Abstract
PNPLA3 was recently associated with the susceptibility to nonalcoholic fatty liver disease, a common cause of chronic liver disease characterized by abnormal triglyceride accumulation. Although it is established that PNPLA3 has both triacylglycerol lipase and acylglycerol O-acyltransferase activities, is still unknown whether the gene has any additional role in the modulation of the human liver metabolome. To uncover the functional role of PNPLA3 on liver metabolism, we performed high-throughput metabolic profiling of PNPLA3 siRNA-silencing and overexpression of wild-type and mutant Ile148Met variants (isoleucine/methionine substitution at codon 148) in Huh-7 cells. Metabolomic analysis was performed by using GC/MS and LC/MS platforms. Silencing of PNPLA3 was associated with a global perturbation of Huh-7 hepatoma cells that resembled a catabolic response associated with protein breakdown. A significant decrease in amino- and γ-glutamyl-amino acids and dipeptides and a significant increase in cysteine sulfinic acid, myo-inositol, lysolipids, sphingolipids, and polyunsaturated fatty acids were observed. Overexpression of the PNPLA3 Met148 variant mirrored many of the metabolic changes observed during gene silencing, but in the opposite direction. These findings were replicated by the exploration of canonical pathways associated with PNPLA3 silencing and Met148 overexpression. Overexpression of the PNPLA3 Met148 variant was associated with a 1.75-fold increase in lactic acid, suggesting a shift to anaerobic metabolism and mitochondrial dysfunction. Together, these results suggest a critical role of PNPLA3 in the modulation of liver metabolism beyond its classical participation in triacylglycerol remodeling.
Keywords: PNPLA3, metabolome, nonalcoholic fatty liver disease, Huh-7 cells, Ile148Met variant, systems biology
the protein encoded by the patatin-like phospholipase domain containing 3 (PNPLA3) gene, also known as adiponutrin, is a multifunctional enzyme that belongs to the IPLA2/lipase (calcium-independent phospholipase A2-epsilon) family, which has both triacylglycerol lipase and acylglycerol O-acyltransferase activities (7).
A common feature of all IPLA2/lipase family enzymes is that they are hormone-sensitive lipases tightly influenced by nutritional needs. In fact, PNPLA3 is highly induced by changes in energy balance (12, 15). Recently, a common nonsynonymous gene variant, the rs738409 C/G encoding an isoleucine to methionine substitution at the amino acid position 148 (I148M), has been associated with the susceptibility to nonalcoholic fatty liver disease (NAFLD) (17), a finding that was widely replicated around the world (21). In addition, the G allele (methionine at position 148, Met148) was significantly associated with the histological disease severity and disease progression (21). Of note, the PNPLA3-Met148 allele does not associate with the body mass index, insulin resistance, or dyslipidemia, the traditional risk factors associated with hepatic steatosis and nonalcoholic steatohepatitis (NASH) (26). Although the clinical and functional impact of Met148 has been extensively studied (4, 6, 8), the focus of these studies have been its role in lipid mobilization and remodeling; it is thus still unknown whether PNPLA3 has a role in the modulation of the global metabolome of the human liver beyond its classical role in lipid remodeling.
Metabolomics allows an unbiased assessment of the global metabolome. To gain additional insights on the metabolic systemwide effects of the PNPLA3 gene, an unbiased metabolomics approach was taken to define the effects of gain of function (overexpression) of wild-type and mutant PNPLA3 as well as loss of function induced by siRNA-silencing of the mutant PNPLA3 gene on the metabolome in Huh-7 cells. Finally, to identify pathways contributing to the observed metabolic derangements, we performed a systems biology exploration by functional enrichment analysis.
MATERIALS AND METHODS
Cell Culture Experiments
Our study design was composed of PNPLA3 silencing and overexpression of the mutant Met148 and wild-type Ile148 in Huh-7 cells. PNPLA3 was silenced in Huh-7 cells by siRNA, with scrambled RNA as a negative control (n = 6 for each). Mutant and wild-type Ile148Met were overexpressed with an empty vector as a control (n = 6 for each) as described below.
PNPLA3 silencing.
Huh-7 cells (2 × 106) were seeded in 100-mm-diameter cell culture dishes and then grown to 50–70% confluence in high-glucose Dulbecco's modified Eagle's medium (Life Technologies) supplemented with 2 mM l-glutamine, 100 U of penicillin/ml, 100 μg of streptomycin/ml (DMEM), and 10% fetal bovine serum. Cells were washed with PBS and 10 ml culture medium were added to each dish and then transfected with a validated siRNA (Santa Cruz Biotechnology), or a negative siRNA control using Lipofectamine transfection reagent (Invitrogen) according to the manufacturer's instructions. The FAM-labeled negative siRNA was used to monitor transfection efficiency by fluorescence microscopy. The optimal concentration of validated PNPLA3 siRNA for transfection was 40 nM giving a transfection efficiency of more than 95%. Cells were incubated for 2 days and then washed three times with PBS (pH 7.4) and then treated accordingly to further experiments.
For detecting silencing efficiency, cells were treated with TRIzol reagent (Invitrogen) to purify total RNA, which was treated with RNase-free DNase to remove genomic DNA. cDNA was synthesized from 4 μg of total RNA by using Moloney murine leukemia virus reverse transcriptase and oligo(dT) primers and subjected to PCR by the manufacturer's instructions. The silencing efficiency was confirmed by real-time PCR using specific PNPLA3 primers (forward: CGT GCG ATA TGT GGA TGG, reverse: CGT GGA CTT GAC TTT AGG G). The quantitative RT-PCR was performed with SYBR Green PCR Master Mix (Bio-Rad, Hercules, CA) on an Bio-Rad C1000 thermal cycler Sequence Detection System as follows: 50°C for 2 min and then at 95°C for 10 min followed by 40 cycles of amplification (95°C for 15 s; 60°C for 30 s; 80°C for 30 s). Glyceraldehyde-3-phosphate dehydrogenase was used as control for loading. All primers were prepared by the Nucleic Acid Core facility at Virginia Commonwealth University.
Ile148Met variant analysis.
To better understand the Ile148Met-PNPLA3 genotype of the cell line used in this experiment, we sequenced the PNPLA3 gene in Huh-7 (hepatocellular carcinoma cells) by using PNPLA3 specific primers; in addition, we also sequenced the HepG2 cell line. DNA sequencing revealed that both the HepG2 and Huh-7 cell lines carry the PNPLA3 Met148 mutation.
