Abstract
Campylobacter, Arcobacter, and Helicobacter species have been isolated from many vertebrate hosts, including birds, mammals, and reptiles. Multiple studies have focused on the prevalence of these Epsilonproteobacteria genera in avian and mammalian species. However, little focus has been given to the presence within reptiles, and their potential zoonotic and pathogenic roles. In this study, occurrence, diversity, and host association of intestinal Epsilonproteobacteria were determined for a large variety of reptiles. From 2011 to 2013, 444 cloacal swabs and fecal samples originating from 417 predominantly captive-held reptiles were screened for Epsilonproteobacteria. Campylobacter, Arcobacter, and Helicobacter genus specific PCRs were performed directly on all samples. All samples were also cultured on selective media and screened for the presence of Epsilonproteobacteria. Using a tiered approach of AFLP, atpA, and 16S rRNA sequencing, 432 Epsilonproteobacteria isolates were characterized at the species level. Based on PCR, Campylobacter, Arcobacter, and Helicobacter were detected in 69.3% of the reptiles; 82.5% of the chelonians, 63.8% of the lizards, and 58.0% of the snakes were positive for one or more of these genera. Epsilonproteobacteria were isolated from 22.1% of the reptiles and were isolated most frequently from chelonians (37.0%), followed by lizards (19.6%) and snakes (3.0%). The most commonly isolated taxa were Arcobacter butzleri, Arcobacter skirrowii, reptile-associated Campylobacter fetus subsp. testudinum, and a putative novel Campylobacter taxon. Furthermore, a clade of seven related putative novel Helicobacter taxa was isolated from lizards and chelonians. This study shows that reptiles carry various intestinal Epsilonproteobacteria taxa, including several putative novel taxa.
Introduction
Campylobacter, Arcobacter, and Helicobacter species occupy a broad vertebrate host range and have been isolated from birds, mammals, and reptiles. These Epsilonproteobacteria genera are often found as commensals in the digestive tract, but some species are associated with disease in specific hosts [1]–[3]. The diverse genus Arcobacter also contains species free-living in the environment and species associated with non-vertebrate hosts [4]. Campylobacter, Helicobacter, and some Arcobacter species are mainly associated with endothermic vertebrate hosts, which is reflected in an optimal growth temperature of 30–42°C for most of the bacterial species [5]. However, Campylobacter, Arcobacter, and Helicobacter species have also been isolated from ectothermic reptiles [4], [6]–[10], which show a large variation in preferred body temperature and usually show considerable fluctuations in their body temperatures, often resulting in a lower average body temperature compared to most endothermic vertebrates [11]. A genetically distinct variant of Campylobacter fetus has been found in reptiles and humans that had direct or indirect contact with reptiles [6]–[13]. This reptile-associated C. fetus has been shown to cause infection in humans with underlying disease [12], [13]. Few bacterial species have been recognized as primary pathogens in reptiles [14], [15], and currently no cases have been reported of Campylobacter or Arcobacter species causing disease in reptiles, although a fatal septicemia in a pancake tortoise (Malacochersus tornieri) associated with an unidentified Helicobacter species has been described [9].
The role of reptiles in the epidemiology of Epsilonproteobacteria is currently unknown. Therefore, the aim of this study was to determine occurrence, diversity, and host association of intestinal Epsilonproteobacteria in reptiles.
Materials and Methods
Ethics statement
The procedures conducted on the reptiles in the study were merely combined with veterinary diagnostic and therapeutic acts and were therefore not considered to cause any additional discomfort. Consequently, the study was not considered an animal experiment under the Dutch Experiments on Animals Act (1977), making an assessment by an animal ethics committee unnecessary. The sampling was performed in strict accordance with the Dutch Law for Practicing Veterinary Medicine (1990). Fecal samples and cloacal swabs were collected as part of routine post mortem examination and diagnostic fecal examination for Campylobacter and other infectious agents. Sampling of live animals was performed by a veterinarian specialized in reptiles and amphibians (MK, Diplomate of the European College of Zoological Medicine, Herpetology). All owners provided consent for use of the samples collected from the reptiles.
Sample collection and preparation
From 2011 to 2013, a total of 444 cloacal swabs (n = 363) and fecal samples (n = 81) were collected from 417 healthy, diseased, or dead reptiles (Table S1). Free-living (n = 2), recently wild-caught (n = 56), and captive-held animals (n = 359) were included. Sampled animals belonged to 97 unique species of 27 families (Table S1). Diversity was high amongst sampled animals and the reptilian orders Squamata (comprising suborders Lacertilia (lizards) and Serpentes (snakes)) and Testudines (chelonians) were covered. Samples were obtained from three zoos (n = 170), a veterinary pathology division (n = 77), a veterinary clinic specialized in reptiles and amphibians (n = 102), a veterinary microbiological diagnostic laboratory (n = 5), a pet shop specialized in reptiles and amphibians (n = 56), and private owners (n = 34), all located in the Netherlands. For 27 animals two samples per animal were included.
Cloacal swabs (Amies charcoal 114C and 116C; Copan, Brescia, Italy) were extracted in 500 µl phosphate buffered saline (PBS); fecal samples were diluted in PBS until a fluid suspension was obtained. Of the each suspension, 240 µl was used directly for culturing and 100 µl was stored at −80°C with 10 mM EDTA for DNA extraction and direct genus specific PCR analysis.
