Abstract
Recent genomic sequencing of the foxtail millet, an abiotic, stress-tolerant crop, has provided a great opportunity for novel gene discovery and functional analysis of this popularly-grown grass. However, few stress-mediated gene families have been studied. Aldehyde dehydrogenases (ALDHs) comprise a gene superfamily encoding NAD (P) +-dependent enzymes that play the role of “aldehyde scavengers”, which indirectly detoxify cellular ROS and reduce the effect of lipid peroxidation meditated cellular toxicity under various environmental stresses. In the current paper, we identified a total of 20 ALDH genes in the foxtail millet genome using a homology search and a phylogenetic analysis and grouped them into ten distinct families based on their amino acid sequence identity. Furthermore, evolutionary analysis of foxtail millet reveals that both tandem and segmental duplication contributed significantly to the expansion of its ALDH genes. The exon-intron structures of members of the same family in foxtail millet or the orthologous genes in rice display highly diverse distributions of their exonic and intronic regions. Also, synteny analysis shows that the majority of foxtail millet and rice ALDH gene homologs exist in the syntenic blocks between the two, implying that these ALDH genes arose before the divergence of cereals. Semi-quantitative and real-time quantitative PCR data reveals that a few SiALDH genes are expressed in an organ-specific manner and that the expression of a number of foxtail millet ALDH genes, such as, SiALDH7B1, SiALDH12A1 and SiALDH18B2 are up-regulated by osmotic stress, cold, H2O2, and phytohormone abscisic acid (ABA). Furthermore, the transformation of SiALDH2B2, SiALDH10A2, SiALDH5F1, SiALDH22A1, and SiALDH3E2 into Escherichia coli (E.coli) was able to improve their salt tolerance. Taken together, our results show that genome-wide identification characteristics and expression analyses provide unique opportunities for assessing the functional roles of foxtail millet ALDH genes in stress responses.
Introduction
As they grow, plants often encounter a wide spectrum of environmental stresses [1], such as drought, salinity, extreme temperatures and oxidative stress. To assure the survival and prosperity of their offspring, plants regulate the expression of a wide range of stress-responsive genes capable of coping with various abiotic stresses, which negatively affect plants by eliciting rapid and excessive accumulation of reactive oxygen species (ROS) that leads to cellular injury (e.g., lipid peroxidation or protein and nucleic acid modification) [2], [3]. Moreover, ROS reacting with lipids and proteins are known to cause an accumulation of toxic products (i.e. aldehydes), which in turn amplify ROS-induced damage. As such, a better understanding of the mechanisms involved in the evolutionary survival of plants to environmental stresses would be of great value.
Aldehyde dehydrogenases (ALDHs) represent an evolutionary conserved gene superfamily encoding NAD (P)+-dependent enzymes that catalyze the irreversible oxidation of a wide range of endogenous and exogenous aromatic and aliphatic aldehydes into corresponding carboxylic acids [4]. Several studies show that many ALDHs protect against various environmental stressors by indirectly detoxifying cellular ROS and/or reducing lipid peroxidation [5]. For instance, overexpression of AtALDH3 reduces lipid peroxidation and increases resistance to osmotic stress, metal toxicity, H2O2 and paraquat treatment [6]. Furthermore, ectopic expression of ALDH7 in both Arabidopsis and tobacco enhances their protection against various osmotic stressors, such as dehydration and high salinity [7]. In addition, overexpression of ALDH22A1 in transgenic tobacco plants increases its stress tolerance and leads to decreased levels of malondialdehyde (MDA) [8]. ALDH genes also play vital roles in many fundamental metabolic pathways under normal conditions. For example, they facilitate in the synthesis and catabolism of a wide of range of biomolecules, such as amino acids, lipids, and vitamins. As such, the ability of ALDHs to facilitate stress responses in plants has made these enzymes the focus of numerous studies on developing stress-resistant crops.
Aldehyde dehydrogenases constitute a diverse protein family found in various organisms. In 1999, the ALDH Gene Nomenclature Committee (AGNC) established criteria for cataloguing deduced ALDH protein sequences [9]. Sequences with more than 40% identity to a previously identified ALDH sequence represent a family and sequences with more than 60% identity within the ALDH family represent a protein subfamily. To date, ALDHs have been classified into 24 distinct families; ALDH10, ALDH12, ALDH21, ALDH22, ALDH23, and ALDH24 are unique to plants [10]–[13]. Completion of the genomic sequencing of various plants has allowed more and more aldehyde dehydrogenase genes to be identified and classified from lower to higher plants, such as Physcomitrella patens, Chlamydomonas reinhardtii, Arabidopsis thaliana, Vitis vinifera, Zea mays, Sorghum bicolor and Glycine max [10], [12]–[15]. However, there have been any systematic investigations of aldehyde dehydrogenase families from the foxtail millet published.
Foxtail millet (Setaria italica L.) is well known for its drought tolerance and is one of the oldest cultivated millet crops and an important food and fodder grain crop in arid and semi-arid regions of Asia and Africa. It is a diploid C4 panicoid crop with a small genome of ∼515 Mb, a short life cycle, and a highly conserved genome structure relative to ancestral grass lineages [16]. As such, foxtail millet has been proposed as an ideal model crop for genetic and molecular studies. Recently, the US Department of Energy Joint Genomic Institute [17] and Beijing Genomics Institute (BGI), China [16] was able to fully sequence its genome, paving the way to additional genome-wide identification studies and the analysis of gene families in foxtail millet.
The SiNAC [18] and SiWD40 [19] gene families have recently been systematically analyzed. However, to date there are no reports on the analysis of the SiALDH gene family. In the current study, we identified 20 ALDH genes from foxtail millet, classified them into 10 different gene families, and investigated their expansion and evolutionary history by examining their duplication, chromosomal distribution, exon-intron structure and synteny map with their rice orthologs. Subsequently, we analyzed the expression profiles of these SiALDH genes in different tissues and under various abiotic stressors and identified the function of ten SiALDH genes expressed in transgenic E.coli in response to salinity-induced stress.
Results and Discussion
The foxtail millet ALDH gene family: nomenclature and phylogenetic analysis
Keywords, HMM profile, and BLAST searches uncovered that approximately twenty ALDH proteins exist in the S.italica genome. Furthermore, based on criteria established by the AGNC, 20 putative ALDH genes were also identified in S.italica genome and grouped into ten families (Table 1). Four families contained multiple members (ALDH2, six members; ALDH3, four members; ALDH10 and ALDH18, two members each), each of the other six families was represented by a single gene (ALDH5, ALDH6, ALDH7, ALDH11, ALDH 12, and ALDH22) and each gene was assigned to different subfamilies (Table 1). A unique identifier was assigned to each of the S.italica ALDH proteins. A number, based on the present location of the ALDH gene on the chromosome, following the subfamily name was used to distinguish multiple members contained in a single family. Subcellular localization predictions revealed that most of S.italica ALDH proteins are located in the cytoplasm and mitochondrion. Moreover, ten S.italica ALDH proteins had splice variant and only primary transcripts were selected for phylogenetic and comparative analyses. Among the 20 foxtail millet ALDH genes identified, eighteen genes were supported by full length protein sequences and cDNA sequences (their corresponding GeneBank Accession are listed in Table 1); the other two genes were not found in GeneBank, but the full length sequences are shown in the Text S1.
Table 1. Foxtail millet ALDH genes and superfamilies.
| Family | Gene Locus ID | Annotation | Accession No. | CDS (bp) | ORF (aa) | Subcellular Localization |
| Family 2 | Si000898m | SiALDH2C2 | XM_004968994 | 1662 | 553 | Cytoplasm |
| Si000743m | SiALDH2C3 | XM_004968990 | 1818 | 605 | Mitochondrion | |
| Si040073m | SiALDH2C4 | No entry | 1359 | 453 | Peroxisome | |
| Si006255m | SiALDH2C1 | XM_004965940 | 1545 | 514 | Cytoplasm | |
| Si006183m | SiALDH2B2 | XM_004965148 | 1650 | 549 | Mitochondrion | |
| Si016807m | SiALDH2B1 | XM_004953741 | 1668 | 555 | Mitochondrion | |
| Family 3 | Si009981m | SiALDH3H1 | XM_004977167 | 1449 | 482 | Cytoplasm |
| Si026307m | SiALDH3H2 | No entry | 1434 | 447 | Cytoplasm | |
| Si009984m | SiALDH3E2 | XM_004976343 | 1446 | 481 | Cytoplasm | |
| Si017050m | SiALDH3E1 | XM_004953227 | 1458 | 485 | Cytoplasm | |
| Family 5 | Si016884m | SiALDH5F1 | XM_004951692 | 1587 | 528 | Mitochondrion |
| Family 6 | Si029327m | SiALDH6B1 | XM_004955659 | 1746 | 581 | Cytoplasm |
| Family 7 | Si029116m | SiALDH7B1 | XM_004956875 | 2055 | 684 | Mitochondrion |
| Family 10 | Si009902m | SiALDH10A2 | XM_004975822 | 1518 | 505 | Mitochondrion |
| Si013592m | SiALDH10A1 | XM_004973405 | 1518 | 505 | Cytoplasm | |
| Family 11 | Si013613m | SiALDH11A1 | XM_004973492 | 1497 | 498 | Cytoplasm |
| Family 12 | Si021508m | SiALDH12A1 | XM_004961322 | 1833 | 610 | Cytoplasm |
| Family 18 | Si000348m | SiALDH18B2 | XM_004970516 | 2412 | 803 | Cytoplasm |
| Si021235m | SiALDH18B1 | XM_004961829 | 2457 | 818 | Cytoplasm | |
| Family 22 | Si029482m | SiALDH22A1 | XM_004958707 | 1581 | 526 | Cytoplasm |
In the present study, we listed the numbers of gene family members for each individual ALDH family in S.italica and ten other plant species (Arabidopsis thaliana, Oryza sativa, Sorghum bicolor, Zea mays, Vitis vinifera, Populus trichocarpa, Physcomitrella patens, Chlamydomonas reinhardtii, Volvox carteri, and Glycine max) and Homo sapiens (Table S1). Of note, there is a lack of ALDH1 and ALDH4 gene family members found in plants due to nomenclature errors made when the genes were originally identified [12]. Plants have 13 various ALDH gene families: ALDH2, ALDH3, ALDH5, ALDH6, ALDH7, ALDH10, ALDH11, ALDH12, ALDH18, ALDH21, ALDH22, ALDH23, and ALDH24; in addition, ALDH19 has only been identified within tomato genome [20]. ALDH21, ALDH23 and ALDH24 are unique to lower plants. Like other monocot/dicot plants, S.italica contain ten common core ALDH families: ALDH2, ALDH3, ALDH5, ALDH6, ALDH7, ALDH10, ALDH11, ALDH12, ALDH18, and ALDH22). Unfortunately, the evolutionary relationships of the ALDH protein in foxtail millet and other plants had never been performed.