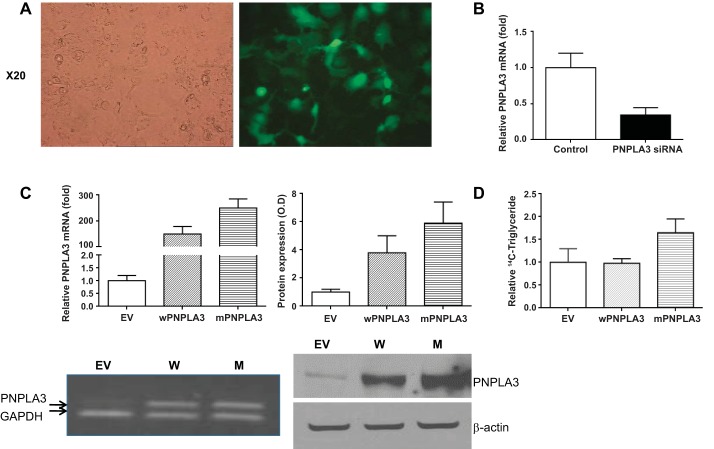
For transfection experiments, Huh7 cell line was grown in DMEM and 10% fetal bovine serum. Cells were cultured at 37°C in an incubator maintained at 6% CO2. Met148 (mutant PNPLA3) was prepared from wild-type PNPLA3 (isoleucine at location 148, Ile148). cDNA was inserted into the pCMV6-XL4 vector (Origene), which was mutated to convert Ile148 to Met148 by using the site-directed mutagenesis protocol recommended by the manufacturer (QuickChange Site-Directed Mutagenesis Kit) by changing the +148 codon from ATC to ATG. Cells were transfected by using Lipofectamine transfection reagent (Invitrogen) according to the manufacturer's instructions and were incubated for 2 days. Cells were washed three times with PBS pH 7.4 and then the pellet was frozen and stored −80°C for further experiments. To determine the transfection efficiency, Huh-7 cells were also transfected with pcDNA-GFP (green fluorescent protein) plasmid DNA, and GFP-positive cells and total cells were counted from five different random areas for each experiment; transfection efficiency was ∼70–80% (Fig. 1A).
Fig. 1.
A: exploration of transfection efficiency: Huh-7 cells transfected with pcDNA-GFP (green fluorescent protein) plasmid DNA. B: PNPLA3 mRNA abundance in siRNA-transfected cells. It demonstrates an ∼65% decrease in PNPLA3 mRNA levels. C: abundance of mRNA and protein levels for both Met148 (mPNPLA3) and Ile148 (wPNPLA3). EV, empty vector. RNA was measured by quantitative real-time PCR, and proteins were identified by Western blotting with visualization using Dura substrate. D. Triglyceride accumulation assay. Cells were exposed to [14C]palmitate for 48 h and then triglyceride fraction in cells transfected with Met148 PNPLA3 was compared with controls exposed to empty vector and cells transfected with Ile148 PNPLA3, indicating that the mutant PNPLA3 was associated with lower triglyceride lipase activity and/or an increase in acyltransferase activity.
For the hydrolysis of triglycerides, Huh-7 cells were cultured and transfected as above with empty vector, wild-type PNPLA3, or Ile148Met PNPLA3. Cells were treated with 1 μCi [14C]palmitate for 4 h, the medium was changed to DMEM plus 10% FCS, and then the cells were harvested after 48 h. The lipid extraction was performed and quantitated by the method of He et al. (4).
High-Throughput Metabolomic Profiling
Gas chromatography (GC)/mass spectometry (MS) and liquid chromatography (LC)/MS/MS platforms were used to generate metabolic profiles by previously published and validated methods (4). All samples were processed by Metabolon (Research Triangle Park, NC) with the methodological details summarized as follows, with more detailed information in the appendix.
Sample preparation.
After cell disruption, total protein amount was measured in the pellet after centrifugation of cell lysate samples. Upon receipt of cell lysate supernatants, aliquots were immediately stored at −80°C until time of analysis. At the time of analysis samples were extracted and prepared for analysis by Metabolon's standard solvent extraction method as previously published (4). The extracted samples were split into equal parts for analysis on the GC/MS and LC/MS/MS platforms. Also included were several technical replicate samples created from a homogeneous pool containing a small amount of all study samples (“Client Matrix”).
LC/MS, LC/MS2.
The LC/MS portion of the platform was based on a Waters ACQUITY UPLC and a Thermo-Finnigan LTQ mass spectrometer, which consisted of an electrospray ionization (ESI) source and linear ion-trap (LIT) mass analyzer. The sample extract was split into two aliquots, dried, then reconstituted in acidic or basic LC-compatible solvents, each of which contained 11 or more injection standards at fixed concentrations. One aliquot was analyzed under acidic positive ion optimized conditions and the other under basic negative ion optimized conditions in two independent injections using separate dedicated columns. Extracts reconstituted in acidic conditions were gradient eluted with water and methanol both containing 0.1% formic acid, whereas the basic extracts, which also used water/methanol, contained 6.5 mM ammonium bicarbonate. The MS analysis alternated between MS and data-dependent MS2 scans using dynamic exclusion (4).
GC/MS analysis.
Samples destined for GC/MS analysis were redried under vacuum desiccation for a minimum of 24 h prior to being derivatized under dried nitrogen with use of bistrimethyl-silyl-triflouroacetamide. The GC column was 5% phenyl and the temperature ramp is from 40° to 300°C in a 16-min period. Samples were analyzed on a Thermo-Finnigan Trace DSQ fast-scanning single-quadrupole mass spectrometer using electron impact ionization. The instrument was tuned and calibrated for mass resolution and mass accuracy on a daily basis.
Metabolite identification.
Compounds were identified by comparison to library entries of purified standards or recurrent unknown entities. As of this writing, more than 1,000 commercially available purified standard compounds had been acquired registered into Laboratory Information Management System for distribution to both the LC and GC platforms for determination of their analytical characteristics. The combination of chromatographic properties and mass spectra gave an indication of a match to the specific compound or an isobaric entity. Additional entities could be identified by virtue of their recurrent nature (both chromatographic and mass spectral). These compounds have the potential to be identified by future acquisition of a matching purified standard or by classical structural analysis.
Data normalization.
Samples were analyzed over the course of 2 wk. Each run day was balanced for different set of samples. Within each run day, samples were completely randomized to avoid group block effects. The raw area counts for each metabolite in each sample were normalized to correct for variation resulting from instrument interday tuning differences. For each metabolite, the raw area counts were divided by its median value for each run day, therefore setting the medians equal to 1 for each day's run. This correctly preserves all variation between samples yet allows metabolites of similar graphical scale. Missing values were assumed to result from areas falling below limits of detection. For each metabolite, missing values were imputed with its observed minimum after the normalization step.
Statistical Analysis
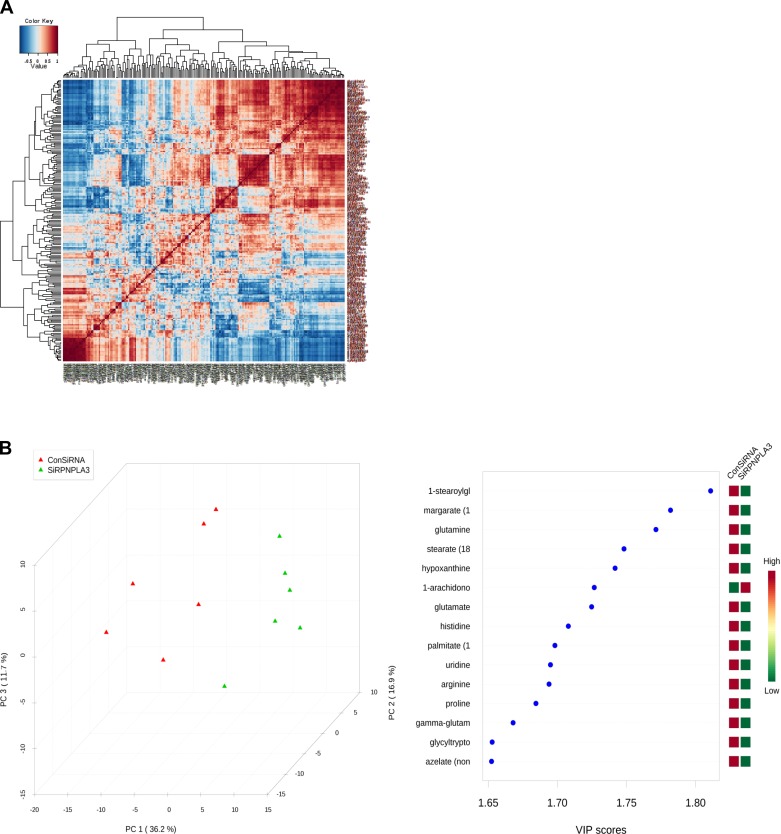
For metabolomics analysis, a variety of univariate analysis (fold change, two-sample t-test, or ANOVA were used with Tukey post hoc tests for pairwise comparisons between groups after log-transformation for normalization of variables, volcano plots, or the nonparametric Wilcoxon test) and multivariate analysis (partial least squares-discriminant analysis, PLS-DA) were performed by using MetaboAnalyst 2.0, a web-based analytical pipeline for high-throughput metabolomics studies (18). For the PNPLA3 knockdown experiment, 233 metabolites were analyzed by using peak intensities normalized by milligrams of protein. False discovery rate (FDR) was used for controlling multiple testing; nevertheless, because the majority of variables are highly correlated among them, within but also between well-defined biological groups (amino acids, urea cycle metabolites, etc.) (Fig. 2A), this correction was very conservative.
