Abstract
Background
Highly abundant proteins in biological fluids such as serum or cerebrospinal fluid (CSF) can hinder the detection of proteins in lower abundance, e.g., potential biomarkers. Commercial products are available for the depletion of albumin and immunoglobulins (Igs), although most of these kits have not been validated for dog samples. The present study therefore examines the use of different types of depletion kits for dog CSF.
Findings
Three kits, with different mechanisms for the depletion of albumin and Igs, were tested with dog CSF specimens. One product significantly decreased the amount of albumin; with all kits, IgG was less efficiently removed than albumin. Mass spectrometry of the fractions eluted from the depletion columns revealed considerable co-depletion of other CSF proteins.
Conclusions
A commercially available depletion kit was identified which depletes albumin and (to a lower extent) immunoglobulins from dog CSF. However, the limited efficacy and the concomitant loss of other proteins from the sample should be taken into account when using this product.
Keywords: Cerebrospinal fluid, Dog, Depletion, Albumin, Mass spectrometry
Findings
Introduction
Biomarkers for an individual biological condition can be of outstanding value for mapping a normal or pathophysiological process or for studying the response to a therapeutic intervention. Specific proteins are of particular interest as biomarkers, as their concentrations can usually be measured cheaply and reliably. Moreover, the enormous progress in the development of proteomic techniques provides highly-specific and sensitive technologies for the identification of individual proteins, including potential biomarkers [1].
Consequently, the protein profiling of body fluids is of great interest for the identification of biomarker candidates. Albumin and immunoglobulins (Igs) are the main protein constituents of plasma or serum and other body fluids. In particular, it has been shown that albumin contributes up to 60% of the protein content of cerebrospinal fluid (CSF), which is considerably more than in plasma [2]. However, potential biomarkers are usually proteins of lower abundance and these are often easier to detect after depletion of highly-abundant species. Various procedures for depleting albumin and/or Igs have been described in the literature (e.g., [3-9]), and diverse “depletion kits” can be obtained from different commercial sources which, in turn, have been tested for several parameters (e.g., [4,10,11]).
Unfortunately, these depletion kits have been tested for only a limited number of donor species (usually human beings and rodents). On the other hand, there is increasing interest in the development of dog models for diseases, such as the Hurler syndrome [12], the Sanfilippo syndrome [13] or Duchenne muscular dystrophy [14]. This interest, together with the first proteomic studies directed at the identification of CSF biomarkers for canine diseases [15,16], prompted us to investigate the applicability of commercial depletion kits for dog CSF.
Materials and methods
CSF samples from eight healthy adult Beagle dogs, four male and four females, obtained by lumbar puncture, were purchased from Seralab (Haywards Heath, UK). The provider guarantees that the specimens were collected at USDA inspected and approved facilities. These samples (pooled pairwise - one male, one female) were centrifuged (2,000 × g for 10 min) to remove cells or other insoluble substances. Proteins were precipitated overnight at 20°C by adding four sample volumes of ice-cold acetone. The proteins were pelleted by centrifugation (10,000 × g, 30 min, 4°C). The supernatant was removed and the pellet was washed with ice-cold acetone (90%). The samples were vortexed and centrifuged again (10,000 × g, 30 min, 4°C). The supernatant was discarded and the pellets were dried at room temperature (30 min). Control experiments using dog serum were performed (see Additional file 1) to confirm the compatibility of this procedure, which has been recommended for proteomic investigations using CSF [17]. The ProteoSeek™ Albumin/Ig Removal Kit (Thermo-Fisher, Bonn, Germany) was applied for albumin and Ig depletion, as based on an affinity resin column. According to the manufacturer, this kit has been positively tested with human, monkey, swine and rabbit samples. Depletion experiments directed at albumin and Ig were also carried out using the ProteoExtract Albumin Removal Kit (Merck Millipore, Darmstadt, Germany, tested by the producer for human, rabbit, rat, mouse, pig and bovine samples, with decreasing activity) and by the antibody-based Vivapure Anti-HSA/Ig Kit (Sartorius, Göttingen, Germany), designed for humans, but also reactive in rat and mouse samples (according to the product information). Proteins obtained from at least three different pairwise pooled CSF samples were applied to each kit according to the manufacturer’s instructions after acetone precipitation. Optimisation procedures suggested by the manufacturers (such as prolonged and/or repeated sample application, extended mixing) did not clearly improve the depletion results for the kits when tested in dog serum. The protein quantities subjected to depletion (100 μg each) were always lower than the upper limits specified by the respective instructions, by a factor of at least 5. Elution from the depletion columns was performed as suggested by manufacturer’s protocols (if available) or by using Laemmli buffer. Flow-through (FT) and elution fractions (E) of CSF were reconstituted in Laemmli buffer. Cysteine residues were reduced using dithiothreitol (10 mM, 15 min, 60°C) and alkylated (55 mM iodoacetamide, 15 min, 20°C) in the dark. All fractions (FT and E of each kit) were separated electrophoretically on precast Novex Tris/Glycine 4%-20% gels (Life Technologies, Darmstadt, Germany). Proteins were visualised using Coomassie Brilliant Blue G-250, and the resulting band patterns obtained from at least three different samples were qualitatively equivalent for the respective kit. All clearly stained bands of eluate fractions (originating from not less than two independent samples for each kit) were excised for subsequent mass spectrometry (MS). After in-gel digestion of proteins and peptide extraction [18], MS was conducted on a LTQ-Orbitrap XL mass spectrometer (Thermo Scientific) equipped with a reverse-phase capillary liquid chromatography system (Eksigent 2D nanoflow LC, Axel Semrau GmbH, Sprockhövel, Germany). The generated peak lists and the MASCOT server (version 2.2, Matrix Science Ltd., London/UK) were used to search against the non-redundant NCBI database (version 131022) restricted to mammals (1,974,848 sequences). A maximum of two missed cleavages was allowed, and the mass tolerance of precursor and sequence ions was set to 10 ppm and 0.35 Da, respectively. The program Scaffold (version 4.2.1, Proteome Software Inc., Portland OR, USA) was used to validate MS/MS-based peptide and protein identifications, which were accepted if established at greater than 99.0% probability with at least two identified peptides.
Results and discussion
Different depletion kits for albumin and Igs were tested using dog CSF samples. Numerous other products for depletion are commercially available [19]. However, data providing an assessment of depletion kits is sparse even for human CSF, and several authors have found that these depletion kits give relatively low reproducibility. Previous work has shown that more than 95% albumin was depleted from human CSF with the ProteoSeek kit, resulting in a 20% increase in the number of detected proteins (from 135 to 163) [20]. The Vivapure anti-HSA/IgG kit was applied to human CSF and depleted albumin less effectively than the Sigma anti-HSA/IgG column or the ProteaPrep HSA/IgG column [21]. The Vivapure kit was found to be superior to the ProteoExtract kit for the analysis of the recovery of the prostate-specific antigen in human serum samples [22]. On the other hand, in studies on human serum, ProteoExtract has been found to give satisfactory reproducibility and specificity [23]. Other authors (e.g., [24]) used the ProteoExtract kit for depleting human CSF, without reporting problems.