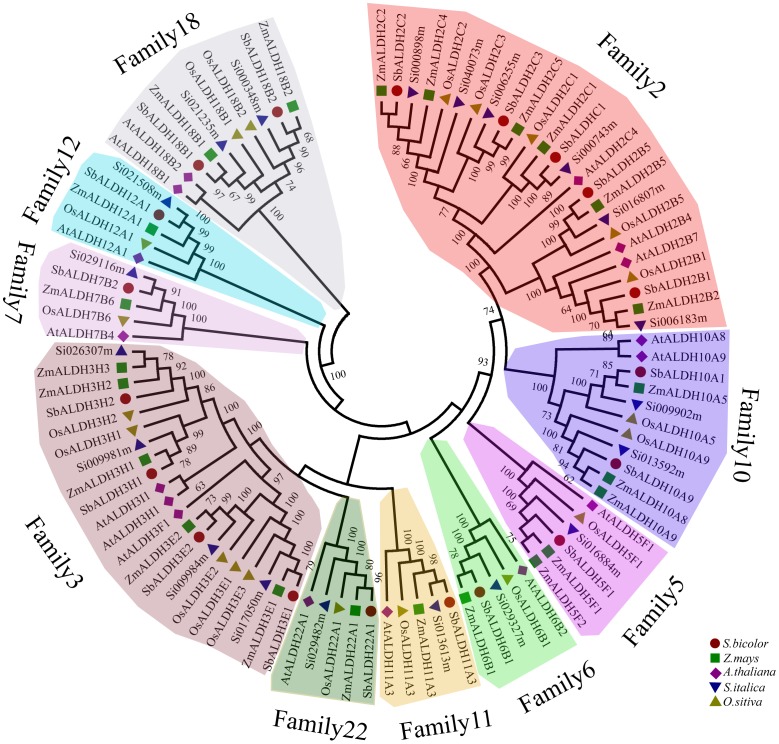
In recent years, the ALDH protein in multiple plant species has been identified. As such, we chose several representational plants for analysis, including Arabidopsis thaliana, Oryza sativa, Sorghum bicolor, and Zea mays and constructed a phylogenetic tree of the ALDH genes in these species, in addition to foxtail millet. The tree was classified into ten major families (Fig. 1 & Table 1) and ALDH proteins from the same families were clustered together (Fig. 1). The phylogenetic tree also revealed that these plant ALDHs split into three clades and share the common core plant ALDH families, listed above. In addition, we found that the majority of foxtail millet ALDH families are more closely related to those in grass species (O. sativa, Z. Mays, and S. Bicolor). These results are consistent with the present understanding of plant evolutionary history [21].
Figure 1. Phylogenetic analysis of foxtail millet and other plant ALDHs.
Phylogenetic tree was constructed with ALDH protein sequences from S.bicolor (Sb), Z.mays (Zm), O.sativa (Os), S.italica (Si), and A. thaliana (At). Members of respective ALDH families are depicted in a specific background color.
Segmental and tandem duplication contribute to superfamily expansion
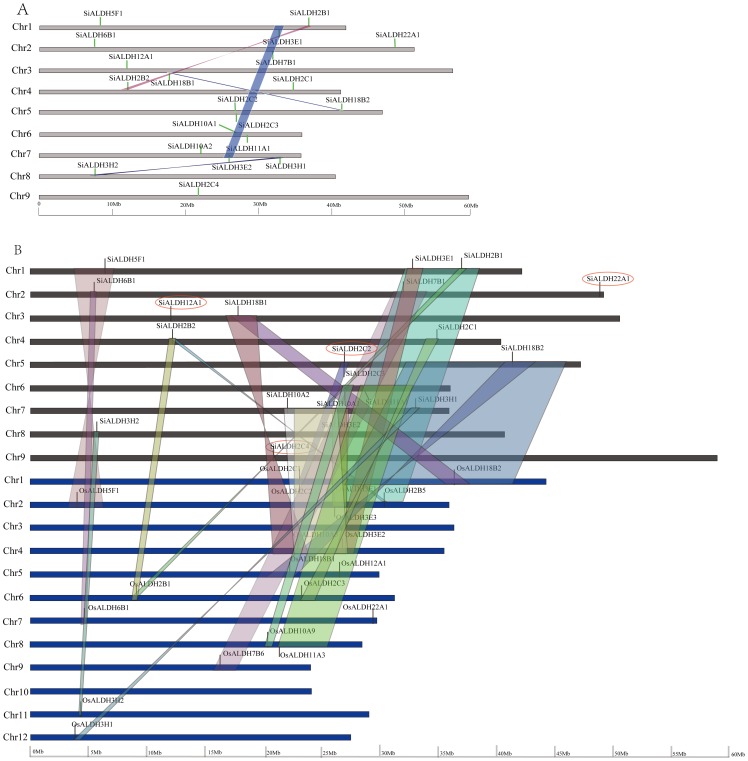
Gene duplication plays an important role in the expansion of gene families. It was recently reported that the foxtail millet genome underwent whole-genome duplication, similar to other grasses, approximately 70 MYA (million year ago) and most of the duplications were generated in the whole genome duplication (WGD) event shared by all grasses [16]. In the present analysis, we found that the 20 foxtail millet ALDH genes are randomly distributed on nine foxtail millet chromosomes (Fig. 2A). Among them, eight genes (SiALDH3E1/SiALDH3E2, SiALDH3H1/SiALDH3H2, SiALDH2B1/SiALDH2B2 and SiALDH18B1/SiALDH18B2) are located in four pairs of segmental duplicated genome regions, likely caused by whole genome duplication. In addition, SiALDH2C2 and SiALDH2C3 genes are identified as a tandem duplication in foxtail millet ALDH family. In summary, four foxtail millet multi-member ALDH gene families are linked with either segmental or tandem duplication events, suggesting that segmental and tandem duplication events play significant roles in the expansion of SiALDH genes. Moreover, the contribution of selection on coding sequences can be quantified by measuring the ratio of nonsynonymous to synonymous substitutions (Ka/Ks). A pair of sequences with Ka/Ks<1 indicates that one sequence has undergone purifying selection while the other has been drifting neutrally. Alternatively, if Ka/Ks = 1 then both sequences have been drifting neutrally. And finally, in the rare event that Ka/Ks>1 specific sites in that sequence have been under positive selection [22]. Our results indicate that the Ka/Ks ratios for five duplicated pairs varied from 0.0024 to 0.1726 with an average of 0.06998 (Table S2), strongly indicating that the SiALDH family underwent strong purifying selection pressure. All five paralogous pairs showed Ks values >1.11, signifying that these duplications might have occurred >85.48 MYA (Table S2). This result suggests that duplications of these 5 paralogous pairs occurred before the whole genome duplication (WGD) of grass. However, we are cautions about asserting that theses SiALDH duplicated before the WGD, considering that the pseudogenization or frame-shift mutation have the potential to inflate Ks value [23].
Figure 2. Distribution and synteny of ALDH genes on foxtail millet chromosomes and synteny analysis of ALDH genes between foxtail millet and rice.
(A) Chromosomes 1–9 are depicted as horizontal gray bars. ALDH genes are indicated by vertical green line. Colored bars denote syntenic of the foxtail millet genome. (B) Foxtail millet and rice chromosomes are depicted as horizontal black and blue bars, respectively. Foxtail millet and rice ALDH gene are indicated by vertical black lines, Colored bars denote syntenic regions between foxtail millet and rice chromosomes. A twisted colored bar indicates that syntenic regions are in opposite orientations. SiALDH genes don't located in the syntenc blocks between rice and foxtail millet chromosomes were marked by red oval.
