Abstract
Two new Penicillium species isolated from plant leaves are reported here, namely, P. fusisporum (type strain AS3.15338T = NRRL 62805T = CBS 137463T) and P. zhuangii (type strain AS3.15341T = NRRL 62806T = CBS 137464T). P. fusisporum is characterized by fast growth rate, apical-swelling monoverticillate penicilli, verrucose stipes, fusiform to oblong conidia about 3.5–4×2–2.5 µm and cinnamon-colored sclerotia. While P. zhuangii presents a moderate growth rate, it also bears apical-swelling monoverticillate penicilli but its stipes are smooth-walled, and produces ovoid to globose smooth-walled conidia about 3–3.5 µm. Both species belong to section Aspergilloides, and P. fusisporum is related to “P. thomii var. flavescens”, while P. zhuangii is morphologically similar to P. lividum. Phylogenetic analyses of sequences of calmodulin and beta-tubulin genes both show that the two new taxa form distinct monophyletic clades.
Introduction
Species of Penicillium Link with vesiculate or apical-swelling monoverticillate conidiophores and moderate to fast growth rate were accommodated in the P. thomii series, P. frequentans series and P. lividum series respectively by Raper and Thom [1]. Pitt [2] included 10 species sharing these characters in series Glabra Pitt of Subgen. Aspergilloides Pitt and regarded P. glabrum as the earlier valid name for the species commonly known as P. frequentans Westling. Moreover, he broadened the concept of P. thomii to encompass 6 synoyms, which was followed by Pitt et al. [3]. Based on ITS1-5.8S-ITS2 and LSU ribosomal DNA (rDNA) sequences, Peterson [4] included 7 members of this series in his Goup 2. Later, Barreto et al. [5] indicated the heterogeneity of the prevailing species concept of P. glabrum and added a new member, P. subericola to series Glabra. Then, Houbraken and Samson [6] established section Aspergilloides Houbraken and Samson based on 4 gene loci to encompass 19 species showing those characters, while other species in Pitt's subgenus Aspergilloides were put in sections Charlesii and Sclerotiora.
In an investigation of phylloplane molds in China, we found many isolates superficially similar to P. glabrum, P. lividum and P. thomii, and thus report here on two new species, P. fusisporum sp. nov. and P. zhuangii sp. nov. belonging to section Aspergilloides.
Materials and Methods
Isolation of strains
Leaf samples were collected from trees and kept in sterilized plastic bags. Isolation of phylloplane fungi followed the methods of Nakase and Takashima [7]. No specific permissions were required for these locations/activities. The field studies did not involve endangered or protected species and the GPS coordinates of the specific locations in our study are 31°45′35″N 110°38′57″E, 29°54′43″N 93°10′43″E and 33°52′54″N 107°47′51″E. Eleven strains with vesciculate or apical-swelling stipes and monoverticillate penicilli were obtained and deposited at the China General Microbiological Culture Collection (CGMCC) of Institute of Microbiology, Chinese Academy of Sciences, Beijing, China. The ex-types culture P. fusisporum AS3.15338T and P. zhuangii AS3.15341T were also deposited at the USDA ARS Culture Collection as NRRL 62805T and NRRL 62806T, CBS-KNAW of The Netherlands as CBS 137463T and CBS 137464T, respectively. All cultures are also maintained at the corresponding author's laboratory and will be supplied upon request for educational or scientific purpose.
Morphological studies
Colony characters were assessed using Czapek agar (Cz, Raper and Thom [1], Czapek yeast autolysate agar (CYA, Pitt [2]), 2% malt extract agar (MEA, malt extract (Difco), Pitt [2), YES (yeast extract sucrose agar, yeast extract (Oxoid), Frisvad and Samson [8], and 25% glycerol nitrate agar (G25N, Pitt [2]). Color names followed Ridgway [9]. Wet mounts were prepared using material from colonies growing on MEA at 25°C after 7 d and mounting in 85% lactic acid without dye. Microscopic examination and photography were performed with a Nikon Eclipse 80i microscope equipped with a Nikon DS-L1 Digital Sight system.
Molecular studies
DNA extraction followed the method of Scott et al. [10]. Partial β-tubulin gene (BenA) sequences were amplified using the sense primers I2 [11] or Bt2a, with the antisense primer Bt2b [12]; the ITS1-5.8S-ITS2 region of rDNA was amplified using the primers ITS5 and ITS4 [13]; the calmodulin gene (CaM) was amplified using the primers of Wang [14]. Polymerase chain reactions (PCR) were carried out in 20 µL reaction mixture containing 0.5 µL of each primer (10 pM/µL), 1.0 µL of genomic DNA (10 ng/µL), 8 µL of 2×PCR MasterMix buffer (0.05 u/µL Taq polymerase, 4 mM MgCl2, 0.4 mM dNTPs), and 10 µL of ultra pure sterile water (Biomed Co. Ltd, Beijng, China). Amplifications were performed in a PTC-150 thermocycler (MJ Research, Watertown, Massachusetts, USA), which was programmed for touch-down PCR (TD PCR) consisting of 94°C for 3 min; 94°C for 30 s, 50°C for 30 s, −0.5°C/cycle, 72°C for 45 s, 20 cycles; 72°C for 5 min; 94°C for 30 s, 40°C for 30 s, 72°C for 45 s, 15 cycles; 72°C for 5 min. After amplification the PCR fragments were electrophoresed in 2.0% agarose gels with a 100 bp DNA ladder (MBI Fermentas) at 80 V for 20 min. Gels were then stained in an aqueous 0.5 µg/mL ethidium bromide water solution for 15 min and examined under 254 nm UV using a portable UV light. Samples showing one single, obvious band of the anticipated length on the gel were then purified and sequenced on both strands with an ABI 3700 DNA analyzer (Tsingke Biotechnologies Co., Ltd., Beijing, China). Raw sequences were proof-read and edited manually with BioEdit 7.0.9 [15]. Edited sequences were aligned using muscle in MEGA version 5 [16]. A sum of 43 strains from section Aspergilloides (Table 1) with validly published sequences from others' work were included. The ITS1-5.8S-ITS2 sequences of P. odoratum NBRC 7741T, “P. trzebinskianum” Abe (nom. inval., Art. 36) NBRC 6038T and P. trzebinskii NBRC 6110T were retrieved from the on-line catalogue of Biological Resource Center (NBRC), NITE of Japan (http://www.nbrc.nite.go.jp/e/), and the ITS1-5.8S-ITS2 sequence of P. trzebinskii CBS 382.48T( = NBRC 6110T) can also be downloaded from the Global Mirror System of DNA Barcode Data (GMS-DBD) (http://nz.boldmirror.net/) as Sample ID KAS3070, Sequence ID PATE046-08. The three sequence matrices of the three loci were analyzed using Maximum Likelihood (ML) method and subjected to 1000 bootstrap replications with Kimura-2 parameter model for CaM and BenA data and General Time Reversible model for ITS1-5.8S-ITS2 datum, and gaps were treated as partial deletion according to Hall [17]. The aligned sequences (Data S1–3) were submitted to TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S14916).
