Abstract
The nitrate anion is a simple, abundant and relatively stable species, yet plays a significant role in global cycling of nitrogen, global climate change, and human health. Although it has been known for quite some time that nitrate is an important species environmentally, recent studies have identified potential medical applications. In this respect the nitrate anion remains an enigmatic species that promises to offer exciting science in years to come. Many bacteria readily reduce nitrate to nitrite via nitrate reductases. Classified into three distinct types – periplasmic nitrate reductase (Nap), respiratory nitrate reductase (Nar) and assimilatory nitrate reductase (Nas), they are defined by their cellular location, operon organization and active site structure. Of these, Nap proteins are the focus of this review. Despite similarities in the catalytic and spectroscopic properties Nap from different Proteobacteria are phylogenetically distinct. This review has two major sections: in the first section, nitrate in the nitrogen cycle and human health, taxonomy of nitrate reductases, assimilatory and dissimilatory nitrate reduction, cellular locations of nitrate reductases, structural and redox chemistry are discussed. The second section focuses on the features of periplasmic nitrate reductase where the catalytic subunit of the Nap and its kinetic properties, auxiliary Nap proteins, operon structure and phylogenetic relationships are discussed.
1. Scope
Periplasmic nitrate reductase (Nap) catalyzes the transformation of nitrate to nitrite in prokaryotes. Isolation and biochemical characterization of the catalytic subunit, NapA, with or without an electron transfer subunit, NapB, from several genera of bacteria has established that NapA’s high affinity for nitrate is conserved. Despite catalytic consistencies and confinement in the periplasmic space, the physiological role of NapA is diverse. This is in contrast to the more commonly studied eukaryotic nitrate reductase (eukNR)1–3 and bacterial respiratory nitrate reductase (Nar).4–6 All nitrate reductases are molybdopterin enzymes, where molybdenum is explicitly ligated by one or two pyranopterin prosthetic groups at the enzyme’s catalytic heart. Herein we examine the similarities and differences in nap gene structure, nap operon regulation, and NapA enzyme biochemistry in an attempt to learn about the physiological function of NapA in prokaryotes. The impact of NapA-catalyzed nitrate reduction on the environment and human health is discussed. Additionally, we review Nap in the context of other molybdopterin containing enzymes, and nitrate reduction in general.
2. Nitrate reduction
2.1 Nitrate in the nitrogen cycle
Nitrogen is distributed throughout Earth’s lithosphere, atmosphere, hydrosphere, and biosphere. The lithosphere contains ~94% of all nitrogen on Earth, ~6% in the atmosphere, and a small fraction (<0.006%) within the hydrosphere and biosphere. 7 Nitrogen is the 4th most abundant element in the biosphere, preceded only by oxygen, carbon and hydrogen, and is a vital component of the collective biomass.
The biosphere’s relative abundance of nitrogen is an indication of the importance of nitrogen for living organisms. The ground state electronic configuration of nitrogen is 1s22s22p3. Nitrogen can form numerous stable compounds, organic or inorganic, with oxidation states ranging from −3 to +5. Organic nitrogen molecules e.g., hormones, amino acids, and nucleic acids, are vital to life on Earth. Additionally, inorganic nitrogen, (e.g., nitrate, nitric oxide), is of paramount importance as it is the keystone of the global nitrogen cycle. Inorganic nitrogen species such as NO and NO2− serves as important signaling molecules. The importance of inorganic nitrogen compounds in the environment and human health will be elaborated later in this review.
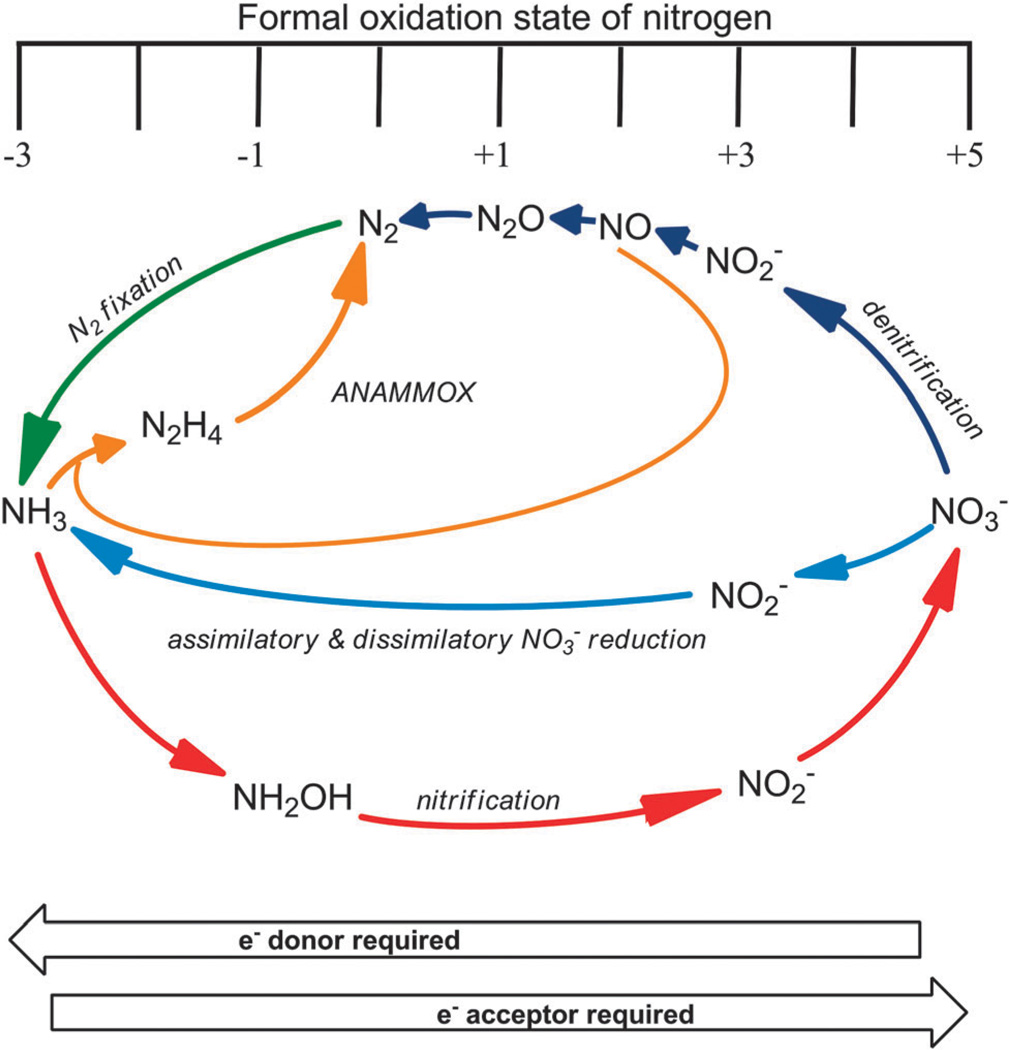
The global nitrogen cycle is an interconnected network of chemical transformations that recycles nitrogen between the atmosphere, biosphere, hydrosphere, and lithosphere. Inorganic nitrogen compounds (i.e., nitrate (NO3−), nitrite (NO2−), nitrous oxide (N2O), nitric oxide (NO), dinitrogen (N2), and ammonia (NH3)) are central to the global nitrogen cycle (Fig. 1). The most oxidized form of nitrogen is NO3−, while the most reduced form is NH3. Microbes are the driving force behind the global nitrogen cycle as they use an assortment of redox reactions to metabolize nitrogen for energy transduction, detoxification, or assimilation.
Fig. 1.
A schematic representation of the bacterial nitrogen cycle.
Both anaerobic ammonium oxidation (ANAMMOX) and nitrification oxidize ammonia, either into N2 (eqn (1)) or NO3− (eqn (2)), respectively. Denitrification is a four-step reductive process, which transforms NO3− to N2, via NO2−, NO, and N2O (eqn (3)). Nitrogen fixation reduces N2 into NH3 (eqn (4)). Nitrate reduction to ammonia is a two-step process with conversion of NO3− to NO2− followed by reduction of NO2− to ammonium (NH4+) (eqn (5)).7,8 Intermediates in NO2− reduction pathways such as
| (1) |
| (2) |
| (3) |
| (4) |
| (5) |
NO and hydroxylamine are not released into the environment.9 Nitrate reduction is designated as assimilatory or dissimilatory depending on the fate of the reduced nitrogen; when incorporated into the biomass it is called assimilatory, and in dissimilatory reduction it is excreted from the cell (vide infra). Microbes encode specialized enzymes, which catalyze the transformations of each nitrogen compound (Table 1). These microbe-driven transformations impact all life, as they are essential to maintain the bioavailability of nitrogen.
Table 1.
Biogenic processes involved in the nitrogen cycle
| Pathway(s) | Reactions catalyzed | Enzyme | Redox potential (mV) | Ref. |
|---|---|---|---|---|
| Assimilatory and dissimilatory nitrate reduction | NO3− + 2H+ + 2e− → NO2− + H2O | Nitrate reductase (NR: eukNR, Nar, Nap and Nas) | +433 | 313 |
| NO2− + 8H+ + 6e− → NH4+ + 2H2O | Nitrite reductase (Nrf) | +340 | 314 | |
| Denitrifieation | NO2− + 2H+ + e− → NO + H2O | Nitrite reductase (NiR) | +350 | 313 |
| 2NO + 2H+ + 2e− → N2O + H2O | Nitric oxide reductase (NoR) | +1175 | 313 | |
| N2O + 2H+ + 2e− → N2 + H2O | Nitrous oxide reductase | +1355 | 313 | |
| Anammox | NO + NH3 + 3H+ + 3e− → N2H4 + H2O | Hydrazine hydrolase (HH) | +340 | 315, 316 |
| N2H4 → N2 + 4H+ + 4e− | Hydrazine dehydrogenase (HDH) | −230 | 313 | |
| Nitrification | NO2− + H2O → NO3− + 2H+ + 2e− | Nitrite oxidase (NO) | +420 | 317, 318 |
| NH2OH + H2O → NO2− + 5H+ + 4e− | Hydroxylamine oxidoreductase (HAO) | +60 | 317 | |
| NH3 + O2 + 2H+ + 2e− → NH2OH + H2O | Ammonia oxidase (AMO) | +730 | 317 | |
| Nitrogen fixation | N2 + 6H+ + 6e− → 2NH3 | Nitrogenase | −92 | 319 |
Anthropogenic activity disrupts the nitrogen cycle through the use of fossil fuels and nitrogen-rich fertilizers. Nitrogen speciation facilitates the movement of nitrogen among the nitrogen-rich lithosphere and atmosphere and the nitrogen-limited biosphere and hydrosphere. In general, volatile nitrogen compounds (e.g., NO, N2O, N2) accumulate in the atmosphere, solid compounds sediment in the lithosphere (e.g., NO3−, NO2−, NH4+), and ionic water-soluble compounds (e.g., NO3−, NO2−, NH4+) are mobile in the hydrosphere. In the biosphere, organisms acquire nitrogen in a species-specific manner; the key compounds are NO3− and NH4+ (elaborated in the following paragraph). Burning fossil fuels releases volatile nitrogen species (e.g., NO2, NO and N2O), which accumulate in the atmosphere. N2O contributes to global warming, it is the 3rd most abundant greenhouse gas, with a heat capacity of 37.2 J mol−1 K−1. This heat capacity is comparable to that of carbon dioxide and methane, 37.1 J mol−1 K−1, and 35.7 J mol−1 K−1, respectively.10 Agricultural run-off contaminates water supplies with NO3− and NH4+ introduced through the disproportionate use of nitrogen-rich fertilizers. Ammonia can exhibit adverse effects on aquatic life (e.g., fish), and NO3− contaminated well water has been associated with undesirable human health effects,11–14 as well as eutrophication.15,16 The use of fossil fuels and nitrogen-rich fertilizers also limits nitrogen-bioavailability, by lowering natural nitrogen content in the sediment and elevating nitrogen content in water supplies.
Despite the diversity of nitrogen species generated in the nitrogen cycle, only a select few inorganic nitrogen compounds can be utilized by higher organisms. Animals require organic nitrogen compounds as a source of nitrogen; amino acids and nucleic acids acquired from diet are recycled for synthesis of new nitrogen compounds. In contrast, bacteria, plants, and fungi can utilize inorganic nitrogen compounds, (e.g., NO3−, NH4+). Ammonia can be directly used to synthesize proteins and nucleic acids, while NO3− can be stored and converted to ammonia via nitrate and nitrite reduction. The lithosphere contains the largest pool of biologically useful nitrogen compounds, in the form of decomposed organic matter or ionic nitrogen compounds. Inorganic nitrogen stays in the lithosphere as a result of nitrogen’s non-rock forming nature. Nitrification can transform NH4+, a by-product of organic decomposition, into NO3−, a nontoxic nitrogen storage molecule. In essence, the microbe-driven nitrogen cycle supplies biologically active forms of nitrogen to the biota.
Although a vast pool of atmospheric N2 covers Earth’s surface, it is essentially unusable. Breaking the N–N bond (distance, 1.097 Å) in N2 is associated with a large heat of dissociation (eqn (6)),17 thus the endothermicity of this reaction lends itself to the inertness of N2. Note that the reduction of
| (6) |
dinitrogen in nitrogen fixation is thermodynamically favorable with a negative free energy (eqn (4)). Denitrification and ANAMMOX pathways replenish atmospheric N2. Nitrogen fixation transforms N2 into NH4+, which then is deposited in the lithosphere and hydrosphere.
Of the nitrogen compounds involved in the nitrogen cycle, nitrate is the most stable and important source of nitrogen that can be retained in soils, sediments, and water providing a biologically useful pool. Enzymatic reduction of NO3− to NO2− can generate a large amount of energy, which organisms can utilize to sustain life. This is evident by the large negative free energy associated with microbial pathways that start with nitrate reduction, denitrification (eqn (3)) and nitrate reduction to ammonia (eqn (5)).
Even though nitrate forms naturally in the environment, it is often found with contaminants that formed either naturally or anthropogenically. For example, naturally occurring nitrate and perchlorate have been reported to co-accumulate in soils from arid environments such as the Mojave Desert in California.18 Perchlorate is a known contaminant that is used as an oxidant in rocket fuel, and has additional uses, such as fireworks, flares and explosives. Perchlorate interferes with iodine uptake, thus impacting the thyroid gland function. Elevated levels of nitrate have also been associated with soils contaminated with heavy metals and radionuclides as nitric acid is used in processing of these metals. Like nitrate, perchlorate can serve as an electron acceptor in biology.19 For example, an elevated nitrate level has been associated with chromium,20,21 and uranium.22,23 Thus successful strategies for the bioremediation of heavy metal contaminated sites must address the influence of nitrate in the process.21
2.2 Nitrate in human health
Humans naturally ingest nitrate through diet as it is found in meat, fruits, vegetables, grains, dairy products, and drinking water.24 Nitrate concentration in meat is low (~5 mg kg−1), although “cured” meats have higher levels (>10 mg kg−1).25 Vegetables contain a high level of nitrate, ranging from a low of 13 mg kg−1 in asparagus to a high of 2500 mg kg−1 in spinach. The USA Environmental Protection Agency (EPA) has set the maximum contamination level (MCL) of nitrate in drinking water at 10 mg L−1 (10 ppm),26 whereas that set by the World Health Organization (WHO) is 50 mg L−1 (50 ppm).27 Despite its natural abundance in food, nitrate has been considered to have negative health effects, due to the fact that it can be transformed into nitrite (which has an MCL of only 1 mg L−1) by bacterial nitrate reductases (NR) in the oral cavity28–30 and gastrointestinal tract.31–33
Nitrate-contaminated drinking water has been known to cause methemoglobinanemia in infants (less than 4 month) and adults with a genetic mutation.34 The disease results from nitrite oxidizing the iron center of hemoglobin from Fe2+ to Fe3+ producing methemoglobin, which cannot bind and transport oxygen, a condition that can have deadly consequences. In healthy adults, methemoglobin reductase, also known as cytochrome b5 reductase, rapidly converts methemoglobin into hemoglobin. Infants do not have a high level of methemoglobin reductase at birth, and as a result, methemoglobin levels can accumulate in the blood stream, leading to hypoxia. Methemoglobinanemia is associated with nitrate and nitrite contaminated well water, although congenital methemoglobinanemia exists.35,36 Genetic mutations in cytochrome b5 reductase or glucose-6-phosphate dehydrogenase have been linked with this condition.
For many years, nitrite has been implicated in human carcinogenesis, because nitrite can form reactive N-nitroso compounds, which have been associated with cancer.37–40 It has been argued that generation of toxic nitrogen compounds occurs at non-physiological concentrations. Dietary intake of such compounds increase the risk of cancer,41 but it is yet to be unequivocally proven that nitrate or nitrite can cause cancer.27 More recently, beneficial applications of nitrate and nitrite have been reported in the treatment of cardiovascular disorders,42–49 pulmonary hypertension,50 ischemia reperfusion induced injury,51 and host defense mechanisms (i.e., mucus generation). 52 The benefits of dietary nitrate are being established and the possible role of nitrate–nitrite therapy in myocardial infarction, stroke, systemic and pulmonary hypertension, and gastric ulceration are now being investigated.53,54
Nitrite can be converted into nitric oxide,53 which then can be oxidized back to nitrite or nitrate.55 Accordingly, nitrite has been described as a storage pool for nitric oxide in mammals.56 Nitric oxide is a pluripotent signaling molecule, an essential component of vasodilation,54 and has numerous physiological roles. The exact nature depends on factors such as cell type, concentration, and time. Sustained pulses of NO can be genotoxic at low concentrations (~100 nM) while higher concentrations (>1 µM) have been linked to increased nitrosation of the DNA repair enzyme and Zn-finger proteins.57–59 Taken together, it is evident that nitrate and nitrite are integral components of NO-biosynthesis and regulation. As a result, their levels are now monitored in certain humans and animal diseases. For example, patients with sickle cell disease have elevated levels of nitrate and nitrite in their plasma,60 with NO affecting vascular function.61
At this time, the effect of nitrate-rich diets on the human microbiota has not been investigated fully. Oral bacteria transform nitrate into nitrite affecting bioavailable NO62 which can influence cardiovascular health.47 Similarly, gastrointestinal bacteria can metabolize nitrate into nitrite, and nitrite into NO as well as ammonia.63–65 Interestingly, NO production increases antibiotic resistance in certain bacteria such as in B. anthracis and S. auresus.66
The involvement of microbes in organ function is a relatively new concept; however there is an increasing body of evidence suggesting microbial activity can affect organ function in mammals.67 It is estimated that there are 1014 bacterial cells present in a human body, which is an order of magnitude more than the human cells present.68 Detailed biochemical and biophysical studies on the pathogenic or communalistic species with specific NRs can provide insight into the effects of bacterial nitrate reduction in the human gastrointestinal tract. Gomez et al. have suggested that in the presence of highly reduced carbon sources, heterotrophic nitrification can occur in the intestine.69 Although this pathway is yet to be fully elucidated, elevated nitrification can lead to a higher concentration of nitrate and nitrite in the intestine that then can support microbial growth on nitrate via nitrate reductase. Nitrate reduction by pathogenic microbes has also been noted. A large fraction of the bacterial pathogens are gram-negative such as the nosocomial multidrug-resistant Acinetobacter baumannii, Klebsiella, Escherichia coli, and Pseudomonas aeruginosa. Mycobacterium and Pseudomonas also utilize nitrate reductases during host (lung) infection, suggesting nitrate reduction is important to the physiology of pathogens.70–72 It has recently been demonstrated that nitrate reduction is an important pathway for Campylobacter during host colonization.73,74 Commonly known pathogenic bacteria, their nitrate–nitrite reductases, and relationship to NO are listed in Table 2. Macrophages utilize nitric oxide to kill invading organisms as part as the innate immune system.75 A correlation between the presence of nitrate and nitrite reductase enzymes and an organism’s insensitivity to nitric oxide (Table 2) implies that nitrate reductase may have a role in microbial survival.
Table 2.
Common pathogenic organisms, nitrate and nitrite reductases and relationship to NO (uk = unknown, nh = no homologue, na = not applicable)
| Organism | Disease | Nitrate reductase |
Nitrite reductase |
End product |
Relationship to NO |
|---|---|---|---|---|---|
| Bacillus anthracis | Anthrax | NarG | NirA, NasB | NH3 | Makes NO by NOS66 |
| Bordetella pertussis | Whooping cough | −nh- | −nh- | −na- | −uk- |
| Borrelia burgdorferi | Lyme disease | −nh- | −nh- | −na- | −uk- |
| Campylobacter jejuni | Gastroenteritis, reactive arthritis | NapA | NrfA | NH3 | Not sensitive to NO320 |
| Chlamydia trachomatis | STD, eye infections | −nh- | −nh- | −na- | NO sensitive321 |
| Clostridium botulinum | Botulism | −nh- | NirA | NH3 | NO sensitive53 |
| Clostridium difficile | Pseudomembranous colitis | −nh- | NirA | NH3 | Makes NO63 |
| Escherichia coli | Diarrhea | NarG, NarZ, NapA | Nrf, Nir | NH3, NO | NO detoxification via Nrf322 |
| Haemophilus influenzae | Respiratory disease | NapA | NrfA | −uk- | −uk- |
| Helicobacter pylori | Gastric cancer and ulcer | −nh- | −nh- | −na- | Killed by nitrite and NO323 |
| Helicobacter hepaticus | Diarrhea | NapA | Nrf | NH3 | −uk- |
| Legionella pneumophila | Legionnaires disease, ventilator-associated pneumonia | −nh- | −nh- | −na- | Killed by NO324 |
| Listeria monocytogenes | Listeriosis | −nh- | −nh- | −na- | −uk- |
| Mycobacterium tuberculosis | Tuberculosis | NarG | NirBD | NH3 | Resistant to NO70,71 |
| Neisseria gonorrhoeae | STD | −nh- | NirB, NirK | NO | Resistant to NO24,325 |
| Neisseria meningitidis | Bacterial meningitis | −nh- | NirB | NO | Makes NO |
| Pseudomonas aeruginosa | Lung infection | NapA, NarG, NasA | NirBD, NasB | NH3, NO | Makes NO326 |
| Rickettsia rickettsii | Spotted fever, typhus | −nh- | −nh- | −na- | −uk- |
| Salmonella typhimurium | Salmonellosis | NapA, NarG, NarZ | Nrf, Nir | NH3, NO | −uk- |
| Shigella dysenteriae | Shigellosis, dysentery | NapA, NarG, NarZ | Nrf, NirB | NH3, NO | −uk- |
| Staphylococcus aureus | Boils sinusitis, food poisoning | NarG | NasD | NH3 | Makes NO |
| Streptococcus pneumoniae | Bacterial meningitis | −nh- | −nh- | −na- | −uk- |
| Vibrio cholera | Cholera, diarrhea | NapA | −nh- | −uk- | Makes NO |
| Yersinia pestis | Pneumonic, septicemia, bubonic plague | NapA | NirBD | NH3 | Sensitive |
2.3 Taxonomy of nitrate reductases
All nitrate reductases are molybdenum dependent enzymes, and have been divided into four major groups: eukNR, Nas, Nap, and Nar.76 Nitrate reductases have been classified according to physiological function, subcellular location (i.e., cytoplasmic, periplasmic, or membrane) of the enzyme, structure of the molybdenum active site (e.g., dioxo-Mo(vi) and monooxo-Mo(vi)). In plants and fungi, eukNR functions in nitrate assimilation, while the three prokaryote nitrate reductases can function in both assimilatory and dissimilatory nitrate reduction: assimilatory nitrate reductase (Nas) or dissimilatory nitrate reductases i.e., Nar and Nap.76 The metabolic roles of both assimilatory and dissimilatory nitrate reduction are discussed in Section 2.4, the cellular localization is discussed in Section 2.5, while the structural aspects of the catalytic subunit are discussed in Section 2.6. Eukaryotic nitrate reductase (eukNR) has been isolated from plants and fungi; animals do not encode a classical nitrate reductase, although bovine xanthine oxidase (XO) can reduce nitrate into nitrite.77–80 Moreover, eukaryotic molybdenum enzymes have been investigated as nitrite reductase; Nas, XO, aldehyde oxidase (AO), and mitochondrial amidoxime reducing component (mARC) are also capable of nitrite reduction to NO.79,81–84 Most of these molybdenum enzymes i.e., sulfite oxidase (SO), XO, and AO, have well-established physiological functions and thus they are not generally considered as classical “nitrate or nitrite reductases”.
2.4 Assimilatory and dissimilatory nitrate reduction
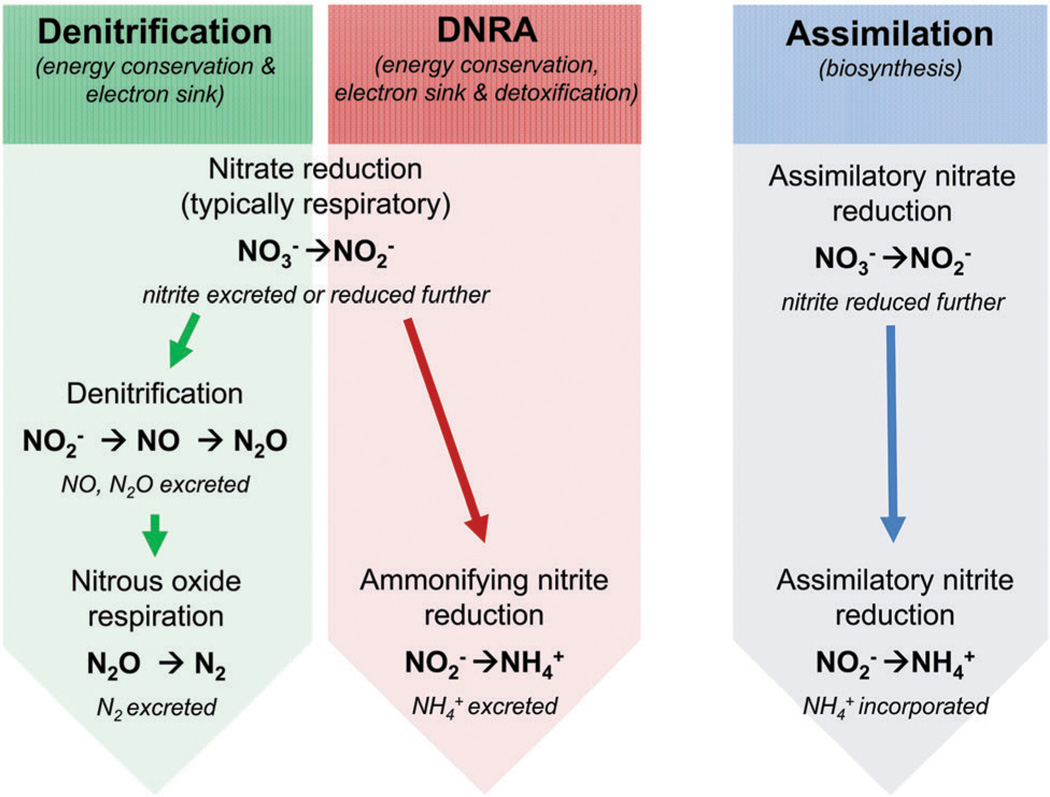
The assimilatory and dissimilatory nitrate reductase enzymes were designated based on the ultimate fate of the nitrogen. Assimilatory nitrate reduction, a form of anabolism, incorporates nitrogen from nitrate into the organism’s biomass. Nitrate is first reduced by nitrate reductases to nitrite, and then nitrite is transformed into ammonia via assimilatory nitrite reductases. Dissimilatory nitrate reduction, a catabolic pathway, does not incorporate nitrogen into the biomass; instead the end products of dissimilatory nitrate reduction are excreted from the cell. Although dissimilatory NR also forms nitrite from nitrate, nitrite can be furthermetabolized into dinitrogen or ammonia as part of denitrification or dissimilatory nitrate reduction to ammonia (DNRA), respectively (Fig. 2). DNRA is an important part of the global nitrogen cycle because ammonia can be stored in the soil. Denitrification generates volatile compounds, which accumulate in the atmosphere. In some cases, dissimilatory nitrate reduction is an energy yielding process; therefore, the term “nitrate respiration” is used to specify nitrate reduction coupled to ATP generation.