Direct genus specific PCR analysis of samples
On all samples Campylobacter, Arcobacter, and Helicobacter genus specific PCRs were performed. For DNA isolation, 50 µl of lysis buffer (Cobas PCR Female Swab Sample Kit, Roche Molecular Systems Inc., Branchburg, USA) was added to 50 µl of each sample, followed by freezing at −80°C for 24 h and immediate heating at 95°C for 15 min in order to release the DNA and to inactivate DNAses. DNA was then extracted from this 100 µl sample on MagNA Pure96 (Roche), using the Roche MagNA Pure96 and Viral NA Small Volume Kit with the Viral NA Universal SV extraction protocol according to the manufacturer’s instructions. Final elution was in 100 µl, and 2 µl of these eluates was used as input for all PCRs. The Arcobacter, Campylobacter, and Helicobacter genus specific PCRs were performed as described previously [16]–[18], with slight adaptations. The Arcobacter genus specific PCR was performed using Go Taq Hot Start Green Master Mix (Promega, Madison, USA); total reaction volume was 25 µl and the PCR was run for 40 cycles on a 2720 Thermal Cycler (Applied Biosystems). Of each reaction, 10 µl was analyzed on a 1% agarose gel for the 1223 bp amplicon. Both Campylobacter and Helicobacter genus specific PCRs were performed using a LightCycler 480 II (Roche) and were run 45 cycles. LightCycler 480 Probes Master (Roche) was used for the Campylobacter genus specific PCR in a total reaction volume of 25 µl; LightCycler 480 SYBR Green I Master (Roche) was used for the Helicobacter genus specific PCR in a total reaction volume of 15 µl. Using the LightCycler 480 software (Roche), the Campylobacter and Helicobacter genus specific PCRs were analyzed based on absolute quantification and melting curves (range 85.5–87°C), respectively. Arcobacter butzleri strain SX-460, Campylobacter fetus strain NCTC 10842, and Helicobacter pylori strain J99 were used as reference strains. A subset of the amplicons was sequenced to validate the PCR results.
Culturing
From each of the sample suspensions, 10 µl was plated on each of four different media: blood agar (Colombia agar with 5% sheep blood), CCD agar, Preston agar and Skirrow agar (Oxoid, Landsmeer, The Netherlands). Additionally, a membrane filtration method was used; 200 µl suspension was applied on a cellulose filter membrane (ME26; 50 mm; pore size 0.6 µm; Whatman, Dassel, Germany) placed on a blood agar plate, incubated at atmospheric conditions at 37°C for one hour, after which the filter membrane was discarded. All agar plates were incubated in a microaerobic atmosphere containing hydrogen (83.3% N2, 7.1% CO2, 3.6% H2, and 6% O2) at 37°C and screened for colonies showing Epsilonproteobacteria-like morphology after 24 h, 48 h, and after a week of incubation. Selected colonies were colorless, whitish, greyish, or brownish, often small, slow growing, and flat, with or without the tendency to spread and showing no or little hemolysis on blood containing media. The number and diversity of colonies showing Epsilonproteobacteria-like morphology varied considerably for each sample and determined the number of selected colonies per sample. On average 9.0 (±6.1) colonies per sample were subcultured onto a fresh blood agar plate, incubated at 37°C under microaerobic conditions until growth was visible, and Gram-stained. Gram-negative bacteria showing helical or curved rod-shaped microscopic morphology typical for most Epsilonproteobacteria were selected and stored at −80°C.
For molecular analysis of the isolates, bacterial genomic DNA of all isolates was extracted following the Gram-negative bacteria protocol of either the High pure PCR template preparation kit (Roche Diagnostics, Almere, The Netherlands) or Gentra Puregene yeast/bacteria kit (Qiagen, Venlo, The Netherlands).
AFLP
Amplified fragment length polymorphism (AFLP) was used to analyze diversity amongst the isolates and for preliminary species identification by inclusion of well-characterized Campylobacter, Arcobacter, and Helicobacter strains. AFLP was performed on all isolates as described by Duim et al. [19], with 20 ng genomic DNA for each 14 µl restriction/ligation reaction mixture. Fingerprint data were created with a 3730 DNA analyzer (Applied Biosystems, Nieuwerkerk aan den IJssel, The Netherlands) and clustered (pairwise similarities; curve-based Pearson correlation; UPGMA clustering) using BioNumerics v.6.6 (Applied Maths, Sint-Martens-Latem, Belgium). For each sample one representative isolate per distinct fingerprint cluster (≥75% similarity) was selected for subsequent atpA and 16S rRNA sequencing.
atpA and 16S rRNA sequencing for species identification of isolates
For further characterization and species identification of the isolates atpA sequencing was performed. The atpA (or uncA) locus, encoding the ATPase synthase α subunit, is part of the MLST-scheme for Campylobacter [20]. In Epsilonproteobacteria, atpA is conserved and has been used to improve Campylobacteraceae and Helicobacteraceae species identification by 16S rRNA sequencing [21], thereby making it a good candidate for (sub)species identification of the isolates. Two degenerate primer sets targeting atpA from a broad range of Campylobacter, Arcobacter, and Helicobacter species were used: atpAF/R [20] and HFatpF/R [22]. For isolates for which atpA could not be sequenced, 16S rRNA encoding DNA sequencing was used using the 16S rDNA targeting primers 27F and 1492R [23]. Each PCR amplification mixture contained 2.5 µM each primer, 1x Go Taq Hot Start Green Master Mix (Promega) or Hot Star Taq Master Mix (Qiagen) and 1 ng/µl genomic DNA. PCRs were performed on a 2720 Thermal Cycler (Applied Biosystems) with the following conditions: 30 s at 94°C, 30 s at 53°C, and 2 min at 72°C (35 (16S rRNA) or 40 cycles (atpA)). DNA sequencing was performed by Macrogen (Amsterdam, The Netherlands), and DNA sequences were assembled and analyzed using BioNumerics v.6.6 (Applied Maths). Species were identified by aligning and clustering DNA sequences (MegAlign; DNASTAR Lasergene v.10.1) with an extensive database containing most currently known Epsilonproteobacteria (atpA) or using NCBI BLASTn (16S rRNA). By combining AFLP and sequence data, the species or genus was identified for all isolates. In addition, for all isolated Helicobacter taxa and most described Helicobacter species, 16S rRNA sequences of one strain per taxon were aligned with Muscle and a 16S rRNA based dendrogram was created using the neighbor-joining method, with bootstrap values determined using 500 repetitions (MEGA v.5.2).
