Abstract
Sex chromosome genes directly influence sex differences in behavior. The discovery of the Sry gene on the Y chromosome (Gubbay et al., 1990; Koopman et al., 1990) substantiated the sex chromosome mechanistic link to sex differences. Moreover, the pronounced connection between X chromosome gene mutations and mental illness produces a strong sex bias in these diseases. Yet, the dominant explanation for sex differences continues to be the gonadal hormones. Here we review progress made on behavioral differences in mouse models that uncouple sex chromosome complement from gonadal sex. We conclude that many social and cognitive behaviors are modified by sex chromosome complement, and discuss the implications for human research. Future directions need to include identification of the genes involved and interactions with these genes and gonadal hormones.
Keywords: Klinefelter Syndrome, Turner Syndrome, Four Core Genotypes, Sex differences, Behavior, Sexual differentiation
Introduction
In species with sexual reproduction males and females must accommodate the demands of both shared and divergent evolutionary pressures. The responses to different demands are reflected in sex differences in morphology and behaviors. Primary sex differences, such as male versus female gonads, are distinctly different between the two sexes, while secondary sexual characteristics like antlers, body hair, and musculature of the larynxes can be completely sexually dimorphic or can vary by degrees. Importantly, sexually dimorphic structures are also present in the central nervous system, and are necessary for the regulation of behavioral sex differences.
Morphological sex differences are primarily orchestrated by developmental gene expression, and, in many vertebrates, sex chromosome genes initiate sex-specific gene expression during early development. These genes, and the proteins they encode, propagate a cascade of signaling events within gene-networks to drive sexual differentiation programs. The best-documented example of this in mammals is the gene of the sex-determining region of the Y chromosome, Sry. When Sry is expressed at the correct time and place it causes the undifferentiated gonads to become testes. In turn, androgens, produced by the male testes during development, organize the brain to respond to male-typical steroid hormones in adulthood, thereby activating sexually dimorphic masculine behaviors.
In addition to steroid hormones, the substantial genetic difference between males (bearing a single X and a Y chromosome) and females (bearing two X chromosomes) plays a significant role in brain development and may lead to neurological disorders (Lyon, 1961; Qureshi and Mehler, 2010; Raymond, 2006; Savic, 2012). Notably, the proportion of genes on the X chromosome that are expressed in brain is higher than any other single chromosome (Xu and Andreassi, 2011), and it is well known that some of these genes play a role in cognitive behavior. Human males with functional mutations in their only allele of these X-genes (i.e. FMR1, JARID1C, ATRX etc.) exhibit mental illnesses (Raymond, 2006). In addition, males with three sex chromosomes (XXY or XYY) can have behavioral problems and low IQs (van Rijn et al., 2006), and XXY (Klinefelter Syndrome) boys are diagnosed with autism spectrum disorders, schizophrenia, affective disorders, and language disabilities more often than XY boys (Savic, 2012). Likewise, XO females (Turner Syndrome) have variable behavioral phenotypes but are often described as socially immature or fearful (Elsheikh et al., 2002; Lesniak-Karpiak et al., 2003). These are very common diseases, ranging in incidence from 1:2,500 (Turner) to 1:600–1,000 (Klinefelter), which highlights the potential clinical applications of research using mouse models of sex chromosome aneuploidy, as well as models that isolate the behavioral actions of sex chromosome genes from actions of hormones.
Over the past ten years, a surge in interest in sex chromosome genes and behavior has been driven by the hypothesis that these genes play a role in sexual differentiation of the brain (Arnold, 2009; Arnold et al., 2003; De Vries et al., 2002). Because the sex chromosomes are intrinsically linked to gonadal development, and thereby hormonal differences, genetically manipulated mice along with spontaneous mutants, have been used as tools for examining direct, genetically derived sex differences that arise independently of hormones. While many non-behavioral phenotypes have been described from these models, this review will focus on and summarize sex chromosome gene effects on sexually dimorphic behaviors. We aim to broaden the readers’ interests in the roles that sex chromosome genes play in sexually dimorphic behavior as well as other behaviors that may relate to human neurodevelopmental disorders with sex-biased incidences.
The Sex Chromosomes
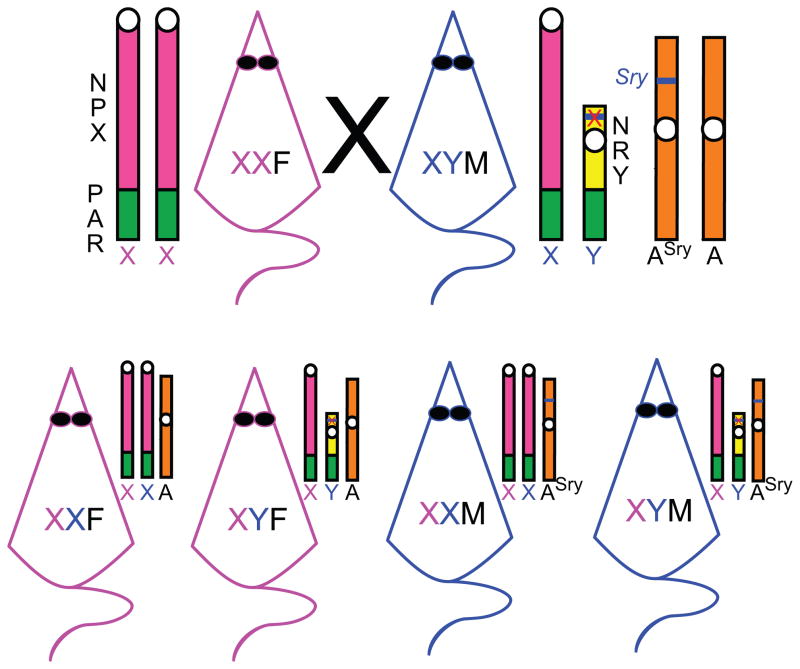
The mammalian sex chromosomes, X and Y, are structurally heteromorphic, and unequally represented in the cells of XX females and XY males. It is hypothesized that the mammalian sex chromosomes started as identical autosomes that diverged once the precursor to the modern day Y chromosome acquired the gene(s) and mutations necessary for genetic inheritance of testes determination (Graves, 1995; Graves et al., 2006). After the Y chromosome captured the testis determining gene(s), three major genetic domains among the two sex chromosomes of mammals evolved (Figure 1; (Ellegren, 2011)). The two chromosomes share a region of identical sequence homology, called the pseudoautosomal region (PAR) (Ellis and Goodfellow, 1989) that, in humans, contains about 29 genes (Ross et al., 2005). Recombination of the sex chromosome PAR region, like recombination in autosomes, is essential for meiosis. In addition to the PAR genes shared with X, the human Y chromosome has approximately 27 known unique genes (429 predicted) in the non-recombining region of Y (NRY; (Skaletsky et al., 2003), NCBI human genome assembly), while the X chromosome contains approximately 1,400 known unique genes (1,672 predicted) in the non-pseudoautosomal region of X (NPX; (Xu and Disteche, 2006); NCBI human genome assembly). In contrast to humans, the single PAR region in mouse sex chromosomes contains only two known genes (Dal Zotto et al., 1998; Perry et al., 2001; Raudsepp and Chowdhary, 2008), the NRY contains 54 predicted Y-unique genes, and the NPX contains 2025 predicted X-unique genes (NCBI mouse genome assembly).
Figure 1. Structural domains of mouse X and Y sex chromosomes.
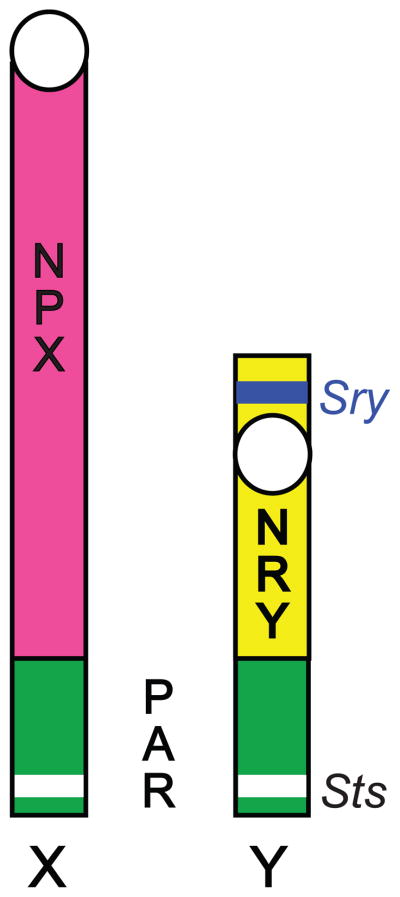
Schematic of the sex chromosomes in mice. PAR (in green), the pseudoautosomal region has sequence homology between X and Y with identical genes that recombine during male meiosis. Sts (in white), the steroid sulfatase gene. NPX (in pink), the non-pseudoautosomal region of X-chromosome genes unique to X. NRY (in yellow), the non-recombining region of Y-chromosome genes unique to Y. Sry (in blue), the sex determining region of Y encoding the testis-determining transcription factor. The centromere is represented as a white circle.
In order to deal with the stoichiometric problem of different doses of sex chromosome genes between the sexes, mammals developed X-inactivation dosage compensation, whereby one of the two X chromosomes is transcriptionally silenced at random in every female cell to match the single X expression levels in XY males (Lyon, 1961; Nguyen and Disteche, 2006; Wutz, 2011). However, in humans, 15% (Carrel and Willard, 2005), and in mice 3% (Berletch et al., 2011; Disteche et al., 2002; Yang et al., 2010), of genes on the silenced X chromosome are capable of escaping inactivation. This can result in a genetic imbalance between males and females such that, besides the unique genes on the Y chromosome that are only expressed in males, some of the genes unique to the X chromosome which escape X-inactivation may be more highly expressed in females. While the number of differentially expressed X chromosome genes is small (Itoh et al., 2007; Johnston et al., 2008), some of these genes are important regulators of downstream gene expression (Xu and Andreassi, 2011). In addition, females, not males, inherit a paternal X chromosome; therefore, any paternally imprinted X genes will be expressed in females only. Dimorphic sex chromosome gene expression is apparent in brain tissue and thus may cause sex differences in behavior (Xu and Andreassi, 2011; Xu et al., 2002; Xu et al., 2008a; Xu et al., 2008b; Xu and Disteche, 2006; Xu et al., 2005). In the case of human sex chromosome aneuploidies, which have more or fewer than two sex chromosomes, variation in expression of sex chromosome genes within the brain may also affect behavior (Knickmeyer, 2012; Savic, 2012). For example, when compared with XX females, females with Turner syndrome (XO) would have decreased expression of any X chromosome genes that escape X-inactivation, while individuals with Klinefelter syndrome (XXY) would have over expression of these same genes when compared to XY males.
The Mice
Before reviewing the behavioral data, we provide detailed descriptions of many of the mouse models that have been used for evaluating sex chromosome effects on behavior. A simplified summary of these models is found in Table 1. A critical consideration when interpreting results in mice is that the background strain influences most behaviors; therefore, we make note of model background strains, and, where appropriate, the Y chromosome strain of origin.
Table 1.
Genetic mouse models for differentiating the contributions of gonads and sex chromosome genes on sex differences
| Copy Number | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
|
Model (Strain) Dam x Sire |
Genotype | Offspring common abbreviations | Gonads | NPX | NRY | Sry | PAR | Suggested uses: |
| Normal (various strains) | ||||||||
| XX x XY | X Y | M | Testes | 1 | 1 | 1 | 2 | Establishing sex differences |
| X X | F | Ovaries | 2 | 0 | 0 | 2 | ||
|
| ||||||||
| FCG (various strains) | ||||||||
| XX x XY− Sry | X Y− Sry | XYM | Testes | 1 | 1 | 1 | 2 | Determining whether sex differences are due to gonadal sex or sex chromosome complement |
| X X Sry | XXM | Testes | 2 | 0 | 1 | 2 | ||
| X Y− | XYF | Ovaries | 1 | 1 | 0 | 2 | ||
| X X | XXF | Ovaries | 2 | 0 | 0 | 2 | ||
|
| ||||||||
| Y* (C57BL/6J or MF1) | ||||||||
| XX x XY* | X Y* | 1XM | Testes | 1 + | 1 | 1 | 2 − | Determining if sex differences correlate with numbers of X- chromosomes and examining effects of Sts |
| X XY* | 2XM | Testes | 2 | 1 | 1 | 2 − | ||
| X Y*X | 1XF | Ovaries | 1 + | 0 | 0 | 2 | ||
| X X | 2XF | Ovaries | 2 | 0 | 0 | 2 | ||
|
| ||||||||
| XY−Y*X Cross (MF1) | ||||||||
| XX x XY− Y*X(Sry+) | XY−(Sry+) | XYM | Testes | 1 | 1 | 1 | 2 | Determining effects of the balance of sex chromosomes when two are present in either gonadal sex |
| XY*X(Sry+) | X0+PAR M | Testes | 1 + | 0 | 1 | 2 | ||
| XX(Sry+) | XXM | Testes | 2 | 0 | 1 | 2 | ||
| XY− | XYF | Ovaries | 1 | 1 | 0 | 2 | ||
| XY*X | X0+PAR F | Ovaries | 1 + | 0 | 0 | 2 | ||
| XX | XXF | Ovaries | 2 | 0 | 0 | 2 | ||
|
| ||||||||
| Y* Aneuploids (MF1 and/or C57) | ||||||||
| XX x XY* | XY* | 39, XY*O | Testes | 1 | 1 | 1 | 2 − | When compared to normal, determining effects of aneuploidy, numbers of PAR and/or Sts and X chromosome origins |
| X O | 39, XO | Ovaries | 1 | 0 | 0 | 1 | ||
|
| ||||||||
| XX x XPafY* | X maternal origin | 39, XmO | Ovaries | 1 | 0 | 0 | 1 | |
| 40, ln(X)1H/XPaf x 40, XY | X paternal origin | 39, XpO | Ovaries | 1 | 0 | 0 | 1 | |
|
| ||||||||
XY*X x XY
 XX x XYY*X XX x XYY*X
|
X X Y | 41, XXY | Testes | 2 | 1 | 1 | 3 | |
|
| ||||||||
| XYSry Aneuploids (MF1) | ||||||||
| XXY− x XYSry | X Y | only male offspring shown | Testes | 1 | 1 | 1 | 2 | Comparing actions of the endogenous Sry versus the Sry transgene. Further analysis of effects of numbers and composition of sex chromosomes (including PAR) |
| X Y Y− | Testes | 1 | 2 | 1 | 3 | |||
| X Y Sry | Testes | 1 | 1 | 2 | 2 | |||
| X Y Y− Sry | Testes | 1 | 2 | 2 | 3 | |||
| X X Sry | Testes | 2 | 0 | 1 | 2 | |||
| X X Y− Sry | Testes | 2 | 1 | 1 | 3 | |||
|
| ||||||||
| Sex Chromosome Trisomy (MF1) | ||||||||
| XXY− x XY−Sry | X X Y− Sry | XXY M | Testes | 2 | 1 | 1 | 3 | Examining effects of the balance of sex chromosomes when three are present in either gonadal sex |
| X Y− Y− Sry | XYY M | Testes | 1 | 2 | 1 | 3 | ||
| X X Y− | XXY F | Ovaries | 2 | 1 | 0 | 3 | ||
| X Y− Y− | XYY F | Ovaries | 1 | 2 | 0 | 3 | ||
| FCG offspring | ||||||||
|
| ||||||||
| POS (C57BL/6J) | ||||||||
| XX x XYPOSI | X Y-poschiavinus | XYPOSI | Ovotestes | 1 | 1 | 1 | 2 | Determining effects of sex chromosome complement in gonadal females |
| X Y-poschiavinus | XYPOSF | Ovaries | 1 | 1 | 1a | 2 | ||
| XX | XXF | Ovaries | 2 | 0 | 0 | 2 | ||
|
| ||||||||
| Odsex (FVB) | ||||||||
| XX x XYMOds | X Y Sox9-overexp. | XYMOds | Testes | 1 | 1 | 1 | 2 | Determining effects of sex chromosome complement in gonadal males |
| X X Sox9-overexp. | XXMOds | Testes | 2 | 0 | 0 | 2 | ||
|
| ||||||||
| SF-1 KO (C57BL/6J) | ||||||||
| XX SF1-het x XY SF1-het | X Y Sf1-het. mutant | XY SF1-het | Testes | 1 | 1 | 1 | 2 | Examining the role for sex chromosomes independent of gonadal sex |
| X Y Sf1-null mutant | XY SF1-KO | None | 1 | 1 | 1 | 2 | ||
| X X Sf1-het. mutant | XX SF1-het | Ovaries | 2 | 0 | 0 | 2 | ||
| X X Sf1-null mutant | XX SF1-KO | None | 2 | 0 | 0 | 2 | ||
|
| ||||||||
| Sex Reversed (C57BL/6J) | ||||||||
| XX x XYSxr | X Y-Sxr | XYSxrM | Testes | 1 | 1+ partial | 1 | 2 | Determining effects of sex chromosome complement in gonadal males |
| X X-Sxr | XXSxrM | Testes | 2 | partial | 1 | 2 | ||
Legend: Model, simplest nomenclature for the offspring produced in these mice; Strain, published strain of models; Dam x Sire, mating pair used to make offspring; Offspring Genotypes, the first X represents the maternally inherited sex chromosome, the second sex chromosome is paternally inherited, and, where applicable, following the paternal sex chromosome are autosomal genetic features that includes transgenic autosomal Sry in the FCG model; Offspring, published names of offspring. NPX, copies of non-PAR X genes; NRY, copies of non-PAR Y genes; Sry, copies of sex-determining region of Y chromosome; PAR, copies of pseudoautosomal region of sex chromosomes. Critical comparisons available within each model are highlighted in gray.
