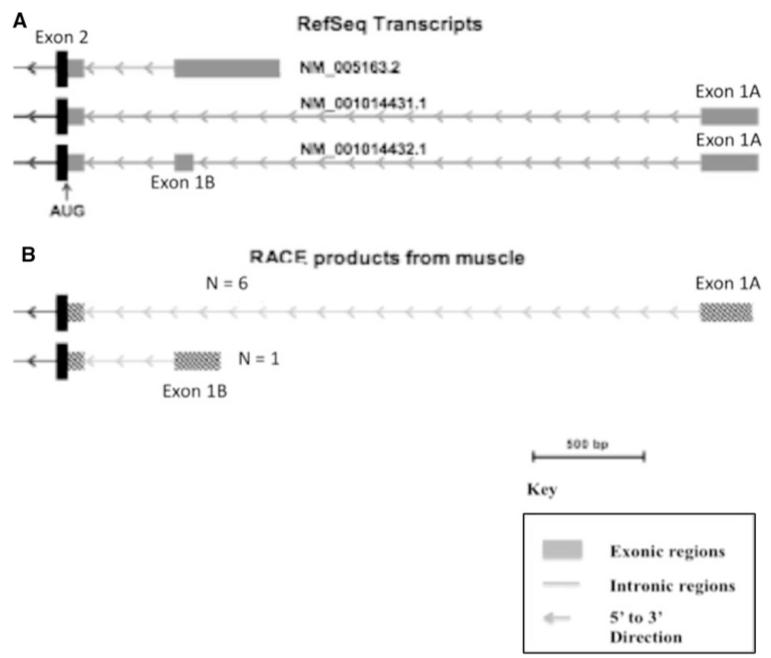
In order to map the 5′ end (exon 1 and promoter) of the human AKT1 gene, we performed 5′ RACE on total RNA isolated from human muscle tissue. Figure 1 shows the RefSeq transcripts for AKT1 from the UCSC genome browser (Fig. 1a) as well as the results of our RACE analysis on the 5′ end of AKT1 (Fig. 1b). Sequence analysis of seven PCR product clones showed two alternative promoters: six clones contained the longer transcript, one contained a shorter transcript with an alternative exon 1, annotated exon 1B. The longer transcript matched RefSeq transcript NM_001014431.1; however, exon 1A was truncated in our transcript. The shorter transcript matched RefSeq transcript NM_005163.2. Both of these transcripts share exon 2, but with distinct 5′ untranslated regions. There were no putative TATA boxes immediately upstream of either of these transcripts discovered via RACE. TATA boxes are often found upstream of the promoter of genes and direct RNA polymerases to the transcription initiation site. Because six of the seven clones contained the longer sequence, we chose to focus on that sequence under the assumption that muscle cells preferentially express the NM_001014431.1 RefSeq transcript. The SNP numbering for this study is relative to exon 1A, which is the major promoter in human skeletal muscle. However, we use the rs number for each SNP (Table 1).
Fig. 1.
AKT1 5′ RACE transcripts in human muscle identify preferential use of upstream promoter. RefSeq transcripts for AKT1 from the UCSC genome browser (a) as well as the results of our RACE analysis on the 5′ end of AKT1 (b). The initial exon of the AKT1 gene in human muscle was identified through 5′ RACE. Seven products were sequenced, with six (N = 6) showing a first exon (Exon 1A) corresponding to RefSeq NM_001014431.1 (a) and one corresponding (N = 1) (b) to an alternative promoter closer to exon 2 (Exon 1B from a)
Table 1.
Conservation of sequence surrounding the three core AKT1 H2 polymorphisms
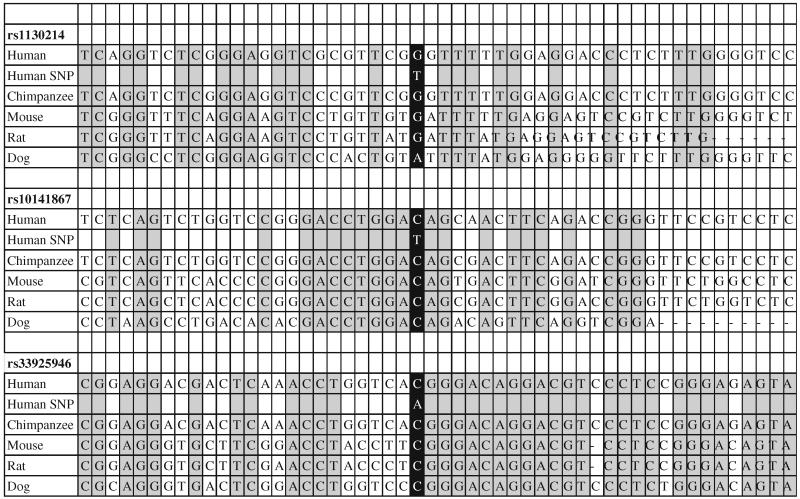
|
Sequences are shown relative to the human reference sequence of chromosome 14. Only human, chimpanzee, mouse, rat, and dog are shown, although there is also strong conservation through fish of the –C6024T polymorphism. The base altered by the H2 polymorphism is shown in black and conserved residues shown in gray highlight
The three core AKT1 H2 polymorphisms are highly conserved through human, chimp, mouse, rat and dog genomes
Conservation of genomic DNA sequences through different species can be indicative of that region having a functional relevance (Frazer et al. 2003; Kellis et al. 2003). We utilized the UCSC genome browser to identify highly conserved sequences in and near the three core loci in the H2 haplotype. This showed that the 12-kb H1 haplotype contained multiple regions of high conservation, including local conservation around each of the three core polymorphisms. To determine if the common alleles at each locus represented the ancestral alleles, we compared the DNA sequences of the human AKT1 gene to chimpanzee, mouse, rat, and dog, immediately bordering the three core loci in the H2 haplotype (Table 1). The ancestral allele of each SNP (haplotype H1) was highly conserved between human, chimpanzee, mouse, rat and dog as shown in the black boxes, which are centered in Table 1. In addition, the regions surrounding the alleles of H1 showed high conservation among the vertebrates as shown in the gray and white boxes to the left and right of the H1 SNPs in Table 1. In each case, the H2 allele changed the polymorphism away from the conserved sequence, suggesting that the polymorphism might have functional significance (Frazer et al. 2003; Kellis et al. 2003). SNP rs10141867 is located within the coding sequence of ZBTB42; however, this is a synonymous change and therefore does not change the amino acid of the gene product.
