Abstract
The 1000 Genomes Project aims to provide a deep characterization of human genome sequence variation by sequencing at a level that should allow the genome-wide detection of most variants with frequencies as low as 1%. However, in the major histocompatibility complex (MHC), only the top 10 most frequent haplotypes are in the 1% frequency range whereas thousands of haplotypes are present at lower frequencies. Given the limitation of both the coverage and the read length of the sequences generated by the 1000 Genomes Project, the highly variable positions that define HLA alleles may be difficult to identify. We used classical Sanger sequencing techniques to type the HLA-A, HLA-B, HLA-C, HLA-DRB1 and HLA-DQB1 genes in the available 1000 Genomes samples and combined the results with the 103,310 variants in the MHC region genotyped by the 1000 Genomes Project. Using pairwise identity-by-descent distances between individuals and principal component analysis, we established the relationship between ancestry and genetic diversity in the MHC region. As expected, both the MHC variants and the HLA phenotype can identify the major ancestry lineage, informed mainly by the most frequent HLA haplotypes. To some extent, regions of the genome with similar genetic or similar recombination rate have similar properties. An MHC-centric analysis underlines departures between the ancestral background of the MHC and the genome-wide picture. Our analysis of linkage disequilibrium (LD) decay in these samples suggests that overestimation of pairwise LD occurs due to a limited sampling of the MHC diversity. This collection of HLA-specific MHC variants, available on the dbMHC portal, is a valuable resource for future analyses of the role of MHC in population and disease studies.
Introduction
30 years of MHC genetics
The human major histocompatibility complex (MHC) is located in the short arm of chromosome 6p21. While the region contains only a small fraction of all human genes [1], it has been extensively studied due to its pivotal role in the immune response and the need for matching the human leukocyte antigen genes (HLA) between donor and recipient in allogeneic tissue and cell transplantation [2], [3]. For example, in addition to HLA typing performed for solid organ transplantation, HLA polymorphisms have been determined in more than 23 million unrelated donors worldwide in order to match patients in need of hematopoietic stem cell transplantation, [4]. Beyond transplantation, polymorphisms in the MHC region have been used as molecular markers for population genetics and studies of diseases and traits. In the past 30 years, no other region in the genome has provided more association signals with multifactorial traits, including autoimmune diseases [5]–[8], inflammatory and infectious diseases [9], cancer [10], adverse drug effects [11], [12], and behavioral traits such as mating [13], [14]. To assess HLA allelic diversity, these studies employed a broad range of methodologies from serology, restriction fragment length polymorphism, and microsatellites up to the latest generation of single nucleotide polymorphism (SNP) genotyping methods. In the most recent genome-wide association studies (GWASs), the high number of MHC-region SNPs included in the arrays and the great complexity of resulting association signals encouraged efforts to impute classical HLA alleles based on SNP profiles [15]. However, the extremely large number of known HLA alleles (unique gene sequences), currently over 8,000 for HLA class I genes and over 2,400 for HLA class II genes [16], [17], creates a formidable challenge when attempting to capture HLA alleles using genotypes derived from common SNPs, such as those typically included on GWAS arrays.
Determining HLA polymorphisms in genomic reference samples
Building on the increasing feasibility of new generation sequencing methods, the 1000 Genomes Project provides a deep characterization of human genome sequence variation as a foundation for investigating the relationship between genotype and phenotype [18]. A goal of this project is to characterize over 95% of variants present (in genomic regions accessible to current high-throughput sequencing technologies) in 14 representative human populations from Europe, East Asia, South Asia, West Africa and the Americas. Whole genome sequencing is performed at low coverage, but at a level that should allow the genome-wide detection of most variants with frequencies as low as 1%, the classical threshold for definition of polymorphisms [18]. However, hundreds of well characterized HLA variants have frequency lower than 1%, and thousands of HLA haplotypes are present at even lower frequencies [19]. Because of the complexity of the exonic polymorphisms, several statistical methods are needed when calling HLA alleles from the sequence data [20], [21]. Higher coverage and longer read length that what the 1000 Genomes Project currently achieve, is required to positively identify all HLA alleles at all loci with an accuracy that compares to classical HLA typing experiments. The 1000 Genomes Project is nevertheless a primary reference dataset for modern genetic studies, including the SNP-based imputation of HLA alleles for disparate population and disease studies. In this report, we used sequence-based techniques to type alleles of the HLA-A, HLA-B, HLA-C, HLA-DRB1 and HLA-DQB1 genes in the available 1000 Genomes samples. This effort allowed the combined analysis of the 103,310 MHC SNPs made publicly available by the 1000 Genomes Project and the HLA alleles of these samples. While making these dataset available, we show that HLA alleles and MHC SNPs are extremely diverse in this dataset and highly specific to ancestral backgrounds. We also demonstrate that gathering HLA and SNP data on large numbers of samples worldwide increases the accuracy of HLA-SNP linkage disequilibrium (LD) estimations, revealing the HLA haplotype specificity of SNP variation. The availability of these HLA genotypes will promote analysis of the genomic architecture and immunobiology of this important super-locus at greater resolution than has heretofore been possible.