Fig. 2.
Schematic representation of dissimilatory and assimilatory nitrate reduction.
The nitrate anion is composed of two elements, nitrogen and oxygen, which can exist in more than one stable isotope (15N and 14N; 18O and 16O). The isotopic ratios of nitrogen and oxygen have been used to assess biological roles in nitrate cycling such as in denitrification and nitrate assimilation.85 Nitrate composed of light isotopes (14N16O3 −) is consumed more rapidly than the nitrate composed of heavy isotopes (15N18O3−).86 Thus, generation of volatile nitrogen compounds, such as N2, via denitrification leaves a pool of the heavy isotope-enriched nitrate for nitrate assimilation. As a result of the disproportionate isotope distribution among the atmosphere and lithosphere, dissimilatory nitrate reduction rates would be slower than rates of nitrate assimilation, hence an observable kinetic isotope effect.86 The periplasmic nitrate reductase (Nap) fractionates with an O-to-N ratio of ~0.6, and is considered to have a limited effect on the overall pool of nitrate reduction in the environment. It was suggested that the O-to-N ratio for respiratory nitrate reductase (Nar) should be similar to that of eukaryotic nitrate reductase (eukNR), both of which play a major role in biogenic nitrate reduction.86
In a recent study, Li et al. have shown that biogenic magnetite formation by Magnetospirillum gryphiswaldense requires periplasmic nitrate reductase (Nap).87 Nap deletion mutants of M. gryphiswaldense exhibit attenuated growth and production of magnetite crystals. Biogenic membrane encapsulated magnetite crystals are favored by low cellular redox potential, although how the cellular redox impacts the formation of the mixed valence iron-oxide material is not clear. It is now believed that the oxygen in the iron-oxide comes from water not molecular oxygen. It is possible that dissimilatory nitrate reduction dominates under microaerobic conditions and is perhaps important in redox balance.
2.5 Cellular locations and functional diversity
The cellular location of nitrate reductases differs in a manner that bears functional significance. The assimilatory NRs (both eukNR and Nas) are soluble, cytoplasmic proteins, while the dissimilatory NRs (i.e., Nap and Nar) are membrane associated. The Nap complex is associated with the periplasmic side of the cell membrane. The Nar complex faces the cytoplasmin bacteria, and the periplasm in Archaea (pNar).88–92 The cytoplasmic location of the assimilatory NR is appropriate, as biosynthesis occurs in the cytoplasm. Conversely, the dissimilatory NRs are associated with the cell membrane, and are typically involved in energy acquisition, detoxification, and redox regulation. Nar, the classical respiratory NR, couples nitrate respiration to proton translocation across the cytoplasmic membrane of bacteria. The resulting electrochemical gradient can then be exploited to drive ATP generation via ATP synthase. In general, the periplasmic-oriented complexes i.e., Nap and pNar are functionally diverse. Unaccompanied Nap and pNar are not capable of producing a proton motive force across the inner membrane; however nitrate reduction when coupled to formate oxidation does indeed generate a proton gradient across the cell membrane (see below).93
The periplasmic nitrate reductase (Nap) was first purified from Paracoccus denitrificans in 1993.94 Initially, it was differentiated from the well-known respiratory nitrate reductases (Nar) not just by its cellular location but also by its insensitivity to azide and its inability to reduce chlorate. Since the first report of Nap several functionally diverse Nap enzymes have been isolated from a variety of prokaryotes (see Section 3.6.1, Table 7).
Table 7.
The nap operon gene content and organization in bacteria
| Quinone oxidase | nap Operon | Phylogenic class | Organism |
|---|---|---|---|
| H | AGHBF_LD | ε | Sulfurovum sp. Autotrophic |
| AGHBFLD | Aquificae | Hydrogenivirga sp. 128-5-R1-1 | |
| Persephonella marina EX-H1 | |||
| Sulfurihydrogenibium sp. YO3AOP1 | |||
| Deinococci | Oceanithermus profundus DSM 14977 | ||
| ε | Arcobacter (butzleri RM4018 and Nitrofigilis) Campylobacter species | ||
| (concisis, curvis, hominis, fetus) | |||
| Helicobacter mustelae 1219 | |||
| Nautilla profundicola | |||
| Sulfuricurvum kujiensedum | |||
| Sulfurospirillum (barnesii and deleyianum) | |||
| Wolinella succinogenes | |||
| AGHBFLDS | ε | Nitratiruptor sp. SB155-2 | |
| Caminibacter mediatlanticus | |||
| AGHBFLSD | ε | Thiomicrospira (formerly Sulfurimonas) denitrificans DSM 125 | |
| AGHBLD | ε | Campylobacter (lari, coli, jejuni (8 strains), upsalensis) Helicobacter hepaticus | |
| DAGHB | β. | Burkholderiales bacterium 1_1_47 | |
| γ | Shewanella amazonensis SB2B*, halifaxensis HAW-EB4, loihica PV-4*, oneidensis MR-1, | ||
| pealeana ATCC 700345*, sediminis HAW-EB3*, sp. ANA-3*, MR-4*, MR-7, W3-18-1* | |||
| DAGHB_F< | β | Candidatus Accumulibacter phophatis | |
| DAGHF | δ | Geobacter lovleyi | |
| DGBAH:G | Clostridia | Desulfitobacterium hafniense strains (2 DCB-2 & 2 DCB-1) | |
| FDAGHB | α | Magnetospirillum magnetotacticum AMB-1 | |
| GBAH:G | Clostridia | Desulfitobacterium hafniense Y51 | |
| MADGH | Chrysiogenetes | Desulfurispirillum indicum | |
| Deferribacteres | Calditerrivibrio nitroreducens | ||
| Deferribacter desulfuricans | |||
| Denitrovibrio acetiphilus | |||
| C | ABC | γ | Photobacterium (damselae CIP 102761 and profundum SS9*) |
| AC | Sphingobacteria | Rhodothermus marinus | |
| CDAG | Clostridia | Symbiobacterium thermophilum | |
| DABC | β | Dechloromonas aromatica RCB | |
| γ | Hahella chejuensis KCTC 2396 | ||
| Photobacterium sp. SKA34* | |||
| Shewanella (piezotolerans WP3*, sediminis HAW-EB3*, violacea DSS12) | |||
| Vibrio vulnificus CMCP6 | |||
| DABCF | Flavobacteria | Maribacter Sp. HTCC2170 | |
| EABC | γ | Photobacterium profundum 3TCK* | |
| EDABC | α | Afipia sp. 1NLS | |
| Azospirillum sp. B510 | |||
| Bradyrhizobium japonicum USDA 110 | |||
| Bradyrhizobium sp. BTAi1 | |||
| Bradyrhizobium sp. ORS278 | |||
| Methylobacterium sp. 4-46 | |||
| Paracoccus (denitrificans PD1222 and pantatrophus) Rhodopseudomonas palustris BisB18 | |||
| β | Achromobacter xylosoxidans A8 | ||
| Azoarcus sp. BH72* | |||
| Burkholderia xenovorans LB400 | |||
| Comamonas testosteroni CNB-2 | |||
| Cupriavidus metallidurans CH34, taiwanensis LMG 19424 and necator | |||
| (formerly Ralstonia eutropha H1 6) | |||
| Ralstonia (eutropha JMP134 and pickettii strains 12D & 12J) | |||
| γ | Marine γ proteobacterium HTCC2148 | ||
| Pseudomonas (mendocina ymp, and stutzeri A1501) | |||
| Saccharophagus degradans 2-40 | |||
| Shewanella amazonensis SB2B*, baltica OS155, baltica OS185*, baltica OS195*, | |||
| baltica OS223, denitrificans OS217, frigidimarina NCIMB 40, loihica PV-4*, | |||
| pealeana ATCC 700345*, ANA-3*, MR-4*, W3-18-1*, woodyi ATCC 51908* | |||
| EFDABC | α | Agrobacterium tumefaciens str. C58 | |
| Pseudomonas sp. G-179 | |||
| Rhizobium leguminosarum bv. trifolii WSM2304 | |||
| Rhodobacterales bacterium Y4I | |||
| Rhodospirillum centenum SW | |||
| Sinorhizobium (fredii NGR234, medicae WSM419, meliloti 1021) | |||
| Starkeya novella | |||
| γ | Colwellia psychrerythraea 34H | ||
| Pseudomonas aeruginosa (strains LESB58, UCBPP-PA14, PAO1 & PA7) | |||
| Reinekea sp. MED297 | |||
| FDABC | γ | Pantoea sp. At-9b, Photobacterium (angustum S14, profundum strains (3TCK* & SS9*)) | |
| Psychromonas ingrahamii 37 | |||
| Serratia odorifera 4Rx13, proteamaculans 568, Vibrio (cholerae str. N16961, | |||
| harveyi ATCC BAA-1116,fischeri strains (ES114, MJ11), parahemeolyticus | |||
| RIMD 2210633, vulnificus YJ016) | |||
| Yersinia (pestis (5 strains), enterocolitica) | |||
| FDABC_ABC | γ | Moritella sp. PE36 | |
| KEFDABC | α | Rhodobacter sphaeroides strains (2.4.1 & ATCC 17025*) | |
| HC | CMADGH | Aquificae | Thermovibrio ammonificans |
| δ | Desulfovibrio desulfuricans strain 27774 | ||
| Desulfovibrio sp. 3 1 syn3 | |||
| DAGHBC | β | Leptothrix cholodnii | |
| γ | Aggregatibacter aphrophilus | ||
| Citrobacter (koseri and rodentium) | |||
| Escherichia coli IAI3 | |||
| Salmonella typhimurium | |||
| Shigella flexneri (2 strains) | |||
| FDABGHBC | α | Rhodobacter sphaeroides ATCC 17025 | |
| FDAGHBC | α | Dinoroseobacter shibae DFL 12 | |
| Laribacter hongkongensis | |||
| β | Azoarcus sp. BH72* | ||
| Thauera sp. MZ1T | |||
| γ | Aeromonas hydrophila | ||
| Edwardsiella (ictaluri, tarda) | |||
| Escherichia coli (16 strains), Grimontia hollisae CIP 101886 | |||
| Haemophilus influenza | |||
| Pectobacterium (atrosepticum, carotovorum (2 strains), wasabiae) | |||
| Providencia (alcalifaciens, rettgeri, rustigiianii, stuartii) | |||
| Salmonella enterica | |||
| Serratia odorifera DSM 4582 | |||
| Shigella (boydii (2 strains) and dysenteriae) | |||
| NA | A | γ | Photobacterium sp. SKA34* |
| ε | Helicobacter felis | ||
| AB | δ | Sorangium cellulosum | |
| DAB | γ | Shewanella (baltica OS185*, baltica OS195*, Piezotolerans WP3*, woodyi ATCC 51908*) | |
| MA | δ | Anaeromyxobacter sp. K and dehalogenans strains (2CP-1 & 2CP-C) |
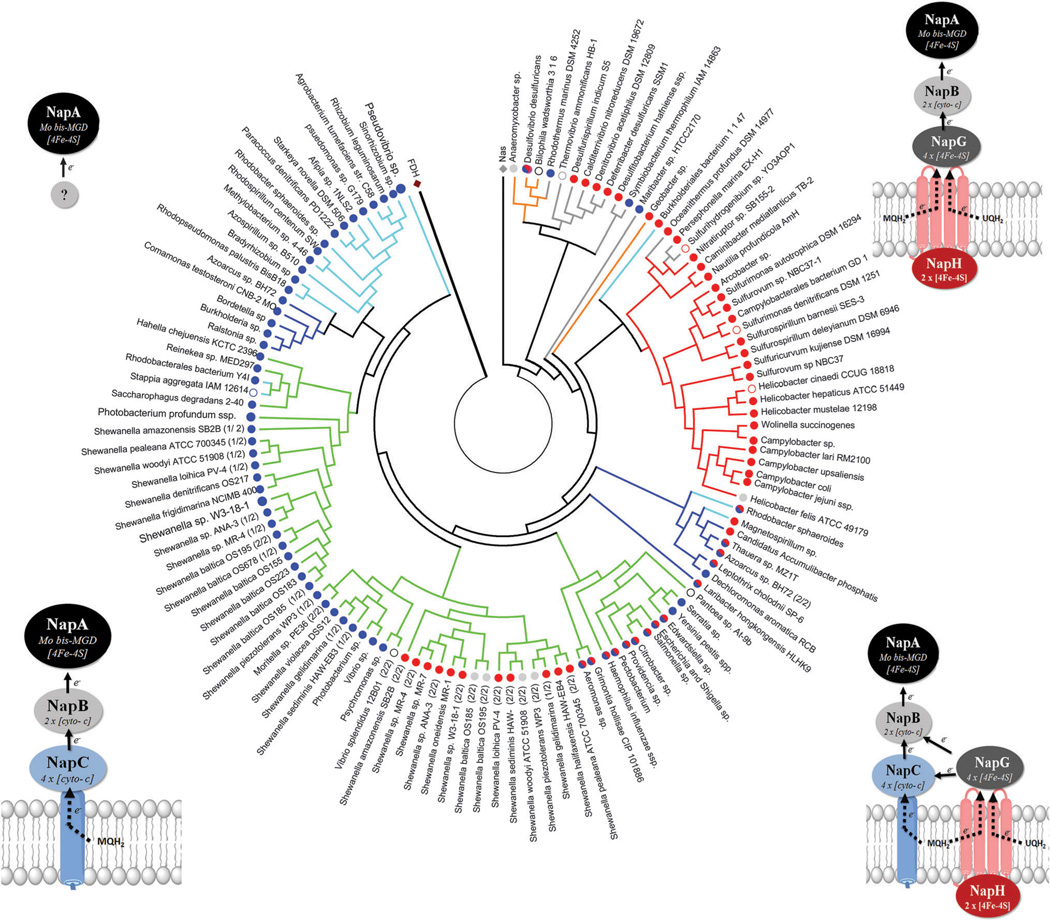
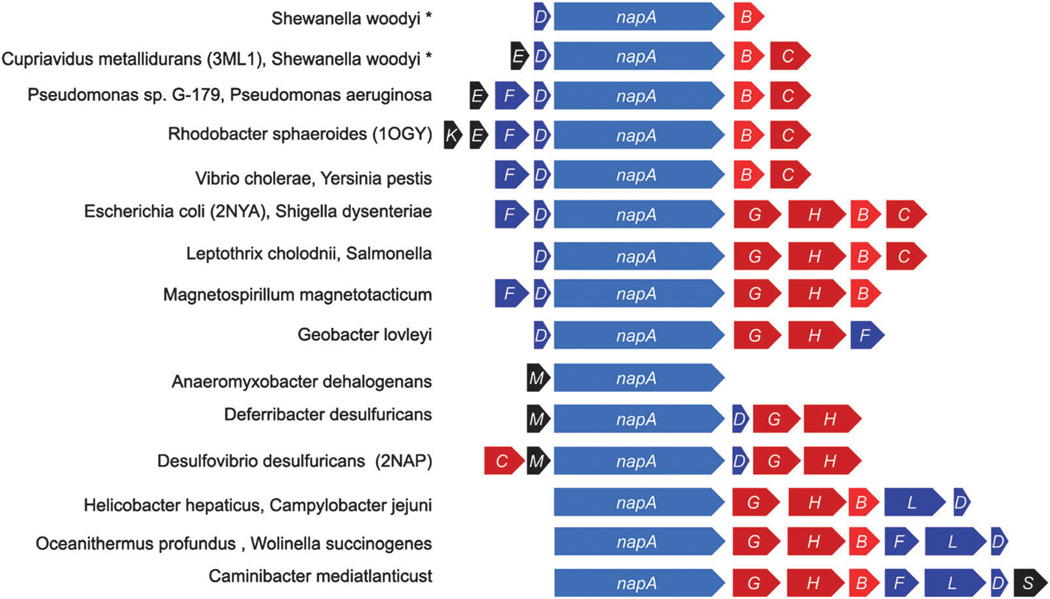
The nap operons categorized into four groups based on the presence or absence of a membrane quinone oxidase (napC or napH). The gene order in each organisms nap operon was provided by the NCBI gene and phenotype mapping database (http://www.ncbi.nlm.nih.gov/gene), and in a few cases the JGI database (http://img.jgi.doe.gov). The identity of each translated Nap protein was confirmed by performing NCBI database blast searches. In each operon the nap genes are listed by the order in which they are found. Organisms that encode two separate nap operons are shown with asterisks (*); fused operons with a colon (:), underscore indicates a gap, and gene encoded on the complementary strand is indicated with an arrow (<). Proteobacteria are denoted as α, β, γ δ, ε.
In some species, periplasmic NRs have been implicated in nitrate respiration; however the validity of this is under debate. The Epsilonproteobacteria only contain Nap yet are capable of anaerobic nitrate respiration.93 For example, Sulfurospirillum barnesii has been shown to couple lactate oxidation to dissimilatory nitrate reduction to ammonia.95 Nevertheless, respiration via the periplasmic nitrate reductase is predicted to generate less ATP than Nar. As both the electron donor complex (e.g., formate dehydrogenase) and the acceptor complex (e.g., Nap) are oriented into the periplasm, the net change in number of protons within the periplasmic space should be zero and thus no proton gradient should be formed across the membrane.96 In reality, when nitrate reduction is coupled to formate oxidation, a proton gradient is indeed generated across the cell membrane.96 Although the gradient may be less than that generated by the cytoplasmic-facing Nar complex, the loss of potential energy may be compensated by the higher affinity for nitrate typically found in periplasmic nitrate reductases.97 Furthermore, the variety of additional subunits associated with the different Nap complexes may further increase this influence.
Unlike other nitrate reductases that have conserved functionality from species to species, Nap is functionally diverse. Nap has been implicated in dissimilatory nitrate reduction, both denitrification and nitrate reduction to ammonia,98,99 maintenance of cellular oxidation–reduction potential i.e., redox poise,100 as well as nitrate scavenging.101,102 Similar to Nar, Nap has the ability to function in nitrate respiration.103 In some organisms, nitrate respiration is dependent on the presence of the nap operon104 e.g., Epsilonproteobacteria.93,105 Nap proteins have also been associated with pathogenicity in Helicobacter and Campylobacter, where Nap genes are up-regulated in response to oxidative stress.73,106–110 The functional diversity of Nap demonstrates that the presence of a napA homolog in an organism’s genome does not necessarily confer functionality. The potential sources of the functional diversity in Nap, and their molecular properties are discussed in subsequent sections.
2.6 Structural aspects of nitrate reductases
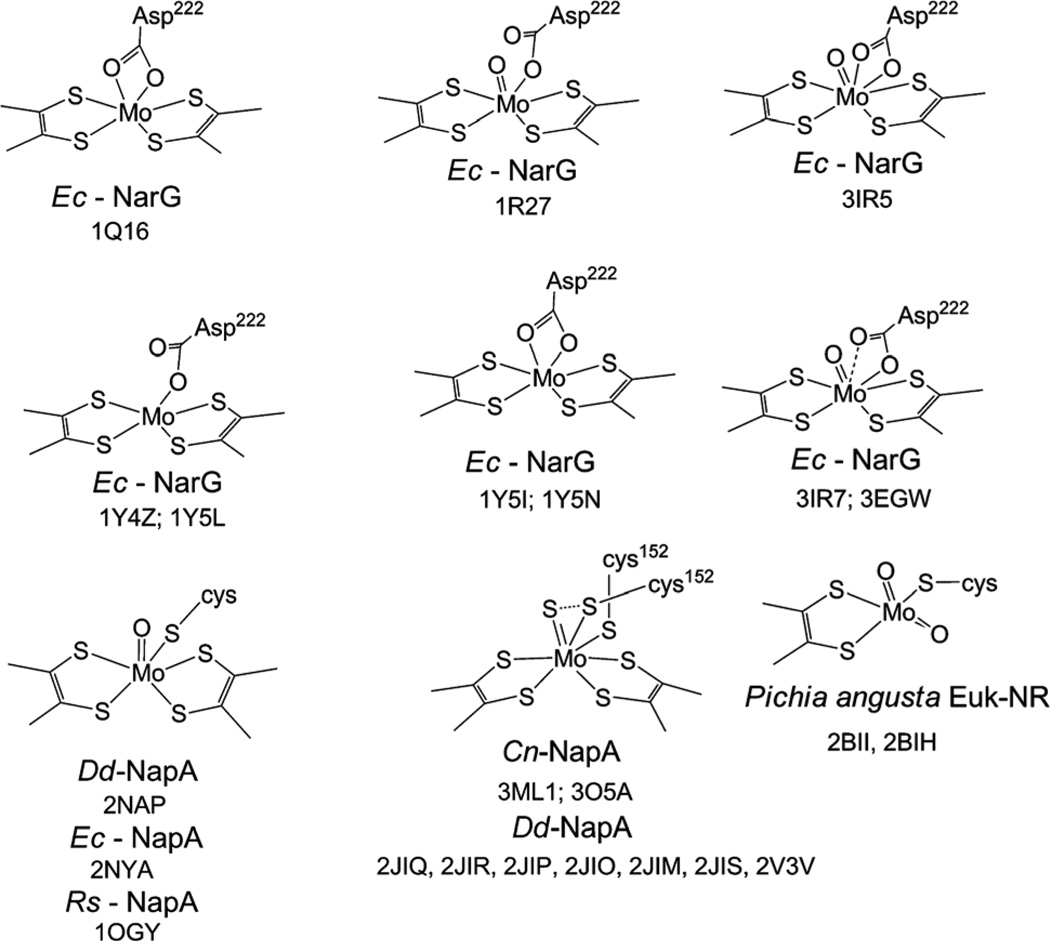
Pterin dependent molybdenum enzymes contain a dithiolene-modified pterin that binds to a molybdenum atom constituting the active site. The basic pterin molecule, (i.e., pyranopterin),111,112 serves as a scaffold, holding molybdenum in the active site. The majority of these enzymes have been classified into three families, dimethyl sulfoxide reductase (DMSOR), sulfite oxidase (SO), and xanthine oxidase (XO).113 Of these, the DMSOR enzymes contain two pyranopterin moieties, while members of the SO and XO families contain a single pterin moiety. Dissimilatory NRs, Nar and Nap, as well as prokaryotic assimilatory nitrate reductase (Nas) belong to the DMSOR family. In the case of Nap and Nar the presence of two pterin cofactors has been unequivocally established, however, no such information for Nas is yet available. Eukaryotic assimilatory NR (eukNR), contains one pyranopterin moiety and is classified as a member of the SO family. In addition to the dithiolene from the pterin, molybdenum is coordinated by additional ligands. A cysteine residue coordinates to molybdenum in eukNR, as it does in Nap and presumably in Nas.114,115 In Nar, an asparate residue coordinates to the molybdenum center.116,117 The eukNR is further distinguished from the other nitrate reductases as it has a dioxo Mo-center. The Mo coordination sites, as determined by crystallography, are shown in Fig. 3. It is important to note that crystallography alone cannot unambiguously define the nature of the coordination spheres, especially, as some of the structures are solved at lower resolution and there is evidence of radiation damage during crystallography.118 A complete picture must therefore include spectroscopic information that is calibrated with well-defined discrete inorganic compounds. However, in the absence of a more robust data set, here we have used crystallographic information as a starting point of discussion.
Fig. 3.
Coordination about the molybdenumcenter of crystallographically characterized nitrate reductases with PDB codes. Ec: Escherichia coli, Dd: Desulfovibrio desulfuricans; Rs: Rhodobacter sphaeroides; Cn: Cupriavidus necator. Conformationally flexible Cys 152 in Cn-NapA structure is shown in two positions.