The Coomassie-stained gels shown in Figure 1 demonstrate that none of the kits gave satisfactory depletion of albumin and Igs from dog CSF samples. The ProteoSeek kit clearly exhibited the highest efficacy with respect to albumin; the amounts depleted from CSF by Vivapure and ProteoExtract kits were lower or negligible, respectively. Proteins identified in bands of the eluate gel lanes are given in Table 1 which lists proteins with substantial abundance, i.e., exponentially modified protein abundance index (emPAI) > 1.0. For approximately 56% of the canine proteins found, at least one isoform coincides with the human CSF proteome previously reported [25]. Albumin was enriched in the eluate fractions obtained from the ProteoSeek and Vivapure kits, but only one low abundant fragment was found in the eluate from the ProteoExtract kit (see Additional file 2 showing all eluted proteins identified by mass spectrometry). In agreement with the protein staining, immunoglobulin fragments were detected with lower emPAI values in the eluate fractions obtained by application of the depletion kits. However, additional proteins were identified by MS in all these fractions. Significant quantities of haemoglobin (obviously a contamination of the CSF samples) were found in the eluate fractions of all kits tested. As expected from the gel staining, the greatest co-depletion was observed for the ProteoSeek kit (115 proteins detected in total), but numerous non-targeted proteins have been also identified in samples obtained with the Vivapure kit (total proteins, 64), whereas only 3 detectable proteins were eluted from the columns of the ProteoExtract kit (see Additional file 2. Table showing all eluted proteins identified by mass spectrometry).
Figure 1.
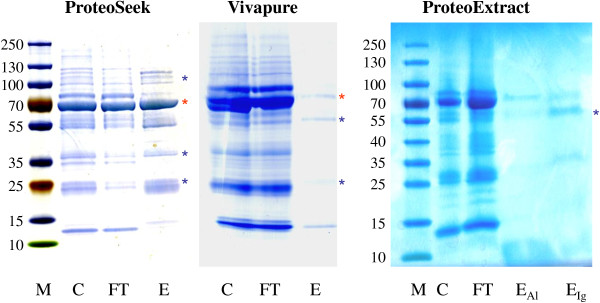
Polyacrylamide gel electrophoresis of Coomassie blue-stained protein fractions obtained by applying different depletion kits for removal of albumin and immunoglobulins from dog cerebrospinal fluid. Protein loads: ProteoSeek, 3 μg each; Vivapure, 3 μg (C, FT), complete eluate fraction (E); ProteoExtract, 3 μg (C) 5 μg (FT), complete eluate fractions (EA/EIg); red/blue asterisks, mass spectrometric identification of albumin/immunoglobulin fragments; M: marker; C: dog cerebrospinal fluid (undepleted fraction); FT: flow-through fraction after depletion procedure; E: eluate fraction(s) obtained from depletion columns.
Table 1.
Proteins identified by mass spectrometry with exponentially modified protein abundance index (emPAI) > 1.0 1
| Protein | Accession No. 2 | M.W. 3 | emPAI max 4 |
|---|---|---|---|
|
ProteoSeek Depletion Kit |
|
|
|
| Serum albumin precursor [Canis lupus familiaris] |
gi|55742764 |
69 |
11.85 |
| Hemoglobin subunit beta-like [Canis lupus familiaris] |
gi|399567834 |
16 |
6.30 |
| PREDICTED: Serum albumin isoform X1 [Canis lupus familiaris] |
gi|545520919 |
69 |
4,41 |
| PREDICTED: Tetranectin [Canis lupus familiaris] |
gi|545535577 |
18 |
4.28 |
| PREDICTED: Apolipoprotein A-I [Canis lupus familiaris] |
gi|73955106 |
30 |
3.34 |
| PREDICTED: apolipoprotein E isoform X5 [Canis lupus familiaris] |
gi|545488191 |
47 |
3.20 |
| Immunoglobulin heavy chain variable region [Mus musculus] |
gi|81237613 |
13 |
2.90 |