Materials and Methods
HLA typing by reference methods
The HLA typing assay was designed to capture the amino acid sequence of the Antigen Recognition Site (ARS) [22]. DNA samples were purchased from the Coriell Institute for Medical Research (Camden, NJ). The HLA typing data of 1,267 individuals related to the 1000 Genomes Project (Table 1) covers 14 populations encompassing 4 major ancestral groups. After specific PCR amplification, exons were sequenced by Sanger technique. The sequences were compared to available sequence information in the HLA allele database on exons 2 and 3 for class I and on exon 2 class II genes, therefore any polymorphism occurring in exon 4 of class I allele or exon 3 of class II gene was not investigated. Typing ambiguities between alleles were allowed since HLA-A, HLA-B, HLA-C gene products have identical sequences in exon 2 and exon 3 antigen recognition sites. Similarly, for class II genes, typing ambiguities occur if HLA-DRB1, HLA-DQB1 gene products have identical sequences in exon 2 antigen recognition sites. (Appendix S1). The Allele Database version used in the report is IMGT 2.26.0 (Jul 2009), effective Feb 2010. Several Hapmap and CEPH samples were previously HLA typed [23], [24]. Confirmatory typing was performed when the typing of the five HLA loci were missing or ambiguous (12 samples). No discrepancies were found. The previously obtained HLA types were otherwise included. The public genotype calls for the 1000 Genomes sequence analysis were downloaded from 1000 Genomes servers (phase 1) for all available samples (See on-line resources [25]–[27]).
Table 1. Overview of the 1000 Genomes project samples typed for HLA genes.
| Code | Ancestry | Description of 1000 Genomes projects project Samples | Size | HLA and genomic variants | variants |
| LWK | African | Luhya from Webuye, Kenya | 90 | 87 | 97 |
| YRI | African | Yoruba from Ibadan, Nigeria | 90 | 38 | 88 |
| ASW | American | African Ancestry from Southwest, USA | 90 | 53 | 61 |
| CLM | American | Colombian from Medellin, Colombia | 70 | 60 | 60 |
| MXL | American | Mexican Ancestry from Los Angeles-California, USA | 89 | 55 | 66 |
| PUR | American | Puerto Rican, Puerto Rico | 70 | 55 | 55 |
| CHB | East Asian | Han Chinese from Beijing, China | 90 | 85 | 97 |
| CHD | East Asian | Chinese from Denver-Colorado, USA | 90 | 0 | NA |
| CHS | East Asian | Han from south, China | 100 | 100 | 100 |
| JPT | East Asian | Japanese from Tokyo, Japan | 91 | 80 | 89 |
| CEU | European | Northern and Western European from Utah, USA | 111 | 47 | 87 |
| FIN | European | Finnish, Finland | 100 | 93 | 93 |
| GBR | European | British from England and Scotland, UK | 96 | 89 | 89 |
| TSI | European | Italian from Tuscany, Italy | 90 | 90 | 98 |
| TOTAL | 4 | 14 | 1267 | 932 | 1080 |
Ibericos from Spain (n = 14) were genotyped in the KGP but were not available for HLA typing at the time of the project. Chinese Han from Denver were typed for HLA they are currently publically available for sequencing data.
SNP genotype data from the 1000 Genomes project
The 103,310 MHC SNPs in the 1000 Genomes were extracted from the MHC (chr6: 28,866,528–33,775,446 See Table S1) [25]–[27]. Similar number of variants was extracted at random throughout the genome [28]. Additional variants were extracted in regions of the genomes with similar density of variants and similar recombination rate to the characteristics of the MHC region. Among the MHC variants, 6,040 MHC SNP previously genotyped in 800 African American controls [29], were used to compute linkage disequilibrium decay with distance by resampling datasets of various sample sizes. All coordinates refer to genome build HG19/GRCh37.
Data availability on-line
The HLA genotype data of the present study is available online: The full specification of HLA alleles in the specified release of the HLA nomenclature [17] is provided on the dbMHC portal (See on-line resources) at NCBI [30]. In addition, allele frequencies can be viewed online using tools developed in the anthropology and cell line components of the Histocompatibility Workshops [31](Figure S2, screen capture of the display [32]). Allele frequency tables are extremely sparse, reflecting the high diversity of HLA alleles for all loci and the limited sampling of the HLA alleles in 1000 Genome Project sample sets (Tables S2 and S3 for HLA allele naming convention used, also available online at dbMHC [32]).
