Abstract
Light chain (LC) amyloidosis (AL) is a fatal disease in which immunoglobulin LC deposit as fibrils. Although the LC amyloid-forming propensity is attributed primarily to the variable region, fibrils also contain full-length LC comprised of variable-joining (VL) and constant (CL) regions. To assess the role of CL in fibrillogenesis, we compared the thermal stability of full-length LC and corresponding VL and CL fragments. Protein unfolding and aggregation were monitored by circular dichroism and light scattering. A full-length λ6 LC purified from urine of a patient with AL amyloidosis showed irreversible unfolding coupled to aggregation. The transition temperature decreased at slower heating rates, indicating kinetic effects. Next, we studied five recombinant λ6 proteins: full-length amyloidogenic LC, its VL, germline LC, germline VL, and CL. Amyloidogenic and germline proteins showed similar rank order of stability, VL < LC < CL; hence, in the full-length LC, VL destabilizes CL. Amyloidogenic proteins were less stable than their germline counterparts, suggesting that reduction in VL stability destabilizes the full-length LC. Thermal unfolding of the full-length amyloidogenic and germline LC required high activation energy and involved irreversible aggregation, yet the unfolding of the isolated VL and CL fragments was partially reversible. Therefore, compared to their fragments, full-length LCs are more likely to initiate aggregation during unfolding and provide a template for the VL deposition. The kinetic barrier for this aggregation is regulated by the stability of the VL region. This represents a paradigm shift in AL fibrillogenesis and suggests CL region as a potential therapeutic target.
Light chain (LC)1 amyloidosis is a rapidly progressing disease in which normally soluble immunoglobulin LC form extracellular fibrillar deposits (AL) in target organs, leading to organ failure and death. The available treatment for AL amyloidosis is limited to cytotoxic agents that kill the monoclonal plasma cells which produce the amyloidogenic LC (1). This treatment helps to extend patients’ lives and improve their quality; however, it is not effective for every patient and, as any chemotherapy, has side effects (2, 3). Understanding the molecular pathway of LC aggregation and fibril formation can help to develop novel therapeutic targets and design new agents that can block this pathogenic pathway and thereby delay the progression of the disease and alleviate its symptoms. To find novel therapeutic targets for AL amyloidosis, it is important to elucidate the pathogenic roles of various LC constituents in amyloid deposits.
Full-length LC is comprised of two similar structural domains of nearly 100 amino acids each, termed variable and constant regions, connected by a short joining region (4). The X-ray crystal structure of LC shows that the variable and constant regions share a similar immunoglobulin-like fold that is rich in β-sheet (4). Despite their structural similarity, the two domains have distinct functions: the variable region mediates interactions with antigens and effector molecules, and the constant region is responsible for binding to cell surface. These functional differences reflect the differences in the protein sequence. As indicated by their names, the constant regions are encoded by the host genome and are nonpolymorphic, while the variable regions are altered by somatic hypermutation during B cell maturation in response to antigenic selection. As a result, each clonal amino acid sequence of the LC variable region is different. This sequence variability of the variable region leads to diversity in aggregating propensity; i.e., some LC sequences are highly prone to aggregation and fibril formation, while others allow the LC to remain water-soluble (for example, LC with variable regions encoded by λ germline genes, which account for nearly 70% of all AL LC cases, are reported to be more amyloidogenic than those with κ genes (5–8)). The molecular determinants for LC aggregation are not clearly understood and are the focus of this work.
Previous reports suggested that AL LC deposits were composed mainly of variable fragments (9, 10). Thus, earlier biophysical studies of LC have focused primarily on the LC variable regions (that were imprecisely referred to as “light chains”) (11–16). These prior analyses showed that thermal and chemical denaturations of variable fragments were well approximated by a reversible two-state transition and revealed a correlation between thermodynamic instability of the LC variable region and its ability to form amyloid. Furthermore, fibril formation was shown to proceed via an aggregation-prone protein unfolding intermediate (17–20). It was proposed that the unfolded intermediate accumulated during the slow kinetic step detected in the unfolding of the 6aJL2 variable region, which led to slow protein aggregation and, eventually, fibril formation (12).
In addition to variable fragments, our recent proteomic studies of amyloid deposits from patient organs detected considerable amounts of full-length LC (21). To test, for the first time, the role of full-length LC in protein aggregation, we carried out thermal denaturation studies of recombinant λ6 LC and its individual fragments, VL comprised of variable and joining regions, and CL corresponding to constant domain. The results, taken together with similar studies of the germline proteins, have revealed that (i) full-length LC is more prone to irreversible aggregation upon unfolding than its VL or CL fragments and (ii) unfolding and aggregation of the full-length LC are subject to kinetic control and are modulated by the stability of the VL region and its interactions with the CL region. These results prompted us to postulate a key role for interdomain interactions in full-length LC at early stages of fibril nucleation.
MATERIALS AND METHODS
Extraction and Analysis of the Whole Protein from the Heart Tissue
The protein was extracted from tissue samples as described (21). Briefly, the samples were washed repeatedly with sterile isotonic saline followed by one wash with double-distilled water. Isoelectric focusing buffer (7 M urea, 2 M thiourea, 4% CHAPS, 65 mM dithiothreitol) was added (200 µL buffer per 100 g of tissue). The samples were crushed with a pestle to remove the protein. Samples were centrifuged at 80000g for 1.5 h at 19 °C The central aqueous layer between the top lipid layer and the cell debris pellet was recovered. Residual lipids were removed by centrifuging at 25000g for 30 min at 4 °C, followed by recovery of the central layer. Total protein was quantified by using the Bio-Rad protein assay. Samples were stored at −80 °C.
Two-dimensional PAGE was carried out as described (21). For each gel, 500 µg of protein was loaded. Extracts were diluted to a final volume of 300 µL with 100 µL of Destreak buffer (Amersham Biosciences), IEF buffer, and pI 3–10 ampholytes (Bio-Rad) to a final concentration of 0.02%. Seventeen centimeter ReadyStrip IPG strips (Bio-Rad) with nonlinear gradients of pH 3–10 were used for the first dimension electrophoresis step, i.e., IEF. Strips were passively rehydrated for 1 h and then actively rehydrated at 50 V for 8 h. IEF was performed using a Protean IEF cell (Bio-Rad) as follows: 120 V for 1 h, 300 V for 30 min, a linear increase up to 3500 V over 3 h, 5000 V for 10 min, and 8000 V steady until a total of 67000 V•3 h had elapsed. After IEF, strips were subjected to disulfide reduction with dithiothreitol and cysteine alkylation with iodoacetamide. Next, second dimension electrophoresis was performed using 9–16% gradient ReadyGels (Bio-Rad). The gels were stained with GelCode colloidal Coomassie Blue (Pierce) and were imaged with an EDAS 290 (Eastman Kodak Co.).
For immunoblotting, the 2D PAGE gels were transferred to Millipore Q PVDF membranes (Millipore). Semidry transfers were conducted using a TransBlot semidry electrophoretic transfer cell (Bio-Rad) and probed with polyclonal rabbit anti-human immunoglobulin λ light chain antibodies. Subsequently, the membranes were probed with goat anti-rabbit polyclonal IgG antibodies coupled with horseradish peroxidase. Membranes were developed using an enhanced chemiluminescence system (Eastman Kodak Co.).
