Abstract
CRC cancer is one of the deadliest diseases in Western countries. In order to develop prognostic biomarkers for CRC (colorectal cancer) aggressiveness, we analyzed retrospectively 267 CRC patients via a novel, multidimensional biomarker platform. Using nanofluidic technology for qPCR analysis and quantitative fluorescent immunohistochemistry for protein analysis, we assessed 33 microRNAs, 124 mRNAs and 9 protein antigens. Analysis was conducted in each single dimension (microRNA, gene or protein) using both the multivariate Cox model and Kaplan-Meier method. Thereafter, we simplified the censored survival data into binary response data (aggressive vs. non aggressive cancer). Subsequently, we integrated the data into a diagnostic score using sliced inverse regression for sufficient dimension reduction. Accuracy was assessed using area under the receiver operating characteristic curve (AUC). Single dimension analysis led to the discovery of individual factors that were significant predictors of outcome. These included seven specific microRNAs, four genes, and one protein. When these factors were quantified individually as predictors of aggressive disease, the highest demonstrable area under the curve (AUC) was 0.68. By contrast, when all results from single dimensions were combined into integrated biomarkers, AUCs were dramatically increased with values approaching and even exceeding 0.9. Single dimension analysis generates statistically significant predictors, but their predictive strengths are suboptimal for clinical utility. A novel, multidimensional integrated approach overcomes these deficiencies. Newly derived integrated biomarkers have the potential to meaningfully guide the selection of therapeutic strategies for individual patients while elucidating molecular mechanisms driving disease progression.
Introduction
CRC is one of the deadliest diseases worldwide. Caucasian patients with local, regional, or metastatic disease exhibit a 5-year survival rate of 66%, 44%, and 4%, respectively [1]. Disease stage at the time of surgery is well established as the most important prognostic factor in CRC. In the last two decades, median overall survival has increased significantly with the introduction of new cytotoxic agents and biologic therapies. The response to such treatments depends on molecular determinants whose elucidation has been the focus of intense and productive research efforts. We now know, for example, that cancers harboring activating KRAS mutations do not respond to anti-EGFR therapy [2]. However, the goal of optimizing treatment protocols based on the unique molecular characteristics of an individual's tumor still remains elusive. Development of novel biomarkers that can reliably identify patients at high risk for disease progression and death would be especially useful in determining the clinical circumstances where adjuvant chemotherapy is warranted. Whereas the use of the antimetabolite 5-Fluorouracil (5FU) is standard therapy for patients with stage III CRC, its potential benefits compared to risks in stage II CRC patients is a matter of controversy and debate [3]. In the absence of a robust clinical predictor of disease outcome, the decision to treat or not to treat stage II patients with 5FU cannot rest on objective and firm criteria. Previously identified predictive biomarkers which had shown great promise in this arena including telomerase, transforming growth factors (TGFα and TGFβ), epidermal growth factors (erbB2 and erbB3) and mucin (MUC1 and MUC2) have disappointed in studies of clinical utility [4].
The traditional approach to biomarker development relies on single dimensional (microRNA, gene or protein) analysis in an attempt to link a single molecular entity to tumor behavior. This method seems to have reached a zenith that is suboptimal for clinical decision-making. Previous multidimensional approaches have demonstrated that through the combination of biomarkers coming from different dimensions a better knowledge of the biology of CRC can be achieved [5], [6], [7]. In an attempt to provide more personalized options, we developed a novel method that further advances the integration and incorporates multiple molecular entities from all three molecular dimensions (microRNA, genes and protein) simultaneously to generate accurate predictors of outcome in patients with CRC. Our results clearly demonstrate the superiority of this novel, multidimensional approach as compared with the traditional tools of single dimension analysis. We are hopeful that newly discovered multidimensional biomarkers will provide a basis for successful triage and stratification of patients in prospective clinical trials while simultaneously revealing molecular agents and pathways playing prominent sinister roles in CRC disease progression.
Materials and Methods
Gene and micro-RNA expression assessed with nanofluidic technology
A clinical cohort of 267 colon cancer patients was analyzed in this retrospective study. After approval of the Danbury Hospital Internal Review Board (DHIRB) and collection of the relevant clinical information, FFPE samples were obtained from colon cancer cases that had been preserved between 2000 and 2008. According to the protocol of the study (DH-17/12) including full de-identification of patient information, DHIRB waived the need of informed consent. FFPE samples were cut to 10 µm thickness and two tissue slices were put into a 1.5 ml tube. To each tube, one milliliter of xylene was added for deparaffinization followed by mixing twice with a high speed vortex for 3 min at room temperature. Total RNA was then automatically extracted with the QIAcube using the miRNeasy FFPE kit (Qiagen, Valencia, CA) following manufacturer's protocol. The RNA from SW837 cells was automatically extracted with the QIAcube using the miRNeasy kit (Qiagen, Valencia, CA) following manufacturer's protocol. RNA quantity and the quality were assessed by Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Analysis was carried out using the 48.48 dynamic array (Fluidigm Corporation, CA, USA) and a Biomark platform following the manufacturer's protocol as previously described [8], [9].
Quantitative fluorescent immunohistochemistry
Quantitative fluorescent immunohistochemistry was performed for protein analysis. Tissue specimens were prepared in a Tissue Micro Array (TMA) format: representative tumor areas were obtained from Formalin Fixed Paraffin Embedded (FFPE) specimens of the primary tumor, and up to three representative replicate 3-mm cores from multiple tumor blocks were taken after review and marking of the hematoxylin and eosin stained slides by board-certified pathologists (SS and PF). In total, 630 cores were taken and distributed over 16 slides from 267 patients. FFPE tissues used as controls of the reaction included normal colon, kidney, liver, brain, breast, lymph nodes, thyroid, skin, tonsil, skeletal muscle and bladder along with breast cancer and non-small cell lung cancer.
TMA slides were deparaffinized in xylene and then rehydrated in sequentially diluted ethanol solutions. Antigen retrieval was conducted by heating the slides in a steamer for 30 minutes in a solution of Tris-EDTA pH 8.0. Endogenous peroxidase activity was blocked by treating the slides in Peroxidazed reagent (Biocare Medical, Concord, CA) for 5 minutes. Non-specific binding was reduced by incubation with Background Sniper (Biocare Medical, Concord, CA) for 10 minutes. Slides were incubated with the primary target antibodies and epithelial and stromal cell mask antibodies diluted in Da Vinci Green antibody diluent (Biocare Medical, Concord, CA) for 1 hour at room temperature. Details of all antibodies used are in Table S1. Cyanine 5 (Cy5) directly conjugated to tyramide (Perkin-Elmer, Boston, MA) at 1∶50 dilution was used as the fluorescent detection for all the target antigens.
Statistical Analysis
For single dimension analysis, overall survival was calculated from the date of diagnosis to the date of death or date last seen. Medians and life tables were computed using the product-limit estimate by the Kaplan and Meier method, and the Log Rank test was employed only to assess the statistical significance. Multivariate analysis assessed the clinical role of each factor matched with other clinical variables (age, stage, grading, type of tumor, and gender), following Cox proportional hazards model. To simplify the issue of censoring, we remove the patients who were censored within 3 years and transformed the survival data into binary response, either aggressive or non-aggressive. For each factor proven to be significant in multivariate analysis (p-value <0.05), the area under the curve (AUC) in the receiver operating characteristic (ROC) curve was utilized to assess discriminatory power.
For multidimensional analysis, the dataset was randomly divided into training and testing subsets, with 125 cases in each subset. Multiple biomarkers were combined to yield a diagnostic score that was used as a predictor of outcome. To generate the score, we first used sliced inverse regression [10], [11] to do the sufficient dimension reduction whereby no information about the conditional distribution of outcome was lost during the dimension reduction. Next, a scalar diagnostic score was computed from the lower dimensional data generated in the first step by likelihood ratio statistic which has been proven to be optimal among all possible functions of multiple markers for binary disease outcomes [12]. This approach enabled utilization of information from multiple markers simultaneously without the need to make assumptions concerning the distributions of the markers. Cox and Kaplan-Meier models were employed to evaluate the statistical significance of multidimensional biomarkers in multivariate analysis as described above.
Results
Expression Analysis of microRNA
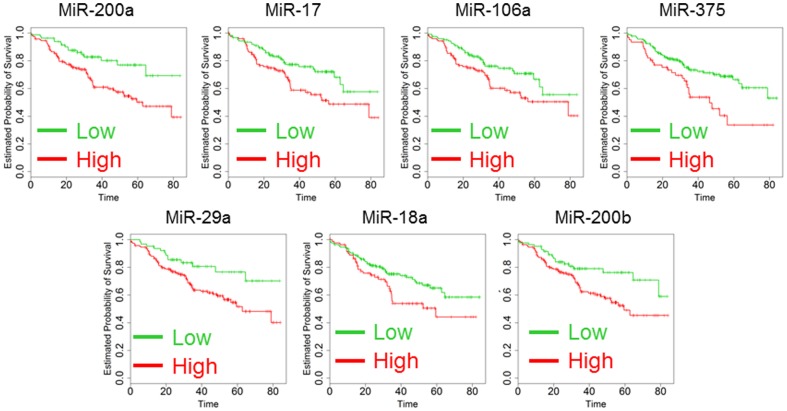
The main clinical parameters of the 267 CRC patients enrolled in this retrospective analysis are illustrated in Table 1. All the specimens were collected at the first surgery before any treatment. As anticipated the most important clinical factor to predict outcome was the stage of the disease. For patients at stage IV the progression was fast with a median survival rate of 11 months, while for patients at earlier stages the outcome was better (Fig. 1). All the patients were then treated with the best available care and this study will focus on pure prognostic predictors and not to predictors of response to specific treatments. As a first step, we screened a series of 33 microRNAs to identify potential predictors of outcome in multivariate analysis including age and stage in the Cox model. MicroRNAs were chosen according to the number of citations in Pubmed using as keywords the terms “colorectal cancer” and “microRNA”. Ten microRNAs (MiR-532-3p, Mir-200a, Mir-17, Mir-106a, MiR-193a-5p, MiR-145, MiR-375, Mir-29a, MiR-18a and Mir-200b) were statistically significant with values of range risk ratio (RR) less than 1 for each, meaning that high expression was related to a good outcome (Table 2). To further support the results of Cox analysis, data were also assessed using the Kaplan-Meier method. Five quintile cutoffs (25, 33, 50, 67 and 75) were used to stratify patients for high and low expression of each microRNA and log-rank test served to detect if differences in the outcome were significant. The quintile cutoff providing the lowest p-value at the log-rank test was used as discriminator (Table 2). Seven microRNAs (Mir-200a, Mir-17, Mir-106a, MiR-375, Mir-29a, MiR-18a and Mir-200b) were confirmed to be significant using the Kaplan-Meier method and the corresponding plots are shown in Fig. 2.