Results
Ancestral diversity of the MHC in the 1000 Genomes samples
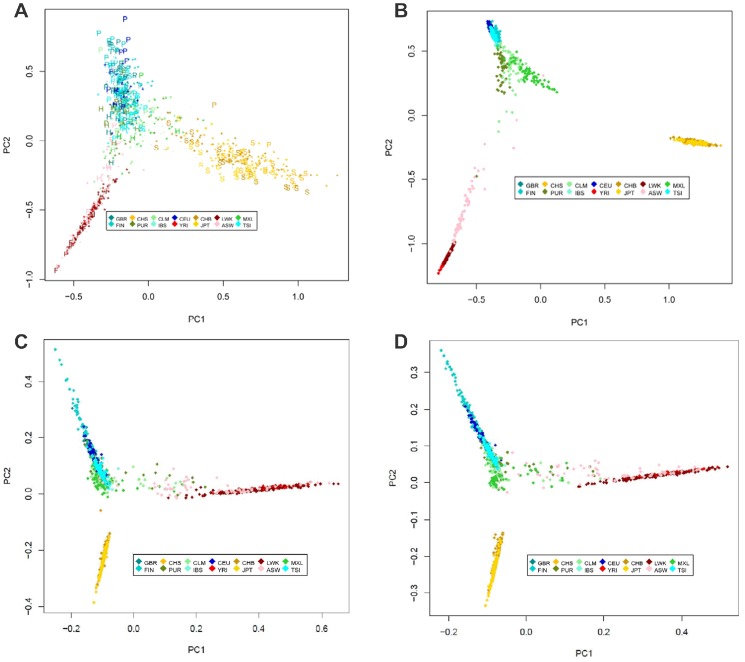
To focus on the ancestral information embedded in SNPs from the MHC, we compared the principal component analyses (PCA) of the Identity by Descent (IBD) distances between all individuals of the 1000 Genomes MHC dataset. IBD distances were computed using Beagle 2 [33] and averaged over ten runs. Both, the variants in the MHC region and an equal number of SNP variants randomly selected throughout the genome were used. The variants' density and recombination rate were computed from 1000 Genome data using Beagle (See web resources). We compared the IBD distances PCA analysis using the SNPs of the MHC region (Figure 1A), using the same number of SNPs randomly selected throughout the genome (Figure 1B). The MHC region has been also compare to other regions of the genomes with similar density of variant (Figure 1C) and similar recombination rate (figure 1D) (Additional examples and information in Figure S1 A–C)). As expected, the analysis shows that distances computed from genome-wide SNPs clearly identify samples of EuropeS1an, Asian and African ancestries as well as the admixed nature of several populations: ASW, PUR, CLM, and MXL (Figure 1B). Discordance between observed IBD ancestry and self-declared ancestry was seen for a handful of samples (Figure 1B legend).
Figure 1. Principal Component analysis of the pairwise IBD distances between 1000 Genomes samples using MHC region marker (A), genome-wide markers (B), and using markers of regions with similar variants' density (C, chr9 : 116,750,000–121,650,000), with a recombination rate (D, chr9:800,000–5,700,000).
(A) The presence of the most frequent ancestry specific HLA haplotype in the samples of the 1000 Genomes project using MHC region markers. Principal component analysis of the 103 K variants from the MHC region in the 1000 Genomes samples. PC1 captures 6.00% of total variance; PC2 captures 5.05%. The PCA analysis is based on publicly available SNPs. In order to integrate the SNP based information to the HLA allele information, individual spots are replaced by letters when a frequent HLA haplotype is predicted when the HLA typing is phased using HLA haplotype frequencies. The so called “frequent” haplotypes are defined in an ancestry specific manner: P for frequent HLA haplotypes in Europeans, S for frequent HLA haplotype in Asians, H for frequent HLA haplotype in Hispanics and F for frequent haplotype in Africans. The detailed list of the frequent haplotypes is presented in supplementary information. Frequent haplotypes and definition of overlap between ancestries were documented in a recent modeling effort for the development of haplobank. (B) Principal Component analysis of the pairwise IBD distances between 1000 Genomes samples using genome-wide markers. Principal component analysis of 100 K variants selected at random throughout of the genome in the 1000 Genomes samples. PC1 captures 55.16% of total variance PC2 captures 41.96%. The representation of distances computed from genome-wide SNPS clearly identifies samples of European, Asian and African ancestries. The results are consistent with self-declared ancestry and the admixed nature of several populations. There are however a few notable exceptions: NA20314 from south west African Americans (ASW) clusters with Mexicans (MXL), NA20291 from ASW clusters with LWK, and HG01108 from the Puerto Rican (PUR) who clusters with the majority of Africans Americans (ASW). In addition, four Columbians (CLM: HG01342, HG01390, HG01462, HG01551) and three African Americans (ASW: NA20278, NA20299, NA20414) cluster together away from their groups. These are also clustering far from their self-declared ancestry in the MHC centered analysis. This most likely reflects their genome-wide ancestry rather than a different ancestry of the MHC. (C) Principal Component analysis of the pairwise IBD distances of 1000 Genomes samples using genome-wide markers of a region (chr9 : 116,750,000–121,650,000) with a variants' density that is similar to the MHC region. Principal component analysis of 100 K variants selected at random throughout of the genome in the 1000 Genomes samples. PC1 captures 2.98% of total variance PC2 captures 1.56%. The representation of distances computed from genome-wide SNPS clearly identifies samples of European, Asian and African ancestries. PC1 and PC2 have been flipped to ease the comparison of the patterns in Figures 1A and 1B. (D) Principal Component analysis of the pairwise IBD distances of 1000 Genomes samples using genome-wide markers of a region (chr9:800,000–5,700,000) with an avergage recombination rate that is similar to the MHC region. Principal component analysis of 100 K variants selected at random throughout of the genome in the 1000 Genomes samples. PC1 captures 2.55% of total variance PC2 captures 1.57%. The representation of distances computed from genome-wide SNPS clearly identifies samples of European, Asian and African ancestries. PC1 and PC2 have been flipped to ease the comparison of the patterns in Figures 1A and 1B.