+, Y* and Y*X chromosomes have a small piece of an NPX consisting of about 8 genes;
−, Y* and XY* chromosomes are missing distal PAR which contains the Sts gene;
congenic Sry does not cause differentiation into testes in XYPos mice; partial, a small number of Y genes surrounding and including Sry (see text).
Four Core Genotypes
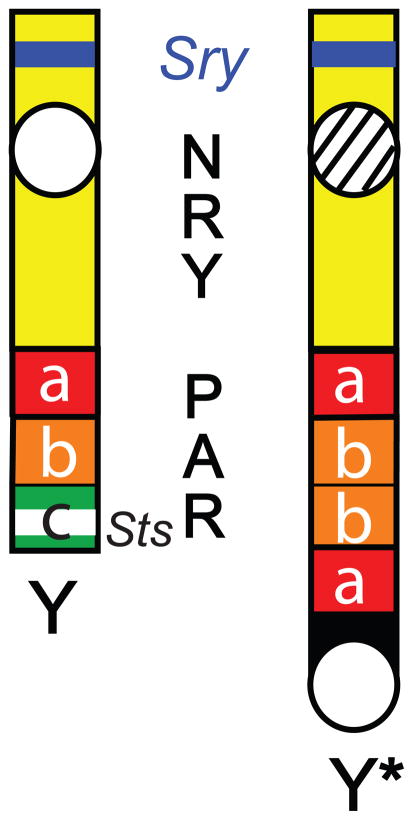
A mouse model now known as the “Four Core Genotypes” (FCG) has been used to directly test the contributions of sex chromosome complement versus gonadal sex (De Vries et al., 2002). This model takes advantage of a spontaneous mutation that resulted in a deletion of the testis determining gene, Sry, on the mouse Y chromosome (Lovell-Badge and Robertson, 1990), which is rescued by a transgenic insertion of Sry within an autosome (Mahadevaiah et al., 1998). An animal with an Sry-deleted Y chromosome (henceforth called ‘Y−’) and an autosomal Sry transgene (XY− Sry male) develops normal testes and is a fertile male. Mating a XY− Sry male with a normal XX female produces offspring of four genotypes (Figure 2): females with two X chromosomes (XXF), females with an X and a Y− chromosome (XYF), males with two X chromosomes and no Y chromosome (XXM), and males with an X and a Y chromosome (XYM). The FCG unlinks gonadal determination from the inheritance of the sex chromosomes, and allows for independent analysis of the two factors.
Figure 2. Cross of XXF and XY−Sry to generate Four Core Genotypes (FCG).
Top line: Breeder mice to generate the FCG offspring. The dam is a normal XX female, and the sire is an XY male with a deletion on Y of testis-determining factor rescued by an autosomal transgene. The color of the mouse body denotes gonadal sex: female (F) is pink, and male (M) is blue. The autosomes (A) are in orange with Sry, the sex determining region of Y encoding the testis-determining transcription factor, in blue. PAR (in green), is the pseudoautosomal region. NPX (in pink), is the non-pseudoautosomal region of X- chromosome genes unique to X. NRY (in yellow), is the non-recombining region of Y chromosome genes unique to Y. The X on this region denotes the Sry gene mutation, and all Y-chromosomes have the Sry deletion. The centromere is represented as a white circle. Bottom Line: FCG offspring consisting of the following: XX females, XXF; XY females, XYF; XX males, XXM; and XY males, XYM. Maternally inherited X chromosomes are labeled with a pink X. Paternally inherited X chromosomes are labeled with a blue X. All other depictions are the same as for the breeders on the top line.
The FCG mice have been bred into several background strains (SJL, MF1, DBA/2 and C57BL/6J), but because of the origin of the Sry deleted Y− chromosome, which was discovered in a 129 male mouse, all of the Y− carrying males and females are Y chromosome congenics with a 129 Y chromosome, while their other chromosomes are from the background strain. This needs to be considered when data from this cross are compared with a normal XY that has both sex chromosomes from the same genetic background. Nonetheless, experiments conducted with the FCG are a good first step to differentiate whether gonadal sex (presence of ovaries or testes) and/or sex chromosome complement (XX or XY genotype) contribute to a sex difference in phenotype.
The Y* Mouse
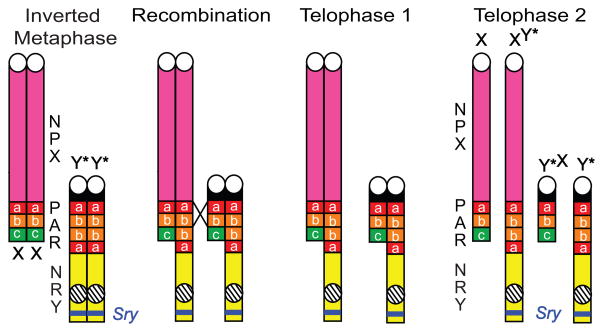
The Y* chromosome was discovered in the LT/Sv inbred mouse line, and was the result of a spontaneous translocation event. During this chromosome rearrangement in a normal male, a portion of the X chromosome consisting of the X-centromere and most of the X-PAR, attached to the end of PAR on the Y chromosome (Eicher et al., 1991); Figure 3). Thus, the Y* chromosome contains all of the Y-specific genes located in NRY, an inverted duplication of the proximal and middle portions of PAR joined end-to-end, and a functional X-centromere on the end of the long arm. In addition, between the duplicated PAR and the functional centromere is a very tiny fragment of NPX containing approximately 8 genes (Burgoyne et al., 1998; Eicher et al., 1991; Isles et al., 2004; Wolstenholme et al., 2013).
Figure 3. Domains of the Y* chromosome.
Represented are both the normal Y (on the left) and the Y* chromosomes. The pseudoautosomal region (PAR) of the normal Y has been separated into three domains based on their locations relative to the normal centromere (depicted here as a white circle on the normal Y): proximal (a = red), middle (b = orange), and distal (c = green) domains. The Y* chromosome contains all of the unique Y-genes located in the non-recombining region of Y (NRY, yellow domain) including the sex determining region of Y (Sry, blue bar). A spontaneous translocation event from the X chromosome duplicated the PAR proximal and middle regions of Y* and attached the duplicate domains in an inverted fashion along with a new centromere. Hatched circle, inactive Y-centromere on the Y* chromosome. Note that the Y* chromosome is missing the Sts gene.
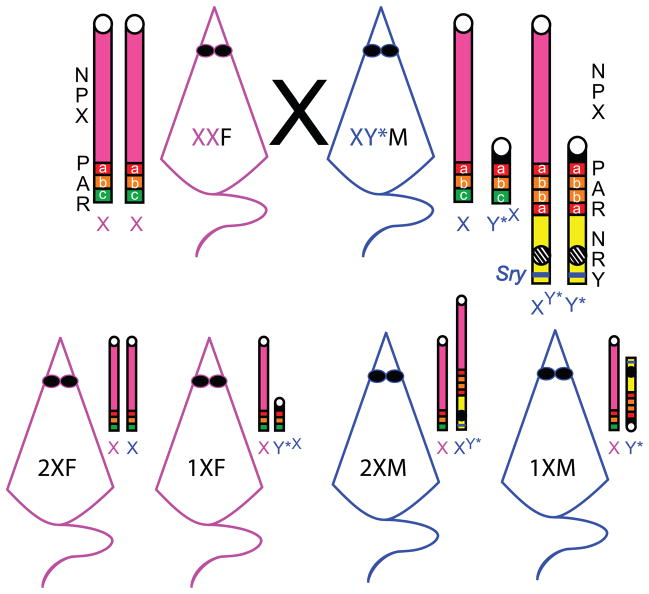
The Y* chromosome has been backcrossed into both MF1 and a substrain of the C57BL/6J background (B6Ei.LT-Y*/EiJ; Eicher et al., 1991). In the C57BL/6J background mating an XY* male to a normal XX female produces litters with four genotypes (Figure 4). In addition to XX female and XY* male offspring, recombination of the Y* chromosome with X yields two additional, unusual genotypes. Half of the females inherit an abnormally small chromosome, consisting of the small (8-gene) NPX fragment of the Y* chromosome and a full PAR, while half of the males inherit an abnormally large chromosome, which consists of an X chromosome (lacking the distal end of the PAR, including the Sts gene) attached to the NRY and shortened PAR (also lacking the distal end with Sts) of the Y* chromosome (Eicher et al., 1991). All of the offspring have a normal, maternal copy of the NPX genes of the X chromosome, and both male genotypes retain all of the NRY chromosome genes. For simplicity’s sake, the offspring are called 1X and 2X, males and females (Table 1). The Y* mouse model is a useful tool for understanding the role of X chromosome dose, including the modulation of behavioral sex differences. It also bears relevance to human disease, as the abnormal female offspring are nearly XO, similar to Turner Syndrome in humans, while the abnormal male offspring are nearly XXY, similar to Klinefelter Syndrome. The Y* mouse model may be particularly useful for understanding Turner syndrome, as this disease is not always caused by complete monosomy (patients can carry a truncated second X chromosome). However, one limitation of the Y* model is that the mice have the normal total number of chromosomes (40) and thus are not aneuploids in the strictest sense.
Figure 4. Y* meiosis.
During metaphase of a 4S gamete, the altered pseudoautosomal region (PAR) of the Y* chromosome pairs with the X PAR in an inverted fashion. To demonstrate the arrangements of the domains of the pseudoautosomal region (PAR) throughout mitosis and recombination of X and Y*, the PAR region has been depicted as three domains based on their locations relative to the normally active centromere (depicted here as a white circle on X): proximal (a = red) domain; middle (b = orange) domain; and distal (c = green) domain. When genetic material is exchanged between X and Y* in the inverted orientation during recombination, the recombined chromatids are different in structure and content of sex chromosome genes compared to the non-recombined chromatids. After recombination and separation of the four unique chromosomes after telophase 1 and 2, four male gametes are produced that differ in content of the following paternal sex chromosomes: X, XY*, Y*X, and Y*. The non-pseudoautosomal region of X (NPX), and the non-recombining region of Y (NRY), are depicted in pink and yellow, respectively. The active Y* centromere is depicted as a white circle, and the inactivate centromere is a circle with stripes. Sry, the sex determining region of Y (blue bar in NRY).
XY−Y*X Cross
Another limitation of the Y* offspring is that they cannot differentiate effects due to Y chromosome genes that are independent of gonadal sex. To address this issue, Chen et al. (Chen et al., 2013a) utilized a cross designed by Paul Burgoyne that mates a normal XX female to an XY−Y*X(Sry+) male in the MF1 background. This cross generates seven sex chromosome genotypes all with and without an autosomal Sry transgene (14 groups). To measure the effects of the Y-chromosome on adiposity and metabolism, Chen and colleagues (Chen et al., 2013a) compared three of the sex chromosome genotypes: XY−, XY*X, and XX (Table 1). Direct comparisons between XY− to XY*X can measure the effects of the presence or absence of NRY genes independent of gonadal sex and the number of PARs.
Aneuploids
The offspring of Y* 1XM and 2XF, in the C57BL/6J background are described above (Table 1). They possess abnormally configured sex chromosomes, and unusual numbers of NPX, NRY and PAR genes, but are not true aneuploids (with more or less than two sex chromosomes). True aneuploids, with more or fewer than two sex chromosomes, can be generated in several ways. In the MF1 background strain, breeding Y* mice can produce a number of true aneuploids with 39 chromosomes (39,XO females) along with the other four Y* genotypes (Table 1). Other strains of mice harboring X chromosome mutations also produce 39,XO offspring at a high frequency. Specifically, patchy fur (XPaf) mice with a mutation in the X-linked Paf gene that resides near PAR, and 40, In(X)1H/XPaf mutants with a large inversion of the X chromosome, display a high frequency of non-disjunction of the sex chromosomes during meiosis (Evans and Phillips, 1975; Lane and Davisson, 1990). When the XPaf mutation is combined with the Y* chromosome rearrangement by mating these two strains, the numbers of aneuploid offspring produced increases even further (Burgoyne and Evans, 2000). In addition, these mice can be used to examine X chromosome parent of origin effects by producing female offspring with either a single maternal or paternal X chromosome. Mating 40,XX females with XPaf Y* males carrying the patchy fur mutation on X, produces XO females with a single maternal X (39,XmO). The reciprocal females with a paternal X (39,XpO) are produced by mating 40, ln(X)1H/XPaf to 40,XY (Evans and Phillips, 1975). Another breeding strategy using the Y* mouse produces 41,XXY aneuploid mice. When XY* males are mated to XX females they produce XY*X females. XY*X females can be mated to normal XY males to obtain XYY*X males. Finally, mating XYY*X to XX females produces 41,XXY males, a model for Klinefelter syndrome (Table 1). 41,XXY males can then be bred to produce more 41,XXY male offspring (Hunt and Eicher, 1991; Hunt et al., 1998); these four generations of crosses can even make 41,XXY mice in the C57BL/6J background (Lue et al., 2005).
A second method of generating aneuploids is mating XY MF1 males that have an additional copy of an Sry transgene (XYSry) inserted onto an autosome with females possessing an XXY− genotype (Table 1). These females have two X chromosomes and an Sry-deleted 129 YTdym1 chromosome (Y−) and, in the MF1 background, they are fertile. The XYSry by XXY− cross can produce the following six testes-bearing males: XY, XYY−, XYSry, XYY− Sry, XXSry, and XXY− Sry. Three of the male genotypes are aneuploids with an extra Y− from the 129Tdym1 strain (XYY−, XYY− Sry, and XXY− Sry) and two genotypes have an extra autosomal copy of Sry from the transgene in addition to the endogenous MF1 copy on Y (XYSry and XYY− Sry). In addition, two males have two X chromosomes (XXSry, and XXY− Sry), and one male has a normal XY genotype and serves as a control (Park et al., 2008).
