Abstract
Aims—We have previously demonstrated the abnormal localisation of expression of the CD44 gene in carcinoma cells in cryostat sections of fresh frozen tumour tissues, using radioactive in situ hybridisation (RISH). In order to facilitate further analysis of the expression of this gene in a wider range of neoplastic and non-neoplastic conditions, we have developed a technique which can visualise its low copy number transcripts in archival paraffin wax embedded specimens.
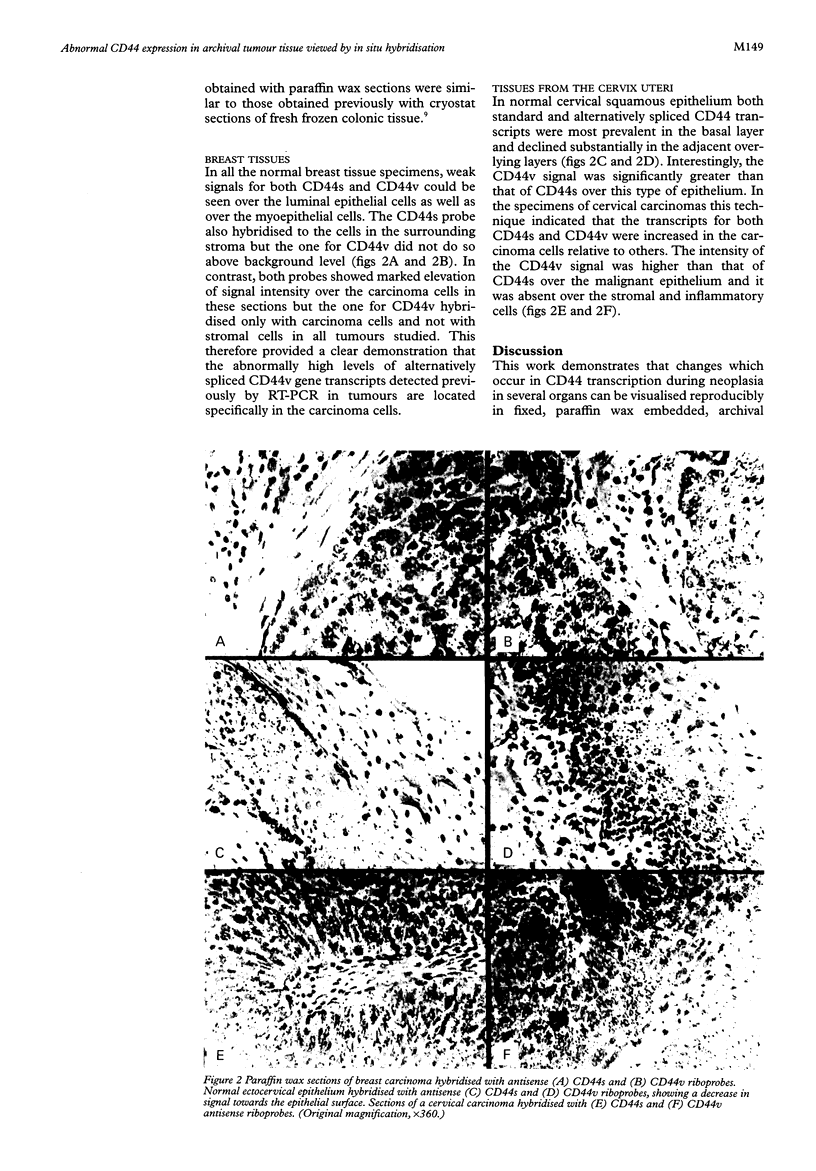
Methods—35S labelled riboprobes complementary to transcripts from the standard (CD44s) and variant (CD44v) regions of the gene were used on paraffin wax embedded sections of tumours and corresponding normal tissues of the colon, breast and uterine cervix.
Results—Elevated levels of signals for CD44s and CD44v transcripts were observed in carcinoma cells relative to their non-neoplastic counterparts in all tissues examined.
Conclusion—This method permits easy access to material which can be selected for suitability, handled at room temperature without degradation and relied upon to show good histological detail. Comparison of the results with those on frozen tissues showed similar distributions of signals. Furthermore, the resolution and morphological detail was improved in paraffin wax sections.
Keywords: CD44
Keywords: in situ hybridisation
Keywords: tumour diagnosis
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bolodeoku J., Yoshida K., Yeomans P., Wells C. A., Tarin D. Demonstration of CD44 gene expression in cells from fine needle aspirates of breast lesions by the polymerase chain reaction. Clin Mol Pathol. 1995 Dec;48(6):M307–M309. doi: 10.1136/mp.48.6.m307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dall P., Heider K. H., Hekele A., von Minckwitz G., Kaufmann M., Ponta H., Herrlich P. Surface protein expression and messenger RNA-splicing analysis of CD44 in uterine cervical cancer and normal cervical epithelium. Cancer Res. 1994 Jul 1;54(13):3337–3341. [PubMed] [Google Scholar]
- Matsumura Y., Tarin D. Significance of CD44 gene products for cancer diagnosis and disease evaluation. Lancet. 1992 Oct 31;340(8827):1053–1058. doi: 10.1016/0140-6736(92)93077-z. [DOI] [PubMed] [Google Scholar]
- Screaton G. R., Bell M. V., Jackson D. G., Cornelis F. B., Gerth U., Bell J. I. Genomic structure of DNA encoding the lymphocyte homing receptor CD44 reveals at least 12 alternatively spliced exons. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):12160–12164. doi: 10.1073/pnas.89.24.12160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tarin D., Bolodeoku J., Hatfill S. J., Sugino T., Woodman A. C., Yoshida K. The clinical significance of malfunction of the CD44 locus in malignancy. J Neurooncol. 1995 Dec;26(3):209–219. doi: 10.1007/BF01052624. [DOI] [PubMed] [Google Scholar]
- Yoshida K., Bolodeoku J., Sugino T., Goodison S., Matsumura Y., Warren B. F., Toge T., Tahara E., Tarin D. Abnormal retention of intron 9 in CD44 gene transcripts in human gastrointestinal tumors. Cancer Res. 1995 Oct 1;55(19):4273–4277. [PubMed] [Google Scholar]




