Abstract
The complex relationship between ontogeny and phylogeny has been the subject of attention and controversy since von Baer’s formulations in the 19th century. The classic concept that embryogenesis progresses from clade general features to species-specific characters has often been revisited. It has become accepted that embryos from a clade show maximum morphological similarity at the so-called phylotypic period (i.e., during mid-embryogenesis). According to the hourglass model, body plan conservation would depend on constrained molecular mechanisms operating at this period. More recently, comparative transcriptomic analyses have provided conclusive evidence that such molecular constraints exist. Examining cis-regulatory architecture during the phylotypic period is essential to understand the evolutionary source of body plan stability. Here we compare transcriptomes and key epigenetic marks (H3K4me3 and H3K27ac) from medaka (Oryzias latipes) and zebrafish (Danio rerio), two distantly related teleosts separated by an evolutionary distance of 115–200 Myr. We show that comparison of transcriptome profiles correlates with anatomical similarities and heterochronies observed at the phylotypic stage. Through comparative epigenomics, we uncover a pool of conserved regulatory regions (≈700), which are active during the vertebrate phylotypic period in both species. Moreover, we show that their neighboring genes encode mainly transcription factors with fundamental roles in tissue specification. We postulate that these regulatory regions, active in both teleost genomes, represent key constrained nodes of the gene networks that sustain the vertebrate body plan.
Behind the broad anatomical diversity observed in vertebrate species rests a common body plan that is established early during embryogenesis and is shared by the entire clade. Central to our modern view of the ontogeny/phylogeny relationship is the concept that basic animal blueprints stand on the evolutionary conservation of key gene regulatory circuits that define tissue and organ identity during embryogenesis (Davidson and Erwin 2006; Carroll 2008). This notion can be traced back to von Baer’s formulations in the 19th century, proposing that embryo development progresses from the more general features of a clade to the specific characters of the species. Or, in other words, that within a particular group the early embryonic forms are more similar than the adults (Gould 1977). During the past few decades, it has become accepted that the window of development at which embryos of a clade show maximum morphological similarity is the phylotypic period (Slack et al. 1993), which does not correspond to the earliest stages of development but rather to mid-embryogenesis once the main body axis has been formed (i.e., pharyngula in vertebrates). However, whether this morphological invariance is also reflected by the conservation of molecular modules has been the subject of debate. According to the egg-timer/hourglass model, conservation of the body plan would depend on constrained molecular mechanisms operating at the phylotypic phase. Among the potential causative mechanisms postulated are the molecular logic imposed by Hox gene colinearity (Duboule 1994) and the low modularity, and therefore high interdependence, of developmental networks during the phylotypic period (Raff 1996; Galis and Metz 2001). Molecular studies in vertebrates based on the ontogenetic analysis of expression for essential genes, as well as protein–protein interactions and signaling pathways, have failed to identify a clear constrained signature during the phylotypic period, thus supporting a funnel-like model (Roux and Robinson-Rechavi 2008; Comte et al. 2010). However, systematic comparative transcriptomic analyses in vertebrates, Drosophila, Caenorhabditis, and even in plants have recently provided conclusive evidence for the existence of molecular constraints during mid-embryogenesis. These studies have reported both the convergence of interspecific gene expression and the prevalence of ancient genes at the phylotypic phase (Domazet-Loso and Tautz 2010; Kalinka et al. 2010; Irie and Kuratani 2011; Levin et al. 2012; Quint et al. 2012).
Examining the cis-regulatory logic is a fundamental step toward understanding the evolutionary sources of the observed developmental constraints imposed on animal body plans. Comparative chromatin immunoprecipitation-sequencing (ChIP-seq) and epigenomics studies have recently opened the possibility of uncovering conserved cis-regulatory modules during development (Schmidt et al. 2010; Woo and Li 2012; Xiao et al. 2012; Cotney et al. 2013). In this sense, recent work in zebrafish indicates that enhancers that become activated at late gastrula and remain active during mid-embryogenesis are evolutionarily more conserved than those activated earlier or later during development (Bogdanovic et al. 2012). The direct comparative analysis of functionally conserved enhancers in related species will now shed some light on the constrained architecture of the regulatory networks operating at the phylotypic phase (Nelson and Wardle 2013). To address this issue, we have compared transcriptomes and epigenetic marks from medaka (Oryzias latipes) and zebrafish (Danio rerio), two distantly related teleosts separated by an evolutionary distance of 115–200 Myr (Furutani-Seiki and Wittbrodt 2004). To complement transcriptomic and epigenomic data sets previously reported in zebrafish (Aday et al. 2011; Bogdanovic et al. 2012; Collins et al. 2012; Pauli et al. 2012; Choudhuri et al. 2013), we have generated RNA-seq and genomic tracks for key histone modifications (H3K4me3 and H3K27ac) from stage 24 (44 hpf) medaka embryos. This embryonic stage in medaka corresponds anatomically to 24-hpf embryos in zebrafish (early pharyngula), that is within the phylotypic period (Kimmel et al. 1995; Iwamatsu 2004). Our comparative analysis of fish transcriptomes shows that expression levels of tissue-specific genes correlate with anatomical similarities and heterochronies between medaka and zebrafish. Furthermore, comparative epigenomic analysis of putative active regulatory regions (PARRs) reveals that only 36% of them (4672 out of 12,938) are conserved at the sequence level between the analyzed teleosts. Among these conserved regions, only 14% (680 out of 4672), here termed shared putative active regulatory regions (SPARRs), are simultaneously active in both species during the phylotypic period. Interestingly, genes associated with this small set of co-acetylated regions show a broader and more complex regulatory landscape. In fact, this collection of genes is highly enriched in transcription factors and signaling molecules with key roles in the control of regulatory circuits involved in the specification of tissues and organs. We propose that SPARRs are evolutionarily constrained nodes that highlight core gene networks involved in the definition of the vertebrate body plan.
Results
Anatomical similarities and heterochronies between zebrafish and medaka phylotypic embryos
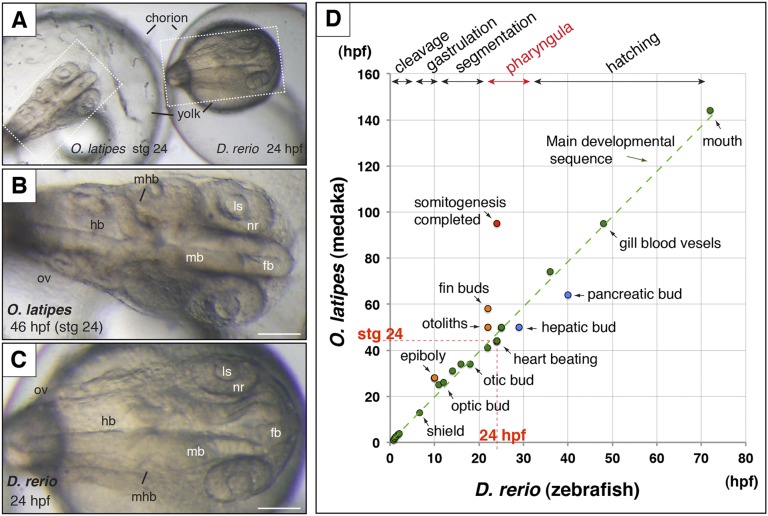
The ontogenetic analysis of the cumulative evolutionary age of the zebrafish transcriptome (i.e., age index) has revealed that the most ancient set of transcripts corresponds to the late segmentation to early pharyngula stages. The onset of heart beating and blood circulation at 24 hpf are two prominent morphological features that characterize this period of maximum evolutionary constraint (Domazet-Loso and Tautz 2010). Conveniently for our comparative study, RNA-seq and genomic tracks for H3K4me3 and H3K27ac were previously obtained for 24-hpf zebrafish embryos (Bogdanovic et al. 2012; Collins et al. 2012; Choudhuri et al. 2013). To determine which developmental stage in medaka shows the highest similarity to this zebrafish stage, we examined anatomical landmarks used as a reference for staging in both species (Kimmel et al. 1995; Iwamatsu 2004). These include, among others, the onset of heart beating and blood circulation, the formation of the optic cup and lens vesicles, or the formation of fin and hepatic buds (Supplemental Table S1). According to most of the features analyzed, medaka embryos show maximum anatomical similarity to 24-hpf zebrafish embryos at ∼44–48 h of development (stage 24) (Supplemental Fig. S1). Despite the relatively large evolutionary distance separating both teleost lineages (115–200 Myr), zebrafish and medaka embryos show a very similar body plan within this developmental window (Fig. 1). Therefore, medaka stage 24 and zebrafish 24 hpf were selected as equivalent reference stages in our comparative study.
Figure 1.