Functional assessment of component loci in the H1 and H2 haplotypes upstream of AKT1
The three component polymorphisms of H2 altered evolutionarily conserved residues yet none were predicted to influence protein-coding sequences, which may indicate a functional change at the protein level. Therefore, we hypothesized that the SNPs of H2 may reside in transcriptional regulatory sequences and thus might alter transcriptional regulation of the AKT1 or ZBTB42 genes. We studied the enhancer/repressor activity of each region in both undifferentiated and differentiated muscle, bone, and fat cells. We chose muscle, fat and bone, because AKT1 has been implicated in skeletal muscle hypertrophy and atrophy (Bodine et al. 2001). In addition, bone can undergo hypertrophy following exercise (Jones et al. 1977) and the role of AKT1 is not well understood in bone health. Subsequent analyses showed strong associations with metabolic phenotypes in a forthcoming paper (J. Devaney et al., in preparation).
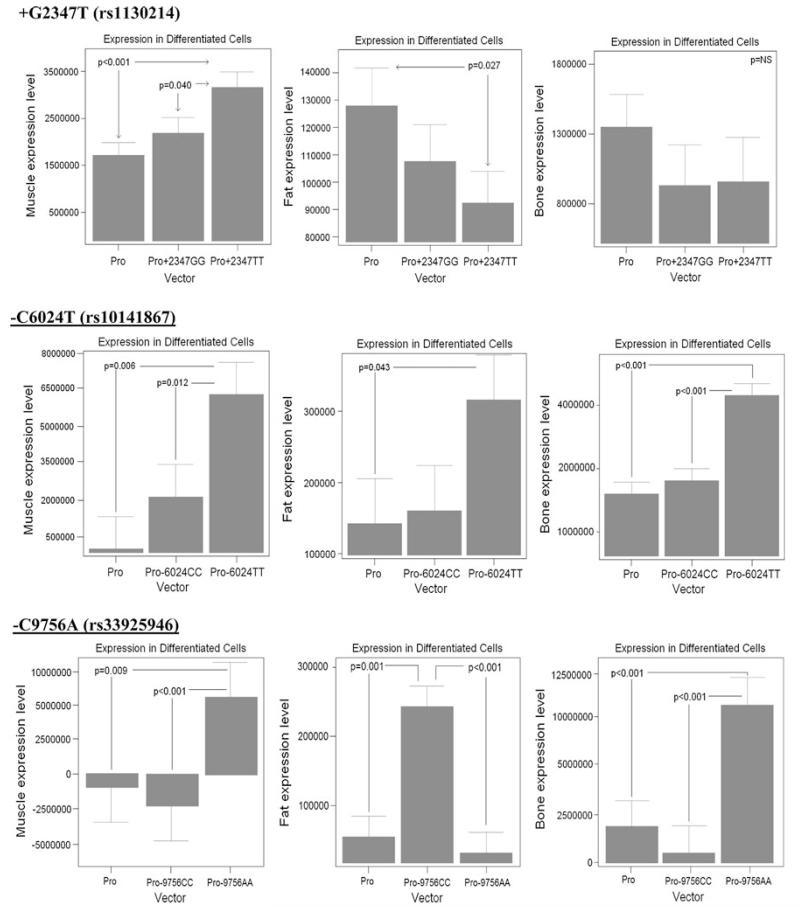
250-bp regions were amplified, with each containing one of the three core loci (Table 1) from human volunteers homozygous for H1, and others homozygous for H2. Each allele-specific region was then cloned into pGL3-Promoter reporter constructs, leading to six constructs (two genotypes each for three regions), as well as empty vector controls. Each of the three constructs was transfected in triplicate (9 transfections) into muscle cells (C2C12), fat cells (3T3-L1), and bone cells (MC3T3-E1). Each construct was transfected into both proliferating undifferentiated cells (70% confluence), and differentiated cells (differentiation media specific for each cell type). A total of 486 transfections were performed, luciferase activity assessed, and repeated measures analysis of variance models computed, with results summarized in Table 2. These data show that each of the three conserved regions functioned as either a transcriptional enhancer or repressor, with the activity often dependent on the specific allele (H1 vs. H2), cell type (bone, muscle, or fat), and differentiation state (Table 2). Results for differentiated cells are shown in Fig. 2.