Fig. 2.
A: PNPLA3 siRNA silencing in Huh-7 cells in vitro: overall correlation pattern among metabolites, with color intensity representing Spearman rank correlation coefficient. B: partial least squares discriminant analysis score (PLS-DA) plot. Left: PLS-DA was performed to identify compounds that contribute most to the separation between groups (PNPLA3 siRNA and negative control). Tridimensional plot of the 3 first principal components shows that the complete separation in clusters of the samples classified in the correct groups. Right: VIP, variable importance in prediction.
Functional Enrichment Analysis
To understand the biological meaning of the observed metabolic changes, we performed a functional enrichment analysis of the experimental data by MetaboAnalyst 2.0, which performs metabolite set enrichment analysis (MSEA) for human and mammalian species. The analysis is based on several libraries containing ∼6,300 groups of biologically meaningful metabolite sets collected primarily from human studies (18).
RESULTS
The Metabolic Signature of In Vitro Liver PNPLA3 siRNA-Silencing Is Associated with Protein Breakdown and an Osmotic Stress Response
Our first approach was to measure the concentrations of the whole metabolite repertory of the liver cells after siRNA silencing of the PNPLA3 gene; siRNA transfection efficiency was high with more than 65% decrease in PNPLA3 mRNA abundance (Fig. 1B). We identified 76 of 233 metabolites that were differentially down- or upregulated in the Huh-7 cells after PNPLA3 knockdown with a FDR lower than 0.15 (Table 1). We observed a dramatic and significant decrease in glutamine-among other amino acids- and dipeptides and γ-glutamyl-amino acids, including γ-glutamyl-glutamine, γ-glutamyl-leucine and γ-glutamyl-isoleucine (Table 1). Interestingly, PLS-DA shows that the first three components perfectly discriminate between samples (Fig. 2B), and glutamine and glutamate are third and seventh in variable importance of prediction. Five of the top 15 such variables are amino acids.
Table 1.
The principal effects of siRNA-PNPLA3 silencing on the global metabolome of Huh-7 hepatoma cells
| Main Pathway | Subpathway | Metabolites | Fold Change | Nominal P Value | Empirical P Value (FDR) |
|---|---|---|---|---|---|
| Amino acid metabolism | Alanine and aspartate metabolism | Asparagine | 0.4 | 0.0008 | 0.01 |
| Alanine | 0.5 | 0.0001 | 0.02 | ||
| α-Amino Acids and Derivatives | γ-Glutamyl-glutamine | 0.4 | 0.0006 | 0.01 | |
| γ-Glutamyl-leucine | 0.6 | 0.004 | 0.03 | ||
| γ-Glutamyl-isoleucine | 0.6 | 0.02 | NS | ||
| γ-Glutamyl-threonine | 0.6 | 0.04 | NS | ||
| Putrescine | 0.5 | 0.04 | NS | ||
| 5-oxoproline | 0.7 | 0.04 | NS | ||
| Cysteine, methionine, SAM, taurine metabolism | Methionine | 0.5 | 0.007 | 0.04 | |
| Cysteine sulfinic acid | 2.5 | 0.007 | 0.04 | ||
| N-acetylmethionine | 0.6 | 0.009 | 0.04 | ||
| Serine | 0.7 | 0.05 | NS | ||
| Dipeptides | Glycyl-tryptophan | 0.1 | 0.0007 | 0.01 | |
| Glycyl-phenylalanine | 0.3 | 0.0007 | 0.01 | ||
| Alanyl-threonine | 0.3 | 0.002 | 0.02 | ||
| Glycyl-tyrosine | 0.3 | 0.003 | 0.03 | ||
| Aspartyl-phenylalanine | 0.5 | 0.005 | 0.03 | ||
| Glycyl-isoleucine | 0.3 | 0.007 | 0.04 | ||
| Glycyl-valine | 0.3 | 0.01 | 0.05 | ||
| Glycyl-leucine | 0.4 | 0.02 | NS | ||
| Alanyl-valine | 0.3 | 0.04 | NS | ||
| Glutamate metabolism | Glutamine | 0.3 | 0.00008 | 0.006 | |
| Glutamate | 0.5 | 0.0002 | 0.007 | ||
| N-acetylglutamate | 1.2 | 0.02 | NS | ||
| Histidine metabolism | Histidine | 0.5 | 0.0003 | 0.007 | |
| Lysine metabolism | Lysine | 0.4 | 0.002 | 0.02 | |
| 2-Aminoadipate | 2.0 | 0.002 | 0.02 | ||
| Other amino acids | Proline | 0.7 | 0.0004 | 0.008 | |
| Leucine | 0.5 | 0.003 | 0.03 | ||
| Isoleucine | 0.6 | 0.009 | 0.04 | ||
| Valine | 0.6 | 0.01 | 0.05 | ||
| Phenylalanine metabolism | Tyrosine | 0.6 | 0.005 | 0.03 | |
| Phenylalanine | 0.6 | 0.005 | 0.03 | ||
| Tryptophan metabolism | Kynurenine | 0.6 | 0.0009 | 0.01 | |
| Tryptophan | 0.5 | 0.004 | 0.03 | ||
| Urea cycle and Arginine metabolism | Arginine | 0.4 | 0.0004 | 0.007 | |
| Assymetricdimethylarginine (ADMA) | 0.6 | 0.03 | NS | ||
| Carbohydrate metabolism | Glycolysis, gluconeogenesis, pyruvate metabolism | Isobar: fructose 1,6-diphosphate, glucose 1,6-diphosphate | 0.4 | 0.005 | 0.03 |
| Sedoheptulose-7-phosphate | 1.8 | 0.007 | 0.04 | ||
| Lactate | 0.7 | 0.01 | 0.05 | ||
| Inositol metabolism | Myo-inositol | 2.1 | 0.003 | 0.02 | |
| Nucleotide, sugars, pentose metabolism | Hypoxanthine | 0.9 | 0.0001 | 0.007 | |
| Uridine | 0.6 | 0.0003 | 0.007 | ||
| Cytidine | 0.5 | 0.006 | 0.04 | ||
| Guanine | 0.6 | 0.02 | NS | ||
| Thymidine 5¢-monophosphate | 1.5 | 0.03 | NS | ||
| Ribose 5-phosphate | 0.5 | 0.03 | NS | ||
| Flavin adenine dinucleotide (FAD) | 1.2 | 0.05 | NS | ||
| Vitamins | Pantothenate | 0.8 | 0.006 | 0.04 | |
| Lipid metabolism | Bile acid metabolism | Taurolithocholate | 0.5 | 0.003 | 0.03 |
| Cholesterol | 0.7 | 0.006 | 0.03 | ||
| Choline metabolism | Choline | 0.7 | 0.002 | 0.02 | |
| Choline phosphate | 0.7 | 0.04 | NS | ||
