Abstract
Genome-wide association studies (GWAS) have strongly implicated MIR137 (the gene encoding the microRNA miR-137) in schizophrenia. A parsimonious hypothesis is that a pathway regulated by miR-137 is important in the etiology of schizophrenia. Full evaluation of this hypothesis requires more definitive knowledge about biological targets of miR-137, which is currently lacking. Our goals were to expand knowledge of the biology of miR-137 by identifying its empirical targets, and to test whether the resulting lists of direct and indirect targets were enriched for genes and pathways involved in risk for schizophrenia. We overexpressed miR-137 in a human neural stem cell line and analyzed gene expression changes at 24 and 48 h using RNA sequencing. Following miR-137 overexpression, 202 and 428 genes were differentially expressed after 24 and 48 h. Genes differentially expressed at 24 h were enriched for transcription factors and cell cycle genes, and differential expression at 48 h affected a wider variety of pathways. Pathways implicated in schizophrenia were upregulated in the 48 h findings (major histocompatibility complex, synapses, FMRP interacting RNAs and calcium channels). Critically, differentially expressed genes at 48 h were enriched for smaller association P-values in the largest published schizophrenia GWAS. This work provides empirical support for a role of miR-137 in the etiology of schizophrenia.
Introduction
Schizophrenia is now known to be highly polygenic with a genetic architecture characterized by contributions from dozens and probably hundreds of different genetic loci. Although rare deleterious exonic and copy number variation (CNVs) have roles, numerous common variants of more subtle effect account for sizable fractions of the heritability of schizophrenia.1, 2, 3, 4
A replicated and robust association with schizophrenia is in MIR137, the gene encoding the microRNA miR-137. This region was initially identified by the Psychiatric Genomics Consortium (P=1.6 × 10−11).5 In the largest genome-wide association study (GWAS) yet conducted (by the Psychiatric Genomics Consortium, 36 989 cases), the second strongest finding was within MIR137 (P=3.4 × 10−19).1 Pathway analyses have suggested that genes containing predicted miR-137 target sites are significantly enriched for smaller P-values for association with schizophrenia.5, 6, 7 This enrichment is not robustly identified in all studies,1 and one possibility is that miR-137 target predictions are not sufficiently accurate.
It is possible that a pathway regulated by miR-137 is important in the etiology of schizophrenia. microRNAs can target large numbers of mRNAs for degradation or translational inhibition, and a single microRNA can have major effects on cellular pathways.8, 9, 10, 11 miR-137 has important roles in neural development12, 13, 14, 15 and has been implicated in autism, intellectual disability, Rett syndrome, Alzheimer's disease, Huntington's disease and multiple cancers.14,16, 17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28
Schizophrenia GWAS results, pathway analyses and the roles of miR-137 in development and disease are consistent with the broad hypothesis that genetic variation in miR-137 and its targets constitute a pathway whose dysfunction is involved in the etiology of schizophrenia. There are several major limitations in evaluating this hypothesis. A key issue is that miR-137 target genes are mostly based on computational algorithms of imperfect sensitivity and specificity. These algorithms predict only the direct targets of miR-137.29,30 Some direct targets of miR-137 have been verified12, 13, 14,18,25, 26, 27, 28,31,32 including genes implicated in schizophrenia (CACNA1C, TCF4, CSMD1, C10orf26 and ZNF804A).33,34 However, many of these targets were identified in cancers, and their neuronal role is unclear. miRNA regulatory activity is complex and can include regulatory loops, post-translational modifications and non-canonical target sequences (for example, in coding regions and not 3' untranslated regions).15,35,36 The 'downstream' effects of miR-137 (that is, its impact beyond a directly targeted transcript) have not been systematically evaluated.
We thus evaluated the effects of miR-137 overexpression in a human neuronal cell line. Our goal was to expand knowledge of the biology of miR-137 by identifying its empirical targets. We transduced neuronal stem cells with vectors expressing miR-137 or a control miRNA mimic and used high-throughput sequencing (RNA-seq) at 24 and 48 h after transduction to assess the effects of miR-137 overexpression. Overexpression of miR-137 had an impact on multiple cell cycle genes, consistent with other findings of effects on proliferation and downregulation in cancer. However, we also found that the indirect effects of miR-137 overexpression at 48 h had strong connections with GWAS findings for schizophrenia. These results provide functional support for a role of miR-137 in the etiology of schizophrenia.
Materials and methods
Cell line
ReNcell-VM (Millipore, Billerica, MA, USA) isa human neural precursor cell line derived from the ventral mesencephalon.37 We chose this cell line because it was readily available, accessible to our methods and not derived from a cancer. Moreover, we genotyped two different clones with Illumina OminExpress arrays and confirmed the expected male sex, European ancestry, the absence of aneuploidy and the absence of CNV larger than >100 kb that affected a gene.
Infection of cells
ReNcell-VMs were grown on laminin-coated plates in ReNcell NSC Maintenance Media (Millipore) with 20 ng ml−1 FGF-2 and 20 ng ml−1 epidermal growth factor. We used pEZX-MR03 lentiviral vectors with miR-137 or a control packaged with an HIV-1-compatible system (GeneCopoeia, Rockville, MD, USA). Infection efficiency was assessed by microscopic evaluation of eGFP expression (contained in the pEZX-MR03 plasmid). To evaluate nonspecific effects because of overloading of the miRNA-processing machinery, we used a control lentiviral vector containing a miRNA mimic (that is, a random miR-like sequence of no similarity to any known miRNA). Flasks of cells were infected in parallel with the miR-137 or control lentivirus. Viral amounts were titrated to achieve ~100% infection rates without evidence of toxicity. For the 48-h time point, the cells were exposed to virus for 24 h and then replaced with fresh media. Cells were collected 24 or 48 h after infection, stored in RNA-stabilizing media (RNAlater, Qiagen, Venlo, The Netherlands) and frozen at −80 °C until analysis. Evaluation of two relatively early time points was consistent with our goal of identifying the immediate effects of miR-137 overexpression as these are likely to be enriched for genes targeted by miR-137.
RNA extraction
Total RNA was extracted using either Maxwell 16 LEV Simply RNA kit (Madison, WI, USA; 24 h RNA-seq) or Norgen Biotek RNA/DNA/Protein Kit (Thorold, ON, Canada; 48 h RNA-seq and mature miR-137 assays). Although both methods yield total RNA, the Norgen kit extracts more mature microRNAs (data not shown) and is better suited for quantification of miR-137.