A third method used to produce aneuploid mice is similar to the strategy just described. Designated by the authors as the “sex chromosome trisomy” model, XXY− females can be bred with XY− Sry males (from the four core genotypes) to make eight genotypes of four males and four females (Chen et al., 2013b). The genotypes include: XX, XY−, XXY−, and XY−Y−; when the Sry transgene is absent the animals have ovaries, and when present they have testes. The virtue of this model is that all of the Y-chromosomes are from the same inbred line (129) and only Sry is a transgene. Like the other two aneuploidy models, these mice cannot be produced in C57BL/6 and, so far, are only available in the MF1 strain.
POS mice
This Y-congenic strain (C57BL/6JEiYPos) was formed when the Y chromosome from wild mice Mus domensticus poschiavinus (YPos) was bred into a C57BL/6Ei background strain (Eicher et al., 1982). After a few backcrosses there were no true male offspring produced, but instead there were normal XX females, XYPos females (XYPosF) and intersexed XYPos (XYPosI) with ovotestes (Canastar et al., 2008; Eicher et al., 1995). The intersex individuals produce sperm that can be extracted from the testicular tissue of the ovotestes to propagate the line. The ovary-bearing XYPosF have normal circulating hormone levels and can become pregnant (Correa et al., 2012; Stavnezer et al., 2000). One caution for interpretation of the data from these mice is that the ovotestes may be transient during embryogenesis (Eicher and Washburn, 2001). If that were the case, normal XYPosF may have been exposed to higher than normal levels of androgen as embryos.
Odsex mice
Odsex mice carry a coat color transgene that was inserted into a regulatory region approximately 1Mb upstream of the Sox9 gene of the FVB/NTacfBR inbred strain. The Sox9 gene is part of the normal testes differentiation cascade, and the transgene insertion causes overexpression of Sox9, which is normally repressed in females. This results in XX males (XXMOds) with small testes that, although capable of producing androgens, are lacking normal spermatogenesis (Bishop et al., 2000).
SF-1 Knockout Mice
Steroidogenic factor 1 (SF-1) is a nuclear receptor that is expressed in the gonads, adrenal glands, pituitary, and brain (Parker et al., 2002; Parker and Schimmer, 1997). SF-1 is essential for gonad development (Ingraham et al., 1994), and, due to the important role of SF-1 in adrenal function, mice lacking the gene (Nr5a1) for SF-1 die shortly after birth unless their adrenals are replaced via organ transplant (Majdic et al., 2002). To keep them alive from birth, SF-1 knockout (SF1 KO) mice typically receive daily injections of mineralocorticoids and glucocorticoids, and at about one week of age they receive adrenal transplants from normal, same age mice. The mice continue to develop in the absence of gonadal hormones, and therefore brain sex differences can be attributed to sex chromosome effects (Budefeld et al., 2008). SF1 KO mice (Table 1) have been backcrossed into the C57BL/6J mouse strain (Grgurevic et al., 2012). Some limitations of this model are that 1) the ventromedial nucleus of the hypothalamus does not form normally, 2) the pituitaries of SF1 KO mice do not produce normal levels of gonadotropins, and 3) the mice have to be exposed to exogenous glucocorticoids during the first week of life, any of which could potentially alter a number of behaviors in these mice.
Sex Reversed Mice
Sex Reversed mice (C57BL/6J-AW-J YSxra), also called Sxr mice (Table 1), are produced by males possessing two X chromosomes, one of which contains a fragment of the Y chromosome that includes the Sry, Zfy2, Kdm5d, Ube1y, and Zfy1 genes (Bishop and Mitchell, 1996). The presence of the Sry on the X chromosome (XSxr) leads to testicular development in these males (Evans et al., 1982; McLaren et al., 1992). However, in the C57BL/6J background, testicular development can be incomplete with this mutation producing ovotestes in the XX males (XXSxr), which complicates the interpretation of behavioral studies (Nagamine et al., 1998).
Sex Chromosomes and Behavior
Now that the relevant mouse models have been described, here we review the literature on behavioral studies conducted using these mice. However, there are some caveats to keep in mind when reviewing the behavioral literature. First, even small procedural differences in behavior tests can have large effects on the outcomes; therefore we have noted experimental differences when applicable. Secondly, since strain differences can also impact behavioral findings, as was done with the description of the mouse models above, we have tried to note differences in background strain and methodology throughout this review as needed. For a quick reference, the studies reviewed below are also summarized in Table 2.
Table 2.
Influence of sex chromosomes on sexually dimorphic behaviors.
| Behavior | Mouse Model | Hormonal Status | Sex Chromosome Phenotype | Refs. |
|---|---|---|---|---|
| Aggression | B10D1F1, D1B10F1 (hybrid) | Intact | Levels match the sire | Selmanoff 1975 |
| D1B10-Y1 F1 (hybrid) | Intact | DBA/1 Y increases aggression in hybrids | Maxson 1979 | |
| CBA/NZB (hybrid) | Intact | NZB Y increases aggression in hybrids | Roubertoux 1988 | |
| FCG (C57BL/6J) | GDX + T | XYF same as male | Gatewood 2006 | |
| POS (C57BL/6JEiYPos) | Intact | No influence of Y or X | Canastar 2008 | |
| Odsex (FVB/NTacfBR) | Intact | No influence of Y or X | Canastar 2008 | |
| Y* (C57BL/6J) | GDX +T | Trend for increased levels in 2XM | Bonthuis 2012 | |
|
| ||||
| Male sexual behavior | DBA/2.DBA/1-Y | Intact | DBA/1-Y reduced mounting in DBA/2 background | Shrenker 1984 |
| FCG (MF1) | GDX + T | No influence of Y or X | De Vries 2002 | |
| XYSry Aneuploids (MF1) | GDX+T | Males with supernumerary Y have increased behaviors. In presence of Sry transgene, males with two X chromosomes have decreased behaviors | Park 2008 | |
| POS (C57BL/6JEiYPos) | Intact | No influence of Y or X | Canastar 2008 | |
| Odsex (FVB/NTacfBR) | Intact | No influence of Y or X | Canastar 2008 | |
| FCG (C57BL/6J) | GDX + T | XXM faster to ejaculate than XYM | Bonthuis 2012 | |
| Y* (C57BL/6J) | GDX + T | Increased behaviors of 2X mice compared to 1X | Bonthuis 2012 | |
|
| ||||
| Female sexual behavior | SF1 KO (C57BL/6J) | No Gonads | XXKOSF1 more receptive and attractive than XYKOSF1 | Grgurevic 2012 |
|
| ||||
| Parental behavior | Sex Reversed (C57BL/6J-AW-J YSxra) | Intact | XXSxrM more parental than XYM | Reisert 2002 |
| FCG (C57BL/6J) | Intact and GDX | XXF more maternal than XYF | Gatewood 2006 | |
|
| ||||
| Adult social interaction | FCG (C57BL/6J) | GDX | XY mice investigate intruder more than XX mice | McPhie-Lalmansingh 2008 |
| Aneuploid (MF1) | Intact | 41, XXY males more sociable than XY | Liu 2010 | |
|
| ||||
| Juvenile social interaction | FCG (C57BL/6J) | Intact | XXF least social with stranger, and most social with sibling | Cox 2011 |
| Y* (C57BL/6J) | Intact | 2X mice more sociable than 1X mice | Cox et al., unpublished | |
|
| ||||
| Partner Preference | Aneuploid (MF1) | GDX Intact |
41, XXY prefers male to OVX female compared to no preference for XY 41, XXY has more of a preference for estrus female odor than XY |
Liu 2010 |
|
| ||||
| Anxiety | Y* (MF1) | Intact | 1XF and 39, XOF more fearful than 2XF | Isles 2004 |
|
| ||||
| Visiospatial attention | Y* (MF1) | Intact | Deficits in X0F and XY*0M | Davies 2007, 2009 |
|
| ||||
| Learning and cognition | Y* (MF1) | Intact | Additional X reduces memory XXY* in novel object recognition | Lewejohann 2009 |
| Aneuploid (C57BL/6EiJ) | Intact | 41, XXY slower to acquire Pavlovian conditioning than XY | Lue 2005 | |
| Aneuploid (MF1) | Intact | 39, XmO show more perseveration than 39, Xp0 | Davies 2005 | |
|
| ||||
| Feeding | FCG (C57BL/6J) | GDX | XX mice consume more food than XY during the light | Chen 2012 |
| XX x XY−Y*X (MF1) | GDX | Mice with 2 or three sex chromosomes eat more high fat diet than mice with a single X | Chen 2013a | |
|
| ||||
| Nociception | FCG (C57BL/6J) FCG (MF1) |
GDX adult Intact P1 |
XY mice less sensitive than XX mice to pain stimuli | Gioiosa 2008a, 2008b |
|
| ||||
| Habit formation | FCG (MF1) | Intact and GDX | XX form stronger habit for food. XY acquire stronger habit for alcohol |
Quinn 2007, Barker 2010 |
Listed is the literature (described in detail in the text) that reveals sex chromosome complement regulation of sexually dimorphic behaviors using the mouse models described in the introduction and Tables 1 and 2. In the mouse model column, the background strain used to acquire the behavioral data with the model is indicated in parentheses. Intact, gonad intact. GDX, gonadectomized. T, testosterone replaced. P1, postnatal day one.
Aggression
Offensive aggressive behavior is highly sexually dimorphic in mice, and largely dependent upon gonadal hormones (Conner et al., 1969). Different mouse strains are also strikingly diverse in the display of aggressive behaviors (Carlier et al., 1990). For example, DBA/1 males engage in acts of aggression (“tail rattling, wrestling, flank biting, chasing, or full attack”) toward male conspecifics more frequently than C57BL/10 males (Selmanoff et al., 1975). Furthermore, the aggression levels of DBA/1 by C57BL/10 F1 hybrid mice depend upon which strain was used for the sire and which was used for the dam. In general, F1 hybrid aggression levels more closely resemble the phenotype of the sire’s background strain than that of the dam’s strain, suggesting a role for gene(s) on the Y chromosome in aggressive behaviors. Early studies backcrossed hybrids to produce Y chromosome congenics with the DBA/1 Y chromosome in the C57BL/10 genetic background strain. Congenic males were then mated to DBA/1 females to generate male hybrid offspring with a DBA/1-Y, despite having paternal autosomes from the C57BL/10 strain (D1B10-Y1 F1). These hybrid mice with a DBA/1 Y were more aggressive than hybrids with a C57BL/10 Y, elegantly demonstrating that the Y chromosome contributes to strain differences in aggression (Maxson et al., 1979). Later, using a positional gene candidate strategy it was determined that the NRY gene, Sry, was most likely involved in establishing these strain differences (Maxson, 1996a; Maxson, 1996b), although these conclusions could not be proven with the methods available at the time.
Further evidence for the importance of the Y chromosome strain origin in aggressive behavior came from F1 hybrids of CBA and NZB mice (Roubertoux and Carlier, 1988), and inbred lines of wild mice selected for aggression (Van Oortmerssen and Sluyter, 1994). These studies showed not only that the Y chromosome was important, but also using cross-fostering that the postnatal maternal environment could alter aggressive behavior (Carlier et al., 1991; Sluyter et al., 1995). In contrast to the studies by Maxson and colleagues, another group suggested that a gene within the PAR of the Y chromosome, rather than an NRY gene, influenced differences in aggression by using different congenic strains (N and H) with substitutions in the PAR regions of the Y chromosome (Roubertoux et al., 1994). Eventually, both groups studying Y chromosome effects on aggressive behavior directly compared their hybrid strains using the same testing procedures, and agreed that an NRY gene (presumably Sry) was the best candidate gene on the Y chromosome associated with aggression (Guillot et al., 1995). Nonetheless, using a different testing paradigm, the PAR located steroid sulfatase gene (Sts) and its enzymatic activity were found to be positively correlated with aggressive behavior (Le Roy et al., 1999; Mortaud et al., 2010); thus, suggesting that both Sts and Sry may be involved in the regulation of aggressive behavior.
These early studies on the genetics of aggressive behavior revealed the importance of the Y chromosome, but due to the function of the Sry gene, could not delineate effects that were caused by differences in androgen secretion. However, research from other animal models (such as birds) suggested that the sex chromosomes might contribute to brain development and behavior directly, not via the actions of gonadal hormones. Thus, while Sry and Sts may be involved in strain differences in male aggression, these and other sex chromosome factors may also influence sex differences in aggression. Following this work, studies with gonadectomized and testosterone replaced FCG mice in the C57BL/6J background showed that the XYF were faster and more likely to attack a submissive male intruder in their home cage as compared to XXF, which virtually never attack (Gatewood et al., 2006). These results suggested that genes on the Y chromosome other than Sry have an effect on aggression. However, as mentioned above, the FCG Y chromosome is from the 129 strain. Since normal males of the 129 strain attack intruders more than C57BL/6J males (Abramov et al., 2008), if the strain difference is due to a Y-genetic component as others have noted with hybrids from other strains (Maxson et al., 1979; Roubertoux et al., 1994), aggression in FCG XYF may depend on the strain origin of the Y chromosome. Moreover, the data in FCG mice was collected prior to the strain being completely backcrossed into a C57BL/6J background (still partially on a MF1 background), and this too could have influenced the aggressive behavior in these mice.
In addition to genes on the Y chromosome, the X chromosome may also influence aggression. To investigate the contribution of the X chromosome in offensive aggression, adult Y* mice of both sexes were gonadectomized, testosterone replaced, and tested in resident-intruder tests. Neither 2X nor 1X females (2XF and 1XF) attacked intruders (neither genotype has a Y chromosome). However, 2X male (2XM) mice showed a trend for increased aggression as measured by attack latency and number of attacks (Bonthuis et al., 2012). Prior to this study, two genes located on the X chromosome had been implicated in aggression: the previously discussed Sts gene which is in the PAR of both sex chromosomes (Le Roy et al., 1999), and Monoamine oxidase A (Maoa), which encodes an enzyme that degrades monoamine neurotransmitters. Transgenic male mice with MAOA deficiency show increased aggressive behavior (Cases et al., 1996). Interestingly, while MAOA levels have not yet been assessed in adult Y* mice, neither genotype of male Y* mice possess the Y chromosome copy of the Sts gene because of the nature of the Y* rearrangement (Davies et al., 2007), suggesting that Sts is not the cause of the X chromosome effect on aggression in Y* males.
Along with the FCG and Y* mice, three additional models, SF1 KO, POS (XYPos) and Odsex mice, have been used to assess the role of the sex chromosomes on aggression, and resulted in conflicting findings. In one study, XY (genetically male) and XX (genetically female) SF1 KO and WT control mice were gonadectomized prior to puberty, treated with testosterone, and tested with intruder males (Grgurevic et al., 2008). The SF1 KO mice of both XX and XY genotypes (XXKOSF1 and XYKOSF1) were far less aggressive than wild type males (XYMSF1), indicating that testosterone given during adulthood was not enough to overcome a deficit in steroid hormones during development, and importantly, that hormone exposure, rather than genetic sex, is needed for proper development of the neural circuits that regulate aggressive behavior. These results point to a potential interplay between sex chromosome genes and hormones during development to establish sexually dimorphic neural circuitry. In another study, ovary-bearing XYPosF were no different than XX females in measures of aggression toward an intruder male, and displayed less aggressive behavior than XY males, while Odsex XX males (XXMOds) did not differ from XY males in aggressive behavior (Canastar et al., 2008). However, in both of these experiments the aggression tests were done in a neutral cage, a significantly different test than resident-intruder aggression, and one that might reduce the amount of aggression since neither mouse is the resident. In addition, the mice did not receive exogenous testosterone to normalize hormone levels, which is in contrast to the FCG and Y* studies. Therefore, the differences between the findings of this study and others may be caused by strain differences and/or discrepancies in testing conditions.
