Abstract
Streptococcus pneumoniae (SP) and nontypeable Haemophilus influenzae (NTHi) are common commensals of the human airway and major bacterial pathogens of otitis media (OM) and other upper airway infections. The interaction between them may play an important role in the pathogenesis of polymicrobial infections. Although previous studies suggested NTHi could promote pneumococcal survival and biofilm formation, how NTHi affects pneumococcal activities has not been defined. Our data in the present studies indicated that the outcome of the interaction between SP and NTHi was in a cell-density-dependent manner and the enhancement of pneumococcal survival happened at the later stages of culturing. Using quantitative PCR, we found that the expression of pneumococcal genes regulating autolysis and fratricide, lytA and cbpD, were significantly down-regulated in co-culture with NTHi. We further observed that influence of NTHi was not on direct cell-to-cell contact, but that this contact may contribute to the interaction between these two microorganisms. These results suggest that pneumococcal survival and biofilm formation can be enhanced by down-regulating pneumococcal cell wall hydrolase production thereby inhibiting pneumococcal autolysis and fratricide in the presence of NTHi.
Keywords: Streptococcus pneumoniae, nontypeable Haemophilus influenzae, survival, inhibit, autolysis, fratricide
1. Introduction
Many bacterial infections are caused by multiple pathogens, termed polymicrobial infection. Traditional treatment strategies for bacterial infections aimed at a single causative agent are ineffective in cases of polymicrobial infection due in part to changing relative abundances of pathogens and/or replacing the major causative agent. It is therefore imperative to develop novel systemic treatment strategies to control polymicrobial infections. The human respiratory tract harbors a diverse population of microbial flora. Many, including Streptococcus pneumoniae (SP) and nontypeable Haemophilus influenzae (NTHi), can cause opportunistic infections. In a healthy individual there is a dynamic balance between the flora and host and between different members of microbial community, which is crucial in determining bacterial carriage. The balance can be broken by shifting the relative abundance of certain species within the community, thus leading to disease. Factors affecting bacterial abundance include microbial interaction within the flora or the consequences of vaccination or antibiotic treatment targeting a particular pathogen. The goal of this study is to discover the interaction between SP and NTHi so that a more comprehensive strategy can be developed to control infectious diseases caused by these microorganisms.
Otitis media (OM) is one of the most common diagnoses in pediatric patients and the leading reason for pediatric office visits and new antibiotic prescriptions to children [1–3]. Epidemiologic studies have shown that OM is naturally involved with multiple pathogens [4–7]. Among known pathogens, SP and NTHi are involved in 30% to 77% of multiple pathogen infection cases of OM [4, 8, 9]. By polymerase chain reaction (PCR) and specific fluorescent in situ hybridization (FISH), coinfections of SP and H. influenzae were detected in about 10% of the middle ear effusions of OM patients and in the biofilm on the middle ear mucosal surface of patients undergoing tympanostomy [10, 11]. Co-colonization and mixed biofilm of multiple microorganisms may enhance pathogenicity by increasing bacterial resistance against host immune clearance and treatment, as well as being a major contributor to chronic OM and recurrent OM. SP and NTHi are also common bacterial pathogens found in chronic rhinosinusitis [12] and the common bacteria contributing to exacerbations of chronic obstructive pulmonary disease (COPD) [13].
There is little information available regarding the mechanics of pathogen/pathogen interactions in polymicrobial OM. SP has potential competitive advantage over H. influenzae in vitro and in vivo by producing hydrogen peroxide and neuraminidase A [14, 15]. Interestingly, using a murine nasopharyngeal colonization model, Lysenko et al. found that H.influenzae had a competitive advantage over SP during colonization by enhancing opsonophagocytic killing of SP [16]. The Swords’ group recently reported that NTHi could enhance pneumococcal biofilm formation in vitro and in vivo [17]. These data and other previous studies [18–20] together suggest a complicated and dynamic interaction between these microorganisms, which predetermines bacterial colonization in the nasopharynx and the associated disease outcomes. However, the molecular and cellular mechanisms directly related to this interspecies interaction have not been defined.
One significant outcome of co-growth of SP and NTHi is the enhancement of pneumococcal survival. SP can spontaneously break down cells by autolysis and fratricide [21–24]. The key molecules mediating autolysis and fratricide include murein hydrolase LytA, peptidoglycan hydrolase LytC and choline-binding protein D (CbpD) [23–26]. Based on that knowledge and the observation of the enhanced pneumococcal survival in vivo in the presence of NTHi, we postulated that the presence of NTHi can down-regulate the expression of pneumococcal autolysis/fratricide-associated enzymes resulting in enhanced SP survival and biofilm formation. In order to address this hypothesis, we investigated pneumococcal survival and gene expression changes in co-culture with NTHi in this study.
2. Materials and Methods
2.1 Strains and growth
SP strains TIGR4, D39, D39lytA− and NTHi strains 86028NP and 2019 were used in this study. NTHi strains were grown on brain heart infusion agar supplemented (sBHI) with hemin (ICN Biochemicals) and nicotinamide adenine dinucleotide (NAD) (Sigma) and SP strains on Todd-Hewitt broth agar plus 0.5% yeast extract (THY) for maintenance. Bacterial cultures were grown in Todd-Hewitt broth plus 0.5% yeast extract liquid medium supplemented with hemin and NAD at 37°C with 5% CO2. For viable bacterial counting, resuspended cultures were serially diluted and plated on BHI agar plates for SP and sBHI agar plates containing 3 µg/mL of vancomycin for NTHi.
2.2 Competition growth
To investigate whether there is competition between SP and NTHi, we set up co-cultures with high or low initial cell densities for each strain to check bacterial survival. Bacteria grown overnight on agar plates were suspended in medium and diluted to an OD600 of 0.15 for 86028NP and 0.5 for TIGR4 (concentration of 108 cfu/ml), respectively. They were further diluted to concentrations of 106 cfu/ml (high) and 104 cfu/ml (low), respectively. Combinations with different concentrations of 86028NP and TIGR4 were made and bacteria were grown statically at 37°C with 5% CO2. Viable cells were counted by diluting the resuspended cultures and plating agar plates at 16 and 24 hour time points after inoculation.
2.3 Co-cultures
In order to evaluate the effect of NTHi on pneumococcal biofilm formation, optimal densities of TIGR4 and 86028NP cells (~104 cfu/ml vs ~106 cfu/ml) were added alone or mixed into wells of chambered cover glasses and were grown statically at 37°C with 5% CO2. Biofilm structures were visualized by confocal laser scanning microscopy (CLSM) following live/dead staining with LIVE/DEAD BacLight Bacterial Viability Kit (Invitrogen, Eugene, OR) at 16 and 24 hour points after inoculation. Five stacks of Z-series images, each representing a different field of view, were taken from biofilm of TIGR4 alone or the mixed culture at 16 and 24 hour points. In order to quantify the total biomass and thickness of biofilm, images were exported into the MATLAB package and analyzed by COMSTATsoftware as described previously [27].
In order to identify and localize SP and NTHi in the biofilm structure, fluorescent Gram staining was performed with ViaGramTM Red+ Bacterial Gram Stain and Viability Kit (Molecular Probe, Eugen, OR) followed by CLSM. Briefly, biofilm was fixed with 4% paraformaldehyde for 6 hours and stained with DAPI/SYTOX for 10 minutes at room temperature. After a PBS wash, the biofilm was stained with 100µg/ml of Texas Red-X conjugate of wheat germ agglutinin (WGA) in 3M KCl [28] for 10 minutes at room temperature. The biofilm was visualized by confocal laser scanning microscopy.
In order to test the universality of the interaction between NTHi and SP, optimal cell densities of NTHi strain 86028NP or 2019 were co-cultured with SP strain TIGR4 or D39, respectively. Viable pneumococcal cells were counted at 16 hours and 24 hours post-inoculation.