| PREDICTED: Complement component C7 isoform 1 [Canis lupus familiaris] |
gi|73953824 |
95 |
2.21 |
| PREDICTED: Immunoglobulin lambda-like polypeptide 5-like isoform X1 [Canis lupus familiaris] |
gi|545544681 |
25 |
2.12 |
| Prostaglandin-H2 D-isomerase precursor [Canis lupus familiaris] |
gi|50978842 |
21 |
2.08 |
| Hemoglobin subunit alpha [Canis lupus familiaris] |
gi|44888810 |
15 |
1.96 |
| PREDICTED: Immunoglobulin lambda-like polypeptide 5-like isoform X2 [Canis lupus familiaris] |
gi|545544683 |
25 |
1.77 |
| PREDICTED: Gelsolin [Canis lupus familiaris] |
gi|545518174 |
94 |
1.70 |
| PREDICTED: Fibulin-1 isoform X1 [Canis lupus familiaris] |
gi|545514459 |
78 |
1.63 |
| Immunoglobulin gamma heavy chain A [Canis lupus familiaris] |
gi|17066524 |
52 |
1.63 |
| Immunoglobulin gamma heavy chain B [Canis lupus familiaris] |
gi|17066526 |
52 |
1.61 |
| PREDICTED: EGF containing fibulin-like extracellular matrix protein 1 isoform X3 [Canis lupus familiaris] |
gi|57092953 |
55 |
1.60 |
| PREDICTED: Angiotensinogen [Canis lupus familiaris] |
gi|545494757 |
52 |
1.53 |
| Immunoglobulin heavy chain constant region CH2 [Canis lupus familiaris] |
gi|124390009 |
12 |
1.45 |
| Ig heavy chain V region GOM [Canis lupus familiaris] |
gi|123768 |
12 |
1.37 |
| Immunoglobulin heavy chain constant region CH4 [Canis lupus familiaris] |
gi|124390013 |
14 |
1.21 |
| PREDICTED: LOW QUALITY PROTEIN: Serotransferrin isoform 1 [Canis lupus familiaris] |
gi|545539001 |
85 |
1.16 |
| Protease serine 4 isoform B [Homo sapiens] |
gi|33126535 |
28 |
1.13 |
| Clusterin precursor [Canis lupus familiaris] |
gi|50979240 |
52 |
1.08 |
| PREDICTED: Plasminogen isoform X1 [Canis lupus familiaris] |
gi|545485785 |
91 |
1.06 |
|
Vivapure Depletion Kit |
|
|
|
| Serum albumin precursor [Canis lupus familiaris] |
gi|55742764 |
69 |
11.30 |
| Hemoglobin subunit beta-like [Canis lupus familiaris] |
gi|399567834 |
16 |
5.69 |
| PREDICTED: Apolipoprotein A-I [Canis lupus familiaris] |
gi|73955106 |
30 |
4.80 |
| PREDICTED: Angiotensinogen [Canis lupus familiaris] |
gi|545494757 |
52 |
3.35 |
| PREDICTED: Serum albumin isoform X1 [Canis lupus familiaris] |
gi|545520919 |
69 |
2.98 |
| Hemoglobin subunit alpha [Canis lupus familiaris] |
gi|44888810 |
15 |
2.91 |
| Immunoglobulin gamma heavy chain B [Canis lupus familiaris] |
gi|17066526 |
52 |
2.58 |
| PREDICTED: Immunoglobulin lambda-like polypeptide 5-like isoform X1 [Canis lupus familiaris] |
gi|545544681 |
25 |
1.62 |
| PREDICTED: Immunoglobulin lambda-like polypeptide 5-like isoform X2 [Canis lupus familiaris] |
gi|545544683 |
25 |
1.62 |
| Prostaglandin-H2 D-isomerase precursor [Canis lupus familiaris] |
gi|50978842 |
21 |
1.33 |
| Albumin, isoform CRA_h [Homo sapiens] |
gi|119626071 |
69 |
1.21 |
| Immunoglobulin heavy chain constant region CH2 [Canis lupus familiaris] |
gi|124390009 |
12 |
1.18 |
| Immunoglobulin gamma heavy chain A [Canis lupus familiaris] |
gi|17066524 |
52 |
1.16 |
| beta-2-Glycoprotein 1 precursor [Canis lupus familiaris] |
gi|54792721 |
38 |
1.00 |
|
ProteoExtract Depletion Kit |
|
|
|
| Immunoglobulin heavy chain variable region [Mus musculus] |
gi|81237613 |
13 |
2.39 |
| Hemoglobin subunit alpha [Canis lupus familiaris] | gi|122508 | 15 | 1.64 |
1The complete list of identified proteins is available at Additional file 2.