Table 1. Forty-four strains included in phylogenetic analyses and the GenBank accession numbers for three genetic markers.
| Species | Strains* | Source | Genetic markers# | ||
| CaM | BenA | ITS | |||
| Penicillium adametzii K. M. Zaleskii | AS3.4470T = CBS 209.28 T = NRRL 737T | Soil under conifers, Poland | AY678540 | JN625957 | AF033401 |
| P.adametzioides Abe ex G. Smith | CBS 313.59T | Unknown source, Japan | JN686387 | JN799642 | JN686433 |
| P. angulare S. W. Peterson, E. M. Bayer & D. T. Wicklow | CBS 130293T | Mount Wheeler Road, Red River, New Mexico, USA | KC773804 | KC773779 | KC773828 |
| P. bilaiae Chalab. | NRRL 3391T | Soil, Kiev, Ukraine | JN626009 | JN625966 | AF033402 |
| P. charlesii G. Smith | NRRL 778T | Moldy corn (Zea mays), Italy | AY741754 | JX091508 | AY742708 |
| P. expansum Link | CBS 325.48T = NRRL 976T | Apple fruit, USA | DQ911134 | AY674400 | FJ463031 |
| P. fellutanum Biourge | NRRL 746T | Unknown source, USA | AY741753 | EF198545 (NRRL 35619) | AF033399 |
| P. fuscum (Sopp) Biourge | CBS 295.62T = NRRL 3008T | Soil, conifer and hardwood forest, Wisconsin, USA | GQ367539 | GQ367513 | AF033411 |
| P. fusisporum L. Wang, sp. nov. | AS3.15338T | Leaves of Rhododendron sp., Nangongshan Forest Park, Shaanxi, China | KF769413 | KF769400 | KF769424 |
| AS3.15372 | Plant leaves, Gongbujiangda, Linzhi, Tibet, China | KF769417 | KF769404 | KF769428 | |
| P. glabrum Thom | CBS 105.11 | Ex-type of P. frequentans; unknown substrate, Germany | GQ367525 | GQ367501 | GU981567 (CBS 125543T) |
| CBS 229.28 | Ex type of P. paczowskii; soil under conifer, Poland | GQ367531 | GQ367506 | N/A | |
| NRRL 35621 | Cork bark, southern Portugal | EF198575 | EF198547 | N/A | |
| NRRL 35684 | Cork bark, southern Portugal | EF198592 | EF198564 | N/A | |
| AS3.15335 | Leaves of Rhododendron sp., Cona County, Tibet, China | KF302640 | KF302630 | KF302650 | |
| AS3.15336 | Plant leaves, Tiantaishan Forest Park, Shaanxi, China | KF769418 | KF769405 | KF769429 | |
| AS3.15337 | Leaves of Rhododendron sp., Nangongshan Forest Park, Shaanxi, China | KF769419 | KF769406 | KF769430 | |
| AS3.15345 | Leaves of Quercus palustris, Nangongshan Forest Park, Shaanxi, China | KF302645 | KF302635 | KF302655 | |
| P. grancanariae C. Ramírez, A.T. Martínez & Ferrer | CBS 687.77T | Air, Gran Canaria, Spain | GQ367533 | GQ367507 | N/A |
| CBS 336.79 | Ex-type of P. palmense, air, Gran Canaria, Spain | GQ367534 | GQ367508 | N/A | |
| P. herquei Bainier & Satory | CBS 336.48T = NRRL 1040T | A leaf of Agauria pyrifolia, France | JN626013 | JN625970 | AF033405 |
| P. hirayamae Udagawa | NRRL 143 T = CBS 229.60T | Milled Thai rice, Japan | EU021691 | JN625955 | JN626095 |
| P. lividum Westling | IMI 39736 T = NRRL 754T | Unrecorded source, Scotland | DQ911124 | FJ004420 | AF033406 |
| AS3.15334 | Cona County, Tibet, China | KF769420 | KF769407 | KF769431 | |
| P. maximae C.M. Visagie, J. Houbraken & R.A. Samson | NRRL 2060T | Cellulose nitrate coated cellophane, Florida, USA | EU427282 | EU427265 | EU427298 |
| P. purpurascens (Sopp) Biourge | IMI 39745T = CBS 366.48T = NRRL 720T | Soil, Canada | DQ911125 | GQ367512 | AF033408 |
| P. saturniforme (L. Wang & W.Y. Zhuang) Houbraken & Samson | AS3.6886T | Soil, Little Peony Forest Reserve, Dunhua, Jilin Province, China | EU644062 | EU644080 | EU644081 |
| P. sclerotiorum van Beyma | NRRL 2074T | Air, Buitenzorg, Java, Indonesia | JN626044 | JN626001 | JN626132 |
| P. spinulosum Thom | IMI 24316iT = AS3.7980T | Hanover, Germany | DQ911126 | KF769408 | KF769432 |
| CBS 223.28 | N/A | GQ367536 | GQ367509 | N/A | |
| CBS 268.35 | Ex-type of P. mediocre; soil, pine forest; Germany | GQ367527 | GQ367499 | N/A | |
| CBS 271.35 | Ex-type of P. tannophilum; leaf litter,Germany | GQ367530 | GQ367505 | N/A | |
| CBS 289.36 | Ex-type of P. tannophagum; tannin solution,Germany | GQ367528 | GQ367503 | N/A | |
| P. subericola Barreto, Frisvad & Samson | CBS 125096T | N/A | GQ367547 | GQ367521 | N/A |
| CBS 125100 | Dried grapes (sultanas, Vitis vinifera), Mildura, Vic, Australia | GQ369760 | GQ369759 | N/A | |
| P. thomii Maire | IMI 189694T = AS3.7982T | Pine cone, unknown country | DQ911127 | KF769409 | KF769433 |
| “P. thomii var. flavescens” S. Abe | CBS 347.59T | from unrecorded substrate, Japan | GQ367535 | GQ367510 | N/A |
| AS3.15339 | Plant leaves, Tiantaishan Forest Park, Shaanxi, China | KF769414 | KF769401 | KF769425 | |
| AS3.15346 | Leaves of Betula utilis, Heihe Forest Park, Shaanxi, China | KF769415 | KF769402 | KF769426 | |
| AS3.15371 | Plant leaves, Gongbujiangda, Linzhi, Tibet, China | KF769416 | KF769403 | KF769427 | |
| “P. yezoense” Hanzawa in Sasaki & Nakane | CBS 350.59T | Butter, Japan | GQ367548 | GQ367517 | N/A |
| P. species (related to “P. yezoense”) | AS3.15349 | Plant leaves, Tiantaishan Forest Park, Shaanxi, China | KF769421 | KF769410 | KF769434 |
| P. zhuangii L. Wang sp. nov. | AS3.15341T | Leaves of Betula utilis , Heihe Forest Park, Shaanxi, China | KF769422 | KF769411 | KF769435 |
| AS3.15347 | Leaves of Rhododendron sp., Nangongshan Forest Park, Shaanxi, China | KF769423 | KF769412 | KF769436 | |
*AS, China General Microbiological Culture Collection, Academia Sinica, Beijing, China; CBS, Centraalbureau voor Schimmelcultures, Utrecht, The Netherlands; IMI, International Mycological Institute, Surrey, UK; NRRL, Agricultural Research Service Culture Collection, Illinois, USA; ex-type strains are indicated with T.
