Abstract
Most vertebrate promoters lie in unmethylated CpG-dense islands, whereas methylation of the more sparsely distributed CpGs in the remainder of the genome is thought to contribute to transcriptional repression. Nonmethylated CG dinucleotides are recognized by CXXC finger protein 1 (CXXC1, also known as CFP1), which recruits SETD1A (also known as Set1) methyltransferase for trimethylation of histone H3 lysine 4, an active promoter mark. Genomic regions enriched for CpGs are thought to be either absent or irrelevant in invertebrates that lack DNA methylation, such as C. elegans; however, a CXXC1 ortholog (CFP-1) is present. Here we demonstrate that C. elegans CFP-1 targets promoters with high CpG density, and these promoters are marked by high levels of H3K4me3. Furthermore, as for mammalian promoters, high CpG content is associated with nucleosome depletion irrespective of transcriptional activity. We further show that highly occupied target (HOT) regions identified by the binding of a large number of transcription factors are CpG-rich promoters in C. elegans and human genomes, suggesting that the unusually high factor association at HOT regions may be a consequence of CpG-linked chromatin accessibility. Our results indicate that nonmethylated CpG-dense sequence is a conserved genomic signal that promotes an open chromatin state, targeting by a CXXC1 ortholog, and H3K4me3 modification in both C. elegans and human genomes.
Transcription is regulated through functional elements in the genome such as promoters and enhancers. Identifying these genomic elements, many of which are recognized by transcription factors (TFs), is a necessary first step in their functional analysis. Toward this goal, the modENCODE and ENCODE Projects have used chromatin immunoprecipitation (ChIP) to map the binding sites for a large number of TFs in C. elegans, Drosophila, and humans (The ENCODE Project Consortium 2007, 2012; Gerstein et al. 2010, 2012; The modENCODE Project Consortium 2010; Nègre et al. 2011; Niu et al. 2011; AP Boyle, CL Araya, C Brdlik, P Cayting, C Cheng, Y Cheng, K Gardner, L Hillier, J Janette, L Jiang, et al., in prep.).
Most of the identified TF-binding regions are “low occupancy,” showing binding by one or a few different TFs. As expected of enhancers, low-occupancy sites are enriched for DNA sequence motifs recognized by the bound factors, suggesting direct DNA binding (Gerstein et al. 2010; The modENCODE Project Consortium 2010). In contrast, these mapping studies also identified an unusual class of sites bound by the majority of mapped TFs, termed HOT (Highly Occupied Target) regions. The large number of factors associated with HOT regions is incompatible with simultaneous occupancy, and the regions usually lack sequence-specific binding motifs of the associated factors, indicating that they are unlikely to be classical enhancers (Gerstein et al. 2010; The modENCODE Project Consortium 2010). Consistent with this idea, Drosophila HOT regions are depleted for annotated enhancers; however, a significant fraction displays enhancer activity in transgenic analyses (Kvon et al. 2012). Drosophila and human HOT regions have features of open chromatin, such as nucleosome depletion and high turnover, and in C. elegans they are usually located near genes with ubiquitous expression (Gerstein et al. 2010; The modENCODE Project Consortium 2010; Yip et al. 2012). These previous reports suggest that HOT regions are active genomic elements but fail to provide a clear picture of their function, possibly because of differences in the definition and characterization of these regions.
Other DNA sequence features are also known to play regulatory roles. For example, a hallmark of many mammalian promoters is enrichment for nonmethylated CpG dinucleotides defined as a “CpG island (CGI)” (Bird et al. 1985; Bird 1986; Gardiner-Garden and Frommer 1987; Illingworth and Bird 2009; Deaton and Bird 2011). In contrast to these promoter regions, CpG dinucleotides are sparsely distributed and cytosine-methylated in most other regions in the genome, and differential DNA methylation is thought to be important for the recognition and function of CGIs (Jones et al. 1998; Nan et al. 1998; Cameron et al. 1999; Klose and Bird 2006; Joulie et al. 2010). Promoter-enriched CpGs have not been observed in invertebrates and are believed to be absent in organisms lacking DNA methylation. Indeed, it is widely considered that the enrichment of CpGs at promoters is a vertebrate-specific phenomenon that requires DNA methylation (Duncan and Miller 1980; Gardiner-Garden and Frommer 1987; Cooper and Krawczak 1989; Ehrlich et al. 1990; Antequera and Bird 1999; Caiafa and Zampieri 2005; Illingworth and Bird 2009; Turner et al. 2010; Deaton and Bird 2011). In mammals, nonmethylated CpGs are associated with active promoter chromatin features and bound by CXXC1 (CFP1), which is part of a SETD1A complex that catalyzes methylation of H3K4 (Lee and Skalnik 2005; Ansari et al. 2008; Butler et al. 2008; Tate et al. 2010; Thomson et al. 2010; Xu et al. 2011). Although C. elegans lacks DNA methylation, the C. elegans genome encodes a CXXC1 ortholog, CFP-1, required for global H3K4 trimethylation (Simonet et al. 2007; Li and Kelly 2011), suggesting a possible similarity of function.
Here we directly compared HOT regions in C. elegans and humans to investigate their functions and ask if these are conserved. We discovered that HOT regions are active promoters enriched for CpG dinucleotides in both organisms. We further show that high CpG density is a feature of many C. elegans promoters, and such promoters are targeted by CFP-1. Our results support a conserved function for promoter-dense CpGs in C. elegans and human genomes.
Results
We collected modENCODE and ENCODE TF mapping data sets for 90 C. elegans factors (across developmental stages) and 159 human factors (from multiple cell types) and determined regions of factor overlap (Supplemental Fig. S1; Methods; The ENCODE Project Consortium 2007, 2012; Gerstein et al. 2010, 2012; The modENCODE Project Consortium 2010; Nègre et al. 2011; Niu et al. 2011; AP Boyle, CL Araya, C Brdlik, P Cayting, C Cheng, Y Cheng, K Gardner, L Hillier, J Janette, L Jiang, et al., in prep). Within each region of TF overlap, we further defined a minimal core region with the highest factor occupancy. This identified 35,062 C. elegans regions bound by 1–87 factors and 737,151 human regions bound by 1–138 factors.