In order to investigate whether NTHi modulates pneumococcal growth pattern, TIGR4 was cultured alone or co-cultured with 86028NP in flasks at 37°C with 5% CO2. The cells were resuspended and viable TIGR4 cells were measured by plating the cultures every 2 hours after inoculation.
2.4 Pneumococcal gene expression
In order to investigate whether NTHi affects pneumococcal gene expression patterns in co-cultures, transcript levels of important genes involved in pneumococcal autolysis and fratricide were tested by quantitative real-time PCR (qPCR). Total bacterial RNAs from TIGR4 alone or TIGR4 with 86028NP cultures were prepared at 16 and 24 hours after inoculation using RiboPure™-bacteria Kit (Ambion, Inc, Austin, TX). cDNAs were synthesized using the SuperScript™ III First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). Taqman qPCR primers/probes for qPCR were designed and synthesized (Applied Biosystems, Carlsbad, CA). The primers/probes are listed in Table 1. The expression levels of pneumococcal genes lytA, lytB, lytC, cbpD and ply were detected by qPCR and normalized to the mRNA level of the housekeeping gene (shikimate 5-dehydrogenase), which is responsible for the biosynthesis of aromatic amino acids in bacteria [29]. The cultures and RNA preparations were repeated 3 times. The relative fold changes of mRNA in the mixed culture to TIGR4 alone were calculated using the 2−ΔΔCt methodology [30]. A fold change of more than 1.5 times up or down regulation, and P value of less than 0.05 was considered significant.
Table 1.
Sequences of primers for pneumococcal quantitative real-time PCR
| Gene Name | Primer Sequence | Probe Sequence |
|---|---|---|
| lytA | GCCGTTCTCAATATCATGCTTAAACTGCGTTGACCCTTATCCATATCTTGCT | CTCACGGCTAATGCC |
| lytB | GTGGTATTTGGGATTCTAACTGTTGGACATTAGTTTGAGCAGGAAAGCGAAA | CAAGGCTCCAATACCG |
| lytC | CGAGTACCATCCATAATCACTTCGTGTGGCAAGGCAACTACTATTTGAC | CATGGCACCACTTCCA |
| cbpD | CTGCCTATAATGGAAGCTATCGTTATGTTCCACCTGTTGAAGAAAGAACAGAATT | CACAGCCTCCAATTGA |
| ply | CCTCAGACAGAGTGGAAACAGATTTCTTGGGTCGCCCCCTAAAATAA | CCGCCTTCACTTCTG |
| Shikimate 5-dehydrogenase | GTGGTGCGGCTAAATCAATCTTGACGAACAAAGACCGAAATCTGACT | ACGCCATCCAAAATAG |
Additionally, we also detected expression of pneumolysin gene (ply) to investigate whether the presence of NTHi could affect pneumococcal pathogenicity since pneumolysin is one of the major virulence factors and plays important roles in SP-causing infection [31].
2.5 Immunostaining
In order to confirm pneumococcal gene expression influenced by NTHi, the lytA gene was cloned and expressed in E. coli followed by anti-LytA rabbit serum preparation. LytA expression and location in different cultures was visualized by immunostaining. Briefly, the lytA gene of TIGR4 was cloned by PCR with primers P33 (5’-gag tag aat atg gat cca att aat gtg ag-3’) and P34 (5’-gat gat atg cgg ccg cga cat tcc att att att tta ctg taa tca agc c-3’). PCR product was digested with BamHI/NotI and inserted into pENTR™2B vector (Invitrogen, Carlsbad, CA) to generate the pENTR2BlytA plasmid. Expression plasmid pDESTlytA was constructed by an LR recombination reaction with pENTR2BlytA and pDEST™17 (Invitrogen, Carlsbad, CA) following the manufacturer’s instructions. Plasmid pDESTlytA was transformed into BL21-AI™ E. coli competent cells (Invitrogen, Carlsbad, CA) to express LytA protein. Purified LytA was used to generate rabbit anti-LytA serum in LAMPIRE Biological Laboratories (Pipersville, PA). Specificity of anti-LytA serum was verified by western blot with cell lysates of SP strain D39 and its isogenic mutant, D39lytA- [32].
LytA protein in biofilm was detected by immunostaining with anti-LytA serum. Biofilms grown in the chambered-cover glasses were fixed with 4% paraformaldehyde and immunostained with rabbit anti-LytA serum. Goat anti-rabbit Alexa Fluor 568 antibody (Invitrogen, Carlsbad, CA) was used as the secondary antibody and DAPI (Invitrogen, Carlsbad, CA) was used for counterstaining. The samples were visualized by CLSM.
2.6 Affect of physical contact
To determine the effect of cell-to-cell contact on pneumococcal survival, a transwell system (Corning Incorporated, Corning, NY) was used for co-cultures. 86028NP cells were grown in the upper compartment and TIGR4 cells in the lower compartment of the system. The two compartments were separated by a membrane with a pore size of 0.4µm, which can block cell migration between the compartments. TIGR4 only, as well as TIGR4 mixed with 86028NP were grown in the wells without transwell inserts as controls. Viable cells were counted at 16, 24 and 48 hour time points after inoculation. Cells were collected at 16 and 24 hour points for gene expression analysis as described above.
2.7 Statistic analysis
The difference between groups for bacterial counts, gene expression levels, and biomass and thickness of biofilm was statistically analyzed using the unpaired Student t test (two tailed). Significance level was determined at P value ≤ 0.05.
3. Results
3.1 Cell density-dependent interaction between SP and NTHi
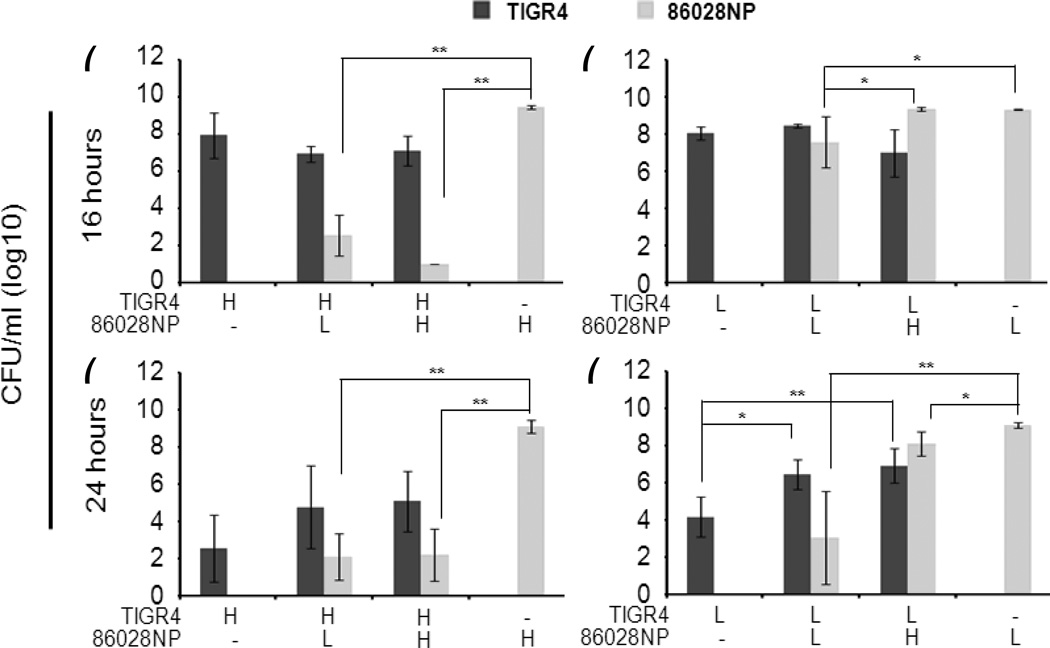
To investigate whether bacterial numbers play a role in the interaction, we set up co-cultures with varying initial amounts of cells of each bacterium. The bacterial counts of both strains were summarized in Fig. 1. The results suggested that the initial densities of each bacterium were important to pneumococcal survival. 86028NP cells didn’t change TIGR4 growth at the 16 hour time point regardless of initial amounts of TIGR4 cells (Fig. 1a and 1b). However, TIGR4 survival was enhanced at the 24 hour time point in the co-culture in combination with low density TIGR4 cells and high or low density 86028NP cells (Fig 1d). When co-cultured with high density TIGR4 cells, 86028NP growth was significantly inhibited at 16 and 24 hour time points regardless of its initial density (Fig 1a and 1c) (P<0.01). When co-cultured with low density TIGR4 cells (Fig. 1b and 1d), growths of 86028NP with low density were significantly inhibited at 16 and 24 hour time points but no inhibition was observed in the co-culture with high density of 86028NP cells at 16 hour time point. Notably, no significant difference was observed between cultures of individual strains with different initial concentrations at each time point. In summary, the data showed that higher cell densities of 86028NP could overcome the inhibition of TIGR4 and enhance TIGR4 survival, while higher cell densities of TIGR4 could inhibit 86028NP growth regardless of how many 86028NP cells there were initially.