Table 2.
The component polymorphisms of H2 of AKT1 show cell type- and differentiation-specific effects on transcriptional activity
|
AKT1 (+G2347T), rs1130214 |
AKT1 (−C6024T), rs10141867 |
AKT1 (−C9756A), rs33925946 |
||||
|---|---|---|---|---|---|---|
| Allele G H1 | Allele T H2 | Allele C H1 | Allele T H2 | Allele C H1 | Allele A H2 | |
| Undifferentiated | ||||||
| Muscle | −1.2§ | − | − | +786.0§ | − | +6.6§ |
| Bone | − | − | −1.4§ | + 1.4§ | − | +9.0# |
| Fat | −4.2 | −3.2 | − | +2.5§ | − | −2.8 |
| Differentiated | ||||||
| Muscle | − | + 1.7§ | − | +15.7# | − | +7.5# |
| Bone | − | − | − | +2.6§ | − | +5.8§ |
| Fat | − | −1.4 | − | +2.2 | +3.9§ | − |
All values shown have p < 0.05;
p < 0.01;
p < 0.001; – not significant
Fig. 2.
H2 component sequences function as enhancers in differentiated muscle and bone cells, and repressors in fat cells. Shown are representative data of allele-specific cell transfections of differentiated cells for muscle (left-most graphs C2C12 myotubes), fat (center 3T3-L1), and bone (right MC3T3-E1). Means and standard errors are shown for a 3 × 3 replicates (total 9 transfections). Shown are the empty promoter construct (pGL3, “Pro”), H1 sequences: rs1130214, rs10141867, rs33925946 (Pro+2347GG, Pro−6024CC, Pro−9756CC), and H2 sequences (Pro+2347TT, Pro−6024TT, Pro−9756AA). Each SNP included approximately 250 bp surrounding the polymorphism. The polymorphisms showed a complex effect on enhancer and repressor activity of the constructs, although H2 generally showed enhancer effects in muscle and bone, and repressor effects in fat, consistent with the human phenotype data
We found that the regions containing the SNPs of the H2 haplotype showed more dramatic effects in enhancer activities, often altering the function of the SNPs of H1. In muscle cells (C2C12), the expression constructs containing H2 sequences showed greater enhancer activity than H1 constructs, both in the undifferentiated and differentiated states, with the exception of the rs1130214 allele in undifferentiated muscle, which shows no significant change (Table 2; Fig. 2). In differentiated muscle cells, the H2 constructs showed a 1.7-fold (rs1130214), 15.7-fold (rs10141867), and 7.5-fold (rs33925946) greater expression than the empty vector. The H1 constructs for each of these same SNPs did not show significant enhancer activity. These data suggest that H2 likely leads to greater expression of AKT1 mRNA in both undifferentiated and differentiated muscle cells.
The results for fat cells were more variable depending on allele and differentiation state of cells. The results for both differentiated and undifferentiated cells are listed in Table 2, while the results for differentiated cells are shown in Fig. 2. Here, we discuss the results of the differentiated fat cells. The H2 rs1130214 construct decreased expression by −1.4-fold; H1 did not significantly alter expression. The H2 rs10141867 construct increased expression by 2.2-fold; H1 did not significantly alter expression. The H1 rs33925946 construct showed the strongest effect and increased expression by 3.9-fold; H2 did not significantly alter expression in differentiated cells (Table 2; Fig. 2). In bone cells, H2 expression constructs showed greater enhancer activity than H1 constructs both in the undifferentiated and differentiated states (Table 2; Fig. 2). In the rs1130214 construct, neither H1 nor H2 caused a significant change in expression in differentiated cells. However, in the rs10141867 construct and the rs33925946 construct, H2 increased expression in differentiated cells by 2.6- and 5.8-fold, respectively. The polymorphisms showed a complex effect on enhancer and repressor activity of the constructs, although H2 generally showed enhancer effects in muscle and bone, and repressor effects in fat, consistent with the human phenotype data.
Overall, the most dramatic enhancer activity was of the derived allele of rs10141867, which is within the coding region of the ZBTB42 gene. This region showed a 786-fold induction of transcription in undifferentiated muscle, and a 15.7-fold induction in differentiated muscle (Fig. 2; Table 2).
Development of a novel technique to measure allele-specific expression
ASE is a novel method of analyzing mRNA expression levels in heterozygotes for a SNP of interest. The SNP of interest has to be located within the coding sequence of a gene, because the template is the cDNA of heterozygotes instead of genomic DNA. This assay exploits the highly sensitive Taqman Assay, which was designed to discriminate between two alleles, by labeling them with one of two reporter dyes, within one DNA sample with high sensitivity in endpoint analysis.
We have named the assay the ASE assay. ASE exploits the highly sensitive Taqman Assay, which is used to discriminate between two alleles, by labeling them with one of two reporter dyes (VIC and FAM), within one DNA sample with high sensitivity in endpoint analysis. We altered some components of the Taqman Allele Discrimination Assay to allow us to get an answer to the specific question of allele-dependent transcript expression level variation. First, the SNP must be located within the coding sequence of a gene, because the template is the cDNA of heterozygotes instead of genomic DNA, which is used in classic genotyping. Second, instead of using genomic DNA, we reverse transcribed a cDNA template from mRNA. A genomic template was used from the same individual to act as a control for equal allelic expression. Finally, ASE is done in real-time (as opposed to endpoint analysis) with measures of fluorescence levels of each probe taken at the end of each 44 amplification cycles. This provides us with 44 snapshots of transcript doubling, plotted as log 2. The linear portion of this scale is then used to set a threshold at which the cycle number necessary to reach that fluorescence level can be used as a relative value of expression in the starting material. The cDNA values are normalized to the genomic values for the two markers providing a fold-difference between the major and minor alleles of the coding SNP in cDNA.
