Abstract
Poxviruses are widespread pathogens, which display extremely different host ranges. Whereas some poxviruses, including variola virus, display narrow host ranges, others such as cowpox viruses naturally infect a wide range of mammals. The molecular basis for differences in host range are poorly understood but apparently depend on the successful manipulation of the host antiviral response. Some poxvirus genes have been shown to confer host tropism in experimental settings and are thus called host range factors. Identified host range genes include vaccinia virus K1L, K3L, E3L, B5R, C7L and SPI-1, cowpox virus CP77/CHOhr, ectromelia virus p28 and 022, and myxoma virus T2, T4, T5, 11L, 13L, 062R and 063R. These genes encode for ankyrin repeat-containing proteins, tumor necrosis factor receptor II homologs, apoptosis inhibitor T4-related proteins, Bcl-2-related proteins, pyrin domain-containing proteins, cellular serine protease inhibitors (serpins), short complement-like repeats containing proteins, KilA-N/RING domain-containing proteins, as well as inhibitors of the double-stranded RNA-activated protein kinase PKR. We conducted a systematic survey for the presence of known host range genes and closely related family members in poxvirus genomes, classified them into subgroups based on their phylogenetic relationship and correlated their presence with the poxvirus phylogeny. Common themes in the evolution of poxvirus host range genes are lineage-specific duplications and multiple independent inactivation events. Our analyses yield new insights into the evolution of poxvirus host range genes. Implications of our findings for poxvirus host range and virulence are discussed.
Keywords: Poxvirus, Host range genes, Virus evolution, Host–pathogen interactions
1. Introduction
Poxviruses are widespread pathogens, which as a family infect a wide range of animals and have been responsible for mass mortalities of animals and humans (Fenner, 2000). They include variola virus (VARV; abbreviations are shown in Table 1), the causative agent of smallpox, which is believed to have caused more fatalities throughout human history than all other infectious diseases combined (McFadden, 2005), and vaccinia virus, which has been successfully used as a vaccine in the eradication of smallpox, one of the biggest achievements in the history of medicine (Fenner, 2000). Despite this success, poxviruses continue to be constant threats for animals, including humans. In fact more than 10 poxviruses are known to infect humans, including monkeypox virus (MPXV), an emerging zoonotic disease, causing high fatality rates (Essbauer et al., 2010; Lewis-Jones, 2004). Strikingly, poxviruses display extremely different host ranges. Whereas some poxviruses such as VARV and camelpox virus (CMLV) have very narrow host ranges, others such as MPXV and cowpox virus (CPXV) naturally infect a broad spectrum of vertebrates (Fenner, 2000; McFadden, 2005).
Table 1.
Names and abbreviations of poxviruses used in this study.
| Name | Abbreviation | Genus |
|---|---|---|
| Amsacta moorei entomopoxvirus | AMV | Entomopoxvirus B |
| Melanoplus sanguinipes entomopoxvirus | MSV | Unassigned |
| Canarypox virus | CNPV | Avipoxvirus |
| Fowlpox virus | FPV | Avipoxvirus |
| Crocodilepox virus | CRV | Unclassified |
| Molluscum contagiosum virus subtype 1 | MOCV | Molluscipoxvirus |
| Bovine papular stomatitis virus | BPSV | Parapoxvirus |
| Orf virus | ORFV | Parapoxvirus |
| Camelpox virus | CMLV | Orthopoxvirus |
| Cowpox virus – Brighton Red | CPXV-BR | Orthopoxvirus |
| Cowpox virus – GRI-90 | CPXV-GRI | Orthopoxvirus |
| Ectromelia virus | ECTV | Orthopoxvirus |
| Horsepox virus | HSPV | Orthopoxvirus |
| Monkeypox virus | MPXV | Orthopoxvirus |
| Rabbitpox virus | RPXV | Orthopoxvirus |
| Taterapox virus | TATV | Orthopoxvirus |
| Vaccinia virus | VACV | Orthopoxvirus |
| Variola virus | VARV | Orthopoxvirus |
| Yaba-like disease virus | YLDV | Yatapoxvirus |
| Yaba monkey tumor virus | YMTV | Yatapoxvirus |
| Tanapox virus | TPV | Yatapoxvirus |
| Myxoma virus | MYXV | Leporipoxvirus |
| Rabbit fibroma virus | RFV | Leporipoxvirus |
| Deerpox virus W-848-83 | DPV-W83 | Cervidpoxvirus |
| Deerpox virus W-1170-84 | DPV-W84 | Cervidpoxvirus |
| Goatpox virus | GTPV | Capripoxvirus |
| Lumpy skin disease virus | LSDV | Capripoxvirus |
| Sheeppox virus | SPPV | Capripoxvirus |
| Swinepox virus | SWPV | Suipoxvirus |
Poxviruses are DNA viruses that exclusively replicate in the cytoplasm of infected cells (Lefkowitz et al., 2006). They are grouped into two subfamilies: Entomopoxvirinae, which infect insects, and Chordopoxvirinae, which infect vertebrates. Currently, nine different genera of Chordopoxvirinae are recognized: avipoxviruses, molluscipoxviruses, parapoxviruses, orthopoxviruses, yatapoxviruses, leporipoxviruses, capripoxviruses, cervidpoxviruses and suipoxviruses. In the past, classification and grouping into different poxvirus genera and species has relied on pathological findings and host range as well as structural and antigenic properties (Fenner, 2000; Lefkowitz et al., 2006). Recent advances in the genomic characterization of poxviruses lay the groundwork for more precise phylogenetic characterization (Bratke and McLysaght, 2008; Lefkowitz et al., 2006; McLysaght et al., 2003). The complete genomes of about 25–29 different poxvirus species, depending on species definition, have been determined to date. For some species multiple strains were sequenced (http:// www.poxvirus.org, Lefkowitz et al., 2005). Poxvirus genomes possess large genomes, ranging from 130 to 365 kb and often contain more than 200 genes (Lefkowitz et al., 2006). A core of 33 gene families are found as single copy genes in all sequenced genomes, and have been used to reconstruct the poxvirus phylogeny (Bratke and McLysaght, 2008). Approximately 90 genes are commonly found in Chordopoxvirinae, with the remaining genes being unique to some lineages or representing lineage-specific gene duplications (Lefkowitz et al., 2006).
Poxviruses are excellent models for studying genome evolution. They display a relatively low rate of accumulation of point mutations (Li et al., 2007), whereas gene duplications, losses, gain by horizontal gene transfer (HGT) and recombination between different species occur frequently (Bratke and McLysaght, 2008; Hughes and Friedman, 2005; McLysaght et al., 2003). These events are important for adapting to their hosts and for subverting the host antiviral response. The terminal regions of poxvirus genomes are especially prone to recombination, important for the assimilation of new genes, and contain identical inverted terminal repeats (ITR), which range in size between 0.1 and 13 kb (Lefkowitz et al., 2006).
Poxviruses contain many genes that are non-essential for viral replication in cell culture but important for modulating and circumventing the host-response and thus influence the course of poxvirus infection and pathology. Such genes are designated as virulence genes (McFadden, 2005). A portion of these genes has been identified that are important for viral replication in only a subset of tissue culture cells, which were derived from different tissues or animal species and are commonly referred to as host range genes or factors and are thought to be responsible for poxvirus-specific differences in tropism and host range (McFadden, 2005; Werden et al., 2008). Unlike many other vertebrate viruses, poxviruses do not rely on specific receptors to enter cells, but harness molecules that are ubiquitously present in many different cell-types in many animal species. Whether a virus can replicate after entry into a cell, depends on the successful manipulation of the cellular antiviral response (McFadden, 2005). Approximately 12 different host range genes or gene families have been identified to-date (reviewed in Werden et al., 2008).
Although some progress has been made in recent years in understanding poxvirus host range, the molecular basis for it is only poorly understood. We conducted systematic analyses on the presence or absence of known poxvirus host range genes. We found evidence for lineage-specific gene inactivation, deletions, duplications and recombination events. Furthermore, we analyzed the evolution and phylogeny of an extended family of host range genes, which often included known virulence factors that have not yet been identified as host range genes. The data presented here fill an important gap in our understanding and knowledge of poxvirus host range, have important implications for disease control and identify threats that might emerge through the assimilation of novel host range factors.
2. Materials and methods
2.1. Construction of poxvirus phylogenetic trees
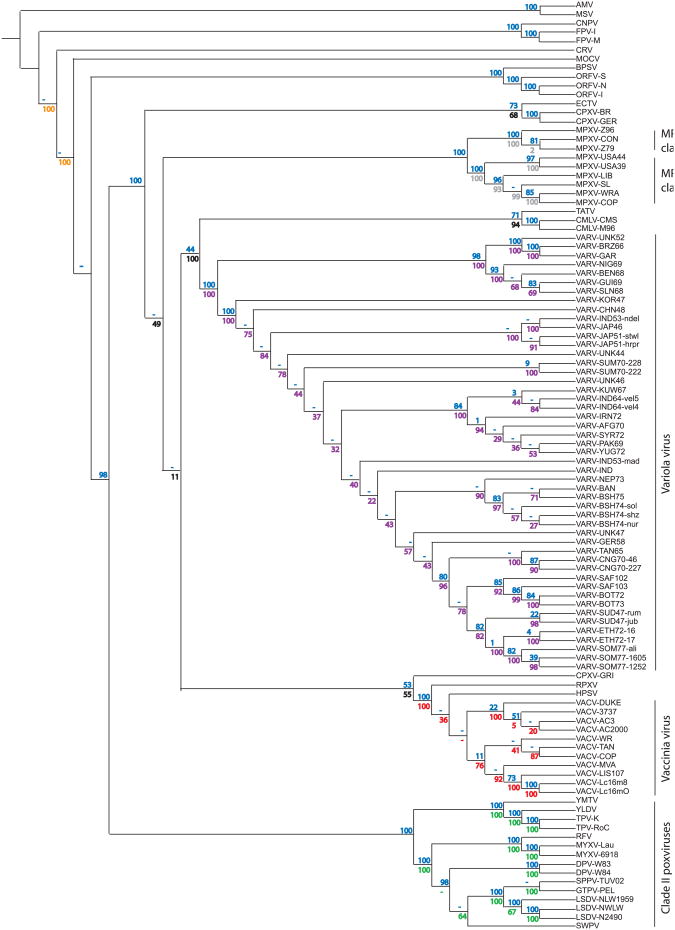
We used a concatenated sequence alignment containing protein sequences of 28 genes (Supplementary Table 1) that are present in single copy (sc) in 104 completely sequenced poxviruses that were used in this study and performed phylogenetic analyses. ClustalW (Thompson et al., 1994) was used to infer a neighbor-joining (NJ) phylogenetic tree based on this alignment. While it provides the backbone for the topology presented in Fig. 1, the large number of genomes and small number of families used to determine their relationship leave some branches where the bootstrap values are low or where the inferred topology is in conflict with the accepted poxvirus phylogeny. We have used concatenated alignments of larger numbers of protein families present in sc in subsets of the 104 genomes to resolve these branches. The tree shown in Fig. 1 is therefore a composite tree. Different colored bootstraps are derived from different subset trees as follows.
Fig. 1.
Phylogenetic tree of completely sequenced poxviruses. A neighbor-joining tree based on 28 core protein families forms the backbone for this tree (blue bootstraps). Concatenated alignments of larger numbers of protein families present in single copy in subsets of the 104 genomes are used to resolve poorly supported branches. Bootstrap values in different colors correspond to these sub-trees. Orange: 36 families in CRV, MOCV, BPSV, FPV-I, AMV. Green: 95 families in clade II. Black: 83 families in orthopoxviruses. Grey: 128 families in MPXV. Purple: 114 families in VARV. Red: 100 families in VACV, HSPV, RPXV. Schematic tree topology is shown and branch lengths are not drawn to scale. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
The inferred monophyly of molluscum contagiosum virus (MOCV) and crocodilepox virus (CRV) is an artifact according to the author of the CRV genome paper (Afonso et al., 2006). When using 36 sc families present in CRV, MOCV, BPSV, FPV-I and AMV to draw a neighbor-joining tree, the topology observed is that in Fig. 1, represented by orange bootstrap values.
The relationship of genera in clade II poxviruses was clarified using a maximum-likelihood (ML) tree based on 95 families from all clade II genomes plus orthopoxvirus outgroup, and this corresponds to the green bootstraps in Fig. 1.
Black bootstraps are derived from a NJ tree of orthopoxvirus representatives plus outgroup based on 83 shared sc gene families.
In agreement with previous analyses containing fewer MPXV genomes (Likos et al., 2005), MPXVs form two clades, which we term clade I (includes MXPV strains Z96, CON, Z79) and clade II (includes strains SL, LIB, WRA, COP, USA39 and USA44) strains. As there was weak bootstrap support for a branch within clade II MPXV, the within-genus topology was resolved with a NJ tree of MPXV plus outgroup based on 128 sc families, and the bootstrap values are noted in gray in Fig. 1.
The 49 VARV isolates that are fully sequenced and were analyzed by (Esposito et al., 2006) are difficult to separate in the poxvirus tree due to their high sequence similarity. A ML tree based on 114 sc families present in all VARV genomes, CMLV, Taterapox virus (TATV) and the ECTV outgroup agrees to a large extent with the topology favored by (Esposito et al., 2006). A NJ tree of the same genomes is presented by purple bootstraps in Fig. 1.
The poorly-supported topology of 11 VACV isolates and closely related genomes horsepox (HSPV) and rabbitpox (RPXV) is resolved by a NJ tree of 100 families present in sc in these genomes and the closest outgroup CPXV-GRI-90. This tree is the source of the red bootstrap values.
All ML trees were inferred with PhyML (Guindon and Gascuel, 2003) using the JTT model of nucleotide substitution.
2.2. Database searches for identifying homologs of poxvirus host range genes
Database searches for poxvirus host range gene homologs were performed using Basic Local Alignments Search Tools (BLAST) (Altschul et al., 1997) (http://BLAST.ncbi.nlm.nih.gov/BLAST.cgi). Protein or nucleotide sequences of previously identified host range genes were used as probes for BLASTP, Position-Specific Iterated (PSI)-BLAST and BLASTN searches. In some cases, searches were limited to poxviridae or specific poxviral species. If no predicted protein ortholog was identified for a given virus species, TBLASTN searches (Gertz et al., 2006) were performed and hits were analyzed for the presence of gene inactivating mutations by comparing nucleotide sequences of probe and target genes.
Protein domains were identified using the conserved domain database (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi).
2.3. Multiple sequence alignments and phylogenetic analyses
Multiple sequence alignments were performed with nucleotide or protein sequences using ClustalX (Thompson et al., 1997), MUSCLE (Edgar, 2004) or MAFFT (Katoh et al., 2002). Alignments for subsequent analyses were chosen after visual inspection using MacClade (Sinauer Associates), taking published data about functional residues and domain architecture of proteins into account. Poorly-aligned and highly divergent parts of multiple sequence alignments were removed using GBlocks (Talavera and Castresana, 2007). Phylogenetic analyses were carried out using the NJ method with nodal support assessed via bootstrapping (1000 replicates) as implemented in PAUP (Swofford, 2002).
Genomic sequences surrounding CPXV-BR and CPXV-GRI ORF203 were compared using PipMaker (http://pipmaker.bx.psu. edu/pipmaker).
3. Results and discussion
3.1. Poxvirus phylogeny
We used sequences of 104 completely sequenced poxvirus genomes to construct the poxvirus phylogeny. To obtain a phylogenetic tree we used a concatenated sequence alignment containing protein sequences of 28 genes that are present in all completely sequenced poxviruses and performed phylogenetic analyses and refined the analyses with additional sequences of poxviruses clades as indicated in materials and methods. The basic phylogeny, as shown in Fig. 1, is comparable to one published previously (Bratke and McLysaght, 2008).
Yatapox-, leporipox-, cervidpox-, capripox- and suipoxviruses formed a sisterclade to orthopoxviruses, which can be referred to as clade II poxviruses (Hughes and Friedman, 2005). MPXV formed two clades, in agreement with previous analyses containing fewer MPXV genomes (Chen et al., 2005; Likos et al., 2005), which we term clade I (includes MPXV Congo Basin strains Z96, Z79 and CON) and clade II (includes West African-derived strains SL, LIB, WRA, COP, USA39 and USA44).
Three completely sequenced CPXV genomes were available for analyses. Whereas CPXV-BR and GER were nested with ECTV, CPXV-GRI formed a clade with RPXV, HSPV and VACV strains, supporting the notion that CPXV-BR and CPXV-GRI represent two different orthopoxvirus species (Gubser et al., 2004; Lefkowitz et al., 2006).
RPXV, HSPV and VACVs formed a monophyletic clade indicating a close relationship as previously described (Tulman et al., 2006). While the branch point between RPXV and HSPV shows only low statistical support, the branches separating the former with all VACV strains showed high bootstrap support. The positioning of RPXV and HSPV as sister clades to the VACV clades indicates that they represent distinct species that shared a common ancestor.
3.2. K3L family
Many poxviruses possess proteins – called K3 in VACV (encoded by K3L) – that are homologous to the S1 domain of the α subunit of eukaryotic translation initiation factor 2 (eIF2). K3 acts as a pseudo-substrate and competitive inhibitor of the antiviral protein kinase PKR by binding to the activated, autophosphorylated form of PKR (Carroll et al., 1993; Dar and Sicheri, 2002; Rothenburg et al., 2011). K3L-deleted VACV (ΔK3L) showed impaired replication in mouse L929 cells, especially after interferon treatment (Beattie et al., 1991). Replication of VACVΔK3L was also severely impaired in hamster BHK cells, whereas it replicated well in human HeLa (Langland and Jacobs, 2002) and rabbit RK13 cells (Shors et al.,1998).
