Abstract
Retrotransposons constitute a major source of genetic variation, and somatic retrotransposon insertions have been reported in cancer. Here, we applied TranspoSeq, a computational framework that identifies retrotransposon insertions from sequencing data, to whole genomes from 200 tumor/normal pairs across 11 tumor types as part of The Cancer Genome Atlas (TCGA) Pan-Cancer Project. In addition to novel germline polymorphisms, we find 810 somatic retrotransposon insertions primarily in lung squamous, head and neck, colorectal, and endometrial carcinomas. Many somatic retrotransposon insertions occur in known cancer genes. We find that high somatic retrotransposition rates in tumors are associated with high rates of genomic rearrangement and somatic mutation. Finally, we developed TranspoSeq-Exome to interrogate an additional 767 tumor samples with hybrid-capture exome data and discovered 35 novel somatic retrotransposon insertions into exonic regions, including an insertion into an exon of the PTEN tumor suppressor gene. The results of this large-scale, comprehensive analysis of retrotransposon movement across tumor types suggest that somatic retrotransposon insertions may represent an important class of structural variation in cancer.
Retrotransposons are genomic elements that mobilize via an RNA intermediate in a copy-and-paste mechanism across the genome. Regarded as “drivers of genome evolution,” retrotransposons comprise nearly half of the human genome and are important vehicles of genomic diversity (Lander et al. 2001; Kazazian 2004). Although the majority of these elements are inactive ancient insertions, a small proportion retains its retrotransposition capacity (Brouha et al. 2003; Beck et al. 2010). The three most active retrotransposon families known are the Long INterspersed Element (LINE-1 or L1), Alu, and SVA (SINE/VNTR/Alu) families (Moran et al. 1996; Kimberland et al. 1999; Xing et al. 2009), specifically the L1HS, AluYa5, and AluYb8 subfamilies in humans (Burns and Boeke 2012). These are thought to retrotranspose via a target-primed reverse transcription (TPRT) mechanism (Luan et al. 1993; Luan and Eickbush 1995; Jurka 1997; Ostertag and Kazazian 2001; Cost et al. 2002), wherein the L1-endonuclease creates two nicks in the genomic DNA followed by insertion of a new copy of the element into the lesion, resulting in short duplicated sequences surrounding the insertion.
Retrotransposon insertions are coming to light as a major source of genetic variation (Ewing and Kazazian 2011; Stewart et al. 2011). It is estimated that one of every 20 live human births exhibits a de novo retrotransposon insertion (Cordaux and Batzer 2009; Xing et al. 2009). A pair of individuals of European origin are believed to differ by ∼500–800 retrotransposon insertion polymorphisms (Stewart et al. 2011). Depending on where they land in the genome, retrotransposon insertions can affect protein function, alter gene expression, and catalyze genomic instability. More than 90 germline retrotransposon insertions have been implicated in disease (Hancks and Kazazian 2012). Specific instances of putative somatic retrotransposon insertions have previously been identified in cancer, including insertions of L1 elements in an exon of the APC tumor-suppressor gene in a case of colorectal cancer (Miki et al. 1992) and within the MYC gene in a breast-carcinoma specimen (Morse et al. 1988), although only the APC event has been verified as a bona fide L1 insertion. Experimental approaches have since identified nine somatic L1 insertions in six primary non–small cell lung tumors (Iskow et al. 2010), numerous L1 insertions in 16 colorectal tumors (Solyom et al. 2012), and a somatic insertion in ST18 in hepatocellular carcinoma (Shukla et al. 2013).
The advent of next-generation sequencing studies of cancer (Stratton et al. 2009; Meyerson et al. 2010) now provides the opportunity to comprehensively investigate the extent of somatic retrotransposon insertions. A recent study identified almost 200 putative somatic retrotransposon insertions from 43 tumor genomes (Lee et al. 2012). Here, we analyze 200 tumor/normal pairs across 11 cancer types using TranspoSeq, a tool we developed to localize retrotransposon insertions from paired-end sequencing data. We find a total of 810 somatic retrotransposon insertions, with 324 in 19 lung squamous cell carcinomas and 206 in 28 head and neck squamous cell carcinomas, while other tumor types appear comparatively quiet. Some of these insertions mobilize to genic regions, including exons, and genes previously implicated in cancer progression. We expand our search to exome data using a modified tool and find additional somatic insertions into exons in endometrial carcinoma.
Results
Whole-genome sequencing reveals numerous nonreference retrotransposon insertions
To identify nonreference somatic retrotransposon insertions computationally from whole-genome sequencing data, we developed TranspoSeq (Helman and Meyerson 2011a; http://cancergenome.nih.gov/newsevents/multimedialibrary/videos/retroseqhelman). Briefly, TranspoSeq locates clusters of unique sequencing reads whose pair-mates align to a database of consensus retrotransposon sequences and predict a genomic fragment length that is nonconcordant with the fragment length distribution of the sample (Supplemental Fig. 1). TranspoSeq classifies putative novel retrotransposon insertion sites as germline, present in both tumor and normal samples but not in the reference, or as somatic, present only in the tumor. We assessed TranspoSeq’s performance using simulated data, determining a sensitivity of 99% with no false-positive calls and a drop in sensitivity at inserted element lengths of <100 bp (Supplemental Fig. 2). We also compared TranspoSeq’s performance to other methods on the same individual and found high concordance (Lee et al. 2012; Keane et al. 2013; Supplemental Material). Finally, we ran TranspoSeq on swapped tumor and normal samples and found no spurious retrotransposon insertions unique to the matched normal tissue (Supplemental Material).
To determine the extent of somatic retrotransposon activity across cancer, we applied TranspoSeq to whole-genome sequencing data from 200 tumor and matched normal samples collected and sequenced through The Cancer Genome Atlas across 11 tumor types: lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian carcinoma (OV), rectal adenocarcinoma (READ), colon adenocarcinoma (COAD), kidney clear-cell carcinoma (KIRC), uterine corpus endometrioid carcinoma (UCEC), head and neck squamous cell carcinoma (HNSC), breast carcinoma (BRCA), acute myeloid leukemia (LAML), and glioblastoma multiforme (GBM). We identified 7724 unique, nonreference germline insertion sites seen in both tumor and matched normal samples (Supplemental Table 2). Of these, 65% are known retrotransposon insertion polymorphisms annotated in previous studies (Xing et al. 2009; Beck et al. 2010; Ewing and Kazazian 2010, 2011; Hormozdiari et al. 2010; Huang et al. 2010; Iskow et al. 2010; Witherspoon et al. 2010; Stewart et al. 2011; Lee et al. 2012). Many of the novel germline retrotransposon insertions identified here represent previously unannotated common polymorphisms, present in as many as 114 individuals (Supplemental Fig. 3D).
We attempted experimental validation on a set of 47 putative somatic retrotransposon insertions, across 21 individuals and four tumor types, including five somatic insertions identified from exome data, as well as four predicted germline transpositions. Validation was carried out via site-specific PCR designed to span the 5′ and 3′ junctions of candidate insertions for tumor and matched normal samples, followed by Illumina sequencing. We found that 39/47 (83%) of predicted somatic insertions have experimental evidence for a transposition event by amplification of either 5′ or 3′ junctions in the tumor, but no junctional amplification from the matched normal sample. Moreover, 32 of 47 (68%) predicted somatic insertions had evidence for amplification of both 5′ and 3′ junctions in the tumor sample and no evidence in the matched normal. Finally, 2/47 putative somatic retrotranspositions had some evidence of the insertion in the matched normal, and 6/47 failed to produce any amplicons in either tumor or matched normal (Supplemental Table 1).
