Abstract
Breast cancer stem-like cells (BCSC) have been implicated in tumor growth, metastasis, drug resistance and relapse but druggable targets in appropriate subsets of this cell population have yet to be identified. Here we identify a fundamental role for the prolyl isomerase Pin1 in driving BCSC expansion, invasiveness and tumorigenicity, defining it as a key target of miR-200c which is known to be a critical regluator in BSCS. Pin1 overexpression expanded the growth and tumorigenicity of BCSC and triggered epithelial-mesenchymal transition (EMT). Conversely, genetic or pharmaacological inhibition of Pin1 reduced the abundance and self-renewal activity of BCSC. Moreover, moderate overexpression of miR-200c-resistant Pin1 rescued the BCSC defect in miR-200c-expressing cells. Genetic deletion of Pin1 also decreased the abundance and repopulating capability of normal mouse mammary stem cells. In human cells freshly isolated from reduction mammoplasty tissues, Pin1 overexpression endowed BCSC traits to normal breast epithelial cells, expanding both luminal and basal/myoepithelial lineages in these cells. In contrast, Pin1 silencing in primary breast cancer cells isolated from clinical samples inhibited the expansion, self-renewal activity and tumorigenesis of BCSC in vitro and in vivo. Overall, our work demonstrated that Pin1 is a pivotal regulator acting downstream of miR-200c to drive BCSC and breast tumorigenicity, highlighting a new therapeutic target to eradicate BCSC.
Keywords: Pin1, Breast Cancer Stem-like Cells, MiR-200c
Introduction
Breast cancer is the second leading cause of cancer-related death in women in the US, and most deaths are due to cancer metastasis or recurrence. Although cells in a tumor have traditionally been regarded to be biologically homogenous and highly proliferative, it has become evident that breast cancer is a genetically and clinically heterogeneous disease (1, 2). Recent studies suggest that breast cancers follow the cancer stem cell model (2), although the topic is of considerable controversy. In this model, cancer is hierarchically organized into tumorigenic and nontumorigenic components, and only a limited, although not necessarily small, number of cancer stem-like cells (CSCs) or tumor-initiating cells (TICs) can proliferate extensively and give rise to both more CSCs as well as nontumorigenic cancer cells (2). Breast cancer stem-like cells (BCSCs) are thought to be responsible for tumor initiation, progression, metastasis, relapse and drug resistance (2, 3). Thus, the elucidation of regulatory mechanisms of BCSCs and identification of druggable targets to eradicate the BCSC compartment in a tumor may be essential to achieve long-term remission of breast cancer (3).
Recently, miRNAs have been identified as major regulators of BCSCs (4, 5). Notably, miR-200c is downregulated in cancers (6) and strongly inhibits the function of both BCSC and normal stem cells (4). Moreover, miR-200c has further been shown to regulate the BCSC stemness and EMT via the downstream transcription factors Bmi1 and Zeb1/Zeb2 (4, 7, 8). However, so far nothing is known whether miR-200c would have any effects on the regulators of upstream signal pathways in BCSCs.
Protein phosphorylation on certain serine or threonine residues preceding a proline (pSer/Thr-Pro) is a central signaling mechanism in diverse cellular processes, especially cell proliferation and transformation (9). We have previously shown that certain pSer/Thr-Pro motifs exist in two distinct conformations, cis and trans, and identified a unique prolyl isomerase Pin1, which binds to and catalyzes cis/trans isomerization of specific pSer/Thr-Pro motifs, catalytically inducing conformational changes following phosphorylation (10). Such conformational changes can have profound effects on phosphorylation signalling by regulating a spectrum of Pin1 substrate activities, thereby playing an important role in many cellular events (10). Importantly, Pin1 is tightly regulated normally at multiple levels and its deregulation contributes to the pathogenesis of human disease, notably cancer (10).
Pin1 is overexpressed and/or activated in human cancers, including breast cancer, with upregulation being correlated with poor prognosis (11, 12). In contrast, the Pin1 polymorphism that reduces Pin1 expression is associated with reduced risk for multiple cancers including human breast cancer (13, 14). Pin1 activates numerous oncogenes and also inactivates many tumor suppressors (10, 15). Notably, whereas Pin1 overexpression causes cell transformation and tumorigenesis, Pin1 knockdown (KD) inhibits breast cancer cell growth in vitro and in vivo (16, 17). Pin1 knockout (KO) mice fail to undergo massive mammary hyperplasia during pregnancy, and develop widespread premature aging phenotypes (18, 19). Moreover, these mice are fully resistant to breast tumorigenesis induced by overexpressing oncogenes, such as MMTV-Neu/ErbB2 or -Ras (20). Thus, Pin1 is pivotal for the development of breast cancer. However, although Pin1 has been shown to increase protein stability of Nanog in embryonic stem cells (21) and Oct4 in induced pluripotent stem cells (22), so far little is known about its role in BCSCs.
In this paper, we show that as an important target of miRNA-200c, Pin1 plays a pivotal role in driving human BCSCs and tumorigenesis as well as regulating normal mouse mammary stem cells (MaSCs). The clinical significance of these novel findings are further substantiated in human primary normal and cancerous breast tissues by the demonstrations that Pin1 overexpression endows BCSC traits to normal breast epithelial cells, whereas Pin1 knockdown potently inhibits the expansion and tumorigenesis of BCSCs in vitro and in vivo. These results have not only provided novel insight into breast cancer development, but also might have novel therapeutic implications because Pin1 inhibitors, which are being developed actively, might be used to overcome cancer resistance to current therapies.
Materials and Methods
Mice
Pin1 KO mice are in the C57/BL6J background. As Pin1+/− heterozygous mice are indistinguishable from Pin1+/+ mice, we focused on the phenotypes on Pin1−/− mice (19). All studies involving mice were approved by the Institutional Animal Care and Use Committee at Beth Israel Deaconess Medical Center and performed in accordance with the relevant protocol.
Cell Culture
The immortalized human mammary epithelial cells (HMLE) and the transformed cells (HMLE-Ras) were kindly provided by Dr. Robert A. Weinberg, and maintained as described (1). MCF10A mammary epithelial cells were cultured as previously described (2). For PiB treatment, cells were exposed to 1 mM PiB for three days. Freshly sorted primary mouse mammary epithelial cells were cultured in DMEM/F12 medium supplemented with 20 ng/ml of EGF, 10 mg/ml of insulin, 0.5 mg/ml of hydrocortisone, 1% bovine serum albumin, and 2% calf serum (3). Freshly isolated primary normal human MEC or breast cancer cells were cultured in MEGM with supplements (4).
Generation of Stable Cell Lines
For overexpression, Pin1 CDS were subcloned into the pBabe retroviral vector or pBybe lentiviral vector. Specific point mutations were introduced using the Quickchange kit (Stratagene) and sequences were verified. All lentiviral shRNA constructs were provided by Dr. William C. Hahn. The production of retroviruses or lentiviruses as well as the infection of target cells was described previously (5). Following infection, the cells were selected using hygromycin or puromycin. Cells were used immediately following selection and for up to three weeks after selection. Fresh stable cell lines were made before each group of experiments and experiments were performed following at least two separate infections.
MicroRNA Related Analysis
Total RNA was isolated from miRNeasy kit (Qiagen) and reversely transcribed by miScript PCR starter kit. Qiagen's miScript PCR system was used to detect miR-200c and miR-15a transcription. MiR-200c was cloned into pLVX-puro (Clonetech) for inducible expression. Cells were exposed to doxycline at final concentration of 4 μg/ml to induce miR-200c expression.
In vitro microRNA binding assay was performed as described (23). In short, Bluescript plasmid containing the Pin1 3' UTR was used as template in PCR. Forward primer located at the vector. Reverse primer is from the DNA sequence of miR-200c. The parameters for the PCR reaction were: one cycle at 95°C for 5 min; 35 cycles at 95°C for 1 min, 37°C for 1 min, 72°C for 1 min; and a final elongation step at 72°C for 10 min. The PCR products were then visualized with a 1.5% agarose gel stained with ethidium bromide.