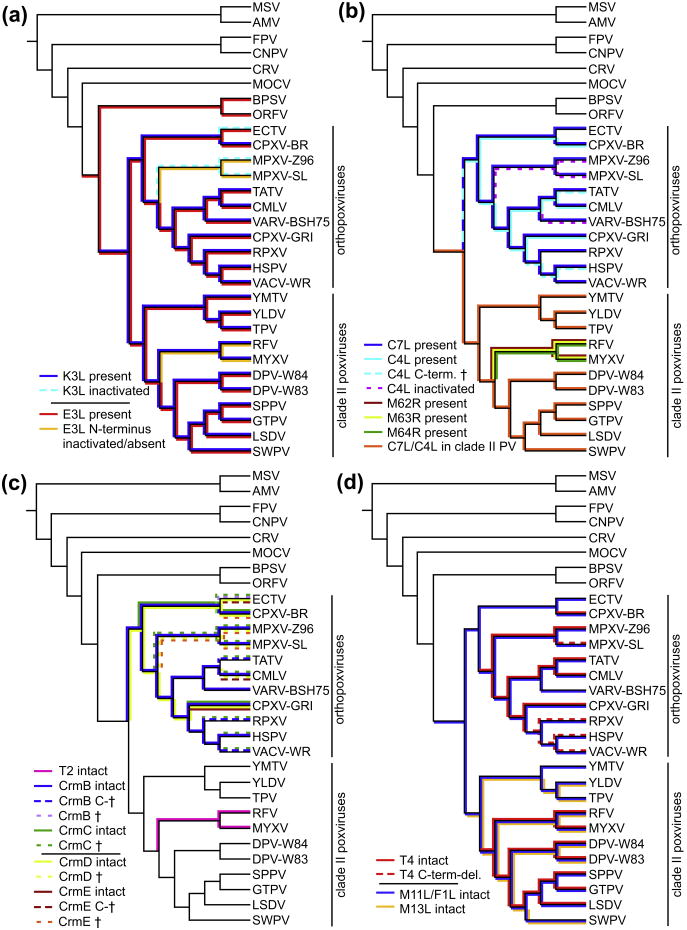
Putative functional K3L orthologs were identified in all clade II poxviruses and orthopoxviruses, except for ectromelia virus (ECTV) and MPXV, in which the open reading frames are interrupted by premature stop codons (Fig. 2a and Supplementary Fig. S1a). In ECTV an insertion of a T at nt position 94, relative to the predicted start codon, leads to the generation of a stop codon (not shown). In all MPXV strains, a stop codon at nt position 130–132 terminates the open reading frame (not shown). Remarkably, a 25 bp deletion is found in both ECTV and MPXV clade I strains at the identical position near the 3′ end of the ORF (Supplementary Fig. S1a and S1b). This would lead to a deletion comprising amino acids YID of the KGYID motif, which is essential for inhibition of PKR (Kawagishi-Kobayashi et al., 1997), and a frame shift. A further 3 bp deletion adjacent to the larger deletion is found in MPXV type II strains. Taking the identical deletion into account, it seems most likely that the K3L orthologs in MPXV and ECTV arose from a common ancestor that acquired this deletion, which was probably non-functional, and that the premature stop codons in ECTV and MPXV arose independently as secondary mutations. Since ECTV and MPXV are found in different clades in all phylogenetic analyses, the presence of the identical deletion in both viruses is suggestive of an intergenomic recombination event. This notion was further supported by phylogenetic analyses using multiple sequence alignments of the K3L genes, which showed that ECTV and MPXV sequences clustered together (not shown). Restriction of K3L orthologs to orthopoxviruses and type II poxviruses indicates that K3L evolved in a common ancestor of those. In addition to VACV K3L, orthologs of VARV and SWPV were shown to inhibit PKR activity (Elde et al., 2009; Kawagishi-Kobayashi et al., 2000; Rothenburg et al., 2009; Seo et al., 2008).
Fig. 2.
Presence of selected poxvirus host range genes projected on trees showing phylogenetic relationships of representative poxvirus strains. (a) Presence of VACV K3L orthologs is indicated above the branches (dark blue for deduced functional genes, dashed light blue lines represent inactivated genes). VACV E3L orthologs are shown below the branches in red (full length genes) and yellow (N-terminus inactivated). (b) Presence of genes of the C7L/M63R family is indicated. Solid blue lines above the branches represent C7L orthologs in orthopoxviruses, C4L orthologs are shown below branches as solid light blue lines (intact ORFs), dashed light blue lines (C-terminus inactivated) or dashed purple lines (ORFs inactivated). C7L/C4L orthologs in clade II poxviruses are shown above the branches, except for leporipoxviruses, for which three homologs (M62R, M63R and M64R) are present. (c) Presence of Tumor necrosis factor receptor II homologs is shown. CrmB orthologs are shown in solid blue lines (intact genes), dashed blue lines (C-terminus inactivated) or dashed light purple lines (ORF inactivated). CrmC orthologs are shown as solid (intact genes) or dashed green lines (ORFs inactivated). CrmD orthologs are represented by solid (intact genes) or dashed yellow lines (ORFs inactivated). CrmE orthologs are shown in solid red lines (intact genes), dashed red lines (C-terminus inactivated) or dashed organge lines (ORF inactivated). Purple lines indicate presence of T4. (d) Occurrence of T4, M11L/F1L and M13L families is shown. Presence of T4 genes is indicated by solid (intact genes) and dashed (C-terminus deleted) red lines. Blue and yellow lines indicate presence of M11L/F1L and M13L orthologs, respectively. Schematic tree topologies are shown and branch lengths are not drawn to scale. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A likely explanation for why K3L is important for virus replication in mouse but not human cells is the recent finding that mouse PKR is much more sensitive to inhibition by VACV K3 than human PKR. Interestingly, mouse and human PKR showed comparable, moderate sensitivity to inhibition by the SWPV K3 ortholog C8 (Rothenburg et al., 2009). Those results indicate that differences in both a cellular protein (PKR), as well as a viral protein (K3 ortholog), could be responsible for distinct poxviral host ranges.
The likely inactivation of a K3L ortholog in the ancestral ECTV and MPXV sequence does not necessarily mean that it was not important for virulence in the respective host(s). In fact, the deduced sequence of this postulated K3L ancestor differs from VACV-WR K3L, which was a very effective inhibitor of mouse PKR, in only two amino acid positions (K22 and F36 in VACV K3L to N and S, respectively).
3.3. E3L family
VACV E3L encodes a protein that contains an N-terminal Z-DNA binding (Zα) domain and a C-terminal double-stranded RNA-binding domain (dsRBD). It has been identified as a host range factor, because its deletion led to severe replication defects in HeLa, Vero and to a lesser extent in L929 cells but not in chicken embryonic fibroblasts, BHK and RK13 cells (Beattie et al., 1996; Langland and Jacobs, 2002). E3 is a well-established inhibitor of PKR, likely by preventing dsRNA-induced PKR activation (reviewed in Langland et al., 2006; Toth et al., 2006). Furthermore, E3 can block activity of other dsRNA-binding proteins including the dsRNA-activated editing enzyme ADAR1 (Liu et al., 2001) and 2′-5′-oligoadenylate synthetase (Rivas et al., 1998). Currently, it is not clear which targets are responsible for the observed host range effects of E3L and what the molecular mechanism for that is. In HeLa cells, it appears that PKR is a main target of E3, because PKR knock-down rescued replication of an E3L deleted VACV (Zhang et al., 2008).
We identified orthologs of E3L containing Zα and dsRBD in all sequenced parapoxviruses, orthopoxviruses and clade II poxviruses, with the exception of MPXV, myxoma virus (MYXV) and rabbit (Shope) fibroma virus (RFV), in which only predicted functional dsRBD were found (Fig. 2a and Table 2). In all MPXV strains, the start codon found in all orthopoxviruses is mutated from ATG to ATT, a triplet that could in principle serve as a translation initiation codon, albeit with lower efficiency (Peabody, 1989). Additionally, there are two 2 bp deletions at nt positions 50/51 and 72/73, relative to the start codon found in VACV E3L, which would lead to frame shifts if the translational start site were functional (Supplementary Fig. S2). The first ATG is found within the Zα domain and is in frame. The ORF is predicted to encode a protein without a functional Zα domain. In MYXV and RFV, no traces of Zα domains can be found.
Table 2.
Summary of presence and absence of host range genes in poxvirus genomes.
| GENE FAMILIES | Serpins | C7L family | B5R- related |
ANK/F-box | TNFR II family | ||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
|
|
|
|
|
|||||||||||||||||||
| HOST RANGE FACTORS | K3L | E3L | SPI-1 | SPI-2 | SPI-3 | p28-like | CL7 | C4L | M62R | M63R | M64R | T4 | B5R | VCP/C3L | M13L | M11L/F1L | K1L | T2 | CrmB | CrmC | CrmD | CrmE | |
| Amsacta moorei EPV | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Melanoplus sanguinipes EPV | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Crocodilepox virus | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Canarypox virus | – | – | [ 4 ] | 2 | – | – | – | – | – | – | – | – | – | – | 35+1 | – | – | – | – | – | – | ||
| Fowlpox virus | – | – | [ 4 ] | 2 | – | – | – | – | – | – | – | – | – | – | 19 | – | – | – | – | – | – | ||
| Molluscum contagiosum virus | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – | – |
| Bovine papular stomatitis virus | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 7 | – | – | – | – | – | – |
| Orf virus NZ2 | – | 1 | – | – | – | – | – | – | – | – | – | – | – | – | – | – | 5 | – | – | – | – | – | – |
| Ectromelia virus Moscow | † | 1 | 1 | 1 | 1 | 1 | 1 | – | – | – | – | – | 1 | 1 | – | 1 | 4+1 | 1 | – | † | † | 1+1 | c† |
| Cowpox virus Brighton Red | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | – | – | – | 1 | 1 | 1 | – | 1 | 10+2 | 1 | – | 1+1 | 1 | 1 | † |
| Monkeypox virus Sierra Leone | † | 1N† | 1 | 1 | 1 | 1 | 1 | 1† | – | – | – | 1C† | 1 | – | – | 1 | 6+1 | 1 | – | 1+1 | † | † | † |
| Monkeypox virus Zaire 96 | † | 1N† | 1 | 1 | 1 | 1 | 1 | 1† | – | – | – | 1 | 1 | 1C† | – | 1 | 6+1 | 1 | – | 1+1 | † | † | † |
| Camelpox virus M-96 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | – | – | – | 1 | 1 | 1 | – | 1 | 4+1 | † | – | 1+1 | † | † | C† |
| Taterapox virus | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1C† | – | – | – | 1 | 1 | 1C† | – | 1 | 3+1 | † | – | C† | † | – | – |
| Variola major virus BSH75 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1† | – | – | – | – | 1 | 1 | – | 1 | 4+1 | † | – | 1 | – | – | – |
| Cowpox virus GRI-90 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | – | – | – | 1 | 1 | 1 | – | 1 | 10+2 | 1 | – | 1+1 | 1 | 1 | 1 |
| Rabbitpox virus | 1 | 1 | 1 | 1 | 1 | 1 | 1 | – | – | – | – | † | 1 | 1 | – | 1 | 4 | 1 | – | 1C† | † | – | – |
| Horsepox virus | 1 | 1 | 1 | 1 | 1 | 1C† | 1 | 1C† | – | – | – | 1C† | 1 | 1 | – | 1 | 6+1 | 1 | – | 1+1 | † | – | – |
| Vaccinia virus WR | 1 | 1 | 1 | 1 | 1 | 2C† | 1 | – | – | – | – | 1C† | 1 | 1 | – | 1 | 3 | 1 | – | 1C† | † | – | – |
| Yaba monkey tumor virus | 1 | 1 | [ 1 ] | – | 1 | – | – | [ 1 ] | – | [ 1 ] | – | 1 | 4 | – | – | – | – | – | – | ||||
| Yaba-like disease virus | 1 | 1 | [ 1 ] | – | 1 | – | – | [ 1 ] | – | [ 1 ] | – | 1 | 4 | – | – | – | – | – | – | ||||
| Tanapox virus | 1 | 1 | [ 1 ] | 1 | 1 | – | – | [ 1 ] | – | [ 1 ] | – | 1 | 4 | – | – | – | – | – | – | ||||
| Myxoma virus | 1 | 1N† | [ 2 ] | 1+1 | 1 | – | – | [ 1 ] | 1+1 | [ 1 ] | – | 1 | 4+1 | – | 1 | – | – | – | – | ||||
| Rabbit fibroma virus | 1+1 | 1N† | [ 1 ] | – | 1 | – | – | [ 1 ] | 1+1 | [ 1 ] | – | 1 | 3+1 | – | 1 | – | – | – | – | ||||
| Deerpox virus W-848-83 | 1 | 1 | [ 2+1 ] | – | 1 | – | – | [ 1 ] | 1+1 | [ 1 ] | – | 1 | 5 | – | – | – | – | – | – | ||||
| Deerpox virus W-1170-84 | 1 | 1 | [ 2+1 ] | – | 1 | – | – | [ 1 ] | 1+1 | [ 1 ] | – | 1 | 5 | – | – | – | – | – | – | ||||
| Sheeppox virus TU-V02127 | 1 | 1 | [ 1 ] | – | 1 | – | – | [ 1 ] | 1+1 | [ 1 ] | – | 1 | 4 | – | – | – | – | – | – | ||||
| Goatpox virus Pellor | 1 | 1 | [ 1 ] | – | 1 | – | – | [ 1 ] | 1+1 | [ 1 ] | – | 1 | 4 | – | – | – | – | – | – | ||||
| Lumpy skin disease virus | 1 | 1 | [ 1 ] | – | 1 | – | – | [ 1 ] | 1+1 | [ 1 ] | – | 1 | 4 | – | – | – | – | – | – | ||||
| Swinepox virus | 1 | 1 | [ 1 ] | – | 1 | – | – | [ 1 ] | – | [ 1 ] | 1 | 1 | 4 | – | – | – | – | – | – | ||||
Numbers indicate quantities of closely related genes based on our database searches and phylogenetic analyses. Brackets [ ] indicate relatedness of (a) gene(s) to closely related genes that evolved after gene duplications.
indicates deletions or stop codons;
N = amino-terminal; C = carboxy-terminal. Superscripts indicate additional identical copies in the inverted terminal repeat region (ITR).
E3L apparently evolved in a virus ancestral to parapoxviruses, orthopoxviruses and clade II poxviruses. The Zα domain probably got lost early in the evolution of leporipoxviruses. The finding that the Zα domain in MPXV is inactivated by mutations in the start-codon and small deletions, but is still present at the genomic level indicates that its inactivation might have occurred more recently than the presumptive Zα loss in leporipoxviruses.
The Zα domain of VACV E3L acts as a virulence factor. It is dispensable for VACV replication in HeLa cells (Chang et al., 1995), but is required for full pathogenesis in a mouse model, in which wild type VACV was approximately 1000 fold more lethal than a Zα domain deleted virus (VACV ΔZα) (Brandt and Jacobs, 2001). Interestingly, after intranasal infection of mice, VACV ΔZα replicated normally in nasal mucosa but failed to efficiently spread to lungs and brain. Neurovirulence was also severely reduced after intracranial infection (Brandt et al., 2005). Remarkably, neurovirulence of VACV was well correlated with the ability of E3 to bind left-handed Z-DNA. Virulence was restored when the Zα domain of E3 was substituted for Zα of ADAR1 and ZBP1, whereas single point mutations that abolished Z-DNA binding dramatically decreased virulence (Kim et al., 2003). These experiments indicate that inactivation of Zα domains in MPXV, MYXV and RFV might also result in reduced virulence and altered tissue tropism in vivo. Interestingly, SWPV and MYXV E3L orthologs were found to functionally complement E3L in HeLa cells, whereas orthologs from sheeppox (SPPV) and yaba monkey tumor virus (YMTV) did not or only poorly complemented E3L, respectively, which correlated well with the ability of the E3 orthologs to inhibit PKR (Myskiw et al., 2011). This finding indicates that virus-specific differences in E3L orthologs might contribute to differences in poxvirus host range.
3.4. The C7L/M063R family
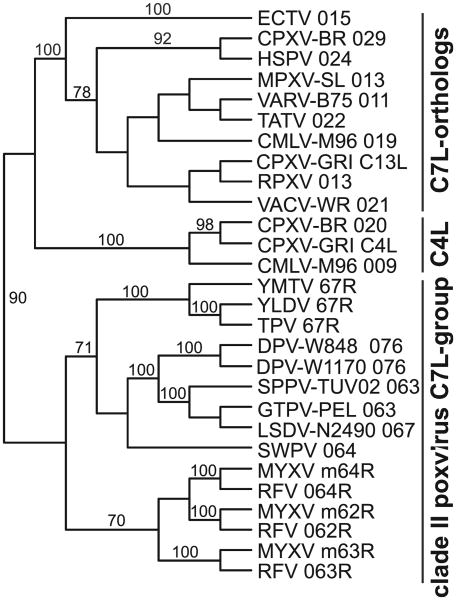
Vaccinia virus C7L encodes a protein with a predicted molecular mass of 18 kDa. Homologs of C7L are only found in poxviruses and it does not seem to be related to any known non-poxviral gene. Some orthopoxviruses possess a second C7L-related gene, called C4L in CPXV-GRI and 020 in CPXV-BR. Clade II poxviruses possess one or three C7L related genes (Table 2).