The catalytic subunit of periplasmic nitrate reductase from Desulfovibrio desulfuricans, NapA, was the first nitrate reductase to be structurally characterized.119 Initially the Mo-center was proposed to contain a terminal oxo-group. Later redetermination of the structure revealed a terminal sulfido group instead of an oxo-group.120 The sulfide group showed S···S interaction with the coordinating S from a disordered cysteine. A similar structure was determined from the same group from Cupriavidus necator.121
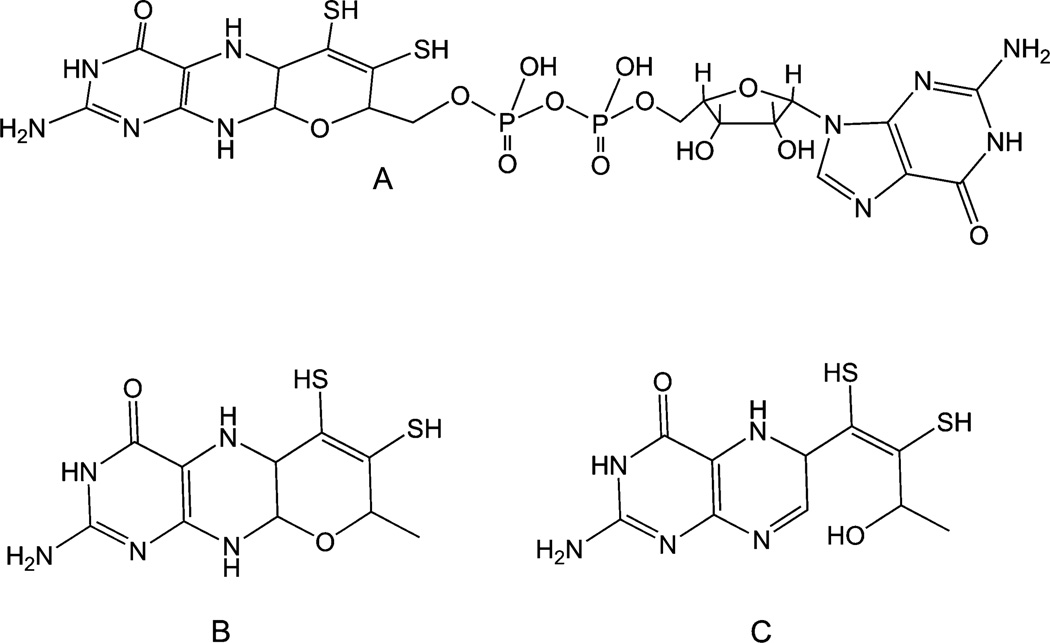
The pterin cofactor of prokaryotic NR is modified with a guanine dinucleotide (Fig. 4). This molecule has been referred to as molybdopterin guanidine dinucleotide (MGD) or pyranopterin guanidine dinucleotide (PGD).112 Themolybdenum center is deeply buried inside the protein milieu. The two pyranopterins, are designated as distal or proximal to the iron–sulfur cluster. It is believed that the proximal pyranopterin participates in the intermolecular electron transfer, from electron donors in the cell to the molybdenum active site. In all NapA and eukNR structures determined to date, the pyran ring of the pyranopterin cofactor is in the closed form; however, the distal pyranopterin ring is open in E. coli Nar (Fig. 4). A similar open pyran ring has also been observed in the crystal structure of ethylbenzene reductase isolated from Aromatoleum aromaticum.122 It has been suggested that the opening and closing of the pyran ring may serve as a means to shuttle proton during the catalytic turnover.123 There are only a handful of synthetic pyran dithiolene molecules available,111,124–128 therefore the fundamental properties are not well defined. However, under oxidative conditions, pyran ring scission has been observed in a pyran dithiolene molecule.129 Whether or not the ring scission and closure has a functional significance remains to be established, but the presence of the ring open structure in two structures raises a question as to what controls the ring-open structure i.e., is it naturally there or an artifact. Future studies should elaborate this aspect more.
Fig. 4.
(A) The pyranopterin cofactor attached to the dinucleotide as found in bacterial nitrate reductases; (B) the basic pyranopterin cofactor found in all nitrate reductases; (C) the ring open form of the cofactor found in the structures of respiratory nitrate reductase, Nar.
The saturated nature of the pyrazine ring in pyranopterin cofactor is reminiscent of the structure of a tetrahydropterin, and in some renditions it has been thought to be in fully reduced state. However, a simple scission of the pyran ring leads to a 5,6-dihydropterin, which then can tautomerize to other dihydro forms, or can be oxidized to a fully oxidized pterin or can be reduced to tetrahydropterin. Thus, the pyranopterin cofactor should be taken as a ‘dihydropterin’. The two electron oxidation of any of the isomeric forms would lead to the fully oxidized ring open structure.112 A detailed discussion can be found in a recent review article.111
The pterin cofactor is expected to adopt different geometries depending on the redox state, and these differences are manifested in their torsion angles. An analysis of the <OCCS and <OCCC torsion angles determined by crystallography of all pterincontaining molybdenum enzymes reveals a linear trend between the two angles. For most structures, the angles linearly correlate with the exception of a few DMSOR and formate dehydrogenase (FDH) structures that may reflect the quality of the structure rather than an intrinsic property of the cofactor. In structures where the pyran ring is open, the observed values of <OCCS angle are lower while <OCCC angles are of higher value, even though they fall on the same line. When looked at closely, the structures from NR (excluding the ring open cofactor) and structures of enzymes belonging to the XO and SO families, it appears that the angles from the DMSOR family cluster at a different position than those from SO, and members of the XO family (Fig. 5). Most representatives of the SO family analyzed here were from mammalian sources, while the NR representatives were from microbial sources. The XO enzymes however, exhibit more heterogeneity regardless of their source – eukaryotic or prokaryotic. The different geometric features are related to different redox states of the pterin, which suggests that redox states of the pterin are fundamentally different in NR and SO. If so, the impact of their redox state on function remains to be established. Based on a detailed analysis of the conformation of the pterin cofactor as determined from crystallography, Rothery et al.130 suggested the redox state of the pterin in different enzymes are as follows: sulfite oxidase in dihydro, xanthine oxidase in tetrahydro, the two pterins in DMSO reductases are in tetrahydro and dihydro forms. Implication of this finding is that the redox state of the pterin allows tuning of the redox state of the metal center and vectorial electron transfer in the case of NapA, which has two pyranopterin cofactors coordinating the Mo-center.
Fig. 5.
Plot of two torsion angles (highlighted in bold lines) in molybdenum cofactors in SO, XO and NR. The linear line is represented by the equation [<OCCC] = 163.9(±7.4) – 0.95(±0.04) × [<OCCS] (N = 49; R2 = 95; P < 0.0001). Note that values in NR exhibit a larger spread, those of XO and SO are tightly clustered, with the exception of four points, which are representing substrate or inhibitor bound form or eukNR.
The pyran ring can not only protect a reduced state of the pterin cofactor but also influence the orientation of the cofactor in the protein milieu, possibly modulating electron transfer. The efficient electron transfer is reminiscent of the ‘entatic state’ described for copper centers in biology.131–138 Whether this is indeed the case in molybdopterin enzymes, needs to be tested experimentally.
In crystallographically characterized pterin-containing molybdenum enzymes, the Mo-center is found to be nearly 15 Å inside the protein and Nap is no exception. This topological feature may be a protective mechanism from dimerization of the molybdenum center, a pervasive reaction in synthetic oxo-transferring molybdenum complexes.139 Model studies have shown that not only does this topological feature protect the Mo-centers from dimerization,140,141 but also changes the electronic structure of the center via modulation of metal–sulfur covalency. This alteration in covalency along with a change in the dielectric properties alters the reduction potential of the molybdenum center, thus providing the driving force of the reaction.142,143
The redox potential of Mo centers in these enzymes is dependent on the electronic structure of the molybdenum center. The terminal oxo-group in oxo-molybdenum complexes dominate the ligand field making the dxy orbital energetically isolated and the lowest in energy among the 4d orbitals. In the desoxo-Mo complexes, the dominating effect of the terminal oxo group is absent,144–148 and the dithiolene ligand effectively dictates the Mo ligand field, again resulting in an isolated dxy orbital in the ground state.149 In the Mo(iv) state the dxy orbital is fully occupied and is empty in the Mo(vi) state. The energy of this orbital can be altered through small perturbations in the geometry around the molybdenum center (e.g., changing the torsion angle of the coordinated amino acid, changing the fold angle of dithiolene ligand) and solvation of the center preferentially stabilizes the higher charges.150–153 Both factors, the electronic structure dictated by the geometric structure and charge stabilization through solvation, are important in the defining the redox potentials in these enzymes. The exact contribution from each component is difficult to assess a priori, and should be taken case by case.
2.7 Substrate level redox chemistry
The reduction of nitrate to nitrite occurs at the molybdenum site of NRs, and it requires two electrons and is coupled with the transfer of two protons. Such reactions are a hallmark of pterin-containing molybdenum enzymes, which catalyze substrate transformations spanning an astonishing 1.8 V range (Table 3). Of the substrates, the redox potential of the nitrate–nitrite couple is highly positive (+433 mV) justifying the ease of its reduction. The oxygen atom in nitrate to nitrite transformation forms a water molecule coupling with the proton mentioned above. In oxidases, such as SO a water molecule serves as a source of the oxygen atom. Because only one oxygen atom is transferred to or from the substrate, these enzymes can be viewed as a monooxygenase. The net transfer of the oxygen atom, a two-electron transfer process, can occur either via a hydroxylation or by a formal atom transfer processes. It is believed that the DMSO reductase and SO family of enzymes function via atom transfer process,154 while XO members function via hydroxylation.155–157 Direct atom transfer processes have been demonstrated with model complexes, and have been summarized elsewhere.126,139,158–163 Such reactions may proceed via multiple transition states involving an associative transition state and formation of a product bound intermediate.164–172 In enzymatic systems, complications with the labeling studies have hindered definitive demonstrations of direct oxygen atom transfer (OAT) reactions in NR.154 The involvement of a proton in the reaction has also been demonstrated with model compounds.173–177 During the two-electron substrate transformation, the metal center changes its redox state by two units, and the catalytically competent center is regenerated in two one-electron steps. In most cases, including nitrate reductase, the intermediate +5 oxidation state of molybdenum has been characterized by electron paramagnetic (EPR) spectroscopy. During the electron regeneration process, the Mo center is reduced via physiological partners such as quinones, or hemes. The redox potentials of representative redox partners are listed in Table 4.
Table 6.
EPR parameters of nitrate reductases
| Enzyme | Signal | Spin (%) | g1 | g2 | g3 | gav | A/MHz | Anisotropy, g1 –g3 |
Rhombieity, (g1 – g2)/(g1 – g3) |
Ref. |
|---|---|---|---|---|---|---|---|---|---|---|
| Dd-NapA | Resting | <5 | 120, 246 | |||||||
| Low-potential | 5–15 | 2.016 | 1.987 | 1.964 | 1.989 | 0.052 | 0.56 | |||
| High potential | NR | 2.019 | 1.988 | 1.960 | 1.989 | 0.059 | 0.53 | |||
| High-g nitrate | ≤40 | 2.000 | 1.990 | 1.981 | 1.990 | 12.9, 14.0, 12.9 | 0.019 | 0.56 | ||
| Turnover | 50 | 1.999 | 1.990 | 1.982 | 1.990 | 14.0, 16.8, 14.0, 7.8 (H) | 0.017 | 0.53 | ||
| 1.999 | 1.992 | 1.982 | 1.991 | 16.3, 18.2, 15.4 | 0.017 | 0.41 | ||||
| Cyanide | 3 | 2.024 | 2.001 | 1.995 | 2.007 | 22.4, 21.0, 16.8 | 0.029 | 0.79 | ||
| Pp-NapAB | High g-split/high g-resting | 2.5–10 | 1.999 | 1.991 | 1.981 | 1.990 | 17.9, 14.6, 14.0, 7.8 (H) | 0.018 | 0.46 | 212, 214 |
| High-g (NO3−) | NR | 1.999 | 1.982 | 1.990 | 1.990 | 17.94, 12.05, 12.89, 14.29 | 0.017 | 0.59 | ||
| Low-g split | 10–20 | 1.996 | 1.969 | 1.961 | 1.975 | 36.4, 38.1, 42.8 | 0.035 | 0.77 | ||
| Low g-unsplit | NR | 1.997 | 1.962 | 1.959 | 1.973 | 0.037 | 0.93 | |||
| Very high-g | 40 | 2.022 | 1.999 | 1.994 | 2.005 | 20.74, 20.74, 18.5, 19.90 (H) | 0.029 | 0.80 | ||
| High-g (N3−) | NR | 2.007 | 1.994 | 1.988 | 1.996 | 19.06, 17.94, 15.97, 17.66 (H) | 0.023 | 0.74 | ||
| High-g (SCN−) | 10 | 2.008 | 1.991 | 1.985 | 1.994 | 18.78, 18.50, 15.41, 17.66 (H) | 0.023 | 0.74 | 206 | |
| Ec NapAB | High-g split | 70–90 | 1.997 | 1.988 | 1.981 | 1.989 | 16.8, 13.4, 13.4 | 0.016 | 0.55 | 96 |
| Rs NapAB | Very High-g | 2.023 | 2.000 | 1.994 | 2.006 | 21.8, 19.9, 16.8 | 0.029 | 0.79 | 186, 205, 248 | |
| High-g split | 10–20 | 1.999 | 1.991 | 1.981 | 1.990 | 17.1, 15.7, 14.0, 8.1, | 0.018 | 0.45 | 186, 213, 247 | |
| Rs NapA | High-g split | 60 | 1.998 | 1.991 | 1.981 | 1.990 | 17.6, 15.1, 14.0 | 0.018 | 0.44 | |
| Sg NapAB | High-g | 1.999 | 1.990 | 1.981 | 1.990 | 17.9, 14.6, 15.4 | 0.018 | 0.50 | 254 |
Table 4.
Redox potentials of partner reactions
| Enzymes | Redox partner reactions | Potentials (mV) |
Ref. | |
|---|---|---|---|---|
| Plant SO |
|
+815 | 335 | |
| XO | O2 + e− → O2− | −330 | 336 | |
| XO | O2 + H+ + e− → HO2 | −80 | 336 | |
| O2 + 2H+ + 2e− → H2O2 | +295 | 336 | ||
| FAD + 2H+ + 2e− → FADH2 | 0 | 335 | ||
| SO | Cytochrome b5: Fe3+ + e− → Fe2+ | 77 | 335 | |
| NR | Fdox + e− → Fdred | −350 to −500 | 335 | |
| Cytochrome c: Fe3+ + e− → Fe2+ | 235 | 335 | ||
| NAD+ + H+ + 2e− → NADH | −320 | 335 | ||
| Ubiquinone: U + 2H+ + 2e− → UH2 | 45 | 335 | ||
| 90 | 262 | |||
| Menaquinone: M + 2H+ + 2e− → MH2 | −70 | 262 | ||
| Aio | Rieske: [(Cys)2Fe2S2(His)2] + e− → [(Cys)2Fe2S2(His)2]− | −140 to +350 | 337 |
It is interesting to note that while the redox potentials of the substrate half reactions (Table 3) can vary significantly, the redox potential of the Mo-centers do not change as much.113 The redox flexibility of molybdenum enzymes involves the redox active pyranopterin i.e., pterin as well as the dithiolene, not just the metal center. The cofactor itself can be viewed as a combination of three distinct redox active components – the Mo center, the dithiolene ligand and the pterin unit. The Mocenter shuttles between +4 and +6 oxidation states, thus can carry out two-electron chemistry. The dithiolene ligand can also carry out two-electron chemistry from fully reduced dithiolene to fully oxidized dithione, and these redox states have been experimentally demonstrated.178–183 A pterin unit in dihydro state can carry out two electron chemistry,112,184 and pterin based radical has been detected in AO.185 Thus, in principle, molybdenum centers coordinated by two pyranopterins can provide up to ten-electron chemistry, making it one of the most electron rich redox active centers in biology. Studying the distribution of redox equivalents through the entire pyranopterin cofactor within the individual molecular components of the enzyme remains an experimental challenge. Representative redox isomers are shown in Fig. 6.
Table 3.
Reactions catalyzed by the pterin-containing molybdenum enzymes. Potentials are with respect to standard hydrogen electrode at pH ~ 7 and 25 °C
| Enzymes | Redox reactions | Potential (mV) | Ref. |
|---|---|---|---|
| Perchlorate reductase | ClO4− + 2H+ + 2e− → ClO3− + H2O | +1200 | 327 |
| +788 | 328 | ||
| Chlorate reductase | ClO3− + 2H+ + 2e− → ClO2− + H2O | +709 | 328 |
| Selenate reductase | SeO42− + 2H+ + 2e− → SeO32− + H2O | +895 | 329 |
| +475 | |||
| Aldehyde oxidase | CH3COO− + 3H+ + 2e− → CH3CHO + H2O | +580 | 330 |
| Arsenate reductase | H3AsO4 + 2H+ + 2e− → H3AsO3 + H2O | +560 | 331 |
| Nitrate reductase | NO3− + 2H+ + 2e− → NO2− + H2O | +420 | 332 |
| +433 | 313 | ||
| DMSO reductase | Me2SO + 2H+ + 2e− → Me2S + H2O | +160 | 332 |
| TMAO reductase | Me3NO + 2H+ + 2e− → Me3N + H2O | +130 | 333 |
| Tetrathionite reductase | S4O62− + 2e− → 2S2O32− | +80 | 329 |
| +24 | 313 | ||
| CO dehydrogenase | CO2 + 2H+ + 2e− → CO + H2O | −106 | 334 |
| Xanthine oxidase | uric acid + 2H+ + 2e− → xanthine + H2O | −360 | 332 |
| Sulfite oxidase | SO42− + 2H+ + 2e− - SO32− + H2O | −380 | 332 |
| Formate dehydrogenase | CO2 + H+ + 2e− → HCOO− | −430 | 313, 332 |
| Formylmethanofuran dehydrogenase | CO2 + MFR− + H+ + 2e− → CHO-MFR + H2O | −497 | 330, 332 |
Fig. 6.
Representative redox isomers of MPT. The Moco is viewed as a dihydropterin (see text for discussion).
3 Periplasmic nitrate reductase
3.1 Structure of the catalytic subunit, NapA
The crystal structures of NapA from several species have been reported: Cupriavidus necator (PDB ID: 3ML1),121 E. coli (PDB ID: 2NYA),96 R. sphaeroides (PDB ID: 1OGY),186 and D. desulfuricans (PDB ID: 2NAP).119 NapA contains two redox active centers: a molybdenum active site and an iron sulfur cluster (Fig. 7). As in other pterin-containing molybdenum enzymes, the Mo-active site of NapA is buried ~15 Å below the surface of the protein and accessible to nitrate via a large funnel shaped cavity. In addition to four dithiolene sulfurs, a fifth sulfur ligand derived from a conserved cysteine residue coordinates the Mo in the active site of NapA.96,119,186 In the initial structure of NapA from D. desulfuricans, the sixth ligand was assigned as a terminal oxo, and in subsequent structures of NapAB from E. coli and R. sphaeroides the sixth ligand to the oxidized molybdenum center was viewed as a coordinated water or hydroxo ligand. These descriptions were consistent with the spectroscopically characterized Mo(v) state where it is generally accepted that a water or hydroxyl group coordinates to the Mo-center. However, reexamination of the D. desulfuricans NapA structure with single-wavelength anomalous diffraction experiments revealed that the sixth ligand is a sulfide, rather than an oxo.120 A high resolution (1.5 Å) structures of NapAB complex from Cupriavidus necator in both the oxidized and partially reduced state support the notion that the sixth ligand is a sulfide group and in the reduced state this sulfide group forms a partial bond with the cysteine sulfur that coordinates to the molybdenum center.121
Fig. 7.
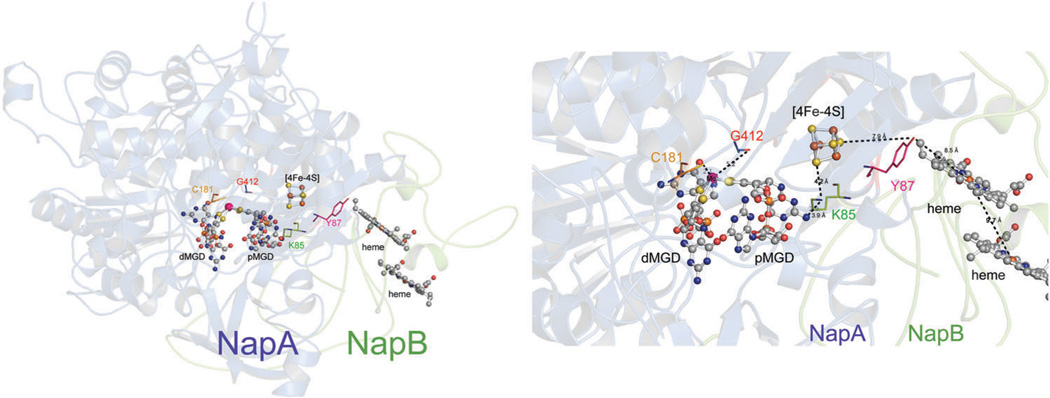
Left: the structure of Rhodobacter sphaeroides NapAB (1OGY) highlighting the metal cofactors along with important amino acid residues. Right: a close up of the metal cofactors and select amino acid residues with distance suggesting a potential electron and or proton transfer path.
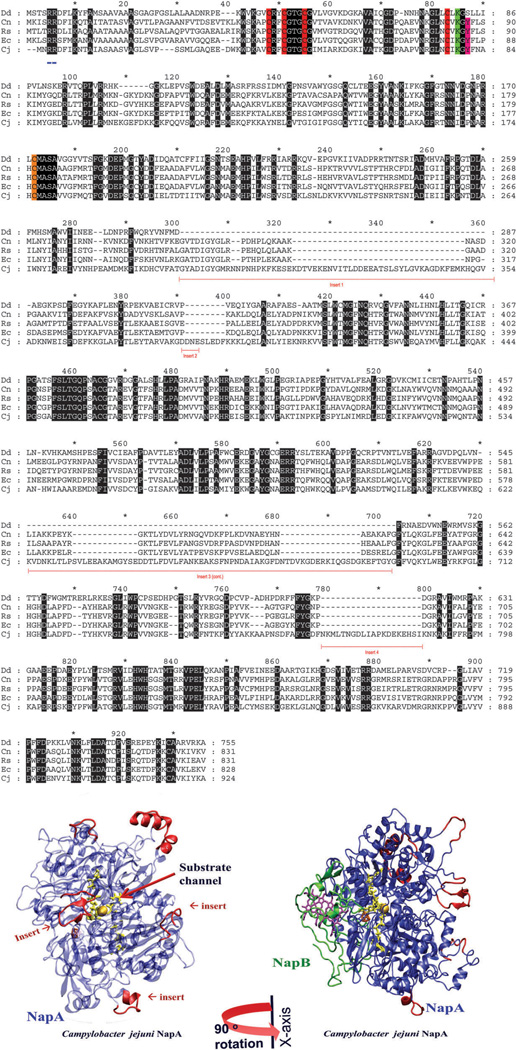
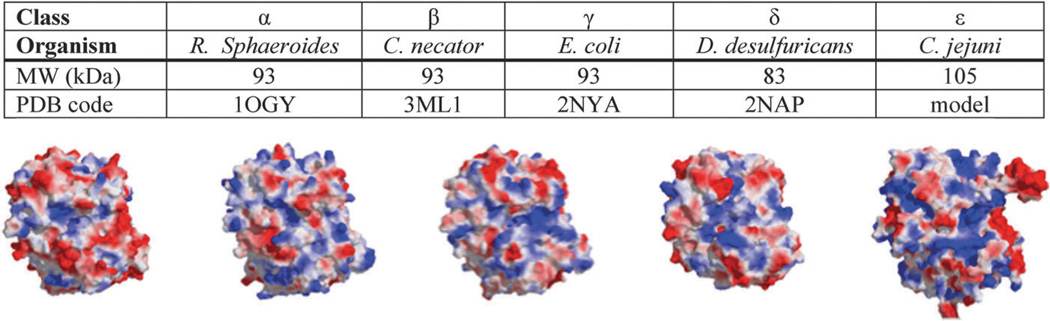
NapA contains an N-terminus twin arginine translocase (TAT) signal sequence (approximately 20 amino acids) and an iron–sulfur ([4Fe–4S]) binding motif (C–X2–C–X3–C–X24–26–C). Additional residues to note include the strictly conserved lysine residue, believed to transport electrons from the [4Fe–4S] cluster to the active site, and the tyrosine-58 in R. sphaeroides participates in electron transfer from NapB to the [4Fe–4S] cluster of NapA, conserved in all heterodimeric NapA, but absent in monomeric NapA such as D. desulfuricans.186 Size is also a prominent difference among NapA enzymes, with some as small as 70 kDa to larger ones of 105 kDa. The additional residues are dispersed throughout the protein rather than extensions at the N- or C-termini. Superimposition of the D. desulfuricans (70 kDa) and R. sphaeroides (90 kDa) NapA structures indicates that the sequence insertions are located on the surface of the protein. This would be expected for molybdopterin proteins that share a high degree of sequence similarity, and the extra residues are distributed over themolecular surface.187 Interestingly, the two larger inserts in the Epsilonproteobacteria NapA expand upon the same polypeptide inserts observed in heterodimeric NapA like R. sphaeroides and E. coli (Fig. 8).96 Jepson et al. described the two inserts as an evolutionary feature, which can be used to distinguish the smaller monomeric NapA (i.e. D. desulfuricans) from larger heterodimer NapA (i.e., R. sphaeroides and E. coli).188 The amino acid composition also varies among species. The isoelectric point of NapA spans from basic (for example, C. jejuni NapA) to acidic (R. sphaeroides NapA) as a result of differences in the amino acid composition (Fig. 9). Whether the differences in the amino acid content and size affect protein function needs to be explored.
Fig. 8.
A sequence alignment, created using GeneDoc, of NapA from D. desulfuricans, Ralstonia eutropha, Rhodobacter sphaeroides, E. coli, and C. jejuni, representing δ, α, β, γ and ε proteobacteria, respectively. Sequence inserts (underlined in red), identical residues (black), [4Fe–4S] binding motif (red), the Mo-coordinated cysteine residue (orange); the lysine (green) and tyrosine (pink) are shown here, the latter two residues are predicted to be involved in electron transfer. The twin arginine translocase (TAT) signal peptide is underlined in blue. Also shown (bottom panel) is the homology models of C. jejuni NapA and NapAB. The insertion sequences, shown in red, are opposite to the NapB binding site. The homology models were created using Molecular Operating Environment (MOE) software heme is purple, [4Fe–4S] is orange, and the MPT is yellow.
Fig. 9.
Properties of NapA from representative organisms from each class of proteobacteria: alpha (α), beta (β), gamma (γ), delta (d), and epsilon (e). One representative organism from each proteobacteria is presented, though multiple organisms were evaluated from each class. C. jejuni NapA structure was generated from the 1OGY structure following standard homology modeling technique using MOE software. Pymol was used in aligning protein structures, calculate and graph electrostatic potential and rendered images.
3.2 Kinetic properties of nitrate reductase
The crystal structure of NapA provided a map of the redox active iron–sulfur and molybdopterin centers, leading to mechanistic proposals. As mentioned, the reduction of nitrate takes place at the molybdenum center. Upon reduction, the molybdenum center gets oxidized and must be reduced to regain catalytic competency. Electrons flow from the partner prosthetic group, i.e., the [4Fe–4S] cluster, to the Mo-center. The Mo atom is 12 Å away from the closest Fe atom of the [4Fe–4S] cluster; a lysine (lys49 in 2NAP) residue is hydrogen bonded to the NH2 group of the pterin cofactor, providing a pathway for electron transfer. Alternatively, electron transfer can follow to the pyrimidine (pterin) nitrogen from the [4Fe–4S] cluster via two water molecules. The oxidized [4Fe–4S] cluster receives electrons from NapB, which in turn receives electrons from the quinone pool completing the electron shuttling.