ASE correctly quantifies the fold-changes in genomic spike-ins
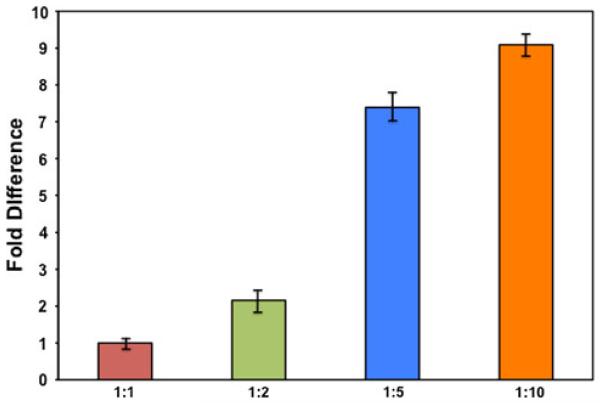
To determine if our modified ASE method showed true differences in allelic expression, we designed a gradient of genomic spike-ins representing different ratios of the major to minor allele for rs10141867. To mimic a heterozygote but also be able to adjust the ratio between the two alleles in a controlled manner, we mixed genomic DNA from homozygote normal (NN) and homozygote mutant (MM) individuals for a SNP of interest at various ratios (N:M): 1:1, 1:2, 1:5 and 1:10. The 1:1 ratio is the control sample. The design of these testing ratios may be useful for the characterization of mitochondrial DNA heteroplasmy using ASE (Tang and Huang 2010). Mean Ct values for the “test” ratios are normalized to this to account for the difference in emission intensities between the two dyes. The test samples represent a gradient of fold differences that may occur physiologically at the transcript level in a heterozygote with a coding sequence SNP that is regulating transcription in one direction.
The fold-changes of the different spike-ins were close to the actual ratio (Fig. 3). The calculated fold-change of our 1:2 (N:M) genomic spike-in was 2.16 of the mutant allele in reference to the normal allele. As the concentration of the mutant compared to the normal increased (ratios 1:5 and 1:10), the assay got less sensitive with fold-changes of 7.39 and 9.09, respectively.
Fig. 3.
ASE correctly identifies the fold-change of a genomic spike-in at ratios 1:1, 1:2, 1:5 and 1:10. Genomic DNA from one individual homozygous for the H1 allele of rs10141867 and one individual homozygous for the H2 allele of rs10141867 were mixed to create four mixtures of varying ratios (H1:H2): 1:1, 1:2, 1:5 and 1:10. ASE analysis was done in triplicate on the four mixtures. Expression levels of the 1:1 and 1:2 ratios showed the fold-change expected (1:1 and 1:2.16, respectively). The 1:5 and 1:10 ratios were not as accurately identified in fold-change by ASE, but the trend was in the correct direction (1:7.39 and 1:9.09, respectively). ASE analysis may be better able to identify smaller fold-changes with higher accuracy
ASE correctly quantifies the fold-change in expression in transcripts carrying nonsense SNPs
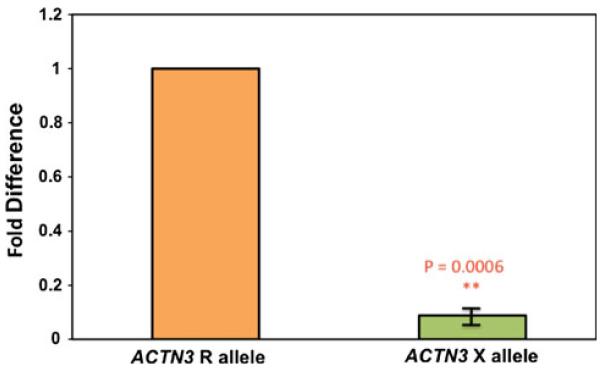
To further test the sensitivity of the ASE method to capture intra-sample variation, we tested the coding sequence SNP R577X of ACTN3. This SNP codes for a nonsense mutation, which initiates nonsense-mediated decay (NMD) of the transcript in vivo resulting in transcript level reduction of ~90% compared to the ancestral allele (North et al. 1999).
Using our new ASE method, we show a tenfold downregulation of the X allele compared to the R allele within heterozygote individuals (Fig. 4). These data correspond with the literature surrounding expression levels of ACTN3 R577X. This verified that the ASE method could detect expression differences in heterozygotes for a coding sequence SNP.
Fig. 4.