Results
Campylobacter, Arcobacter, or Helicobacter were detected by PCR in 69.3% (289/417) of the reptiles; 82.5% (127/154) of Testudines, 63.8% (104/163) of Lacertilia, and 58.0% (58/100) of Serpentes were positive for one or more of these genera (Table 1). The occurrence of each Epsilonproteobacteria genus was highest in Testudines, followed by Lacertilia and Serpentes.
Table 1. Number of animals positive for each Epsilonproteobacteria taxon per reptilian (sub)order.
| Lacertilia (n = 163) | Serpentes (n = 100) | Testudines (n = 154) | Total (n = 417) | |||||
| Animals (%) | Animals (%) | Animals (%) | Animals (%) | |||||
| PCR | Culturing | PCR | Culturing | PCR | Culturing | PCR | Culturing | |
| Arcobacter genus | 23 (14.1) | 6 (3.7) | 13 (13.0) | 2 (2.0) | 47 (30.5) | 25 (16.2) | 83 (19.9) | 33 (7.9) |
| Arcobacter butzleri | 6 (3.7) | 2 (2.0) | 11 (7.1) | 19 (4.6) | ||||
| Arcobacter cryaerophilus | 1 (0.6) | 0 (0.0) | 9 (5.8) | 10 (2.4) | ||||
| Arcobacter skirrowii | 0 (0.0) | 0 (0.0) | 14 (9.1) | 14 (3.4) | ||||
| Campylobacter genus | 62 (38.0) | 18 (11.0) | 32 (32.0) | 3 (3.0) | 93 (60.4) | 39 (25.3) | 187 (44.8) | 60 (14.4) |
| Campylobacter fetus taxon 1* | 9 (5.5) | 3 (3.0) | 11 (7.1) | 23 (5.5) | ||||
| Campylobacter fetus taxon 2* | 0 (0.0) | 0 (0.0) | 1 (0.6) | 1 (0.2) | ||||
| Campylobacter hyointestinalis | 0 (0.0) | 0 (0.0) | 9 (5.8) | 9 (2.2) | ||||
| Campylobacter taxon 1* | 10 (6.1) | 0 (0.0) | 24 (15.6) | 34 (8.2) | ||||
| Campylobacter taxon 2* | 0 (0.0) | 0 (0.0) | 1 (0.6) | 1 (0.2) | ||||
| Helicobacter genus | 50 (30.7) | 12 (7.4) | 26 (26.0) | 0 (0.0) | 87 (56.5) | 8 (5.2) | 163 (39.1) | 20 (4.8) |
| Helicobacter taxon 1* | 1 (0.6) | 0 (0.0) | 0 (0.0) | 1 (0.2) | ||||
| Helicobacter taxon 2* | 0 (0.0) | 0 (0.0) | 8 (5.2) | 8 (1.9) | ||||
| Helicobacter taxon 3* | 4 (2.5) | 0 (0.0) | 0 (0.0) | 4 (1.0) | ||||
| Helicobacter taxon 4* | 1 (0.6) | 0 (0.0) | 0 (0.0) | 1 (0.2) | ||||
| Helicobacter taxon 5* | 1 (0.6) | 0 (0.0) | 0 (0.0) | 1 (0.2) | ||||
| Helicobacter taxon 6* | 1 (0.6) | 0 (0.0) | 0 (0.0) | 1 (0.2) | ||||
| Helicobacter taxon 7* | 4 (2.5) | 0 (0.0) | 0 (0.0) | 4 (1.0) | ||||
| Sulfurospirillum taxon* | 0 (0.0) | 0 (0.0) | 1 (0.6) | 1 (0.2) | ||||
| Total ** | 104 (63.8) | 32 (19.6) | 58 (58.0) | 3 (3.0) | 127 (82.5) | 57 (37.0) | 289 (69.3) | 92 (22.1) |
n, total of sampled animals;
*, putative novel species or subspecies based on 16S rRNA sequence and AFLP fingerprint;
**, not sum of Epsilonproteobacteria positive animals in case of multiple Epsilonproteobacteria taxa per animal.
With an occurrence of 44.8% (187/417) in all animals, Campylobacter was detected with the highest frequency. Compared to Campylobacter, Helicobacter and Arcobacter were detected less frequently: 39.1% (163/417) and 19.9% (83/417), respectively.
Overall, 432 Epsilonproteobacteria isolates were obtained by culturing (Table S1). Of the 92 positive animals, on average 4.7 (±4.1) Epsilonproteobacteria isolates were obtained from each positive animal. Based on AFLP fingerprint data of all isolates, a selection of 188 isolates with unique patterns for each sample was made for species identification with atpA and/or 16S rRNA sequencing. Combining AFLP and sequence data, all isolates could be identified.
Epsilonproteobacteria belonging to the genera Campylobacter, Arcobacter, Helicobacter, and Sulfurospirillum were isolated from 22.1% (92/417) of the animals (Table 1). Examining the occurrence of Epsilonproteobacteria for each reptilian (sub)order included in this study, highest occurrence is observed in Testudines 37.0% (57/154), followed by Lacertilia 19.6% (32/163) and Serpentes 3.0% (3/100). Testudines showed the highest Epsilonproteobacteria diversity with up to four different taxa per animal. Amongst Testudines, terrestrial chelonians of the Testudinidae family showed the highest occurrence of Epsilonproteobacteria: while 49.4% (76/154) of the sampled chelonians and 18.2% (76/417) of the sampled reptiles belonged to the Testudinidae, these animals made up 80.4% (45/56) of the Epsilonproteobacteria positive chelonians (odds ratio 8.8; 95% confidence interval 4.0–19.4; P<0.0001) and 48.9% (45/92) of all Epsilonproteobacteria positive reptiles (odds ratio 9.1; 95% confidence interval 5.2–15.8; P<0.0001).