Sequences KF769400-KF769436 obtained in present study.
Nomenclature
The electronic version of this article in Portable Document Format (PDF) in a work with an ISSN or ISBN will represent a published work according to the International Code of Nomenclature for algae, fungi, and plants, and hence the new names contained in the electronic publication of a PLOS ONE article are effectively published under that Code from the electronic edition alone, so there is no longer any need to provide printed copies.
In addition, new names contained in this work have been submitted to MycoBank from where they will be made available to the Global Names Index. The unique MycoBank number can be resolved and the associated information viewed through any standard web browser by appending the MycoBank number contained in this publication to the prefix http://www.mycobank.org/MB/. The online version of this work is archived and available from the following digital repositories: PubMed Central and LOCKSS.
Results
Phylogenetic delineation of P. fusisporum and P. zhuangii
PCR amplification gave amplicons of CaM about 680 bp, BenA about 680 bp using primers I2 and Bt2b, and ca. 450 bp using primers Bt2a and Bt2b, and ITS1-5.8S-ITS2 about 560 bp. The trimmed alignments of CaM, BenA and ITS1-5.8S-ITS2 sequences were respectively 507, 475 and 536 characters with gaps.
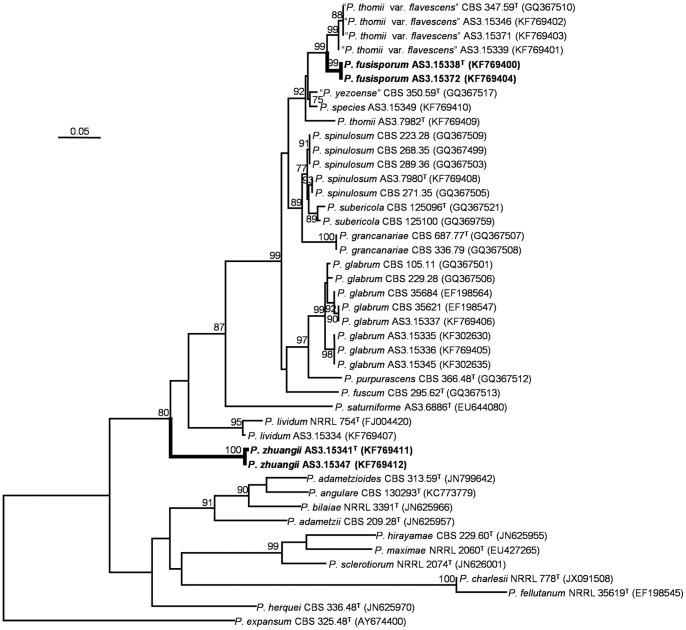
The phylograms resulting from CaM and BenA matrices showed that P. fusisporum was closely related to “P. thomii var. flavescens” with 91% and 99% bootstrap support, respectively. While P. zhuangii was in a clade related to P. lividum with 80% bootstrap support based on the CaM analysis, but in the phylograms based on the BenA and ITS1-5.8S-ITS2 regions, P. zhuangii formed a separate clade without close relatives. The phylogenetic trees generated by the three loci supported them as distinct, monophyletic species (Figures 1–2; Figure S1).
Figure 1. ML phylogram inferred from partial CaM sequences.
Bootstrap percentages over 70% derived from 1000 replicates are indicated at the nodes. Bar = 0.05 substitutions per nucleotide position.
Figure 2. ML phylogram inferred from partial BenA sequences.
Bootstrap percentages over 70% derived from 1000 replicates are indicated at the nodes. Bar = 0.05 substitutions per nucleotide position.
Description of Penicillium fusisporum L. Wang, sp. nov. Figures 3–4
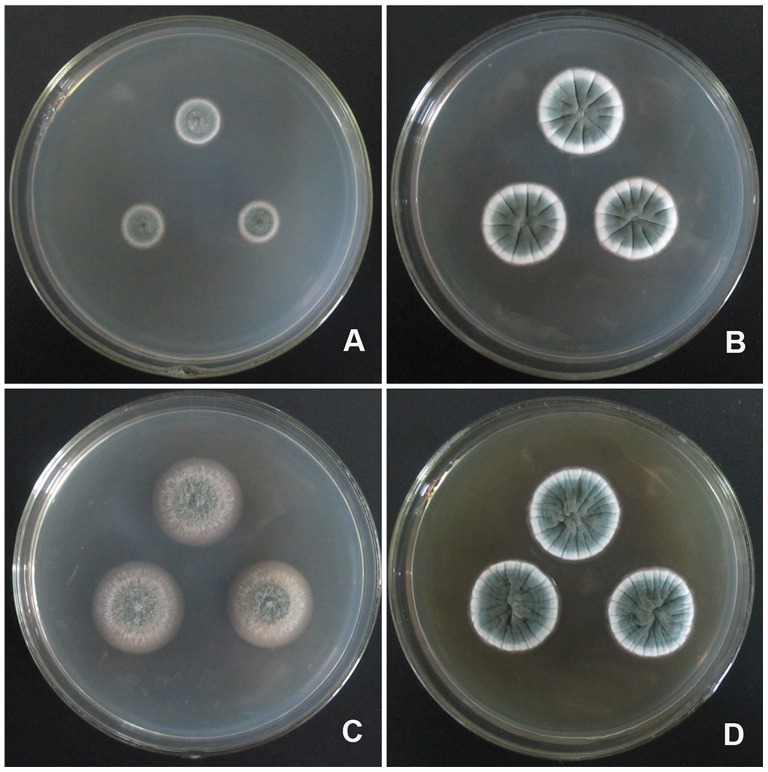
Figure 3. Colonies of P. fusisporum AS3.15338 T incubated 7 d at 25°C.