Cloning of Recombinant Ig LC and Its Fragments
(A) λ6 Amyloidogenic LC
Aλ6 LC from an AL 01–095 patient with multiorgan disease was cloned from bone marrow. The full-length LC-encoding DNA fragment was amplified for insertion into the pQE-1 vector (Qiagen) to allow for expression in Escherichia coli of an N-terminal 6 × His-tagged LC. The primers used to insert appropriate restriction sequences at the 5′ and 3′ ends of the coding sequence of full-length λ6 LC by PCR techniques were 5′ oligonucleotide AATTTTATGCTGACTCAGCCGC and 3′ oligonucleotide GGTAAGCTTATTATGAACATTCTGTAGG-GGC A blunt end was provided by the 5′ oligonucleotide, and a HindIII site (underlined) was engineered into the 3′ oligonucleotide. The DNA sequence of full-length λ6 LC was amplified by PCR with platinum pfx polymerase (Invitrogen). The DNA-encoding λ6 LC was digested with HindIII restriction endonucleases and was ligated into the purified pQE-1 vector that was enzymatically cleaved with PvuII and HindIII. The constructs were verified by DNA sequencing. The resulting plasmid was termed λ6 AL LC/pQE-1.
(B) λ6 Germline LC
The primary sequence of the variable and joining regions of AL 01–095 was aligned against the sequence of its germline donor, IGLV6–57*01 (22), which was obtained from the ALBase database (23). The alignment revealed nine amino acid differences in the variable region, four in the complementarity determining region and five in the framework region (Figure 7). To test the effects of these amino acid substitutions on the protein stability, we have engineered mutations in the variable region of the full-length LC that restore its similarity with the IGLV6–57*01 germline donor sequence, leaving the constant region (encoded by germline donor LC3*04) unchanged. To generate the full-length λ6 Germ LC gene (IGLV6–57*01/LC3*04) from λ6 AL LC, nine point mutations were consecutively introduced using a site-directed mutagenesis kit (Stratagene). The resulting plasmid was termed λ6 Germ LC/pQE-1.
Figure 7.
Amino acid sequence alignment of the variable, joint, and constant regions of AL 01–095 against the sequence of its most probable germline donor, IGLV6-57*01. Dark gray panels show framework regions, and light gray panels show complementarity determining regions according to ref 48.
(C) λ6 Amyloidogenic and Germline VL Regions
To generate the genes corresponding to λ6 amyloid (AL 01–095) and germline (IGLV6–57*01) VL regions, the stop codons (TAG) were incorporated into plasmids λ6 AL LC/pQE-1 and λ6 Germ LC/pQE-1 after the VL sequences using the site-directed mutagenesis kit (Stratagene).
(D) λ6 Constant Region
The DNA fragment encoding λ6 CL region (LC3*04) was amplified by PCR techniques with platinum pfx polymerase (Invitrogen) using the following primers: 5′ oligonucleotide CCCAAGGCTGCCCCCTCG (blunt end) and 3′ oligonucleotide GGTAAGCTTATTATGAACATTCTGTAGGGGC (with an engineered HindIII restriction site, underlined). The DNA-encoding λ6 CL region was digested with HindIII restriction endonucleases and ligated into the purified pQE-1 vector (Qiagen) that had been cut with the PvuII and HindIII enzymes.
Expression and Purification of Recombinant Proteins
The λ 6 LC/pQE-1 expression plasmid was introduced into E. coli Rosetta-gami 2 cells (Novagen) by heat shock; the cells were plated onto Luria broth (LB)-agar media containing 100 µg/mL ampicillin and were incubated overnight at 37 °C A single colony was isolated and cultured overnight at 37 °C in LB media with 100 µg/mL ampicillin (LB/amp). This culture was used to inoculate 500 mL of LB/amp, which was grown at 37°C until OD600 = 0.6 and then cooled to 14 °C Protein expression was induced by adding isopropyl d-thiogalactopyranoside at 0.12 mM and incubating for 20 h at 14°C. The cells were harvested by centrifugation, resuspended in cell lysis buffer (50 mM sodium phosphate, 300 mM NaCl, 10 mM imidazole, pH 8.0) at 3 mL/g wet weight cells, supplemented with hen egg white lysozyme at 1 mg/mL, and incubated for 30 min at 4°C. The cells were lysed by sonication at 4°C, and the supernatant was clarified by centrifugation and passage through 0.22 µm filters. The clarified cell lysate was loaded onto a 3 mL nickel nitrilotriacetic acid agarose column (Qiagen) and rinsed with wash buffer (50 mM sodium phosphate, 300 mM NaCl, 20 mM imidazole, pH 8.0) until OD280 < 0.01. The polyhistidine-tagged rLC was eluted from the column with elution buffer (50 mM sodium phosphate, 300 mM NaCl, 250 mM imidazole, pH 8.0), and fractions with OD280 > 0.05 were collected and pooled. The pooled fractions were exhaustively dialyzed against water.
Circular Dichroism Spectroscopy and Right-Angle Light Scattering
Protein samples (5–20 µM protein in 5 mM sodium phosphate buffer, 15 mM NaCl, pH 7.4) were placed in a 1 mm path length cell, and CD data were collected by using Aviv 400 (Aviv Biomedical, NJ, USA) or Jasco J-815 (Jasco Inc., Japan) spectropolarimeters equipped with thermoelectric temperature controllers. Far-UV CD spectra (190–250 nm) were recorded with 1 nm step size and 1 nm bandwidth. The spectra were smoothed by using the noise reduction routine, the buffer baselines were subtracted, and the spectra were normalized to protein concentration and presented in units of molar ellipticity per residue, [Θ]. Protein secondary structure was estimated from these spectra by using the CDPro program suite (24), which is a modified version of three methods, SELCON3 (25), CONTIN/LL (CONTIN method (26) in locally linearized approximation (27)), and CDSSTR (28).
CD melting data were recorded at 202 nm to monitor β-sheet unfolding upon sample heating and cooling from 25 to 85 °C at a constant rate ranging from 11 to 80 °C/h. Changes in the macromolecular size due to aggregation of the thermally unfolded protein were monitored by right-angle light scattering that was recorded at 202 nm simultaneously with the CD by using fluorescence attachment in an Aviv 400 spectropolarimeter as described (29). The apparent melting temperature Tm at each heating rate was determined from the peak position in the first derivative of the CD or light scattering melting data (30).
The time course of protein unfolding was assessed in kinetic temperature-jump (T-jump) experiments. The unfolding was triggered at t=0 by a rapid temperature increase from 25 °C to a higher constant value and was monitored by CD at 202 nm. The kinetic CD data at each temperature were approximated by a multiexponential decay function; the unfolding rates k(T) were determined at each temperature and were used to obtain the Arrhenius plot, −RT ln k(T) versus 1/T, where R is the universal gas constant and T the temperature in kelvin. The activation energy (enthalpy) Ea ≈ ∆H* of the unfolding was determined from the slope of the Arrhenius plot with an accuracy of about ±10 kcal/mol, which incorporates fitting errors and deviations among different data sets.
Protein Cross-Linking and Size-Exclusion HPLC
Size-exclusion HPLC was used to detect formation of soluble aggregates upon protein heating. Recombinant full-length AL LC and its VL and CL regions, which were intact or heated and cooled from 20 to 85 °C at a rate of 80 °C/h under the same sample conditions as described in CD experiments, were cross-linked with glutaraldehyde following established protocols (31). Briefly, 0.3 µL of 50% glutaraldehyde solution was added to 4 µg of protein in 20 µL of potassium/sodium phosphate buffer at pH 7. After 4 min incubation at room temperature, the reaction was quenched with 1.94 µL of 7% NaBH4 in 0.1 M NaOH. HPLC was performed using a Zorbax Bio Series GF-250 (4 µm, 4.6 × 250 mm) column (Agilent Technologies, Palo Alto, CA), with spectrophotometric monitoring at 280 nm. The proteins were eluted isocratically in a 0.2 M sodium phosphate buffer, pH 7.0, over 20 min at a flow rate of 1 mL/min.