Detection rates in reptiles were lower based on culturing methods (22.1%) than based on genus specific PCRs (69.3%). Animals positive for Epsilonproteobacteria by culturing were all positive by genus specific PCR (Campylobacter and Helicobacter), except one (Arcobacter; 97.0% (32/33)).
The majority of isolated Epsilonproteobacteria belonged to Campylobacter taxa (Table 1). Members of the Campylobacter genus were isolated from 14.4% (60/417) of the reptiles. Based on AFLP, atpA, and 16S rRNA sequence similarity, Campylobacter taxa genetically distinct from described species were identified, likely representing novel species and subspecies. Campylobacter taxon 1 and C. fetus taxon 1 were the most frequently isolated Epsilonproteobacteria taxa. Next to these taxa, C. hyointestinalis, Campylobacter taxon 2, and C. fetus taxon 2 were isolated from Testudines. The latter was distinct from other C. fetus taxa and all isolates were obtained from one animal (Chelonoidis carbonaria). Campylobacter taxon 1 was isolated from Testudines 15.6% (24/154) and Lacertilia 6.1% (10/163). Occurrence of C. fetus taxon 1 was highest in Testudines 7.1% (11/154), followed by Lacertilia 5.5% (9/163) and Serpentes 3.0% (3/100).
Arcobacter species were isolated from 7.9% (33/417) of the reptiles, predominantly from Testudines (Table 1). Species isolated were Arcobacter butzleri, Arcobacter cryaerophilus, and Arcobacter skirrowii. A. butzleri was isolated from Lacertilia, Serpentes, and Testudines while A. skirrowii was exclusively isolated from Testudines.
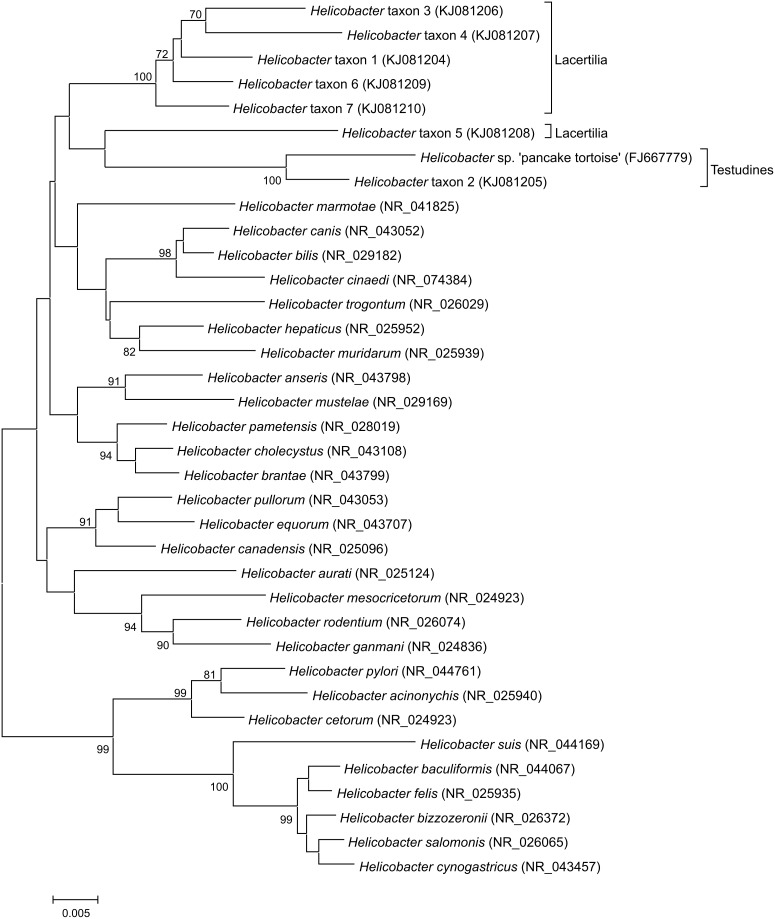
Compared to Arcobacter and Campylobacter, Helicobacter occurrence was low, 4.8% (20/417), but diversity was high. All isolates belonged to seven unknown taxa and were obtained from Lacertilia and Testudines (Table 1). A 16S rRNA based neighbor-joining dendrogram shows that these seven taxa cluster together, but apart from other Helicobacter species isolated from mammals and birds (Figure 1). Within this cluster, three distinct clusters are separated by deep branching. Interestingly, these clusters suggest Helicobacter host association: five out of six taxa obtained from Lacertilia clustered together, whereas Helicobacter taxon 2, obtained from Testudines, clustered with a previously described Helicobacter species isolated from a pancake tortoise (Malacochersus tornieri) [9].
Figure 1. Neighbor-joining dendrogram based on 16S rRNA showing the phylogenetic position of all isolated Helicobacter taxa.
Bootstrap values (≥70%) based on 500 repetitions are shown at the nodes of the dendrogram. The origins of the isolated Helicobacter taxa are indicated: Lacertilia (lizards) or Testudines (chelonians).
In addition to Campylobacter, Arcobacter, and Helicobacter taxa, isolates of an unknown Sulfurospirillum taxon were obtained from a Malaysian giant turtle (Orlitia borneensis) (Table 1).
For each putative novel taxon, 16S rRNA sequences have been deposited in GenBank and accession numbers have been provided (Table S2).
The detection rates for each culturing method based on sampled animals were: filter method 85.9% (79/92), CCD 48.9% (45/92), Preston 46.7% (43/92), Skirrow 40.2% (37/92), and blood agar 13.0% (12/92) (Table 2). All frequently occurring Campylobacter and Arcobacter taxa (≥1 positive animal for each taxon) could be isolated using the filter method, CCD, Preston, and Skirrow agar. Although some Helicobacter isolates were obtained from Preston and Skirrow agar, the majority (53/59) was isolated from blood agar or by the filter method. The Sulfurospirillum isolates were obtained by the filter method and from Preston agar.