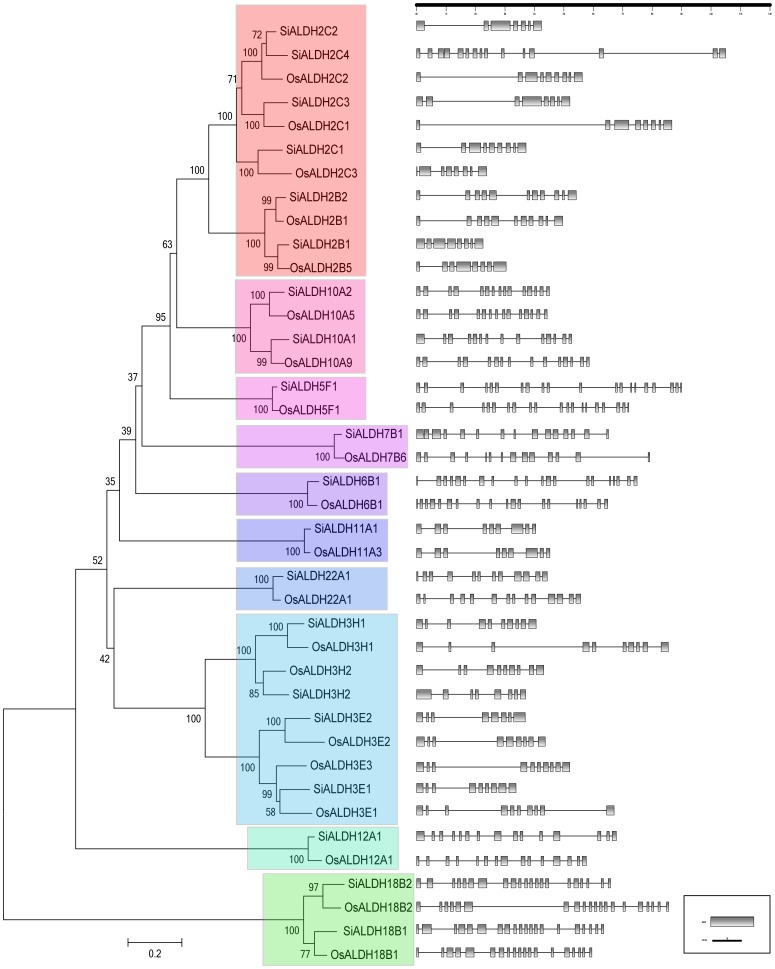
Structural divergences are prevalent in duplicate genes and, in most cases, have led to the generation of functionally distinct paralogs [24]. Investigation into the gene structures of multiple gene families (Fig. S1) shows that duplicate genes of SiALDH have a highly diverse distribution of exons and introns in the foxtail millet genome. In addition, nonduplicate genes of SiALDH also have different gene structures. For example, ALDH10 genes from rice [25] and grape [12] had the same numbers of exons while exhibiting nearly identical lengths, whereas the first exon of SiALDH10A1 and SiALDH10A2 was clearly different in the foxtail millet genome. We hypothesize that this difference is attributed to a rapid evolution of these genes through gene duplications or via their integration into the genome following reverse transcription [26]–[28].
Evolutionary relationship of ALDH gene families between foxtail millet and rice
Researchers can gain a better understanding of the genomic structure/evolution of a lesser-studied taxon via comparative genomic analyses [29]. As such, we were able to infer the probable function of the orthologous genes by comparing the genomes of foxtail millet and rice. Large-scale synteny analysis identified 90% (18/20) of SiALDH genes to have shared synteny with their orthologs of the rice genome. Among them, 10 pairs (SiALDH2C3/OsALDH2C1, SiALDH2C1/OsALDH2C3, SiALDH3H1/OsALDH3H1, SiALDH3H2/OsALDH3H2, SiALDH5F1/OsALDH5F1, SiALDH6B1/OsALDH6B1, SiALDH7B1/OsALDH7B6, SiALDH10A2/OsALDH10A5, SiALDH10A1/OsALDH10A9, and SiALDH11A1/OsALDH11A3) unambiguously existed in both foxtail millet and rice genomes. There were some much stronger cases for a gene/regional duplication where two separate genes of foxtail millet were related to one or two genes in the rice genome, such as SiALDH2B1/SiALDH2B2-OsALDH2B5, SiALDH3E1/SiALDH3E2-OsALDH3E1 and SiALDH18B1/SiALDH18B2-OsALDH18B1/OsALDH18B2. These results suggest that the majority of SiALDH genes share a common ancestor with OsALDH genes counterparts. Certainly, some SiALDH genes were not mapped to any syntenic blocks with rice, such as SiALDH2C2, SiALDH2C4, SiALDH12A1, and SiALDH22A1 (Figure 2B), but it does not mean that these ALDH genes from foxtail millet and rice do not share a common ancestor. Rather, this can be explained by the fact that foxtail millet and rice chromosomes have undergone extensive rearrangements and fusions that possibly lead to selective gene loss [16].
We next want to determine if functional conservations have been maintained between orthologous genes in the two related species. As such, we calculated the ratios of nonsynonymous (Ka) to synonymous (Ks) substitution rate (Ka/Ks) for orthogous gene pairs of foxtail millet SiALDHs with those of rice (22 pairs). We found that the ratios of Ka/Ks for all 22 orthologous gene pairs were less than 1, indicating that functional conservation is likely maintained between these orthologous pairs. We further estimated the approximate divergence time of those orthologous gene pairs. Eight orthologous gene pairs (SiALDH2B2-OsALDH2B1, SiALDH5F1-OsALDH5F1, SiALDH6B1-OsALDH6B1, SiALDH7B1-OsALDH7B6, SiALDH10A1-OsALDH10A9, SiALDH10A2-OsALDH10A9, SiALDH11A1-OsALDH11A3, and SiALDH18B1-OsALDH18B1) have calculated Ks values that vary from 0.3481 to 0.5655 with an average of 0.4467, indicating that the divergence time of these genes was somewhere between 26.77 and 43.50 MYA (Table S2) at a period of time following the divergence between Potidaea and Panicoideae. Additionally, five of the orthologous pairs (SiALDH2C1-OsALDH2C3, SiALDH2B1-OsALDH2B5, SiALDH3H1-OsALDH3H1, SiALDH3H2-OsALDH3H2, SiALDH18B2-OsALDH18B2) had Ks values from 0.6273 to 0.8862 with an average of 0.7487 (Table S2), signifying that their divergence occurred at some point between 48.25 and 68.17 MYA, after the whole genome duplication (WGD) of grass. Taken together, these results are comparable to evolutionary studies of protein-coding genes annotated from a recently released draft genome sequence of foxtail millet. Alternatively, the other nine orthologous pairs showed Ks values larger than 1 (Table S2), demonstrating that these orthologous pairs occurred before he whole genome duplication (WGD) of grass, but we also are cautions about these results and believe that further research is required to be certain of this data.
Orthologous genes conceivably had identical functions, but tended to diverge in regulatory and coding regions which led them to altered the expression patterns and acquire new functions, respectively [24]. In this study, we compared the exon-intron structures of ALDH genes identified in the foxtail millet genome with those found in rice. Our results revealed that a number of exonic losses and gains occurred during the evolution of ALDH genes in both species at the 5′ end or 3′ terminal and even in the middle of their sequences, such as OsALDH18B1/SiALDH18B1, OsALDH2B5/SiALDH2B1 (Fig. 3). Furthermore, SiALDH18B1 have acquired an additional exon between the first and second exon of OsALDH18B1 (Fig. 3), OsALDH2B5 and SiALDH2B1 have the same number of exons but exhibit different exons lengths (Fig. 3), suggesting that these foxtail millet ALDH genes might possess different functions. On the other hand, compared to rice, several SiALDH genes have identical exon/intron structure (OsALDH5F1/SiALDH5F1, OsALDH11A3/SiALDH11A1, OsALDH10A5/SiALDH10A2, OsALDH2B1/SiALDH2B2, and OsALDH3H1/SiALDH3H1) (Fig. 3), indicating that a portion of foxtail millet ALDH genes have similar gene functions to ALDH genes in rice. However, the insertion and deletion of amino acids in the proteins with identical gene structures should not to be disregarded. As such, further investigations are essential in order to illustrate the specifics of any functional divergence between SiALDH genes.
Figure 3. Phylogenetic analysis and exon-intron structures of foxtail millet and rice ALDH genes.
Numbers above or below branches of the tree indicate bootstrap values. Coding exons, represented by ashy, were drawn to scale. Dashed lines connecting two exons represent introns. Members of respective ALDH families are depicted in a specific background color.
