SUMMARY
Since apoptosis of infected cells can limit virus production and spread, some viruses have co-opted prosurvival genes from the host. This includes the Epstein-Barr virus (EBV) gene BHRF1, a homologue of human Bcl-2 proteins that block apoptosis and are associated with cancer. Computational design and experimental optimization were used to generate a novel protein called BINDI that binds BHRF1 with picomolar affinity. BINDI recognizes the hydrophobic cleft of BHRF1 in a manner similar to other Bcl-2 protein interactions, but makes many additional contacts to achieve exceptional affinity and specificity. BINDI induces apoptosis in EBV-infected cancer lines, and when delivered with an antibody-targeted intracellular delivery carrier, BINDI suppressed tumor growth and extended survival in a xenograft disease model of EBV-positive human lymphoma. High specificity designed proteins that selectively kill target cells may provide an advantage over the toxic compounds used in current generation antibody-drug conjugates.
INTRODUCTION
Following virus infection, cells may undergo apoptosis to prevent further virus spread in the host. This has spurred viruses to evolve counteracting mechanisms to prevent host cell death, and during latent infection these factors may contribute to the development of cancer. In particular, there are multiple cancers associated with Epstein-Barr virus (EBV), including Burkitt's lymphoma (BL).
Apoptosis and cell survival are regulated by the homeostatic balance of B cell lymphoma-2 (Bcl-2) family proteins (reviewed in (Martinou and Youle, 2011)), which fall in to three classes. The `executioners', Bak and Bax, initiate apoptosis by increasing mitochondrial outer membrane permeability and facilitating the release of mitochondrial cytochrome c to the cytosol, which activates downstream signaling. Six human prosurvival Bcl-2 proteins (Bcl-2, Bcl-XL, Bcl-B, Mcl-1, Bcl-w and Bfl-1) inhibit this process. Counterbalancing these are numerous proapoptotic BH3-only proteins, including Bim. These factors share a ~26 residue Bcl-2 homology 3 (BH3) motif, an amphipathic α-helical element which binds a hydrophobic groove on the surface of the canonical Bcl-2 fold. Cellular stresses activate proapoptotic BH3-only proteins, which bind and inhibit prosurvival Bcl-2 members, and directly interact with Bak and Bax to favor mitochondrial permeabilization. Conversely, prosurvival Bcl-2 proteins dampen apoptotic triggers and enhance chemoresistance by sequestering BH3-only proteins or directly inhibiting Bak and Bax. Increased expression of prosurvival Bcl-2 proteins is a common feature of many cancers.
Epstein-Barr virus encodes a prosurvival Bcl-2 homologue, BHRF1, which prevents lymphocyte apoptosis during initial infection by sequestering proapoptotic BH3-only proteins (especially Bim), and interacting directly with the executioner Bak (Altmann and Hammerschmidt, 2005; Desbien et al., 2009; Henderson et al., 1993; Kvansakul et al., 2010). Even though BHRF1 is under the control of an early lytic cycle promoter, low levels of constitutive expression have been observed in some cases of EBV-positive BL when the virus is latent, and it has been speculated that BHRF1 may be a necessary viral factor for lymphomagenesis (Kelly et al., 2009; Leao et al., 2007; Watanabe et al., 2010). However, although EBV is one of the earliest viruses to be associated with human cancer, its molecular mechanism of action remains unclear, in part because EBV-positive BLs can have different expression profiles, suggesting the molecular etiology may not be universal (Kelly et al., 2009; Watanabe et al., 2010).
In recent years, computation-based protein design has made considerable progress, including the design of new hyperstable structures (Koga et al., 2012) and the creation of functional sites within existing proteins as scaffolds (Fleishman et al., 2011; Procko et al., 2013). However, except in a few rare instances (Correia et al., 2014; Lanci et al., 2012), the design of new structures that are also functional is a largely unmet challenge. Here, we describe the creation of a picomolar inhibitor of BHRF1 by designing a new protein de novo for optimum interactions. The designed BHRF1 inhibitor triggers apoptosis in several EBV-positive cancer lines, and when delivered with an antibody-targeted carrier system, the inhibitor suppresses tumor progression and extends survival in an animal model of human EBV-positive lymphoma.
RESULTS
BHRF1-binding proteins created by grafting Bim-BH3 side chains to a helical scaffold
Prosurvival Bcl-2 proteins share a similar fold that resembles a cupped hand, with a characteristic hydrophobic surface groove that clasps one side of an amphipathic BH3 motif helix (Kvansakul et al., 2010; Martinou and Youle, 2011). Rigidifying BH3 peptides with hydrocarbon staples, disulfides or lactam bridges on the non-interactive back side of the helix can reduce the entropic cost of a partially-folded peptide acquiring a rigid helical conformation upon binding, and improves BH3 peptide affinity (Azzarito et al., 2013). We reasoned that building a folded structure around a BH3 peptide would similarly pre-stabilize the bound conformation, but also provide additional interactions for increased affinity and specificity. In previous work, interacting residues of the BH3 motif were grafted to the surface of a minimal structured peptide, but after directed evolution these folded peptides displayed only moderate affinity and specificity, and did not always bind to the correct site on the target Bcl-2 protein (Chin and Schepartz, 2001; Gemperli et al., 2005). We instead sought to incorporate the interacting residues of the BH3 motif onto an exposed surface of a larger 3- or 4-helix bundle protein, which would thereby allow additional contacts for increased affinity and specificity extending beyond the BH3 motif.
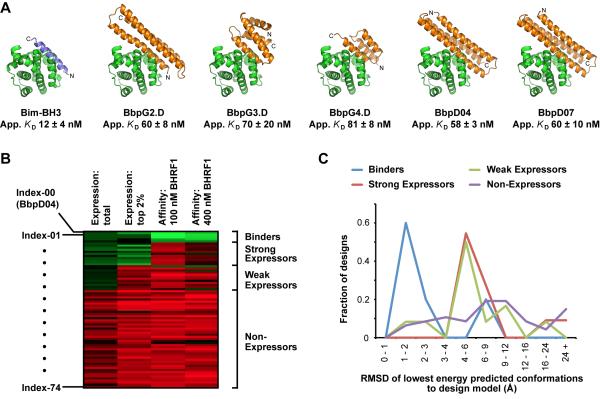
Two computational design strategies were used. The first was a side chain grafting approach onto existing natural proteins (Figure S1A online) (Correia et al., 2010; Ofek et al., 2010). Three helical-bundle proteins were chosen as scaffolds with different sizes and shapes for diversity: the small 3-helix protein Z (PDB 1LP1 (Hogbom et al., 2003)), a larger 3-helix bundle derived from a bacterial ribosome recycling factor (3LHP chain S (Correia et al., 2010)), and a 4-helix viral coat protein (3FBL (Goulet et al., 2009)). Beginning with the crystal structure of BHRF1 bound to a Bim-BH3 peptide (Kvansakul et al., 2010), helical regions within the scaffolds were structurally aligned to the Bim-BH3 helix. The scaffold positions corresponding to Bim-BH3 residues which interact with BHRF1 were then mutated to the Bim-BH3 amino acids. Next, scaffold residues surrounding the graft site were redesigned to minimize the calculated energy of the modeled bound complex (Kuhlman et al., 2003; Leaver-Fay et al., 2011). BHRF1 interface residues, which normally reach over the backside of the Bim-BH3 helix, were simultaneously repacked to alternative low energy rotamers compatible with the new designed interface. From six different graft sites on the three scaffolds, three proteins (BbpG2.D, BbpG3.D and BbpG4.D) were obtained with moderate affinities for BHRF1 (apparent KDs of approximately 60 to 80 nM as evaluated using yeast surface display titrations, Figure 1A, Tables 1 and S1). These affinities are weaker than the wild-type interaction of Bim-BH3 with BHRF1, which has an apparent KD of 12 nM (Table 1), probably reflecting non-optimal backbone conformations and/or slightly unfavorable designed interactions outside the core motif. To evaluate the importance of the additional designed interactions outside the core motif, control designs were produced in which Bim-BH3 binding residues were transplanted to the scaffolds without redesign of the surrounding surface. These control designs did not bind BHRF1 (Table S1), indicating that computational design of novel interactions outside the core motif was necessary.