Sexual Behavior
Like aggression, male sexual behavior may be sensitive to the strain of origin of the Y chromosome. The first demonstration of this compared DBA/2 males to males with a DBA/1 Y chromosome in the DBA/2 genetic background (DBA/2.DBA/1-Y). In this case, the DBA/1 Y chromosome reduced the frequency of males mounting during a 10-minute test to 13%, down from 50% in normal DBA/2 mice possessing their own Y chromosome (Shrenker and Maxson, 1984). These findings may be relevant to the two subsequent sexual behavior studies conducted in FCG mice, in which different results were found using mice on two different background strains.
The first study examining behavior in FCG mice was in the MF1 background strain and did not detect any sex chromosome complement effects on male sex behavior (De Vries et al., 2002). However, this study showed that both XXF and XYF were faster to initiate mounting and thrusting than were XXM or XYM. Furthermore, when the XYM in the FCG cross (harboring a Y from the 129 strain) were compared with normal XY males possessing an MF1 Y chromosome, the normal XY males took longer to commence thrusting and displayed fewer mounts than FCG XYM (XY− Sry males), suggesting a Y chromosome strain-of-origin effect. Alternatively, differences in the prenatal or postnatal environments or effects of the Sry transgene might also contribute to these strain differences.
The second FCG study used mice on the C57BL/6J background (Bonthuis et al., 2012). Again, females were faster to mount and thrust, and engaged in more of these behaviors than males, indicating that these behaviors are not exclusively displayed by males. However, in contrast to the finding from the mice on an MF1 background, there was also a sex chromosome effect: counter intuitively, XX individuals (XXM and XXF) displayed more mounts and thrusts than XY mice in both sexes. Moreover, XXM were significantly faster to ejaculate than XYM. Aside from differences between strains, discrepancies in the data between the two studies using FCG mice may have also been produced by interactions between the autosomes and the 129 Y− chromosome. Regardless, as is true of all studies using only FCG mice, these results do not reveal whether the absence of a Y chromosome, or the presence of the extra X chromosome produces faster ejaculation latencies in XX males.
To differentiate between the effects of multiple X chromosomes versus the presence of the Y chromosome, male sexual behavior in Y* mice of the B6Ei.LT-Y*/EiJ strain was also examined. When gonadectomized and given testosterone implants, both 2XM and 2XF, displayed increased male sex behaviors as compared with mice of both sexes with only one X chromosome (Bonthuis et al., 2012), indicating that dosage of X chromosome genes influences male sexual behavior. These data clarify the interpretations of the FCG data and show that mice with two X chromosomes perform more male sexual behavior. In addition, males with two X chromosomes (2XM) ejaculated more frequently than males with a single X (1XM). In contrast to Bonthuis et al. (2012), a study using XYPosF females, and XXMOds, found that they were no differences in measures of male sex behavior when compared to normal XX females, and XY males or XYMOds, respectively (Canastar et al., 2008). However, XYPosF in this study were gonad intact and not treated with testosterone, and the frequencies and latencies to ejaculation were not reported in XXMOds; therefore, it is difficult to directly compare the results of the two studies.
The experiments conducted in the Y* mice also suggested that aneuploidy influences male sexual behavior. This was tested directly using aneuploids in the MF1 background (Park et al., 2008). In the absence of an Sry transgene, an extra Y− chromosome increased the frequency of ejaculation, and decreased the latency to intromit and ejaculate in XYY− males compared to XY control males. In the presence of the Sry transgene, the extra Y− had no effect on male sex behavior. In contrast to work on the FCG and Y* male mice, the presence of two X chromosomes appeared to increase latencies to mount, thrust and ejaculate, and decreased the number of mounts, intromissions and ejaculations in XXSry and XXY− Sry males as compared to XYSry and XYY− Sry males, respectively. Importantly, these differences were not confounded by genotype differences in testosterone levels at the time of testing, because all animals were gonadectomized and replaced with testosterone in adulthood. However, the direct comparisons of the effects of aneuploidy are complicated by Y-chromosomes from two different inbred lines, and by having either or both copies of an endogenous and transgenic Sry.
Like male aggressive behavior, female sexual behavior is also highly sexually dimorphic, but to date there is only one report on the role of sex chromosomes in the development of female-typical sexual behaviors. Adult SF-1 knockout mice and their wild-type littermates were given injections of estradiol, followed by progesterone, to induce behavioral estrus (Grgurevic et al., 2012). While, as expected, WT females were the most receptive and attractive to a stud male and WT males the least receptive and attractive, a sex chromosome difference persisted in the SF-1 knockout mice with the XXKOSF1 mice receiving more mounts from males than the XYKOSF1. It is important to note that all SF1 KO mice (XXKOSF1 and XYKOSF1) have female external phenotypes. These findings suggest that the development of testes in males is not the only factor important for defeminizing their behavior, and that genes on the Y chromosome may reduce, or genes on the X might increase, receptivity. Moreover, the investigators reported that the sex chromosome effect might be attributable to differences in progesterone receptor expression within the hypothalamus (Grgurevic et al., 2012). However, in FCG neonates sexual dimorphisms in the numbers of progesterone receptor containing cells was not influenced by sex chromosome composition (Wagner et al., 2004). Unpublished data from our laboratory also failed to show any differences in the receptivity scores of FCG females (XXF versus XYF; Rissman et al., unpublished), but it will be important to investigate the role of sex chromosome genes on female sexual behavior using other models. For example, experiments using the Y* model could help to differentiate whether a second X chromosome increases, or Y decreases, receptivity in SF1 KO mice.
Parental Behavior
The influence of sex chromosome genes on parental behavior has been examined in two mouse models. In the FCG mouse on a C57BL/6J background, spontaneous maternal behavior is influenced by sex chromosome complement. When adult, virgin FCG mice of both sexes were exposed to newborn pups, XXF retrieved more pups back to the nest, and began retrieving sooner, than XYF. The XYF were indistinguishable from the males in both behavioral measures (Gatewood et al., 2006). These findings are similar to those found in sex-reversed (Sxr) mice, where, in tests of parental care, Sxr XX males (XXSxrM) displayed less infanticide and more pup retrieval than XY males, but were not quite as good as XX females at these tasks (Reisert et al., 2002). However, in this study the authors did not discriminate XY from XYSxr males, which could have potentially altered their interpretation. Nonetheless, in sum, these studies suggest an inhibitory effect of the Y chromosome on parental behavior, and/or a facilitating effect of X chromosome on these behaviors. However, an advantage of the XX genotype was not noted in the FCG XXM. Alternatively, the amount of androgen exposure during the neonatal period may differ between these strains and this may influence adult parental behaviors.
Social Interactions
Non-aggressive, social interactions have also been examined in the FCG mouse and Y* mouse models, both in the C57BL/6J strain. Adult gonadectomized XY FCG mice of both sexes spend more time engaged in social activities, such as investigating and grooming a strange male, as compared to XX mice (McPhie-Lalmansingh et al., 2008). In these same tests, XXF mice, in comparison to the other three genotypes, also spend more time performing digging, an asocial behavior. Social interactions and play behaviors have been examined in juvenile FCG mice (Cox and Rissman, 2011). When paired with an unfamiliar mouse of the same age and sex; juvenile XXF (21–22 days of age) were less social than all other genotypes; however, when paired with same-sex siblings they were the most social of the FCG genotypes. In play-soliciitng behavior, which is distinguishable from prolonged social contact, XYM were the most likely to solicit play from an unfamiliar mouse, and XXF were the least likely to solicit play from a sibling. Both of these studies suggest a contribution of sex chromosome in social interactions in mice without an indication of which sex chromosome(s) are having this effect.
A recent study using juvenile Y* mice has expanded on these earlier findings in the FCG. It was found that both 2XM and 2XF are more interactive and investigate a novel mouse more than 1X mice (Cox et al., unpublished data). While different from the findings in juvenile FCG mice that XXF are less social in certain contexts, these data are consistent with previous findings in adult Y* mice. Adult 2XM mice spend more time than 1XM investigating a novel mouse than a novel object (Liu et al., 2010). Taken together with the FCG data, these results suggest a role for both Y and X chromosome genes in the development of social behaviors.
Anxiety
The first behavioral data generated using the Y* mouse directly compared anxiety in the 1XF mice (generated by the Y* model on an MF1 background) to normal 2XF. These studies were based on observations made in Turner syndrome patients, which demonstrated a parent of origin effect. Patients with their single X from their mothers had more severe impairments to executive function and social behaviors than patients with a paternal X (Skuse et al., 1997). To further assess the potential contribution of genetic imprinting, 39,XO female mice carrying either maternal (Xm) or paternal (Xp) X chromosomes were necessary for comparative studies (Isles et al., 2004). Because the 1XF from the Y* model retain PAR and a small piece of X chromosome (the Y*X), the authors reasoned that behavioral differences between 1XF and true 39,XO mice could be attributed to genes present on the Y*X (Figures 4 and 5). In initial experiments, mice were tested for anxiety (on the elevated plus maze) and locomotor activity. There were no differences in locomotor activity between groups, and no effect of X chromosome imprinting on either behavior, as 39,XmO and 39,XpO behaved similarly. However, both 39,XO and 1XF (from the Y* cross) displayed increased anxiety on the elevated plus maze as compared to 2XF, indicating that the X gene(s) involved in modulating fear reactivity are not on the Y*X chromosome fragment but reside in NPX.
Figure 5. Offspring of the Y* Cross.
When an XX female (XXF) is mated with a male possessing a Y* chromosome (XY*M), four different genotypes of offspring are produced from the inheritance of the sire’s sex chromosome. Every pup receives a maternal X chromosome, and in combination with the paternal sex chromosome the four genotypes are: XX female, 2XF; XY*X female, 1XF; XXY* male, 2XM; and XY* male, 1XM. NPX, the non-pseudoautosomal region of X (pink). NRY, the non-recombining region of Y (yellow). PAR, the non-pseudoautosomal region of Y broken into proximal (a = red), middle (b = orange), and distal (c = green) domains.
More recent studies using juvenile Y* mice on the C57BL/6J background found the same X chromosome effect of heightened anxiety in 1X mice compared to 2X, and, since the juveniles were pre-pubertal, the findings indicate that circulating gonadal hormones are not responsible for this behavioral difference (Cox et al., unpublished data). Importantly, this study also examined gene candidates responsible for this behavioral phenotype, and found that X chromosome number correlates with the expression of vasopressin in the amygdala. Vasopressin is a neuropeptide implicated in a variety of social behaviors in mammals (De Vries, 2008), and is also involved in the regulation of anxiety (Neumann and Landgraf, 2012).
Visuospatial Attention
In addition to anxiety, subsequent experiments using adult MF1 Y* mice examined visuospatial attention in a 5-choice serial reaction time task. As with the elevated plus maze, there was no effect of X chromosome imprinting on baseline behavior in this task. Yet, when the difficulty of the task was increased, 1XF performed as well as normal 2XF, while the 39,XO females made more errors (Davies et al., 2007). Based on these results, the authors hypothesized that the Sts gene, located within the PAR of the sex chromosomes, may be responsible for the attentional deficit seen in 39,XO females. As previously mentioned, the 1XF from the Y* cross have a copy of Sts on the chromosomal fragment (Y*X) that they inherit, and therefore they, like normal XX females, possess 2 copies of the gene. In contrast, the Y* and XY* chromosomes are lacking this gene, due to the deletion within the PAR; therefore, males of the Y* cross, like true 39,XO females, have only 1 copy of the Sts gene.
To confirm that the dose of the Sts gene is involved in attention, a different breeding strategy was used to generate 39,XY*O male mice (with a single X-attached-to-Y chromosome, and thus no Sts gene). These mice were compared to normal XY males (with 2 copies of Sts) in the same 5-choice serial reaction time task. Results from the 39,XY*O males were similar to those obtained in females, showing deficits in attention, further implicating Sts (Davies et al., 2009). More recent studies have expanded on those findings, by showing that XY*O males are hyperactive as compared to XY males and they display increased anxiety in a light-dark box (Trent et al., 2012). However, no direct comparisons have been made between males and females of the Y* cross to determine which other factors, such as gonadal hormones and Y chromosome effects, may also influence attention.
Learning and Cognition
Many genes on the X chromosome are linked to mental retardation (Zechner et al., 2001), pointing to an important role for the X chromosome in cognition. In addition, data from Y* mice in an MF1 background indicate that X chromosome dosage can influence cognitive behaviors. When Y* males were tested in a novel-object recognition task, 1XM recognized the novel object as shown by their increased investigation, whereas 2XM investigated familiar and novel objects equally (Lewejohann et al., 2009). Studies using 41,XXY aneuploid males (Aneuploids, Table 1), also showed impaired Pavlovian conditioning in males with two X chromosomes (Lue et al., 2005) compared with XY males. Together, these studies indicate that an extra X chromosome in male mice causes deficits in cognitive behavior.
In addition to the number of X chromosomes, the parent of origin of the X chromosomes may also have effects on learning. Although there was no suggestion of X chromosome imprinting noted in the anxiety or attention tasks reviewed above, when 39,XmO and 39,XpO female mice were compared to XX females in a serial reversal learning task, 39,XmO females displayed more errors in this task and showed more perseveration as compared to 39,XpO and XX females (Davies et al., 2005). These results led to the discovery of a novel imprinted gene on the X chromosome, Xlr3b, which was also found to be expressed in a sexually dimorphic manner, with XY females and 39,XmO females showing higher expression levels compared to 39,XpO and XX females (Davies et al., 2005). Thus, there may be another level of complexity when considering X chromosome effects on cognitive tasks.
PAR genes, found on both sex chromosomes, may also be a source of variation in learning. XX females on reciprocal congenic backgrounds that were genetically identical except for the source of their sire’s Y chromosomes displayed differences in abilities to learn in two spatial mazes: the Morris water maze, and the Lashley maze (Hoplight et al., 2000). These results suggested that mere exposure to an unknown genetic difference between the two Y chromosomes could potentially alter offspring behavior, presumably through the exchange of genetic information in the PAR between the Y and X chromosomes during meiosis. However, sires remained with the dams after the litters were born, therefore there may have also been postnatal paternal, environmental effects on offspring behavior.
Feeding and Metabolic Activity
Sex chromosome complement has also been shown to effect food intake (Chen et al., 2012; Chen et al., 2013a). Gonadectomized XX FCG mice from both sexes (in the C56BL/6 line) gained more weight, with an increase in fat deposits, compared with XY mice over a 10-month period. XX mice also had a higher percentage of fat versus lean body mass when fed chow with a normal fat content. When switched to a high fat diet, XX mice gained more weight with higher levels of fat and lean mass, secreted more leptin, and suffered more adverse metabolic consequences of a fatty liver and reduced insulin sensitivity than did XY mice. Importantly, the increase in fat mass correlated with a behavioral difference wherein XX FCG mice consumed more food than XY mice during the light portion of the day, a time when mice are less active and eat less.