RESULTS
Analysis of ex Vivo LC Proteins
The composition of AL amyloid fibrils in tissues was determined by proteomic techniques. Total protein was extracted from autopsied heart tissue, obtained from a patient who succumbed to systemic λ6 AL amyloidosis (AL 01–095). Two-dimensional PAGE analysis was used to produce a proteomic map that revealed extensive heterogeneity of the deposited LC proteins (Figure 1a). While other studies have reported that AL fibrils are composed mainly of VL region fragments and rarely contained full-length LC (10, 32), we found abundant amounts of protein with Mr ≅ 25 kDa and immunoreactive to antihuman LC antibodies, along with smaller sized LC fragments (Figure 1b). Similarly, full-length LC and fragments were found in our recent study of adipose tissue samples from patients with AL including a case featuring a λ6 LC (21). In that study, matrix-assisted laser desorption/ionization mass spectrometry and peptide mass fingerprinting were used to confirm the identity of the full-length and truncated λ6 LC proteins.
Figure 1.
Proteomic analysis of heart tissue from AL patient 01-095. (a) Proteins extracted from the tissue were subjected to 2D PAGE followed by Coomassie staining. The pI range is 3–10; molecular mass range is 10–250 kDa. (b) The enlarged boxed region of the gel from panel a with the results of immunoblotting with primary (rabbit anti-human polyclonal lambda light chains) and secondary (goat anti-rabbit IgG-HRP) antibodies. The pI range is 4.5–5.8; molecular mass range is 10–25 kDa. Immunoreactive spots correspond to the full-length LC and its various proteolytic fragments.
The LC gene from patient AL 01–095 was sequenced from bone marrow plasma cells and was determined to be a member of the λ6 family, IGLV6–57. The LC protein was purified from the urine of the same patient and used for structural stability studies. Mass spectrometric analysis showed that the λ6 LC had no posttranslational modifications and formed a disulfide-linked dimer via the C-terminal Cys215.
The thermal stability of the urinary LC AL 01–095 was studied by circular dichroism (CD) spectroscopy. Figure 2a shows the far-UV CD spectra recorded from the intact protein at 25 °C (solid squares) and from the same sample that was consecutively heated to 85 °C (triangles) and cooled to 25 °C (open squares) at a rate of 80 °C/h. The spectrum of the intact protein had a negative peak at 218 nm and a positive peak at 197 nm characteristic of β-sheet. Heating to 85 °C led to major spectral changes, including the appearance of a negative CD peak near 202 nm, which is characteristic of a predominantly unfolded conformation. This unfolding was largely irreversible, as indicated by the large difference between the spectra recorded at 25 °C before and after heating to 85 °C.
Figure 2.
Thermal denaturation of purified urinary full-length λ6 LC from AL patient 01–095 monitored by circular dichroism and right-angle light scattering. (a) Far-UV CD spectra recorded of a protein sample that was intact at 25 °C (filled squares) or was heated to 85 °C (open squares) and consecutively cooled to 25 °C (filled triangles) at a rate of 80 °C/h. (b) Far-UV CD and (c) 90° light scattering data recorded at 202 nm upon sample heating and cooling at a fast (80 °C/h, filled circles) or a slow rate (11 °C/h, open circles) show concomitant irreversible changes indicative of protein unfolding coupled to aggregation. Vertical arrows show apparent melting temperatures Tm determined from the peak positions in the first derivatives of the heating data. Sample conditions in this and other CD experiments are 10 µM protein in 10 mM sodium phosphate buffer, pH 7.0.
The protein secondary structural content was estimated by using the CDPro program suite (24) with two sets of reference proteins (25). The intact amyloidogenic LC was estimated to have 64% β-sheet structure at 25 °C (Table S1, Supporting Information), similar to the β-sheet content observed in X-ray crystal structures of full-length Bence-Jones proteins (33–35). Upon heating to 85 °C, β-sheet content decreases to 34%; this residual β-structure may result from the anchoring effects of the two disulfide bridges, one in the variable region and another in the constant region of the LC (36). The β-sheet content was not restored to its original value upon heating and cooling, suggesting irreversible unfolding of LC (Figure 2).
Irreversible thermal unfolding often results from aggregation of the heat-denatured protein. To test whether such aggregation occurs upon unfolding of the urinary AL 01–095 LC, we used far-UV CD to report on the secondary structure and right-angle light scattering to report on macromolecular size. Right-angle light scattering was measured by using total fluorescence accessory in AVIV-400 spectropolarimeter (29). Far-UV CD at 202 nm for β-sheet to random coil conversion (Figure 2b) and 90° light scattering for protein aggregation (Figure 2c) were recorded simultaneously during sample heating and cooling at a fast rate (80 °C/h, filled) or a slow rate (11 °C/h, open circles). Heating above 40 °C led to cooperative changes in the CD signal at 202 nm, from positive (β-sheet) to negative (random coil), indicating protein unfolding (Figure 2b). Noncoincidence of the heating and cooling CD curves confirms that the unfolding was irreversible. Furthermore, heating above 40 °C led to an irreversible increase in light scattering, due to protein aggregation (Figure 2c). Importantly, the apparent transition temperatures Tm, which were determined from the peak positions in the first derivatives of the CD and light scattering heating data, agreed within the accuracy of their experimental determination (±1 °C), which indicated a close coupling between protein unfolding and aggregation. We conclude that thermal unfolding of urinary full-length LC AL 01–095 is an irreversible transition coupled to protein aggregation.
Furthermore, decreasing the heating rate from 80 to 11 °C/h led to a low-temperature shift in the CD and light scattering heating data (Figure 2b,c). As a result, the apparent Tm of the protein unfolding and aggregation decreased from 54 ± 1 to 48 ± 1 °C. Such a heating rate effect is a hallmark of slow kinetically controlled transitions with high activation energy (37, 38).
Analysis of Recombinant Full-Length AL LC and Its VL and CL Fragments
To assess the roles of individual LC regions in protein unfolding and aggregation, we generated a recombinant LC that had the same sequence as the urinary AL 01–095 LC. In addition, recombinant proteins corresponding to the VL (comprised of variable and joining) and CL regions of the LC were produced and used to compare thermal unfolding properties of these proteins.
(A) Full-Length LC
Since the LC purified from the AL 01–095 patient urine lacked posttranslational modifications, the only difference between the urinary and recombinant LC was the presence of the N-terminal His tag in the recombinant protein. Far-UV CD spectra of the urinary LC and the recombinant full-length protein closely overlapped (Figure S1, Supporting Information), indicating similar secondary structures. Moreover, the CD melting data recorded for both proteins under the same conditions closely overlapped (Figure S1, Supporting Information) and demonstrated identical heating rate effects, an indication of identical thermal stability (data not shown). These data provide strong evidence that the presence of the His tag had no detectable effects on the secondary structure or stability of the recombinant amyloidogenic LC. Furthermore, our mass spectrometric analysis data showed that urinary LC formed a disulfide-linked dimer via the C-terminal Cys215. An S-S bonded dimer was also formed by the recombinant LC, as indicated by nonreducing gel electrophoresis (Figure S2, Supporting Information). Under mildly reducing conditions (~6% (w/w) DTT), this dimer was converted into monomer (Figure S2, Supporting Information). Importantly, protein secondary structure and thermal stability assessed by far-UV CD remained unaltered upon dimer-to-monomer conversion. Therefore, even though protein dimerization was proposed to play a role in amyloid formation by affecting free light chain clearance and metabolism in vivo (39), it has no detectable effect on the structure, stability, and irreversible unfolding of the LC molecule at early stages of aggregation in vitro.
The formation of soluble protein aggregates upon thermal unfolding was assessed by size-exclusion chromatography. Recombinant LC proteins, which were either intact or heated from 25 to 85 °C at a rate of 80 °C/h and then cooled, were cross-linked with gluteraldehyde before HPLC analysis. Intact protein showed a single peak corresponding to full-length LC (~25 kDa). Heated protein showed an additional peak corresponding to higher order soluble oligomers with an apparent molecular mass of > 200 kDa (Figure 3a). Since fibril formation is a slow process, the protein was incubated for 8 days at 50 °C; negative stain electron microscopy showed formation of unbranched fibrils of varying lengths with diameters of ~10 nm (Figure S3, Supporting Information). Taken together, our CD, light scattering, size-exclusion HPLC, and electron microscopic data show that thermal unfolding of full-length λ6 LC is a slow thermodynamically irreversible transition coupled to aggregation that can lead to amyloid fibril formation over time.