The nonsense SNP R577X within the ACTN3 gene causes a tenfold down-regulation of ACTN3 expression. ASE analysis was done on cDNA (reverse transcribed from total RNA) from frozen human skeletal muscle biopsies from heterozygote individuals for the R577X SNP. The derived allele (the nonsense mutation) shows a tenfold down-regulation of ACTN3 expression compared to the ancestral R allele. Expression levels of the two alleles were normalized to genomic DNA from the respective sample
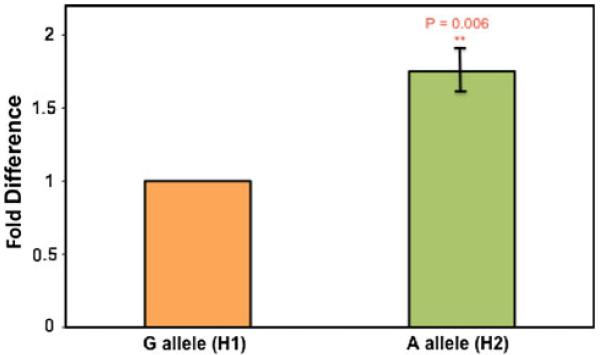
The derived allele of SNP rs10141867 causes a 1.75-fold increase in expression over the ancestral allele in ZBTB42
To test the hypothesis that SNP rs10141867 of our AKT1 haplotype affects transcription levels of the ZBTB42 gene, we used ASE on heterozygote cDNAs. We synthesized cDNAs from mRNA extracted from skeletal muscle from five heterozygote individuals. Genomic DNA from the same individuals served as the controls. When normalized to their genomic match, the derived allele of rs10141867 is expressed at 1.75-fold over the expression of the ancestral allele within the ZBTB42 gene (Fig. 5). This is intriguing as it supports the results of our promoter assay in C2C12 cells, which showed a significant enhancer activity for the derived allele of rs10141867.
Fig. 5.
SNP rs10141867 of H2 causes a 1.75-fold increase in expression of AX721091 in human skeletal muscle. ASE analysis was done on cDNA (reverse transcribed from total RNA) from human skeletal muscle biopsies of heterozygote individuals for SNP rs10141867 (G/A). The derived allele (H2) confers a 1.75-fold increase in transcription of ZBTB42 in vivo. Expression levels of the two alleles were normalized to genomic DNA from the respective sample
Discussion
AKT1 protein function is central to many signaling pathways, and recent publications have begun to show genetic associations with human phenotypes and polymorphisms in and upstream of the AKT1 gene (Emamian et al. 2004; Ludlow et al. 2007; Riska et al. 2007; Schwab et al. 2005). While the AKT1 protein is strongly regulated by phosphorylation state, the association of polymorphisms upstream of the gene suggests that transcriptional regulation may also be important with regard to AKT1 protein function. Transgenic knockouts of AKT1 are consistent with this; in that null mutants for AKT1 are quite runted (Chen et al. 2001).
The H2 haplotype includes polymorphisms previously shown to be linked to muscle strength (Ludlow et al. 2007), body composition (Riska et al. 2007), bone volume, and metabolic syndrome. These data suggest that polymorphisms of functional significance to the regulation of the AKT1 gene are within this 12-kb region. To query the functional significance of the upstream haplotype and component SNPs, we first established promoter usage in human muscle tissue. We carried out 5′ RACE on human muscle RNA, and identified two alternative first exons (Fig. 1). The further upstream promoter (Exon 1A) was preferentially used in muscle (6:1 relative to Exon 1B). Mapping of the upstream haplotype showed that the 12-kb region in strong linkage disequilibrium extended from 10 kb upstream of exon 1A (rs33925946), through Exon 1B (rs1130214), and within the coding sequence of ZBTB42 (rs10141867). All SNP numbering in this current study is relative to Exon 1A, the major promoter in muscle.
Each of the three component SNPs in strong linkage disequilibrium (rs1130214, rs10141867, rs33925946) was amplified from genomic DNA from individuals homozygous for H1, and homozygous for H2, and ~250-bp regions containing these SNPs were studied for transcriptional enhancer/repressor function relative to empty vector. As AKT1 genotypes have been associated with muscle, bone and fat phenotypes (J. Devaney et al., manuscript in preparation), we studied commonly utilized murine cell lines representing each of these tissue types [C2C12 (muscle), 3T3-L1 (fat), MC3T3-E1 (bone)]. H1 includes the ancestral sequence (shared with chimpanzee and other species) in all three regions tested (Table 1). Comparing the enhancer and repressor activities of the three component regions of H1 (ancestral) sequences, we demonstrated that these regions functioned as a repressor in undifferentiated muscle cells (rs1130214), undifferentiated fat cells (rs1130214), and undifferentiated bone cells (rs10141867). This same haplotype showed enhancer activity in differentiated fat (rs33925946). H2 showed more dramatic effects in enhancer activities, often altering the H1 function. Overall, strong enhancer activity was seen for both the derived alleles of rs1130214 and rs33925946 for all cell types and differentiation states, with the strongest enhancer activities in muscle and bone cells (Table 2). Most dramatic was the enhancer activity of the derived allele of rs10141867 within ZBTB42 (Fig. 2). This region showed a 786-fold induction of transcription in undifferentiated muscle, and a 15.7-fold induction in differentiated muscle. The rs33925946 region of H2 also showed strong enhancer activity in both muscle and bone (Table 2).