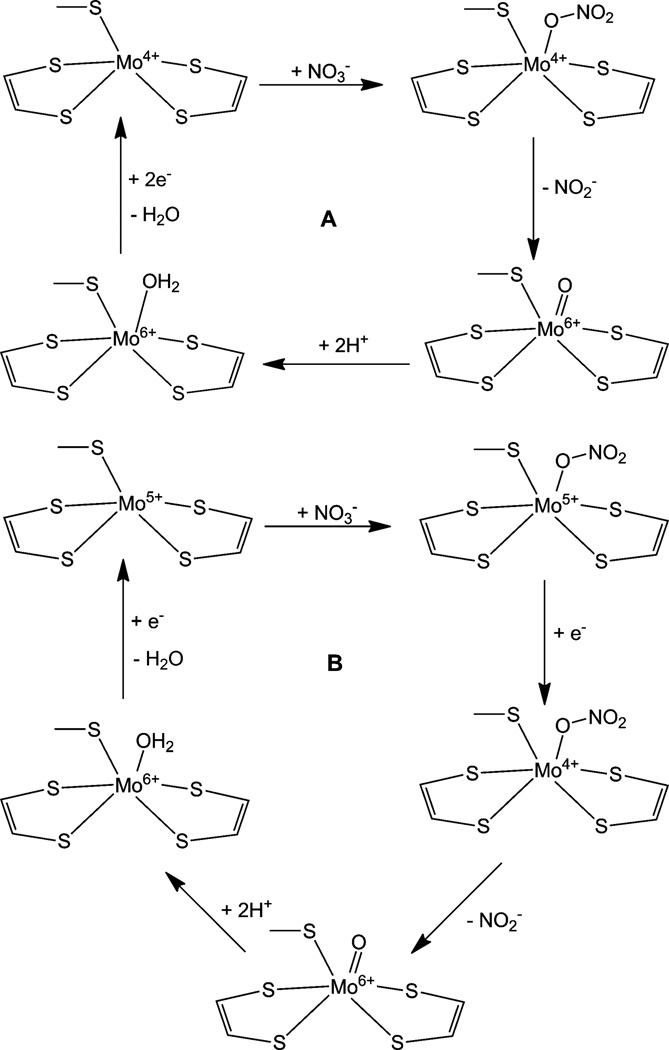
Based on the crystal structure, a catalytic mechanism was proposed for D. desulfuricans NapA (Fig. 10A).119 Accordingly, in the reduced 4+ state, the molybdenum is penta-coordinated, where nitrate binds via one oxygen atom making it a sixcoordinate center. The nitrate is reduced to nitrite and the bound oxygen of nitrate is transferred to the molybdenum as a terminal oxo group. At this time, the molybdenum is oxidized to +6 state, then, the terminal oxygen is protonated making a water molecule, which remains coordinated. The reduction of molybdenum with concomitant loss of the water molecule regenerates the penta-coordinated, reduced Mo(iv) center. Based on the thermodynamic considerations a slightly different mechanism was offered for the E. coli NapA.96 According to the proposed mechanism, nitrate binds to a penta-coordinated Mo(v) center though one of the oxygen atoms, making it a six coordinated species. In this state the molybdenum is reduced by one electron to Mo(iv). The coordinated nitrate is reduced to nitrite and is released. The molybdenum center is oxidized to monooxo-Mo(vi), which is subsequently protonated to give a six-coordinated Mo(vi) state with a coordinated water. This state is reduced by one electron with the loss of a water molecule, regenerating Mo(v) (Fig. 10B). The tenors of these mechanisms are very similar to that proposed for functioning of DMSO reductase where in essence the Mo-center shuttles between a desoxo-Mo(iv) andamonooxo-Mo(vi) centers.154,189 Furthermore, such OAT reactivity has been observed in discrete inorganic molecules,144,161,173,190–194 as well as the proton coupled electron transfer reactions.173–175,177,195–197 Interestingly, stoichiometric nitrate reduction can be achieved by inorganic molybdenum and tungsten compounds with148,190,198,199 or without200 dithiolene coordination.
Fig. 10.
Proposed catalytic mechanism of NapA of D. desulfuricans (A) and E. coli (B).
The revised crystal structure of D. desulfuricans NapA and the structures of C. necator show no terminal oxo-group. Instead a sulfido group was found in the oxidized state and in the reduced state a persulfide bond between the sulfido sulfur and the cysteine sulfur. These structural descriptions were incorporated into a mechanistic proposal whereby significant structural reorganization was proposed.120 Specifically, the persulfide bond was viewed under dynamic control that generates an oxygen-sulfido intermediate and a sulfido species under turnover conditions, in both cases the coordinated cysteine comes off of the Mo-center.120,121,201 The mechanistic proposal was corroborated by density functional theory calculations. 202,203 It is important to point out that initial crystal structures of DMSO reductase showed different geometries about the Mo-center due to sample heterogeneity as revealed by a high resolution structure coupled with spectroscopic investigations.118,204 Such a situation may exist in the case nitrate reductase and indeed has been proposed by Fourmond et al.205 In addition, previous EXAFS studies of Nap have pointed out the presence of a terminal oxo group.206 Kinetics studies of the alpha, beta and gamma-proteobacteria NapA indicated a relatively high affinity for nitrate (Table 5). Steady state kinetic properties have been determined for both the monomeric NapA and heterodimeric NapAB complex, and in most cases the assays were conducted using reduced methyl viologen as a surrogate electron donor (Table 5).
Table 5.
Kinetic parameters of periplasmic nitrate reductases
| Class Organism | Subunit | Substrate | Km (µM) | kcat (s−1) |
Vmax (µmol min−1 mg−1) |
Assay methoda | Ref. | |
|---|---|---|---|---|---|---|---|---|
| α | Magnetospirillum magnetotacticum | NapAB | Nitrate | 3.2 | 2.5 | MV | 338 | |
| Rhodobacter sphaeroides | NapAB | Nitrate | 170 | 25 | MV | 186 | ||
| NapA | Nitrate | 45 | 5.9 | MV | ||||
| NapAB | Nitrate | 30 | PFV | 221 | ||||
| NapAB | Nitrate | 100 (pH 7–8) | PFV | 339 | ||||
| 140 (pH 8.4) | ||||||||
| 185 (pH 9) | ||||||||
| NapAB | Nitrate | 97 | PFV | 340 | ||||
| NapAB (WT) | Nitrate | 410 | 13.1 | MV | 205 | |||
| NapAB (M153A) | Nitrate | 3200 | 13.2 | MV | ||||
| NapAB (R392A) | Nitrate | 65 500 | 11.9 | MV | ||||
| NapAB (Q384N) | Nitrate | 1100 | 9 | MV | ||||
| NapAB | Nitrate | 80 | PFV | 247 | ||||
| Paracoccus pantotrophus (Thiospera | NapAB | Nitrate | 240 | 3.41 (Mo grown cell) | MV | 341 | ||
| pantothropha, Paracoccus denitrificans) | NapAB | Nitrate | 112 | 58 | MV | |||
| NapAB | Nitrate | 45 | PFV | 222 | ||||
| NapAB | 3910 | 0.05 (W grown cell) | MV | 341 | ||||
| NapAB | Nitrate | 11–5.5 ms | 95c | MV | 342 | |||
| NapAB | Nitrate | 1300 | 240 | MV and dithionite | 212 | |||
| NapAB | Azideb | 11 000 (Ki) | MV, dithionite | |||||
| NapAB | Cyanideb | 1300 (Ki) | MV, dithionite | |||||
| NapAB | Thiocyanateb | 4000 (Ki) | MV | 206 | ||||
| γ | Escherichia coli | NapAB | Nitrate | ~ 0.140 (Mo grown cell) | MV | 341 | ||
| NapAB | Nitrate | 0.07 (W grown cell) | MV | |||||
| Aliivibrio (Achromobacter, Vibrio) fischeri | NapAB | Nitrate | 65 | 1.5 | BV | 343 | ||
| β | Alcaligenes eutrophus (Ralstonia | NapAB | Nitrate | 120 | BV | 280 | ||
| eutropha, Cupriavidus necator) | NapAB | Nitrate | 88 | PFV + MV | 344 | |||
| δ | Desulfovibrio desulfuricans | NapA | Nitrate | 32 | 18.6c | MV | 210 | |
In most cases a reduced viologen dye (methyl viologen, MV or benzyl viologen, BV) was used as an electron donor and probed spectrophotometrically.
Inhibitor.
Reported as specific activity.
Tungsten, a group 6 transition metal like molybdenum, often thought of as an antagonist of molybdenum enzymes, can be incorporated into the pyranopterin cofactor, replacing molybdenum, yielding an inefficient enzyme. In the case of DMSOR, the enzyme is functional with Mo or W.207 This is not the case with nitrate reductases. No W-substituted nitrate reductase has been reported in any bacterium. However, a functional W-containing nitrate reductase (Nar) has been isolated from an Achaea, Pyrobaculum aerophilum.208 It has a very high specific activity (Kcat 1162 s−1 and Km 58 µM) and functions optimally at a temperature greater than 95 °C. Even this enzyme, purified from cells grown in the presence of Mo and W, was reported to contain mostly Mo indicating a natural selection of Mo in nitrate reductases.208,209
The detailed kinetic properties of the monomeric NapA from D. desulfuricans have revealed that ionic strength has a peculiar effect on the activity. Monovalent cations (e.g., potassium and sodium) have a stimulatory effect, while divalent cations (e.g., magnesium and calcium) have an inhibitory effect.210 Both ferricyanide and cyanide are inhibitors; however, the presence of phosphate neutralizes the inhibition by cyanide at a lower concentration.210 The optimal pH for nitrate reduction was found to be 8.211 NapA from Paracoccus pantotrophus is competitively inhibited by azide212 and thiocyanate206 with a Ki 11.0 mM and 4.0 mM, respectively. The mechanism of azide inhibition is not clear but it may directly bind to the Mo-center. Thiocyanate inhibition occurs with direct binding to the Mo-center and subsequent displacement of one pyranopterin cofactor. Cyanide also inhibits this enzyme but noncompetitively, with a distinct change in Vmax but no significant change in Km,212 however, crystal structure of the cyanide inhibited form of C. necator NapA (PDB code: 2JIR) clearly show its coordination to Mo. Rhodobacter sphaeroides NapA is noncompetitively inhibited by Zn2+ ions with a Ki of ~1 µM.213
In general, Nap enzymes have a high affinity for nitrate (Table 5), with micromolar Km values (less than 300 µM), and a Vmax ~2 µmol min−1 mg−1. Removal of NapB from the NapAB complex reduces the efficiency of the monomer only 4–6 fold in R. sphaeroides186 but 30 fold in Thiosphaera pantotropha.214 The substrate binding is also affected similarly, and has been suggested to follow a ping-pong type mechanism where nitrate binding is controlled by the reduction of the Mo center.186 Interestingly, napB gene deletion mutants in R. sphaeroides215 and E. coli102 exhibit very little activity, indicating an essential role of NapB. The NapAB complex formation may be oxidation state dependent as well. In the reduced state the calculated Kd is ~0.5 nM while that in the oxidized state is 2.4 µM.186 However, redox modulated binding may control the flow of electrons only in the reduced state.
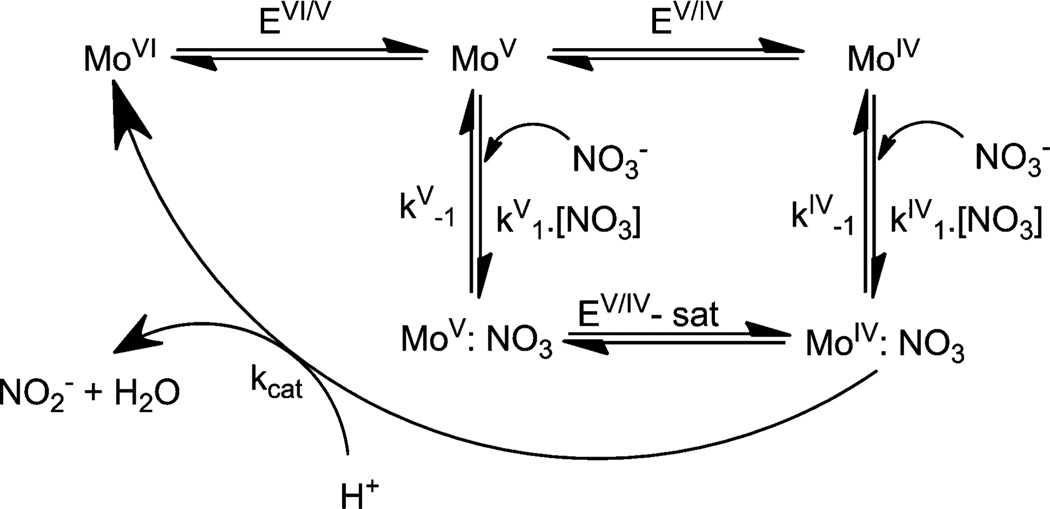
Gates et al. argue that such experiments, in general, do not account for any implicit change in the redox potential of the catalytic metal center, which can be measured by protein film voltammetric (PFV) studies.216 Indeed, PFV experiments on Nap and Nar enzymes reveal different kinetic parameters.217–220 Anderson et al. reported that for the NarGHI complex, there are two states capable of substrate reduction and these two states are coupled by a redox reaction.217 The relative rate of each of the processes then dictates the overall ease of the reaction. Here the Mo(v) state associates more rapidly with nitrate than the Mo(iv) state and the Mo(v):NO3 complex is more stable. A similar conclusion was also reached by Elliot et al.218 At lower nitrate concentrations the substrate is transformed effectively via a slower Mo(iv) pathway. However, at higher substrate concentrations where the catalysis becomes enzyme-limited, the pathway involving the Mo(v) state, which is also faster, becomes more involved. In this scenario, Mo(v) is reduced to Mo(iv) in a substrate bound form. Marangon et al. reported that the NarGHI complex from Marinobacter hydrocarbonoclasticus exhibits a substrate dependent catalysis implying that the enzyme–substrate complex formation precedes electron transfer (Fig. 11).219 There is a redox switch between the two states, termed as “catalytic” and “boost” which are essentially the Mo(iv) state and Mo(v) states, respectively. This redox switch is thought to occur under catalytic conditions, which is independent of the redox potentials of the cofactors present in Nar but dependent on the pH.
Fig. 11.
Proposed catalytic mechanism of respiratory nitrate reductase as suggested by Marangon et al.219
PFV has also been employed in investigating the catalytic mechanism of Nap enzymes. Frangioni et al. reported that the catalytic activity of the R. sphaeroides NapAB complex exhibits catalytic activity close to the reduction potential of the nitrate saturated Mo(v/iv) couple even under non-saturating substrate conditions suggesting a strong binding of the substrate.221 Furthermore, the same group reported the interfacial electron transfer is much faster than the catalytic turnover.205,220 Under turnover conditions, the enzyme has two forms – active and inactive, and they interconvert on a time scale slower than the catalytic turnover. Furthermore, under reducing conditions and in the presence of excess nitrate (high nitrate loading), the inactive form accumulates, causing inhibition of the enzyme.205 The concept of the redox switch as discussed above for Nar has also been invoked in the discussion of the NapAB complex from Paracoccus pantotrophus, which may be attributed to the partner prosthetic group [4Fe–4S] cluster or the Mo center.222 In this case, the activity of the complex has been linked to a single protonation event with a pKa = 7.8; the enzyme exhibits a higher catalytic current at pH~6.222 The authors also conducted experiments with P. pantotrophus cells that express both Nar and Nap. Because the NapAB complex operates at a lower potential than NarGH, the authors argue that electron transfer to Nar through the quinone pool would be thermo-dynamically more favorable; thus is the dominant process. In the presence of high quinone concentrations, reduction of nitrate through NapAB is activated. The exact molecular nature of the components of the redox switch in either enzyme is not clear at this time; it represents catalytically competent but kinetically distinct forms of the enzymes. As mentioned before, it may represent the redox state of the Mo-center, or conformational change that impact redox potential.
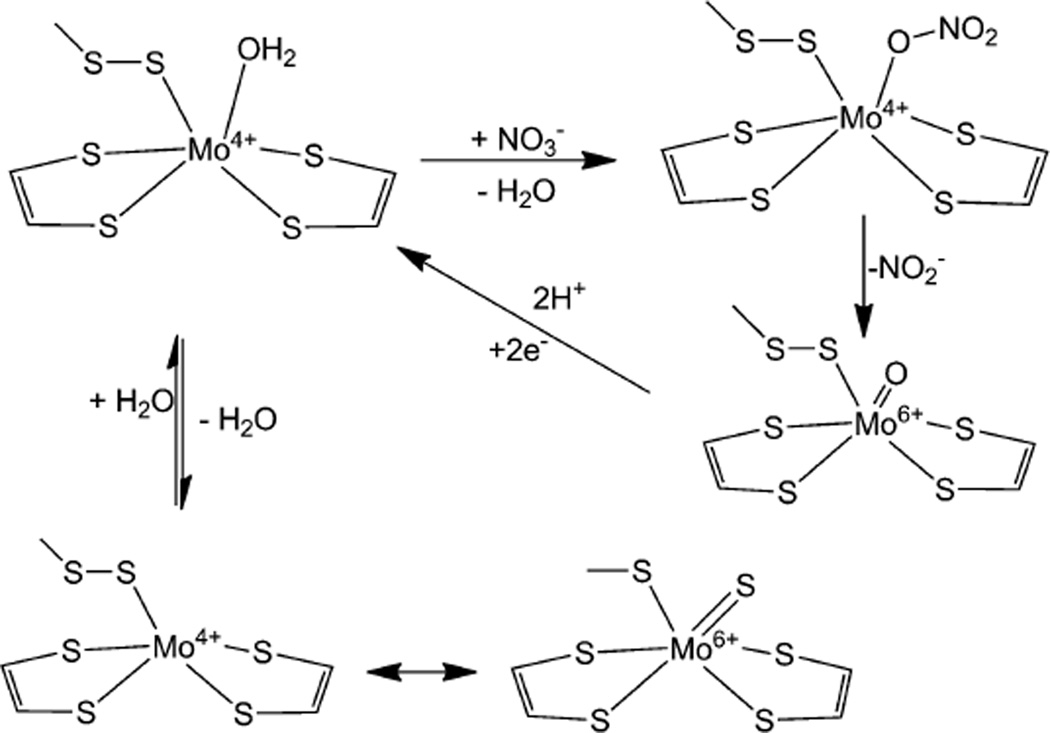
Clearly, the enzymatic mechanism of Nap is complex and fascinating. There are different variables such as pH, substrate and electron source that can influence the mechanism. Further complicating this scenario is the revelation that the Mo center has a sulfide coordination rather than an oxo coordination. Combining the different evidences discussed above, a simplified mechanism can be proposed (Fig. 12). This mechanism takes advantage of the internal redox chemistry of sulfur and assumes that it is operative under low nitrate loading. Here the persulfide bond may remain intact during nitrate binding and transformation, whereby the metal center is converted into a monooxo-Mo(vi) center which can be reduced back to Mo(iv) by two one electron steps associated with proton transfer. In the resting state, the Mo(iv) persulfide moiety undergoes an internal redox change generating a Mo-sulfido species. Noninnocent behavior of the coordinated sulfur involving internal redox has been reported in discrete inorganic compounds.223–231 The proposed mechanism utilizes a direct transfer of an oxygen atomfrom the substrate to themetal. The two forms of catalytically competent enzyme may be related to the redox process mentioned above or a redox process involving pterin cofactor may be possible. The description of the metal redox and details of the Mo(vi) center utilized in the catalytic cycle remain to be fully appreciated. In principle, spectroscopic characterization of different species in the catalytic cycle should help in delineating their electronic structure, which ultimately controls the reactivity.
Fig. 12.
A minimalistic mechanistic proposal for periplasmic nitrate reductase.
From the early days, continuous wave electron paramagnetic resonance (EPR) spectroscopy has played an important role in understanding of pterin-containing molybdenum enzymes. Much of that effort was directed by Bray and his coworkers on XO and SO,232–235 more recently, pulsed EPR spectroscopy on studying sulfite oxidase by Enemark and coworkers.236–241 The naturally abundant, stable molybdenum isotopes (95Mo, I = 5/2, 15.72%; 97Mo, I = 5/2, 9.46% and I = 0 isotopes, 74.82%) give rise to characteristic and unique EPR signals for Mo(v). The nuclear gyromagnetic ratios of the two isotopes with I = 5/2 are very similar to one another, and the signals originating from the two isotopes are normally indistinguishable in continuous wave EPR spectroscopy. Thus, an isotropic Mo(v) EPR signal consists of one large central peak (~75% of total intensity) with 6 satellite peaks (~25% of total intensity). The g-factor falls within a narrow range of 1.8–2.2, yet it is sensitive to small geometric perturbations. EPR spectra of Mo(v) are normally observable at room temperature due to its long electronic relaxation time of Mo(v). Anisotropic EPR spectra can be obtained at liquid nitrogen temperature (77 K). Because of the narrow line-width of the Mo(v) signals, small hyperfine coupling due to the interaction with other magnetic nuclei such as hydrogen and phosphorus, can be observed in CW EPR spectra either directly or by difference spectroscopy.235,242 In the last decade, in addition to measurements at a higher field and with more careful analysis, pulsed EPR techniques have been utilized to understand the finer details about the molybdenum center.118,243,244 Detection of these couplings has played important roles in delineating the mechanistic details. One advantage of this technique is that it can selectively probe the Mo(v) center in the presence of other prosthetic groups. The same can also be viewed as a weakness, as other relevant states are not probed. Like other pterin-containing molybdenum enzymes, nitrate reductases, including periplasmic nitrate reductases, have been investigated by EPR spectroscopy. In most cases, however, the g-values but not the molybdenum hyperfine constants (i.e., A-values), are reported.
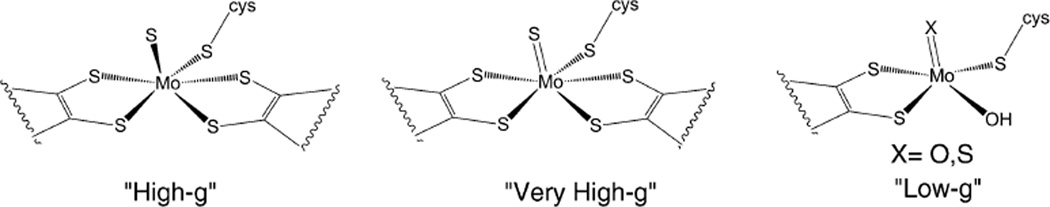
The EPR signals of the Mo-center in Nap have been designated into three broad categories based on their average g values (〈g〉). These are “very high-g”, “low-g” and “high-g” species (Table 6), and all of them are rhombic in nature. The g1 value in “very high-g” is greater than 2 suggesting substantial sulfur contributions in the singly occupied molecular orbital (SOMO). This “very high-g” species is considered to originate from an inactive form of the enzyme. The “low-g” (gav ~ 1.96) is observed when the enzyme is incubated with dithionite, reminiscent of the XO signal. Thus, it suggests the presence of only one dithiolene moiety, as is the case for XO or SO. The species giving this signal is also considered to be inactive. Perhaps the most common signal is the so-called “high-g” species with a gav ~ 1.99, also known as ‘high-g split’ or ‘high-g resting’ signal. It has been suggested that this signal does not represent a catalytically competent species due to its insensitivity to pH and other ligands (i.e., nitrate, nitrite, cyanide, azide, and thiocyanate). 206,212 Under turnover conditions using dithionite as a reductant, another signal called ‘high-g nitrate’ with a lower anisotropy has been reported.206,212 However, subsequent experiments with reduced viologen showed the ‘high-g’ signal during catalysis raising questions about the ‘high-g nitrate’ signal.245 Studies with D. desulfuricans suggest that the ‘nitrate species’ is not catalytically relevant.120,246 Detailed work with R. sphaeroides demonstrated that the ‘high-g’ species is catalytically relevant.247 A recent theoretical (QM/MM) study provided a structural rationale for the different species detected for NapA (Fig. 13).248 Biaso et al. considered different ‘high-g’ signals – ‘high g-resting’, ‘high-g nitrate’ and ‘high-g turnover’ to be magnetically similar to each other indicating little difference in the first coordination sphere. What they suggest is a MoS6 core, and different species arise due to the presence of different ions, and/or a structure distortion. Of these, the ‘high-g’ species is a more reduced state where the disulfide bond is reduced, while the ‘low-g’ signal is attributed to a species coordinated by only one pterin cofactor, as proposed before.
Fig. 13.
Proposed structures of the active sites of ‘high-g’, ‘very-high-g’ and ‘low-g’ species.
