Abstract
Maximal aerobic metabolic rate (MMR) is an important physiological and ecological variable that sets an upper limit to sustained, vigorous activity. How the oxygen cascade from the external environment to the mitochondria may affect MMR has been the subject of much interest, but little is known about the metabolic profiles that underpin variation in MMR. We tested how seven generations of artificial selection for high mass-independent MMR affected metabolite profiles of two skeletal muscles (gastrocnemius and plantaris) and the liver. MMR was 12.3% higher in mass selected for high MMR than in controls. Basal metabolic rate was 3.5% higher in selected mice than in controls. Artificial selection did not lead to detectable changes in the metabolic profiles from plantaris muscle, but in the liver amino acids and tricarboxylic acid cycle (TCA cycle) metabolites were lower in high-MMR mice than in controls. In gastrocnemius, amino acids and TCA cycle metabolites were higher in high-MMR mice than in controls, indicating elevated amino acid and energy metabolism. Moreover, in gastrocnemius free fatty acids and triacylglycerol fatty acids were lower in high-MMR mice than in controls. Because selection for high MMR was associated with changes in the resting metabolic profile of both liver and gastrocnemius, the result suggests a possible mechanistic link between resting metabolism and MMR. In addition, it is well established that diet and exercise affect the composition of fatty acids in muscle. The differences that we found between control lines and lines selected for high MMR demonstrate that the composition of fatty acids in muscle is also affected by genetic factors.
Keywords: Artificial selection, Metabolomics, Fatty acid, Laboratory mouse, Maximal metabolic rate
1. Introduction
Maximal aerobic metabolic rate (MMR), aerobic capacity, or VO2 max is an important physiological variable that sets the upper limit to sustained, vigorous activity. Consequently, MMR may affect the ability to defend territories, gather food, and many other ecologically relevant variables (Stearns, 1992; Zera and Harshman, 2001; Brown et al., 2004; Speakman, 2008; Piersma, 2010). The upper limit to MMR is likely due the ability to supply metabolizable suibstrate, the ability to supply oxygen (via steps in the oxygen cascade), or the ability of the animal to metabolize the available substrate (Taylor and Weibel, 1981; Wagner, 1996; Hammond and Diamond, 1997; Swallow et al., 1998; Koch and Britton, 2001; Rezende et al., 2005; Gębczyński and Konarzewski, 2011). While several decades of studies have attempted to identify the limits to MMR from an organ system perspective (Koteja, 1986; Henderson et al., 2002; Gonzales et al., 2006; Garland and Rose, 2009), the metabolic profiles of animals that vary in MMR have received little study.
A promising approach to identifying metabolic underpinnings to MMR is offered by combining artificial selection experiments on metabolic rates with metabolomics, because artificial selection is a powerful tool for studying physiological variation (Garland and Rose, 2009; Swallow et al., 2009), and metabolomics is a powerful tool for providing insights into what underpins various physiological states (Atherton et al., 2006; 2009; Nelson et al., 2010; Epperson et al., 2011). Indeed, metabolomics is being increasingly used to elucidate biochemical mechanisms that underlie physiological processes (Want et al., 2007; Wikoff et al., 2008; Atherton et al., 2009; Nelson et al., 2010; Epperson et al., 2011).
We used artificial selection to manipulate the metabolic rates of mice. Then we used untargeted global metabolic profiling to test how the metabolic pathways responded to the perturbations caused by selection. An untargeted global approach is less focused than a targeted approach looking at specific hypotheses related to individual metabolites, but given the dearth of data on metabolic profile related to MMR, we chose this more exploratory approach to ensure that we did not miss metabolic changes that may not have been anticipated from a targeted strategy. Morever, an untargeted strategy lays the necessary groundwork for future targeted approaches that might focus on one or a small number of metabolites.
We artificially selected for high mass-independent MMR (i.e., on residuals from regressions of MMR on body mass; high-MMR mice) to generate populations of mice that were genetically differentiated with respect to MMR. Then we use those mice to compare the meta-bolomic profiles of the liver, the plantaris muscle, and the gastrocnemius muscle of control mice with mice resulting from 7 generations (G7) of artificial selection for increased mass-independent MMR (i.e., high-MMR mice). The changes in metabolic profiles should enable us and others to generate hypotheses regarding the physiological function underlying variation in MMR. The liver was chosen because it is one of the main contributors to BMR and because variation in BMR and MMR may be correlated. The gastrocnemius muscle was chosen because it is one of the main contributors to MMR and one of the main locomotor muscles. The gastrocnemius muscle is a mixture of Type I oxidative and Type II glycolytic skeletal muscle. The plantaris muscle was also studied because it, too, is a mixture of Type I oxidative and Type II glycolytic skeletal muscle but is not one of the main locomotor muscles (Gardiner and Olha, 1987; Delp and Duan, 1996; Moore and Dalley, 2006).
2. Materials and methods
2.1. Study organism and metabolic rate measurements
We studied mice derived from an artificial selection experiment on aerobic metabolism. The base population for that selection experiment was HS (heterogeneous stock)/IBG (Institute of Behavioral Genetics) laboratory house mice, Mus musculus, obtained from the Institute of Behavioral Genetics at the University of Colorado, Boulder, CO, USA. While the larger selection experiment had three treatments, mice in this study came from only two of those treatments. Those treatments were: (1) randomly bred controls, and, (2) mice directionally selected for increased mass-independent MMR (i.e., high-MMR mice). Each treatment was replicated four times (i.e., blocks) such that there were 8 lines of mice altogether (4 control lines and 4 high-MMR lines). Herein we studied offspring resulting from 7 generations of selection (for high MMR) or from 7 generations of random breeding (controls). The metabolic rate measurements have been previously described in detail (Wone et al., 2009).
In brief, MMR was measured once using an incremental step test during forced exercise on a motorized treadmill contained within a flow-through respirometry chamber. For the incremental step test, the treadmill rate was increased 8 m min−1 every 2 min. To motivate the mouse to run, a shock was applied to the metal grid at the rear of the treadmill. When the mouse did not move off the grid, this was an indication that the mouse reached MMR and the trial was ended.
BMR was measured at least 2 days after MMR with a flow-through respirometry system with 16 chambers. Twelve chambers were used to measure individual BMR and 4 chambers were used to record baseline concentrations of oxygen in ambient air. Mice were monitored during a 6 h period consisting of 6 cycles of 1 h each. Each mouse was monitored for 16 min out of every hour with an ambient oxygen baseline taken immediately before and immediately after each mouse measurement. BMR measurements were completed under post-absorptive conditions and at 32 °C which is within the animal's thermal neutral zone. All mouse procedures and experimental protocols were approved by the University of Nevada, Reno Institutional Animal Care and Use Committee.