For the reporter assay of Pin1 3'UTR by miR-200c, wildtype and mutant Pin1 3'UTR was cloned into a luciferase construct psiCHECK2 (Promega). Cells were harvested and luciferase activity was measured using the Dual-Luciferase Reporter Assay System (Promega).
Flow Cytometry and the ALDEFLUOR Assay
Freshly isolated mouse mammary cells were incubated with biotinylated anti-CD31, CD45, and Ter119 cocktail and the labeled Lin+ cells were removed by EasySep magnet (StemCell Technologies). The Lin− cells were incubated with fluorescence-conjugated antibodies, including CD24-PE, CD29-APC, and CD49f-FITC antibodies (all from Biolegend), as described (24, 25). For human cell lines, CD24-PE and CD44-APC antibodies (eBioscience) were used to fractionate the BCSC-enriched population, as described (26). Isotype antibodies were used as negative controls. Sorting for BCSCs freshly isolated from human breast cancers was performed with Epics Altra flow cytometer (Beckman Coulter). To deplete non-tumor cells from primary cancer samples, a cocktail of lineage marker antibodies including CD2, CD3, CD10, CD16, CD18, CD31, CD64 and CD140b (PharMingen) were used, while to deplete non-tumor cells from mouse specimens, anti-H2 Kd antibody was used.
The ALDEFLUOR kit (StemCell Technologies) was used to isolate the population with a high ALDH enzymatic activity, as described (27). As negative control, for each sample of cells an aliquot was treated with a specific ALDH inhibitor diethylaminobenzaldehyde (DEAB).
Cleared Fat Pad Transplantation
Fresh Lin− MECs from Pin1 KO and WT mice were isolated and injected into cleared inguinal fat pads of 3-week-old syngeneic mice at limiting dilutions (28). Ten weeks after transplantation mammary glands were harvested and processed for whole mount staining. An outgrowth is defined as a branched structure comprising multiple ducts emanating from a central point, with lobules and terminal end buds (24, 25).
Primary Human Specimens
All studies involving human subjects were approved by the Institutional Review Board at Beth Israel Deaconess Medical Center or Sun Yat-Sen Memorial Hospital, and performed in accordance with the relevant protocol. Normal tissues were from two cases of reduction mammoplasty. Tumors were from surgical resections of eight patients with breast carcinomas (Supplementary Table). All patients received no treatment before surgery.
Tumor Implantation and Serial Transplantation Assay
Aliquots of indicated numbers of HMLER cells were injected into 5-week-old BALB/c nude mice (Jackson Laboratories), as described (28). The tumor incidence was monitored by palpation and determined at two months after injection, with the same tumor incidence at 6 months postinjection. After tumors were detected, tumor size was measured every three days.
Lin−CD24−CD44+ cells were sorted from eight breast cancer specimens and cultured as single cell suspension in ultra-low attachment dishes, and then infected with lentivirus expressing control vector or Pin1 shRNA. After puromycin selection, 2,000 transduced cells from each patient were injected into the mammary fat pads of 5-week-old nude mice. For serial passaging, cells from the primary tumors were sorted again for Lin−CD24−CD44+ cells. Among the 6 primary tumors formed in the shCtrl group, four tumors were randomly selected and passaged into eight mice (two mice per tumor). For the one tumor formed from shPin1 cells, tumor cells were injected into eight mice for serial passaging. The same procedure was applied to the second passage of xenograft cells. The size of tumors was measured every 3 d by calipers, and tumor volumes were calculated as Volume (mm3) = L × W2 × 0.4, as described (5).
Statistical Analysis
All data are presented as the means ± SD, followed by determining significant differences using the two-tailed t test or ANOVA test. Limiting dilution data were analyzed by the single-hit Poisson model using a complementary log-log generalized linear model (29) with L-Calc Software (Stemcell Technologies). All tests of significance were set at P < 0.05.
Results
Pin1 is a major miR-200c downstream target in regulating BCSCs
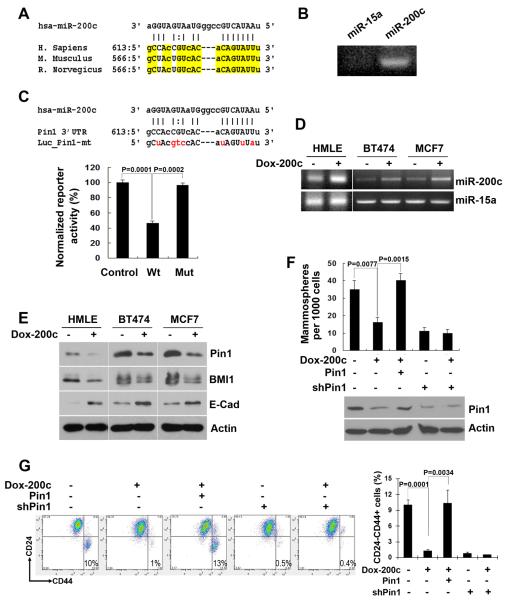
MiR-200c has been reported to regulate the self-renewal and tumorigenicity of BCSCs (4). Interestingly, we found Pin1 might be a potential target for miR-200c, by using the miRNA target prediction programs PicTar (30) and TargetScan (31) to search for miRNA binding sites in the Pin1 mRNA sequence. We found evolutionarily highly conserved binding sites for miR-200c in the 3' untranslated region (UTR) of Pin1 (Fig. 1A), suggesting a possible biological significance. To test if miR-200c would indeed target Pin1, we first performed a PCR-based miRNA binding assay, as described (23). We found that miR-200c bound to Pin1 3'UTR and generated expected PCR products of about 100 bp, while miR-15a, a control miRNA that is not expected to bind to Pin1 3'UTR, did not produce any PCR products (Fig. 1B). Regulation of Pin1 by miR-200c was further evaluated by luciferase reporter assay, as described (4). Luciferase constructs bearing wild-type or mutated miR-200c binding site of Pin1 3'UTR were cotransfected with miR-200c expression construct. We observed that miR-200c suppressed the luciferase activity of the vector with wild-type Pin1 3'UTR by about 50%, while mutation of miR-200c seed region in Pin1 3'UTR abolished the regulating effects of miR-200c on Pin1 3'UTR (Fig. 1C). A random fragment of E-cadherin coding region of approximately 500 bp containing no miR-200c binding site was cloned into luciferase construct to serve as control.
Figure 1. Pin1 is a major miR-200c target in regulating BCSCs.
A, bioinformatic analyses point Pin1 as a promising target of miR-200c. The seed-pairing target sites of miR-200c within the 3'UTR of Pin1 are evolutionary conserved across mammals as highlighted. Capitalized letters are conserved binding nucleotides that directly interact with miR-200c.
B, miR-200c binds to the Pin1 3' UTR. In vitro miRNA binding assay showed that miR-200c binds to Pin1 3'UTR, forming a PCR product of the expected size, while control miR-15a does not generate any visible products.
C, miR-200c reduces expression of the Pin1 3'UTR. Luciferase constructs bearing an unrelated fragment (Control), Pin1 3'UTR (WT), or Pin1 3'UTR containing mutated binding site of miR-200c (Mut) were co-transfected with miR-200c. Results showed that miR-200c reduces luciferase activity by 50%, but its inhibition was abolished when miR-200c binding site on Pin1 3'UTR was mutated. Mutated sequences are shown in red.
D, tetracycline-inducible miR-200c expression in HMLE, BT474 and MCF7 cells. Cells were stably infected with lentiviruses expressing tetracycline-inducible miR-200c, followed by confirming miR-200c expression using RT-PCR after doxycycline treatment for 48 hrs. Expression of miR-15a was monitored to serve as a control.
E, inducible miR-200c expression downregulates Pin1 and BMI1 expression, but upregulates E-Cadherin expression, as detected by immunoblotting analysis. Actin serves as loading control.
F and G, Pin1 is a major miR-200c target in BCSCs. Expression of miR-200c-resistant Pin1 fully rescues the ability of inducible miR-200c expression, to inhibit the mammosphere-forming capability (F) and to reduce the CD24−CD44+ population (G) in HMLE cells. Either Pin1 knockdown or inducible miR-200c expression inhibits the mammosphere-forming capability (F) and reduces the CD24−CD44+ population (G) in HMLE cells, and these effects remain the same when two are combined together, indicating that they both act in the same pathway.
