Summary
The human host has co-evolved with the collective of bacteria species, termed microbiota, in a complex fashion that affects both innate and adaptive immunity. Differential regulation of regulatory T-cell and effector T-cell responses are a direct result of specific microbial species present within the gut, and this relationship is subject is dysregulation during inflammation and disease. The microbiota varies widely between individuals and has a profound effect on how one reacts to various environmental stimuli, particularly if a person is genetically predisposed to an immune-mediated inflammatory disorder such as inflammatory bowel disease (IBD), including Crohn’s disease (CD) and ulcerative colitis (UC). Approximately half of all CD patients have elevated antibodies to CBir1, a microbiota flagellin common to mice and humans, demonstrating flagellins as immunodominant antigens in the intestines. This review focuses on the use of flagellins as probes to study microbiota specific responses in the context of health and disease as well as probes of innate and adaptive responses employed by the host to deal with the overwhelming bacterial presence of the microbiota.
Keywords: microbiota, flagellin, T cells, B cells, IgA, IBD
Introduction
The co-evolution of the microbiota with human has generated complex networks of bacterial reactivity and tolerance, some for the benefit of the host and some for its detriment. For example, the presence of certain Clostridia species in mice have been demonstrated to be protective against dextran sodium sulfate (DSS)-induced colitis, while the presence of Klebsiella pneumoniae and Proteus mirabilis in the murine intestine is associated with inflammation and contributes to colitis in certain immune compromised mice. This complexity is illustrated by the presence of 10-fold more microbial cells than eukaryotic cells in the human body, and these bacterial cells contain 100 times as many genes as the entire human genome (1). Certain clostridia species, most predominantly from cluster XIVa, have been associated with increased numbers of T-regulatory cells (Tregs) in the mouse colon (2), while segmented filamentous bacteria (SFB) has been associated with the development of the T-helper 17 (Th17) cell lineage in the murine small intestine (3, 4). Numerous additional bacterial species have been associated with immune cell development and are discussed further below. Dysregulated responses to the microbiota have been associated with immune-mediated diseases such as Crohn’s disease (CD) (5, 6). CBir1 and related flagellins have been identified as immunodominant antigens in murine colitis and in CD, thus flagellin reactivity has proven to be a valuable tool in understanding microbiota specific responses (6–10). In this review, we update the current understanding of microbiota-specific responses in both innate and adaptive immunity, including microbiota effects on the epithelium, innate lymphoid cells (ILCs), T-cell development, and immunoglobulin A (IgA), as well as recent approaches assisting in understanding how the immune system and the microbiota work in concert.
Innate immune responses to the microbiota
Secretory IgA limits bacterial access to the host
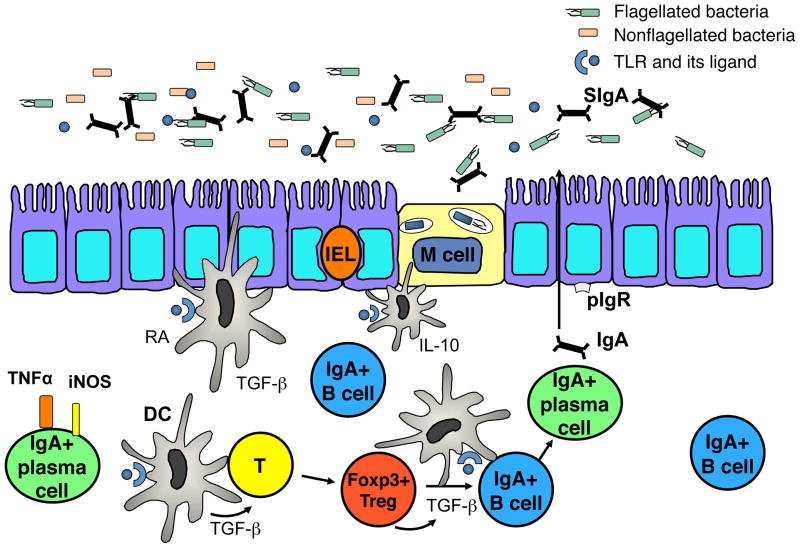
The innate arm of the immune system has critical mechanisms for eliminating pathogenic bacteria and is vital in restricting systemic adaptive responses to microbiota species in order to maintain a homeostatic environment. Secretory IgA (SIgA) is a vital component in communicating the contents of the microbiota to the immune system. After SIgA binds and forms complexes with commensal species, it can subsequently cross from the lumen to the mucosa by binding to a specialized IgA receptor on microfold (M) cells (11) (Fig. 1). SIgA selectively presents the bacterial components to tolerogenic CD11c+CD11b+CD8− dendritic cells (DCs), which produce interleukin-10 (IL-10) and have a propensity to induce IgA class switching (12, 13) in the subepithelial dome (SED) of Peyer’s patches (PPs) (14–16). This process is vital in establishing a constant, albeit nominal, sampling of commensal species by SIgA that ensures effective communication between the microbiota and the immune system. This selective presentation of commensal species to tolerogenic DCs is in line with the anti-inflammatory nature of SIgA and aids in limiting inflammation that could result from the immense load of bacteria in the lumen.
Fig. 1. IgA and gut homeostasis.
SIgA communicates luminal contents to the lamina propria by sampling commensals and presenting them to tolerogenic DCs, which have a propensity to induce IgA class switching, via a specialized receptor on M cells. Foxp3+ Tregs are major T-helper cells involved in IgA production and IgA+ B-cell survival in the lamina propria. IgA+ plasma cells can also express the antimicrobial mediators TNF-α and iNOS to act as an effector cell in the lamina propria. The multifaceted IgA system acts to maintain an anti-inflammatory setting by compartmentalizing microbial responses to the mucosal immune system to circumvent a systemic response to the microbiota.
SIgA is also a critical member of the first line of defense against invading pathogens. Polymeric IgA attaches to the poly-immunoglobulin receptor (pIgR) on the basolateral surface of the epithelium, where it is then transported into the intestinal lumen as SIgA after interacting with secretory component (SC) (17, 18). SIgA blocks adherence of invading bacteria and toxins to the thick mucus layer of the epithelium through broad recognition of pathogenic epitopes on their surface followed by subsequent cross-linking of these antigens in the intestinal lumen, thus preventing the colonization of these species and eliminating the potential for inflammatory responses (14, 18–21). Additional roles for IgA in maintaining mucosal homeostasis are further discussed below.
The role of protective mucus layers and spatial segregation in the intestine An additional mechanism of restricting immune responses to commensal organisms is by spatial segregation at the mucosal interface. This spatial segregation present within the murine colon is in the form of two mucus layers, a firm layer that is in direct contact with epithelium and devoid of bacteria and a loose layer between the firm layer and the lumen that contains limited bacteria (22). Both layers are structurally dependent on goblet cell (GC) production of Muc2 mucin and serve to limit bacterial attachment to the epithelium. In support of this, Muc2−/− mice lack appropriate separation of the microbiota and the colonic epithelium and therefore have bacteria located in colonic crypts and surface epithelium, making them susceptible to intestinal inflammation and colon cancer (22). These findings were recently recapitulated in human studies. Healthy controls were demonstrated to have an impenetrable inner mucus layer in the colon, while ulcerative colitis (UC) patients (and murine models of colitis) were demonstrated to have bacteria present in the inner, normally impenetrable layer (23). These data are in agreement with other reports linking epithelial cell dysregulation and inflammatory bowel disease (IBD) (22, 24, 25).
In the small intestine, epithelial cell secretion of RegIIIγ, a C-type bacterial lectin specific for targeting Gram-positive bacteria, and likely other anti-microbial peptides, is essential in establishing a 50 μm zone devoid of bacteria between the mucus layer and the lumen. The formation of this protective zone is dependent on myeloid differentiation factor 88 (MyD88) signaling and is vital in limiting adaptive immune responses to the microbiota (26, 27). Mice deficient in RegIIIγ have been demonstrated to exhibit increased quantities of Gram-positive mucosa-associated bacteria, heightened intestinal IgA levels, and augmented numbers of CD4+interferon-γ (IFNγ)+ T cells in the intestines (27). The dependence on MyD88 signaling for the induction of this protective zone suggests TLR signals play in a role in generating this response, though the exact trigger or if a threshold of signaling is required for RegIIIγ production is currently unclear.
TLR5 and anti-flagellin responses in mice
Toll-like receptor 5 (TLR5) is expressed on the basolateral surface of intestinal epithelial cells and is the cell surface receptor for flagellin, the structural component of bacteria flagella (28, 29). Both CD11c+CD11b+ and CD11c+CD11b− intestinal DCs express TLR5 to a markedly greater extent than splenic DCs (30). Various microbial ligands including lipopolysaccharide (LPS), FliC, and CpG oligonucleotides downregulate TLR5 expression in bone marrow-derived DCs (BMDCs). In contrast, BMDCs treated with retinoic acid (RA) display increased TLR5 protein, suggesting its expression is differentially regulated in the intestine by various dietary and microbiota constituents (30). Additionally, systemic flagellin administration has been demonstrated to induce significant IL-23 production from CD103+CD11b+ lamina propria (LP) DCs via TLR5 activation (31). IL-23 acts on various cell types and is vital in controlling intestinal inflammation. Mice deficient in TLR5 (TLR5−/−) have elevated levels of pro-inflammatory cytokines compared to wild type mice at baseline and are susceptible to colitis induced by the inflammasome cytokine IL-1β, in the setting of IL-10 neutralization (32). Although inhibiting IL-10 signaling altered the microbiota in both wildtype and TLR5−/− mice, only the TLR5−/− mice were subject to colitis induction, suggesting innate mechanisms may be compensating for the lack of IL-10 in wildtype mice (32). Colitic TLR5−/− mice have transiently increased proportions of proteobacteria, specifically enterobacteria species, compared to wildtype or non-colitic TLR5−/− mice (33).