In gonadectomized Y* mice (MF1) the number of sex chromosomes is also positively correlated with food intake during the first three days when fed a high fat diet. 2XF (with two sex chromosomes) gained more weight than 1XM (with 2 sex chromosome) and were equal to 2XM (with 3 sex chromosomes) after 16 weeks on high fat diet (Chen et al., 2013a). Results from that study, also showed that sex chromosome aneuploidy, in and of itself, influences metabolic measures. The authors crossed FCG and Y* models to make novel males (XY−Y*XSry+), then mated them to XX females (all on an MF1 background) to produce aneuploid offsping (Table 1). The number of sex chromosomes and type (either X or Y) affects body weight and fat mass (Chen et al., 2013a). Mice with two or three sex chromosomes had a higher body weight than mice with a single X chromosome. From these results, it is clear that the influence of sex and sex chromosomes on food intake, metabolic factors, and body mass is complex and will require continued effort to unravel.
Focusing on 11 genes that escape X inactivation in FCG mice in C57BL/6 background, higher expression levels in all three XX peripheral tissues of the liver, and inguinal and gonadal fat, as compared with XY tissues, suggest three candidate: Eif2s3x, Kdm6a, and Ddx3x (Chen et al., 2012). Six of the 11 escapees examined (Kdm5c, Usp9x, Uba1, Rik, Shroom4, and Mid1), were also differentially expressed in at least one of the tissues; all except Mid1 were more highly expressed in XX tissue. Furthermore, systemic signals (i.e. leptin, glucose, insulin) feedback on the brain to affect feeding behavior, and we have found that growth hormone (GH) is sexually dimorphic in several hypothalamic regions (Addison and Rissman, 2011). In addition, we have shown in Y* mice that body weight, which is influenced by the number of X chromosomes, is positively correlated with GH mRNA in the mPOA (Bonthuis and Rissman, 2013). Peripheral and central administration of GH increases appetite (Blissett et al., 2000; Bohlooly et al., 2005; Bonthuis and Rissman, 2013), and GH is peripherally secreted in sexually dimorphic circadian patterns (Jaffe et al., 1998). Our working hypothesis is that part of the impact of sex chromosome complement on body weight is mediated by a central effect on GH, which stimulates appetite particularly during the inactive part of the day/night cycle. Intriguingly, a recent paper on sex differences in circadian rhythms demonstrated a small difference in FCG mice. In the C57BL/6J strain, XX mice were more active in constant dark than XY mice, and XXM mice were more active than XXF. In a light cycle however, no sex chromosome effects were found (Kuljis et al., 2013). Together, these data suggest that sex chromosome genes could be intimately involved in a variety of homeostatic behaviors.
Nociception and Habit Formation
One additional interesting behavioral phenotype is that FCG mice (on either a C57BL/6J or an MF1 background) with an XX genotype are more sensitive to noxious pain than are XY mice, regardless of gonadal sex. Adult XXM and XXF are faster to withdraw their paws from a hotplate, and spend more time licking their paws when injected with formalin, as compared with XY males and females (Gioiosa et al., 2008a). Similarly, when tested the day after birth, XX pups are faster to withdraw their tails from a hot water bath than are XY pups (Gioiosa et al., 2008b). In humans, sex differences in pain perception have been studied for decades, but some of the findings are contentious (Racine et al., 2012); therefore, while the results of the FCG studies are intriguing, it is unclear what role the sex chromosomes are playing in this particular behavioral response.
In contrast to the pain literature, sex differences in addiction are well documented, although not always in the same directions when comparing rats, mice and humans (Anker and Carroll, 2011; Kerstetter and Kippin, 2011). In a test of conditioned habit formation for a food reward (Quinn et al., 2007), XX (all studies in the MF1 FCG) mice regardless of gonadal sex formed the habit more quickly than XY mice. When food was paired with conditioned taste aversion, XY mice showed an aversion while XX mice did not, suggesting that they had acquired an instrumental habit. However, when alcohol was used for conditioning, XY mice did not devalue alcohol when it was paired with conditioned taste aversion, whereas XX mice reduced their consumption (Barker et al., 2010). Whether X or Y sex chromosome genes influence sex differences in habit formation and why natural versus alcohol rewards are percieved differently by XY and XX mice have yet to be determined, but these findings are interesting, nonetheless.
Summary and Conclusions
Y chromosome effects on behavior
Collectively, the literature on aggressive and social behaviors suggests an effect of Y chromosome genes on male social interactions (Gatewood et al., 2006; McPhie-Lalmansingh et al., 2008); but in some cases exposure to androgens may substitute for the actions of Y genes. It is possible that Y chromosome genes “prime” a male to respond to testosterone later, and therefore that females with a Y chromosome are also primed for this response. It is not clear which gene(s) on the Y chromosome are involved, and, since the Sry gene is always coupled with the development of testes, FCG mice cannot be used to assess the direct effects of Sry on these behaviors. However, data from SF1 KO mice, (Grgurevic et al., 2008), suggest that expression of Sry outside of the gonad has no impact on aggressive behaviors. These results are complicated by the fact that, as previously mentioned, the ventromedial nucleus of the hypothalamus (VMN) of SF1 KO mice is disorganized. Indeed, the ventral lateral VMH (vlVMH) is critically important for aggression (Lin et al., 2011; Nelson and Trainor, 2007; Yang et al., 2013). Therefore, their aggressive behavior may, in fact, be related to the unique cellular organization of the VMN, rather than sex chromosome effects.
Effects attributed to the X chromosomes
A role for X chromosome genes in male sexual behavior has been clearly established using both the FCG and Y* mouse models (Bonthuis et al., 2012). In addition, females also display more male copulatory behaviors (indicative of sexual motivation) than males. We need to broaden our examination of the contexts under which these behaviors occur to discover the role of these behaviors when conducted by normal females. One possibility is that females use mounting behavior in social interactions (dominance) with other females (Fang and Clemens, 1999).
In addition to male sexual behavior, there also appears to be an X chromosome effect on female attractivity and receptivity, as shown using the SF1 KO mice (Grgurevic et al., 2012). In our laboratory we attempted to replicate this finding with FCG mice that were gonadectomized prior to puberty, but we did not find a sex chromosome effect on receptivity (unpublished data). However, in contrast to our study, the SF1 KO mice were fed a phytoestrogen-free diet to eliminate the influence of exogenous estrogens on behavior, and they never had ovaries. Thus, as with male aggressive behavior, hormones, both endogenous and in the environment, may interact with sex chromosome genes and block their actions on female sexual behavior.
Aside from social behaviors, X chromosome genes may also impact a variety of other behaviors. The number of X chromosomes influences anxiety behavior as shown in both adult and juvenile Y* mice (Davies et al., 2007)(Cox et al., unpublished data), and there has been an elegant demonstration of the affect of X chromosome genes on food intake and metabolism (Chen et al., 2012). X chromosome copy number is also involved in learning and cognition, as has been shown with the Y* mouse model (Lewejohann et al., 2009; Lue et al., 2005), and, interestingly, parent of origin effects are implicated in learning tasks that are influenced by the X chromosome (Davies et al., 2005). Attempts to uncover the imprinted X chromosome genes led to the discovery of a cluster of imprinted genes on the mouse X (Raefski and O’Neill, 2005), but, unfortunately, none have human homologs.
Future Directions for Behavioral Research
Taken together, the data presented here lead us to conclude that genes on the Y chromosome are playing an important role in the sex difference in male-typical defensive aggression. However, genes on the X chromosome are solidly implicated in several other behaviors, including male sexual behavior, social behavior, anxiety/attention, and feeding. Two of these four behaviors are not sexually dimorphic, and the one behavior that is classically considered sexually dimorphic, male copulatory behavior, is actually not highly dimorphic under conditions of testosterone replacement in C57BL/6J mice, the strain used for many of the behavioral studies discussed. To date, only these few behaviors have been examined for the potential roles of sex chromosome genes. However, it is likely that a variety of other behaviors are influenced by sex chromosome genes, and perhaps include behaviors that are not as strongly influenced by gonadal hormones as those that have been investigated in the literature.
In addition to the need for discovering new behaviors to interrogate, there is still follow-up work to be done on the behaviors that we know are influenced by sex chromosomes. The data on Y chromosome effects on aggression have been available for many years, but so far there has not been direct manipulation of Y chromosome gene candidates. It would be prudent to focus on the genes that are essential for male aggression. Partial Y chromosome deletion (short versus long arm) mice, BAC insertions in XX mice, and CRISPRi (Gilbert et al., 2013; Larson et al., 2013) technologies could be used to test the importance of specific Y genes. Few laboratories have made use of these and other modern genetic techniques that could lead to important information on the interactions between genes on Y and hormone levels, which undoubtedly affect behavior (Abel and Rissman, 2012). Also, studies that manipulate the source of the Y chromosome and neonatal hormones are warranted. We have shown that sex differences in calbindin message in the cerebellum of mice is influenced by both sex chromosome complement and by the estrogen receptor alpha, but how these two factors interact is not known (Abel et al., 2011). The use of classical sexual differentiation paradigms (Phoenix et al., 1959) to investigate such gene-hormone interactions would be a large step ahead for the field. Additionally, environmental variables that may trigger enzymatic signaling, such as low nutrition, can also be modeled in mice with sex chromosome abnormalities.
In combination with behavioral research, more work also needs to be done to map the effects of sex chromosome genes on the development of brain regions involved in behaviors. This will require using not only traditional mapping and immunocytochemical approaches, but also novel technologies to examine brain activity. A recent fascinating experiment used neural imaging of XX, XY and 39,XpO mice in young adult hybrid mice derived from a novel crossing strategy to investigate sex chromosome effects on activity (Raznahan et al., 2013). Genotype differences in activity were noted in the majority of regions examined, but the most striking differences between 39,XO and XX brains were present in the bed nucleus of the stria terminalis, thalamus, and the nucleus accumbens. How these differences are related to behavior has yet to be determined, but point to a tremendous opportunity for research in this field.
Relevance to Human Behaviors
The mouse models we have presented in this review have the potential to inform human research on sex chromosome related disorders. The mice used are not designed to mimic human disease, but rather are engineered to isolate effects of X and/or Y-chromosomes on behavior. However, there are patients that have chromosomal abnormalities similar to these mice. For example, individuals with mutations of regions of the Y chromosome that include the SRY or SOX9 genes have disorders of sexual differentiation (Sim et al., 2008). Phenotypes of these 46,XY individuals include gonadal dysgenesis, an external female phenotype, and in some cases underdeveloped gonads that produce no or low hormone levels. Likewise, translocations of SRY can produce 46,XX individuals, and in some cases the X chromosome with the translocation is preferentially activated (Bouayed Abdelmoula et al., 2003), and the individuals have testes but are infertile. These types of mutations in humans are rare, and so the precise chromosomal complements produced by the FCG, sex reversed mice, or the odsex mutation are not often mirrored in humans. Certainly there are too few patients to determine whether their gonadal and sex chromosome discordance influences any of their behaviors.
Unlike the abovementioned rare sex chromosome mutations, sex chromosome aneuploidy is very common in humans. Klinefelter Syndrome (47,XXY) is the most common sex aneuploidy disorder and occurs in 1 out of every 1,000 to 600 men (Savic, 2012), while Turner Syndrome (45,X) affects approximately 1 in every 2,000 women (Elsheikh et al., 2002). In these syndromes gonadal and chromosomal sex are concordant; however, patients may have cognitive and social behavioral phenotypes (Lesniak-Karpiak et al., 2003; Schmidt et al., 2006). The data collected in mice may lead to predictions about genes on sex chromosomes that affect behavior in humans.
There is a wide range of symptoms found in men with Klinefelter syndrome. Delayed puberty and infertility are the main reasons that these men are diagnosed, but it is likely that more than 80% are undiagnosed (Savic, 2012). Working with what may be the most severe population, a variety of abilities have been assessed in 47,XXY men, and these patients have lower abilities in object and pattern recognition (Bruining et al., 2010), lower scores on cognitive tests (DeLisi et al., 2005), and impaired social processing (Temple and Sanfilippo, 2003; van Rijn et al., 2006) as compared with normal 46,XY men. The aspect of the syndrome that has received the most attention is impaired verbal abilities, including lower verbal IQ scores, in Klinefelter boys (Geschwind et al., 2000), which correlate with differences in brain morphology and function. Specifically, the frontal and temporal lobes and the cerebellum are reduced in volume in Klinefelter males (Steinman et al., 2009), and Klinefelter men have less lateralized processing in language centers than controls (Aleman et al., 2008). One of the key features of autism spectrum disorders (ASD) is disrupted language skills, and in one study 27% of boys with Klinefelter syndrome (Bruining et al., 2009) met the criteria for ASD. In addition, Klinefelter men are more likely than controls to have psychosis symptoms, schizophrenia, and/or depressive disorders.
Sex chromosome trisomy can present in configurations other than 47,XXY and for males 47,XYY chromosome complement may be just as common, along with 47,XXX in females. Because these two conditions do not interfere with gonadal hormone production they are rarely diagnosed. However, there are some similarities between these conditions and Klinefelter syndrome (Bishop et al., 2011). Klinefelter and 47,XXX share similarly disrupted motor skills and language impairments. Boys with 47,XYY also have language deficits and lower than average IQ scores (Leggett et al., 2010). Boys with sex chromosome trisomy also have attention and executive control difficulties and poor motor control (Ross et al., 2009). However, a study using a population identified via prenatal diagnosis found that 37% of 47,XXY, 14% of 47,XYY and 55% of 47,XXX children had no neurodevelopmental difficulties (Bishop et al., 2011). This variability could be caused by several mechanisms described below.
In contrast to sex chromosome trisomy, girls with Turner syndrome, characterized by the loss of all or part of an X chromosome, are recognized at an early age based on short stature and other physical characteristics. Turner syndrome is associated with cognitive deficits, including problems with visuospatial perception and memory, emotionality, and attention (Skuse, 2005; Skuse et al., 2005). Girls with Turner syndrome also have reduced social competence and facial recognition (Hong et al., 2011; Lawrence et al., 2003), and may be at higher risk for autism (Burnett et al., 2010; Marco and Skuse, 2006). Using a tracking task that required increasing amounts of attention, Beaton and colleagues (Beaton et al., 2010) examined BOLD patterns in normal and Turner syndrome girls. When the task was a simple activity, Turner brains were similar to the normal girls. However, as the number of objects tracked increased so did activity in several cortical areas, including prefrontal cortex and in the amygdala. The authors suggest that the Turner girls process information differently from the controls, using less efficient subcortical regions. Interestingly, a variety of phenotypes in Turner patients vary in severity depending on the parent of origin for the single X chromosome (Donnelly et al., 2000; Ko et al., 2010; Lepage et al., 2011). In general, a more pronounced phenotype is noted in girls with a maternally inherited X as compared with the paternal X only (Lepage et al., 2012). While imprinting in mice may be different from humans, data from mice may give more information as to which autosomal genes are downstream of the X chromosome genes responsible for this effect on cognition.
Pre-pubertal girls with Turner syndrome are typically treated with growth hormone and later with estrogens prior to and after puberty. This is done to both increase heights to near average and to ameliorate the low levels of estrogens produced by dysfunctional ovaries. A few, but not all, of the behavioral symptoms of this illness are also reduced by hormone treatment (Ross et al., 1998; Ross et al., 2004; Ross et al., 2002). Klinefelter boys are typically older when their disorder is detected and testosterone replacement is prescribed. Few studies have sorted out effects of hormones on behavior in this population, and the lasting neurobehavioral effects of aneuploidy indicate that this is an area that requires further research. Studies in mice that lead to candidate genes affecting cognitive and social behaviors may well inform treatment of Klinefelter and Turner syndromes. Reciprocally, the few candidate genes that have been suggested based on the human data; STS, NLGN4X, and RPS4X (Pessia et al., 2012; Zinn et al., 2007), could have their roles investigated in mice.