HOT regions are promoters
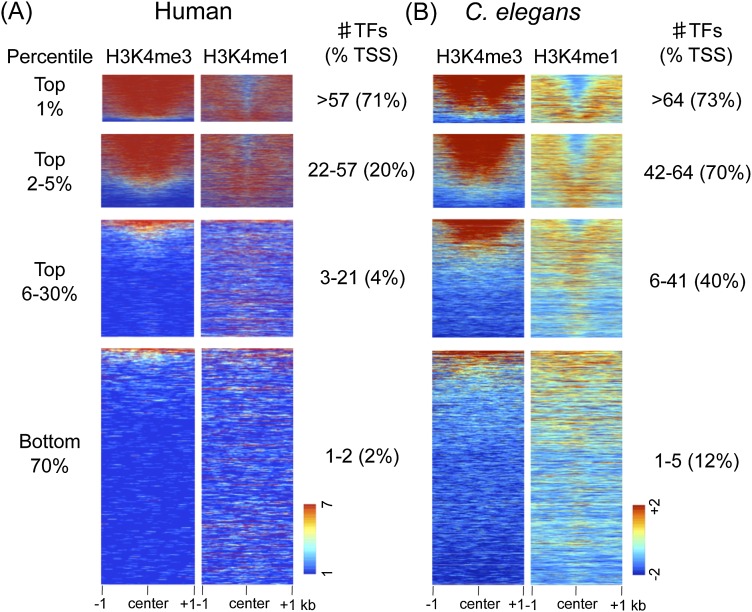
We first examined chromatin modifications at TF binding regions. We ranked TF binding regions by factor occupancy (high to low) and plotted H3K4me3 and H3K4me1 as a heat map. As expected, in both organisms we find that the chromatin of low-occupancy regions has features typical of enhancers (H3K4me3low/H3K4me1high) (Fig. 1). In contrast, as the number of factors increases, the chromatin signature becomes more promoter-like (H3K4me3high/H3K4me1low). The majority of TF binding regions in the top 5% of occupancy have a promoter-like signature in both organisms, suggesting that they are located at promoters. Consistent with this idea, high-occupancy TF binding regions more often overlap with proximal promoter regions compared to low-occupancy regions (Fig. 1).
Figure 1.
HOT regions display promoter features. H3K4me3 and H3K4me1 signals plotted at the centers of core TF overlap regions ranked by the indicated percentile of TF occupancy in humans and C. elegans. (A) For human TF overlap regions, 7419 (top 1%), 30, 945 (top 2%–5%), a random selection of 100,000 of 241,975 (top 6%–30%), and 456,812 (bottom 70%) regions are plotted. (B) For C. elegans TF overlap regions, 376 (top 1%), 1429 (top 2%–5%), 9721 (top 6%–30%), and 23,536 (bottom 70%) regions are plotted. In each TF overlap band, the number of TFs and the percentage of TF core midpoints ±500 bp of a TSS are indicated. Scales show linear (human) or log2 (C. elegans) input normalized signal ranges.
To increase the power of finding similarities between HOT regions within and between organisms, we analyzed HOT regions in the top 1% of occupancy (more than 64 factors in C. elegans and more than 57 factors in humans) since these display the strongest enrichment for promoter-like chromatin features in both organisms. For comparison, we define “COLD” low-occupancy regions as those with single-factor binding (representing 33.6% of regions in C. elegans and 30.8% in humans).
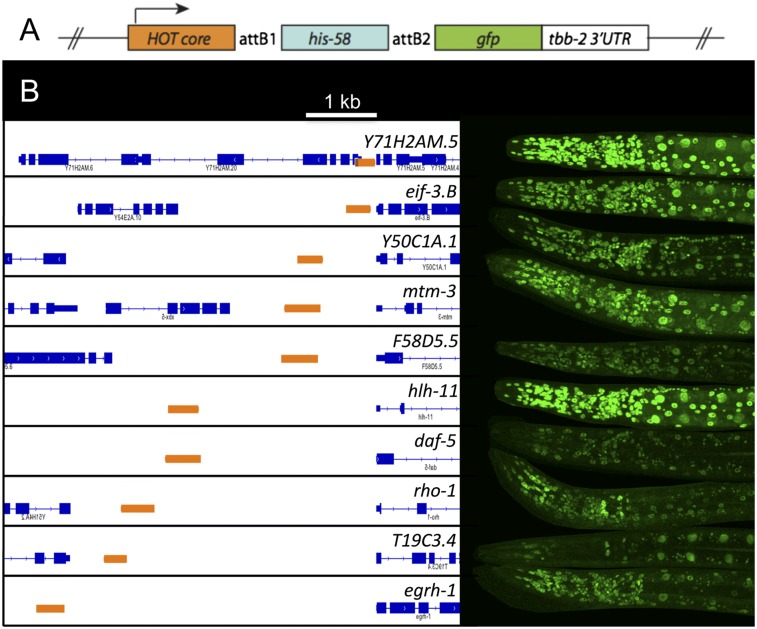
The preceding findings suggest that HOT regions may be active promoters. In C. elegans, the annotated transcript start is often the start of the mature transcript, not the start of transcription, because the primary 5′ end is removed and degraded following trans-splicing (Bektesh and Hirsh 1988; Blumenthal 1995, 2012; Allen et al. 2011). To directly investigate whether core HOT regions are functional promoters, we chose 10 C. elegans core HOT regions of 241–525 bp located at various distances upstream of the nearest annotated transcript start (150 bp to 4.7 kb upstream) and cloned each one directly upstream of a GFP reporter gene (Fig. 2A). All 10 tested HOT regions drove GFP, with most showing widespread expression (Fig. 2B; Supplemental Table S1), indicating that the short tested regions are widely active promoters.
Figure 2.
C. elegans HOT regions are functional promoters. (A) The indicated HOT regions (orange) were cloned directly upstream of a histone∷GFP fusion gene and examined for the expression of GFP using transgenic reporter assays. (B) Ten of ten assayed regions drove GFP expression. The expression in representative larvae stage 4 worms is shown. Coordinates of cloned regions and further information on expression patterns are shown in Supplemental Table S1.
We also found that genes proximal to HOT regions are usually widely expressed. In C. elegans, 78% of genes associated with HOT regions are expressed in all tissues assayed using gene expression profiling (Spencer et al. 2011). Similarly, most (91%) human genes associated with HOT regions are expressed in all examined cell types in gene expression analyses from the ENCODE Project (see Methods). Taken together, these findings support the view that that most C. elegans and human HOT regions are ubiquitously active core promoters.