Quantitative PCR
We used TaqMan assays to measure expression of mature miR-137 (Applied Biosystems, assay ID 001129; Grand Island, NY, USA). Each sample was run in quadruplicate, and StepOne software (v2.1; SAS, Cary, NC, USA) was used to evaluate ΔCt and fold change (RQ) values. The endogenous control was RNU6B. Statistical significance of the ΔCt values was assessed using nested analysis of variance (SAS v9.2, PROC GLM). All assays were performed on the same RNA samples as were used for RNA-seq except for the miR-137 assays at the 24-h time point, which were carried out on replicate samples (using RNA extracted with the Norgen kit).
RNA-seq
RNA-sequencing libraries were generated using Truseq RNA sample prep kits (Illumina, San Diego, CA, USA), which generate libraries from poly-adenylated RNAs. Each library was barcoded, pooled and sequenced using Illumina HiSeq 2000 and single-end 100-bp reads. Single-end sequencing was chosen to maximize read depth and detection of small fold changes. There were three miR-137 overexpressions and three controls at 24 h, and five miR-137 overexpressions and six controls at 48 h. Read depth averaged ~55 million reads per replicate at 24 h and ~160 million reads per replicate at 48 h (see below for explanation of greater numbers of reads at 48 h). All procedures were randomized.
Data processing and analysis
We used the TopHat-Cufflinks-Cuffdiff pipeline to detect differential gene expression between neural stem cells with increased expression of miR-137 compared with controls. First, we used FastQC (see URLs) to evaluate sequencing quality. As one 48-h sample failed, we repeated RNA-seq on all of the 48-h samples. We verified sample identity across all experiments via the creation of a vector of genotypes based on high-confidence heterozygous exonic RNA-seq reads. RNA-seq reads were also aligned to the overexpression sequence (miR-137 or a miR-like mimic) to confirm experimental condition. Second, we mapped RNA-seq reads to the human reference genome (UCSC hg19) using TopHat2 and transcript annotations from the TopHat website (see URLs).38 Third, the mapped reads were assembled into transcripts using Cufflinks (v2.0.1; http://cufflinks.cbcb.umd.edu).39 Fourth, we pooled the Cufflinks output files from multiple RNA-seq samples using Cuffcompare to assemble a comprehensive set of transcripts. Finally, we used Cuffdiff39 to evaluate differential gene expression of miR-137 overexpression versus the control vector separately at 24 and 48 h with correction for multiple comparisons using a false-discovery rate approach (q-value).40 We used the RNA-seq data to evaluate gene expression of transcripts whose protein products are known to characterize neuronal differentiation using a read count approach (Supplementary Methods). A few genes were reported more than once, and the transcript with the smallest P-value was retained.41
Additional analyses
We compared the effects of miR-137 overexpression in neural progenitor cells to computational predictions of genes targeted by miR-137. We used Sylarray (see URLs) to test all known miRNA seed sequences in the RNA-seq gene level data.42 The analyses are performed at varying cutoffs to create a curve of log10(P) values for each miRNA. We also compared differentially expressed genes from miR-137 overexpression to computationally predicted miR-137 targets (TargetScan v6.2; http://www.targetscan.org)43 with significance determined using permutation.
DAPPLE was used to evaluate whether the lists of genes had more protein–protein interactions than predicted by chance.44 We used DAVID45,46 to evaluate whether genes differentially expressed upon miR-137 overexpression tended to occur in specific biological pathways or processes including GO,47 KEGG48 and Biocarta (see URLs). DAVID generated enrichment scores for correlated pathways and Benjamini corrected P-values for individual pathways. PWM scan49,50 was used to predict high-confidence transcription factor-binding sites in gene promoters. We then identified transcription factor-binding site over-represented in the promoters of significantly differentially expressed genes by Monte Carlo simulation analysis.
We compared genes differentially expressed following miR-137 overexpression to an analogous experiment by Hill et al.51 and to a gene expression study in lymphoblastoid cell lines from schizophrenia cases and controls.52 We evaluated the relevance of genes differentially expressed following miR-137 overexpression to pathways previously implicated in the etiology of schizophrenia: expert-curated lists of genes found at synapses (1043 genes),53 genes encoding proteins found at the postsynaptic density (1457 genes),54 genes in neuronal activity-regulated cytoskeleton-associated protein postsynaptic signaling complexes (28 genes),55 N-methyl-D-aspartate receptors (61 genes),55 L-voltage-gated calcium channel subunits (26 genes),6 genes whose proteins interact with calcium channel (Cav2, 207 genes)56 and genes whose mRNAs interact with FMRP (fragile X mental retardation protein, 842 genes).57
Finally, we assessed the overlap of genes differentially expressed following miR-137 overexpression to schizophrenia GWAS results. This analysis is a relatively direct evaluation of the salience of sets of genes directly or indirectly regulated by miR-137 to genetic variation implicated in the etiology of schizophrenia. We used INRICH58 to test whether differentially expressed genes were enriched for smaller GWAS P-values in the largest published schizophrenia GWAS6 (Supplementary Methods).
Results
We transduced human neural precursor cells (ReNcell-VM) with lentiviral vectors expressing either miR-137 or a scrambled miRNA-like control. We verified infection rates >99% via fluorescent imaging of the eGFP tag in the vector. Alignment of RNA-seq reads to the lentiviral sequence confirmed correct expression of the experimental vector in each sample.
Verification
We verified increased miR-137 expression in two ways. qPCR confirmed marked increases in the amounts of mature miR-137 at 24 h (111x increase) and 48 h (548x increase) in cells exposed to lentivirus expressing miR-137 compared with controls (Supplementary Figure S1A). We used RNA-seq to estimate expression of the primary poly-adenylated MIR137 transcript from the miR-137 lentiviral vector and the MIR137 locus. Consistent with the qPCR results, we found 123x increase in the primary transcript at 24 h and 660x increase at 48 h (Supplementary Figure S1B).