While we have focused on abnormal human sex chromosome arrangements in our discussion of human studies, it is also very likely that individual genes identified to affect behavior in mice will also have effects in normal humans. For example, KDM5C (also known as SMCX and JARID1C) is a gene that escapes X-inactivation in mice and in humans. Human males with normal XY sex chromosome complement, and mutations in their single copy of this gene have mental retardation (Akbarian and Huang, 2009; Jensen et al., 2005). This gene codes for a histone demethylase and as such may regulate transcription of multiple autosomal and/or other sex chromosome genes. Other X-inactivation escaping genes (i.e. RPS4X) also conserved in mice and humans may provide new information about the roles of X chromosome gene expression in normal human behavior. The X chromosome genes are highly expressed in brain, and mutations in several lead to mental illnesses. We speculate that in lieu of DNA mutations, epigenetic modifications of these genes, may lead to disease. Thus, the sex chromosomes may do much more than just influence sex differences in behavior: they may be key to the regulation of many neural processes.
Highlights.
Mouse models with sex chromosome aneuploidy are reviewed.
Social and other behaviors correlate with sex chromosome compositions.
Implications for humans with similar sex chromosome conditions are suggested.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Selected Publications
- Abel JL, Rissman EF. Location, location, location: genetic regulation of neural sex differences. Rev Endocr Metab Disord. 2012;13:151–61. doi: 10.1007/s11154-011-9186-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abel JM, Witt DM, Rissman EF. Sex differences in the cerebellum and frontal cortex: roles of estrogen receptor alpha and sex chromosome genes. Neuroendocrinology. 2011;93:230–40. doi: 10.1159/000324402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abramov U, Puussaar T, Raud S, Kurrikoff K, Vasar E. Behavioural differences between C57BL/6 and 129S6/SvEv strains are reinforced by environmental enrichment. Neurosci Lett. 2008;443:223–7. doi: 10.1016/j.neulet.2008.07.075. [DOI] [PubMed] [Google Scholar]
- Addison ML, Rissman EF. Sexual dimorphism of growth hormone in the hypothalamus: regulation by estradiol. Endocrinology. 2011;153:1898–907. doi: 10.1210/en.2011-1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akbarian S, Huang HS. Epigenetic regulation in human brain-focus on histone lysine methylation. Biol Psychiatry. 2009;65:198–203. doi: 10.1016/j.biopsych.2008.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleman A, Swart M, van Rijn S. Brain imaging, genetics and emotion. Biol Psychol. 2008;79:58–69. doi: 10.1016/j.biopsycho.2008.01.009. [DOI] [PubMed] [Google Scholar]
- Anker JJ, Carroll ME. Females are more vulnerable to drug abuse than males: evidence from preclinical studies and the role of ovarian hormones. Curr Top Behav Neurosci. 2011;8:73–96. doi: 10.1007/7854_2010_93. [DOI] [PubMed] [Google Scholar]
- Arnold AP. The organizational-activational hypothesis as the foundation for a unified theory of sexual differentiation of all mammalian tissues. Horm Behav. 2009;55:570–8. doi: 10.1016/j.yhbeh.2009.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold AP, Rissman EF, De Vries GJ. Two perspectives on the origin of sex differences in the brain. Ann N Y Acad Sci. 2003;1007:176–88. doi: 10.1196/annals.1286.018. [DOI] [PubMed] [Google Scholar]
- Barker JM, Torregrossa MM, Arnold AP, Taylor JR. Dissociation of genetic and hormonal influences on sex differences in alcoholism-related behaviors. J Neurosci. 2010;30:9140–4. doi: 10.1523/JNEUROSCI.0548-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beaton EA, Stoddard J, Lai S, Lackey J, Shi J, Ross JL, Simon TJ. Atypical functional brain activation during a multiple object tracking task in girls with Turner syndrome: neurocorrelates of reduced spatiotemporal resolution. Am J Intellect Dev Disabil. 2010;115:140–56. doi: 10.1352/1944-7558-115.2.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berletch JB, Yang F, Xu J, Carrel L, Disteche CM. Genes that escape from X inactivation. Hum Genet. 2011;130:237–45. doi: 10.1007/s00439-011-1011-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bishop CE, Mitchell MJ. Encyclopedia of the mouse genome V. Mouse Y chromosome. Mamm Genome. 1996;6(Spec No):S331–3. [PubMed] [Google Scholar]
- Bishop CE, Whitworth DJ, Qin Y, Agoulnik AI, Agoulnik IU, Harrison WR, Behringer RR, Overbeek PA. A transgenic insertion upstream of sox9 is associated with dominant XX sex reversal in the mouse. Nat Genet. 2000;26:490–4. doi: 10.1038/82652. [DOI] [PubMed] [Google Scholar]
- Bishop DV, Jacobs PA, Lachlan K, Wellesley D, Barnicoat A, Boyd PA, Fryer A, Middlemiss P, Smithson S, Metcalfe K, Shears D, Leggett V, Nation K, Scerif G. Autism, language and communication in children with sex chromosome trisomies. Arch Dis Child. 2011;96:954–9. doi: 10.1136/adc.2009.179747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blissett J, Harris G, Kirk J. Effect of growth hormone therapy on feeding problems and food intake in children with growth disorders. Acta Paediatr. 2000;89:644–9. doi: 10.1080/080352500750043927. [DOI] [PubMed] [Google Scholar]
- Bohlooly YM, Olsson B, Bruder CE, Linden D, Sjogren K, Bjursell M, Egecioglu E, Svensson L, Brodin P, Waterton JC, Isaksson OG, Sundler F, Ahren B, Ohlsson C, Oscarsson J, Tornell J. Growth hormone overexpression in the central nervous system results in hyperphagia-induced obesity associated with insulin resistance and dyslipidemia. Diabetes. 2005;54:51–62. doi: 10.2337/diabetes.54.1.51. [DOI] [PubMed] [Google Scholar]
- Bonthuis PJ, Cox KH, Rissman EF. X-chromosome dosage affects male sexual behavior. Horm Behav. 2012;61:565–72. doi: 10.1016/j.yhbeh.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bonthuis PJ, Rissman EF. Neural growth hormone implicated in body weight sex differences. Endocrinology. 2013;154:3826–35. doi: 10.1210/en.2013-1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouayed Abdelmoula N, Portnoi MF, Keskes L, Recan D, Bahloul A, Boudawara T, Saad A, Rebai T. Skewed X-chromosome inactivation pattern in SRY positive XX maleness: a case report and review of literature. Ann Genet. 2003;46:11–8. doi: 10.1016/s0003-3995(03)00011-x. [DOI] [PubMed] [Google Scholar]
- Bruining H, Swaab H, Kas M, van Engeland H. Psychiatric characteristics in a self-selected sample of boys with Klinefelter syndrome. Pediatrics. 2009;123:e865–70. doi: 10.1542/peds.2008-1954. [DOI] [PubMed] [Google Scholar]
- Bruining H, van Rijn S, Swaab H, Giltay J, Kates W, Kas MJ, van Engeland H, de Sonneville L. The parent-of-origin of the extra X chromosome may differentially affect psychopathology in Klinefelter syndrome. Biol Psychiatry. 2010;68:1156–62. doi: 10.1016/j.biopsych.2010.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Budefeld T, Grgurevic N, Tobet SA, Majdic G. Sex differences in brain developing in the presence or absence of gonads. Dev Neurobiol. 2008;68:981–95. doi: 10.1002/dneu.20638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgoyne PS, Evans EP. A high frequency of XO offspring from X(Paf)Y* male mice: evidence that the Paf mutation involves an inversion spanning the X PAR boundary. Cytogenet Cell Genet. 2000;91:57–61. doi: 10.1159/000056819. [DOI] [PubMed] [Google Scholar]
- Burgoyne PS, Mahadevaiah SK, Perry J, Palmer SJ, Ashworth A. The Y* rearrangement in mice: new insights into a perplexing PAR. Cytogenet Cell Genet. 1998;80:37–40. doi: 10.1159/000014954. [DOI] [PubMed] [Google Scholar]
- Burnett AC, Reutens DC, Wood AG. Social cognition in Turner’s Syndrome. J Clin Neurosci. 2010;17:283–6. doi: 10.1016/j.jocn.2009.09.006. [DOI] [PubMed] [Google Scholar]
- Canastar A, Maxson SC, Bishop CE. Aggressive and mating behaviors in two types of sex reversed mice: XY females and XX males. Arch Sex Behav. 2008;37:2–8. doi: 10.1007/s10508-007-9257-1. [DOI] [PubMed] [Google Scholar]
- Carlier M, Roubertoux PL, Kottler ML, Degrelle H. Y chromosome and aggression in strains of laboratory mice. Behav Genet. 1990;20:137–56. doi: 10.1007/BF01070750. [DOI] [PubMed] [Google Scholar]
- Carlier M, Roubertoux PL, Pastoret C. The Y chromosome effect on intermale aggression in mice depends on the maternal environment. Genetics. 1991;129:231–6. doi: 10.1093/genetics/129.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434:400–4. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- Cases O, Vitalis T, Seif I, De Maeyer E, Sotelo C, Gaspar P. Lack of barrels in the somatosensory cortex of monoamine oxidase A-deficient mice: role of a serotonin excess during the critical period. Neuron. 1996;16:297–307. doi: 10.1016/s0896-6273(00)80048-3. [DOI] [PubMed] [Google Scholar]
- Chen X, McClusky R, Chen J, Beaven SW, Tontonoz P, Arnold AP, Reue K. The number of x chromosomes causes sex differences in adiposity in mice. PLoS Genet. 2012;8:e1002709. doi: 10.1371/journal.pgen.1002709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, McClusky R, Itoh Y, Reue K, Arnold AP. X and Y chromosome complement influence adiposity and metabolism in mice. Endocrinology. 2013a;154:1092–104. doi: 10.1210/en.2012-2098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Williams-Burris SM, McClusky R, Ngun TC, Ghahramani N, Barseghyan H, Reue K, Vilain E, Arnold AP. The Sex Chromosome Trisomy mouse model of XXY and XYY: metabolism and motor performance. Biol Sex Differ. 2013b;4:15. doi: 10.1186/2042-6410-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conner RL, Levine S, Wertheim GA, Cummer JF. Hormonal determinants of aggressive behavior. Ann N Y Acad Sci. 1969;159:760–76. doi: 10.1111/j.1749-6632.1969.tb12977.x. [DOI] [PubMed] [Google Scholar]
- Correa SM, Washburn LL, Kahlon RS, Musson MC, Bouma GJ, Eicher EM, Albrecht KH. Sex reversal in C57BL/6J XY mice caused by increased expression of ovarian genes and insufficient activation of the testis determining pathway. PLoS Genet. 2012;8:e1002569. doi: 10.1371/journal.pgen.1002569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox KH, Rissman EF. Sex differences in juvenile mouse social behavior are influenced by sex chromosomes and social context. Genes Brain Behav. 2011;10:465–72. doi: 10.1111/j.1601-183X.2011.00688.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dal Zotto L, Quaderi NA, Elliott R, Lingerfelter PA, Carrel L, Valsecchi V, Montini E, Yen CH, Chapman V, Kalcheva I, Arrigo G, Zuffardi O, Thomas S, Willard HF, Ballabio A, Disteche CM, Rugarli EI. The mouse Mid1 gene: implications for the pathogenesis of Opitz syndrome and the evolution of the mammalian pseudoautosomal region. Hum Mol Genet. 1998;7:489–99. doi: 10.1093/hmg/7.3.489. [DOI] [PubMed] [Google Scholar]
- Davies W, Humby T, Isles AR, Burgoyne PS, Wilkinson LS. X- monosomy effects on visuospatial attention in mice: a candidate gene and implications for Turner syndrome and attention deficit hyperactivity disorder. Biol Psychiatry. 2007;61:1351–60. doi: 10.1016/j.biopsych.2006.08.011. [DOI] [PubMed] [Google Scholar]
- Davies W, Humby T, Kong W, Otter T, Burgoyne PS, Wilkinson LS. Converging pharmacological and genetic evidence indicates a role for steroid sulfatase in attention. Biol Psychiatry. 2009;66:360–7. doi: 10.1016/j.biopsych.2009.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies W, Isles A, Smith R, Karunadasa D, Burrmann D, Humby T, Ojarikre O, Biggin C, Skuse D, Burgoyne P, Wilkinson L. Xlr3b is a new imprinted candidate for X-linked parent-of-origin effects on cognitive function in mice. Nat Genet. 2005;37:625–9. doi: 10.1038/ng1577. [DOI] [PubMed] [Google Scholar]
- De Vries GJ. Sex differences in vasopressin and oxytocin innervation of the brain. Prog Brain Res. 2008;170:17–27. doi: 10.1016/S0079-6123(08)00402-0. [DOI] [PubMed] [Google Scholar]
- De Vries GJ, Rissman EF, Simerly RB, Yang LY, Scordalakes EM, Auger CJ, Swain A, Lovell-Badge R, Burgoyne PS, Arnold AP. A model system for study of sex chromosome effects on sexually dimorphic neural and behavioral traits. J Neurosci. 2002;22:9005–14. doi: 10.1523/JNEUROSCI.22-20-09005.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisi LE, Maurizio AM, Svetina C, Ardekani B, Szulc K, Nierenberg J, Leonard J, Harvey PD. Klinefelter’s syndrome (XXY) as a genetic model for psychotic disorders. Am J Med Genet B Neuropsychiatr Genet. 2005;135B:15–23. doi: 10.1002/ajmg.b.30163. [DOI] [PubMed] [Google Scholar]
- Disteche CM, Filippova GN, Tsuchiya KD. Escape from X inactivation. Cytogenet Genome Res. 2002;99:36–43. doi: 10.1159/000071572. [DOI] [PubMed] [Google Scholar]
- Donnelly SL, Wolpert CM, Menold MM, Bass MP, Gilbert JR, Cuccaro ML, Delong GR, Pericak-Vance MA. Female with autistic disorder and monosomy X (Turner syndrome): parent-of-origin effect of the X chromosome. Am J Med Genet. 2000;96:312–6. doi: 10.1002/1096-8628(20000612)96:3<312::aid-ajmg16>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- Eicher EM, Hale DW, Hunt PA, Lee BK, Tucker PK, King TR, Eppig JT, Washburn LL. The mouse Y* chromosome involves a complex rearrangement, including interstitial positioning of the pseudoautosomal region. Cytogenet Cell Genet. 1991;57:221–30. doi: 10.1159/000133152. [DOI] [PubMed] [Google Scholar]
- Eicher EM, Shown EP, Washburn LL. Sex reversal in C57BL/6J-YPOS mice corrected by a Sry transgene. Philos Trans R Soc Lond B Biol Sci. 1995;350:263–8. doi: 10.1098/rstb.1995.0160. discussion 268–9. [DOI] [PubMed] [Google Scholar]
- Eicher EM, Washburn LL. Does one gene determine whether a C57BL/6J-Y(POS) mouse will develop as a female or as an hermaphrodite? J Exp Zool. 2001;290:322–6. doi: 10.1002/jez.1072. [DOI] [PubMed] [Google Scholar]