When genetic similarity is computed using MHC SNPs only, the analysis clearly identifies the same three major ancestral lineages (Europeans, Asians, and Africans) (Figure 1A). Like regions with similar variants' density and recombination rate, MHC captures well the major ancestry backgrounds. However, more variability is observed within the MHC of the 3 major ancestries (Figure 1C and 1D), this is consistent with the selection of diversity driven by HLA molecules in a cumulative manner for class I and class II. Some individuals spread across the population hubs and display significant overlap (Figure 1A). This reflects the close relation between MHC polymorphisms and the migratory history of these populations [34], [35]. For example, in contrast to the genome-wide analysis, samples of African ancestry (YRI, LWK, and most of the ASW) overlap fully. African American samples (ASW) appears more split between European and African ancestries. It suggests that intra-group differences can rarely be differentiated from cross-ancestry sequence variation, at least for these populations. Thus, within the major ancestral backgrounds, SNP haplotypes can be shared between individuals of different populations. This observation further supports the empirical HLA compatibility of donor/recipient from distinct populations grounding international exchanges of allogeneic HSC donors. We conclude that the variability of the ancestral MHC signature may not be fully captured by the overall genome ancestral estimation potential, potentially weakening case control analyses due to stratification. For example, several African American samples whose genome-wide IBD distances indicate close relation to Africans cluster with the European groups in the MHC region-based analysis (NA19703, NA19707, NA19904, NA19921).
Frequent HLA haplotypes in the 1000 Genomes samples
In order to integrate these results based on SNPs with the classical HLA typing in Figure 1A, we used HLA haplotype frequencies from the National Marrow Donor Program Registry to infer the phase of the most frequent HLA haplotypes represented in the dataset [19], [36]. HLA information was integrated to the PCA graphical display and HLA genotypes were phased using the haplotype frequencies [36]. Given the sample size, only frequent haplotypes were displayed (frequency >1%, as defined in “frequent” HLA haplotype modeling of haplobank [37]). When the statistical phasing of HLA alleles results in the presence of a frequent haplotype, letters were used as symbol at the PCA coordinate of the individual (“P” for European haplotypes, “H” for haplotypes frequent in Hispanics, “S” for haplotypes frequent in Asians and “F” for haplotypes frequent in Africans Listed in Table S4). Frequent haplotypes are found along the axis drawn by the PCA, which is consistent with frequent HLA haplotypes driving the IBD similarities within an ancestry. Interestingly, the analysis identified the presence of typical European HLA haplotypes in Asians: A*03:01∼B*35:01∼DRB1*01:01 (81% posterior phase probability NA185596 [38]) and A*01:01∼B*57:01∼DRB1*07:01 (88% posterior phase probability HG00708 [9]) Thus, a typical Asian SNP background associated with an HLA type is compatible with a mixed European Asian haploytpe, confirming the SNP background differences of conserved HLA haplotypes because. Therefore, even if only a few copies of the most frequent haplotypes are found in the 1000 Genome samples, and even if chromosomal phase is statistically estimated, it appears that this dataset will rapidly allow the in-depth analysis of haplotype specific variants of interest for both HLA allele imputation and HLA haplotype inference (Sup. Table S5, Imputation analysis limited to tag- SNP is suggestive of the existence of Haplotype specific SNPs).
Linkage disequilibrium decay in the MHC
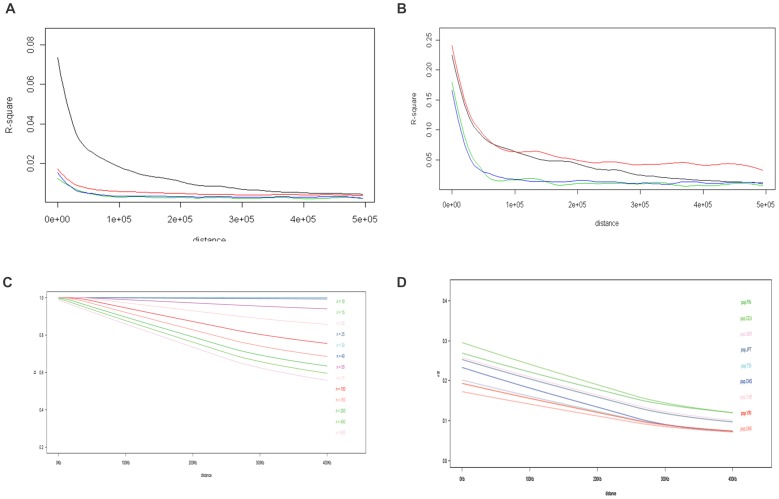
We followed on the analyses presented in Figure 1 in different regions of the genome comparing LD decay in segments with similar characteristics of the MHC (Figure 2A (rare variants) and 2B (variants >with frequent greater than 5%)). The results suggest that the MHC has a strong LD decay, but this decay depends also on the estimated frequencies of the variants affecting the comparison between regions (figure 2B). Then, in order to compare LD MHC configurations between populations we assessed the influence of the sample size on LD decay in the MHC region. By using the 90th percentile of pairwise LD for a given distance between SNP variants, emphasis was placed on the strongest LD components, which are central to both genetic association studies and SNP-based imputation methods of HLA alleles. To evaluate samples sizes larger than those of the 1000 Genomes Project, high-density genotypes of the MHC in a large sample of African Americans from a previously published study were used [29]. Figures 2C and 2D show the 90th percentile of D′ and r2 LD measures respectively for sample sizes ranging from N = 10 to N = 800 as a function of pairwise distance between MHC SNPs. As previously anticipated by Weiss and colleagues [39], sample size influences the estimation of LD: the smaller the sample size, the slower the LD decay with distance between markers. It demonstrates that for a sample size in the order of magnitude of those collected by the 1000 Genomes Project, LD in the MHC region is most likely overestimated. For low sample sizes, sampling fluctuations result in a drastic reduction of the haplotype diversity, which mimics a bottleneck effect. Such effect reduces the sample haplotype diversity compared to the source population haplotype diversity. It tends to inflate the estimation of the frequency of the sampled haplotype as compared to their real frequencies in the population and induces an overestimation of LD that diminishes in higher sample sizes. Such effect makes even more challenging the interpretation of genetic associations hitting the MHC because LD may extends further away from the primary signal than it appears from LD estimated with 1000 genome samples.