Comparative anatomy of stage 24 (44 hpf) medaka (O. latipes) and 24-hpf zebrafish (D. rerio) embryos during the phylotypic window (A). Note that both embryo size and general body plan are comparable between medaka (B) and zebrafish (C) at selected stages. The graph shows the onset of key anatomical landmarks plotted in hpf for zebrafish (x-axis) and medaka (y-axis) throughout development (D). The main developmental sequence is indicated as a green dotted line. Red and blue dots represent heterochronic structures between both species. See also Supplemental Table S1. Bar = 100 µm. (fb) Forebrain, (hb) hindbrain, (ls) lens, (mb) midbrain, (mhb) midbrain-hindbrain boundary, (nr) neural retina, (ov) otic vesicle.
The relative developmental timing of ontogenetic events is largely conserved between zebrafish and medaka during mid-embryogenesis. This is the case for the onset of heart beating, the development of the optic cup and lens vesicle, and the general morphology of the brain (Fig. 1A–C). In addition to the observed similarities, a few heterochronies (i.e., outliers from the main developmental sequence) were also evident (Fig. 1D; Supplemental Table S1). While somitogenesis has only progressed halfway through in medaka at this stage, it is already completed in zebrafish. Furthermore, in contrast to immobile medaka embryos, zebrafish show spontaneous contractions of the trunk and the tail at 24 hpf (Supplemental Fig. S1; Supplemental Movie 1; Kimmel et al. 1995). This is in agreement with previous observations showing that somitogenesis onset and completion, as well as somite number, vary considerably among vertebrate species (Richardson et al. 1998). A second prominent heterochrony was also noticeable for the formation of the fin buds, which happens much earlier in zebrafish (22 hpf) than in medaka embryos (stage 27) (Fig. 1D). Similarly to somitogenesis, fin bud formation has been described as a developmental process frequently uncoupled from the general zootype in vertebrate embryos (Bininda-Emonds et al. 2007; Sakamoto et al. 2009). Interestingly, we could detect only a couple of anatomical traits for which organogenesis progresses earlier in medaka than in zebrafish: the formation of the hepatic and pancreatic buds (Fig. 1D). This observation is consistent with previous descriptions of endoderm derivatives development in both species (Field et al. 2003; Watanabe et al. 2004).
Tissue-specific expression levels resemble anatomical similarities and heterochronies during the phylotypic period
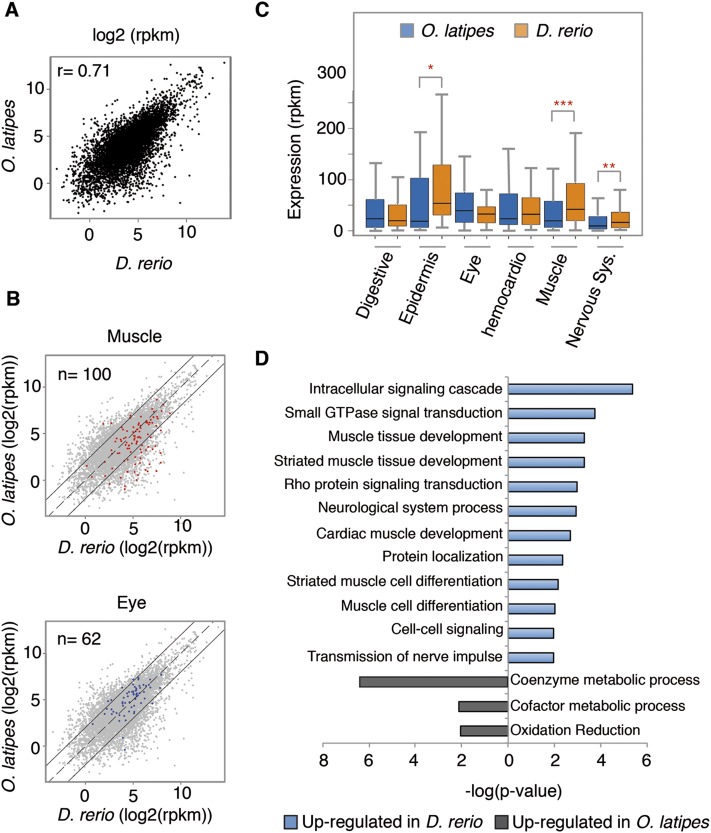
To examine whether the morphological similarities and asynchronies observed correlate with an underlying molecular activity, we performed RNA sequencing (RNA-seq) analyses in medaka at stage 24 (44 hpf) in duplicate and compared RNA levels with the previously published 24-hpf zebrafish transcriptomes (Collins et al. 2012; Choudhuri et al. 2013). The quality of the medaka RNA-seq data was confirmed by the high correlation of the biological replicates (Pearson correlation = 0.99). Although embryo staging is standardized within the zebrafish community (Kimmel et al. 1995), potential differences in the collection and processing of the embryos may be observed. However, the zebrafish 24-hpf data sets used in this study showed a high Pearson correlation coefficient (0.96), despite having been generated in two independent laboratories (Collins et al. 2012; Choudhuri et al. 2013). For interspecies comparisons, we analyzed the expression levels of a set of 9178 orthologous genes, excluding those with reduced RNA expression (counts per million reads [CPM] < 1) (Supplemental Table S2; see Methods). We found a relatively high correlation between the overall transcriptomes of the two species (Pearson correlation = 0.71) (Fig. 2A). This is in agreement with a previous study comparing vertebrate transcriptomes that shows highest correlation coefficients during the pharyngula window (Irie and Kuratani 2011). To compare gene profiles for different structures, a selection of tissue-specific genes was made based on the ZFIN expression database (Sprague et al. 2006). This list was filtered further through the 9178 orthologous list (Supplemental Table S2). First, we compared the expression levels of genes expressed in the eye, an organ for which no anatomical differences were observed between both species; and in the muscles, for which the differences were evident (Fig. 1). Consistent with the morphological data, we observed that 30% of the genes expressed in the muscles were up-regulated (i.e., more than fourfold) in zebrafish (Fig. 2B). On the contrary, 88.7% of the genes expressed in the eye did not show differential expression between the two organisms (Fig. 2B). We next extended the analysis to other tissues and quantified the significance of the observed changes in expression levels. As is shown in Figure 2C, in addition to the muscles, significant differences in RNA levels were also observed in nervous system–specific genes, which are higher in zebrafish than in medaka. This suggests a premature development of the nervous system in zebrafish that may be consistent with the formation of the neuromuscular junctions required for the active twitching of the tail musculature. With the exception of small but significant differences observed for genes expressed in the epidermis, no additional differences were observed for the rest of the tissue-specific genes examined (Fig. 2C; Supplemental Fig. S2).
Figure 2.
Comparison of expression levels between zebrafish and medaka at the phylotypic stage. (A) Correlation plot of zebrafish and medaka expression levels of the 7118 orthologs with expression higher than 1 count per million reads (CPM). The Pearson correlation coefficient (r) is indicated at the upper left corner. (B) Genes expressed in muscles (red) or the eye (blue), according to ZFIN annotations, are drawn over the total number of orthologous genes (gray). Each point represents the expression level of a given gene (log2 RPKM) in both species. Continuous black lines mark a fourfold change in expression. (C) Comparison of the expression levels in different tissues in the two species mentioned above. Bottom and top of boxes indicate 25th and 75th percentiles, respectively, and lines in the boxes indicate medians. Whiskers indicate the lowest and the highest data points within 1.5× interquartile range from the box. (*) P < 0.05, (**) P < 0.01, (***) P < 0.001 (Wilcoxon rank sum test). See also Supplemental Figure S2. (D) Gene ontology categories most significantly overrepresented in differentially expressed genes sorted by P-value. (Blue) Categories up-regulated in zebrafish; (gray) categories up-regulated in medaka.
To examine whether the divergent expression of tissue-specific genes was due to a delay or an advance in the timing of ontogenetic events, we included two other reference vertebrates, mouse (Mus musculus) and Xenopus (X. tropicalis) in our comparative transcriptomics analyses. Based on previous studies, we selected embryos within the pharyngula period for these two organisms (Irie and Kuratani 2011). For Xenopus, previously published data from stage 24–26 embryos were included in our study (Tan et al. 2013). For mouse, we performed a complete RNA sequencing analysis in duplicate using 10.5-d embryos (Pearson correlation between replicates = 0.75). As expected, pairwise correlation between these four vertebrates revealed that the general expression levels of orthologous genes are more similar in evolutionarily related species (Supplemental Fig. S2A). However, when we analyzed gene expression in specific tissues, we found more similarity when either zebrafish vs. Xenopus or medaka vs. mouse were compared (Supplemental Fig. S2C). In particular, transcriptional profiles indicated that specific tissues, such as the muscles and the nervous system, develop comparatively faster in zebrafish and Xenopus than in medaka and mouse. A possible explanation for this observation may be derived from species-specific ecological adaptations during embryogenesis. Zebrafish and Xenopus produce large clutches of eggs (100–300 and 1000–3000, respectively) and, most importantly, hatch as free-swimming larvae after a few days of development. In contrast, embryos are produced in smaller numbers (10–30 and 10–15, respectively) and develop at a slower pace in medaka and in the mouse (Supplemental Table S3). This suggests that, although anatomical similarities are maximal at the phylotypic stage, the developmental timing of individual tissues can be conditioned by adaptive requirements and ecological strategies.