- Eicher EM, Washburn LL, Whitney JB, 3rd, Morrow KE. Mus poschiavinus Y chromosome in the C57BL/6J murine genome causes sex reversal. Science. 1982;217:535–7. doi: 10.1126/science.7089579. [DOI] [PubMed] [Google Scholar]
- Ellegren H. Sex-chromosome evolution: recent progress and the influence of male and female heterogamety. Nat Rev Genet. 2011;12:157–66. doi: 10.1038/nrg2948. [DOI] [PubMed] [Google Scholar]
- Ellis N, Goodfellow PN. The mammalian pseudoautosomal region. Trends Genet. 1989;5:406–10. doi: 10.1016/0168-9525(89)90199-6. [DOI] [PubMed] [Google Scholar]
- Elsheikh M, Dunger DB, Conway GS, Wass JA. Turner’s syndrome in adulthood. Endocr Rev. 2002;23:120–40. doi: 10.1210/edrv.23.1.0457. [DOI] [PubMed] [Google Scholar]
- Evans EP, Burtenshaw MD, Cattanach BM. Meitoic crossing-over between the X and Y chromosomes of male mice carrying the sex-reversing (Sxr) factor. Nature. 1982;300:443–5. doi: 10.1038/300443a0. [DOI] [PubMed] [Google Scholar]
- Evans EP, Phillips RJ. Inversion heterozygosity and the origin of XO daughters of Bpa/+female mice. Nature. 1975;256:40–1. doi: 10.1038/256040a0. [DOI] [PubMed] [Google Scholar]
- Fang J, Clemens LG. Contextual determinants of female-female mounting in laboratory rats. Anim Behav. 1999;57:545–555. doi: 10.1006/anbe.1998.1025. [DOI] [PubMed] [Google Scholar]
- Gatewood JD, Wills A, Shetty S, Xu J, Arnold AP, Burgoyne PS, Rissman EF. Sex chromosome complement and gonadal sex influence aggressive and parental behaviors in mice. J Neurosci. 2006;26:2335–42. doi: 10.1523/JNEUROSCI.3743-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geschwind DH, Boone KB, Miller BL, Swerdloff RS. Neurobehavioral phenotype of Klinefelter syndrome. Ment Retard Dev Disabil Res Rev. 2000;6:107–16. doi: 10.1002/1098-2779(2000)6:2<107::AID-MRDD4>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Gilbert LA, Larson MH, Morsut L, Liu Z, Brar GA, Torres SE, Stern-Ginossar N, Brandman O, Whitehead EH, Doudna JA, Lim WA, Weissman JS, Qi LS. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–51. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioiosa L, Chen X, Watkins R, Klanfer N, Bryant CD, Evans CJ, Arnold AP. Sex chromosome complement affects nociception in tests of acute and chronic exposure to morphine in mice. Horm Behav. 2008a;53:124–30. doi: 10.1016/j.yhbeh.2007.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gioiosa L, Chen X, Watkins R, Umeda EA, Arnold AP. Sex chromosome complement affects nociception and analgesia in newborn mice. J Pain. 2008b;9:962–9. doi: 10.1016/j.jpain.2008.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graves JA. The origin and function of the mammalian Y chromosome and Y-borne genes--an evolving understanding. Bioessays. 1995;17:311–20. doi: 10.1002/bies.950170407. [DOI] [PubMed] [Google Scholar]
- Graves JA, Koina E, Sankovic N. How the gene content of human sex chromosomes evolved. Curr Opin Genet Dev. 2006;16:219–24. doi: 10.1016/j.gde.2006.04.007. [DOI] [PubMed] [Google Scholar]
- Grgurevic N, Budefeld T, Rissman EF, Tobet SA, Majdic G. Aggressive behaviors in adult SF-1 knockout mice that are not exposed to gonadal steroids during development. Behav Neurosci. 2008;122:876–84. doi: 10.1037/0735-7044.122.4.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grgurevic N, Budefeld T, Spanic T, Tobet SA, Majdic G. Evidence that sex chromosome genes affect sexual differentiation of female sexual behavior. Horm Behav. 2012;61:719–24. doi: 10.1016/j.yhbeh.2012.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–50. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- Guillot PV, Carlier M, Maxson SC, Roubertoux PL. Intermale aggression tested in two procedures, using four inbred strains of mice and their reciprocal congenics: Y chromosomal implications. Behav Genet. 1995;25:357–60. doi: 10.1007/BF02197285. [DOI] [PubMed] [Google Scholar]
- Hong DS, Dunkin B, Reiss AL. Psychosocial functioning and social cognitive processing in girls with Turner syndrome. J Dev Behav Pediatr. 2011;32:512–20. doi: 10.1097/DBP.0b013e3182255301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoplight BJ, Mobraaten L, Sherman GF, Hyde LA, Harding S, Denenberg VH. Effects of the non-pseudoautosomal region of the Y-chromosome on behavior in female offspring of two congenic strains of mice. Neuroscience. 2000;96:837–42. doi: 10.1016/s0306-4522(99)00600-4. [DOI] [PubMed] [Google Scholar]
- Hunt PA, Eicher EM. Fertile male mice with three sex chromosomes: evidence that infertility in XYY male mice is an effect of two Y chromosomes. Chromosoma. 1991;100:293–9. doi: 10.1007/BF00360527. [DOI] [PubMed] [Google Scholar]
- Hunt PA, Worthman C, Levinson H, Stallings J, LeMaire R, Mroz K, Park C, Handel MA. Germ cell loss in the XXY male mouse: altered X-chromosome dosage affects prenatal development. Mol Reprod Dev. 1998;49:101–11. doi: 10.1002/(SICI)1098-2795(199802)49:2<101::AID-MRD1>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Ingraham HA, Lala DS, Ikeda Y, Luo X, Shen WH, Nachtigal MW, Abbud R, Nilson JH, Parker KL. The nuclear receptor steroidogenic factor 1 acts at multiple levels of the reproductive axis. Genes Dev. 1994;8:2302–12. doi: 10.1101/gad.8.19.2302. [DOI] [PubMed] [Google Scholar]
- Isles AR, Davies W, Burrmann D, Burgoyne PS, Wilkinson LS. Effects on fear reactivity in XO mice are due to haploinsufficiency of a non-PAR X gene: implications for emotional function in Turner’s syndrome. Hum Mol Genet. 2004;13:1849–55. doi: 10.1093/hmg/ddh203. [DOI] [PubMed] [Google Scholar]
- Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, Schadt EE, Lusis AJ, Arnold AP. Dosage compensation is less effective in birds than in mammals. J Biol. 2007;6:2. doi: 10.1186/jbiol53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaffe CA, Ocampo-Lim B, Guo W, Krueger K, Sugahara I, DeMott-Friberg R, Bermann M, Barkan AL. Regulatory mechanisms of growth hormone secretion are sexually dimorphic. J Clin Invest. 1998;102:153–64. doi: 10.1172/JCI2908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jensen LR, Amende M, Gurok U, Moser B, Gimmel V, Tzschach A, Janecke AR, Tariverdian G, Chelly J, Fryns JP, Van Esch H, Kleefstra T, Hamel B, Moraine C, Gecz J, Turner G, Reinhardt R, Kalscheuer VM, Ropers HH, Lenzner S. Mutations in the JARID1C gene, which is involved in transcriptional regulation and chromatin remodeling, cause X-linked mental retardation. Am J Hum Genet. 2005;76:227–36. doi: 10.1086/427563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston CM, Lovell FL, Leongamornlert DA, Stranger BE, Dermitzakis ET, Ross MT. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS genetics. 2008;4:e9. doi: 10.1371/journal.pgen.0040009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerstetter KA, Kippin TE. Impact of Sex and Gonadal Hormones on Cocaine and Food Reinforcement Paradigms. J Addict Res Ther. 2011:S4. doi: 10.4172/2155-6105.s4-002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knickmeyer RC. Turner syndrome: advances in understanding altered cognition, brain structure and function. Curr Opin Neurol. 2012;25:144–9. doi: 10.1097/WCO.0b013e3283515e9e. [DOI] [PubMed] [Google Scholar]
- Ko JM, Kim JM, Kim GH, Lee BH, Yoo HW. Influence of parental origin of the X chromosome on physical phenotypes and GH responsiveness of patients with Turner syndrome. Clin Endocrinol (Oxf) 2010;73:66–71. doi: 10.1111/j.1365-2265.2010.03782.x. [DOI] [PubMed] [Google Scholar]
- Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348:450–2. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- Kuljis DA, Loh DH, Truong D, Vosko AM, Ong ML, McClusky R, Arnold AP, Colwell CS. Gonadal- and sex-chromosome-dependent sex differences in the circadian system. Endocrinology. 2013;154:1501–12. doi: 10.1210/en.2012-1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane PW, Davisson MT. Patchy fur (Paf), a semidominant X-linked gene associated with a high level of X-Y nondisjunction in male mice. J Hered. 1990;81:43–50. doi: 10.1093/oxfordjournals.jhered.a110923. [DOI] [PubMed] [Google Scholar]
- Larson MH, Gilbert LA, Wang X, Lim WA, Weissman JS, Qi LS. CRISPR interference (CRISPRi) for sequence-specific control of gene expression. Nat Protoc. 2013;8:2180–96. doi: 10.1038/nprot.2013.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence K, Kuntsi J, Coleman M, Campbell R, Skuse D. Face and emotion recognition deficits in Turner syndrome: a possible role for X-linked genes in amygdala development. Neuropsychology. 2003;17:39–49. [PubMed] [Google Scholar]
- Le Roy I, Mortaud S, Tordjman S, Donsez-Darcel E, Carlier M, Degrelle H, Roubertoux PL. Genetic correlation between steroid sulfatase concentration and initiation of attack behavior in mice. Behav Genet. 1999;29:131–6. doi: 10.1023/a:1021664607131. [DOI] [PubMed] [Google Scholar]
- Leggett V, Jacobs P, Nation K, Scerif G, Bishop DV. Neurocognitive outcomes of individuals with a sex chromosome trisomy: XXX, XYY, or XXY: a systematic review. Dev Med Child Neurol. 2010;52:119–29. doi: 10.1111/j.1469-8749.2009.03545.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepage JF, Dunkin B, Hong DS, Reiss AL. Contribution of executive functions to visuospatial difficulties in prepubertal girls with Turner syndrome. Dev Neuropsychol. 2011;36:988–1002. doi: 10.1080/87565641.2011.584356. [DOI] [PubMed] [Google Scholar]
- Lepage JF, Hong DS, Hallmayer J, Reiss AL. Genomic imprinting effects on cognitive and social abilities in prepubertal girls with Turner syndrome. J Clin Endocrinol Metab. 2012;97:E460–4. doi: 10.1210/jc.2011-2916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesniak-Karpiak K, Mazzocco MM, Ross JL. Behavioral assessment of social anxiety in females with Turner or fragile X syndrome. J Autism Dev Disord. 2003;33:55–67. doi: 10.1023/a:1022230504787. [DOI] [PubMed] [Google Scholar]
- Lewejohann L, Damm OS, Luetjens CM, Hamalainen T, Simoni M, Nieschlag E, Gromoll J, Wistuba J. Impaired recognition memory in male mice with a supernumerary X chromosome. Physiol Behav. 2009;96:23–9. doi: 10.1016/j.physbeh.2008.08.007. [DOI] [PubMed] [Google Scholar]
- Lin D, Boyle MP, Dollar P, Lee H, Lein ES, Perona P, Anderson DJ. Functional identification of an aggression locus in the mouse hypothalamus. Nature. 2011;470:221–6. doi: 10.1038/nature09736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu PY, Erkkila K, Lue Y, Jentsch JD, Schwarcz MD, Abuyounes D, Hikim AS, Wang C, Lee PW, Swerdloff RS. Genetic, hormonal, and metabolomic influences on social behavior and sex preference of XXY mice. Am J Physiol Endocrinol Metab. 2010;299:E446–55. doi: 10.1152/ajpendo.00085.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovell-Badge R, Robertson E. XY female mice resulting from a heritable mutation in the primary testis-determining gene, Tdy. Developmental biology. 1990;109:635–46. doi: 10.1242/dev.109.3.635. [DOI] [PubMed] [Google Scholar]
- Lue Y, Jentsch JD, Wang C, Rao PN, Hikim AP, Salameh W, Swerdloff RS. XXY mice exhibit gonadal and behavioral phenotypes similar to Klinefelter syndrome. Endocrinology. 2005;146:4148–54. doi: 10.1210/en.2005-0278. [DOI] [PubMed] [Google Scholar]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.) Nature. 1961;190:372–3. doi: 10.1038/190372a0. [DOI] [PubMed] [Google Scholar]
- Mahadevaiah SK, Odorisio T, Elliott DJ, Rattigan A, Szot M, Laval SH, Washburn LL, McCarrey JR, Cattanach BM, Lovell-Badge R, Burgoyne PS. Mouse homologues of the human AZF candidate gene RBM are expressed in spermatogonia and spermatids, and map to a Y chromosome deletion interval associated with a high incidence of sperm abnormalities. Hum Mol Genet. 1998;7:715–27. doi: 10.1093/hmg/7.4.715. [DOI] [PubMed] [Google Scholar]
- Majdic G, Young M, Gomez-Sanchez E, Anderson P, Szczepaniak LS, Dobbins RL, McGarry JD, Parker KL. Knockout mice lacking steroidogenic factor 1 are a novel genetic model of hypothalamic obesity. Endocrinology. 2002;143:607–14. doi: 10.1210/endo.143.2.8652. [DOI] [PubMed] [Google Scholar]
- Marco EJ, Skuse DH. Autism-lessons from the X chromosome. Soc Cogn Affect Neurosci. 2006;1:183–93. doi: 10.1093/scan/nsl028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxson SC. Issues in the search for candidate genes in mice as potential animal models of human aggression. Ciba Found Symp. 1996a;194:21–30. doi: 10.1002/9780470514825.ch2. discussion 30–5. [DOI] [PubMed] [Google Scholar]
- Maxson SC. Searching for candidate genes with effects on an agonistic behavior, offense, in mice. Behav Genet. 1996b;26:471–6. doi: 10.1007/BF02359751. [DOI] [PubMed] [Google Scholar]
- Maxson SC, Ginsburg BE, Trattner A. Interaction of Y-chromosomal and autosomal gene(s) in the development of intermale aggression in mice. Behav Genet. 1979;9:219–26. doi: 10.1007/BF01071302. [DOI] [PubMed] [Google Scholar]
- McLaren A, Simpson E, Bishop CE, Mitchell MJ, Darling SM. Recombination between the X and Y chromosomes and the Sxr region of the mouse. Genet Res. 1992;60:175–84. doi: 10.1017/s0016672300030925. [DOI] [PubMed] [Google Scholar]
- McPhie-Lalmansingh AA, Tejada LD, Weaver JL, Rissman EF. Sex chromosome complement affects social interactions in mice. Horm Behav. 2008;54:565–70. doi: 10.1016/j.yhbeh.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortaud S, Nicolas L, Pinoteau W, Tordjman S, Carlier M, Roubertoux PL. Brain pathways mediating the pro-aggressive effect of the steroid sulfatase (Sts) gene. Behav Genet. 2010;40:211–9. doi: 10.1007/s10519-010-9340-6. [DOI] [PubMed] [Google Scholar]
- Nagamine CM, Capehart J, Carlisle C, Chang D. Ovotestes in B6-XXSxr sex-reversed mice. Dev Biol. 1998;196:24–32. doi: 10.1006/dbio.1997.8826. [DOI] [PubMed] [Google Scholar]