The strong enhancer activity of the rs10141867 SNP of H2 led to our hypothesis that this SNP is increasing the transcription levels of the ZBTB42 gene. We found that the derived allele of this SNP was in fact causing a 1.75-fold increase in expression of the ZBTB42 gene when compared to the ancestral allele within skeletal muscle of heterozygotes. These data show that each of the three component polymorphisms of H2 upstream of the AKT1 gene and within the coding sequence of ZBTB42 has strong functional consequences on transcriptional activity. Our data also suggest that each of the three regions in strong linkage disequilibrium functions as linked enhancer/repressors.
In conclusion, our study shows that the transcriptional regulation of AKT1 protein expression is likely very complex, with multiple enhancers and repressors showing cell- and differentiation-specific effects. Moreover, polymorphisms of three regions seem to have co-evolved to establish a haplotype that may serve to coordinate metabolic phenotypes of fasting glucose, insulin, BMI and predisposition to metabolic syndrome. These associations coupled with the enhancer activity and higher expression of transcripts carrying H2 implies a deviation from the historic need in humans to have thrifty genes. These are genes or polymorphisms that allow the individual to be more economical with energy so that they will be better able to survive famines and droughts. The advent of agriculture and domestication or herding of game may have relaxed this need to store higher levels of fat and keep muscles smaller. In this relaxation, the H2 haplotype may have developed and grown to high frequency due to its protection against metabolic phenotypes that predispose to comorbidities such as T2D and cardiovascular pathologies.
Abbreviations
- 3T3-L1
Murine fat cells
- AKT1
v-akt murine thymoma viral oncogene homolog 1
- ASE
Allele-specific expression assay
- BMI
Body mass index
- C2C12
Murine muscle cells
- FAMUSS
Functional SNPs associated with muscle size and strength
- FOXO
Forkhead box O
- H1
AKT1 haplotype H1
- H2
AKT1 haplotype H2
- LPH
Lactase-phlorizin hydrolase
- MAF
Minor allele frequency
- MC3T3-E1
Murine bone cells
- MM
Homozygote mutant
- mTOR
Mammalian target of rapamycin
- mZBTB42
Mouse ZBTB42
- NN
Homozygote normal
- PDK
PI-dependent kinases
- PI3K
Phosphatidylinositol 3 kinase
- PKB
Protein kinase B
- PPARg
Peroxisome proliferator-activated receptor gamma
- RACE
Rapid Amplification of cDNA Ends
- RST
Resistin
- S6K1
S6 kinase
- SH2
Src-homology 2
- SNPs
Single nucleotide polymorphisms
- T2D
Type 2 diabetes
- UTR
Untranslated region
- WHO
World Health Organization
- YRI
Yoruban HapMap population
- ZBTB42
Zinc finger and BTB domain containing 42
Contributor Information
Brennan T. Harmon, Department of Integrative Systems Biology, Research Center for Genetic Medicine, Children’s National Medical Center, 111 Michigan Avenue NW, Washington, DC 20010, USA
Stephanie A. Devaney, Department of Integrative Systems Biology, Research Center for Genetic Medicine, Children’s National Medical Center, 111 Michigan Avenue NW, Washington, DC 20010, USA
Heather Gordish-Dressman, Department of Integrative Systems Biology, Research Center for Genetic Medicine, Children’s National Medical Center, 111 Michigan Avenue NW, Washington, DC 20010, USA.
Erica K. Reeves, Department of Integrative Systems Biology, Research Center for Genetic Medicine, Children’s National Medical Center, 111 Michigan Avenue NW, Washington, DC 20010, USA
Po Zhao, Department of Integrative Systems Biology, Research Center for Genetic Medicine, Children’s National Medical Center, 111 Michigan Avenue NW, Washington, DC 20010, USA.
Joseph M. Devaney, Department of Integrative Systems Biology, Research Center for Genetic Medicine, Children’s National Medical Center, 111 Michigan Avenue NW, Washington, DC 20010, USA
Eric P. Hoffman, Department of Integrative Systems Biology, Research Center for Genetic Medicine, Children’s National Medical Center, 111 Michigan Avenue NW, Washington, DC 20010, USA; Department of Integrative Systems Biology, George Washington University School of Medicine and Health Sciences, Washington, DC, USA
References
- Alessi DR, Andjelkovic M, Caudwell B, Cron P, Morrice N, Cohen P, Hemmings BA. Mechanism of activation of protein kinase B by insulin and IGF-1. EMBO J. 1996;15:6541–6551. [PMC free article] [PubMed] [Google Scholar]