Somatic retrotransposon insertion rates vary across tumor types
We detected a total of 810 putative retrotransposon insertions occurring in cancer DNA but not in matched normal DNA from the same patient. These candidate somatic retrotransposition events exhibit the hallmarks of target-primed reverse transcription, such as target site duplications (TSDs) ∼15 bp in length (Fig. 1A), and a canonical L1-endonuclease motif (Feng et al. 1996; Morrish et al. 2002) at the site of insertion (Fig. 1B). There is an additional class of somatic events lacking a TSD, however, suggesting a possible alternative mechanism for somatic insertion (Supplemental Fig. 5). Consistent with previous reports (Iskow et al. 2010; Lee et al. 2012; Solyom et al. 2012), we find that somatic insertions consist primarily (97%) of L1HS elements, specifically L1HS elements that are severely 5′ truncated, differing significantly from germline insertions (Fig. 1C,D). In addition, we find several full-length L1HS somatic insertions, as well as one putative somatic insertion of an SVA element (Supplemental Table 3). It should be noted that, given the 83% validation rate of TranspoSeq, it is likely that roughly 670 of the 810 putative somatic insertions are real events, while the remaining may be false positives due to sequencing artifact or presence in normal tissue.
Figure 1.
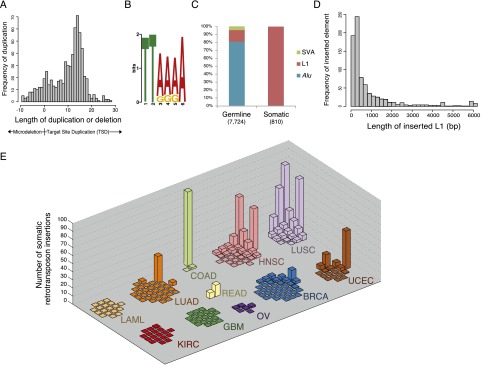
Landscape of retrotransposon insertions across cancer reveals a tumor-type specific pattern. (A) Distribution of duplication or deletion lengths at sites of somatic retrotransposon insertion. Target-site duplication (TSD) lengths are sequence duplications of positive length, while microdeletions at the breakpoint are plotted as negative values according to the length of the deletion. See Supplemental Figure 3A for an analogous plot of germline retrotransposon insertions. (B) A sequence logo of the consensus motif at the predicted breakpoints of somatic retrotransposon insertions. See Supplemental Figure 3B for germline insertion sequence motif. (C) Percentage of each retrotransposon family inserted in both tumor and matched normal (germline) and only in tumor (somatic) across all samples. (D) Length of somatically inserted L1 element (see Supplemental Fig. 3C for germline). (E) Distribution of somatic retrotransposon insertion events per individual across all tumor types. For each tumor type, the vertical axis displays the number of somatic retrotransposon events identified within each individual queried. These data are whole-genome sequences from 200 individuals collected and sequenced through The Cancer Genome Atlas, across 11 tumor types: lung adenocarcinoma (LUAD), lung squamous cell carcinoma (LUSC), ovarian carcinoma (OV), rectal adenocarcinoma (READ), colon adenocarcinoma (COAD), kidney clear cell carcinoma (KIRC), uterine corpus endometrioid carcinoma (UCEC), head and neck squamous cell carcinoma (HNSC), breast carcinoma (BRCA), acute myeloid leukemia (LAML), and glioblastoma multiforme (GBM). See Supplemental Figure 4, A and B, for other representations of these data.
Somatic retrotransposon insertions display a tumor-specific pattern. While GBM, LAML, BRCA, KIRC, OV, and LUAD samples exhibit little or no detected somatic retrotransposition, LUSC, COAD/READ, HNSC, and UCEC show active mobilization of retrotransposons (Fig. 1E; Supplemental Fig. 4; Supplemental Table 3). These findings are in accordance with other studies where L1 insertions were seen in epithelial cancers but not in glioblastomas or hematopoietic cancers (Iskow et al. 2010; Lee et al. 2012; Solyom et al. 2012). Within tumor types, there is wide variation of somatic events among individuals with, for example, a range from 0 up to 79 somatic insertions per sample in squamous cell lung carcinomas.
Earlier studies found enrichment of disease-causing retrotransposon insertions on the X chromosome, possibly due to an ascertainment bias from X-linked disorders. We find cancer-associated somatic events to be evenly distributed across the autosomal and X chromosomes (Supplemental Fig. 4). The distribution of retrotransposon insertions across chromosomal arms significantly differs between germline and somatic events (Wilcoxon P = 3.706 × 10−8). Specifically, the short arm of chromosome 4 has a 1.6-fold enrichment compared with a null distribution of somatic retrotransposon insertions, differing from germline insertions in that arm (Fisher’s P = 0.0087).
Retrotransposons can mobilize into genic regions
Retrotransposons have the capacity to mobilize into genes and surrounding regulatory regions to affect gene expression and disrupt protein function; these insertions have previously been implicated in cancer. Most recently, Shukla et al. (2013) discovered an L1 insertion into ST18 in hepatocellular carcinoma that resulted in overexpression of the gene. We find that the proportion of somatic retrotransposon insertions into genes is similar to that of germline events, where ∼35% of insertions fall in genic regions (coding gene plus 1 kb upstream and downstream) as would be expected from the proportion of the human genome that is comprised of these genic regions. We find several genes that are recurrently disrupted by retrotransposon insertions in multiple samples across tumor types, including CNTNAP2, DLG2, and PDE4B (Fig. 2A). Many of these appear to be known large, common fragile site genes (Fungtammasan et al. 2012; Supplemental Fig. 6A,B). A closer look at the specific genes that contain somatic insertions reveals several known cancer genes, such as RUNX1, a putative tumor suppressor in gastric carcinoma (Silva et al. 2003) that is subject to recurrent loss-of-function inactivation in breast cancer and esophageal adenocarcinoma (Banerji et al. 2012; Dulak et al. 2012; Koboldt et al. 2012), as well as in the exon of REV3L, which has been implicated as a novel tumor suppressor in colorectal and lung cancers, and is involved in maintenance of genomic stability (Brondello et al. 2008; Zhang et al. 2013). One UCEC sample contains an intronic somatic L1 insertion in the ESR1 gene, an important hormone receptor often overexpressed in endometrial and breast cancers (Lebeau et al. 2008). While previous studies found somatic insertion only in intronic regions, we identify 21 somatic events in or within 200 bp of exons of genes such as CYR61 and HSF2, with seven falling in the protein-coding sequence itself (Fig. 2B; Supplemental Table 3). In general, genes with somatic retrotransposon insertions tend to be involved in cell adhesion processes (Supplemental Fig. 7).
Figure 2.