Expression profiles and promoter analysis of foxtail millet ALDH genes
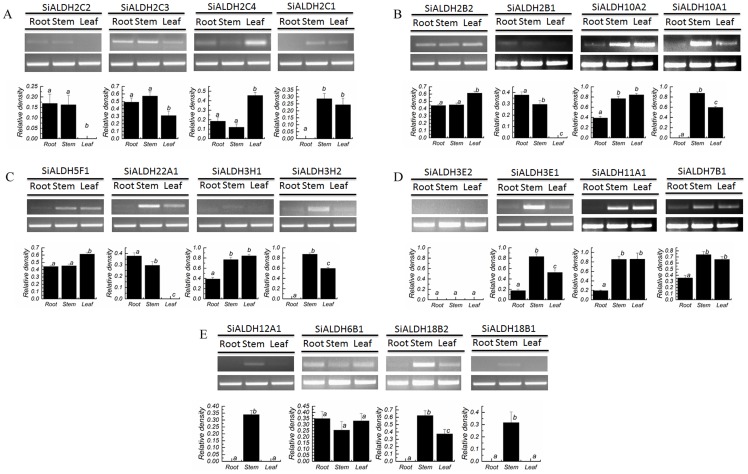
We ran semi-quantitative RT-PCRs to observe the organ-specificity of SiALDH genes using three tissues: young root, stem, and leaf. We found that the majority of SiALDH genes were expressed in the stem and leaf and had significantly lower expression levels (and in some cases were even were non-detectable) in the root, such as SiALDH7B1, SiALDH2C1, SiALDH5F1, and SiALDH22A1. SiALDH12A1 and SiALDH18B1 were mainly expressed in the stem and SiALDH3E1 was expressed in the root, stem and leaf, unlike its sister gene, SiALDH3E2, which was not expressed in the three tissues. Similar results were observed in other duplicated pairs, such as SiALDH18B1/SiALDH18B2 and SiALDH2C2/SiALDH2C3 (Fig. 4).
Figure 4. Semi-quantitative RT-PCR results of 20 foxtail millet ALDH genes in root, stem, and leaf.
(A), (B), (C), (D) and (E): The RT-PCR products, generated with 20 SiALDHs and actin (AF288226.1) gene specific primers, were electrophoresed in a 1.5%agarose gel and densitometric analysis of the corresponding band to SiALDHs. The bars represent the mean ± SD of the results from three separate experiments. One-way ANOVA analysis of variance showed significant differences between group means (P<0.05). Tukey's multiple comparison test showed significant differences between the means of groups depicted by the different letters on the bars (P<0.05). Actin mRNA (AF288226.1) was used as an internal control.
A comprehensive promoter analysis is capable of providing a reference for functional predictions of the 20 stress-related foxtail millet ALDH genes. For this purpose, we identified regulatory elements were identified in the DNA sequences (∼1 kb region upstream of the predicted SiALDH genes) using plantCARE [30]. A number of regulatory elements, including MBS, ARE, LTR and ABRE, known to be involved in responses to either abiotic stresses or hormone levels were found to be overrepresented in the 1kb region upstream of the SiALDH gene (Table S3). The 1 kb region upstream of all SiALDH genes have light or ABA regulatory elements (Table S3). Taken together, this data may indicate that the predicted SiALDH genes have a crucial role in stress-mitigation in the foxtail millet.
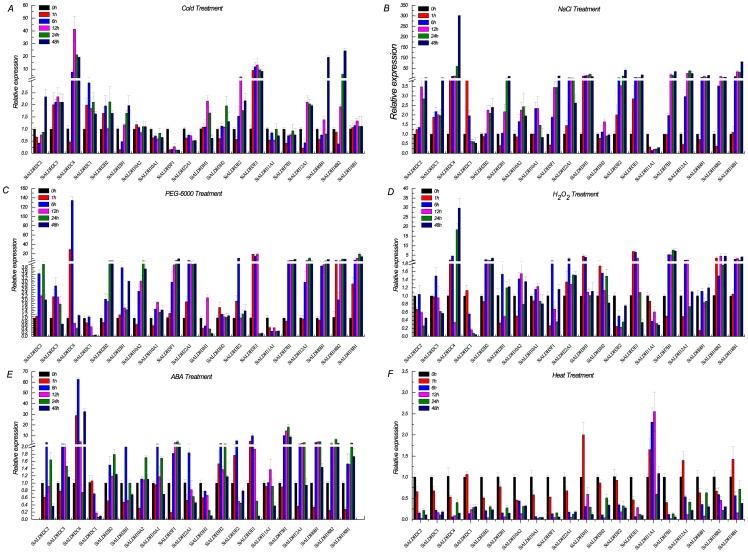
We next subjected the 20 SiALDH genes to quantitative expression analysis in order to decipher the role of SiALDH genes during diverse environmental situations: dehydration (20% PEG-6000), salinity, high temperature, cold, ABA, and H2O2 stress (0, 1, 6, 12, 24, 48 h treatment durations) (Fig. 5 and Table S4). ALDH2 family members required for male plant fertility were first identified in Maize [31], [32]. Soon afterwards, a number of publications showed that plant ALDH family members are associated with cell wall strength [33], [34], aluminum stress [35], and protection against pathogen infection [36]. In the current paper, we found that SiALDH2 genes are induced by abiotic stressors and hormones. Namely, under PEG-6000 and ABA treatment, all of the SiALDH2 genes were up-regulated at least one time points, except for the expression of SiALDH2C1, which was down-regulated under ABA treatment (Fig. 5C and E). Following cold and NaCl treatment, the expression levels of the all SiALDH2 genes increased at a number of time points and after 6 h (Fig. 5A and B). H2O2 treatment, the expression profiles of SiALDH2C4 and SiALDH2B2 both rapidly increased at 1h and 6h post-treatment, respectively, and remained elevated throughout the experiment, while the expression of SiALDH2C2 and SiALDH2C1 decreased or remained at baseline (Fig. 5D). The expression levels of the all other SiALDH2 genes were slightly increased. Following heat treatment, the transcript levels of SiALDH2 genes were down-regulated (Fig. 5F).
Figure 5. QRT-PCR analysis of the 20 foxtail millet ALDH genes.
Time course expression analysis of the 20 foxtail millet genes under various stresses. (A) Cold at 4°C; (B) 250 mM NaCl; (C) 20%PEG-6000; (D) 200 µM H2O2; (E) 100 µM ABA; (F) Heat at 42°C. Actin mRNA (AF288226.1) was used as an internal control. The bars represent the mean± SD of the results from three separate experiments.
ALDH3, ALDH7, ALDH10, ALDH11, ALDH12, and ALDH18 gene families contain several stress inducible ALDH genes that have been identified in various plants. Many ALDH3 genes are believed to be regulated by the ABA stress-response pathway. For example, in Arabidopsis, AtALDH3I1 is induced by ABA exposure, salinity, dehydration, heavy metals, oxidants, and pesticides [6], [37], [38] and AtALDH3H1 also known to be stress-responsive. In this study, we were able to induce SiALDH3H1 via H2O2, cold, and NaCl (Fig. 5A, B and D) whereas the expression levels of SiALDH3H2 displayed no significant difference under NaCl and PEG-6000 treatment (Fig. 5B and C). Furthermore, the transcript levels of SiALDH3E2 and SiALDH3E1 were up-regulated under various stressors, with the exception of SiALDH3E2 following H2O2 and heat treatment, which was down-regulated (Fig. 5A-F), while the expression of ALDH3E genes in rice was down-regulated under both drought and high salinity stresses [25].
A previous study established that ectopic expression of ALDH7 in both Arabidopsis and tobacco enhances their tolerance to drought, salinity and oxidative stress [7]. Rice ALDH7 can also be induced by oxidative and abiotic stresses [39]. Here, we found that SiALDH7B1, which has the closest phylogenetic relationship with OsALDH7, was induced by all various stressors except for low temperature. Moreover, the ALDH10 family, encoding for betaine aldehyde dehydrognases (BADHs; EC 1.2.1.8), are known to participate in the generation of osmolyte and the quaternary ammonium compound glycine betaine [40], [41]. Glycine betaine (GB) is a non-toxic cellular osmolyte that plays a key role during hyperosmolyte conditions and helps to stabilize the protein structure and to maintain the integrity of membranes against the damaging effects of excessive abiotic stresses [42]. In the current research, both SiALDH10A1 and SiALDH10A2 were induced by PEG-6000, NaCl, H2O2 and ABA (Fig. 5B-E). However, neither of the genes responded to cold treatment (Fig. 5A).
The ALDH11 gene family, encodes a cytosolic glyceraldehyde-3-phosphate dehydrogenase that catalyzes the irreversible NADP+-dependent oxidation of GAP to 3-phosphoglycerate and NADPH [43]. The protein plays a role in the response to desiccation in A. thaliana and Craterostigma plantagineum [44], [45]. Likewise, this gene can also be induced by other environmental stress conditions, such as heat shock and anaerobic stress. Here we found that the expression of SiALDH11A1 was down-regulated under cold, PEG-6000, H2O2, and salt stresses (Fig. 5B–E), similar results were previously published on rice under drought stress [25]. Furthermore, heat stress actually increased the expression levels of SiALDH11A1 (Fig. 5F), suggesting potential roles for this gene in mediating cell damage during heat shock.
In plants, ALDH12 family members play a key role in proline (Pro) degradation and their expression levels have been shown to be up-regulated via various stresses [46]. For example, both exogenous proline and salinity can induce the expression levels of AtALDH12A1. Moreover, OsALDH12 gene is induced by drought and salt stresses. Our data ( Fig. 5A–F ) reveals that SiALDH12 was upregulated under all abiotic stress conditions and ABA treatment, suggesting a possible role for it in foxtail millet during periods of environmental hardship.