Table 2. Number of animals positive for each Epsilonproteobacteria taxon per culture medium used.
| Blood agar | Filter | CCD | Preston | Skirrow | Total | |
| Taxon | Animals (%) | Animals (%) | Animals (%) | Animals (%) | Animals (%) | Animals (%) |
| Arcobacter butzleri | 0 (0.0) | 12 (63.2) | 7 (36.8) | 4 (21.1) | 9 (47.4) | 19 (20.2) |
| Arcobacter cryaerophilus | 0 (0.0) | 9 (90.0) | 4 (40.0) | 5 (50.0) | 4 (40.0) | 10 (10.9) |
| Arcobacter skirrowii | 1 (7.1) | 13 (92.9) | 7 (50.0) | 3 (21.4) | 3 (21.4) | 14 (15.2) |
| Campylobacter fetus taxon 1* | 0 (0.0) | 14 (60.9) | 16 (69.6) | 8 (34.8) | 10 (43.5) | 23 (25.0) |
| Campylobacter fetus taxon 2* | 0 (0.0) | 0 (0.0) | 1 (100.0) | 1 (100.0) | 0 (0.0) | 1 (1.1) |
| Campylobacter hyointestinalis | 2 (22.2) | 3 (33.3) | 6 (66.7) | 6 (66.7) | 2 (22.2) | 9 (9.8) |
| Campylobacter taxon 1* | 3 (8.8) | 18 (52.9) | 9 (26.5) | 18 (52.9) | 8 (23.5) | 34 (37.0) |
| Campylobacter taxon 2* | 0 (0.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (100.0) | 1 (1.1) |
| Helicobacter taxon 1* | 0 (0.0) | 1 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.1) |
| Helicobacter taxon 2* | 1 (12.5) | 8 (100.0) | 0 (0.0) | 0 (0.0) | 1 (12.5) | 8 (8.7) |
| Helicobacter taxon 3* | 2 (50.0) | 4 (100.0) | 0 (0.0) | 1 (25.0) | 1 (25.0) | 4 (4.3) |
| Helicobacter taxon 4* | 0 (0.0) | 1 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.1) |
| Helicobacter taxon 5* | 0 (0.0) | 1 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.1) |
| Helicobacter taxon 6* | 0 (0.0) | 1 (100.0) | 0 (0.0) | 0 (0.0) | 0 (0.0) | 1 (1.1) |
| Helicobacter taxon 7* | 3 (75.0) | 4 (100.0) | 0 (0.0) | 0 (0.0) | 2 (50.0) | 4 (4.3) |
| Sulfurospirillum taxon* | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (100.0) | 0 (0.0) | 1 (1.1) |
| Total ** | 12 (13.0) | 79 (85.9) | 45 (48.9) | 43 (46.7) | 37 (40.2) | 92 (100.0) |
*, putative novel species or subspecies based on 16S rRNA sequence and AFLP fingerprint;
**, not sum of Epsilonproteobacteria positive animals in case of multiple Epsilonproteobacteria taxa per animal.
Discussion
Epsilonproteobacteria were present in all reptilian orders examined. Chelonians showed highest Epsilonproteobacteria occurrence and diversity. Based on direct genus specific PCR analysis, Campylobacter occurrence was highest in all reptilian (sub)orders, followed by Helicobacter and Arcobacter. The majority of isolated Epsilonproteobacteria belonged to the genera Campylobacter and Arcobacter. Helicobacter was isolated less frequently. One undescribed Sulfurospirillum taxon was isolated. All currently described Sulfurospirillum species were isolated from environmental sources, potentially making this the first Sulfurospirillum taxon isolated from an animal host. Most commonly isolated taxa were Campylobacter taxon 1 and Campylobacter fetus taxon 1. Campylobacter taxon 1 has been proposed as a novel species, Campylobacter “iguaniorum” (Cig) (unpublished data). The 16S rRNA sequences of Cig were homologous to a recently described Campylobacter strain isolated from a leopard tortoise (Stigmochelys pardalis) [10]. Campylobacter fetus taxon 1 has recently been described as a novel subspecies, Campylobacter fetus subsp. testudinum (Cft) [24]. Cft is homologous to the previously described reptile-associated C. fetus [6]–[8], [12], [13] and similar detection rates in reptiles as found in this study have been reported [8]. A demarcated Campylobacter species distribution was observed in this study: species commonly found in mammals and birds, such as C. jejuni, C. coli, and C. lari, were not found in reptiles. The reverse was observed for Cig and Cft, which were isolated from reptiles, but have not (Cig) or rarely (Cft) been isolated from endothermic vertebrates. This differential distribution of Campylobacter species likely reflects the body temperature of the host, which is fluctuating and on average lower in ectothermic reptiles (with minimum and maximum voluntary temperature ranging from 5–46°C and a mean voluntary temperature ranging from 20–35°C for most families) than in endothermic birds and mammals [11]. Cft and Cig grow well at lower temperatures (20–37°C) and show less or no growth at higher temperatures (42°C), which might be an adaptation to their reptilian hosts. Many Epsilonproteobacteria taxa did not show a strict host association within the examined reptiles, i.e. the same taxon could be isolated from Lacertilia, Serpentes, and Testudines. Especially some of the Arcobacter and Campylobacter taxa displayed broad host ranges. In contrast, the isolated Helicobacter taxa suggest a more confined host association, as based on 16S rRNA phylogeny the taxa were separated in a cluster with isolates originating from lizards and a cluster with isolates originating from chelonians of the Testudinidae family. All Helicobacter taxa isolated from reptiles clustered separate from Helicobacter species isolated from mammals and birds, and, based on 16S rRNA divergence, likely represent novel species.