Fig. 1.
Initial cell densities were critical for the outcomes of interaction between SP and NTHi. Co-cultures were set up with mixtures of different amounts of TIGR4 cells and 86028NP cells. Viable cells were counted at 16 and 24 hour time points after inoculation. (H: high concentration/~106 cfu/ml; L: medium concentration/~104 cfu/ml). High densities of TIGR4 completely inhibited 86028NP survival no matter how many 86028NP cells there were initially. However, higher cell densities of 86028NP could overcome the inhibition of TIGR4 in lower densities and enhance TIGR4 survival. The bacterial cultures and viable cell counts were performed four times independently. *: P<0.05; **: P<0.01.
3.2 NTHi enhanced pneumococcal biofilm formation and survival
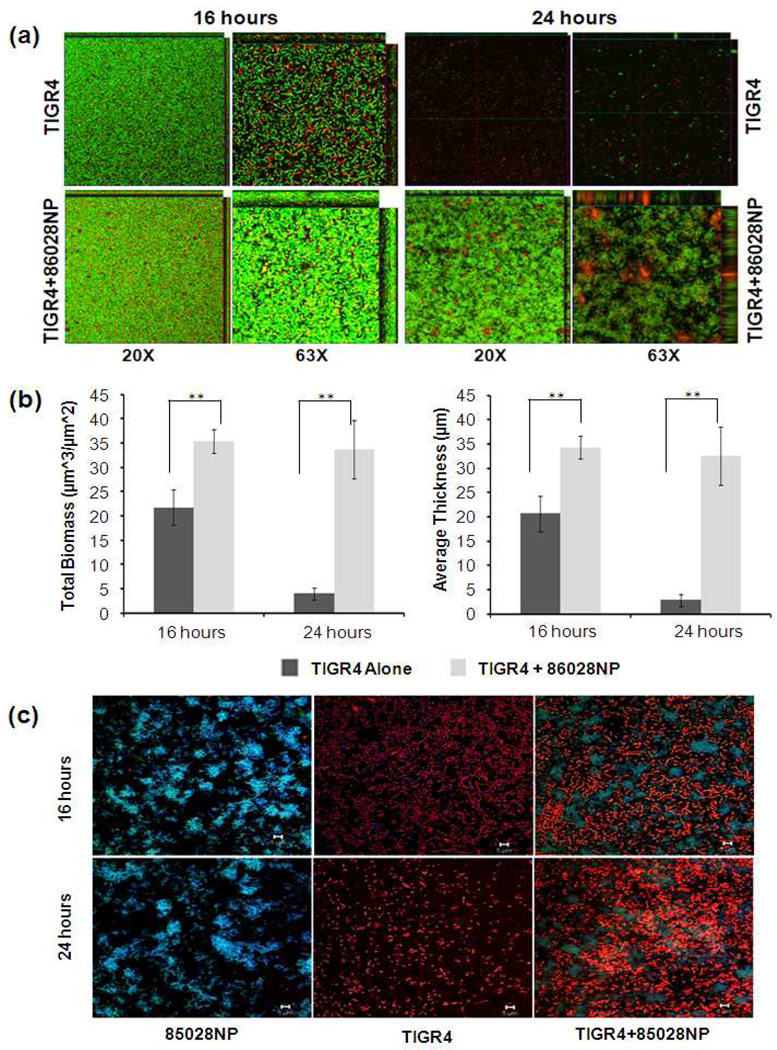
Pneumococcal autolysis starts at stationary phase after inoculation within liquid medium resulting in a rapid decrease of viable cells, leading to the disruption of biofilm structure. Therefore, the pneumococcal biofilm may be maintained because of the enhanced pneumococcal survival. This has been shown by microscopy (Fig. 2a). Using a static biofilm model, we found both TIGR4 alone, and co-cultured with 86028NP formed mature biofilm at the 16 hour point of culture, but a thicker biofilm formed in the co-cultured group due to the presence of 86028NP(P<0.01). The biofilm of TIGR4 alone at the 24 hour time point was dispersed significantly while a sustained biofilm structure was observed in the co-culture group. The biofilm structures were analyzed using COMSTAT software(Fig. 2b). The total biomasses and average thicknesses of the mixed biofilm were significantly higher than those of TIGR4 alone at both time points (P<0.01). TIGR4 and 86028NP cells in the biofilms were identified by fluorescent Gram staining and visualized by CLSM (Fig. 2c). Both TIGR4 and 86028NP cells were shown in the mixed biofilm. There were more intact TIGR4 cells forming chains in the 24 hour mixed biofilm than TIGR4 alone biofilm which contained fewer single cells or debris.
Fig. 2.
NTHi promoted pneumococcal biofilm formation and survival. (a). Biofilms grown in chambered cover glasses were visualized by CLSM following the bacterial live/dead staining process. The images are representatives of a single slice of each biofilm. (b). Total biomasses and average thicknesses of biofilm were analysed using COMSTAT. Mixed biofilms had more biomasses and were thicker than biofilms of TIGR4 alone at both time points. The values of biomass and thickness represented the averages of 5 Z-series images taken from different field of views, respectively. **: P<0.01. (c). TIGR4 and 86028NP cells in the biofilms were identified by fluorescent Gram staining followed by confocal laser scanning microscopy. 86028NP cells (Gram negative) were blue (live) or green (dead). TIGR4 cells (gram positive) were orange. The bar of microscopic image was 10µm.