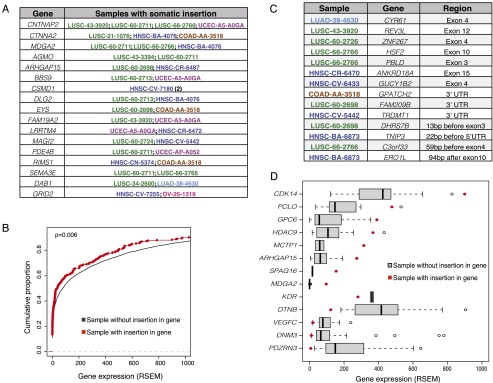
Retrotransposons can mobilize into genic regions. (A) Genes that contain somatic retrotransposon insertions in more than one sample. (B) Empirical cumulative distribution function (ecdf) of gene expression, quantified by RNA-seq by Expectation Maximization (RSEM) values, of genes that contain somatic retrotransposon insertions in a specific sample (red) versus the ecdf of gene expression in genes that do not contain retrotransposon insertions across all other samples (black). (C) Genes that contain somatic retrotransposon insertions in or within 200 bp of exons, 5′, and 3′ UTRs. (D) Gene expression of a selection of genes with somatic retrotransposon insertions; the red dot shows the RSEM value in the particular tumor sample that contained the retrotransposon insertion in that gene, while the gray represents the gene’s expression across all other samples within that tumor type that do not contain a retrotransposon insertion.
We asked whether somatic retrotransposon insertion into a gene impacts the gene’s expression. Using available RNA-seq data across the eight tumor types with retrotransposon insertions in genes (LUSC, LUAD, HNSC, UCEC, BRCA, OV, COAD, and READ), we find that genes with retrotransposon insertions tend to be expressed at a lower level than the same genes in samples of the same tumor type without an insertion (KS-test P = 0.006) (Fig. 2C). When examined individually, some genes with retrotransposition insertions show extreme expression relative to all other samples, in either direction (Fig. 2D).
Somatic 3′-sequence transductions elucidate active retrotransposon elements in cancer
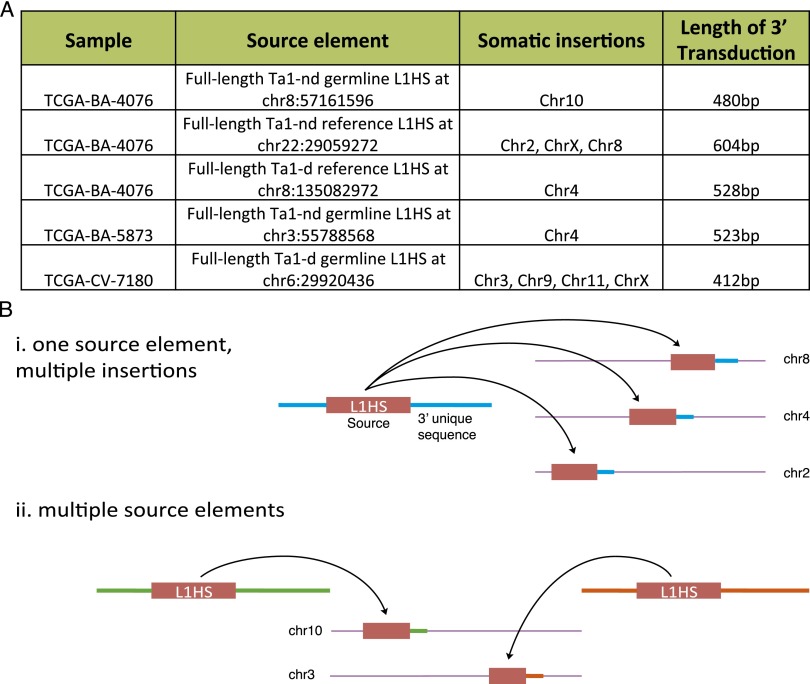
In several samples, we find evidence for the retrotransposition of an L1 along with a short unique genomic sequence. These unique sequences originate from the region downstream from both reference and nonreference germline L1 elements. Known as 3′-transduction, this process is thought to result from the read-through of the weak L1 poly(A) signal and is estimated to occur in 15%–23% of all genomic L1s (Holmes et al. 1994; Moran et al. 1999; Goodier et al. 2000; Pickeral et al. 2000; Szak et al. 2002 ). L1s carrying 3′ transductions have been shown to disrupt several human genes in disease including APC (Miki et al. 1992), DMD (Holmes et al. 1994), CYBB (Meischl et al. 2000), RP2 (Schwahn et al. 1998), and CHM (van den Hurk et al. 2003). 3′-transductions exhibit known TPRT characteristics, including TSDs, the L1 endonuclease motif at the insertion point, and polyadenylation of the 3′-transduced segment. One HNSC sample displayed several such 3′-transduction events from different regions of the genome, suggesting that at least three separate L1HS elements were active in the tumor sample (Supplemental Table 4). In another sample, we find a known nonreference polymorphic full-length germline L1HS element (chr6:29920436) to be highly active and result in at least four separate instances of somatic 5′-truncated L1HS insertions on chromosomes 3, 9, 11, and X (Fig. 3A; Supplemental Table 4). Thus we see evidence for two models of somatic retrotransposon activity in cancer: (1) a single hyperactive source element may insert itself multiple times throughout the genome in the tumor sample, and (2) multiple elements may become active in the tumor sample (Fig. 3B).
Figure 3.
3′-transductions elucidate source retrotransposon element. (A) Select 3′-transduction events, including the sample, the source element location (i.e., genomic origin of the unique sequence), the transposition insertion location, and the length of the transduced sequence. See Supplemental Table 4 for a full list. (B) Schematic of the two models of somatic retrotransposition detected seen in this analysis: (i) one source L1HS element becoming active and inserting multiple times across the tumor sample, and (ii) several source elements becoming active in the tumor sample.
The genomic context of somatic retrotransposition
To evaluate genomic correlates of somatic retrotransposition, cases were binned into two groups by the number of somatic retrotransposon insertions: Retrotransposon-High (RTI-H) tumors have >10 somatic insertions and Retrotransposon-Low (RTI-L) have 10 or fewer insertions. We used the rearrangement detection tool, dRanger (Bass et al. 2011; Chapman et al. 2011) to identify the number of rearrangements in LUSC, LUAD, and HNSC. Samples in the high somatic retrotransposition cluster have more complex genomes in terms of somatic rearrangements (Wilcoxon P = 0.0097) (Fig. 4A). Retrotransposon-high samples also have greater numbers of total somatic substitution mutations per sample than do retrotransposon-low samples (Wilcoxon P = 2.8 × 10−4) (Fig. 4B).
Figure 4.