| Free fatty acids | Margarate (17:0) | 0.8 | 0.00006 | 0.006 | |
| Stearate (18:0) | 0.8 | 0.0001 | 0.007 | ||
| Palmitate (16:0) | 0.8 | 0.0003 | 0.007 | ||
| Azelate (nonanedioate) | 0.6 | 0.0007 | 0.01 | ||
| Docosahexaenoate (DHA; 22:6n3) | 0.8 | 0.001 | 0.01 | ||
| Docosatrienoate (22:3n3) | 1.3 | 0.004 | 0.03 | ||
| Laurate (12:0) | 0.7 | 0.01 | 0.05 | ||
| Eicosapentaenoate (EPA; 20:5n3) | 0.8 | 0.01 | 0.05 | ||
| Glycerol derivatives | Glycerophosphorylcholine (GPC) | 0.5 | 0.006 | 0.03 | |
| Glycerol 3-phosphate (G3P) | 0.8 | 0.010 | 0.05 | ||
| Lysolipids | 1-Stearoyl-glycero-phosphoinositol | 0.6 | 0.00003 | 0.006 | |
| 1-Arachidonoyl-glycero-phosphoethanolamine | 2.3 | 0.0002 | 0.007 | ||
| 1-Oleoyl-glycero-phosphoinositol | 0.7 | 0.001 | 0.01 | ||
| 1-Palmitoyl-glycero-phosphoinositol | 0.6 | 0.003 | 0.03 | ||
| 2-Palmitoyl-glycero-phosphoethanolamine | 3.4 | 0.02 | NS | ||
| 2-Arachidonoyl-glycero-phosphoinositol | 2.0 | 0.02 | NS | ||
| 1-Arachidonoyl-glycero-phosphoinositol | 1.5 | 0.02 | NS | ||
| 2-Linoleoyl-glycero-phosphocholine | 4.2 | 0.03 | NS | ||
| 1-Linoleoyl-glycero-phosphocholine | 3.8 | 0.04 | NS | ||
| 2-Oleoyl-glycero-phosphoethanolamine | 1.6 | 0.04 | NS | ||
| Sphingolipids | Sphinganine | 3.0 | 0.02 | NS |
Using LC/MS and GC/MS analysis, 234 distinct metabolites in the sample matched a named structure in the metabolomic reference library but one was filtered out because of lack of informative variation. Fold change refers to the ratio of the metabolite level between siRNA against PNPLA3-treated cells vs. control cells. After log transformation of the variables with equal variances, unpaired 2-sample t-test was used for comparing silencing vs. control group (nominal P value); similar results were obtained by a nonparametric test. FDR, false discovery rate; NS, not significant; SAM, S-adenosyl-methionine.
PNPLA3 silencing was also associated with a significant decrease in taurolithocholic acid, suggesting a role in the Huh-7 cells in vitro transformation of this secondary bile acid.
In contrast, we observed a significant 2.5-fold increase in cysteine sulfinic acid, which is derived from direct addition of oxygen to cysteine; cysteine oxidation is involved in the modulation of oxidative stress response and in the maintenance of cellular redox homeostasis (14, 19). Furthermore, a significant twofold increase in the polyol myo-inositol was also observed; rise in osmolytes, such as myo-inositol, is a regulatory mechanism to adjust cell volume to functional demands in several pathophysiological disorders, including liver insufficiency, diabetic ketoacidosis, and hypercatabolism (9).
Finally, PNPLA3 silencing was associated with a significant increase of lysolipids [glycerophospholipids primarily but not restricted to those esterified at its carbon 2 (sn-2) position], sphingolipids such as sphinganine, and polyunsaturated fatty acids, such as docosatrienoic acid (docosatrienoate, 22:3n3) (Table 1).
Overexpression of the PNPLA3 Met148 Variant Is Associated with Metabolic Changes in Huh-7 Cells in the Opposite Direction to Gene Silencing and Is Associated with a Shift to Anaerobic Metabolism
To test the specificity of metabolic changes associated with gene silencing, we explored the opposite scenario, a gain of protein function, by transfecting with a plasmid that overexpressed the endogenous mutant protein Met148. Hence, we explored the metabolic profile of the 76 significantly gene-silencing associated metabolites. In addition, we compared the results after transfecting cells with an expression vector carrying the coding sequence for the native Ile148 PNPLA3 protein. Abundance of mRNA and protein levels for both Met148 and Ile148 PNPLA3 were increased by more than 200-fold and 4-fold, respectively (Fig. 1C). To further confirm that there were functional differences between cells overexpressing the Ile148 vs. Met148 PNPLA3 variants, the triglyceride lipase activity was assessed by the triglyceride accumulation assay following a 4 h incubation with [14C]palmitate. The medium was changed to DMEM plus 10% FCS, and then the cells were harvested after 48 h. This demonstrated significantly greater labeled triglyceride accumulation in cells transfected with the Met148 compared with those transfected with Ile148 or exposed to an empty vector (Fig. 1D).
Results of cells transfected with Met148 are shown in Table 2. Of note, highly significant changes in metabolites associated with amino acid metabolism were observed, but in the opposite direction of PNPLA3 silencing, since there was a significant increase of dipeptides, such as glycyl-isoleucine and γ-glutamyl-leucine. In addition to a significant 1.2-fold increase of cholesterol levels and decrease in docosatrienoate, transfected cells with Met148 also showed a significant 1.75-fold increase in lactate levels (Table 2). On the other hand, metabolic profile of cells transfected with Ile148 (Table 2) showed a significant increase in γ-glutamyl-leucine, along with an increase in N-acetyl-glutamate and cholesterol that remained nonsignificant after multiple adjustments.
Table 2.