To further analyze comparatively the transcriptome of medaka and zebrafish in an independent manner, we computed the number of differentially expressed genes using the edgeR package (Robinson et al. 2010). Selecting a false discovery rate threshold (FDR) < 5% and a fold change greater than fourfold (log2 FC > 2), we identified 1085 genes (15.2% of the orthologs list) with higher expression in zebrafish and 600 genes (8.4% of the orthologs) up-regulated in medaka (Supplemental Table S2). Interestingly, the functional categories (Biological process) obtained by DAVID gene ontology (GO) analysis (Huang da et al. 2009) of differentially expressed genes confirmed the up-regulation in zebrafish for muscle tissue development (P = 5.18 × 10−4) and neurological system process- (P = 1.17 × 10−3) related genes. Besides, we found differences in other biological processes not identified through direct morphological observation such as signaling cascade, cardiac muscle tissue development, and protein localization (Fig. 2D; Supplemental Table S2). In the case of medaka up-regulated transcripts, we found that genes related to the cofactor metabolic process (P = 7.51 × 10−3) and oxidation reduction (P = 4.15 × 10−7) were overrepresented with respect to zebrafish. In order to confirm our GO analyses, we decided to use PANTHER, a second bioinformatics tool that has been recently released (Mi et al. 2013). This second analysis corroborated our previous conclusions, for it showed a significant enrichment in GO terms linked to synaptic transmission and muscle development (e.g., neurological system process, synaptic transmission, mesoderm development, muscle organ development, and transmission of nerve impulse) in genes up-regulated in zebrafish (Supplemental Table S2). In the case of medaka up-regulated transcripts, we found less significantly enriched GO terms, and they were child terms linked to metabolic processes (e.g., lipid metabolic process, cellular amino acid metabolic process, carbohydrate metabolic process), as we observed previously in our DAVID analysis.
All together, these results indicate that the correlation level observed upon comparative analysis of tissue-specific genes resembles not only the anatomical similarities, but also the developmental heterochronies identified between both species.
Identification of conserved H3K27ac marks during the phylotypic period
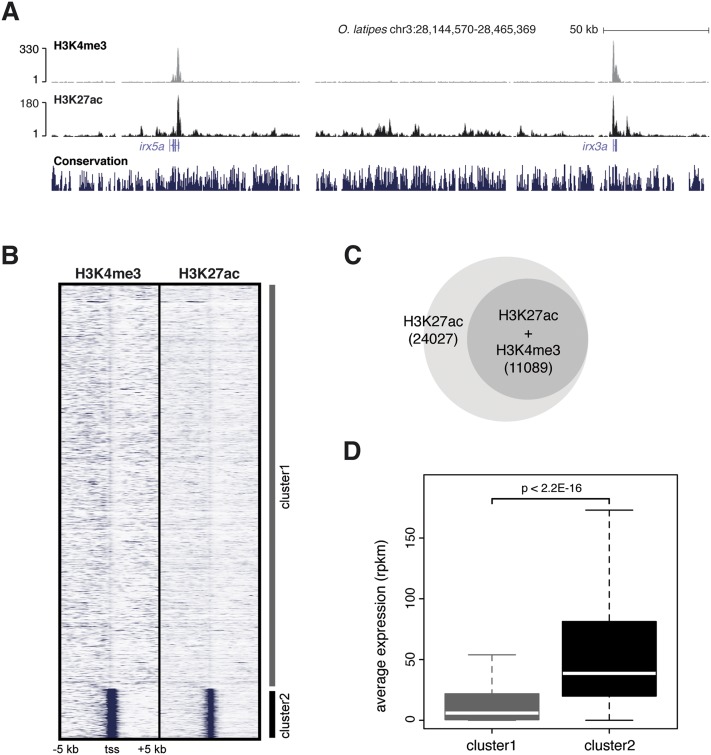
During embryogenesis, transcriptional control is achieved through the coordinated activation of cis-regulatory elements. In recent years, a number of epigenetic marks have been identified as molecular signatures of the activity-state of these regulatory elements (Ong and Corces 2012; Calo and Wysocka 2013). One of them, acetylation of lysine 27 on histone 3 (H3K27ac) has been shown to be a landmark of active transcriptional regulatory elements and promoters in different species (Wang et al. 2008; Heintzman et al. 2009; Creyghton et al. 2010; Rada-Iglesias et al. 2011; Bogdanovic et al. 2012). Although comparative analyses of these marks have been performed in a number of cell types, including stem cells (Goke et al. 2011; Woo and Li 2012; Xiao et al. 2012), no such comparisons have been carried out during embryogenesis in general, and at the phylotypic stage in particular. In order to address this point, first we set out to identify active transcriptional regulatory elements in medaka. To that end, we performed ChIP-seq experiments with specific antibodies against H3K4me3 (histone 3 lysine 4 trimethylation) and H3K27ac (Fig. 3A). The reads obtained from sequencing of immunoprecipitated DNA were aligned to the medaka genome (oryLat2 assembly, Ensembl) (Flicek et al. 2013). Then, we used the H3K4me3 mark to filter out promoters from putative active enhancers, both harboring the H3K27ac mark (Fig. 3A C; Ong and Corces 2012). Of 24,027 H3K27ac peaks obtained, we could identify 12,938 that did not overlap with H3K4me3 domains and therefore represent the subset of putative active regulatory regions (PARRs) at this stage (Fig. 3C). The remaining 11,089 H3K27ac peaks represent those regions occupying active promoters (Fig. 3C). As a validation of our data sets, we found that regions containing both H3K27ac and H3K4me3 marks are associated with transcriptionally active genes, as confirmed by the analysis of our medaka RNA-seq data (Fig. 3D).
Figure 3.
Characterization of epigenetic marks in stage 24 (44 hpf) medaka embryos. (A) UCSC Genome Browser view of H3K4me3 and H3K27ac tracks obtained from medaka ChIP-seq data. As previously described, both epigenomic marks cover the promoter regions of active genes. (B) K-means clustering (k = 2) of H3K4me3 and H3K27ac signals in ±5 kb around the TSS of all the genes annotated in the medaka genome. Cluster2 is enriched in both signals around promoters. (C) Venn diagram showing the fraction of H3K27ac regions overlapping with H3K4me3 regions (promoters). (D) Average expression (in RPKM) of genes grouped in clusters in B. Cluster2 genes show an average expression level higher than genes from Cluster1.