Figure 1. Computationally-designed proteins incorporating elements of the Bim-BH3 motif bind BHRF1.
(A) Computational models of individually-tested designed proteins (orange) that bind BHRF1 (green). The crystal structure of BHRF1 bound to Bim-BH3 (blue) is shown at left for comparison. BbpG designs are from side chain grafting, and BbpD designs are de novo assembled proteins. Apparent affinities (mean ± SE, n = 3–6) are from yeast display titrations. See Figure S1, and Tables S1 and S2. An archive file of designed structures is provided in Supplemental Information online.
(B) Seventy-four computationally designed proteins without human modifications (Indexes-01 to 74) were included in a yeast display library. BbpD04 (Index-00) was included as a positive control. The library was sorted for cells expressing surface protein (lane 1), for the 2% of cells with highest expression (lane 2), and for cells showing binding signal after incubation with 100 nM (lane 3) or 400 nM BHRF1 (lane 4). The gene frequencies in the sorted population were divided by their frequencies in the naive library to calculate a log2 enrichment ratio, plotted from −4 (i.e. depleted, red) to +4 (i.e. enriched, green). See Table S3.
(C) Histogram of the mean RMSD between the ten lowest energy structures found in ab initio structure prediction calculations and the intended designed structure for each of the sets of designs in (B). Designs with computed energy minima near the designed target conformation have a higher probability of binding BHRF1. See Figure S2.
Table 1.
Apparent dissociation constants (nM; mean ± SE, n = 3–6) from yeast surface display titrations
| Protein | BHRF1 | Bcl-2 | Bcl-w | Mcl-1 | Bfl-1 | Bcl-XL | Bcl-B |
|---|---|---|---|---|---|---|---|
| Bim-BH3 | 12 ± 4 | 2.02 ± 0.08 | 2.1 ± 0.1 | 0.6 ± 0.2 | 2.1 ± 0.3 | 3 ± 1 | 12.2 ± 0.1 |
| BbpG2.D | 60 ± 8 | 9 ± 1 | > 100 | 1.5 ± 0.3 | – | 106 ± 9 | > 100 |
| BbpG3.D | 70 ± 20 | 20 ± 2 | 4 ± 2 | 9 ± 2 | – | 31 ± 3 | 79 ± 8 |
| BbpG4.D | 81 ± 8 | > 100 | 35 ± 4 | 20 ± 3 | > 100 | > 100 | 80 ± 4 |
| BbpD07 | 60 ± 10 | 76 ± 7 | – | 3.1 ± 0.3 | > 100 | – | > 100 |
|
| |||||||
| BbpD04 | 58 ± 3 | – | – | 17 ± 7 | > 100 | – | – |
| BbpD04.1 | 8 ± 4 | 110 ± 20 | 14 ± 5 | 30 ± 10 | > 100 | 25 ± 1 | – |
| BbpD04.2 | 0.6 ± 0.2 | 33 ± 4 | 40 ± 10 | 26 ± 4 | 70 ± 20 | 31 ± 2 | – |
| BbpD04.3 | 0.54 ± 0.01 | 20 ± 2 | 34 ± 3 | 19 ± 1 | 32 ± 6 | 34 ± 7 | – |
| BINDI | 0.9 ± 0.2 | 45 ± 7 | 60 ± 10 | 21.6 ± 0.8 | > 100 | > 100 | – |
|
| |||||||
| Accurate dissociation constants (nM; mean ± SD, n = 4–6) measured by BLI | |||||||
|
| |||||||
| Bim-BH3 | 7 ± 3 | 0.75 ± 0.09 | 20 ± 10 | 0.17 ± 0.02 | 0.61 ± 0.04 | 1.56 ± 0.09 | 7 ± 2 |
| BINDI | 0.22 ± 0.05 | 2,100 ± 100 | 870 ± 40 | 40 ± 10 | 2,600 ± 800 | 810 ± 80 | > 10,000 |
| BINDI N62S | 0.16 ± 0.08 | 30,000 ± 10,000 | 4,600 ± 400 | 230 ± 40 | 4,000 ± 2,000 | 8,000 ± 2,000 | 50,000 ± 10,000 |
Creating new proteins for optimized interactions with the BHRF1 ligand-binding groove
Designed proteins from side chain grafting are limited by the assumed rigid backbone of the scaffold. To escape this constraint, a second design strategy was used that builds protein structures de novo using proteins of known structure only as topology guides (Correia et al., 2014). The Bim-BH3 helix from the complex with BHRF1 was taken as a folding nucleus, and helical bundle structures were assembled around it using fragments from the PDB, followed by cycles of sequence design and structure minimization. Cα-Cα atom-pair distances from the topology guide constrained the assembling protein to within 3.0 Å root mean square deviation (RMSD) of the guide. Independent runs of this stochastic design protocol gave an ensemble of structural homologues, roughly centered on the guide scaffold structure (Figure S1B). This protocol was previously applied to the design of an immunogenic protein incorporating a viral epitope, but in that case the incorporated epitope provided all necessary interactions with a target antibody (Correia et al., 2014). Here, the incorporated Bim-BH3 motif provides only a fraction of the interaction surface (approximately 40%), with potential for many additional contacts across an expansive interface.
Each designed protein in the ensemble was docked to the BHRF1 surface via alignment of the incorporated Bim-BH3 motif, and the surrounding interface residues were optimized as described above. The designs were filtered both for stability of the monomer and for interface quality. From thousands of computer-assembled proteins, a small number of designs were selected for further manual modifications, synthetic E. coli codon-optimized genes were constructed, and those proteins that were expressed and soluble in E. coli were tested by yeast surface display for binding to BHRF1 (Table S2; yeast surface display allows rapid measurement of approximate affinities and is a powerful technology for later directed evolution). Two designs with folds based on PDB 3LHP(S) were found to bind BHRF1 with apparent KDs of 58–60 nM (BbpD04 and BbpD07; Figure 1A and Table 1). These designs were `seeded' by a fifteen-residue fragment of the Bim-BH3 motif of which nine side chains contacting the BHRF1 surface were kept fixed. Other residues, primarily on the backside of the motif and buried in the protein core, were designed to minimize the calculated potential energy. The equivalent 3LHP(S) fixed backbone graft (design BbpG1.D, Table S1) had numerous steric clashes between the scaffold backbone and BHRF1 surface in the computational model (described later), and failed to bind BHRF1.
In silico folding probability correlates with binding activity
The success rate for designing functional proteins is low, and design calculations are often followed by substantial human intervention to choose and modify the designs prior to experimental validation (Figure S2A). To investigate the factors distinguishing working from non-working designs and evaluate the importance of human intervention, we characterized BHRF1 binding to a set of computational designs generated as described for BbpD04 and BbpD07, but without any human modifications. Seventy-four designs were displayed on yeast and sorted with a single round of fluorescence-activated cell sorting (FACS) for surface protein expression and BHRF1 binding. Deep sequencing revealed that five designs (Indexes-00 to 04) were highly expressed and enriched after sorting for BHRF1 interaction (Figures 1B and S2A–D, Table S3); BHRF1 binding was confirmed on clonal yeast populations (Figure S2E). Standard computational metrics for assessing interface quality or monomer stability did not distinguish the working designs (Figure S2F–K). Instead, we found that BHRF1 binding strongly correlated with the extent to which the designed sequences folded to the designed structures in ab initio structure prediction calculations (Figure 1C and S2L). Rather than a single calculated metric from a static representation of the structure, ab initio structure prediction generates tens of thousands of conformations from stochastic folding simulations. Amongst the designs that bind BHRF1, the closer the predicted conformations are to the designed structure (i.e. lower RMSD in Figure 1C), the lower their calculated energy and hence the higher the probability of correctly folding. This observation suggests that the primary shortcomings of inactive designs are improper folding and designed state instability, rather than the details of the designed protein-protein interface.