Effects of overexpression of PNPLA3 with methionine (M148) or isoleucine (I148) at position 148 on the Huh-7 cells metabolome
| Main Pathway | Subpathway/Metabolite | Fold Change | Nominal P Value | Empirical P Value (FDR) |
|---|---|---|---|---|
| Overexpression of PNPLA3 with methionine at position 148 | ||||
| Amino acid metabolism | Tryptophan metabolism | |||
| Tryptophan | 0.8 | 0.002 | 0.06 | |
| Urea cycle | ||||
| Assymetricdimethylarginine (ADMA) | 0.6 | 0.009 | 0.05 | |
| Cysteine, methionine, SAM, taurine metabolism | ||||
| Methionine | 0.8 | 0.009 | 0.05 | |
| Valine, leucine, and isoleucine metabolism | ||||
| Valine | 0.8 | 0.009 | 0.06 | |
| Phenylalanine and tyrosine metabolism | ||||
| Phenylalanine | 0.8 | 0.009 | 0.06 | |
| Dipeptides | ||||
| Glycyl-isoleucine | 1.7 | 0.02 | 0.04 | |
| α-Amino Acids and Derivatives | ||||
| γ-Glutamyl-leucine | 1.6 | 0.03 | NS | |
| Lipid metabolism | Long-chain fatty acid | |||
| Docosatrienoate (22:3n3) | 0.8 | 0.002 | 0.02 | |
| Sterol/Steroid | ||||
| Cholesterol | 1.2 | 0.03 | NS | |
| Carbohydrate metabolism | Glycolysis, gluconeogenesis, pyruvate metabolism | |||
| Lactate | 1.8 | 0.002 | 0.02 | |
| Overexpression of PNPLA3 with isoleucine at position 148 | ||||
| Amino acid metabolism | α-Amino Acids and Derivatives | |||
| γ-Glutamyl-leucine | 1.9 | 0.006 | 0.007 | |
| Glutamate metabolism | ||||
| N-acetylglutamate | 1.3 | 0.06 | NS | |
| Lipid metabolism | Long-chain fatty acid | |||
| Docosatrienoate (22:3n3) | 0.8 | 0.02 | NS | |
| Sterol/Steroid | ||||
| Cholesterol | 1.3 | 0.03 | NS | |
| Nucleotides | Pyrimidine, cytidine metabolism | |||
| Cytidine | 0.8 | 0.02 | NS | |
Fold change refers to the ratio of the metabolite levels between M148 or Ile148 PNPLA3 expression vector-treated cells vs. empty vector- treated control cells, respectively. Nominal P value stands for test T after log transformation of variables with equal variances. Similar results were obtained by a nonparametric test.
Systems Biology Exploration by Functional Enrichment Analysis Shows That PNPLA3 Silencing and Met148 Modify Amino Acid Metabolic
To explore whether the set of significantly associated metabolites were enriched with a particular biological pathway we performed a metabolite set enrichment analysis. Thus the list of metabolites shown in Tables 1 and 2 was contrasted with a library that contains 88 metabolite sets based on normal metabolic pathways (27); enrichment analysis was performed independently for each experimental condition.
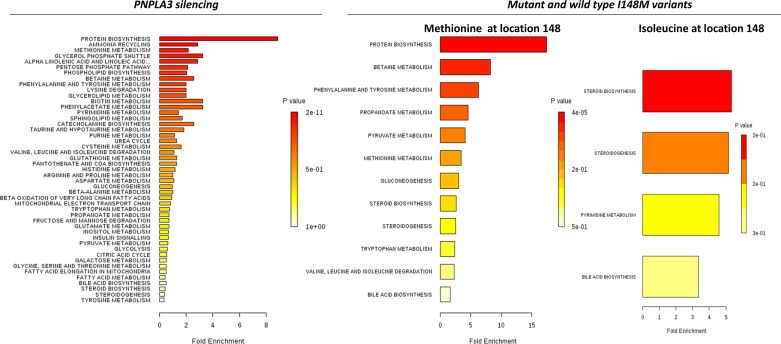
Notably, there was a remarkable overlapping between the pathway-associated metabolite sets of PNPLA3 silencing and Met148 (Fig. 3); protein biosynthesis, betaine metabolism (betaine is closely related with choline and S-adenosyl methionine and both of these compounds function as methyl donors), phenylalanine, and tyrosine metabolism, among others, were significantly associated. On the contrary, enrichment analysis of terms associated with Ile148 was restricted to few biological pathways, including steroid, cholesterol, and bile acids biosynthesis (Fig. 3).
Fig. 3.
Metabolite set enrichment analysis of PNPLA3 silencing and the PNPLA3 Ile148Met variant overexpression. Figure shows a graphical representation of the pathway-associated metabolite sets by enrichment analysis in each experimental condition (PNPLA3 siRNA silencing and overexpression of PNPLA3 with Met148 or Ile148).
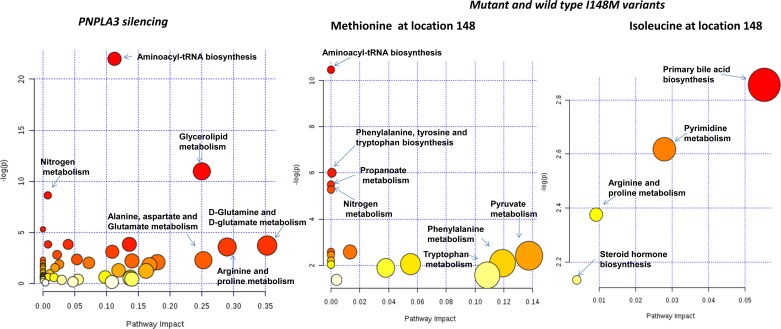
To gain further insight into the underlying biological mechanisms associated with PNPLA3 silencing, we performed overrepresentation analysis; the statistical test performed was hypergeometric distribution and the P value represented the enrichment of certain metabolites in a pathway (P value ≤0.05 is indicative of significant enrichment). Interestingly, pathway impact values from pathway topology analysis showed that 10 canonical pathways were significantly perturbed during PNPLA3 silencing (Fig. 4 and Table 3); the two top ones were aminoacyl-tRNA biosynthesis and glycerophospholipid metabolism and most of the remaining ones were related with amino acid metabolism (Fig. 4 and Table 3). Notably, three of four of the significantly perturbed canonical pathways during Met148 overexpression were overlapped with the ones perturbed during gene silencing (Fig. 4 and Table 3).
Fig. 4.
Identification of perturbed pathways by overrepresentation analysis (ORA) in the list of significantly altered metabolites. The analysis was done by using a pathway library restricted to Homo sapiens, and P values for ORA stand for hypergeometric test. All the experimental conditions were analyzed (PNPLA3 siRNA silencing and overexpression of PNPLA3 with Met148 or Ile148).
Table 3.
Results of overrepresentation analysis: canonical pathways affected by differential regulation of metabolites during PNPLA3 silencing and overexpression of PNPLA3 with the I148M variants
| Canonical Pathway | P Value |
|---|---|
| PNPLA3 silencing | |
| Aminoacyl-tRNA biosynthesis | 2.9 E−10 |
| Glycerophospholipid metabolism | 0.00002 |
| Nitrogen metabolism | 0.0002 |
| Cyanoamino acid metabolism | 0.005 |
| Glycine, serine and threonine metabolism | 0.02 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 0.02 |
| Valine, leucine and isoleucine biosynthesis | 0.02 |
| d-Glutamine and d-glutamate metabolism | 0.02 |
| Arginine and proline metabolism | 0.03 |
| Pyrimidine metabolism | 0.04 |
| Overexpression of PNPLA3 with methionine at position 148 (M148) | |
| Aminoacyl-tRNA biosynthesis | 0.00003 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 0.002 |
| Propanoate metabolism | 0.004 |
| Nitrogen metabolism | 0.005 |
| Overexpression of PNPLA3 with isoleucine at position 148 (I148) | |
| Primary bile acid biosynthesis | 0.06 |
Comparison between Met148 vs. Ile148 showed increased levels of γ-glutamyl-glutamine [fold change (FC): 1.845, nominal P value: 0.001385, FDR: 0.086] and lactate (FC: 1.4896, nominal P value: 0.00982, FDR: 0.20), and decreased levels of tryptophan (FC: 0.7951, nominal P value: 0.004872, FDR: 0.151) with no changes in carbohydrate or lipid metabolism.