C7L was identified as a host range factor by its ability to restore viral replication of a VACV-WR strain that had K1L inactivated in human MRC-5 and porcine LLC-PK1 cells but not in rabbit RK13 cells (Perkus et al., 1990). Three tandemly arranged C7L homologs are found in MYXV: M062R, M063R and M064R. Deletion of MYXV 063R led to severe replication defects in various rabbit cell lines, whereas replication in other cell lines was not (monkey BGMK and BSC-40 cells) or only moderately (human HOS and murine 3T3 cells) impaired (Barrett et al., 2007). A MYXV 062R deficient virus was replication competent in BSC-40 cells, but showed replication defects in BGMK, RK13, RL-5 and in 18 of 21 tested human cell lines (Liu et al., 2011). The molecular mechanism by which C7L acts as a virulence- and host range factor is currently unknown.
Our analyses show that C7L/M63R-related genes are only present in orthopoxviruses and clade II poxviruses (Fig. 2b and Table 2). All orthopoxviruses possess a direct C7L ortholog, whereas full length C4L-orthologs are only found in CPXV-GRI, CPXV-BR (020) and CMLV (009). In all sequenced VARV strains the putative start-codon of the C4L/020 ortholog is mutated from ATG to TTG. Even if translation was initiated at this codon efficiently, a 1 bp deletion at nt position 139, in 42 out of 49 strains (clade I), including VARV-BSH75, would lead to a frame shift resulting in premature termination of the ORF (Fig. 2b). Three (BEN68, GUI69 and SLN68) of the other 7 strains which all form VARV clade II in the phylogenetic tree harbor a 1 bp insertion, that would lead to a frame shift resulting in premature termination. The presence of three out of frame start codons following the above mentioned putative start codon would make the translation of a downstream ORF, which corresponds to amino acids 48–170 of CPXV-GRI C4L and which is annotated as ORF2 in VARV-BSH75, unlikely if translation was not initiated at the TTG codon.
A similar situation is found in all MPXV strains, in which the start-codon of the C4L-ortholog is mutated from ATG to TTG. Several insertions and deletions (indels) lead to frame shifts resulting in premature stop-codons. Moreover several short out of frame ORFs are present in the 5′ region. In clade II MPXV strains only the first 389 nt are present, whereas complete C4L orthologs (514 bp) are found in clade I MPXV strains. A TATV virus C4L-ortholog contains an intact start codon but has a two bp deletion at position 313 relative to the putative start codon leading to a frame-shift and premature termination codon. A comparable situation is found for HSPV in which a one bp deletion leads to the disruption and premature termination of the ORF. No traces of C4L-orthologs are found in any VACV, RPXV and ECTV strains. Since C4L orthologs are found in closely related viruses, it appears likely that C4L orthologs got lost in those viruses in multiple independent events.
In yatapoxviruses, capripoxviruses, DPV and SWPV one copy of a C7L-homolog is present, while three copies are found in leporipoxviruses. Phylogenetic analysis indicates that the 3 C7L-related genes M062R, M063R and M064R arose during leporipoxvirus evolution from two duplication events (Fig. 3). Another duplication event during orthopoxvirus evolution led to the emergence of the C7L and C4L/020 clades, the latter of which got lost or inactivated in some viruses. Inclusion of C-terminally deleted or inactivated C4L/020 orthologs in phylogenetic analyses showed clustering within the C4L clade (not shown).
Fig. 3.
Phylogenetic relationship of C7L/M063R family genes. The phylogenetic relationship between C7L/M063R homologs is shown. A mid-point rooted tree was generated from a multiple protein sequence alignment of the indicated sequences using the neighbor-joining method. Bootstrap support of ≥70 is indicated above the branches.
Interestingly, MYXV 062R and YLDV 67R were able to rescue VACV replication of a strain that had both C7L and K1L deleted in mouse 3T3 and human HeLa and A431 cells, whereas MYXV 063R, 064R and CPXV-BR 020 failed to rescue VACV replication (Meng et al., 2008). In a related study, SPPV 063 rescued VACV replication in HeLa cells but not in mouse 3T3 and LA-4 cells. Substitution of amino acid residues 134 and 135 with residues found in VACV C7L and other homologs rescued virus replication in mouse cells (Meng et al., 2012).
To date only VACV C7L, SPPV 063 and MYXV M063R and M062R have been identified as host range factors. However, it is possible that the other homologs also contribute to host range in settings not yet tested.
3.5. Tumor necrosis factor receptor II homologs – the T2/Crm family
Some poxviruses contain homologs of cellular receptors for tumor necrosis factor (TNFR), which play important roles in the antiviral response and inflammatory processes. RFV and MYXV T2 proteins were the first identified viral TNFR homologs (Smith et al., 1990; Smith et al., 1991; Upton et al., 1991). T2 are secreted proteins that contain 4 N-terminal cysteine rich domains (CRDs), which show greatest sequence identity with cellular TNFR-2 and confer TNF binding, and a C-terminal domain (CTD), which is unique to poxviruses and is involved in efficient T2 secretion (Schreiber and McFadden, 1996). In addition to binding and neutralizing TNF, an intracellular form of T2 can block apoptosis independently of its TNF binding abilities. This activity is mediated by the pre-ligand assembly domain (PLAD), which is also found in TNFRs and is located at the N-terminus and is partly overlapping with the first CRD (Sedger et al., 2006). A T2-deleted MYXV strain displayed strongly decreased virulence in European rabbits and showed a replication defect in rabbit RL-5 but not in RK13 cells.
Multiple copies of T2 homologs are found in some orthopoxviruses, e.g. in CPXV, in which they are called cytokine response modifiers (Crm) B (Hu et al., 1994), C (Smith et al., 1996), D (Loparev et al., 1998) and E (Saraiva and Alcami, 2001). CrmB and CrmD contain both CRDs and CTDs, whereas CrmC and CrmE contain only the N-terminal CRDs. The CTD of VARV CrmB has been shown to confer binding and inactivation of several chemokines independent of the CRDs (Alejo et al., 2006).
We extended the analysis described in the aforementioned papers for the presence of Crm B-E genes in poxviruses (Fig. 2c and Table 2). Full-length CrmB orthologs are encoded by CPXV-GRI, CPXV-BR, CMLV, VARV, HSPV and MPXV. In all of these viruses except VARV, two identical copies are found in the ITR. Interestingly, in RPXV and VACV strains at least five identical deletions, accounting for 96 bp in total, disrupt the ORFs, which show 99% sequence identity to HSPV CrmB without taking the deletions into account. The deletions result in premature stop codons leading to predicted 86 amino acids (aa) C-terminally truncated proteins. This indicates that HSPV CrmB is ancestral to the RPXV and VACV orthologs and that deletions arose in a common ancestor of the latter. In VACV Ankara and derivative strains the first 287 bp are deleted and in VACV TAN the first 540 bp relative to HSPV CrmB are absent. In TATV a 2 bp deletion at nt position 285 relative to the predicted translational start site, in the otherwise completely present CrmB ortholog, leads to a frame shift incorporating a premature stop codon and results in a predicted C-terminally truncated protein. The truncated CrmB orthologs of VACV, RPXV and TATV contain only CRD1 and part of CRD2. Since truncation mutants of MYXV T2 that lack CRD3 and CDR4 were unable to bind TNF (Xu et al., 2000), the naturally occurring C-terminal deleted proteins are also predicted to be incapable of binding TNF. In all ECTV strains only an 89 bp fragment of a CrmB ortholog was identified. This fragment displays approximately 90% sequence identity to other orthopoxvirus CrmB genes.
Full length CrmD orthologs are only found in CPXV-GRI, CPXV-BR and ECTV, the latter of which contains two identical copies in the ITR. In CPXV-GER, which is closely related to CPXV-BR, a 4 bp deletion at nt position 203 leads to a frame shift resulting in premature termination. In all MPXV strains the first 528 bp are missing. Due to the absence of in-frame start codons and the presence of deletions the remaining fragment is not predicted to encode for a protein. In CMLV the first 728 bp are deleted. CrmD orthologs are absent in VARV, TATV, HSPV and RPXV and most VACV strains. In VACV-Lister derived strains a 90 bp fragment showing 97% sequence identity to the 3′ end of CPXV CrmD is found.
Full-length CrmC orthologs are present in CPXV-GRI, CPXV-BR and VACV strains DUKE, 3737, AC3 and AC2000. In the other VACV strains as well as in RPXV and HSPV the CrmC ORFs are disrupted by short and partly independent deletions. In ECTV the CrmC ortholog is disrupted by two 1 bp deletions and one 1 bp insertion. Multiple independent small indels also destroy the ORF in all MPXV, TATV and CMLV strains. It is striking that the gene-inactivating mutations apparently arose independently in ECTV, MPXV, TATV, CMLV and the VACV/RPXV/HSPV complex. No trace of a CrmC ortholog was found in VARV.
CrmE has been described in CPXV-GRI (Saraiva and Alcami, 2001). Orthologs displaying 99% sequence identity are also found in CPXV-GER, VACV-LIS, Vac-Lc16m8, VACV-Lc16mO and VACV-USSR strains, but are absent in all other VACV strains as well as in HSPV, RPXV, VARV and TATV. C-terminal truncated CrmE orthologs are found in CMLV, in which a 2 bp deletion at nt position 206 leads to a premature stop codon at nt position 208, and in ECTV, where in addition to a 24 bp “in-frame” deletion at nt position 74, a 26 bp deletion at nt position 220 leads to premature termination at nt position 244 resulting in a predicted 81 aa protein (Supplementary Fig. S3). In MPXV clade I strains a 2 bp insertion at nt position 56 leads to a premature stop codon at nt position 61. A 209 nt ORF, annotated as K3L, would only be translated if there were leaky scanning of the first start codon or a transcriptional start 3′ of the first start codon (Supplementary Fig. S3). A 26 bp deletion at nt position 244 leads to premature termination of this ORF at nt position 270. Such a protein would lack the leader peptide. In MPXV clade II strains an additional 1 bp deletion at nt position 124 would destroy the aforementioned ORF (not shown). The CPXV-BR CrmE ortholog contains a stop-codon at nt position 25, a 1 bp insertion at position 170 and a 26 bp deletion at nt position 237 (Supplementary Fig. S3). In CPXV-BR the complete 3′ region corresponding nt 369–504 is missing from the genome. Another CPXV-BR CrmE-like gene is found in BLAST searches (accession no.: AJ272007). This gene is 100% identical to ECTV CrmE and absent in the CPXV-BR genome. We therefore suspect that AJ272007 is the result of an annotation error or PCR artifact. Multiple sequence alignment of the different CrmE orthologs shows that the 26 bp deletions in MPXV, ECTV and CPXV-BR are found at the identical position (Supplementary Fig. S3). Thus, it is likely that this deletion arose in a CrmE gene that was ancestral to those found in these viruses, either in a common ancestor or as a result of recombination events. It is striking that CPXV-GER, which is closely related to CPXV-BR, contains an intact CrmE ortholog. Moreover VACV strains derived from VACV-Lis, but not the other closely related VACV, HSPV and RPXV strains, carry CrmE orthologs. These patterns are suggestive of inter-species recombination events.
Another TNFR-related gene has been identified in CPXV. This protein, which is most closely related to cellular CD30, can bind the CD30 ligand CD153 and inhibit the ligand from binding CD30 (Panus et al., 2002). CD30 homologs are found in CPXV-BR (015), CPXV-GRI (D13L), ECTV (009), HSPV (010) and DPV-W84 (005), but not in other poxviruses including DPV-W83 in which the largest part of this gene is deleted. An additional TNFR-related protein is found in CNPV (ORF086) (Tulman et al., 2004), which appears to be most closely related to TNFR superfamily member 11b. Additionally we identified genes encoding putative proteins that lack the CRDs and display only homology to the CrmB/D CTD in CPXV-BR (014), CPXV-GRI (D12L), ECTV (008) and DPV (016). Interestingly, in orthopoxviruses the stop codon of the CD30 homolog CPXV-BR 015 overlaps with the start codon of the neighboring CrmB/D-related gene, CPXV-BR 014 (Supplementary Fig. S4). A 1 bp deletion or 2 bp insertion between CPXV ORFs 015 (vCD30) and 014, or a frame shift during translation, would create a fusion protein. It is possible that CPXV-015 and 014 resulted from a mutation of an ancestral gene that combined both domains. The presence of a CPXV-014 ortholog in the distantly related DPV (016), which is found at a different location than vCD30 (005), indicates a function for this ORF.
We performed phylogenetic analyses with different fragments from vTNFR homologs to determine the relationship between these genes. Analysis using the complete ORF showed that T2 formed a sister clade to the CrmB and CrmD groups (Supplementary Fig. S5a). The same topology was also observed when the N-terminus was analyzed and flounder (Po) TNFR2 was used as outgroup (Supplementary Fig. S5b). In this setting, the vTNFR family was a sister clade to human and mouse TNFR2. Inclusion of more vertebrate TNFR2 sequences and the more distantly related TNFR1 sequences confirmed the previous finding that the vTNFR might have evolved after horizontal gene transfer (HGT) from an early mammalian TNFR2 (Supplementary Fig. S5c). Inclusion of CrmE and CrmC genes showed CrmC as a sister clade to mammalian TNFR2 and the other vTNFRs, the latter of which appeared to be monophyletic, with CrmE being a sister clade to the T2, CrmB and CrmD groups (Supplementary Fig. S5d). Taken together, the analyses confirm the previous notion that poxviral TNFR2s probably evolved from a cellular TNFR2 (Smith et al., 1990). Phylogenetic analysis of the CTDs of CrmB, CrmD and T2 homologs with the CTDs found among the predicted proteins showed that the former are more closely related to one another. T2 formed a sister clade to CrmB and CrmD, which can thus be considered as orthologs of T2 and are likely to have evolved by a gene duplication early in ortho-poxvirus evolution.
Interestingly, MYXV T2 was shown to specifically inhibit the activity of rabbit, but not human or mouse, TNF (Schreiber and McFadden, 1994). Moreover, orthopoxvirus TNFR2 homologs have been reported to display both virus and species-specific differences in binding and inactivating TNF and TNF-induced cell death, respectively (Alejo et al., 2006; Gileva et al., 2006). Thus, they represent good candidates for being host range factors. It is noteworthy that CPXVs, which show the greatest host range among orthopoxviruses, also possess the most intact TNFR2 homologs, i.e. 4 in CPXV-GRI and 3 in CPXV-BR (CrmB, CrmC and CrmD) and CPXV-GER (CrmB, CrmC and CrmE).
3.6. Apoptosis inhibitor T4 family
T4 is an endoplasmic reticulum-localized apoptosis inhibitor that has been best characterized in MYXV. T4 encodes a 25 kDa protein, which shows no apparent sequence homology with any non-poxvirus protein (Barry et al., 1997; Hnatiuk et al., 1999). A T4 deleted MYXV replicated normally in RK13 cells, but showed a replication defect in rabbit RL5 and peripheral blood lymphocytes due to the induction of apoptosis. Moreover, the T4-deleted virus showed decreased virulence in European rabbits, in which it produced fewer secondary lesions and showed a reduced replication rate (Barry et al., 1997). The molecular mechanism of how T4 prevents apoptosis in infected cells is unknown. In a screen for attenuated CPXV-BR on the chorioallantoic membrane of chicken embryos, a recombination event in the T4 ortholog (ORF203) (Pickup et al., 1984), led to a truncated protein and could thus contribute, along with a larger deletion, to attenuation.
T4 orthologs were identified in all leporipoxviruses, capripoxviruses and DPV, which all contain two identical copies in the ITR, but are absent in yatapoxviruses and SWPV. Intact T4 orthologs are also found in the following orthopoxviruses as single copy genes: CPXV-BR (203), CMLV (188), TATV (195), CPXV-GRI (B8R) and all clade I MPXV strains (B10R) (Table 2 and Fig. 2d). In clade II MPXV strains LIB, SIE, WRA and COP a two bp deletion at position 218/219, relative to the start codon, results in a premature stop codon and disruption of the ORF leaving only the first 72 aa intact. In the USA39 and USA44 strains a large deletion covering bp 142 to 622 is found. This confirms and extends a previous analysis (Likos et al., 2005). T4 orthologs of CPXV-BR, CMLV and TATV are very similar, showing 95-96% sequence identity on the protein level with one another, whereas sequence identity between the former group and CPXV-GRI and MPXV, which are 97% identical to each other, is only around 65%. Because the two groups do not form phylogenetically linked clades, this striking finding suggests recombination or gene transfer events. Sequence comparison of the genomic region surrounding ORF203/B8R (T4) between CPXV-BR and CPXV-GRI revealed that ORF203 is more divergent than the surrounding ORFs 201, 202, 204 and 205, which show much higher sequence identity levels (Supplementary Fig. S6).
No traces of T4 orthologs were found in all sequenced ECTV and VARV strains. In HSPV, RPXV and VACV strains only the first 206 nucleotides of the 663 bp ORF, with respect to CPXV-GRI, are present. A 1579 bp deletion truncates the T4 ortholog and also affects the neighboring gene (WR192 in VACV-WR); however, the retained sequence displays 97% sequence identity with CPXV-GRI B8R. In addition to the truncation RPXV contains a 1 nt deletion at position 15, which is predicted to result in premature translation termination. This mutation likely arose after the split of RPXV, HSPV and VACV from a common ancestor. T4 orthologs are absent in all other poxviruses. A T4-like gene probably evolved in a common ancestor of orthopoxviruses and clade II poxviruses, and subsequently got lost in yatapoxviruses, SWPV, ECTV and VARV. Alternatively, gene transfer events between poxviruses, like those evident in ortho-poxviruses, could explain the observed distribution patterns.
3.7. The Bcl-2-related M11L/F1L family
Many poxviruses possess mitochondria-localized anti-apoptotic proteins, encoded by 11L in MYXV (Everett et al., 2000) and F1L in VACV (Wasilenko et al., 2003), which block apoptosis by sequestering apoptosis mediators Bax and Bak (Su et al., 2006; Taylor et al., 2006; Wang et al., 2004; Wasilenko et al., 2005; Postigo et al., 2006). M11L and F1L encoded proteins are structurally related to cellular prosurvival Bcl-2 family proteins and adopt a very similar protein fold as BclXl, despite little if any primary sequence similarity (Douglas et al., 2007; Kvansakul et al., 2007, 2008).