Enhanced BHRF1 binding affinity and specificity through improved electrostatic complementarity
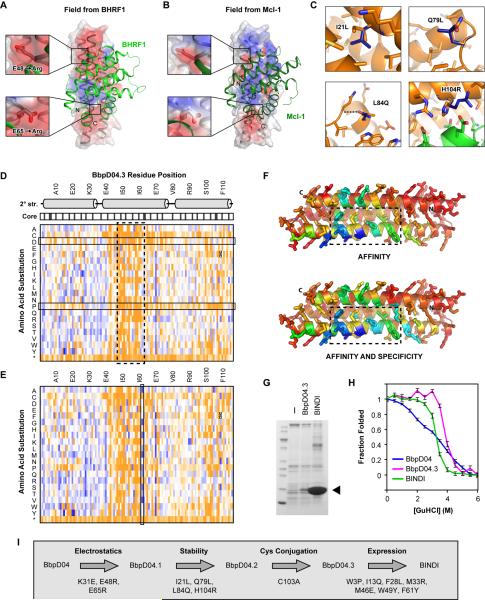
To illuminate BHRF1 biology, the designed protein must not only bind with high affinity, but do so specifically. Design BbpD04, a de novo designed protein without sequence homologues identified by BLAST (Altschul et al., 1997), bound BHRF1 with higher affinity (apparent KD = 58 ± 3 nM) than all human prosurvival Bcl-2 proteins except Mcl-1 (Table 1), and was chosen for further optimization. The computed electric field (Baker et al., 2001) experienced by BbpD04 when bound to BHRF1 is markedly more negative than when bound to Mcl-1 (Figure 2A–B). Nine point substitutions were made to eight residues of BbpD04 to specifically increase electrostatic complementarity for BHRF1. Six decreased the KD(BHRF1)/KD(Mcl-1) ratio as predicted (Table S4). The variant BbpD04.1 containing the best two point mutations (E48R and E65R), together with a third compensatory mutation (K31E) to preserve a putative stabilizing salt-bridge, bound BHRF1 with tighter affinity (Table 1).
Figure 2. Affinity maturation of BbpD04.
(A) Computational model of BHRF1 (light green ribbon) bound to design BbpD04 (transparent surface over a grey ribbon). The electrostatic potential from BHRF1 is shown on the BbpD04 surface (red, −2 kT/e to blue, +2 kT/e; calculated using the Adaptive Poisson-Boltzmann Solver built in ROSETTA). Mutations E48R and E65R (insets) decrease electrostatic repulsion in regions where the field from BHRF1 is negative.
(B) Same as in (A), with Mcl-1 in place of BHRF1. E48 and E65 are now in regions that border positive potential from Mcl-1. The Mcl-1•BbpD04 model was generated by superposition of the BbpD04 binding site to Bim-BH3 bound to Mcl-1 in crystal structure 2PQK, followed by rotamer repacking and side chain/backbone minimization in ROSETTA. See Table S4.
(C) The four additional mutations in BbpD04.2 (dark blue sticks) are shown on the computational model of BbpD04.1 (orange) bound to BHRF1 (green). Three mutations are distant from the interface. See Figure S3 and S4.
(D) Sequence-fitness landscape. All single amino acid substitutions of BbpD04.3 were expressed in a yeast display library. The 1% of cells with highest binding signal for 400 pM biotinylated BHRF1 relative to surface expression were collected by FACS. Plotted for each substitution is the log2 enrichment ratio from −3.5 (depleted, orange) to +3.5 (enriched, blue). Stop codons, *. The region of the incorporated Bim-BH3 motif is boxed with a broken line. Secondary structure and core residues (shaded grey) are indicated above. Substitutions to aspartate (tend to be depleted for core residues) and to proline (depleted for helical residues) are boxed.
(E) As in (D), except the library was sorted for binding to 400 pM biotinylated BHRF1 in the presence of 8 nM competitor Bcl-2 proteins for specificity. Substitutions of N62 are boxed.
(F) The modeled structure of BbpD04.3 is colored by sequence Shannon entropy from 2.8 (highly conserved, dark blue) to 4.3 (variable, red), based on the sequence-fitness landscapes. A broken line boxes the incorporated Bim-BH3 motif.
(G) BbpD04.3 and its derivative BINDI were expressed as 6his-tagged proteins in E. coli, precipitated from cleared lysate with NiNTA-agarose and analyzed on a Coomassie-stained SDS-polyacrylamide electrophoretic gel. An arrowhead indicates the expected MW at 15 kD. See Figure S5.
(H) The fraction of protein folded in the presence of guanidinium hydrochloride, based on the change in CD signal at 222 nm.
(I) Summary of all mutations made to BbpD04 during affinity maturation.
Optimization of affinity, specificity and stability
Error-prone PCR mutagenesis of BbpD04.1 followed by FACS for BHRF1 binding in the presence of unlabeled human Bcl-2 proteins to favor specificity identified two mutations at the designed interface (H104R, predicted to enhance electrostatic complementarity, and N62S, predicted to improve specificity based on sequence-fitness landscape mapping described below), and three mutations distal from the interface that might alter protein stability. I21L slightly alters packing in the hydrophobic core, Q79L increases hydrophobic interactions buttressing the second connecting loop, and L84Q forms a stabilizing hydrogen bond to the loop backbone. The substitutions were recombined in a combinatorial library, and FACS followed by analysis of expression in E. coli identified a clone, BbpD04.2 (with four mutations, see Figure 2C), that was monodisperse and monomeric by size exclusion chromatography (SEC) after protein purification from E. coli (Figure S3). To enable site-specific chemical coupling for later characterization, the single internal cysteine of BbpD04.2 was removed by mutagenesis (mutant C103A, called BbpD04.3; Table 1 and Figure S4).
To probe the sequence-fitness landscape of the designed protein, site-specific saturation mutagenesis was used to independently diversify every codon of the BbpD04.3 gene to NNK (N is any base, K is G or T), producing a library of (116 positions) × (20 amino acids + stop codon) = 2,436 protein variants. The variants were expressed on yeast and the library was sorted by a single round of FACS for BHRF1 affinity (Figure 2D) or affinity-and-specificity (unlabeled human Bcl-2 proteins were included as competitors; Figure 2E). DNA was extracted from the naive and post-sorted yeast populations, and the BbpD04.3 coding region was deep sequenced. The ratio of the frequencies of each protein variant in the pre-sorted and post-sorted populations reports on the effect of each substitution on affinity/specificity fitness.
The comprehensive BbpD04.3 affinity sequence-fitness landscape reveals the critical nature of the incorporated Bim-BH3 motif, with most substitutions of interface residues depleted (Figure 2D). In addition, substitutions to proline, which can break regular helical secondary structure, are depleted across the first, second and third helical spans of the designed helical bundle fold. Substitutions to aspartate, a short and charged amino acid, are depleted within the hydrophobic core as anticipated. The BbpD04.3 affinity-specificity sequence-fitness landscape, in which unlabeled Bcl-2 proteins were included as competitors for BHRF1 binding, is similar (Figure 2E). Notably, several mutations of Asn62, including N62S described in detail later, were found to selectively increase BHRF1 specificity.
Bacterial expression of BbpD04.3 was very low, limiting the quantity and purity that could be purified for biochemical applications. To improve expression, twenty-nine BbpD04.3 point mutants with positive enrichment ratios in either the affinity or affinity-specificity sequence-fitness landscapes were expressed in E. coli and analyzed for increased soluble protein levels (Figure S5A). Nine mutations were identified: W3A and W3P increase helical propensity of the initiating residue in the starting helix; and I13Q, F28L, M33R, F61Y, W49E/Y and M46E decrease surface hydrophobicity. These substitutions perhaps reduce misfolding or aggregation, thereby reducing clearance of the protein by the quality control machinery. Because the mutations are generally surface-exposed at distinct sites on a long helical bundle, we reasoned they could likely be combined without negative interference (Figure S5B). A BbpD04.3 variant with seven mutations (Figure S5C) had significantly increased bacterial expression and improved specificity (Figure 2G and Table 1). This variant is named BHRF1-INhibiting Design acting Intracellularly (BINDI). A summary of all mutations introduced in the original design is provided in Figure 2I.
The increased affinity for BHRF1 following in vitro evolution correlates with enhanced protein stability. Both BbpD04.3 and BINDI undergo cooperative unfolding at high concentrations (> 3 M) of the chemical denaturant guanidinium hydrochloride measured by circular dichroism (CD) spectroscopy (Figure 2H). However, the original design, BbpD04, has nearly linear loss of CD signal over a wide concentration range of guanidinium hydrochloride (Figure 2H). The absence of a cooperative melting transition is associated with molten globules that lack a rigid core or single native conformation. While BbpD04, BbpD04.3 and BINDI have high thermostability and retain partly α-helical CD spectra at 95°C, only evolved BbpD04.3 and BINDI fully renature when the heated proteins are cooled (Figure S5D–G). Further, the original BbpD04 design is sensitive to rapid hydrolysis by proteases, which require unfolded substrate backbone to access the enzyme active site, whereas BINDI and BbpD04.3 are resistant (Figure S5H).