Figure 2. Across genomic region comparison of the Linkage Disequilibrium (LD) for variants with frequency lower than 5% (A), greater than 5% (B), and 90th percentile of LD by D′ (C) and R2 (D) as a function of distance (kb) for various sample size as measure.
(A) Across genomic region comparison of the LD decay (R-Square) in the 1000 genome samples for variants whose frequency is lower than 5%. (B) Across genomic regions comparison of the LD decay (R-Square) in the 1000 genome samples for variants whose frequency is greater than 5%. Chr6:28,850,000:33,750,000 (black) representing the MHC; Chr9:116,750,000:121,650,000 (green with similar variants' density as MHC used in Fig. 1C); chr9:800,000:5,700,000 (blue with similar recombination rate as MHC used in FIG. 1D), an additional control with similar variants' density chr8:9,400,000 = red (with similar variants' density as MHC), The plot is presented for 0–500 Kbp. In 2A, all markers are included in 2B only markers whose frequencies are greater than 5% are included, showing that the analysis is affected by low frequency variants which requires large sample size for accurate estimation. (C) Average 90th Percentile of pairwise linkage disequilibrium (D′) as a function of distance (kb) for various sample size. (D) Average 90th Percentile of pairwise linkage disequilibrium (R2) as a function of distance (kb ranging from 0–400 Kb) for various sample sizes. (C and D) The AAMS dataset consists of 405 African American controls and 594 African American individuals with multiple sclerosis (MS) typed at 6040 MHC SNPs using Infinium iSelect HD Custom Genotyping BeadChip (Illumina). After strict quality control for missingness <0.1% and minor allele frequency >5%, 3224 markers remained for analysis. A subset of n = 10 random control individuals was selected. Linkage disequilibrium (r2 and D′) was calculated between all pairs of SNPs (5,195,476 unique pairs) using Haploview software. All r2 and d′ estimates were sorted by distance between markers, and grouped into bins of 500 bases. The 90th percentile r2 and d′ were calculated within each bin. Locally weighted regression (Cleveland, W. S. (1981) LOWESS: A program for smoothing scatterplots by robust locally weighted regression (The American Statistician, 35, 54) was used to create a smooth regression line across the 90th percentile r2 and d′ measures. The line in the figure represents the median across 10 trials of re-sampling the n = 10 individuals. The same procedure was repeated for larger sample sizes (n = 15, 20, 25, 30, 40, 50, 75, 100, 150, and 200). For the largest sample sizes (n = 400 and n = 800), MS cases were included in the analysis. The Correlation between sample size and average LD measure at a distance of 400 kb is shown in Figure S3A and S3B in Supplementary material.
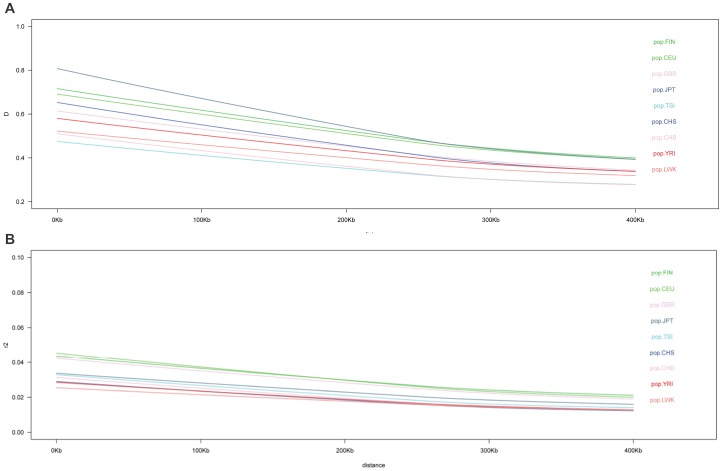
Next, we randomly sampled 85 unrelated individuals from nine of the1000 Genomes Project populations to directly compare the LD decay across the samples. Figure 3 displays the 90th percentile of D′ (Figure 3A) and r2 (Figure 3B) LD measures (Y-axis) according to the distance between markers (X-axis) for nine populations of the 1000 Genomes project. The northern European populations (FIN and CEU) exhibit the highest LD along the MHC; British and Japanese samples have an intermediate LD. The Chinese (CHS and CHB), African (LWK and YRI), and surprisingly the Tuscani (TSI) samples have the lowest LD levels. These curves are influenced by both the genetic diversity of the most frequent haplotypes and the amount of recombination/drift occurring in the population history. In Tuscani and Chinese populations the most frequent HLA haplotypes are composed of frequent alleles. In Africans, HLA haplotypes are on average less frequent and more diverse. Given the density of genes in the MHC region and their functional relevance, long-range LD components can be involved in disease association signals; it also shows that using the analysis of samples of non-European ancestry can refine variants that may be causally involved.