Table 1. Distribution of clinico-pathological characteristics of patients.
| Characteristics | All Cases (%) | Training Set (%) | Testing Set (%) | p-value* |
| Number | 267 | 125 | 125 | 0.9469 |
| Age, yrs Median (range) | 70 (22–96) | 72 (22–96) | 70 (25–95) | |
| AJCC Stage | ||||
| I–II | 176 (65.9) | 82 (65.6) | 80 (64.0) | 0.8947 |
| III–IV | 91 (34.1) | 43 (34.4) | 45 (36.0) | |
| Gender | ||||
| Male | 122 (45.6) | 55 (44.0) | 59 (47.2) | 0.7033 |
| Female | 145 (54.3) | 70 (56.0) | 66 (52.8) | |
| Histotype | ||||
| Adenocarcinoma | 243 (91.0) | 114 (91.2) | 115 (92.0) | 0.9114 |
| Mucinous | 24 (9.0) | 11 (8.8) | 10 (8.0) | |
| Grade | ||||
| G1-2 | 212 (78.2) | 99 (79.2) | 101 (80.8) | 0.6091 |
| G3 | 31 (20.9) | 18 (14.4) | 13 (10.4) | |
| N/A | 24 (3.8) | 8 (6.4) | 11 (8.8) |
*testing the difference between training and testing set.
Figure 1. Kaplan-Meier analysis according to stage of the clinical group of 267 patients enrolled in this retrospective analysis.
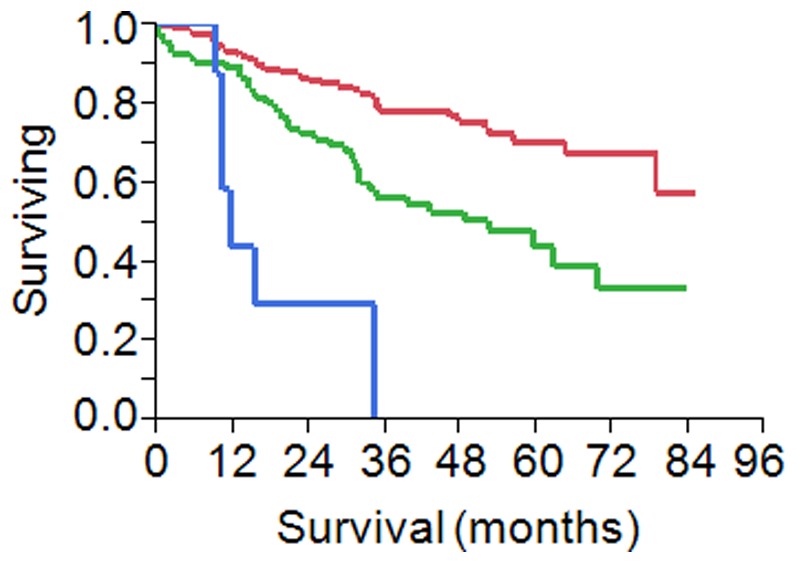
In red stage I–II patients (n = 176), in green stage III patients (n = 82) and in blue stage IV (n = 8).
Table 2. Results of microRNA expression analysis in the clinical setting of 267 patients with multivariate Cox and Kaplan-Meier method.
| P-value from KM | P-value from Cox | Range risk ratio | Range risk ratio lower limit | Range risk ratio upper limit | |
| MiR-532-3p | 0.064771 | 0.008339 | 0.094842 | 0.01648 | 0.545822 |
| Mir-200a | 0.0029 | 0.010557 | 0.077561 | 0.010928 | 0.550463 |
| Mir-17 | 0.019042 | 0.011139 | 0.079489 | 0.011251 | 0.561619 |
| Mir-106a | 0.020048 | 0.015244 | 0.099816 | 0.015518 | 0.642047 |
| MiR-193a-5p | 0.071498 | 0.016423 | 0.108225 | 0.0176 | 0.665506 |
| MiR-145 | 0.107096 | 0.019985 | 0.193244 | 0.048386 | 0.771785 |
| MiR-375 | 0.000434 | 0.020366 | 0.21165 | 0.056987 | 0.786067 |
| Mir-29a | 0.001772 | 0.024357 | 0.135351 | 0.023734 | 0.771891 |
| MiR-18a | 0.020165 | 0.027317 | 0.224383 | 0.059514 | 0.845988 |
| Mir-200b | 0.02183 | 0.047943 | 0.144104 | 0.021133 | 0.98261 |
| Mir-200c | 0.015671 | 0.053802 | 0.193341 | 0.03639 | 1.027235 |
| Mir-429 | 0.009123 | 0.057033 | 0.180053 | 0.030799 | 1.052607 |
| Let-7c | 0.122339 | 0.064338 | 0.218802 | 0.043733 | 1.094688 |
| Mir-126 | 0.013457 | 0.07168 | 0.198886 | 0.034305 | 1.15305 |
| Mir-92a | 0.015478 | 0.088624 | 0.311977 | 0.081623 | 1.192433 |
| MiR-128 | 0.074133 | 0.091335 | 0.322118 | 0.086476 | 1.199867 |
| MiR-18b | 0.057048 | 0.093705 | 0.350199 | 0.102678 | 1.194406 |
| Mir-141 | 0.030167 | 0.100176 | 0.252985 | 0.049144 | 1.302326 |
| Mir-27a | 0.03627 | 0.103385 | 0.259099 | 0.051005 | 1.316174 |
| Let-7g | 0.11726 | 0.104305 | 0.295949 | 0.068104 | 1.286059 |
| Mir-221 | 0.063595 | 0.119559 | 0.322513 | 0.077581 | 1.340726 |
| Mir-21 | 0.103007 | 0.13283 | 0.341332 | 0.084031 | 1.386486 |
| MiR-320 | 0.005064 | 0.155964 | 0.299032 | 0.056422 | 1.584841 |
| MiR-642 | 0.039954 | 0.185248 | 0.280021 | 0.042591 | 1.841022 |
| Mir-34a | 0.042544 | 0.237098 | 0.395142 | 0.084772 | 1.84185 |
| Let-7e | 0.03418 | 0.328038 | 0.514886 | 0.136147 | 1.947215 |
| Mir-20a | 0.321134 | 0.331775 | 0.509387 | 0.130451 | 1.989068 |
| MiR-328 | 0.02598 | 0.383902 | 0.551185 | 0.144202 | 2.106798 |
| Mir-31 | 0.343731 | 0.507135 | 0.570242 | 0.108455 | 2.998244 |
| Mir-183 | 0.31913 | 0.572282 | 0.5731 | 0.083001 | 3.95708 |
| MiR-30c | 0.291601 | 0.882707 | 0.893081 | 0.198839 | 4.011246 |
| MiR-125b | 0.12562 | 0.941408 | 0.947918 | 0.227684 | 3.946476 |
| Mir-203 | 0.033555 | 0.954696 | 0.959546 | 0.230859 | 3.988278 |
Figure 2. Kaplan-Maier analysis of 267 patients according to the analysis of Mir-200a, Mir-17, Mir-106a, MiR-375, Mir-29a, MiR-18a and Mir-200b expression.
Kaplan-Maier analysis was performed dividing the patients as high (green) and low (red) setting. Survival time scale is in months. All the differences were significant and p-values are reported in Table 2 (Log-rank test).
Expression Analysis of genes
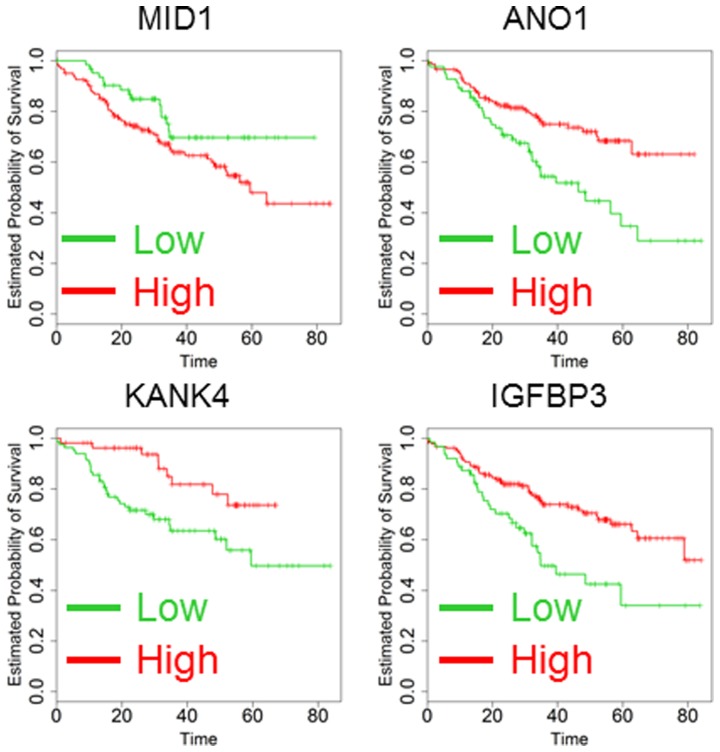
Nanofluidic technology offers the advantage of allowing analysis of microRNAs and their target genes (targetome) in the same RNA sample due to the low volume of each individual qPCR analysis. To perform this analysis, we employed multiple software applications (www.miRbase.org)[13] to prepare a list of genes that might be targetable by the 33 microRNAs investigated in this study. The list was prioritized according to a functional network obtained with the DAVID software (http://david.abcc.ncifcrf.gov) [14] in order to enrich the pool with actionable targets and master regulators of gene expression and apoptosis. After an initial analysis of 180 candidates, we focused on 79 genes whose expression was detectable in a large number of CRC cancer patients. Six genes (MID1, INHBA, OSBPL3, BGN, DICER1 and FAP) were predictors in univariate analysis (data not shown), but only MID1 remained significant after multivariate correction and Kaplan-Meier analysis (Table 3 and Fig. 3). As a second measure to possibly increase the number of candidate genes, we analyzed the public dataset GSE14333 reporting transcriptome analysis of 290 CRC cancer patients [15]. For each individual gene, data were analyzed and computed in a multivariate Cox model as described above (Table 4). Predictive capability was confirmed by Kaplan-Maier analysis using a multiple procedure of quintile selection for the cutoff as described above. The 45 genes with the lowest p-values in multivariate analysis were assessed in our platform of nanofluidic gene expression. Only 3 out of 45 (7%) genes were confirmed as predictors of outcome in both GSE14333 and our clinical setting in multivariate Cox regression and Kaplan-Meier analysis (ANO1, KANK4 and IGFBP3, Table 4 and Fig. 3).