2.2. Tissue collection and extraction
Tissues were collected from 10 male mice of each line after 7 generations of artificial selection for increased mass-independent MMR. Mice did not have access to exercise wheels, and they were collected from their home cages during the day (their inactive phase). Hence, the metabolomic profiles should be reflective of mice that were at rest or nearly so (Refinetti, 2006). We did not sample female mice because of potential confounding of metabolic profiles resulting from pre- and post-breeding conditions. Mice were injected subcutaneously with a 0.3-mL mixture of Dormitor (10%; medetomidine hydrochloride; Orion Corp, Espoo, Finland), Ketaset (10%; ketamine hydrochloride; Fort Dodge Animal Health, Fort Dodge, Iowa, USA), and sterile water (80%), and tissue collection was performed after cervical dislocation. The liver, gastrocnemius muscle, and plantaris muscle were rapidly dissected (<90 s post-mortem time before freezing), snap frozen in liquid nitrogen, and stored at −80 °C until extraction.
Approximately 50 mg of tissue was pulverized with a BioPulverizer (BioSpec Products Inc., Bartlesville, OK, USA) under dry ice and liquid nitrogen, with the exception of the plantaris muscle where less than 20 mg were collected. Metabolites and lipids were then extracted from the tissues using methanol-chloroform (Le Belle et al., 2002; Atherton et al., 2006). Ice cold methanol-chloroform (2:1, 600 μL) was added and tissue samples were placed in a sonicating bath for 15 min. After sonication of the samples, chloroform-water (1:1) was added (198 μL of each) to the tissue samples. Two microliters of each of the internal standards containing 15 mg/L of phenanthrene and 25 mg/L of ribitol were also added to the samples. Samples were centrifuged at 13,500 g for 20 min, and the aqueous (i.e., extracted metabolites) and organic layers (i.e., extracted lipids) were stored separately at −80 °C.
2.3. Metabolomic and fatty acid profiling by GC–MS
For the aqueous layer (i.e., metabolites), 150 μL samples were transferred to a 2 mL glass auto-sampler vials and dried for 30 min at high setting in an evacuated centrifuge before derivatization (Gullberg et al., 2004; Atherton et al., 2006). To derivatize the aqueous samples, we added 40 μL of methoxyamine HCL (20 mg/mL in pyridine). The samples were vortex mixed for 1 min and then incubated at 30 °C for 1 h. Afterwards, samples were silylated at room temperature with 70 μL of N-methyl-N-trimethylsilyltrifluoroacetamid (MSTFA) for 30 min. Samples were then transferred to auto-sampler vials with 1.5 mL glass inserts for GC–MS analysis.
For the organic layer, 150 μL were transferred to activated silica Sep-Pak cartridges (Waters Corp, Milford, MA, USA) to separate phosphoand neutral lipids. Sep-Pak cartridges were activated with 1 mL of chloroform. Neutral lipids (i.e., triacylglycerol fatty acids, or TGA) were eluted with 2×1 mL of ethyl acetate under gentle pressure. Neutral lipid extracts were stored at −80 °C or dried under a stream of nitrogen and derivatized. The samples were reconstituted with 750 μL of chloroform-methanol (1:1 vol/vol). We converted the neutral lipid samples to fatty acid methyl esters (FAMEs) by incubating with 150 μL of BF3-methanol at 80 °C for 90 min. Samples were room cooled and a methylated C19 internal standard (25 mg/L) dissolved in chloroform was added. We then added 300 μL of LC-MS grade water and 600 μL of hexane (1:2 ratio) to the samples. Samples were vortex mixed for 1 min and allowed to separate over night. The organic layer was transferred into a 2 mL auto-sampler vial and dried under a stream of nitrogen. Samples were reconstituted with 150 μL of hexane and transferred to auto-sampler vials with 1.5 mL glass inserts for GC–MS analysis. FAMEs analyses profile the total fatty acids found within the tissues (i.e., intramuscular TGA). To analyze the free fatty acids (FFA), we derivatized the free fatty acids and cholesterol derivatives following the derivatization protocol for the aqueous samples described earlier (Atherton et al., 2009).
One microliter of the derivatized metabolites was injected into a Thermo Finnigan gas chromatography (GC; Thermo Scientific, Waltham, MA, USA) equipped with a 30 m×0.32 mm-internal diameter column, part number 123-3832 DB-35MS (Agilent Technologies, Santa Clara, CA, USA). The initial column temperature of 80 °C was held for 2 min and then increased 5 °C/min to 330 °C, which was held for 6 min. Similarly, 1 μL of the FAMEs was injected into the Thermo Finnigan GC equipped with a HP INNOWAX column (60 m×0.25 mm-internal diameter column, part number 1909IN-136; Agilent Technologies). The initial column temperature was 200 °C. The temperature was increased 5 °C/min until the temperature reached 240 °C, which was held for 30 min. For the derivatized free fatty acids, 2 μL was injected into the Thermo Finnigan GC equipped with a 60 m×0.25 mm-internal diameter column, part number 122-5562 DB-5MS (Agilent Technologies). The initial column temperature of 80 °C was held for 2 min. The temperature was then increased 5 °C/min until the temperature reached 315 °C, which was held for 17 min. All column effluent was introduced into a Polaris Q trap mass spectrometer (Thermo Scientific) for mass analysis.
2.4. Data analysis
To analyze the response of MMR and BMR to selection, we used a mixed-model approach where sex and selection treatment were fitted as fixed effects. In addition, block and line nested within treatment were fitted as random effects. For BMR analysis, body mass, and age were included as fixed effects. For MMR analysis, body mass, age, treadmill, and observer were included as fixed effects.
GC–MS chromatograms were analyzed using Xcalibur v.1.3 (Thermo Scientific). An individual metabolite or FAME peak was identified in the chromatogram by comparing its mass spectra to the National Institute of Standards and Technology (NIST) Mass Spectral Library (NIST 08; Gaithersburg, MD, USA), the Golm Metabolite Database (Max Planck Institute of Molecular Plant Physiology, Potsdam; Germany), and the University of Nevada, Reno standard databases. We used MET-IDEA for semi-quantitation of each peak in the GC–MS chromatograms (Broeckling et al., 2006). Deconvolution and semi-quantitation of peaks and overlapping peaks were achieved by directed extraction of ion intensity values based on quantifier ion-retention time for the metabolite (Broeckling et al., 2006). A 0.1-min threshold window was used for the deviation of peaks away from the predicted retention time across the data set. All peaks were normalized by an internal standard with the following equation.
AN = (AO)/AIS, where AN – normalized area, AO – not normalized area, AIS — area of internal standard.