A, Cz; B, CYA; C, MEA; D, YES.
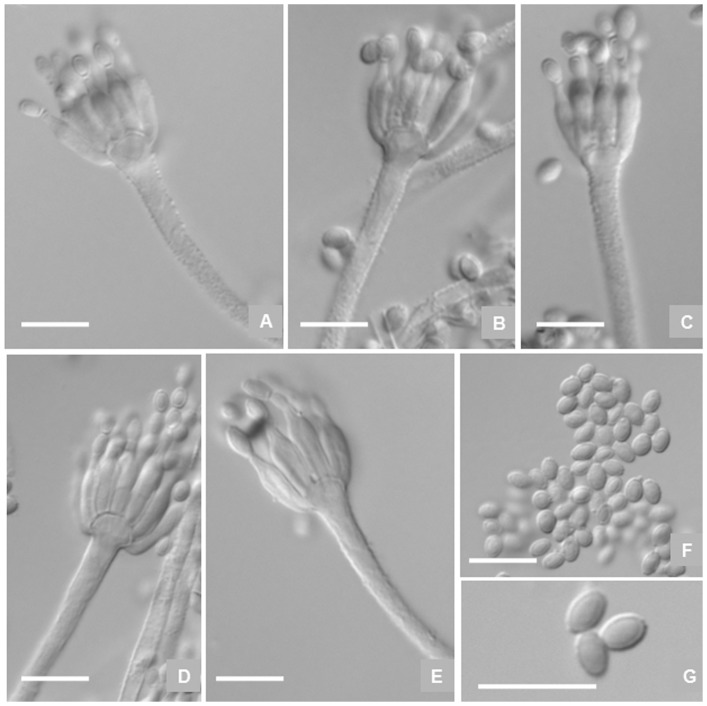
Figure 4. Microscopic characters of P. fusisporum AS3.15338 T.
A–E, Conidiophores; F–G, Conidia. Bar = 10 µm.
[urn:lsid:indexfungorum.org:names: 806119], MycoBank MB 806119
Etymology: The specific epithet is derived from the fusiform shape of its conidia.
Holotype: HMAS 244961
Colonies 33–34 mm diam on Cz at 25°C after 7 d, thin, radially sulcate in central areas, umbonate in centers; sclerotia moderate in centers, white when young, then Cinnamon (R. Pl. XXIX) when mature; velutinous; conidiogenesis moderate to abundant, near Pea Green to Sage Green (R. Pl. XLVII); mycelia white; clear exudate limited; soluble pigment absent; reverse Cinnamon to Verona Brown or Warm Sepia in centers, but showing Pale Pinkish Buff at periphery (R. Pl. XXIX). Colonies 50–53 mm diam on CYA at 25°C after 7 d, thin, umbonate or protuberant in centers, irregularly sulcate; sclerotia moderate in centers, white when immature, then Cinnamon (R. Pl. XXIX); velutinous; conidiogenesis abundant, near Pea Green to Sage Green (R. Pl. XLVII); mycelium white; clear exudate moderate in central areas and no soluble pigment; reverse Verona Brown in centers but Pinkish Buff at periphery (R. Pl. XXIX). Colonies 58–60 mm diam on MEA at 25°C after 7 d, low, plane, slightly protuberant in centers; velutinous; conidiogenesis abundant, near Sage Green (R. Pl. XLVII); no exudate and soluble pigment; reverse Deep Olive-Buff (R. Pl. XL). Colonies 55–57 mm diam on YES at 25°C after 7 d, thin, protuberant or convolute in centers, irregularly sulcate; Cinnamon (R. Pl. XXIX) sclerotia abundant in centers; velutinous; conidiogenesis abundant, Grayish Olive (R. Pl. XLVI); exudate and soluble pigment absent; reverse Colonial Buff (R. Pl. XXX). On G25N at 25°C after 7 d, colonies 7–8 mm diam, thin, radially sulcate moderate; velutinous; with moderate conidiogenesis colored Light Grayish Olive (R. Pl. XLVI); no exudate and soluble pigment; reverse Plae Ochraceous-Salmon ((R. Pl. XV). On CYA at 37°C after 7 d, no growth. On CYA at 5°C after 7 d, germination only.
Sclerotia commonly produced on Cz, CYA and YES, first colored white then Cinnamon when mature, irregular in shape, up to 600 µm in the long axis. Conidiophores arising from agar surface; stipes (90–) 120–200 (−250)×2.5–3.5 µm, verrucose, seldom smooth-walled, apically swelling up to 4–6 µm diam; penicilli monoverticillate, but occasionally with one branch about 12–20 µm long on CYA; phialides (8–) 12–16 per verticil, ampulliform with distinguishable collula, 9–11×2–3 µm; conidia fusiform to oblong, 3.5–4×2–2.5 µm, walls smooth, conidial chains irregularly tangled about 120–180 µm long.
Strains examined
China, Shaanxi, Nangongshan Forest Park, 31°45′35″N 110°38′57″E, 1500 m, from leaves of Rhododendron sp., 6 Oct 2012, coll. P-J Han, leaf samples no. NGS13E, ex-type culture AS3.15338 (Holotype: HMAS 244961 from dried culture of ex-type AS3.15338 on CYA). Tibet, Linzhi, Gongbujiangda, 29°54′43″N 93°10′43″E, 3400 m, from unidentified plant leaves, 12 Aug 2012, coll. P-J Han, leaf samples no. GBJD3D, additional culture AS3.15372.
Notes
This new species is characterized by its fast growth rate, cinnamon-colored sclerotia, apical-swelling monoverticillate conidiophores,verrucose stipes, and fusiform to oblong smooth-walled conidia.