Interestingly, all snakes carrying Cft were sick or deceased, while most lizards and chelonians carrying Cft appeared clinically healthy. Of all isolates obtained from snakes, 88.0% (22/25) were Cft, and no other Epsilonproteobacteria taxa except three A. butzleri isolates were obtained from these animals. Necropsy showed that all Cft infected snakes had severe infections in the colon or abdominal cavity. It is tempting to speculate that Cft is associated with disease, but due to the low number of animals this should be interpreted with care. Little is known about pathogenicity in reptiles of other Epsilonproteobacteria taxa, although a fatal septicemia in a pancake tortoise (Malacochersus tornieri), associated with a Helicobacter species related to Helicobacter taxon 2, isolated from spur-thighed tortoises (Testudo graeca) and Russian tortoises (Agrionemys horsfieldii), has been described [9]. The majority of Epsilonproteobacteria species isolated from reptiles during this study (C. fetus, C. hyointestinalis, A. butzleri, A. cryaerophilus, and A. skirrowii) have been shown to be potentially pathogenic in humans [1], [25], [26]. Although for reptilian C. fetus there appears to be an association between contact with reptiles and infection in humans [6], [12], [13], to this date, no such association has been described for the other Epsilonproteobacteria species. The growing number of reptiles kept as pets in some European countries [27] and increased farming of reptiles, predominantly freshwater turtles for human consumption, in Asian countries [28], might increase the risk of reptilian derived Epsilonproteobacteria infections in humans.
It has to be noted that most samples were obtained from captive-held reptiles, which may lead to other transmission dynamics and an intestinal microbiota composition distinct from free-living reptiles, as has been shown for other vertebrates [29]. Indeed, AFLP indicated that unrelated reptile species having direct or indirect contact (same housing, same zoo, or same shipment) can harbor the same specific Epsilonproteobacteria variants, suggesting horizontal transmission (data not shown). Fecal-oral transmission is likely, especially in captive animals and species displaying coprophagy, although a common feed related origin cannot be excluded. Particularly chelonians of the Testudinidae family commonly display coprophagy, presumably to supplement their intestinal microbiota and optimize digestion of their herbivorous diet. This behavior could partly explain the frequent occurrence and high diversity of intestinal Epsilonproteobacteria in these chelonians. Nevertheless, the majority of Epsilonproteobacteria species were also isolated from recently caught free-living reptiles. Studies comparing Salmonella carriage in captive-held and free-living reptiles showed that occurrence was lower in the latter [30], [31], but additional sampling of free-living reptiles would be necessary to elucidate the native Epsilonproteobacteria carrier status. In addition to this, analysis of additional samples obtained from species of the excluded orders Rhynchocephalia and Crocodilia is required to obtain a more precise estimation of the intestinal Epsilonproteobacteria occurrence in reptiles.
The methods used in this study might have influenced the observed intestinal Epsilonproteobacteria occurrence, as described below. A total of 69.1% (PCR) and 22.3% (culturing) of the cloacal swabs, and 55.6% (PCR) and 16.0% (culturing) of the fecal samples was positive for Epsilonproteobacteria. Although there was no statistical difference between the detection rates for cloacal swabs and fecal samples (two tailed Fisher exact test; P = 0.03 (PCR) and P = 0.23 (culturing)), it is uncertain whether these detection rate differences are inherent to the sample type or merely a result of stochastic processes. Of 27 animals two samples were included, which potentially increased the detection rate: 12 (PCR) and 22 (culturing) cases contained two negative samples; eight (PCR) and three (culturing) cases contained one positive sample; seven (PCR) and two (culturing) cases contained two positive samples. The culturing temperature was 37°C, which may have been too high for culturing potential Campylobacter, Arcobacter, and Helicobacter from reptiles with low preferred body temperatures. A subset of samples was cultured at both 30°C and 37°C to determine the effect of temperature on growth rate and species composition. Although a lower growth rate was observed at 30°C, colony composition appeared similar at both temperatures (data not shown). The culture media were selected to promote Epsilonproteobacteria growth and inhibit growth of non-Epsilonproteobacteria. Although most of the antimicrobial containing media used are optimized for C. jejuni and C. coli, the results of the filter method showed high agreement with media containing antimicrobials for all isolated Campylobacter and Arcobacter taxa (positive animals >1). Most Helicobacter taxa were isolated using media without antimicrobials. The discrepancy in detection rates between culturing and direct genus specific PCR was most prominent in the Helicobacter genus, suggesting that the culturing methods were suboptimal for Helicobacter. Also, non-motile Epsilonproteobacteria species, such as C. gracilis, will likely not pass through the filter. Microaerobic conditions are preferred by most culturable Epsilonproteobacteria species [5], although some species, such as C. gracilis and C. rectus, prefer anaerobic conditions. The presence of hydrogen, formate, or succinate as an electron source is essential for growth of some Campylobacter species. Furthermore, only 10 µl (≤2%) of the fecal suspension was plated on selective media, and a selection of colonies displaying Epsilonproteobacteria-like morphology was isolated. Since not all Epsilonproteobacteria display the typical helical or curved rod shape, screening microscopically for this feature might lead to an underestimation of the occurrence.