To visualize and model how well the metabolite signatures classified the treatments, we used partial least squares-discriminant analysis (PLS-DA; Ramadan et al., 2006; Atherton et al., 2009). We used the chemometrics software Solo (Eigenvector Research, Inc., Wenatchee, WA, USA) to discriminate group classification of the metabolite signatures. PLS-DA is a regression extension of PCA and is considered a supervised method to model separation of independent samples and group classification (Goodacre et al., 2004). R2X, R2Y, and Q2Y are used as measures for the robustness of a PLS-DA model, where R2X is the cumulative variance explained by the metabolite signatures, and R2Y is the cumulative variance explained by the PLS-DA components. Cross-validation of R2Y estimates Q2Y, which explains the cumulative variation predicted by the model. Thus, R2Y and Q2Y values indicate how well the overall model discriminates group membership in a data set. The range of these values is 0 to 1, and the more they approach 1, the better they represent good discrimination. For all PLSD-DA models, we used a 3-fold random sample cross-validation method. In addition, we used the PLS-DA class grouping to build the models (Eigenvector Research), where we grouped the lines of control mice as one class and the lines of high-MMR mice as the other. In addition, the identified metabolites were mapped onto general biochemical pathways according to the Kyoto Encyclopedia of Genes and Genomes (KEGG).
Concentrations of identified metabolites and fatty acids were compared between high-MMR and control mice. The individual concentrations were compared using separate one-way analysis of variance (ANOVA), where the block and line nested within treatment are random effects. Recall that for our experimental design, the block was the unit of replication. In each block there was a treatment (i.e., high-MMR) line and a control line of mice. We had a total of 4 blocks of mice, hence there were 4 lines of high-MMR mice and 4 lines of control mice. Lines of mice are the units of replication in a selection experiment (Henderson, 1989). The error term used for the ANOVAs was between-replication variation. To account for multiple ANOVA comparisons, we estimated the false discovery rate as the maximum q value (Storey, 2002). All statistical analyses were performed using SAS, v. 9.2 (SAS Institute, Cary, NC, USA).
3. Results
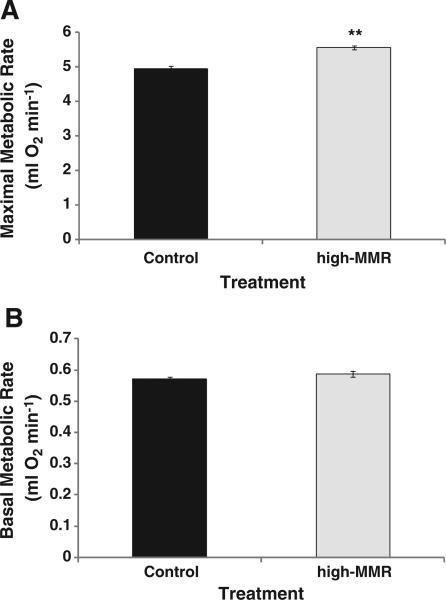
After seven generations of selection, high-MMR mice had 12.3% higher mass-adjusted MMR than control mice (n=278, df=1, 3, F=48.45, p=0.006; Fig. 1A). Similarly, mass-adjusted BMR was also higher in high-MMR mice than controls, but that difference was only borderline significant (n=240, df=1, 3, F=9.07, p=0.057; Fig. 1B).
Fig. 1.
A. Response to selection for high maximal metabolic rate (MMR) at generation 7 (**significantly different from control; n=278, df=1, 3, F =48.45, p=0.006). V. Correlated response of mass independent basal metabolite rate (BMR) to selection for high MMR at generation 7 (n=240, df=1, 3, F=9.07, p=0.057). Values presented are mean (±sem).
The untargeted GC–MS screen of tissue samples resolved approximately 95 putative metabolites. Subsequent analyses focused on those metabolites we were able to unambiguously identify. We unambiguously identified 30 metabolites in the gastrocnemius muscle, 42 metabolites in the liver, and 34 metabolites in the plantaris muscle.
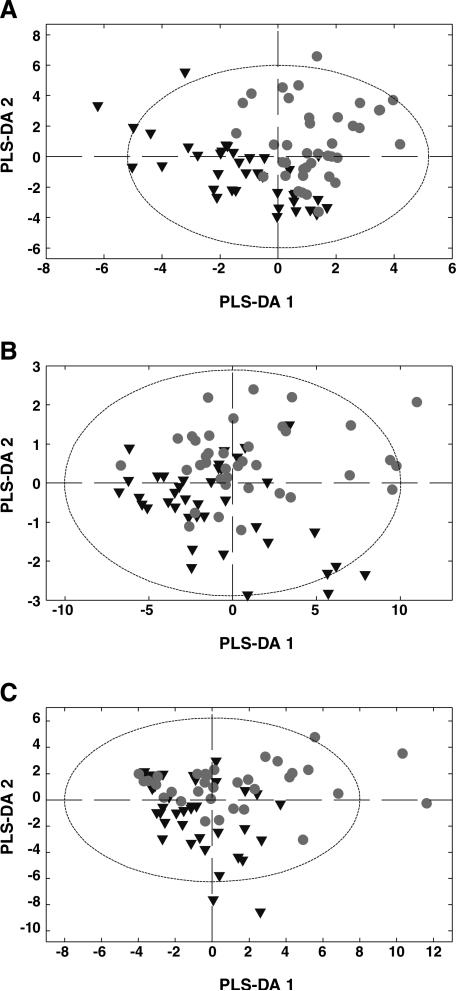
PLS-DA models showed some clustering of mice as a function of selection treatment (Fig. 2). These models revealed metabolic alterations in high-MMR mice compared to control mice, but only the gastrocnemius muscle model performed well in terms of classification (i.e., R2Y or the percentage of the samples in the training set successfully classified) and prediction (i.e., Q2Y or the percentage of the test set correctly classified by using R2Y) abilities. The gastrocnemius muscle model explained 58.9% (R2Y) and predicted 27.8% (Q2Y) of the data based on the cross-validation (Fig. 2A). The gastrocnemius model also explained 38.1% of the metabolite signatures (R2X). Similarly, the liver model explained 32.7% (R2Y) and predicted 4.1% (Q2Y) of the data based on the cross-validation (Fig. 2B). The liver model also explained 41.3% of the metabolite signatures (R2X). In contrast, the plantaris muscle model explained 27.5% (R2Y), and predicted only 0.9% (Q2Y) of the data based on the cross-validation (Fig. 2C). The plan-taris model also explained 50.5% of the metabolite signatures (R2X).
Fig. 2.
