Abstract
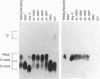
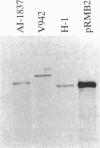
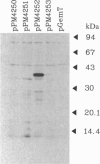
The recent emergence of a pathogenic new non-O1 serotype (O139) of Vibrio cholerae has led to numerous studies in an attempt to identify the origins of this new strain. Our studies indicate that O139 strains have clear differences in the surface polysaccharides when compared with O1 strains: the lipopolysaccharide can be described as semi-rough. Southern hybridization with the O1 rfb region demonstrates that O139 strains no longer contain any of the rfb genes required for the synthesis of the O1 O-antigen or its modification and also lack at least 6 kb of additional contiguous DNA. However, O139 strains have retained rfaD and have a single open reading frame closely related to three small open reading frames of the O1 rfb region. This region is closely related to the H-repeat of Escherichia coli and to the transposases of a number of insertion sequence elements and has all the features of an insertion sequence element that has been designated VcIS1. Transposon insertion mutants defective in O139 O-antigen (and capsule) biosynthesis map to the same fragment as VcIS1. Preliminary sequence data of complementing clones indicate that this DNA encodes a galactosyl-transferase and other enzymes for the utilization of galactose in polysaccharide biosynthesis. We propose a mechanism by which both the Ogawa serotype of O1 strains and the O139 serotype strains may have evolved.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albert M. J., Siddique A. K., Islam M. S., Faruque A. S., Ansaruzzaman M., Faruque S. M., Sack R. B. Large outbreak of clinical cholera due to Vibrio cholerae non-O1 in Bangladesh. Lancet. 1993 Mar 13;341(8846):704–704. doi: 10.1016/0140-6736(93)90481-u. [DOI] [PubMed] [Google Scholar]
- Bhattacharya M. K., Bhattacharya S. K., Garg S., Saha P. K., Dutta D., Nair G. B., Deb B. C., Das K. P. Outbreak of Vibrio cholerae non-O1 in India and Bangladesh. Lancet. 1993 May 22;341(8856):1346–1347. doi: 10.1016/0140-6736(93)90855-b. [DOI] [PubMed] [Google Scholar]
- Calia K. E., Murtagh M., Ferraro M. J., Calderwood S. B. Comparison of Vibrio cholerae O139 with V. cholerae O1 classical and El Tor biotypes. Infect Immun. 1994 Apr;62(4):1504–1506. doi: 10.1128/iai.62.4.1504-1506.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das B., Ghosh R. K., Sharma C., Vasin N., Ghosh A. Tandem repeats of cholera toxin gene in Vibrio cholerae O139. Lancet. 1993 Nov 6;342(8880):1173–1174. doi: 10.1016/0140-6736(93)92157-o. [DOI] [PubMed] [Google Scholar]
- Davis M. A., Simons R. W., Kleckner N. Tn10 protects itself at two levels from fortuitous activation by external promoters. Cell. 1985 Nov;43(1):379–387. doi: 10.1016/0092-8674(85)90043-1. [DOI] [PubMed] [Google Scholar]