Defensin regulation of mucosal homeostasis
α-defensins contribute to intestinal homeostasis by regulating IL-1β. Although overproduction of IL-1β leads to overt inflammation, a complete lack of this signaling molecule can contribute to an increased susceptibility to various infections. Matrix metalloproteinase-7-deficient (MMP7−/−) mice, which are deficient in mature α-defensins, have augmented basal levels of IL-1β, demonstrating a role for defensins as negative regulators of IL-1β. MMP7−/− mice also have increased sensitivity to DSS-induced colitis, revealing a role for defensins in controlling intestinal epithelial health (34). Loss of function mutations in NOD2, a risk allele associated with CD, are associated with decreased α-defensin production from Paneth cells, further establishing a link between altered defensin responses and intestinal inflammation (35, 36).
Autophagy as a mechanism of innate immunity
Recent work has demonstrated autophagy as a vital innate immune mechanism to combat opportunistic commensals and pathogenic bacteria at the epithelial surface (37). This epithelial cell-intrinsic mechanism functions by ingesting invasive bacteria in a MyD88-dependent manner, limiting the spread of bacteria to sites beyond the epithelial layer. This observation is intriguing, considering that NOD1 and NOD2 have been shown to regulate autophagy, and NOD2 mutations are linked with CD in the same pathway as ATG16L1, an autophagy-associated gene (36, 38–41). Therefore, dysregulation of autophagy due to a genetic defect could play a role in the onset of CD by altered innate, protective responses against invading bacteria as well as the previously mentioned defect in Paneth cells (37, 41, 42). Furthermore, ATG16L1 may function as a suppressor of inflammatory cytokine induction by NOD1 and NOD2 (43). ATG16L1 hinders poly-ubiquitination of the Rip2 adapter and subsequent recruitment and activation of Rip2. This function is independent of the autophagy defect associated with the CD risk allele and reveals an additional role for defects in ATG16L1 in contributing to intestinal inflammation and predisposition to CD (43).
Inflammasomes as mediators of innate immunity
Inflammasomes are critical innate mediators of the microbiota-host dialog that sense pathogen-associated molecular patterns (PAMPs) and damage-associated molecular patterns (DAMPs). Recent work has demonstrated that mice deficient in the NLRP6 inflammasome signaling complex, a member of the NOD-like receptor (NLR) family, have reduced levels of IL-18, a cytokine important in epithelial turnover and barrier function (44). NLRP6−/− mice also had a dysbiosis of the intestinal microbiota including elevated levels of Prevotellaceae and TM7 bacterial family members and reciprocal decreases in the Lactobacillus genus and Firmicute phylum. This dysbiosis was both vertically transferred through progeny and horizontally transferred to co-housed pups and adult mice. NLRP6−/− mice as well as wildtype mice with this altered flora had increased sensitivity to DSS-induced colitis due to upregulation of CCL5, a T-cell chemokine (44). These data support the hypothesis that genetic defects in inflammasome activation and signaling can contribute to an increased susceptibility to intestinal inflammation by inducing dysbiosis that is ‘colitigenic’ in nature.
Ipaf, a member of the NOD-LRR (leucine-rich repeat) family of proteins, has been demonstrated to recognize intracellular flagellin and is capable of inflammasome activation, stimulation of caspase-1, and activation of pro-inflammatory IL-1β in a TLR5-independent manner in Salmonella infected macrophages (45). Flagellin stimulation of IL-1β via ipaf occurs in naïve as well as LPS-tolerized macrophages, indicating this activation is independent of NFκB and mitogen-activated protein kinase (MAPK) signaling (45).
ILCs: bridging innate and adaptive immunity
Innate lymphoid cells (ILCs) have emerged as important regulators of adaptive immunity. ILCs have a vital role in barrier function and integrity (46) and can be categorized into three groups (ILC1, ILC2, ILC3) according to their dependence on specific transcription factors (47). The heterogeneous group 3 ILCs, which are dependent on the canonical Th17 lineage transcription factor retinoic-acid-receptor related orphan receptor- γt (RORγt), have been recently shown to contribute to microbiota-specific responses independently of the classical Th17 lineage-associated cytokines, IL17A, IL-22, and IL-23 (48). The CCR6+RORγt+Tbet−IFNγ− subset of the group 3 ILCs express major histocompatibility complex class II (MHCII) and are able to present antigen in vivo. This subset lacks costimulatory molecules and therefore does not induce T-cell proliferation in the gut but instead functions to limit CD4+ mucosal T-cell responses. ILCs pulsed with CBir1 flagellin peptide and transferred into a naïve congenic mouse along with CBir1-transgenic T cells resulted in a decreased expansion of CBir1-specific T cells and IFNγ production, compared to transfer of CBir1-specific T cells only, after systemic challenge with CBir1 peptide (48). These data implicate a significant role for ILCs in limiting CD4+ T-cell responses to the microbiota by interacting with T cells through an MHCII-dependent mechanism. While this mechanism is independent of IL-23, group 3 ILCs have recently been demonstrated to be dependent on IL-23 production from DCs to produce IL-22, an important mechanism maintaining epithelial integrity (31). Thus, ILCs have a multifaceted role in regulating intestinal homeostasis.
Innate regulation of adaptive CD4+ T-cell responses to microbiota antigens T-cell transfer models, most notably the transfer on naive (CD45RBhi) cells into immunodeficient mice, have long been used as experimental models of colitis (49, 50). Co-transfer of memory T cells (CD45RBlo) along with naive T cells, as well as neutralization of specific pro-inflammatory cytokines, are effective at inhibiting intestinal inflammation (50). Following transfer, the CD4+ T cells expand in vivo, but the mechanism of colitogenic CD4+ T-cell proliferation and whether this proliferation was necessary for colitis development has been clarified. The induction of colitis by transfer of naive CBir1 flagellin TCR transgenic CD4+ T cells into an immunocompromised mouse requires both innate stimulation via TLR signals for spontaneous proliferation of T cells, as well as antigen-specific CD4+ T-cell activation (51). Adoptive transfer of CD4+ CBir1- specific T cells but not CD4+ OT-II cells, which are specific for ovalbumin, into B6.Rag−/− specific pathogen-free (SPF) mice resulted in colitis. Colonizing the B6.Rag−/− mice with an OVA-expressing E. coli before transfer of OT-II CD4+ T cells resulted in colitis induction, demonstrating that the lack of colitis was not due to an insufficiency of T cells but rather a lack of cognate antigen (51). While transgenic T (OT-II and CBir1) cells were able to proliferate and cause colitis when transferred into Rag−/− (SPF) mice in the presence of cognate antigen, these cells did not proliferate or cause intestinal inflammation when transferred into germ-free (GF) Rag−/− mice, indicating a role for the microbiota in spontaneous T-cell proliferation and colitis induction. Co-transfer of DCs pulsed with mouse cecal contents with CD4+ CBir1 T cells but not IL6−/− or MyD88−/− DCs were able to restore spontaneous T-cell proliferation in GF mice. Therefore, microbiota-derived ligands are vital in stimulating DCs to produce IL-6 via the MyD88 pathway, an essential cytokine in spontaneous T-cell proliferation (51). These data are in agreement with a two-hit model of colitis in which microbial stimulation of the innate intestinal system and activation of T cells by their cognate microbial antigen are both required for colitis to occur (51).
Microbiota-specific adaptive responses
IgA maintains a mutualistic relationship with the microbiota
The immune system has multiple strategies for dealing with the immense bacterial load of the intestinal system, one of the most prominent being IgA. In contrast to the classical prime-boost responses characteristic of IgG reactivity, IgA responses are additive in nature and dependent on the bacterial species that are contemporaneously present, suggesting an intimate crosstalk between IgA and the microbiota (52). Using a reversible microbial colonization system, microbiota-specific IgA responses were found to be long-lived and capable of reaching high titers but subject to attrition, and therefore directed at the bacterial species currently present within the intestinal tract (52). IgA responses in the intestine also require a high threshold for induction, approximately 109 bacteria, and have a relatively long half-life, greater than 16 weeks. These characteristics, coupled with a slow onset of response, implicate IgA as an anti-inflammatory antibody that acts to maintain mutualism with the microbiota rather than mount an immune response against it (52). Dysbiosis of the microbiota during inflammation can contribute to the breakdown of this mutualistic phenotype and lead to unregulated bacterial growth contributing to systemic microbiota responses.
To generate commensal-specific IgA, DCs sample bacteria in close proximity to the epithelium and subsequently interact with B and T cells in the PPs to generate IgA-producing B cells (53). Commensal-specific IgA can also be produced in a T-independent fashion that is not dependent on follicular lymphoid tissue organization, largely through B1 B cells (17, 54). The commensal-specific IgA generated is confined to the mucosal immune system, and therefore circumvents systemic responses against the resident bacteria. This compartmentalization of murine responses to the microbiota has been demonstrated by the homeostatic response to 20 random recombinant intestinal bacterial proteins (rIBs) cloned from the cecum of a C3H/HeJ mouse (55). In contrast to strong IgA responses to the majority of the 20 rIBs, there were no splenic CD4+ T-cell responses or baseline serum IgG reactivity to any of these microbiota antigens in normal C3H/HeJ mice. However, all 20 rIBs were able to induce strong IgG responses after parental immunization, equivalent to those induced with ovalbumin (OVA) (55). This demonstrates ignorance, rather than a tolerance, of commensal bacteria by the systemic immune system. In agreement, adoptively transferred bacteria-specific T cells do not proliferate in the spleen or mesenteric lymph nodes in wildtype mice with normal IgA levels after oral challenge with antigen (7). This is due to endogenous IgA preventing uptake of antigen into the mesenteric lymph node DCs, because T cells readily proliferate in mice deficient in IgA or the in the poly-immunoglobulin receptor (pIgR) after oral challenge with antigen (7). These data reveal a role for IgA in promoting homeostasis with the microbiota by limiting exposure to systemic immune responses.