2Accession number in NCBI database.
3M.W., Protein molecular weight in kDa.
4The emPAI value expresses mass spectrometry-based quantitation of protein abundance and is calculated according to , (N observed , N observable , number of observed/observable peptides); cf. http://www.matrixscience.com/help/quant_empai_help.html for more details.
Unspecific binding, either to the matrix or to the bait material, is a well-known phenomenon occurring in affinity purification approaches. In addition, one has to expect that proteins bound to albumin will recover in the eluate fractions. All of the proteins identified in the eluate were also present in the recently characterised CSF “albuminome” [21] for ProteoExtract, compared to 44% for Vivapure and 30% for ProteoSeek, although this comparison does not consider different experimental conditions and species. Since albuminome data were obtained in human CSF samples, it is likely that the co-depletion observed in our experiments results from both albumin binding and unspecific association.
In conclusion, our results do not support the use of these kits for depletion of albumin and Igs from dog CSF samples. Even with the most active kit tested, albumin and Igs were only incompletely removed. This was accompanied by co-depletion of numerous other proteins, a particular drawback for the identification of potential biomarkers.
Abbreviations
CSF: Cerebrospinal fluid; E: Elution; emPAI: Exponentially modified protein abundance index; F: Flow-through; Ig: Immunoglobulin; MS: Mass spectrometry.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
RG carried out the biochemical experiments and participated in drafting the manuscript. EK and MS planned and performed mass spectrometric experiments. JA, JH and IB participated in the design of the study, the evaluation of the data and in drafting the manuscript. RH conceived the study and participated in its design and coordination and in drafting the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Gel electrophoresis of dog serum before and after albumin depletion using the ProteoSeek kit. The flow-through fraction and the column eluate were compared with and without prior acetone precipitation.
Table showing all eluted proteins identified by mass spectrometry.
Contributor Information
Ramona Günther, Email: rguenthr@fmp-berlin.de.
Eberhard Krause, Email: ekrause@fmp-berlin.de.
Michael Schümann, Email: schueman@fmp-berlin.de.
Jérome Ausseil, Email: ausseil.jerome@chu-amiens.fr.
Jean-Michel Heard, Email: jean-michel.heard@pasteur.fr.
Ingolf E Blasig, Email: iblasig@fmp-berlin.de.
Reiner F Haseloff, Email: haseloff@fmp-berlin.de.
Acknowledgement
RG was supported by the Pasteur Institute (Paris, France) in a collaborative project.
References
- Kroksveen AC, Opsahl JA, Aye TT, Ulvik RJ, Berven FS. Proteomics of human cerebrospinal fluid: discovery and verification of biomarker candidates in neurodegenerative diseases using quantitative proteomics. J Proteomics. 2011;74:371–388. doi: 10.1016/j.jprot.2010.11.010. [DOI] [PubMed] [Google Scholar]
- Roche S, Gabelle A, Lehmann S. Clinical proteomics of the cerebrospinal fluid: towards the discovery of new biomarkers. Proteomics Clin Appl. 2008;2:428–436. doi: 10.1002/prca.200780040. [DOI] [PubMed] [Google Scholar]
- da Silva MAO, Arruda MAZ. An aqueous two-phase system as a strategy for serum albumin depletion. Talanta. 2009;77:985–990. doi: 10.1016/j.talanta.2008.07.055. [DOI] [PubMed] [Google Scholar]