CpG dinucleotides are enriched in C. elegans and human HOT regions
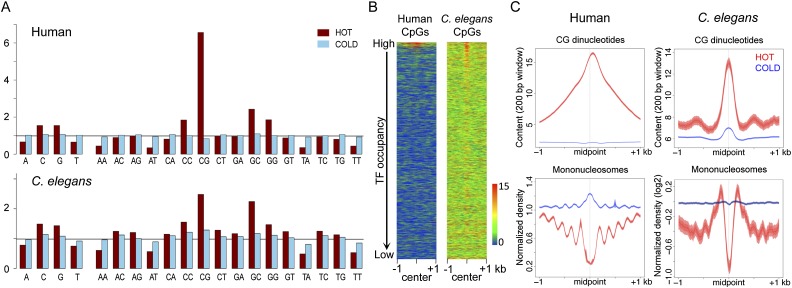
We next searched for common sequence characteristics of C. elegans and human HOT regions by analyzing their composition of mono- and dinucleotides. In both organisms, HOT regions have high GC content (Fig. 3A). Remarkably, C. elegans and human HOT regions show similar patterns of dinucleotide enrichment and depletion, with CG dinucleotides showing the highest enrichment in both organisms (Fig. 3A). To further investigate the distribution of CG dinucleotides at HOT and COLD regions, we plotted CpG density across these regions, which showed prominent peaks around the midpoints of HOT regions (Fig. 3B,C, upper panel). In contrast, COLD regions do not display these patterns (Fig. 3B,C, upper panel). In addition, we find that HOT regions but not COLD regions show strong nucleosome depletion (Fig. 3C, bottom panel).
Figure 3.
HOT regions are enriched for CpG dinucleotides and depleted for nucleosomes. (A) Frequency of the indicated mono- and dinucleotides in HOT (red) and COLD (blue) regions is shown relative to the genome-wide frequency scaled to one (black horizontal line). (B) Heat maps showing the distribution of CpG density in ranked TF overlap regions in human and C. elegans. The color scheme shows the scale (0 to 15) for CpG content in a 200-bp window. (C) The distribution of CpG density (top) and nucleosomes (bottom) was plotted for HOT (red) and COLD (blue) regions in the human and C. elegans genomes. Lines show mean signal, darker filled areas show standard error, and lighter filled areas are 95% confidence intervals. All plots show 2-kb regions centered at the midpoint of core regions.
The finding that HOT regions are at core promoters raised the possibility that CpG-dense sequences may be a shared promoter feature in C. elegans and human genomes. As described above, CpG enrichment is a known property of the majority of human promoters (Gardiner-Garden and Frommer 1987; Illingworth and Bird 2009). In these “CpG islands,” the cytosines of CpGs are typically nonmethylated; whereas CpGs are broadly depleted from most regions of the human genome, and nonpromoter CpGs are usually cytosine methylated. Since DNA methylation differences are thought to underlie the recognition and function of CpGs as well as the mutation of cytosine that drives depletion of bulk CpGs, it was unexpected to observe enrichment of CpGs at promoters in the C. elegans genome, where DNA methylation and CpG depletion are both absent.
The preceding analyses only examined CpG enrichment at narrowly defined HOT regions. To ask whether CpG dinucleotides might be more widely found at C. elegans promoters, we plotted CpG density and observed/expected CpG content (normalizing for local GC content) across protein-coding promoters and found clear CpG enrichment just upstream of the transcription start site (TSS) in many C. elegans promoters, with a narrower distribution in C. elegans than in human (Supplemental Figs. S2, S3; heat map in Fig. 5D, see below). In both humans and C. elegans, genes with CpG-rich promoters more frequently show ubiquitous expression compared to those with CpG-poor promoters (in human 68.6% of high CpG and 2.3% of low CpG; in C. elegans 56.3% of high CpG and 21.8% of low CpG; see Methods) (Schug et al. 2005; Ramskold et al. 2009). Taken together, these findings suggest that CpG dinucleotides are a feature of widely active C. elegans promoters, as they are in vertebrates.
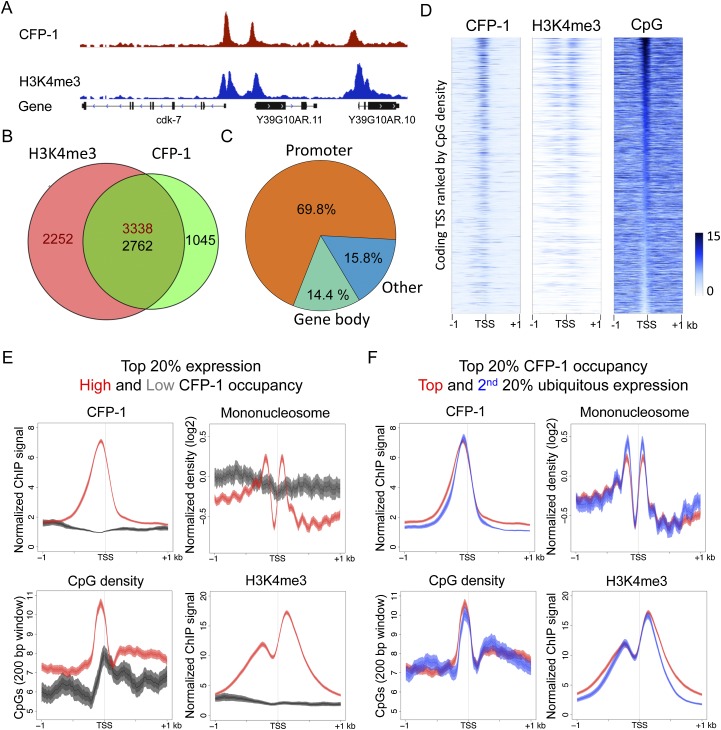
Figure 5.
C. elegans CFP-1 is targeted to CpG-rich promoters marked by H3K4me3. (A) A representative screen shot for the distribution of CFP-1 (red) and H3K4me3 (blue) normalized ChIP signals in late embryos. (B) Venn diagram showing the overlap between CFP-1 and H3K4me3 ChIP ChIP-seq peaks. Numbers in the overlap region are not equal because single peaks in one data set may overlap more than one peak in the other data set. (C) Percentage of CFP-1 peaks overlapping promoter regions (orange; ±500 bp of coding TSSs) (Chen et al. 2013; Kruesi et al. 2013), gene bodies (green), and the remaining genomic regions (blue). (D) Signal distributions for CFP-1, H3K4me3, and CpG density plotted in 2-kb windows centered by coding TSSs identified in Chen et al. (2013) in heat maps ranked by the density of promoter CpGs. The color scheme shows the scale (0–15) for the indicated signal. (E) Plots of CFP-1 ChIP signal, CpG content, mononucleosome pattern, and H3K4me3 signal across promoters of active ubiquitous genes in the top 20% of expression with high (red, top 20%), or low (dark gray, bottom 20%) CFP-1 occupancy. (F) Plots of CFP-1 ChIP signal, CpG content, mononucleosome pattern, and H3K4me3 ChIP signal across ubiquitous promoters highly targeted by CFP-1 (top 20%) and with high (red, top 20%) or low (blue, second 20%) expression levels. All plots display the indicated features in 2-kb windows centered at TSSs. Lines show mean signal, darker filled areas show standard error, and lighter filled areas are 95% confidence intervals.