In all panels, bar graphs present mean±SD of three independent experiments.
To confirm that miR-200c indeed regulates Pin1 expression, we generated tetracyclineinducible miR-200c-expressing lentiviruses, followed by stable infection of immortalized human breast epithelial HMLE cells and breast cancer cell lines BT474 and MCF7. The expression of miR-200c with or without the inducer doxycycline was confirmed by RT-PCR, with miR-15a being using as a control (Fig. 1D). Protein expression of Pin1 was monitored together with BMI1 and E-cadherin, which are downstream molecules of miR-200c (4, 8), by western blot. Both Pin1 and BMI1 level decreased whereas E-cadherin expression increased specifically after the induction of miR-200c by doxycycline treatment (Fig. 1E). These results together indicate that Pin1 is indeed a miR-200c downstream target.
Given that miR-200c regulates BCSCs (4), we next examined whether Pin1 is a key mediator for miR-200c in this function. We stably and moderately overexpressed a miR-200c-resistant Pin1 coding sequence using a retroviral construct in HMLE cells that already stably expressed tetracycline-inducible miR-200c. As expected, induction of miR-200c expression by addition of tetracycline reduced BCSC-enriched CD24−CD44+ population by ~ 10 folds and mammosphere formation, a property associated with mammary stem cells, by about 50%. These stem cell phenotypes were fully restored by expression of miR-200c-resistant Pin1 (Fig. 1F and 1G). Moreover, miR-200c expression in Pin1 KD cells could not further decrease CD24−CD44+ population or mammosphere forming capacity, suggesting that Pin1 is a functionally critical target of miR-200c for conferring stem cell traits. Thus, Pin1 is a key miRNA-200c downstream gene in regulating BCSCs.
Pin1 overexpression potently drives the expansion and tumorigenicity of BCSCs
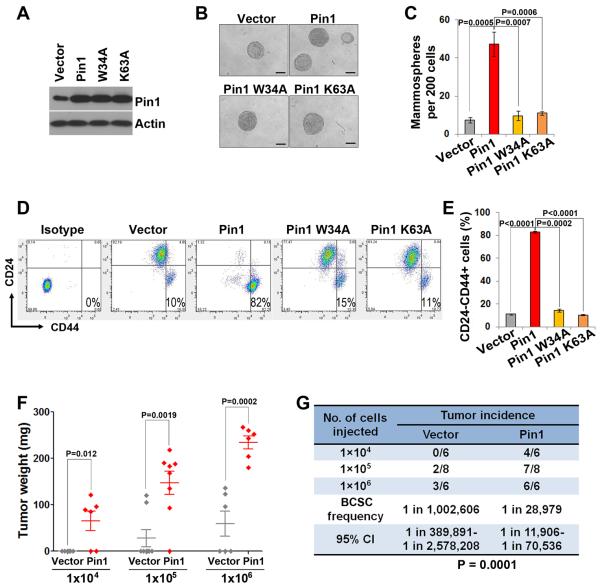
We have previously shown that Pin1 is commonly overexpressed in human breast cancer tissues and cell lines (11, 12). To directly examine the role of Pin1 in BCSCs, we first carried out gain-of-function experiments in HMLEs, which have been immortalized by serial transfection with hTERT and SV40 (32) and widely used to study BCSCs. We stably infected HMLE cells with retroviruses expressing Pin1, which moderately overexpressed Pin1 at 2–3 times above the endogenous level in HMLE cells (Fig. 2A). Comparing to HMLEs expressing empty vector, Pin1-overexpressing cells formed more and bigger mammospheres (Fig. 2B and 2C). Moreover, Pin1 overexpression drastically increased the population of BCSC-enriched CD24−CD44+ cells by 8–9 folds above that of the vector control infected HMLE cells (Fig. 2D and 2E). To confirm the BCSC properties of the CD24−CD44+ population in Pin1-expressing HMLE cells, we sorted the CD24−CD44+ and non-CD24−CD44+ fractions from Pin1-expressing HMLE cells. The CD24−CD44+ cells formed mammospheres efficiently, whereas the non-CD24−CD44+ fraction barely formed mammospheres (data not shown). Importantly, the promoting effects of Pin1 on BCSCs were dependent upon its prolyl isomerase function because the Pin1 point mutants either in the WW domain (W34A) or the PPIase domain (K63A), which cannot bind to or isomerize pSer/Thr-Pro motifs, respectively (17), failed to increase the mammosphere formation or the CD24−CD44+ population (Fig. 2B–2E), as shown for many other known Pin1 cellular functions (10). Thus, moderate Pin1 overexpression in HMLE cells results in enrichment of cells with BCSC properties.
Figure 2. Pin1 overexpression potently drives the expansion and tumorigenicity of BCSCs in HMLEs.
A, moderate and stable overexpression of Pin1, and its W34A mutant and K63A mutant in HMLE cells using retrovirus-mediated gene transfer, assessed by immunoblot.
B and C, overexpression of Pin1, but not its W34A or K63A mutant increased mammosphere-forming activity in HMLE cells. Scale bars, 100 μm.
D and E, overexpression of Pin1, but not its mutants, in HMLE cells potently induced expansion of BCSCs, as assayed by FACS analysis of the BCSC-enriched CD24−CD44+ population.
F and G, Pin1 overexpression increases tumorigenicity of BCSCs. Transformed HMLE (HMLE-Ras) cells stably infected with control vector and Pin1 were injected into subcutaneous sites of nude mice in limiting dilutions. Two months later, mice were sacrificed and evaluated for tumor weight (F) and frequency (G). Pin1-overexpressing HMLE-Ras cells exhibited significantly much higher tumor incidence and grew much faster than control cells.
In all panels, error bars represent SD.
To assess whether the gain of BCSC properties of Pin1-overexpressing HMLE cells could enhance tumorigenicity, we performed tumor-seeding experiments at limiting dilutions in nude mice. We overexpressed Pin1 in HMLER cells, HMLE cells transformed with V12H-Ras, which is needed to enable Snail or Twist-overexpressing HMLE cells to form tumors in nude mice (28, 32). Pin1-overexpressing and control HMLER cells were injected in limiting dilutions subcutaneously in nude mice. 1×104 Pin1-expressing HMLER cells formed tumors in 4 out of 6 mice, while no tumors formed when an equal number of cells expressing a control vector were injected into mice (Fig. 2F and 2G). In fact, 105 of control cells were required to initiate tumors, and even then, only 2 of 8 injected hosts develop tumors. However, 105 of Pin1-overexpressing cells formed tumors in 7 out of 8 mice. Hence, Pin1 overexpression potently increases the BCSC frequency over 30 folds in HMLER cells (P= 0.0001). Thus, moderate Pin1 overexpression potently drives the expansion and tumorigenicity of BCSCs in vitro and in vivo.
Moderate Pin1 overexpression in HMLEs induces epithelial-mesenchymal transition
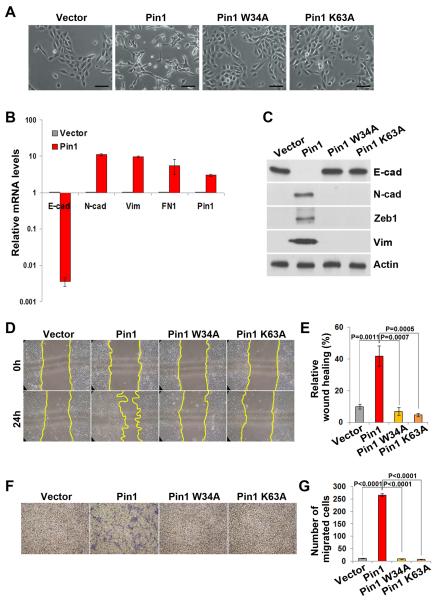
Overexpression of Twist or Snail in HMLE cells also induces an epithelial to mesenchymal transition (EMT), which may be linked to BCSC properties (28). Given the effects of Pin1 overexpression on BCSCs, we examined whether Pin1 might activate EMT. Pin1-overexpressing cells, which had a much higher percentage of the CD24−CD44+ population and mammosphere-forming activity, developed a fibroblast-like appearance, suggesting a transition to a mesenchymal phenotype (Fig. 3A). Neither W34A nor K63A Pin1 point mutant induced such morphological changes, consistent with their failure to promote BCSC properties (Fig. 3A). To confirm that Pin1-overexpressing cells have undergone EMT, we analyzed epithelial and mesenchymal markers using qRT-PCR and western blot. Indeed, Pin1 overexpression drastically downregulated mRNA levels of epithelial markers, such as E-Cadherin, but upregulated expression of mesenchymal markers, such as N-Cadherin, vimentin, and fibronectin (Fig. 3B). These results were further confirmed by the findings that Pin1-overexpressed cells had decreased protein levels of E-Cadherin, increased protein levels of N-Cadherin, Zeb1 and vimentin, whereas W34A or K63A mutants had no effect (Fig. 3C)(28). Moreover, ectopic Pin1, but not W34A or K63A mutant expression caused an increase in cell migration, a property associated with EMT, as measured by wound healing assay (Fig. 3D and 3E) and transwell assay (Fig. 3F and 3G). Thus, moderate overexpression of Pin1, but not its inactive mutants, potently co-induces BCSC and EMT properties, as did Twist or Snail (28).