Replication of an 11L-deficient MYXV was normal in RK13 cells, but impaired in rabbit splenocytes, and showed a markedly reduced virulence in rabbits coupled with an increased inflammatory response (Opgenorth et al., 1992).
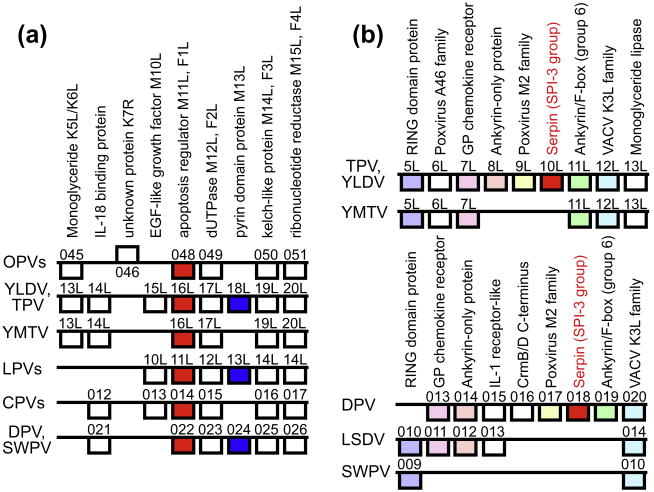
Using PSI-BLAST, MYXV 11L orthologs were identified as single copy genes in all clade II poxviruses but not in other poxviruses (Table 2 and Fig. 2d). Sequence identity on the protein level between the M11L orthologs of different clade II poxvirus genera is between 20% and 35%, which is remarkably low. PSI-BLAST searches using the M11L DPV ortholog 16L as a probe resulted in multiple significant hits in orthopoxviruses, including VACV F1L as a representative family member. All orthopoxviruses possess an M11L ortholog, whereas it is absent in all other completely sequenced poxviruses (Table 2 and Fig. 2d). However, an ortholog (C14R) is present in the partially sequenced squirrelpox virus (McInnes et al., 2006).
Since orthopoxvirus F1L-like proteins and clade II virus M11L orthologs can be connected by PSI-BLAST searches and no other homologs were identified in these searches, we consider them as true orthologs, probably originating after capture of a cellular Bcl-2 family gene. This notion is further supported by the conserved synteny of M11L/F1L genes in orthopoxviruses and clade II poxviruses (Fig. 4a).
Fig. 4.
Synteny conservation around F1L/M11L, M13L and SPI-3 genes in poxvirus genomes. (a) Synteny conservation around F1L/M11L and M13L in selected poxvirus genomes is shown. Boxes above and below horizontal lines represent genes, transcribed from left to right or right to left, respectively. Homologous genes in different genomes are aligned vertically, with any functional information and the name of the gene (in vaccinia virus Copenhagen and/or myxoma virus as a leporipoxvirus (LPV) representative listed at the top. F1L/M11L orthologs are highlighted in red; M13L orthologs are shown in blue. Gene names are given for representative family members: for orthopoxviruses (OPVs), gene names for CPXV-BR are shown. Note that CPXV-BR ORF 045 encoding a lipase is disrupted in VACV (K5L/K6L) and CMLV by multiple in/dels, and 95% of it is deleted in VARV. For capripoxviruses (CPVs), gene names for GTPV and SSPV are shown. The same genes are also present in LSDV but are represented by ORFs 015-020. Identical gene content for this region is found in DPV and SWPV. ORFs (021-026) found in DPV are shown here, and correspond to ORFs 011-016 in SWPV. (b) Synteny conservation around SPI-3 orthologs in selected clade II poxvirus genomes is shown. SPI-3 orthologs are highlighted in red. Identical colors in the upper and lower part of this figure represent orthologous genes in respective genomes. The gene that corresponds to LSDV ORF13 is disrupted in SPPV and GTPV by multiple indels. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
VACV N1L, A52 and B14 (B15 in VACV-COP) are additional Bcl-2 related proteins that adopt a Bcl-2 fold despite the apparent lack of sequence similarity to M11L and F1L (Aoyagi et al., 2007; Cooray et al., 2007; Graham et al., 2008; Kvansakul et al., 2008). Orthologs of these proteins are found in many orthopoxviruses and clade II poxviruses.
Interestingly, unrelated Bcl-2-like proteins, which are most closely related to vertebrate MCL1 and lack apparent sequence similarity to other poxvirus Bcl-2-related proteins, are found in FPV (039) and CNPV (058) (Afonso et al., 2000; Tulman et al., 2004). FPV039 was shown to inhibit apoptosis in chicken and human cells and could functionally replace VACV F1L in the context of VACV infection (Banadyga et al., 2007; Turner and Moyer, 2002).
3.8. Pyrin domain-containing M13L family
MYXV 13L encodes a small 127 aa protein that contains a pyrin domain (PYD/PAAD) (Johnston et al., 2005). The family of cellular PYD-containing proteins comprises 19 members in humans, which are involved in the processing of proinflammatory cytokines and are localized in the inflammasome (Reed et al., 2003). M13L was shown to inhibit caspase-1 activation and thus processing of mature IL-1β and IL-18 in human THP-1 cells. An M13L deficient virus showed normal replication in rabbit RK13 cells but showed impaired replication in RL5 cells and in primary blood monocytes and lymphocytes. Moreover, this virus was attenuated in a rabbit model for MYXV infection (Johnston et al., 2005). Both MYXV and RFV 13L were shown to physically interact with the human PYD-containing protein ASC-1, a caspase-1 activator (Johnston et al., 2005; Dorfleutner et al., 2007).
M13L orthologs are only found in the clade II poxviruses RFV (13L), SWPV (014), DPV (024), YLDV (18L) and TPV (18L). In contrast, M13L orthologs are missing in capripoxviruses and YMTV, whereas adjacent ORFs are present (Figs. 2d and 4a). Since YLDV and capripoxviruses are not immediately related, this indicates that deletions occurred independently in both lineages, or alternatively that a recombination event took place. In the latter case, higher sequence identity between the neighboring genes of these viruses than with other clade II viruses could be expected, which is not the case, making this scenario more unlikely. Independent deletion of 13L orthologs is further supported by the observation that the distance between adjacent genes differs between capripoxviruses (∼45 nt) and YMTV (134 nt).
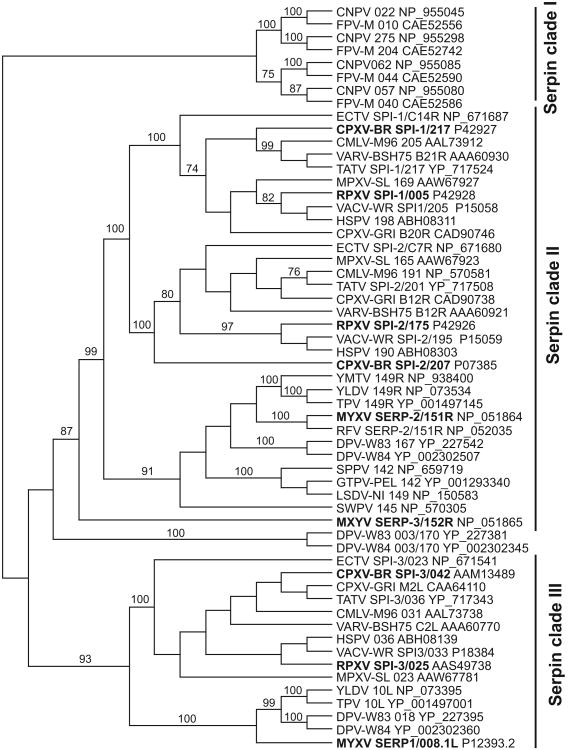
3.9. The Serpin family
Cellular serine protease inhibitors (serpins) play important roles in the regulation of biological networks that include inflammation, apoptosis, coagulation and complement activation (Silverman et al., 2001; van Gent et al., 2003). Many poxviruses also encode serpins, which counteract the host-response to infection. Three serpins are found in orthopoxviruses: SPI-1, SPI-2 (crmA) and SPI-3. They display different target specificity that includes cathepsin G (SPI-1), caspases 1, 8 and 10 and granzyme B (SPI-2) and plasminogen activator, plasmin, thrombin and factor Xa (SPI-3) (reviewed in Silverman et al., 2001)
SPI-1 deficient RPXV showed a replication defect in porcine PK-15 and human A549 cells, but grew well in other cells, in contrast to SPI-2 deficient RPXV, which behaved like wild type in those cells (Ali et al., 1994). Similarly, an SPI-1 deficient VACV-WR strain showed a replication defect in A549 and human keratinocytes but not in African green monkey BS-C-1 cells (Shisler et al., 1999). In MYXV three serpins called SERP1, SERP2 and SERP3 have been described, the numbering of which does not directly reflect a relationship with orthopoxvirus serpins.
Serpins are absent in entomopoxviruses, CRV, MOCV and para-poxviruses. All orthopoxviruses included in this study contain orthologs of SPI-1, SPI-2 and SPI-3. Clade II poxviruses possess one (YMTV, RFV, SWPV and capripoxviruses), two (YLDV and TPV) or three (MYXV and DPV) serpin genes. Phylogenetic analysis showed 3 major clades of poxvirus serpin genes (Fig. 5). Four serpins that are present in both FPV and CNPV clustered in one clade (termed clade I). Orthopoxvirus SPI-1 and SPI-2 orthologs were found in another clade (II) nested with clade II poxvirus serpins, including MYXV 151R/SERP2 and 152R/SERP3 genes. Positioning of M152R/SERP3 outside other clade II poxvirus serpins might be explained by small internal deletions. It is noteworthy that all clade II poxviruses possess at least one clade II serpin, which indicates essential roles for these. Our analyses support the notion that orthopoxvirus SPI-1 and SPI-2 genes probably originated from a duplication during orthopoxvirus evolution (Irving et al., 2000). A third clade is formed by SPI-3 orthologs, which includes MYXV 008.1/SERP-1 and other clade II poxvirus genes. Clade III serpins are absent in YMTV, RFV, SWPV and capripoxviruses. Deletions of clade III serpins likely occurred independently at least three times in YMTV, RFV and SWPV/capripoxviruses, because they are present in other more closely related clade II poxviruses and deletions appear to be lineage-specific. In YMTV the respective ORF 10L is deleted along with 9L and 8L, as compared to YLDV and TPV (Fig. 4b). In RFV more than 80% of the SPI-3 ortholog is deleted, while surrounding genes are conserved between RFV and MYXV (Fig. 4b). In capripoxviruses (e.g. LSDV), the SPI-3 ortholog is deleted along with three neighboring genes. In SWPV three additional neighboring genes are deleted (Fig. 4b). DPV products from duplicated 003/170 genes could not be assigned to one of the serpin clades with high confidence using phylogenetic analyses, however higher sequence identities with clade II serpins supports a closer relationship to those.
Fig. 5.
Phylogenetic relationship of poxvirus serpin proteins. The midpoint-rooted phylogenetic tree was constructed using the neighbor-joining method with a multiple protein sequence alignment of the indicated sequences. Bootstrap support of ≥70 is indicated above the branches. Prototypic poxvirus serpins are highlighted (bold letters). Names of proteins/ORFs and accession numbers are indicated.
Although host range activity has only been reported for ortho-poxvirus SPI-1 genes so far, it is possible that other serpins also con-tributetopoxvirus host range.Inthis context itis noteworthy that an SPI-2/CrmA deleted CPXV, while attenuated in both intratracheal and intradermal infection models, only showed a replication defect in the latter. This indicates that CPXV SPI-2/CrmA plays distinct roles in different paths of infection in mice (MacNeill et al., 2009).
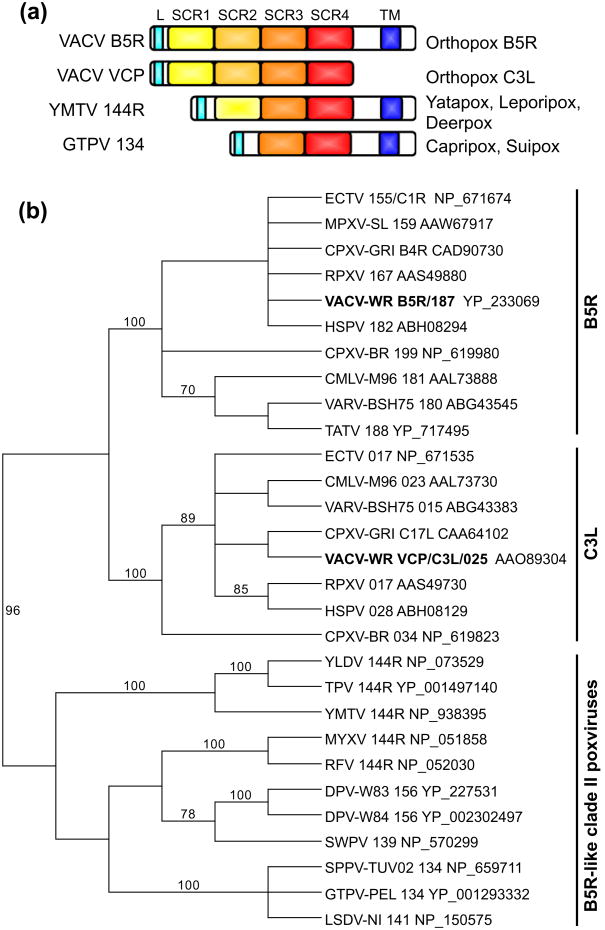
3.10. Short complement-like repeats containing B5R/VCP family
The B5R family comprises genes coding for proteins containing short complement-like repeats (SCRs), which consist of approximately 60 amino acids and are also found in complement system proteins (Engelstad et al., 1992; Takahashi-Nishimaki et al., 1991). B5R-orthologs in orthopoxviruses consist of four tandemly arranged extracellular SCRs and a carboxy-terminal transmembrane domain. B5R is a component of the extracellular virus envelope (Engelstad et al., 1992; Isaacs et al., 1992b) and its deletion in VACV severely impaired production of extracellular enveloped virus (EEV) and virulence in mice (Engelstad and Smith, 1993; Wolffe et al., 1993). Interaction of B5R with a yet to be identified cellular surface molecule activates the cellular Src kinase, leading to actin polymerization and subsequent enhancement of cell-to-cell virus spread (Newsome et al., 2004).
B5R was identified as a host range factor because it contains a premature termination codon in the VACV strain LC16m8, an attenuated derivative of the Lister strain that shows impaired virus production in African green monkey Vero but not in rabbit RK13 cells. Restoration of wild type B5R in VACV-LC16m8 restored plaque size and host range (Takahashi-Nishimaki et al., 1991). A RPXV strain, in which the B5R ortholog was deleted showed either reduced plaque formation in rabbit RK13, rat Rat2 and African green monkey CV-1 cells or no plaques in African green monkey Vero, pig PK-15, chicken embryonic fibroblasts (CEFs) and quail QT-6 cells (Stern et al., 1997). At low-multiplicity of infection, the mutant virus produced titers comparable to wild-type virus in RK13 and Vero cells; however, production of infective virus was severely impaired in CEFs (Stern et al., 1997).
The related VACV complement control protein (VCP/C3L) is a secreted protein that contains four SCRs but lacks a transmembrane domain (Kotwal and Moss, 1988). It acts as a virulence factor by inhibiting complement activation (Isaacs et al., 1992a; Kotwal et al., 1990).
Our analyses showed that B5R-related genes are absent in entomopoxviruses, avipoxviruses, parapoxviruses, CRV and MOCV. In all clade II poxviruses, a single B5R-related gene is found, which encodes a protein with two predicted functional SCRs and a carboxy-terminal transmembrane domain. In yatapoxviruses, leporipoxviruses and DPVs the N-terminal part is distantly related to SCR1 of orthopoxvirus B5R and VCP. However, as it was not recognized as a SCR using the conserved domain database, it might not be functional. Multiple sequence alignments and BLAST searches showed that the two SCRs of clade II poxvirus B5R-orthologs are most closely related to SCR3 and SCR4 of orthopoxvirus B5R and C3L genes (Fig. 6a). Full-length VCP/C3L orthologs are found in most orthopoxviruses except for MPXV and TATV. In clade II MPXV strains the whole C3L ortholog is deleted along with 3 other adjacent genes. In clade I MPXVs a premature stop-codon terminates the open reading frame at the beginning of SCR4. The truncated MPXV VCP ortholog, also called MOPICE, was at least partially functional in in vitro assays. MOPICE binding of human complement factors was weaker than that of the VARV ortholog SPICE but stronger than VACV VCP, whereas decay-accelerating activity was only identified for the VARV and VACV orthologs (Liszewski et al., 2006). In TATV a single nt substitution at position 584 results in a premature stop codon within SCR3. Additionally, a 2 bp insertion at position 668 leads to premature ORF termination. Moreover, the hexanucleotide sequence GCTAAT at nt position 118 encoding Ala-Asn is duplicated in other orthopoxviruses, and repeated 9 times in TATV. This Ala-Asn expansion, which is localized within SCR1, prevents its detection as an SCR in conserved domain searches and might compromise complement factor binding.
Fig. 6.