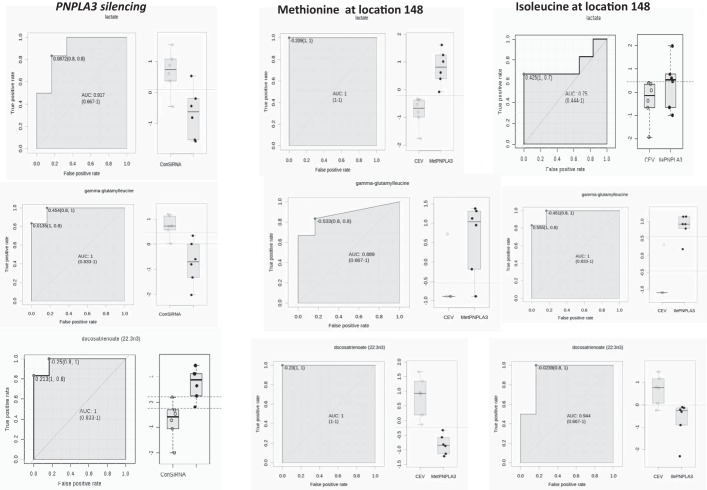
Together, the metabolotype associated with the amino acid substitution at position 148 seems to be clearly restricted to few metabolites, including polyunsaturated fatty acids, such as docosatrienoic acid, and γ-glutamyl-amino acids (Fig. 5); components of the glutathione biosynthetic pathway such as γ-glutamyl-leucine were significantly increased in both the Met148 and Ile148 variant. Surprisingly, a distinctive metabolite that fit for almost perfect discrimination between Met148 vs. Ile148 was lactic acid (area under the curve: 0.917, 95% confidence interval: 0.667–1), being higher (FC: 1.5, P value < 0.016) in the Met148 vs. Ile148-transfected cells (Fig. 5).
Fig. 5.
Receiver Operating Characteristic (ROC) Curve analysis for the most significantly associated metabolites in the overexpression of the PNPLA3 with Met148 or Ile148 in comparison with PNPLA3 silencing. ROC-based analysis on the data sets was performed by the web-based tool ROCCET (http://www.roccet.ca/). As an example, comparisons were performed by Lactate, gamma-glutamyl-leucine and docosatrienoic acid.
DISCUSSION
In this study we performed nontargeted metabolomics to explore the role of PNPLA3 in the regulation of Huh-7 cells in vitro metabolome. This approach revealed several novel findings and demonstrated that PNPLA3 induces widespread effects on the global Huh-7 cells in vitro metabolome beyond its classical role as triacylglycerol lipase and acylglycerol O-acyltransferase. Silencing of PNPLA3 was associated with a global perturbation of Huh-7 cells in vitro metabolome from amino acid, lipid, and carbohydrate metabolism that resembles a catabolic response with extensive protein breakdown. These observations are consistent with the significant decrease in a variety of amino acids, including glutamate, and dipeptides that mimics a fasting/starvation condition. Overexpression of the PNPLA3 Met148 variant mirrored the metabolic changes observed during gene silencing but in the opposite direction, which along with the comparison with the effect of the transfection with the Ile148 variant. These results were also replicated by the exploration of associated canonical pathways that showed a remarkable concordance between PNPLA3 silencing and overexpression of the PNPLA3 Met148 variant.
Another hitherto unrecognized finding from this study is that γ-glutamyl-amino acids, direct markers of oxidative stress, were decreased during gene silencing and increased during both wild-type and mutant overexpression, indicating that PNPLA3 may increase oxidative stress (11). Cells that overexpressed Met148 also showed an approximately twofold increase in lactic acid that suggests a shift to anaerobic metabolism and mitochondrial dysfunction, a finding that was previously associated with NAFLD disease progression (13, 19, 24). This finding might also explain the role of the rs738409 in the transition from fatty liver to NASH because NASH is more frequently observed in Met148 homozygous carriers who have 3.24-fold more risk of higher necroinflammatory scores (21).
Our results also showed an interesting pathway associated with the PNPLA3 function in the Huh-7 cells in vitro, such as alanine, aspartate, and glutamate metabolism. Indeed, our findings may explain, at least in part, the previous observation of the first genomewide association study on the loci influencing plasma levels of liver enzymes (18). Yuan and coworkers (27) reported that variants in PNPLA3-SAMM50 on chromosome 22, including the rs738409, significantly influence the plasma levels of alanine aminotransferase (ALT); follow-up studies confirmed this initial finding, showing that the rs738409 GG genotype vs. the CC genotype was associated with a 28% increase in serum ALT levels (21).
The present data raise the possibility that this observation could reflect a role for PNPLA3 with amino acid metabolism. Actually, much of the deregulated pathways associated with either gene silencing or overexpression of the PNPLA3 Met148 variant are consistent with the hypothesis that alanine and aspartate aminotransferases might reflect high levels of hepatic transamination of amino acids in the liver (22).
Similarly, PNPLA3 also significantly affected the metabolic pathway associated with phenylalanine and tyrosine metabolism, which also involves the transamination of α-ketoglutarate. Of note, it was previously observed that metabolic risk factors (central obesity, insulin resistance, hypertension, and dyslipidemia) are associated with multiple metabolites, including branched-chain amino acids, tryptophan breakdown products, nucleotide metabolites, and particularly glutamine and glutamate changes (2). High glutamine-to-glutamate ratio was associated with lower risk of incident diabetes mellitus. Interestingly, the glutamine-to-glutamate ratio is decreased by PNPLA3 knocking down (data not shown).
We also report here that PNPLA3 silencing in the Huh-7 cells involves changes in myo-inositol, an organic osmolyte associated with osmotic stress and a plethora of cell-signaling and transcriptional events (5). This observation is consistent with the finding of a metabolotype associated with extreme protein catabolism that causes cell “shrinkage” because of amino acid starvation (1).
Furthermore, the metabolic signature of PNPLA3 silencing was associated with a dramatic increase in lysolipids, including glycerophospholipids with a glycerol-3-phosphate molecule esterified at its sn-2 position, such as 2-linoleoylglycerophosphocholine, 2-palmitoylglycero-phosphoethanolamine, and 2-arachidonoylglycerophosphoinositol. This finding might be partially explained by the lack of enzymatic activity with an ester bond specificity of the phospholipase A2 (PLA2) domain, which mediates the acyl ester hydrolysis at the sn-2 position of phospholipids to produce free fatty acids and lysophospholipids (10). Nevertheless, the significant accumulation of some other glycerophospholipids, such as 1-arachidonoylglycerophospho-inositol, 1-linoleoylglycerophosphocholine, and 1-arachidonoyl-glycerophosphoethanolamine, might suggest a novel finding regarding the physiological role of the PNPLA3 as phospholipase A1, which might have also specificity for some sn-1 positions of the glycerol backbones of phospholipids, for example those arachidonoyl or linoleoyl-containing glycerophospholipids, suggesting that at least in the liver the enzyme may be able to hydrolyze a broader spectrum of fatty acids. More experiments are necessary to rule out that other putative PLA1-enzymes were induced during PNPLA3 silencing and that this effect is not dependent of the adaptation of the Lands cycle (16) and, certainly, to further explore its relation with proinflammatory arachidonic acid derivatives (23).