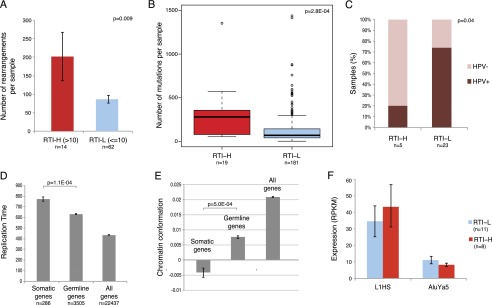
Retrotransposon load is correlated with genomic instability, late-replication, and closed chromatin. (A) Number of somatic rearrangements in LUSC, LUAD, and HNSC samples with high retrotransposon load (>10 somatic retrotransposon insertions, RTI-H) and with low retrotransposon load (≤10 somatic insertion, RTI-L). (B) Number of somatic mutations in RTI-H and RTI-L samples across all 11 tumor types. (C) HPV status of RTI-H and RTI-L HNSC samples. (D) Replication timing of genes that contain somatic retrotransposon insertions versus genes that contain germline insertions, and all RefSeq genes. Later replicating genes have higher values of replication time on the y-axis. (E) Chromatin conformation of genes that contain somatic retrotransposon insertions versus genes that contain germline insertions, and all RefSeq genes. The y-scale represents relative chromatin “openness,” the lower the y-value, the more closed the chromatin state. (F) Expression (RPKM) of consensus L1HS and AluYa5 sequences in RTI-H and RTI-L LUSC samples. All error bars represent standard error of the distribution.
Within each tumor type we correlated somatic mutation, methylation, copy number, and microRNA data where available to retrotransposon clusters. We find that in HNSC samples, both TP53 mutation and CDKN2A (also known as p16) focal deletion are significantly correlated to high retrotransposition activity (Fisher’s P = 0.01481, data not shown). Since HPV-positive HNSC tumors are less likely to have TP53 mutation (Gillison et al. 2000), we looked at somatic retrotransposition versus HPV status in the 28 HNSC samples and found that, accordingly, samples with high retrotransposition are disproportionately HPV negative (Fisher’s exact P = 0.041) (Fig. 4C).
Furthermore, we find that retrotransposons tend to insert somatically in late-replicating genes, as compared to germline insertions (Wilcoxon P = 1.1 × 10−4) and the null distribution of genic replication times (Wilcoxon P < 2 × 10−16) (Fig. 4D). This is in agreement with a recent report that somatically mutated genes are biased toward later replication time (Lawrence et al. 2013). Interestingly, chromatin conformation as assessed by Hi-C long-range interaction data (Lieberman-Aiden et al. 2009) shows that somatic retrotransposon insertions are targeted at regions of the genome that have a more closed conformation (Wilcoxon P = 5 × 10−4) (Fig. 4E).
Finally, using RNA-seq data from LUSC tumor samples, we assessed the expression levels of two active subfamilies of retrotransposons, L1HS and AluYa5, and found that retrotransposon expression does not appear to correlate with retrotransposition activity in this tumor type (Fig. 4F).
Retrotransposon insertions identified in exome capture data
Since we find somatic retrotransposon insertions into exonic regions, we modified TranspoSeq to interrogate the large number of exome sequencing data available through TCGA. TranspoSeq-Exome locates clusters of split reads, where one portion of the read aligns uniquely to the genome and the other portion aligns to the database of consensus retrotransposon sequences, in effect spanning the junction between unique genome and retrotransposon (Supplemental Fig. 8). This method is effective because the sequencing reads used in this study are 100 bp in length and provide adequate split read sequence. We applied TranspoSeq-Exome to whole-exome sequence data of 199 LUSC, 327 HNSC, and 241 UCEC samples, focusing our analysis on L1HS retotransposition. We were able to recapitulate four of the exonic insertions detected in the whole-genome sequences of the same samples, and also found 22 novel somatic L1HS insertions in LUSC, five in UCEC, and eight in HNSC (Fig. 5A; Supplemental Table 5). Exome data reveals somatic retrotransposon insertions in exons of several of the same genes that have intronic somatic insertions in the whole-genome sequencing data, as well as new exonic insertions. Thus, we add 35 novel somatic retrotransposon events and show that somatic retrotransposon insertions into exons can be detected from hybrid-capture exome sequencing.
Figure 5.
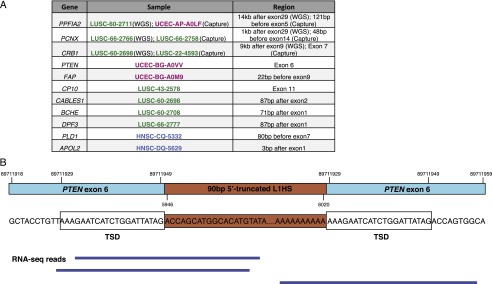
Exome sequencing identifies novel retrotransposon insertions into exons. (A) Genes with somatic retrotransposon insertions into exons as detected by TranspoSeq-Exome. Somatic insertions in PPFIA2, PCNX, and CRB1 were identified in the whole-genome sequencing cohort as well as in separate samples in the exome sequencing set. (B) Diagram of a 90-bp 5′-truncated L1HS element inserted into exon 6 of PTEN. In dark blue are RNA-seq reads that span the reference–transposon junction, supporting its expression.
Notably, we find an 112-bp 5′-truncated L1HS element in exon 6 of the PTEN tumor suppressor in DNA from an endometrial carcinoma (Fig. 5B). RNA-seq reads spanning the insertion at both ends confirm the expression of a chimeric mRNA containing a somatically inserted L1HS sequence. While its 3′ end inserted at the canonical L1-endonuclease cleavage motif, this retrotransposition is likely the result of a 5′ microhomology-mediated end-joining (Zingler 2005) with a 12-bp overlap between reference sequences at the 5′-end integration site and the 5′-truncated L1HS element. We experimentally validated the presence and sequence of this insertion in the endometrial sample and not-matched normal tissue (Supplemental Fig. 9).
Discussion
We present here a large-scale comprehensive analysis of somatic retrotransposon movement in cancer. We find that not only colorectal cancer (Lee et al. 2012; Solyom et al. 2012), but also lung squamous cell, head and neck squamous cell, and endometrial carcinomas exhibit considerable L1 retrotransposition. Other cancer types, including glioblastoma multiforme, acute myeloid leukemia, and kidney clear-cell carcinoma, remain quiet. We demonstrate the novel insertion of L1HS into known and putative tumor-suppressor genes, such as RUNX1 and REV3L, and identify genes that undergo recurrent insertion across samples and tumor types, such as CNTNAP2. We also present the first analysis of retrotransposon insertions using exome-capture data, revealing several interesting exonic insertions, including one into PTEN. Our findings suggest that somatic retrotransposon insertions are an important class of cancer-associated structural variation with the potential to play a role in the tumorigenesis of certain cancers.
A small set of active L1s accounts for most of the L1 activity in humans (Brouha et al. 2002; Beck et al. 2010). We find that the majority of somatically inserted L1s are severely 5′-truncated, and are thus rendered inactive upon insertion. Nonetheless, we do identify several full-length L1HS somatic insertions, as well as common full-length germline polymorphisms that mobilize in the tumor sample, as evidenced by their transduction of unique 3′-sequences. This raises the possibility that polymorphic transposable elements in the germline may predispose to increased somatic retotransposon activity.
The typical mechanism of retrotransposition, TPRT, leads to double-stranded breaks (DSBs), and so it is thought that L1 transposition has genome-destabilizing effects (Belgnaoui et al. 2006). Whether it is the L1 that is causing these DSBs or rather contributing to L1-mediated repair of preexisting DSBs (Morrish et al. 2002) is an open question; we do, however, see some evidence of somatic L1-endonuclease-independent insertions lacking the canonical endonuclease motif and TPRT TSDs (Supplemental Fig. 5), suggesting a possible alternate mechanism of L1 insertion in tumor genomes.