The SiALDH18 family, encoding a Δ-1-pyrroline-5-carboxylate synthetases (P5CSs; EC 1.2.1.41 and EC 2.7.2.11), is a bifunctional protein that contains an N-terminal amino-acid kinase domain and a C-terminal aldehyde dehydrogenase domain. These enzymes play an important role in the biosynthesis of proline [47]. Furthermore, the expression levels of ALDH18 in a number of plants have been shown to be up-regulated in response to osmotic stress [48], [49]. Similarly, we also showed that SiALDH18 was up-regulated following PEG-6000, NaCl, H2O2, low temperature, and ABA ( Fig. 5A–E ).
The ALDH5 gene family is comprised of succinic semialdehyde dehydrogenases (SSADH; EC 1.2.1.24), which participates in GABA ‘shunt’ pathway in bacteria, plants and animals. In plants, ALDH5 mutations cause enhanced accumulation of reactive oxygen intermediates and cell death in response to light and heat stress [50]. Our results ( Fig. 5A–F ) indicate an upregulation of SiALDH5 by all of the investigated stressors except for low temperature.
The ALDH6 gene family is composed of methylmalonyl semialdehyde dehydrogenases (EC 1.2.1.27). Thus far, the function of these genes in plants is not clear. Here we found an upregulation in the expression of SiALDH6B1 under PEG-6000, NaCl, and ABA treatment ( Fig.5 B, C and E). However, we believe their exact functions should be elucidated further. Moreover, ZmALDH22A1 was recently found to be up-regulated in response to a variety of stressors such as dehydration, high salinity, and ABA treatment [51]; the same consequence in foxtail millet is displayed in Fig. 5A–F . Nonetheless, ALDH22 is not known to be induced by osmotic stress in rice and Arabidopsis [25], [52]. As such, the role of ALDH22 in plants still needs further study.
In summary, the overall variability in gene expression patterns implies that SiALDHs participate in a complex network of pathways in order to perform different physiological functions in response to different challenges. This comprehensive expression profile provides a clue to the role of SiALDHs in imparting stress tolerance.
Enhancement of salt stress tolerance of recombinant E.coli harboring foxtail millet ALDH genes
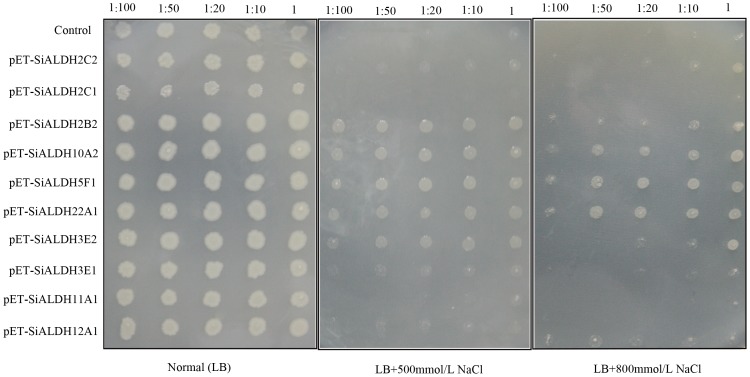
The expression of foreign plant genes can directly contribute to increasing stress tolerance in bacteria host cells [53], [54]. In plants, abiotic stress can trigger the generation of reactive oxygen species (ROS) that disrupts cellular homeostasis and induce the expression of genes involved in defense mechanisms [55]. Moreover, aldehyde dehydrogenases play a key role in the detoxification of various aldehyde molecules produced in response to abiotic stress. Thus, we expressed ten SiALDH proteins (SiALDH2C2, SiALDH2C1, SiALDH2B2, SiALDH10A2, SiALDH5F1, SiALDH22A1, SiALDH3E1, SiALDH3E2, SiALDH11A1, and SiALDH12A1) in E.coli in an attempt to determine the function of foxtail millet ALDH proteins to salt stress condition, SDS-PAGE analysis results demonstrated that the molecular weights of the ten recombinant proteins agreed with the predicted molecular weights (Fig. S2). Subsequently, we spotted aliquots of recombinant strains harboring the empty vector pET-28a (Control) and 10 recombinant vectors (pET-SiALDH2C2, pET-SiALDH2C1, pET-SiALDH2B2, pET-SiALDH10A2, pET-SiALDH5F1, pET-SiALDH22A1, pET-SiALDH3E1, pET-SiALDH3E2, pET-SiALDH11A1, and pET-SiALDH12A1) onto LB plates and supplemented them with either 500 mmol/L or 800 mmol/L NaCl. The growth status of Rosetta cells (transformed with Control and recombinant vectors) indicated that the recombinants and control cells showed similar growth on LB medium, suggesting that exogenous SiALDHs do not restrain the cell growth.
Results show that high salt severely inhibits the growth of the control strain. Amazingly, the five recombinants (pET-SiALDH2B2, pET-SiALDH10A2, pET-SiALDH5F1, pET-SiALDH22A1, and pET-SiALDH3E2) were able to grow normally at different dilution gradients under 500 mmol/L NaCl (Fig. 6). Similarly, under 800 mmol/L NaCl, we can clearly see that Rosetta cells harboring SiALDH10A1, SiALDH22A1, and SiALDH5F1 were able to still grow normally at high bacterial concentrations (Fig. 6), revealing that these five SiALDH genes (SiALDH2B2, SiALDH10A2, SiALDH5F1, SiALDH22A1, and SiALDH3E2) are associated with high salt tolerance. This enhancement of salt tolerance further indicates that the expression of these foxtail millet ALDH proteins in host cells is able to confer their protective function against protein damage, cellular membrane disruption, and cellular apoptosis; their exact function in plants needs to be further examined.
Figure 6. Spot assay of E. coli growth.
The viabilities of the strains were detected on different LB plated with additional 500/L NaCl (B), 800 mmol/L NaCl and normal LB plates (A). Strains were Control: strain harboring plasmid pET-28(a), pET-ALDHs: strain harboring different SiALDH proteins. 1.5 microliters of the serially diluted E. coli cells cultured for 16 h at 16°C with 0.5 mmol/L IPTG were dotted onto LB agar plates containing NaCl at the marked concentration.
Summary
Aldehyde dehydrogenases (ALDHs) are a set of NAD (P)+-dependent enzymes that oxidize a wide range of aldehydes into their corresponding carboxylic acids. The identification and characterization of ALDH gene families has been well studied in different plant species. However, currently that are no reports investigating ALDH gene families in the foxtail millet. In the current study, we identified 20 ALDH genes in the foxtail millet genome, grouped them into ten families, and then assigned each a unique identifier to each of the predicted S.italica ALDH proteins based on the criteria established by the ALDH Gene Nomenclature Committee (AGNC). Importantly, evolutionary analysis reveals that both tandem and segmental duplications have contributed significantly to the expansion of the foxtail millet ALDH genes. Our gene structure analysis shows that ALDHs from the same family in foxtail millet or the orthologous genes in rice display highly diverse distributions of their exonic and intronic regions. Comparative synteny analysis between foxtail millet and rice genomes show that the majority of SiALDH and OsALDH gene homologs are located in syntenic regions between the two, indicating that these ALDH genes share common ancestors. In addition, we found that analysis of the Ka/Ks ratios of paralogous and orthologous gene pairs facilitates in a deeper understanding of the evolutionary significance of gene-duplications and divergence. Furthermore, we show for the first time that a few SiALDH genes extremely organ-specific and that the expression pattern of a number of SiALDH genes is affected by several stressors, including dehydration, salinity, cold, ABA treatment, and heat stress. Finally, we find that the transformation of SiALDH2B2, SiALDH10A2, SiALDH5F1, SiALDH22A1, and SiALDH3E1 into E.coli was able to improve its salt tolerance. Taken together, this work lays a solid foundation for future studies on the function of ALDH genes in foxtail millet and many other grasses, as well.
Methods and Materials
Identification and annotation of foxtail millet ALDH genes
Rice [15] and A. thaliana [10] ALDH amino acid sequences were retrieved from the PHYTOZOME v9.1 database (http://www.phytozome.org/) and utilized to identify homologous peptides from foxtail millet by BLASTP search using default settings. The keywords “ALDH”, “Aldehyde dehydrogenases”, and the HMM profiles of the ALDH domain PF00171, KOG2450 (aldehyde dehydrogenase), KOG2451 (aldehyde dehydrogenase), KOG2453 (aldehyde dehydrogenase) and KOG2456 (aldehyde dehydrogenase) were all used as queries for searching the foxtail millet genomic database to identify ALDH and ALDH-like sequences [56]. Similarity searches were also performed through BLASTP in GeneBank non-redundant protein database to eliminate possible exclusions of any additional ALDH member. All hits with the excepted values <1.0 were retrieved and all redundant sequences were removed using the decrease redundancy tool (http://web.expasy.org/decrease_redundancy/). Each non-redundant sequence was monitored for the presence of the conserved ALDH domain (PF00171) by SMART (http://smart.embl-heidelberg.de/) [57], Pfam (http://pfam.sanger.ac.uk/) and CDD (Conserved Domain Database) [58] searches. Likewise, the two active sites (PS00070 (cysteine active site) and PS00687 (glutamic acid active site)) were checked by PROSITE (http://prosite.expasy.org/). The deduced ALDH polypeptides were annotated using criteria established by the ALDH Gene Nomenclature Committee (AGNC) [3], [9]. In brief, deduced amino acid sequences greater than 40% identical to other previously identified ALDH sequences were defined as a family, while sequences with greater than 60% identity were defined as a protein subfamily. Deduced amino acid sequences with less than 40% identity described a new ALDH protein family. YLoc [59], [60] was used to predict the foxtail millet ALDH gene subcellular localization.