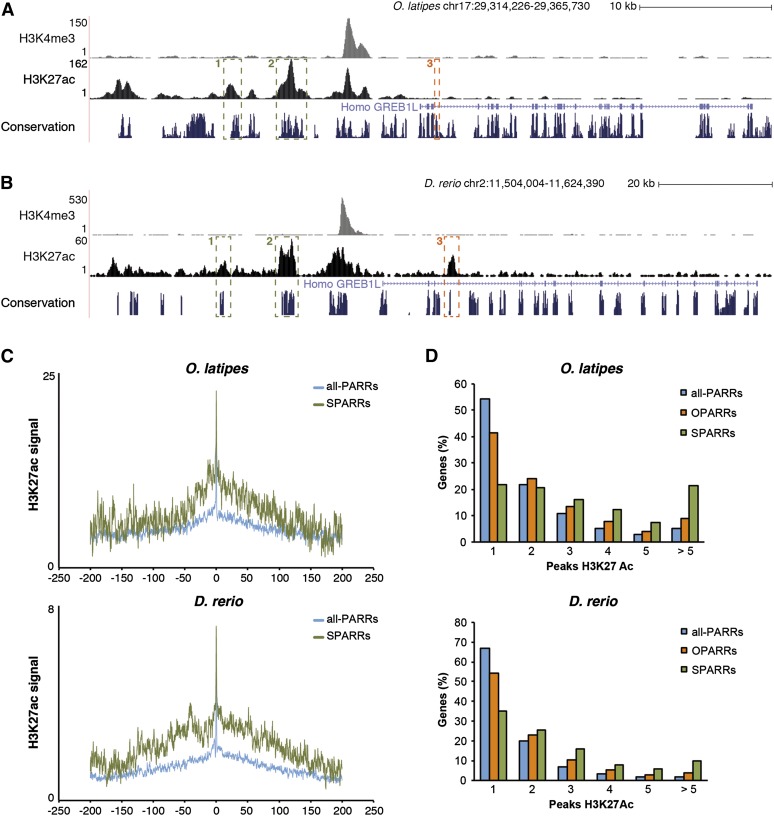
Once we identified the putative cis-regulatory elements at the phylotypic stage in medaka, we proceeded to analyze their evolutionary conservation, using zebrafish as a reference, a distantly related teleost species. To that end, published ChIP-seq data (Bogdanovic et al. 2012) were used to identify an equivalent set of 8892 PARRs in zebrafish (Supplemental Table S4). For our analyses, we compared these two data sets from medaka and zebrafish, together with a list of conserved regions between both species, as obtained from the UCSC Genome Browser (Meyer et al. 2013). Based on this information, we defined two kinds of conserved DNA domains: Only-one-species PARRs (OPARRs—peaks conserved but putatively active only in one of the two species) and Shared PARRs (SPARRs—peaks conserved and putatively active in both species) (Fig. 4A,B; see peaks validation by qPCR in Methods section). The medaka data set was compared against the zebrafish one, and vice versa. As a result, we obtained 3992 OPARRs and 680 SPARRs in medaka and 2032 OPARRs and 701 SPARRs in zebrafish (Supplemental Table S4). The small discrepancy observed among species in the number of SPARRs is due to both the presence of duplicated regions and the occasional incomplete overlap between SPARRs and conserved regions. To explore the functional significance of these results, we assigned the nearest gene to each PARR to further study their features. Independent of whether gene assignation was examined in medaka or zebrafish, we found that the genomic landscape of SPARR-associated genes had a much wider and higher H3K27ac mean profile than the average of all PARRs-associated genes (Fig. 4C). This might correspond to genes with a high number of cis-regulatory regions, many of them located far away from the promoter, which would result in a more complex transcriptional regulation. In fact, when we calculated the number of peaks associated with OPARRs, SPARRs, and all PARRs-related genes, we found that the proportion of genes including several H3K27ac peaks was significantly higher in the SPARRs subset (Fig. 4D). To minimize potential errors caused by inaccuracies in the assignment of SPARRs to neighbor genes (i.e., due both to local assembly mistakes and to chromosomal rearrangements), we refined our analysis by focusing in on SPARRs associated with the same gene in both species. This refined list of SPARRs, here named as cSPARRs, are associated with genes that showed an even higher number of H3K27ac regulatory regions than the original SPARR-associated genes (Supplemental Fig. S3A,B). Interestingly, a significant fraction of genes associated with SPARRs also include OPARRs in their vicinity, thus indicating that their regulation is more complex (Supplemental Fig. S3C). Taken together, these results indicate that genes with a more complex regulation are also those harboring conserved active enhancers.
Figure 4.
Analysis of the regulatory landscape of phylotypic genes associated with SPARRs and OPARRs. (A,B) Two examples of SPARR (1, 2) and one of OPARR (3) in the medaka genome (A) and their orthologous regions in zebrafish (B) are shown. (C) Average profiles of H3K27ac signal covering a 400-kb landscape for genes associated with all-PARRs (blue line) and SPARRs (green line) in both species: medaka and zebrafish (upper and lower panels, respectively). The average of reads in each bin of 200 bp is represented in log scale on the x-axis. The y-axis shows the position around the gene TSS, in kb. (D) Frequency distribution of orthologous genes (in percent) associated with either all H3K27ac peaks (all-PARRs), OPARRs, or SPARRs, according to the number of H3K27ac regulatory regions included in their vicinity; medaka and zebrafish (upper and lower panels, respectively).
To determine whether this conservation is restricted to teleosts or is also maintained in other vertebrates, we compared our data with that of the VISTA Enhancer Browser, a resource including experimentally validated human and mouse noncoding fragments with gene enhancer activity (Visel et al. 2007). In this project, 1857 noncoding human regions selected by means of phylogenetic foot-printing analyses and tissue-specific ChIP-seq assays of epigenomic marks have been tested in mouse transgenic assays. Of these sequences, 982 are able to drive consistent expression patterns and, therefore, are considered as active regulatory regions. Of the 12,938 PARRs found in medaka, 2157 are conserved with the human genome, and from them 115 overlap with regions analyzed in the VISTA Enhancer Browser collection. A high proportion of these conserved regions (82, 71.3%) were found active in mouse transgenic assays (Supplemental Table S5). When we compared the SPARRs (n = 680) from medaka, 253 were conserved in humans. Interestingly, an even higher percentage (88.6%) of the SPARRs were experimentally confirmed as active enhancers in transgenic mice (31 out of the 35 regions found in the VISTA Enhancer Browser database) (Supplemental Fig. S4). Similar results were obtained using the zebrafish data as a reference (Supplemental Table S5). This significantly higher percentage (hypergeometric test, P = 0.00095) of positive regulatory regions within the SPARRs suggests that regions putatively active in both teleost species are also active in other vertebrates as well. To test this hypothesis, we crossed the VISTA Enhancer Browser information of elements tested in transgenesis assays with H3K27ac tracks obtained from human ES cells differentiated into distinct cell types representing the basic embryonic cell layers (Xie et al. 2013). Approximately 2/3 (21 out of 31 in medaka and 18 out of 27 in zebrafish) of the regions that showed regulatory activity in mouse transgenesis assays were also acetylated in at least one human differentiated cell type. In contrast, most of the regions (three out of four in medaka and five out of five in zebrafish) that were negative in transgenesis assays were also negative for the acetylation mark in differentiated human ES cells (Supplemental Table S5).
To further validate the functional conservation of SPARRs across vertebrates, we carried out transient transgenesis assays in zebrafish by injecting the corresponding fish regions (n = 6) orthologous to the tested mammalian enhancers (VISTA Enhancer Browser). Interestingly, the six regions tested (hs73, hs619, hs625, hs969, hs1315, and hs1327) directed the expression of the reporter GFP in a similar manner (i.e., to the same tissues) as the homologous regions in mice (Supplemental Fig. S5). Moreover, we tested three of these regions (hs73, hs1315, and hs1327) in transient transgenesis assays in medaka, obtaining very similar results (Supplemental Fig. S5). These results further confirmed that regions active in both teleost species are also functionally conserved in other vertebrates.
Conserved transcriptional control of genes associated with shared regulatory regions
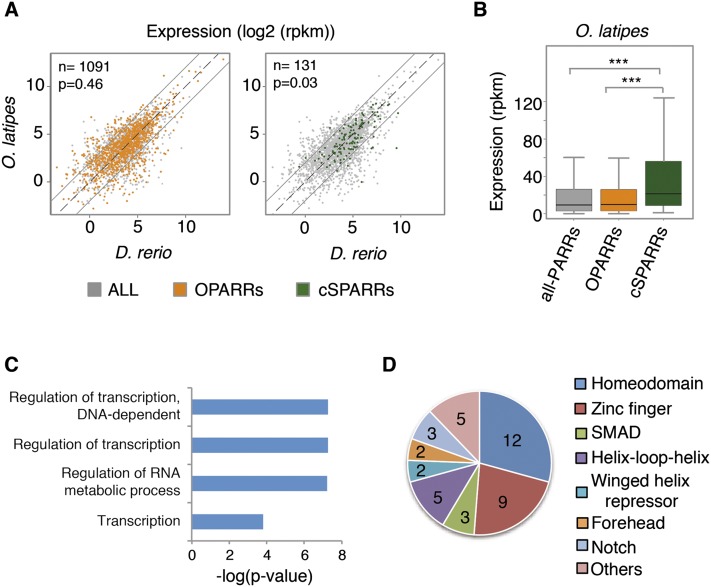
To integrate the information we obtained from the analysis of chromatin epigenetic marks with our gene expression data, we examined the expression levels of genes associated with OPARR and cSPARRs regions. Expression analysis of medaka genes in the vicinity of H3K27ac regions showed that whereas OPARRs-associated genes display very variable expression levels between species, cSPARR-associated genes were significantly enriched in nondifferentially expressed genes (P = 0.46 and P = 0.03 for OPARR and cSPARRs, respectively; hypergeometric test) (Fig. 5A). Similar results were derived from the analysis of zebrafish genes associated with H3K27ac regions (data not shown). Moreover, we observed that the median expression level of cSPARR-associated genes was significantly higher than the expression average of both genes containing OPARRs and the overall transcriptome (Fig. 5B). These results indicate that the expression control of genes associated with shared regulatory regions is significantly conserved through evolution.
Figure 5.