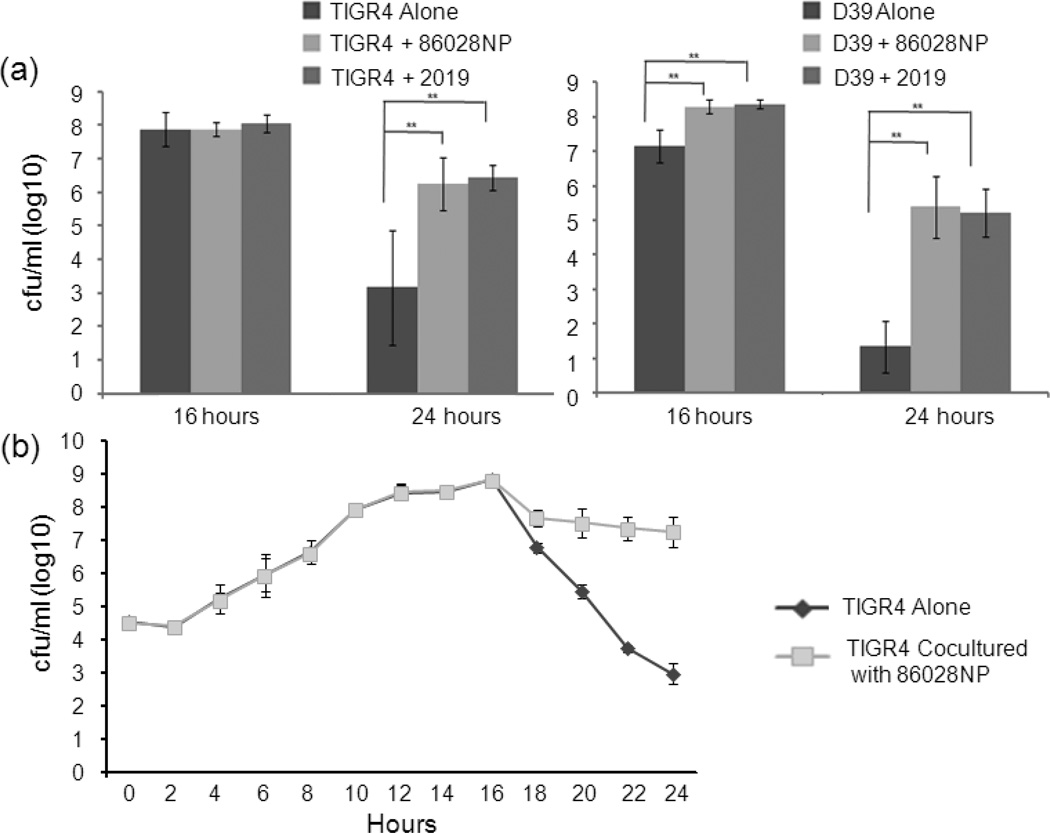
Enhancing pneumococcal survival at the late stage of culture was observed in the mixed cultures with different strains (Fig. 3a), indicating that it is a universal interaction rather than strain-specific between S. pneumoniae and H. influenzae. Both NTHi strains 86028NP and 2019 significantly enhanced TIGR4 survival at 24 hours after inoculation (P<0.01). Interestingly, enhanced survivals of pneumococcal strain D39 by NTHi were observed as early as 16 hours post-inoculation (P<0.01).
Fig. 3.
NTHi enhanced pneumococcal survival. (a). Enhanced pneumococcal survivals were observed in co-cultures with different strains. Both NTHi strains 86028NP and 2019 enhanced TIGR4 survival at 24 hour point. NTHi strains also enhanced other pneumococci D39 survival. The cfu counts represented total viable cells including planktonic and biofilm cells. The bacterial culture and viable cell counting were performed four times independently. **: P<0.01. (b). Co-culturing with NTHi didn’t alter the growth pattern of SP up to the stationary phase. The amount of viable TIGR4 cells of TIGR4 alone culture declined rapidly after 16 hours due to the autolysis and/or fratricide but there were much more viable SP cells in the co-cultured group than SP alone (P<0.01). The cfu counts were average results of 4 independent experiments.
We also observed that NTHi didn’t change pneumococcal growth patterns during the early stages of growth based on the amounts of viable cells, which indicated SP alone and SP co-cultured with NTHi entered stationary phase similarly (Fig. 3b). However, more viable TIGR4 cells were counted in the co-culture of TIGR4 and 86028NP than TIGR4 alone after the stationary phase (P<0.01).
These data together suggest that NTHi enhanced pneumococcal survival and biofilm formation. The enhancement may be due to arresting pneumococcal autolysis, thus promoting its survival at a late stage of culture since the enhancement was only observed after stationary phase.
3.3 NTHi down regulated transcription of pneumococcal genes which were involved in autolysis and fratricide
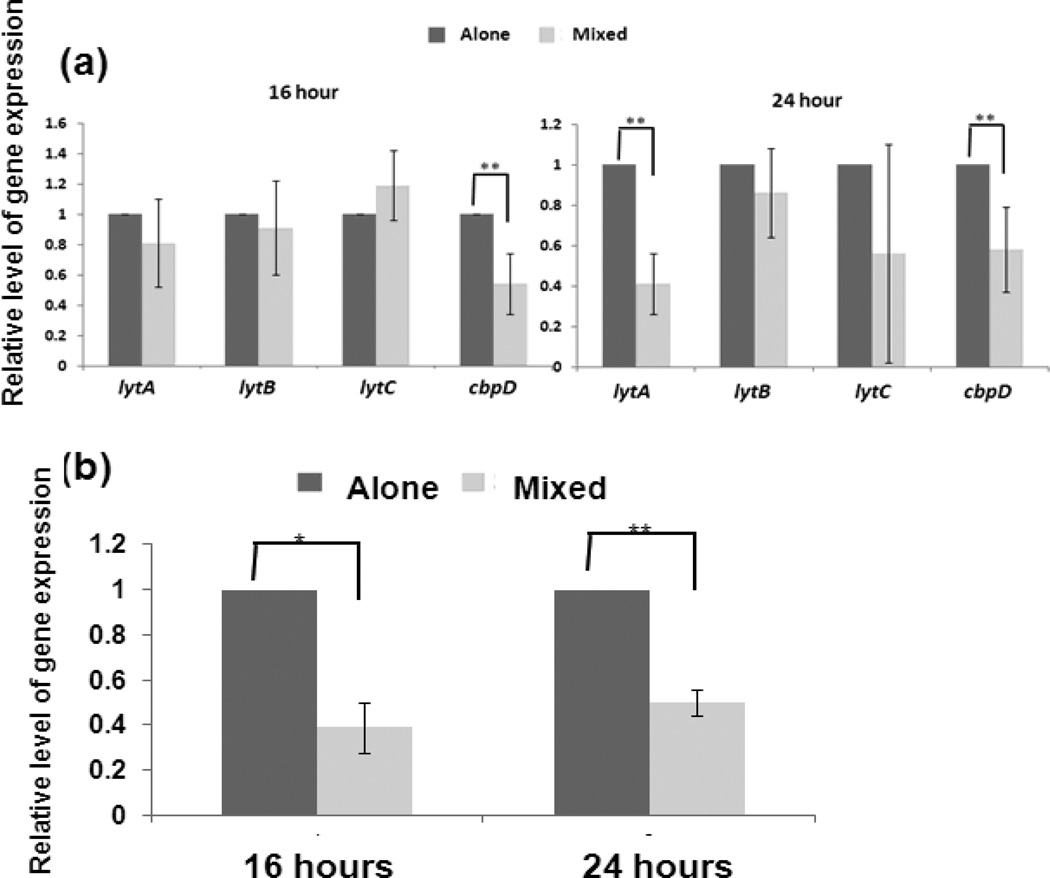
Since pneumococcal survival and biofilm formation could be regulated by autolysis and fratricide, we postulate that co-culture with NTHi inhibits the expression of genes involved in pneumococcal autolysis and fratricide. Using qPCR, we found that co-culture with 86028NP modified these gene’s expression patterns after stationary phase (Fig. 4a). The transcription levels of lytA and lytC were not significantly different at 16 hour cultures between TIGR4 alone and co-culture with 86028NP, but cbpD was significantly down-regulated in the co-culture group. Both lytA and cbpD gene expression were significantly down-regulated in the co-culture group compared to TIGR4 alone at the 24 hour time point. LytC gene expression was down-regulated in the co-culture group with high variation in repeated experiments. Expression of another pneumococcal gene lytB, which is essential for pneumococcal cell separation but not involved in autolysis or fratricide [33], didn’t change significantly in all conditions.
Fig. 4.
Changes of pneumococcal gene expression in the presence of NTHi. Relative mRNA levels of pneumococcal lytA, lytB, lytC, cbpD and ply were measured by qPCR, normalized to a housekeeping gene (shikimate 5-dehydrogenase). The data represented the folds of relative mRNA levels of genes in the co-culture group to those in TIGR4 alone group. (a). Pneumococcal genes involved in autolysis and fratricide were significantly down-regulated in the presence of NTHi. The expression levels of lytA, lytB, lytC genes were not significantly different in 16 hour cultures of TIGR4 alone and mixed culture with 86028NP but cbpD was down-regulated significantly in the mixed culture compared to TIGR4 alone. Both lytA and cbpD gene expressions were significantly down-regulated at the 24 hour time point in the mixed culture compared to TIGR4 alone culture. Another gene, lytB, which isn’t involved in fratricide, didn’t change significantly either between different cultures or different time points. The bacterial cultures and qPCR were performed four times independently. (b). Expression of pneumococcal ply gene was down-regulated in the presence of NTHi. *: P<0.05; **: P<0.01.