Description of Penicillium zhuangii L. Wang, sp. nov. Figures 5–6
Figure 5. Colonies of P. zhuangii AS3.15341 T incubated 7 d at 25°C.
A, Cz; B, CYA; C, MEA; D, YES.
Figure 6. Microscopic characters of P. zhuangii AS3.15341 T.
A–D, Conidiophores; E–F, Conidia. Bar = 10 µm.
[urn:lsid:indexfungorum.org:names: 805945], MycoBank MB 805945
Etymology: named after Prof. Jian-Yun Zhuang, who made great contribution to the taxonomy of Pucciniales in China.
Holotype: HMAS 244922
Colonies 12–14 mm diam on Cz at 25°C after 7 d, thin, plane, protuberant in central areas; velutinous; conidiogenesis moderate, near Pea Green to Celandine Green (R. Pl. XLVII); mycelium white; no exudate and soluble pigment; reverse Light Cinnamon-Drab to Cinnamon Drab (R. Pl. XLVI). Colonies 23–27 mm diam on CYA at 25°C after 7 d, thin, radially sulcate; velutinous; conidiogenesis moderate, near Pea Green to Andover Green (R. Pl. XLVII); mycelium white; no exudate or soluble pigment; reverse Pale Ochraceous-Buff (R. Pl. XV). Colonies 24–26 mm diam on MEA at 25°C after 7 d, low, plane, margins submerged in agar; velutinous, with sparsely floccose white mycelia overlaid; conidiogenesis limited to moderate, near Grayish Olive (R. Pl. XLVI); no exudate or soluble pigment; reverse Tawny-Olive (R. Pl. XXIX). Colonies 24–26 mm diam on YES at 25°C after 7 d, thin, convolute in centers, radially sulcate and lightly annually plicate; velutinous; conidiogenesis abundant, Andover Green or Pea Green to Celandine Green (R. Pl. XLVII); exudate absent; slightly Ochraceous-Tawny (R. Pl. XV) soluble pigment limited; reverse Tawny to Ochraceous-Tawny (R. Pl. XV). On G25N at 25°C after 7 d, colonies 2–5 mm diam, with limited conidiogenesis colored Light Grayish Olive (R. Pl. XLVI). On CYA at 37°C after 7 d, no growth. On CYA at 5°C after 7 d, colonies 2–6 mm diam, with white mycelium only.
Conidiophores arising from agar surface; stipes (120–) 140–180 (−220)×2.5–3.6 µm, smooth-walled, apically swelling up to 3.5–5.5 µm diam; penicilli exclusively monoverticillate; phialides 8–12 per verticil, ampulliform with distinguishable collula, (7–) 9–13×2.5–3.5 µm; conidia ovoid to globose, 3–3.5 µm, walls smooth, born in irregularly tangled chains about 100–140 µm long.
Strains examined
China, Shaanxi, Zhouzhi Heihe Forest Park, 33°52′54″N 107°47′51″E, 1200 m, from leaves of Betula utilis, 28 Sep 2012, coll. P-J Han, leaf samples no. ZHH20, ex-type culture AS3.15341 (Holotype: HMAS 244922, from dried culture of ex-type AS3.15341 on CYA). Shaanxi: Nangongshan Forest Park, 31°45′35″N 110°38′57″E, 1500 m, from leaves of Rhododendron sp., 6 Oct 2012, coll. P-J Han, leaf samples no. NGS13D, additional culture AS3.15347.
Notes
This new taxon is characterized by its moderate growth rate, apical-swelling monoverticillate conidiophores, smooth-walled stipes, and ovoid to subglobose smooth-walled conidia.
Discussion
The two new taxa reported here should be placed in Penicillium sensu stricto section Aspergilloides in the classification of the most recent phylogenetic scheme by Houbraken and Samson [6].
Raper and Thom regarded the strains with monoverticillate penicilli and hard but brittle sclerotia in pink color as P. thomii. Abe [18] reported a new variety of P. thomii, namely “P. thomii var. flavescens” (nom. inval., Art. 36), which differs from P. thomii by its fast growth rate, abundant Pinkish Cinnamon sclerotia and the yellow-green colony color. Pitt [2] and Pitt et al. [3] broadened the concept of P. thomii to include P. aurantioviolaceum, P. crocicola, P. roseoviride, “P. thomii var. flavescens”, and “P. yezoense” (nom. inval., Art. 36). However in our phylogenies based on CaM and BenA, the type culture CBS 347.59 of “P. thomii var. flavescens” together with our three strains forms a monophyletic lineage well separated from the type culture IMI 189694 of P. thomii, which means that “P. thomii var. flavescens” represents a different species other than P. thomii (Figures 1–2). In addition, “P. thomii var. flavescens” is different from P. thomii in many morphological aspects. For instance, the isolates of “P. thomii var. flavescens” produce both smooth-walled and sparsely granular conidiophores, while those of P. thomii are finely roughened. Moreover, the conidia of “P. thomii var. flavescens” also show subglobose to ovoid shape in addition to ellipsoidal shape, but those of P. thomii are ellipsoidal.
Although isolates of P. fusisporum and “P. thomii var. flavescens” grow rapidly on standard media and produce sclerotia colored near Cinnamon, isolates of “P. thomii var. flavescens” excretes pale yellow-green soluble pigment on Cz and its colony reverse also shows the same tint, but P. fusisporum does not. Besides, the conidial of P. fusisporum colored near Pea Green to Sage Green, instead of Bottle Green or Russian Green in”P. thomii var. flavescens”. Still, the stipe walls of P. fusisporum are commonly verrucose and conidia are fusiform to oblong, whereas, stipe walls of isolates in “P. thomii var. flavescens” are delicately or sparsely granular to smooth and its conidial shape is subglobose or ovoid to ellipsoidal. Our phylogenies based on CaM and BenA both show the kinship between them (91% and 99% bootstrap support respectively), but also indicate that P. fusisporum forms a distinct clade basal to those isolates “P. thomii var. flavescens” (Figures 1–2).
