Abstract
Randomization and blocking have the potential to prevent the negative impacts of non-biological effects on molecular biomarker discovery. Their use in practice, however, has been scarce. To demonstrate the logistic feasibility and scientific benefits of randomization and blocking, we conducted a microRNA study of endometrial tumors (n=96) and ovarian tumors (n=96) using a blocked randomization design to control for non-biological effects; we profiled the same set of tumors for a second time using no blocking or randomization. We assessed empirical evidence of differential expression in the two studies. We performed simulations through virtual re-hybridizations to further evaluate the effects of blocking and randomization. There was moderate and asymmetric differential expression (351/3523, 10%) between endometrial and ovarian tumors in the randomized dataset. Non-biological effects were observed in the non-randomized dataset and 1934 markers (55%) were called differentially expressed (DE). Among them, 185 were deemed DE (185/351, 53%) and 1749 non-DE (1749/3172, 55%) in the randomized dataset. In simulations, when randomization was applied to all samples at once or within batches of samples balanced in tumor groups, blocking improved the true positive rate (TPR) from 0.95 to 0.97 and the false positive rate (FPR) from 0.02 to 0.002; when sample batches were unbalanced, randomization had a worse TPR (0.92) and FPR (0.10) regardless of blocking. Normalization improved the detection of true positive markers but still retained sizeable false positive markers. Randomization and blocking should be used in practice to more fully reap the benefits of genomics technologies.
Introduction
Technological advances in genomics profiling provide a plethora of molecular data to discover potential biomarkers for cancer diagnosis and treatment (1, 2). Discovering molecular biomarkers that are accurate and reproducible – the ‘needles in a haystack’ – still remains challenging (3–5). A major source of the challenge comes from the non-biological effects in the data that are resulted from the experimental process (6, 7). Non-biological effects can be introduced into the data at numerous steps of the experiment such as sample preparation, array hybridization, and image scanning (8). Previous efforts to remove non-biological effects have been mainly focused on post-hoc data adjustments through a ‘normalization’ step (9, 10).
Careful study planning and sound experimental design offer a preventive opportunity to reduce the level of non-biological effects and mitigate their negative impact on biomarker discovery (11, 12). Two basic principles of experimental design are randomization and blocking (13, 14). Random assignment of experimental units to comparison groups eliminates bias due to known and unknown confounders. Randomization has been widely used in many scientific fields including clinical studies to determine the efficacy of experimental treatments (15). Arranging experimental units in blocks of similar units can reduce the variance and hence increase the power to detect differences between comparison groups (16). Many profiling platforms come with natural blocks. For example, the Agilent human microRNA (miRNA) arrays have eight arrays on each glass slide; the Illumina human gene expression BeadChips have twelve bead-chips on each slide; the Illumina sequencer has eight lanes on each flow cell. Randomization and blocking have been previously suggested for use in genomic studies (17–19). However, their applications in practice are scarce possibly due to the lack of awareness and the conceived difficulties in logistic planning.
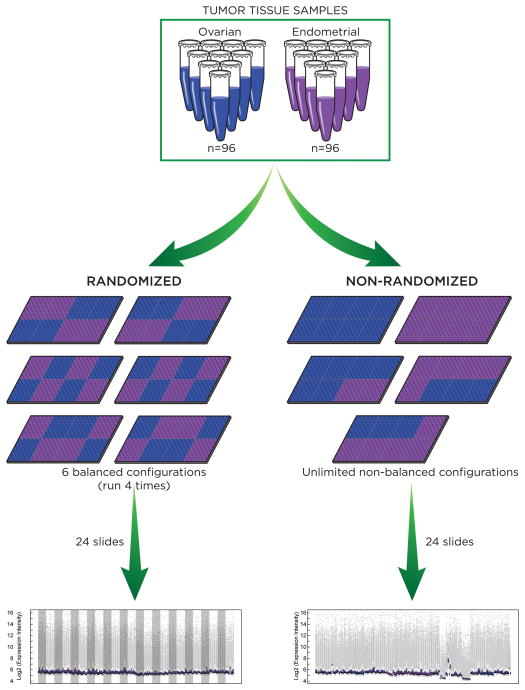
We set out to demonstrate the logistic feasibility and scientific benefits of randomization and blocking in molecular biomarker discovery so as to facilitate their adoption in cancer genomic studies. Towards this end, we profiled the same set of tumor samples twice with different experimental designs, once using the blocked randomization design and uniform array handling and a second time using no blocking, randomization, or uniform handling. Empirical evidence of differential expression was assessed in both the randomized study and the non-randomized study. The overall design of our study is illustrated in Figure 1. This design is general and can be applied to many genomics platform. In this study we profiled miRNA expression for a set of 96 endometrial tumors and 96 ovarian tumors using Agilent miRNA arrays (20, 21).