Global metabolite comparison of tissues sampled. The figures show the lines of control mice as one group (filled circles) and the lines of high-MMR mice another group (filled upside-down triangles) to make the data easier to visualize. (A) Gastrocnmeius muscle PLS-DA model is constructed from 30 metabolites. (B) Liver PLS-DA model is constructed from 42 metabolites. (C) Plantaris muscle PLS-DA model is constructed from 34 metabolites. Ellipse represents the 95% confidence interval.
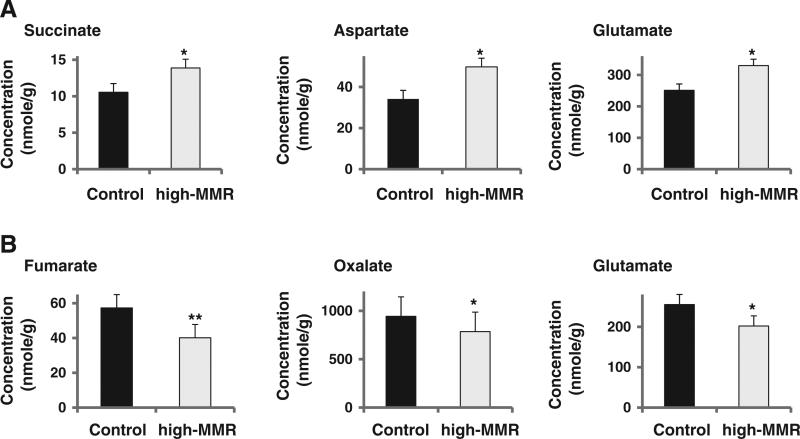
Significant changes in metabolomic profile were detected in the gastrocnemius muscle and liver of high-MMR mice. Of the 30 metabolites identified in the gastrocnemius muscle, 5 metabolites differed significantly between high-MMR and control mice (Table S1). Three of these metabolites are involved in the energy and amino acid metabolism (Fig. 3A), and the other two are by-products of amino acid metabolism. Of the 34 metabolites identified in the plantaris muscle, none differed significantly between high-MMR and control mice. Of the 42 metabolites identified in the liver, 3 metabolites differed significantly between high-MMR and control mice (Table S1). Two of these metabolites are involved in energy metabolism and the other one is involved in the amino acid metabolism (Fig. 3B).
Fig. 3.
Significant changes in detected metabolites involved in the glycolysis/TCA cycle and amino acids metabolism; (A) gastrocnemius muscle, (B) liver. Values presented are mean (± sem) concentrations of metabolites detected. * p and q<0.05. ** p<0.01 and q<0.05 compared to control.
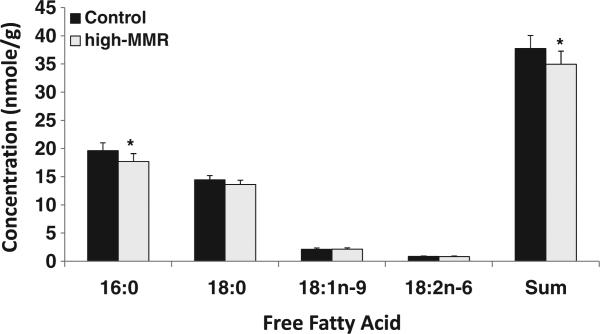
Glucose concentration was elevated in high-MMR lines of mice but controls and high-MMR were not significantly different. To clarify whether the elevated resting metabolic rate of high-MMR mice was associated with fatty acid metabolism, we profiled the free fatty acids and triacylglycerol fatty acids of the gastrocnemius muscle. High-MMR mice had significantly less total free fatty acids in the gastrocnemius muscle than control mice (Fig. 4). In particular, the free fatty acid concentration of palmitic acid (16:0) was markedly decreased. Although the statistical significance was borderline (p= 0.06) stearic acid (18:0) concentration was also lower in high-MMR lines of mice compared to control lines of mice. High-MMR lines of mice had lower triacylglycerol fatty acids concentrations in the gastrocnemius muscle (Fig. 5). Notably, triacylglycerol fatty acids concentrations of stearic acid, eicosadienoic acid (20:2 n-6), dihomo-gamma-linolenic acid (20:3 n-6), and docosahexaenoic acid (22:6 n-3) were decreased.
Fig. 4.
Free fatty acid profile of gastrocnemius muscle. Values presented are mean (±sem) concentrations of free fatty acid detected; *p and q<0.05 compared to control.
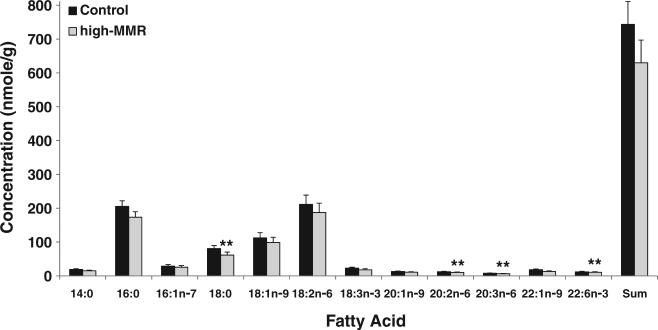
Fig. 5.
Triacylglyceride fatty acid profile of gastrocnemius muscle. Values presented are mean (±sem) concentrations of triacylglycerides detected; *p and q<0.05. **p<0.01 and q<0.05 compared to control.
4. Discussion
Selection for high mass-independent MMR led to a significant increase in mass-adjusted MMR and a borderline significant (p=0.057) increase in mass-adjusted BMR. Selection for high MMR also resulted in metabolomic changes in the gastrocnemius muscle and liver, but not in the plantaris mucle. The overall pattern for high-MMR mice was increased energy and amino acid metabolism in the gastrocnemius muscle. Specifically, elevated TCA cycle intermediates suggest that selection for increased MMR elevated aerobic metabolism in the gastrocnemius muscle. Intererstingly, these differences were shown even in mice that were in or near at resting state. Most TCA cycle intermediates were elevated in mice selected for high MMR compared to controls, but not all of the differences were statistically significant (Fig. 1SA). Besides the elevated TCA cycle intermediates, many amino acids or by products of amino acid metabolism, such as creatinine derivatives, were elevated in mice selected for high MMR compared to controls, but not all of the differences were statistically significant (Fig. 1S A). The functional relevance of the elevated amino acid metabolism in the gastrocnemius muscle is not entirely clear, but one possible explanation is that dietary proteins are being used as an energy substrate (Cherel and Le Maho, 1988; Lumeij and Remple, 1991; Landys et al., 2005). Another possible explanation is that the elevated amino acid metabolism maintains the TCA cycle intermediates needed for fatty acid metabolism (Veiga et al., 1978; Dohm, 1986; Bauchinger and Biebach, 2001). The non-significant elevated amino acid detected in the gastrocnemius muscle was β-alanine. Elevated levels of β-alanine appear to influence maximal (sprint and endurance performance) metabolic functions. That is, β-alanine increases sprint performance in humans (Derave et al., 2007; Hill et al., 2007; Thienen et al., 2009). In addition, because fatty acid oxidation is hampered by low pH levels in skeletal muscles (Derave et al., 2007; Hill et al., 2007), β-alanine, when converted to carnosine, can have a buffering effect aerobic metabolism of fatty acids in skeletal muscle during endurance performance (Hill et al., 2007; Baguet et al., 2010).