The distribution of retrotransposon insertions may depend on the accessibility of the chromosome to the transposition machinery. L1-endonuclease, however, shows preference for supercoiled DNA (Feng et al. 1996), and although L1-endonuclease nicking of histone-bound DNA was found to be repressed, some sites were enhanced for L1 nicking when nucleosomal (Cost et al. 2001). We find a disproportionate amount of somatic retrotransposon insertions occurring in closed chromatin regions of the genome. Although we used chromatin open/closed states derived from a normal human lymphoblastoid cell line, Lieberman-Aiden et al. (2009) found high reproducibility between cell lines of different origin and tissue type. Because genes within closed chromatin states are expressed at lower rates, it is conceivable that somatic insertions are tolerated in these regions, despite the difficulty in access.
Some limitations of TranspoSeq and TranspoSeq-Exome for identifying novel nonreference somatic retrotransposon insertions include inherent problems associated with read lengths of 100 bp, fragment-length dispersion, alignment uncertainty, and disparate sequencing coverage. Additionally, the accuracy of retrotransposon subfamily calling is limited by the corresponding fragment length of the sequencing library. Future integration of sequencing technologies that enable long fragments and longer reads (Carneiro et al. 2012) will aid in the precise identification of the inserted elements.
In addition to identifying novel somatic retrotransposon insertions across multiple tumor and sequencing data types, we also sought to answer the question: What does somatic retrotransposition target? We show here that somatic retrotransposition recurrently targets large, common-fragile site genes that are late replicating and tend to be located in regions of closed chromatin. Whether these regions are specifically targeted by L1 or whether negative selection eliminated the cells with insertions into other areas remains to be elucidated. L1 insertions are frequent genomic passenger events in cancer, and their ability to act as drivers has yet to be demonstrated (Rodić and Burns 2013). Thus, somatic retrotransposition should continue to be investigated in large sequencing studies such as TCGA and may provide insight into tumor biology, clinically viable targets, and potential biomarkers for patient stratification.
Methods
Data
Sequencing data
Sequencing data were downloaded from the TCGA CGHub repository (https://cghub.ucsc.edu/). Primary and processed data for TCGA can be downloaded by registered users at https://tcga-data.nci.nih.gov/tcga/tcgaDownload.jsp, https://cghub.ucsc.edu/. The downloaded BAM file for each tumor and normal sample (aligned to genome build hg18 for LAML, COAD, READ, and two OV samples, and hg19 for all others using BWA) were used as input to TranspoSeq.
Processed RNA-seq data, in the form of RNA-seq by Expectation Maximization (RSEM) values, and mutation data were downloaded from Synapse (syn300013, https://www.synapse.org/#!Synapse:syn300013).
Retrotransposon data
Consensus retrotransposon sequences were downloaded from GIRI Repbase (http://www.girinst.org/repbase/). All elements in the L1 (n = 117) and SINE1/7SL (n = 55) families, as well as SVA were included in this analysis. See Supplemental Table 6 for the sequences used. Reference retrotransposon identities were downloaded from RepeatMasker on January 12, 2013 (http://www.repeatmasker.org/).
Retrotransposon insertion polymorphism data
dbRIP (Wang et al. 2006) was accessed on May 22, 2012. At the time of download, it contained 2086 Alu, 598 L1, and 77 SVA annotated elements. We also include data from 10 other previous studies reporting germline retrotransposon insertions (Xing et al. 2009; Beck et al. 2010; Ewing and Kazazian 2010, 2011; Hormozdiari et al. 2010; Huang et al. 2010; Iskow et al. 2010; Witherspoon et al. 2010; Stewart et al. 2011; Burns and Boeke 2012; Lee et al. 2012).
Gene annotation data
RefSeq annotation files for both hg18 and hg19 were downloaded from UCSC Genome Browser (genome.ucsc.edu/cgi-bin/hgTables?command=start) on Oct 19, 2011.
Computational analysis
TranspoSeq was first presented in 2011 as RetroSeq (Helman and Meyerson 2011a,b; http://cancergenome.nih.gov/newsevents/multimedialibrary/videos/retroseqhelman). It uses both paired and split read information to identify and characterize nonreference retrotransposon insertion events from tumor and matched normal BAM files. It is functionally similar to other read-anchored and split-read mobile element insertion tools such as Tea (Lee et al. 2012) and the Sanger Institute’s RetroSeq (Keane et al. 2013), but includes additional de novo assembly and contig alignment procedures. TranspoSeq consists of three main steps: (1) Get Reads, (2) Process Reads, and (3) Assemble Reads. See Supplemental Figure 1 for a detailed schematic of the process.
Get reads
Beginning with the input BAM file, TranspoSeq parses out all discordant read-pairs, defined as pair-mates whose aligned positions are nonconcordant with the fragment length distribution. We use a threshold of 1 kb to call a nonconcordant read-pair order to balance the desired sensitivity and specificity given an average fragment length of about 400 bp. These read-pairs are then aligned to a database of consensus retrotransposon sequences using NCBI’s blastn algorithm. Reads that align with either a predefined minimal percent identity and number of consecutive bases, or a predefined maximal BLAST e-value are kept for further processing. In this analysis, we use a BLAST e-value threshold of 2 × 10−7, which is equivalent to ∼30 consecutive nucleotides with 85% identity to the consensus retrotransposon sequence. For each read that successfully aligns, we locate its pair-mate: If this mate also aligns to the retrotransposon database, the pair is discarded; if not, and the mate aligns to the genome with adequate mapping quality (MAPQ>0), the pair is collected for further processing.
Process reads
Unique reads whose pair-mates align to a retrotransposon consensus sequence are grouped by read orientation (forward or reverse) and each set is clustered separately. Clusters are defined by the distance between the start positions of two adjacent reads as no larger than 200 bp. Forward and reverse clusters are then overlapped—allowing for an overlap of up to 60 bp and a gap of up to 500 bp between a forward and reverse cluster in order to account for target sequence duplications (TSDs) and variable coverage. Parameter values were chosen based on prior knowledge as well as empirically, and tested on simulated data sets. One-sided events, clusters without an overlapping cluster in the opposing orientation are set aside for future investigation.
Events supported by clusters in both directions are annotated based on: presence in matched normal sample, proximity (within a 200-bp window) to a reference retrotransposon, known RIP (dbRIP) (Wang et al. 2006) and 1000 Genomes Project (Ewing and Kazazian 2011; Stewart et al. 2011), known gene (RefSeq track of UCSC Genome Browser), and known CNV (Beroukhim et al. 2007). Events are also annotated with information pertaining to alignment to the retrotransposon database: identity, inferred length, and inversion status of inserted retrotransposon element.
Inferred length of an inserted element is determined computationally via the alignment positions at either end of the insertion, i.e., if the 5′ junction read aligns to position 1000 of a consensus L1HS element and the 3′ junction read aligns to position 6020, an inserted element of 4020 bp is postulated at that site.