Figure 3. Pin1 overexpression potently induced EMT in HMLEs.
A, HMLE cells overexpressing Pin1, but not its mutants, showed a fibroblast-like, mesenchymal morphology. Scale bars, 100 μm.
B, overexpression of Pin1 induced the downregulation of E-cadherin mRNA and upregulation of N-cadherin, fibronectin and vimentin mRNA, determined by real-time RT-PCR. GAPDH expression was used to normalize the variability in template loading.
C, overexpression of Pin1, but not its mutants, downregulated E-cadherin protein expression and upregulated N-cadherin, Zeb1 and vimentin protein expression.
D and E, overexpression of Pin1, but not its mutants, increased cell migration capacity of HMLEs, as determined by wound-healing migration assay. Cells migrating into wounds were monitored by time-lapse microscopy, with images captured at the indicated times after wounding (C).
F and G, overexpression of Pin1, but not its mutants, increased cell migration in the transwell assay. Quantified were the numbers of cells that transversed the transwell membranes (F).
In all panels, bar graphs present mean±SD of three independent experiments.
To further confirm the role of Pin1 in EMT, we silenced Pin1 expression using shRNA in MCF7 cells (Supplementary Fig. S2). The control MCF7 cells displayed an elongated spindle shape, whereas Pin1 KD cells exhibited more epithelial-like cobblestone morphology (Supplementary Fig. S2A). Pin1 KD also increased the expression of E-Cadherin and decreased the expression of Zeb1 and Vimentin (Supplementary Fig. S2B). Therefore, Pin1 is critical for the EMT process in breast cancer cells.
Pin1 inhibition by chemical compound, shRNA or miRNA suppresses the BCSC-enriched population
Given the dramatic effects of Pin1 overexpression on promoting BCSC expansion, we wondered whether endogenous Pin1 is required for maintaining the BCSC population. To address this question, we first inhibited Pin1 function by treating HMLE cells with PiB, a chemical compound that selectively inhibits the parvulin family of prolyl isomerases that include Pin1 (33). A 4~5 time reduction of the CD24−CD44+ population was detected 72 h after PiB treatment (Fig. 4A and 4C). We then evaluated the ALDH+ population, which is enriched in normal stem cells, luminal progenitor cells and/or BCSCs, using the ALDEFLUOR assay (27). PiB treatment also significantly reduced ALDH+ population by 4 times in HMLE (Supplementary Fig. S1A). We also treated MCF10A cells and found PiB treatment led to about 4 times decrease of CD24−CD44+ population and over one half lost of ALDH+ population (data not shown). The ability of PiB to inhibit the BCSC population is consistent with the above findings that the catalytically inactive Pin1 mutant fails to regulate BCSCs (Fig. 2B–2E).
Figure 4. Pin1 inhibition suppresses the expansion of BCSC-enriched population.
A, the treatment of HMLE cells with PiB, a Pin1 chemical inhibitor, reduced the CD24−CD44+ population.
B, Pin1 KD in HMLE cells, as confirmed by immunoblotting analysis, reduced the CD24−CD44+population.
C, bar graph showed decreased percentage of CD24−CD44+ in PiB-treated and Pin1 KD HMLEs shown in A and B.
D and E, Pin1 KD in HMLE cells reduced mammosphere-forming activity, showing smaller and fewer mammospheres than the controls. Data present mammospheres formed by 1000 cells. Scale bars, 100 μm.
F and G, doxycycline-induced expression of miR-200c or Pin1 shRNA decreased the abundance of CD24−CD44+ cells in BT474 and MCF7, which was rescued by expression of miR-200c-resistant Pin1.
H, doxycycline-induced expression of miR-200c or Pin1 shRNA decreased the mammosphere-forming activity in BT474 and MCF7, which was rescued by expression of miR-200c-resistant Pin1.
In all panels, bar graphs present mean±SD of three independent experiments.
Given that PiB is not a very potent or specific Pin1 inhibitor; it can also inhibit Par14, another member of the parvulin family (33), we used Pin1 KD to confirm the role of endogenous Pin1 in sustaining the population of BCSCs. Pin1 was effectively and stably silenced using lentiviruses expressing a validated Pin1 shRNA in HMLE, BT474 and MCF7 cells (Fig. 4B and Supplementary Fig. S1B), as shown (34). Importantly, silencing Pin1 reduced the size of CD24−CD44+ population about 30~50 times in HMLE (Fig. 4B and 4C) and about 2~6 times in BT474 and MCF7 cells (Fig. 4F and 4G). Similar results were observed in ALDH+ population as well (Supplementary Fig. S1A). Consistently, Pin1 KD cells formed fewer and smaller mammospheres than the control cells in HMLE (Fig. 4D and 4E) and BT474 and MCF7 cells (Fig. 4H). Thus, either chemical or genetic inhibition of Pin1 potently decreases the BCSC-enriched population.
To investigate the effects of Pin1 inhibition by miR-200c in breast cancer cells, we examined the CD24−CD44+ population and mammosphere formation in BT474 and MCF7 cells infected with tetracycline-inducible miR-200c lentiviruses. As shown in HMLE cells (Fig. 1F and 1G), induction of miR-200c expression by adding tetracycline decreased Pin1 expression (Supplementary Fig. S1B) and reduced CD24−CD44+ population and mammosphere formation in these breast cancer cell lines (Fig. 4F–4H). To further confirm that Pin1 mediates the BCSC effects of miR-200c in breast cancer cells, we moderately expressed the miR-200c-resistant Pin1 coding sequence using a retroviral construct in BT474 and MCF7 cells that already stably expressed tetracycline-inducible miR-200c (Supplementary Fig. S1B), as did in HMLEs (Fig. 1F). Indeed, moderate expression of miR-200c-resistant Pin1 fully rescued the BCSC phenotypes inhibited by miR-200c expression in both BT474 and MCF7 cells (Fig. 4F–4H). Thus, Pin1 is a critical mediator of miRNA-200c to regulate BCSCs in breast cancer.
Pin1 knockout decreases the abundance and repopulating capability of normal mouse mammary stem cells
BCSCs share some characteristics and regulatory pathways with normal mammary stem cells (MaSCs) (1, 4, 27). Expression of genes that modulate stem cells is also associated with poor prognosis in cancer, suggesting that CSCs may require stem cell functions for tumor initiation, growth, and/or metastasis (35, 36). Therefore, we evaluated whether Pin1 regulates normal MaSCs.