As miR-137 can effect neuronal proliferation and differentiation,12, 13, 14,59 we evaluated cell density, morphology and expression of differentiation markers in the same flasks used for RNA-seq. There were no significant differences in cell densities between miR-137 overexpression and control flasks at 24 (P=0.53) or 48 h (P=0.78), although the 48-h flasks had higher cell densities than at 24 h (P=7.6 × 10−9, as expected because of additional growth time). We noted no qualitative differences in cellular morphology between the experimental conditions at either time point. We used RNA-seq to compare expression of 12 genes whose protein products are commonly used to classify neuronal cell types (Supplementary Table S1). There were no significant differences between the miR-137 overexpression and control conditions. The overall pattern of results is consistent with that of a neuronal stem cell. Thus, large increases in expression of miR-137 did not lead to clear alterations in cell fate after 24 or 48 h.
We verified the downregulation of predicted targets of miR-137 in two ways. First, we applied Sylarray42 to the RNA-seq data to evaluate fold change differences associated with all known miRNA seed sequences. Genes with the miR-137 seed sequence (AGCAATTA) in the 3'-untranslated repeat evidenced the greatest downregulation of any seed sequence (Supplementary Figure S1C, minimum P=1.1 × 10−22 in the 24-h condition and minimum P=2.4 × 10−45 in the 48-h condition). As a complementary test, we used permutation to evaluate differential gene expression in 1115 genes with predicted miR-137 target sites (TargetScan v6.2).43 Genes with a predicted miR-137 target site were significantly more likely to be differentially expressed (Supplementary Figure S1D, P<10−200 at both 24 and 48 h), and most were downregulated at 24 (82%) and 48 h (87%). These results are consistent with profound and relatively specific downregulation resulting from miR-137 overexpression.
General expression patterns
Supplementary Table S2 shows patterns of RNA-seq analyzability at 24 and 48 h. After removing transcripts that had insufficient sequencing depth, low expression levels or technical faults and resolving genes reported more than once, there were 14 682 transcripts analyzable at both 24 and 48 h. The correlations between miR-137 overexpression and control conditions were 0.999 at both 24 and 48 h, and the correlations between the 24 and 48 h conditions were 0.980–0.982 (Supplementary Figures S2a and S2b). We applied principal component analysis to the expression data matrix (Supplementary Figure S3). The first principal component accounted for 97% of the variance and completely distinguished the replicates from 24 and 48 h. These descriptive results suggest the comparability of the RNA-seq data within and between time points. The results also suggest that only a fraction of transcripts was altered by the miR-137 experimental manipulation.
Differential gene expression
Supplementary Figure S4 shows descriptive statistics for log2 fold changes, P-values and q-values for the 24 and 48 h analyses. These analyses compare the miR-137 overexpression to a control vector within each time point. At 24 h, 14 869 unique genes were analyzable and 202 (1.36%) had evidence of differential gene expression (q<0.05, Supplementary Table S3). Of the differentially expressed genes, 130 genes (64.4%) had evidence of inhibition of gene expression following exposure to markedly elevated amounts of miR-137 and 72 genes (35.6%) showed enhancement. At 48 h, 428 (2.71%) of 15 780 analyzable and unique genes had evidence of differential expression (q<0.05, Supplementary Table S4). Of these, 157 genes (36.7%) showed inhibition after miR-137 overexpression and 271 genes (63.3%) showed enhancement.
Review of the read count data showed no instances where miR-137 overexpression acted as a switch at either time point (that is, high expression in the control condition and very low expression with miR-137 overexpression). Differentially expressed genes had subtler effects with transcriptional changes ranging from 1.8-fold downregulation to 1.8-fold upregulation at 24 h and from 2.9-fold downregulation to 2.8-fold upregulation at 48 h (only 12 differentially expressed transcripts had fold changes in excess of 2). Thus, despite substantial increases in miR-137 concentrations, the effects were quantitative not absolute and consistent with a role for miR-137 acting to fine-tune gene expression.
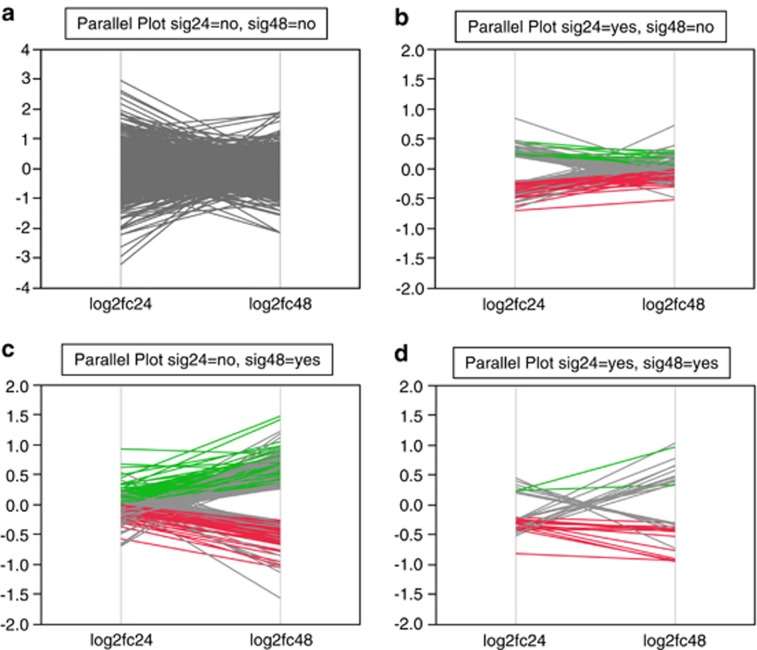
Figure 1 depicts full results from this study and lists of differentially expressed genes are in Supplementary Tables S3 and S4. The results indicate the complexity of the effects of miR-137 overexpression with time. For example, of transcripts that were differentially expressed at both 24 and 48 h, only 44.1% (15/34) had the same direction of effect each time.
Figure 1.
Parallel plots of fold change values for the 14 694 analyzable genes at both 24 and 48 h. The four graphs are for each combination of significance at 24 and 48 h time points (q<0.05). In each graph, the x axis shows fold changes (FCs) at 24 and 48 h time points, log2 (FC24) and log2 (FC48). The y axis is log2 FC with positive values indicating upregulation upon overexpression of miR-137 and negative values the opposite. Each line connects the log2 FC values for a gene. Large FC values may not be statistically significant (can occur when expression levels are low). All lines in a are colored gray. For b–d, red lines indicate inhibition at both 24 and 48 h, green lines increased expression at both 24 and 48 h and grey lines show genes whose log2 FC values switched signs between 24 and 48 h.