Figure 3.
Heat-induced aggregation of recombinant AL proteins assessed by size-exclusion high-performance liquid chromatography. The proteins that were intact (lower curve) or heated and cooled from 25 to 85 °C at a rate of 80 °C/h (upper curve) were cross-linked by gluteraldehyde and analyzed by HPLC. Upon heating and cooling, full-length LC forms soluble aggregates with an apparent molecular mass of >200 kDa (the true molecular mass is hard to estimate since the aggregated protein is largely unfolded). (a) VL fragment shows no detectable aggregation (b) and CL-fragment forms lower order oligomers (c)
(B) VL Fragment
Figure 4a shows far-UV CD spectra recorded from a VL sample containing intact protein (filled squares) that was consecutively heated to 65 °C (triangles) and cooled to 25 °C (open squares) at a rate of 80 °C/h. The spectrum of the intact VL λ6 protein was similar to that reported for other VL proteins (17,19, 40, 41). This spectrum differed from that of the full-length LC at short wavelengths (Figures 2a and 4a, solid circles) but showed a similar negative peak near 218 nm, indicating a predominantly β-sheet conformation. The estimated β-sheet content in VL was ~62%, similar to that in full-length LC. This is comparable to the β-sheet content observed in the X-ray crystal structures of other lambda VL proteins (14, 42, 43). Heating to 65 °C led to a substantial unfolding of this β-structure (Figure 4a, open squares), with a residual β-sheet content of ~30% (Table S1, Supporting Information). Importantly, in contrast to full-length protein, the CD spectra recorded at 25 °C of intact VL and of VL that was heated and cooled at a fast rate from 25 to 65 °C largely overlapped (Figure 4a, filled and open squares), suggesting that VL unfolding was largely reversible. This is consistent with many earlier studies reporting reversible unfolding of various VL fragments of LC (ref 41 and references therein).
Figure 4.
Thermal unfolding and refolding of recombinant VL fragment of AL LC. (a) Far-UV CD spectra of intact protein recorded at 25 °C (filled squares) and of the same protein that was heated to 65 °C (filled triangles) followed by cooling to 25 °C (open squares) at a rate of 80 °C/h. (b) CD melting data recorded at 202 nm during protein heating and cooling at a fast (80 °C/h, filled circles) or a slow rate (11 °C/h, open circles)
To test the thermodynamic reversibility of VL unfolding, CD melting data were recorded during heating and cooling at two different rates (Figure 4b). At a fast rate of 80 °C/h (filled circles), the heating and cooling data of VL largely overlapped, confirming the reversible nature of unfolding. At a slow rate of 11 °C/h (open circles), only partial protein refolding upon heating and cooling was observed. This is consistent with the findings from a recent study of a similar V protein, λ6 germline 6aJL2; this protein showed reversible thermal unfolding on a fast time scale but formed an irreversible soluble oligomeric unfolding intermediate on a slow time scale (41). Importantly, the CD melting data recorded for VL at a fast and a slow rate closely overlapped (Figure 4b, open and filled circles). This contrasts with the hysteresis and scan rate effects observed in the melting data of full-length LC. We conclude that, in contrast to full-length LC that undergoes irreversible unfolding coupled to aggregation, the unfolding of VL is inherently reversible on a fast time scale.
(C) CL Fragment
The far-UV CD spectrum of the C fragment (Figure 5a, filled squares) was similar to that of the full-length protein and indicated a similar β-sheet content of about 64% (Table S1, Supporting Information). Comparison of far-UV CD spectra recorded before and after heating to 85 °C at a fast rate (80 °C/h) showed a partially irreversible unfolding (Figure 5a, filled and open squares). Furthermore, comparison of such CD spectra in Figures 2a, 4a, and 5a suggests that the unfolding of the full-length LC was less reversible than that of its individual VL or CL fragments. This was confirmed by the comparison of the heating and cooling data recorded for the full-length LC and its fragments: fast unfolding was irreversible in LC, partially reversible in CL, and fully reversible in VL (Figures 2b, 4b, and 5b, filled symbols). Consequently, irreversible unfolding of full-length LC results from the higher aggregating propensity of the partially unfolded CL region and/or from interactions between the CL and VL regions during their unfolding.
Figure 5.
Thermal unfolding and refolding of recombinant CL fragment of AL LC. (a) Far-UV CD spectra recorded from intact protein at 25 °C (filled squares) and from the same protein that was heated to 85 °C (filled triangles) and cooled to 25 °C (open squares) at a rate of 80 °C/h. (b) CD melting data recorded at 202 nm during sample heating and cooling at a fast (80 °C/h, filled circles) or a slow rate (11 °C/h, open circles).
Evidence for Interdomain Interactions in LC
Comparison of far-UV CD spectra of recombinant proteins in their intact folded state shows that the spectrum of full-length LC cannot be represented by a weighted average of the spectra of the isolated VL and CL fragments (filled squares in Figures 2a, 4a, and 5a; Figure S4a, Supporting Information). This effect cannot result from the His tag as it provides no detectable contribution to the far-UV CD of the LC. Hence, these data must reflect conformational differences between the full-length LC and its individual VL and CL domains. The nonaddititive nature of the far-UV CD spectra of the full-length LC and its fragments indicates that interdomain interaction affects protein conformation in the folded state.
The importance of interdomain interactions during LC unfolding is further supported by the melting data in Figure 6a. The CD heating data were recorded at a rate of 80 °C/h for the full-length LC and its VL and CL fragments (filled symbols). The sigmoidal unfolding transition of the full-length protein cannot be represented as a sum of two consecutive transitions of its VL and CL fragments (Figure S4b, Supporting Information). This is illustrated by the first derivatives of the CD melting data, d[Θ](T)/dT, which show a well-resolved peak corresponding to the unfolding of full-length LC (Figure 6b). The peak is located between those of the VL and CL fragments and cannot be represented by their weighted average. Therefore, the VL and CL regions unfold together rather than independently, which implies extensive interactions between these regions during the unfolding of the full-length LC. Furthermore, the heating data in Figure 6 demonstrate clearly that the rank order of the protein stability inferred from their apparent melting temperatures is VL < LC < CL. Consequently, in the full-length protein, the constant region stabilizes the variable region while the variable destabilizes the constant region.
Figure 6.
Thermal unfolding of recombinant amyloidogenic and germline LC proteins compared. (a) CD data were recorded of full-length LC proteins (circles), their VL fragments (triangles), and their common CL fragment (filled squares) at 202 nm during heating at a rate of 80 °C/h. AL proteins are in filled symbols and germline proteins are in open symbols. The apparent transition temperatures Tm, which were determined from the peak position in the dΘ/dT functions, are 60 °C(C-region) > 53 °C (full-length germline LC) > 51 °C (full-length amyloidogenic LC) > 47 °C (germline VL) > 43 °C (amyloidogenic VL). (b) First derivatives of the CD heating data of AL proteins (selected from panel a).
Comparison of AL and Germline Proteins
To understand whether mutations acquired during the development of amyloid-producing plasma cells affect LC stability and aggregation, we compared the AL 01–095 protein with a protein expressed from the sequence of its most probable germline donor, IGLV6–57*01, which was obtained from the ALBase database (23). Sequence alignment revealed nine amino acid differences in the variable region, four in the complementarity determining regions and five in the framework region (Figure 7). To test the effects of these substitutions on the protein stability, we engineered mutations in the VL region of the full-length AL 01–095 LC to restore its identity with the IGLV6–57*01 germline donor sequence, leaving the CL region unchanged. The resulting full-length LC and its VL fragment (termed germline proteins) were produced recombinantly, and their secondary structure and stability were analyzed by CD spectroscopy.