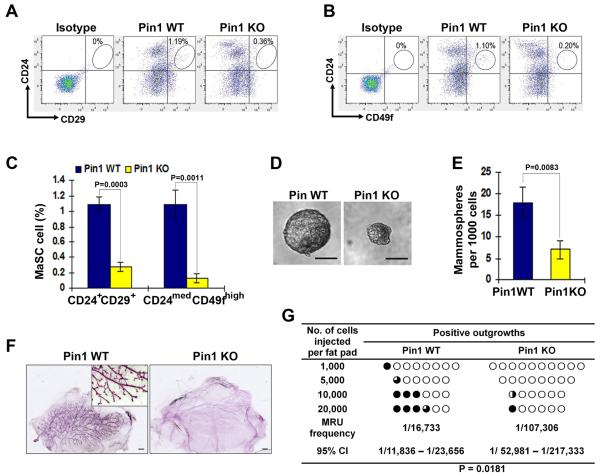
To address this question, we first performed flow cytometry analysis of mammary epithelial cells (MECs) isolated from 10-week old Pin1 KO and wild-type (WT) littermates at the proestrus stage of the estrous cycle for cell surface markers, Lin−CD24+CD29+ and Lin−CD24medCD49fhi, which have been widely used to enrich for stem cell populations in mammary tissues (24, 25). MaSC-enriched Lin−CD24+CD29+ and Lin−CD24medCD49fhi populations were dramatically reduced in mammary glands in Pin1 KO mice, as compared with those in Pin1 WT littermates (Fig. 5A–5C). Moreover, mammosphere formation assays also showed that Pin1 KO Lin− MECs formed fewer and smaller mammospheres than WT controls (Fig. 5D and 5E).
Figure 5. Pin1 knockout decreases the abundance and repopulating capability of mouse MaSCs.
A and B, Pin1 KO in mice reduced the MaSCs-enriched population, as assayed by FACS analyses of the CD24+CD29+ (A) and CD24medCD49fhi (B) fraction in the Lin− MECs isolated from WT and Pin1 KO littermates.
C, bar graph showed decreased percentage of CD24+CD29+ and CD24medCD49fhi cells in three Pin1 KO mice, comparing to WT littermates.
D and E, Pin1 KO in mice reduced mammosphere-forming activity, showing smaller (D) and fewer (E) mammospheres than the controls. Scale bars, 100 μm.
F and G, Pin1 KO in mice reduced the repopulating capability of MaSCs. Representative whole mount image showed carmine-stained outgrowths formed by 1,000 Pin1WT or KO Lin− MECs transplanted into cleared fat pads of virgin recipient mice (K). The frequency of mammary repopulating unit (MRU) in Lin− MECs from Pin1WT or KO littermates was measured by the limiting dilution analysis (G). The circles represent the transplanted fat pads. The dark areas in circles represent the percentages of the reconstitute outgrowth within fat pads. Scale bars, 1 mm. In all panels, bar graphs present mean±SD of three independent experiments.
Although mammosphere assays are a powerful surrogate method to evaluate stem cells, not all mouse-derived mammosphere-forming cells contain regenerative stem cell activity. Therefore, we performed functional limiting dilution transplantation experiments to determine the effects of Pin1 deletion on the repopulating capability of MaSCs, as described (24, 25). Freshly dissociated Lin− Pin1 WT or KO MECs were transplanted into cleared fat pads of syngeneic mice at decreasing cell numbers. Pin1 KO had dramatically decreased repopulating capability (Fig. 5F and 5G). Based on a single-hit Poisson distribution, the mammary repopulating unit (MRU) was determined to be one MRU per 16,733 cells in Pin1 WT MECs, while the frequency decreased about 6-fold in Pin1 KO cells (Fig. 5G). Importantly, mammary fat pad reconstitution was severely impaired in outgrowths of Pin1 KO MECs (Fig. 5F and 5G). These in vitro and in vivo results together indicate that Pin1 deletion in MECs leads to reduced MaSC frequency and repopulating activity. These results are consistent with our previous findings that mammary epithelial cells in Pin1 KO MECs fail to undergo massive proliferation during pregnancy in mice (19), a major function of MaSCs (24, 25).
Pin1 promotes the expansion of BCSC-enriched populations, as well as basal/myoepithelial and luminal progenitors in primary normal human MECs
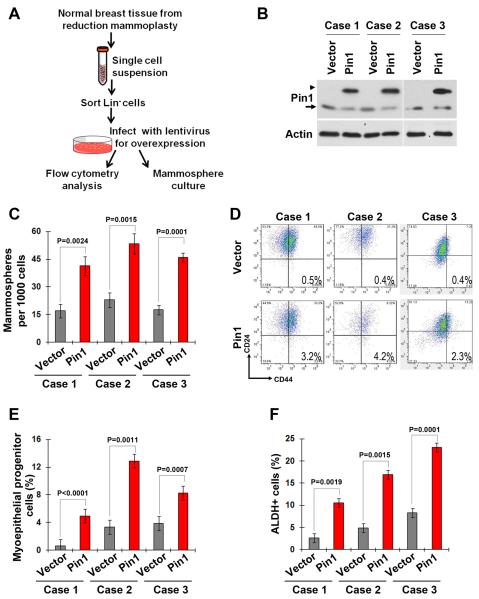
To extend our findings in cell lines and animal models, we test the influence of Pin1 expression on the stem cell-enriched population in normal primary human MECs. We first sorted Lin−MECs isolated from reduction mammoplasty tissues, and then infected them with lentiviruses expressing Flag-Pin1 or control vector (Fig. 6A and 6B). Pin1-overexpressing cells showed increased mammosphere formation in all of the three cases tested (Fig. 6C). Moreover, Pin1 overexpression led to 6–10 fold increase in the CD24−CD44+ population (Fig. 6D and Supplementary Fig. S3A). Thus, Pin1 confers stem cell-related properties on normal human MECs.
Figure 6. Pin1 promotes the expansion of BCSC-enriched populations, as well as basal/myoepithelial and luminal progenitors in primary human MECs.
A, schematic of the experiments on normal human MECs from reduction mammoplasty tissues.
B, western blot showed lentivirus-mediated overexpression of Flag-Pin1 in three cases of human normal Lin-MECs. Arrowhead, exogenous Flag tagged protein; Arrow, endogenous protein.
C, Pin1 overexpression increased the mammosphere formation in primary human MECs. Bar graph presents mean±SD of three independent experiments.
D, Pin1 overexpression increased the CD24−CD44+ population in primary human MECs.
E, Pin1 overexpression increased the basal/myoepithelial progenitor-enriched population in primary human MECs, as fractionated as MUC1-EpCAM-CD10+CD49f+ population.
F, Pin1 overexpression increased the luminal progenitor-enriched population in primary human MECs, as analyzed by the ALDH assay.
Bar graphs present mean±SD of three independent experiments.
Increasing evidence shows that human mammary epithelium is organized in a hierarchical manner. Taking advantage of the fact that primary MECs may contain the cell types of both luminal and basal/myoepithelial lineages, we attempt to determine the effects of Pin1 overexpression on these two lineages. We found that Pin1-overexpressing cells have higher percentage of the EpCAM− MUC1− CD10+ CD49f+ population (Fig. 6E and Supplementary Fig. S3B), which enriched basal/myoepithelial progenitors (37). As ALDH activity is also a marker of luminal progenitors (37), we measured the ALDH+ population in these primary MECs and found that Pin1 overexpression increased the abundance of ALDH+ cells (Fig. 6F and Supplementary Fig. S3C). These data suggest that Pin1 not only promotes the expansion of stem cell populations, but also the basal/myoepithelial and luminal progenitors.
Pin1 is required to sustain tumorigenic potential of human primary BCSCs
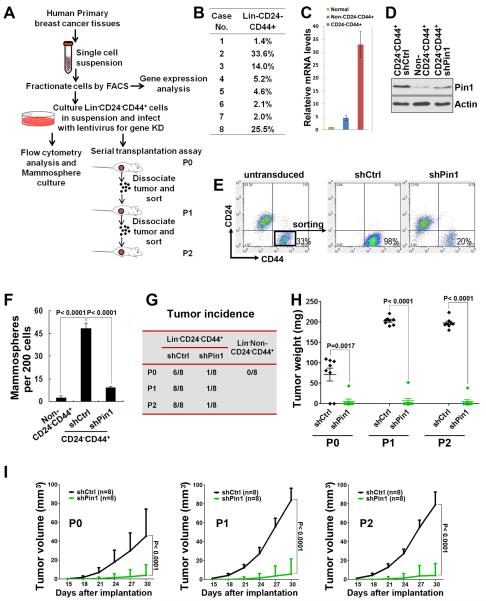
In above experiments, we have demonstrated that endogenous Pin1 is required for the BCSC maintenance in cell lines (Fig. 4). To further assess whether Pin1 is critical for the tumorigenesis of BCSCs in primary breast cancers, we sorted Lin−CD24−CD44+ cells from freshly isolated human breast cancer cells of eight patients (Fig. 7A, 7B and Supplementary Table), and then evaluated the impact of Pin1 on BCSCs in vitro and in vivo.