Table 3. Results of gene expression analysis in the clinical setting of 267 patients with multivariate Cox and Kaplan-Meier method.
| P-value from KM | P-value from Cox | Range risk ratio | Range risk ratio lower limit | Range risk ratio upper limit | |
| ANO1 | 0.000455 | 0.001999 | 251.7049 | 7.554459 | 8386.487 |
| MID1 | 0.022369 | 0.00219 | 0.061488 | 0.010323 | 0.366259 |
| KANK4 | 0.00051 | 0.014574 | 29.03691 | 1.946079 | 433.2518 |
| IGFBP3 | 0.000968 | 0.021595 | 14.17555 | 1.476178 | 136.1259 |
| KLF6 | 0.239788 | 0.02687 | 0.130799 | 0.021595 | 0.792224 |
| GLI3 | 0.114457 | 0.038389 | 0.115389 | 0.014944 | 0.890962 |
| SMCR7L | 0.037868 | 0.04224 | 9.413194 | 1.081742 | 81.91252 |
| NEK6 | 0.066407 | 0.050178 | 133.963 | 0.996207 | 18014.43 |
| NAT2 | 0.356728 | 0.065965 | 7.387218 | 0.876405 | 62.26683 |
| PBX3 | 0.140048 | 0.072855 | 0.076392 | 0.004598 | 1.26913 |
| ANGPT2 | 0.000343 | 0.073683 | 21.83904 | 0.744166 | 640.9105 |
| AIMP2 | 0.431665 | 0.07928 | 16.0743 | 0.722844 | 357.4535 |
| OSBPL3 | 8.41E-05 | 0.08083 | 4.974909 | 0.821414 | 30.13061 |
| ATXN1 | 0.020848 | 0.102364 | 8.834232 | 0.646996 | 120.6247 |
| TUBB3 | 0.019182 | 0.106898 | 3.208144 | 0.777738 | 13.2335 |
| TPBG | 0.009543 | 0.109652 | 3.902565 | 0.735954 | 20.69425 |
| IGF1R | 0.134023 | 0.110324 | 9.739804 | 0.595836 | 159.2113 |
| EZR | 0.016156 | 0.117108 | 17.77504 | 0.485969 | 650.1487 |
| SAV1 | 0.011384 | 0.118547 | 14.03151 | 0.509015 | 386.7929 |
| AR | 0.348534 | 0.121365 | 0.161848 | 0.01616 | 1.621013 |
| RAI14 | 0.00433 | 0.131537 | 3.122865 | 0.710897 | 13.71828 |
| CXCR4 | 0.05891 | 0.140109 | 0.269459 | 0.047199 | 1.53833 |
| UST | 0.013135 | 0.140332 | 7.706586 | 0.510569 | 116.324 |
| INHBA | 0.002297 | 0.145151 | 3.199909 | 0.669214 | 15.30066 |
| CCL5 | 0.07661 | 0.146705 | 0.218204 | 0.027924 | 1.705063 |
| STC1 | 0.047976 | 0.154726 | 11.71039 | 0.395198 | 346.9982 |
| CALU | 0.540614 | 0.173061 | 0.215761 | 0.02376 | 1.959315 |
| CD109 | 0.159253 | 0.178425 | 5.255192 | 0.46883 | 58.90633 |
| DICER1 | 0.166057 | 0.19096 | 0.255369 | 0.033012 | 1.975454 |
| HIF1A | 0.779411 | 0.208162 | 0.362001 | 0.074392 | 1.76154 |
| MKI67 | 0.007381 | 0.224242 | 3.396451 | 0.472755 | 24.40137 |
| VAV3 | 0.024025 | 0.224528 | 0.47273 | 0.141103 | 1.583763 |
| TFAM | 0.482992 | 0.240308 | 10.34319 | 0.209424 | 510.8378 |
| BGN | 0.013917 | 0.24456 | 2.629649 | 0.516015 | 13.40088 |
| UGCG | 0.133881 | 0.245772 | 18.61169 | 0.133532 | 2594.106 |
| PLK2 | 0.121792 | 0.271772 | 7.080799 | 0.215788 | 232.3469 |
| CAV2 | 0.095172 | 0.276778 | 2.660236 | 0.456206 | 15.51242 |
| BBX | 0.374652 | 0.279638 | 12.47811 | 0.128517 | 1211.536 |
| SNAI1 | 0.183131 | 0.282896 | 0.363617 | 0.057377 | 2.304384 |
| MCL1 | 0.231297 | 0.283847 | 0.295541 | 0.031804 | 2.746347 |
| COX7A2L | 0.122784 | 0.285949 | 0.277405 | 0.026316 | 2.924187 |
| CEP170 | 0.018982 | 0.299886 | 4.728032 | 0.250678 | 89.17529 |
| CCND1 | 0.171718 | 0.300918 | 3.251198 | 0.348254 | 30.35228 |
| COMMD2 | 0.077355 | 0.303629 | 5.395563 | 0.217408 | 133.9053 |
| ADAMTS5 | 0.123317 | 0.305862 | 2.314392 | 0.464361 | 11.535 |
| ESR2 | 0.153752 | 0.311968 | 2.66113 | 0.399116 | 17.74326 |
| KLF12 | 0.465026 | 0.319069 | 2.64606 | 0.390215 | 17.943 |
| CCL2 | 0.441674 | 0.319525 | 0.438755 | 0.086651 | 2.22162 |
| EPAS1 | 0.490625 | 0.331663 | 0.418505 | 0.072098 | 2.429288 |
| RTN2 | 0.249843 | 0.334748 | 2.735875 | 0.353963 | 21.14633 |
| CHST13 | 0.039054 | 0.336699 | 2.876949 | 0.33316 | 24.84341 |
| PIM1 | 0.105052 | 0.367117 | 3.552454 | 0.226011 | 55.83778 |
| PPP2CA | 0.19979 | 0.373204 | 0.317213 | 0.025341 | 3.970806 |
| CLOCK | 0.209762 | 0.374067 | 0.492709 | 0.103457 | 2.346506 |
| FAM84A | 0.107645 | 0.37997 | 0.496851 | 0.104249 | 2.367992 |
| TM4SF1 | 0.158023 | 0.392541 | 3.496384 | 0.198371 | 61.62552 |
| CAMK2D | 0.703007 | 0.396634 | 1.968245 | 0.411242 | 9.420213 |
| HIPK1 | 0.368779 | 0.397418 | 0.322743 | 0.023514 | 4.429794 |
| KLF5 | 0.089271 | 0.408022 | 0.523601 | 0.113069 | 2.42469 |
| PTEN | 0.554417 | 0.413923 | 0.412612 | 0.049346 | 3.450134 |
| MYOF | 0.275264 | 0.417654 | 5.561907 | 0.087722 | 352.6461 |
| PNRC1 | 0.318134 | 0.420618 | 3.398165 | 0.173127 | 66.69977 |
| IGF2R | 0.096886 | 0.429115 | 2.60505 | 0.242724 | 27.95882 |
| HGF | 0.003672 | 0.429973 | 2.099431 | 0.332818 | 13.24333 |
| CDKN1A | 0.059386 | 0.430643 | 0.388964 | 0.037156 | 4.071878 |
| TGFB1 | 0.006336 | 0.446801 | 2.421279 | 0.248095 | 23.63038 |
| KDR | 0.18116 | 0.449974 | 1.72842 | 0.417918 | 7.14838 |
| EPHA3 | 0.567195 | 0.472304 | 0.55043 | 0.108045 | 2.804128 |
| CFTR | 0.384598 | 0.47535 | 2.360475 | 0.223261 | 24.95658 |
| ARNT2 | 0.06533 | 0.481856 | 0.413958 | 0.035442 | 4.834986 |
| MITF | 0.110527 | 0.487799 | 1.829958 | 0.331946 | 10.08821 |
| AHNAK2 | 0.188641 | 0.495026 | 1.774388 | 0.341724 | 9.213451 |
| KLF7 | 0.049811 | 0.506272 | 1.612536 | 0.39409 | 6.598172 |
| JAK2 | 0.394445 | 0.516395 | 0.591344 | 0.120979 | 2.890488 |
| SOX2 | 0.150192 | 0.541916 | 0.530877 | 0.069385 | 4.061828 |
| MAPRE1 | 0.357987 | 0.543075 | 0.557944 | 0.085098 | 3.658149 |
| CDH1 | 0.03582 | 0.552333 | 1.615003 | 0.332349 | 7.847868 |
| IGSF5 | 0.019559 | 0.555833 | 1.679289 | 0.29926 | 9.423273 |
| ANTXR2 | 0.546413 | 0.565429 | 3.671851 | 0.043523 | 309.7803 |
| PLK1 | 0.416995 | 0.572943 | 0.671878 | 0.168582 | 2.677758 |
| MECP2 | 0.462499 | 0.581138 | 0.626129 | 0.118665 | 3.303723 |
| VAMP2 | 0.161211 | 0.58226 | 0.628316 | 0.119973 | 3.290569 |
| COL1A1 | 0.02623 | 0.584364 | 0.529685 | 0.054348 | 5.162451 |
| FES | 0.382467 | 0.593556 | 1.591399 | 0.288902 | 8.766134 |
| PTK2 | 0.060876 | 0.601408 | 1.649351 | 0.25232 | 10.78138 |
| BCL2 | 0.338186 | 0.610589 | 0.626785 | 0.103814 | 3.784251 |
| HOXB5 | 0.27284 | 0.62191 | 1.486459 | 0.307571 | 7.183917 |
| CYP39A1 | 0.486591 | 0.642143 | 1.671365 | 0.191529 | 14.58506 |
| CDX2 | 0.002486 | 0.683233 | 0.651889 | 0.083486 | 5.090169 |
| PDGFRB | 0.042645 | 0.69718 | 1.42049 | 0.242459 | 8.322194 |
| CD59 | 0.005198 | 0.700335 | 1.635662 | 0.133489 | 20.04205 |
| EPHB2 | 0.042245 | 0.744099 | 0.655446 | 0.051878 | 8.281188 |
| FAM1738 | 0.112702 | 0.744395 | 1.849962 | 0.045828 | 74.67826 |
| SHMT2 | 0.040828 | 0.745961 | 0.73874 | 0.118273 | 4.614223 |
| SLC12A2 | 0.434188 | 0.771848 | 0.776719 | 0.140768 | 4.285722 |
| HOXB7 | 0.195627 | 0.788445 | 0.798834 | 0.154871 | 4.120429 |
| PBK | 0.445747 | 0.790426 | 0.821722 | 0.193118 | 3.496454 |
| DUSP10 | 0.13842 | 0.794739 | 1.669082 | 0.03519 | 79.16626 |
| CISH | 0.090655 | 0.810002 | 0.754679 | 0.076079 | 7.486208 |
| FAP | 0.030751 | 0.811833 | 1.191001 | 0.282441 | 5.022236 |
| MAFF | 0.255153 | 0.814144 | 0.748927 | 0.067236 | 8.342135 |
| MET | 0.13437 | 0.823224 | 1.374103 | 0.084554 | 22.33087 |
| CHEK1 | 0.163926 | 0.825053 | 0.822722 | 0.145827 | 4.641614 |
| ESR1 | 0.056936 | 0.846116 | 1.3123 | 0.084339 | 20.41915 |
| CDX1 | 0.010579 | 0.848595 | 1.30181 | 0.086808 | 19.52242 |
| ADM | 0.262582 | 0.853341 | 1.156516 | 0.247503 | 5.40409 |
| HECTD2 | 0.353439 | 0.855316 | 0.860183 | 0.170415 | 4.341829 |
| PPARGC1B | 0.37968 | 0.855611 | 1.222648 | 0.14027 | 10.65707 |
| LDLR | 0.355447 | 0.858784 | 1.222084 | 0.134155 | 11.13256 |
| HIC2 | 0.176202 | 0.862587 | 0.815525 | 0.081015 | 8.209401 |
| CHEK2 | 0.155676 | 0.866904 | 1.172525 | 0.182286 | 7.542064 |
| AXL | 0.039057 | 0.882521 | 1.167514 | 0.149675 | 9.107027 |
| SRC | 0.503974 | 0.883268 | 0.895666 | 0.205758 | 3.898842 |
| FGFR1 | 0.416422 | 0.892855 | 0.896111 | 0.181618 | 4.421444 |
| CXCL10 | 0.090745 | 0.898925 | 0.868597 | 0.098801 | 7.636207 |
| ETS2 | 0.135175 | 0.904279 | 0.80061 | 0.021349 | 30.02398 |
| KIF11 | 0.243986 | 0.91388 | 0.918496 | 0.196749 | 4.287873 |
| ROCK1 | 0.302625 | 0.915522 | 1.123317 | 0.131032 | 9.630014 |
| DROSHA | 0.25995 | 0.924716 | 1.086434 | 0.194645 | 6.064063 |
| JAK1 | 0.113129 | 0.930365 | 0.905077 | 0.096647 | 8.47581 |
| ERBB2 | 0.302875 | 0.962123 | 1.048409 | 0.14901 | 7.376446 |
| CTNNB1 | 0.099735 | 0.963275 | 1.03343 | 0.254908 | 4.18966 |
| GBP1 | 0.040014 | 0.969138 | 1.043144 | 0.122758 | 8.864152 |
| SLITRK4 | 0.238082 | 0.973046 | 0.978316 | 0.274291 | 3.489367 |
Figure 3. Kaplan-Maier analysis of 267 patients according to the analysis of MID1, ANO1, KANK4 and IGFBP3 expression.