Figure 1.
Illustration of the overall design for a randomized miRNA array study paired with a non-randomized study using the same set of tumor samples.
We present empirical evidence of confounding non-biological effects in the non-randomized study. We compare the results of differential expression analysis for the randomized study versus that for the non-randomized study. We consider the impact of a post-hoc normalization step to remove non-biological effects. In addition to the empirical evaluations, we perform simulation studies to separately examine the effect of randomization and blocking when the true biomarker status is known and array handling is non-uniform. We also use simulations to assess the effect of randomization when samples come in multiple batches and each batch is randomized individually. Finally, we make recommendations on study design for cancer molecular biomarker studies.
Materials and Methods
Patient samples
Ninety-six high-grade serous ovarian cancer samples and 96 endometrioid endometrial cancer samples were used in our study. These samples were all newly diagnosed, previously untreated, and collected at Memorial Sloan Kettering Cancer Center (MSKCC) between 2000 and 2012. Their use in our study was approved by the MSKCC Institutional Review Board.
Tumor microdissection and RNA extraction
Sample preparation used strict quality control on the specimens. Freshly harvested tissue was snap frozen for eventual cryomold embedding and RNA extraction for microarray analysis. Prior to RNA extraction a 5 μm H&E histologic section was cut from the top of the cryomold to assess for tumor content and percent necrosis. For any specimens with less than 60% tumor cell nuclei we used macrodissection to obtain enriched areas so as to make the specimen usable for RNA extraction and as uniform as possible. All specimens had less than 20% necrosis. All pathology specimens used for molecular study were diagnosed by dedicated gynecologic pathologists to establish histologic cell type, grade of malignancy, and site of origin. RNA was extracted from 30–100mg of macrodissected cryomold tissue using the Ambion mirVana miRNA Isolation kit (Ambion) according to the manufacturer’s protocol. Total RNA yield and quality were assessed using a nanodrop spectrophotometer and an Agilent Bioanalyzer.
MiRNA microarray hybridization
Human miRNA microarrays (Agilent Technologies), containing 3523 markers that represent 1,205 human and 142 human viral miRNAs, were used for miRNA expression profiling according to the manufacturer protocol. For samples meeting the quality control standards, 200 ng of total RNA was labeled using the miRNA Complete Labeling and Hybridization Kit (Agilent). Labeled RNA was hybridized in Agilent Human miRNA Microarray Release 16.0. Slides were washed and scanned according to the manufacturer’s instructions. Images were quantified using Feature Extraction 10.7.3.1 (Agilent).
Study design for the randomized study
Agilent miRNA arrays have an 8-plex design, where each array slide contains eight individual arrays arranged as two rows and four columns. Therefore each array slide represents an experimental block. We used the blocked randomization design when assigning arrays to sample groups in order to remove confounding non-biological effects. To achieve the best balance, we further required the blocks to be row and column balanced. That is, there were equal numbers of cases and controls on any row and any column of a slide. There is a total of six possible configurations that achieve row and column balance for a 2 by 4 block such as the Agilent 8-plex array (Figure 1). The 192 tumor samples used 192 arrays corresponding to 24 array slides. When implementing the blocked randomization design, we assigned the 24 slides to one of the six configurations with equal probabilities and consequently dedicated the arrays to one of the two sample groups. We then assigned each group of arrays to a random permutation of the samples in the corresponding group. We carefully planned our randomized study. All 24 slides were ordered from the same manufacture batch. Their hybridization and production were processed in one run by a single experienced technician.