Comparison of the far-UV CD data of the amyloidogenic and germline proteins showed several important features. First, CD spectra of the germline LC and VL proteins were very similar to their amyloidogenic counterparts, indicating that the germline mutations had no significant effect on the secondary structure (Figure S5, Supporting Information). Second, similar to amyloidogenic proteins, the germline proteins showed nonadditive far-UV CD spectra, Θ(full-length LC) ≠ Θ(VL) + Θ(CL), indicating that the protein conformation in the folded state is affected by the interdomain interactions in full-length LC. Third, similar to amyloidogenic proteins, the melting data of the germline proteins showed an irreversible transition coupled to aggregation for full-length LC and an inherently reversible transition for the VL fragment (Figure S5, Supporting Information). Fourth, similar to amyloidogenic proteins, the CD heating data of the germline proteins were nonadditive, indicating interdomain interactions during protein unfolding (Figure 6a). Finally, the melting data of amyloidogenic and germline proteins showed similar rank order of stability, VL < LC < CL (Figure 6a). The only distinction between the amyloidogenic and germline proteins observed by CD was that, compared to amyloidogenic proteins, the melting data of the germline LC and VL proteins were shifted to higher temperatures by about 2–4 °C (Figure 6a, open and filled circles and triangles). This implies higher stability of the germline proteins as compared to their pathogenic counterparts, which is consistent with exhibiting a less amyloidogenic nature of the germline proteins.
Analysis of LC Unfolding Kinetics
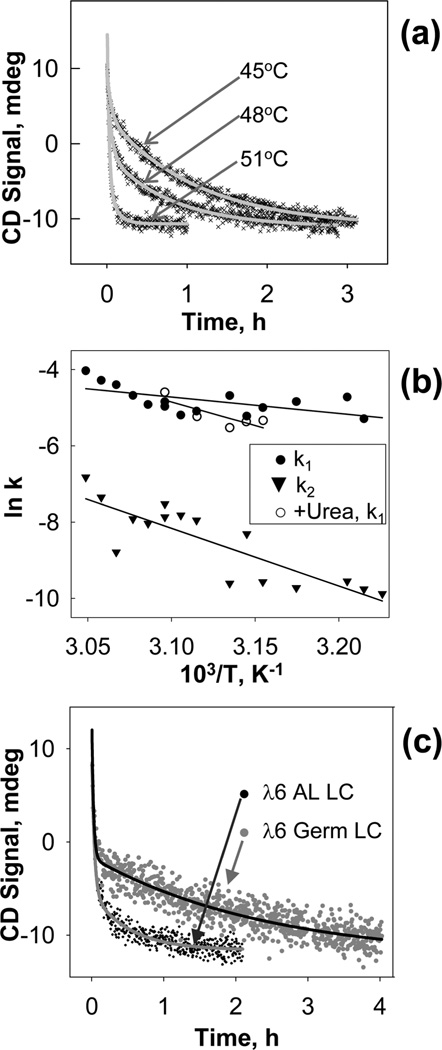
The hysteresis and scan rate effects in the melting data in Figure 2 indicate that thermal unfolding of full-length LC is a kinetically controlled transition with high activation energy. To quantify this unfolding kinetics, we performed temperature-jump (T-jump) experiments using recombinant amyloidogenic and germline LC proteins. Protein unfolding was triggered at time t = 0 by a rapid temperature increase from 25 °C to a higher constant value (37–55 °C), and the time course of the β-sheet to random coil conversion was monitored by CD at 202 nm, Θ](t). Figure 8a shows selected T-jump data recorded from full-length AL LC at 10 µM. No effects of protein concentration on the transition kinetics were detected in the concentration range explored (5–20 µM protein). The T-jump data were approximated with multiexponential decay functions, [Θ](t)=A1 exp(−k1t) + A2 exp(−k2t) + …, where Ai is the amplitude and ki is the temperature-dependent rate constant of the ith kinetic phase. Two exponents were required for adequate data fitting (solid lines in Figure 8a), suggesting two kinetic phases: the first faster phase occurs on a time scale of minutes, and the second slower phase takes hours to complete. Figure 8b shows Arrhenius plots, ln k(T) versus 1/T, for the two kinetic phases of LC unfolding (filled symbols). Linear approximation of these plots suggests comparable slopes, implying comparable activation energies (enthalpies) for these phases, Ea=30 ± 10 kcal/mol. The relatively large uncertainly in this estimate, which incorporates fitting errors and deviations among different data sets, precludes more detailed quantification of the LC unfolding kinetics.
Figure 8.
Unfolding kinetics of recombinant full-length LC proteins. (a) Representative CD data recorded at 202 nm of AL LC in temperature jumps from 25 °C to 45, 48, or 51 °C. Gray lines show two-exponential data fitting, [Θ](t) = A1 exp(−k1t)+A2 exp(ℒk2t), indicating two kinetic phases with rate constants k1 and k2. (b) Arrhenius plots, ln(k) versus 1/T, for the two kinetic phases in the unfolding of recombinant AL LC: a fast phase (filled circles) and a slow phase (filled triangles). Solid lines show least-squares fitting of all data points by linear functions; the slopes of these linear plots correspond to the Arrhenius activation energies for the two phases of about Ea~ 30 ± 10 kcal/mol. The accuracy of this estimate reflects the fitting errors and the deviations among fitting three different data sets. Open circles show the Arrhenius plot for the single unfolding phase observed in AL LC in the presence of 1 M urea. (c) Representative kinetic CD data of amyloidogenic and germline full-length LC recorded in a T-jump from 25 to 50 °C. Solid lines show two-exponential data fitting.
What is the physical origin of the two kinetic phases in LC unfolding? The answer is suggested by comparing our LC data to that from the LC VL fragment, 6aJL2, the only LC protein whose unfolding kinetics has been reported (41). The rate of the first unfolding phase observed in our LC studies was comparable to the unfolding rate reported for the 6aJL2 VL fragment and was in the range reported for other immunoglobuline-like proteins (41, 44). Furthermore, on a time scale of minutes, the VL fragment 6aJL2 was shown to undergo reversible two-state unfolding without aggregation, but on a slower time scale this protein reportedly formed soluble aggregates (41). This raises the possibility that the first faster phase seen in Figure 8 involves reversible LC unfolding without aggregation, while the second slower phase is linked to aggregation. This interpretation is strongly supported by the irreversible heat-induced aggregation of LC that was detected by light scattering in our melting experiments (Figure 2) and confirmed by size-exclusion chromatography (Figure 3). Therefore, we propose that the faster kinetic phase in Figure 8b reflects the reversible unfolding of full-length AL LC while the slower phase involves irreversible protein aggregation.
To test this hypothesis, we carried out kinetic analysis of AL LC in the presence of low concentrations of urea to disrupt the protein oligomers without altering the secondary structure. In 1 M urea, full-length AL LC had an intact secondary structure as indicated by far-UV CD spectra but showed faster thermal unfolding in T-jumps (data not shown) as compared to that in denaturant-free buffer. The T-jump data recorded in 1 Murea were well approximated by monoexponentials, suggesting one unfolding phase. Arrhenius analysis showed that this phase was similar to the fast phase detected in denaturant-free buffer (Figure 8b, overlapping open and filled circles). Therefore, the addition of urea had little effect on the fast unfolding phase but eliminated the slow unfolding phase. This further supports the conclusion that the fast phase reflects unfolding of one protein molecule while the slow phase involves aggregation of full-length LC.