Figure 7. Pin1 regulates the expansion and tumorigenicity of human primary BCSCs.
A, schematic of the experiments on freshly isolated primary human BCSCs.
B, Lin−CD24−CD44+ cells were sorted from breast cancer tissues of eight patients, with percentage ranged from 1.4–33.6%.
C, real-time PCR showed that expression of Pin1 mRNA was markedly increased in the Lin−CD24−CD44+ population, comparing to the Lin−Non-CD24−CD44+ or normal epithelial cells.
D, western blot showed upregulated Pin1 expression in the BCSC-enriched population and the knockdown of Pin1 in the Lin−CD24−CD44+ population isolated from primary human breast cancer cells.
E, Pin1 KD decreased the CD24−CD44+ population. Lin−CD24−CD44+cells sorted from primary human breast cancers were infected with lentivirus expressing control or Pin1 shRNA, and then analyzed for the CD24−CD44+ population.
F, Pin1 KD decreased the mammosphere formation in Lin−CD24−CD44+ cells isolated from primary human breast cancer.
G–I, Pin1 KD interfered with both tumor initiation and growth of primary BCSCs in vivo, as shown by tumor incidence (G), tumor weights (H) and growth curve (I). 2,000 lentivirus transduced Lin−CD24−CD44+ cells freshly isolated from eight breast cancer patients were serially transplanted as xenografts into eight nude mice. P0, freshly isolated primary cells; P1, passage 1; P2, passage 2.
In all panels, error bars represent SD.
We first examined the expression of Pin1 in Lin−CD24−CD44+, Lin−non-CD24−CD44+ cancer cells and normal MECs from patients. Comparing with those in normal MECs, Pin1 mRNA levels were ~ 5 times higher in Lin-non-CD24-CD44 + cancer cells and over ~30 times higher in BCSC-enriched Lin−CD24−CD44+cells in case 2 (Fig. 7C). Pin1 protein was also markedly upregulated in the Lin−CD24−CD44+cells infected with shCtrl, compared to non-CD24−CD44+cells in this case (Fig. 7D). This upregulation in BCSC-enriched population was consistent with the role of Pin1 in promoting the BCSC expansion.
Given that Pin1 is highly expressed in the BCSC-enriched population, next we tested whether endogenous Pin1 was required to maintain the BCSC population in the primary breast cancer by transducing Lin−CD24−CD44+ primary breast cancer cells with lentivirus expressing Pin1 or control shRNA. Pin1 was efficiently silenced after three days of puromycin selection (Fig. 7D). As we cultured the sorted CD24−CD44+ cells in ultra-low attachment dishes, the cells infected with control shRNA still had a high percentage of CD24−CD44+ cells after selection (Fig. 7E), as shown (5). However, this population was significantly reduced in Pin1 KD cells, being only 1/5 of the control cells in case 2 (Fig. 7E). Pin1 KD also significantly decreased the mammosphere-forming activity of the CD24−CD44+ cells in this case (Fig. 7F). Thus, Pin1 plays an important role in sustaining the BCSC properties in human primary breast cancer cells.
We lastly investigated whether Pin1 was required for the tumorigenicity of the BCSC-enriched Lin−CD24−CD44+ population. We injected 2,000 control and Pin1 shRNA transduced Lin−CD24−CD44+ cells, or Lin−non-CD24−CD44+ cells isolated from eight breast cancer patients into eight nude mice, using the same procedure as that described previously (5). While no tumors developed in mice injected with the cells that were not CD24−CD44+, 2,000 control cells from same patients generated six tumors in eight injected mice (P0 tumors) (Fig. 7G). However, lentivirus-mediated KD of Pin1 not only drastically reduced tumor incidence (Fig. 7G), but also potently reduced tumor growth, as measured by tumor volumes and weights (Fig. 7H and 7I). We further dissociated the tumors and sorted again for CD24−CD44+ cells for the serial transplantation. We randomly selected four P0 shCtrl tumors, and passaged each one into two nude mice for the next generation of xenografts (P1). For the one P0 shPin1 tumor, we passaged it into eight mice for P1 xenografts. When 2,000 control cells were passaged in nude mice, they could be serially transplanted at least for two more passages (P1 and P2) without reduced tumorigenicity (Fig. 7G), as described (5). However, 2,000 Pin1 KD cells had substantially decreased frequency of tumor formation and reduced tumor growth through passages (Fig. 7H and 7I). Thus, expression of Pin1 is highly enriched in primary human BCSCs and silencing Pin1 strongly interferes with the expansion and tumorigenesis of human primary BCSCs in vitro and in vivo.
Discussion
We uncover that as an important target suppressed by miR-200c, Pin1drives the expansion, invasiveness and tumorigenicity of human BCSCs, as well as enhances the abundance and repopulating capacity of normal mouse MaSCs. Expression of Pin1 is highly enriched in human primary BCSCs, and overexpression of Pin1 endows BCSC traits normal human primary breast epithelial cells, whereas KD of either gene potently inhibits human primary BCSCs and tumorigenesis. Thus, miR-200c/Pin1 signals a pivotal pathway that regulates BCSCs and offers promising new drug targets in BCSCs.
MiR-200c has been shown to regulate CSC function and EMT through repressing Bmi1, Zeb1/Zeb2 (4, 7, 8). Here we showed that miR-200c directly bound to a highly conserved region in the 3' UTR of Pin1. Mutations in this region abolished the repressing effect of miR-200c on Pin1 transcription. Moreover, in the presence of miR-200c overexpression, recovering Pin1 expression by a miR-200c-resistant construct could fully rescue the BCSC phenotypes repressed by miR-200c, such as mammosphere formation and CD24−CD44+ population size. Consistently, Pin1 KD could completely mimic the BCSC phenotypes, which were induced by miR-200c overexpression. In primary human breast cancer samples, we have shown that Pin1 expression markedly increased in the BCSC-enriched population, which is consistent with the finding that miR-200c is specifically downregulated in BCSCs in comparison to non-tumorigenic cancer cells (4). These data demonstrate that Pin1 is a functionally important target of miR-200c in BCSC maintenance.
Pin1 is overexpressed and plays a critical role in the development of breast cancer, but its role in BCSCs is unknown. We found that Pin1 overexpression dramatically increased the BCSC-enriched CD24−CD44+ population and mammosphere formation, as well as induced EMT and enhanced tumorigenic potential in human breast cell lines, whereas inhibiting Pin1 by chemical inhibitor or gene silencing potently decreased the BCSC-enriched population and mammosphere formation. The significance of these findings is further substantiated by the demonstrations that Pin1 expression was highest in BCSCs among human primary breast tumor cells or normal breast epithelial cells, that Pin1 overexpression in primary human normal MECs increased the CD24−CD44+ population and mammosphere formation, and that Pin1 KD in primary human breast cancer cells reduced the population, mammosphere formation and tumorigenesis of BCSCs. Thus, Pin1 drives the expansion and tumorigenesis of BCSCs not only in human breast cell lines, but also in primary human normal and breast cancer cells.
We have demonstrated that Pin1 KO reduced the abundance and repopulating activity of normal MaSC-enriched populations in mice, indicating that Pin1 is also required to sustain MaSCs in normal mammary glands. It's worth noting that CSCs share many characteristics of normal stem cells (35, 36). The tightly regulated process of normal stem cell expansion is dysregulated in CSCs due to transforming events, resulting in an unrestricted expansion of self-renewing cells in cancer. Emerging evidence suggests that BCSCs are likely regulated by some important components present in normal stem cells (4, 27). The miRNA expression profile of BCSCs and normal mammary stem cells is remarkably similar (4). MiR-200c is poorly expressed in both normal and tumorigenic stem cells and inhibits the function of both normal MaSCs and BCSCs (4), consistent with our findings that Pin1 promote the expansion of normal MaSCs and BCSCs but is inhibited by MiR-200c. Moreover, expression of normal human MaSC signature correlates with high grade breast cancers, which also have higher frequencies of BCSCs (1). Our data suggest that Pin1 is a pivotal regulator shared by both MaSCs and BCSCs.