Study design for the non-randomized study
RNAs for the same 192 tumors used for the randomized study were re-arrayed for a second study that used no blocking or randomization. Care was taken to ensure consistent sample handling for the two studies, and RNAs used for two arrays of the same tumor sample were taken from the same master stock. The 192 arrays for the non-randomized study were ordered from multiple manufacturer batches. In order to mimic what would be done in the clinical setting, the arrays in the non-randomized study were assigned to samples in the order of sample collection (Supplementary Table S3), and were processed in multiple runs by two technicians.
Statistical analysis of miRNA array data
Each marker on the Agilent array has multiple replicates (ranging from 10 to 40) on the array. Our data show minimum variation among replicates for the same marker (Supplementary Fig. S1). Hence we summarized data from replicates for the same marker using the median. Evidence against the null hypothesis of equivalent expression was assessed using the t statistic comparing the endometrial and ovarian sample groups (22). A separate t-test was performed for each of the 3,523 markers on the Agilent array and a two-sided p-value was calculated. The p-values were used to derive a marker set at a given significance level: markers with p-values smaller than the significance level were declared differentially expressed (DE), and those having larger p-values were declared non-differentially-expressed (non-DE). The resulting differential expression status of markers was compared between the randomized study and the non-randomized study using a venn diagram. Statistical analyses were conducted using R and Bioconductor.
Simulation study
Our goal for the simulation study was three-fold: (1) to examine the effect of randomization and blocking when non-uniform handling is used and when the true biomarker status is known; (2) to assess the effect of randomization and blocking separately; and (3) to evaluate the effect of randomization when samples come in batches and randomization can only be done within each batch.
Toward this goal, we simulated data for the below scenarios of experimental designs for array-to- sample assignment, each combined with non-uniform array handling.
blocking, randomization of all samples together;
blocking, randomization within balanced batches of samples;
blocking, randomization within unbalanced batches of samples;
no blocking, randomization of all samples together;
no blocking, randomization within balanced batches of samples;
no blocking, randomization within unbalanced batches of samples;
blocking, no randomization;
no blocking, no randomization;
complete confounding of sample groups and array slides.
In order to mimic the characteristics of miRNA array data, we estimated the biological effects of each sample and the non-biological effects of each array using the empirical data from the paired array studies and then simulated data through virtual re-hybridizations of samples to arrays. Specific steps of the simulation study are as follows.
Biological effects estimation. We approximated the biological effects of each sample as its measurements in the randomized data, except that for non-DE markers in the ovarian samples mean differences between the two tumor groups were subtracted from the observed data.
Non-biological effects estimation. We estimated the non-biological effects for each array in the non-randomized data as the difference between this array and its paired array in the randomized data.
-
Array-to-sample assignment. For each simulation, samples are randomly shuffled, and arrays are re-assigned to samples according to an experimental design:
Blocking, randomization of all samples together: 4 arrays from each slide are randomly selected and assigned to one tumor group, and the rest to another group.
Blocking, randomization within balanced batches: 24 slides are randomly allocated to 5 batches each with equal number of samples from the two tumor groups; 4 arrays of each slide are randomly selected and assigned to one group, and the rest to another.
Blocking, randomization within unbalanced batches: 24 slides are randomly allocated to 5 batches each with unequal number of samples from the two tumor groups (with the ratio of the two tumor types 1:3 or 3:1); 4 arrays of each slide are randomly selected and assigned to one group, and the rest to another.
No blocking, randomization of all samples together: 96 arrays across all array slides are randomly selected and assigned to one tumor group, and the rest to another.
No blocking, randomization within balanced batches: 24 slides are randomly allocated to 5 batches each with equal number of samples from the two tumor groups; half arrays across slides in each batch are randomly selected and assigned to one group, and the rest to another.
No blocking, randomization within unbalanced batches: 24 slides are randomly allocated to 5 batches each with unequal number of samples from the two tumor groups; half arrays across slides in each batch are randomly selected and assigned to one group, and the rest to another.