Finally, the unfolding kinetics of the full-length amyloidogenic and germline proteins were compared. Figure 8c shows selected T-jump data recorded by CD at 202 nm. These and other kinetic data reveal that the germline LC undergoes slower unfolding, particularly in the second kinetic phase, and, hence, is more stable and less prone to aggregation when compared to the AL LC. This is consistent with the melting data in Figure 6 showing that, under identical experimental conditions, germ-line proteins unfold at higher temperatures and, consequently, are more stable than their AL counterparts. The difference in the unfolding of the amyloidogenic and germline proteins is more pronounced in the T-jump than in the melting data (Figures 6 and 8), suggesting that, similar to other reports, kinetic experiments provide a particularly sensitive tool for elucidating small differences in the stability of proteins and their assemblies (45). Taken together, our results clearly demonstrate lower kinetic stability of the AL LC, which is consistent with a higher amyloid-forming propensity compared to germline LC.
DISCUSSION
Proteomic analysis carried out in this and our previous study (21) has revealed that fibrillar deposits in the tissues of patients with λ6 LC systemic amyloidosis contain not only variable fragments but also full-length LC. Here, we report the first detailed biophysical study of full-length amyloidogenic λ6 LC. The results suggest an important role of the full-length LC in fibrillogenesis. Our CD data reveal that full-length LC undergoes slow irreversible thermal unfolding with a high activation energy, Ea~30 kcal/mol. Analysis of the CD and light scattering melting data, together with the size-exclusion chromatography results, shows clearly that the unfolding of the full-length LC is coupled to irreversible aggregation. Such aggregation apparently corresponds to the slow phase in protein unfolding. Importantly, prolonged incubation of full-length LC at high temperatures leads to formation of fibrils, detected by electron microscopy. We speculate that the residual β-sheet structure in the heat-unfolded protein (~30%) facilitates conversion of the thermally unfolded LC aggregates into the cross-β-sheet conformation in amyloid fibrils (17).
Taken together, our results reveal that full-length LC undergoes a slow irreversible thermal unfolding coupled to aggregation that leads to fibril formation over time. This contrasts with the largely reversible unfolding of the λ6 VL fragment observed on a fast time scale in this and other studies (ref 41 and references therein) and with partially reversible unfolding of the CL region (amyloid fibril formation by κ CL domain in vitro was recently reported (46)). Moreover, similar to AL and germline LC from the λ6 family, an AL LC protein from the κ1 family also showed irreversible unfolding of full-length LC and partially reversible unfolding of its VL and CL fragments (Figure S6, Supporting Information). This suggests that irreversible unfolding and aggregation of the full-length LC, which contrasts starkly with at least partially reversible unfolding and reduced aggregating propensity of its fragments, may be a common feature of immunoglobulin LCs. It is consistent with our analysis of partially irreversible unfolding of full-length κ1 purified from the urine of the patient with multiple myeloma showing that low thermo-dynamic stability is not sufficient for amyloid formation and suggesting the importance of kinetic factors (47).
Our data suggest that the enhanced aggregation of the full-length protein results not only from the combined aggregating propensities of its VL and CL regions but also from the intramolecular interactions between these regions. Such interactions are inferred from the nonadditive far-UV CD spectra and from the nonadditive melting data recorded from these proteins (Figures 2, 4, and 5; Figure S4, Supporting Information). Furthermore, far-UV CD spectra and the melting data of the synthetic germline proteins show similar nonadditive effects (Figure 7; Figure S5, Supporting Information), indicating that these effects are not limited to one particular LC sequence. Taken together, our results prompt us to postulate that kinetic stability and aggregating propensity of full-length LC depend upon the structural stability of the VL and CL regions, as well as upon their interactions.
Interestingly, our CD studies of the recombinant λ6 AL and germline proteins show that the rank order of protein stability is VL<LC<CL. Consequently, in the full-length LC, the VL region destabilizes the CL region while the CL region stabilizes the VL region. What intramolecular interactions between the VL and CL regions can cause this effect? The answer is suggested by the comparison of AL proteins to their germline counterparts: lower thermodynamic stability of the VL fragment corresponds to lower kinetic stability of the full-length protein (Figures 6a and 8c). This implies that, in the full-length protein, increasing the population of the unfolded VL region destabilizes the CL region, possibly via the direct interactions between these partially unfolded regions facilitated by their linkage via the joining region. These interdomain interactions shift the conformation of the full-length protein toward the partially unfolded state that is prone to irreversible aggregation.
The results reported here prompt us to propose a new model for the early events in AL LC aggregation. Our model differs from the existing paradigm that is centered solely on the polymorphic variable region, and it invokes not only thermodynamic but also kinetic instability. We postulate that, compared to the VL region, full-length LC has a higher propensity to aggregate upon partial unfolding. This high aggregating propensity is counterbalanced by the high kinetic barrier that separates the native soluble state of the full-length LC from its partially unfolded aggregation-prone state. The height of this kinetic barrier determines the aggregation rate of full-length LC and is regulated, in part, by the thermodynamic stability of its variable region: the less stable the VL region, the lower the kinetic stability of the full-length LC and the faster it unfolds and aggregates. (For a relationship between thermodynamic and kinetic stability see a recent review by Sanchez-Ruiz (45).) Once the full-length LC has nucleated the aggregate, it provides a template for aggregation of the variable regions, whose rapid deposition is facilitated by their low thermodynamic stability. Thus, the aggregation-prone full-length LC initiates the nucleation, while the relatively unstable variable regions are central to protein deposition at the propagation stage.
This model is consistent with the predominance of variable region fragments found in amyloid deposits in patients with AL amyloidosis. Our model is also consistent with the reports that the amyloid-forming propensity of λ6 LC correlates with, but cannot be entirely attributed to, thermodynamic instability of the VL fragment (11–13). We propose that, in addition to VL instability, another essential and previously unrecognized factor for amyloid nucleation is the interdomain interactions between VL and CL regions in full-length LC.
Our model suggests two complementary strategies in developing small molecules to reduce protein aggregation for the treatment of AL amyloidosis. One is the conventional approach aimed at raising the structural stability of the variable region. This will not only hinder the unfolding and eventual aggregation of the VL fragment but will also diminish its destabilizing effects on the full-length LC, which, according to our model, will decelerate nucleation. This strategy is complicated by the polymorphic nature of the target protein. An alternative strategy is to search for molecules that bind to the constant region, possibly at the interface with the variable region and/or at the joining linker, thereby inhibiting unfavorable interactions between the variable and constant regions in their partially unfolded aggregation-prone states and/or increasing the kinetic stability of full-length LC. This strategy would be expected to slow down the early nucleation stage, i.e., increase the lag time of irreversible aggregation of LC, and perhaps preventing or retarding fibrillogenesis.
Supplementary Material
ACKNOWLEDGMENT
We thank Dr. J. Ward for providing cDNA which was used to make a λ6 AL LC/pQE-1 plasmid. We also thank Dr. T. Prokaeva and Mr. B. Spencer for cloning and DNA sequence analysis of AL 01-095 LC and Dr. C. Costello and Mrs. M. Budnik for mass spectrometry analysis of urinary purified AL01-095 .LC.
Footnotes
This work was supported by National Institutes of Health Grant HL 68705, The Young Family Amyloid Research Fund, and the Amyloid Research Fund at Boston University School of Medicine. CD instruments were supported by NIH Grants HL026355 and GM067260 and by HRSA Grant C76HF 16067.
Abbreviations: AL, amyloid deposits, light chain type; LC, light chain(s); CL region, constant region of light chain; VL region, variable and joined region of light chain; LB, Luria broth; CD, circular dichroism; T-jump, temperature jump.