The concept of CSCs has important therapeutic implications because current therapies have been developed to decrease tumor size and, albeit they may produce dramatic responses, are not likely to result in stable, long-lasting remission if the rare CSCs are not targeted too. In this regard, Pin1 may offer a promising target for cancer therapy, because Pin1 not only promote the growth of regular breast cancer cells, but also the expansion and tumorigenicity of BCSCs. The following properties make Pin1 a particularly attractive candidate as a new anticancer target (10). Firstly, it is an enzyme with high substrate specificity and a well-defined active site structure (38). Secondly, Pin1 is often overexpressed and/or activated in human cancers and its expression strongly correlates with poor patient outcome (12, 39). Reducing Pin1 expression by SNP is associated with reduced cancer risk for a wide range of cancers, including for breast cancer (13, 14, 40, 41). Whereas Pin1 overexpression causes cell transformation and tumorigenesis (16, 17), Pin1 KD inhibits tumor growth in vitro and in vivo (42). Moreover, Pin1 KO mice, which develop normally, are fully resistant to tumorigenesis induced by MMTV-Neu/ErbB2 or –Ras (20). Thirdly, Pin1 activates numerous oncogenes and also inactivates a large number of tumor suppressors (10). As a results, inhibiting Pin1 may have the unique and desired feature to block many other oncogenic pathways as well as to restore the function of tumor suppressors. Lastly and most importantly, Pin1 drives the expansion, invasiveness and tumorigenicity of BCSCs. The inhibitory effects of Pin1 inhibition on BCSCs were not only demonstrated in cell lines, but also verified in freshly isolated primary human breast cancer cells, suggesting that Pin1 inhibitors may have the potential to restrict or even eradicate CSCs. Thus, Pin1 inhibitors, which are under active development (33, 38, 43–50), might have the unique properties to inhibit the growth of regular breast cancer cells, and also to suppress the expansion and tumorigenicity of BCSCs, which are resistant to current therapies.
Supplementary Material
Acknowledgments
We are very grateful to Robert Weinberg for providing expert advice and critical reagents, and sending his lab members to perform mouse transplantation experiments, and also thank William C. Hahn for reagents and/or advice, and members of Lu/Zhou laboratories for constructive discussions.
Grant Support C.H.C. is a DOD Breast Cancer Research Program Postdoctoral Fellow and a NIH T32 training grant awardee. D.Y.L. is a Human Frontier Science Program Long Term Fellow. This work was supported by a Komen for Cure grant to X.Z.Z., Ministry of Science and Technology of China 973 project to E.S., and NIH grants (CA167677, AG039405, DA031663) and National Natural Science Foundation of China grant (U1205024) to K.P.L.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Authors' Contributions Conception and design: M.L. Luo, X. Z. Zhou, K.P. Lu
Development of methodology: M.L. Luo, C. Gong, W. Guo, F. Reinhardt
Acquisition of data (provided animals, acquired and managed patients, provided facilities, etc.): M.L. Luo, C. Gong, C.H. Chen, D.Y. Lee, P. Huang, Y. Yao, G. Wulf, E. Song
Analysis and interpretation of data (e.g., statistical analysis, biostatistics, computational analysis): M.L. Luo, H. Hu, D.Y. Lee, P. Huang
Writing, review, and/or revision of the manuscript: M.L. Luo, H. Hu, J. Lieberman, K.P. Lu
Administrative, technical, or material support (i.e., reporting or organizing data, constructing databases): X. Z. Zhou
Study supervision: X. Z. Zhou, K.P. Lu
Supplementary Information Supplementary Information includes Supplementary Methods, Supplementary References, Supplementary Tables and two Supplementary Figures.
Reference
- 1.Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, et al. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 2010;140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- 2.Stingl J, Caldas C. Molecular heterogeneity of breast carcinomas and the cancer stem cell hypothesis. Nat Rev Cancer. 2007;7:791–9. doi: 10.1038/nrc2212. [DOI] [PubMed] [Google Scholar]
- 3.Liu S, Wicha MS. Targeting breast cancer stem cells. J Clin Oncol. 2010;28:4006–12. doi: 10.1200/JCO.2009.27.5388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shimono Y, Zabala M, Cho RW, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138:592–603. doi: 10.1016/j.cell.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yu F, Yao H, Zhu P, Zhang X, Pan Q, Gong C, et al. let-7 regulates self renewal and tumorigenicity of breast cancer cells. Cell. 2007;131:1109–23. doi: 10.1016/j.cell.2007.10.054. [DOI] [PubMed] [Google Scholar]
- 6.Adam L, Zhong M, Choi W, Qi W, Nicoloso M, Arora A, et al. miR-200 expression regulates epithelial-to-mesenchymal transition in bladder cancer cells and reverses resistance to epidermal growth factor receptor therapy. Clin Cancer Res. 2009;15:5060–72. doi: 10.1158/1078-0432.CCR-08-2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol. 2008;10:593–601. doi: 10.1038/ncb1722. [DOI] [PubMed] [Google Scholar]
- 8.Park SM, Gaur AB, Lengyel E, Peter ME. The miR-200 family determines the epithelial phenotype of cancer cells by targeting the E-cadherin repressors ZEB1 and ZEB2. Genes Dev. 2008;22:894–907. doi: 10.1101/gad.1640608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Blume-Jensen P, Hunter T. Oncogenic kinase signalling. Nature. 2001;411:355–65. doi: 10.1038/35077225. [DOI] [PubMed] [Google Scholar]
- 10.Lu KP, Zhou XZ. The prolyl isomerase PIN1: a pivotal new twist in phosphorylation signalling and disease. Nat Rev Mol Cell Biol. 2007;8:904–16. doi: 10.1038/nrm2261. [DOI] [PubMed] [Google Scholar]
- 11.Bao L, Kimzey A, Sauter G, Sowadski JM, Lu KP, Wang DG. Prevalent overexpression of prolyl isomerase Pin1 in human cancers. Am J Pathol. 2004;164:1727–37. doi: 10.1016/S0002-9440(10)63731-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wulf GM, Ryo A, Wulf GG, Lee SW, Niu T, Petkova V, et al. Pin1 is overexpressed in breast cancer and cooperates with Ras signaling in increasing the transcriptional activity of c-Jun towards cyclin D1. EMBO J. 2001;20:3459–72. doi: 10.1093/emboj/20.13.3459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Han CH, Lu J, Wei Q, Bondy ML, Brewster AM, Yu TK, et al. The functional promoter polymorphism (−842G>C) in the PIN1 gene is associated with decreased risk of breast cancer in non-Hispanic white women 55 years and younger. Breast Cancer Res Treat. 2010;122:243–9. doi: 10.1007/s10549-009-0682-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu HR, Xu ZF, Sun YL, Han JJ, Li ZJ. The −842G/C Polymorphisms of PIN1 Contributes to Cancer Risk: A Meta-Analysis of 10 Case-Control Studies. PLoS ONE. 2013;8:e71516. doi: 10.1371/journal.pone.0071516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Finn G, Luo ML, Zhou XZ, Lu KP. Pin1: A promising novel diagnostic and therapeutic target that acts on numerous cancer-driving pathways. In: Atta-ur-Rahman, editor. Curr Cancer Drug Targets. 2013. pp. 146–71. [Google Scholar]