Induction of IgA responses by T-regulatory cells
Treg cells in the intestine provide critical survival factors for mucosal IgA+ B cells in the intestine (7, 17). CD4+Foxp3+ and CD4+CD25+ Tregs are able to induce B-cell class switch recombination (CSR) and IgA production in vitro, and CBir1-specific as well as total IgA production in the intestine is severely impaired after Treg depletion. Intestinal IgA is fully restored after adoptive transfer of CD4+CD25+ or CD4+Foxp3+ Tregs compared to transfer of CD4+CD25− or CD4+Foxp3− T cells into B6.TCRβδ−/− mice. The substantial decrease in IgA+ B cells and reciprocal increase in Annexin V staining after selective Treg depletion indicates that Tregs serve a survival function for IgA+ B cells in the LP (7). These data support a model in which Tregs act in concert with mucosal IgA+ B cells to both inhibit host inflammatory responses and maintain the gut microbiota, which in turn protects the host against intestinal pathogens (Fig. 1).
IgA+ B-cell effector functions
An additional role for the multifaceted IgA antibody producing B cells was recently elucidated by the finding that a subset of IgA+ plasma cells in the LP express both inducible nitric oxide synthase (iNOS), an antimicrobial factor, and tumor necrosis factor α (TNF-α) (56). Interestingly, the production of iNOS by this IgA+ B-cell subset is dependent on microbial colonization. GF mice are completely deficient in this subset, but iNOS expression by IgA+ plasma cells is successfully restored after colonization with a limited flora (56). Mice deficient in iNOS+TNFα+ IgA+ plasma cells have diminished IgA production and are unable to effectively clear infection with Citrobacter rodentium. These data establish a new effector function for IgA+ plasma cells in producing antimicrobial factors in vivo, further broadening role of IgA+ B cells in maintaining gut homeostasis in addition to compartmentalizing microbiota-specific responses and limiting uptake of microbiota antigens (Fig. 1).
Colitogenic flagellin-specific T-cell responses and dependence on IL-23
Augmented production of the pro-inflammatory cytokines, IL-17A as well as IFN-γ and TNF-α, is a defining characteristic of experimental colitis induction in mice (50, 57, 58). To determine the most prominent cytokines triggered by the microbiota during colitis, a T-cell line specific for intestinal antigens was developed by repeated stimulation of splenic CD4+ cells from colitic mice with cecal bacteria preparations from a C3H mouse. Transfer of the bacteria-reactive CD4+ cells into C3H.SCID mice resulted in colitis within four weeks (57, 59). Although the majority of CD4+ T cells expressed IL-17 after transfer, there was still a substantial population of CD4+ IFN-γ-expressing cells. To determine which T-helper cell type was most associated with inducing colitis, these bacteria-reactive cells were differentiated into Th17 or Th1 phenotypes prior to transfer into a C3H.SCID mouse. Interestingly, the IL-17-producing Th17 cells reactive to cecal bacterial antigens were decidedly more potent in inducing colitis and immunopathology than the Th1 cells expressing IFNγ (57), an outcome likely due to Th17 plasticity. The Th17 subset has been demonstrated to have the potential to differentiate into Th1+ Th17+ and Th1+ effectors, whereas Th1 cell types are less fluid and retain traditional Th1 phenotypes in vivo (60). The murine Th17 lineage requires IL-6 and TGF-β for differentiation, and IL-23 has been shown to interact with and stabilize this pathway (57, 61–63). IL-23 is heterodimeric member of the IL-12 family of cytokines, with a unique subunit, IL-23p19, and shared subunit with IL-12, IL-12p40 (64). Mutations in the IL-23 receptor are associated with various immune-mediated disorders including CD, UC, psoriasis, and ankylosing spondylitis (36). A monoclonal antibody to the IL23p19 subunit of IL-23 was tested in this bacterial-specific T-cell transfer model of colitis to determine the function of IL-23 in intestinal inflammation. Neutralization of IL-23 with the anti-IL-23p19 antibody at the time of T-cell transfer was sufficient to inhibit the development of colitis in immunocompromised mice. In addition, the IL-23p19 antibody was effective at treating established colitis as well as downregulating multiple inflammatory genes such as IL-6, IFN-γ, TNF-α, IL-1β, and IL-17 (57). Increased apoptosis of Th17 cells was observed after administration of the IL-23p19 antibody, supporting a role for IL-23 as a survival factor for Th17 cells. These data reveal the importance of IL-23 in fostering the sustainability of Th17 cells during intestinal inflammation. IL-23 also plays an important protective role in the intestine and whether IL-23 targeted therapy will be beneficial in the treatment of IBD in humans is unknown.
Activation of systemic microbiota reactive T cells by gut infection
Infection with the gastrointestinal pathogen, Toxoplasma gondii, activated microbiota-specific T-cell responses through a breech in the compartmentalization of the mucosal immune system (65). Naive CBir1 TCR transgenic T cells proliferated abundantly and expressed the classical Th1 transcription factor, T-bet, when transferred into mice subsequently infected with T.gondii. This is in contrast to the transfer of OT-II TCR transgenic T cells, which did not undergo similar activation, demonstrating that commensal-specific T cells are capable of activation and differentiation into an effector phenotype during a heterologous mucosal infection (65). Interestingly, these cells persisted as effector memory cells with increased expression of CD27 and low expression of Ly6c. Memory CBir1 TCR Tg T cells were able to effectively proliferate after a secondary systemic challenge with CBir1 peptide 35 days post oral infection with T.gondii. In addition, these long-lived cells maintained Tbet expression and low levels of IL-7Rα, consistent with a memory cell recall response. CBir1 flagellin-specific cells could be reactivated during a gastrointestinal infection (GI), when CBir1 Tg T cells were stimulated with CBir1 peptide and LPS prior to a later oral challenge with T. gondii, demonstrating that commensal-reactive T cells can respond to subsequent pathogenic infections after initial antigen-specific stimulation (65). These data provide insight into how dysregulated responses to commensals may arise in the setting of infection or epithelial barrier disruption, potentially leading to inflammation and disease.
Adaptive immunity maintains a healthy microbiota
Lack of competent adaptive immune responses can result in deficient responses to the microbiota and shifts in bacterial composition. Deficiency in the T-box transcription factor family member Tbet (Tbx21) in mice lacking adaptive immunity (Rag2−/−) is capable of causing spontaneous colitis and epithelial layer malfunction (66). The colitis induced in the T-bet−/−Rag2−/− (TRUC) mice is dependent on the microbiota dysbiosis that is vertically transmissible to progeny as well as to wildtype and Rag2−/− pups crossfostered with TRUC mothers. The colitis is marked by increased production of TNF-α from colonic DCs, for which Tbet is a negative regulator but is sufficiently controlled by transfer of CD4+CD25+ regulatory T cells or treatment with antibiotics (66). 16S rRNA-based analysis of the microbiota constituents of the TRUC mice revealed the presence of both Klebsiella pneumoniae and Proteus mirabilis (67). While colonization of GF re-derived TRUC mice with K. pneumoniae and P. mirabilis was not sufficient to induce colitis, the two bacterial species are able to elicit intestinal inflammation, albeit to a lower degree than that see in TRUC mice, in both WT and Rag2−/− mice in the presence of an endogenous flora (67). These data indicate that certain microbial species, while not pathogenic on their own, may be able to interact with endogenous members of the microbiota to induce disease under specific circumstances. Indeed, numerous murine models of colitis have varying species of bacteria contributing to inflammation depending on the experimental model (Table 1).
Table 1. Multiple bacterial strains contribute to colitis.
Multiple bacterial species are capable of contributing to or inducing colitis in mice depending on the experimental conditions and background strain. There is no single common species across all models, indicating that the resulting IBD depends on not only on genotype but also on the local environment, particularly the microbiota composition in different facilities.
| Model | Organism | Mouse Strain | Publication |
|---|---|---|---|
| CD45RBhi | SFB + others | BALB/cJHanHsd-SCID | Stepankova et al. 2007 |
| CD4-dnTgfβ × IL- 10R2−/− | Bacteroides | C57BL/6 | Bloom et al. 2011 |
| IL-10−/− | Clostridia XIVa Lachnospiraceae |
C3H/HeJBir | Duck et al. 2007 |
| TRUC | K. pneunomiae P. mirabilis |
Balb/c × Rag2−/− × Tbet−/− | Garrett et al. 2007 |
| DSS NLRP6−/− | Prevotellaceae TM7 |
C57BL/6 | Elinav et al. 2011 |
| IL-10−/− | E.coli E. faecalis |
129S6/SvEv | Kim et al. 2005 |
Microbiota effects on CD4+ T cells
Recent advances in sequencing technology has allowed for a more in-depth look at the collection of genes, termed the microbiome, that make up the microbiota. Current estimates are that the microbiome contains approximately 3×106 genes, and considering that the microbiota has co-evolved with their host, it is logical that many of these genes encode antigens that function in maintaining mutualism with the host (68). Considering the extensive population of bacterial species in the intestine, gnotobiotic mice have been useful in understanding the effects of specific bacterial strains on host immunity. Monocolonization of GF mice with Bacteroides thetaiotaomicron, a strain of bacteria known to colonize both human and mouse intestine, is sufficient to induce multiple alterations in host genes affecting intestinal functions including nutrient absorption, barrier function, xenobiotic metabolism, angiogenesis, and maturation of immune cells. These changes in gene expression are bacterial species specific because monocolonization with other species such as Escherichia coli K12 and Bifidobacterium infantis induce contrasting effects compared to B. thetaiotaomicron monocolonization (69). These data demonstrate that disparate bacterial species have targeted and varying effects on the host and highlight the need to investigate specific functions of dominant bacterial organisms on host immunity.