Kaplan-Maier analysis was performed dividing the patients as high (green) and low (red) setting. Survival time scale is in months. All the differences were significant and p-values are reported in Table 3 (Log-rank test).
Table 4. Results of gene expression analysis in the public dataset GSE14333 with multivariate Cox and Kaplan-Meier method.
| Probe | Gene | P-value from KM | P-value from Cox | Range risk ratio | Range risk ratio lower limit | Range risk ratio upper limit |
| 229357_at | ADAMTS5 | 6.04E-06 | 0.000201 | 34.0864 | 5.305339 | 219.0025 |
| 235368_at | ADAMTS5 | 1.32E-06 | 8.75E-05 | 42.64678 | 6.539883 | 278.101 |
| 219935_at | ADAMTS5 | 8.92E-05 | 0.000265 | 22.25267 | 4.200208 | 117.8945 |
| 1558636_s_at | ADAMTS5 | 0.001966 | 0.005793 | 100.589 | 3.802689 | 2660.788 |
| 220726_at | AHNAK2 | 0.26889 | 0.690171 | 0.697388 | 0.118539 | 4.102851 |
| 1558378_a_at | AHNAK2 | 0.000362 | 0.00308 | 27.08616 | 3.047499 | 240.7416 |
| 212992_at | AHNAK2 | 1.55E-06 | 3.46E-05 | 483.5156 | 25.93489 | 9014.392 |
| 209971_x_at | AIMP2 | 0.000178 | 0.000173 | 0.056117 | 0.01248 | 0.252328 |
| 202138_x_at | AIMP2 | 0.000483 | 5.44E-05 | 0.0415 | 0.00885 | 0.194598 |
| 209972_s_at | AIMP2 | 0.012259 | 0.457615 | 0.415432 | 0.040911 | 4.218525 |
| 205572_at | ANGPT2 | 0.007138 | 0.027838 | 11.73135 | 1.307621 | 105.248 |
| 236034_at | ANGPT2 | 0.003749 | 0.000137 | 102.5228 | 9.496737 | 1106.794 |
| 211148_s_at | ANGPT2 | 0.000103 | 0.066738 | 26.07117 | 0.798486 | 851.2436 |
| 237261_at | ANGPT2 | 0.032343 | 0.153008 | 12.34905 | 0.392984 | 388.0543 |
| 1555269_a_at | ANO1 | 0.005692 | 0.02366 | 23.37811 | 1.52436 | 358.5346 |
| 218804_at | ANO1 | 0.001514 | 0.000249 | 88.27612 | 8.031295 | 970.2886 |
| 1555536_at | ANTXR2 | 0.165872 | 0.448339 | 1.992017 | 0.33543 | 11.82999 |
| 225524_at | ANTXR2 | 0.002183 | 0.00047 | 44.01017 | 5.278853 | 366.916 |
| 228573_at | ANTXR2 | 0.005047 | 0.000316 | 19.32602 | 3.857263 | 96.82903 |
| 213015_at | BBX | 0.003728 | 0.006287 | 20.99437 | 2.364569 | 186.4033 |
| 1557239_at | BBX | 0.291266 | 0.980533 | 1.036403 | 0.058639 | 18.31775 |
| 223134_at | BBX | 6.95E-05 | 0.00274 | 30.21056 | 3.248415 | 280.9611 |
| 226331_at | BBX | 2.81E-05 | 8.81E-07 | 359.268 | 34.41053 | 3750.987 |
| 213016_at | BBX | 0.004471 | 0.000586 | 102.7534 | 7.326352 | 1441.135 |
| 223135_s_at | BBX | 6.36E-08 | 1.22E-05 | 45.64845 | 8.239986 | 252.8865 |
| 232008_s_at | BBX | 0.000531 | 9.24E-05 | 102.4825 | 10.06176 | 1043.82 |
| 1557240_a_at | BBX | 0.169963 | 0.299311 | 2.43368 | 0.453789 | 13.05188 |
| 213426_s_at | CAV2 | 0.188766 | 0.031867 | 7.132542 | 1.18575 | 42.90378 |
| 203324_s_at | CAV2 | 3.97E-07 | 4.18E-05 | 41.99252 | 7.026959 | 250.9438 |
| 203323_at | CAV2 | 1.60E-09 | 2.04E-06 | 77.00721 | 12.82191 | 462.4983 |
| 229900_at | CD109 | 0.041603 | 0.004916 | 26.16536 | 2.689981 | 254.5096 |
| 226545_at | CD109 | 0.000325 | 0.00023 | 20.46414 | 4.105505 | 102.0048 |
| 200984_s_at | CD59 | 0.000362 | 0.000149 | 55.54448 | 6.96658 | 442.8556 |
| 200983_x_at | CD59 | 1.14E-06 | 3.41E-05 | 70.30158 | 9.407096 | 525.3813 |
| 200985_s_at | CD59 | 9.51E-06 | 3.67E-05 | 52.14801 | 7.975754 | 340.9602 |
| 212463_at | CD59 | 6.84E-07 | 1.22E-06 | 175.8647 | 21.78629 | 1419.626 |
| 228748_at | CD59 | 0.004633 | 0.041237 | 7.88942 | 1.085611 | 57.33449 |
| 206430_at | CDX1 | 6.36E-07 | 4.45E-05 | 0.030374 | 0.005675 | 0.162561 |
| 206387_at | CDX2 | 0.0014 | 3.67E-05 | 0.073955 | 0.021472 | 0.254721 |
| 231606_at | CDX2 | 0.000568 | 0.095068 | 0.303118 | 0.074635 | 1.231068 |
| 212746_s_at | CEP170 | 0.000299 | 0.009884 | 23.82942 | 2.142353 | 265.055 |
| 207719_x_at | CEP170 | 0.000131 | 6.49E-05 | 31.19191 | 5.766271 | 168.7287 |
| 234702_x_at | CFTR | 0.01694 | 0.27293 | 0.461545 | 0.115856 | 1.838699 |
| 215703_at | CFTR | 0.037325 | 1.37E-05 | 0.053111 | 0.014147 | 0.199386 |
| 217026_at | CFTR | 0.385981 | 0.252799 | 0.168605 | 0.007976 | 3.563921 |
| 215702_s_at | CFTR | 0.012329 | 0.000782 | 0.02279 | 0.002509 | 0.206981 |
| 234706_x_at | CFTR | 0.371555 | 0.578887 | 0.511618 | 0.047986 | 5.454732 |
| 205043_at | CFTR | 0.021692 | 3.17E-05 | 0.065349 | 0.018078 | 0.236224 |
| 239647_at | CHST13 | 0.000319 | 3.61E-05 | 0.07416 | 0.021584 | 0.254811 |
| 242503_at | CHST13 | 0.246465 | 0.263741 | 0.463753 | 0.120515 | 1.784564 |
| 223377_x_at | CISH | 0.01124 | 0.030594 | 0.153875 | 0.02821 | 0.839335 |
| 223961_s_at | CISH | 4.08E-05 | 5.85E-05 | 0.033215 | 0.006312 | 0.174775 |
| 221223_x_at | CISH | 0.002772 | 0.001846 | 0.036573 | 0.004558 | 0.293441 |
| 226910_at | COMMD2 | 0.000633 | 0.000126 | 37.40157 | 5.872115 | 238.2237 |
| 223491_at | COMMD2 | 3.60E-05 | 0.019365 | 10.47272 | 1.462487 | 74.99413 |
| 221563_at | DUSP10 | 5.91E-05 | 0.001099 | 37.05756 | 4.233756 | 324.3604 |
| 215501_s_at | DUSP10 | 1.84E-05 | 0.000388 | 26.5064 | 4.335402 | 162.0586 |
| 209588_at | EPHB2 | 0.00033 | 9.89E-05 | 0.05185 | 0.011687 | 0.230028 |
| 209589_s_at | EPHB2 | 0.010517 | 0.00193 | 0.082811 | 0.017148 | 0.399908 |
| 211165_x_at | EPHB2 | 2.20E-05 | 0.000149 | 0.046433 | 0.0095 | 0.226942 |
| 210651_s_at | EPHB2 | 0.027564 | 0.000968 | 0.060346 | 0.011386 | 0.319845 |
| 234158_at | EPHB2 | 0.031839 | 0.169435 | 0.412742 | 0.116806 | 1.45845 |
| 233699_at | EPHB2 | 0.231256 | 0.453665 | 1.688996 | 0.428775 | 6.653159 |
| 222303_at | ETS2 | 0.006806 | 0.005887 | 0.114387 | 0.024448 | 0.535198 |
| 201328_at | ETS2 | 0.000411 | 8.08E-05 | 0.081369 | 0.023375 | 0.283247 |
| 201329_s_at | ETS2 | 0.000328 | 8.38E-05 | 0.065016 | 0.016656 | 0.253792 |
| 208621_s_at | EZR | 0.034737 | 0.097102 | 7.073765 | 0.7013 | 71.35055 |
| 217234_s_at | EZR | 0.044295 | 0.041498 | 9.910504 | 1.092409 | 89.90964 |
| 208622_s_at | EZR | 0.000622 | 0.001303 | 99.00849 | 6.013656 | 1630.07 |
| 208623_s_at | EZR | 4.02E-07 | 2.11E-05 | 1207.412 | 45.89005 | 31768.2 |
| 217230_at | EZR | 0.174403 | 0.140414 | 3.361179 | 0.670718 | 16.84393 |
| 238645_at | EZR | 0.021088 | 0.042168 | 11.38081 | 1.089858 | 118.8439 |
| 215200_x_at | EZR | 0.286994 | 0.318044 | 2.823012 | 0.368122 | 21.64879 |
| 225670_at | FAM173B | 2.61E-05 | 0.000117 | 0.033282 | 0.005894 | 0.187931 |
| 225668_at | FAM173B | 0.063779 | 0.343216 | 0.331004 | 0.033645 | 3.256445 |
| 234335_s_at | FAM84A | 1.84E-05 | 2.34E-05 | 0.021597 | 0.003653 | 0.127692 |