Promoter-dense CpG regions are associated with nucleosome depletion
Mammalian CpG-rich promoters show strong nucleosome depletion that appears at least partially independent of transcriptional activity, suggesting that CpG density plays a role in promoter accessibility (Fenouil et al. 2012; Vavouri and Lehner 2012). We therefore investigated the relationship between CpG density and chromatin accessibility at C. elegans promoters.
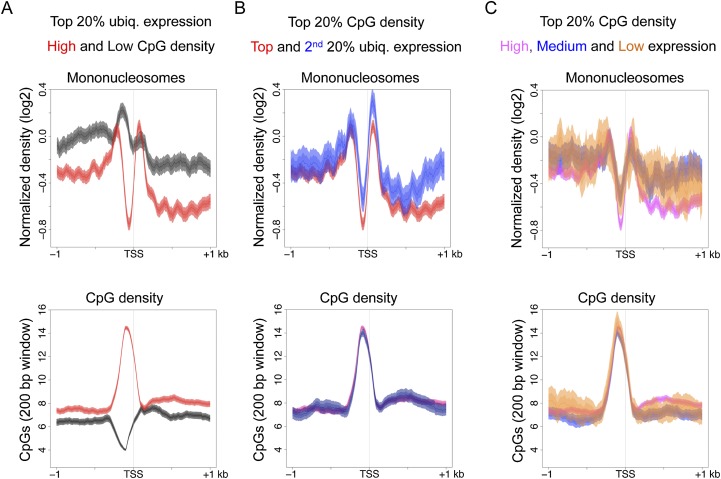
We first separated promoters of highly expressed genes into those with high and low CpG density and compared their nucleosome distribution patterns. For this analysis, we used ubiquitously active C. elegans promoters to avoid effects due to tissue-specific regulation. We found that among these highly active promoters, those with high CpG density are strongly nucleosome depleted compared to those with low CpG density (Fig. 4A). We obtained similar results after normalizing CpG content to local GC content (Supplemental Fig. S4A). Therefore, high CpG density but not high transcriptional activity is associated with promoter accessibility. To test whether high GC content rather than high CpG density was related to nucleosome depletion, we separated high GC content promoters into those with high or low CpG content and plotted their nucleosome distributions. Promoters with high CpG content show stronger nucleosome depletion than those with low CpG content, indicating that accessibility is linked with high CpG rather than generally high GC content (Supplemental Fig. S4B,C).
Figure 4.
A promoter harboring a CpG dense region is associated with nucleosome depletion in C. elegans. (A) Plots of mononucleosome and CpG distributions across promoters of ubiquitously expressed genes in the top 20% of expression band, separated by high CpG density (red, top 20%) or low CpG density (dark gray, bottom 20%). (B) Mononucleosome and CpG distributions were analyzed for ubiquitously active promoters within 20% of CpG density (protein coding promoters) and separated into the top 20% (red) or second 20% (blue) of expression. (C) The distribution of mononucleosomes and CpGs across promoters with the top 20% CpG density separated into those with high (top 20%, pink), medium (middle 20%, blue), or low/no expression (bottom 40%, orange). Lines show mean signal, darker filled areas show standard error, and lighter filled areas are 95% confidence intervals.
To investigate whether the level of transcriptional activity affects the nucleosome distribution pattern at promoters, we separated ubiquitously active promoters with high CpG density into different gene expression bands. Ubiquitously active genes are relatively highly expressed, with most (94%) falling into the top 40% of expression when considering all genes. We separated these genes into those in the top and bottom of this range (top 20% and second 20% of expression) and compared their nucleosome distributions. The two gene expression classes both show strong nucleosome depletion (Fig. 4B), suggesting that among ubiquitously expressed genes, the level of transcriptional activity has little effect on the level of nucleosome depletion at promoters with high CpG density.
Finally, to investigate whether high CpG density per se can drive nucleosome depletion, we separated all high CpG density promoters into different expression bands. Genes in the middle and bottom expression bands are enriched for tissue-specific expression, whereas those in the top expression band are enriched for being ubiquitously expressed. Promoters of all three expression bands display clear nucleosome depletion. However, we found that the level of depletion is lower at promoters of genes in the low/no and middle expression bands compared to those of high expression (Fig. 4C). This difference suggests that high CpG density alone is not sufficient for strong nucleosome depletion, and that additional regulation can counteract the effects of CpG-rich sequences in promoting chromatin accessibility.
C. elegans CFP-1 is targeted to CpG-rich promoters marked by H3K4me3
In higher vertebrates, nonmethylated CpGs are directly recognized by the CXXC domain of CXXC1 (CFP1), which leads to trimethylation of lysine 4 of histone H3 through recruitment of the SETD1A histone methyltransferase (Lee and Skalnik 2005; Tate et al. 2010; Thomson et al. 2010). Despite the lack of DNA methylation in C. elegans (Simpson et al. 1986), a CXXC1 ortholog (CFP-1) containing a conserved CXXC domain exists (highlighted in Supplemental Fig. S5). In addition, it was previously demonstrated that C. elegans CFP-1 is required for global H3K4me3 levels (Simonet et al. 2007; Li and Kelly 2011). These findings raise the possibility that C. elegans CFP-1 might target promoter regions of high CpG density, as in mammals.
To test this hypothesis, we mapped the binding sites of CFP-1 by chromatin immunoprecipitation (ChIP) assays of GFP-tagged CFP-1. We find that CpG di-nucleotides are enriched at CFP-1 binding sites and are densely distributed at CFP-1 peaks (Supplemental Fig. S6). Most C. elegans CFP-1 binding sites are at promoters and overlap regions marked by H3K4me3 (Fig. 5A–C), consistent with the requirement for CFP-1 in generating H3K4me3. Furthermore, the signal intensities of both CFP-1 and H3K4me3 at promoters show a striking concordance with CpG density (Fig. 5D,E). The CFP-1 peaks not near annotated 5′ ends also harbor high CpG density and promoter-like chromatin signatures (H3K4me3high/H3K4me1low), suggesting that they might identify unknown promoters (Supplemental Fig. S7A). In contrast, little CFP-1 signal is found at COLD regions, which usually have an enhancer-like chromatin signature (Supplemental Fig. S7B). We also find that highly expressed genes with high levels of CFP-1 (top 20%) show H3K4me3 marking and nucleosome depletion, but highly expressed genes with little CFP1 (bottom 20%) do not, suggesting that CFP-1 binding is important for these patterns (Fig. 5E). Furthermore, as with high CpG density, we found that promoters with high CFP-1 levels show nucleosome depletion and high H3K4me3 marking, which was observed in promoters linked with genes in both the top 20 and second 20% expression bands (Fig. 5F). We conclude that targeting of CXXC1 orthologs to nonmethylated CpG-dense promoters and the relationship with H3K4me3 marking is conserved between C. elegans and humans.