We asserted above that the widely used, computationally derived TargetScan (v6.2) list of miR-137 targets is imperfect. The presence of a predicted TargetScan-binding site was associated with significant (q<0.05) differential gene expression at 48 h (odds ratio 4.0, 95% confidence interval (CI) 3.2–5.1, P=2.7 × 10−33) and, as expected from the analyses above, this was mainly because of inhibitory effects of miR-137 overexpression (odds ratio=10.6, 95% CI 7.6–14.6, P=6.4 × 10−69) rather than enhancement (odds ratio=1.2, 95% CI 0.7–2.27, P=0.52). However, these strong statistical associations were not robustly predictive as only 8.9% (87/1002) of analyzable genes with predicted miR-137 target sites were differentially expressed and only 20.3% (87/428) of differentially expressed transcripts had predicted miR-137 target sites. The direct and indirect effects of miR-137 thus appear to be complex and not robustly captured using TargetScan.
Pathway analyses
As the effects of miR-137 overexpression influenced multiple genes and never acted as a switch, we attempted to identify biological themes that could unify the effects of miR-137 across many different transcripts. We used a more liberal definition of differential expression in these analyses (q<0.1 instead of 0.05).
We used DAPPLE44 to assess whether the protein products of genes with significant differential expression had greater protein–protein interaction than expected by chance. We found significantly more interactions at 24 h (P=0.01) and at 48 h (P=0.001), and many of these genes formed a single network at each time point (Supplementary Figure S5). This result is consistent with the hypothesis that miR-137 influences multicomponent functional networks rather than targeting of unrelated individual genes.
We applied pathway analysis to lists of differentially expressed genes using DAVID (Table 1).45,46 At 24 h, genes with inhibition after exposure to markedly increased amounts of miR-137 (presumably enriched for direct targets of miR-137) were enriched for pathways related to extracellular matrix, positive regulation of transcription, neuronal differentiation/projection and extracellular signaling (Supplementary Table S5a). Genes with increased expression at 24 h (presumptive indirect targets of miR-137) were enriched for cell cycle consistent with prior reports (Supplementary Table S5b).15,27,59 In the 48-h data, pathway analysis of differentially expressed genes yielded more changes. Interestingly, cell cycle pathways (upregulated at 24 h) were strongly downregulated at 48 h (Supplementary Table S6a). Pathways involved in DNA replication and cell cycle processes were also downregulated at 48 h. Upregulated pathways at 48 h included strong enrichment for genes involved in multiple types of cell–cell interactions, the major histocompatibility complex class II antigens (a region containing the most significant GWAS association for schizophrenia),5,6,60,61 synapses and ion channels/calcium binding (Supplementary Table S6b).
Table 1. DAVID pathway analysis of differentially expressed genes (q<0.1).
| Time | Direction of effect | Enrichment | Annotation cluster description |
|---|---|---|---|
| 24 h | Downregulation (183 genes) | 3.04 | Extracellular matrix |
| 2.92 | Positive regulation of transcription | ||
| 2.61 | Neuron differentiation/projection | ||
| 2.55 | Extracellular signaling | ||
| Upregulation (126 genes) | 2.75 | Cell cycle | |
| 48 h | Downregulation (239 genes) | 10.75 | Cell cycle |
| 7.16 | DNA replication | ||
| 4.74 | Lumen/membrane enclosed | ||
| 4.42 | Cell cycle checkpoint | ||
| 3.31 | Chromosome segregation | ||
| 3.04 | Mitotic checkpoint/chromosome | ||
| 3.01 | Microtubule/spindle organization | ||
| 2.79 | DNA binding | ||
| Upregulation (376 genes) | 15.98 | Signaling | |
| 12.07 | Transmembrane | ||
| 10.47 | Cell adhesion | ||
| 5.87 | Plasma membrane | ||
| 4.41 | EGF-like | ||
| 4.18 | MHC class II/antigen processing | ||
| 3.89 | Ion transport/gated channels | ||
| 3.79 | Immunoglobulin-like | ||
| 3.63 | Calcium ion binding/ion binding | ||
| 3.21 | Angiogenesis | ||
| 2.80 | Cell junction/synapse | ||
| 2.58 | Synaptic transmission | ||
| 2.55 | Matrix adhesion/integrins | ||
| 2.52 | Extracellular matrix |
Abbreviations: EGF, epidermal growth factor; MHC, major histocompatibility complex. Pathway analysis from DAVID. As many annotated pathways contain overlapping genes, DAVID combines such pathways into clusters. Column 4 shows our labels for these clusters. Individual pathways are reported in Supplementary Tables S5a and S6b.
Positive regulators of transcription were the second most enriched cluster from the pathway analyses (Table 1), and four of the most significantly enriched individual pathways from DAVID involved transcriptional regulation (Supplementary Table S5a). One of the top associations arising from schizophrenia GWAS is TCF4, a transcription factor that contains two known miR-137-binding sites.33 These results suggest that the effects of miR-137 can be extended markedly by altering the regulation of transcription factors. To explore this possibility, we obtained a list of 1745 genes encoding transcription factors from Transfac.62 At 24 h, 29 different transcription factors were downregulated (q<0.1), slightly more than chance expectations (P=0.01), whereas there was no over-representation of transcription factors in the upregulated genes (P=0.61). At 48 h, genes encoding transcription factors were less likely to be upregulated data (P=8.2 × 10−4). These results are consistent with miR-137 acting as an amplifier via directly fine-tuning the regulation of transcription factors, which then leads to secondary regulatory effects. TCF4 is a known direct target of miR-13733 and has been implicated in schizophrenia GWAS.5,6 Although TCF4 was not itself differentially expressed, we found enrichment for TCF4-bindings sites in the promoters of genes with significant differential expression at 48 h (P=0.003), including both up- (P=0.007) and downregulation (P=0.01). These results highlight the evident complexity of miR-137 gene regulation and that effects on true direct targets may not be detectable (due to transcriptional repression or subtle effects) and yet influence transcription for multiple genes.