Integration of genome-wide epigenetic and expression data. (A) Comparison of zebrafish and medaka expression levels (log2 RPKM) for genes associated with OPARR (orange) and cSPARR (green) regions, as identified in medaka. These genes are plotted over the total number of genes associated with H3K27ac regions identified in medaka (gray). The number of genes included in each group is indicated in the upper left corner. Only genes with CPM > 1 are plotted. Note that genes associated with cSPARRs are significantly enriched in nondifferentially expressed transcripts (P = 0.46 and P = 0.03 for OPARRs and cSPARRs, respectively; hypergeometric test). (B) Box plot indicating the average expression levels (RPKM) of genes associated with cSPARR, OPARR, and all H3K27ac regions identified in medaka. (***) P < 0.001 (Wilcoxon rank sum test). (C) Gene ontology analysis of the total number of genes associated with cSPARRs. Significant biological process categories (P < 0.05) revealed that genes associated with cSPARRs are involved mainly in transcriptional regulation. (D) Representative InterPro domains present in transcription factors identified within the total collection of genes associated with cSPARRs. Numbers inside the chart indicate the number of genes present in each category.
A general DAVID analysis of GO term enrichment in the general list of genes associated with PARRs both in zebrafish and medaka reflected the transcriptionally active state of a broad set of genes related to diverse developmental processes. A number of GO terms involved in tissue patterning (e.g., regionalization, P = 6.08 × 10−7; or pattern specification process, P = 1.17 × 10−6), cellular and epithelial morphogenesis (e.g., tissue morphogenesis, P = 5.04 × 10−8; or cell motion, P = 3.24 × 10−6), or precursor differentiation (e.g., neuron differentiation, P = 2.13 × 10−5) were derived from these analyses (Supplemental Table S6). In contrast, when GO terms were analyzed only for the list of cSPARR-associated genes, all the significantly enriched terms were related to transcriptional categories such as regulation of transcription: P = 5.65 × 10−8; regulation of RNA metabolism process: P = 6.04 × 10−8; or transcription: P = 1.57 × 10−4 (Fig. 5C; Supplemental Table S6). Moreover, the enrichment analysis of InterPro protein domains related to these cSPARR-associated genes showed the overrepresentation of important transcriptional domains for developmental processes, such as the homeodomain (P = 7.45 × 10−5), zinc finger C2H2 (P = 8.47 × 10−4), SMAD domain (P = 5.10 × 10−3), or winged helix repressor DNA-binding (P = 9.48 × 10−3) (Supplemental Table S6). A detailed analysis of the occurrence of the InterPro domains present in the transcription factors identified within the collection of 145 cSPARR-associated genes is shown in Figure 5D.
Taken together, these results indicate that not only are developmental genes conserved at the vertebrate phylotypic stage (Domazet-Loso and Tautz 2010) but the key regulatory regions responsible for their tight and complex modulation are also conserved. Our data suggest that the shared regulatory elements identified in our study constitute essential nodes of the constrained transcriptional network operating at the phylotypic stage.
Discussion
In this work, we compared zebrafish and medaka pharyngula embryos morphologically and molecularly. We have examined both their transcriptomes and predictive epigenetic marks for conserved active enhancers during the phylotypic window. Whereas in closely related vertebrates, the high overall genome similarity masks the identification of noncoding conserved elements, only a few of them can be identified outside the vertebrate group and even less show enough transphyletic conservation to be tracked beyond the Cambrian horizon (McEwen et al. 2009; Royo et al. 2011; Clarke et al. 2012). The evolutionary distance between zebrafish and medaka (115–200 Myr) is suitable for the identification and analysis of conserved regulatory elements in vertebrates (Furutani-Seiki and Wittbrodt 2004).
Despite their evolutionary distance, zebrafish and medaka share a very similar anatomy, which is particularly noticeable when embryos are compared at the phylotypic stage. Nevertheless, a number of heterochronies are observed during this developmental window (here described in Supplemental Table S1). In fact, the observation of such conspicuous heterochronies between vertebrate phylotypic embryos has been an argument raised against the hourglass model (Richardson et al. 1998). In agreement with the hourglass hypothesis, comparative transcriptomics in vertebrates have revealed that interspecies correlation in gene expression levels is maximal within this phylotypic window (Irie and Kuratani 2011). Our comparative analysis of tissue-specific genes shows that there is a high concordance of expression levels in synchronously developing tissues, and thus is in line with previous comparative transcriptomic analyses (Domazet-Loso and Tautz 2010; Irie and Kuratani 2011). In addition, our work shows that this concordance drops when gene expression is compared for heterochronic structures (e.g., muscles and nervous system). This observation fits under the umbrella of the general notion that changes in gene regulatory networks (GRNs) play a prevalent role in the evolution of animal form (Davidson 2006; Carroll 2008).
The objective of this study is not to provide additional evidence showing molecular constraints at the phylotypic period; this has been sufficiently addressed by others. We rather aim to have a first look at the nature of such constraints. Cis-regulatory modules (CRMs) have been considered not only the units of input information in GRNs but also the fundamental units of evolutionary change (Davidson 2006). In this report, we have performed a comparative epigenomics study to identify a subset of ∼700 putative CRMs (here termed SPARRs) that are both conserved and active in zebrafish and medaka pharyngula embryos. Here we have associated each CRM to the nearest gene. Provided that enhancers for a particular gene could even lie in a neighbor gene intron (Lettice et al. 2003; Smemo et al. 2014), this assumption may lead to potential errors. However, assignment by nearest gene model is the most widely used method, and it has been shown that patterns of enhancer activity correlate strongly with patterns of nearest-gene expression (Ernst et al. 2011; Shen et al. 2012). Our analysis of molecular domains for genes associated with SPARRs reveals that a large proportion of them provide regulatory input to genes encoding transcription factors. This finding suggests that these regulatory regions represent constrained nodes from essential GRNs operating at the phylotypic period in the teleost group. Furthermore, it is likely that the core set of nodes responsible for the evolutionary stability of the vertebrate body plan is, to a large extent, comprised within these regions conserved in teleosts. In agreement with this, a large proportion (88%) of the human SPARRs homologs included in the tested (i.e., in transgenesis assays) collection of CRMs at the VISTA Enhancer Browser behave as tissue-specific active enhancers.
There are a number of reasons to think that the collection of 700 SPARRs identified here represents an underestimate of the actual number of core CRMs responsible for the architecture of the vertebrate phylotype. First, in our analysis we have considered only regulatory regions conserved between the two teleosts, which roughly correspond to a third of the acetylated regions (PARRs) identified in each species. This approach, however, may have excluded a number of elements that still share similar functional logic (i.e., a similar composition of transcription factor binding sites) but whose overall sequence conservation is beyond the detection limits of conventional alignment tools (Fisher et al. 2006; He et al. 2011; Taher et al. 2011). In addition, comparative ChIP-seq studies have also pointed to the existence of pervasive species-specific gene regulation in a number of tissues, including ES cells (The ENCODE Project Consortium 2007; Schmidt et al. 2010; The ENCODE Project Consortium 2012). To what extent this also applies to complex CRMs regulating master developmental genes is currently unclear. Finally, the intrinsic technical limitations imposed by ChIP-seq approaches applied to whole embryos might result in false negatives and hence in an underestimate of the total number of co-acetylated regions in teleost genomes. This may partially explain why a large proportion of the conserved acetylated regions identified in our study appear to be active only in one species at the phylotypic period (OPARRs). Although we show that, collectively, gene regulatory features associated with SPARRs and OPARRs are significantly different, we cannot rule out the possibility that a fraction of the regions classified here as OPARRs is in fact active below the detection level in one of the species (i.e., due to different regulatory weight). Alternatively, the differential and complex activation timing of these regions in the teleost genomes could also account for the observed prevalence of OPARRs versus SPARRs during the narrow developmental window under study.
Among vertebrate regulatory sequences, evolutionary divergence has been proposed to occur faster in fish genomes. The partitioning of regulatory elements between duplicate gene loci after fish-specific whole-genome duplication (FSGD) has been suggested as a causative mechanism driving their divergence and hence the extensive adaptive radiation observed in teleosts (Taylor et al. 2001; Christoffels et al. 2004; Hoegg et al. 2004; Meyer and Van de Peer 2005). Thus, it has been shown that more than twice as many noncoding elements are conserved between elephant shark and human genomes than between teleost fish and human genomes (Venkatesh et al. 2006). Moreover, comparative genomics studies have shown that conserved noncoding elements have been evolving rapidly in teleost fishes (Wang et al. 2009; Lee et al. 2011). Comparative analyses of epigenetic marks in other vertebrates including tetrapods, cartilaginous fish, and agnates will complement our study and help to define more precisely the ancestral set of CRMs in vertebrates. The analysis of these marks in basal ray-finned fish that diverged from teleosts before the FSGD, such as the spotted gar (Lepisosteous oculatus) (Amores et al. 2011), may be also important to determine the degree of regulatory divergence in the teleost group. However, even if our comparative analysis in teleosts overlooks a fraction of the ancestral set of vertebrate CRMs, our approach would be biased toward the identification of “essential nodes,” precisely those enhancers more resilient to evolutionary change due to their central role in the definition of the vertebrate body plan.