Blocking, no randomization: 4 arrays on the first row of each slide are assigned to one tumor group, and the rest to another group.
No blocking, no randomization: the allocation of arrays to the tumor groups is kept the same as the non-randomized study.
Complete confounding of sample groups and array slides: 12 array slides are randomly assigned to one tumor group, and the rest slides to another group.
Virtual hybridization. Simulated data were generated by summing the biological effects of each sample and the non-biological effects of its re-assigned array.
Analysis of each simulated dataset. Simulated data were analyzed for differential expression similar to the analysis of the non-randomized empirical study.
Summary of simulated datasets. Steps 3–5 were repeated to generate 100 datasets for each experimental design. True positive rate (TPR) and false negative rate (FNR) are calculated and averaged across the 100 simulated datasets.
Results and Discussion
Empirical evaluation of molecular biomarkers in the randomized study
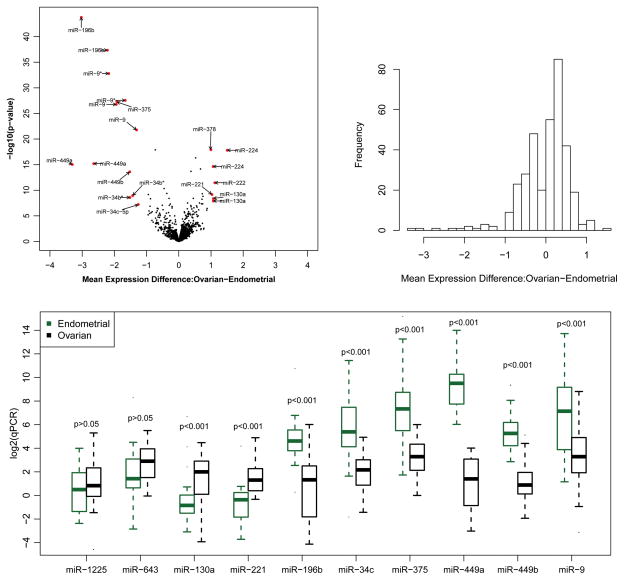
We compared miRNA expression levels between endometrial and ovarian tumors for each of the 3,523 markers on the Agilent array using the randomized data. Among them, 351 markers (10%) were differentially expressed at a p-value cutoff of 0.01 (Figure 2a, Supplementary Table S1). Among the 351 markers, 210 are over-expressed in ovarian tumors, 6 of which have a log2-fold-change (L2FC) greater than 1 with a maximum of 1.5; 141 are under-expressed, 13 of which have a L2FC greater than 1 with a maximum of 3.3. By chance, 35 markers are expected to have a p-value less than 0.01. The level of differential expression is moderately abundant in our data and the pattern of differential expression is not symmetric around 0 (Figure 2b).
Figure 2.
Figure 2a. Volcano plot for comparing miRNA expression between ovarian tumors versus endometrial tumors using the randomized data.
Figure 2b. Histogram of mean expression differences between ovarian tumors versus endometrial tumors among markers that are differentially expressed in the randomized data.
Figure 2c. Boxplot of the qPCR data for ten selected miRNAs. The data were derived by the delta-delta-Ct method and displayed on the log2 scale.
The 6 markers that are up-regulated in ovarian tumors and have a L2FC greater than 1 represent 4 unique miRNAs, and the 13 down-regulated markers represent 8 unique miRNAs (Supplementary Table S2). MiR-224, a gene located on the X-chromosome and thought to be active in mammalian ovaries, is the most up-regulated in ovarian tumors with a L2FC of 1.5 (23). The other three up-regulated miRNAs in ovarian tumors are miR-130a, miR-221, and its paralogue miR-222, which all have been previously reported to be oncogenic (24–26). MiR-449a, which has been shown to be involved in vertebrate multiciliogenesis, is the most up-regulated in endometrial tumors with a L2FC of 3.3 (27, 28). The other seven down-regulated miRNAs are miR-9, miR-34a/b/c, miR-196b, miR-375, and miR-449b, which have been implicated in a number of cancer types (28–34).