Sequence alignments, phylogenetic and promoter analysis
The protein sequences of ALDH genes in A. thaliana, O. sativa, S. Bicolor, Z. Mays [10] and S.italica were also retrieved from the PHYTOZOME v9.1 database (http://www.phytozome.org/). Subsequently, we performed multiple alignments of ALDH protein sequences using ClustalX2.0 [61]. The alignments results was adjusted using BioEdit V7.0.5.3 [62] and eliminated the portions of the sequences that could not be reliably aligned. Phylogenetic trees were constructed with the MEGA5.10 Beta4 [63] software utilizing the neighbor-joining (NJ) method and the bootstrap test was replicated 1000 times. The entire alignment has been provided as Text S2.
In order to investigate the promoter regions of the SiALDH gene family, the 1 kb upstream regions (based on the position of the genes provided by the S.italica annotation information) were selected and analyzed using plantCARE [30]. Known as stress-mediated regulatory elements are listed in Table S3.
Physical Mapping, Gene Duplication and Gene Structure Analysis
Specific chromosomal location and segment duplication of SiALDH genes were determined by FeatView (http://genomevolution.org/CoGe/FeatView.pl) tool. The genes were plotted separately on to all nine foxtail millet chromosomes according to their ascending orders of physical position (bp) and then displayed using Adobe Illustrator CS6. Duplications in the foxtail millet genome and synteny between foxtail millet and rice were established utilizing the SynMap tool (detailed settings are as following: Blast Algorithm: Last, DAGChainer options and Merge syntenic Blocks use the recommended parameters, and the synonymous (Ks) and non-synonymous (Ka) substitution rates were calculated using the CODEML program [64]) and confirmed with the GEvo tool at the CoGe Web site (http://synteny.cnr.berkeley.edu/CoGe/). The formulas T = Ks/2λ (λ = 6.5×10−9) [65], [66] was used to calculate time of duplication and divergence of each SiALDH genes. Next, we identified tandem duplications of ALDH genes in the foxtail millet genome by checking their physical locations in individual chromosomes. Tandem duplicated genes were defined as adjacent homologous ALDH genes on the foxtail millet chromosomes, with no more than one intervening gene. All paralogous and orthologous gene pairs are listed in Table S5 and displayed using Adobe Illustrator CS6. Finally, The exon/intron structures of foxtail millet ALDH genes were determined from alignments of their coding sequences with corresponding genomic sequences using the est2genome program [67]. The diagrams of exon-intron structures were obtained using the online program FancyGene [68].
Plant materials, growth conditions and treatments
The foxtail millet variety, Tie-8396, was used for all experiments. The seeds were germinated on wet filter paper at room temperature for 2 days and planted in pots with a mixture of peat/forest and vermiculite (1∶1 v/v). Plants were grown in a greenhouse (28°Cday/20°Cnight, 16 h photoperiod, natural lighting, 70% relative humidity) and when the seedlings were five weeks old they were chronologically plunged into a 250 mM NaCl solution, a 100 µM ABA solution, a 20%PEG-6000 solution, and 100 µM H2O2 solutions for multiple periods of time (0, 1, 6, 12, 24, and 48 h). Moreover, we also subjugated the seedlings to 4 h of 42°C heat stress in an incubator followed recovery (0, 1, 6, 12, 24, and 48 h) at room temperature. Similarly, for cold stress tests, seedlings were placed in a 4°C freezer; also sampled at 0, 1, 6, 12, 24, and 48 h. Concurrently, the roots, stems, leaves of the untreatment seedlings were also harvested for RNA extraction. These samples were put in liquid nitrogen and stored in −80°C. The above experiments were repeated three times to ensure precision and reproducibility.
RNA extraction, Semi-quantitative and quantitative RT-PCR analysis
Total RNA was extracted using the RNAprep Pure Plant Kit (TIANGEN, Beijing) according to the manufacturer's instructions. Prime Script RT reagent Kit (TaKaRa, Dalian) was used for reverse transcription. The condition for Semi-quantitative PCR amplification of cDNA was as follows: denature at 94°C for 10 min and 32 (actin) or 35(gene) cycles for 94°C 30 s; 58°C (gene) or 60°C (actin) 30 s; 72°C 30 s, followed by further incubation for 5 min at 72°C (1 cycle). Furthermore, densitometry and band quantization was performed utilizing Bio-Rad Quantity One software. Quantitative PCR was performed on the Applied Biosystems 7500 real-time PCR system using Super Real PreMix Plus (SYBR Green) (TIANGEN, Beijing), as per instructions. The PCR thermal cycle conditions were as follows: denature at 95°C for 15 min and 40 cycles for 95°C, 10 s; 60°C, 20 s; 72°C, 32 s. The specificity of the PCR reactions was determined by analyzing the melting curve. The constitutive actin gene (AF288226.1) was used as the endogenous control; quantification of the 20 SiALDH gene transcript was analyzed using the 2−ΔΔCt method [69]. The primers used for real-time PCR analysis were designed with primique [70] and listed in Table S6. The each experiment repeat three times.
Construction of expression vector pET- ALDH and the expression of foxtail millet ALDH genes in response to salinity
Primers in Table S6 were subsequently used for cloning SiALDH genes from cDNA. Amplification was performed as follows: 10 min at 95°C; 32 cycles of 40 s at 94°C, 2 min at 68°C; followed by a final extension for 10 min at 72°C. PET-28a (+) was linearized via BamHI and SacI restriction digestion. Then according to the In-Fusion HD Cloning System instructions, the cloned ALDH genes were sub-cloned into the PET-28a (+). For functional expression, E.coli Rosetta competent cells (CoWin Bioscience Co., Ltd, Beijing) were transformed with either the recombinant plasmid (PET-ALDH/Rosetta) or with PET-28a (+) vector lacking SiALDH genes (Control). The PET-ALDH/Rosetta and control were both allowed to grow overnight with shaking at 37°C. We next transferred 400 µl of each culture mixture into a fresh 40 ml liquid LB with kanamycin and shaken at 37°C for about 3 h. β-D-thiogalactopyranoside (IPTG) was then added to a final concentration of 0.5 mmol/L in order to induce the expression of the inserted gene. After a 16 h induction (16°C), the samples (adjusting the original A600 value of all E.coli groups to the same value) were spotted onto the LB agar plates supplemented with NaCl (500 mmol/L, 800 mmol/L). The experiment was repeated three times with similar results.
Data Analysis
Statistical analyses were completed by One-way ANOVA followed by Tukey's test utilizing Origin 9.1 software. P values of less than 0.05 were considered statistically significant.
Supporting Information
Phylogenetic analysis and exon-intron structures of foxtail millet ALDH from the same family. Numbers above or below branches of the tree indicate bootstrap values. Coding exons, represented by ashy, were drawn to scale. Lines connecting two exons represent introns.
(TIF)
The SDS-PAGE analysis for recombinant PET-ALDH. Total proteins from SiALDH2C1, SiALDH2C2, SiALDH2B2, SiALDH10A2, SiALDH5F1, SiALDH22A1, SiALDH3E1, SiALDH3E2, SiALDH11A1, and SiALDH12A1 were separated by SDS-PAGE. The differential protein bands near the calculated molecular mass of polypeptides expressed by SiALDH2C1, SiALDH2C2, SiALDH2B2, SiALDH10A2, SiALDH5F1, SiALDH22A1, SiALDH3E1, SiALDH3E2, SiALDH11A1, and SiALDH12A1 were about 61 kDa, 64 kDa, 63 kDa, 60 kDa, 62 kDa, 62 kDa, 58 kDa, 57 kDa, 59 kDa, and 71 kDa, respectively.
(TIF)
Numbers of ALDH family members identified in various organisms.
(DOCX)
The Ka/Ks ratios and estimated divergence time for orthologous ALDH proteins between foxtail millet and rice and paralogous ALDH proteins in foxtail millet.
(DOCX)
The promoter analysis of the 20 foxtail millet ALDH genes.
(XLSX)
Relative expression values of 20 foxtail millet ALDH genes in response to various stress and hormone treatments. One-way ANOVA analysis of variance showed significant differences between group means (P<0.05). Tukey's multiple comparison test showed significant differences between the means of groups depicted by the different letters (P<0.05).
(XLSX)
Paralogous gene pairs within foxtail millet and orthologous gene pairs between foxtail millet and rice.
(DOCX)
A list of Primers sequences of 20 foxtail millet ALDH genes for real-time PCR and gene clone.
(DOCX)
The information of 20 SiALDH genes. 20 SiALDH gene CDS and protein sequenced were completely listed.
(TXT)
Multiple alignments of ALDH amino acid sequences from A. thaliana , O. sativa , S. Bicolor , Z. Mays and S.italica .
(TXT)
Acknowledgments
Grateful thanks are due to Prof. Diao Xian-Min, Institute of Crop sciences, Chinese Academy of Agricultural Sciences, Beijing, China for providing the foxtail millet variety (Tie-8396). We are especially grateful to Prof. Shuijin Hu, North Carolina State University, Raleigh, North Carolina, USA for helpful critical reading of the manuscript.