Discussion
Our results support the view that promoter CpG dinucleotides may function as an ancient conserved promoter signal. Many C. elegans and human promoters show local CpG enrichment and are targeted by a nonmethyl CpG binding protein 1 ortholog (Lee and Skalnik 2005 and this work), and extreme HOT regions in both organisms are CpG-rich promoters. Furthermore, C. elegans CpG-dense promoters are strongly nucleosome depleted in vivo, as in mammals (Fenouil et al. 2012; Vavouri and Lehner 2012).
How CpG-rich sequences function in promoter accessibility is unclear. One possibility is that the DNA sequence itself initiates accessibility, as it has been observed that selected human CpG-rich promoter sequences poorly assemble nucleosomes in vitro (Ramirez-Carrozzi et al. 2009), and large CGIs show low in vitro occupancy (Fenouil et al. 2012). If CpG-dense sequence does intrinsically disfavor nucleosome assembly, this could make the regions more available for binding to factors such as CXXC1 (CFP1). However, in contrast to the findings of Ramirez-Carrozzi et al. (2009), global in vitro analyses that mapped intrinsically favored nucleosome positions in genomic DNA showed that human CpG-rich promoters are on average enriched for nucleosome occupancy (Valouev et al. 2011). Similarly, we also find that C. elegans CpG-rich promoters display strong nucleosome occupancy in in vitro assembly data sets (Supplemental Fig. S8; Locke et al. 2013).
These conflicting in vitro results regarding CpG-rich sequences could be due to differences in the assay conditions used for nucleosome assembly in vitro. Ramirez-Carrozzi et al. (2009) directly tested nucleosome assembly on 300-bp fragments, whereas the global in vitro studies (Valouev et al. 2011; Locke et al. 2013) assembled nucleosomes on larger fragmented genomic DNAs before micrococcal nuclease treatment to isolate mononucleosomes. Another study (Zhang et al. 2011) showed that conditions used for nucleosome reconstitution can strongly affect the results obtained, as more in vivo-like nucleosome positions are observed when a salt dialysis method is applied during nucleosome reassembly. Interestingly, the addition of whole-cell extract and ATP in the absence of transcription nearly reconstituted in vivo patterns (Zhang et al. 2011), supporting a role for external factors in nucleosome positioning and occupancy. Further investigations will be required to elucidate the roles of CpG sequences and non-nucleosome factors in determining nucleosome patterns at promoters.
Irrespective of such future studies, our findings suggest a paradigm whereby CpG-rich sequences promote chromatin accessibility, either intrinsically or through the activities of other factors, which facilitates the formation of an active promoter with an open chromatin state. Our results also provide a plausible explanation for the high occupancy of transcription factors at HOT regions. High CpG density at these regions might create and/or maintain highly accessible regions that are available for interactions with TFs and other factors.
The finding that C. elegans and human HOT regions are active promoters may seem at odds with the finding that Drosophila HOT regions can function as enhancers in transgenic assays (Kvon et al. 2012). This variance may be partly explained by differences in how HOT regions were defined. We analyzed extreme HOT regions in the top 1% of occupancy, whereas Kvon et al. (2012) studied those in the top 5% of occupancy. Nevertheless, we find that Drosophila TF binding regions in the top 5% of occupancy are enriched for having promoter-like chromatin signatures and frequently overlap transcription start sites (Supplemental Fig. S9A), similar to C. elegans and human HOT regions. We also note that the HOT regions assayed in the Drosophila transgenic study (Kvon et al. 2012) were relatively large (a median length of 2.1 kb) and therefore may harbor multiple regulatory elements. As activity assays were not conducted in the absence of a basal promoter, it is possible that the tested Drosophila regions might contain active promoters as well as enhancers. Interestingly, in contrast to C. elegans and human, we observed only a very small enrichment for CpG dinucleotides in Drosophila HOT regions, with similar enrichment in COLD regions (Supplemental Fig. S9B), suggesting that CpG dinucleotides may not be relevant at Drosophila HOT regions.
After CpG, the GC dinucleotide shows the second highest enrichment in human and C. elegans HOT regions (Fig. 3), suggesting that GpC might play an uncharacterized role at promoters. CpG shows much higher enrichment in humans than in C. elegans, whereas GpC enrichment is similar in the two organisms. This difference might be a consequence of the global depletion of CpG in the nonpromoter regions of the human genome, which is not seen in C. elegans. A possible role for GpC dinucleotides at promoters remains to be characterized.
Computational CpG island predictions, which identify DNA sequences with the highest relative CpG density, do not highly overlap promoters in cold-blooded vertebrates (Long et al. 2013) or in C. elegans (Supplemental Table S4; Wu et al. 2010). Therefore, CpG density alone is unlikely to be the sole signal for an open chromatin state at promoters. Consistent with this, purification of nonmethylated DNA via CXXC binding identifies promoters across a range of vertebrate genomes (Long et al. 2013), suggesting that differential DNA methylation may be important for recognition of these regions. As C. elegans lacks DNA methylation, recognition of nonmethylated CpG-rich sequence must involve alternative mechanisms in this organism. An important goal for the future will be to understand how these regions are identified. Given the conservation of CpG promoter enrichment, it seems plausible that mechanisms not involving DNA methylation may also operate in animals harboring DNA methylation.
Methods
Defining the overlaps of transcription factor binding sites
We used modENCODE and ENCODE collections of TF mapping data for 90 C. elegans factors, 54 D. melanogaster factors, and 159 human factors (The ENCODE Project Consortium 2007, 2012; Gerstein et al. 2010, 2012; The modENCODE Project Consortium 2010; Nègre et al. 2011; Niu et al. 2011; AP Boyle, CL Araya, C Brdlik, P Cayting, C Cheng, Y Cheng, K Gardner, L Hillier, J Janette, L Jiang, et al., in prep). Data sets for a given factor were merged into single files to prevent double counting of factors; then at every base, the number of factors where a peak was present was counted. The occupancy of each region is the number of unique factors found inside. For each region of TF overlap, the “core region” (Supplemental Fig. S1) was defined as the range with local maxima of peak call coverage. This process identified 35,062 C. elegans regions bound by 1–87 factors, 32,168 D. melanogaster by 1–37 factors, and 73,7151 human regions bound by 1–138 factors (Supplemental File S1). For all analyses, we defined HOT regions as those in the top 1% of occupancy (376 for C. elegans, 341 for D. melanogaster, and 7419 for humans) and “COLD” regions as single factor binding regions (13,425 for C. elegans, 18,177 for D. melanogaster, and 314,323 for humans) (Supplemental File S2). The following genome versions were used: hg19 for human, ce10/WS220 for C. elegans, and dm3 for Drosophila.