We experimentally validated 2 non-DE miRNAs and 8 DE miRNAs (2 up-regulated in ovarian tumors and 6 down-regulated) using qRT-PCR. The differential expression status was confirmed for all 10 miRNAs (Figure 2c).
Empirical evaluation of molecular biomarkers in the non-randomized study
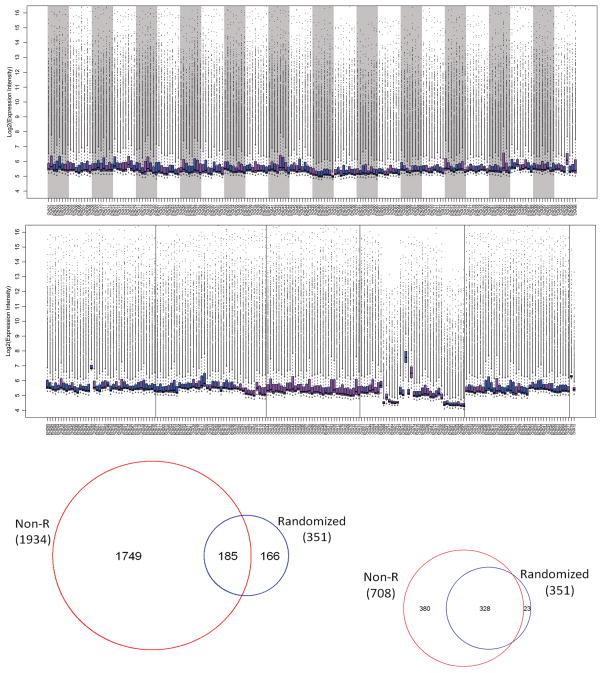
Figures 3a and 3b shows the distribution of the non-randomized data without normalization in comparison with the distribution of the randomized data. It indicates clear non-biological effects in the non-randomized data. When comparing miRNA expression between endometrial and ovarian tumors in the non-randomized data, 1934 markers (55%) were called DE at a p-value cutoff of 0.01. Among these markers, 181 were called DE in the randomized data (185/351, 53%); 1749 were called non-DE in the randomized data (1749/3172, 55%) (Figure 3c). With quantile normalization, the number of DE markers identified in the non-randomized data was reduced to 708. Among them, 328 were DE (328/351, 93%) and 380 were non-DE (380/3172, 12%) in the randomized data (Figure 3d).
Figure 3.
Figure 3a. Boxplot of the randomized data.
Figure 3b. Boxplot of the non-randomized data before normalization.
Figure 3c. Venn diagram comparing differentially expressed markers identified by the non-randomized data before normalization and those by the randomized data.
Figure 3d. Venn diagram comparing differentially expressed markers identified by the non-randomized data after quantile normalization and those by the randomized data.
Simulation study
The results of the simulation study are summarized in Table 1 and graphically displayed in Supplementary Fig. S2.
Table 1.
Results of the simulation study. Mean and standard deviation (in parenthesis) of true positive rate (TPR) and false positive rate (FPR) are reported. Simulation settings with the highest TPR and the lowest FPR are indicated in bold font. Simulation settings with the second highest TPR and the second lowest FPR are indicated in bold italic font.