| 229546_at | FAM84A | 0.000894 | 2.59E-05 | 0.034042 | 0.007048 | 0.164431 |
| 225667_s_at | FAM84A | 0.000701 | 1.19E-05 | 0.013224 | 0.001908 | 0.091637 |
| 234331_s_at | FAM84A | 0.000489 | 3.23E-07 | 0.010521 | 0.001834 | 0.06036 |
| 231439_at | FAM84A | 0.002881 | 0.00016 | 0.043208 | 0.008456 | 0.220772 |
| 228459_at | FAM84A | 0.000124 | 0.00285 | 0.104682 | 0.023768 | 0.461058 |
| 228319_at | FAM84A | 0.088904 | 0.688981 | 0.609438 | 0.053918 | 6.888528 |
| 210095_s_at | IGFBP3 | 0.00038 | 0.000122 | 34.81351 | 5.693815 | 212.8592 |
| 212143_s_at | IGFBP3 | 3.87E-06 | 6.58E-05 | 40.75663 | 6.598764 | 251.7294 |
| 243027_at | IGSF5 | 0.000113 | 0.001432 | 55.30184 | 4.691637 | 651.8607 |
| 229125_at | KANK4 | 8.48E-06 | 0.000276 | 12.86927 | 3.247498 | 50.99869 |
| 217173_s_at | LDLR | 0.011169 | 0.16538 | 4.576761 | 0.533656 | 39.25138 |
| 202067_s_at | LDLR | 0.002542 | 0.120351 | 14.71872 | 0.494593 | 438.0182 |
| 202068_s_at | LDLR | 0.002236 | 0.000255 | 51.66079 | 6.236509 | 427.9376 |
| 217103_at | LDLR | 0.340416 | 0.568331 | 0.54143 | 0.065787 | 4.456002 |
| 217183_at | LDLR | 0.129907 | 0.480234 | 1.708543 | 0.386148 | 7.559592 |
| 217005_at | LDLR | 0.074924 | 0.228696 | 2.675823 | 0.538828 | 13.28814 |
| 205193_at | MAFF | 0.000158 | 0.000102 | 44.54421 | 6.562634 | 302.3461 |
| 211864_s_at | MYOF | 1.04E-05 | 0.000138 | 62.98781 | 7.483162 | 530.1855 |
| 201798_s_at | MYOF | 0.000208 | 2.47E-05 | 188.5189 | 16.51764 | 2151.602 |
| 217518_at | MYOF | 0.020004 | 0.166863 | 4.972052 | 0.511631 | 48.31859 |
| 206797_at | NAT2 | 0.000112 | 2.13E-05 | 0.062607 | 0.017448 | 0.224656 |
| 201939_at | PLK2 | 0.003791 | 7.69E-05 | 14.06108 | 3.792438 | 52.13371 |
| 209034_at | PNRC1 | 1.45E-05 | 4.87E-05 | 31.43427 | 5.954339 | 165.9484 |
| 1555282_a_at | PPARGC1B | 0.060267 | 0.352001 | 0.505394 | 0.120092 | 2.126897 |
| 1553639_a_at | PPARGC1B | 3.01E-06 | 2.18E-05 | 0.03799 | 0.008394 | 0.171939 |
| 1563943_at | PPARGC1B | 0.458245 | 0.985653 | 1.014717 | 0.206439 | 4.98768 |
| 202052_s_at | RAI14 | 5.41E-07 | 0.000287 | 2081.501 | 33.49958 | 129334.4 |
| 204217_s_at | RTN2 | 0.000905 | 0.007263 | 165.478 | 3.970497 | 6896.612 |
| 222573_s_at | SAV1 | 0.002144 | 0.027685 | 13.52864 | 1.331125 | 137.4957 |
| 218276_s_at | SAV1 | 1.68E-06 | 4.00E-05 | 39.62951 | 6.846883 | 229.3742 |
| 234491_s_at | SAV1 | 0.023935 | 0.012655 | 15.70887 | 1.802495 | 136.9039 |
| 236606_at | SAV1 | 0.028297 | 0.161562 | 149.8623 | 0.134718 | 166709.5 |
| 204404_at | SLC12A2 | 3.73E-06 | 0.000236 | 0.05294 | 0.011053 | 0.253562 |
| 225835_at | SLC12A2 | 0.001286 | 0.007522 | 0.111752 | 0.022405 | 0.557401 |
| 232636_at | SLITRK4 | 8.73E-06 | 0.000342 | 1081.219 | 23.6349 | 49462.24 |
| 204596_s_at | STC1 | 0.001429 | 0.002766 | 128.3408 | 5.339502 | 3084.814 |
| 204595_s_at | STC1 | 9.53E-06 | 0.000234 | 26.79181 | 4.6472 | 154.4589 |
| 230746_s_at | STC1 | 0.000866 | 0.000953 | 17.63863 | 3.214301 | 96.79288 |
| 204597_x_at | STC1 | 2.33E-06 | 0.002335 | 76.65634 | 4.689406 | 1253.079 |
| 238443_at | TFAM | 0.059819 | 0.225228 | 0.371229 | 0.074838 | 1.841456 |
| 203177_x_at | TFAM | 1.71E-05 | 0.000113 | 0.022829 | 0.00335 | 0.155556 |
| 208541_x_at | TFAM | 0.002744 | 0.054875 | 0.216047 | 0.045206 | 1.032524 |
| 203176_s_at | TFAM | 0.000342 | 0.003043 | 0.115476 | 0.027697 | 0.481457 |
| 238168_at | TM4SF1 | 0.030768 | 0.014898 | 614.9803 | 3.498935 | 108090.2 |
| 209387_s_at | TM4SF1 | 2.56E-08 | 2.02E-06 | 800.6651 | 50.79976 | 12619.44 |
| 215034_s_at | TM4SF1 | 3.95E-07 | 1.61E-07 | 1067.398 | 78.60081 | 14495.26 |
| 209386_at | TM4SF1 | 5.87E-10 | 8.06E-08 | 1440.194 | 101.0956 | 20516.8 |
| 215033_at | TM4SF1 | 0.000849 | 0.024728 | 76.44376 | 1.736114 | 3365.936 |
| 203476_at | TPBG | 6.74E-05 | 0.000102 | 53.32757 | 7.179862 | 396.0842 |
| 224967_at | UGCG | 0.000167 | 0.000189 | 25.07686 | 4.619337 | 136.1341 |
| 204881_s_at | UGCG | 0.030888 | 0.25574 | 2.664385 | 0.491631 | 14.43957 |
| 221765_at | UGCG | 6.94E-05 | 0.01459 | 18.46403 | 1.778815 | 191.6559 |
| 205138_s_at | UST | 0.039899 | 0.095085 | 3.788817 | 0.792891 | 18.10481 |
| 205139_s_at | UST | 0.000152 | 8.32E-05 | 39.8298 | 6.355425 | 249.6155 |
| 214792_x_at | VAMP2 | 0.00019 | 0.000615 | 45.85393 | 5.13567 | 409.4078 |
| 201556_s_at | VAMP2 | 0.00036 | 0.000231 | 51.11359 | 6.295788 | 414.9757 |
| 201557_at | VAMP2 | 0.001365 | 0.001694 | 1058.693 | 13.68399 | 81908.26 |
Expression Analysis of proteins
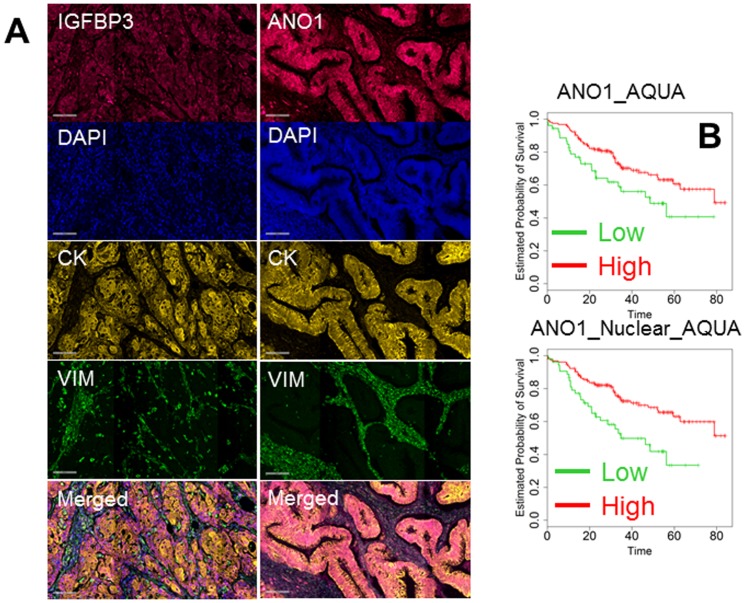
To conduct analysis at the protein level, we chose 9 factors (TUBB3, ELAVL1, OSBPL3, IGFBP3, ANO1, HGF, GLI3, PPP2CA and ARNT2). TMAs were prepared from the same paraffin blocks used for gene and microRNA analysis. Triplicate cores of each case were included in the TMAs to capture clonal heterogeneity, and each TMA was analyzed in triplicate by multiplexed, quantitative fluorescent immunohistochemistry. Nuclei were stained with DAPI (blue channel), and stromal and epithelial cells were stained with anti-vimentin (green channel) and anti-cytokeratin (yellow channel), respectively. Antigens of interest were acquired in the red channel, and a representative image of the analysis for IGFBP3 and ANO1 is shown in Fig. 4A. For each protein, expression was quantified with AQUA software which utilizes an unsupervised algorithm to quantify expression in defined subcellular compartments or “masks”. In our study, we selected four masks: tumor (cytokeratin+), stromal (vimentin+), tumor nuclei (DAPI+/cytokeratin+) and tumor cytoplasm (DAPI-/cytokeratin+). For each 3 mm core, at least three electronic subsegments (histospots) were analyzed. Because of replicate analysis, we collected up to 18 AQUA scores for each patient which were then averaged. GLI3, ARNT2 and HGF showed predominantly nuclear staining in some cancer cells, while in others, the staining was predominantly cytoplasmic. To exploit this phenomenon, an index was created by dividing the nuclear over the cytoplasmic expression. A value >1 was typical of a strong nuclear staining, while a value <1 indicated a predominantly cytoplasmic pattern of expression. Expression of all proteins and the index were analyzed with multivariate Cox regression analysis. Only expression of ANO1 (in cancer cells and in the nuclei of cancer cells) was significant in multivariate analysis (Table 5 and Fig. 4B).