- Gustafson C. E., Chu S., Trust T. J. Mutagenesis of the paracrystalline surface protein array of Aeromonas salmonicida by endogenous insertion elements. J Mol Biol. 1994 Apr 8;237(4):452–463. doi: 10.1006/jmbi.1994.1247. [DOI] [PubMed] [Google Scholar]
- Hall R. H., Khambaty F. M., Kothary M., Keasler S. P. Non-O1 Vibrio cholerae. Lancet. 1993 Aug 14;342(8868):430–430. doi: 10.1016/0140-6736(93)92839-l. [DOI] [PubMed] [Google Scholar]
- Hitchcock P. J., Brown T. M. Morphological heterogeneity among Salmonella lipopolysaccharide chemotypes in silver-stained polyacrylamide gels. J Bacteriol. 1983 Apr;154(1):269–277. doi: 10.1128/jb.154.1.269-277.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. A., Salles C. A., Panigrahi P., Albert M. J., Wright A. C., Johnson R. J., Morris J. G., Jr Vibrio cholerae O139 synonym bengal is closely related to Vibrio cholerae El Tor but has important differences. Infect Immun. 1994 May;62(5):2108–2110. doi: 10.1128/iai.62.5.2108-2110.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin R. J., Capage M., Hill C. W. A repetitive DNA sequence, rhs, responsible for duplications within the Escherichia coli K-12 chromosome. J Mol Biol. 1984 Jul 25;177(1):1–18. doi: 10.1016/0022-2836(84)90054-8. [DOI] [PubMed] [Google Scholar]
- Macpherson D. F., Morona R., Beger D. W., Cheah K. C., Manning P. A. Genetic analysis of the rfb region of Shigella flexneri encoding the Y serotype O-antigen specificity. Mol Microbiol. 1991 Jun;5(6):1491–1499. doi: 10.1111/j.1365-2958.1991.tb00795.x. [DOI] [PubMed] [Google Scholar]
- Manning P. A., Heuzenroeder M. W., Yeadon J., Leavesley D. I., Reeves P. R., Rowley D. Molecular cloning and expression in Escherichia coli K-12 of the O antigens of the Inaba and Ogawa serotypes of the Vibrio cholerae O1 lipopolysaccharides and their potential for vaccine development. Infect Immun. 1986 Aug;53(2):272–277. doi: 10.1128/iai.53.2.272-277.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris J. G., Jr Non-O group 1 Vibrio cholerae: a look at the epidemiology of an occasional pathogen. Epidemiol Rev. 1990;12:179–191. doi: 10.1093/oxfordjournals.epirev.a036052. [DOI] [PubMed] [Google Scholar]
- Pearson G. D., Woods A., Chiang S. L., Mekalanos J. J. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qadri F., Chowdhury A., Hossain J., Chowdhury K., Azim T., Shimada T., Islam K. M., Sack R. B., Albert M. J. Development and evaluation of rapid monoclonal antibody-based coagglutination test for direct detection of Vibrio cholerae O139 synonym Bengal in stool samples. J Clin Microbiol. 1994 Jun;32(6):1589–1590. doi: 10.1128/jcm.32.6.1589-1590.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramamurthy T., Garg S., Sharma R., Bhattacharya S. K., Nair G. B., Shimada T., Takeda T., Karasawa T., Kurazano H., Pal A. Emergence of novel strain of Vibrio cholerae with epidemic potential in southern and eastern India. Lancet. 1993 Mar 13;341(8846):703–704. doi: 10.1016/0140-6736(93)90480-5. [DOI] [PubMed] [Google Scholar]