Genomic studies of schizophrenia have implicated biological pathways using multiple types of genomic data (common variation, rare CNVs and rare exonic variation).3,6,55,63 It is possible that genes influenced by miR-137 alter risk by directly or indirectly targeting pathways central to pathogenesis. We thus evaluated whether pathways implicated in schizophrenia were enriched for transcripts with differential expression consequent to exposure to increased amounts of miR-137. First, pathways including sets of synaptic genes have been implicated in schizophrenia. Expert-curated lists of synaptic genes are enriched for smaller GWAS P-values,6,63 and genes encoding postsynaptic density proteins, N-methyl-D-aspartate receptors and activity-regulated cytoskeleton-associated protein complexes (activity-regulated cytoskeleton-associated protein) have been implicated in schizophrenia by studies of de novo CNV, de novo deleterious exome variation and deleterious exome variation in cases and controls.3,4,55 We tested for associations of these pathways with genes with differential expression at 24 and 48 h (Table 2). None of these pathways were associated in the 24-h data but three showed significant associations in the 48-h data (all but N-methyl-D-aspartate receptor genes). The associations for synaptic genes were particularly strong. Intriguingly, the 48-h associations tended to be in differentially expressed genes showing upregulation. Second, genes encoding RNAs that interact with FMRP (the protein product of FMR1)57 are enriched for smaller GWAS P-values and deleterious exon mutations in schizophrenia.3 We found significant association between genes whose RNAs interact with FMRP with upregulated genes at 48 h, supporting the idea that RNAs bound by FMRP are influenced by miR-137. Third, neuronal calcium channel signaling has been implicated in schizophrenia and bipolar disorder.6,6,64 We found significant associations between calcium channel subunits with genes upregulated at 48 h following exposure to markedly increased amounts of miR-137. The analogous association with genes whose protein products were identified as interacting with Cav2 (voltage-gated calcium channel) was nominally significant.56 Overall, there was little evidence of association between genes differentially regulated by miR-137 at 24 h but relatively consistent associations with upregulation at 48 h. It is plausible that indirect/upregulation effects require longer exposure to increased levels of miR-137; this supports a role for miR-137 as an effector that indirectly regulates pathways critical to schizophrenia.
Table 2. Analyses of pathways implicated by schizophrenia genome-wide studies.
| Pathway | DE genes (q<0.1) |
24 h |
48 h |
||
|---|---|---|---|---|---|
| P | OR (95% CI) | P | OR (95% CI) | ||
| Synaptic genes | All | 0.90 | 0.94 (0.57–1.56) | 2.8 × 10−7 | 2.13 (1.63–2.78) |
| (1043 genes) | Downregulated | 0.86 | 0.85 (0.41–1.74) | 1.0 | 0.95 (0.5–1.81) |
| Upregulated | 0.70 | 1.11 (0.54–2.28) | 3.2 × 10−8 | 2.54 (1.88–3.44) | |
| Postsynaptic | All | 0.78 | 0.88 (0.49–1.58) | 0.06 | 1.4 (0.99–1.99) |
| density | Downregulated | 1.0 | 0.86 (0.38–1.97) | 0.39 | 0.63 (0.28–1.44) |
| (1457 genes) | Upregulated | 1.0 | 0.98 (0.43–2.24) | 0.0049 | 1.82 (1.23–2.71) |
| ARC | All | 1.0 | N/A | 0.0096 | 5.51 (1.86–16.32) |
| (28 genes) | Downregulated | 1.0 | N/A | 0.20 | 4.96 (0.61–40.5) |
| Upregulated | 1.0 | N/A | 0.03 | 5.03 (1.4–18.09) | |
| NMDA receptor | All | 1.0 | 0.89 (0.12–6.45) | 0.17 | 1.94 (0.7–5.38) |
| (61 genes) | Downregulated | 1.0 | N/A | 0.24 | 2.24 (0.53–9.43) |
| Upregulated | 0.41 | 1.94 (0.26–14.27) | 0.32 | 1.84 (0.43–7.88) | |
| FMRP interactors | All | 0.08 | 1.49 (0.96–2.32) | 4.7 × 10−5 | 1.92 (1.43–2.57) |
| (842 genes) | Downregulated | 0.07 | 1.61 (0.98–2.65) | 0.71 | 1.13 (0.57–2.21) |
| Upregulated | 1.0 | 0.84 (0.31–2.28) | 1.5 × 10−4 | 1.98 (1.42–2.75) | |
| Calcium channel | All | 1.0 | N/A | 0.0056 | 6.61 (2.19–19.97) |
| subunits | Downregulated | 1.0 | N/A | 1.0 | N/A |
| (26 genes) | Upregulated | 1.0 | N/A | 0.0098 | 5.68 (1.84–17.52) |
| Cav2 interactors | All | 0.08 | N/A | 0.11 | 1.68 (0.9–3.1) |
| (207 genes) | Downregulated | 0.27 | N/A | 0.73 | 0.47 (0.06–3.36) |
| Upregulated | 0.65 | N/A | 0.04 | 2.09 (1.08–4.05) | |
Abbreviations: ARC, neuronal activity-regulated cytoskeleton-associated protein postsynaptic signaling complexes; Cav2, voltage-gated calcium channel; CI, confidence interval; DE, differentially expressed; FMRP, fragile X mental retardation protein; NMDA, N-methyl-D-aspartate; OR, odds ratio. Boldface shows P<0.01.
We compared these results with those of a similar experiment by Hill et al.51 (~1000-fold increase in miR-137 expression in the neural cell line CTXOE03, gene expression measured at 72 h using Illumina HT-12v4 arrays). For the genes present in both studies, there was a significant negative correlation in the log2 fold change values at 24 h (Spearman ρ=−0.33, P=5 × 10−19) and a significant positive correlation at 48 h (Spearman ρ=0.25, P=3 × 10−11). Despite the use of a different vector, cell line and expression technology, these results are surprisingly comparable to these in our experiment. However, there was no significant overlap between our results and genes that were differentially expressing in lymphoblastoid cell lines from schizophrenia cases and controls (data not shown).52
Schizophrenia GWAS results
Previous work has shown significant enrichment of smaller schizophrenia GWAS P-values for predicted targets of miR-137.5,6 However, these analyses were based on TargetScan-predicted direct effects of miR-137 which, as described above, have incomplete overlap with the biological direct and indirect effects of miR-137 overexpression in human neural precursor cells. We thus compared the empirical findings from this report with results from the largest published schizophrenia GWAS (Table 3).7 There was no significant enrichment for genes differentially expressed at 24 h. However, there was significant enrichment of smaller GWAS P-values for downregulated targets of miR-137.
Table 3. Enrichment of schizophrenia GWAS results and genes with differential expression after overexpression of miR-137.
| Time | DE genes (q<0.1) | Enrichment P-value |
|---|---|---|
| 24 h | All | >0.1 |
| Downregulation | >0.1 | |
| Upregulation | >0.1 | |
| 48 h | All | 0.014 |
| Downregulation | 0.005 | |
| Upregulation | 0.011 |
Abbreviation: GWAS, genome-wide association study.