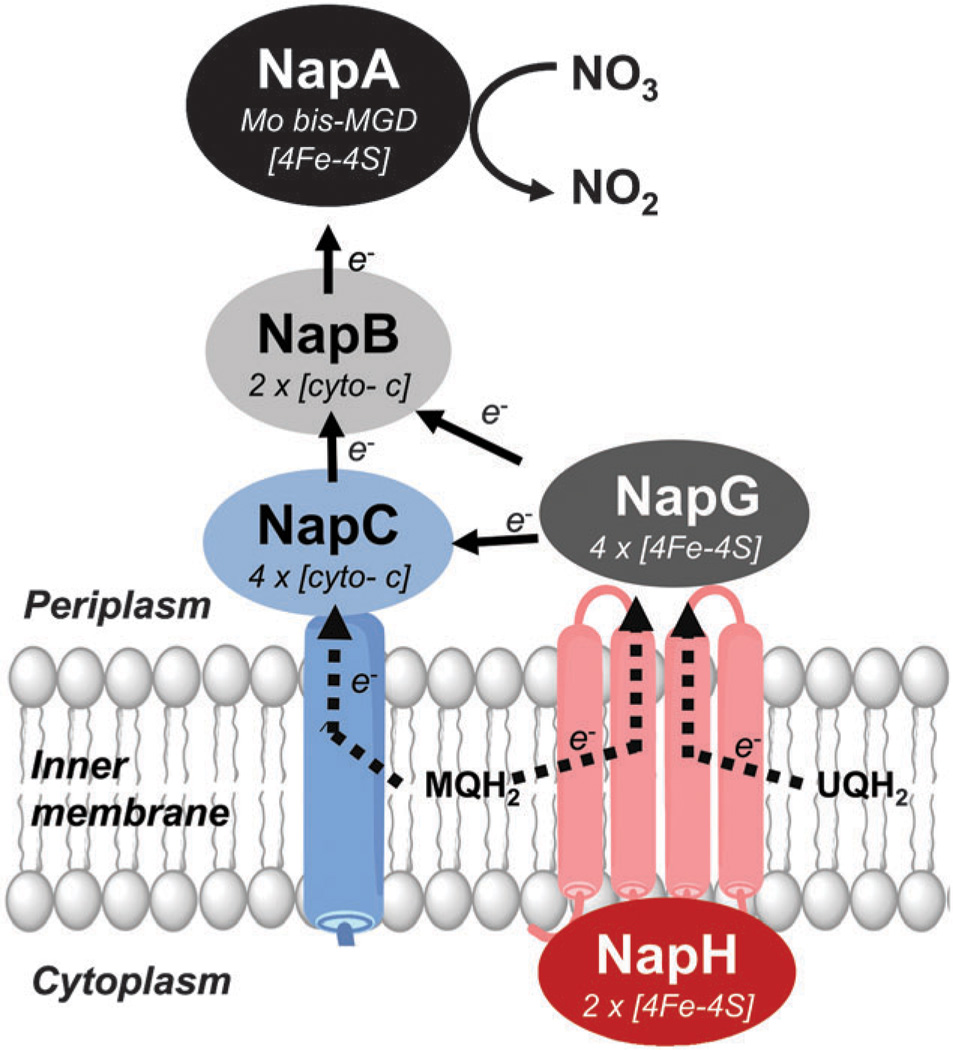
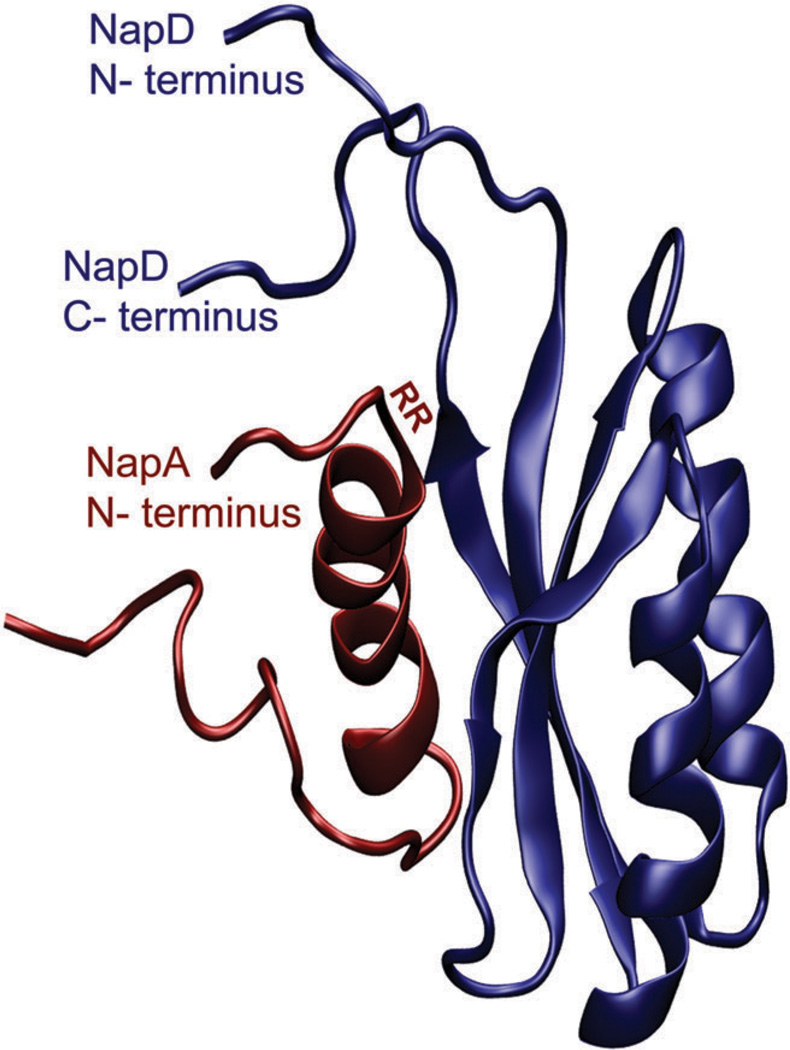
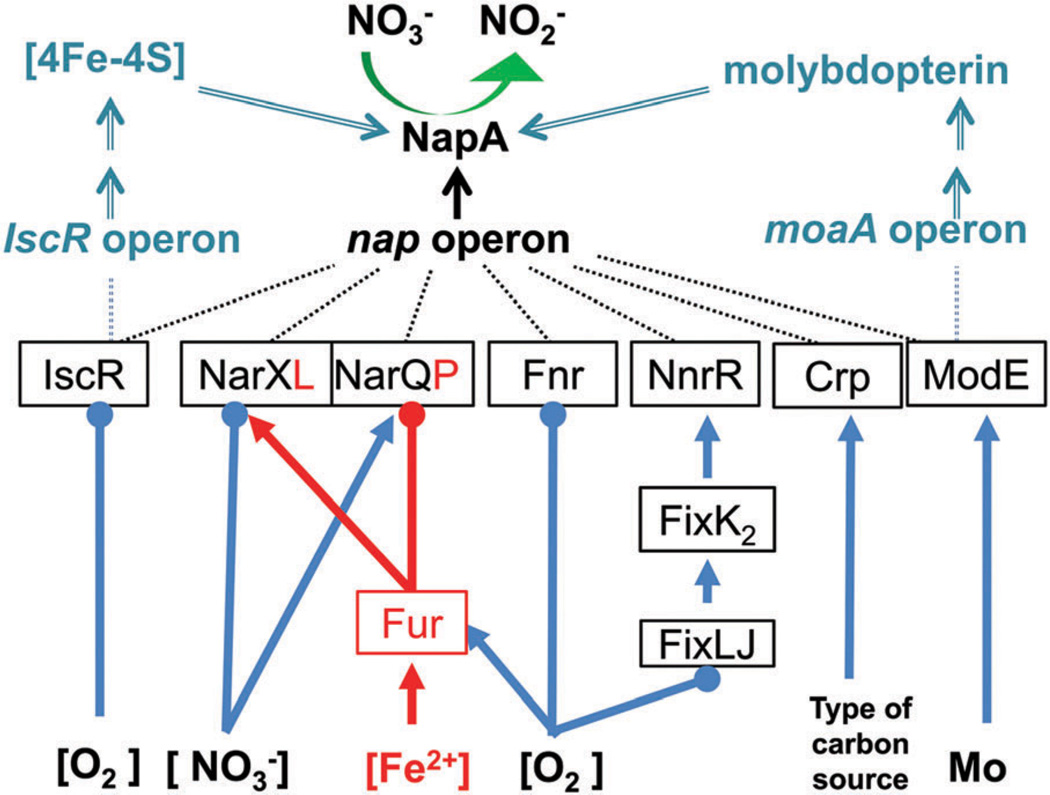
3.3 Redox active auxiliary Nap proteins (NapB, NapC, NapG and NapH)
The genes in the nap operon can be divided into two groups, redox active or maturation, based on the physiological functions of the translated proteins. NapB, NapC, NapG, NapH, and NapM are electron transport proteins involved in the transfer electrons to or from the terminal reductase, NapA. Thematuration proteins NapD, NapL and NapF are involved in post-translational modification of NapA (discussed later). The presence of these genes varies among organisms and no solitary nap gene can be considered ubiquitous to all napA encoding organisms. Mutational studies conducted on R. sphaeroides,215 E. coli,102 and Wolinella succinogenes105 have given insight into the significance of each auxiliary nap gene in nitrate reduction via Nap.
Of the redox active Nap proteins other than NapA, NapB is the most extensively studied. This small c-type cytochrome is an essential component of electron transport in organisms where it is present.101,105 Organisms lacking the napB gene, such as Desulfovibrio desulfuricans, encode alternative iron cofactor redox partners, such as NapM or NapC.249,250 In most organisms, the functional enzyme has been isolated as a NapAB heterodimer. The Rhodobacter sphaeroides and Cupriavidus necator NapAB heterodimer structures have been determined using X-ray crystallography.121,186 Additionally, the crystal structures of two monomeric NapAs are available.96,119 In E. coli the functional enzyme is also a heterodimer, however NapB is not present in the crystal structure because it dissociated from the complex during purification. A tyrosine residue in NapA is proposed to aid in electron transfer from the heme of NapB to the [4Fe–4S] cluster of NapA. This tyrosine residue is conserved in all heterodimeric NapA proteins and absent in monomeric NapA proteins such as D. desulfuricans.188
The crystal structure of the recombinant Haemophilus influenza NapB has revealed that the two heme molecules are stacked parallel and hydrogen bonded to each other via propionic acid side chains.251 In this structure, the conserved residues of NapB are located in close proximity to the heme binding motifs (HXnCXnCH). The N- and C-termini are poorly conserved, and are not involved in electron transfer to NapA.251 The N-terminus encodes a hydrophobic signal sequence, roughly 30 amino acids long, which targets the unfolded NapB for periplasmic export via the sec dependent pathway.186,252,253 Arnoux et al. suggest that the N- and C-termini of NapB are involved in NapA binding.186 The crystal structures of NapAB heterodimers from C. necator and R. sphaeroides underscore this aspect.121,186 The poor conservation of the NapA binding sites on NapB proteins could be the source of the different binding affinities observed in the active enzyme. E. coli and R. sphaeroides NapAB exhibit different dissociation constants, 32 µM ref. 96 and 5 × 10−4 µM ref. 186 respectively, and are reflective of the purified form. This could explain why the E. coli NapA was isolated as a monomer, while the R. sphaeroides homologue was isolated as a heterodimer.254
Similar to NapA, the post-translational modification for functional NapB requires maturation proteins. In this case cytochrome c maturation (Ccm) proteins are involved in NapB cytochrome c biosynthesis and insertion. While the ccmABC-DEFGHI operon is located directly adjacent to the nap operon in E. coli,255 this is not the situation in most other organisms. The ccm operon is missing completely from the genomes of Epsilonproteobacteria, suggesting that these organisms have alternative methods for iron uptake and heme synthesis. C. jejuni encodes a large variety of genes associated with iron scavenging, uptake, and storage, one of which (chu) can transport intact heme into the cell.256 The ability of C. jejuni to utilize intact heme could make cytochrome c biogenesis redundant, and thus energetically unfavorable.
The nap operons can also encode one or both of the membrane quinone oxidases NapC and NapH (Table 4). The presence of NapC and/or NapH may be an indication that the organism can catalyze nitrate respiration via Nap. This variability and diversity may indicate diverging functional roles of Nap as well. The presence of and type of quinone oxidase to classify nap operons in bacteria is shown in Table 7.
NapC, a tetra-heme transmembrane protein, is also considered to be an essential component of electron transport to NapA in organisms in which it is found.102 NapC transports electrons via the heme cofactors from the membrane quinone pool to the periplasmic NapB.257 In E. coli, electron transfer from ubiquinone and menaquinone to NapAB is dependent on NapC. Ubiquinone electron transfer from NapGH to NapABC is more efficient than ubiquinol transfer directly from NapC to NapAB.258 Like napB, napC is not present in all nap operons. NapC belongs to a large group of tetra heme proteins, the NapC/NirT family.259 The possibility that other members of the NapC/NirT family, NrfH and Cym, could fulfill the function of NapC (i.e., electron transport to NapA) in organisms which lack napC has been postulated,259–261 although supporting experimental evidence is not available.
The genes napG and napH encode two putative ferredoxins. NapH is an integral membrane protein. The cytoplasmic portion of NapH contains two iron sulfur binding motifs. NapG contains four iron sulfur binding motifs and is associated with the periplasmic face of the membrane. In organisms which do not encode a NapC homologue, NapG and NapH are predicted to transfer electrons from the quinone pool directly to NapAB.262 Campylobacter only has menaquinone, but does not have a NapC homologue. In Campylobacter, NapGH are believed to transport electrons from the menaquinone pool to NapAB.93 The MQH2 couple has a lower midpoint redox potential than UQH2, −70 mV compared to +90 mV,262 indicating that MQH2 would be a less efficient electron acceptor. The proposed electron transfer pathway from the quinone pool to the substrate nitrate is shown in Fig. 14 as well as in Fig. 18 (vide infra).
Fig. 14.
The proposed routes of electron transport to NapAB from quinone pool during nitrate reduction. Electrons can be transferred from menaquinone (MQH2) or ubiquinone (UQH2) with in the inner membrane by a quinone oxidase (i.e., NapC or NapH). Electrons are transported to NapAB via NapC, NapGH or NapCGH.
Fig. 18.
Phylogenic tree of NapA using 292 protein sequences. Positions containing gaps were eliminated, yielding a total of 600 amino acid positions in the final dataset. MEGA5 software was used for alignments and phylogenetic analyses. Formate dehydrogenase (FDH) was included in the analysis as an outlier. Three assimilatory nitrate reductases (Nas) were also included for reference. NapA sequences were collected from the NCBI database. Branches corresponding to similar species (sp.) or subspecies (ssp.) are collapsed. Branches are colored according to the phyla of proteobacteria: alpha (aqua); beta (navy blue); gamma (green); delta (orange); epsilon (red); non-proteobacteria (grey). Branch nodes are colored red (NapH); blue (NapC); red/blue (NapH + NapC); grey (no quinone oxidase). Diagrams at the four corners display the topology of the different Nap forms with respect to the periplasmic membrane and correspond to the node colors. Open circles represent NapA sequences from organisms where complete nap operon content is unknown.
3.4 Non redox-active auxiliary Nap proteins (NapD, NapF, and NapL)
Of the auxiliary Nap proteins, the napD gene is the most common and appears to be the most vital. The R. sphaeroides, E. coli, and W. succinogenes mutational studies are in agreement, napD is an essential component of nitrate reduction, as in all napD mutants nitrate reduction is inhibited.102,105,215 Translated NapD is a dedicated chaperone protein as it has a defined role in NapA maturation.263 NapD is a member of the well-studied TorD chaperone protein family.264–266 It interacts with the twin arginine translocase (TAT) leader sequence of NapA and possibly other maturation proteins, such as NapF, in the biosynthesis of NapA.267
NapD modulates the periplasmic export of NapA by binding to the N-terminal TAT leader sequence of NapA. Structural characterization of the E. coli NapD267 indicates (Fig. 15) that the non-polar residues in the β-sheet of NapD bind, via protein–protein interactions, to the TAT leader-sequence of NapA (Fig. 8). It is conceivable that NapD delays export of NapA until the protein is properly folded and the cofactors are inserted.267 While the purpose of the NapA-D interaction is predicted to be comparable in all organisms, the chemical basis of the interaction is species-specific, thus may be considered as a redox enzyme maturation protein (REMP).268,269 NapD lacks a conserved NapA binding motif.
Fig. 15.
The protein structure of E. coli NapD in complex with the N-terminus of NapA. The protein backbone, of NapD (blue) and NapA (red), is displayed as a ribbon. The location of the two arginine residues (RR), for which the TAT leader sequence is named, is on the N-terminus of NapA. The figure the NMR solution structure (PDB ID: 2PQ4 was created using VMD (version 1.8.7) software.
Despite the importance of NapD, some organisms do not possess a napD homologue (e.g., Sorangium cellulosum, Photobacterium damselae, Rhodothermus marinus, and Anaeromyxobacter species). In general, nap operons which lack napD have relatively few genes. Its absence may be an indication of reduced functionality as a result of the incapacity to synthesize a functional NapA. For example, neither Photobacterium damselae,270 or Rhodothermus marinusas271 are capable of reducing nitrate despite having a napA homologue. Sorangium cellulosum and Anaeromyxobacter species can reduce nitrate,272,273 though this activity cannot be attributed to NapA without a more detailed investigation as the genomes of these organisms encode multiple nitrate reductases (i.e., Nar, Nap and Nas). A better understanding of how essential NapD is may come from studies of the organisms without the gene or those lacking the twin arginine motif (e.g., S. cellulosum, P. damselae).
NapF, a cytoplasmic, non-heme iron sulfur protein also plays a role in NapA maturation. While mutation of the napF gene does not completely abolish nitrate reductase activity in E. coli,102 a drastic decrease in activity is observed in R. sphaeroides and W. succinogenes.105,215 NapF is not considered to be essential for NapA maturation, although it does aid in [4Fe–4S] insertion. 105,258,274 NapF contains four iron sulfur binding motifs. Mutation of each motif demonstrated that one specific motif is responsible for transferring the [4Fe–4S] cluster into NapA.103 Moreover mutation of the W. succinogenes NapF led to the accumulation of the inactive precursor NapA in the cytoplasm.103 The ability of NapF to reconstitute the [4Fe–4S] cluster of the apo-NapA has also been demonstrated in vitro.275 The association of NapF and NapA has also been confirmed.274 Interestingly, recombinant affinity-tagged NapF has been used to purify NapA from E. coli.274 NapA maturation is more efficient in the presence of NapF. Mature NapA is able to accumulate in the periplasmic space with time, as indicated by nitrate reductase activity assays, growth curves and SDS-PAGE data. Thus, NapF is possibly an evolutionary advantage to organisms that possess it.
A third putative maturation protein, NapL, is found primarily in the Epsilonproteobacteria. The gene napL encodes a 35 kDa periplasmic chaperone protein,103,105 with no known metal binding motifs.103 While the function of NapL is unknown, mutation of napL in W. succinogenes and C. jejuni delays respiratory growth on nitrate media.105,276
3.5 Nap protein of unconfirmed function (NapS)
No experimental evidence is available to confirm if NapS is translated, and if so, what the function may be. The napS gene, found in Arcobacter butzleri (YP_001489295.1 or ZP_07891038.1), Sulfurovum sp. (YP_004049825.1), Nitratiruptor sp. SB155-2 (YP_001357263.1) and Sulfurimonas denitrificans (YP_003892305.1), is annotated as a methyl-accepting chemotaxis sensor protein, part of the PAS (Per-ARNT-Sim) superfamily. The putative NapS proteins exhibit significant homology with the structurally characterized region of Azotobacter vinelandii NifL. NifL is the redox sensor domain of the NifLF two-component regulatory system, which, along with NifA, regulates nitrogen fixation genes in response to oxygen. 277,278 Thus NapS may be involved in regulation of the nap operon, however, this has yet to be experimentally determined. The variety of nap genes indicates that the nap operon evolved by gaining or losing genes in an effort to fit the functional needs of the organism.
3.6 The Nap operon
3.6.1 Operon structure and organization
The nap operon is typically found on the chromosome, although examples of plasmid borne nap operons are known e.g., Paracoccus denitrificans, Cupriavidus metallidurans, Sinorhizobium meliloti Sinorhizobium medicae Sulfuricurvum kujiense, Rhodobacter sphaeroides,279 Ralstonia eutropha (also known as Cupriavidus necator).280,281 In some organisms (e.g. Shewanella species) multiple copies of the nap operon have been found.254,282 The nap operon exhibits significant variability in gene organization and composition (Fig. 16). In addition to the napA gene, eleven nap genes have been identified to date.4,249,283 While different operon structures have been observed (Table 7), the most common structure of the nap operon is napEDABC, which is found primarily in alpha-, beta-, and gamma-proteobacteria.
Fig. 16.
Representative examples of the nap operon gene content and organization. Each nap gene (arrow) was identified by performing a blast search of the NCBI database, (*) indicates organisms that encode two nap operons.
3.6.2 Regulation of the nap operon
The functional diversity of Nap has a foundation in transcriptional regulation. The nap operon can be regulated in response to multiple environmental cues e.g., the absence or presence of nitrate, oxygen, molybdenum and/or iron, the type of carbon source, or redox potential in the cell. As a result mechanisms of nap transcriptional regulation are not conserved, considerable variations exist.
Microbes utilize multiple terminal reductases during respiratory growth, thus regulating translation of each reductase, under the appropriate conditions, is a concerted endeavor. During aerobic growth, oxygen is typically the preferred terminal electron acceptor, a result of the high redox potential (E0′ = +840 mV). Under anaerobic conditions, nitrate takes precedence over other terminal electron acceptors, again due to the relatively high redox potential (E0′ = +433 mV). Not surprisingly, the presence and absence of both oxygen and nitrate can affect transcription of Nap (Fig. 17).
Fig. 17.
Transcriptional regulation of the nap operon, and the pyranopterin and [4Fe–4S] cofactor biogenesis operons. Note that not all regulatory effects are present in the same organism. Proteins directly regulating the nap operon transcription are connected to “NapA” with a dashed line (–), and those that affect the cofactor biogenesis operons are connected to the specific operons by a double dashed line (==). The operons for pyranopterin and [4Fe–4S] cofactor biogenesis (blue) are moaABCDE and iscRSUA-hscBA-fdx, respectively. Lines with arrow (→) denote a positive regulatory effect, i.e., up-regulation, in the presence of the associated environmental signal; negative effects, i.e., down-regulation, is indicated with circle lines (−•). Concentration dependent regulation is indicated with brackets surrounding the chemical, for example [O2].
Multiple global transcription regulators have been identified for nitrate reductases although no unique nap regulator has been identified (Fig. 17). Each regulatory protein binds to a DNA consensus sequence upstream from the regulated genes. In response to oxygen, the fumarate and nitrate reductase (FNR) protein can control transcription of the nap operon.284,285 Additionally, the NarPQ/NarLX dual interacting two-component regulatory system is employed to regulate transcription of nap in the presence of nitrate.286 The dualistic constituents, a membrane bound sensor (i.e., NarX and NarQ), and a DNA binding response regulator, (i.e., NarL, NarP), work together to control expression. E. coli uses both the Fnr and NarPQ/NarLX transcriptional regulators to sense nitrate and oxygen levels and coordinate expression of nitrate reductases, Nap and Nar (both narG and narZ operons) as well as nitrite reductases, Nrf and Nir.287 Nap is maximally expressed anaerobically in the presence of low nitrate concentrations, possibly providing a selective advantage over Nar during colonization of the human GI tract.101,255,288,289
In Bradyrhizobium japonicum napEDABC expression is also up-regulated in response to micro-aerophillic growth and the presence of nitrate. In B. japonicum the FixLJ-FixK2-NnrR regulatory cascade controls nap expression in response to oxygen.290,291 FixLJ, a two-component regulatory system, down-regulates translation of the nap operon, when an elevated concentration of oxygen is present. However translation of the nap operon is not always repressed in the presence of oxygen. In P. pantotrophus Nap expression occurs aerobically. The P. pantotrophus nap operon does not have a FNR-binding site upstream.292 R. capsulatus and R. sphaeroides express nap both in the presence and absence of oxygen.293
The cellular redox state can also regulate nap expression. P. pantotrophus utilizes the ferric uptake regulator (Fur) as a transcriptional regulator of NarP and NarL. Fur is a global regulator, which controls iron (Fe2+) uptake and detoxification. 294,295 Fur controls nap expression and the response to oxygen and cellular redox status as it is dependent on free Fe2+. Consequently oxidants (i.e., O2) can decrease Fur activity.287 In E. coli, the cyclic AMP receptor protein (Crp) has also been demonstrated to up-regulate nap operon translation in response to carbon source. Less favorable sugars, such as mannose, increased nap translation compared to the typical fermentative sugars, such as glucose.296 In Shewanella oneidensis it has been shown than NapA is responsive to nitrate but nitrite reductase (NrfA) is responsive to both nitrate and nitrite in a Crp dependent way.297
The translation of the nap operon is also coordinated with the biosynthesis of the iron sulfur cluster and the pyranopterin cofactors required in active NapA. An iron–sulfur clustercontaining transcription factor, the iron–sulfur cluster regulator (IscR), represses nap transcription under aerobic conditions.298 IscR also regulates iron–sulfur cluster biogenesis.298 The IscR binding motif is typically located up-stream from the nap operon. The IscR binding region overlaps with an additional regulatory protein-binding site, the molybdate responsive (ModE) transcriptional regulator protein. In response to molybdenum, ModE up-regulates the nap operon translation. ModE also regulates transcription of the pyranopterin biosynthesis genes (moaABCDE)299 and a high affinity molybdate transporter (modABC).300 The ModE and IcsR regulatory sites are located further up-stream from the nap operon than the FnrR, NarP, NarL binding sites.301 Thus ModE and IscR can supersede transcription of the nap operon, dominating FnrR, NarP, or NarL regulatory elements, if molybdate (MoO42−) and iron are limited.302
The Epsilonproteobacterium C. jejuni appears to employ analogous regulatory mechanisms for nitrate respiration. Unlike E. coli, which encodes three dissimilatory nitrate reductases (i.e. NapA, NarG and NarZ), C. jejuni only encodes NapA. In C. jejuni NapABGH are expressed in response to iron303–305 and molybdenum.109,305 This suggests that the IscR and ModE regulatory proteins are involved in transcriptional regulation of the C. jejuni nap operon. Additionally, C. jejuni preferably up-regulates Nap expression anaerobically. The napABG expression is up-regulated during colonization in chickens306 and napABGH in elevated growth temperatures,307 possibly an indication of Fnr regulation. The alignment of the C. jejuni pre-nap operon DNA with the corresponding E. coli sequence indicates that all of the regulatory elements binding sites are conserved.
3.7 Phylogenetic relationship
In 2002, we (JFS and PB) explored the possible evolutionary relationships among the nitrate reductases;76 and more recently, the evolutionary aspects of the different sub families of nitrate reductases have been discussed.308,309 In our original study, using available sequence data at the time (13 NarG, 12 NasA and 11 NapA), we found that bacterial nitrate reductases formed two major clades, the NarG clade and the NasA/NapA clade. We postulated76 that NapA might have evolved from the catalytic subunit of the assimilatory nitrate reductases; Jepson and colleagues further explored this idea.188 Bacterial assimilatory nitrate reductases have been further divided into three types based on their protein sequence and the biochemical properties of the catalytic subunits, namely NasA, NasC and NarB.4 NarB is the closest phylogenetic relative of NapA.188 Although the structural aspects of NapA are conserved among the different microbial species, (e.g., the molybdenum center, iron sulfur cluster, and TAT leader sequence), they may also be subdivided into distinct subclades based on differences in amino acid sequence and operon organization.