SUPPORTING INFORMATION AVAILABLE
(1) Secondary structural content in recombinant AL and germline LC proteins; (2) comparison of secondary structure and thermal unfolding of full-length urinary λ6 LC and its recombinant counterpart; (3) SDS gels showing S-S linked dimer and S-S reduced monomer of urinary purified and recombinant full-length LC; (4) electron micrographs showing fibril formation upon prolonged incubation of AL LC at elevated temperatures; (5) comparison of CD data of full-length AL LC and a mixture of its VL and CL fragments; (6) thermal unfolding and refolding of recombinant λ6 germline LC proteins and of recombinant amyloidogenic κ1 LC proteins. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- 1.Sanchorawala V, Seldin DC. An overview of high-dose melphalan and stem cell transplantation in the treatment of AL amyloidosis. Amyloid. 2007;14:261–269. doi: 10.1080/13506120701613984. [DOI] [PubMed] [Google Scholar]
- 2.Skinner M, Sanchorawala V, Seldin DC, Dember LM, Falk RH, Berk JL, Anderson JJ, O’Hara C, Finn KT, Libbey CA, Wiesman J, Quillen K, Swan N, Wright DG. High-dose melphalan and autologous stem-cell transplantation in patients with AL amyloidosis: an 8-year study. Ann. Intern. Med. 2004;140:85–93. doi: 10.7326/0003-4819-140-2-200401200-00008. [DOI] [PubMed] [Google Scholar]
- 3.Seldin DC, Anderson JJ, Sanchorawala V, Malek K, Wright DG, Quillen K, Finn KT, Berk JL, Dember LM, Falk RH, Skinner M. Improvement in quality of life of patients with AL amyloidosis treated with high-dose melphalan andautologous stem cell transplantation. Blood. 2004;104:1888–1893. doi: 10.1182/blood-2004-01-0089. [DOI] [PubMed] [Google Scholar]
- 4.Amzel LM, Poljak RJ. Three-dimensional structure of immunoglobulins. Annu. Rev. Biochem. 1979;48:961–997. doi: 10.1146/annurev.bi.48.070179.004525. [DOI] [PubMed] [Google Scholar]
- 5.Abraham RS, Geyer SM, Price-Troska TL, Allmer C, Kyle RA, Gertz MA, Fonseca R. Immunoglobulin light chain variable (V) region genes influence clinical presentation and outcome in light chain-associated amyloidosis (AL) Blood. 2003;101:3801–3808. doi: 10.1182/blood-2002-09-2707. [DOI] [PubMed] [Google Scholar]
- 6.Comenzo RL, Wally J, Kica G, Murray J, Ericsson T, Skinner M, Zhang Y. Clonal immunoglobulin light chain variable region germline gene use in AL amyloidosis: association with dominant amyloid-related organ involvement and survival after stem cell transplantation. Br. J. Haematol. 1999;106:744–751. doi: 10.1046/j.1365-2141.1999.01591.x. [DOI] [PubMed] [Google Scholar]
- 7.Comenzo RL, Zhang Y, Martinez C, Osman K, Herrera GA. The tropism of organ involvement in primary systemic amyloidosis: contributions of Ig V(L) germ line gene use and clonal plasma cell burden. Blood. 2001;98:714–720. doi: 10.1182/blood.v98.3.714. [DOI] [PubMed] [Google Scholar]
- 8.Solomon A, Frangione B, Franklin EC. Bence Jones proteins and light chains of immunoglobulins. Preferential association of the V lambda VI subgroup of human light chains with amyloidosis AL (lambda) J. Clin. Invest. 1982;70:453–460. doi: 10.1172/JCI110635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Solomon A, Weiss DT. Protein and host factors implicated in the pathogenesis of light chain amyloidosis (AL amyloidosis) Amyloid. 1995;2:269–279. [Google Scholar]
- 10.Olsen KE, Sletten K, Westermark P. Fragments of the constant region of immunoglobulin light chains are constituents of AL-amyloid proteins. Biochem. Biophys. Res. Commun. 1998;251:642–647. doi: 10.1006/bbrc.1998.9508. [DOI] [PubMed] [Google Scholar]
- 11.Wall J, Schell M, Murphy C, Hrncic R, Stevens FJ, Solomon A. Thermodynamic instability of human lambda 6 light chains: correlation with fibrillogenicity. Biochemistry. 1999;38:14101–14108. doi: 10.1021/bi991131j. [DOI] [PubMed] [Google Scholar]
- 12.del Pozo Yauner L, Ortiz E, Sanchez R, Sanchez-Lopez R, Guereca L, Murphy CL, Allen A, Wall JS, Fernandez-Velasco DA, Solomon A, Becerril B. Influence of the germline sequence on the thermodynamic stability and fibrillogenicity of human lambda 6 light chains. Proteins. 2008;72:684–692. doi: 10.1002/prot.21934. [DOI] [PubMed] [Google Scholar]
- 13.Kim Y, Wall JS, Meyer J, Murphy C, Randolph TW, Manning MC, Solomon A, Carpenter JF. Thermo-dynamic modulation of light chain amyloid fibril formation. J. Biol. Chem. 2000;275:1570–1574. doi: 10.1074/jbc.275.3.1570. [DOI] [PubMed] [Google Scholar]
- 14.Wall JS, Gupta V, Wilkerson M, Schell M, Loris R, Adams P, Solomon A, Stevens F, Dealwis C. Structural basis of light chain amyloidogenicity: comparison of the thermodynamic properties, fibrillogenic potential and tertiary structural features of four Vlambda6 proteins. J. Mol. Recognit. 2004;17:323–331. doi: 10.1002/jmr.681. [DOI] [PubMed] [Google Scholar]
- 15.McLaughlin RW, De Stigter JK, Sikkink LA, Baden EM, Ramirez-Alvarado M. The effects of sodium sulfate, glycosaminoglycans, and Congo red on the structure, stability, and amyloid formation of an immunoglobulin light-chain protein. Protein Sci. 2006;15:1710–1722. doi: 10.1110/ps.051997606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arendt BK, Ramirez-Alvarado M, Sikkink LA, Keats JJ, Ahmann GJ, Dispenzieri A, Fonseca R, Ketterling RP, Knudson RA, Mulvihill EM, Tschumper RC, Wu X, Zeldenrust SR, Jelinek DF. Biologic and genetic characterization of the novel amyloidogenic lambda light chain-secreting human cell lines, ALMC-1 and ALMC-2. Blood. 2008;112:1931–1941. doi: 10.1182/blood-2008-03-143040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khurana R, Gillespie JR, Talapatra A, Minert LJ, Ionescu-Zanetti C, Millett I, Fink AL. Partially folded intermediates as critical precursors of light chain amyloid fibrils and amorphous aggregates. Biochemistry. 2001;40:3525–3535. doi: 10.1021/bi001782b. [DOI] [PubMed] [Google Scholar]
- 18.Kim YS, Randolph TW, Stevens FJ, Carpenter JF. Kinetics and energetics of assembly, nucleation, and growth of aggregates and fibrils for an amyloidogenic protein. Insights into transition states from pressure, temperature, and co-solute studies. J. Biol. Chem. 2002;277:27240–27246. doi: 10.1074/jbc.M202492200. [DOI] [PubMed] [Google Scholar]
- 19.Qin Z, Hu D, Zhu M, Fink AL. Structural characterization of the partially folded intermediates of an immunoglobulin light chain leading to amyloid fibrillation and amorphous aggregation. Biochemistry. 2007;46:3521–3531. doi: 10.1021/bi061716v. [DOI] [PubMed] [Google Scholar]
- 20.Souillac PO, Uversky VN, Fink AL. Structural transformations of oligomeric intermediates in the fibrillation of the immunoglobulin light chain LEN. Biochemistry. 2003;42:8094–8104. doi: 10.1021/bi034652m. [DOI] [PubMed] [Google Scholar]
- 21.Lavatelli F, Perlman DH, Spencer B, Prokaeva T, McComb ME, Theberge R, Connors LH, Bellotti V, Seldin D, Merlini G, Skinner M, Costello CE. Amyloidogenic and associated proteins in systemic amyloidosis proteome of adipose tissue. Mol. Cell. Proteomics. 2008;7:1570–1583. doi: 10.1074/mcp.M700545-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ch′ang L-Y, Yen C-P, Besl L, Schell M, Solomon A. Identification and characterization of a functional human Ig VlVI germline gene. Mol. Immunol. 1994;31:531–536. doi: 10.1016/0161-5890(94)90040-x. [DOI] [PubMed] [Google Scholar]