Funding Statement
This work was supported by the National Key Project for Research on Transgenic Biology (2013ZX08002-002) and the National 863 High-tech Project (2012AA10A309). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Knight H, Knight MR (2001) Abiotic stress signalling pathways: specificity and cross-talk. Trends in Plant Science 6: 262–267. [DOI] [PubMed] [Google Scholar]
- 2. Bartels D (2001) Targeting detoxification pathways: an efficient approach to obtain plants with multiple stress tolerance? Trends in plant science 6: 284–286. [DOI] [PubMed] [Google Scholar]
- 3. Gill SS, Tuteja N (2010) Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiology and Biochemistry 48: 909–930. [DOI] [PubMed] [Google Scholar]
- 4. Yoshida A, Rzhetsky A, Hsu LC, Chang C (1998) Human aldehyde dehydrogenase gene family. European Journal of Biochemistry 251: 549–557. [DOI] [PubMed] [Google Scholar]
- 5. Singh S, Brocker C, Koppaka V, Chen Y, Jackson BC, et al. (2013) Aldehyde dehydrogenases in cellular responses to oxidative/electrophilic stress. Free Radical Biology and Medicine 56: 89–101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sunkar R, Bartels D, Kirch HH (2003) Overexpression of a stress-inducible aldehyde dehydrogenase gene from Arabidopsis thaliana in transgenic plants improves stress tolerance. The Plant Journal 35: 452–464. [DOI] [PubMed] [Google Scholar]
- 7. Rodrigues SM, Andrade MO, Gomes APS, DaMatta FM, Baracat-Pereira MC, et al. (2006) Arabidopsis and tobacco plants ectopically expressing the soybean antiquitin-like ALDH7 gene display enhanced tolerance to drought, salinity, and oxidative stress. Journal of experimental botany 57: 1909–1918. [DOI] [PubMed] [Google Scholar]
- 8. Huang W, Ma X, Wang Q, Gao Y, Xue Y, et al. (2008) Significant improvement of stress tolerance in tobacco plants by overexpressing a stress-responsive aldehyde dehydrogenase gene from maize (Zea mays). Plant Molecular Biology 68: 451–463. [DOI] [PubMed] [Google Scholar]
- 9. Vasiliou V, Bairoch A, Tipton KF, Nebert DW (1999) Eukaryotic aldehyde dehydrogenase (ALDH) genes: human polymorphisms, and recommended nomenclature based on divergent evolution and chromosomal mapping. Pharmacogenetics 9: 421–434. [PubMed] [Google Scholar]
- 10. Kirch HH, Bartels D, Wei YL, Schnable PS, Wood AJ (2004) The ALDH gene superfamily of Arabidopsis. Trends in Plant Science 9: 371–377. [DOI] [PubMed] [Google Scholar]
- 11. Kotchoni SO, Jimenez-Lopez JC, Kayode APP, Gachomo EW, Baba-Moussa L (2012) The soybean aldehyde dehydrogenase (ALDH) protein superfamily. Gene 495: 128–133. [DOI] [PubMed] [Google Scholar]
- 12.Zhang YC, Mao LY, Wang H, Brocker C, Yin XJ, et al. (2012) Genome-Wide Identification and Analysis of Grape Aldehyde Dehydrogenase (ALDH) Gene Superfamily. PLoS One 7. [DOI] [PMC free article] [PubMed]
- 13. Brocker C, Vasiliou M, Carpenter S, Carpenter C, Zhang YC, et al. (2013) Aldehyde dehydrogenase (ALDH) superfamily in plants: gene nomenclature and comparative genomics. Planta 237: 189–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jimenez-Lopez JC, Gachomo EW, Seufferheld MJ, Kotchoni SO (2010) The maize ALDH protein superfamily: linking structural features to functional specificities. Bmc Structural Biology 10. [DOI] [PMC free article] [PubMed]
- 15. Kotchoni SO, Jimenez-Lopez JC, Gao D, Edwards V, Gachomo EW, et al. (2010) Modeling-dependent protein characterization of the rice aldehyde dehydrogenase (ALDH) superfamily reveals distinct functional and structural features. PLoS One 5: e11516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Zhang G, Liu X, Quan Z, Cheng S, Xu X, et al. (2012) Genome sequence of foxtail millet (Setaria italica) provides insights into grass evolution and biofuel potential. Nature biotechnology 30: 549–554. [DOI] [PubMed] [Google Scholar]
- 17. Bennetzen JL, Schmutz J, Wang H, Percifield R, Hawkins J, et al. (2012) Reference genome sequence of the model plant Setaria. Nature biotechnology 30: 555–561. [DOI] [PubMed] [Google Scholar]
- 18. Puranik S, Sahu PP, Mandal SN, Parida SK, Prasad M (2013) Comprehensive genome-wide survey, genomic constitution and expression profiling of the NAC transcription factor family in foxtail millet (Setaria italica L.). PloS one 8: e64594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mishra AK, Muthamilarasan M, Khan Y, Parida SK, Prasad M (2014) Genome-Wide Investigation and Expression Analyses of WD40 Protein Family in the Model Plant Foxtail Millet (Setaria italica L.). PloS one 9: e86852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. García-Ríos M, Fujita T, LaRosa PC, Locy RD, Clithero JM, et al. (1997) Cloning of a polycistronic cDNA from tomato encoding γ-glutamyl kinase and γ-glutamyl phosphate reductase. Proceedings of the National Academy of Sciences 94: 8249–8254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kellogg EA (2001) Evolutionary history of the grasses. Plant physiology 125: 1198–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Juretic N, Hoen DR, Huynh ML, Harrison PM, Bureau TE (2005) The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome research 15: 1292–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Wang X, Tang H, Paterson AH (2011) Seventy million years of concerted evolution of a homoeologous chromosome pair, in parallel, in major Poaceae lineages. Plant Cell 23: 27–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Xu G, Guo C, Shan H, Kong H (2012) Divergence of duplicate genes in exon-intron structure. Proceedings of the National Academy of Sciences 109: 1187–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gao CX, Han B (2009) Evolutionary and expression study of the aldehyde dehydrogenase (ALDH) gene superfamily in rice (Oryza sativa). Gene 431: 86–94. [DOI] [PubMed] [Google Scholar]
- 26. Lecharny A, Boudet N, Gy I, Aubourg S, Kreis M (2003) Introns in, introns out in plant gene families: a genomic approach of the dynamics of gene structure. J Struct Funct Genomics 3: 111–116. [PubMed] [Google Scholar]
- 27. Lurin C, Andres C, Aubourg S, Bellaoui M, Bitton F, et al. (2004) Genome-wide analysis of Arabidopsis pentatricopeptide repeat proteins reveals their essential role in organelle biogenesis. Plant Cell 16: 2089–2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Jain M, Khurana P, Tyagi AK, Khurana JP (2008) Genome-wide analysis of intronless genes in rice and Arabidopsis. Functional & Integrative Genomics 8: 69–78. [DOI] [PubMed] [Google Scholar]
- 29.Paterson A, Wang X, Tang H, Lee T (2012) Synteny and genomic rearrangements. Plant Genome Diversity Volume 1: Springer. pp. 195–207.