A neighbor joining tree has been constructed from multiple sequence alignments of 288 NapA sequences with three assimilatory nitrate reductases and one formate dehydrogenase out-group taxa (Fig. 18). The monomeric NapA, exemplified by D. desulfuricans NapA, is clearly the most similar to the assimilatory nitrate reductase (Nas). The deepest branch in the NapA tree (most similar to assimilatory nitrate reductases) is comprised mainly of Deltaproteobacteria, along with some non-proteobacteria (i.e., Firmicutes, Deferribacteres, Bacteroidetes and Chrysiogenetes). A second, deep branch in the NapA tree is comprised of mainly Epsilonproteobacteria, although Aquificae and one Deinococcus also cluster into this branch. This unique Epsilonproteobacterial subclade was hinted to by the sole Campylobacter species in the original phylogenetic tree.76 Two additional subclades include NapA from alpha-, beta- and gamma-proteobacteria possibly indicating more heterogeneity and a greater divergence from assimilatory nitrate reductase. A clearer picture emerges of this divergence with further examination of the distribution of Nap isoforms.
There are at least 11 nap genes (napA,B,C,D,E,F,G,H,L,M, and S) and at least 4 distinct nap operons (EDABC, DAGHBC, CMADGH, and AGHBFLD) (Fig. 16 and Table 7). Thus the 4 major subclades evident in the NapA tree in Fig. 15 underscore not only differences in protein sequence, but also differences in operon organization, with DAGHBC defining the subclade predominated by Gammaproteobacteria, EDABC defining the subclade with alpha-, beta-, and gamma-proteobacteria, AGHBFLD defining the Epsilonproteobacterial subclade, CMDAGH found in the Deltaproteobacteria subclade. Only the Epsilonproteobacterial subclade appears to be highly conserved but even so has exceptions with some operons having an additional gene (e.g., napS). A further investigation of Shewanella species provides interesting insight into the distribution of the napEDABC and napDAGHBC isoforms as many species have both.282 The Nap from two operons have been isolated and characterized, and one of them (Nap-α) was found to be more stable.254 Jepson et al., based on their examination of gene order in 19 Shewanella species, postulated that the different isoforms might serve different functions and/or could be under different regulatory control.254 However, a closer look at not just the presence of the two isoforms, but the actual number of genes in the operons reveals another more fascinating phenomenon. Table 7 shows the gene orders for a total of 26 strains representing 19 species of Shewanella. Nine strains have both complete EDABC and DAGHB operons. Two species (S. oneidensis, S. halifaxensis) have only DAGHB. S. denitrificans is the only known species with EDABC. The rest have one complete operon with the other isoform missing crucial genes. Even more intriguing is the presence of a unique isoform of napA found in the genomes of both S. benthica and S. violacea. In addition to a DABC (missing napE) there is a napA homolog with an accompanying gene annotated as dsmC, a membrane-anchored quinone electron shuttle. Furthermore, amino acid sequence alignment revealed that the – CMAS – amino acid sequence of the core of the catalytic site is conserved whilst other residues are not. We suggest that the nap operon is rapidly evolving and inmost strains only one complete operon is functional as the periplasmic nitrate reductase. Thus the NapA phylogeny can be explained by nap operon gene content and organization. It suggests an evolutionary progression of the nap operon, possibly away from the monomeric assimilatory nitrate reductase operon of NarB, into more complex operons with horizontal gene transfer widespread among the alpha-, beta-, and gamma-proteobacteria. In that context Shewanella may represent an intermediate stage in evolution as both types of operon are present, and it has been suggested that nap-β may have been acquired by horizontal gene transfer.310
Shewanella strains are not the only organisms where multiple copies of the nap operon can be found. In Rhodobacter sphaeroides 2.4.3 two copies of nap operon were also found. These two nap operons are napFDABC that encodes for Nap-α and napDFAGHBC that encodes for Nap-β. Activity from Nap-α is only detected when the cells are grown in solid media, perhaps it’s synthesis have different regulatory elements and functionally it is involved in redox poise (homeostasis). The Nap-β has a negative oxygen control, and does not contribute to nitrite production.310
Another example of gene modification (e.g., truncation or loss) can be found in Campylobacter jejuni. There are two subspecies of C. jejuni, C. jejuni subs. jejuni and C. jejuni subs. doylei. The latter is often found in patients with bacteremia and gastroenteritis and can be differentiated from C. jejuni subs. jejuni by its inability to respire nitrate. It was found to have a deletion of the napA gene and mutations in napB.311
Interestingly, the phylogeny of NapA correlates with the nap operon structure. Examining the presence, or absence, of nap genes, which encode proteins involved in post-translational modification of NapA (i.e. NapD, NapF, and NapL) and electron transport to NapA (i.e. NapB, NapC, NapG, and NapH), can provide fundamental information on how the mechanism of NapA maturation and electron transport differs in each organism. Whilst the presence of dedicated nap chaperone proteins and electron transport proteins would increase the overall efficiency of nitrate reduction via NapA, it is possible that, in the absence of these dedicated genes, the organism can use more undefined proteins to fulfill the same role. For example, in Shewanella oneidensis uses CymA, a c-type cytochrome containing membrane protein similar to NapC, to transfer electrons from the quinone pool to NapA312 It will be important to explore the strength as well as time scale of the interaction between different subunits of Nap. Comparison with those of the membrane bound complexes such formate dehydrogenase (FDH-GHI) or respiratory nitrate reductase (NarGHI) should provide a map of the physiological function.
4 Summary
This review has tried to convey important aspects of nitrogen cycling in the environment and in health as well as call attention to the remarkable forms and functions of nitrate reductase. There have been significant advances in our understanding of the molybdopterin cofactor and its role in nitrate reduction as well as insights into the impact of the protein milieu on the reactivity and specificity. While catalyzing a single reaction, nitrate reduction to nitrite, periplasmic nitrate reductase has been linked to a number of cellular functions including the maintenance of redox poise, nitrate scavenging, nitrate respiration and pathogenicity. In addition to this functional diversity, genetic variability is also common as revealed by the variety of nap genes and operon structure. The transcriptional level regulatory mechanisms vary among organisms with as many as three different promoters and seven regulatoryprotein binding sites. Clearly, in the last decade many exciting results informing the structure, genetics, and kinetics were reported and it is hoped the same excitement will continue in the next decade. An important aspect would be how Nap enzymes play a role in pathogenicity and whether therapeutic intervention would be possible utilizing the chemistry of Nap. One cautionary aspect is that many of the organisms described herein have been renamed, some several times, posing a challenge to keep track of just what has been studied on which organism. We look forward to future studies enhanced by the ever-increasing volume of genomic data, advances in instrumentation, and new isolates.
Acknowledgements
Work in authors’ laboratories has been supported by grants from the National Institutes of Health, the Department of Energy, and the Commonwealth of Pennsylvania. We also appreciate numerous stimulating discussions with our present and former collaborators and coworkers. Special thanks to Mr Andrew K. Adams, Mr Samih Nassif, Mr John Thomas, and Mr Benjamin Mogesa for their assistance in preparation of this manuscript. We also thank Dr Mark Gladwin for insightful discussion on NO in biology, and Dr Jesus Tejero for helpful discussion on enzyme functions.
Abbreviations
- Aio
Arsenite oxidase
- SO
Sulfite oxidase
- XO
Xanthine oxidase
- DMSOR
Dimethyl sulfoxide reductase
- mARC
Mitochondrial amidoxime reducing component
- PGD
Pyranopterin guanidine dinucleotide
- Nap
Periplasmic nitrate reductase
- pNar
Periplasmic respiratory nitrate reductase
- Nar
Respiratory nitrate reductase
- eukNR
Eukaryotic nitrate reductase
- NasA
Assimilatory nitrate reductase A
- NarB
Assimilatory nitrate reductase B
- Fdh
Formate dehydrogenase
- DNRA
Dissimilatory nitrate reduction to ammonia
- TAT
Twin arginine translocase
- MQH2
Menaquinone
- UQH2
Ubiquinone
- Crp
Cyclic AMP receptor protein
- IscR
Iron–sulfur cluster regulator
- FNR
Fumarate and nitrate reductase
- ModE
Molybdate-responsive transcriptional regulator protein
- AMP
Adenosine monophosphate
- MOE
Molecular operating environment
- PDB
Protein data bank
Biographies

Courtney Sparacino-Watkins
Courtney Sparacino-Watkins earned her BS in Chemistry and Biochemistry from Slippery Rock University, and her PhD from Duquesne University, under the advisement of Partha Basu. She is currently a postdoctoral fellow at the University of Pittsburgh, School of Medicine, Division of Pulmonary, Allergy and Critical Care Medicine, and in the Vascular Medicine Institute under the mentorship of Mark T. Gladwin. Her research interests center on metabolism and signaling of nitrogen oxyanions (nitrate, nitrite, and nitric oxide) in human vascular diseases and pathogenic bacteria.

John F. Stolz
John Stolz earned his BSc (Biology) from Fordham University and a PhD (Microbial Ecology and Evolution) from Boston University. He was an NRC Post Doctoral Fellow at NASA Jet Propulsion Laboratory and a Visiting Associate at California Institute of Technology (with Joseph Kirschvink). Following an NSF Post Doctoral Fellowship in Plant Biology at the University of Massachusetts, Amherst (with R. Clinton Fuller), he joined the faculty at Duquesne University in 1990. His research interests are the ecophysiology, biochemistry and molecular biology of phototrophic and metal(loid) reducing bacteria and include the microbial transformation of arsenic, the environmental impact of non-conventional shale gas extraction, and community structure in modern marine stromatolites.

Partha Basu
Partha Basu earned his BSc (Honors) in Chemistry and MSc in Inorganic Chemistry from Calcutta University, and a PhD from Jadavpur University (with Animesh Chakravorty). After a post-doctoral work at the University of Arizona (with John Enemark) he joined the faculty of Duquesne University in 1998. His research interest is in metals in biology specifically the role of metallobiomolecules in nitrate and arsenic metabolism. His group designs and develops new molecules for detailed kinetic, spectroscopic and structural studies, and as sensors of heavy metal ions. Another aspect of his research is proteomics and protein biochemistry of molybdenum enzymes.
References
- 1.Campbell WH. Adv. Photosynth. Respir. 2002;12:35–48. [Google Scholar]
- 2.Campbell WH. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1999;50:277–303. doi: 10.1146/annurev.arplant.50.1.277. [DOI] [PubMed] [Google Scholar]
- 3.Guerrero MG, Vega JM, Losada M. Annu. Rev. Plant Physiol. 1981;32:169–204. [Google Scholar]
- 4.Richardson DJ, Berks BC, Russell DA, Spiro S, Taylor CJ. Cell. Mol. Life Sci. 2001;58:165–178. doi: 10.1007/PL00000845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zumft WG. Microbiol. Mol. Biol. Rev. 1997;61:533–616. doi: 10.1128/mmbr.61.4.533-616.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin JT, Stewart V. Adv. Microb. Physiol. 1998;39:1–30. doi: 10.1016/s0065-2911(08)60014-4. [DOI] [PubMed] [Google Scholar]
- 7.Rosswall T. In: Some perspectives of the major biogeochemical cycles. Likens GE, editor. Vol. 2. New York, USA: John Wiley & Sons Ithaca; 1981. p. 199. [Google Scholar]
- 8.Stolz JF, Oremland RS. FEMS Microbiol. Rev. 1999;23:615–627. doi: 10.1111/j.1574-6976.1999.tb00416.x. [DOI] [PubMed] [Google Scholar]
- 9.Einsle O, Messerschmidt A, Huber R, Kroneck PMH, Neese F. J. Am. Chem. Soc. 2002;124:11737–11745. doi: 10.1021/ja0206487. [DOI] [PubMed] [Google Scholar]
- 10.Lide D, editor. CRC. 75 edn. Cleveland, OH: CRC Press; 1994-1995. pp. 5–16.pp. 15–20. [Google Scholar]
- 11.Moga M, Vasile I, Bigiu N. J. Environ. Prot. Ecol. 2013;14:341–349. [Google Scholar]
- 12.Ward MH, de Kok TM, Levallois P, Brender J, Gulis G, Nolan BT, VanDerslice J. Environ. Health Perspect. 2005;113:1607–1614. doi: 10.1289/ehp.8043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Powlson DS, Addiscott TM, Benjamin N, Cassman KG, de Kok TM, van Grinsven H, L’Hirondel J-L, Avery AA, van KC. J. Environ. Qual. 2008;37:291–295. doi: 10.2134/jeq2007.0177. [DOI] [PubMed] [Google Scholar]
- 14.Knuckles TL, Buntz JG, Paffett M, Channell M, Harmon M, Cherng T, Lucas SN, McDonald JD, Kanagy NL, Campen MJ. J. Toxicol. Environ. Health, Part A. 2011;74:828–837. doi: 10.1080/15287394.2011.570225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schindler DW. Science. 1974;184:897–899. doi: 10.1126/science.184.4139.897. [DOI] [PubMed] [Google Scholar]
- 16.Holloway JM, Dahlgren RA, Hansen B, Casey WH. Nature. 1998;395:785–788. [Google Scholar]
- 17.Cotton FA, Wilkinson G, Murillo CA, Bochmann M. Advanced Inorganic Chemistry. 6th edn. New York: John Wiley & Sons, Inc; 1999. [Google Scholar]
- 18.Lybrand RA, Michalski G, Graham RC, Parker DR. Geochim. Cosmochim. Acta. 2013;104:136–147. [Google Scholar]
- 19.Liebensteiner MG, Pinkse MWH, Schaap PJ, Stams AJM, Lomans BP. Science. 2013;340:85–87. doi: 10.1126/science.1233957. [DOI] [PubMed] [Google Scholar]
- 20.Kourtev PS, Nakatsu CH, Konopka A. Appl. Environ. Microbiol. 2009;75:6249–6257. doi: 10.1128/AEM.00347-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chovanec P, Sparacino-Watkins C, Zhang N, Basu P, Stolz JF. Front. Microbiol. 2012;3:416. doi: 10.3389/fmicb.2012.00416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ganesh R, Robinson KG, Chu L, Kucsmas D, Reed GD. Water Res. 1999;33:3447–3458. [Google Scholar]
- 23.Wu Q, Sanford RA, Loffler FE. Appl. Environ. Microbiol. 2006;72:3608–3614. doi: 10.1128/AEM.72.5.3608-3614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lundberg JO, Weitzberg E, Cole JA, Benjamin N. Nat. Rev. Microbiol. 2004;2:593–602. doi: 10.1038/nrmicro929. [DOI] [PubMed] [Google Scholar]
- 25.Dennis MJ, Wilson LA. Nitrates and nitrites. Academic Press; 2003. [Google Scholar]
- 26.Environmental Protetion Agency, US EPA. 2007. [Google Scholar]
- 27.World Health Organization. Nitrate and nitrite in Drinkingwater. Geneva: 2011. [Google Scholar]
- 28.Doel JJ, Benjamin N, Hector MP, Rogers M, Allaker RP. Eur. J. Oral Sci. 2005;113:14–19. doi: 10.1111/j.1600-0722.2004.00184.x. [DOI] [PubMed] [Google Scholar]
- 29.Takahama U, Hirota S. Chem. Res. Toxicol. 2010;23:1067–1075. doi: 10.1021/tx1000464. [DOI] [PubMed] [Google Scholar]
- 30.Li H, Thompson I, Carter P, Whiteley A, Bailey M, Leifert C, Killham K. Oral Microbiol. Immunol. 2007;22:67–71. doi: 10.1111/j.1399-302X.2007.00313.x. [DOI] [PubMed] [Google Scholar]
- 31.Parham NJ, Gibson GR. FEMS Microbiol. Ecol. 2000;31:21–28. doi: 10.1111/j.1574-6941.2000.tb00667.x. [DOI] [PubMed] [Google Scholar]
- 32.Allison C, Macfarlane GT. J. Gen. Microbiol. 1988;134:1397–1405. doi: 10.1099/00221287-134-6-1397. [DOI] [PubMed] [Google Scholar]
- 33.Forsythe SJ, Dolby JM, Webster ADB, Cole JA. J. Med. Microbiol. 1988;25:253–259. doi: 10.1099/00222615-25-4-253. [DOI] [PubMed] [Google Scholar]
- 34.Knobeloch L, Salna B, Hogan A, Postle J, Anderson H. Environ. Health Perspect. 2000;108:675–678. doi: 10.1289/ehp.00108675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lorenzo FRV, Phillips JD, Nussenzveig R, Lingam B, Koul PA, Schrier SL, Prchal JT. Blood Cells, Mol., Dis. 2011;46:277–281. doi: 10.1016/j.bcmd.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Percy MJ, Aslan D. J. Clin. Pathol. 2008;61:1122–1123. doi: 10.1136/jcp.2008.058701. [DOI] [PubMed] [Google Scholar]
- 37.Ward MH. Rev. Environ. Health. 2009;24:357–363. doi: 10.1515/reveh.2009.24.4.357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward MH, Mark SD, Cantor KP, Weisenburger DD, Correa-Villaseor A, Zahn SH. Epidemiology. 1996;7:465–471. [PubMed] [Google Scholar]
- 39.Hughes R, Rowland IR. Microb. Ecol. Health Dis. 2000;12:179–185. [Google Scholar]
- 40.Griesenbeck J, Brender J, Sharkey J, Steck M, Huber J, Rene A, McDonald T, Romitti P, Canfield M, Langlois P, Suarez L. Environ. Health. 2010;9:1–17. doi: 10.1186/1476-069X-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Loh YH, Jakszyn P, Luben RN, Mulligan AA, Mitrou P, Khaw K-T. Am. J. Clin. Nutr. 2011;93:1053–1061. doi: 10.3945/ajcn.111.012377. [DOI] [PubMed] [Google Scholar]
- 42.Dejam A, Hunter CJ, Gladwin MT. N. Engl. J. Med. 2007;356:1590. doi: 10.1056/NEJMc070163. [DOI] [PubMed] [Google Scholar]
- 43.Dejam A, Hunter CJ, Schechter AN, Gladwin MT. Blood Cells, Mol., Dis. 2004;32:423–429. doi: 10.1016/j.bcmd.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 44.Gladwin MT. J. Clin. Invest. 2004;113:19–21. doi: 10.1172/JCI200420664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Alef MJ, Vallabhaneni R, Carchman E, Morris SM, Shiva S, Wang Y, Kelley EE, Tarpey MM, Gladwin MT, Tzeng E, Zuckerbraun BS. J. Clin. Invest. 2011;121:1646–1656. doi: 10.1172/JCI44079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gilchrist M, Winyard PG, Aizawa K, Anning C, Shore A, Benjamin N. Free Radical Biol. Med. 2013;60:89–97. doi: 10.1016/j.freeradbiomed.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 47.Kapil V, Haydar SMA, Pearl V, Lundberg JO, Weitzberg E, Ahluwalia A. Free Radical Biol. Med. 2013;55:93–100. doi: 10.1016/j.freeradbiomed.2012.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelly J, Fulford J, Vanhatalo A, Blackwell JR, French O, Bailey SJ, Gilchrist M, Winyard PG, Jones AM. Am. J. Physiol. 2013;304:R73–R83. doi: 10.1152/ajpregu.00406.2012. [DOI] [PubMed] [Google Scholar]
- 49.Lidder S, Webb AJ. Br. J. Clin. Pharmacol. 2013;75:677–696. doi: 10.1111/j.1365-2125.2012.04420.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zuckerbraun BS, George P, Gladwin MT. Cardiovasc. Res. 2011;89:542–552. doi: 10.1093/cvr/cvq370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Raat NJH, Noguchi AC, Liu VB, Raghavachari N, Liu D, Xu X, Shiva S, Munson PJ, Gladwin MT. Free Radical Biol. Med. 2009;47:510–517. doi: 10.1016/j.freeradbiomed.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bjorne HH, Petersson J, Phillipson M, Weitzberg E, Holm L, Lundberg JO. J. Clin. Invest. 2004;113:106–114. doi: 10.1172/JCI200419019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lundberg JO, Weitzberg E, Gladwin MT. Nat. Rev. Drug Discovery. 2008;7:156–167. doi: 10.1038/nrd2466. [DOI] [PubMed] [Google Scholar]
- 54.Gladwin MT, Raat NJH, Shiva S, Dezfulian C, Hogg N, Kim-Shapiro DB, Patel RP. Am. J. Physiol.: Heart Circ. Physiol. 2006;291:H2026–H2035. doi: 10.1152/ajpheart.00407.2006. [DOI] [PubMed] [Google Scholar]
- 55.Sparacino-Watkins CE, Lai Y-C, Gladwin MT. Circulation. 2012;125:2824–2826. doi: 10.1161/CIRCULATIONAHA.112.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gladwin MT, Shelhamer JH, Schechter AN, Pease-Fye ME, Waclawiw MA, Panza JA, Ognibene FP, Cannon RO. Proc. Natl. Acad. Sci. U. S. A. 2000;97:11482–11487. doi: 10.1073/pnas.97.21.11482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thomas DD, Ridnour LA, Isenberg JS, Flores- Santana W, Switzer CH, Donzelli S, Hussain P, Vecoli C, Paolocci N, Ambs S, Colton CA, Harris CC, Roberts DD, Wink DA. Free Radical Biol. Med. 2008;45:18–31. doi: 10.1016/j.freeradbiomed.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wink DA, Hines HB, Cheng RY, Switzer CH, Flores-Santana W, Vitek MP, Ridnour LA, Colton CA. J. Leukocyte Biol. 2011;89:873–891. doi: 10.1189/jlb.1010550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ridnour LA, Thomas DD, Switzer C, Flores-Santana W, Isenberg JS, Ambs S, Roberts DD, Wink DA. Nitric Oxide. 2008;19:73–76. doi: 10.1016/j.niox.2008.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Rusanova I, Escames G, Cossio G, de Borace RG, Moreno B, Chahboune M, Lopez LC, Diez T, Acuna-Castroviejo D. Eur. J. Haematol. 2010;85:529–537. doi: 10.1111/j.1600-0609.2010.01528.x. [DOI] [PubMed] [Google Scholar]
- 61.Aslan M, Freeman BA. Free Radical Biol. Med. 2007;43:1469–1483. doi: 10.1016/j.freeradbiomed.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Duncan C, Dougall H, Johnston P, Green S, Brogan R, Leifert C, Smith L, Golden M, Benjamin N. Nat. Med. 1995;1:546–551. doi: 10.1038/nm0695-546. [DOI] [PubMed] [Google Scholar]