BINDI binds BHRF1 with high affinity and specificity
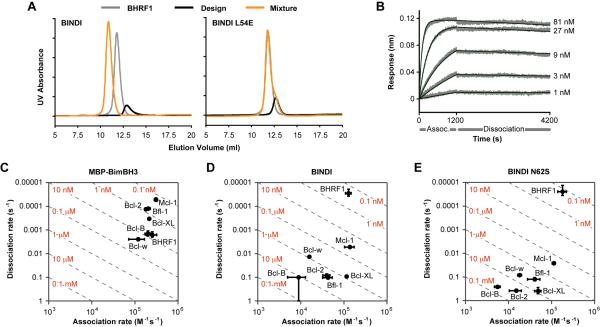
Apparent dissociation constants by yeast surface display are useful approximations, but may be artificially tight due to avidity effects or ligand rebinding to a dense receptor surface, or may be artificially weak if binding equilibrium is not reached. The BINDI•BHRF1 interaction was therefore further characterized by alternative methods. BINDI eluted as a higher molecular weight complex by SEC when mixed with BHRF1 in solution, whereas BINDI L54E with a knockout mutation in the designed interface did not (Figures 3A and S3). Using bio-layer interferometry (BLI) to measure the kinetic rate constants, BINDI•BHRF1 was found to form a very tight complex (KD 220 ± 50 pM) with a slow dissociation rate (koff = [2.8 ± 0.9] × 10−5 s−1) (Figure 3B and 3D). BINDI has high specificity for BHRF1; the dissociation constants for human Bcl-2 family members are much weaker. BINDI bound human Mcl-1 with KD 40 ± 10 nM (180-fold increase compared to BHRF1), Bcl-2 with KD 2.1 ± 0.1 μM (10,000-fold increase), Bcl-w with KD 870 ± 40 nM (4,000-fold increase), Bfl-1 with KD 2.6 ± 0.8 μM (12,000-fold increase), Bcl-B with KD > 10 μM (> 45,000-fold increase) and Bcl-XL with KD 810 ± 80 nM (4,000-fold increase). The N62S variant of BINDI was found to have even higher specificity for BHRF1 versus Mcl-1, the closest competitor (Figure 3E). Compared to the measured affinities of Bcl-2 proteins for Bim-BH3 (Figure 3C) and to other published values (Caria et al., 2012; Dutta et al., 2013; Dutta et al., 2010; Gemperli et al., 2005; Kvansakul et al., 2010; Lessene et al., 2013; Tse et al., 2008), the affinity and specificity of BINDI for BHRF1 is considerably greater than any previously described BHRF1 ligand, and is similar to or exceeds that of any other protein, peptide or drug designed to specifically bind a Bcl-2 family protein.
Figure 3. BINDI binds BHRF1 with high affinity and specificity.
(A) BINDI or knockout mutant BINDI L54E were mixed with BHRF1 and separated by SEC.
(B) Biotinylated BHRF1 was immobilized to a BLI sensor and the interaction with BINDI was measured at the indicated concentrations.
(C–E) BLI kinetic analysis of interactions between immobilized Bcl-2 proteins and soluble Bim-BH3 fused to the C-terminus of maltose-binding protein (C), BINDI (D) and BINDI N62S (E). Red labels on broken diagonal lines indicate the corresponding affinities / KD. Plotted are means ± SD from 4–6 experiments.
The crystal structure of BINDI•BHRF1
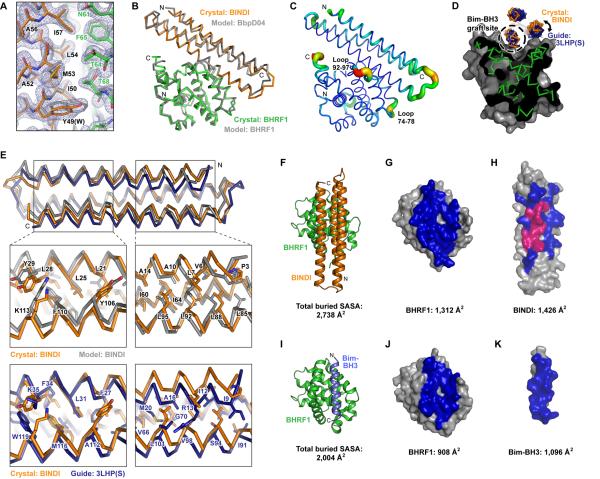
The crystal structure of the BINDI•BHRF1 complex was determined at 2.05 Å resolution (Figure 4A and Table S5) and found to closely match the design model. The asymmetric unit contains two nearly identical copies of BINDI-bound BHRF1; the analysis presented here corresponds to one copy of the complex (Figure S6A–B). The backbones of the crystal structure and the original BbpD04•BHRF1 model align with a Cα-Cα RMSD of 1.06 Å (Figure 4B). The designed core and interface regions were modeled accurately, while connecting loops at the ends of the helical bundle deviate slightly from the computational prediction (Figure 4C). Excluding flexible termini, only two regions differ substantially from the model; BHRF1 loop 74–78 and BHRF1 loop 92–97 are located further away from the interface in the crystal structure by 3 to 5 Å (Figure 4C and S6C–D). Our computational modeling assumed that the target BHRF1 backbone was rigid, and therefore changes to the target structure could not be anticipated. These regions have been found to be flexible in BHRF1 and other Bcl-2 family members to accommodate the binding of different BH3 motif ligands (Fire et al., 2010; Kvansakul et al., 2010; Lee et al., 2009).
Figure 4. The crystal structure of BINDI-bound BHRF1.
(A) Electron density at 2.0 σ (blue mesh) in the region of the Bim-BH3 incorporation site. The crystallographic model of BINDI (orange) bound to BHRF1 (green) is superimposed on the original starting computational model of BbpD04•BHRF1 (grey). Residues of BINDI and BHRF1 are labeled black and green, respectively, and residues of BbpD04 prior to affinity maturation that differ are indicated in parentheses. See Figure S6.
(B) The crystal structure of BINDI bound to BHRF1 superimposed on the original BbpD04•BHRF1 model.
(C) Agreement between the crystal structure and computational model is represented from low (thin blue tubing) to higher Cα-Cα RMSD (thick red tubing).
(D) Slice through the crystal structure of BINDI (orange ribbon) bound to BHRF1 (green ribbon with grey surface). The guiding scaffold 3LHP(S) (dark blue) is aligned to BINDI at the Bim-BH3 incorporation site. A direct graft of the BH3 motif into 3LHP(S) at this position causes clashes elsewhere with the BHRF1 surface.
(E) Aligned ribbon traces of BINDI from the BHRF1•BINDI crystal structure in orange; the computational model of BINDI in grey; and the crystal structure of the guiding scaffold 3LHP(S) in dark blue. The three structures are aligned at the Bim-BH3 incorporation site on the middle helix in the back. The upper magnified insets show the agreement between core residue conformations in the BINDI computational model versus the crystal structure. The lower magnified insets show that the core residues of BINDI are distinct in both identity and position from those in the guiding scaffold. When aligned at the Bim-BH3 incorporation site, the crystal structure is closer to the design model than the guiding scaffold.
(F) Crystal structure of BINDI (orange) bound to BHRF1 (green).
(G) The surface of BHRF1, in the same orientation as in (F), with the buried contact surface in BHRF1•BINDI colored blue.
(H) The surface of BINDI, rotated 180º compared to the orientation in (F), with the buried contact surface in BHRF1•BINDI colored. Buried residues whose identities were taken from the incorporated Bim-BH3 fragment are magenta. Buried residues that were designed are blue.
(I) The crystal structure (PDB 2WH6) of Bim-BH3 (blue) bound to BHRF1 (green). Buried surface areas are calculated based on the 22 ordered residues of Bim built into the electron density (a 26-residue Bim-BH3 peptide was crystallized with BHRF1 (Kvansakul et al., 2010)).