| Index | Blocking, Randomization | No Normalization | Quantile Normalization | ||
|---|---|---|---|---|---|
| TPR | FPR | TPR | FPR | ||
| a | yes, across all samples | 0.97 (0.02) | 0.002 (0.005) | 1 (0) | 0.19 (0.03) |
| b | yes, within balanced batches | 0.97 (0.02) | 0.001 (0.003) | 1 (0) | 0.18 (0.03) |
| c | yes, within unbalanced batches | 0.92 (0.04) | 0.10 (0.17) | 1 (0) | 0.24 (0.07) |
| d | no, across all samples | 0.95 (0.03) | 0.02 (0.05) | 1 (0) | 0.19 (0.04) |
| e | no, within balanced batches | 0.95 (0.03) | 0.01 (0.02) | 1 (0) | 0.18 (0.04) |
| f | no, within unbalanced batches | 0.92 (0.05) | 0.17 (0.26) | 1 (0) | 0.24 (0.07) |
|
| |||||
| g | yes, no | 0.97 (0.01) | 0.001 (0) | 1 (0) | 0.24 (0.02) |
| h | no, no | 0.84 (0.01) | 0.07 (0.03) | 1 (0) | 0.17 (0.02) |
| i | completely confounded | 0.89 (0.08) | 0.20 (0.30) | 0.999 (0.001) | 0.26 (0.09) |
Not surprisingly, blocking and randomization applied to all samples at once led to a best accuracy of biomarker detection with the highest TPR (0.97, 340/351) and lowest FPR (0.002, 6/3172). This level of accuracy was equally, if not slightly better, achieved by blocking and randomization applied to sample batches that were balanced in sample groups (TPR=0.97, FPR=0.001=3/3172). The next best level of biomarker detection accuracy was achieved when the array-to-sample assignment used randomization across all samples (TPR=0.95=333/351, FPR=0.02=63/3172) or randomization within balanced batches of samples (TPR=0.95, FPR=0.01) but with no blocking. When randomization was used for samples that came in batches unbalanced in sample groups, the accuracy of biomarker detection had a slightly worse TPR (0.92, 323/351) and a much worse FPR (0.10–0.17, 312–539/3172) regardless of whether blocking was used within each batch. As expected, complete confounding of sample group and array slide resulted in a low TPR (0.89=312/351) and a high FPR (0.20=634/3172).
When no randomization was used, our simulation showed a high accuracy of biomarker detection (TPR=0.97, FPR=0.001) with blocking, and a low detection accuracy without blocking (TPR=0.84=295/351, FPR=0.07=222/3172). We note that this result depends on the actual level of confounding between sample group and array slide in the data. In this simulation, we used the array-to-sample assignment in the empirical non-randomized study, where the sample group was partly confounded with array slides with the number of endometrial tumors being 0, 1, 2, 3, 5, 6, and 7 for 4, 2, 1, 4, 2, 7, and 4 array slides, respectively.
Regardless of the experimental design, the post-hoc normalization step, using quantile normalization in our study, improved the detection of true positive markers to an almost perfect level and at the same time still had false positive markers with a FPR often around 0.20, corresponding to a false discovery rate of about 64%.
We used p-values at a cutoff of 0.01 to call significance in our study. We tested other p-value cutoffs and observed similar results (results not shown). We also repeated our analysis using the false discovery rate to adjust for multiple comparisons when calling significant markers, and observed similar results on the relative ranking of the experimental designs (results not shown).
Taken together, our simulation study further underlines the importance of blocking and randomization for accurate detection of disease relevant markers. Even blocking alone, or randomization alone, or randomization within balanced batches alone offers significant benefits in accurate biomarker detection. Post-hoc data adjustments through normalization improve the identification of true positive markers, but still possess a large number of false positive markers.
Conclusions
We have demonstrated that blocking and randomization are valuable for miRNA array studies of tumor samples. We also showed, through both empirical and simulated studies, that blocking, randomization across all samples, or randomization within balanced sample batches can effectively remove confounding non-biological effects and ensure the accuracy of detecting disease relevant markers with both a high TPR and a low FPR. We observed that post-hoc normalization to the data improves the detection of true positive markers and at the same time retains a large number of false positive markers in our study.
In practice, we recommend use of the block-type design of profiling platforms, placing equal numbers of samples from each comparison group in each block to the extent possible. If blocking cannot be done, one should randomize the samples altogether if they are available (for example, in a small scale study or a retrospective study using banked tissues), or batch samples so that each batch is balanced in comparison groups and then randomize within each batch.
Supplementary Material
Acknowledgments
Financial Support: This work was supported by NIH R01 grant CA151947 (L.-X. Qin, Q. Zhou, F. Bogomolniy, N. Olvera, J.M. Satagopan, C.B. Begg, and D.A. Levine), the Chia Family Foundation (F. Bogomolniy, N. Olvera, and D.A. Levine), and the Entertainment Industry Foundation (F. Bogomolniy, N. Olvera, and D.A. Levine).