Additionally, the presence of NTHi also decreased ply gene expression (Fig. 4b). This down-regulation occurred as early as 16 hours after inoculation.
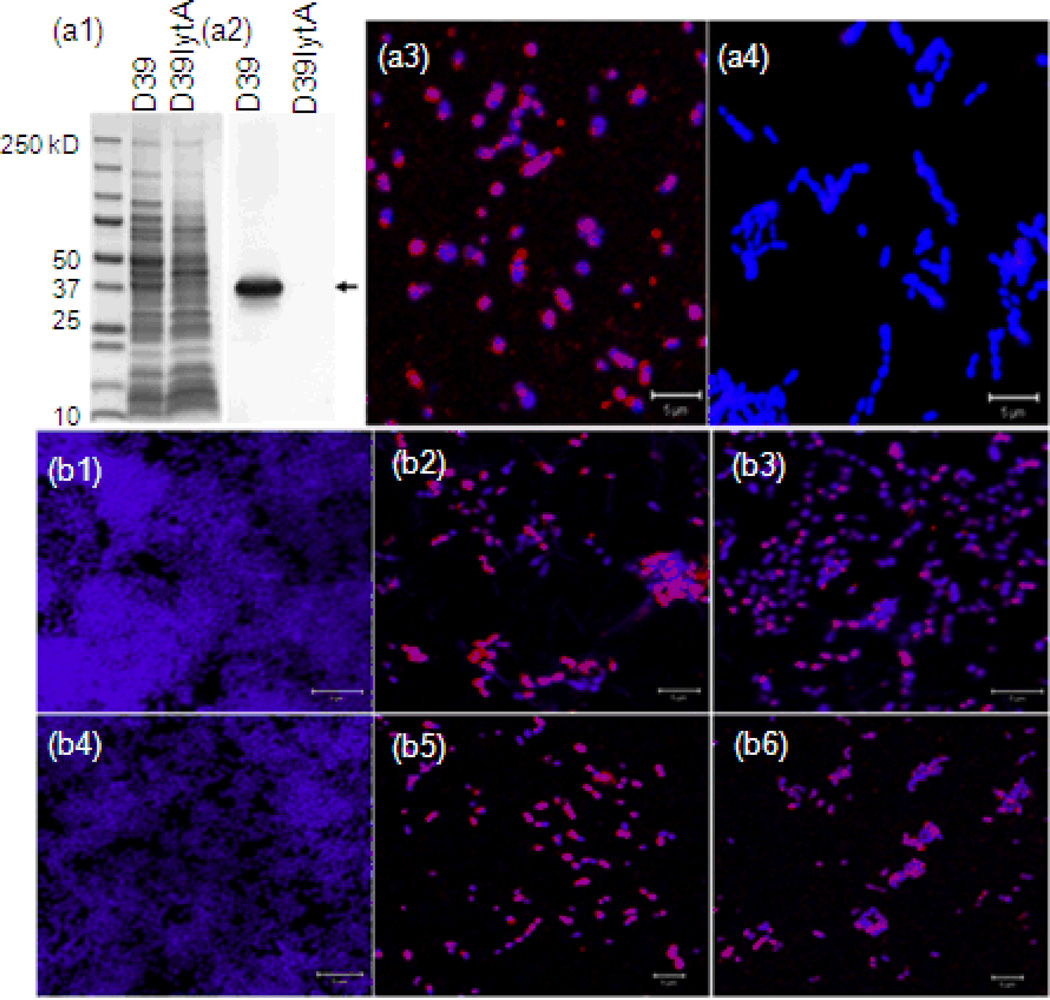
Decreased LytA protein was also confirmed by immunostaining. The specific binding activity of anti-LytA serum and LytA in biofilm cells was confirmed using a D39lytA-mutant [32] (Fig. 5a1–a4). Subsequently, LytA in the biofilms of TIGR4 alone and co-cultures was confirmed by immunostaining. There was more LytA protein detected in the biofilms of TIGR4 alone cultures than that of the co-cultures at both 16 and 24 hour time points (Fig. 5b1–b6). The correlation of down-regulated autolysin/fratricide-related gene expression and more viable pneumococcal cells in the co-culture group suggests that NTHi may jeopardize autolysis and fratricide directly or indirectly to promote pneumococcal survival and biofilm formation.
Fig. 5.
Expression and binding of pneumococcal autolysin in biofilm were decreased in the presence of NTHi. Pneumococcal LytA proteins produced (red) in the biofilm were detected by immunostaining with anti-LytA serum followed by confocal laser scanning microscopy. Biofilms were counterstained with DAPI (purple). a1–4: binding–specificities of anti-LytA serum were confirmed by western blot and immunostaining with D39 and its isogenic mutant D39lytA-. a1, results of SDS-PAGE; a2, results of western blot; a3, immunostaining of D39 biofilm; a4, immunostaining of D39lytA- biofilm. b1–6: results of immunostaining. b1, 16 hour biofilm of 86028NP; b2, 16 hour biofilm of TIGR4; b3, 16 hour biofilm of TIGR4 mixed with 86028NP; b4, 24 hour biofilm of 86028NP; b5, 24 hour biofilm of TIGR4; b6, 24 hour biofilm of TIGR4 mixed with 86028NP. At both time points, there were more LytA proteins in the biofilm of TIGR4 alone than the mixed biofilm cultures. The bar of microscopic image was 10 µm.
3.4 Cell-to-cell contact was not necessary to enhance pneumococcal survival by NTHi
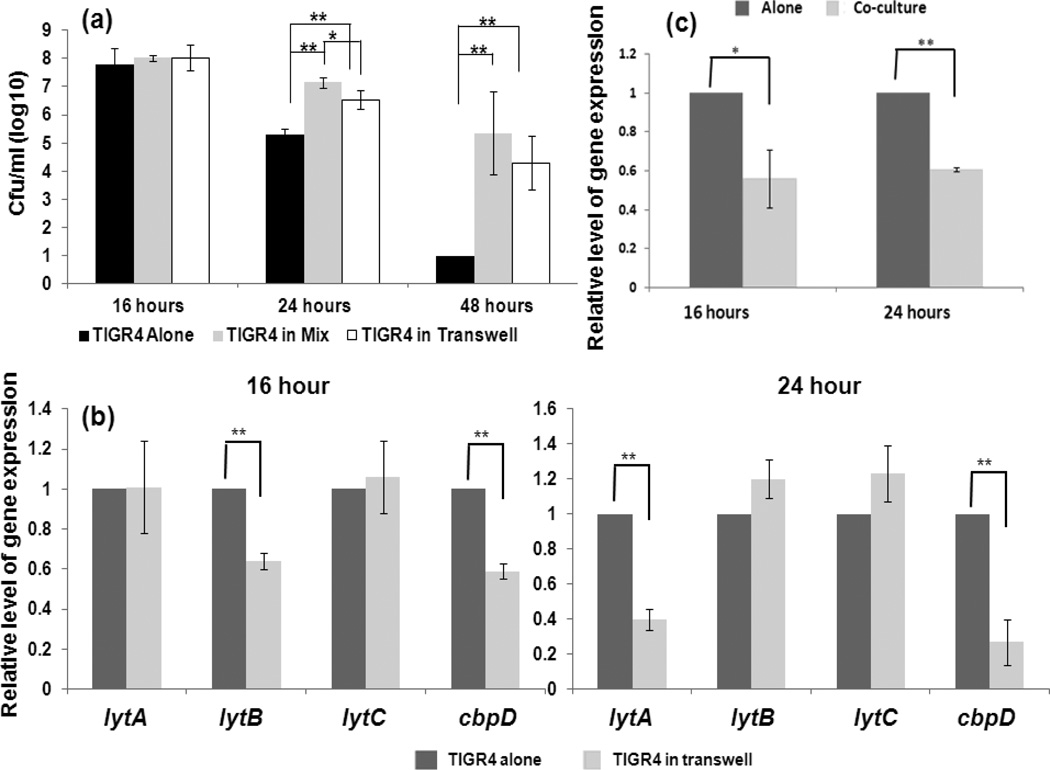
To assess whether direct cell-to-cell contact plays a role in the interaction, we set up co-cultures within a transwell system followed by viable cell counting and pneumococcal gene expression analysis. The results are summarized in Fig. 6. At 16 hours, there was no significant difference in viable TIGR4 cells between TIGR4 alone and the mixed culture in wells or co-culture with 86028NP in a transwell system. At 24 hours, the amount of viable TIGR4 cells of the mixed culture and co-culture in the transwell systems was significantly more than that of TIGR4 alone (P<0.01). In addition, the amount of viable TIGR4 cells in the mixed culture was significantly more than that of co-culture in the transwell system (P<0.05). At 48 hours, there were not viable TIGR4 cells detected in the monoculture but there were more TIGR4 cells detected in the mixed culture and co-culture in the transwell systems (P<0.01) (Fig. 6a).