P-values from INRICH evaluating the enrichment of lower GWAS P-values (P<0.01)6 to differentially expressed genes (q<0.1) from miR-137 overexpression. The extended major histocompatibility complex region (chr6:25–34 mb) was excluded due to its high gene density, extended linkage disequilibrium and strong GWAS signal in schizophrenia. Boldface shows P<0.01.
Discussion
miR-137 is a particularly intriguing association for schizophrenia, given the strength of its association and the co-occurrence of additional associations in many of its predicted targets. These empirical findings from unbiased genomic screens are consistent with the hypothesis that a biological or developmental process orchestrated by miR-137 contributes to the etiology of schizophrenia. As we demonstrated, a key limitation of our current knowledge is that we have only an approximate idea of the genes influenced by miR-137 (computational prediction of direct targets using TargetScan and almost nothing on its indirect targets).
As an initial approach to understanding the reach of miR-137, we overexpressed miR-137 in human neuronal stem cells to detect genes whose transcripts showed differential expression upon exposure to large increases in amounts of miR-137. Notably, our results had approximate overlap with a similar study.51 Using a number of verification analyses, we demonstrated that we successfully increased the amounts of miR-137 and that these effects were relatively specific. Our approach allowed detection of direct targets influenced by RNA degradation plus secondary effects in the form of increased expression. Via identification of individual genes and larger-scale alterations in biological pathways, our findings provide a window into the cellular mechanisms regulated by miR-137 in neuronal stem cells. Moreover, the different patterns of findings at 24 and 48 h highlight the dynamic regulation of miR-137, and emphasize that findings at a single time point may not encompass all targets of a given miRNA. In aggregate, these admittedly circumstantial results increase support for a role of miR-137 in schizophrenia.
First, the direct and indirect targets of miR-137 that we identified had relatively small changes in expression, typically less than twofold. We identified no instances where miR-137 acted as an on- or off-switch. These findings are consistent with prior studies showing that microRNAs yield subtle changes in many genes.10,11 Moreover, many differentially expressed genes were not predicted targets of miR-137, and many predicted targets are not altered. We do not believe that this was be due to off target effects or secondary effects on different microRNAs, given that Sylarray detected only enrichment in miR-137 seed sequences (Supplementary Figure S1C).
Second, miR-137 has been shown to affect proliferation and differentiation.14,25,26,59 We found that cell density, morphology and neuronal fate markers were not altered by miR-137 at 24 or 48 h. Given that cell cycle genes showed differential expression (Table 1), it is possible that longer periods of observation would have detected alterations in morphology or cell fate.
Third, our results were consistent with the coordinated influence of miR-137 on multiple sets of functionally interconnected genes (for example, via the significant enrichment of protein–protein interactions in the differentially expressed genes and the pathway analyses). These results support the hypothesis that miR-137 causes subtle differential regulation of multiple genes in interrelated pathways, leading to larger physiological changes to a cell or organism.
Fourth, as summarized above, highly significant schizophrenia GWAS findings near MIR137 and enrichment of its predicted targets support a role of MIR137 in schizophrenia. However, the region of association includes other candidate genes (DPYD and MIR2682). Critically, genes influenced by miR-137 in a human neural stem cell line (particularly downregulated genes) were enriched for smaller GWAS P-values (Table 3). Moreover, pathway analyses provide supporting data. Genes upregulated at 48 h (presumptive indirect targets) were consistently enriched for genes and pathways related to schizophrenia (Tables 1 and 2). These included significant overlap with major histocompatibility complex class II genes (encompassed by the strongest schizophrenia finding to date), genes with TCF4 (a schizophrenia risk locus and known target of miR-137) promoter-binding sites and pathways previously implicated in schizophrenia (synaptic genes, genes whose mRNAs interact with FMRP and L-type voltage-gated calcium channels).
These results should be interpreted in the context of several limitations. First, we are cautious in our comparison of results between 24 and 48 h, given the change to a different method of RNA extraction. Second, we attempted to sequester miR-137 as an inhibition control but were unable to obtain technically satisfactory results. Third, the degree of overexpression of miR-137 was continual and far greater the naturally occurring levels of expression.
In summary, our results have identified genes and pathways regulated through miR-137, supporting a role of miR-137 in fine-tuning neuronally important pathways including cell cycle regulation, L-type voltage-gated calcium channels, cell–cell interactions and synaptic function. These results provide empirical data supporting the hypothesis that direct and indirect miR-137 targets have roles in the etiology of schizophrenia.
Acknowledgments
We offer special thanks to Dr Wei Sun for statistical advice and direction, Peter S. Chines for assistance with the transcription factor-binding site enrichment analysis pipeline and to Dr David Rubinow for his support. The project was funded by the Foundation For Hope (Raleigh, NC, http://www.walkforhope.com).