- 23.Bodi K, Prokaeva T, Spencer B, Eberhard M, Connors LH, Seldin DC. AL-Base: a visual platform analysis tool for the study of amyloidogenic immunoglobulin light chain sequences. Amyloid. 2009;16:1–8. doi: 10.1080/13506120802676781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sreerama N, Woody RW. Estimation of protein secondary structure from circular dichroism spectra: comparison of CONTIN, SELCON, and CDSSTR methods with an expanded reference set. Anal. Biochem. 2000;287:252–260. doi: 10.1006/abio.2000.4880. [DOI] [PubMed] [Google Scholar]
- 25.Sreerama N, Venyaminov SYu, Woody RW. Estimation of the number of R-helical and β-strand segments in proteins using circular dichroism spectroscopy. Protein Sci. 1999;8:370–380. doi: 10.1110/ps.8.2.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Provencher SW, Glöckner J. Estimation of protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 27.van Stokkum IHM, Spoelder HJW, Bloemendal MR, van Grondelle R, Groen FCA. Estimation of protein secondary structure and error analysis from circular dichroism. Anal. Biochem. 1991;291:110–118. doi: 10.1016/0003-2697(90)90396-q. [DOI] [PubMed] [Google Scholar]
- 28.Johnson WC., Jr Analyzing protein circular dichroism spectra for accurate secondary structures. Proteins. 1999;35:307–312. [PubMed] [Google Scholar]
- 29.Benjwal S, Verma S, Rohm KH, Gursky O. Monitoring protein aggregation during thermal unfolding in circular dichroism experiments. Protein Sci. 2006;15:635–639. doi: 10.1110/ps.051917406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.John DM, Weeks KM. Van’t Hoff enthalpies without baselines. Protein Sci. 2000;9:1416–1419. doi: 10.1110/ps.9.7.1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tojo K, Sekijima Y, Kelly JW, Ikeda S. Diflunisal stabilizes familial amyloid polyneuropathy-associated transthyretin variant tetramers in serum against dissociation required for amyloidogenesis. Neurosci. Res. 2006;56:441–449. doi: 10.1016/j.neures.2006.08.014. [DOI] [PubMed] [Google Scholar]
- 32.Solomon A, Weiss DT. Structural and functional properties of human lambda-light-chain variable-region subgroups. Clin. Diagn. Lab. Immunol. 1995;2:387–394. doi: 10.1128/cdli.2.4.387-394.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ely KR, Herron JN, Harker M, Edmundson AB. Three-dimensional structure of a light chain dimer crystallized in water. Conformational flexibility of a molecule in two crystal forms. J. Mol. Biol. 1989;210:601–615. doi: 10.1016/0022-2836(89)90135-6. [DOI] [PubMed] [Google Scholar]
- 34.Huang DB, Ainsworth CF, Stevens FJ, Schiffer M. Three quaternary structures for a single protein. Proc. Natl. Acad. Sci. U.S.A. 1996;93:7017–7021. doi: 10.1073/pnas.93.14.7017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bourne PC, Ramsland PA, Shan L, Fan ZC, DeWitt CR, Shultz BB, Terzyan SS, Moomaw CR, Slaughter CA, Guddat LW, Edmundson AB. Three-dimensional structure of an immunoglobulin light-chain dimer with amyloidogenic properties. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2002;58:815–823. doi: 10.1107/s0907444902004183. [DOI] [PubMed] [Google Scholar]
- 36.Titani K, Wikler M, Shinoda T, Putnam FW. The amino acid sequence of a λ type Bence-Jones protein. III. The complete amino acid sequence and the location of the disulfide bridges. J. Biol. Chem. 1970;245:2171–2176. [PubMed] [Google Scholar]
- 37.Sanchez-Ruiz J, Lopez-Lacomba JL, Cortijo M, Mateo PL. Differential scanning calorimetry of the irreversible thermal denaturation of thermolysin. Biochemistry. 1988;27:1648–1652. doi: 10.1021/bi00405a039. [DOI] [PubMed] [Google Scholar]
- 38.Gursky O, Ranjana Gantz DL. Complex of human apolipoprotein C-1 with phospholipid: thermodynamic or kinetic stability? Biochemistry. 2002;41:7373–7384. doi: 10.1021/bi025588w. [DOI] [PubMed] [Google Scholar]
- 39.Kaplan B, Ramirez-Alvarado M, Sikkink L, Golderman S, Dispenzieri A, Livneh A, Gallo G. Free light chains in plasma of patients with light chain amyloidosis and non-amyloid light chain deposition disease. High proportion and heterogeneity of disulfide-linked monoclonal free light chains as pathogenic features of amyloid disease. Br. J. Haematol. 2009;144:705–715. doi: 10.1111/j.1365-2141.2008.07522.x. [DOI] [PubMed] [Google Scholar]
- 40.Souillac PO, Uversky VN, Millett IS, Khurana R, Doniach S, Fink AL. Effect of association state and conformational stability on the kinetics of immunoglobulin light chain amyloid fibril formation at physiological pH. J. Biol. Chem. 2002;277:12657–12665. doi: 10.1074/jbc.M109230200. [DOI] [PubMed] [Google Scholar]
- 41.Blancas-Mejia LM, Tellez LA, del Pozo-Yauner L, Becerril B, Sanchez-Ruiz JM, Fernandez-Velasco DA. Thermo-dynamic and kinetic characterization of a germ line human lambda6 light-chain protein: the relation between unfolding and fibrillogenesis. J. Mol. Biol. 2009;386:1153–1166. doi: 10.1016/j.jmb.2008.12.069. [DOI] [PubMed] [Google Scholar]
- 42.Pokkuluri PR, Solomon A, Weiss DT, Stevens FJ, Schiffer M. Tertiary structure of human lambda 6 light chains. Amyloid. 1999;6:165–171. doi: 10.3109/13506129909007322. [DOI] [PubMed] [Google Scholar]
- 43.Hernandez-Santoyo A, Del Pozo Yauner L, Fuentes-Silva D, Ortiz E, Rudino-Pinera E, Sanchez-Lopez R, Horjales E, Becerril B, Rodriguez-Romero A. A single mutation at the sheet switch region results in conformational changes favoring lambda6 light-chain fibrillogenesis. J. Mol. Biol. 2010;396:280–292. doi: 10.1016/j.jmb.2009.11.038. [DOI] [PubMed] [Google Scholar]
- 44.Clarke J, Cota E, Fowler SB, Hamill SJ. Folding studies of immunoglobulin-like beta-sandwich proteins suggest that they share a common folding pathway. Structure. 1999;7:1145–1153. doi: 10.1016/s0969-2126(99)80181-6. [DOI] [PubMed] [Google Scholar]
- 45.Sanchez-Ruiz JM. Protein kinetic stability. Biophys. Chem. 2010;148:1–15. doi: 10.1016/j.bpc.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 46.Yamamoto K, Yagi H, Lee YH, Kardos J, Hagihara Y, Naiki H, Goto Y. The amyloid fibrils of the constant domain of immunoglobulin light chain. FEBS Lett. 2010;584:3348–3353. doi: 10.1016/j.febslet.2010.06.019. [DOI] [PubMed] [Google Scholar]
- 47.Chung CM, Chiu JD, Connors LH, Gursky O, Lim A, Dykstra AB, Liepnieks J, Benson MD, Costello CE, Skinner M, Walsh MT. Thermodynamic stability of a kappaI immunoglobulin light chain: relevance to multiple myeloma. Biophys. J. 2005;88:4232–4242. doi: 10.1529/biophysj.105.061317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kabat E, Wu TT, Perry HM, Gottesman KS, Foeller C. Sequences of proteins of immunological interest, Vol. 5, National Institutes of Health. Bethesda, MD: 1991. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.