As expected, direct genus specific PCRs resulted in higher detection rates compared to culturing. Nearly all samples positive for Epsilonproteobacteria by culturing were positive by PCR as well. Members of the Campylobacter genus were most frequently detected by both PCR and culturing. Remarkably, members of the Arcobacter genus were detected least frequently by PCR, whereas members of the Helicobacter genus were detected least frequently by culturing. This discrepancy indicates that the Arcobacter and Helicobacter PCRs likely have different sensitivities and that both genera show differential growth on the media used. Indeed, many members of the Helicobacter genus are considered fastidious organisms [2]. Since Epsilonproteobacteria were detected on genus level by PCR and higher detection rates were observed by PCR than by culturing, fastidious or non-culturable species might not have been identified, which could have led to an underestimation of the true Epsilonproteobacteria diversity. Ideally, one PCR detecting all Epsilonproteobacteria directly in crude samples would overcome the potential differences in sensitivities between PCRs. Although a PCR-RFLP method targeting 16S rRNA of most Campylobacter, Arcobacter, and Helicobacter species has been described [32], this assay is designed for species identification of isolates, and not for direct assessment of samples. While molecular methods are useful, isolates obtained by culturing are preferred for further analysis, such as examination of epidemiology, diversity, and phenotypic characters, making both methods complementary.
The polyphasic approach based on direct genus specific PCR and culturing was useful to determine intestinal Epsilonproteobacteria occurrence and diversity from genus to subspecies level. Our results show that reptiles comprise a significant reservoir for Epsilonproteobacteria. A clade of related putative novel Helicobacter taxa appears to be specifically associated with reptiles. Arcobacter butzleri, Arcobacter skirrowii, Campylobacter fetus subsp. testudinum, and Campylobacter “iguaniorum” represent a major portion of the Epsilonproteobacteria isolated from reptiles, chelonians in particular. The Campylobacter taxa are thus far predominantly found in reptiles, which may constitute their primary reservoir.
Supporting Information
Number of samples, animals, and detected Epsilonproteobacteria for all reptilian species included in this study. Epsilon, Epsilonproteobacteria; A, Arcobacter genus; Ab, Arcobacter butzleri; Ac, Arcobacter cryaerophilus; As, Arcobacter skirrowii; C, Campylobacter genus; Cf1, Campylobacter fetus taxon 1; Cf2, Campylobacter fetus taxon 2; Ch, Campylobacter hyointestinalis; C1, Campylobacter taxon 1; C2, Campylobacter taxon 2; H, Helicobacter genus; H1, Helicobacter taxon 1; H2, Helicobacter taxon 2; H3, Helicobacter taxon 3; H4, Helicobacter taxon 4; H5, Helicobacter taxon 5; H6, Helicobacter taxon 6; H7, Helicobacter taxon 7; S, Sulfurospirillum taxon.
(XLSX)
GenBank accession numbers of the 16S rRNA sequences for all putative novel Epsilonproteobacteria taxa isolated during this study. *, 16S rRNA sequence deposited at GenBank prior to this study.
(XLSX)
Acknowledgments
We kindly thank Mark de Boer (Rotterdam Zoo), Dennis de Haan, Warren Spencer (Artis Royal Zoo), Arendo Flipse (Herpetological Center Holland), Job Stumpel, and Carlos Voogdt for their valuable contribution to sample collection, Abigaël Koulen and Linda van der Graaf-van Bloois (Utrecht University) for technical support, and Rebekah Keesler (Utrecht University) for proof reading.
Funding Statement
The authors have no support or funding to report.
References
- 1.Nachamkin I, Szymanski CM, Blaser MJ, editors (2008) Campylobacter. Washington D.C.: ASM Press. [Google Scholar]
- 2. Solnick JV, Schauer DB (2001) Emergence of diverse Helicobacter species in the pathogenesis of gastric and enterohepatic diseases. Clin Microbiol Rev 14: 59–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Collado L, Figueras MJ (2011) Taxonomy, epidemiology, and clinical relevance of the genus Arcobacter . Clin Microbiol Rev 24: 174–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Debruyne L, Gevers D, Vandamme P (2008) Taxonomy of the family Campylobacteraceae. In: Nachamkin I, Szymanski CM, Blaser MJ, editors. Campylobacter. Washington D.C.: ASM Press. 3–25. [Google Scholar]
- 5. On SL, Holmes B, Sackin MJ (1996) A probability matrix for the identification of campylobacters, helicobacters and allied taxa. J Appl Bacteriol 81: 425–432. [DOI] [PubMed] [Google Scholar]
- 6. Harvey S, Greenwood JR (1985) Isolation of Campylobacter fetus from a pet turtle. J Clin Microbiol 21: 260–261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Dingle KE, Blaser MJ, Tu ZC, Pruckler J, Fitzgerald C, et al. (2010) Genetic relationships among reptilian and mammalian Campylobacter fetus strains determined by multilocus sequence typing. J Clin Microbiol 48: 977–980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Wang CM, Shia WY, Jhou YJ, Shyu CL (2013) Occurrence and molecular characterization of reptilian Campylobacter fetus strains isolated in Taiwan. Vet Microbiol 164: 67–76. [DOI] [PubMed] [Google Scholar]
- 9. Stacy BA, Wellehan JF Jr (2010) Fatal septicemia caused by Helicobacter infection in a pancake tortoise (Malacochersus tornieri). J Vet Diagn Invest 22: 660–662. [DOI] [PubMed] [Google Scholar]
- 10. Benejat L, Gravet A, Sifré E, Ben Amor S, Quintard B, et al. (2014) Characterization of a Campylobacter fetus-like strain isolated from the faeces of a sick leopard tortoise (Stigmochelys pardalis) using matrix-assisted laser desorption/ionization time of flight as an alternative to bacterial 16S rDNA phylogeny. Lett Appl Microbiol 58: 338–343. [DOI] [PubMed] [Google Scholar]
- 11.Vitt LJ, Caldwell JP (2013) Herpetology: An introductory biology of amphibians and reptiles. San Diego: Academic Press. [Google Scholar]
- 12. Tu ZC, Zeitlin G, Gagner JP, Keo T, Hanna BA, et al. (2004) Campylobacter fetus of reptile origin as a human pathogen. J Clin Microbiol 42: 4405–4407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Patrick ME, Gilbert MJ, Blaser MJ, Tauxe RV, Wagenaar JA, et al. (2013) Human infections with new subspecies of Campylobacter fetus . Emerg Infect Dis 19: 1678–1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Pasmans F, Blahak S, Martel A, Pantchev N (2008) Introducing reptiles into a captive collection: The role of the veterinarian. The Veterinary Journal 175: 53–68. [DOI] [PubMed] [Google Scholar]
- 15.Girling S, Raiti P (2004) BSAVA manual of reptiles. Quedgeley, Gloucester: British Small Animal Veterinary Association.