Figure 3. Across sample comparison linkage disequilibrium as a function of pairwise distance between SNPs for similar number of individual (n = 85) as measured by D′ (A) and R2 (B).
(A) Across sample comparison of Median of LD (D′) as a function of pairwise distance between SNPs for similar number of individual (n = 85). (B) Across sample comparison of Median of LD (R2′) as a function of pairwise distance between SNPs for similar number of individual (n = 85). We resampled 85 unrelated individuals from the various populations of the 1000 Genomes in order to compare the LD decay pattern for a similar sample size. The figure shows the relation between the median percentile of pairwise LD measures according to the distance between the two markers between 0 and 400 Kb.
Discussion
We report the public availability of high resolution HLA typing in the samples of the 1000 Genomes Project and describe the ancestry specific content of HLA allele and SNP variant haplotypes of the MHC. The data complements the resource made available by the 1000 genomes project and other collaborative effort on those samples [23], [40]. The MHC region can be described as a “genome within the genome,” able to identify the ancestral history of the individual. However, the relative low sample size of the 1000 Genomes Project fails to properly reflect the full range of haplotype diversity and, consequently, the SNP-based analysis can overestimate the extent of LD patterns. Furthermore, the effect of sample size on LD depends on the baseline haplotype diversity and frequency distribution of the source population. While the difference observed between European and African populations is conservatively estimated, larger sample sizes would reduce this haplotype diversity truncation effect. The sample size effect is particularly strong on D′ measures of LD due to the apparently complete LD (D′ = 1 R2<1) that may generate sampling fluctuations that prevent interpretation of the D′ based LD decay comparison between populations.
Large sample size are required to capture the haplotypic diversity of the MHC region
The availability of high resolution HLA typing information for the 1000 Genomes project dataset opens an array of possibilities for studying MHC polymorphisms and HLA alleles. It contributes to reducing the gap between large HLA registries, as illustrated by the recent publication of haplotype frequencies estimated from 2.9 million individuals [41], and the deep characterization of the human genome sequence diversity of the 1000 genomes project [18]. In addition to evolutionary studies of MHC haplotypes and HLA alleles, this HLA data will facilitate the training of the various SNP-based HLA imputation algorithms and the possibility to use the 1000 genome as reference samples for next-generation capture and sequencing of HLA genes. In order to illustrate the potential use of the public availability of the HLA gene typing with the 1000 Genomes sequencing data, we have explored the existence of SNP variants that can be used to indicate the presence of common HLA haplotypes (Sup. Table 2). Interestingly, many such variants seem to occur in the most common European HLA haplotype HLA-A*01:01∼HLA-B*08:01∼HLA-DRB1*03:01. The HLA haplotype HLA-A*3303∼HLA-B*4403∼HLA-DRB1*1302, which is common in Asians, also shows a high number of associated variants (r2>0.6, Sup. Table S4 and S5). Although HLA haplotype statistical phasing does not allow us to conclude that these are “tag-SNPs”, it adds further support to the examination of rare SNP variations embedded in long HLA haplotypes. Finally, we expect that the data will help to define the best reference and strategies for the use of SNPs to impute HLA alleles for population and disease studies.
Supporting Information Legends
Supplemental data consists in: Figure S1 MHC region definition (Table S1), HLA Allele frequencies in the samples of the 1000 Genomes (Tables S2), HLA alleles grouped by similarities in the antigen recognition site (Table S3), Screen capture of the display of allelic frequencies in dbMHC for the 1000 genome populations (Figure S2), The most frequent ancestry specific HLA haplotypes (Tables S4), Please note that the V2 ‘old’ style HLA nomenclature and ARS “g” code were used in supplementary material, please refer to website for more up to date information and specification of HLA allele ambiguities strings. Variants associated with frequent haplotypes in Europeans (Tables S5), Correlation between sample size and r2 90th percentile in African American samples for marker of a pairwise distance of 1000 kb (Figure S3).
Supporting Information
MHC region definition. A, Selection of the region by recombination rate et variants' density. B, chr16:86,750,000–91,650,000, a region on chromosome 16 with similar recombination rate as MHC shown in Figure 1D. C, chr16:74,200,000–79,100,000, a region on chromosome 16 with similar variants' density as MHC shown in Figure 1C.
(TIF)
Screen capture of the display of allelic frequencies in dbMHC for the 1000 genome populations. A, Homepage. B, Population selection. C, Data download.
(TIF)
Correlation between sample size and the 90th percentile of D′ (S3-A) and r2 (S3B) in African American samples for markers' pairs at a distance of 400 kb.
(TIF)
Additional details for HLA typing protocol.
(DOCX)
MHC region definitions.
(DOCX)
HLA Allele frequencies in the samples of the 1000 Genomes.