- 63.Sobko T, Reinders CI, Jansson EA, Norin E, Midtvedt T, Lundberg JO. Nitric Oxide. 2005;13:272–278. doi: 10.1016/j.niox.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 64.Sobko T, Huang L, Midtvedt T, Norin E, Gustafsson LE, Norman M, Jansson EA, Lundberg JO. Free Radical Biol. Med. 2006;41:985–991. doi: 10.1016/j.freeradbiomed.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 65.Lundberg JO, Weitzberg E. Gut. 2013;62:616–629. doi: 10.1136/gutjnl-2011-301649. [DOI] [PubMed] [Google Scholar]
- 66.Gusarov I, Shatalin K, Starodubtseva M, Nudler E. Science. 2009;325:1380–1384. doi: 10.1126/science.1175439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Claus SP, Ellero SL, Berger B, Krause L, Bruttin A, Molina J, Paris A, Want EJ, de Waziers I, Cloarec O, Richards SE, Wang Y, Dumas M-E, Ross A, Rezzi S, Kochhar S, Van Bladeren P, Lindon JC, Holmes E, Nicholson JK. mBio. 2011;2:e00271–10. doi: 10.1128/mBio.00271-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.O’Hara AM, Shanahan F. EMBO Rep. 2006;7:688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gomez RF, Tannenbaum SR, Savoca J, Ralt D, Rockowitz N. Cancer. 1980;45:1066–1067. doi: 10.1002/1097-0142(19800315)45:5+<1066::aid-cncr2820451306>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 70.Khan A, Sarkar D. Microbiology. 2012;158:301–307. doi: 10.1099/mic.0.054759-0. [DOI] [PubMed] [Google Scholar]
- 71.Weber I, Fritz C, Ruttkowski S, Kreft A, Bange F-C. Mol. Microbiol. 2000;35:1017. doi: 10.1046/j.1365-2958.2000.01794.x. [DOI] [PubMed] [Google Scholar]
- 72.Palmer KL, Brown SA, Whiteley M. J. Bacteriol. 2007;189:4449–4455. doi: 10.1128/JB.00162-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Weingarten RA, Grimes JL, Olson JW. Appl. Environ. Microbiol. 2008;74:1367–1375. doi: 10.1128/AEM.02261-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Liu X, Gao B, Novik V, Galan JE. PLoS Pathog. 2012;8:e1002562. doi: 10.1371/journal.ppat.1002562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.MacMicking J, Xie Q-w, Nathan C. Annu. Rev. Immunol. 1997;15:323–350. doi: 10.1146/annurev.immunol.15.1.323. [DOI] [PubMed] [Google Scholar]
- 76.Stolz JF, Basu P. ChemBioChem. 2002;3:198–206. doi: 10.1002/1439-7633(20020301)3:2/3<198::AID-CBIC198>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 77.Li H, Samouilov A, Liu X, Zweier JL. Biochemistry. 2003;42:1150–1159. doi: 10.1021/bi026385a. [DOI] [PubMed] [Google Scholar]
- 78.Millar TM, Stevens CR, Benjamin N, Eisenthal R, Harrison R, Blake DR. FEBS Lett. 1998;427:225–228. doi: 10.1016/s0014-5793(98)00430-x. [DOI] [PubMed] [Google Scholar]
- 79.Li H, Samouilov A, Liu X, Zweier JL. J. Biol. Chem. 2001;276:24482–24489. doi: 10.1074/jbc.M011648200. [DOI] [PubMed] [Google Scholar]
- 80.Jansson EA, Huang L, Malkey R, Govoni M, Nihlen C, Olsson A, Stensdotter M, Petersson J, Holm L, Weitzberg E, Lundberg JO. Nat. Chem. Biol. 2008;4:411–417. doi: 10.1038/nchembio.92. [DOI] [PubMed] [Google Scholar]
- 81.Meyer C, Lea US, Provan F, Kaiser WM, Lillo C. Photosynth. Res. 2005;83:181–189. doi: 10.1007/s11120-004-3548-3. [DOI] [PubMed] [Google Scholar]
- 82.Maia LB, Moura JJG. JBIC, J. Biol. Inorg. Chem. 2011;16:443–460. doi: 10.1007/s00775-010-0741-z. [DOI] [PubMed] [Google Scholar]
- 83.Sparacino-Watkins C, Tejero J, Wang J, Ragireddy VP, Gauthier M, Thomas J, Bueno M, Azarov I, Lai Y-C, Sun B, Frizzell S, Basu P, Mora A, Gladwin MT. Nitric Oxide. 2013;31(S1):S45–S46. [Google Scholar]
- 84.Wang J, Krizowski S, Fischer K, Niks D, Tejero J, Wang L, Sparacino-Watkins C, Ragireddy P, Frizzell S, Kelley EE, Shiva S, Zhang Y, Basu P, Hille R, Schwarz G, Gladwin MT. Nitric Oxide. 2013;31(S1):S39–S40. [Google Scholar]
- 85.Kelley CJ, Keller CK, Evans RD, Orr CH, Smith JL, Harlow BA. Soil Biol. Biochem. 2013;57:731–738. [Google Scholar]
- 86.Karsh KL, Granger J, Kritee K, Sigman DM. Environ. Sci. Technol. 2012;46:5727–5735. doi: 10.1021/es204593q. [DOI] [PubMed] [Google Scholar]
- 87.Li Y, Katzmann E, Borg S, Schueler D. J. Bacteriol. 2012;194:4847–4856. doi: 10.1128/JB.00903-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Martinez-Espinosa RM, Dridge EJ, Bonete MJ, Butt JN, Butler CS, Sargent F, Richardson DJ. FEMS Microbiol. Lett. 2007;276:129–139. doi: 10.1111/j.1574-6968.2007.00887.x. [DOI] [PubMed] [Google Scholar]
- 89.Lowe EC, Bydder S, Hartshorne RS, Tape HL, Dridge EJ, Debieux CM, Paszkiewicz K, Singleton I, Lewis RJ, Santini JM, Richardson DJ, Butler CS. J. Biol. Chem. 2010;285:18433–18442. doi: 10.1074/jbc.M110.115873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kraft B, Strous M, Tegetmeyer HE. J. Biotechnol. 2011;155:104–117. doi: 10.1016/j.jbiotec.2010.12.025. [DOI] [PubMed] [Google Scholar]
- 91.Magalon A, Fedor JG, Walburger A, Weiner JH. Coord. Chem. Rev. 2011;255:1159–1178. [Google Scholar]
- 92.Cabello P, Roldán MD, Moreno-Vivián C. Microbiology. 2004;150:3527–3546. doi: 10.1099/mic.0.27303-0. [DOI] [PubMed] [Google Scholar]
- 93.Kern M, Simon J. Biochim. Biophys. Acta, Bioenerg. 2009;1787:646–656. doi: 10.1016/j.bbabio.2008.12.010. [DOI] [PubMed] [Google Scholar]
- 94.Sears HJ, Ferguson SJ, Richardson DJ, Spiro S. FEMS Microbiol. Lett. 1993;113:107. [Google Scholar]
- 95.Oremland RS, Blum JS, Culbertson CW, Visscher PT, Miller LG, Dowdle P, Strohmaier FE. Appl. Environ. Microbiol. 1994;60:3011–3019. doi: 10.1128/aem.60.8.3011-3019.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Jepson BJN, Mohan S, Clarke TA, Gates AJ, Cole JA, Butler CS, Butt JN, Hemmings AM, Richardson DJ. J. Biol. Chem. 2007;282:6425–6437. doi: 10.1074/jbc.M607353200. [DOI] [PubMed] [Google Scholar]
- 97.Potter L, Angove H, Richardson D, Cole J. Adv. Microb. Physiol. 2001;45:51–112. doi: 10.1016/s0065-2911(01)45002-8. [DOI] [PubMed] [Google Scholar]
- 98.Tamegai H, Ikeda E, Kato C, Horikoshi K. Biosci., Biotechnol., Biochem. 2007;71:2041–2045. doi: 10.1271/bbb.70256. [DOI] [PubMed] [Google Scholar]
- 99.Cruz-García C, Murray AE, Klappenbach JA, Stewart V, Tiedje JM. J. Bacteriol. 2007;189:656–662. doi: 10.1128/JB.01194-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Richardson DJ. Microbiology. 2000;146:551–571. doi: 10.1099/00221287-146-3-551. [DOI] [PubMed] [Google Scholar]
- 101.Potter LC, Millington P, Griffiths L, Thomas GH, Cole JA. Biochem. J. 1999;344(Pt 1):77–84. [PMC free article] [PubMed] [Google Scholar]
- 102.Potter LC, Cole JA. Biochem. J. 1999;344:69–76. doi: 10.1042/0264-6021:3440069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kern M, Simon J. Microbiology. 2009;155:2784–2794. doi: 10.1099/mic.0.029983-0. [DOI] [PubMed] [Google Scholar]
- 104.Delgado MJ, Bonnard N, Tresierra-Ayala A, Bedmar EJ, Müller P. Microbiology. 2003;149:3395–3403. doi: 10.1099/mic.0.26620-0. [DOI] [PubMed] [Google Scholar]
- 105.Kern M, Mager AM, Simon J. Microbiology. 2007;153:3739–3747. doi: 10.1099/mic.0.2007/009928-0. [DOI] [PubMed] [Google Scholar]
- 106.Cooksley C, Jenks PJ, Green A, Cockayne A, Logan RPH, Hardie KR. J. Med. Microbiol. 2003;52:461–469. doi: 10.1099/jmm.0.05070-0. [DOI] [PubMed] [Google Scholar]
- 107.Vegge CS, Brondsted L, Li Y-P, Bang DD, Ingmer H. Appl. Environ. Microbiol. 2009;75:5308–5314. doi: 10.1128/AEM.00287-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Spiro S. FEMS Microbiol. Rev. 2007;31:193–211. doi: 10.1111/j.1574-6976.2006.00061.x. [DOI] [PubMed] [Google Scholar]
- 109.Taveirne ME, Sikes ML, Olson JW. Mol. Microbiol. 2009;74:758–771. doi: 10.1111/j.1365-2958.2009.06901.x. [DOI] [PubMed] [Google Scholar]
- 110.Pittman MS, Kelly DJ. Biochem. Soc. Trans. 2005;33:190–192. doi: 10.1042/BST0330190. [DOI] [PubMed] [Google Scholar]
- 111.Basu P, Burgmayer SJN. Coord. Chem. Rev. 2011;255:1016–1038. doi: 10.1016/j.ccr.2011.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Fischer B, Enemark JH, Basu P. J. Inorg. Biochem. 1998;72:13–21. doi: 10.1016/s0162-0134(98)10054-5. [DOI] [PubMed] [Google Scholar]
- 113.Hille R. Chem. Rev. 1996;96:2757–2816. doi: 10.1021/cr950061t. [DOI] [PubMed] [Google Scholar]
- 114.Fischer K, Barbier GG, Hecht H-J, Mendel RR, Campbell WH, Schwarz G. Plant Cell. 2005;17:1167–1179. doi: 10.1105/tpc.104.029694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.George GN, Mertens JA, Campbell WH. J. Am. Chem. Soc. 1999;121:9730–9731. [Google Scholar]
- 116.Bertero MG, Rothery RA, Palak M, Hou C, Lim D, Blasco F, Weiner JH, Strynadka NCJ. Nat. Struct. Biol. 2003;10:681–687. doi: 10.1038/nsb969. [DOI] [PubMed] [Google Scholar]
- 117.Jormakka M, Richardson D, Byrne B, Iwata S. Structure. 2004;12:95–104. doi: 10.1016/j.str.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 118.Pushie MJ, George GN. Coord. Chem. Rev. 2011;255:1055–1084. [Google Scholar]
- 119.Dias J, Than M, Humm A, Bourenkov GP, Bartunik HD, Bursakov S, Calvete J, Caldeira J, Carneiro C, Moura JJ, Moura I, Romao MJ. Structure. 1999;7:65–79. doi: 10.1016/s0969-2126(99)80010-0. [DOI] [PubMed] [Google Scholar]
- 120.Najmudin S, Gonzalez PJ, Trincao J, Coelho C, Mukhopadhyay A, Cerqueira NMFSA, Romao CC, Moura I, Moura JJG, Brondino CD, Romao MJ. JBIC, J. Biol. Inorg. Chem. 2008;13:737–753. doi: 10.1007/s00775-008-0359-6. [DOI] [PubMed] [Google Scholar]
- 121.Coelho C, Gonzalez PJ, Moura JG, Moura I, Trincao J, Romao MJ. J. Mol. Biol. 2011;408:932–948. doi: 10.1016/j.jmb.2011.03.016. [DOI] [PubMed] [Google Scholar]
- 122.Kloer DP, Hagel C, Heider J, Schulz GE. Structure. 2006;14:1377–1388. doi: 10.1016/j.str.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 123.Enemark JH, Garner CD. JBIC, J. Biol. Inorg. Chem. 1997;2:817–822. [Google Scholar]
- 124.Hine FJ, Taylor AJ, Garner CD. Coord. Chem. Rev. 2010;254:1570–1579. [Google Scholar]
- 125.Schulzke C. Eur. J. Inorg. Chem. 2011:1189–1199. [Google Scholar]
- 126.Sugimoto H, Tsukube H. Chem. Soc. Rev. 2008;37:2609–2619. doi: 10.1039/b610235m. [DOI] [PubMed] [Google Scholar]
- 127.Marbella L, Serli-Mitasev B, Basu P. Angew. Chem., Int. Ed. 2009;48:3996–3998. doi: 10.1002/anie.200806297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Williams BR, Fu Y, Yap GPA, Burgmayer SJN. J. Am. Chem. Soc. 2012;134:19584–19587. doi: 10.1021/ja310018e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pimkov IV, Nigam A, Venna K, Fleming FF, Solntsev PV, Nemykin VN, Basu P. J. Heterocycl. Chem. 2013;50:879–886. doi: 10.1002/jhet.1715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Rothery RA, Stein B, Solomonson M, Kirk ML, Weiner JH. Proc. Natl. Acad. Sci. U. S. A. 2012;109:14773–14778. doi: 10.1073/pnas.1200671109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Williams RJP. Inorg. Chim. Acta, Rev. 1971;5:137–155. [Google Scholar]
- 132.Lane RH, Pantaleo NS, Farr JK, Coney WM, Newton MG. Adv. Chem. Ser. 1979;173:358–363. [Google Scholar]
- 133.Williams RJP. Eur. J. Biochem. 1995;234:363–381. doi: 10.1111/j.1432-1033.1995.363_b.x. [DOI] [PubMed] [Google Scholar]
- 134.LaCroix LB, Shadle SE, Wang Y, Averill BA, Hedman B, Hodgson KO, Solomon EI. J. Am. Chem. Soc. 1996;118:7755–7768. [Google Scholar]
- 135.Comba P. Coord. Chem. Rev. 2000:200–202. 217–245. [Google Scholar]
- 136.Chaka G, Sonnenberg JL, Schlegel HB, Heeg MJ, Jaeger G, Nelson TJ, Ochrymowycz LA, Rorabacher DB. J. Am. Chem. Soc. 2007;129:5217–5227. doi: 10.1021/ja068960u. [DOI] [PubMed] [Google Scholar]
- 137.Zong C, Wilson CJ, Shen T, Wittung-Stafshede P, Mayo SL, Wolynes PG. Proc. Natl. Acad. Sci. U. S. A. 2007;104:3159–3164. doi: 10.1073/pnas.0611149104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Solomon EI, Hadt RG. Coord. Chem. Rev. 2011;255:774–789. [Google Scholar]
- 139.Holm RH. Coord. Chem. Rev. 1990;100:183–221. [Google Scholar]
- 140.Mondal S, Basu P. Inorg. Chem. 2001;40:192–193. doi: 10.1021/ic0011866. [DOI] [PubMed] [Google Scholar]
- 141.Sengar RS, Basu P. Inorg. Chim. Acta. 2007;360:2092–2099. doi: 10.1016/j.ica.2006.10.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Basu P, Nemykin VN, Sengar RS. Inorg. Chem. 2003;42:7489–7501. doi: 10.1021/ic034821r. [DOI] [PubMed] [Google Scholar]
- 143.McNaughton RL, Mondal S, Nemykin VN, Basu P, Kirk ML. Inorg. Chem. 2005;44:8216–8222. doi: 10.1021/ic0482281. [DOI] [PubMed] [Google Scholar]
- 144.Lim BS, Holm RH. J. Am. Chem. Soc. 2001;123:1920–1930. doi: 10.1021/ja003546u. [DOI] [PubMed] [Google Scholar]
- 145.Lorber C, Donahue JP, Goddard CA, Nordlander E, Holm RH. J. Am. Chem. Soc. 1998;120:8102–8112. [Google Scholar]
- 146.Lim BS, Sung K-M, Holm RH. J. Am. Chem. Soc. 2000;122:7410–7411. [Google Scholar]
- 147.Majumdar A, Pal K, Nagarajan K, Sarkar S. Inorg. Chem. 2007;46:6136–6147. doi: 10.1021/ic7006777. [DOI] [PubMed] [Google Scholar]
- 148.Majumdar A, Pal K, Sarkar S. Dalton Trans. 2009:1927–1938. doi: 10.1039/b815436h. [DOI] [PubMed] [Google Scholar]
- 149.McNaughton RL, Lim BS, Knottenbelt SZ, Holm RH, Kirk ML. J. Am. Chem. Soc. 2008;130:4628–4636. doi: 10.1021/ja074691b. [DOI] [PubMed] [Google Scholar]
- 150.Nemykin VN, Basu P. Inorg. Chem. 2003;42:4046–4056. doi: 10.1021/ic0262942. [DOI] [PubMed] [Google Scholar]
- 151.Kail B, Nemykin VN, Davie SR, Carrano CJ, Hammes B, Basu P. Inorg. Chem. 2002;41:1281–1291. doi: 10.1021/ic011169w. [DOI] [PubMed] [Google Scholar]
- 152.Davie SR, Rubie ND, Hammes BS, Carrano CJ, Kirk ML, Basu P. Inorg. Chem. 2001;40:2632–2633. doi: 10.1021/ic0012017. [DOI] [PubMed] [Google Scholar]
- 153.Inscore FE, Knottenbelt SZ, Rubie ND, Joshi HK, Kirk ML, Enemark JH. Inorg. Chem. 2006;45:967–976. doi: 10.1021/ic0506815. [DOI] [PubMed] [Google Scholar]
- 154.Schultz BE, Hille R, Holm RH. J. Am. Chem. Soc. 1995;117:827–828. [Google Scholar]
- 155.Choi E-Y, Stockert AL, Leimkuhler S, Hille R. J. Inorg. Biochem. 2004;98:841–848. doi: 10.1016/j.jinorgbio.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 156.Hille R. Biochemistry. 1991;30:8522–8529. doi: 10.1021/bi00099a004. [DOI] [PubMed] [Google Scholar]
- 157.Kim JH, Ryan MG, Knaut H, Hille R. J. Biol. Chem. 1996;271:6771–6780. doi: 10.1074/jbc.271.12.6771. [DOI] [PubMed] [Google Scholar]
- 158.Holm RH, Solomon EI, Majumdar A, Tenderholt A. Coord. Chem. Rev. 2011;255:993–1015. [Google Scholar]
- 159.Young CG. Biomimetic Oxidations Catalyzed by Transition Metal Complexes. Imperial College Press; 2000. pp. 415–459. [Google Scholar]
- 160.McMaster J, Tunney JM, Garner CD. Prog. Inorg. Chem. 2003;52:539–583. [Google Scholar]
- 161.Enemark JH, Cooney JJA, Wang J-J, Holm RH. Chem. Rev. 2004;104:1175–1200. doi: 10.1021/cr020609d. [DOI] [PubMed] [Google Scholar]
- 162.Enemark JH, Young CG. Adv. Inorg. Chem. 1994;40:1–88. [Google Scholar]
- 163.Holm RH. Chem. Rev. 1987;87:1401–1449. [Google Scholar]
- 164.Basu P, Kail BW, Young CG. Inorg. Chem. 2010;49:4895–4900. doi: 10.1021/ic902500h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Basu P, Nemykin VN, Sengar RS. Inorg. Chem. 2009;48:6303–6313. doi: 10.1021/ic900579s. [DOI] [PubMed] [Google Scholar]
- 166.Kail BW, Perez LM, Zaric SD, Millar AJ, Young CG, Hall MB, Basu P. Chem.-Eur. J. 2006;12:7501–7509. doi: 10.1002/chem.200600269. [DOI] [PubMed] [Google Scholar]
- 167.Millar AJ, Doonan CJ, Smith PD, Nemykin VN, Basu P, Young CG. Chem.-Eur. J. 2005;11:3255–3267. doi: 10.1002/chem.200401101. [DOI] [PubMed] [Google Scholar]
- 168.Nemykin VN, Basu P. Inorg. Chem. 2005;44:7494–7502. doi: 10.1021/ic0508324. [DOI] [PubMed] [Google Scholar]
- 169.Nemykin VN, Laskin J, Basu P. J. Am. Chem. Soc. 2004;126:8604–8605. doi: 10.1021/ja049121f. [DOI] [PubMed] [Google Scholar]
- 170.Sengar RS, Nemykin VN, Basu P. J. Inorg. Biochem. 2008;102:748–756. doi: 10.1016/j.jinorgbio.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Smith PD, Millar AJ, Young CG, Ghosh A, Basu P. J. Am. Chem. Soc. 2000;122:9298–9299. [Google Scholar]
- 172.Basu P, Kail BW, Adams AK, Nemykin VN. Dalton Trans. 2013;42:3071–3081. doi: 10.1039/c2dt32349d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Nemykin VN, Davie SR, Mondal S, Rubie N, Kirk ML, Somogyi A, Basu P. J. Am. Chem. Soc. 2002;124:756–757. doi: 10.1021/ja017178l. [DOI] [PubMed] [Google Scholar]
- 174.Dutta SK, McConville DB, Youngs WJ, Chaudhury M. Inorg. Chem. 1997;36:2517–2522. [Google Scholar]
- 175.Laughlin LJ, Young CG. Inorg. Chem. 1996;35:1050–1058. doi: 10.1021/ic950815m. [DOI] [PubMed] [Google Scholar]
- 176.Subramanian P, Spence JT, Ortega R, Enemark JH. Inorg. Chem. 1984;23:2564–2572. [Google Scholar]
- 177.Sugimoto H, Tano H, Miyake H, Itoh S. Dalton Trans. 2011;40:2358–2365. doi: 10.1039/c0dt00763c. [DOI] [PubMed] [Google Scholar]
- 178.Perera E, Basu P. Dalton Trans. 2009:5023–5028. doi: 10.1039/b904113c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Nemykin VN, Olsen JG, Perera E, Basu P. Inorg. Chem. 2006;45:3557–3568. doi: 10.1021/ic051653p. [DOI] [PubMed] [Google Scholar]
- 180.Sproules S, Banerjee P, Weyhermuller T, Yan Y, Donahue JP, Wieghardt K. Inorg. Chem. 2011;50:7106–7122. doi: 10.1021/ic2006265. [DOI] [PubMed] [Google Scholar]
- 181.Sproules S, Wieghardt K. Coord. Chem. Rev. 2011;255:837–860. [Google Scholar]
- 182.Mtei RP, Perera E, Mogesa B, Stein B, Basu P, Kirk ML. Eur. J. Inorg. Chem. 2011:5467–5470. doi: 10.1002/ejic.201101084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Matz KG, Mtei RP, Rothstein R, Kirk ML, Burgmayer SJN. Inorg. Chem. 2011;50:9804–9815. doi: 10.1021/ic200783a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Burgmayer SNJ, Pearsall DL, Blaney SM, Moore EM, Sauk-Schubert C. JBIC, J. Biol. Inorg. Chem. 2004;9:59–66. doi: 10.1007/s00775-003-0496-x. [DOI] [PubMed] [Google Scholar]
- 185.Luykx DMAM, Duine JA, de Vries S. Biochemistry. 1998;37:11366–11375. doi: 10.1021/bi972972y. [DOI] [PubMed] [Google Scholar]
- 186.Arnoux P, Sabaty M, Alric J, Frangioni B, Guigliarelli B, Adriano J-M, Pignol D. Nat. Struct. Biol. 2003;10:928–934. doi: 10.1038/nsb994. [DOI] [PubMed] [Google Scholar]
- 187.Moura JJG, Brondino CD, Trincao J, Romao MJ. JBIC, J. Biol. Inorg. Chem. 2004;9:791–799. doi: 10.1007/s00775-004-0573-9. [DOI] [PubMed] [Google Scholar]
- 188.Jepson BJN, Marietout A, Mohan S, Cole JA, Butler CS, Richardson DJ. Biochem. Soc. Trans. 2006;36:122–126. doi: 10.1042/BST0340122. [DOI] [PubMed] [Google Scholar]
- 189.Schindelin H, Kisker C, Hilton J, Rajagopalan KV, Rees DC. Science. 1996;272:1615–1621. doi: 10.1126/science.272.5268.1615. [DOI] [PubMed] [Google Scholar]
- 190.Jiang J, Holm RH. Inorg. Chem. 2005;44:1068–1072. doi: 10.1021/ic040109y. [DOI] [PubMed] [Google Scholar]
- 191.Wang J-J, Tessier C, Holm RH. Inorg. Chem. 2006;45:2979–2988. doi: 10.1021/ic0521630. [DOI] [PubMed] [Google Scholar]
- 192.Sugimoto H, Tatemoto S, Suyama K, Miyake H, Mtei RP, Itoh S, Kirk ML. Inorg. Chem. 2010;49:5368–5370. doi: 10.1021/ic100825x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Donahue JP, Goldsmith CR, Nadiminti U, Holm RH. J. Am. Chem. Soc. 1998;120:12869–12881. [Google Scholar]
- 194.Majumdar A, Sarkar S. Coord. Chem. Rev. 2011;255:1039–1054. [Google Scholar]
- 195.Stiefel EI, Gardner JK. J. Less-Common Met. 1974;36:521–533. [Google Scholar]
- 196.Sugimoto H, Tarumizu M, Miyake H, Tsukube H. Eur. J. Inorg. Chem. 2006:4494–4497. [Google Scholar]
- 197.Das SK, Chaudhury PK, Biswas D, Sarkar S. J. Am. Chem. Soc. 1994;116:9061–9070. [Google Scholar]
- 198.Majumdar A, Pal K, Sarkar S. Inorg. Chem. 2008;47:3393–3401. doi: 10.1021/ic7024268. [DOI] [PubMed] [Google Scholar]
- 199.Majumdar A, Pal K, Sarkar S. J. Am. Chem. Soc. 2006;128:4196–4197. doi: 10.1021/ja0586135. [DOI] [PubMed] [Google Scholar]
- 200.Craig JA, Holm RH. J. Am. Chem. Soc. 1989;111:2111–2115. [Google Scholar]
- 201.Gonzalez PJ, Rivas MG, Mota CS, Brondino CD, Moura I, Moura JJG. Coord. Chem. Rev. 2013;257:315–331. [Google Scholar]
- 202.Hofmann M. JBIC, J. Biol. Inorg. Chem. 2009;14:1023–1035. doi: 10.1007/s00775-009-0545-1. [DOI] [PubMed] [Google Scholar]
- 203.Cerqueira NMFSA, Gonzalez PJ, Brondino CD, Romao MJ, Romao CC, Moura I, Moura JJG. J. Comput. Chem. 2009;30:2466–2484. doi: 10.1002/jcc.21280. [DOI] [PubMed] [Google Scholar]
- 204.Cotelesage JJH, Pushie MJ, Grochulski P, Pickering IJ, George GN. J. Inorg. Biochem. 2012;115:127–137. doi: 10.1016/j.jinorgbio.2012.06.019. [DOI] [PubMed] [Google Scholar]
- 205.Fourmond V, Sabaty M, Arnoux P, Bertrand P, Pignol D, Leger C. J. Phys. Chem. B. 2010;114:3341–3347. doi: 10.1021/jp911443y. [DOI] [PubMed] [Google Scholar]
- 206.Butler CS, Charnock JM, Garner CD, Thomson AJ, Ferguson SJ, Berks BC, Richardson DJ. Biochem. J. 2000;352:859–864. [PMC free article] [PubMed] [Google Scholar]
- 207.Stewart LJ, Bailey S, Bennett B, Charnock JM, Garner CD, McAlpine AS. J. Mol. Biol. 2000;299:593–600. doi: 10.1006/jmbi.2000.3702. [DOI] [PubMed] [Google Scholar]
- 208.Afshar S, Johnson E, de Vries S, Schröder I. J. Bacteriol. 2001;183:5491–5495. doi: 10.1128/JB.183.19.5491-5495.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.de Vries S, Momcilovic M, Strampraad MJF, Whitelegge JP, Baghai A, Schroeder I. Biochemistry. 2010;49:9911–9921. doi: 10.1021/bi100974v. [DOI] [PubMed] [Google Scholar]
- 210.Bursakov SA, Carneiro C, Almendra MJ, Duarte RO, Caldeira J, Moura I, Moura JJG. Biochem. Biophys. Res. Commun. 1997;239:816–822. doi: 10.1006/bbrc.1997.7560. [DOI] [PubMed] [Google Scholar]
- 211.Bursakov S, Liu M-Y, Payne WJ, LeGall J, Moura I, Moura JJG. Anaerobe. 1995;1:55–60. doi: 10.1016/s1075-9964(95)80444-7. [DOI] [PubMed] [Google Scholar]