It is also interesting to note the effects of overexpression of the wild-type PNPLA3 protein and mutant protein on triglyceride accumulation following exposure to [14C]palmitate. The failure of overexpression of wild-type PNPLA3 to enhance triglyceride hydrolysis indicates that it is a weak lipase and is in line with previously published data (4). It is also possible that, as recently described (25), the mutant protein remodels triglyceride by increased labeled palmitate incorporation at carbon 1 and 2 of triacylglycerol via its acyltransferase activity. Both possibilities would lead to labeled triglyceride accumulation in our experimental conditions.
In summary, the encoded protein by PNPLA3 is so far regarded as a triacylglycerol lipase/acyltransferase that mediates triacylglycerol hydrolysis in both adipocytes and hepatocytes. Notably, the present study provides evidence for a pivotal role of PNPLA3 in the regulation of the liver metabolome by controlling major biochemical pathways associated with the energy usage/storage cycle. An unexpected involvement of PNPLA3 in bile acid metabolism was also uncovered by the gene silencing experiment. We also demonstrated a novel metabolotype associated with the Met148 variant that not only impacts on cholesterol synthesis in the Huh-7 cells but also is associated with a metabolic signature of anaerobic metabolism and disturbances in mitochondrial oxidative phosphorylation associated with a cellular hypoxic condition. This observation might explain the importance of PNPLA3 in modifying the natural history and disease phenotype of NAFLD and the magnitude of the effect of the rs738409 on the histological severity of NAFLD, which is far beyond the small magnitude observed for common variants on complex traits. In addition, the Met148 variant might either show a loss of lipase function or a gaining of acyltransferase activity, as recently shown (18).
GRANTS
This research was supported by grants NIDDK 5RO1DK081410-05 to A. J. Sanyal, NIDDK T32 DK 007150-35 to A. J. Sanyal, NIDDK 5R01AA020758-03 to A. J. Sanyal, and Scientific Exchange in the context of International Cooperation Agreements (NSF-CONICET), Res. 1038 to S. Sookoian.
DISCLOSURES
A. J. Sanyal holds financial interest in or has received remuneration from Abbott, Exhalenz, Genentech, GenFit, Gilead, Gore, Ikaria, Intercept, Merck, Norgine, Roche, Salix, Uptodate, Takeda, Astellas, Novartis, Galectin, Nitto Denko, and Bristol Myers. Dr. Sanyal consulted with Genentech re NASH and fibrosis without remuneration as of March 2013. Bayer-Onyx's Gideon trial is a global study, for which Dr. Sanyal is the US PI; no personal remuneration has been received. Dr. Sanyal will be the PI of the Immuron upcoming trial for alcoholic hepatitis as part of the NIAAA-funded TREAT consortium; Immuron will provide drug and no additional funding. He has provided advice to Novartis and Galectin but not taken personal remuneration. Research grants for Salix, Gore, Gilead, and Exhalenz represent the site budgets for VCU clinical trials involving these companies and do not support Dr. Sanyal directly. Research collaborations without any funding from the commercial entity: Regulus, CSL Behring, Ferring, Zora Lipidomics, and Metabolon.
AUTHOR CONTRIBUTIONS
H.-K.M. and A.J.S. conception and design of research; H.-K.M., J.C., and F.M. performed experiments; H.-K.M., S.C.S., C.J.P., and A.J.S. analyzed data; H.-K.M., S.C.S., C.J.P., J.C., and A.J.S. interpreted results of experiments; H.-K.M., S.C.S., C.J.P., J.C., F.M., and A.J.S. prepared figures; H.-K.M., S.C.S., C.J.P., J.C., and A.J.S. drafted manuscript; H.-K.M., S.C.S., C.J.P., and A.J.S. edited and revised manuscript; H.-K.M., S.C.S., C.J.P., and A.J.S. approved final version of manuscript.
Appendix
Methodological Details About Metabolomic Profiling
Sample preparation.
The sample preparation process was carried out with the automated MicroLab STAR system (Hamilton, Salt Lake City, UT). Recovery standards were added prior to the first step in the extraction process for quality control (QC) purposes. Sample preparation was conducted by using a proprietary series of organic and aqueous extractions to remove the protein fraction while allowing maximum recovery of small molecules. The resulting extract was divided into two fractions: one for analysis by LC and one for analysis by GC. Samples were placed briefly on a TurboVap (Zymark) to remove the organic solvent. Each sample was then frozen and dried under vacuum. Samples were then prepared for the appropriate instrument, either LC/MS or GC/MS.
Accurate mass determination and MS/MS fragmentation.
The LC/MS portion of the platform was based on a Waters ACQUITY UPLC and a Thermo-Finnigan LTQ-FT mass spectrometer, which had a LIT front end and a Fourier transform ion cyclotron resonance (FT-ICR) mass spectrometer back end. For ions with counts greater than two million, an accurate mass measurement could be performed. Accurate mass measurements could be made on the parent ion as well as fragments. The typical mass error was less than 5 ppm. Ions with less than two million counts require a greater amount of effort to characterize. Fragmentation spectra (MS/MS) were typically generated in data-dependent manner, but, if necessary, targeted MS/MS could be employed, such as in the case of lower level signals (3).
Data extraction and QA.
The data extraction of the raw MS data files yielded information that could loaded into a relational database and manipulated without resorting to binary large object (BLOB) manipulation. Once in the database the information was examined and appropriate QC limits were imposed. Peaks were identified by using Metabolon's proprietary peak integration software, and component parts were stored in a separate and specifically designed complex data structure. The median relative standard deviation (MRSD), a quality assurance (QA) metric of quantification and measure of instrument variability, was determined to be 5% for a panel of 30 internal standards. Overall process variability (i.e., extraction, recovery, resuspension, and instrument performance) for endogenous biochemicals (i.e., noninstrument standards) within technical replicate cell lysate samples was calculated to be 12% MRSD. These SD values reflected acceptable levels of variability for overall process and instrumentation of the analytical platform (3).
A variety of curation procedures were carried out to ensure that a high-quality data set was made available for statistical analysis and data interpretation. The QC and curation processes were designed to ensure accurate and consistent identification of true chemical entities and to remove those representing system artifacts, misassignments, and background noise. Metabolon data analysts used proprietary visualization and interpretation software to confirm the consistency of peak identification among the various samples. Library matches for each compound were checked for each sample and corrected if necessary.
For QA/QC purposes, a number of additional samples were included with each day's analysis. Furthermore, a selection of QC compounds was added to every sample, including those under test. These compounds were carefully chosen so as not to interfere with the measurement of the endogenous compounds. These QC samples were primarily used to evaluate the process control for each study as well as aiding in the data curation.