Raper and Thom [1] distinguished P. aurantioviolaceum Biourge from P. thomii for its nonsclerotigenic characters and narrow- ellipsoidal to fusiform conidia, and treated P. roseoviride Stapp and Bortels as the synonym of it. Although Pitt [2] and Pitt et al. [3] did not follow this practice, from the characters described by Raper and Thom [1], we think P. aurantioviolaceum would be regarded as a separate species. Though both P. fusisporum and P. aurantioviolaceum have monoverticillate penicilli with occasionally branched stipes and narrowly ellipsoidal to fusiform conidia, P. fusisporum shows a velutinous colony texture on all standard media and bears abundant sclerotia, while P. aurantioviolaceum gives a loosely velutinous to floccose texture and never produces sclerotia. Moreover, the stipe length of P. fusisporum is much shorter (less than 250 µm), nearly one half of those of P. aurantioviolaceum, which are at least 400 µm, and the stipe walls of P. fusisporum are commonly verrucose, but those of P. aurantioviolaceum are closely and finely echinulate. Furthermore, P. fusisporum produces smooth-walled conidia, whereas, those of P. aurantioviolaceum are predominantly delicately roughened.
Also, Pitt [2] and Pitt et al. [3] relegated P. crocicola as a synonym of P. thomii, but Houbraken and Samson [6] confirmed its species status, and showing its outgroup position to P. patens and “P. thomii” CBS 347.59. While P. fusisporum is unlikely conspecific with P. crocicola, because in our phylogram of ITS1-5.8S-ITS2 (Figure S1) P. fusisporum is much more closely related to “P. thomii var. flavescens” than P. crocicola is, and above all, P. crocicola is the outgroup of P. aurantioviolaceum, “P. thomii var. flavescens”, P. fusisporum, P. purpurascens, P. thomii, and P. trzebinskii Zaleski as well as P. glabrum. Given the fact that ITS1-5.8S-ITS2 and the DNA-dependent RNA polymerase II second largest subunit gene (RPB2) are more evolutionarily conserved than CaM and BenA in penicillia, it could be inferred that P. fusisporum would be much distant to P. crocicola. In addition to the molecular evidence, there are clear differences between P. fusisporum and P. crocicola referring to the translated diagnosis of by Kulik [20]. For example, P. fusisporum produces longer stipes up to 250 µm than those of P. crocicola which are about 106 µm at most. Moreover, the stipe walls of P. fusisporum are verrucose but those of P. crocicola are smooth. The penicilli of P. crocicola are monoverticillate, while P. fusisporum bears sub-terminal branches on CYA. Furthermore, the conidia of P. fusisporum are fusiform to oblong, whereas, those of P. crocicola are subglobose to globose. Still more, the sclerotia of P. fusisporum are about twice longer (up to 600 µm) as those of P. crocicola which are about 320 µm in the long axis.
The Pea Green conidial color, abundant sclerotia and vesiculate monoverticillate penicilli of P. fusisporum also resemble those of P. thomii, however, the difference between them is obvious. Firstly, P. fusisporum grows faster and produces larger sclerotia up to 600 µm long and in a darker color near Cinnamon, while the growth rate of P. thomii is slower, the sclerotia are less than 350 µm long and in Apricot or Salmon shade. Secondly, P. fusisporum occasionally bears one sub-terminal branch on CYA, but those of P. thomii are strictly monoverticillate. Thirdly, the stipe walls of P. fusisporum are apparently verrucose whereas, those of P. thomii are delicately roughened. Still, the new taxon produces fusiform to oblong conidia, nonetheless, P. thomii bears ellipsoidal conidia.
Pit and Hocking [19] reported a new species producing sclerotia and long, roughened stipes bearing monoverticillate penicilli, P. patens Pitt and Hocking. In the phylogram of Houbraken and Samson [6], P. patens was in the clade with”P. thomii” CBS 347.59 with a significant bootstrap support, just like the relationship of P. fusisporum with the four strains of “P. thomii var. flavescens” including CBS 347.59 in our phylograms. Although P. fusisporum and P. patens are alike in colony appearance, P. fusisporum can be readily distinguished from P. patens in many aspects. First, the sclerotia of P. fusisporum are much larger (up to ca. 600 µm of the long axis) than those of P. patens (up to 300 µm in the long axis). Second, P. fusisporum produces much shorter stipes less than 250 µm long, than those of P. patens, which are up to 500–800 µm. Third, the stipe walls of P. fusisporum are verrucose, while those of P. patens are finely to conspicuously roughened, and the stipes are apical-swelling in P. fusisporum but P. patens bears nonvesiculate stipes. Still, though P. fusisporum produces predominantly monoverticillate penicilli, it occasionally bears sub-terminal branches on CYA, whereas, P. patens produces strictly monoverticillate penicilli. Furthermore, the conidial shapes of P. fusisporum are fusiform to oblong, but those of P. patens are ellipsoidal [19]. All the evidence discussed above supported the new species status of P. fusisporum. The comparisons of the key characters of the above species are summarized in Table 2.
Table 2. Comparisons of microscopic characters among P. aurantioviolaceum, P. crocicola, P. fusisporum, P. patens, P. thomii and “P. thomii var. flavescens” *.
| P. aurantioviolaceum | P. crocicola | P. fusisporum | P. patens | P. thomii | “P. thomii var. flavescens” | |
| Sclerotia (µm) | None | Yellow-brown to dark brown, 140–320 | Cinnamon, up to 600 | Colorless to pale brown, 130–200×130–150 | Pinkish to apricot color, 250–350 | Pinkish cinnamon to salmon orange, 190–510×150–410 |
| Conidiophores | ||||||
| Stipes (µm) | Closely and finely echinulate, 400 or more ×2.5–3.5, | Smooth, 32–106×3–4 | Verrucose, seldom smooth-walled, (90–) 120–200 (−250)×2.5–3.5 | Finely to conspicuously roughened, 500–800×2.5–3 | Delicately echinulate, 200–400×2.8–4 | Delicately or sparsely granular, 90–300×2.1–3.4 |
| Apical swellings or vesicles (diam., µm) | 4.5–5 | N/A | 4–6 | None | 4–6 | 3.7×6.2 |
| Penicilli | Strictly monoverticillate | Monoverticillate | Monoverticillate, occasionally with one subterminal branch on CYA | Strictly monoverticillate | Monoverticillate, seldom branched | Strictly monoverticillate |
| Phialides (µm) | 8–10×2.0–2.5 | 7–14×2.5–3.5 | 9–11×2–3 | 8–10×2.5–3 | 8–12×2–3 | 9.3–10.6×1.7–2.5 |
| Conidia (µm) | Strongly ellipsoidal to fusiform, 3–3.5×2–2.5 | Subglobose to globosel, 2.3–3.5×2–3 | Fusiform to oblong, 3.5–4×2–2.5 | Ellipsoidal, 3–3.5×2–2.5 | Ellipsoidal to subglobose, 3–3.5(−4)×2.5–2.8 | Ellipsoidal to subglobose, 2.8–3.6×1.7–2.1 |
| Conidial walls | Delicately roughened | Smooth | Smooth | Smooth | Smooth or finely to conspicuously oughend | Smooth |
*These data were integrated from our observations, Raper and Thom (1949), Abe (1956), Kulik (1968), Pitt (1979), Pitt and Hocking (1985).