In contrast to the gastrocnemius muscle, the overall pattern for high-MMR mice was decreased energy and amino acid metabolism in the liver. That is, the decreased levels of TCA cycle metabolites suggest that selection for increased MMR decreased aerobic metabolism in the liver. Most TCA cycle metabolites were decreased in mice selected for high MMR compared to controls but not all of the differences were statistically significant (Fig. 1SB). The functional significance of these decreases in amino acid and energy metabolism is not clear at this point, but it makes sense that we found elevated energy and amino acid metabolism in the gastrocnemius muscle and not the liver. High MMR is the character we selected on and the liver is not thought to be strongly linked to MMR, while skeletal muscle is thought to be the main organ system contributing to MMR (Bishop, 1999; Weibel et al., 2004; Weibel and Hoppeler, 2005). If high-MMR mice stored more glycogen in their livers, which might be another factor contributing to reduced concentrations of metabolites in the liver. If enough glycogen were stored then that could reduce the concentrations of all metabolites in the livers of high-MMR mice. However, while we did not measure liver glycogen levels, we suspect that they may not have been strongly affected by selection because if they were strongly affected then we might also have expected to detect significant differences in lactate or glucose levels, and we did not detect significant differences in lactate or glucose levels (Jacobs, 1981). Our findings suggest that the metabolic composition of the gastrocnemius muscle reflects its being primed to meet the elevated metabolic capacity during MMR. Recall that high- MMR mice had a small (3.5%) and borderline significant increase in BMR compared to controls and that the metabolomic profiles of the mice were obtained during their inactive phase (i.e., RMR). So, the metabolite changes were associated with selection for increased MMR but these changes are reflected in the resting or near resting state. This finding is consistent with the idea that the capacity for high MMR is associated with changes in the resting state (Hammond et al., 1994; Speakman and Johnson, 2000; Johnson et al., 2001; Piersma, 2010).
MMR and BMR are predictably correlated at the inter-specific level (Koteja, 1987; Bozinovic, 1992; Hayes and Garland, 1995; Dutenhoffer and Swanson, 1996; Rezende et al., 2002), but why this is so is unclear. MMR is largely determined by metabolic output by muscle, and in contrast, visceral organs have been thought to be the primary determinants of BMR. However, a recent inter-specific analysis of variation in muscle mass and BMR in mammals suggests that muscle mass explains more of the variation in BMR than previously thought (Raichlen et al., 2010). In addition to the visceral organs, muscle mass is indeed an important contributor to BMR (Daan et al., 1990; Konarzewski and Diamond, 1995; Even et al., 2001; Selman et al., 2001), then our findings may help explain why (a) skeletal muscle metabolism in rats accounts for nearly 50% of total tissue metabolism (Field et al., 1939), (b) muscle metabolism in humans accounts for 20% of the oxygen consumption at rest (Rolfe and Brown, 1997), (c) muscle mass is correlated with BMR in birds (McNab, 1994; McNab and Ellis, 2006), or (d) muscle mass appears to explain variation in BMR in mammals (McNab, 1978; 2000; 2007; Raichlen et al., 2010).
Our findings suggest that high-MMR lines of mice are utilizing fatty acids (in addition to carbohydrates) to fuel their elevated metabolic capacity. Changes in the free fatty acids and muscle triacylglycerol fatty acid composition are generally linked to exercise training and to dietary fat intake (Andersson et al., 2002; Szabo et al., 2002; Petridou et al., 2005; Dimopoulos et al., 2006; Ehrenborg and Krook, 2009). Our mice were not exercised and standard laboratory rat chow was available ad libitum. Hence, selection for high-MMR in our lines of mice altered the composition of free fatty acids and triacylglycerol fatty acids in skeletal muscle.
The correlated response of free fatty acids and triacylglycerol fatty acids to selection for high-MMR indicates that there is a genetic component to muscle free fatty acids and triacylglycerol fatty acid composition. Interestingly, free fatty acid levels and triacylglycerol fatty acid composition have a central role in determining membrane properties and cell signaling, and they are ligands that bind to the peroxisome proliferator-activated receptors (PPAR) α and γ in skeletal muscle and other tissues, such as liver (Borkman et al., 1993; Ehrenborg and Krook, 2009). PPARs are nuclear receptor proteins that function as transcription factors regulating expression of genes for fatty acid catabolism (Ehrenborg and Krook, 2009). Hence, free fatty acid concentrations and triacylglycerol fatty acid composition affect skeletal muscle lipid metabolism.
5. Conclusions
In this study, we explored the metabolites of gastrocnemius and plantaris muscles, and of liver in inactive mice that have been selected for increased MMR. Metabolic profiling indicated elevated amino acids and energy metabolism in the gastrocnemius muscle, but not in the liver or plantaris muscle. Notably, free fatty acids and triacylglycerol fatty acids are energy sources that could increase metabolic rate. Collectively, our findings suggest that MMR and BMR may be mechanistically linked via elevated amino acid and energy metabolism in the musculature. Most importantly, our results should enable others to generate working hypotheses regarding the physiological function underlying variation in MMR. Lastly, it is well established that diet and exercise affect the composition of fatty acids in muscle. The differences that we found between control lines and lines selected for high MMR demonstrate that the composition of fatty acids in muscle is also affected by genetic factors.
Supplementary Material
Acknowledgments
Study was funded by NSF IOS 0344994 to Jack P. Hayes and by University of Nevada, Reno Graduate Student Association Research Grants to Bernard Wone. We thank Beate Wone for the help with the tissue extractions. We thank John Cushman and Dave Shintani for the use of their lab facilities. We thank Angelica Adrian, Cynthia Downs, Amber Huleva, Andy Hicks, Else Huerta, Marta Labocha, Kim Mclean, Mike Sears, and Alexandra Watson for their help with the selection experiment. The UNR Proteomics Center acknowledges support from NIH Grant P20 RR-016464 from the INBRE Program of the National Center for Research Resources.
Footnotes
Supplementary materials related to this article can be found online at doi:10.1016/j.cbd.2011.09.003.