It has been shown that although epigenomic conservation does not always correlate with genomic sequence conservation, it can provide an additional layer of information that is necessary to interpret genome regulation (Xiao et al. 2012). Hence, the collection of shared enhancers identified here represents a powerful resource to investigate the architecture of the GRNs operating during the phylotypic window. It has been postulated that the developmental programs controlling different organ primordia may be interdependent in a way that cannot be resolved into individual modules. This lack of modularity may have functioned as an evolutionary constraint to stabilize the vertebrate body plan (Raff 1996). Some of the data presented here are in line with this hypothesis. A large proportion of the SPARR-associated genes encode transcription factors and components of signaling pathways that, in turn, may act as upstream regulators of other conserved nodes of the GRNs. In addition, SPARR-associated genes show a complex regulatory profile, often including multiple CRMs, which suggests that they represent “hub” genes with high connectivity within the GRNs. In fact, an important proportion (between 44%–53%) of the SPARR-associated genes also include in their neighborhood conserved regions that are acetylated only in one of the two species (here termed OPARRs). Whether these putative enhancers act as “shadow enhancers” providing functional robustness to a “primary” enhancer (Hong et al. 2008; Frankel et al. 2010) or, alternatively, bring independent regulatory input needs to be determined. Furthermore, detailed analyses of predicted connectivity focused in the nodes of phylotypic and nonphylotypic GRNs will be required to formally prove Raff’s lack-of-modularity hypothesis.
Methods
Fish stocks and genomes
Medaka (O. latipes) and zebrafish (D. rerio) wild-type strains were kept as closed stocks, and embryos were staged as previously described (Kimmel et al. 1995; Iwamatsu 2004). The genome assemblies for medaka and zebrafish genomes have been released (Flicek et al. 2013). The zebrafish genome (Zv9) has a size of 1505 Mb, and 26,206 protein-coding genes have been annotated (Collins et al. 2012; Howe et al. 2013). The medaka genome (HdrR-2005) has a size of 700 Mb, and a total of 20,141 coding genes have been predicted (Kasahara et al. 2007).
Embryo collection and RNA samples
Whole medaka and mouse embryos (without extra-embryonic membranes) were collected according to standard procedures. All the animal experiments were carried out in accordance with the guidelines of our Institutional Animal Ethics Committee. For medaka experiments, a total of 60 embryos were suspended in TRIzol reagent (Intron Biotechnology) with chloroform. Two replicates for each sample were used for RNA-seq analyses. Total RNA was isolated from the aqueous phase, purified by isopropanol precipitation, and cleaned using the RNeasy MinElute Cleanup kit (Qiagen). For mouse samples, four day 10.5 embryos were pooled and homogenized, and total RNA was extracted similarly. Two replicates for each sample were also used for mouse RNA-seq analyses. Subsequent processes, including preparation of sequencing libraries, were performed by standard TruSeqTM RNA sample preparation (Illumina) with the following changes: Purifications were carried out using Qiagen clean-up columns, and e-gels were used for size selection. Samples were sequenced using HiSeq 2000 at the EMBL Genomics Core Facility (EMBL-Heidelberg).
Criteria for orthologous genes identification
Basic Local Alignment Search Tool (BLAST) searching (E-value < 1 × 10−20) was applied to the nonredundant proteome of each organism downloaded from the EMBL Ensembl website (http://www.ensembl.org/). Pairs of genes with reciprocal best BLAST hits (RBBHs) were defined to be orthologs.
RNA-seq data processing
Raw RNA sequence data from medaka and mouse, and previously published Xenopus (Tan et al. 2013) and zebrafish (Collins et al. 2012; Choudhuri et al. 2013) data, were aligned with the oryLat2 (October 2005), mm9 (July 2007), xenTro2 (August 2005), and danRer7 (July 2010) genome versions, respectively, using TopHat (Trapnell et al. 2009). To minimize errors due to the variability between species annotations, we took into account only the number of mapped reads that overlapped with the Ensembl coding sequences of those genes present in our orthologs list. Expression values were obtained by calculating the sum of all the expression hits from distinct exons annotated to a single locus using RSeQC software (Wang et al. 2012). To filter low-expressed genes, loci with counts per million reads (CPM) <1 in at least two samples were discarded. For differential expression analyses, raw count data were processed using the edgeR package under default conditions (Robinson and Smyth 2008), and genes with FDR < 5% and fold change > 4 were considered significant. For analyses of expression levels in different tissues, data were normalized by scaling read counts to reads per kilobase per million reads (RPKM), followed by quantile normalization to reduce variability between samples. Data were log2 transformed and the mean of the replicates was used in further analyses. Genes expressed in specific tissues were obtained from Ensembl filtered by expression in ZFIN (Sprague et al. 2006) anatomical system data: “digestive,” “epidermis,” “eye,” “hemocardio,” “muscle,” and “nervous,” and filtered to obtain genes that only are expressed in one of the tissues (Supplemental Table S2).
Gene ontology analyses
Gene ontology analyses were performed using DAVID (Huang da et al. 2009) and PANTHER (Mi et al. 2013). Only the “Biological Process” tree was used in the study. As the medaka genome was not represented in DAVID, only zebrafish Ensembl gene names were used for the analysis. For GO analyses of differentially expressed genes, we used as a reference background the list of orthologous genes with CPM > 1 (Supplemental Table S2). We considered significant GO categories with a P-value < 0.05 and more than seven genes. For GO analyses of ChIP-seq data, the complete list of orthologous genes was used as background. GO categories with a P-value < 0.05 were considered significant. P-values were corrected by multiple testing.
Medaka ChIP-seq
Chromatin immunoprecipitation (ChIP) was performed following a protocol reported for zebrafish (Bogdanovic et al. 2013) with minor modifications. Per ChIP, we used 600 dechorionated embryos at stage 24. Samples were sonicated using the Diagenode Bioruptor device with the following cycling conditions: 12 min high–30 sec on, 30 sec off; 12 min on ice; 12 min high–30 sec on, 30 sec off. The size of sonicated DNA was in the range of 100–500 bp. The anti-H3K4me3 (pAB-033-050) antibody was obtained from Diagenode. The anti-H3K27ac (ab4729) antibody was purchased from Abcam. Immunoprecipitated DNA was purified with QIAquick columns (Qiagen). DNA ends were repaired and the adaptors ligated. The size-selected (300 bp) library was then amplified in a PCR reaction and sequenced using the Genome Analyzer (Illumina). The sequenced reads were mapped to the reference medaka genome (oryLat2 assembly) with Bowtie software (Langmead et al. 2009). Peak callings were performed with MACS (Zhang et al. 2008) using default parameters. Peaks were independently validated by qPCR, using specific primers for 12 regions, four of them acetylated in medaka but not in zebrafish, four acetylated in zebrafish but not in medaka, and four acetylated in both species (Supplemental Fig. S6). To further test the reproducibility of the ChIP-seq experiment for H3K27ac marks, a second biological replicate was analyzed. Reads from both replicates (grouped in windows of 1 kb over the genome) show a Pearson correlation coefficient of 0.97 (Supplemental Fig. S7).
Comparison between zebrafish and medaka ChIP-seq data
In order to compare acetylation peaks obtained from ChIP-seq analyses in both species, chained and netted alignments (axt format) between danRer7 and oryLat2 assemblies were downloaded from the UCSC Genome Browser downloads web page (http://hgdownload.cse.ucsc.edu/downloads.html). Minus-strand coordinates were transformed to plus-strand coordinates. To obtain epigenomic marks corresponding to enhancers, only those H3K27-acetylated peaks that do not overlap with an H3K4-trimethylated peak were considered for each species. This subtraction was performed using BEDTools software (Quinlan and Hall 2010). Putative active regulatory regions identified in one species were crossed with the axt alignment file to map the orthologous positions in the second species. The resulting list was then compared with the mapped PARRs in the second species. A fraction of the PARRs in each species overlapped with conserved regions from the UCSC Genome Browser. For zebrafish PARRs, this overlapping was determined as 86.5% of the total length of the conserved region, on average. For medaka, the mean overlap was 80.5% of the conserved region.