We thank Dr. Agnes Viale and Jeffrey Zhao at the MSKCC Genomics Core Laboratory for their help with the microarray experiments.
Footnotes
Disclosure of Potential Conflicts of Interest: No potential conflicts of interest were disclosed.
Note: Supplementary data for this article are available at Clinical Cancer Research Online (http://clincancerres.aacrjournals.org/).
References
- 1.Cancer Genome Atlas Research N. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–8. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Zhang J, Baran J, Cros A, Guberman JM, Haider S, Hsu J, et al. International Cancer Genome Consortium Data Portal--a one-stop shop for cancer genomics data. Database: the journal of biological databases and curation. 2011:2011:bar026. doi: 10.1093/database/bar026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Diamandis EP. Cancer biomarkers: can we turn recent failures into success? Journal of the National Cancer Institute. 2010;102:1462–7. doi: 10.1093/jnci/djq306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ferte C, Trister AD, Huang E, Bot BM, Guinney J, Commo F, et al. Impact of bioinformatic procedures in the development and translation of high-throughput molecular classifiers in oncology. Clinical cancer research: an official journal of the American Association for Cancer Research. 2013;19:4315–25. doi: 10.1158/1078-0432.CCR-12-3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ransohoff DF. Bias as a threat to the validity of cancer molecular-marker research. Nat Rev Cancer. 2005;5:142–9. doi: 10.1038/nrc1550. [DOI] [PubMed] [Google Scholar]
- 6.Owzar K, Barry WT, Jung SH, Sohn I, George SL. Statistical challenges in preprocessing in microarray experiments in cancer. Clinical cancer research: an official journal of the American Association for Cancer Research. 2008;14:5959–66. doi: 10.1158/1078-0432.CCR-07-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Schadt EE, Li C, Su C, Wong WH. Analyzing high-density oligonucleotide gene expression array data. Journal of cellular biochemistry. 2000;80:192–202. [PubMed] [Google Scholar]
- 8.Speed T. Statistical Analysis of Gene Expression Microarray Data. CRC Pr I Llc; 2003. [Google Scholar]
- 9.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003b;4:249–64. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 10.Qin LX, Satagopan JM. Normalization method for transcriptional studies of heterogeneous samples--simultaneous array normalization and identification of equivalent expression. Statistical applications in genetics and molecular biology. 2009;8:Article 10. doi: 10.2202/1544-6115.1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Churchill GA. Fundamentals of experimental design for cDNA microarrays. Nat Genet. 2002;32 (Suppl):490–5. doi: 10.1038/ng1031. [DOI] [PubMed] [Google Scholar]
- 12.Ransohoff DF. How to improve reliability and efficiency of research about molecular markers: roles of phases, guidelines, and study design. J Clin Epidemiol. 2007;60:1205–19. doi: 10.1016/j.jclinepi.2007.04.020. [DOI] [PubMed] [Google Scholar]
- 13.Cochran Cox. Experimental Designs. Wiley, John & Sons Incorporated; 1992. [Google Scholar]
- 14.Fisher RA, Prance GT. The Design of Experiments. Hafner Press; 1935. [Google Scholar]
- 15.Rosenberger WF, Lachin JM. Randomization in Clinical Trials: Theory and Practice. Wiley; 2004. [Google Scholar]
- 16.Bailey RA. Design of Comparative Experiments. Cambridge University Press; 2008. [Google Scholar]
- 17.Kerr MK, Churchill GA. Statistical design and the analysis of gene expression microarray data. Genet Res. 2001;77:123–8. doi: 10.1017/s0016672301005055. [DOI] [PubMed] [Google Scholar]