Dr Sullivan was on the SAB of Expression Analysis (Durham, NC, USA). The remaining authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Translational Psychiatry website (http://www.nature.com/tp)
Supplementary Material
References
- Schizophrenia Working Group of the Psychiatric Genomics Consortium Biological Insights From 108 Schizophrenia-Associated Genetic Loci. Nature. 2014. [DOI] [PMC free article] [PubMed]
- Levinson DF, Duan J, Oh S, Wang K, Sanders AR, Shi J, et al. Copy number variants in schizophrenia: confirmation of five previous findings and new evidence for 3q29 microdeletions and VIPR2 duplications. Am J Psychiatry. 2011;168:302–316. doi: 10.1176/appi.ajp.2010.10060876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell SM, Moran JL, Fromer M, Ruderfer D, Solovieff N, Roussos P, et al. A burden of ultra-rare disruptive mutations concentrated in synaptic gene networks increases risk of schizophrenia. Nature. 2014;506:185–190. doi: 10.1038/nature12975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromer M, Pocklington AJ, Kavanagh D, Williams H, Dwyer S, Gormley P, et al. De novo mutations in schizophrenia identify pathogenic gene networks regulating synaptic strength and overlap with autism and intellectual disability. Nature. 2014;506:179–184. doi: 10.1038/nature12929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, Sanders AR, Kendler KS, Levinson DF, Sklar P, Holmans PA, et al. Genome-wide association study identifies five new schizophrenia loci. Nat Genet. 2011;43:969–976. doi: 10.1038/ng.940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripke S, O'Dushlaine C, Chambert K, Moran JL, Kahler AK, Akterin S, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–1159. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guella I, Sequeira A, Rollins B, Morgan L, Torri F, van Erp TG, et al. Analysis of miR-137 expression and rs1625579 in dorsolateral prefrontal cortex. J Psychiatr Res. 2013;47:1215–1221. doi: 10.1016/j.jpsychires.2013.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Carrington JC, Ambros V. Role of microRNAs in plant and animal development. Science. 2003;301:336–338. doi: 10.1126/science.1085242. [DOI] [PubMed] [Google Scholar]
- Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, Castle J, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–773. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455:58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- Tarantino C, Paolella G, Cozzuto L, Minopoli G, Pastore L, Parisi S, et al. miRNA 34a, 100, and 137 modulate differentiation of mouse embryonic stem cells. FASEB J. 2010;24:3255–3263. doi: 10.1096/fj.09-152207. [DOI] [PubMed] [Google Scholar]
- Smrt RD, Szulwach KE, Pfeiffer RL, Li X, Guo W, Pathania M, et al. MicroRNA miR-137 regulates neuronal maturation by targeting ubiquitin ligase mind bomb-1. Stem Cells. 2010;28:1060–1070. doi: 10.1002/stem.431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szulwach KE, Li X, Smrt RD, Li Y, Luo Y, Lin L, et al. Cross talk between microRNA and epigenetic regulation in adult neurogenesis. J Cell Biol. 2010;189:127–141. doi: 10.1083/jcb.200908151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun G, Ye P, Murai K, Lang MF, Li S, Zhang H, et al. miR-137 forms a regulatory loop with nuclear receptor TLX and LSD1 in neural stem cells. Nat Commun. 2011;2:529. doi: 10.1038/ncomms1532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devanna P, Vernes SC. A direct molecular link between the autism candidate gene RORa and the schizophrenia candidate MIR137. Sci Rep. 2014;4:3994. doi: 10.1038/srep03994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen MH, Valles A, Kirkels LA, Mastebroek M, Olde Loohuis N, Kos A, et al. Chromosome 1p21.3 microdeletions comprising DPYD and MIR137 are associated with intellectual disability. J Med Genet. 2011;48:810–818. doi: 10.1136/jmedgenet-2011-100294. [DOI] [PubMed] [Google Scholar]
- Wu H, Tao J, Chen PJ, Shahab A, Ge W, Hart RP, et al. Genome-wide analysis reveals methyl-CpG-binding protein 2-dependent regulation of microRNAs in a mouse model of Rett syndrome. Proc Natl Acad Sci USA. 2010;107:18161–18166. doi: 10.1073/pnas.1005595107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geekiyanage H, Chan C. MicroRNA-137/181c regulates serine palmitoyltransferase and in turn amyloid {beta}, novel targets in sporadic Alzheimer's disease. J Neurosci. 2011;31:14820–14830. doi: 10.1523/JNEUROSCI.3883-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soldati C, Bithell A, Johnston C, Wong KY, Stanton LW, Buckley NJ. Dysregulation of REST-regulated coding and non-coding RNAs in a cellular model of Huntington's disease. J Neurochem. 2013;124:418–430. doi: 10.1111/jnc.12090. [DOI] [PubMed] [Google Scholar]
- Langevin SM, Stone RA, Bunker CH, Grandis JR, Sobol RW, Taioli E. MicroRNA-137 promoter methylation in oral rinses from patients with squamous cell carcinoma of the head and neck is associated with gender and body mass index. Carcinogenesis. 2010;31:864–870. doi: 10.1093/carcin/bgq051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandres E, Agirre X, Bitarte N, Ramirez N, Zarate R, Roman-Gomez J, et al. Epigenetic regulation of microRNA expression in colorectal cancer. Int J Cancer. 2009;125:2737–2743. doi: 10.1002/ijc.24638. [DOI] [PubMed] [Google Scholar]
- Dacic S, Kelly L, Shuai Y, Nikiforova MN. miRNA expression profiling of lung adenocarcinomas: correlation with mutational status. Mod Pathol. 2010;23:1577–1582. doi: 10.1038/modpathol.2010.152. [DOI] [PubMed] [Google Scholar]
- Zhi F, Chen X, Wang S, Xia X, Shi Y, Guan W, et al. The use of hsa-miR-21, hsa-miR-181b and hsa-miR-106a as prognostic indicators of astrocytoma. Eur J Cancer. 2010;46:1640–1649. doi: 10.1016/j.ejca.2010.02.003. [DOI] [PubMed] [Google Scholar]
- Liu M, Lang N, Qiu M, Xu F, Li Q, Tang Q, et al. miR-137 targets Cdc42 expression, induces cell cycle G1 arrest and inhibits invasion in colorectal cancer cells. Int J Cancer. 2011;128:1269–1279. doi: 10.1002/ijc.25452. [DOI] [PubMed] [Google Scholar]
- Balaguer F, Link A, Lozano JJ, Cuatrecasas M, Nagasaka T, Boland CR, et al. Epigenetic silencing of miR-137 is an early event in colorectal carcinogenesis. Cancer Res. 2010;70:6609–6618. doi: 10.1158/0008-5472.CAN-10-0622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wang J, Shen H, Lu J, Li C, Hu D, et al. Epigenetics, microRNAs, and carcinogenesis: functional role of microRNA-137 in uveal melanoma. Invest Ophthalmol Vis Sci. 2010;52:1193–1199. doi: 10.1167/iovs.10-5272. [DOI] [PubMed] [Google Scholar]
- Chen Q, Chen X, Zhang M, Fan Q, Luo S, Cao X. miR-137 is frequently down-regulated in gastric cancer and is a negative regulator of Cdc42. Dig Dis Sci. 2011;56:2009–2016. doi: 10.1007/s10620-010-1536-3. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Megraw M, Hatzigeorgiou AG. A guide through present computational approaches for the identification of mammalian microRNA targets. Nat Methods. 2006;3:881–886. doi: 10.1038/nmeth954. [DOI] [PubMed] [Google Scholar]