Structure and phylogeny of the B5R/VCP family. (a) Schematic representation of the domain organization of B5R and VCP (C3L) family proteins. Leader sequences (light blue), transmembrane domains (dark blue) and short complement-like repeats (SCR1-4: yellow, light orange, orange and red, respectively) are highlighted. (b) Mid-point rooted neighbor-joining tree of a protein multiple sequence alignment of SCR3 and SCR4 from indicated sequences. Bootstrap support of ≥70 is indicated above the branches. Names of proteins/ORFs and accession numbers are indicated. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Phylogenetic analysis of the orthologous SCR3 and SCR4 domains revealed 3 major clades (Fig. 6b): orthopoxvirus B5R and C3L orthologs, as well as B5R-like proteins from clade II poxviruses. Orthopoxvirus B5R and C3L clades were nested with one another and separation from clade II poxvirus B5R orthologs showed high bootstrap support.
Taking the phylogenetic analysis and the same number of SCRs in orthopoxvirus B5R and C3L orthologs into account, it appears likely that an ancestral B5R-like gene was duplicated early in orthopoxvirus evolution. While one copy (B5R) retained the transmembrane domain, the other copy (VCP/C3L) lost it and adopted a new function as a different virulence factor by inhibiting host complement factors. This apparently provides an example of neofunctionalization during poxvirus evolution.
It is unknown how B5R functions as a host range factor, but species- and maybe allele-specific differences in the cellular membrane-associated factor interacting with B5R might play an important role. Whether VCP can also function as a host range factor in cell lines or animals is unknown, but seems possible given the high species-specific variation of the host complement system. Along this line it is particularly interesting that the VARV VCP ortholog SPICE/D15L, which displays 95% identity to VACV VCP on the protein level, was a better inhibitor of human and baboon than of guinea pig and dog complement in in vitro assays, whereas the reverse was observed for VACV VCP. Moreover, SPICE was shown to be more than 100- and 6-fold more potent than VACV VCP in inhibiting human complement factors C3b and C4b, respectively (Rosengard et al., 2002).
3.11. The KilA-N/RING domain-containing p28/N1R protein family
ECTV p28 (012) and RFV N1R (143) are prototypic members of a family of poxvirus proteins that combine N-terminal KilA-N and C-terminal RING domains, a combination unique to poxviruses (Iyer et al., 2002). Disruption of the p28 gene had no effect on ECTV replication in BS-C-1 and RAW264.7 cells as well as in primary MEFs and primary mouse ovary cells, but resulted in a replication defect in mouse peritoneal macrophages and decreased virulence in mice (Senkevich et al., 1994, 1995). ECTV p28 as well as orthologs from VARV, VACV-IHD and MYXV were shown to possess ubiquitin ligase activity, which was associated with their RING domain (Huang et al., 2004; Nerenberg et al., 2005).
Single copy genes of p28 orthologs are found in all clade II poxviruses and most orthopoxviruses (Table 2). Genes coding for predicted truncated p28 orthologs are found in HSPV and in all VACV strains, except for strains LC16m8 and LC16mO, in which they are full-length. An intact p28 encoding gene has also been reported for the not completely sequenced VACV strain IHD-W (Upton et al., 1994). In VACV-WR and LIS107, an 11 bp deletion at nt position 537, leads to premature termination of the ORF at amino acid position 184. This deletion is found in all sequences of the VACV complex except for LC16m8, LC16mO and RPXV (intact ORF) and HSPV, in the latter of which the ORF is terminated as a result of a 1 nt deletion at nt position 278. Additional, presumably secondary, mutations are found in the other VACV strains that lead to the incorporation of earlier stop-codons. In VACV-WR the p28 gene is duplicated in the ITR. One copy (011) contains a deletion of 9 nt encoding amino acids T139, P140 and N141 relative to the other (208). Since the deduced proteins are different, which is unusual for ITR located genes, we consider them as two distinct genes (Table 2). The p28 gene is completely or largely deleted in the genomes of VACV-COP and MVA strains, respectively.
Most VARV strains contain intact p28 orthologs, except for IND64-vel4, JAP51-hpr, SUM70-222 and SUM70-228, the three latter of which contain the same 1 bp A insertion at nt position 278. Since JAP51-hpr is not directly related to SUM70-222 and SUM70-228, this indicates a recombination event between strains, or alternatively independent mutations or sequencing errors. The insertion is part of an A7 stretch. Since it is hard to imagine a recombination event between strains, which are well separated in the phylogenetic tree and were isolated 19 years apart, independent mutations or sequencing artifacts may be more likely.
Two closely related p28 orthologs are found in FPV (150 and 157) and CNPV (197 and 205), which contain the poxvirus-specific combination of KilA-N and RING domains and likely emerged through a duplication in a common ancestor. Presence of p28 homologs in avipoxviruses and orthopox/clade II poxviruses indicates that p28 evolved in an early chordopoxvirus ancestor and got lost in CRV, MOCV and parapoxvirus lineages, or alternatively that either a recombination event between an avipoxvirus ancestor and an orthopox/clade II poxvirus ancestor occurred or that p28 evolved independently in both lineages. Absence of p28 orthologs in CRV, MOCV and parapoxviruses indicates that they were lost in these lineages, or alternatively that a recombination event between an avipoxvirus ancestor and an orthopox/clade II poxvirus occurred. Genes encoding proteins containing only KilA-N domains or in which the KilA-N domain is found in combination with other protein domains are present in some other poxviruses, e.g. 022 in CNPV, which are sometimes annotated as N1R/p28 gene family proteins. Some poxviruses possess other RING domain containing proteins. A p28 gene ancestor probably evolved early in poxvirus evolution by the fusion of a KilA-N domain-encoding gene with either a viral or cellular RING containing gene. In the latter scenario, this gene might have been closely related to Makorin genes as suggested (Nicholls and Gray, 2004). The mechanism of how p28 contributes to viral host range is currently not understood but might be related to its ubiquitin ligase activity, which was linked to the RING domain (Huang et al., 2004; Nerenberg et al., 2005). However, the presence of genes encoding only KilA-N domains in other poxviruses makes it possible that this domain exerts an ubiquitin ligase-independent function that might contribute to host range and virulence.
3.12. ANK/F-box proteins (CP77/T5)
Many cellular proteins contain multiple copies of ankyrin repeats (ANK) that mediate protein-protein interactions (Mosavi et al., 2004). ANK-containing proteins form the largest family of poxvirus proteins most of which contain at their carboxy-termini F-box-like domains, which are also known as Pox protein repeats of ankyrin C-terminal (PRANC) domains (Mercer et al., 2005; Sonnberg et al., 2008). Two ANK/F-box proteins have been identified as poxvirus host range factors: CP77 (CHOhr/CPXV-BR025) in CPXV-BR (Spehner et al., 1988) and MYXV-T5 (Mossman et al., 1996).
CP77 was identified as a host range factor based on its ability to rescue viral replication of VACV in CHO cells, in which it can normally not replicate due to a shut-off of both viral and host protein synthesis early after infection. Moreover, CP77 also enabled ECTV, in which most of an orthologous gene is deleted, to efficiently replicate in otherwise poorly permissive hamster (CHO, CCL 14, CCL 16 and CCL 39) and rabbit (SIRC and RK13) cells (Chen et al., 1992). On the molecular level CP77 was shown to inhibit TNFα mediated nuclear factor-κB (NF-κB) activation, likely by the binding of the ANK repeats to the NF-κB subunit p65 and of the F-box to the cellular SCF ligase complex (Chang et al., 2009).
MYXV contains two identical copies of a gene in the ITRs coding for the ANK/F-box protein T5, which is a virulence factor and is essential for myxomatosis in rabbits. Whereas a T5 deleted virus replicated well in RK13 cells, it was replication deficient in RL5 and primary rabbit peripheral mononuclear cells due to the induction of apoptosis (Mossman et al., 1996). Interestingly, the T5 deleted MYXV also showed a replication defect in a subset of human tumor cells that were permissive for the wild type virus (Wang et al., 2006). T5 bound to the serine/threonine kinase Akt and promoted its phosphorylation and activation. The replication defect of the T5 deficient virus correlated with decreased phosphorylation levels of Akt in nonpermissive cells (Wang et al., 2006).
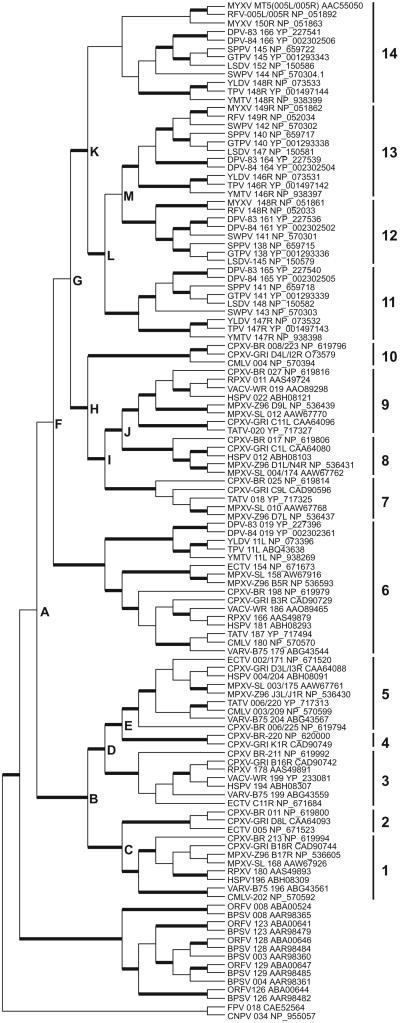
We extended the analysis of ANK/F-box proteins previously reported for selected poxviruses (Sonnberg et al., 2008) (Table 2). ANK/F-box proteins were not identified in entomopoxviruses, MOCV and CRV. The largest numbers of intact ANK/F-box proteins were identified in avipoxviruses CNPV (n = 35) and FPV (=19). In parapoxviruses, 5 (ORFV) and 7 (BPSV) ANK/F-box proteins were identified. In clade II poxviruses 3 to 5 different ANK/F-box proteins were found and between 3 and 10 ANK/F-box proteins were identified in orthopoxviruses.
Phylogenetic analyses showed that all avipoxviruses and parapoxviruses ANK/F-box proteins formed separate clades, indicating that independent duplications occurred in these lineages. To reduce the number of branches displayed in the phylogenetic tree, only one of the ANK/F-box proteins of FPV and CNPV was used for the analysis shown in Fig. 7. The phylogenetic tree is representative of neighbor joining analyses performed with alignments generated with different alignment programs. Bold lines mark branches that showed bootstrap values of ≥80 in all analyses. Bootstrap values for other branches were below 70 in some analyses and can thus be viewed as not significant.
Fig. 7.
Phylogeny of poxvirus ANK/F-box proteins. Neighbor-joining analyses performed with multiple protein sequence alignments generated using Muscle, from which non-alignable portions were excised using GBlocks. The tree was rooted to FPV 018 and CNPV 034. Bold lines mark branches with bootstrap values of ≥80 in all analyses performed. Letters A–M indicate inferred duplication events. Orthopoxvirus and clade II PV ANK/F-box proteins were classified into 14 different groups depending on phylogenetic clades.
The phylogenetic relationship among parapoxviruses genes is comparable to that recently published in two independent studies (Hughes et al., 2010; Sonnberg et al., 2011). For orthopoxviruses and clade II poxviruses, 14 different clades were identified, which show highest sequence identity among each other and likely arose from at least 13 independent duplications, (Table 3 and Fig. 7). These clades were arbitrarily designated as clades 1-14 and inferred duplication events are labeled by capital letters in the phylogenetic tree (Fig. 7). Duplication steps (letter code) that led to the emergence of groups 1–14 were added in a hierarchical manner and similar codes indicate a close relationship (Table 3). Ten ANK/F-box groups were identified in orthopoxviruses. Members of all of these groups were found in CPXV-BR and CXPV-GRI.
Table 3.
Presence and absence of ANK/F-Box genes in poxvirus genomes.
| #1 (ABC) CPXV-BR213 | #2 (ABC) CPXV-BR011 | #3 (ABC) CPXV-BR211 | #4 (ABCD) CPXV-BR220 | #5 (ABCD) CPXV-BR006/225 | #6 (AF) CPXV-BR198/DPV019 | #7 (AFGHI) CPXV-BR025(CPXV77) | #8 (AFGHIJ) CPXV-BR017 | #9 (AFGH) CPXV-BR027 | #10 (AFGH) CPXV-BR008/223 | #11 (AFGKL) DPV165 | #12 (AFGKLM) MYXV148/DPV161 | #13 (AFGKLM) MYXV149/DPV164 | #14 (AFGK) MYXV-T5/DPV166 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ectromelia virus Moscow | † | 1 | 1 | † | 1+1 | 1 | † | † | † | † | – | – | – | – |
| Cowpox virus - Brighton Red | 1 | 1 | 1 | 1 | 1+1 | 1 | 1 | 1 | 1 | 1+1 | – | – | – | – |
| Monkeypox virus Sierra Leone | 1 | – | – | † | 1+1 | 1 | 1 | 1+1 | 1 | – | – | – | – | – |
| Monkeypox virus Zaire 96 | 1 | – | – | † | 1+1 | 1 | 1 | 1+1 | 1 | – | – | – | – | – |
| Camelpox virus M-96 | 1 | – | † | – | 1+1 | 1 | † | – | † | 1 | – | – | – | – |
| Taterapox virus | † | – | † | – | 1+1 | 1 | 1 | – | 1 | N† | – | – | – | – |
| Variola major virus-BSH75 | 1 | – | 1 | – | 1 | 1 | 1C† | – | † | † | – | – | – | – |
| Cowpox virus - GRI-90 | 1 | 1 | 1 | 1 | 1+1 | 1 | 1 | 1 | 1 | 1+1 | – | – | – | – |
| Rabbitpox virus | 1 | – | 1 | † | † | 1 | † | – | 1 | 1+1C† | – | – | – | – |
| Horsepox virus | 1 | – | 1 | † | 1+1 | 1 | † | 1 | 1 | 1+1C† | – | – | – | – |
| Vaccinia virus WR | † | – | 1 | † | † | 1 | † | – | 1 | – | – | – | – | – |
| Yaba monkey tumor virus | – | – | – | – | – | 1 | – | – | – | – | 1 | – | 1 | 1 |
| Yaba-like disease virus | – | – | – | – | – | 1 | – | – | – | – | 1 | – | 1 | 1 |
| Tanapox virus | – | – | – | – | – | 1 | – | – | – | – | 1 | – | – | 1 |
| Myxoma virus | – | – | – | – | – | – | – | – | – | – | – | 1 | 1 | 2+1 |
| Rabbit fibroma virus | – | – | – | – | – | – | – | – | – | – | – | 1 | 1 | 1+1 |
| Deerpox virus W-848-83 | – | – | – | – | – | 1 | – | – | – | – | 1 | 1 | 1 | 1 |
| Deerpox virus W-1170-84 | – | – | – | – | – | 1 | – | – | – | – | 1 | 1 | 1 | 1 |
| Sheeppox virus TU-V02127 | – | – | – | – | – | – | – | – | – | – | 1 | 1 | 1 | 1 |
| Goatpox virus Pellor | – | – | – | – | – | – | – | – | – | – | 1 | 1 | 1 | 1 |
| Lumpy skin disease virus | – | – | – | – | – | – | – | – | – | – | 1 | 1 | 1 | 1 |
| Swinepox virus | – | – | – | – | – | – | – | – | – | – | 1 | 1 | 1 | 1 |
ANK/F-Box genes were grouped according to their phylogenetic relationships. Letters indicate inferred duplication events.
indicates deletion or stop codons;
N = amino-terminal; C = carboxy-terminal; - = gene not present. Numbers indicate quantities of closely related genes.
Intact group 1 (CPXV-BR 213) orthologs were found in all orthopoxviruses, except for TATV and ECTV in which multiple gene-inactivating indels are found. VACV-Acambis is the only VACV strain analyzed in this study encoding a full-length group 1 ortholog, whereas orthologous ORFs are disrupted in all other VACV strains by multiple independent indels.
Group 2 (CPXV-BR 011) orthologs were only identified in CPXV-BR, CPXV-GRI and ECTV, which display about 94% sequence identity with each other. No direct orthologs were found in all other orthopoxviruses. In these latter cases the closest relatives are the group 1 genes, which display about 46% sequence identity.
Intact orthologs of group 3 genes (CPXV-BR 211) are found in ECTV, CPXV-BR, VARV, CPXV-GRI, RPXV, HSPV and most VACV strains, except LIS107, Lc1608 and Lc16m0 in which the ORF is C-terminally truncated due to a deletion. In CMLV and TATV multiple indels disrupt the ORF of the group 3 ortholog, whereas no trace was found in MPXV.
The only full-length group 4 ANK/F-box (CPXV-BR 220) genes were found in CPXV-BR and CPXV-GRI. Multiple indels in group 4 orthologs of ECTV, RPXV, HSPV and VACV disrupt the ORFs in these viruses. In MPXV only a short fragment (125 bp) displaying 92% sequence identity to CPXV-BR 220 was identified, whereas no traces could be found in CMLV, TATV and VARV.
Intact group 5 ANK/F-box genes (CPXV-BR 006/225) are found in most orthopoxviruses as duplicated genes in the ITR, except for VARV in which only one intact copy is found, and in RPXV and VACVs in which group 5 ORFs are disrupted by multiple indels.
Group 6 ANK/F-box genes (CPX-BR198) are found in all orthopoxviruses except for VACV-ANK, -MVA and derivative strains in which multiple indels destroy the ORF. Interestingly, group 6 ANK/F-box genes are also present in DPV and yatapoxviruses.
Group 7 ANK/F-box genes include CP77 (CPXV-BR 025). Intact orthologs are found in CPXV-GRI, MPXV and TATV. In VARV strains, different deletions lead to predicted truncated proteins lacking the F-box region and encoding only the first 355 to 451 N-terminal amino acids depending on the strain. In ECTV, RPXV, HSPV and VACV multiple indels destroy the ORF. In CMLV only a 160 bp fragment showing 98% sequence identity to an internal part of TATV 018 is found.