(J) The surface of BHRF1, in the same orientation as in (I), with the buried contact surface in BHRF1•Bim-BH3 colored blue.
(K) The surface of Bim-BH3, rotated 180º compared to the orientation in (I), with the buried contact surface in BHRF1•Bim-BH3 colored blue.
BINDI incorporates the Bim-BH3 motif within a de novo designed fold guided by the topology of PDB 3LHP chain S. As noted earlier, the direct graft of Bim-BH3 interaction residues to the equivalent site within the 3LHP(S) scaffold (design BbpG1) failed to bind BHRF1, even after extensive design of the surrounding interaction surface (design BbpG1.D; Table S1). While 3LHP(S) is structurally similar to BINDI, it is nonetheless a poor steric fit for the BHRF1 binding groove in this design protocol. Aligning the graft site within 3LHP(S) to the Bim-BH3 motif of BINDI in the BINDI•BHRF1 crystal structure shows that the C-terminal helix of the grafted design comes too close to the BHRF1 surface, such that side chains would clash (Figure 4D). An alignment of the crystal structure, design model and guide scaffold over the Bim-BH3 incorporation site shows that the crystal structure is closer to the design model than to the guide scaffold (Figure 4E). The RMSD between the crystal and design models is 1.37 Å, and between the crystal and guide scaffold is 1.86 Å. Thus, the design process succeeded in identifying a sequence that folds more closely to the designed model than to the structure of the initial guide scaffold.
Compared to the native Bim-BH3 interaction, BINDI contacts an additional 404 Å2 on the surface of BHRF1 (Figure 4F–K). Residues from the Bim fragment that seeded design account for just 587 Å2 of the BINDI surface buried in the complex, whereas designed residues account for 839 Å2 (Figure 4H). Only two residues at the periphery of the incorporated Bim-BH3 motif changed during affinity maturation (the conservative W49Y and F61Y substitutions), while all residues in the core of the motif remained unchanged (Figure S6E–G). Introducing these two mutations into a Bim-BH3 peptide, or mutating the Bim-BH3 peptide at all five positions within the BH3 region that distinguish BbpD04 from specific BINDI, failed to achieve the high affinity and specificity of BINDI (Figure S6H–I). Likewise, excised peptides from BbpD04 and BINDI that could be considered equivalent to a 26-residue BH3 motif, while partially specific, again lack the affinity and specificity of BINDI (Figure S6H–I). The extraordinary specificity of BINDI is therefore accomplished through interactions across an expansive interface, extending well beyond the central Bim-BH3 residues used to seed design.
BINDI triggers apoptosis preferentially in EBV-infected cells
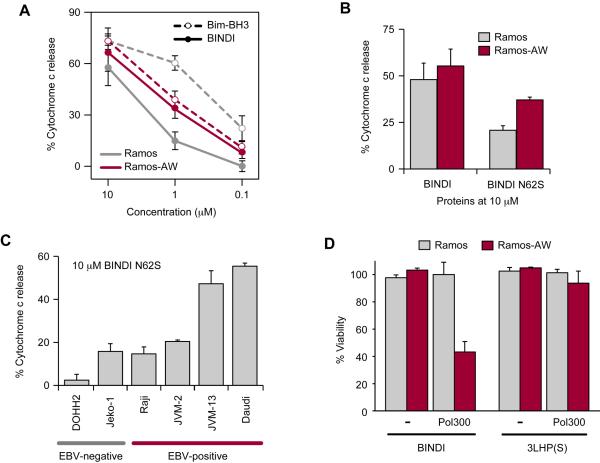
We tested whether inhibition of BHRF1 via steric occlusion of the BH3-binding groove with BINDI could induce mitochondrial cytochrome c release in the EBV-positive BL cell line Ramos-AW. Ramos-AW expresses BHRF1 at very low levels (Leao et al., 2007), and therefore presents a challenging biological target that likely expresses much higher levels of off-target endogenous Bcl-2 family proteins. BINDI was applied to mitochondria isolated from both Ramos-AW and the EBV-negative parental line Ramos (Andersson and Lindahl, 1976). BINDI elicited greater cytochrome c release from Ramos-AW mitochondria (Figure 5A), indicating an EBV-associated factor is likely a BINDI target. Strikingly, the non-specific Bim-BH3 peptide had opposite behavior; mitochondria from EBV-negative Ramos cells were more sensitive to Bim-BH3 treatment than those from EBV-positive Ramos-AW cells. Indeed, EBV-positive cell lines are widely reported as more resistant to nonselective apoptotic stimuli (Ishii et al., 1995; Kvansakul et al., 2010; Leao et al., 2007), making the enhanced activity of BINDI against Ramos-AW cells all the more significant.
Figure 5. BINDI triggers apoptosis in EBV-positive B cell lines.
(A) Cytochrome c release from mitochondria harvested from Ramos (EBV-negative, grey) or Ramos-AW cells (EBV-positive, crimson) treated with Bim-BH3 peptide (broken line) or BINDI (solid line). Mean ± SD, n = 4, for panels A–C. See Figure S7.
(B) Cytochrome c release from Ramos and Ramos-AW mitochondria treated with 10 μM BINDI or BINDI N62S.
(C) Mitochondria were harvested from EBV-negative and positive lines and cytochrome c release was measured after treatment with 10 μM BINDI N62S.
(D) Cells were incubated with sub-lethal doses (2 μM) of antennapedia peptide-fused BINDI or 3LHP(S). Diblock copolymer Pol300 was conjugated to the proteins for enhanced endosomal escape. Cell viability (mean ± SD, n = 3) was measured after 24 hours.
While weaker than the picomolar affinity of BINDI for BHRF1, the moderate affinity for Mcl-1 is likely the reason BINDI still triggers apoptosis in the EBV-negative Ramos cell line. It is possible that the enhanced toxicity of BINDI towards Ramos-AW reflects increased Mcl-1-dependency in this line, rather than expression of EBV BHRF1. To rule out this possibility, we tested variant BINDI N62S with even greater specificity. Asn62 of BINDI hydrogen bonds to the N-terminus of BHRF1 helix α6, and serine at this position is predicted to similarly interact at the interface (Figure S7B). BINDI N62S still binds BHRF1 with tight affinity (KD 160 ± 80 pM), but with better specificity (Table 1 and Figure 3E); in particular, the affinity for Mcl-1 is diminished six-fold (KD 230 ± 40 nM). BINDI N62S has even greater discrimination between Ramos and Ramos-AW cells (Figure 5B). The enhanced activity of BINDI to initiate cytochrome c release preferentially in EBV-positive cells is therefore almost certainly due to BHRF1 inhibition.
Expression profiling of EBV-positive BLs has revealed distinct subgroups (Kelly et al., 2013; Watanabe et al., 2010), and BHRF1 may not be important for cell survival in all cases. Including Ramos and Ramos-AW, mitochondria were isolated from five EBV-positive and three EBV-negative B cell lines. Bim-BH3 peptide triggered cytochrome c release, whereas the inactive guide scaffold 3LHP(S) had no effect (Figure S7C). Incubation with BINDI N62S induced high cytochrome c release in three EBV-positive lines (Figure 5B–C): BL lines Ramos-AW and Daudi, and B-prolymphocytic leukemia JVM-13. Two of the EBV-positive lines had low levels of cytochrome c release similar to EBV-negative cells, despite transcribing the BHRF1 gene (Figure S7C): BL line Raji and mantle cell lymphoma line JVM-2. Hence only some EBV-positive cancer lines appear to be dependent on BHRF1 for survival.
BINDI was genetically fused with a C-terminal antennapedia peptide for non-specific cellular uptake and intracellular delivery in vitro. BINDI-antennapedia applied to the growth medium at 4 μM selectively killed 40% of EBV-positive Ramos-AW cells, with no measurable death of EBV-negative Ramos cells (Figure S7D). Antennapedia-fused proteins concentrate in endocytic organelles and escape to the cytosol with low efficiency (Duvall et al., 2010). To enhance endosomal escape, BINDI-antennapedia was conjugated via a terminal cysteine to a diblock copolymer carrier, Pol300, comprising a hydrophilic first block and a pH-responsive endosomolytic second block (Convertine et al., 2010; Duvall et al., 2010; Manganiello et al., 2012). A lower 2 μM dose of BINDI-antennapedia induced 60% cell death preferentially in Ramos-AW cells when conjugated to the Pol300 polymeric carrier for enhanced cytosolic delivery (Figure 5D). Our data suggest inhibition of BHRF1 may effectively kill EBV-positive BL.