Assemble reads
Read-pairs supporting a candidate insertion as well as split reads spanning the putative insertion breakpoint are then assembled de novo using Inchworm (Grabherr et al. 2011) to form contigs in the forward and reverse directions separately. Contigs in each direction are aligned back to the database of retrotransposon consensus sequences with BLAST (blastn) and to the reference genome using BLAT. The longest contig containing a retrotransposon-aligned region and a reference-aligned region with minimal overlap is returned along with the specific retrotransposon subfamily and alignment properties. If such a contig cannot be constructed, TranspoSeq uses the alignment properties of the discordant reads themselves. Split reads are used, when available, to determine the forward and reverse breakpoints as well as the putative TSD sequence defined as the region between these forward and reverse breakpoints.
Filtering
Post-processing filtering was performed to remove regions with >30% poor quality reads (MAPQ = 0), <0.005 allelic fraction, and >25 discordant reads within the candidate region in the normal sample, as well as regions that did not produce at least one substantial contig (>14 bp) from de novo assembly. Allelic fraction is calculated by (number of split reads supporting insertion)/(number of total reads spanning breakpoint). Candidate insertions of a retrotransposon into the same reference element subfamily were also filtered out. Only events with at least 10 read-pairs, including at least two in each direction, supporting the insertion were maintained. Events consistent with microsatellite instability or ancient retrotransposons were filtered out. Finally, we manually reviewed each putative somatic insertion region using the Broad Institute’s Integrative Genome Viewer (Robinson et al. 2011) and only those events that passed manual inspection were retained for further analysis.
TranspoSeq uses the SAMtools netsf java toolkit to parse BAM files and R for data processing. Pipelines are run with the reference assembly corresponding to the input BAMs, and the resulting calls are then converted to hg19 when necessary using UCSC Genome Browser Database liftOver (Meyer et al. 2012).
TranspoSeq-Exome
We modified TranspoSeq to search for novel junctions between retrotransposons and unique genomic sequence using split reads. Instead of aligning all discordant read-pairs to the database of consensus retrotransposon sequences, TranspoSeq-Exome first parses out all clipped reads identified by BWA and aligns the clipped sequence to the database of retrotransposons. Split reads that have >10 bp aligning to a retrotransposon with an E-value of 2 × 10−7 or lower are then clustered, processed, and annotated as in TranspoSeq (see Supplemental Fig. 8 for a schematic of the method). A limitation of this technique is that we are only able to identify inserted L1s where the 5′ end (even if truncated) of the L1HS is captured, because the poly(A)-containing 3′ end does not align significantly to the database. Additionally, the exact base-pair location of a clip can be misidentified by BWA.
Experimental validation
Validations were carried out via site-specific PCR designed to span the 5′ and 3′ junctions of candidate insertions for tumor and matched normal samples. Primers, designed using Primer3 (Rozen and Skaletsky 2000), and target information is listed in Supplemental Table 1. PCRs were performed with 3 μL of 2.5 ng/μL DNA, 5 μL of 1-μM mixed primers, 0.08 μL of 100 mM dNTPs, 0.04 μL Hot Start Taq DNA polymerase, 0.4 μL of 25 mM MgCL2 and 1 μL of 10× buffer, with 1.47 μL of dH20 for a total reaction volume of 11 μL. The reactions were run with a hot start of 95°C for 5 min, then 30 cycles of 94° for 30 sec, 60° for 30 sec, and 72° for 1 min, followed by a final cool-down at 72° for 3 min. A total of 2 μL of each PCR reaction was run on a caliper to visualize PCR amplicons. Initial PCRs underwent eight cycles of a tailing reaction to add adapters and indexes for sequencing and run on Illumina MiSeq with a single 8-bp index and standard Illumina sequencing primers, resulting in 250-bp paired-end reads and insert size ∼320 bp and a coverage of ∼200×. See Supplemental Figure 9 and accompanying text for further information regarding validation experiments.
Statistical analysis
Correlations with other genomic features
Data for replication timing and chromatin conformation were collected from Chen et al. (2010) and Lieberman-Aiden et al. (2009), respectively, and relationships with retrotransposon insertions were assessed using a two-sided Wilcoxon rank-sum test. HPV status for 28 HNSC samples were derived from the paper freeze analysis set provided by the TCGA HNSC AWG (http://www.broadinstitute.org/collaboration/gcc/samples/hnsc). These calls were based on review of several data types including detection of HPV by RNA and DNA sequencing, mass spectrometry, and available clinical data. Association between HPV status and retrotransposon insertions was quantified with a two-sided Fisher’s exact test.
Retrotransposon high and low clusters were correlated to somatic mutation, arm-level, and focal copy-number changes, miRNA levels, and methylation data when available, using The Broad Institute’s TCGA GDAC (https://confluence.broadinstitute.org/display/GDAC/Home). Fisher’s exact tests are used to assess significance of association.
Sequence motif
Sequence motifs at insertion breakpoints were computed using the MEME Suite (Bailey et al. 2009).
Retrotransposon element expression
Raw RNA-seq FASTQ files were aligned to consensus L1HS and AluYa5 sequences using Bowtie 2 (Langmead et al. 2009) allowing for one mismatch. Values were then converted to reads per kilobase per million (RPKM) by the formula: number of mapped reads/length of transcript in kilobase/total number of reads in Mb.
3′-transductions
Short transductions were identified when reads on one side spanned the transduction and therefore the event maintained evidence for a retrotransposon insertion on both sides. A transduction was called when the 3′ end junction of the insertion spanned across the poly(A) sequence into a region 3′ of an active (either reference or germline/somatic) L1 element. Element characteristics were assessed using L1Base (Penzkofer 2004).
Correlation with gene expression
To assess overall gene expression changes across all tumor types: We compared gene expression in the sample in which the insertion is present to the distribution of RSEM across all other samples investigated. We used a two-tailed Wilcoxon-Mann Whitney test in R to test for the hypothesis that a gene with a retrotransposon insertion is transcribed at a significantly lower level in samples with this insertion.
To assess individual expression changes: For each gene containing a retrotransposon insertion, we compared the RSEM for the sample in which the insertion is present to the empirical cumulative distribution of the RSEM values of that gene across all samples within that tumor type. We used a one-sample, two-sided Kolmogorov-Smirnov test in R (ks.test) to assess the hypothesis that a gene with a retrotransposon insertion is expressed at a significantly different level than in samples without this insertion. P-values were corrected for multiple testing using Bonferroni correction.
Data access
Source code for TranspoSeq and TranspoSeq-Exome is available at http://www.broadinstitute.org/cancer/cga/transposeq. Retrotransposon insertion positions have been submitted to NCBI dbVar (http://www.ncbi.nlm.nih.gov/dbvar/) under accession nstd94.
Acknowledgments
This work was conducted as part of The Cancer Genome Atlas (TCGA), a project of the National Cancer Institute (NCI) and National Human Genome Research Institute (NHGRI). We thank Peter Hammerman, Chandra Pedamallu, Josh Francis, Fujiko Duke, Luc de Waal, Joonil Jung, Angela Brooks, and Alal Eran for their tremendous help. Funding was supported by National Cancer Institute grants U24CA143867 and U24CA126546 to M.M.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.163659.113.