- Bajestan SN, Sabouri AH, Nakamura M, Takashima H, Keikhaee MR, Behdani F, Fayyazi MR, Sargolzaee MR, Bajestan MN, Sabouri Z, Khayami E, Haghighi S, Hashemi SB, Eiraku N, Tufani H, Najmabadi H, Arimura K, Sano A, Osame M. Association of AKT1 haplotype with the risk of schizophrenia in Iranian population. Am J Med Genet B Neuropsychiatr Genet. 2006;141:383–386. doi: 10.1002/ajmg.b.30291. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, Zlotchenko E, Scrimgeour A, Lawrence JC, Glass DJ, Yancopoulos GD. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–1019. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Bouzakri K, Zachrisson A, Al-Khalili L, Zhang BB, Koistinen HA, Krook A, Zierath JR. siRNA-based gene silencing reveals specialized roles of IRS-1/Akt2 and IRS-2/Akt1 in glucose and lipid metabolism in human skeletal muscle. Cell Metab. 2006;4:89–96. doi: 10.1016/j.cmet.2006.04.008. [DOI] [PubMed] [Google Scholar]
- Brunn GJ, Hudson CC, Sekulic A, Williams JM, Hosoi H, Houghton PJ, Lawrence JC, Jr, Abraham RT. Phosphorylation of the translational repressor PHAS-I by the mammalian target of rapamycin. Science. 1997;277:99–101. doi: 10.1126/science.277.5322.99. [DOI] [PubMed] [Google Scholar]
- Burgering BM, Coffer PJ. Protein kinase B (c-Akt) in phosphatidylinositol-3-OH kinase signal transduction. Nature. 1995;376:599–602. doi: 10.1038/376599a0. [DOI] [PubMed] [Google Scholar]
- Chen WS, Xu PZ, Gottlob K, Chen ML, Sokol K, Shiyanova T, Roninson I, Weng W, Suzuki R, Tobe K, Kadowaki T, Hay N. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev. 2001;15:2203–2208. doi: 10.1101/gad.913901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng JQ, Godwin AK, Bellacosa A, Taguchi T, Franke TF, Hamilton TC, Tsichlis PN, Testa JR. AKT2, a putative oncogene encoding a member of a subfamily of protein-serine/threonine kinases, is amplified in human ovarian carcinomas. Proc Natl Acad Sci USA. 1992;89:9267–9271. doi: 10.1073/pnas.89.19.9267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condorelli G, Drusco A, Stassi G, Bellacosa A, Roncarati R, Iaccarino G, Russo MA, Gu Y, Dalton N, Chung C, Latronico MVG, Napoli C, Sadoshima J, Croce CM, Ross J., Jr Akt induces enhanced myocardial contractility and cell size in vivo in transgenic mice. Proc Natl Acad Sci. 2002;99:12333–12338. doi: 10.1073/pnas.172376399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature. 1995;378:785–789. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- DeBosch B, Treskov I, Lupu TS, Weinheimer C, Kovacs A, Courtois M, Muslin AJ. Akt1 is required for physiological cardiac growth. Circulation. 2006;113:2097–2104. doi: 10.1161/CIRCULATIONAHA.105.595231. [DOI] [PubMed] [Google Scholar]
- Devaney J, Gordish-Dressman H, Harmon B, Bradbury M, Devaney S, Harris T, Thompson P, Clarkson P, Price T, Angelopoulos T, Gordon P, Moyna N, Pescatello L, Visich P, Zoeller R, Seip R, Seo J, Kim B, Tosi L, Garcia M, Li R, Zmuda J, Delmonico M, Lindsay R, Howard B, Kraus W, Hoffman E. AKT1 polymorphisms are associated with risk for metabolic syndrome. Hum Genet. 2010 doi: 10.1007/s00439-010-0910-8. submitted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emamian ES, Hall D, Birnbaum MJ, Karayiorgou M, Gogos JA. Convergent evidence for impaired AKT1-GSK3beta signaling in schizophrenia. Nat Genet. 2004;36:131–137. doi: 10.1038/ng1296. [DOI] [PubMed] [Google Scholar]
- Frazer KA, Elnitski L, Church DM, Dubchak I, Hardison RC. Cross-species sequence comparisons: a review of methods and available resources. Genome Res. 2003;13:1–12. doi: 10.1101/gr.222003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda M, Iwata N, Suzuki T, Kitajima T, Yamanouchi Y, Kinoshita Y, Inada T, Ozaki N. Association of AKT1 with schizophrenia confirmed in a Japanese population. Biol Psychiatry. 2004;56:698–700. doi: 10.1016/j.biopsych.2004.07.023. [DOI] [PubMed] [Google Scholar]
- Jones HH, Priest JD, Hayes WC, Tichenor CC, Nagel DA. Humeral hypertrophy in response to exercise. J Bone Joint Surg Am. 1977;59:204–208. [PubMed] [Google Scholar]
- Jones PF, Jakubowicz T, Hemmings BA. Molecular cloning of a second form of rac protein kinase. Cell Regul. 1991a;2:1001–1009. doi: 10.1091/mbc.2.12.1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones PF, Jakubowicz T, Pitossi FJ, Maurer F, Hemmings BA. Molecular cloning and identification of a serine/threonine protein kinase of the second-messenger subfamily. Proc Natl Acad Sci USA. 1991b;88:4171–4175. doi: 10.1073/pnas.88.10.4171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kellis M, Patterson N, Endrizzi M, Birren B, Lander ES. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- Konishi H, Kuroda S, Tanaka M, Matsuzaki H, Ono Y, Kameyama K, Haga T, Kikkawa U. Molecular cloning and characterization of a new member of the RAC protein kinase family: association of the pleckstrin homology domain of three types of RAC protein kinase with protein kinase C subspecies and beta gamma subunits of G proteins. Biochem Biophys Res Commun. 1995;216:526–534. doi: 10.1006/bbrc.1995.2654. [DOI] [PubMed] [Google Scholar]