Treatment of EBV-positive B cell lymphoma in a xenograft mouse model by intracellular delivery of BINDI
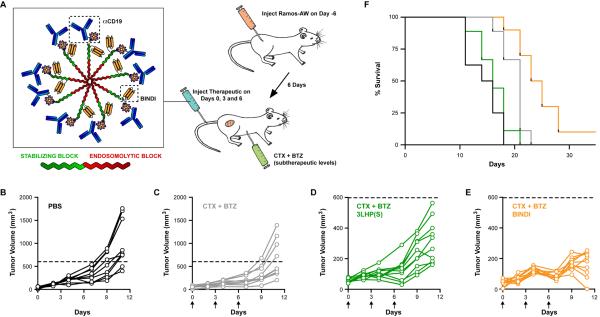
Intracellular delivery of proteins in vivo is challenging, and there are no current intracellular protein drugs available clinically. We developed an antibody-micelle formulation to deliver BINDI to intracellular targets in an EBV-positive human lymphoma disease model. BINDI was coupled via a C-terminal cysteine to a diblock copolymer (Pol950); similar copolymers have been shown to facilitate cytosolic release of fluorescent peptides (Duvall et al., 2010) and have been coupled to antibodies for cell-targeting (Berguig et al., 2012). The copolymer's hydrophilic first block was composed of polyethylene glycol methacrylate (MA) to improve safety and pharmacokinetic properties, pyridyldisulfide MA for cysteine conjugation to BINDI, and biotin-hydroxylethyl MA for coupling to streptavidin-antiCD19 (αCD19; human monoclonal CAT-13.1E10-SA). The endosomolytic pH-responsive second block was composed of diethylaminoethyl MA and butyl MA. Copolymer:αCD19:BINDI forms micelles that disassociate at endosomal pH to expose the membrane-destabilizing segments (Figure 6A). CD19 is a rapidly internalizing surface antigen on B cells, and the αCD19-complex is thought to be endocytosed followed by endosomal escape and protein drug release in the reducing environment of the cytosol.
Figure 6. Treatment of EBV-positive B cell lymphoma xenograft tumors by intracellular delivery of BINDI in vivo.
(A) Schematic representation of the copolymer-based treatment. Pol950 has stabilizing (green) and endosomolytic (red) blocks and forms a micelle at physiological pH. The stabilizing block couples to αCD19 and BINDI. Nude mice with subcutaneous Ramos-AW xenografts were treated on days 0, 3 and 6 with Pol950 (300 mg/kg) : αCD19 (15 mg/kg) : BINDI or 3LHP(S) (105 mg/kg). A maximum tolerated dose study determined this level of BINDI was nontoxic. Mice were injected 30 minutes prior to each treatment with CTX (35 mg/ml) and BTZ (0.5 mg/ml).
(B–E) Tumor growth is plotted for each individual mouse until day 11 when the first mice are euthanized. (B) PBS control treatment, black, n = 8; (C) chemo-only, grey, n = 9; (D) 3LHP(S)-copolymer treatment, green, n = 9; (E) BINDI-copolymer treatment, orange, n = 10.
(F) Kaplan-Meier survival plot. There is a significant increase in survival with treatment (log-rank test χ2 = 46, P < 0.0001). See Figure S7E.
Subcutaneous Ramos-AW xenograft tumors were established in nude BALB/c mice. The mice were treated intravenously on days 0, 3 and 6 with antibody-micelles coupled to the inactive scaffold 3LHP(S) or to BINDI. Thirty minutes prior to each treatment, cyclophosphamide (CTX) and bortezomib (BTZ) were injected intraperitoneally at subtherapeutic doses to prime cells for apoptosis (O'Connor et al., 2006). The treatments were nontoxic, with no substantial change in mouse body weight (Figure S7E).
The intracellular delivery of BINDI to the B cell lymphoma xenograft slowed tumor progression and prolonged survival. Tumors grew rapidly in the untreated/PBS and chemo-only control groups (Figure 6B–C), with mean tumor sizes of 1080 ± 500 mm3 and 680 ± 410 mm3, respectively, at day 11 when the first mice were euthanized due to excessive tumor burden. Both scaffold 3LHP(S) and BINDI treatment groups had reduced tumor sizes due to the therapeutic effects of αCD19 coupled to the micelle carrier, but BINDI substantially slowed tumor growth compared to the scaffold control, especially during the treatment cycle (140 ± 60 mm3 vs. 330 ± 140 mm3 at day 11, unpaired t test P = 0.003, Figure 6D–E). Lifespan was extended in the BINDI-treated mice compared to the scaffold treatment (log-rank test P = 0.006), with median survival of 15 days for PBS treatment, 16 days for chemo-only, and 21 days for 3LHP(S) treatment, extending to 24 days following BINDI treatment (Figure 6F). Future work is needed to determine the fraction of proteins delivered with this new delivery carrier that are internalized and directly engage intracellular targets. Nonetheless, our data represent the first demonstration that a de novo computationally designed protein can reduce tumor growth in a preclinical model.
DISCUSSION
By breaking free of the conformational constraints imposed by repurposing pre-existing scaffolds and instead building a new protein with structure tailored for the target surface, a remarkably tight and specific binder of the EBV apoptosis regulator BHRF1 was designed. The elevated toxicity of the engineered BINDI protein towards EBV-positive cancer lines supports the hypothesis that BHRF1 is necessary for survival in at least some EBV-associated cancers.
BINDI has a structure and amino acid sequence found after computationally filtering thousands of potential designed conformations for optimum interactions with BHRF1. The crystal structure is very close to the BINDI computational model, with the orientations of helices differing from the guide scaffold in ways critical for activity. The ability to custom-tailor the backbone conformation to the challenge at hand was critical to our success in achieving very high affinity and specificity.
Protein structure and function are closely coupled, and our results demonstrate the importance of designing for stability in addition to optimizing the binding interface. Designs with BHRF1 binding activity had energy minima near the intended target structure in ab initio structure calculations, and directed evolution of BbpD04 to enhance affinity introduced mutations far from the interface that increased protein stability. Direct in silico screening for a deep energy minimum around the desired conformation could considerably increase the success rate of protein design efforts.
Finally, we demonstrate that BINDI can slow progression of EBV-positive B cell lymphoma and prolong survival in a human xenograft mouse model. The suppression of tumor growth during treatment with BINDI provides proof-of-principle results that a designer protein toxin can be antibody-targeted and effective in vivo. Further optimization of dosing, targeting and the carrier may all increase therapeutic efficacy, and alternative designed toxins with destructive enzymatic activity would boost potency. Designer toxins with high potency and specificity could provide the basis for a next generation of antibody-drug conjugates for the treatment of cancer.
EXPERIMENTAL PROCEDURES
Protein Design and Purification
Proteins were designed using the ROSETTA software suite, and designed protein genes were synthesized by oligo assembly or by commercial suppliers. Designed proteins (cloned into pET29b (Novagen)) were purified with C-terminal 6His-tags from E. coli BL21(DE3) by NiNTA-affinity chromatography (Qiagen) and SEC (GE Healthcare). Target Bcl-2 proteins had C-terminal avi-6His tags and were similarly purified from E. coli, followed by enzymatic biotinylation using BirA (Avidity). Biotinylated proteins were further purified by additional rounds of NiNTA-affinity chromatography and SEC. All purified proteins were concentrated with ultra-filtration centrifugal devices (Sartorius), snap frozen in liquid nitrogen and stored at −80 °C.
Yeast Surface Display
Design optimization was by yeast surface display. Designed protein genes were cloned into plasmid pETCON (Fleishman et al., 2011), and expressed as fusions with N-terminal Aga2p for surface attachment and a C-terminal myc-tag in the EBY100 strain. Surface expression was detected with anti-myc-FITC (Immunology Consultants Laboratory), while binding to biotinylated Bcl-2 proteins after co-incubation for 1–4 h at 22 °C in phosphate-buffered saline was detected with phycoerythrin-streptavidin (Invitrogen). Yeast were sorted with a BD Influx cell sorter. Gene sequences were diversified either by assembly of degenerate oligos, by over-lapping PCR for generating the site-specific saturation mutagenesis library (Procko et al., 2013), or by error-prone PCR with an average error rate of 1.3 a.a. substitutions per clone using GeneMorph II Random Mutagenesis (Agilent Technologies).