Microbiota influence on T-effector cells
Microbial stimulation of the innate and adaptive immune systems has been demonstrated as indispensable for intestinal health, and recent data have also highlighted the microbiota’s pivotal function in fostering the development of T-cell subsets including Tregs and Th17 cells. Recent work demonstrated that approximately half of all genes affected by microbial colonization of GF mice are associated with immune responses pathways (4). This establishes a major role for bacteria in shaping immune responses. Monocolonization with one bacterium, Segmented Filamentous Bacteria (SFB), is capable of inducing multiple T-cell lineages in the large and small intestine of mice as well as augmenting various ileal mRNA transcripts, including RegIII-γ (4). SFB is a clostridia-related organism that forms filament-like structures and is a potent inducer of polyclonal IgA responses, and to a lesser extent, antigen-specific IgA responses (70). SFB filaments attach to the intestinal epithelium of the distal ilieum, as well as the epithelial dome region of PPs (71). Further data on SFB has elucidated a specific role in the induction of Th17 cells (3). Bacterial composition of mice from two different facilities, Jackson Laboratory and Taconic Farms, were analyzed due to their divergent levels of Th17 cells in the small intestine LP. Mice from Jackson Laboratory had very low numbers of intestinal Th17 cells and a lack of SFB colonization, while the mice from Taconic Farms had sufficient intestinal Th17 numbers and displayed robust adherence of SFB to the ilieum (3, 72). Co-housing of nice from Taconic Farms and Jackson Laboratory was sufficient to establish SFB colonization in Jackson Laboratory mice and was accompanied by a reciprocal increase in intestinal Th17 cells (3). These data support the hypothesis that SFB is a major inducer and regulator of Th17 cell induction in mice. Additionally, the combination of RegIII-γ upregulation in SFB monocolonized mice and concomitant increase in IL-17 production is consistent with a protective role for SFB in regards to epithelial integrity (3, 4). Indeed, mice colonized with SFB demonstrate are more resistant to Citrobacter rodentium colonization and infection, most likely due to the increased production of the Th17-associated cytokines, IL-17, IL-22, and IL-23 (3). Although the exact mechanism of induction of Th17 cells is unclear, SFB does stimulate the epithelial cell production of serum amyloid A (SAA), which can induce Th17 responses in a concentration dependent manner in vitro (3). The lack of overt inflammation in SFB monocolonized mice suggests the major effect of SFB is to maintain homeostasis with the host and prevent the colonization of pathogenic species. The protective role of Th17 cells in the context of mucosal stress is consistent with recent data demonstrating that mice deficient in the IL-17 receptor (IL-17R−/−) are more susceptible to DSS-induced colitis (73). IL-17R−/− mice were also shown to have decreased intestinal pIgR and IgA levels, demonstrating a role for Th17 cells in their regulation and a broad role in overall mucosal homeostasis (73).
Microbiota influence on T-regulatory cells
Multiple bacterial species have been associated with the induction and regulation of CD4+Foxp3+ and CD4+IL10+ regulatory T cells in the intestine. GF mice have low numbers of CD4+Foxp3+ Tregs in the colonic LP, as opposed to other tissues such as the inguinal lymph nodes (iLN), small intestine LP, mesenteric lymph nodes (mLNs), and PPs of SPF mice, indicating a role for microbial stimulation in colonic Treg induction (2). Reconstitution of GF mice with a mix of 46 strains of Clostridium species, predominately from Clostridium clusters IV (Clostridium leptum) and XIV (Clostridium coccoides), both of which have been associated with mucosal health, (74) induced a vigorous accumulation of Foxp3+ Tregs, to levels seen in SPF mice, in the colon of these gnotobiotic mice (2). This increase in Tregs was present only in the colon and cecum of these mice, where these bacteria typically reside, but not in the small intestine. The effect was also specific to Clostridium species because colonization of GF mice with a mix of 16 Bacteroides species, SFB, or a mix of three Lactobacillus strains had minimal effects on colonic Foxp3+ Treg accumulation (2). The increase in Foxp3+ Tregs was accompanied by an increase in TGF-β production by the intestinal epithelial cells (IECs) of the Clostridium-colonized mice. A majority of the Foxp3+ cells were Helios negative, indicating that Clostridium species are primarily augmenting the induction of the induced Foxp3+ Tregs (iTregs) in the colon through increased TGF-β and TGF-β activator production by IECs (2). Additionally, the newly generated Tregs expressed high amounts of cytotoxic T-lymphocyte antigen 4 (CTLA-4) as well as IL-10, and demonstrated similar suppression kinetics to Tregs derived from SPF mice. Early colonization of SPF mice with Clostridia (2 weeks of age) efficiently generated a Clostridia-rich mucosal environment that was less susceptible to DSS and oxazolone-induced colitis, further demonstrating the functionality of these induced Tregs as well as the importance of Clostridia in mucosal homeostasis (2).
Subsequent data have demonstrated that colonization of GF mice with a mix of 17 Clostridia species isolated from a human fecal sample is capable of inducing CD4+Foxp3+ Tregs in the colon (75). The collective of 17 strains was from Clostridia clusters IV, XIVa and XVIII, and all strains lacked overt virulence factors. The increase in colon Tregs was both vertically and horizontally transmissible and marked by an increase in IL-10+ and/or ICOS+ CD4+ T cells within the Treg population (75). Colonic Tregs isolated from Clostridia treated mice were able to suppress OT-I TCR transgenic CD8+ T-cell antigen-specific proliferation in vitro, and their suppressive capacity was increased with the addition of autoclaved cecal contents from Clostridia treated mice, suggesting a portion of the induced Tregs are antigen specific (75). GF mice colonized with a single member or a mix of 3–5 Clostridia species from the group of 17 demonstrated increased colon Tregs, though not to the same extent seen with colonization of all 17 strains. Additionally, cecal extracts from the 17-strain mix were capable of eliciting increased TGF-β1 production from an intestinal epithelial line compared to extracts from mice colonized with a single isolate or a mix of 5 strains. Lastly, SPF mice orally inoculated with the 17-strain mix were less susceptible to TNBS-induced colitis and more protected in a naive T-cell transfer model of colitis (75). These data demonstrate that bacterial species most likely work in concert to induce protective responses and combination therapy may be a more realistic therapeutic for patients with intestinal inflammation.
Although the exact mechanism of how Clostridia species specifically induce Tregs is unclear, it is logical to speculate that bacterial metabolites such as short chain fatty acids (SCFAs) are mediating this response. Administration of acetate to the drinking water of mice protected from DSS colitis and was mediated by engagement of the SCFA receptor, GPR43 (encoded by Ffar2), expressed on innate immune cells (76). SCFAs, including acetic acid, propionic acid and butyric acid, have been shown to induce the generation of mucosal Tregs (77–79). Butyrate, a metabolite of bacterial fermentation, particularly by Clostridia, is capable in augmenting extrathymic Foxp3+ Treg induction in vivo in GF, SPF, as well as antibiotic treated SPF mice when administered systemically (77, 78). Furthermore, local administration of butyrate to the colon has been demonstrated to induce extrathymic de novo generation of Tregs (78). Propionate and acetate were equally capable of augmenting colonic Treg induction after local administration, independent of CNS-1, a conserved non-coding DNA sequence on Foxp3 that acts as an enhancer element responsible for extrathymic Treg induction, suggesting that these SCFAs support expansion of the existing pool of colonic Tregs (78). While systemic administration of butyrate was sufficient to induce extrathymic Tregs, the induction of colonic Tregs was dependent on local administration. Other work has demonstrated increased colonic Tregs in GF and SPF mice after systemic administration of butyrate, acetate, propionate, or a mix of all three SCFAs (77). In these studies, systemically administered propionate was demonstrated to induce colonic Treg generation via the SCFA receptor GPR43, in agreement with previous work (76). GPR43 engagement by SCFAs was sufficient to induce histone deacetylase (HDAC) inhibition, a potential mechanism for the increased colonic Treg proliferation and suppressive capacity. While propionate treatment did not ameliorate intestinal inflammation in lymphopenic mice in a naive CD4+ T-cell transfer model of colitis, it was capable of decreasing disease severity when Tregs were co-transferred along with T naive cells into lymphopenic mice, indicating that propionate acts directly to expand and strengthen the existing pool of regulatory T cells (77, 78). Although SCFA systemic administration was capable of inducing extrathymic Tregs in both studies, their effect on colonic Treg generation varied. The contrasting effects of SCFAs on colonic and extra-intestinal Treg generation may be due to resident microflora differences between different facilities. Further research is needed to clarify which specific strains of bacteria produce SCFAs, although certain clusters of Clostridia have already been demonstrated to produce enhanced levels of SCFAs compared to other bacterial species. It would also be interesting to determine how these SCFAs are differentially regulated during inflammation and disease, considering SCFAs have been shown to be depleted in UC.
Clostrida’s effect on mucosal homeostasis is not restricted to Tregs alone. Recent data have implicated Clostridium butyricum, a probiotic member of Clostridia cluster I, as a potent inducer of IL-10 production from F4/80+CD11b+CD11cint macrophages (Mϕ) in the colon under inflammatory conditions (80). Treatment of mice with C.butyricum was not sufficient to induce IL-10 production from Mϕ under steady state conditions, but rather required an inflammatory milieu, such as one supplied by DSS-induced colitis. Interestingly, increased IL-10 production occurred through a TLR2/MyD88 signaling pathway and was not dependent on T cells, as similar results were seen in Rag−/− mice (80). This T-cell-independent protective effect is different from previous data in which B. fragilis polysaccharide A stimulation induced homeostatic conditions through TLR2 signaling directly on Treg cells (81). These data demonstrate novel methods of how treatment with a microbiota member as a probiotic can contribute to a stabilized microbiota and homeostasis through IL-10-dependent mechanisms.