Fig. 6.
Cell-to-cell contact was not required to modify pneumococcal activities by NTHi. Co-culture of 86028NP and TIGR4 without cell-to-cell contact were performed in a transwell system. Viable pneumococcal cells and gene expression were measured at different time points. a. Viable pneumococcal cell counts. Compared to TIGR4 alone culture, there were significantly more cells when TIGR4 co-grown with 86028NP either in the mixed culture (co-culturing 86028NP and TIGR4 in a same well) or co-cultured in the transwell system. Notably, there were significantly more viable TIGR4 cells in the mixed culture than co-cultured with 86028NP in the transwell system at the 24 hour point after inoculation. b. Expression of pneumococcal genes involved in autolysis and fratricide were influenced by NTHi co-cultured in transwell system. Similar to these in mixed cultures, lytA and cbpD were significantly down-regulated by NTHi. Notably, lytB gene was also down-regulated at 16 hour point which was not observed in mixed culture condition. c. NTHi down-regulated pneumococcal ply gene expression when co-cultured in transwell system. The bacterial cultures, viable cell counts and qPCR were performed four times independently. *: P<0.05; **: P<0.01.
Similar to mixed cultures, co-cultures with 86028NP in a transwell system also affected TIGR4 gene expression (Fig. 6b and 6c). Without cell-to-cell contact, 86028NP down regulated TIGR4 cbpD expression at 16 and 24 hour points and lytA expression at 24 hours after inoculation (P<0.01). Notably, co-cultures of 86028NP and TIGR4 also decreased TIGR4 lytB gene expression at the 16 hour point (P<0.01), which was not observed in the mixed cultures.
4. Discussion
Although a heptavalent conjugate pneumococcal vaccine has been incorporated into vaccination schedules for children in the US since the year 2000 [34], the overall incidence of OM hasn’t declined. Clinical studies indicate that the reasons for this tepid success include the replacement of pneumococcal strains and switching predomination of pathogenic species [35,36]. This fact along with the high incidence of polymicrobial infection highlights an urgent need for a more thorough understanding of bacterial pathogenesis of OM, in the hope that novel and more efficacious strategies can be developed to manage this disease. The results of our work clearly suggest that the presence of NTHi modulates the pneumococcal activities, which may impact the pathogenicity of pneumococcus and disease occurrence in polymicrobial infections.
It has been shown that SP inhibits the growth of H. influenzae by producing hydrogen peroxide. However, survivals of H. influenzae in the presence of SP in in vitro [37] and in vivo [17] models suggest a complicated and dynamic interference between these two organisms. Initiating the cultures with combinations of different concentrations of two bacteria, we found that the outcome of the interaction is in a cell-density dependent manner, i.e., a high density of SP cells would completely inhibit NTHi growth no matter what the initial densities of NTHi were, but a high density of NTHi cells could withstand pneumococcal killing when co-cultured with lower density TIGR4 cells (Figure 1). The potential mechanism leading to different destines of SP and NTHi in co-culturing is yet to be dtermined, but may be due to the catalase removal of hydrogen peroxide produced by SP and a reduction of pneumococcal autolysis/fratricide upon encountering increased competition in the culture. Further studies including the use a hydrogen peroxide negative pneumococcal mutant in co-culture and profiling global gene expression patterns of both bacteria in co-culture are ongoing in our laboratory.
Although SP and H. influenzae could be identified in the same biofilm structure formed on the middle ear mucosa of patients with chronic OM [10], contradictory data of SP and H. influenzae association in human patients have been reported by different groups [5, 38,39]. Our data provides a potential explanation for these varied results. When SP was in dominance over H. influenzae in the nasopharynx, it would inhibit H. influenzae colonization and growth. As a result, an acute OM would be more likely to develop caused by SP. However, if H. influenzae was dominant in the nasopharynx, it would promote SP colonization and compromise the pathogenicity of SP. As such, a mixed biofilm and chronic OM would be more likely to occur. This needs to be verified with more clinical observations and in vivo studies with animal models, which is under way in our laboratory.
Our studies indicate that the impact of NTHi on pneumococcal survival happens at post stationary phase. Notably, SP produces autolysin to suicide at this phase, resulting in the number of viable cells declining rapidly [40–42]. In addition to autolysis, SP also produces cell wall hydrolases to kill sibling cells, known as fratricide. Although both autolysis and fratricide lead to decreasing numbers of viable cells, they are regulated by different mechanisms and involve different factors. Pneumococcal autolysis is mediated by LytA [43] and fratricide in liquid medium is tightly mediated by a combination of CbpD, LytA and LytC [23–26]. Since autolysis and fratricide action are dependent on these proteins, both would be undermined when expression of one or all of them was down-regulated. Counts of viable TIGR4 cells shown in Fig. 3b clearly suggest that the enhanced SP survival in the presence of 86028NP is due to delayed bacterial death rather than augmented growth. Our results generate a straightforward conjecture that the presence of NTHi might down regulate the expression of these enzymes involved in pneumococcal autolysis and fratricide, thus decreasing pneumococcal autolysis/fratricide and delaying its death. Our qPCR results support our hypothesis that the expressions of cbpD and lytA genes were all down-regulated after stationary phase in the presence of NTHi. Using anti-pneumococcal LytA serum, we confirmed that the amounts of LytA protein located on the cell surface in the biofilm of TIGR4 alone were higher than that of TIGR4 co-cultured with 86028NP, which is in accordance with the results of qPCR. These observations correspond to fluctuations in the amount of viable pneumococcal cells in the cultures, suggesting that pneumococcal autolysis and fratricide can be down-regulated by the presence of NTHi. The phenomenon observed in our study suggests, for the first time, that pneumococcal autolysis and fratricide could be altered by other bacterial species in a polymicrobial community.
A critical question in assessing the interaction between different microorganisms is whether there is significant effect on pathogenicity. Ply is an important virulence factor of SP that breaks down the primary defense line of the host and induces inflammation [44, 45]. Cope and colleagues recently reported that co-cultures with NTHi did down-regulate pneumococcal ply gene expression at 24 hour culture [37], which was confirmed in our studies. In fact, the down-regulation of ply expression was observed as early as 16 hours post-inoculation. Down-regulated ply gene expression along with inhibited autolysis and fratricide may lead to decreased concentrations of extracellular Ply by SP in co-infection situations. Since Ply plays critical roles in the pathogenicity of SP, co-existing with H. influenzae in the human respiratory tract could compromise the tissue damage and inflammation caused by Ply. This result may explain the phenomenon observed in our previous studies showing that co-infection with NTHi in chinchilla middle ear decreased systemic infection of pneumococci [17].