Integration of ChIP-seq and RNA-seq data
RNA-seq profiles were integrated with ChIP-seq data by assigning each acetylated region to its nearest gene using BEDTools. The expression levels of genes associated with PARRs were obtained from our medaka RNA-seq or from the reported zebrafish data sets (Collins et al. 2012; Choudhuri et al. 2013)
Data access
The ChIP-seq and RNA-seq data included in this work have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under the following accession numbers: GSE46351 (medaka stage 24 ChIP-seq tracks), GSE46484 (medaka stage 24 RNA-seq tracks), and GSE47033 (mouse E10.5 RNA-seq tracks).
Acknowledgments
We thank Gert-Jan Veenstra and Simon van Heeringen for their critical input, Rocío Polvillo and María Nicolás-Pérez for their excellent technical help, and Iwanka Kozarewa and Lina Chen for their help on the mouse RNA-seq. The Andalusian government (JA) supported A.F-.M. as scientific manager of the Aquatic Vertebrates Platform at CABD. J.W.C. was supported by a studentship from The Institute of Cancer Research. Spanish and Andalusian government grants BFU2010-14839, CSD2007-00008, and P08-CVI-3488 to J.L.G-.S.; and BFU2011-22916 and P11-CVI-7256 to J.R.M-.M. supported this work.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.163915.113.
References
- Aday AW, Zhu LJ, Lakshmanan A, Wang J, Lawson ND 2011. Identification of cis regulatory features in the embryonic zebrafish genome through large-scale profiling of H3K4me1 and H3K4me3 binding sites. Dev Biol 357: 450–462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amores A, Catchen J, Ferrara A, Fontenot Q, Postlethwait JH 2011. Genome evolution and meiotic maps by massively parallel DNA sequencing: spotted gar, an outgroup for the teleost genome duplication. Genetics 188: 799–808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bininda-Emonds OR, Jeffery JE, Sanchez-Villagra MR, Hanken J, Colbert M, Pieau C, Selwood L, Ten Cate C, Raynaud A, Osabutey CK, et al. 2007. Forelimb-hindlimb developmental timing changes across tetrapod phylogeny. BMC Evol Biol 7: 182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Hidalgo C, van Kruysbergen I, van Heeringen SJ, Veenstra GJ, Gomez-Skarmeta JL 2012. Dynamics of enhancer chromatin signatures mark the transition from pluripotency to cell specification during embryogenesis. Genome Res 22: 2043–2053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanovic O, Fernandez-Minan A, Tena JJ, de la Calle-Mustienes E, Gomez-Skarmeta JL 2013. The developmental epigenomics toolbox: ChIP-seq and MethylCap-seq profiling of early zebrafish embryos. Methods 62: 207–215 [DOI] [PubMed] [Google Scholar]
- Calo E, Wysocka J 2013. Modification of enhancer chromatin: what, how, and why? Mol Cell 49: 825–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB 2008. Evo-devo and an expanding evolutionary synthesis: a genetic theory of morphological evolution. Cell 134: 25–36 [DOI] [PubMed] [Google Scholar]
- Choudhuri A, Maitra U, Evans T 2013. Translation initiation factor eIF3h targets specific transcripts to polysomes during embryogenesis. Proc Natl Acad Sci 110: 9818–9823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christoffels A, Koh EG, Chia JM, Brenner S, Aparicio S, Venkatesh B 2004. Fugu genome analysis provides evidence for a whole-genome duplication early during the evolution of ray-finned fishes. Mol Biol Evol 21: 1146–1151 [DOI] [PubMed] [Google Scholar]
- Clarke SL, VanderMeer JE, Wenger AM, Schaar BT, Ahituv N, Bejerano G 2012. Human developmental enhancers conserved between deuterostomes and protostomes. PLoS Genet 8: e1002852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins JE, White S, Searle SM, Stemple DL 2012. Incorporating RNA-seq data into the zebrafish Ensembl genebuild. Genome Res 22: 2067–2078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comte A, Roux J, Robinson-Rechavi M 2010. Molecular signaling in zebrafish development and the vertebrate phylotypic period. Evol Dev 12: 144–156 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cotney J, Leng J, Yin J, Reilly SK, Demare LE, Emera D, Ayoub AE, Rakic P, Noonan JP 2013. The evolution of lineage-specific regulatory activities in the human embryonic limb. Cell 154: 185–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creyghton MP, Cheng AW, Welstead GG, Kooistra T, Carey BW, Steine EJ, Hanna J, Lodato MA, Frampton GM, Sharp PA, et al. 2010. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci 107: 21931–21936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH. 2006. The regulatory genome: gene regulatory networks in development and evolution. Academic Press, Amsterdam, Netherlands. [Google Scholar]
- Davidson EH, Erwin DH 2006. Gene regulatory networks and the evolution of animal body plans. Science 311: 796–800 [DOI] [PubMed] [Google Scholar]
- Domazet-Loso T, Tautz D 2010. A phylogenetically based transcriptome age index mirrors ontogenetic divergence patterns. Nature 468: 815–818 [DOI] [PubMed] [Google Scholar]
- Duboule D 1994. Temporal colinearity and the phylotypic progression: a basis for the stability of a vertebrate Bauplan and the evolution of morphologies through heterochrony. Dev Suppl 1994: 135–142 [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernst J, Kheradpour P, Mikkelsen TS, Shoresh N, Ward LD, Epstein CB, Zhang X, Wang L, Issner R, Coyne M, et al. 2011. Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473: 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field HA, Ober EA, Roeser T, Stainier DY 2003. Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev Biol 253: 279–290 [DOI] [PubMed] [Google Scholar]
- Fisher S, Grice EA, Vinton RM, Bessling SL, McCallion AS 2006. Conservation of RET regulatory function from human to zebrafish without sequence similarity. Science 312: 276–279 [DOI] [PubMed] [Google Scholar]
- Flicek P, Ahmed I, Amode MR, Barrell D, Beal K, Brent S, Carvalho-Silva D, Clapham P, Coates G, Fairley S, et al. 2013. Ensembl 2013. Nucleic Acids Res 41: D48–D55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel N, Davis GK, Vargas D, Wang S, Payre F, Stern DL 2010. Phenotypic robustness conferred by apparently redundant transcriptional enhancers. Nature 466: 490–493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furutani-Seiki M, Wittbrodt J 2004. Medaka and zebrafish, an evolutionary twin study. Mech Dev 121: 629–637 [DOI] [PubMed] [Google Scholar]
- Galis F, Metz JA 2001. Testing the vulnerability of the phylotypic stage: on modularity and evolutionary conservation. J Exp Zool 291: 195–204 [DOI] [PubMed] [Google Scholar]
- Goke J, Jung M, Behrens S, Chavez L, O’Keeffe S, Timmermann B, Lehrach H, Adjaye J, Vingron M 2011. Combinatorial binding in human and mouse embryonic stem cells identifies conserved enhancers active in early embryonic development. PLoS Comput Biol 7: e1002304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gould SJ. 1977. Ontogeny and phylogeny. Belknap Press of Harvard University Press, Cambridge, MA. [Google Scholar]
- He Q, Bardet AF, Patton B, Purvis J, Johnston J, Paulson A, Gogol M, Stark A, Zeitlinger J 2011. High conservation of transcription factor binding and evidence for combinatorial regulation across six Drosophila species. Nat Genet 43: 414–420 [DOI] [PubMed] [Google Scholar]
- Heintzman ND, Hon GC, Hawkins RD, Kheradpour P, Stark A, Harp LF, Ye Z, Lee LK, Stuart RK, Ching CW, et al. 2009. Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459: 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoegg S, Brinkmann H, Taylor JS, Meyer A 2004. Phylogenetic timing of the fish-specific genome duplication correlates with the diversification of teleost fish. J Mol Evol 59: 190–203 [DOI] [PubMed] [Google Scholar]
- Hong JW, Hendrix DA, Levine MS 2008. Shadow enhancers as a source of evolutionary novelty. Science 321: 1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe K, Clark MD, Torroja CF, Torrance J, Berthelot C, Muffato M, Collins JE, Humphray S, McLaren K, Matthews L, et al. 2013. The zebrafish reference genome sequence and its relationship to the human genome. Nature 496: 498–503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA 2009. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4: 44–57 [DOI] [PubMed] [Google Scholar]
- Irie N, Kuratani S 2011. Comparative transcriptome analysis reveals vertebrate phylotypic period during organogenesis. Nat Commun 2: 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwamatsu T 2004. Stages of normal development in the medaka Oryzias latipes. Mech Dev 121: 605–618 [DOI] [PubMed] [Google Scholar]