(DOCX)
HLA alleles grouped by similarities in the antigen recognition site.
(DOCX)
The most frequent ancestry specific HLA haplotypes.
(DOCX)
Variants associated with frequent haplotypes in Europeans.
(DOCX)
Funding Statement
This work was supported by grants from the National Institute of Health U19AI067152 (ARRA administrative supplement), RO1NS076492, RO1NS046297, and from the Office of Naval Research N00014-11-1-0339. PAG is a recipient of the Race to erase MS Junior Investigator Award and the European Federation for Immunogenetics Julia Bodmer Award. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, et al. (2004) Gene map of the extended human MHC. Nat Rev Genet 5: 889–899. [DOI] [PubMed] [Google Scholar]
- 2. Petersdorf EW (2008) Optimal HLA matching in hematopoietic cell transplantation. Curr Opin Immunol 20: 588–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Susal C, Opelz G (2012) Impact of HLA Matching and HLA Antibodies in Organ Transplantation: A Collaborative Transplant Study View. Methods Mol Biol 882: 267–277. [DOI] [PubMed] [Google Scholar]
- 4. van Rood JJ, Oudshoorn M (2008) Eleven million donors in Bone Marrow Donors Worldwide! Time for reassessment? Bone Marrow Transplant 41: 1–9. [DOI] [PubMed] [Google Scholar]
- 5. Cotsapas C, Voight BF, Rossin E, Lage K, Neale BM, et al. (2011) Pervasive sharing of genetic effects in autoimmune disease. PLoS Genet 7: e1002254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rioux JD, Goyette P, Vyse TJ, Hammarstrom L, Fernando MM, et al. (2009) Mapping of multiple susceptibility variants within the MHC region for 7 immune-mediated diseases. Proc Natl Acad Sci U S A 106: 18680–18685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sawcer S, Hellenthal G, Pirinen M, Spencer CC, Patsopoulos NA, et al. (2011) Genetic risk and a primary role for cell-mediated immune mechanisms in multiple sclerosis. Nature 476: 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Raychaudhuri S, Sandor C, Stahl EA, Freudenberg J, Lee HS, et al. (2012) Five amino acids in three HLA proteins explain most of the association between MHC and seropositive rheumatoid arthritis. Nat Genet 44: 291–296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Pereyra F, Jia X, McLaren PJ, Telenti A, de Bakker PI, et al. (2010) The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science 330: 1551–1557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Cozen W, Li D, Best T, Van Den Berg DJ, Gourraud PA, et al. (2012) A genome-wide meta-analysis of nodular sclerosing Hodgkin lymphoma identifies risk loci at 6p21.32. Blood 119: 469–475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. McCormack M, Alfirevic A, Bourgeois S, Farrell JJ, Kasperaviciute D, et al. (2011) HLA-A*3101 and carbamazepine-induced hypersensitivity reactions in Europeans. N Engl J Med 364: 1134–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Illing PT, Vivian JP, Dudek NL, Kostenko L, Chen Z, et al. (2012) Immune self-reactivity triggered by drug-modified HLA-peptide repertoire. Nature 486: 554–558. [DOI] [PubMed] [Google Scholar]
- 13. Chaix R, Cao C, Donnelly P (2008) Is mate choice in humans MHC-dependent? PLoS Genet 4: e1000184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Khankhanian P, Gourraud PA, Caillier SJ, Santaniello A, Hauser SL, et al. (2010) Genetic variation in the odorant receptors family 13 and the mhc loci influence mate selection in a multiple sclerosis dataset. BMC Genomics 11: 626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Dilthey AT, Moutsianas L, Leslie S, McVean G (2011) HLA*IMP–an integrated framework for imputing classical HLA alleles from SNP genotypes. Bioinformatics 27: 968–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Marsh SG (2012) Nomenclature for factors of the HLA system, update February 2012. Tissue Antigens 80: 72–77. [DOI] [PubMed] [Google Scholar]
- 17. Robinson J, Mistry K, McWilliam H, Lopez R, Parham P, et al. (2011) The IMGT/HLA database. Nucleic Acids Res 39: D1171–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. A map of human genome variation from population-scale sequencing. Nature 467: 1061–1073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maiers M, Gragert L, Klitz W (2007) High-resolution HLA alleles and haplotypes in the United States population. Hum Immunol 68: 779–788. [DOI] [PubMed] [Google Scholar]
- 20. Iqbal Z, Caccamo M, Turner I, Flicek P, McVean G (2012) De novo assembly and genotyping of variants using colored de Bruijn graphs. Nat Genet 44: 226–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Major E, Rigo K, Hague T, Berces A, Juhos S (2013) HLA Typing from 1000 Genomes Whole Genome and Whole Exome Illumina Data. PLoS One 8: e78410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cano P, Klitz W, Mack SJ, Maiers M, Marsh SG, et al. (2007) Common and well-documented HLA alleles: report of the Ad-Hoc committee of the american society for histocompatiblity and immunogenetics. Hum Immunol 68: 392–417. [DOI] [PubMed] [Google Scholar]
- 23. Bugawan TL, Klitz W, Blair A, Erlich HA (2000) High-resolution HLA class I typing in the CEPH families: analysis of linkage disequilibrium among HLA loci. Tissue Antigens 56: 392–404. [DOI] [PubMed] [Google Scholar]
- 24. de Bakker PI, McVean G, Sabeti PC, Miretti MM, Green T, et al. (2006) A high-resolution HLA and SNP haplotype map for disease association studies in the extended human MHC. Nat Genet 38: 1166–1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Genome C (2014) Reference Genome Sequence 1000 Genome. Available: ftp://ftp.1000genomes.ebi.ac.uk/vol1/ftp/technical/reference/human_g1k_v37.fasta.gz.