- 30. Lescot M, Déhais P, Thijs G, Marchal K, Moreau Y, et al. (2002) PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic acids research 30: 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Liu F, Cui X, Horner HT, Weiner H, Schnable PS (2001) Mitochondrial aldehyde dehydrogenase activity is required for male fertility in maize. The Plant Cell Online 13: 1063–1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cui X, Wise RP, Schnable PS (1996) The rf2 nuclear restorer gene of male-sterile T-cytoplasm maize. SCIENCE-NEW YORK THEN WASHINGTON-: 1334–1335. [DOI] [PubMed]
- 33. Nair RB, Bastress KL, Ruegger MO, Denault JW, Chapple C (2004) The Arabidopsis thaliana REDUCED EPIDERMAL FLUORESCENCE1 gene encodes an aldehyde dehydrogenase involved in ferulic acid and sinapic acid biosynthesis. The Plant Cell Online 16: 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Grabber JH, Ralph J, Hatfield RD (2000) Cross-linking of maize walls by ferulate dimerization and incorporation into lignin. Journal of agricultural and food chemistry 48: 6106–6113. [DOI] [PubMed] [Google Scholar]
- 35. Mohan Murali Achary V, Jena S, Panda KK, Panda BB (2008) Aluminium induced oxidative stress and DNA damage in root cells of Allium cepa L. Ecotoxicology and Environmental Safety. 70: 300–310. [DOI] [PubMed] [Google Scholar]
- 36. Wen Y, Wang X, Xiao S, Wang Y (2012) Ectopic expression of VpALDH2B4, a novel aldehyde dehydrogenase gene from Chinese wild grapevine (Vitis pseudoreticulata), enhances resistance to mildew pathogens and salt stress in Arabidopsis. Planta 236: 525–539. [DOI] [PubMed] [Google Scholar]
- 37. Kirch HH, Nair A, Bartels D (2001) Novel ABA-and dehydration-inducible aldehyde dehydrogenase genes isolated from the resurrection plant Craterostigma plantagineum and Arabidopsis thaliana. The Plant Journal 28: 555–567. [DOI] [PubMed] [Google Scholar]
- 38.Stiti N, Missihoun TD, Kotchoni SO, Kirch HH, Bartels D (2011) Aldehyde dehydrogenases in Arabidopsis thaliana: biochemical requirements, metabolic pathways, and functional analysis. Frontiers in plant science 2. [DOI] [PMC free article] [PubMed]
- 39. Shin J-H, Kim S-R, An G (2009) Rice aldehyde dehydrogenase7 is needed for seed maturation and viability. Plant physiology 149: 905–915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rathinasabapathi B, Burnet M, Russell BL, Gage DA, Liao PC, et al. (1997) Choline monooxygenase, an unusual iron-sulfur enzyme catalyzing the first step of glycine betaine synthesis in plants: prosthetic group characterization and cDNA cloning. Proc Natl Acad Sci U S A 94: 3454–3458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Nakamura T, Yokota S, Muramoto Y, Tsutsui K, Oguri Y, et al. (1997) Expression of a betaine aldehyde dehydrogenase gene in rice, a glycinebetaine nonaccumulator, and possible localization of its protein in peroxisomes. Plant J 11: 1115–1120. [DOI] [PubMed] [Google Scholar]
- 42. Fitzgerald TL, Waters DL, Henry RJ (2009) Betaine aldehyde dehydrogenase in plants. Plant Biology 11: 119–130. [DOI] [PubMed] [Google Scholar]
- 43. Valverde F, Losada M, Serrano A (1999) Engineering a central metabolic pathway: glycolysis with no net phosphorylation in an Escherichia coli gap mutant complemented with a plant GapN gene. FEBS letters 449: 153–158. [DOI] [PubMed] [Google Scholar]
- 44. Gallardo K, Job C, Groot SP, Puype M, Demol H, et al. (2001) Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiology 126: 835–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Bartels D, Salamini F (2001) Desiccation Tolerance in the Resurrection PlantCraterostigma plantagineum. A Contribution to the Study of Drought Tolerance at the Molecular Level. Plant Physiology 127: 1346–1353. [PMC free article] [PubMed] [Google Scholar]
- 46. Miller G, Honig A, Stein H, Suzuki N, Mittler R, et al. (2009) Unraveling Δ1-pyrroline-5-carboxylate-proline cycle in plants by uncoupled expression of proline oxidation enzymes. Journal of Biological Chemistry 284: 26482–26492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Igarashi Y, Yoshiba Y, Sanada Y, Yamaguchi-Shinozaki K, Wada K, et al. (1997) Characterization of the gene for Δ1-pyrroline-5-carboxylate synthetase and correlation between the expression of the gene and salt tolerance in Oryza sativa L. Plant molecular biology. 33: 857–865. [DOI] [PubMed] [Google Scholar]
- 48.Razavizadeh R, Ehsanpour A (2009) Effects of salt stress on proline content, expression of delta-1-pyrroline-5-carboxylate synthetase, and activities of catalase and ascorbate peroxidase in transgenic tobacco plants.
- 49. Yoshiba Y, Kiyosue T, Nakashima K, Yamaguchi-Shinozaki K, Shinozaki K (1997) Regulation of levels of proline as an osmolyte in plants under water stress. Plant and Cell Physiology 38: 1095–1102. [DOI] [PubMed] [Google Scholar]
- 50. Bouché N, Fait A, Bouchez D, Møller SG, Fromm H (2003) Mitochondrial succinic-semialdehyde dehydrogenase of the γ-aminobutyrate shunt is required to restrict levels of reactive oxygen intermediates in plants. Proceedings of the National Academy of Sciences 100: 6843–6848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Huang W, Ma X, Wang Q, Gao Y, Xue Y, et al. (2008) Significant improvement of stress tolerance in tobacco plants by overexpressing a stress-responsive aldehyde dehydrogenase gene from maize (Zea mays). Plant molecular biology 68: 451–463. [DOI] [PubMed] [Google Scholar]
- 52. Kirch H-H, Schlingensiepen S, Kotchoni S, Sunkar R, Bartels D (2005) Detailed expression analysis of selected genes of the aldehyde dehydrogenase (ALDH) gene superfamily in Arabidopsis thaliana. Plant molecular biology 57: 315–332. [DOI] [PubMed] [Google Scholar]
- 53. Yamada A, Sekiguchi M, Mimura T, Ozeki Y (2002) The role of plant CCTα in salt-and osmotic-stress tolerance. Plant and cell physiology 43: 1043–1048. [DOI] [PubMed] [Google Scholar]
- 54. Liu Y, Zheng Y (2005) PM2, a group 3 LEA protein from soybean, and its 22-mer repeating region confer salt tolerance in Escherichia coli . Biochemical and biophysical research communications 331: 325–332. [DOI] [PubMed] [Google Scholar]
- 55. Noctor G, Foyer CH (1998) Ascorbate and glutathione: keeping active oxygen under control. Annual review of plant biology 49: 249–279. [DOI] [PubMed] [Google Scholar]
- 56. Altschul SF, Madden TL, Schaffer AA, Zhang JH, Zhang Z, et al. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Letunic I, Doerks T, Bork P (2012) SMART 7: recent updates to the protein domain annotation resource. Nucleic acids research 40: D302–D305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Marchler-Bauer A, Anderson JB, Cherukuri PF, DeWweese-Scott C, Geer LY, et al. (2005) CDD: a conserved domain database for protein classification. Nucleic Acids Research 33: D192–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Briesemeister S, Rahnenfuhrer J, Kohlbacher O (2010) Going from where to why-interpretable prediction of protein subcellular localization. Bioinformatics 26: 1232–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Briesemeister S, Rahnenfuhrer J, Kohlbacher O (2010) YLoc-an interpretable web server for predicting subcellular localization. Nucleic Acids Research 38: W497–W502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. (2007) Clustal W and clustal X version 2.0. Bioinformatics 23: 2947–2948. [DOI] [PubMed] [Google Scholar]
- 62.Hall TA. BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT; 1999. pp. 95–98.
- 63. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Molecular biology and evolution 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Yang Z (2007) PAML 4: phylogenetic analysis by maximum likelihood. Molecular biology and evolution 24: 1586–1591. [DOI] [PubMed] [Google Scholar]
- 65. Lynch M, Conery JS (2000) The evolutionary fate and consequences of duplicate genes. Science 290: 1151–1155. [DOI] [PubMed] [Google Scholar]
- 66. Yang Z, Gu S, Wang X, Li W, Tang Z, et al. (2008) Molecular evolution of the CPP-like gene family in plants: insights from comparative genomics of Arabidopsis and rice. Journal of molecular evolution 67: 266–277. [DOI] [PubMed] [Google Scholar]
- 67. Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends in genetics 16: 276–277. [DOI] [PubMed] [Google Scholar]
- 68. Rambaldi D, Ciccarelli FD (2009) FancyGene: dynamic visualization of gene structures and protein domain architectures on genomic loci. Bioinformatics 25: 2281–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Livak KJ, Schmittgen TD (2001) Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- 70. Fredslund J, Lange M (2007) Primique: automatic design of specific PCR primers for each sequence in a family. BMC bioinformatics 8: 369. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Phylogenetic analysis and exon-intron structures of foxtail millet ALDH from the same family. Numbers above or below branches of the tree indicate bootstrap values. Coding exons, represented by ashy, were drawn to scale. Lines connecting two exons represent introns.
(TIF)
The SDS-PAGE analysis for recombinant PET-ALDH. Total proteins from SiALDH2C1, SiALDH2C2, SiALDH2B2, SiALDH10A2, SiALDH5F1, SiALDH22A1, SiALDH3E1, SiALDH3E2, SiALDH11A1, and SiALDH12A1 were separated by SDS-PAGE. The differential protein bands near the calculated molecular mass of polypeptides expressed by SiALDH2C1, SiALDH2C2, SiALDH2B2, SiALDH10A2, SiALDH5F1, SiALDH22A1, SiALDH3E1, SiALDH3E2, SiALDH11A1, and SiALDH12A1 were about 61 kDa, 64 kDa, 63 kDa, 60 kDa, 62 kDa, 62 kDa, 58 kDa, 57 kDa, 59 kDa, and 71 kDa, respectively.
(TIF)
Numbers of ALDH family members identified in various organisms.
(DOCX)
The Ka/Ks ratios and estimated divergence time for orthologous ALDH proteins between foxtail millet and rice and paralogous ALDH proteins in foxtail millet.
(DOCX)
The promoter analysis of the 20 foxtail millet ALDH genes.
(XLSX)
Relative expression values of 20 foxtail millet ALDH genes in response to various stress and hormone treatments. One-way ANOVA analysis of variance showed significant differences between group means (P<0.05). Tukey's multiple comparison test showed significant differences between the means of groups depicted by the different letters (P<0.05).
(XLSX)
Paralogous gene pairs within foxtail millet and orthologous gene pairs between foxtail millet and rice.
(DOCX)
A list of Primers sequences of 20 foxtail millet ALDH genes for real-time PCR and gene clone.
(DOCX)
The information of 20 SiALDH genes. 20 SiALDH gene CDS and protein sequenced were completely listed.
(TXT)
Multiple alignments of ALDH amino acid sequences from A. thaliana , O. sativa , S. Bicolor , Z. Mays and S.italica .
(TXT)