Crystallization and Structure Determination
Untagged BINDI and C-terminal 6his-BHRF1 were co-purified from E. coli lysate via NiNTA-affinity chromatography and SEC. Crystals of the complex were grown by vapor diffusion after mixing 1:1 drops of protein with reservoir (8–12% PEG8000, 100 mM Tris-Cl pH 8.5, 200 mM MgCl2), and were cryopreserved with 18–20 % glycerol in mother liquid prior to flash freezing. Data sets were collected with a Rigaku MicroMax-007HF rotating anode home source and at beam line BL 5.0.2 at the Advanced Light Source at Lawrence Berkeley National Laboratory. The structure was determined by molecular replacement with chain A of PDB 2WH6 and a computational model of BINDI.
Apoptosis Assays
Cytochrome c release was measured by ELISA (Life Technologies) from isolated mitochondria after treating with the indicated peptides/proteins for 0.5 h at room temperature. For cell viability studies, protein and protein-polymer conjugates were incubated with Ramos or Ramos-AW cells for 24 h, and viability measured using a CellTiter 96 Aqueous One Solution Cell Proliferation Assay, MTS (Promega). For these studies, Bim-BH3 peptide (sequence Ac-CMRPEIWIAQELRRIGDEFNAYYARR-ON) was synthesized by solid phase synthesis and purified.
Diblock Copolymers
Pol950 and Pol300 diblock copolymers were synthesized by reversible addition-fragmentation chain transfer. Pol300 is a 25,000 Da diblock copolymer composed of 95% polyethylene glycol MA (300 Da) and 5% pyridyl disulfide MA in the first block, and 60% diethylaminoethyl MA and 40% butyl MA in the second block. Pol950 is a 44,000 Da diblock copolymer composed of 80% polyethylene glycol MA (950 Da), 10% pyridyl disulfide MA, and 10 % biotin-hydroxyethyl MA in the first block, and 60% diethylaminoethyl MA and 40% butyl MA in the second block.
Xenograft Disease Model
BALB/c nu/nu mice (6 to 8 weeks old) were housed under protocols approved by the FHCRC Institutional Animal Care and Use Committee. Ramos-AW cells (107 cells / 200 μl) were injected in the right flank (day -6) and tumors grew for 6 days to a volume of 50 mm3. Mice with similar sized tumors were sorted randomly into treatment groups (n = 8 to 10). On days 0, 3, and 6, mice were injected intraperitoneally with CTX (35 mg/kg) and BTZ (0.5 mg/kg). After 0.5 h, mice were injected via tail vein with conjugates at a dose of 15 mg/kg (αCD19; clone CAT-13.1E10 produced in the monoclonal production facility at FHCRC), 300 mg/kg (Pol950) and 105 mg/kg (BINDI or 3LHP). Body weight was monitored for toxicity and tumor sizes were measured while blinded to treatment groups. Mice were euthanized when tumors reached a volume of 1250 mm3.
Supplementary Material
Highlights.
a specific inhibitor of an EBV prosurvival Bcl-2 protein was computationally designed
the likelihood of designed proteins correctly folding correlates with activity
the designed inhibitor can initiate apoptosis in EBV-infected cancer cells
intracellular delivery with a micelle carrier suppresses xenograft tumor growth
ACKNOWLEDGEMENTS
This work was supported by the National Institute of General Medical Studies of the National Institutes of Health (NIH) under award numbers P41 GM103533 and R01 GM49857, NIH grant R21EB014572, Washington State Life Sciences Discovery Fund grant 2496490 to the Center for Intracellular Delivery of Biologics, and the Defense Threat Reduction Agency. Computational resources were provided by BOINC, supported by the National Science Foundation through awards SCI-0221529, SCI-0438443, SCI-0506411, PHY/0555655 and OCI-0721124.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
ACCESSION NUMBERS The crystal structure factors and coordinates of BINDI-bound BHRF1 have been deposited in the PDB (ID code 4OYD).
SUPPLEMENTAL INFORMATION Supplemental Information includes Extended Experimental Procedures, seven figures, five tables and structure files of designed proteins.
AUTHOR CONTRIBUTIONS E.P. computationally designed proteins with assistance from Y.S. and B.E.C.; E.P. optimized, purified and characterized proteins with assistance from Y.C.; G.Y.B. and E.P. measured mitochondrial cytochrome c release with assistance from D.M.; G.Y.B. developed the micelle carrier with assistance from A.J.C. and performed animal studies with assistance from S.F. and G.B.; B.W.S. crystallized BINDI-bound BHRF1 and determined the structure; E.P. drafted the manuscript with edits and input from all authors; and W.R.S., D.M.H., O.W.P., B.L.S., P.S.S. and D.B. supervised research.
FINANCIAL INTERESTS DISCLOSURE P.S.S. and O.W.P. are co-founders of PhaseRx Pharmaceuticals Inc., which has licensed drug delivery technology from UW that encompasses aspects of the polymeric carriers used in this study. All the work reported in this study was completely independent of the company.
REFERENCES
- Altmann M, Hammerschmidt W. Epstein-Barr virus provides a new paradigm: a requirement for the immediate inhibition of apoptosis. PLoS Biol. 2005;3:e404. doi: 10.1371/journal.pbio.0030404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersson M, Lindahl T. Epstein-Barr virus DNA in human lymphoid cell lines: in vitro conversion. Virology. 1976;73:96–105. doi: 10.1016/0042-6822(76)90064-7. [DOI] [PubMed] [Google Scholar]
- Azzarito V, Long K, Murphy NS, Wilson AJ. Inhibition of alpha-helix-mediated protein-protein interactions using designed molecules. Nat Chem. 2013;5:161–173. doi: 10.1038/nchem.1568. [DOI] [PubMed] [Google Scholar]
- Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berguig GY, Convertine AJ, Shi J, Palanca-Wessels MC, Duvall CL, Pun SH, Press OW, Stayton PS. Intracellular delivery and trafficking dynamics of a lymphoma-targeting antibody-polymer conjugate. Mol Pharm. 2012;9:3506–3514. doi: 10.1021/mp300338s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caria S, Chugh S, Nhu D, Lessene G, Kvansakul M. Crystallization and preliminary X-ray characterization of Epstein-Barr virus BHRF1 in complex with benzoylurea peptidomimetic. Acta Crystallogr F Struct Biol Cryst Commun. 2012;1:1521–1524. doi: 10.1107/S1744309112043333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin JW, Schepartz A. Design and Evolution of a Miniature Bcl-2 Binding Protein. Angew Chem Int Ed Engl. 2001;40:3806–3809. doi: 10.1002/1521-3773(20011015)40:20<3806::AID-ANIE3806>3.0.CO;2-B. [DOI] [PubMed] [Google Scholar]
- Convertine AJ, Diab C, Prieve M, Paschal A, Hoffman AS, Johnson PH, Stayton PS. pH-Responsive Polymeric Micelle Carriers for siRNA Drugs. Biomacromolecules. 2010 doi: 10.1021/bm100652w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Correia BE, Ban YE, Holmes MA, Xu H, Ellingson K, Kraft Z, Carrico C, Boni E, Sather DN, Zenobia C, et al. Computational design of epitope-scaffolds allows induction of antibodies specific for a poorly immunogenic HIV vaccine epitope. Structure. 2010;18:1116–1126. doi: 10.1016/j.str.2010.06.010. [DOI] [PubMed] [Google Scholar]