- Witkos TM, Koscianska E, Krzyzosiak WJ. Practical aspects of microRNA target prediction. Curr Mol Med. 2011;11:93–109. doi: 10.2174/156652411794859250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haflidadottir BS, Bergsteinsdottir K, Praetorius C, Steingrimsson E. miR-148 regulates Mitf in melanoma cells. PLoS ONE. 2010;5:e11574. doi: 10.1371/journal.pone.0011574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bemis LT, Chen R, Amato CM, Classen EH, Robinson SE, Coffey DG, et al. MicroRNA-137 targets microphthalmia-associated transcription factor in melanoma cell lines. Cancer Res. 2008;68:1362–1368. doi: 10.1158/0008-5472.CAN-07-2912. [DOI] [PubMed] [Google Scholar]
- Kwon E, Wang W, Tsai LH. Validation of schizophrenia-associated genes CSMD1, C10orf26, CACNA1C and TCF4 as miR-137 targets. Mol Psychiatry. 2013;18:11–12. doi: 10.1038/mp.2011.170. [DOI] [PubMed] [Google Scholar]
- Kim AH, Parker EK, Williamson V, McMichael GO, Fanous AH, Vladimirov VI. Experimental validation of candidate schizophrenia gene ZNF804A as target for hsa-miR-137. Schizophr Res. 2012;141:60–64. doi: 10.1016/j.schres.2012.06.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tay Y, Zhang J, Thomson AM, Lim B, Rigoutsos I. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature. 2008;455:1124–1128. doi: 10.1038/nature07299. [DOI] [PubMed] [Google Scholar]
- Schnall-Levin M, Zhao Y, Perrimon N, Berger B. Conserved microRNA targeting in Drosophila is as widespread in coding regions as in 3′UTRs. Proc Natl Acad Sci USA. 2010;107:15751–15756. doi: 10.1073/pnas.1006172107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donato R, Miljan EA, Hines SJ, Aouabdi S, Pollock K, Patel S, et al. Differential development of neuronal physiological responsiveness in two human neural stem cell lines. BMC Neurosci. 2007;8:36. doi: 10.1186/1471-2202-8-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci USA. 2003;100:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright F, Sullivan PF, Brooks AI, Zou F, Sun W, Xia K, et al. Heritability and genomics of gene expression in periferal blood. Nat Genet. 2014;46:430–437. doi: 10.1038/ng.2951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartonicek N, Enright AJ. SylArray: a web server for automated detection of miRNA effects from expression data. Bioinformatics. 2010;26:2900–2901. doi: 10.1093/bioinformatics/btq545. [DOI] [PubMed] [Google Scholar]
- Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, et al. Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gene Ontology Consortium The gene ontology: enhancements for 2011. Nucleic Acids Res. 2012;40:D559–D564. doi: 10.1093/nar/gkr1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M. KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res. 2012;40:D109–D114. doi: 10.1093/nar/gkr988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levy S, Hannenhalli S. Identification of transcription factor binding sites in the human genome sequence. Mamm Genome. 2002;13:510–514. doi: 10.1007/s00335-002-2175-6. [DOI] [PubMed] [Google Scholar]
- Sethupathy P, Giang H, Plotkin JB, Hannenhalli S. Genome-wide analysis of natural selection on human cis-elements. PLoS ONE. 2008;3:e3137. doi: 10.1371/journal.pone.0003137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill MJ, Donocik JG, Nuamah RA, Mein CA, Sainz-Fuertes R, Bray NJ. Transcriptional consequences of schizophrenia candidate miR-137 manipulation in human neural progenitor cells. Schizophr Res. 2014;153:225–230. doi: 10.1016/j.schres.2014.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders AR, Goring HH, Duan J, Drigalenko EI, Moy W, Freda J, et al. Transcriptome study of differential expression in schizophrenia. Hum Mol Genet. 2013;22:5001–5014. doi: 10.1093/hmg/ddt350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruano D, Abecasis GR, Glaser B, Lips ES, Cornelisse LN, de Jong AP, et al. Functional gene group analysis reveals a role of synaptic heterotrimeric G proteins in cognitive ability. Am J Hum Genet. 2010;86:113–125. doi: 10.1016/j.ajhg.2009.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croning MD, Marshall MC, McLaren P, Armstrong JD, Grant SG. G2Cdb: the genes to cognition database. Nucleic Acids Res. 2009;37:D846–D851. doi: 10.1093/nar/gkn700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirov G, Pocklington AJ, Holmans P, Ivanov D, Ikeda M, Ruderfer D, et al. De novo CNV analysis implicates specific abnormalities of postsynaptic signalling complexes in the pathogenesis of schizophrenia. Mol Psychiatry. 2012;17:142–153. doi: 10.1038/mp.2011.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller CS, Haupt A, Bildl W, Schindler J, Knaus HG, Meissner M, et al. Quantitative proteomics of the Cav2 channel nano-environments in the mammalian brain. Proc Natl Acad Sci USA. 2010;107:14950–14957. doi: 10.1073/pnas.1005940107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darnell JC, Van Driesche SJ, Zhang C, Hung KY, Mele A, Fraser CE, et al. FMRP stalls ribosomal translocation on mRNAs linked to synaptic function and autism. Cell. 2011;146:247–261. doi: 10.1016/j.cell.2011.06.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee PH, O'Dushlaine C, Thomas B, Purcell S. InRich: Interval-based enrichment analysis for genome-wide association studies. Bioinformatics. 2012;28:1797–1799. doi: 10.1093/bioinformatics/bts191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silber J, Lim DA, Petritsch C, Persson AI, Maunakea AK, Yu M, et al. miR-124 and miR-137 inhibit proliferation of glioblastoma multiforme cells and induce differentiation of brain tumor stem cells. BMC Med. 2008;6:14. doi: 10.1186/1741-7015-6-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Ophoff RA, Steinberg S, Andreassen OA, Cichon S, Rujescu D, et al. Common variants conferring risk of schizophrenia. Nature. 2009;460:744–747. doi: 10.1038/nature08186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irish Schizophrenia Genomics Consortium, Wellcome Trust Case Control Consortium Genome-wide association study implicates HLA-C*01:02 as a risk factor at the MHC locus in schizophrenia. Biol Psychiatry. 2012;72:620–628. doi: 10.1016/j.biopsych.2012.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lips ES, Cornelisse LN, Toonen RF, Min JL, Hultman CM, Holmans PA, et al. Functional gene group analysis identifies synaptic gene groups as risk factor for schizophrenia. Mol Psychiatry. 2012;17:996–1006. doi: 10.1038/mp.2011.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. 2008;40:1056–1058. doi: 10.1038/ng.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.