HOT and COLD core regions were scored as overlapping a promoter defined by ±500 bp of a transcript start site. Coding transcript start sites were downloaded from Ensembl genes 71 (human (GRCh37.p10) and D. melanogaster (BDGP5). For C. elegans, we collected and pooled all coding TSSs recently identified based on capped RNA sequencing (Chen et al. 2013; Kruesi et al. 2013).
Generation and analysis of HOT-region transgenic lines
Ten HOT core sites were PCR amplified from N2 genomic DNA using Phusion polymerase (Finnzymes) and Gateway cloned (Invitrogen) into pDONR221 (regions given in Supplemental Table S1). To create transgenes to test whether HOT regions could function as promoters, MultiSite Gateway cloning (Invitrogen) was used to recombine the HOT regions upstream of his-58 (pJA272) and gfp∷tbb-2 3′UTR (pJA256) sequences on the MosSCI compatible vector pCFJ150, which targets Mos site Mos1(ttTi5605) chrII (Zeiser et al. 2011). C. elegans MosSCI lines were generated as described (Frøkjær-Jensen et al. 2008), injecting strain EG6699 with injection mixes that contained pCFJ103(40 ng/µL), pCFJ90(5 ng/µL), pCFJ104(2.5 ng/µL), and expression clones at 40 ng/µL. Supplemental Table S2 lists all strains generated in this study. All strains were used and cultured using standard methods (Brenner 1974). Transgenic strains were grown at 25°C prior to microscopic examinations. Young adult or L4 stage worms were sedated in 5 mM Tetramisole, aligned, and scanned in groups at controlled laser and scanning settings. A full list of primers used for generation of pDONR221 promoter constructs can be found in Supplemental Table S3.
Generation of GFP-tagged CFP-1 (JA1597)
The coding region of F52B11.1a (cfp-1) was PCR amplified from N2 genomic DNA using Phusion polymerase (Finnzymes) and Gateway cloned into pDONR221. The cfp-1 coding region was then recombined into the MosSCI compatible vector pCFJ201 (which targets Mos site Mos1[cxTi10882] chrIV) downstream from the dpy-30 promoter and upstream of gfp∷tbb-2 3′UTR (Zeiser et al. 2011).
Chromatin immunoprecipitation (ChIP)
Late embryos were obtained as in Vielle et al. (2012) by aging embryos collected by hypochlorite treatment 3.5 h prior to flash freezing in liquid nitrogen. Formaldehyde-fixed chromatin extracts and chromatin immunoprecipitations were as in Kolasinska-Zwierz et al. (2009) except that DNA was sonicated to a size range of 200–400 bp. ChIP assays were performed in 1 mL extract (1 mg protein) in FA buffer with 10 µg of anti-GFP rabbit serum (Abcam ab290) or anti-H3K4me3 (Abcam ab8580); two biological replicates were performed for each antibody. DNA sequencing libraries were constructed using the Illumina TruSeq sequencing kit and were sequenced on the Illumina platform.
Data sets, processing, and visualization
C. elegans ChIP-seq data H3K4me3 (modENCODE_5166) and H3K4me1 (modENCODE_5158) and Drosophila ChIP-seq data H3K4me3 (modENCODE_789) and H3K4me1 (modENCODE_777) were obtained from modENCODE (http://www.modencode.org/). MNase digested mononucleosome data for C. elegans embryos (GSM468574) (Ooi et al. 2010) and human K562 cells (GSM920557) (The ENCODE Project Consortium 2012) were downloaded from the Gene Expression Omnibus. H3K4me3 (wgEncodeBroadHistoneHepg2H3k4me3StdSig.bigWig) and H3K4me1 (wgEncodeBroadHistoneHepg2H3k04me1StdSig.bigWig) ChIP-seq data in HepG2 cells were obtained from the ENCODE Project (The ENCODE Project Consortium 2012; AP Boyle, CL Araya, C Brdlik, P Cayting, C Cheng, Y Cheng, K Gardner, L Hillier, J Janette, L Jiang, et al., in prep) (http://genome.ucsc.edu/ENCODE/downloads.html). TF ChIP-seq data sets used in the HOT region study are listed in Supplemental File S3 and can be downloaded from http://anshul.kundaje.net/projects/modencode (for C. elegans and Drosophila) and http://hgdownload.cse.ucsc.edu/goldenPath/hg19/encodeDCC/wgEncodeAwgTfbsUniform/ (for human).
CpG density, GC content, and observed over expected CpG ratio tracks were calculated in sliding windows of 200-bp size and 1-bp shift, reporting calculated values in the middle of the corresponding range. CpG promoter content was calculated at −200 bp to C. elegans TSSs and ±500 bp of human TSSs. These calculated CpG values for all protein coding promoters—n = 10,106 for C. elegans from Chen et al. (2013) and n = 129,604 for human from Ensembl GRCh37.p12—were ranked to extract promoters harboring the indicated percentile of CpG content.
Ubiquitously expressed genes in C. elegans were defined as those showing expression (FDR <0.05) in embryonic gut, pan neuron, body wall muscle, germline, and hypodermis in tissue-specific RNA-seq profiling data sets (Spencer et al. 2011). Human ubiquitously expressed genes were defined as those detectably expressed in all examined cell lines (n = 34) in human RNA-seq data sets from the ENCODE Project (The ENCODE Project Consortium 2011) data from http://genome.crg.es/encode_RNA_dashboard/hg19/. Genes with high CpG promoters were those having a promoter in the top 20% CpG band but no promoter in the bottom 80% CpG band (n = 2215 for C. elegans and n = 7710 for human). Genes with low CpG promoters are those having a promoter in the bottom 20% CpG band but no promoter in the top 80% CpG band (n = 1764 for C. elegans and n = 1834 for human genes).
CFP-1 and H3K4me3 ChIP-seq data sets used in Figure 5 were aligned using BWA with default settings (Li and Durbin 2009), normalized using BEADS (Cheung et al. 2011), then converted to ratios of BEADS scores (enrichment relative to input) and then Z-scored. Peaks were called using MACS v. 2.0.10 software (Feng et al. 2011) with 1 × 10−10 P-value cutoff. Peak calls from each replicate were intersected, and regions present in both were kept (Supplemental File S4). Heat maps were generated using the Cistrome heatmap function (Liu et al. 2011). The IGV Genome Browser was applied for visualization (Robinson et al. 2011).