References
- Andersson A, Nalsen C, Tengblad S, Vessby B. Fatty acid composition of skeletal muscle reflects dietary fat composition in humans. Am. J. Clin. Nutr. 2002;76:1222–1229. doi: 10.1093/ajcn/76.6.1222. [DOI] [PubMed] [Google Scholar]
- Atherton HJ, Bailey NJ, Zhang W, Taylor J, Major H, Shockcor J, Clarke K, Griffin JL. A combined 1H-NMR spectroscopy- and mass spectrometry-based metabolomic study of the PPAR-α null mutant mouse defines profound systemic changes in metabolism linked to the metabolic syndrome. Physiol. Genomics. 2006;27:178–186. doi: 10.1152/physiolgenomics.00060.2006. [DOI] [PubMed] [Google Scholar]
- Atherton HJ, Gulston MK, Bailey NJ, Cheng KK, Zhang W, Clarke K, Griffin JL. Metabolomics of the interaction between PPAR-α and age in the PPAR-α-null mouse. Mol. Syst. Biol. 2009;5:1–10. doi: 10.1038/msb.2009.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baguet A, Koppo K, Pottier A, Derave W. β-Alanine supplementation reduces acidosis but not oxygen uptake response during high-intensity cycling exercise. Eur. J. Appl. Physiol. 2010;108:495–503. doi: 10.1007/s00421-009-1225-0. [DOI] [PubMed] [Google Scholar]
- Bauchinger U, Biebach H. Differential catabolism of muscle protein in garden warblers (Sylvia borin): flight and leg muscle act as a protein source during long distance migration. J. Comp. Physiol. B. 2001;171:293–301. doi: 10.1007/s003600100176. [DOI] [PubMed] [Google Scholar]
- Bishop CM. The maximum oxygen consumption and aerobic scope of birds and mammals: getting to the heart of the matter. Proc. Biol. Sci. Wash. 1999;266:2275–2281. doi: 10.1098/rspb.1999.0919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borkman M, Storlien LH, Pan DA, Jenkins AB, Chisholm DJ, Campbell LV. The relation between insulin sensitivity and the fatty-acid composition of skeletal-muscle phospholipids. N Engl J. Med. 1993;28:238–244. doi: 10.1056/NEJM199301283280404. [DOI] [PubMed] [Google Scholar]
- Bozinovic F. Scaling basal and maximum metabolic rate in rodents and the aerobic capacity model for the evolution of endothermy. Physiol. Zool. 1992;65:921–932. [Google Scholar]
- Broeckling CD, Reddy IR, Duran AL, Zhao X, Sumner LW. MET-IDEA: a data extraction tool for mass spectrometry-based metabolomics. Anal. Chem. 2006;78:4334–4431. doi: 10.1021/ac0521596. [DOI] [PubMed] [Google Scholar]
- Brown JH, Gillooly JF, Allen AP, Savage VM, West GB. Toward a metabolic theory of ecology. Ecology. 2004;85:1771–1789. [Google Scholar]
- Cherel Y, Le Maho Y. Changes in body mass and plasma metabolites during short-term fasting in the king penguin. Condor. 1988;90:257–258. [Google Scholar]
- Daan S, Masman D, Groenewold A. Avian basal metabolic rates: their association with body composition and energy expenditure. Am. J. Physiol. 1990;259:R333–R340. doi: 10.1152/ajpregu.1990.259.2.R333. [DOI] [PubMed] [Google Scholar]
- Delp MD, Duan C. Composition and size of type I, IIA, IID/X, and IIB fibers and citrate synthase activity of rat muscle. J. Appl. Physiol. 1996;80:261–270. doi: 10.1152/jappl.1996.80.1.261. [DOI] [PubMed] [Google Scholar]
- Derave W, Özdemir MS, Harris RC, Pottier A, Reyngoudt H, Koppo K, Wise JA, Achten E. β-Alanine supplementation augments muscle carnosine content and attenuates fatigue during repeated isokinetic contraction bouts in trained sprinters. J. Appl. Physiol. 2007;103:1736–1743. doi: 10.1152/japplphysiol.00397.2007. [DOI] [PubMed] [Google Scholar]
- Dimopoulos N, Watson M, Sakamoto K, Hundal HS. Differential effects of palmitate and palmitoleate on insulin action and glucose utilization in rat L6 skeletal muscle cells. Biochem. J. 2006;399:473–481. doi: 10.1042/BJ20060244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dohm GL. Protein as a fuel for endurance exercise. Exerc. Sport Sci. Rev. 1986;14:143–173. [PubMed] [Google Scholar]
- Dutenhoffer MS, Swanson DL. Relationship of basal to summit metabolic rate in passerine birds and the aerobic capacity model for the evolution of endothermy. Physiol. Zool. 1996;69:1232–1254. [Google Scholar]
- Ehrenborg E, Krook A. Regulation of skeletal muscle physiology and metabolism by peroxisome proliferator-activated receptor delta. Pharmacol. Rev. 2009;61:373–393. doi: 10.1124/pr.109.001560. [DOI] [PubMed] [Google Scholar]
- Epperson EL, Karimpour-Fard A, Hunter LE, Martin SL. Metabolic cycles in a circannual hibernator. Physiol. Genomics. 2011;43:799–807. doi: 10.1152/physiolgenomics.00028.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Even PC, Rolland V, Roseau S, Bouthegourd JC, Tome D. Prediction of basal metabolism from organ size in the rat: relationship to strain, feeding, age, and obesity. Am. J. Physiol. 2001;280:R1887–R1896. doi: 10.1152/ajpregu.2001.280.6.R1887. [DOI] [PubMed] [Google Scholar]
- Field J, Belding HS, Martin AW. An analysis of the relation between basal metabolism and summated tissue respiration in the rat. I. The post-pubertal albino rat. J. Cell Comp Physiology. 1939;14:143–157. [Google Scholar]
- Gardiner PF, Olha AE. Contractile and electromyographic characteristics of rat plantaris motor unit types during fatigue in situ. J. Physiol. 1987;385:13–34. doi: 10.1113/jphysiol.1987.sp016481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garland T, Jr., Rose MR. Experimental Evolution: Concepts, Methods, and Applications of Selection Experiments. University of California Press; Berkeley, CA: 2009. [Google Scholar]
- Gębczyński AK, Konarzewski M. Effects of oxygen availability on maximum aerobic performance in Mus musculus selected for basal metabolic rate or aerobic capacity. J. Exp. Biol. 2011;214:1714–1720. doi: 10.1242/jeb.051680. [DOI] [PubMed] [Google Scholar]