Figure 4. Representative quantitative immunofluorescence for IGFBP3 (left column) and ANO1 (right column).
A: From top to bottom the following signals are represented: antigen of interest (red channel), cell nuclei (DAPI), tumor cells (cytokeratin), stromal cells (vimentin) and merged image. B: Kaplan-Maier analysis of 267 patients according to the expression of AQUA scores of ANO1 inside the tumor mask (ANO1_AQUA) and in the nucleus of cancer cells (ANO1_Nuclear_AQUA). Kaplan-Maier analysis was performed dividing the patients as high (green) and low (red) setting. All the differences were significant and p-values are reported in Table 5 (Log-rank test).
Table 5. Results of quantitative fluorescent immunohistochemistry quantified with AQUA in the clinical setting of 267 patients with multivariate Cox and Kaplan-Meier method.
| P-value from KM | P-value from Cox | Range risk ratio | Range risk ratio lower limit | Range risk ratio upper limit | |
| ANO1_Nuclear_AQUA | 0.019325 | 0.0163 | 5.334706 | 1.360938 | 20.91138 |
| ANO1_AQUA | 0.000898 | 0.047176 | 3.933242 | 1.017225 | 15.20842 |
| ANO1_Cyto_AQUA | 0.000466 | 0.064055 | 3.455312 | 0.930134 | 12.83598 |
| OSBPL3_Cyto.AQUA | 0.123325 | 0.07897 | 0.188465 | 0.029282 | 1.213009 |
| OSBPL3_Nuclear_AQUA | 0.109894 | 0.080147 | 0.19641 | 0.031728 | 1.21585 |
| ARNT2_Cyto_AQUA | 0.167487 | 0.140698 | 3.728865 | 0.647353 | 21.47889 |
| ANO1_Stromal_AQUA | 0.196194 | 0.168063 | 2.759558 | 0.651671 | 11.68558 |
| ARNT2_Nuclear_AQUA | 0.216922 | 0.190952 | 3.466272 | 0.537979 | 22.33368 |
| HGF_Index | 0.072421 | 0.256612 | 0.421516 | 0.094756 | 1.875095 |
| IGFBP3_Nuclear_AQUA | 0.103525 | 0.280396 | 2.469812 | 0.478288 | 12.75378 |
| IGFBP3_AQUA | 0.03215 | 0.334276 | 2.071344 | 0.472373 | 9.082782 |
| IGFBP3_Cyto_AQUA | 0.022362 | 0.355887 | 1.91389 | 0.482418 | 7.592945 |
| OSBPL3_AQUA | 0.253341 | 0.414725 | 3.737845 | 0.157242 | 88.85318 |
| IGFBP3_Stroma_AQUA | 0.256147 | 0.43221 | 1.819391 | 0.408595 | 8.101375 |
| Gli3_Cytoplasm_AQUA | 0.630287 | 0.440849 | 1.603306 | 0.482704 | 5.3254 |
| HGF_Cyto_AQUA | 0.460081 | 0.504868 | 0.586884 | 0.122545 | 2.810666 |
| Gli3_Index | 0.282345 | 0.566901 | 0.656363 | 0.155334 | 2.773461 |
| ARNT2_Index | 0.419295 | 0.615751 | 0.562225 | 0.059327 | 5.328067 |
| Gli3_Nuclear_AQUA | 0.472219 | 0.621925 | 1.339116 | 0.419535 | 4.274326 |
| ELAVL1_AQUA | 0.185824 | 0.642386 | 1.911476 | 0.124104 | 29.44091 |
| PPP2CA_AQUA | 0.431568 | 0.706374 | 0.667826 | 0.081745 | 5.455859 |
| TUBB3_Aqua | 0.142229 | 0.752119 | 1.238951 | 0.327804 | 4.682667 |
| HGF_AQUA | 0.244703 | 0.75816 | 0.76795 | 0.143024 | 4.12343 |
| HGF_Nuclear_AQUA | 0.403127 | 0.790899 | 0.785888 | 0.13239 | 4.665154 |
| ARNT2_AQUA | 0.137421 | 0.793627 | 1.212292 | 0.286559 | 5.128612 |
| Gli3_AQUA | 0.405624 | 0.813404 | 0.833907 | 0.184549 | 3.768124 |
| TUBB3_Cyto_AQUA | 0.253091 | 0.857207 | 0.884385 | 0.23196 | 3.371855 |
| ELAVL1_Cyto_AQUA | 0.375018 | 0.900491 | 1.157521 | 0.116881 | 11.4634 |
| ELAVL1_Index | 0.296773 | 0.915789 | 0.90653 | 0.147036 | 5.58907 |
| TUBB3_Nuclear_AQUA | 0.443265 | 0.919185 | 0.947465 | 0.334061 | 2.68721 |
| ELAVL1_Nuclear_AQUA | 0.037163 | 0.958367 | 1.07847 | 0.063246 | 18.39011 |
Nuclear and Cyto indicate expression of the antigen in nucleus and cytoplasm, respectively. If no specified expression was assessed inside the cancer cells. Stromal expression refers to vimentin-positive cells. Index was created dividing the nuclear over the cytoplasmic expression.
Calculation of Predictive Accuracy
We divided the patients into two clinical groups of interest to allow simplification of censored data into a binary response. Those surviving less than three years from diagnosis were labeled as having aggressive disease, while those surviving for greater than three years were considered to have more indolent, non-aggressive disease. Each of the individual factors from the three dimensions above (microRNA, gene or protein) was tested as a predictor of disease aggressiveness using ROC curves with AUC calculation. Although some factors were statistically significant in multivariate analysis, the maximum AUC obtained from any single biomarker in a single dimension was only 0.68 (ADAMTS5). Utilizing such a weak predictor for patient care would be unacceptable as it is inaccurate (either falsely positive or falsely negative) in approximately one third of the cases.
Generation of multidimensional biomarkers
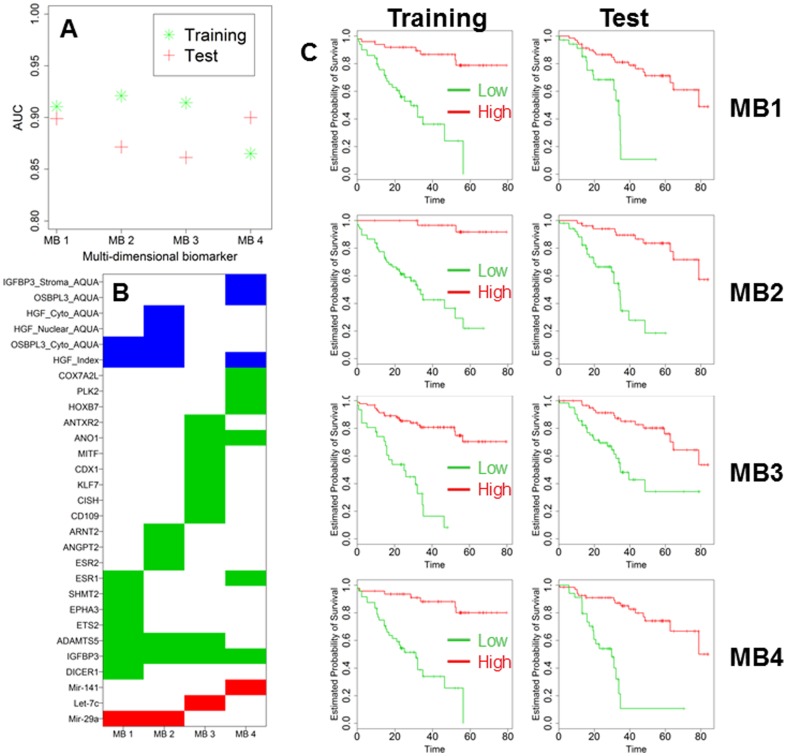
We speculated that, by combining the information from different dimensions (microRNA, gene and protein), we could substantially increase predictive accuracy. However, multidimensionality engenders significant computational complexities and challenges. Whereas in single dimension analysis the number of considered variables in our case is relatively limited at 188, multidimensional analysis of two and three variables yields 17,578 and 1,089,836 combinations, respectively. Controlling for type 1 errors using cross-validation becomes critically important as the number of variables rises. For this reason, after excluding 17 patients due to incomplete data, we randomly assigned the remaining 250 patients to either training or testing set (Tab. 1). As a first step, we randomly chose either two or three variables from all the microRNAs, genes and proteins that we considered. After sufficient dimension reduction, variables were combined into a new diagnostic score, which included all the information of the parental factors. Computation clearly demonstrated that by increasing the amount of data from different dimensions, the calculated AUCs increased in the training set (Fig. 5A). After computation of all the 1,089,836 multidimensional predictors, we selected the combinations with the highest ranking of AUCs. We then added one additional biomarker at a time, a microRNA, gene or protein, into the existing combinations while considering all possible combinations, and calculated the AUCs again in the training set (Fig. 5B). This process was repeated until AUCs reached a maximum and failed to increase significantly by adding additional predictors into the existing combinations. These maximums were reached when number of variables inside each combination reached 10 in the training set (Fig. 5B). Thereafter, we analyzed the top combinations in the testing set and we found 15 multidimensional biomarkers (MB) which showed AUC values >0.83 in the training and testing set (composition is reported in Table 6, 7 and 8) supporting the notion that multidimensional biomarkers are more accurate than any individual single dimension predictor. From this list, we selected the 4 most accurate multidimensional biomarkers (MB1 to MB4), each with AUCs of approximately 0.9 in both training and test sets (Fig. 6A). Their composition is graphically depicted in Fig. 6B. These biomarkers were also outstanding predictors of outcome in Kaplan-Meier analysis (Fig. 6C).