Innate immune mechanisms are vital in limiting bacterial adherence to intestinal tissues, but various bacteria are able to penetrate and maintain close contact with the host epithelium, including SFB and Bacteroides fragilis (3, 4, 81). B. fragilis is a common human commensal organism that is capable of inducing IL-10-producing Foxp3+ Tregs through TLR2-dependent recognition of polysaccharide A (PSA) (81–83). Instead of inducing TLR-mediated inflammatory responses, PSA acts to suppress IL-17 production to successfully colonize the colonic crypts. PSA’s actions occur exclusively through TLR2 signals on Foxp3+ regulatory T cells, even in the absence of antigen-presenting cells (APCs) (81). B.fragilis is an example of a mechanism by which a microbiota species has evolved with its host to circumvent inflammatory responses, typically resulting from TLR signaling, to maintain mutualism. B. fragilis’s recognition by Tregs, instead of innate immune cells, most likely aids in its intimate colonization niche with its host. This symbiotic relationship may have evolved over time to restrict inflammatory IL-17 production and colonization of the colonic crypts with pathogenic species.
Treg/Teff balance in the intestine
Regulatory T cells and effector T cells are dynamically regulated by the microbiota (Fig. 2). Data demonstrated that commensal DNA modulates the T-effector/Treg cell balance in the intestine via a TLR9-dependent mechanism (84). TLR9−/− mice have an increased frequency of Tregs and reduced production of IFN-γ and IL-17 by T-effector cells in the small intestinal LP compared to wildtype mice. This difference is not present in the colonic LP, demonstrating a site-specific effect of TLR9 signaling on T-cell regulation. Treatment of wildtype mice with an antibiotic cocktail diminished host immune response against oral infection with a mucosal pathogen, demonstrating a need for microbial stimulation to mount a successful mucosal immune response. Administration of commensal DNA or CpG, a natural TLR9 ligand, was able to revert this phenotype, further indicating that commensal DNA is able to prime a mucosal response to infection in a TLR9-dependent manner. Additionally, LP DCs treated with CpG are impaired in their ability to induce Tregs in vitro under polarized conditions, but this phenotype is reversed by the inhibition of IL-6, IFN-γ, and IL-4 (84). Therefore, commensal DNA’s regulation of the T-effector/Treg cell balance through TLR9 is most likely site-specific as well as dependent on the cytokine milieu of the affected site.
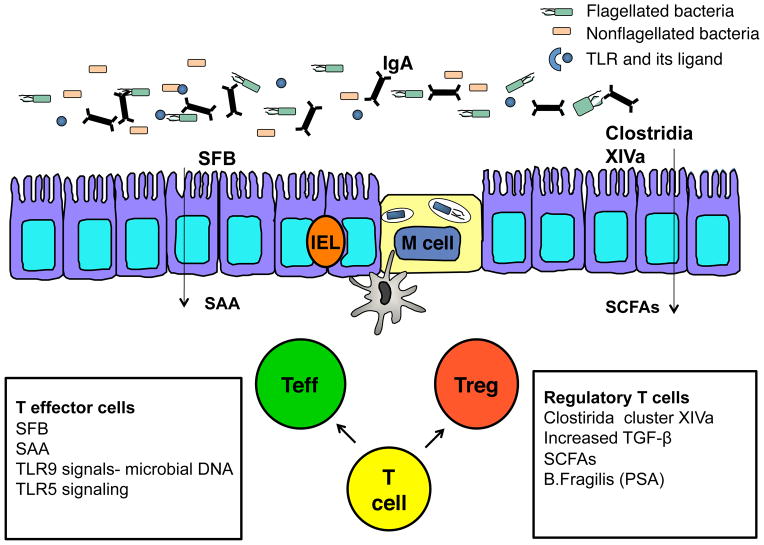
Fig. 2. T-regulatory/T-effector cell balance in the intestine.
Differential expression of microbial species and components contribute to the balance of T-effector cells (Teff) and T-regulatory cells (Tregs) in the lamina propria. SFB colonization is marked by increased SAA production and augmented Th17 cells. Colonization of the colon with Clostridia XIVa family members is associated with increased TGF-β from the epithelium and augmented Tregs. Numerous additional signals contribute to the balance of Teff/Treg including levels of SCFAs and TLR ligand interactions.
In a similar manner, TL5−/− mice have augmented Foxp3+ Tregs compared to wildtype mice (30). TLR5 ligation promotes T-effector cells while opposing Foxp3+ Treg generation in vitro, suggesting that TLR5 signaling affects the Treg/Teff balance. Considering that intestinal DCs express TLR5 to a much greater extent than splenic DCs, TLR5 is most likely an important modulator of microbial signals and the balance of immune cells in the gut (30). TLR5+ CD103+CD11b+ LP DC induction of IL-23 upon flagellin recognition could also contribute to the balance of Treg/Teff in the gut given the known role of IL-23 in supporting the Th17 effector cell pathway and that IL-23 has been shown to negatively regulate Foxp3+ Tregs (31, 61, 85).
Clearly, the bacterial composition of the host most plays a major role in the balance of Tregs and Teffs in the intestine whether by production of SCFAs, colonization by SFB or related organism, (3, 4, 77, 78) or through various innate signals including recognition via TLRs, especially TLR5 and TLR9.
Microbiota flagellin responses in humans
Genetic links to IBD
With more than 160 gene variants encoding susceptibility to IBD identified in genome-wide association studies (GWAS), strong genetic predispositions exist for the development of IBD, most notably CD (86–89). These loci contain variants affecting both the innate and adaptive immune systems and are shared with multiple other immune mediated inflammatory diseases. For example, a polymorphism in the IL-23R gene is common to CD, ulcerative colitis (UC), psoriasis, and ankylosing spondylitis (90). The IL-23R is a heterodimer composed of an α-chain, IL-23R, coupled with a β-chain, IL-12Rβ1, which is shared with the IL-12R (91). The IL-23R gene variant linked to IBD encodes a splice variant resulting in a soluble receptor antagonist of IL-23 (92), and confers protection from IBD (93, 94). The effect of decreased IL-23 signaling could be wide-ranging, because IL-23 acts on innate cells, ILCs, and T cells. While yet to be shown in humans, neutralization of IL-23 is sufficient to ameliorate colitis in mice, demonstrating the importance of this cytokine in modulating intestinal inflammation (57). In mice, systemic administration of flagellin has been demonstrated to induce IL-23 production from CD103+CD11b+ intestinal DCs through the flagellin receptor TLR5, stimulating IL-22 production from RORγt+ ILCs, and providing protective epithelial responses to microbial stimulation (31). Therefore, IL-23 appears to play an important role in immune homeostasis and host protection.
Polymorphisms in NOD2, the gene encoding nucleotide-binding oligomerization domain protein 2, are strongly associated with CD but not UC (36). NOD2 recognizes muramyl dipeptide (MDP), a component of peptidoglycan present on Gram-positive and Gram-negative bacteria, and is therefore a vital receptor for pathogen recognition (36, 95, 96). The most common alterations in NOD2 associated with CD result in decreased activation of NFκB after MDP stimulation, which may contribute to altered bacterial colonization and improper innate immune responses to the microbiota (36, 97). Indeed, CD patients carrying at least one variant in NOD2 have been demonstrated to express quantitatively higher levels of serum IgG anti-flagellin compared to CD patients without any NOD2 variants, suggesting these mutations lead to augmented adaptive antimicrobial responses (9, 98). Recent studies have also linked NOD2 and ATG16L1, an additional risk allele for CD, in an autophagy pathway (38, 40). Human DCs with mutations in NOD2 or ATG16L1 were impaired in autophagy induction, bacterial trafficking and antigen presentation, further demonstrating the need for bacterial recognition and efficient intracellular processing in controlling responses to the microbes (36, 38, 40, 41).
GWAS studies have identified associations between polymorphisms in TL1A and IBD in multiple populations (99). Expression of the cytokine, TL1A (TNFSF15) and its receptor, death domain containing receptor (DR3), is upregulated in T cells and macrophages of IBD patients (100, 101). Bacterial stimulation from both aerobic and anaerobic strains, including the flagellated Lachnospiraceae A4, are capable of inducing TL1A mRNA expression in CD14+ human macrophages and monocyte-derived DCs (MoDCs) (99). Lachnospiraceae A4 was shown to be a potent inducer of membrane-bound TL1A protein expression in human macrophages. This bacterial stimulation is dependent on p38 MAPK signaling and NFκB activation, and results in elevated IFN-γ expression in T cells activated by the TL1A-producing APCs (99). Although genetics play a role in susceptibility to IBD, gene variants are not shared by all CD or UC patients. The current hypothesis for the development of IBD is that chronic inflammation is the result of a dysregulated response to the microbiota in a genetically susceptible individual (102).
Flagellin-specific responses in IBD
A related group of flagellins, most similar to those of Clostridium subphylum cluster XIVa, have been identified as a novel class of immunodominant antigens in multiple models of murine colitis (8). Anti-flagellin IgG2a responses are prominent in the sera of colitic mice, and adoptive transfer of flagellin-specific CD4+ T cells into immunodeficient mice is capable of inducing colitis. Interestingly, serum IgG antibodies to CBir1, a flagellin with significant reactivity in murine colitis, are elevated in approximately half of all CD patients but not in healthy controls or UC patients (8). Recent work characterizing CBir1 flagellin and related flagellins have affirmed their place in Clostridium cluster XIVa and further placed them into the Lachnospiraceae family (103). While the organism responsible for producing CBir1 flagellin has not been isolated, as anaerobes from this cluster are notoriously difficult to culture, a closely related organism, A4, which produces Fla2 in addition to four other flagellins, has been isolated (103) (Fig. 3). Interestingly, IgG reactivity to the Clostridium cluster XIVa related organisms in CD patient sera is specific for flagellins and not to any other potential antigen of the organisms (103). This solidifies flagellins as immunodominant antigens in the intestine and suggests they are potent mediators of innate immune activation and response to the microbiota.