Intact group 8 ANK/F-box genes (CPXV-BR 017) orthologs were identified in MPXV, CPXV-GRI and HSPV. T-BLASTN searches of the whole genomes identified a 225 bp region in ECTV with 96% sequence identity with the 3′ region of CPXV-BR017 that lacks an in frame start codon, whereas no traces of group 8 genes were found in all other orthopoxviruses.
Intact group 9 ANK/F-box (CPXV-BR 027) orthologs were identified in all orthopoxviruses except for all VARV, CMLV and ECTV strains. Multiple independent indels disrupt the ORF in these latter viruses, which are thus not predicted to encode functional proteins. Interestingly CPXV-GRI (C11L) and TATV (020) genes are most closely related to one another, sharing 96% sequence identity on the protein level, whereas sequence identity to other group 9 proteins is below 70%. Disrupted VARV and CMLV group 9 genes are most similar to the TATV/CPXV-GRI subgroup. This is indicative of a recombination event between CPXV-GRI and TATV/VARV/CMLV group. Alternatively, the two sub-groups might result from independent duplications in orthopoxviruses with subsequent deletion of one or the other copy from the respective sub-group.
Intact orthologs of group 10 ANK/F-box (CPXV-BR 008/223) genes are present in CPXV-BR, CPXV-GRI (both contain 2 copies in the ITR) and CMLV (1 full length and 1 truncated copy at the ITR boundary). Indels lead to predicted truncated proteins in HSPV (368 amino acids) and RPXV (385 amino acids) in comparison to the intact CPXV gene. Several indels are also found in some VACV strains, e.g. VACV-COP, whereas the gene is completely deleted in other strains, including VACV-WR. Similarly, no evidence of a group 10 gene was found in MPXV. In TATV the largest part of the gene is deleted leaving only the F-Box encoding region (91 aa) intact. Large parts of group 10 genes are deleted in all VARV and ECTV strains.
Four additional ANK/F-box gene groups (11-14) were identified solely in clade II poxviruses. These genes form a monophyletic clade and thus probably resulted from three duplication events early in the evolution of these viruses (Fig. 7 and Table 3). Group 13 (MYXV-149/DPV164) and group 14 (MYXV-T5/DPV166) orthologs are present in all clade II poxviruses, whereas group 11 (DPV165) genes are absent in leporipoxviruses and group 12 (MYXV-148/DPV161) orthologs are absent in yatapoxviruses.
The phylogenetic analyses indicate that only a few ANK/F-box proteins were present in early poxviruses and that independent duplications of their genes occurred in several lineages. The presence of group 6 ANK/F-box genes in orthopoxviruses and some clade II poxviruses indicates that an ancestral virus possessed one related ANK/F-box gene that underwent independent duplication events in both lineages. Many viruses or lineages subsequently lost the gene or acquired gene-inactivating mutations. Whereas both complete deletions and gene inactivating mutations, which often arose independently of one another, occurred in orthopoxviruses, only lineage-specific gene losses are found in the analyzed clade II poxviruses. Multiple duplications are also evident in parapoxviruses. Duplications of one copy of an ANK/F-box gene likely led to the emergence of 5 and 7 copies in ORF and BPSV, respectively, and even more copies in avipoxviruses in which the largest number of ANK/F-box genes is found.
A striking observation is that all 10 orthopoxviruses ANK/F-box proteins are found in CPXV-BR and CPXV-GRI despite their relatively large phylogenetic distance within orthopoxviruses. Traces of most ANK/F-box genes can be found in some orthopoxviruses that lost intact coding ANK/F-box genes, indicating that most ANK/F-box genes were present in a common orthopoxvirus ancestor. Alternatively, orthopoxviruses might have acquired these genes from other orthopoxviruses during gene transfer events in some instances. Gene loss of ANK/F-box encoding genes is a common theme in poxvirus evolution. In fact, none of the analyzed orthopoxviruses except for CPXV-BR and CPXV-GRI display the same pattern of ANK/F-box genes (Table 3). Since different poxvirus ANK/F-box proteins likely carry out unique biological functions (Werden et al., 2008), and show unique distribution patterns (Table 3), they are good candidates to explain at least in part differences in poxvirus host range. It is particularly interesting that CPXVs, which have the broadest host range among poxviruses, contain the largest number of ANK/F-box encoding genes found in mammalian poxviruses.
3.13. The orthopoxvirus K1L family
Orthopoxviruses possess another ANK-containing protein with a predicted molecular mass of 32 kDa, which is encoded by K1L (WR032) in VACV. It is the only ANK protein in orthopoxviruses that lacks an F-box. Deletion of K1L from VACV-COP resulted in a replication defect in rabbit RK13 but not in human MRC-5, monkey VERO or porcine LLC-PK1 cells (Perkus et al., 1990). The modified vaccinia virus Ankara (MVA) strain is highly attenuated and contains multiple genomic deletions, including a partially deleted K1L gene. Insertion of K1L into the MVA genome restored replication in RK13 cells, but not in other non-permissive cells (Meyer et al., 1991). Moreover, RK13 cells that were stably transfected with K1L were permissive for both MVA and a VACV-WR strain that had K1L deleted (Sutter et al., 1994). The replication defect of K1L-deleted VACV in RK13 cells was associated with a rapid shutdown of viral and host protein synthesis (Ramsey-Ewing and Moss, 1996). Reconstitution of K1L in MVA resulted in inhibition of the NF-κB pathway, presumably by inhibition of IκBα degradation; whereas deletion of K1L in VACV-WR resulted in IκBα degradation and NF-κB activation (Shisler and Jin, 2004). Interestingly, VACV K1L and its ECTV ortholog 022 had different affects in RK13 and human HEp-2 cells when integrated into the genomes of both VACV and ECTV. Whereas K1L and ECTV022 had comparable stimulatory effects on viral replication in HEp-2 cells, ECTV022 had only a weak stimulatory effect on viral replication in RK13 cells. In contrast K1L exerted a strong stimulatory effect in RK13 cells (Chen et al., 1993). VACV-COP K1L and ECTV022 display 96% sequence identity, differing at 11 out of 285 amino acid positions. It seems possible that the different effects of VACV-K1L and ECTV022 reflect host species-specific differences in inhibition of cellular targets.
Intact K1L orthologs are found in all orthopoxviruses with the exception of VARV, CMLV and TATV in which premature stop-codons and short indels destroy the ORFs (Table 2). Since these latter viruses form a monophyletic clade, it seemed possible that a gene-inactivating mutation occurred in a common ancestor. Surprisingly, close inspection of the nucleotide sequences revealed that none of the gene-inactivating mutations is common to all three or even two of these viruses (Fig. 8a). Thus, it appears most likely that gene-inactivating mutations occurred independently in the three species. Since it is possible that a mutation in a regulatory sequence occurred in an ancestor of VARV, CMLV and TATV, we compared the 5′ region of K1L orthologs in different orthopoxviruses (Fig. 8b). An early promoter core motif is found at position –42 to –28, relative to the predicted start codon, which in VACV-WR is identical to the core motif for early poxvirus genes as described (Yang et al., 2010). This motif is deleted in all VARV strains, but is well conserved in CMLV and TATV, which excludes the deletion of this motif in a common ancestor of these viruses. The K1L ORF is intact in all sequenced VACV strains except for VACV-MVA in which only the first 263 nt out of 853 are found and VACV-TAN, in which a 1 bp insertion at nt position 542 leads to a frame shift resulting in the incorporation of a premature stop codon.
Fig. 8.
Gene-inactivating mutations in K1L orthologs of variola, camelpox and taterapox viruses. (a) Horizontal lines represent genes of VARV, CMLV and TATV that are orthologous to vaccinia virus K1L. Inactivating mutations are indicated by red circles (stop codons), deletions (blue triangles) and insertions (inverse green triangles). Numbers indicate deleted or inserted nucleotides, which lead to frame shifts. (b) Comparison of the 5′ region of K1L orthologs in different orthopoxviruses. The box highlights an early promoter core motif that is found at position −42 to −28, relative to the predicted start codon (asterisk). Dots indicate nucleotides that are identical to VACV-WR. Dashes indicate deletions. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
K1L does not appear to have orthologs in the other poxviruses included in this study based on two observations: (1) Some poxviruses (leporipoxviruses, YMTV, SWPV, parapoxviruses, MOCV, CRV and entomopoxviruses) do not possess any ANK proteins besides the aforementioned ANK/F-box proteins; and (2) Some poxviruses contain F-box-less ANK proteins (e.g. YLDV8L, TPV8L, LSDV012, GTPV10, SPPV10 and DPV014) that are only distantly related to K1L, showing not significantly more sequence identity to K1L than to other ANK proteins. However, it cannot be ruled out that some of these latter proteins might possess similar biological activities as K1L.
3.14. General observations
The poxvirus host range gene families analyzed in this study display a strikingly dissimilar distribution among poxvirus genera. None of the analyzed family genes are found in entomopoxviruses, CRV, and MOCV. Only three and two families are present in avipoxviruses and parapoxviruses, respectively. These families comprise ANK/F-Box genes, found in both avipoxviruses and para-poxviruses, serpin genes and p28-like proteins, present in avi-poxviruses, and E3L orthologs, found in parapoxviruses. In contrast, at least 11 of the 12 analyzed family members are found in orthopoxviruses and clade II poxviruses. In the latter groups 11 (leporipoxviruses), 10 (DPVs) and 9 (yatapoxviruses, capripoxviruses and SWPV) host range families were identified. The different distribution of host range families in poxvirus genera might be partially attributed to the fact that members of orthopoxviruses and clade II poxviruses comprise viruses that are important human and animal pathogens. Some of these viruses serve as models for understanding poxvirus biology, and are thus most thoroughly studied. More distantly related poxviruses likely contain different genes that are important for their host ranges but have not yet been identified. The absence of specific host range genes might also contribute to host restriction of poxviruses. Along this line are experiments performed with an experimental HIV vaccine. Inclusion of VACV K3L and E3L genes in CNPV greatly stimulated HIV p24 production in infected human and monkey cell lines, which correlated with PKR inhibition (Fang et al., 2001).
There is also evidence for the evolution of viral genes that target similar antiviral pathways but that evolved independently in distantly related poxviruses. Examples include the Bcl-2-like genes and the TNFR superfamily. Bcl-2-related M11/F1L orthologs were only identified in orthopoxviruses and clade II poxviruses, whereas other Bcl-2-like genes are present in avipoxviruses as discussed above. Similarly, many poxviruses encode TNFR related genes that likely evolved independently by the capture of different cellular TNFR genes. Although host range function has not been described for all of these genes, it appears likely that they evolved to specifically antagonize the antiviral response in their natural hosts.
3.15. Host range genes in orthopoxviruses
Comparison of host range family genes within genera of closely related poxviruses might give valuable insights into a connection between the presence of specific genes and host range. It is striking that CPXVs, which have the broadest host range among poxviruses, possess the largest number of intact host range family genes, i.e. 27 in CPXV-GRI and 26 in CPXV-BR, as opposed to 13 in TATV and VACV, and 15 in VARV, ECTV, RPXV and MPXV-SL on the other side of the spectrum. Since CPXVs possess the largest genomes among orthopoxviruses, the high number of host range family genes is not too surprising. However, it is interesting to note that for the vast majority of host range family genes inactivated genes or traces of orthologous genes could be detected in the available orthopox-virus genomes. Thus, it appears likely that most host range family genes were already present in a common ancestor of the completely sequenced orthopoxviruses and subsequently were inactivated or lost in many orthopoxvirus species.
Only 7 of the 27 host range gene family members are intact in all orthopoxviruses. These are: SPI-1, SPI-2, SPI-3, C7L, F1L and ANK/F-box group 6 genes. Conservation of these genes indicates an essential role for viral replication in a wide variety of hosts.
3.16. Implications of gene inactivations and duplications for poxvirus host range and virulence
A common explanation for gene losses in viruses is that lost genes are not important for infection of their respective host species and have therefore been lost (loss of function). While it is likely that the presence or absence of host range genes contributes to host range, gene losses could also be viewed as an adaptation of the virus to the host that the virus becomes less virulent and detrimental to its host. Attenuated viruses might have advantages over their progenitors in that the infected host becomes less sick, survives longer and stays more mobile, all of which could lead to better virus transmission. Inactivation of VACV K3L orthologs in ECTV and MPXV might be an example of this proposed “attenuation (adaptation) by gene inactivation”: the deduced ancestral K3L ortholog in ECTV and MPXV differs in only two residues from VACV-WR K3L, the latter of which has been shown to be a very potent inhibitor of mouse but not human PKR. The deduced ancestral K3L variant is in fact identical with that found in CPXV-GRI. Since all these viruses probably have rodent reservoirs, it seems possible that they are all good inhibitors of rodent PKRs. However, this has to be verified experimentally.
Another scenario is that viral genes may not be effective in inhibiting the antiviral response and therefore might become non-essential and subsequently lost after the virus becomes established in a new host species. Such a viral specification might result in the emergence of a new virus species and might constrain the virus to infect a limited number or perhaps only one species. This might explain the inactivation or loss of the 11 of the 27 host range family genes analyzed in this study during evolution of the human-specific VARV.
While some host range genes evidently got lost or inactivated in ancestors of extant closely related poxviruses, e.g. an M13L gene in capripoxviruses, the independent loss or inactivation of host range genes apparently occurred frequently in different lineages. For example C4L orthologs in orthopoxviruses experienced partial gene inactivations (TATV and HSPV), complete gene inactivations (MPXV and VARV), or total gene losses (ECTV and RPXV/VACV). Likewise T4 orthologs experienced C-terminal deletions (RPXV/HSPV/VACV and clade II MPXV) or complete gene losses (VARV, ECTV, SWPV and likely yatapoxviruses).
Another remarkable theme in poxvirus genome evolution is the lineage-specific duplication of host range genes. Striking examples for this include ANK/F-Box genes, for which multiple duplication events were observed for orthopoxviruses /clade II poxviruses (13 events), parapoxviruses (6 events) and avipoxviruses (34 events). Viral gene duplications might result in several advantages:
Duplications might allow the adaptation of copies to protein orthologs found in different hosts, thereby extending host range. This notion is supported by the perpetuation of all duplicated ANK/F-box gene copies in CPXVs, which have the broadest host range of poxviruses, and loss or inactivation of ANK/F-box genes in orthopoxviruses that have narrower host ranges.
Products of duplicated genes can lead to the targeting of different but related antiviral host pathways. Examples include the duplicated serpins and TNFR-2 homologs, which preferentially target different host proteases and cytokines, respectively (Alejo et al., 2006; Gileva et al., 2006; Silverman et al., 2001; Turner and Moyer, 2002).
Duplication of host range genes can also lead to gene neofunctionalization. An example of this can be found in the SCR-containing B5R/VCP family. Genes encoding SCR proteins containing a transmembrane domain are found in all orthopoxviruses and clade II viruses. A duplication that likely occurred in an ancestor of orthopoxviruses led to the perpetuation of the transmembrane domain and function in one copy (B5R family). In contrast the second family member lost the transmembrane domain producing instead a secreted protein that evolved to become an inhibitor of the host complement system.
In this study we analyzed the presence of genes belonging to 12 poxvirus gene families, which contain genes with described host range function. Some of these 12 families comprise multiple genes, most of which probably evolved due to lineage-specific duplications. While only a subset of these genes have described host range function, other genes in these families are likely to possess host range function. Taking into account that detection of host range gene function can be complicated and relies on the appropriate detection system, and that many poxviral genes have not been tested for host range function yet, it can be anticipated that more genes will be identified as having host range function. Virulence factors are especially good hostrange candidates, because in many cases adaptation to specific host(s) can be expected. Determining the distribution patterns of host range genes among poxviruses and correlating this with viral host ranges will constitute important steps in better understanding both viral host range and virulence. These analyses will also provide valuable insights in the risk assessment for the emergence of new human and animal poxviruses.
Supplementary Material
Acknowledgments
We are grateful to Thomas E. Dever for helpful discussions and for helpful suggestions on the manuscript. This work was supported in part by NIH grant P20 RR016475 from the INBRE Program of the National Center for Research Resources to Stefan Rothenburg.
Footnotes
Appendix A. Supplementary data: Supplementary data associated with this article can be found, in the online version, at http://dx.doi.org/10.1016/j.meegid.2012.12.002.