Figure 5. Box-whisker plot representing the values of AUC in the training set.

In the boxplot, from bottom to top, they are Q1-1.5*IQR, Q1, median, Q3, and Q3+1.5*Q3 where Q1 is first quartile (25th percentile), Q3 is the third quartile (75th percentile), and IQR is the interquartile (namely, Q3-Q1). In A the analysis is made with a single variable, with all the possible combination of two (n = 17,578) and three variables (n = 1,089,836). In B the analysis is performed by adding one new variable (gene, microRNA or protein) to the previous top combinations.
Table 6. AUC analysis for the top 15 multidimensional biomarkers in the training and testing set.
| AUC Combinations | MB | Training | Test |
| DICER1+IGFBP3+Mir−29a+HGF_Index +ADAMTS5+OSBPL3_Cyto_AQUA+ETS2+EPHA3+SHMT2+ESR1 | 1 | 0.91059 | 0.89881 |
| ADAMTS5+OSBPL3_Cyto_AQUA+HGF_Index+ESR2+ANGPT2+HGF_Nuclear_AQUA+ARNT2+IGFBP3+Mir−29a+HGF_Cyto_AQUA | 2 | 0.921007 | 0.871599 |
| CD109+IGFBP3+Let−7c+CISH+KLF7+CDX1+MITF+ADAMTS5+ANO1+ANTXR2 | 3 | 0.914251 | 0.861305 |
| HOXB7+IGFBP3+Mir−141+HGF_Index+ESR1+ANO1+PLK2+OSBPL3_AQUA+COX7A2L+IGFBP3_Stroma_AQUA | 4 | 0.865103 | 0.899916 |
| HOXB7+IGFBP3+MiR−320+HGF_Index+DROSHA+KLF6+Mir−200a+FGFR1+TM4SF1+Mir−200b | 5 | 0.916084 | 0.847826 |
| IGFBP3+Mir−29a+PNRC1+HGF_Index+CFTR+SRC+KLF5+KIF11+ANO1+CAV2 | 6 | 0.903652 | 0.855797 |
| EZR+Mir−17+HGF_Index+ANO1+IGFBP3_AQUA+OSBPL3_AQUA+Mir−200b+PIM1+ANO1_AQUA+PPP2CA | 7 | 0.827957 | 0.930193 |
| EZR+Mir−200a+HGF_Index+IGFBP3+Mir−17+ESR1+Mir−92a+ERBB2+DUSP10+PLK2 | 8 | 0.868687 | 0.881884 |
| DICER1+IGFBP3+MiR−193a−5p+CDX1+DROSHA+ANTXR2+HGF_Index+KIF11+OSBPL3_Cyto_AQUA+PLK2 | 9 | 0.875868 | 0.872449 |
| RAI14+HGF_Index+STC1+MiR−18a+COX7A2L+SOX2+SMCR7L+NAT2+MCL1+ANTXR2 | 10 | 0.845377 | 0.88913 |
| HOXB7+IGFBP3+Mir−141+HGF_Index+ESR1+ANO1+PLK2+OSBPL3_AQUA+COX7A2L | 11 | 0.844618 | 0.885606 |
| IGFBP3+Mir−29a+PNRC1+HGF_Index+CFTR+SRC+KLF5+KIF11+ANO1 | 12 | 0.885781 | 0.843478 |
| HOXB7+IGFBP3+MiR−320+HGF_Index+DROSHA+KLF6+Mir−200a+FGFR1 | 13 | 0.862471 | 0.845652 |
| DICER1+IGFBP3+Let−7g+CDX1+ANTXR2+ADM+DROSHA+KLF6+TUBB3_Cyto_AQUA+HGF_Cyto_AQUA | 14 | 0.86748 | 0.837662 |
| EZR+Mir17+HGF_Index+ANO1+IGFBP3_AQUA+OSBPL3_AQUA+Mir−200b+PIM1+ANO1_AQUA | 15 | 0.839687 | 0.849453 |
Table 7. KM and Cox information in training set for all the top multidimensional biomarkers.
| MB | P-value from KM | P-value from Cox | Range risk ratio | Range risk ratio lower limit | Range risk ratio upper limit |
| 1 | 2.00E-07 | 1.51E-06 | 653.0181 | 46.55881 | 9159.011 |
| 2 | 1.28E-07 | 3.80E-06 | 258.9312 | 24.54175 | 2731.889 |
| 3 | 1.48E-09 | 7.80E-07 | 327.9436 | 32.9403 | 3264.907 |
| 4 | 6.92E-08 | 1.15E-05 | 331.7012 | 24.79851 | 4436.787 |
| 5 | 1.07E-09 | 1.24E-05 | 40807.52 | 349.2252 | 4768423 |
| 6 | 9.46E-08 | 2.72E-06 | 112.5715 | 15.63995 | 810.2548 |
| 7 | 1.09E-06 | 0.000413 | 195.6531 | 10.46789 | 3656.91 |
| 8 | 1.75E-05 | 2.97E-05 | 266.2441 | 19.35674 | 3662.079 |
| 9 | 9.17E-06 | 0.000101 | 192.0735 | 13.57356 | 2717.948 |
| 10 | 1.67E-06 | 6.71E-05 | 12784.75 | 122.3606 | 1335805 |
| 11 | 1.81E-05 | 5.65E-06 | 102.7509 | 13.90232 | 759.4232 |
| 12 | 7.40E-08 | 3.94E-06 | 560.1007 | 38.10062 | 8233.798 |
| 13 | 3.96E-08 | 6.38E-05 | 32337.23 | 199.1264 | 5251420 |
| 14 | 2.69E-05 | 0.000165 | 402.8112 | 17.78357 | 9123.973 |
| 15 | 0.000179 | 0.000116 | 89.50607 | 9.105603 | 879.8249 |
Table 8. KM and Cox information in testing set for all the top multidimensional biomarkers.
| MB | P-value from KM | P-value from Cox | Range risk ratio | Range risk ratio lower limit | Range risk ratio upper limit |
| 1 | 1.08E-07 | 0.000175 | 149.4211 | 10.9347 | 2041.818 |
| 2 | 8.10E-07 | 0.000655 | 135.4344 | 8.046889 | 2279.45 |
| 3 | 1.00E-05 | 2.96E-07 | 134.664 | 20.66007 | 877.7509 |
| 4 | 1.02E-06 | 0.000173 | 174.0681 | 11.78516 | 2571.004 |
| 5 | 0.000107 | 0.000499 | 64.97979 | 6.196342 | 681.43 |
| 6 | 1.51E-06 | 0.000702 | 75.67874 | 6.197425 | 924.1375 |
| 7 | 1.21E-09 | 6.37E-07 | 301.1185 | 31.85225 | 2846.654 |
| 8 | 2.45E-08 | 7.39E-06 | 1150.195 | 52.76299 | 25073.42 |
| 9 | 1.17E-06 | 2.19E-05 | 165.1841 | 15.62799 | 1745.956 |
| 10 | 0.003986 | 0.008584 | 33.05786 | 2.43376 | 449.0262 |
| 11 | 0.000976 | 0.000481 | 171.8073 | 9.553675 | 3089.673 |
| 12 | 0.000404 | 0.003664 | 16.68279 | 2.499333 | 111.3559 |
| 13 | 0.00047 | 0.002143 | 657810 | 126.8559 | 3.41E+09 |
| 14 | 2.47E-05 | 2.90E-05 | 143.1154 | 13.97101 | 1466.037 |
| 15 | 0.000149 | 4.68E-06 | 647.3492 | 40.53591 | 10338.02 |
Figure 6. Bar Chart reporting AUC of the top MB obtained in the training and testing set (A).
Graphical chart of the composition of MB1-4 (B). In blue, green and red protein, genes and microRNA are reported. Kaplan-Maier analysis of the training and testing set according to the expression of the top 4 MB (C). All the differences were highly significant (Log-rank test) and are reported in Table 7 and 8 for training and testing set, respectively.
Discussion
CRC cancer remains among the deadliest malignancies. For clinical management, particularly in stage II and III disease, multiple therapeutic options are now available. As observed also in our clinical study the outcome is mostly driven by clinical stage at diagnosis with stage IV patients presenting a severe prognosis. However, even in patients with earlier stages the outcome is not only favorable with a significant relapse rate. Discoveries of effective biomarkers that can guide therapeutic decisions are ambitiously sought in the hopes of achieving the best possible outcomes, minimizing not necessary and toxic procedures. A host of studies have been conducted toward this end [2], [16], [17]. The ideal biomarker to drive clinical treatments should be significant in multivariate analysis while having robust predictive accuracy with few false positive and false negative results. Some limited successes have been obtained with regard to the selection of specific therapeutic regimens according to toxicity and efficacy [2]. However, most of these promising individual biomarkers have fallen short in clinical trials [18]. More complex biomarkers have been created, albeit in a single dimension. One 12-gene panel was effective in predicting risk of recurrence and response to treatment in a large clinical study of 1436 patients of stage II and III CRC patients [19]. However, later validation studies did not reproduce the same results [20], since the Achilles' heel of this technology remains the lack of accuracy in independent validation studies. In our study we intend dimension as the nature of the variable, being microRNA, a gene or a protein. We believe that the lack of accuracy should be dependent at least in part from the fact that the 12-gene signature was obtained only in the gene dimension, thus downsizing the possible role that other factors such as microRNA and protein may have in the predictive capability of genes. This past experience prompted us to revisit the way predictive biomarkers are built. Cancer aggressiveness is a complex trait in most of the cases. It is like a multifactorial equation. To make the pattern more complex, such multiple factors are coming from different dimensions such as genes, proteins, DNA sequences and different subset of cells (cancer and stromal). Our idea was centering the prediction on an integrated method of analysis including more dimensions and more factors at the same time. We believe that only an integrated approach can get closer to the solution of a multifactorial equation. The results we present in this study support our hypothesis. In our cohort of CRC patients, we first analyzed a large panel of possible individual predictors coming from each of three single dimensions (microRNA, gene or protein). We were indeed able to identify statistically significant predictors of outcome as determined by multivariate Cox analysis and Kaplan-Meier method. Some of these predictors have not been extensively investigated in CRC to date. As an example, expression of ANO1 (anoctamine 1) was found to be statistically significant at the gene and protein level, re-enforcing recent data coming from analysis of the dataset GSE14333 [15]. However, AUC of ANO1 in our analysis was not greater than 0.65, meaning that as a driver of clinical decisions, ANO1 would misclassify a consistent number of patients. Thus, statistical significance does not necessarily translate into clinical utility. Failure to recognize this fact can account for much of the disappointment with individual biomarkers derived from a single dimension [4], [20].