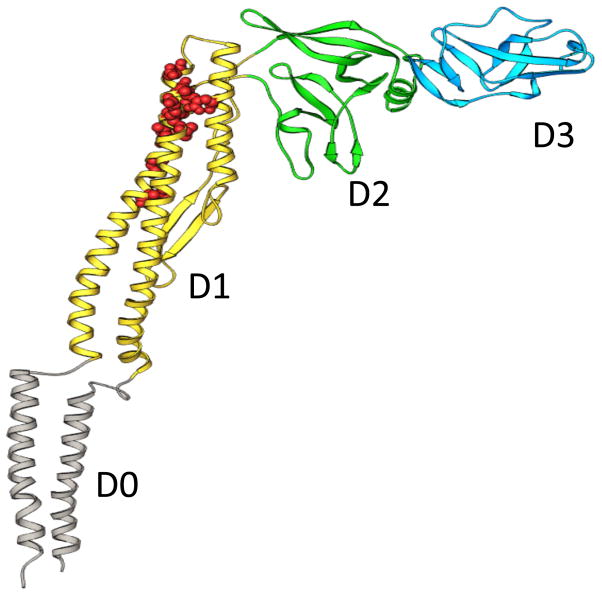
Fig. 3. Structure of CBir1 and related flagellins.
The predicted structure of CBir1 and Lachnospriaceae A4 Fla2 flagellins is similar the previously defined fliC of S. typhimurium, including a TLR5 binding site, depicted in red. Figure used with permission (103).
Previous work has classified CD patients into the following phenotypic groups based on seroreactivity to microbial antigens: (i) oligomannan [anti-Saccharomyces cerevisiae antibody (ASCA)] IgA and IgG reactive, (ii) I2 (CD-related protein from Pseudomonas fluorescens) and OmpC (E. coli outer membrance porin C) IgA reactive, (iii) pANCA (perinuclear antineutrophil cytoplasmic antibody) non-reactive, and (iv) little to no response to any of these microbial antigens (104). Intriguingly, CBir1 flagellin antibody reactivity does not associate with any of the above groups in CD but rather independently marks a subset of patients with a complicated course of CD (9, 10). Anti-CBir1 flagellin positive patients have increased incidence of small bowel internal-penetrating and fibrostenosing disease features and surgery (9, 10). Similarly, antibodies to CBir1 are associated with a more rapid and aggressive disease phenotype in pediatric CD patients (6). Monocytes isolated from peripheral blood (PB) from CD patients produce significantly more IL-6 after stimulation with CBir1 flagellin than do PB monocytes from UC patients or healthy controls (HC) (105). CBir1 flagellin is also capable of inducing proliferation in human PB T cells isolated from CD patients, demonstrating an adaptive T-cell response (105). Additionally, LP mononuclear cells (LPMCs) isolated from CD patients produce increased IFN-γ upon CBir1 flagellin stimulation compared to LPMCs from UC patients or healthy controls, supporting a pathogenic role for CBir1-specific responses in CD (105).
Previous work has identified the cytokine deficiency-induced colitis susceptibility 1 (Cdcs1) locus as the regulator of colitis induction in C3H/HeJBir mice deficient in IL-10 (106). The Cdcs locus was demonstrated to contribute to decreased innate responsiveness to microbiota products and TLR ligands, including flagellin, coupled with exaggerated CD4+ T-cell adaptive responses resulting in colonic inflammation (106, 107). The Cdcs1 locus has been mapped to mouse Chromosome 3 (106), more specifically to an interval containing the NFκB1 gene (107). In support of this, macrophages from C3H/HeJBir mice constitutively express augmented levels of NFκB p50 compared to C57BL/6J macrophages (107). Anti-CBir1 reactivity has recently been linked to chromosome 4 in humans, to an area including the NFκB1 gene and syntenic to the Cdcs1 locus on Chromosome 3 in mice (107, 108). Furthermore, NFκB1 haplotypes have been demonstrated to be associated with anti-CBir1 and anti-ASCA reactivity in CD patients (108). NFκB is composed of two subunits, p50 from NFKB1 and p65 from ReIA. The NFκB1 haplotype (H1) associated with expression of anti-CBir1 antibodies in CD patients is marked by lower expression of the NFκB1 p50 subunit and p105, the inactive precursor of p50, and leads to decreased function and activation of NFκB. This decreased activation of NFκB functions to influence the innate response to the microbiota and contributes to excessive adaptive responses (108). In agreement with these data, innate production of IL-6 is lower in CD patients with quantitatively higher levels of anti-CBir1 antibodies, suggesting an adaptive immune compensatory mechanism (105). These findings are in agreement with the hypothesis that altered innate responses to the microbiota could result in unregulated adaptive microbiota-specific responses in CD.
Concluding remarks
Recent work has characterized specific effects of the microbiota on adaptive immunity, including induction of regulatory and effector CD4+ T cells, production of cytokines and anti-microbial factors, and protection or susceptibility to colitis. However, these events are intimately intertwined with stimulation of the innate immune system, blurring the distinction between the two arms of the immune system. Innate signals via microbial stimulation are necessary for spontaneous proliferation of CD4+ T cells and colitis induction in a T-cell transfer model of colitis, supporting a model in which colitis induction is dependent on signals from both the innate and adaptive immune system. This is in agreement with murine models of colitis in which dampened innate signaling results in aberrant adaptive CD4+ T-cell responses. GWAS findings have elucidated multiple gene variants within the innate as well as the adaptive immune system associating with IBD and by inference mucosal homeostasis. The importance of IgA in the mucosal system is paramount and multifaceted. Protective SIgA aids in resistance to colonization by pathogens while IgA+ plasma cells can secrete iNOS and TNF-α to act as effector cells. Altered innate responses to bacteria most likely favor an environment more susceptible to exaggerated adaptive responses to components of the microbiota, particularly flagellins, and play a role in the onset of IBD. Flagellins as immunodominant microbiota antigens have been useful as probes to investigate both innate and adaptive immune responses.
Acknowledgments
This work is supported by National Institutes of Health grants DK 071176 and RR20136.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Gill SR, et al. Metagenomic analysis of the human distal gut microbiome. Science. 2006;312:1355–1359. doi: 10.1126/science.1124234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Atarashi K, et al. Induction of colonic regulatory T cells by indigenous Clostridium species. Science. 2011;331:337–341. doi: 10.1126/science.1198469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ivanov, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaboriau-Routhiau V, et al. The key role of segmented filamentous bacteria in the coordinated maturation of gut helper T cell responses. Immunity. 2009;31:677–689. doi: 10.1016/j.immuni.2009.08.020. [DOI] [PubMed] [Google Scholar]
- 5.Elson C, Cong Y, Qi F, Hershberg R, Targan S. Molecular approaches to the role of the microbiota in inflammatory bowel disease. Ann NY Acad Sci. 2006;1072:39–51. doi: 10.1196/annals.1326.010. [DOI] [PubMed] [Google Scholar]
- 6.Dubinsky M, et al. Serum immune responses predict rapid disease progression among children with Crohn’s disease: immune responses predict disease progression. Am J Gastroenterol. 2006;101:360–367. doi: 10.1111/j.1572-0241.2006.00456.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cong Y, Feng T, Fujihashi K, Schoeb T, Elson C. A dominant, coordinated T regulatory cell-IgA response to the intestinal microbiota. Proc Natl Acad Sci USA. 2009;106:19256–19261. doi: 10.1073/pnas.0812681106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lodes M, et al. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Papadakis K, et al. Anti-flagellin (CBir1) phenotypic and genetic Crohn’s disease associations. Inflamm Bowel Dis. 2007;13:524–530. doi: 10.1002/ibd.20106. [DOI] [PubMed] [Google Scholar]
- 10.Targan S, et al. Antibodies to CBir1 flagellin define a unique response that is associated independently with complicated Crohn’s disease. Gastroenterol. 2005;128:2020–2028. doi: 10.1053/j.gastro.2005.03.046. [DOI] [PubMed] [Google Scholar]
- 11.Mantis NJ, Cheung MC, Chintalacharuvu KR, Rey J, Corthesy B, Neutra MR. Selective adherence of IgA to murine Peyer’s patch M cells: evidence for a novel IgA receptor. J Immunol. 2002;169:1844–1851. doi: 10.4049/jimmunol.169.4.1844. [DOI] [PubMed] [Google Scholar]
- 12.Iwasaki A, Kelsall BL. Unique functions of CD11b+, CD8 alpha+, and double-negative Peyer’s patch dendritic cells. J Immunol. 2001;166:4884–4890. doi: 10.4049/jimmunol.166.8.4884. [DOI] [PubMed] [Google Scholar]
- 13.Cerutti A. Location, location, location: B-cell differentiation in the gut lamina propria. Mucosal Immunol. 2008;1:8–10. doi: 10.1038/mi.2007.8. [DOI] [PubMed] [Google Scholar]
- 14.Mantis N, Rol N, Corthésy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011;4:603–611. doi: 10.1038/mi.2011.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rol N, Favre L, Benyacoub J, Corthesy B. The role of secretory immunoglobulin A in the natural sensing of commensal bacteria by mouse Peyer’s patch dendritic cells. J Biol Chem. 2012;287:40074–40082. doi: 10.1074/jbc.M112.405001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kadaoui KA, Corthesy B. Secretory IgA mediates bacterial translocation to dendritic cells in mouse Peyer’s patches with restriction to mucosal compartment. J Immunol. 2007;179:7751–7757. doi: 10.4049/jimmunol.179.11.7751. [DOI] [PubMed] [Google Scholar]
- 17.Cerutti A, Rescigno M. The biology of intestinal immunoglobulin A responses. Immunity. 2008;28:740–750. doi: 10.1016/j.immuni.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaetzel CS. The polymeric immunoglobulin receptor: bridging innate and adaptive immune responses at mucosal surfaces. Immunol Rev. 2005;206:83–99. doi: 10.1111/j.0105-2896.2005.00278.x. [DOI] [PubMed] [Google Scholar]