The designation of P. zhuangii as a new species is supported by our all phylograms inferred from CaM, BenA and ITS1-5.8S-ITS2 loci (Figures 1–2; Figure S1). The phylogenies inferred from the three loci indicate that P. zhuangii is a well-separated species with no closely related kins. Although CaM sequence datum slightly show that P. zhuangii might be a sibling of P. lividum (with only 80% bootstrap support), it can be discriminated from P. lividum by many morphological characters such as growth rate, conidiophore elements and conidia. The growth rate of P. zhuangii is characteristically slower than those of typical isolates of P. lividum (commonly faster than 30 mm on standard media) and stipe length of P. zhuangii is much shorter, less than 250 µm, than most of P. lividum isolates, which are 400–600 µm. In addition, the stipe walls of the P. zhuangii are exclusively smooth, while some strains of P. lividum show distinctively rough walls. Moreover, P. zhuangii bears only monoverticillate conidiophores, whereas certain strains of P. lividum produce short branches reminiscent of metulae. Furthermore, the conidia of P. zhuangii are smooth-walled, while, those of P. lividum are characteristically roughened.
Pitt [2] and Pitt et al. [3] listed P. odoratum Christensen and Backus and “P. trzebinskianum” as the synonyms of P. lividum, but in the phylogenetic tree of Houbraken and Samson [6], the sister species to P. lividum is P. odoratum. And our phylogeny based on ITS1-5.8S-ITS2 does not show any close relationship between P. zhuangii and P. odoratum and “P. trzebinskianum” (Fig. S1). In addition, P. zhuangii can be distinguished from P. odoratum due to many distinctive characters. First, P. zhuangii shows moderate conidiogenesis and does not emit fruity odor, while P. odoratum generates conidia tardily and giving off strong aromatic odor like apples, though this was regarded as an ephemeral character by Pitt [2]. Second, the conidiophores of P. zhuangii are much shorter (less than 220 µm) than those of P. odoratum which are about 560 µm long, and the stipe walls of P. zhuangii are smooth but those of P. odoratum are roughened. Third, the apical swellings of stipes in P. zhuangii are much smaller (about 5.5 µm) than those of P. odoratum (up to 7–9 µm in diam.). Still, though both species bear ovoid to globose conidia, the conidial walls of P. zhuangii are smooth, yet those of P. odoratum are roughened and showing faintly banded [21]. In addition, P. zhuangii can also be readily distinguished from “P. trzebinskianum” morphologically in that P. zhuangii grows slower and its conidial color is near Pea Green, however “P. trzebinskianum” grows much faster and its conidial color is near Dusky Dull Green. Moreover, P. zhuangii produces smooth-walled conidiopores but those of “P. trzebinskianum” are punctate or granular. Further, the conidia of P. zhuangii are smooth-walled whereas, those of “P. trzebinskianum” are echinulate [20].
Thom and Raper [1] also treated P. trzebinskii, a slowly-growing species with short, monoverticillate conidiophores as a valid taxon other than P. lividum. However, Pitt (1979), relegated it to one synonym of P. spinulosum. The phylogram inferred from ITS1-5.8S-ITS2 in our studies does not only show that P. trzebinskii and P. spinulosum are different species, but also shows that P. zhuangii is far distant to P. trzebinskii (Fig. S1). In addition to this phylogenetic evidence, P. zhuangii can be readily differentiated from P. trzebinskii in that the walls of stipes and conidia are both smooth in P. zhuangii, while those of P. trzebinskii are conspicuously echinulate. Besides, the colony appearance on Cz of P. zhuangii shows a velutinous texture without any overlaid mycelia appearing floccose, but P. trzebinskii does, and the reverse color of colonies in P. zhuangii shows Cinnamon Drab, but that of P. trzebinskii gives deep dull violet to dark fuscous color.
Pitt et al. [3] regarded P. quercetorum Baghdadi as the synonym of P. thomii, but in the phylogram of Houbraken and Samson [6], P. quercetorum is an outgroup species to P. lividum and P. odoratum, and our ITS1-5.8S-ITS2 phylogram does not only indicates that P. quercetorum is much distant to P. thomii but also shows its outgroup position to P. zhuangii (Fig. S1). Though the growth rate and certain microscopic characters of P. quercetorum and P. zhuangii are overlapped, the most striking difference between them is that P. quercetorum produces orange-brown sclerotia, while no sclerotia were produced by P. zhuangii. Additionally, P. quercetorum produces much longer stipes (200–400 µm) than P. zhuangii (less than 220 µm), and it also produces smaller conidia (less than 3.0 µm) than P. zhuangii (3–3.5 µm). According to the above evidence, P. zhuangii should be regarded as a valid new taxon. The comparisons of the key characters of the above species are summarized in Table 3.
Table 3. Comparisons of microscopic characters among P. lividum, P. odoratum, P. quercetorum, “P. trzebinskianum”, P. trzebinskii and P. zhuangii *.
| P. lividum | P. odoratum | P. quercetorum | “P. trzebinskianum” | P. trzebinskii | P. zhuangii | |
| Sclerotia (µm) | None | None | Orange brown, 200–300 | None | None | None |
| Conidiophores | ||||||
| Stipes (µm) | Smooth, 400–600 or more ×2.5–4 | Coarsely roughened, 100–560×3–4 | Smooth, 200–400×3–3.5 | Punctate or granullar, 60–280×2.5–4 | Delicately and conspicuously echinulate, 150–200×1.8–2.5 | Smooth, (120–) 140–180 (−220)×2.5–3.6 |
| Apical swellings or vesicles (diam., µm) | 5–6 | 7–9 | 6 | 3.7–6.9 | N/A | 3.5–5.5 |
| Penicilli | Monoverticillate, occasionally with a branch | Monoverticillate | Strictly monoverticillate | Strictly monoverticillate | Monoverticillate | Exclusively monoverticillate |
| Phialides (µm) | 8–12×2–3 | 9–12×3–4 | 8–12×2.8–3 | 8.7–12.5×2.1–3.2 | 8–10×1.8–2.2 | (7–) 9–13×2.5–3.5 |
| Conidia (µm) | Ellipsoidal to ovoid or subglobose, 3–4×2.6–3 | Ellipsoidal to ovoid or subglobose, 3–4.1×2.2–4 | Spheroidal, 2.8–3 | Ellipsoidal to ovoid or sometimes subglobose, 2.5–3.8×2.3–3.1 | Broadly ellipsoidal to subglobose, 2.5–3.3 | Ovoid to globose, 3–3.5 |
| Conidial walls | Clearly roughened showing spiral banding | Roughened appearing faintly banded | Smooth | Echinulate | Conspicuously echinulate | Smooth |
*These data were integrated from our observations, Raper and Thom (1949), Christensen and Backus (1956), Kulik (1968), Pitt (1979).