- 18.Verdugo RA, Deschepper CF, Munoz G, Pomp D, Churchill GA. Importance of randomization in microarray experimental designs with Illumina platforms. Nucleic Acids Res. 2009;37:5610–8. doi: 10.1093/nar/gkp573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Auer PL, Doerge RW. Statistical design and analysis of RNA sequencing data. Genetics. 2010;185:405–16. doi: 10.1534/genetics.110.114983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cummins JM, Velculescu VE. Implications of micro-RNA profiling for cancer diagnosis. Oncogene. 2006;25:6220–7. doi: 10.1038/sj.onc.1209914. [DOI] [PubMed] [Google Scholar]
- 21.Wang H, Ach RA, Curry B. Direct and sensitive miRNA profiling from low-input total RNA. Rna. 2007;13:151–9. doi: 10.1261/rna.234507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Statistical applications in genetics and molecular biology. 2004;3:Article3. doi: 10.2202/1544-6115.1027. [DOI] [PubMed] [Google Scholar]
- 23.Christenson LK. MicroRNA control of ovarian function. Anim Reprod. 2010;7:129–33. [PMC free article] [PubMed] [Google Scholar]
- 24.Acunzo M, Visone R, Romano G, Veronese A, Lovat F, Palmieri D, et al. miR-130a targets MET and induces TRAIL-sensitivity in NSCLC by downregulating miR-221 and 222. Oncogene. 2012;31:634–42. doi: 10.1038/onc.2011.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feng M, Li Z, Aau M, Wong CH, Yang X, Yu Q. Myc/miR-378/TOB2/cyclin D1 functional module regulates oncogenic transformation. Oncogene. 2011;30:2242–51. doi: 10.1038/onc.2010.602. [DOI] [PubMed] [Google Scholar]
- 26.Pang Y, Young CY, Yuan H. MicroRNAs and prostate cancer. Acta Biochim Biophys Sin (Shanghai) 2010;42:363–9. doi: 10.1093/abbs/gmq038. [DOI] [PubMed] [Google Scholar]
- 27.Marcet B, Chevalier B, Luxardi G, Coraux C, Zaragosi LE, Cibois M, et al. Control of vertebrate multiciliogenesis by miR-449 through direct repression of the Delta/Notch pathway. Nat Cell Biol. 2011;13:693–9. doi: 10.1038/ncb2241. [DOI] [PubMed] [Google Scholar]
- 28.Yang X, Feng M, Jiang X, Wu Z, Li Z, Aau M, et al. miR-449a and miR-449b are direct transcriptional targets of E2F1 and negatively regulate pRb-E2F1 activity through a feedback loop by targeting CDK6 and CDC25A. Genes Dev. 2009;23:2388–93. doi: 10.1101/gad.1819009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Corney DC, Hwang CI, Matoso A, Vogt M, Flesken-Nikitin A, Godwin AK, et al. Frequent downregulation of miR-34 family in human ovarian cancers. Clinical cancer research: an official journal of the American Association for Cancer Research. 2010;16:1119–28. doi: 10.1158/1078-0432.CCR-09-2642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70:9175–84. doi: 10.1158/0008-5472.CAN-10-1318. [DOI] [PubMed] [Google Scholar]
- 31.Guo LM, Pu Y, Han Z, Liu T, Li YX, Liu M, et al. MicroRNA-9 inhibits ovarian cancer cell growth through regulation of NF-kappaB1. FEBS J. 2009;276:5537–46. doi: 10.1111/j.1742-4658.2009.07237.x. [DOI] [PubMed] [Google Scholar]
- 32.Lehmann U, Hasemeier B, Christgen M, Muller M, Romermann D, Langer F, et al. Epigenetic inactivation of microRNA gene hsa-mir-9-1 in human breast cancer. J Pathol. 2008;214:17–24. doi: 10.1002/path.2251. [DOI] [PubMed] [Google Scholar]
- 33.Li Z, Huang H, Chen P, He M, Li Y, Arnovitz S, et al. miR-196b directly targets both HOXA9/MEIS1 oncogenes and FAS tumour suppressor in MLL-rearranged leukaemia. Nat Commun. 2012;3:688. doi: 10.1038/ncomms1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peurala H, Greco D, Heikkinen T, Kaur S, Bartkova J, Jamshidi M, et al. MiR-34a expression has an effect for lower risk of metastasis and associates with expression patterns predicting clinical outcome in breast cancer. PloS one. 2011;6:e26122. doi: 10.1371/journal.pone.0026122. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.