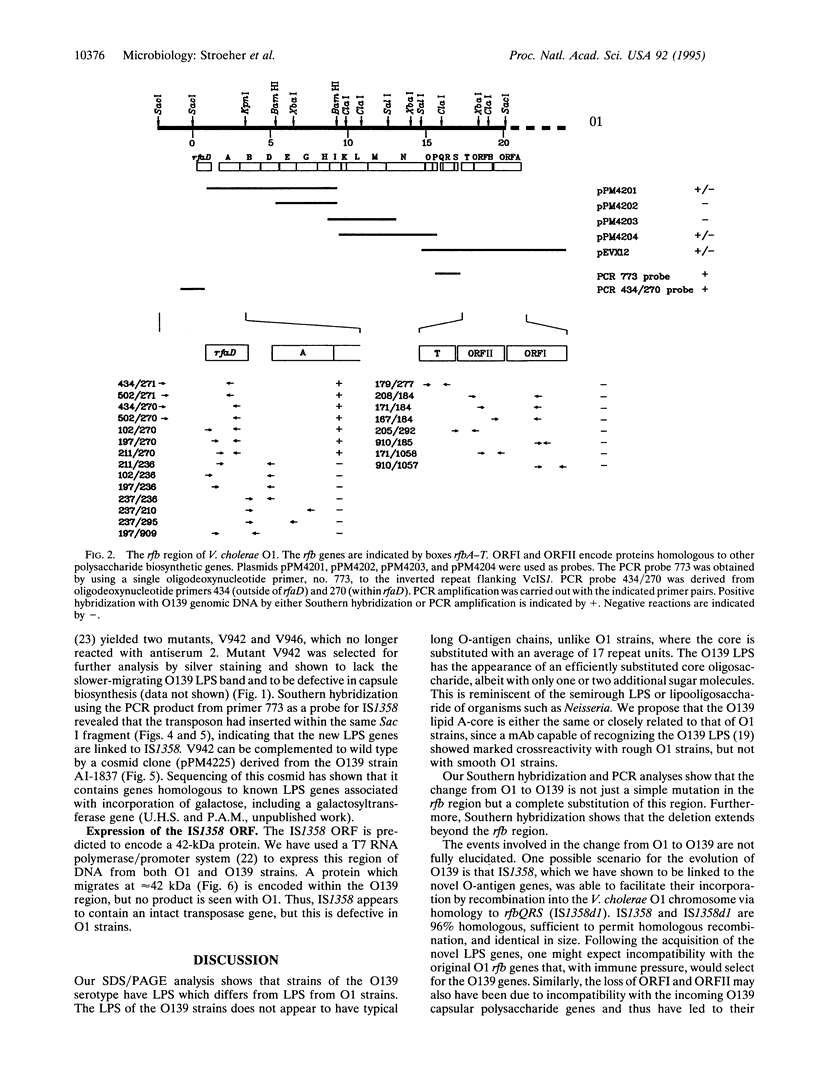
- Rivas M., Toma C., Miliwebsky E., Caffer M. I., Galas M., Varela P., Tous M., Bru A. M., Binsztein N. Cholera isolates in relation to the "eighth pandemic". Lancet. 1993 Oct 9;342(8876):926–927. [PubMed] [Google Scholar]
- Schulz V. P., Reznikoff W. S. Translation initiation of IS50R read-through transcripts. J Mol Biol. 1991 Sep 5;221(1):65–80. doi: 10.1016/0022-2836(91)80205-9. [DOI] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Stroeher U. H., Karageorgos L. E., Morona R., Manning P. A. Serotype conversion in Vibrio cholerae O1. Proc Natl Acad Sci U S A. 1992 Apr 1;89(7):2566–2570. doi: 10.1073/pnas.89.7.2566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor R. K., Manoil C., Mekalanos J. J. Broad-host-range vectors for delivery of TnphoA: use in genetic analysis of secreted virulence determinants of Vibrio cholerae. J Bacteriol. 1989 Apr;171(4):1870–1878. doi: 10.1128/jb.171.4.1870-1878.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waldor M. K., Mekalanos J. J. ToxR regulates virulence gene expression in non-O1 strains of Vibrio cholerae that cause epidemic cholera. Infect Immun. 1994 Jan;62(1):72–78. doi: 10.1128/iai.62.1.72-78.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward H. M., Manning P. A. Mapping of chromosomal loci associated with lipopolysaccharide synthesis and serotype specificity in Vibrio cholerae 01 by transposon mutagenesis using Tn5 and Tn2680. Mol Gen Genet. 1989 Aug;218(2):367–370. doi: 10.1007/BF00331294. [DOI] [PubMed] [Google Scholar]
- Ward H. M., Morelli G., Kamke M., Morona R., Yeadon J., Hackett J. A., Manning P. A. A physical map of the chromosomal region determining O-antigen biosynthesis in Vibrio cholerae O1. Gene. 1987;55(2-3):197–204. doi: 10.1016/0378-1119(87)90280-0. [DOI] [PubMed] [Google Scholar]
- Xiang S. H., Hobbs M., Reeves P. R. Molecular analysis of the rfb gene cluster of a group D2 Salmonella enterica strain: evidence for its origin from an insertion sequence-mediated recombination event between group E and D1 strains. J Bacteriol. 1994 Jul;176(14):4357–4365. doi: 10.1128/jb.176.14.4357-4365.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao S., Sandt C. H., Feulner G., Vlazny D. A., Gray J. A., Hill C. W. Rhs elements of Escherichia coli K-12: complex composites of shared and unique components that have different evolutionary histories. J Bacteriol. 1993 May;175(10):2799–2808. doi: 10.1128/jb.175.10.2799-2808.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]