Supporting Information
ML phylogram inferred from the ITS1-5.8S-ITS2 sequences. Bootstrap percentages over 70% derived from 1000 replicates are indicated at the nodes. Bar = 0.02 substitutions per nucleotide position. The sequences of P. odoratum NBRC 7741 T, “P. trzebinskianum” NBRC 6038 T and P. trzebinskii NBRC 6110 T were retrieved from the on-line catalogue of Biological Resource Center (NBRC), NITE of Japan (http://www.nbrc.nite.go.jp/e/).
(TIF)
The alignment of CaM sequences.
(FAS)
The alignment of BenA sequences.
(FAS)
The alignment of ITS1-5.8S-ITS2 sequences.
(FAS)
Acknowledgments
Prof. Jian-Yun Zhuang identified the plant samples. Prof. Yi-Jian Yao gave advice on nomenclature.
Data Availability
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. The three aligned sequence data sets are available from TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S14916).
Funding Statement
This work is supported by National Natural Science Foundation of China (NSFC no. 31270539 to LW, http://www.nsfc.gov.cn/). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Raper KB, Thom C (1949) A Manual of the Penicillia. Baltimore: Williams and Wilkins. 875 p. [Google Scholar]
- 2.Pitt JI (1979) The genus Penicillium and its teleomorphic states Eupenicillium and Talaromyces. London: Academic Press. 634 p. [Google Scholar]
- 3.Pitt JI, Samson RA, Frisvad JC (2000) List of accepted species and their synonyms in the family Trichocomaceae. In: Samson RA, Pitt JI, editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Amsterdam: Harwood Academic Publishers. pp. 9–49.
- 4.Peterson SW (2000) Phylogenetic analysis of Penicillium species based on ITS and LSU-rDNA nucleotide sequences. In: Samson RA, Pitt JI, editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Amsterdam: Harwood Academic Publishers. pp. 163–178.
- 5. Barreto MC, Houbraken J, Samson RA, Frisvad JC, San-Romão MV (2011) Taxonomic studies of the Penicillium glabrum complex and the description of a new species P. subericola . Fungal Div 49: 23–33. [Google Scholar]
- 6. Houbraken J, Samson RA (2011) Phylogeny of Penicillium and the segregation of Trichocomaceae into three families. Stud Mycol 70: 1–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Nakase T, Takashima M (1993) A simple procedure for the high frequency isolation of new taxa of ballistosporous yeasts living on the surfaces of plants. RIKEN Rev 3: 33–34. [Google Scholar]
- 8. Frisvad JC, Samson RA (2004) Polyphasic taxonomy of Penicillium subgen. Penicillium. A guide to identification of food and air-borne terverticillate Penicillia and their mycotoxins. Stud Mycol 49: 1–173. [Google Scholar]
- 9.Ridgway R (1912) Color standards and color nomenclature. Washington DC: Published by the author. 53 p. [Google Scholar]
- 10.Scott J, Malloch D, Wong B, Sawa T, Straus N (2000) DNA heteroduplex fingerprinting in Penicillium In: Samson RA, Pitt JI, editors. Integration of modern taxonomic methods for Penicillium and Aspergillus classification. Amsterdam: Harwood Academic Publishers. pp. 225–236.
- 11. Wang B, Wang L (2013) Penicillium kongii, a new terverticillate species isolated from plant leaves in China. Mycologia 105: 1547–1554. [DOI] [PubMed] [Google Scholar]
- 12. Glass NL, Donaldson GC (1995) Development of primer sets designed for use with the PCR to amplify conserved genes from filamentous ascomycetes. App Environ Microbiol 61: 1323–1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MS, Gelfand DH, editors. PCR protocols: a guide to methods and applications. New York: Academic Press. pp. 315–322.
- 14. Wang L (2012) Four new records of Aspergillus section Usti from Shandong Province, China. Mycotaxon 120: 373–384. [Google Scholar]
- 15. Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acids Symp Ser 41: 95–98. [Google Scholar]
- 16. Tamura K, Peterson D, Peterson N, Stecher G, Nei M, et al. (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28: 2731–2739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hall BG (2013) Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol 30: 1229–1235. [DOI] [PubMed] [Google Scholar]
- 18. Abe S (1956) Studies on the classification of the penicillia. J Gen Appl Microbiol 2: 50–51. [Google Scholar]
- 19. Pitt JI, Hocking AD (1985) New species of fungi from Indonesian dried fish. Mycotaxon 22: 197–208. [Google Scholar]
- 20.Kulik MM (1968) A compilation of descriptions of new Penicillium species. Agriculture handbook No. 351. Washington DC: U.S. Agricultural Research Service (U.S. Government Printing Office). 80 p. [Google Scholar]
- 21. Christensen M, Backus MP (1961) New or noteworthy penicillia from Wisconsin soils. Mycologia 53: 451–463. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ML phylogram inferred from the ITS1-5.8S-ITS2 sequences. Bootstrap percentages over 70% derived from 1000 replicates are indicated at the nodes. Bar = 0.02 substitutions per nucleotide position. The sequences of P. odoratum NBRC 7741 T, “P. trzebinskianum” NBRC 6038 T and P. trzebinskii NBRC 6110 T were retrieved from the on-line catalogue of Biological Resource Center (NBRC), NITE of Japan (http://www.nbrc.nite.go.jp/e/).
(TIF)
The alignment of CaM sequences.
(FAS)
The alignment of BenA sequences.
(FAS)
The alignment of ITS1-5.8S-ITS2 sequences.
(FAS)
Data Availability Statement
The authors confirm that all data underlying the findings are fully available without restriction. All relevant data are within the paper and its Supporting Information files. The three aligned sequence data sets are available from TreeBase (http://purl.org/phylo/treebase/phylows/study/TB2:S14916).