References
- Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS 2009. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res 37: W202–W208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerji S, Cibulskis K, Rangel-Escareno C, Brown KK, Carter SL, Frederick AM, Lawrence MS, Sivachenko AY, Sougnez C, Zou L, et al. 2012. Sequence analysis of mutations and translocations across breast cancer subtypes. Nature 486: 405–409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bass AJ, Lawrence MS, Brace LE, Ramos AH, Drier Y, Cibulskis K, Sougnez C, Voet D, Saksena G, Sivachenko A, et al. 2011. Genomic sequencing of colorectal adenocarcinomas identifies a recurrent VTI1A-TCF7L2 fusion. Nat Genet 43: 964–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck CR, Collier P, Macfarlane C, Malig M, Kidd JM, Eichler EE, Badge RM, Moran JV 2010. LINE-1 retrotransposition activity in human genomes. Cell 141: 1159–1170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belgnaoui SM, Gosden RG, Semmes OJ, Haoudi A 2006. Human LINE-1 retrotransposon induces DNA damage and apoptosis in cancer cells. Cancer Cell Int 6: 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beroukhim R, Getz G, Nghiemphu L, Barretina J, Hsueh T, Linhart D, Vivanco I, Lee JC, Huang JH, Alexander S, et al. 2007. Assessing the significance of chromosomal aberrations in cancer: methodology and application to glioma. Proc Natl Acad Sci 104: 20007–20012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brondello J-M, Pillaire MJ, Rodriguez C, Gourraud P-A, Selves J, Cazaux C, Piette J 2008. Novel evidences for a tumor suppressor role of Rev3, the catalytic subunit of Pol ζ. Oncogene 27: 6093–6101 [DOI] [PubMed] [Google Scholar]
- Brouha B, Meischl C, Ostertag E, de Boer M, Zhang Y, Neijens H, Roos D, Kazazian HH 2002. Evidence consistent with human L1 retrotransposition in maternal meiosis I. Am J Hum Genet 71: 327–336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brouha B, Schustak J, Badge RM, Lutz-Prigge S, Farley AH, Moran JV, Kazazian HH 2003. Hot L1s account for the bulk of retrotransposition in the human population. Proc Natl Acad Sci 100: 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns KH, Boeke JD 2012. Human Transposon Tectonics. Cell 149: 740–752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carneiro MO, Russ C, Ross MG, Gabriel SB, Nusbaum C, DePristo MA 2012. Pacific biosciences sequencing technology for genotyping and variation discovery in human data. BMC Genomics 13: 375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapman MA, Lawrence MS, Keats JJ, Cibulskis K, Sougnez C, Schinzel AC, Harview CL, Brunet J-P, Ahmann GJ, Adli M, et al. 2011. Initial genome sequencing and analysis of multiple myeloma. Nature 471: 467–472 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CL, Rappailles A, Duquenne L, Huvet M, Guilbaud G, Farinelli L, Audit B, d’Aubenton-Carafa Y, Arneodo A, Hyrien O, et al. 2010. Impact of replication timing on non-CpG and CpG substitution rates in mammalian genomes. Genome Res 20: 447–457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordaux R, Batzer MA 2009. The impact of retrotransposons on human genome evolution. Nat Rev Genet 10: 691–703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Golding A, Schlissel MS, Boeke JD 2001. Target DNA chromatinization modulates nicking by L1 endonuclease. Nucleic Acids Res 29: 573–577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cost GJ, Feng Q, Jacquier A, Boeke JD 2002. Human L1 element target-primed reverse transcription in vitro. EMBO J 21: 5899–5910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulak AM, Schumacher SE, van Lieshout J, Imamura Y, Fox C, Shim B, Ramos AH, Saksena G, Baca SC, Baselga J, et al. 2012. Gastrointestinal adenocarcinomas of the esophagus, stomach, and colon exhibit distinct patterns of genome instability and oncogenesis. Cancer Res 72: 4383–4393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH 2010. High-throughput sequencing reveals extensive variation in human-specific L1 content in individual human genomes. Genome Res 20: 1262–1270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing AD, Kazazian HH 2011. Whole-genome resequencing allows detection of many rare LINE-1 insertion alleles in humans. Genome Res 21: 985–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Q, Moran JV, Kazazian HH, Boeke JD 1996. Human L1 retrotransposon encodes a conserved endonuclease required for retrotransposition. Cell 87: 905–916 [DOI] [PubMed] [Google Scholar]
- Fungtammasan A, Walsh E, Chiaromonte F, Eckert KA, Makova KD 2012. A genome-wide analysis of common fragile sites: What features determine chromosomal instability in the human genome? Genome Res 22: 993–1005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, et al. 2000. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 92: 709–720 [DOI] [PubMed] [Google Scholar]
- Goodier JL, Ostertag EM, Kazazian HH 2000. Transduction of 3′-flanking sequences is common in L1 retrotransposition. Hum Mol Genet 9: 653–657 [DOI] [PubMed] [Google Scholar]
- Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q, et al. 2011. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29: 644–652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancks DC, Kazazian HH Jr 2012. Active human retrotransposons: variation and disease. Curr Opin Genet Dev 22: 191–203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helman E, Meyerson M 2011a. RetroSeq: A tool to discover somatic insertion of retrotransposons, Elena Helman, TCGA Scientific Symposium. http://cancergenome.nih.gov/newsevents/multimedialibrary/videos/retroseqhelman (Accessed June 4, 2013)
- Helman E, Meyerson M 2011b. Translation of the Cancer Genome Program. http://www.aacr.org/home/scientists/meetings–workshops/special-conferences/previous-special-conferences/2011—2012-special-conferences/translation-of-the-cancer-genome/program.aspx (Accessed June 7, 2013)
- Holmes SE, Dombroski BA, Krebs CM, Boehm CD, Kazazian HH 1994. A new retrotransposable human L1 element from the LRE2 locus on chromosome 1q produces a chimaeric insertion. Nat Genet 7: 143–148 [DOI] [PubMed] [Google Scholar]
- Hormozdiari F, Hajirasouliha I, Dao P, Hach F, Yorukoglu D, Alkan C, Eichler EE, Sahinalp SC 2010. Next-generation VariationHunter: combinatorial algorithms for transposon insertion discovery. Bioinformatics 26: i350–i357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang CRL, Schneider AM, Lu Y, Niranjan T, Shen P, Robinson MA, Steranka JP, Valle D, Civin CI, Wang T, et al. 2010. Mobile interspersed repeats are major structural variants in the human genome. Cell 141: 1171–1182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskow RC, McCabe MT, Mills RE, Torene S, Pittard WS, Neuwald AF, Van Meir EG, Vertino PM, Devine SE 2010. Natural mutagenesis of human genomes by endogenous retrotransposons. Cell 141: 1253–1261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jurka J 1997. Sequence patterns indicate an enzymatic involvement in integration of mammalian retroposons. Proc Natl Acad Sci 94: 1872–1877 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazazian HH 2004. Mobile elements: Drivers of genome evolution. Science 303: 1626–1632 [DOI] [PubMed] [Google Scholar]