The universality of the influence of NTHi on enhancing pneumococcal survival was confirmed by co-culturing different strains of SP and NTHi (Figure 3). Co-culturing with Moraxella catarrhalis, an additional important pathogen of OM, we didn’t observe enhanced pneumococcal survival and biofilm formation (data not shown), which suggested a specific cross-talking between SP and NTHi. However, there is much unknown regarding the mechanism regulating this interspecies conversation. Our studies suggested that both physical interaction and chemical-mediated cross-talking could be involved in facilitating the interaction between SP and H. influenzae. Using a transwell system, we found that physical interaction was not crucial for NTHi to enhance pneumococcal survival. This suggests that NTHi was able to influence pneumococcal behaviors via some undefined resoluble factors. That observation was consistent with the result of Cope’s study [37], which indicated that the supernatant of co-culture biofilm of SP and H. influenzae influenced pneumococcal spxB gene expression. However, the differentiated expression of lytB, as well as the higher amounts of viable pneumococcal cells in the mixed cultures compared to the co-cultures in a transwell system, clearly suggests that a physical mechanism also contributes to this interspecies interaction. Furthermore, using spent medium and cell lysate of NTHi we observed both enhanced pneumococcal survival but showed different potencies (data not shown). These data, taken together, suggest both physical and chemical mechanisms are involved in the microbial interaction in a polymicrobial community.
Acknowledgements
We thank Dr. Bing Pang at the Department of Microbiology of Wake Forest University for assistance with COMSTAT. Drs. WE Swords, Dara Frank, and David Friedland kindly reviewed our manuscript. Drs. Briles and Paton kindly provided D39lytA- mutant. This work was supported by the National Institute on Deafness and Other Communication Disorders at the National Institutes of Health [grant number DC007903] (JEK) and funding provided by the Department of Otolaryngology and Communication Sciences, Medical College of Wisconsin to WH.
References
- 1.Klein JO. The burden of otitis media. Vaccine. 2000;19(Suppl 1):2–8. doi: 10.1016/s0264-410x(00)00271-1. [DOI] [PubMed] [Google Scholar]
- 2.Paradise JL, Rockette HE, Colborn DK, Bernard BS, Smith CG, Kurs-Lasky M, Janosky JE. Otitis media in 2253 Pittsburgh-area infants: prevalence and risk factors during the first two years of life. Pediatrics. 1997;99:318–333. doi: 10.1542/peds.99.3.318. [DOI] [PubMed] [Google Scholar]
- 3.Finkelstein JA, Davis RL, Dowell SF, Metlay JP, Soumerai SB, Rifas-Shiman SL, Higham M, Miller Z, Miroshnik I, Pedan A, Platt R. Reducing antibiotic use in children: a randomized trial in 12 practices. Pediatrics. 2001;108:1–7. doi: 10.1542/peds.108.1.1. [DOI] [PubMed] [Google Scholar]
- 4.Post JC, Preston RA, Aul JJ, Larkins-Pettigrew M, Rydquist-White J, Anderson KW, Wadowsky RM, Reagan DR, Walker ES, Kingsley LA, Magit AE, Ehrlich GD. Molecular analysis of bacterial pathogens in otitis media with effusion. JAMA. 1995;273:1598–1604. [PubMed] [Google Scholar]
- 5.Pettigrew MM, Gent JF, Revai K, Patel JA, Chonmaitree T. Microbial interactions during upper respiratory tract infections. Emerg Infect Dis. 2008;14:1584–1591. doi: 10.3201/eid1410.080119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hendolin PH, Markkanen A, Ylikoski J, Wahlfors JJ. Use of multiplex PCR for simultaneous detection of four bacterial species in middle ear effusions. J Clin Microbiol. 1997;35:2854–2858. doi: 10.1128/jcm.35.11.2854-2858.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaur R, Adlowitz DG, Casey JR, Zeng M, Pichichero ME. Simultaneous assay for four bacterial species including Alloiococcus otitidis using multiplex-PCR in children with culture negative acute otitis media. Pediatr Infect Dis J. 2010;29:741–745. doi: 10.1097/INF.0b013e3181d9e639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pichichero ME, Casey JR. Evolving microbiology and molecular epidemiology of acute otitis media in the pneumococcal conjugate vaccine era. Pediatr Infect Dis J. 2007;26(Suppl 10):12–16. doi: 10.1097/INF.0b013e318154b25d. [DOI] [PubMed] [Google Scholar]
- 9.Gok U, Bulut Y, Keles E, Yalcin S, Doymaz MZ. Bacteriological and PCR analysis of clinical material aspirated from otitis media with effusions. Int J Pediatr Otorhinolaryngol. 2001;60:49–54. doi: 10.1016/s0165-5876(01)00510-9. [DOI] [PubMed] [Google Scholar]
- 10.Hall-Stoodley L, Hu FZ, Gieseke A, Nistico L, Nguyen D, Hayes J, Forbes M, Greenberg DP, Dice B, Burrows A, Wackym PA, Stoodley P, Post JC, Ehrlich GD, Kerschner JE. Direct detection of bacterial biofilms on the middle-ear mucosa of children with chronic otitis media. JAMA. 2006;296:202–211. doi: 10.1001/jama.296.2.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ruohola A, Meurman O, Nikkari S, Skottman T, Salmi A, Waris M, Osterback R, Eerola E, Allander T, Niesters H, Heikkinen T, Ruuskanen O. Microbiology of acute otitis media in children with tympanostomy tubes: prevalences of bacteria and viruses. Clin Infect Dis. 2006;43:1417–1422. doi: 10.1086/509332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sanderson AR, Leid JG, Hunsaker D. Bacterial biofilms on the sinus mucosa of human subjects with chronic rhinosinusitis. Laryngoscope. 2006;116:1121–1126. doi: 10.1097/01.mlg.0000221954.05467.54. [DOI] [PubMed] [Google Scholar]
- 13.Sethi S. Bacteria in exacerbations of chronic obstructive pulmonary disease: phenomenon or epiphenomenon? Proc Am Thorac Soc. 2004;1:109–114. doi: 10.1513/pats.2306029. [DOI] [PubMed] [Google Scholar]
- 14.Shakhnovich EA, King SJ, Weiser JN. Neuraminidase expressed by Streptococcus pneumoniae desialylates the lipopolysaccharide of Neisseria meningitidis and Haemophilus influenzae: a paradigm for interbacterial competition among pathogens of the human respiratory tract. Infect Immun. 2002;70:7161–7164. doi: 10.1128/IAI.70.12.7161-7164.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pericone CD, Overweg K, Hermans PW, Weiser JN. Inhibitory and bactericidal effects of hydrogen peroxide production by Streptococcus pneumoniae on other inhabitants of the upper respiratory tract. Infect Immun. 2000;68:3990–3997. doi: 10.1128/iai.68.7.3990-3997.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lysenko ES, Ratner AJ, Nelson AL, Weiser JN. The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog. 2005;1:e1. doi: 10.1371/journal.ppat.0010001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weimer KE, Armbruster CE, Juneau RA, Hong W, Pang B, Swords WE. Coinfection with Haemophilus influenzae promotes pneumococcal biofilm formation during experimental otitis media and impedes the progression of pneumococcal disease. J Infect Dis. 2010;202:1068–1075. doi: 10.1086/656046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kweon SM, Wang B, Rixter D, Lim JH, Koga T, Ishinaga H, Chen LF, Jono H, Xu H, Li JD. Synergistic activation of NF-kappaB by nontypeable H. influenzae and S. pneumoniae is mediated by CK2, IKKbeta-IkappaBalpha, and p38 MAPK. Biochem Biophys Res Commun. 2006;351:368–375. doi: 10.1016/j.bbrc.2006.10.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shen H, Yoshida H, Yan F, Li W, Xu F, Huang H, Jono H, Li JD. Synergistic induction of MUC5AC mucin by nontypeable Haemophilus influenzae and Streptococcus pneumoniae. Biochem Biophys Res Commun. 2008;365:795–800. doi: 10.1016/j.bbrc.2007.11.060. [DOI] [PubMed] [Google Scholar]