- Gonzales NC, Kirkton DS, Howlett RA, Britton SL, Koch LG, Wagner HE, Wagner PD. Continued divergence in VO2, max of rats artificially selected for running endurance is mediated by greater convective blood O2 delivery. J. Appl. Physiol. 2006;101:1288–1296. doi: 10.1152/japplphysiol.01527.2005. [DOI] [PubMed] [Google Scholar]
- Goodacre R, Vaidyanathan S, Dunn WB, Harrigan GG, Kell DB. Metabolomics by numbers: acquiring and understanding global metabolite data. Trends Biotechnol. 2004;22:245–252. doi: 10.1016/j.tibtech.2004.03.007. [DOI] [PubMed] [Google Scholar]
- Gullberg J, Jonsson P, Nordstrom A, Sjostrom M, Moritz T. Design of experiments: an efficient strategy to identify factors influencing extraction and derivatization of Arabidopsi thaliana samples in metabolomic studies with gas chromatography/mass spectrometry. Anal. Biochem. 2004;331:283–295. doi: 10.1016/j.ab.2004.04.037. [DOI] [PubMed] [Google Scholar]
- Hammond KA, Diamond JM. Maximal sustained energy budgets in humans and animals. Nature. 1997;386:457–462. doi: 10.1038/386457a0. [DOI] [PubMed] [Google Scholar]
- Hammond KA, Konarzewski M, Torres R, Diamond JM. Metabolic ceilings under a combination of peak energy demands. Physiol. Zool. 1994;67:1479–1506. [Google Scholar]
- Hayes JP, Garland T. The evolution of endothermy: testing the aerobic capacity model. Evolution. 1995;49:836–847. doi: 10.1111/j.1558-5646.1995.tb02320.x. [DOI] [PubMed] [Google Scholar]
- Henderson ND. Interpreting studies that compare high- and low-selected lines on new characters. Behav. Genet. 1989;19:473–502. doi: 10.1007/BF01066250. [DOI] [PubMed] [Google Scholar]
- Henderson KK, Wagner H, Favret F, Britton SL, Koch LG, Wagner PD, Gonzalez NC. Determinants of maximal O2 uptake in rats selectively bred for endurance running capacity. J. Appl. Physiol. 2002;93:1265–1274. doi: 10.1152/japplphysiol.00809.2001. [DOI] [PubMed] [Google Scholar]
- Hill CA, Harris RC, Kim HJ, Harris BD, Sale C, Boobis LH, Kim CK, Wise JA. Influence of beta-alanine supplementation on skeletal muscle carnosine concentrations and high intensity cycling capacity. Amino Acids. 2007;32:225–233. doi: 10.1007/s00726-006-0364-4. [DOI] [PubMed] [Google Scholar]
- Jacobs I. Lactate concentrations after short, maximal exercise. Acta Physiol. Scand. 1981;111:465–469. doi: 10.1111/j.1748-1716.1981.tb06764.x. [DOI] [PubMed] [Google Scholar]
- Johnson MS, Thomson SC, Speakman JR. Limits to sustained intake. II. Interrelationships between resting metabolic rate, life-history traits and morphology in Mus musculus. J. Exp. Biol. 2001;204:1937–1946. doi: 10.1242/jeb.204.11.1937. [DOI] [PubMed] [Google Scholar]
- Koch LG, Britton SL. Artificial selection for intrinsic aerobic endurance running capacity in rats. Physiol. Genomics. 2001;5:45–52. doi: 10.1152/physiolgenomics.2001.5.1.45. [DOI] [PubMed] [Google Scholar]
- Konarzewski M, Diamond J. Evolution of basal metabolic rate and organ masses in laboratory mice. Evolution. 1995;49:1239–1248. doi: 10.1111/j.1558-5646.1995.tb04450.x. [DOI] [PubMed] [Google Scholar]
- Koteja P. Maximum cold-induced oxygen consumption in the house sparrow Passer domesticus L. Physiol. Zool. 1986;59:43–48. [Google Scholar]
- Koteja P. On the relation between basal and maximum metabolic rate in mammals. Comp. Biochem. Physiol. 1987;87A:205–208. doi: 10.1016/0300-9629(87)90447-6. [DOI] [PubMed] [Google Scholar]
- Landys MM, Piersma T, Guglielmo CG, Jukema J, Ramenofsky M, Wingfield JC. Metabolic profile of long-distance migratory flight and stopover in a shore-bird. Proc. R. Soc. B. 2005;272:295–302. doi: 10.1098/rspb.2004.2952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Belle JE, Harris NG, Williams SR, Bhakoo KK. A comparison of cell and tissue extraction techniques using high-resolution 1H-NMR spectroscopy. NMR Biomed. 2002;15:37–44. doi: 10.1002/nbm.740. [DOI] [PubMed] [Google Scholar]
- Lumeij JT, Remple JD. Plasma urea, creatinine and uric acid concentrations in relation to feeding in peregrine falcons (Falco peregrinus). Avian Pathol. 1991;20:79–83. doi: 10.1080/03079459108418743. [DOI] [PubMed] [Google Scholar]
- McNab BK. Energetics of arboreal folivores: physiological problems and ecological consequences of feeding on an ubiquitous food supply. In: Montgomery GG, editor. The ecology of arboreal folivores. Smithsonian Institution Press; Washington: 1978. pp. 153–162. [Google Scholar]
- McNab BK. Energy conservation and the evolution of fightlessness in birds. Am. Nat. 1994;144:628–642. [Google Scholar]
- McNab BK. The standard energetics of mammalian carnivores: Felidae and Hyaenidae. Can. J. Zool. 2000;78:2227–2239. [Google Scholar]
- McNab BK. The evolution of energetics in birds and mammals. Univ. Calif. Publ. Zool. 2007;137:67–110. [Google Scholar]
- McNab BK, Ellis HI. Flightless rails endemic to islands have lower energy expenditures and clutch sizes than flighted rails on islands and continents. Comp. Biochem. Physiol. A. 2006;145:295–311. doi: 10.1016/j.cbpa.2006.02.025. [DOI] [PubMed] [Google Scholar]
- Moore KL, Dalley AF. Clinically Oriented Anatomy. 5th ed. Lippincott Williams & Wilkins; Philadelphia: 2006. [Google Scholar]
- Nelson CJ, Otis JP, Carey HV. Global analysis of circulating metabolites in hibernating ground squirrels. Comp. Biochem. Physiol. D. 2010;5:265–273. doi: 10.1016/j.cbd.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Petridou A, Nikolaidis MG, Matsakas A, Schulz T, Michna H, Mougios V. Effect of exercise training on the fatty acid composition of lipid classes in rat liver, skeletal muscle, and adipose tissue. Eur. J. Appl. Physiol. 2005;94:84–92. doi: 10.1007/s00421-004-1294-z. [DOI] [PubMed] [Google Scholar]