Data access
The ChIP-seq data for C. elegans CFP-1 and H3K4me3 tracks used in this study have been submitted to the NCBI Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE49870.
Acknowledgments
We thank M. Chesney for helpful comments on the manuscript. R.A.-J.C., P.S., E.Z., and J.A. were supported by a Wellcome Trust Senior Research Fellowship to J.A. (054523) and T.A.D. by a Wellcome Trust Research Career Development Fellowship. S.K.F. was supported by a Gates Cambridge Scholarship. J.A. and T.A.D. also acknowledge support by core funding from the Wellcome Trust and Cancer Research, UK.
Footnotes
[Supplemental material is available for this article.]
Article published online before print. Article, supplemental material, and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.161992.113.
Freely available online through the Genome Research Open Access option.
References
- Allen MA, Hillier LW, Waterston RH, Blumenthal T 2011. A global analysis of C. elegans trans-splicing. Genome Res 21: 255–264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari KI, Mishra BP, Mandal SS 2008. Human CpG binding protein interacts with MLL1, MLL2 and hSet1 and regulates Hox gene expression. Biochim Biophys Acta 1779: 66–73 [DOI] [PubMed] [Google Scholar]
- Antequera F, Bird A 1999. CpG islands as genomic footprints of promoters that are associated with replication origins. Curr Biol 9: R661–R667 [DOI] [PubMed] [Google Scholar]
- Bektesh SL, Hirsh DI 1988. C. elegans mRNAs acquire a spliced leader through a trans-splicing mechanism. Nucleic Acids Res 16: 5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird AP 1986. CpG-rich islands and the function of DNA methylation. Nature 321: 209–213 [DOI] [PubMed] [Google Scholar]
- Bird A, Taggart M, Frommer M, Miller OJ, Macleod D 1985. A fraction of the mouse genome that is derived from islands of nonmethylated, CpG-rich DNA. Cell 40: 91–99 [DOI] [PubMed] [Google Scholar]
- Blumenthal T 1995. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet 11: 132–136 [DOI] [PubMed] [Google Scholar]
- Blumenthal T 2012. Trans-splicing and operons in C. elegans. In WormBook (ed. The C. elegans Research Community), pp. 1–11. http://www.wormbook.org [DOI] [PubMed] [Google Scholar]
- Brenner S 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler JS, Lee JH, Skalnik DG 2008. CFP1 interacts with DNMT1 independently of association with the Setd1 Histone H3K4 methyltransferase complexes. DNA Cell Biol 27: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caiafa P, Zampieri M 2005. DNA methylation and chromatin structure: the puzzling CpG islands. J Cell Biochem 94: 257–265 [DOI] [PubMed] [Google Scholar]
- Cameron EE, Bachman KE, Myohanen S, Herman JG, Baylin SB 1999. Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 21: 103–107 [DOI] [PubMed] [Google Scholar]
- Chen RA, Down TA, Stempor P, Chen QB, Egelhofer TA, Hillier LW, Jeffers TE, Ahringer J 2013. The landscape of RNA polymerase II transcription initiation in C. elegans reveals promoter and enhancer architectures. Genome Res 23: 1339–1347 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung MS, Down TA, Latorre I, Ahringer J 2011. Systematic bias in high-throughput sequencing data and its correction by BEADS. Nucleic Acids Res 39: e103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M 1989. Cytosine methylation and the fate of CpG dinucleotides in vertebrate genomes. Hum Genet 83: 181–188 [DOI] [PubMed] [Google Scholar]
- Deaton AM, Bird A 2011. CpG islands and the regulation of transcription. Genes Dev 25: 1010–1022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan BK, Miller JH 1980. Mutagenic deamination of cytosine residues in DNA. Nature 287: 560–561 [DOI] [PubMed] [Google Scholar]
- Ehrlich M, Zhang XY, Inamdar NM 1990. Spontaneous deamination of cytosine and 5-methylcytosine residues in DNA and replacement of 5-methylcytosine residues with cytosine residues. Mutat Res 238: 277–286 [DOI] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447: 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2011. A user’s guide to the encyclopedia of DNA elements (ENCODE). PLoS Biol 9: e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The ENCODE Project Consortium. 2012. An integrated encyclopedia of DNA elements in the human genome. Nature 489: 57–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J, Liu T, Zhang Y 2011. Using MACS to identify peaks from ChIP-Seq data. Curr Protoc Bioinformatics 34: 2.14.1–2.14.14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenouil R, Cauchy P, Koch F, Descostes N, Cabeza JZ, Innocenti C, Ferrier P, Spicuglia S, Gut M, Gut I, et al. 2012. CpG islands and GC content dictate nucleosome depletion in a transcription-independent manner at mammalian promoters. Genome Res 22: 2399–2408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frøkjær-Jensen C, Davis MW, Hopkins CE, Newman BJ, Thummel JM, Olesen SP, Grunnet M, Jorgensen EM 2008. Single-copy insertion of transgenes in Caenorhabditis elegans. Nat Genet 40: 1375–1383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M, Frommer M 1987. CpG islands in vertebrate genomes. J Mol Biol 196: 261–282 [DOI] [PubMed] [Google Scholar]
- Gerstein MB, Lu ZJ, Van Nostrand EL, Cheng C, Arshinoff BI, Liu T, Yip KY, Robilotto R, Rechtsteiner A, Ikegami K, et al. 2010. Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330: 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstein MB, Kundaje A, Hariharan M, Landt SG, Yan KK, Cheng C, Mu XJ, Khurana E, Rozowsky J, Alexander R, et al. 2012. Architecture of the human regulatory network derived from ENCODE data. Nature 489: 91–100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illingworth RS, Bird AP 2009. CpG islands–‘a rough guide’. FEBS Lett 583: 1713–1720 [DOI] [PubMed] [Google Scholar]
- Jones PL, Veenstra GJ, Wade PA, Vermaak D, Kass SU, Landsberger N, Strouboulis J, Wolffe AP 1998. Methylated DNA and MeCP2 recruit histone deacetylase to repress transcription. Nat Genet 19: 187–191 [DOI] [PubMed] [Google Scholar]
- Joulie M, Miotto B, Defossez PA 2010. Mammalian methyl-binding proteins: what might they do? Bioessays 32: 1025–1032 [DOI] [PubMed] [Google Scholar]
- Klose RJ, Bird AP 2006. Genomic DNA methylation: the mark and its mediators. Trends Biochem Sci 31: 89–97 [DOI] [PubMed] [Google Scholar]