- Kalinka AT, Varga KM, Gerrard DT, Preibisch S, Corcoran DL, Jarrells J, Ohler U, Bergman CM, Tomancak P 2010. Gene expression divergence recapitulates the developmental hourglass model. Nature 468: 811–814 [DOI] [PubMed] [Google Scholar]
- Kasahara M, Naruse K, Sasaki S, Nakatani Y, Qu W, Ahsan B, Yamada T, Nagayasu Y, Doi K, Kasai Y, et al. 2007. The medaka draft genome and insights into vertebrate genome evolution. Nature 447: 714–719 [DOI] [PubMed] [Google Scholar]
- Kimmel CB, Ballard WW, Kimmel SR, Ullmann B, Schilling TF 1995. Stages of embryonic development of the zebrafish. Dev Dyn 203: 253–310 [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL 2009. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10: R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee AP, Kerk SY, Tan YY, Brenner S, Venkatesh B 2011. Ancient vertebrate conserved noncoding elements have been evolving rapidly in teleost fishes. Mol Biol Evol 28: 1205–1215 [DOI] [PubMed] [Google Scholar]
- Lettice LA, Heaney SJ, Purdie LA, Li L, de Beer P, Oostra BA, Goode D, Elgar G, Hill RE, de Graaff E 2003. A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum Mol Genet 12: 1725–1735 [DOI] [PubMed] [Google Scholar]
- Levin M, Hashimshony T, Wagner F, Yanai I 2012. Developmental milestones punctuate gene expression in the Caenorhabditis embryo. Dev Cell 22: 1101–1108 [DOI] [PubMed] [Google Scholar]
- McEwen GK, Goode DK, Parker HJ, Woolfe A, Callaway H, Elgar G 2009. Early evolution of conserved regulatory sequences associated with development in vertebrates. PLoS Genet 5: e1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer A, Van de Peer Y 2005. From 2R to 3R: evidence for a fish-specific genome duplication (FSGD). BioEssays 27: 937–945 [DOI] [PubMed] [Google Scholar]
- Meyer LR, Zweig AS, Hinrichs AS, Karolchik D, Kuhn RM, Wong M, Sloan CA, Rosenbloom KR, Roe G, Rhead B, et al. 2013. The UCSC Genome Browser database: extensions and updates 2013. Nucleic Acids Res 41: D64–D69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi H, Muruganujan A, Casagrande JT, Thomas PD 2013. Large-scale gene function analysis with the PANTHER classification system. Nat Protoc 8: 1551–1566 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson AC, Wardle FC 2013. Conserved non-coding elements and cis regulation: actions speak louder than words. Development 140: 1385–1395 [DOI] [PubMed] [Google Scholar]
- Ong CT, Corces VG 2012. Enhancers: emerging roles in cell fate specification. EMBO Rep 13: 423–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pauli A, Valen E, Lin MF, Garber M, Vastenhouw NL, Levin JZ, Fan L, Sandelin A, Rinn JL, Regev A, et al. 2012. Systematic identification of long noncoding RNAs expressed during zebrafish embryogenesis. Genome Res 22: 577–591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quint M, Drost HG, Gabel A, Ullrich KK, Bonn M, Grosse I 2012. A transcriptomic hourglass in plant embryogenesis. Nature 490: 98–101 [DOI] [PubMed] [Google Scholar]
- Rada-Iglesias A, Bajpai R, Swigut T, Brugmann SA, Flynn RA, Wysocka J 2011. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470: 279–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raff RA. 1996. The shape of life: genes, development, and the evolution of animal form. University of Chicago Press, Chicago, IL. [Google Scholar]
- Richardson MK, Allen SP, Wright GM, Raynaud A, Hanken J 1998. Somite number and vertebrate evolution. Development 125: 151–160 [DOI] [PubMed] [Google Scholar]
- Robinson MD, Smyth GK 2008. Small-sample estimation of negative binomial dispersion, with applications to SAGE data. Biostatistics 9: 321–332 [DOI] [PubMed] [Google Scholar]
- Robinson MD, McCarthy DJ, Smyth GK 2010. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26: 139–140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roux J, Robinson-Rechavi M 2008. Developmental constraints on vertebrate genome evolution. PLoS Genet 4: e1000311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo JL, Maeso I, Irimia M, Gao F, Peter IS, Lopes CS, D'Aniello S, Casares F, Davidson EH, Garcia-Fernández J, Gómez-Skarmeta JL 2011. Transphyletic conservation of developmental regulatory state in animal evolution. Proc Natl Acad Sci 108: 14186–14191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto K, Onimaru K, Munakata K, Suda N, Tamura M, Ochi H, Tanaka M 2009. Heterochronic shift in Hox-mediated activation of sonic hedgehog leads to morphological changes during fin development. PLoS ONE 4: e5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt D, Wilson MD, Ballester B, Schwalie PC, Brown GD, Marshall A, Kutter C, Watt S, Martinez-Jimenez CP, Mackay S, et al. 2010. Five-vertebrate ChIP-seq reveals the evolutionary dynamics of transcription factor binding. Science 328: 1036–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y, Yue F, McCleary DF, Ye Z, Edsall L, Kuan S, Wagner U, Dixon J, Lee L, Lobanenkov VV, et al. 2012. A map of the cis-regulatory sequences in the mouse genome. Nature 488: 116–120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slack JM, Holland PW, Graham CF 1993. The zootype and the phylotypic stage. Nature 361: 490–492 [DOI] [PubMed] [Google Scholar]
- Smemo S, Tena JJ, Kim KH, Gamazon ER, Sakabe NJ, Gomez-Marin C, Aneas I, Credidio FL, Sobreira DR, Wasserman NF, et al. 2014. Obesity-associated variants within FTO form long-range functional connections with IRX3. Nature 507: 371–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprague J, Bayraktaroglu L, Clements D, Conlin T, Fashena D, Frazer K, Haendel M, Howe DG, Mani P, Ramachandran S, et al. 2006. The Zebrafish Information Network: the zebrafish model organism database. Nucleic Acids Res 34: D581–D585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taher L, McGaughey DM, Maragh S, Aneas I, Bessling SL, Miller W, Nobrega MA, McCallion AS, Ovcharenko I 2011. Genome-wide identification of conserved regulatory function in diverged sequences. Genome Res 21: 1139–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan MH, Au KF, Yablonovitch AL, Wills AE, Chuang J, Baker JC, Wong WH, Li JB 2013. RNA sequencing reveals a diverse and dynamic repertoire of the Xenopus tropicalis transcriptome over development. Genome Res 23: 201–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor JS, Van de Peer Y, Braasch I, Meyer A 2001. Comparative genomics provides evidence for an ancient genome duplication event in fish. Philos Trans R Soc Lond B Biol Sci 356: 1661–1679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL 2009. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25: 1105–1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh B, Kirkness EF, Loh YH, Halpern AL, Lee AP, Johnson J, Dandona N, Viswanathan LD, Tay A, Venter JC, et al. 2006. Ancient noncoding elements conserved in the human genome. Science 314: 1892. [DOI] [PubMed] [Google Scholar]
- Visel A, Minovitsky S, Dubchak I, Pennacchio LA 2007. VISTA Enhancer Browser–a database of tissue-specific human enhancers. Nucleic Acids Res 35: D88–D92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Zang C, Rosenfeld JA, Schones DE, Barski A, Cuddapah S, Cui K, Roh TY, Peng W, Zhang MQ, et al. 2008. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40: 897–903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Lee AP, Kodzius R, Brenner S, Venkatesh B 2009. Large number of ultraconserved elements were already present in the jawed vertebrate ancestor. Mol Biol Evol 26: 487–490 [DOI] [PubMed] [Google Scholar]
- Wang L, Wang S, Li W 2012. RSeQC: quality control of RNA-seq experiments. Bioinformatics 28: 2184–2185 [DOI] [PubMed] [Google Scholar]
- Watanabe T, Asaka S, Kitagawa D, Saito K, Kurashige R, Sasado T, Morinaga C, Suwa H, Niwa K, Henrich T, et al. 2004. Mutations affecting liver development and function in Medaka, Oryzias latipes, screened by multiple criteria. Mech Dev 121: 791–802 [DOI] [PubMed] [Google Scholar]
- Woo YH, Li WH 2012. Evolutionary conservation of histone modifications in mammals. Mol Biol Evol 29: 1757–1767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao S, Xie D, Cao X, Yu P, Xing X, Chen CC, Musselman M, Xie M, West FD, Lewin HA, et al. 2012. Comparative epigenomic annotation of regulatory DNA. Cell 149: 1381–1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie W, Schultz MD, Lister R, Hou Z, Rajagopal N, Ray P, Whitaker JW, Tian S, Hawkins RD, Leung D, et al. 2013. Epigenomic analysis of multilineage differentiation of human embryonic stem cells. Cell 153: 1134–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W, et al. 2008. Model-based analysis of ChIP-Seq (MACS). Genome Biol 9: R137. [DOI] [PMC free article] [PubMed] [Google Scholar]