- Piersma T. Why marathon migrants get away with high metabolic ceilings: towards an ecology of physiological restraint. J. Exp. Biol. 2010;214:295–302. doi: 10.1242/jeb.046748. [DOI] [PubMed] [Google Scholar]
- Raichlen DA, Gordon AD, Muchlinski MN, Snodgrass JJ. Causes and significance of variation in mammalian basal metabolism. J. Comp. Physiol. B. 2010;180:301–311. doi: 10.1007/s00360-009-0399-4. [DOI] [PubMed] [Google Scholar]
- Ramadan Z, Jacobs D, Grigorov M, Kochhar S. Metabolic profiling using principal component analysis, discriminant partial least squares, and genetic algorithms. Talanta. 2006;68:1683–1691. doi: 10.1016/j.talanta.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Refinetti R. Variability of diurnality in laboratory rodents. J. Comp. Physiol. A. 2006;192:701–714. doi: 10.1007/s00359-006-0093-x. [DOI] [PubMed] [Google Scholar]
- Rezende EL, Swanson DL, Novoa FF, Bozinovic F. Passerines versus nonpasserines: so far, no statistical differences in avian energetics. J. Exp. Biol. 2002;205:101–107. doi: 10.1242/jeb.205.1.101. [DOI] [PubMed] [Google Scholar]
- Rezende EL, Chappell MA, Gomes FR, Malisch JL, Garland T., Jr. Maximal metabolic rates during voluntary exercise, forced exercise, and cold exposure in house mice selectively bred for high wheel-running. J. Exp. Biol. 2005;208:2447–2458. doi: 10.1242/jeb.01631. [DOI] [PubMed] [Google Scholar]
- Rolfe D, Brown G. Cellular energy utilization and molecular origin of standard metabolic rate in mammals. Physiol. Rev. 1997;77:731–758. doi: 10.1152/physrev.1997.77.3.731. [DOI] [PubMed] [Google Scholar]
- Selman C, Lumsden S, Bünger L, Hill WG, Speakman JR. Resting metabolic rate and morphology in mice (Mus musculus) selected for high and low food intake. J. Exp. Biol. 2001;204:777–784. doi: 10.1242/jeb.204.4.777. [DOI] [PubMed] [Google Scholar]
- Speakman JR. The physiological costs of reproduction in small mammals. Phil. Trans. R. Soc. B-Biol. Sci. 2008;363:375–398. doi: 10.1098/rstb.2007.2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman JR, Johnson MS. Relationships between resting metabolic rate and morphology in lactating mice: what tissues are the major contributors to resting metabolism? In: Heldmaier G, Klingenspor M, editors. Living in the Cold. Vol. 3. Springer-Verlag; Berlin: 2000. pp. 479–486. [Google Scholar]
- Stearns SC. The Evolution of Life Histories. Oxford University Press; Oxford: 1992. [Google Scholar]
- Storey JD. A direct approach to false discovery rates. J. R. Stat. Soc. B. 2002;64:479–498. [Google Scholar]
- Swallow JG, Garland T, Jr., Carter PA, Zhan W-Z, Sieck GC. Effects of voluntary activity and genetic selection on aerobic capacity in house mice (Mus domesticus). J. Appl. Physiol. 1998;84:69–76. doi: 10.1152/jappl.1998.84.1.69. [DOI] [PubMed] [Google Scholar]
- Swallow JG, Hayes JP, Koteja P, Garland T., Jr. Selection experiments and experimental evolution of performance and physiology. In: Garland T Jr., Rose MR, editors. Experimental evolution: concepts, methods, and applications of selection experiments. University of California Press; Berkeley, California: 2009. pp. 301–351. [Google Scholar]
- Szabo A, Romvari R, Febel H, Bogner P, Szendro Z. Training-induced alterations of the fatty acid profile of rabbit muscles. Acta Vet. Hung. 2002;50:357–364. doi: 10.1556/AVet.50.2002.3.11. [DOI] [PubMed] [Google Scholar]
- Taylor CR, Weibel ER. Design of the mammalian respiratory system. I: problem and strategy. Respir. Physiol. 1981;44:1–10. [PubMed] [Google Scholar]
- Thienen RV, Proeyen KV, Eynde BV, Puype J, Lefere T, Hespel P. β-alanine improves sprint performance in endurance cycling. Med. Sci. Sport Exerc. 2009;41:898–903. doi: 10.1249/MSS.0b013e31818db708. [DOI] [PubMed] [Google Scholar]
- Veiga JAS, Roselino ES, Migliorini RH. Fasting, adrenalectomy, and gluconeo-genesis in the chicken and a carnivorous bird. Am. J. Physiol. 1978;234:R115–R121. doi: 10.1152/ajpregu.1978.234.3.R115. [DOI] [PubMed] [Google Scholar]
- Wagner PD. Determinants of maximum oxygen transport and utilization. Annu. Rev. Physiol. 1996;58:21–50. doi: 10.1146/annurev.ph.58.030196.000321. [DOI] [PubMed] [Google Scholar]
- Want EJ, Nordstrom A, Morita H, Siuzdak G. From exogenous to endogenous: the inevitable imprint of mass spectrometry in metabolomics. J. Proteome Res. 2007;6:459–468. doi: 10.1021/pr060505+. [DOI] [PubMed] [Google Scholar]
- Weibel ER, Hoppeler H. Exercise-induced maximal metabolic rate scales with muscle aerobic capacity. J. Exp. Biol. 2005;208:1635–1644. doi: 10.1242/jeb.01548. [DOI] [PubMed] [Google Scholar]
- Weibel ER, Bacigalupe LD, Schmitt B, Hoppeler H. Allometric scaling of maximal metabolic rate in mammals: muscle aerobic capacity as determinant factor. Respir. Physiol. Neurobiol. 2004;140:115–132. doi: 10.1016/j.resp.2004.01.006. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Pendyala G, Siuzdak G, Fox HS. Metabolomic analysis of the cerebrospinal fluid reveals changes in phospholipase expression in the CNS of SIV-infected macaques. J. Clin. Invest. 2008;118:2661–2669. doi: 10.1172/JCI34138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wone B, Sears MW, Labocha MK, Donovan ER, Hayes JP. Genetic variances and covariances of aerobic metabolic rates in laboratory mice. Proc. R. Soc. B. 2009;276:3695–3704. doi: 10.1098/rspb.2009.0980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zera AJ, Harshman LG. The physiology of life history trade-offs in animals. Annu. Rev. Ecol. Syst. 2001;32:95–126. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.