REFERENCES
- 1.Alfieri RR, Petronini PG. Hyperosmotic stress response: comparison with other cellular stresses. Pflügers Arch 454: 173–185, 2007 [DOI] [PubMed] [Google Scholar]
- 2.Cheng S, Rhee EP, Larson MG, Lewis GD, McCabe EL, Shen D, Palma MJ, Roberts LD, Dejam A, Souza AL, Deik AA, Magnusson M, Fox CS, O'Donnell CJ, Vasan RS, Melander O, Clish CB, Gerszten RE, Wang TJ. Metabolite profiling identifies pathways associated with metabolic risk in humans. Circulation 125: 2222–2231, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Evans AM, DeHaven CD, Barrett T, Mitchell M, Milgram E. Integrated, nontargeted ultrahigh performance liquid chromatography/electrospray ionization tandem mass spectrometry platform for the identification and relative quantification of the small-molecule complement of biological systems. Anal Chem 81: 6656–6667, 2009 [DOI] [PubMed] [Google Scholar]
- 4.He S, McPhaul C, Li JZ, Garuti R, Kinch L, Grishin NV, Cohen JC, Hobbs HH. A sequence variation (I148M) in PNPLA3 associated with nonalcoholic fatty liver disease disrupts triglyceride hydrolysis. J Biol Chem 285: 6706–6715, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holub BJ. Metabolism and function of myo-inositol and inositol phospholipids. Annu Rev Nutr 6: 563–597, 1986 [DOI] [PubMed] [Google Scholar]
- 6.Huang Y, He S, Li JZ, Seo YK, Osborne TF, Cohen JC, Hobbs HH. A feed-forward loop amplifies nutritional regulation of PNPLA3. Proc Natl Acad Sci USA 107: 7892–7897, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenkins CM, Mancuso DJ, Yan W, Sims HF, Gibson B, Gross RW. Identification, cloning, expression, and purification of three novel human calcium-independent phospholipase A2 family members possessing triacylglycerol lipase and acylglycerol transacylase activities. J Biol Chem 279: 48968–48975, 2004 [DOI] [PubMed] [Google Scholar]
- 8.Kumari M, Schoiswohl G, Chitraju C, Paar M, Cornaciu I, Rangrez AY, Wongsiriroj N, Nagy HM, Ivanova PT, Scott SA, Knittelfelder O, Rechberger GN, Birner-Gruenberger R, Eder S, Brown HA, Haemmerle G, Oberer M, Lass A, Kershaw EE, Zimmermann R, Zechner R. Adiponutrin functions as a nutritionally regulated lysophosphatidic acid acyltransferase. Cell Metab 15: 691–702, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lang F. Mechanisms and significance of cell volume regulation. J Am Coll Nutr 26: 613S–623S, 2007 [DOI] [PubMed] [Google Scholar]
- 10.Linderoth L, Andresen TL, Jorgensen K, Madsen R, Peters GH. Molecular basis of phospholipase A2 activity toward phospholipids with sn-1 substitutions. Biophys J 94: 14–26, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Martin HL, Teismann P. Glutathione—a review on its role and significance in Parkinson's disease. FASEB J 23: 3263–3272, 2009 [DOI] [PubMed] [Google Scholar]
- 12.Moldes M, Beauregard G, Faraj M, Peretti N, Ducluzeau PH, Laville M, Rabasa-Lhoret R, Vidal H, Clement K. Adiponutrin gene is regulated by insulin and glucose in human adipose tissue. Eur J Endocrinol 155: 461–468, 2006 [DOI] [PubMed] [Google Scholar]
- 13.Pirola CJ, Gianotti TF, Burgueno AL, Rey-Funes M, Loidl CF, Mallardi P, Martino JS, Castano GO, Sookoian S. Epigenetic modification of liver mitochondrial DNA is associated with histological severity of nonalcoholic fatty liver disease. Gut 62: 1356–1363, 2013 [DOI] [PubMed] [Google Scholar]
- 14.Poole LB, Karplus PA, Claiborne A. Protein sulfenic acids in redox signaling. Annu Rev Pharmacol Toxicol 44: 325–347, 2004 [DOI] [PubMed] [Google Scholar]
- 15.Rae-Whitcombe SM, Kennedy D, Voyles M, Thompson MP. Regulation of the promoter region of the human adiponutrin/PNPLA3 gene by glucose and insulin. Biochem Biophys Res Commun 402: 767–772, 2010 [DOI] [PubMed] [Google Scholar]
- 16.Richmond GS, Smith TK. Phospholipases a(1). Int J Mol Sci 12: 588–612, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Romeo S, Kozlitina J, Xing C, Pertsemlidis A, Cox D, Pennacchio LA, Boerwinkle E, Cohen JC, Hobbs HH. Genetic variation in PNPLA3 confers susceptibility to nonalcoholic fatty liver disease. Nat Genet 40: 1461–1465, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruhanen H, Perttilä J, Hölttä-Vuori M, Zhou Y, Yki-Järvinen H, Ikonen E, Käkelä R, Olkkonen VM. PNPLA3 mediates hepatocyte triacylglycerol remodeling. J Lipid Res 55: 739–746, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sanyal AJ, Campbell-Sargent C, Mirshahi F, Rizzo WB, Contos MJ, Sterling RK, Luketic VA, Shiffman ML, Clore JN. Nonalcoholic steatohepatitis: association of insulin resistance and mitochondrial abnormalities. Gastroenterology 120: 1183–1192, 2001 [DOI] [PubMed] [Google Scholar]
- 20.Saurin AT, Neubert H, Brennan JP, Eaton P. Widespread sulfenic acid formation in tissues in response to hydrogen peroxide. Proc Natl Acad Sci USA 101: 17982–17987, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sookoian S, Pirola CJ. Meta-analysis of the influence of I148M variant of patatin-like phospholipase domain containing 3 gene (PNPLA3) on the susceptibility and histological severity of nonalcoholic fatty liver disease. Hepatology 53: 1883–1894, 2011 [DOI] [PubMed] [Google Scholar]
- 22.Sookoian S, Pirola CJ. Alanine and aspartate aminotransferase and glutamine-cycling pathway: their roles in pathogenesis of metabolic syndrome. World J Gastroenterol 18: 3775–3781, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sookoian S, Pirola CJ. The genetic epidemiology of nonalcoholic fatty liver disease: toward a personalized medicine. Clin Liver Dis 16: 467–485, 2012 [DOI] [PubMed] [Google Scholar]
- 24.Sookoian S, Rosselli MS, Gemma C, Burgueno AL, Fernandez GT, Castano GO, Pirola CJ. Epigenetic regulation of insulin resistance in nonalcoholic fatty liver disease: impact of liver methylation of the peroxisome proliferator-activated receptor gamma coactivator 1alpha promoter. Hepatology 52: 1992–2000, 2010 [DOI] [PubMed] [Google Scholar]
- 25.Speliotes EK, Yerges-Armstrong LM, Wu J, Hernaez R, Kim LJ, Palmer CD, Gudnason V, Eiriksdottir G, Garcia ME, Launer LJ, Nalls MA, Clark JM, Mitchell BD, Shuldiner AR, Butler JL, Tomas M, Hoffmann U, Hwang SJ, Massaro JM, O'Donnell CJ, Sahani DV, Salomaa V, Schadt EE, Schwartz SM, Siscovick DS, Voight BF, Carr JJ, Feitosa MF, Harris TB, Fox CS, Smith AV, Kao WH, Hirschhorn JN, Borecki IB. Genome-wide association analysis identifies variants associated with nonalcoholic fatty liver disease that have distinct effects on metabolic traits. PLoS Genet 7: e1001324, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS. MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res 40: W127–W133, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yuan X, Waterworth D, Perry JR, Lim N, Song K, Chambers JC, Zhang W, Vollenweider P, Stirnadel H, Johnson T, Bergmann S, Beckmann ND, Li Y, Ferrucci L, Melzer D, Hernandez D, Singleton A, Scott J, Elliott P, Waeber G, Cardon L, Frayling TM, Kooner JS, Mooser V. Population-based genome-wide association studies reveal six loci influencing plasma levels of liver enzymes. Am J Hum Genet 83: 520–528, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]