- 19.Corthésy B. Multi-faceted functions of secretory IgA at mucosal surfaces. Front Immunol. 2013;4:185. doi: 10.3389/fimmu.2013.00185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Apter FM, Lencer WI, Finkelstein RA, Mekalanos JJ, Neutra MR. Monoclonal immunoglobulin A antibodies directed against cholera toxin prevent the toxin-induced chloride secretory response and block toxin binding to intestinal epithelial cells in vitro. Infect Immun. 1993;61:5271–5278. doi: 10.1128/iai.61.12.5271-5278.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mantis NJ, McGuinness CR, Sonuyi O, Edwards G, Farrant SA. Immunoglobulin A antibodies against ricin A and B subunits protect epithelial cells from ricin intoxication. Infect Immun. 2006;74:3455–3462. doi: 10.1128/IAI.02088-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johansson M, Phillipson M, Petersson J, Velcich A, Holm L, Hansson G. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proc Natl Acad Sci USA. 2008;105:15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johansson M, et al. Bacteria penetrate the normally impenetrable inner colon mucus layer in both murine colitis models and patients with ulcerative colitis. Gut. 2014;63:281–291. doi: 10.1136/gutjnl-2012-303207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Swidsinski A, Weber J, Loening-Baucke V, Hale LP, Lochs H. Spatial organization and composition of the mucosal flora in patients with inflammatory bowel disease. J Clin Microbiol. 2005;43:3380–3389. doi: 10.1128/JCM.43.7.3380-3389.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336:1268–1273. doi: 10.1126/science.1223490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cash H, Whitham C, Behrendt C, Hooper L. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313:1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vaishnava S, et al. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science. 2011;334:255–258. doi: 10.1126/science.1209791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gewirtz AT, Navas TA, Lyons S, Godowski PJ, Madara JL. Cutting edge: bacterial flagellin activates basolaterally expressed TLR5 to induce epithelial proinflammatory gene expression. J Immunol. 2001;167:1882–1885. doi: 10.4049/jimmunol.167.4.1882. [DOI] [PubMed] [Google Scholar]
- 29.Smith KD, et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4:1247–1253. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 30.Feng T, Cong Y, Alexander K, Elson C. Regulation of Toll-like receptor 5 gene expression and function on mucosal dendritic cells. PloS One. 2012;7:e35918. doi: 10.1371/journal.pone.0035918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kinnebrew MA, et al. Interleukin 23 production by intestinal CD103(+)CD11b(+) dendritic cells in response to bacterial flagellin enhances mucosal innate immune defense. Immunity. 2012;36:276–287. doi: 10.1016/j.immuni.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Carvalho FA, et al. Interleukin-1beta (IL-1beta) promotes susceptibility of Toll-like receptor 5 (TLR5) deficient mice to colitis. Gut. 2012;61:373–384. doi: 10.1136/gut.2011.240556. [DOI] [PubMed] [Google Scholar]
- 33.Carvalho FA, et al. Transient inability to manage proteobacteria promotes chronic gut inflammation in TLR5-deficient mice. Cell Host Microbe. 2012;12:139–152. doi: 10.1016/j.chom.2012.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shi J, et al. A novel role for defensins in intestinal homeostasis: regulation of IL-1beta secretion. J Immunol. 2007;179:1245–1253. doi: 10.4049/jimmunol.179.2.1245. [DOI] [PubMed] [Google Scholar]
- 35.Wehkamp J, et al. NOD2 (CARD15) mutations in Crohn’s disease are associated with diminished mucosal alpha-defensin expression. Gut. 2004;53:1658–1664. doi: 10.1136/gut.2003.032805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cho JH. The genetics and immunopathogenesis of inflammatory bowel disease. Nat Rev Immunol. 2008;8:458–466. doi: 10.1038/nri2340. [DOI] [PubMed] [Google Scholar]
- 37.Benjamin JL, Sumpter R, Jr, Levine B, Hooper LV. Intestinal epithelial autophagy is essential for host defense against invasive bacteria. Cell Host Microbe. 2013;13:723–734. doi: 10.1016/j.chom.2013.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cooney R, et al. NOD2 stimulation induces autophagy in dendritic cells influencing bacterial handling and antigen presentation. Nat Med. 2010;16:90–97. doi: 10.1038/nm.2069. [DOI] [PubMed] [Google Scholar]
- 39.Travassos LH, Carneiro LA, Girardin S, Philpott DJ. Nod proteins link bacterial sensing and autophagy. Autophagy. 2010;6:409–411. doi: 10.4161/auto.6.3.11305. [DOI] [PubMed] [Google Scholar]
- 40.Travassos LH, et al. Nod1 and Nod2 direct autophagy by recruiting ATG16L1 to the plasma membrane at the site of bacterial entry. Nat Immunol. 2010;11:55–62. doi: 10.1038/ni.1823. [DOI] [PubMed] [Google Scholar]
- 41.Homer CR, Richmond AL, Rebert NA, Achkar JP, McDonald C. ATG16L1 and NOD2 interact in an autophagy-dependent antibacterial pathway implicated in Crohn’s disease pathogenesis. Gastroenterology. 2010;139:1630–1641. doi: 10.1053/j.gastro.2010.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cadwell K, et al. A key role for autophagy and the autophagy gene Atg16l1 in mouse and human intestinal Paneth cells. Nature. 2008;456:259–263. doi: 10.1038/nature07416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sorbara MT, et al. The protein ATG16L1 suppresses inflammatory cytokines induced by the intracellular sensors Nod1 and Nod2 in an autophagy-independent manner. Immunity. 2013;39:858–873. doi: 10.1016/j.immuni.2013.10.013. [DOI] [PubMed] [Google Scholar]
- 44.Elinav E, et al. NLRP6 inflammasome regulates colonic microbial ecology and risk for colitis. Cell. 2011;145:745–757. doi: 10.1016/j.cell.2011.04.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Franchi L, et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat Immunol. 2006;7:576–582. doi: 10.1038/ni1346. [DOI] [PubMed] [Google Scholar]
- 46.Sonnenberg G, Monticelli L, Elloso M, Fouser L, Artis D. CD4(+) lymphoid tissue-inducer cells promote innate immunity in the gut. Immunity. 2011;34:122–134. doi: 10.1016/j.immuni.2010.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sonnenberg G, Artis D. Innate lymphoid cell interactions with microbiota: implications for intestinal health and disease. Immunity. 2012;37:601–610. doi: 10.1016/j.immuni.2012.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hepworth M, et al. Innate lymphoid cells regulate CD4+ T-cell responses to intestinal commensal bacteria. Nature. 2013;498:113–117. doi: 10.1038/nature12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol. 1993;5:1461–1471. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 50.Powrie F, Leach MW, Mauze S, Menon S, Caddle LB, Coffman RL. Inhibition of Th1 responses prevents inflammatory bowel disease in scid mice reconstituted with CD45RBhi CD4+ T cells. Immunity. 1994;1:553–562. doi: 10.1016/1074-7613(94)90045-0. [DOI] [PubMed] [Google Scholar]
- 51.Feng T, Wang L, Schoeb T, Elson C, Cong Y. Microbiota innate stimulation is a prerequisite for T cell spontaneous proliferation and induction of experimental colitis. The J Exp Med. 2010;207:1321–1332. doi: 10.1084/jem.20092253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hapfelmeier S, et al. Reversible microbial colonization of germ-free mice reveals the dynamics of IgA immune responses. Science. 2010;328:1705–1709. doi: 10.1126/science.1188454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Macpherson A, Uhr T. Induction of protective IgA by intestinal dendritic cells carrying commensal bacteria. Science. 2004;303:1662–1665. doi: 10.1126/science.1091334. [DOI] [PubMed] [Google Scholar]
- 54.Macpherson AJ, Gatto D, Sainsbury E, Harriman GR, Hengartner H, Zinkernagel RM. A primitive T cell-independent mechanism of intestinal mucosal IgA responses to commensal bacteria. Science. 2000;288:2222–2226. doi: 10.1126/science.288.5474.2222. [DOI] [PubMed] [Google Scholar]
- 55.Konrad A, Cong Y, Duck W, Borlaza R, Elson C. Tight mucosal compartmentation of the murine immune response to antigens of the enteric microbiota. Gastroenterology. 2006;130:2050–2059. doi: 10.1053/j.gastro.2006.02.055. [DOI] [PubMed] [Google Scholar]
- 56.Fritz JH, et al. Acquisition of a multifunctional IgA+ plasma cell phenotype in the gut. Nature. 2012;481:199–203. doi: 10.1038/nature10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Elson C, et al. Monoclonal anti-interleukin 23 reverses active colitis in a T cell-mediated model in mice. Gastroenterology. 2007;132:2359–2370. doi: 10.1053/j.gastro.2007.03.104. [DOI] [PubMed] [Google Scholar]
- 58.Yen D, et al. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cong Y, Weaver CT, Lazenby A, Elson CO. Colitis induced by enteric bacterial antigen-specific CD4+ T cells requires CD40-CD40 ligand interactions for a sustained increase in mucosal IL-12. J Immunol. 2000;165:2173–2182. doi: 10.4049/jimmunol.165.4.2173. [DOI] [PubMed] [Google Scholar]
- 60.Lee Y, et al. Late developmental plasticity in the T helper 17 lineage. Immunity. 2009;30:92–107. doi: 10.1016/j.immuni.2008.11.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mangan P, et al. Transforming growth factor-beta induces development of the T(H)17 lineage. Nature. 2006;441:231–234. doi: 10.1038/nature04754. [DOI] [PubMed] [Google Scholar]
- 62.Harrington LE, et al. Interleukin 17-producing CD4+ effector T cells develop via a lineage distinct from the T helper type 1 and 2 lineages. Nat Immunol. 2005;6:1123–1132. doi: 10.1038/ni1254. [DOI] [PubMed] [Google Scholar]