Not satisfied with AUCs below 0.7 and in the hopes of developing more robust predictors, we sought to combine our data in novel ways. In this manuscript, we provide details of a multidimensional platform which combines nanofluidic technology with quantitative fluorescent immunohistochemistry to create biomarkers with AUCs approaching and even exceeding 0.9. While the number of variables that need to be analyzed is immense, this potent toolset can collect multidimensional data at a reasonable reagent cost for FFPE samples ($0.20 for gene/microRNA analysis, $0.85 per protein).
Beyond predicting clinical outcome, our assay can highlight molecular drivers of aggressiveness. For example, IGFBP3 appears in all of the four top multidimensional biomarkers. This antigen is well known to researchers in CRC, although conflicting data are present in the literature regarding its effects [21]. At the gene expression level in both GSE14333 and our data set, high expression of IGFBP3 was related to poor outcome. This is in keeping with other previous studies [21], [22], [23]. The weight of evidence surely implicates this gene as a prominent driver of CRC cancer aggressiveness despite its being at odds with older studies connecting IGFBP3 expression to an anti-proliferative effect on the growth of colon cancer cells (reviewed in [24]).
Only two variables were present in 3 out of the 4 top multidimensional biomarkers: ADAMTS5 and HGF index. ADAMTS5 is a member of the ADAMTS (a disintegrin and metalloproteinase with thrombospondin motifs) protein family. The enzyme encoded by this gene contains two C-terminal TS motifs and functions as aggrecanase to cleave aggrecan, a major proteoglycan of cartilage [25]. As single factor, in the dataset GSE14333, high expression was associated with poor outcome in multiple probes. However, in our analysis, this factor did not show a significant trend in multivariate analysis as single element. Literature on ADAMTS5 in CRC cancer is extremely limited with only one study reporting this gene as one of the most hypermethylated in tumor as compared with the surrounding normal colonic mucosa [26]. The other variable, HGF (hepatocyte growth factor) index, represents a pathway that is known to be activated in aggressive CRC. HGF has been extensively investigated as a potential new target (reviewed in [27]). Although HGF expression in immunoperoxidase staining appears with a clear cytoplasmic pattern in CRC cancer cells [28], our immunofluorescence assay demonstrated a nuclear pattern that was of clinical significance. A similar nuclear localization of the receptor of HGF c-Met has been reported in breast cancer cells, where such overexpression was related to increased metastatic potential and aggressive disease [29].
In summary, CRC cancer aggressiveness is a complex trait that cannot be predicted with suitable accuracy by the use of an individual, single dimensional factor (microRNA, gene or protein). In contrast, a multidimensional integrated approach which utilizes data from microRNA, gene and protein analysis can generate accurate predictors of biological behavior, foster better clinical management of CRC, and shine a spotlight on molecules and molecular pathways which are associated with and potentially the cause of poor outcome.
Supporting Information
List of antibodies, suppliers and final concentration used.
(DOCX)
Acknowledgments
This work is dedicated to the memory of Renato Andreoli who passed away for colorectal cancer at the age of 57.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.
Funding Statement
This work was partially supported by a Grant from the Ruth C. Donovan Cancer Research Program and by a liberal donation from Mr. and Mrs. Ruggles. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Wingo PA, Ries LA, Parker SL, Heath CW Jr (1998) Long-term cancer patient survival in the United States. Cancer Epidemiol Biomarkers Prev 7: 271–282. [PubMed] [Google Scholar]
- 2. Newton KF, Newman W, Hill J (2012) Review of biomarkers in colorectal cancer. Colorectal Dis 14: 3–17. [DOI] [PubMed] [Google Scholar]
- 3. Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, et al. (2010) Colorectal cancer. Lancet 375: 1030–1047. [DOI] [PubMed] [Google Scholar]
- 4. Srivastava S, Gopal-Srivastava R (2002) Biomarkers in cancer screening: a public health perspective. J Nutr 132: 2471S–2475S. [DOI] [PubMed] [Google Scholar]
- 5. Comprehensive molecular characterization of human colon and rectal cancer. Nature 487: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lee H, Flaherty P, Ji HP (2013) Systematic genomic identification of colorectal cancer genes delineating advanced from early clinical stage and metastasis. BMC Med Genomics 6: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ma Y, Zhang P, Wang F, Liu W, Yang J, et al. (2012) An integrated proteomics and metabolomics approach for defining oncofetal biomarkers in the colorectal cancer. Ann Surg 255: 720–730. [DOI] [PubMed] [Google Scholar]
- 8.Mariani M, Zannoni GF, Sioletic S, Sieber S, Martino C, et al. (2012) Gender influences the Class III and V beta-tubulin ability to predict poor outcome in colorectal cancer. Clin Cancer Res. [DOI] [PubMed]
- 9. Prislei S, Martinelli E, Mariani M, Raspaglio G, Sieber S, et al. (2013) MiR-200c and HuR in ovarian cancer. BMC Cancer 13: 72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Li L, Li H (2004) Dimension reduction methods for microarrays with application to censored survival data. Bioinformatics 20: 3406–3412. [DOI] [PubMed] [Google Scholar]
- 11. Li L, Yin X (2008) Sliced inverse regression with regularizations. Biometrics 64: 124–131. [DOI] [PubMed] [Google Scholar]
- 12. Pfeiffer RM, Bur E (2008) A model free approach to combining biomarkers. Biom J 50: 558–570. [DOI] [PubMed] [Google Scholar]
- 13. Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39: D152–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Huang da W, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57. [DOI] [PubMed] [Google Scholar]
- 15. Jorissen RN, Gibbs P, Christie M, Prakash S, Lipton L, et al. (2009) Metastasis-Associated Gene Expression Changes Predict Poor Outcomes in Patients with Dukes Stage B and C Colorectal Cancer. Clin Cancer Res 15: 7642–7651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hogan NM, Joyce MR, Kerin MJ (2012) MicroRNA expression in colorectal cancer. Cancer Biomark 11: 239–243. [DOI] [PubMed] [Google Scholar]
- 17. Gonzalez-Gonzalez M, Garcia JG, Montero JA, Fernandez LM, Bengoechea O, et al. (2013) Genomics and proteomics approaches for biomarker discovery in sporadic colorectal cancer with metastasis. Cancer Genomics Proteomics 10: 19–25. [PubMed] [Google Scholar]
- 18. Tejpar S, Bertagnolli M, Bosman F, Lenz HJ, Garraway L, et al. (2010) Prognostic and predictive biomarkers in resected colon cancer: current status and future perspectives for integrating genomics into biomarker discovery. Oncologist 15: 390–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Gray RG, Quirke P, Handley K, Lopatin M, Magill L, et al. (2011) Validation study of a quantitative multigene reverse transcriptase-polymerase chain reaction assay for assessment of recurrence risk in patients with stage II colon cancer. J Clin Oncol 29: 4611–4619. [DOI] [PubMed] [Google Scholar]
- 20. Sanz-Pamplona R, Berenguer A, Cordero D, Riccadonna S, Sole X, et al. (2012) Clinical value of prognosis gene expression signatures in colorectal cancer: a systematic review. PLoS One 7: e48877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Georges RB, Adwan H, Hamdi H, Hielscher T, Linnemann U, et al. (2011) The insulin-like growth factor binding proteins 3 and 7 are associated with colorectal cancer and liver metastasis. Cancer Biol Ther 12: 69–79. [DOI] [PubMed] [Google Scholar]
- 22. Kansra S, Ewton DZ, Wang J, Friedman E (2000) IGFBP-3 mediates TGF beta 1 proliferative response in colon cancer cells. Int J Cancer 87: 373–378. [DOI] [PubMed] [Google Scholar]
- 23. Baxter RC (2013) Insulin-like growth factor binding protein-3 (IGFBP-3): Novel ligands mediate unexpected functions. J Cell Commun Signal 7: 179–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Baxter RC (2001) Signalling pathways involved in antiproliferative effects of IGFBP-3: a review. Mol Pathol 54: 145–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Held-Feindt J, Paredes EB, Blomer U, Seidenbecher C, Stark AM, et al. (2006) Matrix-degrading proteases ADAMTS4 and ADAMTS5 (disintegrins and metalloproteinases with thrombospondin motifs 4 and 5) are expressed in human glioblastomas. Int J Cancer 118: 55–61. [DOI] [PubMed] [Google Scholar]
- 26. Kim YH, Lee HC, Kim SY, Yeom YI, Ryu KJ, et al. (2011) Epigenomic analysis of aberrantly methylated genes in colorectal cancer identifies genes commonly affected by epigenetic alterations. Ann Surg Oncol 18: 2338–2347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lemos C, Sack U, Schmid F, Juneja M, Stein U (2013) Anti-metastatic treatment in colorectal cancer: targeting signaling pathways. Curr Pharm Des 19: 841–863. [PubMed] [Google Scholar]
- 28. Trovato M, Vitarelli E, Grosso M, Alesci S, Benvenga S, et al. (2004) Immunohistochemical expression of HGF, c-MET and transcription factor STAT3 in colorectal tumors. Eur J Histochem 48: 291–297. [PubMed] [Google Scholar]
- 29. Matteucci E, Bendinelli P, Desiderio MA (2009) Nuclear localization of active HGF receptor Met in aggressive MDA-MB231 breast carcinoma cells. Carcinogenesis 30: 937–945. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
List of antibodies, suppliers and final concentration used.
(DOCX)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files.