- Kolasinska-Zwierz P, Down T, Latorre I, Liu T, Liu XS, Ahringer J 2009. Differential chromatin marking of introns and expressed exons by H3K36me3. Nat Genet 41: 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruesi WS, Core LJ, Waters CT, Lis JT, Meyer BJ 2013. Condensin controls recruitment of RNA polymerase II to achieve nematode X-chromosome dosage compensation. Elife 2: e00808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kvon EZ, Stampfel G, Yáñez-Cuna JO, Dickson BJ, Stark A 2012. HOT regions function as patterned developmental enhancers and have a distinct cis-regulatory signature. Genes Dev 26: 908–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JH, Skalnik DG 2005. CpG-binding protein (CXXC finger protein 1) is a component of the mammalian Set1 histone H3-Lys4 methyltransferase complex, the analogue of the yeast Set1/COMPASS complex. J Biol Chem 280: 41725–41731 [DOI] [PubMed] [Google Scholar]
- Li H, Durbin R 2009. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25: 1754–1760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li T, Kelly WG 2011. A role for Set1/MLL-related components in epigenetic regulation of the Caenorhabditis elegans germ line. PLoS Genet 7: e1001349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T, Ortiz JA, Taing L, Meyer CA, Lee B, Zhang Y, Shin H, Wong SS, Ma J, Lei Y, et al. 2011. Cistrome: an integrative platform for transcriptional regulation studies. Genome Biol 12: R83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Locke G, Haberman D, Johnson SM, Morozov AV 2013. Global remodeling of nucleosome positions in C. elegans. BMC Genomics 14: 284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long HK, Sims D, Heger A, Blackledge NP, Kutter C, Wright ML, Grutzner F, Odom DT, Patient R, Ponting CP, et al. 2013. Epigenetic conservation at gene regulatory elements revealed by non-methylated DNA profiling in seven vertebrates. Elife 2: e00348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The modENCODE Project Consortium. 2010. Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330: 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nan X, Ng HH, Johnson CA, Laherty CD, Turner BM, Eisenman RN, Bird A 1998. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature 393: 386–389 [DOI] [PubMed] [Google Scholar]
- Nègre N, Brown CD, Ma L, Bristow CA, Miller SW, Wagner U, Kheradpour P, Eaton ML, Loriaux P, Sealfon R, et al. 2011. A cis-regulatory map of the Drosophila genome. Nature 471: 527–531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niu W, Lu ZJ, Zhong M, Sarov M, Murray JI, Brdlik CM, Janette J, Chen C, Alves P, Preston E, et al. 2011. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res 21: 245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooi SL, Henikoff JG, Henikoff S 2010. A native chromatin purification system for epigenomic profiling in Caenorhabditis elegans. Nucleic Acids Res 38: e26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramirez-Carrozzi VR, Braas D, Bhatt DM, Cheng CS, Hong C, Doty KR, Black JC, Hoffmann A, Carey M, Smale ST 2009. A unifying model for the selective regulation of inducible transcription by CpG islands and nucleosome remodeling. Cell 138: 114–128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramskold D, Wang ET, Burge CB, Sandberg R 2009. An abundance of ubiquitously expressed genes revealed by tissue transcriptome sequence data. PLoS Comput Biol 5: e1000598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schug J, Schuller WP, Kappen C, Salbaum JM, Bucan M, Stoeckert CJ Jr 2005. Promoter features related to tissue specificity as measured by Shannon entropy. Genome Biol 6: R33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonet T, Dulermo R, Schott S, Palladino F 2007. Antagonistic functions of SET-2/SET1 and HPL/HP1 proteins in C. elegans development. Dev Biol 312: 367–383 [DOI] [PubMed] [Google Scholar]
- Simpson VJ, Johnson TE, Hammen RF 1986. Caenorhabditis elegans DNA does not contain 5-methylcytosine at any time during development or aging. Nucleic Acids Res 14: 6711–6719 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer WC, Zeller G, Watson JD, Henz SR, Watkins KL, McWhirter RD, Petersen S, Sreedharan VT, Widmer C, Jo J, et al. 2011. A spatial and temporal map of C. elegans gene expression. Genome Res 21: 325–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tate CM, Lee JH, Skalnik DG 2010. CXXC finger protein 1 restricts the Setd1A histone H3K4 methyltransferase complex to euchromatin. FEBS J 277: 210–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomson JP, Skene PJ, Selfridge J, Clouaire T, Guy J, Webb S, Kerr AR, Deaton A, Andrews R, James KD, et al. 2010. CpG islands influence chromatin structure via the CpG-binding protein Cfp1. Nature 464: 1082–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner JD, Alt SR, Cao L, Vernocchi S, Trifonova S, Battello N, Muller CP 2010. Transcriptional control of the glucocorticoid receptor: CpG islands, epigenetics and more. Biochem Pharmacol 80: 1860–1868 [DOI] [PubMed] [Google Scholar]
- Valouev A, Johnson SM, Boyd SD, Smith CL, Fire AZ, Sidow A 2011. Determinants of nucleosome organization in primary human cells. Nature 474: 516–520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vavouri T, Lehner B 2012. Human genes with CpG island promoters have a distinct transcription-associated chromatin organization. Genome Biol 13: R110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vielle A, Lang J, Dong Y, Ercan S, Kotwaliwale C, Rechtsteiner A, Appert A, Chen QB, Dose A, Egelhofer T, et al. 2012. H4K20me1 contributes to downregulation of X-linked genes for C. elegans dosage compensation. PLoS Genet 8: e1002933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Caffo B, Jaffee HA, Irizarry RA, Feinberg AP 2010. Redefining CpG islands using hidden Markov models. Biostatistics 11: 499–514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Bian C, Lam R, Dong A, Min J 2011. The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain. Nat Commun 2: 227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yip KY, Cheng C, Bhardwaj N, Brown JB, Leng J, Kundaje A, Rozowsky J, Birney E, Bickel P, Snyder M, et al. 2012. Classification of human genomic regions based on experimentally determined binding sites of more than 100 transcription-related factors. Genome Biol 13: R48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeiser E, Frøkjær-Jensen C, Jorgensen E, Ahringer J 2011. MosSCI and gateway compatible plasmid toolkit for constitutive and inducible expression of transgenes in the C. elegans germline. PLoS ONE 6: e20082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Wippo CJ, Wal M, Ward E, Korber P, Pugh BF 2011. A packing mechanism for nucleosome organization reconstituted across a eukaryotic genome. Science 332: 977–980 [DOI] [PMC free article] [PubMed] [Google Scholar]