- 63.Weaver CT, Hatton RD, Mangan PR, Harrington LE. IL-17 family cytokines and the expanding diversity of effector T cell lineages. Annu Rev Immunol. 2007;25:821–852. doi: 10.1146/annurev.immunol.25.022106.141557. [DOI] [PubMed] [Google Scholar]
- 64.Langrish CL, McKenzie BS, Wilson NJ, de Waal Malefyt R, Kastelein RA, Cua DJ. IL-12 and IL-23: master regulators of innate and adaptive immunity. Immunol Rev. 2004;202:96–105. doi: 10.1111/j.0105-2896.2004.00214.x. [DOI] [PubMed] [Google Scholar]
- 65.Hand T, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science. 2012;337:1553–1556. doi: 10.1126/science.1220961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garrett WS, et al. Communicable ulcerative colitis induced by T-bet deficiency in the innate immune system. Cell. 2007;131:33–45. doi: 10.1016/j.cell.2007.08.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Garrett WS, et al. Enterobacteriaceae act in concert with the gut microbiota to induce spontaneous and maternally transmitted colitis. Cell Host Microbe. 2010;8:292–300. doi: 10.1016/j.chom.2010.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291:881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 70.Talham GL, Jiang HQ, Bos NA, Cebra JJ. Segmented filamentous bacteria are potent stimuli of a physiologically normal state of the murine gut mucosal immune system. Infect Immunol. 1999;67:1992–2000. doi: 10.1128/iai.67.4.1992-2000.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Klaasen HL, Koopman JP, Poelma FG, Beynen AC. Intestinal, segmented, filamentous bacteria. FEMS Microbiol Rev. 1992;8:165–180. doi: 10.1111/j.1574-6968.1992.tb04986.x. [DOI] [PubMed] [Google Scholar]
- 72.Ivanov, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host Microbe. 2008;4:337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cao A, Yao S, Gong B, Elson C, Cong Y. Th17 cells upregulate polymeric Ig receptor and intestinal IgA and contribute to intestinal homeostasis. J Immunol. 2012;189:4666–4673. doi: 10.4049/jimmunol.1200955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Frank DN, St Amand AL, Feldman RA, Boedeker EC, Harpaz N, Pace NR. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc Natl Acad Sci USA. 2007;104:13780–13785. doi: 10.1073/pnas.0706625104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Atarashi K, et al. Treg induction by a rationally selected mixture of Clostridia strains from the human microbiota. Nature. 2013;500:232–236. doi: 10.1038/nature12331. [DOI] [PubMed] [Google Scholar]
- 76.Maslowski KM, et al. Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature. 2009;461:1282–1286. doi: 10.1038/nature08530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Smith PM, et al. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science. 2013;341:569–573. doi: 10.1126/science.1241165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Arpaia N, et al. Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature. 2013;504:451–455. doi: 10.1038/nature12726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Furusawa Y, et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature. 2013;504:446–450. doi: 10.1038/nature12721. [DOI] [PubMed] [Google Scholar]
- 80.Hayashi A, et al. A single strain of Clostridium butyricum induces intestinal IL-10-producing macrophages to suppress acute experimental colitis in mice. Cell Host Microbe. 2013;13:711–722. doi: 10.1016/j.chom.2013.05.013. [DOI] [PubMed] [Google Scholar]
- 81.Round JL, et al. The Toll-like receptor 2 pathway establishes colonization by a commensal of the human microbiota. Science. 2011;332:974–977. doi: 10.1126/science.1206095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mazmanian SK, Round JL, Kasper DL. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature. 2008;453:620–625. doi: 10.1038/nature07008. [DOI] [PubMed] [Google Scholar]
- 83.Round JL, Mazmanian SK. Inducible Foxp3+ regulatory T-cell development by a commensal bacterium of the intestinal microbiota. Proc Natl Acad Sci USA. 2010;107:12204–12209. doi: 10.1073/pnas.0909122107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hall JA, et al. Commensal DNA limits regulatory T cell conversion and is a natural adjuvant of intestinal immune responses. Immunity. 2008;29:637–649. doi: 10.1016/j.immuni.2008.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Izcue A, et al. Interleukin-23 restrains regulatory T cell activity to drive T cell-dependent colitis. Immunity. 2008;28:559–570. doi: 10.1016/j.immuni.2008.02.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Mokry M, et al. Many inflammatory bowel disease risk loci include regions that regulate gene expression in immune cells and the intestinal epithelium. Gastroenterology. 2013 doi: 10.1053/j.gastro.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 87.Franke A, et al. Genome-wide meta-analysis increases to 71 the number of confirmed Crohn’s disease susceptibility loci. Nat Genet. 2010;42:1118–1125. doi: 10.1038/ng.717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Franke A, et al. Genome-wide association study for ulcerative colitis identifies risk loci at 7q22 and 22q13 (IL17REL) Nat Genet. 2010;42:292–294. doi: 10.1038/ng.553. [DOI] [PubMed] [Google Scholar]
- 89.Anderson CA, et al. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nat Genet. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Barrett JC, et al. Genome-wide association defines more than 30 distinct susceptibility loci for Crohn’s disease. Nat Genet. 2008;40:955–962. doi: 10.1038/NG.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Parham C, et al. A receptor for the heterodimeric cytokine IL-23 is composed of IL-12Rbeta1 and a novel cytokine receptor subunit, IL-23R. J Immunol. 2002;168:5699–5708. doi: 10.4049/jimmunol.168.11.5699. [DOI] [PubMed] [Google Scholar]
- 92.Kan SH, Mancini G, Gallagher G. Identification and characterization of multiple splice forms of the human interleukin-23 receptor alpha chain in mitogen-activated leukocytes. Genes Immun. 2008;9:631–639. doi: 10.1038/gene.2008.64. [DOI] [PubMed] [Google Scholar]
- 93.Duerr RH, et al. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Momozawa Y, et al. Resequencing of positional candidates identifies low frequency IL23R coding variants protecting against inflammatory bowel disease. Nat Genet. 2011;43:43–47. doi: 10.1038/ng.733. [DOI] [PubMed] [Google Scholar]
- 95.Girardin SE, et al. Nod2 is a general sensor of peptidoglycan through muramyl dipeptide (MDP) detection. J Biol Chem. 2003;278:8869–8872. doi: 10.1074/jbc.C200651200. [DOI] [PubMed] [Google Scholar]
- 96.Girardin SE, et al. Peptidoglycan molecular requirements allowing detection by Nod1 and Nod2. J Biol Chem. 2003;278:41702–41708. doi: 10.1074/jbc.M307198200. [DOI] [PubMed] [Google Scholar]
- 97.Abraham C, Cho JH. Functional consequences of NOD2 (CARD15) mutations. Inflamm Bowel Dis. 2006;12:641–650. doi: 10.1097/01.MIB.0000225332.83861.5f. [DOI] [PubMed] [Google Scholar]
- 98.Devlin SM, et al. NOD2 variants and antibody response to microbial antigens in Crohn’s disease patients and their unaffected relatives. Gastroenterology. 2007;132:576–586. doi: 10.1053/j.gastro.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 99.Shih D, et al. Microbial induction of inflammatory bowel disease associated gene TL1A (TNFSF15) in antigen presenting cells. Eur J Immunol. 2009;39:3239–3250. doi: 10.1002/eji.200839087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bamias G, et al. Expression, localization, and functional activity of TL1A, a novel Th1-polarizing cytokine in inflammatory bowel disease. J Immunol. 2003;171:4868–4874. doi: 10.4049/jimmunol.171.9.4868. [DOI] [PubMed] [Google Scholar]
- 101.Prehn JL, et al. Potential role for TL1A, the new TNF-family member and potent costimulator of IFN-gamma, in mucosal inflammation. Clin Immunol. 2004;112:66–77. doi: 10.1016/j.clim.2004.02.007. [DOI] [PubMed] [Google Scholar]
- 102.Podolsky DK. Inflammatory bowel disease. N Engl J Med. 2002;347:417–429. doi: 10.1056/NEJMra020831. [DOI] [PubMed] [Google Scholar]
- 103.Duck L, et al. Isolation of flagellated bacteria implicated in Crohn’s disease. Inflamm Bowel Dis. 2007;13:1191–1201. doi: 10.1002/ibd.20237. [DOI] [PubMed] [Google Scholar]
- 104.Landers CJ, et al. Selected loss of tolerance evidenced by Crohn’s disease-associated immune responses to auto-and microbial antigens. Gastroenterol. 2002;123:689–699. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- 105.Shen C, Landers C, Derkowski C, Elson C, Targan S. Enhanced CBir1-specific innate and adaptive immune responses in Crohn’s disease. Inflamm Bowel Dis. 2008;14:1641–1651. doi: 10.1002/ibd.20645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Farmer M, et al. A major quantitative trait locus on chromosome 3 controls colitis severity in IL-10-deficient mice. Proc Natl Acad Sci. 2001;98:13820–13825. doi: 10.1073/pnas.241258698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Beckwith J, Cong Y, Sundberg JP, Elson CO, Leiter EH. Cdcs1, a major colitogenic locus in mice, regulates innate and adaptive immune response to enteric bacterial antigens. Gastroenterology. 2005;129:1473–1484. doi: 10.1053/j.gastro.2005.07.057. [DOI] [PubMed] [Google Scholar]
- 108.Takedatsu H, et al. Linkage of Crohn’s disease-related serological phenotypes: NFKB1 haplotypes are associated with anti-CBir1 and ASCA, and show reduced NF-kappaB activation. Gut. 2009;58:60–67. doi: 10.1136/gut.2008.156422. [DOI] [PMC free article] [PubMed] [Google Scholar]