- 16.Ryo A, Liou YC, Wulf G, Nakamura M, Lee SW, Lu KP. PIN1 is an E2F target gene essential for Neu/Ras-induced transformation of mammary epithelial cells. Mol Cell Biol. 2002;22:5281–95. doi: 10.1128/MCB.22.15.5281-5295.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suizu F, Ryo A, Wulf G, Lim J, Lu KP. Pin1 regulates centrosome duplication, and its overexpression induces centrosome amplification, chromosome instability, and oncogenesis. Mol Cell Biol. 2006;26:1463–79. doi: 10.1128/MCB.26.4.1463-1479.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee TH, Tun-Kyi A, Shi R, Lim J, Soohoo C, Finn G, et al. Essential role of Pin1 in the regulation of TRF1 stability and telomere maintenance. Nat Cell Biol. 2009;11:97–105. doi: 10.1038/ncb1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liou YC, Ryo A, Huang HK, Lu PJ, Bronson R, Fujimori F, et al. Loss of Pin1 function in the mouse causes phenotypes resembling cyclin D1-null phenotypes. Proc Natl Acad Sci U S A. 2002;99:1335–40. doi: 10.1073/pnas.032404099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. EMBO J. 2004;23:3397–407. doi: 10.1038/sj.emboj.7600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Moretto-Zita M, Jin H, Shen Z, Zhao T, Briggs SP, Xu Y. Phosphorylation stabilizes Nanog by promoting its interaction with Pin1. Proc Natl Acad Sci U S A. 2010;107:13312–7. doi: 10.1073/pnas.1005847107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nishi M, Akutsu H, Masui S, Kondo A, Nagashima Y, Kimura H, et al. A distinct role for Pin1 in the induction and maintenance of pluripotency. J Biol Chem. 2011;286:11593–603. doi: 10.1074/jbc.M110.187989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee DY, Shatseva T, Jeyapalan Z, Du WW, Deng Z, Yang BB. A 3'-untranslated region (3'UTR) induces organ adhesion by regulating miR-199a* functions. PLoS ONE. 2009;4:e4527. doi: 10.1371/journal.pone.0004527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shackleton M, Vaillant F, Simpson KJ, Stingl J, Smyth GK, Asselin-Labat ML, et al. Generation of a functional mammary gland from a single stem cell. Nature. 2006;439:84–8. doi: 10.1038/nature04372. [DOI] [PubMed] [Google Scholar]
- 25.Stingl J, Eirew P, Ricketson I, Shackleton M, Vaillant F, Choi D, et al. Purification and unique properties of mammary epithelial stem cells. Nature. 2006;439:993–7. doi: 10.1038/nature04496. [DOI] [PubMed] [Google Scholar]
- 26.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ginestier C, Hur MH, Charafe-Jauffret E, Monville F, Dutcher J, Brown M, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–67. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, et al. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133:704–15. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bonnefoix T, Bonnefoix P, Callanan M, Verdiel P, Sotto JJ. Graphical representation of a generalized linear model-based statistical test estimating the fit of the single-hit Poisson model to limiting dilution assays. J Immunol. 2001;167:5725–30. doi: 10.4049/jimmunol.167.10.5725. [DOI] [PubMed] [Google Scholar]
- 30.Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, et al. Combinatorial microRNA target predictions. Nat Genet. 2005;37:495–500. doi: 10.1038/ng1536. [DOI] [PubMed] [Google Scholar]
- 31.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 32.Elenbaas B, Spirio L, Koerner F, Fleming MD, Zimonjic DB, Donaher JL, et al. Human breast cancer cells generated by oncogenic transformation of primary mammary epithelial cells. Genes Dev. 2001;15:50–65. doi: 10.1101/gad.828901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Uchida T, Takamiya M, Takahashi M, Miyashita H, Ikeda H, Terada T, et al. Pin1 and Par14 peptidyl prolyl isomerase inhibitors block cell proliferation. Chem Biol. 2003;10:15–24. doi: 10.1016/s1074-5521(02)00310-1. [DOI] [PubMed] [Google Scholar]
- 34.Nakamura K, Greenwood A, Binder L, Bigio EH, Denial S, Nicholson L, et al. Proline isomer-specific antibodies reveal the early pathogenic tau conformation in Alzheimer's disease. Cell. 2012;149:232–44. doi: 10.1016/j.cell.2012.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A, et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet. 2008;40:499–507. doi: 10.1038/ng.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merlos-Suarez A, Barriga FM, Jung P, Iglesias M, Cespedes MV, Rossell D, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–24. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 37.Visvader JE. Keeping abreast of the mammary epithelial hierarchy and breast tumorigenesis. Genes Dev. 2009;23:2563–77. doi: 10.1101/gad.1849509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, Daum S, Wildemann D, Zhou XZ, Verdecia MA, Bowman ME, et al. Structural basis for high-affinity peptide inhibition of human Pin1. ACS Chem Biol. 2007;2:320–8. doi: 10.1021/cb7000044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ayala G, Wang D, Wulf G, Frolov A, Li R, Sowadski J, et al. The prolyl isomerase Pin1 is a novel prognostic marker in human prostate cancer. Cancer Res. 2003;63:6244–51. [PubMed] [Google Scholar]
- 40.Lu J, Hu Z, Wei S, Wang LE, Liu Z, El-Naggar AK, et al. A novel functional variant (−842G>C) in the PIN1 promoter contributes to decreased risk of squamous cell carcinoma of the head and neck by diminishing the promoter activity. Carcinogenesis. 2009;30:1717–21. doi: 10.1093/carcin/bgp171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lu J, Yang L, Zhao H, Liu B, Li Y, Wu H, et al. The polymorphism and haplotypes of PIN1 gene are associated with the risk of lung cancer in Southern and Eastern Chinese populations. Hum Mutat. 2011;32:1299–308. doi: 10.1002/humu.21574. [DOI] [PubMed] [Google Scholar]
- 42.Ryo A, Uemura H, Ishiguro H, Saitoh T, Yamaguchi A, Perrem K, et al. Stable suppression of tumorigenicity by Pin1-targeted RNA interference in prostate cancer. Clin Cancer Res. 2005;11:7523–31. doi: 10.1158/1078-0432.CCR-05-0457. [DOI] [PubMed] [Google Scholar]
- 43.Dong L, Marakovits J, Hou X, Guo C, Greasley S, Dagostino E, et al. Structure-based design of novel human Pin1 inhibitors (II) Bioorg Med Chem Lett. 2010;20:2210–4. doi: 10.1016/j.bmcl.2010.02.033. [DOI] [PubMed] [Google Scholar]
- 44.Guo C, Hou X, Dong L, Dagostino E, Greasley S, Ferre R, et al. Structure-based design of novel human Pin1 inhibitors (I) Bioorg Med Chem Lett. 2009;19:5613–6. doi: 10.1016/j.bmcl.2009.08.034. [DOI] [PubMed] [Google Scholar]
- 45.Liu T, Liu Y, Kao HY, Pei D. Membrane permeable cyclic peptidyl inhibitors against human Peptidylprolyl Isomerase Pin1. J Med Chem. 2010;53:2494–501. doi: 10.1021/jm901778v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Potter A, Oldfield V, Nunns C, Fromont C, Ray S, Northfield CJ, et al. Discovery of cell-active phenyl-imidazole Pin1 inhibitors by structure-guided fragment evolution. Bioorg Med Chem Lett. 2010;20:6483–8. doi: 10.1016/j.bmcl.2010.09.063. [DOI] [PubMed] [Google Scholar]
- 47.Potter AJ, Ray S, Gueritz L, Nunns CL, Bryant CJ, Scrace SF, et al. Structure-guided design of alpha-amino acid-derived Pin1 inhibitors. Bioorg Med Chem Lett. 2010;20:586–90. doi: 10.1016/j.bmcl.2009.11.090. [DOI] [PubMed] [Google Scholar]
- 48.Wildemann D, Erdmann F, Alvarez BH, Stoller G, Zhou XZ, Fanghanel J, et al. Nanomolar inhibitors of the peptidyl prolyl cis/trans isomerase Pin1 from combinatorial peptide libraries. J Med Chem. 2006;49:2147–50. doi: 10.1021/jm060036n. [DOI] [PubMed] [Google Scholar]
- 49.Zhang Y, Fussel S, Reimer U, Schutkowski M, Fischer G. Substrate-based design of reversible Pin1 inhibitors. Biochemistry (Mosc) 2002;41:11868–77. doi: 10.1021/bi0262395. [DOI] [PubMed] [Google Scholar]
- 50.Moore JD, Potter A. Pin1 inhibitors: Pitfalls, progress and cellular pharmacology. Bioorg Med Chem Lett. 2013;23:4283–91. doi: 10.1016/j.bmcl.2013.05.088. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.