- 20.Ratner AJ, Lysenko ES, Paul MN, Weiser JN. Synergistic proinflammatory responses induced by polymicrobial colonization of epithelial surfaces. Proc Natl Acad Sci USA. 2005;102:3429–3434. doi: 10.1073/pnas.0500599102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Steinmoen H, Teigen A, Havarstein LS. Competence-induced cells of Streptococcus pneumoniae lyse competence-deficient cells of the same strain during cocultivation. J Bacteriol. 2003;185:7176–7183. doi: 10.1128/JB.185.24.7176-7183.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Claverys JP, Martin B, Havarstein LS. Competence-induced fratricide in streptococci. Mol Microbiol. 2007;64:1423–1433. doi: 10.1111/j.1365-2958.2007.05757.x. [DOI] [PubMed] [Google Scholar]
- 23.Eldholm V, Johnsborg O, Haugen K, Ohnstad HS, Havarstein LS. Fratricide in Streptococcus pneumoniae: contributions and role of the cell wall hydrolases CbpD, LytA and LytC. Microbiology. 2009;155:2223–2234. doi: 10.1099/mic.0.026328-0. [DOI] [PubMed] [Google Scholar]
- 24.Guiral S, Mitchell TJ, Martin B, Claverys JP. Competence-programmed predation of noncompetent cells in the human pathogen Streptococcus pneumoniae: genetic requirements. Proc Natl Acad Sci USA. 2005;102:8710–8715. doi: 10.1073/pnas.0500879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kausmally L, Johnsborg O, Lunde M, Knutsen E, Havarstein LS. Choline-binding protein D (CbpD) in Streptococcus pneumoniae is essential for competence-induced cell lysis. J Bacteriol. 2005;187:4338–4345. doi: 10.1128/JB.187.13.4338-4345.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Eldholm V, Johnsborg O, Straume D, Ohnstad HS, Berg KH, Hermoso JA, Håvarstein LS. Pneumococcal CbpD is a murein hydrolase that requires a dual cell envelope binding specificity to kill target cells during fratricide. Mol Microbiol. 2010;76:905–917. doi: 10.1111/j.1365-2958.2010.07143.x. [DOI] [PubMed] [Google Scholar]
- 27.Heydorn A, Nielsen AT, Hentzer M, Sternberg C, Givskov M, Ersbøll BK, Molin S. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology. 2000;146:2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 28.Holm C, Jespersen L. A Flow-Cytometric Gram-Staining Technique for Milk-Associated Bacteria. Appl Environ Microbial. 2003;69:2857–2863. doi: 10.1128/AEM.69.5.2857-2863.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bentley R. The shikimate pathway--a metabolic tree with many branches. Crit Rev Biochem Mol Biol. 1990;25:307–384. doi: 10.3109/10409239009090615. [DOI] [PubMed] [Google Scholar]
- 30.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 31.Cockeran R, Anderson R, Feldman C. The role of pneumolysin in the pathogenesis of Streptococcus pneumoniae infection. Curr Opin Infect Dis. 2002;15:235–239. doi: 10.1097/00001432-200206000-00004. [DOI] [PubMed] [Google Scholar]
- 32.Berry AM, Lock RA, Hansman D, Paton JC. Contribution of autolysin to virulence of. Streptococcus pneumoniae, Infect Immun. 1989;57:2324–2330. doi: 10.1128/iai.57.8.2324-2330.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Garcia P, Gonzalez MP, Garcia E, Lopez R, Garcia JL. LytB, a novel pneumococcal murein hydrolase essential for cell separation. Mol Microbiol. 1999;31:1275–1281. doi: 10.1046/j.1365-2958.1999.01238.x. [DOI] [PubMed] [Google Scholar]
- 34.Black S, Shinefield H, Fireman B, Lewis E, Ray P, Hansen JR, Elvin L, Ensor KM, Hackell J, Siber G, Malinoski F, Madore D, Chang I, Kohberger R, Watson W, Austrian R, Edwards K. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Northern California Kaiser Permanente Vaccine Study Center Group. Pediatr Infect Dis J. 2000;19:187–195. doi: 10.1097/00006454-200003000-00003. [DOI] [PubMed] [Google Scholar]
- 35.Block SL, Hedrick J, Harrison CJ, Tyler R, Smith A, Findlay R, Keegan E. Community-wide vaccination with the heptavalent pneumococcal conjugate significantly alters the microbiology of acute otitis media. Pediatr Infect Dis J. 2004;23:829–833. doi: 10.1097/01.inf.0000136868.91756.80. [DOI] [PubMed] [Google Scholar]
- 36.Black S, Shinefield H, Baxter R, Austrian R, Elvin L, Hansen J, Lewis E, Fireman B. Impact of the use of heptavalent pneumococcal conjugate vaccine on disease epidemiology in children and adults. Vaccine. 2006;24(Suppl 2):79–80. doi: 10.1016/j.vaccine.2005.01.132. [DOI] [PubMed] [Google Scholar]
- 37.Cope EK, Goldstein-Daruech N, Kofonow JM, Christensen L, McDermott B, Monroy F, Palmer JN, Chiu AG, Shirtliff ME, Cohen NA, Leid JG. Regulation of virulence gene expression resulting from Streptococcus pneumoniae and nontypeable Haemophilus influenzae interactions in chronic disease. PLoS One. 2011;6:e28523. doi: 10.1371/journal.pone.0028523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jacoby P, Watson K, Bowman J, Taylor A, Riley TV, Smith DW, Lehmann D. Kalgoorlie Otitis Media Research Project Team. Modelling the co-occurrence of Streptococcus pneumoniae with other bacterial and viral pathogens in the upper respiratory tract. Vaccine. 2007;25:2458–2464. doi: 10.1016/j.vaccine.2006.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jourdain S, Smeesters PR, Denis O, Dramaix M, Sputael V, Malaviolle X, Van Melderen L, Vergison A. Differences in nasopharyngeal bacterial carriage in preschool children from different socio-economic origins. Clin Microbiol Infect. 2011;17:907–914. doi: 10.1111/j.1469-0691.2010.03410.x. [DOI] [PubMed] [Google Scholar]
- 40.Moscoso M, Claverys JP. Release of DNA into the medium by competent Streptococcus pneumoniae: kinetics, mechanism and stability of the liberated DNA. Mol Microbiol. 2004;54:783–794. doi: 10.1111/j.1365-2958.2004.04305.x. [DOI] [PubMed] [Google Scholar]
- 41.Morrison DA, Baker MF. Competence for genetic transformation in pneumococcus depends on synthesis of a small set of proteins. Nature. 1979;282:215–217. doi: 10.1038/282215a0. [DOI] [PubMed] [Google Scholar]
- 42.Tomasz A, Hotchkiss RD. Regulation of the Transformability of Pheumococcal Cultures by Macromolecular Cell Products. Proc Natl Acad Sci USA. 1964;51:480–487. doi: 10.1073/pnas.51.3.480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lopez R, Garcia JL, Garcia E, Ronda C, Garcia P. Structural analysis and biological significance of the cell wall lytic enzymes of Streptococcus pneumoniae and its bacteriophage. FEMS Microbiol Lett. 1992;79:439–447. doi: 10.1111/j.1574-6968.1992.tb14074.x. [DOI] [PubMed] [Google Scholar]
- 44.Jedrzejas MJ. Pneumococcal virulence factors: structure and function. Microbiol Mol Biol Rev. 2001;65:187–207. doi: 10.1128/MMBR.65.2.187-207.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoo IH, Shin HS, Kim YJ, Kim HB, Jin S, Ha UH. Role of pneumococcal pneumolysin in the induction of an inflammatory response in human epithelial cells. FEMS Immunol Med Microbiol. 2010;60:28–35. doi: 10.1111/j.1574-695X.2010.00699.x. [DOI] [PubMed] [Google Scholar]