- Ichibangase T, Moriya K, Koike K, Imai K. Limitation of immunoaffinity column for the removal of abundant proteins from plasma in quantitative plasma proteomics. Biomed Chromatogr. 2009;23:480–487. doi: 10.1002/bmc.1139. [DOI] [PubMed] [Google Scholar]
- Mahn A, Reyes A, Zamorano M, Cifuentes W, Ismail M. Depletion of highly abundant proteins in blood plasma by hydrophobic interaction chromatography for proteomic analysis. J Chromatogr B. 2010;878:1038–1044. doi: 10.1016/j.jchromb.2010.03.006. [DOI] [PubMed] [Google Scholar]
- Ramstrom M, Zuberovic A, Gronwall C, Hanrieder J, Bergquist J, Hober S. Development of affinity columns for the removal of high-abundance proteins in cerebrospinal fluid. Biotechnol Appl Biochem. 2009;52:159–166. doi: 10.1042/BA20080015. [DOI] [PubMed] [Google Scholar]
- Urbas L, Brne P, Gabor B, Barut M, Strlic M, Petric TC, Strancar A. Depletion of high-abundance proteins from human plasma using a combination of an affinity and pseudo-affinity column. J Chromatogr A. 2009;1216:2689–2694. doi: 10.1016/j.chroma.2008.10.104. [DOI] [PubMed] [Google Scholar]
- Wetterhall M, Zuberovic A, Hanrieder J, Bergquist J. Assessment of the partitioning capacity of high abundant proteins in human cerebrospinal fluid using affinity and immunoaffinity subtraction spin columns. J Chromatogr B. 2010;878:1519–1530. doi: 10.1016/j.jchromb.2010.04.003. [DOI] [PubMed] [Google Scholar]
- Mouton-Barbosa E, Roux-Dalvai F, Bouyssie D, Berger F, Schmidt E, Righetti PG, Guerrier L, Boschetti E, Burlet-Schiltz O, Monsarrat B, de Peredo AG. In-depth exploration of cerebrospinal fluid by combining peptide ligand library treatment and label-free protein quantification. Mol Cell Proteomics. 2010;9:1006–1021. doi: 10.1074/mcp.M900513-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gundry RL, White MY, Nogee J, Tchernyshyov I, Van Eyk JE. Assessment of albumin removal from an immunoaffinity spin column: critical implications for proteomic examination of the albuminome and albumin-depleted samples. Proteomics. 2009;9:2021–2028. doi: 10.1002/pmic.200800686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roche S, Tiers L, Provansal M, Seveno M, Piva MT, Jouin P, Lehmann S. Depletion of one, six, twelve or twenty major blood proteins before proteomic analysis: the more the better? J Proteomics. 2009;72:945–951. doi: 10.1016/j.jprot.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Ciron C, Desmaris N, Colle MA, Raoul S, Joussemet B, Verot L, Ausseil J, Froissart R, Roux F, Cherel Y, Lajat Y, Schwartz B, Vanier MT, Maire I, Tardieu M, Moullier P, Heard JM. Gene therapy of the brain in the dog model of Hurler’s syndrome. Ann Neurol. 2006;60:204–213. doi: 10.1002/ana.20870. [DOI] [PubMed] [Google Scholar]
- Ellinwood NM, Ausseil J, Desmaris N, Bigou S, Liu S, Jens JK, Snella EM, Mohammed EEA, Thomson CB, Raoul S, Joussemet B, Roux F, Cherel Y, Lajat Y, Piraud M, Benchaouir R, Hermening S, Petry H, Froissart R, Tardieu M, Ciron C, Moullier P, Parkes J, Kline KL, Maire I, Vanier MT, Heard JM, Colle MA. Safe, efficient, and reproducible gene therapy of the brain in the dog models of sanfilippo and hurler syndromes. Mol Ther. 2011;19:251–259. doi: 10.1038/mt.2010.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelemy I, Uriarte A, Drougard C, Unterfinger Y, Thibaud JL, Blot S. Effects of an immunosuppressive treatment in the GRMD dog model of duchenne muscular dystrophy. PLoS One. 2012;7(11):e48478. doi: 10.1371/journal.pone.0048478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Miyasho T, Nomura S, Yokota H, Nakade T. Proteome analysis of cerebrospinal fluid in healthy beagles and canine encephalitis. J Vet Med Sci. 2012;74:751–756. doi: 10.1292/jvms.11-0474. [DOI] [PubMed] [Google Scholar]