- Correia BE, Bates JT, Loomis RJ, Baneyx G, Carrico C, Jardine JG, Rupert P, Correnti C, Kalyuzhniy O, Vittal V, et al. Proof of principle for epitope-focused vaccine design. Nature. 2014;507:201–206. doi: 10.1038/nature12966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desbien AL, Kappler JW, Marrack P. The Epstein-Barr virus Bcl-2 homolog, BHRF1, blocks apoptosis by binding to a limited amount of Bim. Proc Natl Acad Sci U S A. 2009;106:5663–5668. doi: 10.1073/pnas.0901036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Chen TS, Keating AE. Peptide ligands for pro-survival protein Bfl-1 from computationally guided library screening. ACS Chem Biol. 2013;8:778–788. doi: 10.1021/cb300679a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dutta S, Gulla S, Chen TS, Fire E, Grant RA, Keating AE. Determinants of BH3 binding specificity for Mcl-1 versus Bcl-xL. J Mol Biol. 2010;398:747–762. doi: 10.1016/j.jmb.2010.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvall CL, Convertine AJ, Benoit DS, Hoffman AS, Stayton PS. Intracellular delivery of a proapoptotic peptide via conjugation to a RAFT synthesized endosomolytic polymer. Mol Pharm. 2010;7:468–476. doi: 10.1021/mp9002267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire E, Gulla SV, Grant RA, Keating AE. Mcl-1-Bim complexes accommodate surprising point mutations via minor structural changes. Protein Sci. 2010;19:507–519. doi: 10.1002/pro.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleishman SJ, Whitehead TA, Ekiert DC, Dreyfus C, Corn JE, Strauch EM, Wilson IA, Baker D. Computational design of proteins targeting the conserved stem region of influenza hemagglutinin. Science. 2011;332:816–821. doi: 10.1126/science.1202617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gemperli AC, Rutledge SE, Maranda A, Schepartz A. Paralog-selective ligands for bcl-2 proteins. J Am Chem Soc. 2005;127:1596–1597. doi: 10.1021/ja0441211. [DOI] [PubMed] [Google Scholar]
- Goulet A, Blangy S, Redder P, Prangishvili D, Felisberto-Rodrigues C, Forterre P, Campanacci V, Cambillau C. Acidianus filamentous virus 1 coat proteins display a helical fold spanning the filamentous archaeal viruses lineage. Proc Natl Acad Sci U S A. 2009;106:21155–21160. doi: 10.1073/pnas.0909893106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson S, Huen D, Rowe M, Dawson C, Johnson G, Rickinson A. Epstein-Barr virus-coded BHRF1 protein, a viral homologue of Bcl-2, protects human B cells from programmed cell death. Proc Natl Acad Sci U S A. 1993;90:8479–8483. doi: 10.1073/pnas.90.18.8479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogbom M, Eklund M, Nygren PA, Nordlund P. Structural basis for recognition by an in vitro evolved affibody. Proc Natl Acad Sci U S A. 2003;100:3191–3196. doi: 10.1073/pnas.0436100100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii HH, Etheridge MR, Gobe GC. Cycloheximide-induced apoptosis in Burkitt lymphoma (BJA-B) cells with and without Epstein-Barr virus infection. Immunol Cell Biol. 1995;73:463–468. doi: 10.1038/icb.1995.72. [DOI] [PubMed] [Google Scholar]
- Kelly GL, Long HM, Stylianou J, Thomas WA, Leese A, Bell AI, Bornkamm GW, Mautner J, Rickinson AB, Rowe M. An Epstein-Barr virus anti-apoptotic protein constitutively expressed in transformed cells and implicated in burkitt lymphomagenesis: the Wp/BHRF1 link. PLoS Pathog. 2009;5:e1000341. doi: 10.1371/journal.ppat.1000341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly GL, Stylianou J, Rasaiyaah J, Wei W, Thomas W, Croom-Carter D, Kohler C, Spang R, Woodman C, Kellam P, et al. Different patterns of Epstein-Barr virus latency in endemic Burkitt lymphoma (BL) lead to distinct variants within the BL-associated gene expression signature. J Virol. 2013;87:2882–2894. doi: 10.1128/JVI.03003-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koga N, Tatsumi-Koga R, Liu G, Xiao R, Acton TB, Montelione GT, Baker D. Principles for designing ideal protein structures. Nature. 2012;491:222–227. doi: 10.1038/nature11600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlman B, Dantas G, Ireton GC, Varani G, Stoddard BL, Baker D. Design of a novel globular protein fold with atomic-level accuracy. Science. 2003;302:1364–1368. doi: 10.1126/science.1089427. [DOI] [PubMed] [Google Scholar]
- Kvansakul M, Wei AH, Fletcher JI, Willis SN, Chen L, Roberts AW, Huang DC, Colman PM. Structural basis for apoptosis inhibition by Epstein-Barr virus BHRF1. PLoS Pathog. 2010;6:e1001236. doi: 10.1371/journal.ppat.1001236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lanci CJ, MacDermaid CM, Kang SG, Acharya R, North B, Yang X, Qiu XJ, DeGrado WF, Saven JG. Computational design of a protein crystal. Proc Natl Acad Sci U S A. 2012;109:7304–7309. doi: 10.1073/pnas.1112595109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leao M, Anderton E, Wade M, Meekings K, Allday MJ. Epstein-barr virus-induced resistance to drugs that activate the mitotic spindle assembly checkpoint in Burkitt's lymphoma cells. J Virol. 2007;81:248–260. doi: 10.1128/JVI.01096-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leaver-Fay A, Tyka M, Lewis SM, Lange OF, Thompson J, Jacak R, Kaufman K, Renfrew PD, Smith CA, Sheffler W, et al. ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 2011;487:545–574. doi: 10.1016/B978-0-12-381270-4.00019-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee EF, Czabotar PE, Yang H, Sleebs BE, Lessene G, Colman PM, Smith BJ, Fairlie WD. Conformational changes in Bcl-2 pro-survival proteins determine their capacity to bind ligands. J Biol Chem. 2009;284:30508–30517. doi: 10.1074/jbc.M109.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lessene G, Czabotar PE, Sleebs BE, Zobel K, Lowes KN, Adams JM, Baell JB, Colman PM, Deshayes K, Fairbrother WJ, et al. Structure-guided design of a selective BCL-X(L) inhibitor. Nat Chem Biol. 2013;9:390–397. doi: 10.1038/nchembio.1246. [DOI] [PubMed] [Google Scholar]
- Manganiello MJ, Cheng C, Convertine AJ, Bryers JD, Stayton PS. Diblock copolymers with tunable pH transitions for gene delivery. Biomaterials. 2012;33:2301–2309. doi: 10.1016/j.biomaterials.2011.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinou JC, Youle RJ. Mitochondria in apoptosis: Bcl-2 family members and mitochondrial dynamics. Dev Cell. 2011;21:92–101. doi: 10.1016/j.devcel.2011.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor OA, Smith EA, Toner LE, Teruya-Feldstein J, Frankel S, Rolfe M, Wei X, Liu S, Marcucci G, Chan KK, Chanan-Khan A. The combination of the proteasome inhibitor bortezomib and the bcl-2 antisense molecule oblimersen sensitizes human B-cell lymphomas to cyclophosphamide. Clin Cancer Res. 2006;12:2902–2911. doi: 10.1158/1078-0432.CCR-05-0308. [DOI] [PubMed] [Google Scholar]
- Ofek G, Guenaga FJ, Schief WR, Skinner J, Baker D, Wyatt R, Kwong PD. Elicitation of structure-specific antibodies by epitope scaffolds. Proc Natl Acad Sci U S A. 2010;107:17880–17887. doi: 10.1073/pnas.1004728107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Procko E, Hedman R, Hamilton K, Seetharaman J, Fleishman SJ, Su M, Aramini J, Kornhaber G, Hunt JF, Tong L, et al. Computational Design of a Protein-Based Enzyme Inhibitor. J Mol Biol. 2013;425:3563–3575. doi: 10.1016/j.jmb.2013.06.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse C, Shoemaker AR, Adickes J, Anderson MG, Chen J, Jin S, Johnson EF, Marsh KC, Mitten MJ, Nimmer P, et al. ABT-263: a potent and orally bioavailable Bcl-2 family inhibitor. Cancer Res. 2008;68:3421–3428. doi: 10.1158/0008-5472.CAN-07-5836. [DOI] [PubMed] [Google Scholar]
- Watanabe A, Maruo S, Ito T, Ito M, Katsumura KR, Takada K. Epstein-Barr virus-encoded Bcl-2 homologue functions as a survival factor in Wp-restricted Burkitt lymphoma cell line P3HR-1. J Virol. 2010;84:2893–2901. doi: 10.1128/JVI.01616-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.