References
- Afonso CL, Tulman ER, Delhon G, Lu Z, Viljoen GJ, Wallace DB, Kutish GF, Rock DL. Genome of crocodilepox virus. J Virol. 2006;80:4978–4991. doi: 10.1128/JVI.80.10.4978-4991.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Afonso CL, Tulman ER, Lu Z, Zsak L, Kutish GF, Rock DL. The genome of fowlpox virus. J Virol. 2000;74:3815–3831. doi: 10.1128/jvi.74.8.3815-3831.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alejo A, Ruiz-Arguello MB, Ho Y, Smith VP, Saraiva M, Alcami A. A chemokine-binding domain in the tumor necrosis factor receptor from variola (smallpox) virus. Proc Natl Acad Sci USA. 2006;103:5995–6000. doi: 10.1073/pnas.0510462103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali AN, Turner PC, Brooks MA, Moyer RW. The SPI-1 gene of rabbitpox virus determines host range and is required for hemorrhagic pock formation. Virology. 1994;202:305–314. doi: 10.1006/viro.1994.1347. [DOI] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aoyagi M, Zhai D, Jin C, Aleshin AE, Stec B, Reed JC, Liddington RC. Vaccinia virus N1L protein resembles a B cell lymphoma-2 (Bcl-2) family protein. Protein Sci. 2007;16:118–124. doi: 10.1110/ps.062454707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banadyga L, Gerig J, Stewart T, Barry M. Fowlpox virus encodes a Bcl-2 homologue that protects cells from apoptotic death through interaction with the proapoptotic protein Bak. J Virol. 2007;81:11032–11045. doi: 10.1128/JVI.00734-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett JW, Shun Chang C, Wang G, Werden SJ, Shao Z, Barrett C, Gao X, Belsito TA, Villenevue D, McFadden G. Myxoma virus M063R is a host range gene essential for virus replication in rabbit cells. Virology. 2007;361:123–132. doi: 10.1016/j.virol.2006.11.015. [DOI] [PubMed] [Google Scholar]
- Barry M, Hnatiuk S, Mossman K, Lee SF, Boshkov L, McFadden G. The myxoma virus M-T4 gene encodes a novel RDEL-containing protein that is retained within the endoplasmic reticulum and is important for the productive infection of lymphocytes. Virology. 1997;239:360–377. doi: 10.1006/viro.1997.8894. [DOI] [PubMed] [Google Scholar]
- Beattie E, Kauffman EB, Martinez H, Perkus ME, Jacobs BL, Paoletti E, Tartaglia J. Host-range restriction of vaccinia virus E3L-specific deletion mutants. Virus Genes. 1996;12:89–94. doi: 10.1007/BF00370005. [DOI] [PubMed] [Google Scholar]
- Beattie E, Tartaglia J, Paoletti E. Vaccinia virus-encoded eIF-2α homolog abrogates the antiviral effect of interferon. Virology. 1991;183:419–422. doi: 10.1016/0042-6822(91)90158-8. [DOI] [PubMed] [Google Scholar]
- Brandt T, Heck MC, Vijaysri S, Jentarra GM, Cameron JM, Jacobs BL. The N-terminal domain of the vaccinia virus E3L-protein is required for neurovirulence, but not induction of a protective immune response. Virology. 2005;333:263–270. doi: 10.1016/j.virol.2005.01.006. [DOI] [PubMed] [Google Scholar]
- Brandt TA, Jacobs BL. Both carboxy- and amino-terminal domains of the vaccinia virus interferon resistance gene, E3L, are required for pathogenesis in a mouse model. J Virol. 2001;75:850–856. doi: 10.1128/JVI.75.2.850-856.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratke KA, McLysaght A. Identification of multiple independent horizontal gene transfers into poxviruses using a comparative genomics approach. BMC Evol Biol. 2008;8:67. doi: 10.1186/1471-2148-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll K, Elroy-Stein O, Moss B, Jagus R. Recombinant vaccinia virus K3L gene product prevents activation of double-stranded RNA-dependent, initiation factor 2 alpha-specific protein kinase. J Biol Chem. 1993;268:12837–12842. [PubMed] [Google Scholar]
- Chang HW, Uribe LH, Jacobs BL. Rescue of vaccinia virus lacking the E3L gene by mutants of E3L. J Virol. 1995;69:6605–6608. doi: 10.1128/jvi.69.10.6605-6608.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SJ, Hsiao JC, Sonnberg S, Chiang CT, Yang MH, Tzou DL, Mercer AA, Chang W. Poxviral host range protein CP77 contains a F-box-like domain that is necessary to suppress NF-{kappa}B activation by TNF-{alpha} but is independent of its host range function. J Virol. 2009;83:4140–4152. doi: 10.1128/JVI.01835-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen N, Li G, Liszewski MK, Atkinson JP, Jahrling PB, Feng Z, Schriewer J, Buck C, Wang C, Lefkowitz EJ, Esposito JJ, Harms T, Damon IK, Roper RL, Upton C, Buller RM. Virulence differences between monkeypox virus isolates from West Africa and the Congo basin. Virology. 2005;340:46–63. doi: 10.1016/j.virol.2005.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Drillien R, Spehner D, Buller RM. Restricted replication of ectromelia virus in cell culture correlates with mutations in virus-encoded host range gene. Virology. 1992;187:433–442. doi: 10.1016/0042-6822(92)90445-u. [DOI] [PubMed] [Google Scholar]
- Chen W, Drillien R, Spehner D, Buller RM. In vitro and in vivo study of the ectromelia virus homolog of the vaccinia virus K1L host range gene. Virology. 1993;196:682–693. doi: 10.1006/viro.1993.1525. [DOI] [PubMed] [Google Scholar]
- Cooray S, Bahar MW, Abrescia NG, McVey CE, Bartlett NW, Chen RA, Stuart DI, Grimes JM, Smith GL. Functional and structural studies of the vaccinia virus virulence factor N1 reveal a Bcl-2-like anti-apoptotic protein. J Gen Virol. 2007;88:1656–1666. doi: 10.1099/vir.0.82772-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dar AC, Sicheri F. X-ray crystal structure and functional analysis of vaccinia virus K3L reveals molecular determinants for PKR subversion and substrate recognition. Mol Cell. 2002;10:295–305. doi: 10.1016/s1097-2765(02)00590-7. [DOI] [PubMed] [Google Scholar]
- Dorfleutner A, Talbott SJ, Bryan NB, Funya KN, Rellick SL, Reed JC, Shi X, Rojanasakul Y, Flynn DC, Stehlik C. A Shope Fibroma virus PYRIN-only protein modulates the host immune response. Virus Genes. 2007;35:685–694. doi: 10.1007/s11262-007-0141-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas AE, Corbett KD, Berger JM, McFadden G, Handel TM. Structure of M11L: A myxoma virus structural homolog of the apoptosis inhibitor, Bcl-2. Protein Sci. 2007;16:695–703. doi: 10.1110/ps.062720107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elde NC, Child SJ, Geballe AP, Malik HS. Protein kinase R reveals an evolutionary model for defeating viral mimicry. Nature. 2009;457:485–489. doi: 10.1038/nature07529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelstad M, Howard ST, Smith GL. A constitutively expressed vaccinia gene encodes a 42-kDa glycoprotein related to complement control factors that forms part of the extracellular virus envelope. Virology. 1992;188:801–810. doi: 10.1016/0042-6822(92)90535-w. [DOI] [PubMed] [Google Scholar]
- Engelstad M, Smith GL. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology. 1993;194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- Esposito JJ, Sammons SA, Frace AM, Osborne JD, Olsen-Rasmussen M, Zhang M, Govil D, Damon IK, Kline R, Laker M, Li Y, Smith GL, Meyer H, Leduc JW, Wohlhueter RM. Genome sequence diversity and clues to the evolution of variola (smallpox) virus. Science. 2006;313:807–812. doi: 10.1126/science.1125134. [DOI] [PubMed] [Google Scholar]
- Essbauer S, Pfeffer M, Meyer H. Zoonotic poxviruses. Vet Microbiol. 2010;140:229–236. doi: 10.1016/j.vetmic.2009.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Everett H, Barry M, Lee SF, Sun X, Graham K, Stone J, Bleackley RC, McFadden G. M11L: a novel mitochondria-localized protein of myxoma virus that blocks apoptosis of infected leukocytes. J Exp Med. 2000;191:1487–1498. doi: 10.1084/jem.191.9.1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang ZY, Limbach K, Tartaglia J, Hammonds J, Chen X, Spearman P. Expression of vaccinia E3L and K3L genes by a novel recombinant canarypox HIV vaccine vector enhances HIV-1 pseudovirion production and inhibits apoptosis in human cells. Virology. 2001;291:272–284. doi: 10.1006/viro.2001.1209. [DOI] [PubMed] [Google Scholar]
- Fenner F. Adventures with poxviruses of vertebrates. FEMS Microbiol Rev. 2000;24:123–133. doi: 10.1016/S0168-6445(00)00027-9. [DOI] [PubMed] [Google Scholar]
- Gertz EM, Yu YK, Agarwala R, Schaffer AA, Altschul SF. Composition-based statistics and translated nucleotide searches: improving the TBLASTN module of BLAST. BMC Biol. 2006;4:41. doi: 10.1186/1741-7007-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gileva IP, Nepomnyashchikh TS, Antonets DV, Lebedev LR, Kochneva GV, Grazhdantseva AV, Shchelkunov SN. Properties of the recombinant TNF-binding proteins from variola, monkeypox, and cowpox viruses are different. Biochim Biophys Acta. 2006;1764:1710–1718. doi: 10.1016/j.bbapap.2006.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham SC, Bahar MW, Cooray S, Chen RA, Whalen DM, Abrescia NG, Alderton D, Owens RJ, Stuart DI, Smith GL, Grimes JM. Vaccinia virus proteins A52 and B14 Share a Bcl-2-like fold but have evolved to inhibit NF-kappaB rather than apoptosis. PLoS Pathog. 2008;4:e1000128. doi: 10.1371/journal.ppat.1000128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gubser C, Hue S, Kellam P, Smith GL. Poxvirus genomes: a phylogenetic analysis. J Gen Virol. 2004;85:105–117. doi: 10.1099/vir.0.19565-0. [DOI] [PubMed] [Google Scholar]
- Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- Hnatiuk S, Barry M, Zeng W, Liu L, Lucas A, Percy D, McFadden G. Role of the C-terminal RDEL motif of the myxoma virus M-T4 protein in terms of apoptosis regulation and viral pathogenesis. Virology. 1999;263:290–306. doi: 10.1006/viro.1999.9946. [DOI] [PubMed] [Google Scholar]
- Hu FQ, Smith CA, Pickup DJ. Cowpox virus contains two copies of an early gene encoding a soluble secreted form of the type II TNF receptor. Virology. 1994;204:343–356. doi: 10.1006/viro.1994.1539. [DOI] [PubMed] [Google Scholar]
- Huang J, Huang Q, Zhou X, Shen MM, Yen A, Yu SX, Dong G, Qu K, Huang P, Anderson EM, Daniel-Issakani S, Buller RM, Payan DG, Lu HH. The poxvirus p28 virulence factor is an E3 ubiquitin ligase. J Biol Chem. 2004;279:54110–54116. doi: 10.1074/jbc.M410583200. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Friedman R. Poxvirus genome evolution by gene gain and loss. Mol Phylogenet Evol. 2005;35:186–195. doi: 10.1016/j.ympev.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Hughes AL, Irausquin S, Friedman R. The evolutionary biology of poxviruses. Infect Genet Evol. 2010;10:50–59. doi: 10.1016/j.meegid.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irving JA, Pike RN, Lesk AM, Whisstock JC. Phylogeny of the serpin superfamily: implications of patterns of amino acid conservation for structure and function. Genome Res. 2000;10:1845–1864. doi: 10.1101/gr.gr-1478r. [DOI] [PubMed] [Google Scholar]
- Isaacs SN, Kotwal GJ, Moss B. Vaccinia virus complement-control protein prevents antibody-dependent complement-enhanced neutralization of infectivity and contributes to virulence. Proc Natl Acad Sci USA. 1992a;89:628–632. doi: 10.1073/pnas.89.2.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isaacs SN, Wolffe EJ, Payne LG, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol. 1992b;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer LM, Koonin EV, Aravind L. Extensive domain shuffling in transcription regulators of DNA viruses and implications for the origin of fungal APSES transcription factors. Genome Biol. 2002;3:RESEARCH0012. doi: 10.1186/gb-2002-3-3-research0012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston JB, Barrett JW, Nazarian SH, Goodwin M, Ricciuto D, Wang G, McFadden G. A poxvirus-encoded pyrin domain protein interacts with ASC-1 to inhibit host inflammatory and apoptotic responses to infection. Immunity. 2005;23:587–598. doi: 10.1016/j.immuni.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawagishi-Kobayashi M, Cao C, Lu J, Ozato K, Dever TE. Pseudosubstrate inhibition of protein kinase PKR by swine pox virus C8L gene product. Virology. 2000;276:424–434. doi: 10.1006/viro.2000.0561. [DOI] [PubMed] [Google Scholar]
- Kawagishi-Kobayashi M, Silverman JB, Ung TL, Dever TE. Regulation of the protein kinase PKR by the vaccinia virus pseudosubstrate inhibitor K3L is dependent on residues conserved between the K3L protein and the PKR substrate eIF2a. Mol Cell Biol. 1997;17:4146–4158. doi: 10.1128/mcb.17.7.4146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim YG, Muralinath M, Brandt T, Pearcy M, Hauns K, Lowenhaupt K, Jacobs BL, Rich A. A role for Z-DNA binding in vaccinia virus pathogenesis. Proc Natl Acad Sci USA. 2003;100:6974–6979. doi: 10.1073/pnas.0431131100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotwal GJ, Isaacs SN, McKenzie R, Frank MM, Moss B. Inhibition of the complement cascade by the major secretory protein of vaccinia virus. Science. 1990;250:827–830. doi: 10.1126/science.2237434. [DOI] [PubMed] [Google Scholar]
- Kotwal GJ, Moss B. Vaccinia virus encodes a secretory polypeptide structurally related to complement control proteins. Nature. 1988;335:176–178. doi: 10.1038/335176a0. [DOI] [PubMed] [Google Scholar]
- Kvansakul M, van Delft MF, Lee EF, Gulbis JM, Fairlie WD, Huang DC, Colman PM. A structural viral mimic of prosurvival Bcl-2: a pivotal role for sequestering proapoptotic Bax and Bak. Mol Cell. 2007;25:933–942. doi: 10.1016/j.molcel.2007.02.004. [DOI] [PubMed] [Google Scholar]
- Kvansakul M, Yang H, Fairlie WD, Czabotar PE, Fischer SF, Perugini MA, Huang DC, Colman PM. Vaccinia virus anti-apoptotic F1L is a novel Bcl-2-like domain-swapped dimer that binds a highly selective subset of BH3-containing death ligands. Cell Death Differ. 2008;15:1564–1571. doi: 10.1038/cdd.2008.83. [DOI] [PubMed] [Google Scholar]
- Langland JO, Jacobs BL. The role of the PKR-inhibitory genes, E3L and K3L, in determining vaccinia virus host range. Virology. 2002;299:133–141. doi: 10.1006/viro.2002.1479. [DOI] [PubMed] [Google Scholar]
- Langland JO, Cameron JM, Heck MC, Jancovich JK, Jacobs BL. Inhibition of PKR by RNA and DNA viruses. Virus Res. 2006;119:100–110. doi: 10.1016/j.virusres.2005.10.014. [DOI] [PubMed] [Google Scholar]
- Lefkowitz EJ, Upton C, Changayil SS, Buck C, Traktman P, Buller RM. Poxvirus Bioinformatics Resource Center: a comprehensive Poxviridae informational and analytical resource. Nucleic Acids Res. 2005;33:D311–D316. doi: 10.1093/nar/gki110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefkowitz EJ, Wang C, Upton C. Poxviruses: past, present and future. Virus Res. 2006;117:105–118. doi: 10.1016/j.virusres.2006.01.016. [DOI] [PubMed] [Google Scholar]
- Lewis-Jones S. Zoonotic poxvirus infections in humans. Curr Opin Infect Dis. 2004;17:81–89. doi: 10.1097/00001432-200404000-00003. [DOI] [PubMed] [Google Scholar]
- Li Y, Carroll DS, Gardner SN, Walsh MC, Vitalis EA, Damon IK. On the origin of smallpox: correlating variola phylogenics with historical smallpox records. Proc Natl Acad Sci USA. 2007;104:15787–15792. doi: 10.1073/pnas.0609268104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Likos AM, Sammons SA, Olson VA, Frace AM, Li Y, Olsen-Rasmussen M, Davidson W, Galloway R, Khristova ML, Reynolds MG, Zhao H, Carroll DS, Curns A, Formenty P, Esposito JJ, Regnery RL, Damon IK. A tale of two clades: monkeypox viruses. J Gen Virol. 2005;86:2661–2672. doi: 10.1099/vir.0.81215-0. [DOI] [PubMed] [Google Scholar]
- Liszewski MK, Leung MK, Hauhart R, Buller RM, Bertram P, Wang X, Rosengard AM, Kotwal GJ, Atkinson JP. Structure and regulatory profile of the monkeypox inhibitor of complement: comparison to homologs in vaccinia and variola and evidence for dimer formation. J Immunol. 2006;176:3725–3734. doi: 10.4049/jimmunol.176.6.3725. [DOI] [PubMed] [Google Scholar]
- Liu J, Wennier S, Zhang L, McFadden G. M062 is a host range factor essential for myxoma virus pathogenesis and functions as an antagonist of host SAMD9 in human cells. J Virol. 2011;85:3270–3282. doi: 10.1128/JVI.02243-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Wolff KC, Jacobs BL, Samuel CE. Vaccinia virus E3L interferon resistance protein inhibits the interferon-induced adenosine deaminase A-to-I editing activity. Virology. 2001;289:378–387. doi: 10.1006/viro.2001.1154. [DOI] [PubMed] [Google Scholar]
- Loparev VN, Parsons JM, Knight JC, Panus JF, Ray CA, Buller RM, Pickup DJ, Esposito JJ. A third distinct tumor necrosis factor receptor of orthopoxviruses. Proc Natl Acad Sci USA. 1998;95:3786–3791. doi: 10.1073/pnas.95.7.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacNeill AL, Moldawer LL, Moyer RW. The role of the cowpox virus crmA gene during intratracheal and intradermal infection of C57BL/6 mice. Virology. 2009;384:151–160. doi: 10.1016/j.virol.2008.10.041. [DOI] [PubMed] [Google Scholar]