- 16. Harmon K, Wesley I (1996) Identification of Arcobacter isolates by PCR. Lett Appl Microbiol 23: 241–244. [DOI] [PubMed] [Google Scholar]
- 17. de Boer RF, Ott A, Guren P, van Zanten E, van Belkum A, et al. (2013) Detection of Campylobacter species and Arcobacter butzleri in stool samples by use of real-time multiplex PCR. J Clin Microbiol 51: 253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Coldham T, Rose K, O'Rourke J, Neilan BA, Dalton H, et al. (2011) Detection, isolation, and characterization of Helicobacter species from the gastrointestinal tract of the brushtail possum. Appl Environ Microbiol 77: 1581–1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Duim B, Wassenaar TM, Rigter A, Wagenaar J (1999) High-resolution genotyping of Campylobacter strains isolated from poultry and humans with amplified fragment length polymorphism fingerprinting. Appl Environ Microbiol 65: 2369–2375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Miller WG, On SL, Wang G, Fontanoz S, Lastovica AJ, et al. (2005) Extended multilocus sequence typing system for Campylobacter coli, C. lari, C. upsaliensis, and C. helveticus . J Clin Microbiol 43: 2315–2329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Miller WG, Yee E, Jolley KA, Chapman MH (2014) Use of an improved atpA amplification and sequencing method to identify members of the Campylobacteraceae and Helicobacteraceae. Lett Appl Microbiol. DOI:10.1111. [DOI] [PubMed]
- 22. Miller WG, Chapman MH, Yee E, On SL, McNulty DK, et al. (2012) Multilocus sequence typing methods for the emerging Campylobacter species C. hyointestinalis, C. lanienae, C. sputorum, C. concisus, and C. curvus . Front Cell Infect Microbiol 2: 45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Maiwald M (2004) Broad-range PCR for detection and identification of bacteria. In: Persing DH, Tenover FC, Versalovic J, Tang Y-W, Relman D, et al., editors. Molecular Microbiology: Diagnostics Principles and Practice. Washington D.C.: ASM press. 379–390. [Google Scholar]
- 24.Fitzgerald C, Tu ZC, Patrick M, Stiles T, Lawson AJ, et al. (2014) Description of Campylobacter fetus subsp. testudinum subsp. nov., isolated from humans and reptiles. Int J Syst Evol Microbiol. DOI:10.1099. [DOI] [PubMed]
- 25. Edmonds P, Patton CM, Griffin PM, Barrett TJ, Schmid GP, et al. (1987) Campylobacter hyointestinalis associated with human gastrointestinal disease in the United States. J Clin Microbiol 25: 685–691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Vandenberg O, Dediste A, Houf K, Ibekwem S, Souayah H, et al. (2004) Arcobacter species in humans. Emerg Infect Dis 10: 1863–1867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Editorial team, Bertrand S, Rimhanen-Finne R, Weill FX, Rabsch W, et al. (2008) Salmonella infections associated with reptiles: The current situation in Europe. Euro Surveill 13: 18902. [PubMed] [Google Scholar]
- 28. Haitao S, Parham JF, Zhiyong F, Meiling H, Feng Y (2008) Evidence for the massive scale of turtle farming in China. Oryx 42: 147–150. [Google Scholar]
- 29. Ochman H, Worobey M, Kuo CH, Ndjango JB, Peeters M, et al. (2010) Evolutionary relationships of wild hominids recapitulated by gut microbial communities. PLoS Biol 8: e1000546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Scheelings TF, Lightfoot D, Holz P (2011) Prevalence of Salmonella in Australian reptiles. J Wildl Dis 47: 1–11. [DOI] [PubMed] [Google Scholar]
- 31. Richards JM, Brown JD, Kelly TR, Fountain AL, Sleeman JM (2004) Absence of detectable Salmonella cloacal shedding in free-living reptiles on admission to the wildlife center of Virginia. J Zoo Wildl Med 35: 562–563. [DOI] [PubMed] [Google Scholar]
- 32. Marshall SM, Melito PL, Woodward DL, Johnson WM, Rodgers FG, et al. (1999) Rapid identification of Campylobacter, Arcobacter, and Helicobacter isolates by PCR-restriction fragment length polymorphism analysis of the 16S rRNA gene. J Clin Microbiol 37: 4158–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Number of samples, animals, and detected Epsilonproteobacteria for all reptilian species included in this study. Epsilon, Epsilonproteobacteria; A, Arcobacter genus; Ab, Arcobacter butzleri; Ac, Arcobacter cryaerophilus; As, Arcobacter skirrowii; C, Campylobacter genus; Cf1, Campylobacter fetus taxon 1; Cf2, Campylobacter fetus taxon 2; Ch, Campylobacter hyointestinalis; C1, Campylobacter taxon 1; C2, Campylobacter taxon 2; H, Helicobacter genus; H1, Helicobacter taxon 1; H2, Helicobacter taxon 2; H3, Helicobacter taxon 3; H4, Helicobacter taxon 4; H5, Helicobacter taxon 5; H6, Helicobacter taxon 6; H7, Helicobacter taxon 7; S, Sulfurospirillum taxon.
(XLSX)
GenBank accession numbers of the 16S rRNA sequences for all putative novel Epsilonproteobacteria taxa isolated during this study. *, 16S rRNA sequence deposited at GenBank prior to this study.
(XLSX)