- 26.Genome C (2014) 1000 Genome FTP data release. Available: ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/.
- 27.Genome C (2014) List of samples in phase 1 integrated calls. Available: ftp://ftp-trace.ncbi.nih.gov/1000genomes/ftp/release/20110521/phase1_integrated_calls.20101123.ALL.panel.
- 28. Software B (2014) Beagle formatted datasets. [Google Scholar]
- 29. McElroy JP, Cree BA, Caillier SJ, Gregersen PK, Herbert J, et al. (2010) Refining the association of MHC with multiple sclerosis in African Americans. Hum Mol Genet 19: 3080–3088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, et al. (2012) Database resources of the National Center for Biotechnology Information. Nucleic Acids Res 40: D13–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Meyer D, Single RM, Mack SJ, Lancaster A, Nelson MP, et al. (2007) Single Locus Polymorphism of Classical HLA Genes. In: Hansen JA, editor. Immunobiology of the Human MHC: Proceedings of the 13th International Histocompatibility Workshop and Conference. Seattle, WA: IHWG press. pp. 653–704. [Google Scholar]
- 32.NCBI (2014) DbMHC Immunogenetic portal. Available: http://www.ncbi.nlm.nih.gov/projects/gv/mhc/.
- 33. Browning SR, Browning BL (2010) High-resolution detection of identity by descent in unrelated individuals. Am J Hum Genet 86: 526–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sanchez-Mazas A, Fernandez-Vina M, Middleton D, Hollenbach JA, Buhler S, et al. (2011) Immunogenetics as a tool in anthropological studies. Immunology 133: 143–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Meyer D, Single RM, Mack SJ, Erlich HA, Thomson G (2006) Signatures of demographic history and natural selection in the human major histocompatibility complex Loci. Genetics 173: 2121–2142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gourraud PA, Lamiraux P, El-Kadhi N, Raffoux C, Cambon-Thomsen A (2005) Inferred HLA haplotype information for donors from hematopoietic stem cells donor registries. Hum Immunol 66: 563–570. [DOI] [PubMed] [Google Scholar]
- 37. Gourraud PA, Gilson L, Girard M, Peschanski M (2012) The role of human leukocyte antigen matching in the development of multiethnic “haplobank” of induced pluripotent stem cell lines. Stem Cells 30: 180–186. [DOI] [PubMed] [Google Scholar]
- 38. Thomas R, Apps R, Qi Y, Gao X, Male V, et al. (2009) HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat Genet 41: 1290–1294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Weiss KM, Clark AG (2002) Linkage disequilibrium and the mapping of complex human traits. Trends Genet 18: 19–24. [DOI] [PubMed] [Google Scholar]
- 40. Marchini J, Cutler D, Patterson N, Stephens M, Eskin E, et al. (2006) A comparison of phasing algorithms for trios and unrelated individuals. Am J Hum Genet 78: 437–450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Gragert L, Madbouly A, Freeman J, Maiers M (2013) Six-locus high resolution HLA haplotype frequencies derived from mixed-resolution DNA typing for the entire US donor registry. Hum Immunol 74: 1313–1320. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
MHC region definition. A, Selection of the region by recombination rate et variants' density. B, chr16:86,750,000–91,650,000, a region on chromosome 16 with similar recombination rate as MHC shown in Figure 1D. C, chr16:74,200,000–79,100,000, a region on chromosome 16 with similar variants' density as MHC shown in Figure 1C.
(TIF)
Screen capture of the display of allelic frequencies in dbMHC for the 1000 genome populations. A, Homepage. B, Population selection. C, Data download.
(TIF)
Correlation between sample size and the 90th percentile of D′ (S3-A) and r2 (S3B) in African American samples for markers' pairs at a distance of 400 kb.
(TIF)
Additional details for HLA typing protocol.
(DOCX)
MHC region definitions.
(DOCX)
HLA Allele frequencies in the samples of the 1000 Genomes.
(DOCX)
HLA alleles grouped by similarities in the antigen recognition site.
(DOCX)
The most frequent ancestry specific HLA haplotypes.
(DOCX)
Variants associated with frequent haplotypes in Europeans.
(DOCX)
Data Availability Statement
The HLA genotype data of the present study is available online: The full specification of HLA alleles in the specified release of the HLA nomenclature [17] is provided on the dbMHC portal (See on-line resources) at NCBI [30]. In addition, allele frequencies can be viewed online using tools developed in the anthropology and cell line components of the Histocompatibility Workshops [31](Figure S2, screen capture of the display [32]). Allele frequency tables are extremely sparse, reflecting the high diversity of HLA alleles for all loci and the limited sampling of the HLA alleles in 1000 Genome Project sample sets (Tables S2 and S3 for HLA allele naming convention used, also available online at dbMHC [32]).