- 212.Butler CS, Charnock JM, Bennett B, Sears HJ, Reilly AJ, Ferguson SJ, Garner CD, Lowe DJ, Thomson AJ, Berks BC, Richardson DJ. Biochemistry. 1999;38:9000–9012. doi: 10.1021/bi990402n. [DOI] [PubMed] [Google Scholar]
- 213.Dementin S, Arnoux P, Frangioni B, Grosse S, Leger C, Burlat B, Guigliarelli B, Sabaty M, Pignol D. Biochemistry. 2007;46:9713–9721. doi: 10.1021/bi700928m. [DOI] [PubMed] [Google Scholar]
- 214.Bennett B, Berks BC, Ferguson SJ, Thomson AJ, Richardson DJ. Eur. J. Biochem. 1994;226:789–798. doi: 10.1111/j.1432-1033.1994.00789.x. [DOI] [PubMed] [Google Scholar]
- 215.Reyes F, Gavira M, Castillo F, Moreno-Vivián C. Biochem. J. 1998;331:897–904. doi: 10.1042/bj3310897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Gates AJ, Kemp GL, To CY, Mann J, Marritt SJ, Mayes AG, Richardson DJ, Butt JN. Phys. Chem. Chem. Phys. 2011;13:7720–7731. doi: 10.1039/c0cp02887h. [DOI] [PubMed] [Google Scholar]
- 217.Anderson LJ, Richardson DJ, Butt JN. Biochemistry. 2001;40:11294–11307. doi: 10.1021/bi002706b. [DOI] [PubMed] [Google Scholar]
- 218.Elliott SJ, Hoke KR, Heffron K, Palak M, Rothery RA, Weiner JH, Armstrong FA. Biochemistry. 2004;43:799–807. doi: 10.1021/bi035869j. [DOI] [PubMed] [Google Scholar]
- 219.Marangon J, Paes dSPM, Moura I, Brondino CD, Moura JJG, Gonzalez PJ. Biochim. Biophys. Acta, Bioenerg. 2012;1817:1072–1082. doi: 10.1016/j.bbabio.2012.04.011. [DOI] [PubMed] [Google Scholar]
- 220.Fourmond V, Burlat B, Dementin S, Sabaty M, Arnoux P, Etienne E, Guigliarelli B, Bertrand P, Pignol D, Leger C. Biochemistry. 2010;49:2424–2432. doi: 10.1021/bi902140e. [DOI] [PubMed] [Google Scholar]
- 221.Frangioni B, Arnoux P, Sabaty M, Pignol D, Bertrand P, Guigliarelli B, Leger C. J. Am. Chem. Soc. 2004;126:1328–1329. doi: 10.1021/ja0384072. [DOI] [PubMed] [Google Scholar]
- 222.Gates AJ, Richardson DJ, Butt JN. Biochem. J. 2008;409:159–168. doi: 10.1042/BJ20071088. [DOI] [PubMed] [Google Scholar]
- 223.Doonan CJ, Nielsen DJ, Smith PD, White JM, George GN, Young CG. J. Am. Chem. Soc. 2006;128:305–316. doi: 10.1021/ja056109u. [DOI] [PubMed] [Google Scholar]
- 224.Eagle AA, Laughlin LJ, Young CG, Tiekink ERT. J. Am. Chem. Soc. 1992;114:9195–9197. [Google Scholar]
- 225.Harmer MA, Halbert TR, Pan WH, Coyle CL, Cohen SA, Stiefel EI. Polyhedron. 1986;5:341–347. [Google Scholar]
- 226.Llusar R, Polo V, Velez E, Vicent C. Inorg. Chem. 2010;49:8045–8055. doi: 10.1021/ic1010693. [DOI] [PubMed] [Google Scholar]
- 227.Thomas S, Eagle AA, Sproules SA, Hill JP, White JM, Tiekink ERT, George GN, Young CG. Inorg. Chem. 2003;42:5909–5916. doi: 10.1021/ic030095g. [DOI] [PubMed] [Google Scholar]
- 228.Wang K, McConnachie JM, Stiefel EI. Inorg. Chem. 1999;38:4334–4341. doi: 10.1021/ic980674z. [DOI] [PubMed] [Google Scholar]
- 229.McConnachie CA, Stiefel EI. Inorg. Chem. 1999;38:964–972. doi: 10.1021/ic980674z. [DOI] [PubMed] [Google Scholar]
- 230.Coucouvanis D. Adv. Inorg. Chem. 1998;45:1–73. [Google Scholar]
- 231.Mayer JM. Chemtracts. 1997;10:350–353. [Google Scholar]
- 232.Bray RC. Flavins Flavoproteins, Proc. Int. Symp. 1984 [Google Scholar]
- 233.Bray RC. Biol. Magn. Reson. 1980;2:45–84. [Google Scholar]
- 234.Bray RC, Swann JC. Struct. Bonding. 1972;11:107–144. [Google Scholar]
- 235.Bray RC, Lowe DJ. Biochem. Soc. Trans. 1997;25:762–768. doi: 10.1042/bst0250762. [DOI] [PubMed] [Google Scholar]
- 236.Enemark JH, Astashkin AV, Raitsimring AM. Dalton Trans. 2006:3501–3514. doi: 10.1039/b602919a. [DOI] [PubMed] [Google Scholar]
- 237.Enemark JH, Astashkin AV, Raitsimring AM. Biochem. Soc. Trans. 2008;36:1129–1133. doi: 10.1042/BST0361129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Enemark JH, Raitsimring AM, Astashkin AV, Klein EL. Faraday Discuss. 2011;148:249–267. doi: 10.1039/c004404k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 239.Rajapakshe A, Astashkin AV, Klein EL, Reichmann D, Mendel RR, Bittner F, Enemark JH. Biochemistry. 2011;50:8813–8822. doi: 10.1021/bi2005762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Astashkin AV, Rajapakshe A, Cornelison MJ, Johnson-Winters K, Enemark JH. J. Phys. Chem. B. 2012;116:1942–1950. doi: 10.1021/jp210578f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Pacheco A, Basu P, Borbat P, Raitsimring AM, Enemark JH. Inorg. Chem. 1996;35:7001–7008. doi: 10.1021/ic9607806. [DOI] [PubMed] [Google Scholar]
- 242.George GN, Bray RC, Morpeth FF, Boxer DH. Biochem. J. 1985;227:925–931. doi: 10.1042/bj2270925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 243.Johnson-Winters K, Tollin G, Enemark JH. Biochemistry. 2010;49:7242–7254. doi: 10.1021/bi1008485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Hille R. Biol. Magn. Reson. 2010;29:91–120. [Google Scholar]
- 245.Butler CS, Fairhurst SA, Ferguson SJ, Thomson AJ, Berks BC, Richardson DJ, Lowe DJ. Biochem. J. 2002;363:817–823. doi: 10.1042/0264-6021:3630817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Gonzalez PJ, Rivas MG, Brondino CD, Bursakov SA, Moura I, Moura JJG. JBIC, J. Biol. Inorg. Chem. 2006;11:609–616. doi: 10.1007/s00775-006-0110-0. [DOI] [PubMed] [Google Scholar]
- 247.Fourmond V, Burlat B, Dementin S, Arnoux P, Sabaty M, Boiry S, Guigliarelli B, Bertrand P, Pignol D, Leger C. J. Phys. Chem. B. 2008;112:15478–15486. doi: 10.1021/jp807092y. [DOI] [PubMed] [Google Scholar]
- 248.Biaso F, Burlat B, Guigliarelli B. Inorg. Chem. 2012;51:3409–3419. doi: 10.1021/ic201533p. [DOI] [PubMed] [Google Scholar]
- 249.Marietou A, Richardson D, Cole J, Mohan S. FEMS Microbiol. Lett. 2005;248:217–225. doi: 10.1016/j.femsle.2005.05.042. [DOI] [PubMed] [Google Scholar]
- 250.Marietou A, Griffiths L, Cole J. J. Bacteriol. 2009;191:882–889. doi: 10.1128/JB.01171-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Brigé A, Leys D, Meyer TE, Cusanovich MA, Van Beeumen JJ. Biochemistry. 2002;41:4827–4836. doi: 10.1021/bi012144b. [DOI] [PubMed] [Google Scholar]
- 252.Brigé A, Cole JA, Hagen WR, Guisez Y, Van Beeumen JJ. Biochem. J. 2001;356:851–858. doi: 10.1042/0264-6021:3560851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253.Berks BC, Ferguson SJ, Moir JWB, Richardson DJ. Biochim. Biophys. Acta, Bioenerg. 1995;1232:97–173. doi: 10.1016/0005-2728(95)00092-5. [DOI] [PubMed] [Google Scholar]
- 254.Simpson PJL, McKinzie AA, Codd R. Biochem. Biophys. Res. Commun. 2010;398:13–18. doi: 10.1016/j.bbrc.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 255.Stewart V, Lu Y, Darwin AJ. J. Bacteriol. 2002;184:1314–1323. doi: 10.1128/JB.184.5.1314-1323.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 256.Wooldridge KG, van Vlietn AHM. In: Campylobacter: molecular and cellular biology. Konkel ME, Ketley JM, editors. Wymondham, UK: Horizon Scientific Press; 2005. [Google Scholar]
- 257.Cartron ML, Roldán MD, Ferguson SJ, Berks BC, Richardson DJ. Biochem. J. 2002;368:425–432. doi: 10.1042/BJ20020865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258.Brondijk TH, Fiegen D, Richardson DJ, Cole JA. Mol. Microbiol. 2002;44:245–255. doi: 10.1046/j.1365-2958.2002.02875.x. [DOI] [PubMed] [Google Scholar]
- 259.Gross R, Eichler R, Simon J. Biochem. J. 2005;390:689–693. doi: 10.1042/BJ20050448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 260.Simon J, Gross R, Einsle O, Kroneck PMH, Kröger A, Klimmek O. Mol. Microbiol. 2000;35:686–696. doi: 10.1046/j.1365-2958.2000.01742.x. [DOI] [PubMed] [Google Scholar]
- 261.Myers C, Myers J. J. Bacteriol. 1997;179:1143–1152. doi: 10.1128/jb.179.4.1143-1152.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Brondijk THC, Nilavongse A, Filenko N, Richardson DJ, Cole JA. Biochem. J. 2004;379:47–55. doi: 10.1042/BJ20031115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 263.Arias-Cartin R, Grimaldi S, Pommier J, Lanciano P, Schaefer C, Arnoux P, Giordano G, Guigliarelli B, Magalon A. Proc. Natl. Acad. Sci. U. S. A. 2011;108:7781–7786. doi: 10.1073/pnas.1010427108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Ilbert M, Méjean V, Giudici-Orticoni M-T, Samama J-P, Iobbi-Nivol C. J. Biol. Chem. 2003;278:28787–28792. doi: 10.1074/jbc.M302730200. [DOI] [PubMed] [Google Scholar]
- 265.Hatzixanthis K, Clarke TA, Oubrie A, Richardson DJ, Turner RJ, Sargent F. Proc. Natl. Acad. Sci. U. S. A. 2005;102:8460–8465. doi: 10.1073/pnas.0500737102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Buchanan G, Maillard J, Nabuurs SB, Richardson DJ, Palmer T, Sargent F. FEBS Lett. 2008;582:3979–3984. doi: 10.1016/j.febslet.2008.10.049. [DOI] [PubMed] [Google Scholar]
- 267.Maillard J, Spronk CAEM, Buchanan G, Lyall V, Richardson DJ, Palmer T, Vuister GW, Sargent F. Proc. Natl. Acad. Sci. U. S. A. 2007;104:15641–15646. doi: 10.1073/pnas.0703967104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Sargent F. Biochem. Soc. Trans. 2007;35:835–847. doi: 10.1042/BST0350835. [DOI] [PubMed] [Google Scholar]
- 269.Shanmugham A, Bakayan A, Voller P, Grosveld J, Lill H, Bollen YJ. PLoS One. 2012;7:e34159. doi: 10.1371/journal.pone.0034159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270.Thyssen A, Grisez L, Van Houdt R, Ollevier F. Int. J. Syst. Bacteriol. 1998;48:1145–1151. doi: 10.1099/00207713-48-4-1145. [DOI] [PubMed] [Google Scholar]
- 271.Alfredsson GA, Kristjansson JK, Hjorleifsdottir S, Stetter KO. J. Gen. Microbiol. 1988;134:299–306. [Google Scholar]
- 272.Knauber T, Doss SD, Gerth K, Perlova O, Müller R, Treuner-Lange A. Mol. Microbiol. 2008;69:254–266. doi: 10.1111/j.1365-2958.2008.06285.x. [DOI] [PubMed] [Google Scholar]
- 273.Sanford RA, Cole JR, Tiedje JM. Appl. Environ. Microbiol. 2002;68:893–900. doi: 10.1128/AEM.68.2.893-900.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Nilavongse A, Brondijk THC, Overton TW, Richardson DJ, Leach ER, Cole JA. Microbiology. 2006;152:3227–3237. doi: 10.1099/mic.0.29157-0. [DOI] [PubMed] [Google Scholar]
- 275.Olmo-Mira MF, Gavira M, Richardson DJ, Castillo F, Moreno-Vivian C, Roldan MD. J. Biol. Chem. 2004;279:49727–49735. doi: 10.1074/jbc.M406502200. [DOI] [PubMed] [Google Scholar]
- 276.Pittman MS, Elvers KT, Lee L, Jones MA, Poole RK, Park SF, Kelly DJ. Mol. Microbiol. 2007;63:575–590. doi: 10.1111/j.1365-2958.2006.05532.x. [DOI] [PubMed] [Google Scholar]
- 277.Dixon R, Kahn D. Nat. Rev. Microbiol. 2004;2:621–631. doi: 10.1038/nrmicro954. [DOI] [PubMed] [Google Scholar]
- 278.Martinez-Argudo I, Little R, Shearer N, Johnson P, Dixon R. J. Bacteriol. 2004;186:601–610. doi: 10.1128/JB.186.3.601-610.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279.Schwintner C, Sabaty M, Berna B, Cahors S, Richaud P. FEMS Microbiol. Lett. 1998;165:313–321. doi: 10.1111/j.1574-6968.1998.tb13163.x. [DOI] [PubMed] [Google Scholar]
- 280.Siddiqui RA, Warnecke-Eberz U, Hengsberger A, Schneider B, Kostka S, Friedrich B. J. Bacteriol. 1993;175:5867–5876. doi: 10.1128/jb.175.18.5867-5876.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 281.Coelho C, Gonzalez PJ, Trincao J, Carvalho AL, Najmudin S, Hettman T, Dieckman S, Moura JJG, Moura I, Romao MJ. Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun. 2007;63:516–519. doi: 10.1107/S1744309107022129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Simpson PJL, Richardson DJ, Codd R. Microbiology. 2010;156:302–312. doi: 10.1099/mic.0.034421-0. [DOI] [PubMed] [Google Scholar]
- 283.Gonzalez PJ, Correia C, Moura I, Brondino CD, Moura JJG. J. Inorg. Biochem. 2006;100:1015–1023. doi: 10.1016/j.jinorgbio.2005.11.024. [DOI] [PubMed] [Google Scholar]
- 284.Stewart V. Antonie Van Leeuwenhoek. 1994;66:37–45. doi: 10.1007/BF00871631. [DOI] [PubMed] [Google Scholar]
- 285.Stewart V. Mol. Mircobiol. 1993;9:425–434. doi: 10.1111/j.1365-2958.1993.tb01704.x. [DOI] [PubMed] [Google Scholar]
- 286.Stewart V, Rabin RS. In: Two-Component Signal Transduction. James TJS, Hoch A, editors. ASM Press; 1995. [Google Scholar]
- 287.Teixidó L, Cortés P, Bigas A, Àlvarez G, Barbé J, Campo S. Int. Microbiol. 2010;13:33–39. doi: 10.2436/20.1501.01.108. [DOI] [PubMed] [Google Scholar]
- 288.Kumar A, Brar R, Wang P, Dee L, Skorupa G, Khadour F, Schulz R, Parrillo JE. Am. J. Physiol. 1999;276:R265–R276. doi: 10.1152/ajpregu.1999.276.1.R265. [DOI] [PubMed] [Google Scholar]
- 289.Fukada T, Hiscock KM, Dennis PF, Grischek T. Water Res. 2003;37:3070–3078. doi: 10.1016/S0043-1354(03)00176-3. [DOI] [PubMed] [Google Scholar]
- 290.Robles EF, Sanchez C, Bonnard N, Delgado MJ, Bedmar EJ. Biochem. Soc. Trans. 2006;34:108–110. doi: 10.1042/BST0340108. [DOI] [PubMed] [Google Scholar]
- 291.Mesa S, Hauser F, Friberg M, Malaguti E, Fischer H-M, Hennecke H. J. Bacteriol. 2008;190:12. doi: 10.1128/JB.00748-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292.Sears HJ, Sawers G, Berks BC, Ferguson SJ, Richardson DJ. Microbiology. 2000;146(Pt 11):2977–2985. doi: 10.1099/00221287-146-11-2977. [DOI] [PubMed] [Google Scholar]
- 293.Gavira M, Roldan MD, Castillo F, Moreno-Vivian C. J. Bacteriol. 2002;184:1693–1702. doi: 10.1128/JB.184.6.1693-1702.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Ellington MJ, Bhakoo KK, Sawers G, Richardson DJ, Ferguson SJ. J. Bacteriol. 2002;184:4767–4774. doi: 10.1128/JB.184.17.4767-4774.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Ellington MJ, Fosdike WL, Sawers RG, Richardson DJ, Ferguson SJ. Arch. Microbiol. 2006;184:298–304. doi: 10.1007/s00203-005-0044-x. [DOI] [PubMed] [Google Scholar]
- 296.Stewart V, Bledsoe PJ, Chen L-L, Cai A. J. Bacteriol. 2009;191:996–1005. doi: 10.1128/JB.00873-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297.Dong Y, Wang J, Fu H, Zhou G, Shi M, Gao H. PLoS One. 2012;7:e51643. doi: 10.1371/journal.pone.0051643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Giel JL, Rodionov D, Liu M, Blattner FR, Kiley PJ. Mol. Microbiol. 2006;60:1058–1075. doi: 10.1111/j.1365-2958.2006.05160.x. [DOI] [PubMed] [Google Scholar]
- 299.Anderson LA, McNairn E, Leubke T, Pau RN, Boxer DH. J. Bacteriol. 2000;182:7035–7043. doi: 10.1128/jb.182.24.7035-7043.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300.Mouncey NJ, Mitchenall LA, Pau RN. Microbiology. 1996;142:1997–2004. doi: 10.1099/13500872-142-8-1997. [DOI] [PubMed] [Google Scholar]
- 301.Stewart V. Biochem. Soc. Trans. 2003;31:1–10. doi: 10.1042/bst0310001. [DOI] [PubMed] [Google Scholar]
- 302.McNicholas PM, Gunsalus RP. J. Bacteriol. 2002;184:3253–3259. doi: 10.1128/JB.184.12.3253-3259.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303.Palyada K, Threadgill D, Stintzi A. J. Bacteriol. 2004;186:4714–4729. doi: 10.1128/JB.186.14.4714-4729.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Holmes K, Mulholland F, Pearson BM, Pin C, McNicholl-Kennedy J, Ketley JM, Wells JM. Microbiology. 2005;151:243–257. doi: 10.1099/mic.0.27412-0. [DOI] [PubMed] [Google Scholar]
- 305.Sampathkumar B, Napper S, Carrillo CD, Willson P, Taboada E, Nash JHE, Potter AA, Babiuk LA, Allan BJ. Microbiology. 2006;152:567–577. doi: 10.1099/mic.0.28405-0. [DOI] [PubMed] [Google Scholar]
- 306.Woodall CA, Jones AK, Barrow PA, Hinds J, Marsden GL, Kelly D, Dorrell N, Wren BW, Maskell DJ. Infect. Immun. 2005;73:5278–5285. doi: 10.1128/IAI.73.8.5278-5285.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Stintzi A. J. Bacteriol. 2003;185:2009–2016. doi: 10.1128/JB.185.6.2009-2016.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 308.Glass JB, Wolfe-Simon F, Anbar AD. Geobiology. 2009;7:100–123. doi: 10.1111/j.1472-4669.2009.00190.x. [DOI] [PubMed] [Google Scholar]
- 309.Chen Y, Wang F, Xu J, Mehmood MA, Xiao X. ISME J. 2011;5:843–855. doi: 10.1038/ismej.2010.182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Hartsock A, Shapleigh JP. J. Bacteriol. 2011;193:6483–6489. doi: 10.1128/JB.05324-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311.Miller WG, Parker CT, Health S, Lastovica AJ. BMC Microbiol. 2007;7:11. doi: 10.1186/1471-2180-7-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Gao H, Yang ZK, Barua S, Reed SB, Romine MF, Nealson KH, Fredrickson JK, Tiedje JM, Zhou J. ISME J. 2009;3:966–976. doi: 10.1038/ismej.2009.40. [DOI] [PubMed] [Google Scholar]
- 313.Thauer RK, Jungermann K, Decker K. Bacteriol. Rev. 1977;41:100–180. doi: 10.1128/br.41.1.100-180.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Simon J. FEMS Microbiol. Rev. 2002;26:285–309. doi: 10.1111/j.1574-6976.2002.tb00616.x. [DOI] [PubMed] [Google Scholar]
- 315.Karlsson R, Karlsson A, Baeckman O, Johansson BR, Hulth S. FEMS Microbiol. Lett. 2009;297:87–94. doi: 10.1111/j.1574-6968.2009.01677.x. [DOI] [PubMed] [Google Scholar]
- 316.Strous M, Pelletier E, Mangenot S, Rattei T, Lehner A, Taylor MW, Horn M, Daims H, Bartol-Mavel D, Wincker P, Barbe V, Fonknechten N, Vallenet D, Segurens B, Schenowitz-Truong C, Medigue C, Collingro A, Snel B, Dutilh BE, Op den Camp HJ, van der Drift C, Cirpus I, van de Pas-Schoonen KT, Harhangi HR, van Niftrik L, Schmid M, Keltjens J, van de Vossenberg J, Kartal B, Meier H, Frishman D, Huynen MA, Mewes HW, Weissenbach J, Jetten MS, Wagner M, Le Paslier D. Nature. 2006;440:790–794. doi: 10.1038/nature04647. [DOI] [PubMed] [Google Scholar]
- 317.Barton LL. Structural and Functional Relationships in Prokaryotes. Springer; 2004. [Google Scholar]
- 318.Luecker S, Wagner M, Maixner F, Pelletier E, Koch H, Vacherie B, Rattei T, Damst JSS, Spieck E, Le PD, Daims H. Proc. Natl. Acad. Sci. U. S. A. 2010;107:13479–13484. doi: 10.1073/pnas.1003860107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 319.Maloy JT. In: Standard Potentials in Aqueous Solution. Bard AJ, Parsons R, Jordan J, editors. New York: Mercel Dekker; 1985. pp. 127–139. [Google Scholar]
- 320.Poole R. Biochem. Soc. Trans. 2005;33:176–180. doi: 10.1042/BST0330176. [DOI] [PubMed] [Google Scholar]
- 321.De Groote MA, Fang FC. Clin. Infect. Dis. 1995;21:S162–S165. doi: 10.1093/clinids/21.supplement_2.s162. [DOI] [PubMed] [Google Scholar]
- 322.Poock SR, Leach ER, Moir JW, Cole JA, Richardson DJ. J. Biol. Chem. 2002;277:23664–23669. doi: 10.1074/jbc.M200731200. [DOI] [PubMed] [Google Scholar]
- 323.Dykhuizen R, Fraser A, McKenzie H, Golden M, Leifert C, Benjamin N. Gut. 1998;42:334–337. doi: 10.1136/gut.42.3.334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 324.Zumft WG. Arch. Microbiol. 1993;160:253–264. doi: 10.1007/BF00292074. [DOI] [PubMed] [Google Scholar]
- 325.Overton TW, Whitehead R, Li Y, Snyder LA, Saunders NJ, Smith H, Cole JA. J. Biol. Chem. 2006;281:33115–33126. doi: 10.1074/jbc.M607056200. [DOI] [PubMed] [Google Scholar]
- 326.Van Alst NE, Picardo KF, Iglewski BH, Haidaris CG. Infect. Immun. 2007;75:3780–3790. doi: 10.1128/IAI.00201-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Mussini T, Longhi P. The Halogens: Chlorine. New York: Marcel Dekker, Inc; 1985. [Google Scholar]
- 328.Wolterink AFWM. Wageningen University, PhD thesis. 2004. [Google Scholar]
- 329.Zhdanov SI. In: Standard Potentials in Aquesous Solution. Bard AJ, Parsons R, Jordan J, editors. New York: Marcel Dekker; 1985. pp. 93–125. [Google Scholar]
- 330.Kletzin A, Adams MW. FEMS Microbiol. Rev. 1996;18:5–63. doi: 10.1016/0168-6445(95)00025-9. [DOI] [PubMed] [Google Scholar]
- 331.Santhanam KSV, Sundaresan NS. In: Standard Potentials in Aqueous Solutions. Bard AJ, Parsons R, Jordan J, editors. New York: Mercel Dekker; 1985. pp. 162–172. [Google Scholar]
- 332.Pilato RS, Stiefel EI. In: Bioinorganic Catalysis. Reedijk J, Bouwman E, editors. New York: Marcel Dekker; 1999. pp. 81–152. [Google Scholar]
- 333.Castell CH. J. Fish. Res. Board Can. 1950;7:567–575. [Google Scholar]
- 334.Galus Z. In: Standard Potentials in Aqueous Solution. Bard AJ, Parsons R, Jordan J, editors. New York: Mercel Dekker; 1985. pp. 189–235. [Google Scholar]
- 335.Cheetham NWH. Introducing Biological Energetics. New York: Oxford University Press; 2011. [Google Scholar]
- 336.Brett CMA, Brett AMO. Electrochemistry Principles, Methods, and Applications. Oxford University Press; 1996. [Google Scholar]
- 337.Mason JR, Cammack R. Annu. Rev. Microbiol. 1992;46:277–305. doi: 10.1146/annurev.mi.46.100192.001425. [DOI] [PubMed] [Google Scholar]
- 338.Taoka A, Yoshimatsu K, Kanemori M, Fukumori Y. Can. J. Microbiol. 2003;49:197–206. doi: 10.1139/w03-028. [DOI] [PubMed] [Google Scholar]
- 339.Bertrand P, Frangioni B, Dementin S, Sabaty M, Arnoux P, Guigliarelli B, Pignol D, Leger C. J. Phys. Chem. B. 2007;111:10300–10311. doi: 10.1021/jp074340j. [DOI] [PubMed] [Google Scholar]
- 340.Fourmond V, Lautier T, Baffert C, Leroux F, Liebgott P-P, Dementin S, Rousset M, Arnoux P, Pignol D, Meynial-Salles I, Soucaille P, Bertrand P, Leger C. Anal. Chem. 2009;81:2962–2968. doi: 10.1021/ac8025702. [DOI] [PubMed] [Google Scholar]
- 341.Gates AJ, Hughes RO, Sharp SR, Millington PD, Nilavongse A, Cole JA, Leach E-R, Jepson B, Richardson DJ, Butler CS. FEMS Microbiol. Lett. 2003;220:261–269. doi: 10.1016/S0378-1097(03)00122-8. [DOI] [PubMed] [Google Scholar]
- 342.Berks BC, Richardson DJ, Robinson,M. P. Reilly C, Aplin RT, Ferguson SJ. Eur. J. Biochem. 1994;220:117–124. doi: 10.1111/j.1432-1033.1994.tb18605.x. [DOI] [PubMed] [Google Scholar]
- 343.Sadana JC, McElroy WD. Arch. Biochem. Biophys. 1957;67:16–34. doi: 10.1016/0003-9861(57)90242-4. [DOI] [PubMed] [Google Scholar]
- 344.Coelho C, Marangon J, Rodrigues D, Moura JJG, Romao MJ, Paes de Sousa PM, Correia dos Santos MM. J. Electroanal. Chem. 2013;693:105–113. [Google Scholar]