- Nelson RJ, Trainor BC. Neural mechanisms of aggression. Nat Rev Neurosci. 2007;8:536–46. doi: 10.1038/nrn2174. [DOI] [PubMed] [Google Scholar]
- Neumann ID, Landgraf R. Balance of brain oxytocin and vasopressin: implications for anxiety, depression, and social behaviors. Trends Neurosci. 2012;35:649–59. doi: 10.1016/j.tins.2012.08.004. [DOI] [PubMed] [Google Scholar]
- Nguyen DK, Disteche CM. Dosage compensation of the active X chromosome in mammals. Nat Genet. 2006;38:47–53. doi: 10.1038/ng1705. [DOI] [PubMed] [Google Scholar]
- Park JH, Burns-Cusato M, Dominguez-Salazar E, Riggan A, Shetty S, Arnold AP, Rissman EF. Effects of sex chromosome aneuploidy on male sexual behavior. Genes Brain Behav. 2008;7:609–17. doi: 10.1111/j.1601-183X.2008.00397.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker KL, Rice DA, Lala DS, Ikeda Y, Luo X, Wong M, Bakke M, Zhao L, Frigeri C, Hanley NA, Stallings N, Schimmer BP. Steroidogenic factor 1: an essential mediator of endocrine development. Recent Prog Horm Res. 2002;57:19–36. doi: 10.1210/rp.57.1.19. [DOI] [PubMed] [Google Scholar]
- Parker KL, Schimmer BP. Steroidogenic factor 1: a key determinant of endocrine development and function. Endocr Rev. 1997;18:361–77. doi: 10.1210/edrv.18.3.0301. [DOI] [PubMed] [Google Scholar]
- Perry J, Palmer S, Gabriel A, Ashworth A. A short pseudoautosomal region in laboratory mice. Genome Res. 2001;11:1826–32. doi: 10.1101/gr.203001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GA. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci U S A. 2012;109:5346–51. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phoenix CH, Goy RW, Gerall AA, Young WC. Organizing action of prenatally administered testosterone propionate on the tissues mediating mating behavior in the female guinea pig. Endocrinology. 1959;65:369–82. doi: 10.1210/endo-65-3-369. [DOI] [PubMed] [Google Scholar]
- Quinn JJ, Hitchcott PK, Umeda EA, Arnold AP, Taylor JR. Sex chromosome complement regulates habit formation. Nat Neurosci. 2007;10:1398–400. doi: 10.1038/nn1994. [DOI] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF. Genetic and epigenetic underpinnings of sex differences in the brain and in neurological and psychiatric disease susceptibility. Prog Brain Res. 2010;186:77–95. doi: 10.1016/B978-0-444-53630-3.00006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racine M, Tousignant-Laflamme Y, Kloda LA, Dion D, Dupuis G, Choiniere M. A systematic literature review of 10 years of research on sex/gender and experimental pain perception - part 1: are there really differences between women and men? Pain. 2012;153:602–18. doi: 10.1016/j.pain.2011.11.025. [DOI] [PubMed] [Google Scholar]
- Raefski AS, O’Neill MJ. Identification of a cluster of X-linked imprinted genes in mice. Nat Genet. 2005;37:620–4. doi: 10.1038/ng1567. [DOI] [PubMed] [Google Scholar]
- Raudsepp T, Chowdhary BP. The horse pseudoautosomal region (PAR): characterization and comparison with the human, chimp and mouse PARs. Cytogenet Genome Res. 2008;121:102–9. doi: 10.1159/000125835. [DOI] [PubMed] [Google Scholar]
- Raymond FL. X linked mental retardation: a clinical guide. Journal of medical genetics. 2006;43:193–200. doi: 10.1136/jmg.2005.033043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raznahan A, Probst F, Palmert MR, Giedd JN, Lerch JP. High resolution whole brain imaging of anatomical variation in XO, XX, and XY mice. NeuroImage. 2013;83:962–8. doi: 10.1016/j.neuroimage.2013.07.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reisert I, Karolczak M, Beyer C, Just W, Maxson SC, Ehret G. Sry does not fully sex-reverse female into male behavior towards pups. Behav Genet. 2002;32:103–11. doi: 10.1023/a:1015297622509. [DOI] [PubMed] [Google Scholar]
- Ross JL, Roeltgen D, Feuillan P, Kushner H, Cutler GB., Jr Effects of estrogen on nonverbal processing speed and motor function in girls with Turner’s syndrome. J Clin Endocrinol Metab. 1998;83:3198–204. doi: 10.1210/jcem.83.9.5087. [DOI] [PubMed] [Google Scholar]
- Ross JL, Stefanatos GA, Kushner H, Bondy C, Nelson L, Zinn A, Roeltgen D. The effect of genetic differences and ovarian failure: intact cognitive function in adult women with premature ovarian failure versus turner syndrome. J Clin Endocrinol Metab. 2004;89:1817–22. doi: 10.1210/jc.2003-031463. [DOI] [PubMed] [Google Scholar]
- Ross JL, Stefanatos GA, Kushner H, Zinn A, Bondy C, Roeltgen D. Persistent cognitive deficits in adult women with Turner syndrome. Neurology. 2002;58:218–25. doi: 10.1212/wnl.58.2.218. [DOI] [PubMed] [Google Scholar]
- Ross JL, Zeger MP, Kushner H, Zinn AR, Roeltgen DP. An extra X or Y chromosome: contrasting the cognitive and motor phenotypes in childhood in boys with 47,XYY syndrome or 47,XXY Klinefelter syndrome. Dev Disabil Res Rev. 2009;15:309–17. doi: 10.1002/ddrr.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross MT, Grafham DV, Coffey AJ, Scherer S, McLay K, Muzny D, Platzer M, Howell GR, Burrows C, Bird CP, Frankish A, Lovell FL, Howe KL, Ashurst JL, Fulton RS, Sudbrak R, Wen G, Jones MC, Hurles ME, Andrews TD, Scott CE, Searle S, Ramser J, Whittaker A, Deadman R, Carter NP, Hunt SE, Chen R, Cree A, Gunaratne P, Havlak P, Hodgson A, Metzker ML, Richards S, Scott G, Steffen D, Sodergren E, Wheeler DA, Worley KC, Ainscough R, Ambrose KD, Ansari-Lari MA, Aradhya S, Ashwell RI, Babbage AK, Bagguley CL, Ballabio A, Banerjee R, Barker GE, Barlow KF, Barrett IP, Bates KN, Beare DM, Beasley H, Beasley O, Beck A, Bethel G, Blechschmidt K, Brady N, Bray-Allen S, Bridgeman AM, Brown AJ, Brown MJ, Bonnin D, Bruford EA, Buhay C, Burch P, Burford D, Burgess J, Burrill W, Burton J, Bye JM, Carder C, Carrel L, Chako J, Chapman JC, Chavez D, Chen E, Chen G, Chen Y, Chen Z, Chinault C, Ciccodicola A, Clark SY, Clarke G, Clee CM, Clegg S, Clerc-Blankenburg K, Clifford K, Cobley V, Cole CG, Conquer JS, Corby N, Connor RE, David R, Davies J, Davis C, Davis J, Delgado O, Deshazo D, et al. The DNA sequence of the human X chromosome. Nature. 2005;434:325–37. doi: 10.1038/nature03440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roubertoux PL, Carlier M. Differences between CBA/H and NZB mice on intermale aggression. II. Maternal effects. Behav Genet. 1988;18:175–84. doi: 10.1007/BF01067839. [DOI] [PubMed] [Google Scholar]
- Roubertoux PL, Carlier M, Degrelle H, Haas-Dupertuis MC, Phillips J, Moutier R. Co-segregation of intermale aggression with the pseudoautosomal region of the Y chromosome in mice. Genetics. 1994;136:225–30. doi: 10.1093/genetics/136.1.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savic I. Advances in research on the neurological and neuropsychiatric phenotype of Klinefelter syndrome. Curr Opin Neurol. 2012;25:138–43. doi: 10.1097/WCO.0b013e32835181a0. [DOI] [PubMed] [Google Scholar]
- Schmidt PJ, Cardoso GM, Ross JL, Haq N, Rubinow DR, Bondy CA. Shyness, social anxiety, and impaired self-esteem in Turner syndrome and premature ovarian failure. Jama. 2006;295:1374–6. doi: 10.1001/jama.295.12.1374. [DOI] [PubMed] [Google Scholar]
- Selmanoff MK, Jumonville JE, Maxson SC, Ginsburg BE. Evidence for a Y chromosomal contribution to an aggressive phenotype in inbred mice. Nature. 1975;253:529–30. doi: 10.1038/253529a0. [DOI] [PubMed] [Google Scholar]
- Shrenker P, Maxson SC. The DBA/1Bg and DBA/2Bg Y chromosomes compared for their effects on male sexual behavior. Behav Neural Biol. 1984;42:33–7. doi: 10.1016/s0163-1047(84)90400-x. [DOI] [PubMed] [Google Scholar]
- Sim H, Argentaro A, Harley VR. Boys, girls and shuttling of SRY and SOX9. Trends Endocrinol Metab. 2008;19:213–22. doi: 10.1016/j.tem.2008.04.002. [DOI] [PubMed] [Google Scholar]
- Skaletsky H, Kuroda-Kawaguchi T, Minx PJ, Cordum HS, Hillier L, Brown LG, Repping S, Pyntikova T, Ali J, Bieri T, Chinwalla A, Delehaunty A, Delehaunty K, Du H, Fewell G, Fulton L, Fulton R, Graves T, Hou SF, Latrielle P, Leonard S, Mardis E, Maupin R, McPherson J, Miner T, Nash W, Nguyen C, Ozersky P, Pepin K, Rock S, Rohlfing T, Scott K, Schultz B, Strong C, Tin-Wollam A, Yang SP, Waterston RH, Wilson RK, Rozen S, Page DC. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature. 2003;423:825–37. doi: 10.1038/nature01722. [DOI] [PubMed] [Google Scholar]
- Skuse DH. X-linked genes and mental functioning. Hum Mol Genet. 2005;14(Spec No 1):R27–32. doi: 10.1093/hmg/ddi112. [DOI] [PubMed] [Google Scholar]
- Skuse DH, James RS, Bishop DV, Coppin B, Dalton P, Aamodt-Leeper G, Bacarese-Hamilton M, Creswell C, McGurk R, Jacobs PA. Evidence from Turner’s syndrome of an imprinted X-linked locus affecting cognitive function. Nature. 1997;387:705–8. doi: 10.1038/42706. [DOI] [PubMed] [Google Scholar]
- Skuse DH, Morris JS, Dolan RJ. Functional dissociation of amygdala-modulated arousal and cognitive appraisal, in Turner syndrome. Brain. 2005;128:2084–96. doi: 10.1093/brain/awh562. [DOI] [PubMed] [Google Scholar]
- Sluyter F, Bohus B, Beldhuis HJ, van Oortmerssen GA. Autosomal and Y chromosomal effects on the stereotyped response to apomorphine in wild house mice. Pharmacol Biochem Behav. 1995;52:17–22. doi: 10.1016/0091-3057(95)00092-b. [DOI] [PubMed] [Google Scholar]
- Stavnezer AJ, McDowell CS, Hyde LA, Bimonte HA, Balogh SA, Hoplight BJ, Denenberg VH. Spatial ability of XY sex-reversed female mice. Behav Brain Res. 2000;112:135–43. doi: 10.1016/s0166-4328(00)00174-1. [DOI] [PubMed] [Google Scholar]
- Steinman K, Ross J, Lai S, Reiss A, Hoeft F. Structural and functional neuroimaging in Klinefelter (47,XXY) syndrome: a review of the literature and preliminary results from a functional magnetic resonance imaging study of language. Dev Disabil Res Rev. 2009;15:295–308. doi: 10.1002/ddrr.84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Temple CM, Sanfilippo PM. Executive skills in Klinefelter’s syndrome. Neuropsychologia. 2003;41:1547–59. doi: 10.1016/s0028-3932(03)00061-7. [DOI] [PubMed] [Google Scholar]
- Trent S, Dennehy A, Richardson H, Ojarikre OA, Burgoyne PS, Humby T, Davies W. Steroid sulfatase-deficient mice exhibit endophenotypes relevant to attention deficit hyperactivity disorder. Psychoneuroendocrinology. 2012;37:221–9. doi: 10.1016/j.psyneuen.2011.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oortmerssen GA, Sluyter F. Studies on wild house mice. V. Aggression in lines selected for attack latency and their Y-chromosomal congenics. Behav Genet. 1994;24:73–8. doi: 10.1007/BF01067930. [DOI] [PubMed] [Google Scholar]
- van Rijn S, Aleman A, Swaab H, Kahn R. Klinefelter’s syndrome (karyotype 47,XXY) and schizophrenia-spectrum pathology. Br J Psychiatry. 2006;189:459–60. doi: 10.1192/bjp.bp.105.008961. [DOI] [PubMed] [Google Scholar]
- Wagner CK, Xu J, Pfau JL, Quadros PS, De Vries GJ, Arnold AP. Neonatal mice possessing an Sry transgene show a masculinized pattern of progesterone receptor expression in the brain independent of sex chromosome status. Endocrinology. 2004;145:1046–9. doi: 10.1210/en.2003-1219. [DOI] [PubMed] [Google Scholar]
- Wolstenholme JT, Rissman EF, Bekiranov S. Sexual differentiation in the developing mouse brain: contributions of sex chromosome genes. Genes Brain Behav. 2013;12:166–80. doi: 10.1111/gbb.12010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wutz A. Gene silencing in X-chromosome inactivation: advances in understanding facultative heterochromatin formation. Nat Rev Genet. 2011;12:542–53. doi: 10.1038/nrg3035. [DOI] [PubMed] [Google Scholar]
- Xu J, Andreassi M. Reversible histone methylation regulates brain gene expression and behavior. Horm Behav. 2011;59:383–92. doi: 10.1016/j.yhbeh.2010.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Burgoyne PS, Arnold AP. Sex differences in sex chromosome gene expression in mouse brain. Hum Mol Genet. 2002;11:1409–19. doi: 10.1093/hmg/11.12.1409. [DOI] [PubMed] [Google Scholar]
- Xu J, Deng X, Disteche CM. Sex-specific expression of the X-linked histone demethylase gene Jarid1c in brain. PloS one. 2008a;3:e2553. doi: 10.1371/journal.pone.0002553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Deng X, Watkins R, Disteche CM. Sex-specific differences in expression of histone demethylases Utx and Uty in mouse brain and neurons. J Neurosci. 2008b;28:4521–7. doi: 10.1523/JNEUROSCI.5382-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Disteche CM. Sex differences in brain expression of X- and Y-linked genes. Brain Res Mol Brain Res. 2006;1126:50–5. doi: 10.1016/j.brainres.2006.08.049. [DOI] [PubMed] [Google Scholar]
- Xu J, Taya S, Kaibuchi K, Arnold AP. Sexually dimorphic expression of Usp9x is related to sex chromosome complement in adult mouse brain. Eur J Neurosci. 2005;21:3017–22. doi: 10.1111/j.1460-9568.2005.04134.x. [DOI] [PubMed] [Google Scholar]
- Yang CF, Chiang MC, Gray DC, Prabhakaran M, Alvarado M, Juntti SA, Unger EK, Wells JA, Shah NM. Sexually dimorphic neurons in the ventromedial hypothalamus govern mating in both sexes and aggression in males. Cell. 2013;153:896–909. doi: 10.1016/j.cell.2013.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang F, Babak T, Shendure J, Disteche CM. Global survey of escape from X inactivation by RNA-sequencing in mouse. Genome Res. 2010;20:614–22. doi: 10.1101/gr.103200.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zechner U, Wilda M, Kehrer-Sawatzki H, Vogel W, Fundele R, Hameister H. A high density of X-linked genes for general cognitive ability: a run-away process shaping human evolution? Trends Genet. 2001;17:697–701. doi: 10.1016/s0168-9525(01)02446-5. [DOI] [PubMed] [Google Scholar]
- Zinn AR, Roeltgen D, Stefanatos G, Ramos P, Elder FF, Kushner H, Kowal K, Ross JL. A Turner syndrome neurocognitive phenotype maps to Xp22.3. Behav Brain Funct. 2007;3:24. doi: 10.1186/1744-9081-3-24. [DOI] [PMC free article] [PubMed] [Google Scholar]