- Krook A, Roth RA, Jiang XJ, Zierath JR, Wallberg-Henriksson H. Insulin-stimulated Akt kinase activity is reduced in skeletal muscle from NIDDM subjects. Diabetes. 1998;47:1281–1286. doi: 10.2337/diab.47.8.1281. [DOI] [PubMed] [Google Scholar]
- Lin T-A, Lawrence JC., Jr Control of the translational regulators PHAS-I and PHAS-II by insulin and cAMP in 3T3-L1 adipocytes. J Biol Chem. 1996;271:30199–30204. doi: 10.1074/jbc.271.47.30199. [DOI] [PubMed] [Google Scholar]
- Liu AX, Testa JR, Hamilton TC, Jove R, Nicosia SV, Cheng JQ. AKT2, a member of the protein kinase B family, is activated by growth factors, v-Ha-ras, and v-src through phosphatidylinositol 3-kinase in human ovarian epithelial cancer cells. Cancer Res. 1998;58:2973–2977. [PubMed] [Google Scholar]
- Ludlow AT, Lui D, Metter J, Ferrucci L, Roth SM. Akt1 G205T polymorphism is associated with muscle strength. Med Sci Sports Exerc. 2007;39(5):S13. [Google Scholar]
- Matsui T, Li L, Wu JC, Cook SA, Nagoshi T, Picard MH, Liao R, Rosenzweig A. Phenotypic spectrum caused by transgenic overexpression of activated Akt in the heart. J Biol Chem. 2002;277:22896–22901. doi: 10.1074/jbc.M200347200. [DOI] [PubMed] [Google Scholar]
- North KN, Yang N, Wattanasirichaigoon D, Mills M, Easteal S, Beggs AH. A common nonsense mutation results in alpha-actinin-3 deficiency in the general population. Nat Genet. 1999;21:353–354. doi: 10.1038/7675. [DOI] [PubMed] [Google Scholar]
- Peng X-d, Xu P-Z, Chen M-L, Hahn-Windgassen A, Skeen J, Jacobs J, Sundararajan D, Chen WS, Crawford SE, Coleman KG, Hay N. Dwarfism, impaired skin development, skeletal muscle atrophy, delayed bone development, and impeded adipogenesis in mice lacking Akt1 and Akt2. Genes Dev. 2003;17:1352–1365. doi: 10.1101/gad.1089403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riska KL, Devaney J, Chen TC, Matos M, Hoffman EP, Clarkson PM. AKT1 association with body composition. Med Sci Sports Exerc. 2007;39(5):S14. [Google Scholar]
- Rondinone CM, Carvalho E, Wesslau C, Smith UP. Impaired glucose transport and protein kinase B activation by insulin, but not okadaic acid, in adipocytes from subjects with Type II diabetes mellitus. Diabetologia. 1999;42:819–825. doi: 10.1007/s001250051232. [DOI] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, Walsh K, Schiaffino S, Lecker SH, Goldberg AL. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab SG, Hoefgen B, Hanses C, Hassenbach MB, Albus M, Lerer B, Trixler M, Maier W, Wildenauer DB. Further evidence for association of variants in the AKT1 gene with schizophrenia in a sample of European sib-pair families. Biol Psychiatry. 2005;58:446–450. doi: 10.1016/j.biopsych.2005.05.005. [DOI] [PubMed] [Google Scholar]
- Shaw M, Cohen P, Alessi DR. The activation of protein kinase B by H2O2 or heat shock is mediated by phosphoinositide 3-kinase and not by mitogen-activated protein kinase-activated protein kinase-2. Biochem J. 1998;336(Pt 1):241–246. doi: 10.1042/bj3360241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiojima I, Sato K, Izumiya Y, Schiekofer S, Ito M, Liao R, Colucci WS, Walsh K. Disruption of coordinated cardiac hypertrophy and angiogenesis contributes to the transition to heart failure. J Clin Invest. 2005;115:2108–2118. doi: 10.1172/JCI24682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stitt TN, Drujan D, Clarke BA, Panaro F, Timofeyva Y, Kline WO, Gonzalez M, Yancopoulos GD, Glass DJ. The IGF-1/PI3K/Akt pathway prevents expression of muscle atrophyinduced ubiquitin ligases by inhibiting FOXO transcription factors. Mol Cell. 2004;14:395–403. doi: 10.1016/s1097-2765(04)00211-4. [DOI] [PubMed] [Google Scholar]
- Tang S, Huang T. Characterization of mitochondrial DNA heteroplasmy using a parallel sequencing system. Biotechniques. 2010;48:287–296. doi: 10.2144/000113389. [DOI] [PubMed] [Google Scholar]
- Ueki K, Yamamoto-Honda R, Kaburagi Y, Yamauchi T, Tobe K, Burgering BM, Coffer PJ, Komuro I, Akanuma Y, Yazaki Y, Kadowaki T. Potential role of protein kinase B in insulininduced glucose transport, glycogen synthesis, and protein synthesis. J Biol Chem. 1998;273:5315–5322. doi: 10.1074/jbc.273.9.5315. [DOI] [PubMed] [Google Scholar]
- Vanhaesebroeck B, Alessi DR. The PI3K-PDK1 connection: more than just a road to PKB. Biochem J. 2000;346(Pt 3):561–576. [PMC free article] [PubMed] [Google Scholar]
- Yang ZZ, Tschopp O, Di-Poi N, Bruder E, Baudry A, Dummler B, Wahli W, Hemmings BA. Dosage-dependent effects of Akt1/protein kinase Balpha (PKBalpha) and Akt3/PKBgamma on thymus, skin, and cardiovascular and nervous system development in mice. Mol Cell Biol. 2005;25:10407–10418. doi: 10.1128/MCB.25.23.10407-10418.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]