- McFadden G. Poxvirus tropism. Nat Rev. 2005;3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McInnes CJ, Wood AR, Thomas K, Sainsbury AW, Gurnell J, Dein FJ, Nettleton PF. Genomic characterization of a novel poxvirus contributing to the decline of the red squirrel (Sciurus vulgaris) in the UK. J Gen Virol. 2006;87:2115–2125. doi: 10.1099/vir.0.81966-0. [DOI] [PubMed] [Google Scholar]
- McLysaght A, Baldi PF, Gaut BS. Extensive gene gain associated with adaptive evolution of poxviruses. Proc Natl Acad Sci USA. 2003;100:15655–15660. doi: 10.1073/pnas.2136653100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Chao J, Xiang Y. Identification from diverse mammalian poxviruses of host-range regulatory genes functioning equivalently to vaccinia virus C7L. Virology. 2008;372:372–383. doi: 10.1016/j.virol.2007.10.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meng X, Schoggins J, Rose L, Cao J, Ploss A, Rice CM, Xiang Y. C7L family of poxvirus host range genes inhibits antiviral activities induced by type I interferons and interferon regulatory factor 1. J Virol. 2012;86:4538–4547. doi: 10.1128/JVI.06140-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer AA, Fleming SB, Ueda N. F-box-like domains are present in most poxvirus ankyrin repeat proteins. Virus Genes. 2005;31:127–133. doi: 10.1007/s11262-005-1784-z. [DOI] [PubMed] [Google Scholar]
- Meyer H, Sutter G, Mayr A. Mapping of deletions in the genome of the highly attenuated vaccinia virus MVA and their influence on virulence. J Gen Virol. 1991;72(Pt 5):1031–1038. doi: 10.1099/0022-1317-72-5-1031. [DOI] [PubMed] [Google Scholar]
- Mosavi LK, Cammett TJ, Desrosiers DC, Peng ZY. The ankyrin repeat as molecular architecture for protein recognition. Protein Sci. 2004;13:1435–1448. doi: 10.1110/ps.03554604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mossman K, Lee SF, Barry M, Boshkov L, McFadden G. Disruption of M-T5, a novel myxoma virus gene member of poxvirus host range superfamily, results in dramatic attenuation of myxomatosis in infected European rabbits. J Virol. 1996;70:4394–4410. doi: 10.1128/jvi.70.7.4394-4410.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myskiw C, Arsenio J, Hammett C, van Bruggen R, Deschambault Y, Beausoleil N, Babiuk S, Cao J. Comparative analysis of poxvirus orthologues of the vaccinia virus E3 protein: modulation of protein kinase R activity, cytokine responses, and virus pathogenicity. J Virol. 2011;85:12280–12291. doi: 10.1128/JVI.05505-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nerenberg BT, Taylor J, Bartee E, Gouveia K, Barry M, Fruh K. The poxviral RING protein p28 is a ubiquitin ligase that targets ubiquitin to viral replication factories. J Virol. 2005;79:597–601. doi: 10.1128/JVI.79.1.597-601.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newsome TP, Scaplehorn N, Way M. SRC mediates a switch from microtubule- to actin-based motility of vaccinia virus. Science. 2004;306:124–129. doi: 10.1126/science.1101509. [DOI] [PubMed] [Google Scholar]
- Nicholls RD, Gray TA. Cellular source of the poxviral N1R/p28 gene family. Virus Genes. 2004;29:359–364. doi: 10.1007/s11262-004-7440-1. [DOI] [PubMed] [Google Scholar]
- Opgenorth A, Graham K, Nation N, Strayer D, McFadden G. Deletion analysis of two tandemly arranged virulence genes in myxoma virus, M11L and myxoma growth factor. J Virol. 1992;66:4720–4731. doi: 10.1128/jvi.66.8.4720-4731.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panus JF, Smith CA, Ray CA, Smith TD, Patel DD, Pickup DJ. Cowpox virus encodes a fifth member of the tumor necrosis factor receptor family: a soluble, secreted CD30 homologue. Proc Natl Acad Sci USA. 2002;99:8348–8353. doi: 10.1073/pnas.122238599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peabody DS. Translation initiation at non-AUG triplets in mammalian cells. J Biol Chem. 1989;264:5031–5035. [PubMed] [Google Scholar]
- Perkus ME, Goebel SJ, Davis SW, Johnson GP, Limbach K, Norton EK, Paoletti E. Vaccinia virus host range genes. Virology. 1990;179:276–286. doi: 10.1016/0042-6822(90)90296-4. [DOI] [PubMed] [Google Scholar]
- Pickup DJ, Ink BS, Parsons BL, Hu W, Joklik WK. Spontaneous deletions and duplications of sequences in the genome of cowpox virus. Proc Natl Acad Sci USA. 1984;81:6817–6821. doi: 10.1073/pnas.81.21.6817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Postigo A, Cross JR, Downward J, Way M. Interaction of F1L with the BH3 domain of Bak is responsible for inhibiting vaccinia-induced apoptosis. Cell Death Differ. 2006;13:1651–1662. doi: 10.1038/sj.cdd.4401853. [DOI] [PubMed] [Google Scholar]
- Ramsey-Ewing AL, Moss B. Complementation of a vaccinia virus host-range K1L gene deletion by the nonhomologous CP77 gene. Virology. 1996;222:75–86. doi: 10.1006/viro.1996.0399. [DOI] [PubMed] [Google Scholar]
- Reed JC, Doctor K, Rojas A, Zapata JM, Stehlik C, Fiorentino L, Damiano J, Roth W, Matsuzawa S, Newman R, Takayama S, Marusawa H, Xu F, Salvesen G, Godzik A. Comparative analysis of apoptosis and inflammation genes of mice and humans. Genome Res. 2003;13:1376–1388. doi: 10.1101/gr.1053803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivas C, Gil J, Melkova Z, Esteban M, Diaz-Guerra M. Vaccinia virus E3L protein is an inhibitor of the interferon (i.f.n.)-induced 2–5A synthetase enzyme. Virology. 1998;243:406–414. doi: 10.1006/viro.1998.9072. [DOI] [PubMed] [Google Scholar]
- Rosengard AM, Liu Y, Nie Z, Jimenez R. Variola virus immune evasion design: expression of a highly efficient inhibitor of human complement. Proc Natl Acad Sci USA. 2002;99:8808–8013. doi: 10.1073/pnas.112220499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburg S, Chinchar VG, Dever TE. Characterization of a ranavirus inhibitor of the antiviral protein kinase PKR. BMC Microbiol. 2011;11:56. doi: 10.1186/1471-2180-11-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothenburg S, Seo EJ, Gibbs JS, Dever TE, Dittmar K. Rapid evolution of protein kinase PKR alters sensitivity to viral inhibitors. Nat Struct Mol Biol. 2009;16:63–70. doi: 10.1038/nsmb.1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saraiva M, Alcami A. CrmE, a novel soluble tumor necrosis factor receptor encoded by poxviruses. J Virol. 2001;75:226–233. doi: 10.1128/JVI.75.1.226-233.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber M, McFadden G. The myxoma virus TNF-receptor homologue (T2) inhibits tumor necrosis factor-alpha in a species-specific fashion. Virology. 1994;204:692–705. doi: 10.1006/viro.1994.1585. [DOI] [PubMed] [Google Scholar]
- Schreiber M, McFadden G. Mutational analysis of the ligand-binding domain of M-T2 protein, the tumor necrosis factor receptor homologue of myxoma virus. J Immunol. 1996;157:4486–4495. [PubMed] [Google Scholar]
- Sedger LM, Osvath SR, Xu XM, Li G, Chan FK, Barrett JW, McFadden G. Poxvirus tumor necrosis factor receptor (TNFR)-like T2 proteins contain a conserved preligand assembly domain that inhibits cellular TNFR1-induced cell death. J Virol. 2006;80:9300–9309. doi: 10.1128/JVI.02449-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senkevich TG, Koonin EV, Buller RM. A poxvirus protein with a RING zinc finger motif is of crucial importance for virulence. Virology. 1994;198:118–128. doi: 10.1006/viro.1994.1014. [DOI] [PubMed] [Google Scholar]
- Senkevich TG, Wolffe EJ, Buller RM. Ectromelia virus RING finger protein is localized in virus factories and is required for virus replication in macrophages. J Virol. 1995;69:4103–4111. doi: 10.1128/jvi.69.7.4103-4111.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo EJ, Liu F, Kawagishi-Kobayashi M, Ung TL, Cao C, Dar AC, Sicheri F, Dever TE. Protein kinase PKR mutants resistant to the poxvirus pseudosubstrate K3L protein. Proc Natl Acad Sci USA. 2008;105:16894–16899. doi: 10.1073/pnas.0805524105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shisler JL, Isaacs SN, Moss B. Vaccinia virus serpin-1 deletion mutant exhibits a host range defect characterized by low levels of intermediate and late mRNAs. Virology. 1999;262:298–311. doi: 10.1006/viro.1999.9884. [DOI] [PubMed] [Google Scholar]
- Shisler JL, Jin XL. The vaccinia virus K1L gene product inhibits host NF-κB activation by preventing IκBα degradation. J Virol. 2004;78:3553–3560. doi: 10.1128/JVI.78.7.3553-3560.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman GA, Bird PI, Carrell RW, Church FC, Coughlin PB, Gettins PG, Irving JA, Lomas DA, Luke CJ, Moyer RW, Pemberton PA, Remold-O'Donnell E, Salvesen GS, Travis J, Whisstock JC. The serpins are an expanding superfamily of structurally similar but functionally diverse proteins. Evolution, mechanism of inhibition, novel functions, and a revised nomenclature. J Biol Chem. 2001;276:33293–33296. doi: 10.1074/jbc.R100016200. [DOI] [PubMed] [Google Scholar]
- Shors ST, Beattie E, Paoletti E, Tartaglia J, Jacobs BL. Role of the vaccinia virus E3L and K3L gene products in rescue of VSV and EMCV from the effects of IFN-alpha. J Interferon Cytokine Res. 1998;18:721–729. doi: 10.1089/jir.1998.18.721. [DOI] [PubMed] [Google Scholar]
- Smith CA, Davis T, Anderson D, Solam L, Beckmann MP, Jerzy R, Dower SK, Cosman D, Goodwin RG. A receptor for tumor necrosis factor defines an unusual family of cellular and viral proteins. Science. 1990;248:1019–1023. doi: 10.1126/science.2160731. [DOI] [PubMed] [Google Scholar]
- Smith CA, Davis T, Wignall JM, Din WS, Farrah T, Upton C, McFadden G, Goodwin RG. T2 open reading frame from the Shope fibroma virus encodes a soluble form of the TNF receptor. Biochem Biophys Res Commun. 1991;176:335–342. doi: 10.1016/0006-291x(91)90929-2. [DOI] [PubMed] [Google Scholar]
- Smith CA, Hu FQ, Smith TD, Richards CL, Smolak P, Goodwin RG, Pickup DJ. Cowpox virus genome encodes a second soluble homologue of cellular TNF receptors, distinct from CrmB, that binds TNF but not LT alpha. Virology. 1996;223:132–147. doi: 10.1006/viro.1996.0462. [DOI] [PubMed] [Google Scholar]
- Sonnberg S, Fleming SB, Mercer AA. Phylogenetic analysis of the large family of poxvirus ankyrin-repeat proteins reveals orthologue groups within and across chordopoxvirus genera. J Gen Virol. 2011;92:2596–2607. doi: 10.1099/vir.0.033654-0. [DOI] [PubMed] [Google Scholar]
- Sonnberg S, Seet BT, Pawson T, Fleming SB, Mercer AA. Poxvirus ankyrin repeat proteins are a unique class of F-box proteins that associate with cellular SCF1 ubiquitin ligase complexes. Proc Natl Acad Sci USA. 2008;105:10955–10960. doi: 10.1073/pnas.0802042105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spehner D, Gillard S, Drillien R, Kirn A. A cowpox virus gene required for multiplication in Chinese hamster ovary cells. J Virol. 1988;62:1297–1304. doi: 10.1128/jvi.62.4.1297-1304.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern RJ, Thompson JP, Moyer RW. Attenuation of B5R mutants of rabbitpox virus in vivo is related to impaired growth and not an enhanced host inflammatory response. Virology. 1997;233:118–129. doi: 10.1006/viro.1997.8556. [DOI] [PubMed] [Google Scholar]
- Su J, Wang G, Barrett JW, Irvine TS, Gao X, McFadden G. Myxoma virus M11L blocks apoptosis through inhibition of conformational activation of Bax at the mitochondria. J Virol. 2006;80:1140–1151. doi: 10.1128/JVI.80.3.1140-1151.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter G, Ramsey-Ewing A, Rosales R, Moss B. Stable expression of the vaccinia virus K1L gene in rabbit cells complements the host range defect of a vaccinia virus mutant. J Virol. 1994;68:4109–41016. doi: 10.1128/jvi.68.7.4109-4116.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swofford DL. Phylogenetic Analysis Using Parsimony (and other methods) version 4. Sinauer Associates Inc.; Sunderland, Massachusetts: 2002. [Google Scholar]
- Takahashi-Nishimaki F, Funahashi S, Miki K, Hashizume S, Sugimoto M. Regulation of plaque size and host range by a vaccinia virus gene related to complement system proteins. Virology. 1991;181:158–164. doi: 10.1016/0042-6822(91)90480-y. [DOI] [PubMed] [Google Scholar]
- Talavera G, Castresana J. Improvement of phylogenies after removing divergent and ambiguously aligned blocks from protein sequence alignments. Syst Biol. 2007;56:564–577. doi: 10.1080/10635150701472164. [DOI] [PubMed] [Google Scholar]
- Taylor JM, Quilty D, Banadyga L, Barry M. The vaccinia virus protein F1L interacts with Bim and inhibits activation of the pro-apoptotic protein Bax. J Biol Chem. 2006;281:39728–39739. doi: 10.1074/jbc.M607465200. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface. flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toth AM, Zhang P, Das S, George CX, Samuel CE. Interferon action and the double-stranded RNA-dependent enzymes ADAR1 adenosine deaminase and PKR protein kinase. Prog Nucleic Acid Res Mol Biol. 2006;81:369–434. doi: 10.1016/S0079-6603(06)81010-X. [DOI] [PubMed] [Google Scholar]
- Tulman ER, Afonso CL, Lu Z, Zsak L, Kutish GF, Rock DL. The genome of canarypox virus. J Virol. 2004;78:353–366. doi: 10.1128/JVI.78.1.353-366.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulman ER, Delhon G, Afonso CL, Lu Z, Zsak L, Sandybaev NT, Kerembekova UZ, Zaitsev VL, Kutish GF, Rock DL. Genome of horsepox virus. J Virol. 2006;80:9244–9258. doi: 10.1128/JVI.00945-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner PC, Moyer RW. Poxvirus immune modulators: functional insights from animal models. Virus Res. 2002;88:35–53. doi: 10.1016/s0168-1702(02)00119-3. [DOI] [PubMed] [Google Scholar]
- Upton C, Macen JL, Schreiber M, McFadden G. Myxoma virus expresses a secreted protein with homology to the tumor necrosis factor receptor gene family that contributes to viral virulence. Virology. 1991;184:370–382. doi: 10.1016/0042-6822(91)90853-4. [DOI] [PubMed] [Google Scholar]
- Upton C, Schiff L, Rice SA, Dowdeswell T, Yang X, McFadden G. A poxvirus protein with a RING finger motif binds zinc and localizes in virus factories. J Virol. 1994;68:4186–4195. doi: 10.1128/jvi.68.7.4186-4195.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Gent D, Sharp P, Morgan K, Kalsheker N. Serpins: structure, function and molecular evolution. Int J Biochem Cell Biol. 2003;35:1536–1547. doi: 10.1016/s1357-2725(03)00134-1. [DOI] [PubMed] [Google Scholar]
- Wang G, Barrett JW, Nazarian SH, Everett H, Gao X, Bleackley C, Colwill K, Moran MF, McFadden G. Myxoma virus M11L prevents apoptosis through constitutive interaction with Bak. J Virol. 2004;78:7097–7111. doi: 10.1128/JVI.78.13.7097-7111.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G, Barrett JW, Stanford M, Werden SJ, Johnston JB, Gao X, Sun M, Cheng JQ, McFadden G. Infection of human cancer cells with myxoma virus requires Akt activation via interaction with a viral ankyrin-repeat host range factor. Proc Natl Acad Sci USA. 2006;103:4640–4645. doi: 10.1073/pnas.0509341103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilenko ST, Banadyga L, Bond D, Barry M. The vaccinia virus F1L protein interacts with the proapoptotic protein Bak and inhibits Bak activation. J Virol. 2005;79:14031–14043. doi: 10.1128/JVI.79.22.14031-14043.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasilenko ST, Stewart TL, Meyers AF, Barry M. Vaccinia virus encodes a previously uncharacterized mitochondrial-associated inhibitor of apoptosis. Proc Natl Acad Sci USA. 2003;100:14345–14350. doi: 10.1073/pnas.2235583100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werden SJ, Rahman MM, McFadden G. Poxvirus host range genes. Adv Virus Res. 2008;71:135–171. doi: 10.1016/S0065-3527(08)00003-1. [DOI] [PubMed] [Google Scholar]
- Wolffe EJ, Isaacs SN, Moss B. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol. 1993;67:4732–4741. doi: 10.1128/jvi.67.8.4732-4741.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X, Nash P, McFadden G. Myxoma virus expresses a TNF receptor homolog with two distinct functions. Virus Genes. 2000;21:97–109. [PubMed] [Google Scholar]
- Yang Z, Bruno DP, Martens CA, Porcella SF, Moss B. Simultaneous high-resolution analysis of vaccinia virus and host cell transcriptomes by deep RNA sequencing. Proc Natl Acad Sci USA. 2010;107:11513–11518. doi: 10.1073/pnas.1006594107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Jacobs BL, Samuel CE. Loss of protein kinase PKR expression in human HeLa cells complements the vaccinia virus E3L deletion mutant phenotype by restoration of viral protein synthesis. J Virol. 2008;82:840–848. doi: 10.1128/JVI.01891-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.