- Shafie INF, Anderson TJ, Penderis J, Eckersall PD, McLaughlin M. A protocol for the management of canine cerebrospinal fluid for the proteomic assessment of putative biomarkers. Vet J. 2013;197:836–841. doi: 10.1016/j.tvjl.2013.05.039. [DOI] [PubMed] [Google Scholar]
- Chen RL, Sage EA, Dunn MJ, Wait R, Preston JE. Optimising ovine cerebrospinal fluid preparation for two-dimensional gel electrophoresis. Proteomics. 2006;6:3170–3175. doi: 10.1002/pmic.200500623. [DOI] [PubMed] [Google Scholar]
- Lange S, Sylvester M, Schumann M, Freund C, Krause E. Identification of phosphorylation-dependent interaction partners of the adapter protein ADAP using quantitative mass spectrometry: SILAC vs O-18-labeling. J Proteome Res. 2010;9:4113–4122. doi: 10.1021/pr1003054. [DOI] [PubMed] [Google Scholar]
- Pottiez G, Ciborowski P. In: Expression profiling in neuroscience. Karamanos Y, editor. New York, NY, USA: Humana Press; 2012. Proteomic profiling of cerebrospinal fluid; pp. 245–270. [Walz W (Series Editor): Neuromethods, vol 13.] [Google Scholar]
- Shores KS, Knapp DR. Assessment approach for evaluating high abundance protein depletion methods for cerebrospinal fluid (CSF) proteomic analysis. J Proteome Res. 2007;6:3739–3751. doi: 10.1021/pr070293w. [DOI] [PubMed] [Google Scholar]
- Holewinski RJ, Jin ZC, Powell MJ, Maust MD, Van Eyk JE. A fast and reproducible method for albumin isolation and depletion from serum and cerebrospinal fluid. Proteomics. 2013;13:743–750. doi: 10.1002/pmic.201200192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fortin T, Salvador A, Charrier JP, Lenz C, Lacoux X, Morla A, Choquet-Kastylevsky G, Lemoine J. Clinical quantitation of prostate-specific antigen biomarker in the Low nanogram/milliliter range by conventional bore liquid chromatography-tandem mass spectrometry (multiple reaction monitoring) coupling and correlation with ELISA tests. Mol Cell Proteomics. 2009;8:1006–1015. doi: 10.1074/mcp.M800238-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorhall K, Miliotis T, Davidsson P. Comparison of different depletion strategies for improved resolution in proteomic analysis of human serum samples. Proteomics. 2005;5:307–317. doi: 10.1002/pmic.200400900. [DOI] [PubMed] [Google Scholar]
- Higuchi M, Iwata N, Matsuba Y, Takano J, Suemoto T, Maeda J, Ji B, Ono M, Staufenbiel M, Suhara T, Saido TC. Mechanistic involvement of the calpain-calpastatin system in Alzheimer neuropathology. FASEB J. 2012;26:1204–1217. doi: 10.1096/fj.11-187740. [DOI] [PubMed] [Google Scholar]
- Schutzer SE, Liu T, Natelson BH, Angel TE, Schepmoes AA, Purvine SO, Hixson KK, Lipton MS, Camp DG, Coyle PK, Smith RD, Bergquist J. Establishing the proteome of normal human cerebrospinal fluid. PLoS One. 2010;5(6):e10980. doi: 10.1371/journal.pone.0010980. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Gel electrophoresis of dog serum before and after albumin depletion using the ProteoSeek kit. The flow-through fraction and the column eluate were compared with and without prior acetone precipitation.
Table showing all eluted proteins identified by mass spectrometry.