- Keane TM, Wong K, Adams DJ 2013. RetroSeq: transposable element discovery from next-generation sequencing data. Bioinformatics 29: 389–390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimberland ML, Divoky V, Prchal J, Schwahn U, Berger W, Kazazian HH 1999. Full-length human L1 insertions retain the capacity for high frequency retrotransposition in cultured cells. Hum Mol Genet 8: 1557–1560 [DOI] [PubMed] [Google Scholar]
- Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF, Fulton LL, Dooling DJ, Ding L, Mardis ER, et al. 2012. Comprehensive molecular portraits of human breast tumours. Nature 490: 61–70 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. 2001. Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, et al. 2013. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499: 214–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebeau A, Grob TJ, Holst F, Seyedi-Fazlollahi N, Moch H, Terracciano L, Turzynski A, Choschzick M, Sauter G, Simon R 2008. Oestrogen receptor gene (ESR1) amplification is frequent in endometrial carcinoma and its precursor lesions. J Pathol 216: 151–157 [DOI] [PubMed] [Google Scholar]
- Lee E, Iskow R, Yang L, Gokcumen O, Haseley P, Luquette LJ, Lohr JG, Harris CC, Ding L, Wilson RK, et al. 2012. Landscape of somatic retrotransposition in human cancers. Science 337: 967–971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. 2009. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326: 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Eickbush TH 1995. RNA template requirements for target DNA-primed reverse transcription by the R2 retrotransposable element. Mol Cell Biol 15: 3882–3891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH 1993. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell 72: 595–605 [DOI] [PubMed] [Google Scholar]
- Meischl C, Boer M, Ahlin A, Roos D 2000. A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur J Hum Genet 8: 697–703 [DOI] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. 2012. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 41: D64–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerson M, Gabriel S, Getz G 2010. Advances in understanding cancer genomes through second-generation sequencing. Nat Rev Genet 11: 685–696 [DOI] [PubMed] [Google Scholar]
- Miki Y, Nishisho I, Horii A, Miyoshi Y, Utsunomiya J, Kinzler KW, Vogelstein B, Nakamura Y 1992. Disruption of the APC gene by a retrotransposal insertion of L1 sequence in a colon cancer. Cancer Res 52: 643–645 [PubMed] [Google Scholar]
- Moran JV, Holmes SE, Naas TP, DeBerardinis RJ, Boeke JD, Kazazian HH 1996. High frequency retrotransposition in cultured mammalian cells. Cell 87: 917–927 [DOI] [PubMed] [Google Scholar]
- Moran JV, DeBerardinis RJ, Kazazian HH 1999. Exon shuffling by L1 retrotransposition. Science 283: 1530–1534 [DOI] [PubMed] [Google Scholar]
- Morrish TA, Gilbert N, Myers JS, Vincent BJ, Stamato TD, Taccioli GE, Batzer MA, Moran JV 2002. DNA repair mediated by endonuclease-independent LINE-1 retrotransposition. Nat Genet 31: 159–165 [DOI] [PubMed] [Google Scholar]
- Morse B, Rotherg PG, South VJ, Spandorfer JM, Astrin SM 1988. Insertional mutagenesis of the myc locus by a LINE-1 sequence in a human breast carcinoma. Nature 333: 87–90 [DOI] [PubMed] [Google Scholar]
- Ostertag EM, Kazazian HH 2001. Biology of mammalian L1 retrotransposons. Annu Rev Genet 35: 501–538 [DOI] [PubMed] [Google Scholar]
- Penzkofer T 2004. L1Base: from functional annotation to prediction of active LINE-1 elements. Nucleic Acids Res 33: D498–D500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickeral OK, Makałowski W, Boguski MS, Boeke JD 2000. Frequent human genomic DNA transduction driven by LINE-1 retrotransposition. Genome Res 10: 411–415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodić N, Burns KH 2013. Long interspersed element–1 (LINE-1): passenger or driver in human neoplasms? PLoS Genet 9: e1003402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H 2000. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Schwahn U, Lenzner S, Dong J, Feil S, Hinzmann B, van Duijnhoven G, Kirschner R, Hemberger M, Bergen AA, Rosenberg T, et al. 1998. Positional cloning of the gene for X-linked retinitis pigmentosa 2. Nat Genet 19: 327–332 [DOI] [PubMed] [Google Scholar]
- Shukla R, Upton KR, Muñoz-Lopez M, Gerhardt DJ, Fisher ME, Nguyen T, Brennan PM, Baillie JK, Collino A, Ghisletti S, et al. 2013. Endogenous retrotransposition activates oncogenic pathways in hepatocellular carcinoma. Cell 153: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silva FPG, Morolli B, Storlazzi CT, Anelli L, Wessels H, Bezrookove V, Kluin-Nelemans HC, Giphart-Gassler M 2003. Identification of RUNX1/AML1 as a classical tumor suppressor gene. Oncogene 22: 538–547 [DOI] [PubMed] [Google Scholar]
- Solyom S, Ewing AD, Rahrmann EP, Doucet TT, Nelson HH, Burns MB, Harris RS, Sigmon DF, Casella A, Erlanger B, et al. 2012. Extensive somatic L1 retrotransposition in colorectal tumors. Genome Res 22: 2328–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart C, Kural D, Strömberg MP, Walker JA, Konkel MK, Stütz AM, Urban AE, Grubert F, Lam HYK, Lee W-P, et al. 2011. A comprehensive map of mobile element insertion polymorphisms in humans. PLoS Genet 7: e1002236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stratton MR, Campbell PJ, Futreal PA 2009. The cancer genome. Nature 458: 719–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szak ST, Pickeral OK, Makalowski W, Boguski MS, Landsman D, Boeke JD 2002. Molecular archeology of L1 insertions in the human genome. Genome Biol 3: research0052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van den Hurk JAJM, van de Pol DJR, Wissinger B, van Driel MA, Hoefsloot LH, de Wijs IJ, van den Born LI, Heckenlively JR, Brunner HG, Zrenner E, et al. 2003. Novel types of mutation in the choroideremia (CHM) gene: a full-length L1 insertion and an intronic mutation activating a cryptic exon. Hum Genet 113: 268–275 [DOI] [PubMed] [Google Scholar]
- Wang J, Song L, Grover D, Azrak S, Batzer MA, Liang P 2006. dbRIP: a highly integrated database of retrotransposon insertion polymorphisms in humans. Hum Mutat 27: 323–329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Witherspoon DJ, Xing J, Zhang Y, Watkins WS, Batzer MA, Jorde LB 2010. Mobile element scanning (ME-Scan) by targeted high-throughput sequencing. BMC Genomics 11: 410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing J, Zhang Y, Han K, Salem AH, Sen SK, Huff CD, Zhou Q, Kirkness EF, Levy S, Batzer MA, et al. 2009. Mobile elements create structural variation: analysis of a complete human genome. Genome Res 19: 1516–1526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S, Chen H, Zhao X, Cao J, Tong J, Lu J, Wu W, Shen H, Wei Q, Lu D 2013. REV3L 3′UTR 460 T>C polymorphism in microRNA target sites contributes to lung cancer susceptibility. Oncogene 32: 242–250 [DOI] [PubMed] [Google Scholar]
- Zingler N 2005. Analysis of 5′ junctions of human LINE-1 and Alu retrotransposons suggests an alternative model for 5′-end attachment requiring microhomology-mediated end-joining. Genome Res 15: 780–789 [DOI] [PMC free article] [PubMed] [Google Scholar]